Abstract
Osteoarthritis is the most common musculoskeletal disease and the leading cause of disability globally. Here, we perform a genome-wide association study for osteoarthritis (77,052 cases and 378,169 controls), analysing 4 phenotypes: knee osteoarthritis, hip osteoarthritis, knee and/or hip osteoarthritis, and any osteoarthritis. We discover 64 signals, 52 of them novel, more than doubling the number of established disease loci. Six signals fine map to a single variant. We identify putative effector genes by integrating eQTL colocalization, fine-mapping, human rare disease, animal model, and osteoarthritis tissue expression data. We find enrichment for genes underlying monogenic forms of bone development diseases, and for the collagen formation and extracellular matrix organisation biological pathways. Ten of the likely effector genes, including TGFB1, FGF18, CTSK and IL11 have therapeutics approved or in clinical trials, with mechanisms of action supportive of evaluation for efficacy in osteoarthritis.
Osteoarthritis affects 40% of individuals over the age of 701, is a major cause of pain, comorbidity and mortality2. Ten million people in the UK alone suffer from osteoarthritis, with a total indirect cost to the economy of £14.8 billion per annum2. Disease management targets the main symptom (pain) and culminates in joint replacement surgery (1.76 million per year in the EU) with variable outcomes3. There is a clear and urgent need to translate genomic evidence into druggable mechanisms of disease aetiology and progression, to support the development of disease-modifying therapies for osteoarthritis.
Here, we leverage the UK Biobank and arcOGEN resources to perform a genome-wide meta-analysis for osteoarthritis across ~17.5 million single nucleotide variants in up to 455,221 individuals (Supplementary Figure 1). We identify 65 genome-wide significant variants at 64 loci (P≤3x10-8; Online Methods, Supplementary Table 1), 52 of which are novel, thus increasing the number of established loci from 344 to 86: 24 novel signals for osteoarthritis at any site (77,052 cases), 15 for hip osteoarthritis (15,704 cases), 7 for knee osteoarthritis (24,955 cases), and 6 for osteoarthritis of the hip and/or knee (39,427 cases) (Table 1, Supplementary Figures 2-6, Supplementary Tables 2 and 3). We find that 25 of 34 previously-reported loci show association (P<0.05) with at least one of the four osteoarthritis traits we evaluate (Supplementary Table 4).
Table 1. Independent variants with P<3x10-8 in an inverse-variance weighted fixed effects meta-analysis of UK Biobank and arcOGEN.
Variant positions are reported according to build 37 and their alleles are coded based on the positive strand.
| rsID | Trait | Other Traits | EA/NEA | WEAF | OR | OR_95CI | PV | q_pv | i2 |
|---|---|---|---|---|---|---|---|---|---|
| Newly Identified Loci | |||||||||
| rs4338381 | OA_hip | OA OA_kneehip | A/G | 0.63 | 1.1 | 1.07, 1.13 | 4.37E-15 | 0.93 | 0 |
| 1:150214028 | OA | OA_hip | C/CT | 0.37 | 1.03 | 1.02, 1.05 | 2.54E-08 | 1.00 | 0 |
