Abstract
RASGRP1 is an important guanine nucleotide exchange factor and activator of the RAS-MAPK pathway following T cell antigen receptor (TCR) signaling. The consequences of RASGRP1 mutations in humans are unknown. In a patient with recurrent bacterial and viral infections, born to healthy consanguineous parents, we used homozygosity mapping and exome sequencing to identify a biallelic stop-gain variant in RASGRP1. This variant segregated perfectly with the disease and has not been reported in genetic databases. RASGRP1 deficiency was associated in T cells and B cells with decreased phosphorylation of the extracellular-signal-regulated serine kinase ERK, which was restored following expression of wild-type RASGRP1. RASGRP1 deficiency also resulted in defective proliferation, activation and motility of T cells and B cells. RASGRP1-deficient natural killer (NK) cells exhibited impaired cytotoxicity with defective granule convergence and actin accumulation. Interaction proteomics identified the dynein light chain DYNLL1 as interacting with RASGRP1, which links RASGRP1 to cytoskeletal dynamics. RASGRP1-deficient cells showed decreased activation of the GTPase RhoA. Treatment with lenalidomide increased RhoA activity and reversed the migration and activation defects of RASGRP1-deficient lymphocytes.
RASGRP1 is a guanine-nucleotide-exchange factor that is involved in lymphocyte development and function1. The catalytic region of RASGRP1 consists of a RAS exchange motif and a CDC25 domain. RASGRP1 contains two atypical calcium-binding EF hands, a C1 diacylglycerol (DAG)-binding domain and a protein kinase C phosphorylation site that is necessary for activation of the protein2. Although the protein has high expression in lymphoid cells, its role is best studied in T cells following activation of the TCR2. Following increased DAG expression and/or phosphorylation by protein kinase C, RASGRP1 translocates to the membrane and converts the small GTPase RAS from an inactive GDP-bound to an active GTP-bound state. This process mediates activation of the MAP-kinase (RAS-RAF-MEK-ERK) cascade3.
Although RAS signaling is well known for its association with malignant transformation, RAS GTPases control a variety of cellular processes under physiological conditions, including cell growth, differentiation, apoptosis, neuronal activity and cell migration. There are three main RAS isoforms (K, H and N) that all share a conserved core GTPase domain but have distinct biological functions. Depending on the upstream stimulus, RASGRP1 can activate all three Ras isoforms4. Studies of Rasgrp1−/− mice have revealed substantial defects in thymocyte development and function as a result of defective TCR-mediated ERK phosphorylation. Accordingly, Rasgrp1−/− mice have considerably fewer CD4+ and CD8+ T cells compared with wild-type mice5.
Studies of patients with inborn errors of their immune system (primary immunodeficiency disorders) have considerably shaped the molecular understanding of key nodes of human immunity, sometimes revealing surprising differences between patients and mouse models with deficiency in corresponding gene products6–8. In humans, the subgroup of quantitative or qualitative defects of T lymphocytes that result in combined immunodeficiencies comprise a heterogeneous subgroup of primary immunodeficiencies9,10. Often, these defects are associated with primary or secondary B cell defects11,12. Despite some heterogeneity, impairment of adaptive immunity in these diseases increases susceptibility to recurrent, earlyonset and severe infections with a variety of pathogens, including viruses, bacteria, fungi and parasites9.
We found that deficiency in human RASGRP1, a major regulator of RAS signaling in T lymphocytes, resulted in a previously unknown primary immunodeficiency disease. Our findings indicate a major role for RASGRP1 in immune cell signaling and function in T cells, B cells and NK cells. We also decipher a previously unknown role for RASGRP1 in the dynamic regulation of the cytoskeleton and identify lenalidomide as a potential treatment option for this immunodeficiency.
Results
Identification of human RASGRP1 deficiency
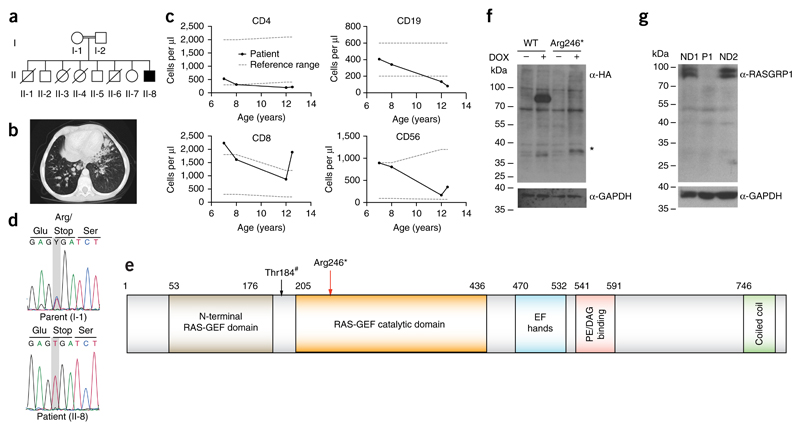
We studied a 12-year-old index patient of healthy consanguineous parents (Fig. 1a). Three older siblings of the patient (II/1, II/3 and II/4; Fig. 1a) died in the first 2 years of life, possibly as a result of immunodeficiency-related complications. However, no material from these siblings was available for further genetic testing. The index patient presented with recurrent infections, including multiple episodes of pneumonia that resulted in bronchiectasis and collapse of the right upper pulmonary lobe (Fig. 1b). Over time, severe failure to thrive, with height and weight below the third percentile, as well as finger clubbing became evident (data not shown). At the age of 12 years, after another episode of pneumonia, monthly intravenous immunoglobulin therapy was started. Prophylactic antibiotic therapy was recommended, which the patient did not use regularly. Further episodes of pneumonia and herpetic lesions necessitated hospitalization. Serial immunological analyses revealed normal numbers of total lymphocytes, but a progressive decrease in the number of CD4+ T cells, an inverted ratio of CD4+ T cells to CD8+ T cells, an increased relative proportion of TCRγδ cells, and progressively decreasing B cell counts compared with healthy individuals (Fig. 1c and Supplementary Table 1). Routine adenotomy was performed on this patient at the age of 15 and, unexpectedly, histology revealed a low-grade Epstein-Barr virus (EBV)-associated B cell lymphoma (Supplementary Fig. 1a). Given the immunodeficiency-related complications and the development of lymphoma, chemotherapy was performed, followed by myeloablative conditioning and allogeneic hematopoietic stem cell transplantation.
Figure 1. Identification of human RASGRP1 deficiency.
(a) Pedigree of the index family: double lines indicate consanguinity; filled black indicates the index patient; diagonal lines indicate deceased siblings. (b) Computed tomography scan of the index patient showing bronchiectatic segments, centrilobular nodular opacities, and paratracheal, subcarinal and bilateral hilar lymphadenopathy. (c) Longitudinal peripheral blood lymphocyte counts (exact measurements, Supplementary Table 1); gray dotted lines represent reference values. (d) Capillary sequencing confirmation of the stop-gain mutation in RASGRP1 (c. C726T, p. Arg246*). (e) RASGRP1 protein domains: red arrow indicates position of the stop-gain mutation; black arrow indicates the Thr184 PKC-phosphorylation site (Thr184#). (f) Cropped immunoblot analysis of wild-type (WT) and mutant (Arg246*) RASGRP1 tagged at the amino terminus with Strep-HA-in doxycyclin-inducible HEK293T cells. *, expected size of the truncated protein. (g) Cropped immunoblot analysis of full-length RASGRP1 in EBV-immortalized B cells from the patient (P1) and two normal donors (ND). Data are representative of four (f) or two (g) independent experiments.
Given the known consanguinity in the family and the absence of clinical signs of immunodeficiency in either parent, paralleled by unremarkable leukocyte immunophenotyping (Supplementary Tables 2 and 3), we suspected an autosomal-recessive disease trait. We performed homozygosity mapping and exome sequencing to identify the underlying genetic disease etiology. Although we detected no variants in genes previously associated with primary immunodeficiencies, we found a homozygous nonsense variant on Chr15: 38805097: G>A (hg19 build 137) (c. C726T, p. Arg246*) in RASGRP1 (Fig. 1d,e). This variant has not been observed in heterozygous or homozygous form in several human genetic variation databases including the 1000 Genomes Project and the Exome Aggregation Consortium (ExAC) (Supplementary Tables 4 and 5a). It was predicted to be highly damaging for the corresponding protein function, with a combined annotation-dependent depletion (CADD) score of 35 (Supplementary Table 5b). The RASGRP1 gene has a negative residual variation intolerance score and is among the 11.77% most intolerant genes, indicative of the evolutionary conservation of RASGRP1. Accordingly, the number of ‘observed’ nonsense variants was significantly lower than the number of ‘expected’ nonsense variants (Supplementary Table 5b). Of note, no other homozygous variants in RASGRP1 leading to premature stop codons or frameshift mutations have been observed with higher population frequencies (Supplementary Table 5a). The variant showed perfect co-segregation with the disease under the assumption of autosomal-recessive inheritance, with both parents and all three available healthy siblings being heterozygous for the detected variant (Supplementary Fig. 1). In addition to the identified stop codon variant in RASGRP1, a variant in UROC1 (p.G313R), which encodes urocanase enzyme, segregated with the disease in the core family (data not shown). Biallelic loss-of-function mutations in this gene have been shown to cause urocanic aciduria, mental retardation and ataxia13. As we could not reconcile this variant with the immunodeficiency in our patient, we disregarded this gene as being causative of the immunological phenotype in the index patient.
To assess the effect of the variant in RASGRP1, we expressed wild-type or mutant streptavidin-hemagglutinin (Strep-HA)-tagged RASGRP1 in doxycycline-inducible HEK293T human embryonic kidney cells and observed markedly reduced expression of the truncated protein product (Fig. 1f). Consistent with these observations, immunoblot analysis with an antibody to RASGRP1 (anti-RASGRP1) did not detect endogenous full-length RASGRP1 protein in cells from the patient (Fig. 1g).
Profound multifaceted peripheral blood T cell defects
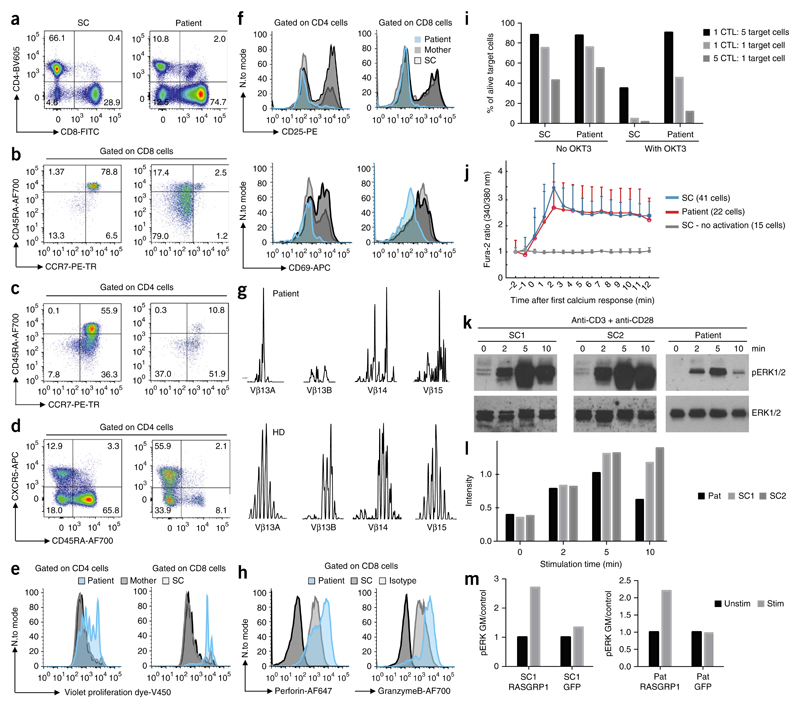
Mouse studies have identified RASGRP1 as a guanine-nucleotide-exchange factor that acts as an essential activator of the RAS-RAF-MEK-ERK signaling in T cells; it thus serves as a major regulator of T cell development14, survival and proliferation15. Activation of RASGRP1 is dependent on DAG and occurs following activation of the signaling protein PLCγ1 in T cells16. RASGRP1 has high expression in all lymphoid cells and in a variety of other cell types17. However, so far the function of mouse RASGRP1 beyond its function in T cells has remained largely elusive (comparison, Supplementary Table 6). We performed detailed T cell immunophenotyping to characterize deficiency in human RASGRP1 and observed persistent CD4+ T cell lymphopenia together with an increase in the number of CD8+ T cells compared with healthy controls (Fig. 2a). Evaluation of the distribution of TCRαβT cells versus that of TCRγδ T cells revealed increased numbers of TCRγδ+ CD8+ T cells compared to reference values (Supplementary Table 1). We further investigated the compartment of non-conventional T cells and observed reduced numbers of mucosal-associated invariant T cells (Supplementary Table 3). Furthermore, the number of invariant NKT cells was very low, bordering on absent (Supplementary Tables 3 and 6, and Supplementary Fig. 2), reminiscent of studies of mice18. Although a prominent population expressing the TCR α-chain variable region 24 (Vα24) was detectable19, these cells expressed the surface markers CD8 and CD45RA (Supplementary Fig. 2a), which suggested that they might belong to oligoclonally expanded and exhausted memory CD8+ T cells.
Figure 2. RASGRP1deficiency causes defective TCR signaling and an aberrant immunophenotype.
(a–d) T cell immunophenotyping displaying relative proportions of CD4+ cells and CD8+ cells amongst CD3+ cells in lymphocyte gate (a), proportions of naive (CD45RA+CCR7+), central memory (CD45RA−CCR7+)T cells, effector memory (CD45RA−CCR7−) T cells and exhausted effector memory (CD45RA+CCR7−) T cells among CD8+ T cells (b) and CD4+ T cells (c), and the relative abundance of CXCR5+CD45RA− cells and naive (CXCR5−CD45RA+) cells among CD4+ cells (d). Full gating strategy, Supplementary Figure 3. (e) Proliferation of CD4+ and CD8+ T cells at day 4 after stimulation with anti-CD3 and anti-CD28. (f) Expression of CD25 and CD69 24 h after stimulation with anti-CD3 and anti-CD28. (g) TCR Vβ spectra-typing of the patient’s peripheral blood T cells. (h) Expression of perforin and granzyme B on the patient’s CD8+ T cells. (i) Killing of target cells by CD8+ T cells with (right) or without (left) OKT3. CTL, cytotoxic T lymphocytes. (j) Microscopy of intracellular Ca2+ flux in immortalized T cell lines following stimulation with anti-CD3, assessed by detection of the Fura-2 excitation ratio. (k) Cropped immunoblot analysis of total and phosphorylated (p-) ERK in primary T cells after stimulation with anti-CD3 and anti-CD28. (l) Quantification of the results in k. (m) Geometric mean (GM) of phosphorylated ERK in the patient’s CD8+ T cells and shipment control CD8+ T cells transfected to express a vector encoding wild-type RASGRP1 or a vector encoding green fluorescent protein (GFP) as control. Data are representative of eight (a), four (b,c), three (e,f,h,i,m) or two (d,g,j,k) independent experiments.
Both CD4+ T cells and CD8+ T cells from the patient exhibited an exhausted phenotype, as indicated by expansion of CCR7−CD45RA+ effector memory T cell populations, which represented up to 80% of CD8+ T cells (Fig. 2b,c, Supplementary Table 3 and Supplementary Fig. 3). In the CD4+ T cell subset, the majority of cells were CD45RA−CXCR5+ (Fig. 2d), suggestive of an increased frequency in peripheral blood follicular helper T cells compared with healthy controls5,20,21. In the CD4+ compartment of the patient, normal proportions of CD25+FOXP3+CD4+ regulatory T cells and recent thymic emigrants were present (Supplementary Table 3).
Functionally, both RASGRP1-deficient CD4+ T cells and RASGRP1-deficient CD8+ T cells showed defective proliferation after stimulation with antibody to the TCR invariant chain CD3 (anti-CD3) and antibody to the costimulatory molecule CD28 (anti-CD28) (Fig. 2e and Supplementary Fig. 3), accompanied by impaired expression of the activation markers CD25 and CD69 (Fig. 2f). Defective T cell proliferation was recapitulated in Jurkat human T cells with doxycycline-inducible short hairpin RNA-based knockdown of RASGRP1 (shRASGRP1) (Supplementary Fig. 2b). In addition, proliferation assays using thymidine incorporation also showed defective cellular proliferation after nonspecific stimulation with various mitogens (Supplementary Fig. 2c). Consistent with the above-mentioned proliferation defects, TCR Vβ spectratyping of patient peripheral blood T cells revealed a restricted TCR repertoire (Fig. 2g and Supplementary Fig. 2d). CD8+ cytotoxic T cell populations obtained from the patient and expanded ex vivo showed increased expression of perforin and granzyme B compared to healthy donors (Fig. 2h), as well as defective killing of targets cells following activation with the monoclonal antibody against CD3 (OKT-3) (Fig. 2i).
Published studies have shown that RASGRP1 is critical for TCR-induced signal transduction in mouse T cells5,16. Here we sought to investigate the functional consequences of RASGRP1 deficiency on the activation of human T cells. In both inducible shRASGRP1-Jurkat cells and in a patient T cell line, immortalized using human telomerase reverse transcriptase (hTERT) we did not observe defects in initial TCR signaling events upstream of RASGRP1, including intercellular adhesion molecule 1 (ICAM1) ring formation, exclusion from the central supramolecular activation complex (cSMAC), or calcium flux (Fig. 2j and Supplementary Fig. 2e–g). Thus, we focused on potential signaling defects downstream of RASGRP1 in populations of primary peripheral blood mononuclear cells (PBMCs) obtained from patient and expanded ex vivo using anti-CD3 and anti-CD28. Cells were serum- and antibody-starved and re-stimulated with anti-CD3 and anti-CD28. We observed reduced, but not absent, phosphorylation of ERK1/2 (at Thr202 and Tyr204) in RASGRP1-deficient cells compared to healthy donor expanded T cells (Fig. 2k,l and Supplementary Fig. 2h). Transient transfection of the cells with vector encoding wild-type RASGRP1 normalized impaired activation of ERK in the patient’s CD8+ T cells (Fig. 2m).
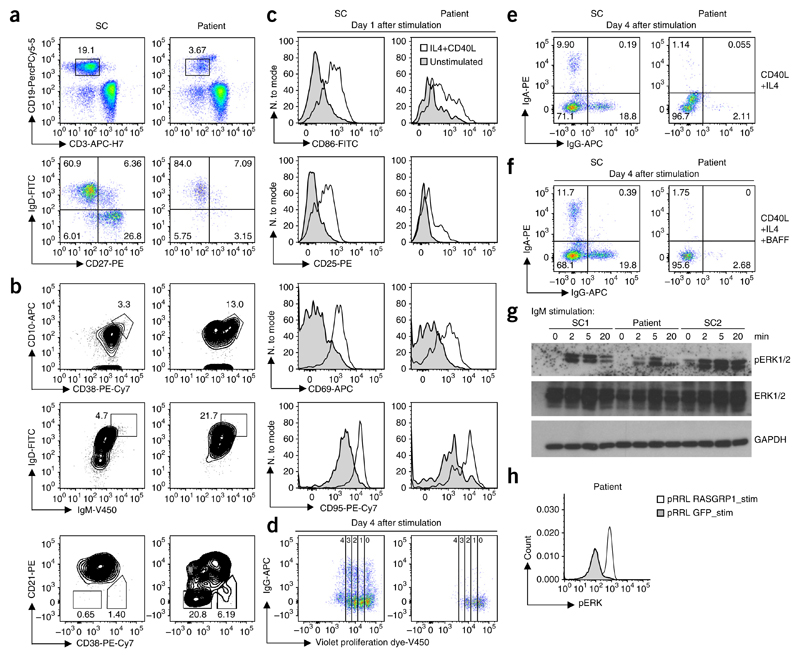
Deficient B cell activation and proliferation
Studies of the role of RASGRP1 in mouse B cells have remained contradictory22,23. We performed detailed immunophenotypic analysis of the patient’s B cell subsets and observed decreased proportions of memory B cells (CD19+CD27+) and increased proportions of both transitional B cells and CD21lo B cells, the latter of which consituted up to 20% of all CD19+ B cells, compared to healthy donor B cell subsets (Fig. 3a,b and Supplementary Fig. 3). The concentrations of total immunoglobulin G (IgG) and IgM were normal in the patient, but IgA concentrations were elevated (Supplementary Table 1). Although findings from mouse studies have suggested a role for RASGRP1 in the development of autoimmunity22 and genome-wide association studies have identified an association of single-nucleotide variants in the gene encoding RASGRP1 with systemic lupus erythematodes24, no clinical signs of autoimmunity were apparent, consistent with the absence of detectable autoantibodies (Supplementary Table 7). However, we observed insufficient vaccination responses, including absent antibodies to hepatitis B surface antigen and decreased pneumococcusspecific antibodies following vaccination (data not shown).