| 1:174192402 | OA | TAAAAAAAAAAAAAAAAA/T | 0.57 | 1.03 | 1.02, 1.05 | 1.05E-08 | 1.00 | 0 | |
| rs11583641 | OA_hip | C/T | 0.72 | 1.08 | 1.06, 1.11 | 5.58E-10 | 0.63 | 0 | |
| rs10218792 | OA | G/T | 0.27 | 1.04 | 1.02, 1.05 | 2.03E-08 | 0.77 | 0 | |
| rs2061027 | OA | OA_knee OA_kneehip | A/G | 0.51 | 1.04 | 1.03, 1.05 | 3.16E-13 | 0.25 | 25.8 |
| rs12470967 | OA_knee | OA_kneehip | A/G | 0.43 | 1.06 | 1.04, 1.08 | 1.50E-08 | 1.00 | 0 |
| rs62182810 | OA | A/G | 0.55 | 1.03 | 1.02, 1.05 | 1.65E-09 | 0.84 | 0 | |
| rs62262139 | OA | A/G | 0.54 | 1.04 | 1.03, 1.05 | 9.09E-11 | 1.00 | 0 | |
| rs11732213 | OA_kneehip | OA OA_hip | T/C | 0.81 | 1.06 | 1.04, 1.08 | 8.81E-10 | 0.60 | 0 |
| rs1913707 | OA_hip | OA | A/G | 0.61 | 1.08 | 1.06, 1.11 | 2.96E-11 | 0.03 | 79 |
| rs34811474 | OA | G/A | 0.77 | 1.04 | 1.03, 1.05 | 2.17E-09 | 0.76 | 0 | |
| rs13107325 | OA | T/C | 0.08 | 1.1 | 1.07, 1.12 | 8.29E-19 | 0.70 | 0 | |
| rs35611929 | OA_knee | A/G | 0.34 | 1.06 | 1.04, 1.08 | 1.21E-08 | 0.96 | 0 | |
| rs3884606 | OA_kneehip | G/A | 0.49 | 1.04 | 1.03, 1.06 | 8.25E-09 | 0.42 | 0 | |
| rs115740542 | OA | OA_hip OA_kneehip | C/T | 0.07 | 1.06 | 1.04, 1.08 | 8.59E-09 | 0.95 | 0 |
| rs9277552 | OA_kneehip | OA_knee OA | C/T | 0.79 | 1.06 | 1.04, 1.08 | 2.37E-10 | 0.78 | 0 |
| rs12154055 | OA | G/A | 0.61 | 1.03 | 1.02, 1.04 | 2.71E-08 | 0.10 | 63.5 | |
| rs80287694 | OA_hip | G/A | 0.11 | 1.12 | 1.08, 1.16 | 2.66E-09 | 0.20 | 39.1 | |
| rs11409738 | OA | OA_kneehip | TA/T | 0.37 | 1.04 | 1.03, 1.05 | 2.13E-10 | 1.00 | 0 |
| rs330050 | OA | OA_kneehip OA_hip | G/C | 0.51 | 1.04 | 1.03, 1.05 | 1.93E-11 | 0.35 | 0 |
| rs60890741 | OA_hip | C/CA | 0.86 | 1.11 | 1.08, 1.16 | 4.50E-09 | 1.00 | 0 | |
| rs919642 | OA | OA_kneehip OA_knee | T/A | 0.27 | 1.05 | 1.04, 1.06 | 8.55E-15 | 0.41 | 0 |
| rs1330349 | OA_hip | C/G | 0.58 | 1.08 | 1.06, 1.11 | 4.10E-11 | 0.71 | 0 | |
| rs62578127 | OA_hip | C/T | 0.63 | 1.09 | 1.06, 1.11 | 2.77E-12 | 0.54 | 0 | |
| rs17659798 | OA_kneehip | A/C | 0.71 | 1.06 | 1.04, 1.07 | 2.06E-10 | 0.86 | 0 | |
| rs11031191 | OA | T/G | 0.35 | 1.03 | 1.02, 1.05 | 1.42E-08 | 0.95 | 0 | |
| rs10896015 | OA_hip | G/A | 0.73 | 1.08 | 1.05, 1.11 | 2.74E-09 | 0.36 | 0 | |
| rs34419890 | OA_hip | T/C | 0.93 | 1.13 | 1.09, 1.18 | 1.99E-08 | 0.75 | 0 | |
| rs1149620 | OA | T/A | 0.57 | 1.04 | 1.02, 1.05 | 6.93E-10 | 0.90 | 0 | |
| rs79056043 | OA_hip | G/A | 0.05 | 1.18 | 1.12, 1.24 | 1.33E-09 | 0.14 | 53 | |
| rs317630 | OA | T/C | 0.27 | 1.04 | 1.02, 1.05 | 1.97E-08 | 0.75 | 0 | |
| rs11105466 | OA_kneehip | A/G | 0.42 | 1.04 | 1.03, 1.06 | 2.15E-08 | 0.26 | 22.6 | |
| rs2171126 | OA | OA_kneehip | T/C | 0.51 | 1.03 | 1.02, 1.05 | 9.07E-10 | 0.26 | 21.5 |
| rs11059094 | OA_hip | T/C | 0.48 | 1.08 | 1.05, 1.1 | 7.38E-11 | 0.44 | 0 | |
| rs56116847 | OA_knee | OA OA_kneehip | A/G | 0.36 | 1.06 | 1.04, 1.08 | 3.19E-10 | 0.05 | 74.2 |
| rs35912128 | OA_knee | AT/A | 0.17 | 1.08 | 1.05, 1.11 | 2.18E-08 | 1.00 | 0 | |
| rs35206230 | OA | OA_kneehip | T/C | 0.67 | 1.04 | 1.03, 1.05 | 1.48E-12 | 0.86 | 0 |
| rs6499244 | OA_knee | OA_kneehip | A/T | 0.56 | 1.06 | 1.04, 1.08 | 3.88E-11 | 0.74 | 0 |
| rs1126464 | OA | G/C | 0.76 | 1.04 | 1.03, 1.06 | 1.56E-10 | 0.07 | 69.3 | |
| rs35087650 | OA_knee | ATT/A | 0.26 | 1.07 | 1.05, 1.1 | 1.18E-09 | 1.00 | 0 | |
| rs2953013 | OA_kneehip | C/A | 0.3 | 1.05 | 1.04, 1.07 | 3.07E-10 | 0.87 | 0 | |
| rs62063281 | OA_hip | G/A | 0.22 | 1.1 | 1.07, 1.13 | 5.30E-12 | 0.91 | 0 | |
| rs547116051 | OA | AC/A | 0.001 | 1.83 | 1.49, 2.26 | 1.50E-08 | 1.00 | 0 | |
| rs7222178 | OA_hip | A/T | 0.2 | 1.1 | 1.07, 1.13 | 3.78E-11 | 0.59 | 0 | |
| rs8067763 | OA_knee | G/A | 0.41 | 1.06 | 1.04, 1.08 | 2.39E-09 | 0.35 | 0 | |
| rs10502437 | OA | G/A | 0.6 | 1.03 | 1.02, 1.04 | 2.50E-08 | 0.69 | 0 | |
| rs1560707 | OA | T/G | 0.37 | 1.04 | 1.03, 1.05 | 1.35E-13 | 0.45 | 0 | |
| rs75621460 | OA | OA_kneehip | A/G | 0.03 | 1.16 | 1.12, 1.2 | 1.62E-15 | 0.58 | 0 |
| rs4252548 | OA_hip | T/C | 0.02 | 1.32 | 1.22, 1.43 | 1.96E-12 | 0.05 | 73 | |
| rs2836618 | OA_hip | OA_kneehip | A/G | 0.26 | 1.09 | 1.06, 1.12 | 3.20E-11 | 0.02 | 82.6 |
| rs528981060 | OA | A/G | 0.001 | 1.68 | 1.4, 2.02 | 2.37E-08 | 1.00 | 0 | |
| Previously reported loci | |||||||||
| rs2820443 | OA_kneehip | OA_hip OA | C/T | 0.3 | 1.06 | 1.04, 1.07 | 6.01E-11 | 0.82 | 0 |
| rs3771501 | OA | OA_hip OA_kneehip | A/G | 0.47 | 1.05 | 1.03, 1.06 | 4.24E-16 | 0.67 | 0 |
| rs3774355 | OA_hip | OA_kneehip | A/G | 0.36 | 1.09 | 1.07, 1.12 | 8.20E-14 | 0.12 | 59 |
| rs2396502 | OA_hip | OA_kneehip | C/A | 0.6 | 1.09 | 1.06, 1.11 | 2.12E-12 | 0.74 | 0 |
| rs12209223 | OA_hip | A/C | 0.1 | 1.17 | 1.13, 1.21 | 3.88E-16 | 0.26 | 22 | |
| rs10974438 | OA | OA_kneehip | A/C | 0.65 | 1.03 | 1.02, 1.05 | 1.34E-08 | 0.36 | 0 |
| rs34687269 | OA_hip | A/T | 0.53 | 1.09 | 1.06, 1.11 | 1.67E-12 | 0.70 | 0 | |
| rs10492367 | OA_hip | OA_kneehip | T/G | 0.19 | 1.16 | 1.13, 1.2 | 1.25E-24 | 0.25 | 24.5 |
| rs4775006 | OA_knee | A/C | 0.41 | 1.06 | 1.04, 1.08 | 8.40E-10 | 0.08 | 68.3 | |
| rs12901372 | OA_hip | C/G | 0.53 | 1.08 | 1.06, 1.11 | 3.46E-11 | 0.13 | 56.2 | |
| rs9930333 | OA_kneehip | G/T | 0.42 | 1.05 | 1.03, 1.06 | 1.52E-09 | 0.04 | 75.8 | |
| rs143384 | OA_knee | OA_kneehip OA | A/G | 0.6 | 1.1 | 1.08, 1.12 | 4.77E-23 | 0.37 | 0 |
Trait: Osteoarthritis trait most significantly associated with variant in the meta-analysis stage; Other Traits: Other osteoarthritis traits with genome-wide significant association following meta-analysis; EA/NEA: Effect allele/non-effect allele; WEAF: Weighted effect allele frequency between UK Biobank and arcOGEN; OR: Odds ratio; OR_95CI: Lower bound of the 95% credible interval of the odds ratio, upper bound of the 95% credible interval of the odds ratio; PV: P value (two-sided); q_pv: P value of Cochran’s Q measure of heterogeneity; i2: I2 statistic describing the percentage of variation across studies that is due to heterogeneity rather than chance; OA: osteoarthritis; OA_hip: Hip osteoarthritis; OA_knee: Knee osteoarthritis; OA_kneehip: Knee and/or hip osteoarthritis.