Figure 3. RASGRP1 deficiency results in a B cell proliferation and activation defect.
(a) Showing relative proportion of naive (IgD+CD27−) B cells, memory non-switched (IgD+CD27+) B cells and memory-switched (IgD−CD27+) B cells. Full gating strategy, Supplementary Figure 3. (b) Flow cytometry of gated CD19+ B cells showing transitional T1 (CD10+CD38+) B cells, transitional (IgD+IgM+) B cells and activated (CD21−CD38lo) B cells. (c) Flow cytometric expression of CD86, CD95, CD25 and CD69 of CD19+ cells 1 d after stimulation with anti-CD40L and IL-4. (d) Proliferating B cells at day 4 after stimulation as in c. (e) IgA+ and IgG+ switched B cells at day 4 stimulation. (f) IgA+ and IgG+ switched B cells at day 4 stimulation as in c with (bottom) or without (top) the addition of BAFF. (g) Cropped immunoblot analysis of total and phosphorylated ERK in EBV-transformed B cells from the patient, assessed after stimulation with IgM. (h) Phosphorylated ERK after gene transfer using a lentiviral backbone (pRRL) with sequence encoding wild-type RASGRP1 or GFP in EBV-transformed B cells from the patient, after stimulation with IgM. Data are representative of four (a,b), two (c,d,g,h) or one (e,f) independent experiment(s).
To assess B cell function, we stimulated the patient’s PBMCs with antibody to the costimulatory receptor CD40 and interleukin 4 (IL-4)25. The patient’s B cells, compared to healthy donor B cells, showed reduced upregulation of expression of the activation markers CD86, CD25, CD95 and CD69 (Fig. 3c) that resulted in decreased B cell proliferation (Fig. 3d) and reduced class-switch recombination following stimulation (Fig. 3e). The observed defects could not be reversed by the addition of the mature B cell survival factor BAFF (Fig. 3f), which suggested that self-reactive transitional B cells were absent from the PBMC compartment26.
Both quantitative defects and qualitative defects in B cells in patients with inherited deficiencies of T cells result from either an intrinsic B cell defect (primary defect) or deficient co-stimulation11,12. To assess our hypothesis of a B cell–intrinsic phenotype, we evaluated B cell antigen receptor (BCR) signaling in EBV-transformed B cell lines from the patient. The EBV-immortalized B cells showed reduced activation of the MAPK pathway following IgM stimulation, similar to the T cell defects that we observed (Fig. 3g). Defective ERK phosphorylation in patient B cells was reversed after lentiviral transduction of wild-type RASGRP1 (Fig. 3h). Collectively, the patient’s B cells showed defective MAPK activation, with defective BCR signaling resulting in reduced proliferation and class switching.
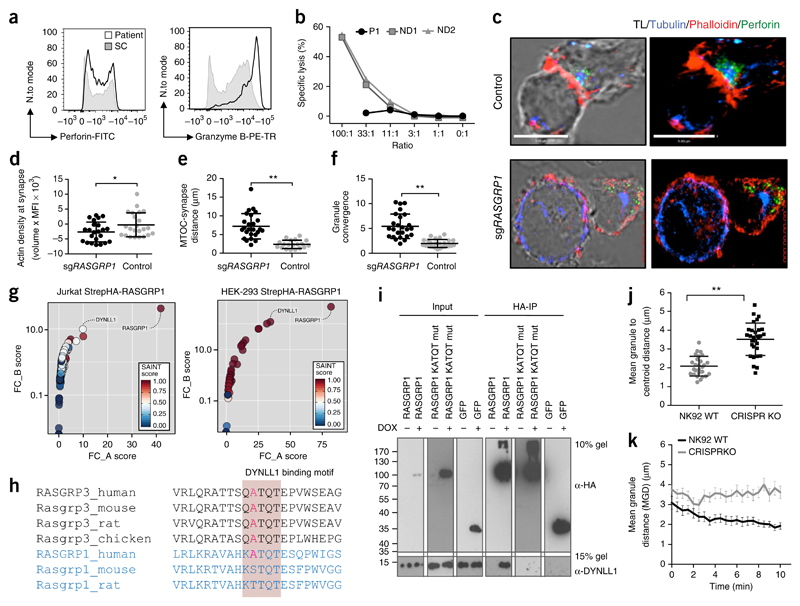
RASGRP1 deficiency causes impaired NK-cell cytotoxicity
Mouse studies have not yet addressed the role of RASGRP1 in NK cells, to the best of our knowledge. We observed unaltered numbers of the patient’s NK cells with normal CD56dim and CD56bright subset distributions (Supplementary Table 1). However, the CD56bright NK cells had high expression of intracellular IL-5 (Supplementary Fig. 4a). These data might suggest that there is a developmental bias toward the production of T helper type 2 cytokines at an intermediate differentiation state, as production of these cytokines is lost following terminal differentiation27. Despite having high protein expression levels of intracellular granzyme B and perforin (Fig. 4a), the patient’s NK cells exhibited severely impaired cytolytic function (Fig. 4b). This impairment was associated with defective formation of a mature NK cell immunological synapse in CRISPR/Cas9-edited NK-92 cell lines deficient in RASGRP1 (sgRASGRP1) (Fig. 4c) with decreased F-actin accumulation, increased distance from the microtubule-organizing center (MTOC) to the synapse and impaired granule convergence (Fig. 4d–f), consistent with impaired formation of the NK cell immunological synapse in primary cells from the patient (Supplementary Fig. 4b).
Figure 4. Aberrant cytoskeletal dynamics in NK cells.
(a) Flow cytometry analyzing the intracellular accumulation of perforin and granzyme B in NK cells from the patient and control donors. Full gating strategy, Supplementary Figure 4. (b) Chromium (51Cr)-release assay of NK cell cytotoxicity. E/T, effector cell/target cell. (c) Confocal microscopy of a fixed RASGRP1 wild-type NK-92 control cell (top) and an NK-92 cell with CRISPR-mediated editing of RASGRP1 (sgRASGRP1) (bottom) conjugated to K562 cells, detected with anti-α-tubulin (blue), anti-perforin (green) and phalloidin-stained F-actin (red). TL, transmitted light. (d–f) Actin density at the synapse (d), distance from lytic granule to MTOC (e), and granule convergence to MTOC (f) of RASGRP1 wild-type NK-92 control cells and sgRASGRP1 NK-92 cells. (g) Tandem affinity purification of RASGRP1 in Jurkat cells (left) and HEK cells (right), presented as FC-A scores versus FC-B scores. (h) Conservation of the KATQT motif in RASGRP1 and RASGRP3 across various vertebrate species. (i) HA-based co-immunoprecipitation of Strep-HA–tagged wild-type (WT) RASGRP1, KATQT-mutant (mut) RASGRP1 or Strep-HA–tagged-GFP with endogenous DYNLL1. (j,k) Video microscopy of granule convergence in sgRAGSRP1 and control NK cells. *P < 0.05 and **P < 0.0001 (unpaired Student’s t test). Data are representative of four (c), three (a,c–f,i), two (g,j,k) or one (b) independent experiment(s) (mean and s.d. in d–f,j,k).
Proteomics links RASGRP1 to cytoplasmic dynein
The patient’s CD8+ T cells displayed normal expression of ICAM1 and did not show any defects in ICAM1 ring formation or exclusion from the central supramolecular activation complex following TCR stimulation (Supplementary Fig. 2f), which suggested that the cells retained the required elements for formation of the immunological synapse. In addition, the patient’s NK cells showed normal expression of NK cell–activating receptors, as well as similar levels of granzyme B and perforin protein expression compared to healthy donor NK cells (Fig. 4a). However, we observed an unusual combination of defective granule convergence together with defects in the accumulation of F actin in both the patient’s NK cells and sgRASGRP1 NK-92 cells. Assuming a similar role for MAPK activation in actin polymerization in both T cells and NK cells28, we hypothesized that the defect in granule convergence might be linked to an as-yet-unknown interaction partner. Thus, we performed tandem affinity purification of RASGRP1 tagged at the amino terminus with Strep-HA plus subsequent mass-spectrometry analysis of bound interactors in HEK293T cells and Jurkat T cells (Fig. 4g). In both HEK293T cells and Jurkat T cells which were engineered for doxycycline-inducible RASGRP1 expression, the dynein light chain DYNLL1 was one of the top-ranking interacting proteins according to both SAINT and FC-scoring (Fig. 4g). Notably, published studies of mice and rats have reported binding of RASGRP3, but not of RASGRP1, to DYNLL1 (ref. 29). Such cross-species differences might be explained by the fact only human RASGRP1 contains a binding motif similar to that of mouse RASGRP3 (ref. 30) (Fig. 4h). Co-immunoprecipitation of RASGRP1 tagged at the amino terminus with Strep-HA confirmed the interaction of RASGRP1 with endogenous DYNLL1 in both Jurkat T cells and HEK293T cells (Fig. 4i, Supplementary Table 8a,b and Supplementary Fig. 4d). Substitution of the assumed DYNLL1-binding motif with alanine residues resulted in abrogation of the binding of DYNLL1 (Fig. 4i), which confirming the critical role of this motif in the RASGRP1-DYNLL1 interaction.
Cytoplasmic dynein can mediate retrograde transport over the microtubule cytoskeleton toward the MTOC in T cells, which results in centrosome polarization following TCR activation31. In NK cells, dynein-driven transport of lytic granules along microtubules is required for convergence to the MTOC preceding formation of the immunological synapse32. Visualization of sgRASGRP1 NK cells by video microscopy revealed markedly defective granule motility (Fig. 4j,k). Consistent with a putative role for RASGRP1 in regulating the microtubule cytoskeleton, accumulation of dynein before centrosome polarization has been shown to depend on the RASGRP1 activator DAG33. Moreover, lytic granule convergence occurs rapidly after target-cell engagement and is independent of actin dynamics32. Thus, RASGRP1 deficiency markedly affects NK cell cytotoxicity by impairing both actin polymerization and the convergence of lytic granules to the MTOC, which is possibly explained by an abrogated interaction with DYNLL1.
Reversal of cellular defects by lenalidomide
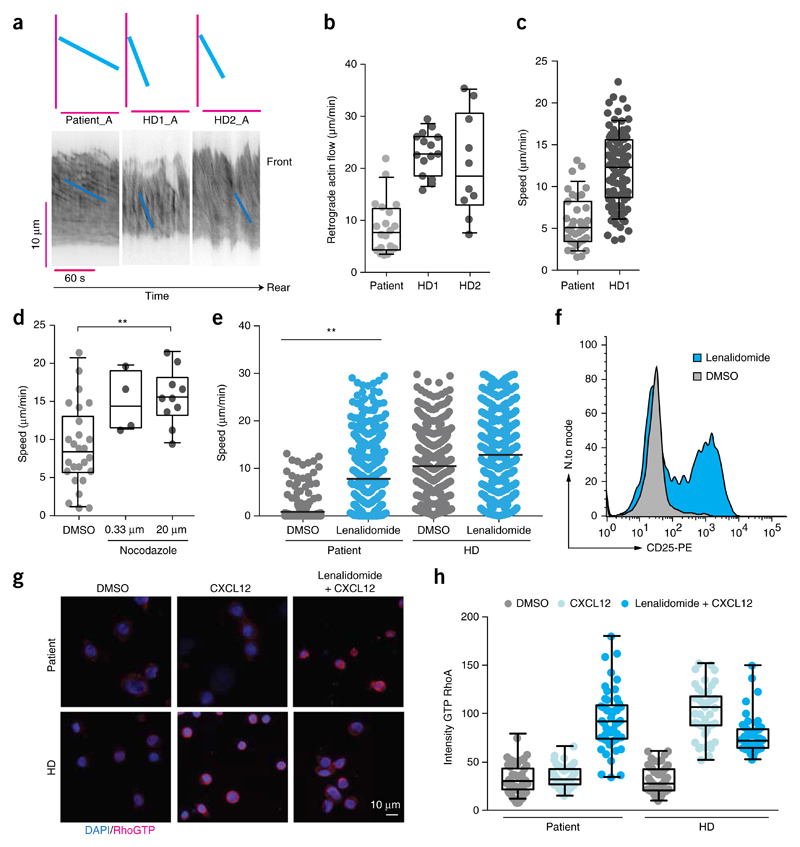
Apart from NK-cell cytotoxicity, microtubules and actin also depend on precisely coordinated interactions to establish cell polarity34 and the coordinated cell migration that is essential for lymphocyte function34. To visualize cytoskeletal dynamics and cell migration in primary cells from the patient, we transiently transfected CD8+ T cells with the actin marker Lifeact™. We observed significantly slower actin turnover in RASGRP1-deficient CD8+ T cells than in CD8+ T cells from healthy donors (Fig. 5a,b). To assess whether slow actin turnover would also ‘translate’ into a low cell-migration speed, we performed cell-migration assays with CD8+ T cells stimulated with the chemokine CXCL12 on ICAM-1-coated plates. Indeed, RASGRP1-deficient cells migrated at lower speeds than did CD8+ T cells from healthy donors (Fig. 5c).
Figure 5. RASGRP1 deficiency leads to cell-migration defects that are reversed following treatment with lenalidomide.
(a) Kymographs of retrograde actin flow in CD8+ T cells from the patient and healthy donors after transfection with Lifeact. Each kymograph represents one cell. (b) Quantification of retrograde actin flow in CD8+ T cells from the patient and two healthy donors. Each symbol represents one cell. (c) Migration speed of CD8+ T cells from the patient and healthy donors. (d) Migration speed of CD8+ T cells from the patient and healthy donors following the addition of nocodazole. (e) Migration speed of PBMCs from the patient and healthy donors after the addition of lenalidomide. (f) Flow cytometry analyzing the expression of CD25 on proliferating T cells obtained from the patient and incubated with lenalidomide (blue) or DMSO (gray). (g) Immunofluorescence microscopy of CD8+ T cells from the patient and a healthy control that underwent population expansion and were pre-treated for 16 h with either lenalidomide or DMSO and were activated for 5 min with CXCL12, then were stained for active RhoA-GTP (red) and counterstained with the DNA-binding dye DAPI (blue). (h) Quantification of immunofluorescence images as in g (50 cells per condition) by measurement of the mean intensity of total RhoA and RhoA-GTP. (P > 0.05), *P ≤ 0.05 (e), **P < 0.01 (e) and ***P ≤ 0.0001 (ANOVA (e,d)) Data are representative of four (c), or two (a,b,e,f,g) independent experiments.
RASGRP1-deficient mouse mast cells display decreased RhoA activity35. We found that levels of active RhoA were markedly lower in the patient’s T cells following CXCL12 stimulation compared with healthy donor T cells, whereas total RhoA levels differed only mildly (Supplementary Fig. 5). Thus, we hypothesized that activation of RhoA signaling might reverse the migration defect of RASGRP1-deficient CD8+ T cells. Indeed, the known RhoA activator nocodazole normalized the observed migration defect (Fig. 5d). However, as nocodazole is not amenable for clinical use, we sought to identify an alternative compound that could induce similar functional effects. In patients with chronic lymphocytic leukemia, T cells exhibit an exhausted (immunodeficient) phenotype that is similar to that of the RASGRP1-deficient patient36,37. The thalidomide-derived drug lenalidomide can modulate the activity of key transcription factors that are important for restoring T cell function, which results in increased RhoA activity38. Indeed, treatment with lenalidomide significantly increased cell velocity and thus normalized the overt migration defect observed in RASGRP1-deficient PBMCs (Fig. 5e); thus, this might represent a potential treatment option for RASGRP1-deficient patients. Consistent with the published positive effect of lenalidomide on IL-2 secretion39, we observed increased CD25 expression in T cells following lenalidomide treatment (Fig. 5f). Given that lenalidomide modifies multiple targets of the Cullin–E3 ligase complex by binding to its intracellular target cereblon, we aimed to investigate whether the effect of lenalidomide on the cellular phenotype could be linked to normalization of RhoA signaling. In CD8+ T cells from healthy control donors, stimulation with CXCL12 activated RhoA, as detected with an antibody specific for the GTP-bound state of RhoA (Fig. 5g,h). In contrast, CD8+ T cells from the RASGRP1-deficient patient failed to activate RhoA following stimulation with CXCL12 (Fig. 5g,h). Notably, treatment of the patient’s CD8+ T cells with additional lenalidomide restored active RhoA to amounts comparable to those measured in the control CD8+ T cells (Fig. 5g,h).
Discussion
We identified human RASGRP1 deficiency as a previously unknown primary immunodeficiency with notable defects in the lymphoid compartment that were amenable to partial reversal with lenalidomide. Although our study was performed on a pedigree with a single affected patient, our results demonstrated that the loss of RASGRP1 caused the phenotype, consistent with outlined criteria as published40: the patient’s candidate genotype was monogenic and did not occur in subjects without the clinical phenotype; we found that the detected mutation caused the absence of full-length RASGRP1 protein and reduced ERK phosphorylation; we observed restoration of the defective phosphorylation of ERK following expression of wild-type RASGRP1 in the patient’s cells; and shRNA and CRISPR-based studies of Jurkat cells and NK-92 cell lines reproduced the phenotype observed in primary cells from the patient. The phenotype of human RASGRP1 deficiency differs substantially from previously described mouse models5,22 and suggests a previously unknown role for RASGRP1 in cellular responses beyond TCR signaling. Although RASGRP1-deficient mice exhibit decreased numbers of both CD4+ and CD8+ single-positive thymocytes5, the patient’s immunological phenotype showed an inverted ratio of CD4+ T cells to CD8+ T cells with population expansion of exhausted effector memory cells, consistent with a partial T cell defect9. Partial T cell defects are often associated with autoimmunity9. The role of RASGRP1 in autoimmunity has remained controversial. Both RASGRP1-deficient mice22 with an autoreactive BCR and Rasgrp1−/− mice overexpressing a truncated form of RASGRP1 that lacks the carboxy-terminal tail domain41 develop a lymphoproliferative disorder with features reminiscent of human systemic lupus erythematosus. We did not observe overt autoimmunity in human RASGRP1 deficiency. Future studies, including more RASGRP1-deficient subjects, will enable delineation of the full phenotypic spectrum of human RASGRP1 deficiency.