To identify putative effector genes at the 64 genome-wide significant regions, we integrated results from several strands of investigation, including transcriptomic/proteomic characterisation of primary tissue from osteoarthritis patients undergoing joint replacement surgery, coupled with statistical fine-mapping, annotation of predicted consequences of variants in the credible sets, eQTL colocalization, and relevant rare human disease and animal model evidence (Online Methods, Supplementary Table 5 and 6). We observe evidence of colocalization in at least one tissue for 49 out of the 64 loci, 44 of which are at newly-associated osteoarthritis signals (Supplementary Table 7 and Supplementary Figure 7). Using MetaXcan, we identify 11 genes with additional evidence of colocalization at loci not reaching genome-wide significance in SNV analyses (Supplementary Figure 8, Supplementary Tables 8 and 9).
Pathway analyses (Online Methods and Supplementary Note) identify 64 biological processes associated with osteoarthritis, of which 46 are bone-, cartilage- and chondrocyte- morphology related (Supplementary Table 10). The collagen formation and extracellular matrix organisation biological pathways are consistently identified by different pathway analysis methods. Genome-wide linkage disequilibrium (LD) score regression analysis5,6 unveils significant correlation between osteoarthritis and traits within the obesity, cognition, smoking, bone mineral density and reproductive trait categories (Figure 1; Supplementary Table 11 and 12). Mendelian randomization analyses (Online Methods) support a role for higher body mass index (BMI) and adiposity in osteoarthritis risk, and identify a potential protective effect of LDL cholesterol, and of higher level of education against osteoarthritis (Supplementary Tables 13-15 and Supplementary Note). Two of the BMI loci (SLC39A8 and FTO) show genome-wide significant associations with osteoarthritis, with SLC39A8 showing much larger effects on osteoarthritis than expected given the BMI-raising effects (Supplementary Figure 9). Apparent causal associations of knee pain with osteoarthritis (Supplementary Table 13 and 16) are potentially attributable to reverse causality (Supplementary Note). We estimate the proportion of the total narrow sense heritability explained by osteoarthritis loci to be 14.7 % for knee osteoarthritis, 51.9 % for hip osteoarthritis, 24.2% for osteoarthritis of the hip and/or knee, and 22.5% of osteoarthritis at any site (Supplementary Table 17). We do not find evidence for a role of low-frequency or rare variation of large effect in osteoarthritis susceptibility, and have limited power to detect smaller effects at lower-frequency variants (Figure 2). In the future, meta-analyses of osteoarthritis studies in global populations will help further deconvolute the genetic underpinning of this disabling disease.
Figure 1. Genetic correlations between osteoarthritis and other traits and diseases.
Genetic correlations (rg) between osteoarthritis and other publicly available GWAS results, based on LD score regression as implemented in LDHub. The diagram shows traits with significant correlation (P<0.05) and 95% confidence intervals across all osteoarthritis definitions. The red outline of the bars denotes negative correlation and the blue outline denotes positive correlation. The upper right legend shows the categories of the traits. OA: osteoarthritis; OA_hip: Hip osteoarthritis; OA_knee: Knee osteoarthritis; OA_kneehip: Knee and/or hip osteoarthritis. Lumbar spine bone mineral density 1 and 2 relate to two different published studies.
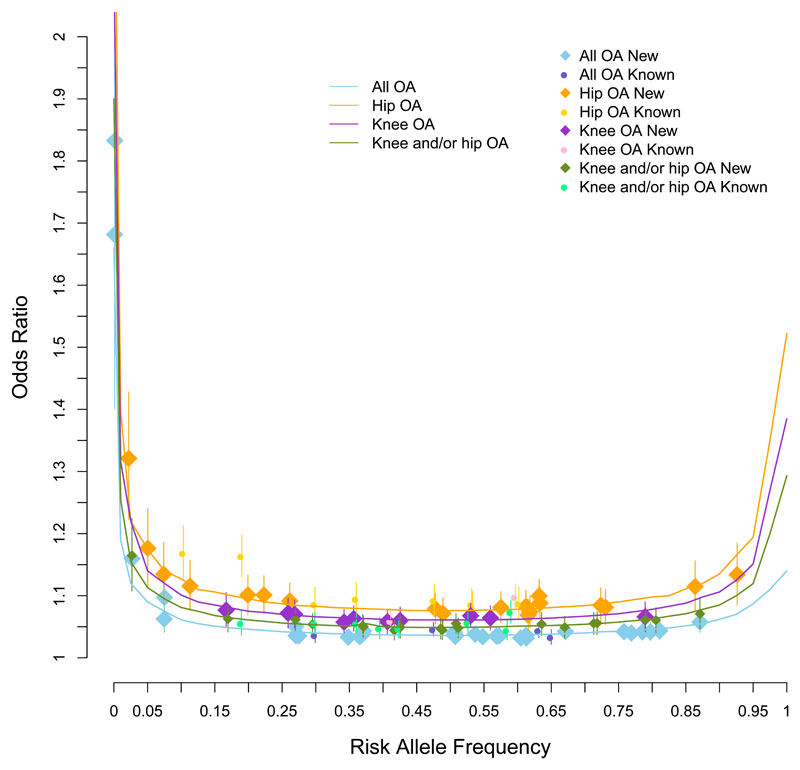
Figure 2. Allelic architecture of index variants.
Meta-analysis based odds ratio with its 95% confidence interval of 99 variants (previously-reported denoted as circles and newly-reported denoted as diamonds) with UK Biobank and arcOGEN meta-analysis P<3.0x10-8 (two-sided) as a function of their weighted allele frequency. The curves indicate 80% power at the genome-wide significance threshold of P≤3.0x10-8, for the four sample sizes of the meta-analyses. We have 80% power to detect an association at genome-wide significance for a variant with 1% MAF and allelic odds ratio of 1.19, 1.40, 1.32 and 1.25 for all osteoarthritis, hip osteoarthritis, knee osteoarthritis and knee and/or hip osteoarthritis, respectively. For 0.1% MAF the corresponding odds ratios are 1.66, 2.43, 2.12 and 1.90.
We used a combination of conditional analyses7 followed by asymptotic Bayes’ factor fine-mapping8 (Online Methods) of conditionally distinct association signals to identify causal variants. In six of the novel loci, a single variant could be postulated as causal with more than 95% posterior probability: missense variants in SLC39A8, IL11 and ANAPC4 (rs13107325, rs4252548 and rs34811474, respectively), rs75621460 near TGFB1, rs547116051 near MAPT and rs528981060 near SCUBE1 (Supplementary Table 18 and Supplementary Note).
We observe strong enrichment for genes known to cause monogenic bone development diseases and forms of early-onset osteoarthritis, in the vicinity of osteoarthritis signals (odds ratio [OR] 8.87, P=1.8x10-4, and OR 8.83, P=8x10-3, respectively) (Supplementary Table 19 and 20). This finding highlights bone development as an important physiological process in osteoarthritis aetiology. Several genes identified as likely causal in our study are also linked to osteoarthritis aetiology in animal models. In eight out of the ten cases where we can unequivocally define directionality of association, we observe concordance between our results and those from animal models (i.e. that reduced expression or loss-of-function mutations increase osteoarthritis risk both in humans and in animal models) (Supplementary Table 21). Some of these genes code for structural bone/cartilage proteins (COL11A1, COL11A2) or play a critical role in bone/cartilage development (FGFR3, GDF5). These consistent observations in human and animal models provide compelling evidence for a causal role of these genes in osteoarthritis and point to an agonist strategy as the desired mechanism of action for new osteoarthritis drugs targeting these eight genes.