The role of RASGRP1 in NK cells has not been addressed in detail in mouse models. NK cells are innate immune cells best known for their ability to mediate cytotoxicity after the ligation of germline-encoded activation receptors42. A variety of human primary immunodeficiencies are associated with defective accumulation of actin at the immunological synapse, particularly actin-polymerization deficiencies such as Wiskott-Aldrich syndrome43 or deficiency in the actin regulators WIP44 or DOCK8 (ref. 45). However, the combination of defective polymerization of actin, together with the considerable granule-convergence defect seen in cells from the RASGRP-deficient patient has not been seen in human immunodeficiencies before, to our knowledge.
Accumulation of dynein before centrosome polarization in lymphocytes depends on the known RASGRP1 activator DAG46. Evidence has revealed a RAS-binding domain in the dynein intermediate chain that probably mediates activation of the complex47. Our discovery of an interaction between human RASGRP1 and cytosolic DYNLL1 provides evidence that RASGRP1 mediates granule convergence following activation, a dynein-dependent process32. To our knowledge, RASGRP1 deficiency represents the first association of impaired dynein function with a human primary immunodeficiency. Of note, several viral proteins, including the HIV-1 integrase, interact with DYNLL1, and their interaction has been shown to be involved in efficient reverse transcription of HIV-1 (ref. 48).
Our study has also identified a targeted treatment strategy with lenalidomide, providing proof-of-concept for such approaches in primary (and potentially secondary) immunodeficiencies with aberrant cytoskeletal dynamics. Thus far, the thalidomide derivative lenalidomide has been approved for various lymphoid malignancies, such as multiple myeloma37 and chronic lymphocytic leukemia36. In these diseases, T cells are in a state of pseudo-exhaustion and show defects in activation, proliferation and cytotoxicity49, similar to those observed in RASGRP1 deficiency. The action of lenalidomide has been linked to cereblon which, together with the scaffolding protein CUL4 and the catalytic subunit ROC1, forms the E3 ubiquitin ligase cereblon-CRL4 complex that mediates degradation of the transcription factors Ikaros and Aiolos and may alter other targets of the E3 ubiquitin ligase38. These processes result in, among other effects, the upregulation of RhoA activity, which might be the factor responsible for the rescue of RASGRP1-deficient lymphocytes36; we found that treatment with lenalidomide significantly increased RhoA activity in patient CD8+ T cells. In sum, we have identified RASGRP1 deficiency as a previously unknown type of partial T cell defect, which revealed a previously unknown link between RAS signaling and cytosolic dynein dynamics and which was amenable to improvement following RhoA activation and lenalidomide treatment.
Online Methods
Patient and ethics
The patient was included with informed written consent by the parents and approval from the Institutional Review Boards of the Medical University of Vienna and Ankara Hacettepe University Medical School.
Homozygosity mapping
Affymetrix 6.0 single nucleotide polymorphism (SNP)-based homozygosity mapping was performed as previously described50.
Exome sequencing
DNA extraction, exome sequencing and data analysis were performed as described previously50.
Sanger sequencing
Primers for the variants detected with exome sequencing were designed using PrimerZ. PCR amplification and capillary sequencing was performed as previously described51.
Statistics
Statistical tests used in this study include Student’s t test, with or without Welch’s correction, as appropriate and detailed in the respective methods sections.
Mutational profile and population genetics
The Combined Annotation Dependent Depletion (CADD) score was determined using CADD version v1.3 (ref. 52). The mutational and population genetics profile of RASGRP1 CCDS (ENSG00000172575) was obtained from ExAC (http://exac.broadinstitute.org/gene/ENSG00000172575, accessed 29th April, 2016). The Residual Variation Intolerance Score was obtained from Genic Intolerance (http://genic-intolerance.org, accessed April 29, 2016). A binomial test was used to assess statistical significance of the deviation of the theoretically expected distribution of nonsense variants (expected) from the observed nonsense variants (observed) based on the ExAC database. Loss-of-function carrier frequency (q) was calculated as: q = (SUM of putative loss-of-function population alleles)/(Total population alleles). Expected rate of nullizygous (1 in X non-consanguineous conceptions) variants was calculated as: X = 1/q2. If the Sum of putative loss of function alleles was 0, q⋆ was calculated as: q⋆ = 1/(Total population alleles), and expected rate of nullizygous variants: X = 1/(q_⋆2).
Flow cytometry
All flow cytometric analyses were recorded on a BD LSR Fortessa except NK-cell immunophenotyping (recorded on both BD LSR Fortessa and FACS Canto) and cell proliferation assays upon shRNA-mediated knockdown (recorded on BD FACS Calibur). All data were analyzed using FlowJo X (TreeStar) and data was graphed with Prism 6.0 (GraphPad Software).
Immunophenotyping
PBMCs immunophenotyping was performed on an LSR-Fortessa (BD Biosciences). Antibodies were used as described previously51 apart from: GranzymeB-AF700, TCRαβ-FITC, TCRαβ-PE, TCRγδ-APC, TCRγδ-PE, TCR-PE, all from BD Biosciences; TCR Vα7.2-APC, Ig-PE both from Miltenyi Biotec; Perforin-AF647 from BioLegend; CXCR5-APC from Biomedica.
Cell lines
Feeder cell-mediated T-cell expansion
Feeder T-cell generation was done as described53,54. Obtained expanded T cells were evaluated by) flow cytometry for expression of CD4, CD8, perforin and granzyme B and purified as necessary using a MagniSort CD8 cell separation kit (eBioscience).
Generation of EBV-immortalized B-cell lines
Patient and healthy donor PBMCs were isolated using Ficoll density centrifugation. 106 freshly isolated PBMCs were incubated with EBV virus supernatant and on the following day 1 μg/ml cyclosporin A was added to the cells. Cells were maintained in complete RPMI medium (Gibco) with 10% FCS, 1% L-glutamine and 1% PenStrep (all from Invitrogen).
HEK293T cells
Human RASGRP1 cDNA and the mutant version thereof were cloned into pTO-SII-HA-GW vectors using gateway recombination to generate N-terminal Streptavidin-Hemagglutinin-tagged fusion protein and transfected into HEK293 Flp-In-TREx cells (Thermo Fisher Scientific) as described55,56.
Jurkat cells with inducible overexpression of RASGRP1
Jurkat E6.1 cells (ATCC) were retrovirally infected with a retroviral vector (MSCV-rtTA3-IRES-EcoR-PGK-Puro) as described previously57,58. A codon-optimized version of RASGRP1 cDNA (IDT systems) was cloned into pSIN-TREtight-SH-gw-N-IRES-mCherry-PGK-BlastR as described57.
Inducible shRNA mediated knock down of RASGRP1 in Jurkat cells
Short hairpin RNAs were designed according to published recommendations57. Jurkat T-cell line expressing an rtTA3 element with an ecotropic receptor as well as PlatE cells for virus production were kindly provided by Johannes Zuber (IMP). Cell lines were tested for protein expression 24h after induction with 1 μg/ml doxycycline using flow cytometric analyses. GFP marks successful transfection whereas dsRed expression indicates protein expression after doxycycline addition. The following short hairpin RNAs were used:sh_RGRP1:UGAAACACUAACUACAAGCUA;sh_RGRP2: UUAACUUGUAUACUAUACCUC. Each shRNA has 2 off-targets sites that correspond to known RASGRP1 isoforms and no XhoI/EcoRI restriction sites.
CRISPR-mediated gene deletion in NK-92 cell lines
CRISPRs targeting RASGRP1 were designed as described59 and cloned into a lentiCRISPRv2 vector60. NK-92 cells (ATCC) were cultivated according to the supplier’s instructions. Lentiviral virus production and cell infection were performed according to standard procedures. Limited dilution was performed and single cell clones were selected. Editing at genomic position was confirmed using capillary sequencing and subsequent upload into tide (http://tide.nki.nl/, accessed November, 2014)61. Only clones with frameshift mutations in both alleles of more than 90% were used for further experiments. Following CRISPR targeting sequences were used for cloning: C_RASGRP1_E2_p1_A_t:CACCgAGACACCATCATTCGGAACT; C_RASGRP1_E2_p1_A_b:AAACAGTTCCGAATGATGGTGTCTc; C_RASGRP1_E2_p2_A_t:CACCgGAGACACCATCATTCGGAAC; C_RASGRP1_E2_p2_A_b:AAACGTTCCGAATGATGGTGTCTCc.
Immunoblot experiments
RASGRP1 immunoblot experiments
4 × 106 Patient and healthy donor EBV-immortalized B cells were lysed in 50 μl Laemmli Buffer, ran on a 10% acrylamide gel at 110 V and blotted overnight at 120 mA at 4 °C on a PVDF membrane. Membranes were incubated with anti-human RASGRP1 (Merck Millipore, Clone 10.1) in Tris-buffered saline with 0.5% Tween and 5% milk overnight at 4 °C and a secondary anti-mouse antibody at ~21 °C for 2 h. Blots were developed using ECL (Thermo Scientific) and films (GE healthcare).
TCR and BCR signaling analysis
Patient or healthy donor derived T-cell lines were starved for up to 4 h in RPMI medium (Invitrogen) containing 1% FCS (Sigma-Aldrich), 0.2 mM L-glutamine (Sigma-Aldrich), 10 μg/ml penicillin/streptomycin (Sigma-Aldrich and 20 mM HEPES (Gibco). Cells were put on ice for at least 20 min and ice-cold stimulation mix containing mAb to CD3 (clone OKT-3, purified, 2 μg/ml) and mAb against CD28 (clone Leu28, purified, 1 μg/ml, BD Bioscience). Cells were stimulated for indicated time points, washed in ice-cold PBS and lysed in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA (all from Roth) and complete protease inhibitor tablets (Roche), 1 μM orthovanadate, 50 mM NaF, 1% Triton X-100 (Thermo Scientific). Upon addition of Laemmli buffer samples were boiled for 5 min and loaded on 8 or 10% acrylamide gels. Gels were run at constant voltage and blotted over night with constant 120 mA at 4 °C using a mini-protean wet blotting system (BioRad). Immunoblotting was performed according to standard conditions with Tween-buffered saline and 5% bovine serum albumin (Roth) according to the manufacturer’s instructions. Antibodies are summarized below.