Ten genes have a therapeutic approved or in clinical trials (Table 2), with mechanisms of action that are not inconsistent with potential for efficacy in osteoarthritis, based on eQTL, functional genomics, rare disease and animal model data. Four of these genes, TGFB1, GDF5, FGF18 and CTSK, currently have therapeutics in clinical development for osteoarthritis/cartilage regeneration indications. Of these, only GDF5 has been previously published as genetically associated with osteoarthritis susceptibility9. Two of the genes, IL11 and DPEP1, have approved therapeutics for unrelated indications, opening the possibility for repositioning.
Table 2. Translational context for selected osteoarthritis-associated genes.
| Gene | OA phenotype | OA locus Chr: index variant | MOA needed for OA, if known† | Drug targeting OA gene | Dev. Phase | Molecule type | Drug MOA | Current Indication(s) |
|---|---|---|---|---|---|---|---|---|
| TGFB1 | OA; OA_kneehip | Chr19: rs75621460 | Agonist / upregulator | INVOSSA | Registered | Cell therapy | ↑expression | Knee osteoarthritis |
| GDF5 | OA_knee; OA_kneehip; OA | Chr20: rs143384 | Agonist / upregulator | HMR-4052 | Clinical development | Protein | ↑signalling | Regeneration, cartilage, intervertebral disc |
| FGF18 | OA_kneehip | Chr5: rs3884606 | Agonist / upregulator | AS-902330 | Clinical development | Protein | ↑signalling | Osteoarthritis, cartilage regeneration |
| CTSK | OA; OA_hip | 1:150214028 _CT_C | Unknown | CTSK inhibitor | Clinical development | SM | Inhibitor | Osteoarthritis |
| IL11 | OA_hip | chr19: rs4252548 | ↑ IL11 signalling? | Oprelvekin | Approved | Protein | ↑ IL11 signalling | Thrombocytopenia |
| DPEP1 | OA | Chr16: rs1126464 | Unknown | CILASTATIN | Approved | SM | Inhibitor | Co-administered with imipenem (antibiotic) to prolong effective dose |
| DIABLO | OA_hip | Chr12: rs11059094 |
Unknown | LCL-161 | Clinical development | SM | SMAC mimetic and IAP inhibitor | Breast cancer, leukemia, myeloma |
| CRHR1 | OA_hip | Chr17: rs62063281 |
Inhibitor | NBI-74788 | Clinical development | SM | Antagonist | Adrenal insufficiency, primary, congenital |
| MAPT | OA_hip | Chr17: rs62063281 |
Inhibitor | flortaucipir F 18, Leuco-methylthioninium | Clinical development | SM | Tau aggregation inhibitor | Alzheimer's disease |
| TNFSF15 | OA; OA_hip; OA_kneehip; OA_knee | Chr9: rs919642, rs1330349 | Unknown | PF-06480605 | Clinical development | Ab | Inhibitor | Ulcerative colitis, Wet AMD |
OA: osteoarthritis; OA_hip: Hip osteoarthritis; OA_knee: Knee osteoarthritis; OA_kneehip: Knee and/or hip osteoarthritis; SM: small molecule; Ab: Antibody; MOA: mechanism of action of a therapeutic;
Based on functional evidence supporting the gene as an osteoarthritis risk factor. Criteria for inclusion of “OA locus” gene: target has a therapeutic approved or in clinical development; therapeutic with OA indication and/or target eQTL colocalization with index variant and/or target missense variant with posterior probability of colocalization >0.5. Drug data compiled from Chembl (URLs) and Clinical trials.gov (URLs).
rs4252548 (hip osteoarthritis, posterior probability of causality [PPC] 0.99), is a predicted deleterious missense variant (Arg112His) in IL11 (interleukin 11), associated with increased risk of hip osteoarthritis. Using RNA sequencing (Online Methods), we find that IL11 shows increased expression in degraded compared to intact cartilage (log2 fold change [logFC]=0.787, false discovery rate [FDR]=4.82x10-3). This cytokine is a potent stimulator of bone formation10, is required for normal bone turnover11 and has been previously found to be up-regulated in osteoarthritis knee tissue and to be associated with disease progression12. The rs4252548 osteoarthritis risk allele is also associated with decreased adult height13. A recombinant human IL11 molecule (NEUMEGA) with three-fold-enhanced affinity for IL11RA, compared to IL1114, is approved for the treatment of chemotherapy-induced thrombocytopenia (Table 2). The likely effects of increased IL11 signalling in osteoarthritis joints are currently not well understood, and it is worth evaluating this therapeutic for potential efficacy in disease models.
The rs1126464 (osteoarthritis, PPC 0.89) locus index signal is a missense variant (Glu351Gln) in DPEP1, predicted to be tolerated. DPEP1 hydrolyses a wide range of dipeptides, and is implicated in the renal metabolism of glutathione and its conjugates. A DPEP1 inhibitor, cilastatin, is approved and used in combination with the antibiotic imipenem, in order to protect it from dehydropeptidase and prolong its antibacterial effect15. We suggest investigating the effects of cilastatin in osteoarthritis models to determine whether this has potential as a therapeutic, or whether an agonist may be efficacious.
rs75621460 (hip and/or knee osteoarthritis, single variant in the 95% credible set) is an intergenic variant residing downstream of CCDC97 and TGFB1 (Table 1, Supplementary Table 3), and is colocalised with a TGFB1 eQTL in sun-exposed skin (GTEx) (Supplementary Table 6). Mutations in TGFB1 cause Camurati–Engelmann disease characterised by diaphyseal dysplasia with thickening and fluctuating bone volume giving rise to bone pain, muscle weakness, gait issues and tiredness16,17. TGFB1 plays a critical role in skeletal development and adult bone homeostasis18, including bone remodelling19, osteoclast/osteoblast differentiation20,21 and chondrogenesis22. INVOSSA™, a TGFB1 cell and gene therapy in chondrocytes, was associated with significant improvements in function and pain in patients with knee osteoarthritis23.
The importance of TGFB1 signalling for osteoarthritis is supported by significant enrichment for “TGF Beta Signalling Pathway” genes (Supplementary table 10), including: LTBP1, LTBP3, SMAD3 and RUNX2. LTBP1, at the novel rs4671010 locus, encodes Latent-transforming growth factor beta-binding protein 1 which directly interacts with TGFB1, is involved in the assembly, secretion and targeting of TGFB1 to sites at which it is stored and/or activated, and may contribute to controlling the activity of TGFB124. LTBP3 (novel locus: rs10896015) encodes Latent-transforming growth factor beta-binding protein 3 which directly interacts with and activates TGFB1 in the early proliferative phase of osteogenic differentiation25. SMAD3 (known locus: rs12901372) encodes a transcriptional modulator and plays a critical role in chondrogenic differentiation, and regulates TGFB1 expression26; and the TGFB1/SMAD3 pathway regulates the expression of miR-140 in osteoarthritis27. The directionality of the colocalized eQTL and animal model data suggest that agonism/up-regulation of LTBP1, LTBP3 and SMAD3 may be therapeutic for osteoarthritis (Supplementary Table 21). RUNX2 (known locus: rs2064630) encodes a transcription factor essential for the osteoblast differentiation and chondrocyte maturation28, and is down-regulated by TGFB129. Given the genetic and biological support for the importance of TGFB1 in osteoarthritis aetiology and treatment, there may be scope for the development of simpler osteoarthritis therapeutics which target this mechanism, such as a small molecule or antibody.
Although not a current drug target, the novel SLC39A8 association is noteworthy. rs13107325 (osteoarthritis, PPC 0.99) is a missense variant located in SLC39A8 and demonstrates significantly increased expression in degraded compared to intact articular cartilage (logFC=0.522, FDR=5.80x10-5) (Table1; Supplementary Table 2), consistent with previously-reported increased levels of SLC39A8 in osteoarthritis compared to healthy chondrocytes30,31. rs13107325 is also associated with obesity32, hypertension33, Crohn's disease and altered microbiome composition34. SLC39A8 encodes a zinc transporter ZIP8 which functions in the cellular import of zinc at the onset of inflammation. Suppression of SLC39A8 has been shown to reduce cartilage degradation in osteoarthritis animal models30. The zinc-SLC39A8-MTF1 axis has been proposed to be an essential catabolic regulator of osteoarthritis pathogenesis31.