Prior to stimulation, B cells were serum starved for 3–4 h and stimulated with anti-IgM 500 ng/ml (Southern Biologicals) for the indicated time points. Subsequent handling was performed as described for TCR signaling analyses: p44/42 MAPK (ERK1/2) (clone 137F5), Phospho-p44/42 (Thr202/Tyr204) MAPK (ERK1/2), (clone D13.14.4E; all from Cell Signaling), GAPDH (clone 86C5, Santa Cruz Biotechnology).
Tandem affinity purification and mass spectrometry analyses
Tandem affinity purification and mass spectrometry analyses were performed as described62. Data were uploaded to CRAPome database for FCA, FCB and Saint score calculations63.
Co-immunoprecipitation of RASGRP1 and DYNLL1 in HEK293 cells and Jurkat T cells
Immunoprecipitation was performed by lysing induced cells in IP buffer (50 mM HEPES pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 1 mM Na3VO4, protease inhibitors from Sigma-Aldrich). Lysates were precleared with Protein G–Sepharose beads (GE Healthcare) and tagged RASGRP1 protein purified using anti-HA agarose beads (Sigma-Aldrich). Immunoprecipitates were washed with wash buffer (50 mM HEPES pH 8.0, 300 mM NaCl, 5 mM EDTA, 1% NP-40, 50 mM NaF, 1 mM Na3VO4, protease inhibitors from Sigma-Aldrich), and analyzed by SDS-PAGE separation, blotting and immunostaining using antibodies against DYNLL1, GAPDH (both Santa Cruz Biotechnology) and HA tag (Sigma-Aldrich; 1:1,000 dilution for DYNLL1 antibody and 1:2,000 dilution for others). 1/40 of the lysate used for immunoprecipitation was used as input control.
T-cell proliferation
Thymidine incorporation assays were performed as described51. Flow cytometry-based proliferation assays were performed using Violet proliferation dye 450 (BD Biosciences). In brief, cells were labeled with proliferation dye according to manufacturer’s instructions and incubated with stimulating antibodies 1 μg/ml anti-CD3 (clone OKT3/Leu-4, T3, eBiosciences) and 0.5 μg/ml anti-CD28 (clone T44, CD28.2, eBiosciences) for up to 4 d. Proliferation of shRNA-transfected Jurkat T-cell lines was performed by labeling cells with V450 proliferation dye and subsequent knockdown induction with 1 μg/ml doxycycline. For proliferation studies samples were taken on consecutive days and analyzed by flow cytometry after staining with additional markers (see Immunophenotyping section).
Flow-cytometry-based T-cell cytotoxicity
Assays to quantify the cytotoxic activity of CD8+ T cells were done as previously described with minor modifications64. In brief, GFP-expressing P815 target cells were treated with aphidicolin to inhibit proliferation and pre-incubated either with or without OKT-3 (1 μg/ml). Different ratios of target cells and CD8+ T cells were distributed in 96-well U-bottom low-cell-binding plates (Nunc). Following 6 or 24 h of incubation, cells were stained with 7-AAD and residual alive target cells were evaluated.
T-cell and B-cell reconstitution experiments
T cells
Patient and healthy donor PBMCs were thawn and transfected with a GFP control or wild-type N-terminally GFP-tagged RASGRP1 using the Amaxa human T-cell nucleofector kit (Lonza) and program V024 (unstimulated T cells). Cells were rested for 1 d in complete IMDM, starved for 4 h in serum-free medium and stimulated for 30–40 min with stimulating antibodies 1 μg/ml anti-CD3 (clone OKT3/Leu-4, eBiosciences) and 0.5 μg/ml anti-CD28 (Clone T44, CD28.2, eBiosciences). Cells were fixed in 100 μl IC-Fixation solution (eBioscience) for 20–60 min, washed 2x in Fixation/Permeabilization buffer (eBioscience) and stained with antibodies against pERK, CD4, CD8 and CD3 (see Flow Cytometry section) for 1 h ~21 °C. Samples were washed in 1× in Fixation/Permeabilization buffer and 1× in PBS and analyzed by flow cytometry gating on GFP-positive cells.
B cells
Patient and healthy donor B-cell lines were transduced using a lentiviral vector (pRRL_EF1a_StrepHA_IRES_GFP), with second generation packaging plasmids (PAX2 and MD2G) kindly provided by the Giulio Superti-Furga laboratory (CeMM). For phospho-ERK analyses, transduced B-EBVs were serum-starved for 2–3 h and stimulated using IgM antibody (Southern biologicals) for 30–40 min. Staining was performed as described for T cells with pERK and CD19.
B-cell class switch recombination
B-cell class switch recombination (CSR) experiments were performed on freshly isolated PBMCs. Cells were labeled with Violet proliferation dye 450 (BD Biosciences) according to manufacturer’s instructions and seeded into a 96-well plate at 2 × 105 cells/well in IMDM medium supplemented with 10% FCS, 5% L-Glutamine, penicillin/streptomycin, 1% HEPES and beta-mercaptoethanol. B-cell proliferation and CSR were induced using CD40L (1/800 dilution of the titrated supernatant and IL-4 (30 ng/ml) or CD40L/IL-4/BAFF (1/1,000 dilution of the titrated supernatant; both kindly provided by H. Eibel, Center for Chronic Immunodeficiency) for 4 d. The activation status of the cultured B cells was measured 1 d upon induction by flow cytometry using CD19, CD86, CD25, CD69 and CD95 antibodies. Cell proliferation was measured by monitoring the dilution of the proliferation dye, and CSR by monitoring IgG expression on the surface on CD19+ B cells using flow cytometry after 4 d in culture.
Cell migration assays following lenalidomide treatment
Agarose based assays
Human CD8+ T cells were purified using MagniSort Human CD8+ T-cell Enrichment Kit (eBioscience) and labeled with 5 μM CMTMR (Invitrogen) 1 d before imaging and incubated with 1 μM lenalidomide or DMSO for 24 h, and recorded migrating confined under agarose block. Briefly, home-made glass bottom dishes were coated with 2 μg/ml human ICAM-1 Fc (R&D) at 4 °C overnight and afterwards rinsed with PBS to remove residual, unbound protein. 500 μl of 0.5% agarose (Biozym Gold Agarose) solution containing 100 ng/ml human CXCL12 (R&D), 50 μM ascorbic acid and fully supplemented R10 medium (RPMI 1640, 10% FCS, glutamine, non-essential amino-acids, β-mercaptoethanol) were poured onto the coated dishes to form approximately 3-mm thick layer and allowed to solidify for 1 h at 4 °C. Finally, dishes were equilibrated for 30 min in a humidified incubator at 37 °C, 5% CO2 and cells were injected between the glass and the agarose layer. Cells were recorded at 37 °C on Nikon Eclipse Ti inverted microscope equipped with Hamamatsu EMCCD C9100-02 camera and tracked using Volocity software (PerkinElmer).
Video microscopy without agarose
The experiment was carried out as described36. Stimulation of PBMCs with 1 μM lenalidomide or DMSO was performed for 20 h before microscopy.
Actin retrograde flow assay
Isolated human CD8+ T cells were nucleofected with plasmid expressing Lifeact-GFP using Nucleofector II and Mouse T-cell Nucleofector Kit (Lonza) 1 d before imaging. Glass bottom dishes (MatTek) were coated with 0.2 mg/mL PLL-PEG at 4 °C overnight and afterwards rinsed with PBS to remove residual PLL-PEG. 500 μl of 0.5% agarose (Biozym Gold Agarose) solution containing 100 ng/ml human CXCL12 (R&D) were poured onto the coated dishes. Cells were injected between the glass and the solidified agarose layer as described above.
Total internal reflection (TIRF) microscopy was performed at 37 °C with an inverted Axiovert 200 (Zeiss) microscope, a TIRF 488/561-nm laser system (Visitron systems) and an EvolveTM EMCCD camera (Photometrics) triggered by VisiView software (Visitron). FIJI image processing package was used for image and kymograph analysis.
Protein expression and lipid bilayers
DNA constructs for production of monovalent streptavidin (MonoSAv) were a gift from A. Ting (Massachusetts Institute of Technology). A 3C protease-recognition sequence was inserted between the alive streptavidin subunit and the six-glutamate tag. The gene encoding the dead streptavidin subunit was prolonged with a six-histidine tagged tail mediating binding to supported lipid bilayers containing 18:1 DGS-NTA(Ni) (1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid)succinyl] (nickel salt)65. MonoSAv was produced by an in vitro refolding protocol as described66,67 and subsequently purified by MonoQ (MonoQ 5/50, GE Healthcare) and size-exclusion chromatography (Superdex 200, GE Healthcare). The six-glutamate tagged tail was cleaved with the 3C protease (GE Healthcare).
Calcium flux experiments
T cells (0.5−1 × 106) were loaded with 5 μM Fura-2 (Invitrogen) in complete RPMI-1640 medium for 30 min at 25 °C and then washed twice with 10 ml imaging buffer (1 × HBSS, Gibco, supplemented with 2% FCS, Sigma, 10 mM HEPES pH 7.4, Gibco, 1 mM MgCl2 and 1 mM CaCl2). For imaging, cells were spotted on a supported lipid bilayer functionalized with MonoSAv (50 ng/well) linked to CD3-biotin antibodies (OKT-3, eBioscience) at 25 °C. Image acquisition was performed with a DMI4000B microscope (Leica Microsystems) equipped with a 40× immersion objective (Leica HCX PL Apo 40×, NA 1.25) and an Andor iXon Ultra-8871 EM-CCD camera (Andor Technologies) controlled by the Leica Application Suite Advanced Fluorescence software (version AF6000LX). Imaging of Fura-2 (excitation, 340 nm and 380 nm; emission, 510 nm) was achieved through a fast external filter wheel equipped with excitation filers at 340 nm and 380 nm (Leica Fura EX) and a filter cube consisting of a dichroic beamsplitter and an emission band pass filter at 520 nm (Fura-2 Leica EM520/36, DC:409). DIC and Fura-2 images were collected at intervals of 1 min over 15 min. Intracellular calcium dynamics were determined from the Fura-2 excitation ratio (excitation at 340 nm/excitation at 380 nm) with the open source image analysis software package Fiji and Matlab.
Calcium flux measurement in shRNA cell lines was performed using Indo-1 Calcium Indicator (Life-Technologies). 24 h after doxycycline induction cells were labeled with Indo-1 Calcium Indicator stimulated with OKT3 (eBiosciences) and analyzed on a BD LSR-Fortessa. Data were analyzed using FlowJo Software.
NK-cell experiments on patient cells and CRISPR edited NK-92 cell lines
Conjugates of NK-92 CRISPR edited cells and K562 cells were formed for 60 min at 37 °C on silane coated slides (Electron Microscopy Services) and then permeabilized, fixed and stained for F-actin with phalloidin Alexa-Fluor568, anti-perforin Alexa-Fluor488 (clone G9, BioLegend) and anti-α-tubulin biotin (Life Technologies) followed by streptavidin Alexa-Fluor647 (Life Technologies). Conjugates were imaged as z stacks of 0.2-μm thickness to cover the entire volume of the immunological synapse, determined individually for each conjugate, on a Zeiss Axio-Observer Z1 equipped with a Yokogawa CSU10 spinning disc, Zeiss 63× 1.43 NA objective, and Hamamatsu Orca-AG camera. Images were acquired and analyzed with Volocity software (PerkinElmer) as described68.
For measurement of F-actin mesh in patient cells, PBMC were activated for 25 min on #1.5 coverslips coated with 5 μg/ml anti-CD18 (clone IB4) and anti-NKp30 (BioLegend) then fixed, permeabilized and stained with phalloidin Alexa-Fluor532 (Life Technologies) and anti-perforin Alexa-Fluor488 (clone δG9, BioLegend). Slides were mounted with ProLong Gold (Life Technologies). Stimulation emission depletion (STED) images were acquired on a Leica SP8 confocal microscope with excitation by white light laser, STED depletion at 660 nm with time-gating, and emission detected by HyD (GaAs) detectors. Data was exported to Volocity software (PerkinElmer) for analysis following deconvolution with Huygens software (Scientific Volume Imaging). All data was graphed using Prism 6.0 (GraphPad). Statistical analysis was performed using the Student’s two-tailed unpaired t test. P < 0.05 was considered significant.
Analysis of RhoA activation by immunofluorescence
Untransformed CD8+ T cells were expanded with a feeder mixture and a supplement of IL-2 and IL-15, as described above. Cells (2 × 105) were treated with lenalidomide (1 μM) or DMSO (1:1,000) in complete medium for 16 h. Half of the cells were subsequently stimulated with CXCL12 (100 ng/ml) for 5 min. Cells (105 per condition) were then deposited onto poly-L-lysine-coated coverslips. Following a sedimentation of 5 min at 37 °C, cells were spun down for 2 min at 350 rpm. Cells were fixed with ice-cold 10% TCA/30 mM glycine onto coverslips as described69. Following incubation on ice for 15 min, coverslips were washed three times with PBS/glycine. Cells were permeabilized with 0.2% Triton X-100 in PBS/glycine, washed and blocked for 30 min with 3% BSA, 0.01% Triton X-100 in PBS/glycine. Cells were stained with a rabbit mAb recognizing total RhoA (catalog number: 67B9, Cell Signaling) or active RhoA-GTP (catalog number: 26904, NewEast Biosciences) in a 1:50 dilution in 0.01% Triton X-100 in PBS/glycine and incubated overnight at 4 °C. Cells were incubated with anti-rabbit Alexa-Fluor555 or anti-mouse Alexa-Fluor488 secondary Abs at 1/400 dilution. Following nuclear staining with DAPI, slides were mounted with Prolong Gold antifade mounting medium (Life Technologies). Randomly selected fields were examined with a Leica DMI 6000B fluorescence microscope equipped with a 40× objective. The mean intensity of total RhoA and RhoA-GTP was calculated from 8-bit images in 50 cells per condition using cell masks created with ImageJ software.
SiR-tubulin staining and live convergence imaging
For imaging NK cells on activated glass surface, 3 × 105 NK cells were incubated with 500 nm of the live cell fluorogenic microtubule labeling probe SiR-tubulin (Spirochrome) and 5 μm Verapamil for 2 h at 37 °C. For imaging lytic granules, cells were incubated with 1 μM LysoTracker Red DND-99 (Thermo Fisher Scientific) for 30 min at 37 °C, washed once, and resuspended in dye-free R10 (RPMI 1640 (Gibco), 10% FBS (Atlanta Biologicals), 2 mM L-glutamine (Gibco), 1 mM sodium pyruvate (CellGro), 10 mM HEPES (Gibco), 100 μM MEM nonessential amino acids (Gibco), and 100 U/ml penicillin and streptomycin), supplemented with 5 μm Verapamil. ΔT dishes (Bioptechs) were coated with 5 μg/ml anti-NKp30 (BioLegend) and 5 μg/ml anti-CD18 (Clone IB4) for 1 h at 37 °C, washed with PBS, and pre-warmed before imaging with 300 μl dye-free R10.
For imaging of NK cells with K562 target cells, 105 LysoTracker Red-loaded NK cells (effectors) were stained with 500 nm SiR-tubulin and 5 μm Verapamil for 2 h at 37 °C, washed once, resuspended in dye-free R10, and then mixed at a 1:2 ratio with target cells that had been pre-incubated for 5 min with 2.5 μM CellTrace CFSE (Thermo Fisher Scientific). Conjugates were incubated in 1 ml microcentrifuge tubes in the presence of 5 μm Verapamil for 30 min at 37 °C and plated in Lab-Tek #1.0 Borosilicate chamber slides (Nunc) pre-coated with 5 μg/ml anti-CD58 (BD Pharmingen) for 30 min at 37° and washed with PBS. Target cells were allowed to adhere to glass for 15 min and conjugates were imaged at 30s per frame for 30–45 min on a Leica SP8 laser scanning confocal microscope with 100× objective. Excitation was provided by a tunable white light laser at 488, 561 and 647 nm.
For evaluation of lytic granule convergence, raw images were analyzed in Volocity (PerkinElmer) and mean granule distance (MGD) to the MTOC was calculated as described previously32. Number of replicates performed = 3 for both types of experiments. Graphs were plotted in Prism (GraphPad) showing mean values with s.e.m. Statistical test used is unpaired t test with Welch’s correction.
Supplementary Material
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Acknowledgments
We thank the patient and his family for participating in this study; B. Fleckenstein and M. Schmidt for generating and providing patient-derived T cell lines; and G. Superti-Furga, J. Bigenzahn for providing the inducible protein expression system used for Jurkat T cells and together with N. Serwas, C.D. Conde, A. Kalinichenko, K. Ackerman and R. Martins for critically reviewing the manuscript and providing comments. The research leading to these results was funded by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement 310857 (K.B.), the Vienna Science and Technology Fund (WWTF) through project LS14-031 (J.B.H. and K.B.), an unrestricted research grant from Celgene Austria (U.J.), the National Institutes of Health (R01AI067946 to J.S.O.), Boehringer Ingelheim Fonds (R.P.), the Austrian Science Fund (FWF): Project M1809-B19 (K.L.W.), and the French Agence Nationale de la Recherche (ANR-13-BSV1-0031 to L.D.).
Footnotes
Author Contributions
E.S. performed most of the experiments, analyzed data, interpreted results, and, together with K.B., wrote the initial draft and revised version of the manuscript. D.C., I.T. and Ö.S. cared for the patient, and provided and interpreted clinical and immunological data. M.H. and E.S. performed migration and Lifeact experiments. M.S. provided critical input to the content of the manuscript. E.M.M., P.S., M.M., P.P.B., H.T.H. and J.S.O. performed and interpreted NK-cell immunological synapse experiments and detailed flow-cytometry-based NK-cell immunophenotyping. S.A.B. and E.S. identified the RASGRP1 mutation and performed initial experiments. R.P. and J.B.H. performed lipid bilayer calcium-flux experiments, and analyzed and interpreted data. L.P. generated an untransformed CD8+ T cell line from the patient and performed the CD8+ T cell cytotoxic assays. H.S. and V.S. performed proliferation analyses together with E.S. and provided critical input. Ö.Y.P. and L.D. performed the immunofluorescence experiments to quantify RhoA activation. K.L.W., W.G. and I.B. performed experiments and provided critical input. F.M., J.G.G. and U.J. helped with migration analyses. K.L.B. performed mass spectrometry analyses. W.F.P. and G.J.Z. performed thymidine incorporation assays, chromium release assays and analyses of autoantibody titers. K.B. conceived of and coordinated the study, provided laboratory resources, interpreted data, supervised E.S., W.G., Ö.Y.P., I.B., S.B. and K.W., wrote the manuscript together with E.S. and took overall responsibility for the study. All of the authors provided critical input and agreed to this publication.
Competing Financial Interests
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Roose J, Weiss A. T cells: getting a GRP on Ras. Nat Immunol. 2000;1:275–276. doi: 10.1038/79713. [DOI] [PubMed] [Google Scholar]
- 2.Stone JC. Regulation and function of the RasGRP family of Ras activators in blood cells. Genes Cancer. 2011;2:320–334. doi: 10.1177/1947601911408082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downward J, Graves JD, Warne PH, Rayter S, Cantrell DA. Stimulation of p21ras upon T-cell activation. Nature. 1990;346:719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- 4.Kremer KN, Kumar A, Hedin KE. G alpha i2 and ZAP-70 mediate RasGRP1 membrane localization and activation of SDF-1-induced T cell functions. J Immunol. 2011;187:3177–3185. doi: 10.4049/jimmunol.1100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dower NA, et al. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 6.Khanna R, Burrows SR. Human immunology: a case for the ascent of non-furry immunology. Immunol Cell Biol. 2011;89:330–331. doi: 10.1038/icb.2010.173. [DOI] [PubMed] [Google Scholar]
- 7.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 8.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 9.Notarangelo LD. Functional T cell immunodeficiencies (with T cells present) Annu Rev Immunol. 2013;31:195–225. doi: 10.1146/annurev-immunol-032712-095927. [DOI] [PubMed] [Google Scholar]
- 10.Notarangelo LD. Partial defects of T-cell development associated with poor T-cell function. J Allergy Clin Immunol. 2013;131:1297–1305. doi: 10.1016/j.jaci.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picard C, et al. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J Clin Immunol. 2015;35:696–726. doi: 10.1007/s10875-015-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bousfiha A, et al. The 2015 IUIS phenotypic classification for primary immunodeficiencies. J Clin Immunol. 2015;35:727–738. doi: 10.1007/s10875-015-0198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinós C, et al. Mutations in the urocanase gene UROC1 are associated with urocanic aciduria. J Med Genet. 2009;46:407–411. doi: 10.1136/jmg.2008.060632. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, et al. Differential requirement of RasGRP1 for γδ T cell development and activation. J Immunol. 2012;189:61–71. doi: 10.4049/jimmunol.1103272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warnecke N, et al. TCR-mediated Erk activation does not depend on Sos and Grb2 in peripheral human T cells. EMBO Rep. 2012;13:386–391. doi: 10.1038/embor.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jun JE, Rubio I, Roose JP. Regulation of ras exchange factors and cellular localization of ras activation by lipid messengers in T cells. Front Immunol. 2013;4:239. doi: 10.3389/fimmu.2013.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Depeille P, et al. RasGRP1 opposes proliferative EGFR-SOS1-Ras signals and restricts intestinal epithelial cell growth. Nat Cell Biol. 2015;17:804–815. doi: 10.1038/ncb3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen S, et al. Critical roles of RasGRP1 for invariant NKT cell development. J Immunol. 2011;187:4467–4473. doi: 10.4049/jimmunol.1003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoya CJ, et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology. 2007;122:1–14. doi: 10.1111/j.1365-2567.2007.02647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priatel JJ, et al. Chronic immunodeficiency in mice lacking RasGRP1 results in CD4 T cell immune activation and exhaustion. J Immunol. 2007;179:2143–2152. doi: 10.4049/jimmunol.179.4.2143. [DOI] [PubMed] [Google Scholar]
- 21.Ma CS, Deenick EK. Human T follicular helper (Tfh) cells and disease. Immunol Cell Biol. 2014;92:64–71. doi: 10.1038/icb.2013.55. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett A, Buhlmann JE, Stone J, Lim B, Barrington RA. Multiple checkpoint breach of B cell tolerance in Rasgrp1-deficient mice. J Immunol. 2013;191:3605–3613. doi: 10.4049/jimmunol.1202892. [DOI] [PubMed] [Google Scholar]
- 23.Coughlin JJ, Stang SL, Dower NA, Stone JC. RasGRP1 and RasGRP3 regulate B cell proliferation by facilitating B cell receptor-Ras signaling. J Immunol. 2005;175:7179–7184. doi: 10.4049/jimmunol.175.11.7179. [DOI] [PubMed] [Google Scholar]
- 24.Sun C, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet. 2016;48:323–330. doi: 10.1038/ng.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tangye SG, Ferguson A, Avery DT, Ma CS, Hodgkin PD. Isotype switching by human B cells is division-associated and regulated by cytokines. J Immunol. 2002;169:4298–4306. doi: 10.4049/jimmunol.169.8.4298. [DOI] [PubMed] [Google Scholar]
- 26.Thien M, et al. Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity. 2004;20:785–798. doi: 10.1016/j.immuni.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Perussia B, Chen Y, Loza MJ. Peripheral NK cell phenotypes: multiple changing of faces of an adapting, developing cell. Mol Immunol. 2005;42:385–395. doi: 10.1016/j.molimm.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Navarro MN, Cantrell DA. Serine-threonine kinases in TCR signaling. Nat Immunol. 2014;15:808–814. doi: 10.1038/ni.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamura SM, Oki-Idouchi CE, Lorenzo PS. The exchange factor and diacylglycerol receptor RasGRP3 interacts with dynein light chain 1 through its C-terminal domain. J Biol Chem. 2006;281:36132–36139. doi: 10.1074/jbc.M605093200. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Crespo I, et al. Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett. 2001;503:135–141. doi: 10.1016/s0014-5793(01)02718-1. [DOI] [PubMed] [Google Scholar]
- 31.Huse M. Microtubule-organizing center polarity and the immunological synapse: protein kinase C and beyond. Front Immunol. 2012;3:235. doi: 10.3389/fimmu.2012.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mentlik AN, Sanborn KB, Holzbaur EL, Orange JS. Rapid lytic granule convergence to the MTOC in natural killer cells is dependent on dynein but not cytolytic commitment. Mol Biol Cell. 2010;21:2241–2256. doi: 10.1091/mbc.E09-11-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roose JP, Mollenauer M, Gupta VA, Stone J, Weiss A. A diacylglycerol-protein kinase C-RasGRP1 pathway directs Ras activation upon antigen receptor stimulation of T cells. Mol Cell Biol. 2005;25:4426–4441. doi: 10.1128/MCB.25.11.4426-4441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Zhu M, Nishida K, Hirano T, Zhang W. An essential role for RasGRP1 in mast cell function and IgE-mediated allergic response. J Exp Med. 2007;204:93–103. doi: 10.1084/jem.20061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsay AG, et al. Chronic lymphocytic leukemia cells induce defective LFA-1-directed T-cell motility by altering Rho GTPase signaling that is reversible with lenalidomide. Blood. 2013;121:2704–2714. doi: 10.1182/blood-2012-08-448332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson PG, et al. Lenalidomide, bortezomib, and dexamethasone combination therapy in patients with newly diagnosed multiple myeloma. Blood. 2010;116:679–686. doi: 10.1182/blood-2010-02-268862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krönke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandhi AK, et al. Immunomodulatory agents lenalidomide and pomalidomide costimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) Br J Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med. 2014;211:2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuller DM, et al. Regulation of RasGRP1 function in T cell development and activation by its unique tail domain. PLoS One. 2012;7:e38796. doi: 10.1371/journal.pone.0038796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 43.Thrasher AJ, Burns SO. WASP: a key immunological multitasker. Nat Rev Immunol. 2010;10:182–192. doi: 10.1038/nri2724. [DOI] [PubMed] [Google Scholar]
- 44.Lanzi G, et al. A novel primary human immunodeficiency due to deficiency in the WASP-interacting protein WIP. J Exp Med. 2012;209:29–34. doi: 10.1084/jem.20110896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGhee SA, Chatila TA. DOCK8 immune deficiency as a model for primary cytoskeletal dysfunction. Dis Markers. 2010;29:151–156. doi: 10.3233/DMA-2010-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Kapoor TM, Chen JK, Huse M. Diacylglycerol promotes centrosome polarization in T cells via reciprocal localization of dynein and myosin II. Proc Natl Acad Sci USA. 2013;110:11976–11981. doi: 10.1073/pnas.1306180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder CM, Ostrem JM, Hertz NT, Vale RD. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. eLife. 2014;3:e03351. doi: 10.7554/eLife.03351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jayappa KD, et al. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. J Virol. 2015;89:3497–3511. doi: 10.1128/JVI.03347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zenz T. Exhausting T cells in CLL. Blood. 2013;121:1485–1486. doi: 10.1182/blood-2013-01-475939. [DOI] [PubMed] [Google Scholar]
- 50.Salzer E, et al. Combined immunodeficiency with life-threatening EBV-associated lymphoproliferative disorder in patients lacking functional CD27. Haematologica. 2013;98:473–478. doi: 10.3324/haematol.2012.068791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willmann KL, et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat Commun. 2014;5:5360. doi: 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dupré L, et al. Efficacy of gene therapy for Wiskott-Aldrich syndrome using a WAS promoter/cDNA-containing lentiviral vector and nonlethal irradiation. Hum Gene Ther. 2006;17:303–313. doi: 10.1089/hum.2006.17.303. [DOI] [PubMed] [Google Scholar]
- 54.Dupré L, et al. Lentiviral vector-mediated gene transfer in T cells from Wiskott-Aldrich syndrome patients leads to functional correction. Mol Ther. 2004;10:903–915. doi: 10.1016/j.ymthe.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 55.Pichlmair A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 56.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dow LE, et al. A pipeline for the generation of shRNA transgenic mice. Nat Protoc. 2012;7:374–393. doi: 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuber J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boztug K, et al. JAGN1 deficiency causes aberrant myeloid cell homeostasis and congenital neutropenia. Nat Genet. 2014;46:1021–1027. doi: 10.1038/ng.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vasconcelos Z, et al. Individual human cytotoxic T lymphocytes exhibit intraclonal heterogeneity during sustained killing. Cell Rep. 2015;11:1474–1485. doi: 10.1016/j.celrep.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fairhead M, Krndija D, Lowe ED, Howarth M. Plug-and-play pairing via defined divalent streptavidins. J Mol Biol. 2014;426:199–214. doi: 10.1016/j.jmb.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howarth M, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee PP, Orange JS. Quantitative measurement of F-actin accumulation at the NK cell immunological synapse. J Immunol Methods. 2010;355:1–13. doi: 10.1016/j.jim.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trifari S, et al. Defective Th1 cytokine gene transcription in CD4+ and CD8+ T cells from Wiskott-Aldrich syndrome patients. J Immunol. 2006;177:7451–7461. doi: 10.4049/jimmunol.177.10.7451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.