In this study, we have more than doubled the number of osteoarthritis risk loci, supported by integrated eQTL colocalization, fine-mapping, Mendelian bone disease, animal model and differential osteoarthritis joint expression data, to reveal putative effector genes. In addition to identifying chondrocyte and osteoblast biological mechanisms implicated in osteoarthritis susceptibility, we have revealed biological mechanisms that represent attractive targets for osteoarthritis drug discovery, and highlight approved therapeutics which represent viable considerations for repositioning as osteoarthritis therapies. We anticipate that this advance in basic understanding of osteoarthritis risk factors and mechanisms will stimulate the evaluation of novel drug targets for osteoarthritis.
Online Methods
Studies
UK Biobank
UK Biobank is a cohort of 500,000 participants aged 40-69 years recruited between 2006 and 2010 in 22 assessment centres throughout the UK35. The assessment visit included electronic signed consent; a self-completed touch-screen questionnaire; brief computer-assisted interview; physical and functional measures; and collection of biological samples and genetic data. This work was based on the third UK Biobank release, which includes the full set of the 500,000 genotypes imputed on the Haplotype Reference Consortium36 and the 1000 Genomes Consortium37.
The case and control definition, genotyping, imputation, and association testing are described in Supplementary Note.
Arthritis Research UK Osteoarthritis Genetics (arcOGEN) – cases
arcOGEN is a collection of unrelated, UK-based individuals of European ancestry with knee and/or hip osteoarthritis from the arcOGEN Consortium9,38. Cases were ascertained based on clinical evidence of disease to a level requiring joint replacement or radiographic evidence of disease (Kellgren–Lawrence grade ≥2). The arcOGEN study was ethically approved, and all subjects used in this study provided written, informed consent.
United Kingdom Household Longitudinal Study (UKHLS) – controls
The UKHLS, also known as Understanding Society, is a longitudinal panel survey of 40,000 UK households (England, Scotland, Wales and Northern Ireland) representative of the UK population. Participants are surveyed annually since 2009 and contribute information relating to their socioeconomic circumstances, attitudes, and behaviours via a computer assisted interview. The study includes phenotypical data for a representative sample of participants for a wide range of social and economic indicators as well as a biological sample collection encompassing biometric, physiological, biochemical, and haematological measurements and self-reported medical history and medication use. The UKHLS has been approved by the University of Essex Ethics Committee and informed consent was obtained from every participant. The genotyping, imputation and association testing have been previously described9,39,40 (Supplementary Note).
Meta-analysis
We meta-analysed the UK Biobank and arcOGEN datasets using fixed effects inverse-variance weighted meta-analysis in METAL41. We performed meta-analyses across osteoarthritis definitions using summary statistics from the UK Biobank and arcOGEN cohorts, and defined genome-wide significance based on the meta-analysis combined P value as outlined below.
Significance threshold
The osteoarthritis traits analysed in this study are highly correlated. To calculate M_eff the effective number of independent traits, we estimated the genetic correlation matrix between the 4 osteoarthritis traits (Supplementary Table 1) using LDscore42 with genome-wide summary statistics of common-frequency variants in the UK Biobank dataset. We then calculated M_eff from the eigenvalues λ_i of the correlation matrix43:
For the M = 4 osteoarthritis phenotypes in this study, M_eff = 1.6046. We therefore use P≤3x10-8 as the threshold corrected for the effective number of traits to report genome wide significance.
Statistical independence
To define independent signals within a GWAS, we performed physical clumping using a simple iterative procedure. We rank all variants that reach a P value threshold according to their P value. The variant with the smallest P value is considered the index variant of that signal and any variants within 1MB region either side of that index variant that reach the pre-defined P value threshold are clumped with that variant. We repeat the procedure until no more variants that reach the pre-defined P value threshold exist that have not been assigned to a physical clump. To test that the index variants defined by this procedure are statistically independent, we performed an approximate stepwise model selection procedure, as implemented by COJO in GCTA7. An independent signal in a region is declared if its P value of association in the stepwise regression is less than 3x10-8. LD calculations were based on the full UK Biobank imputed set.
To define independent signals across the four osteoarthritis GWAS, we performed reciprocal approximate conditional analyses, as implemented by COJO in GCTA7, of each index variant of one GWAS conditioned on each index variant of the other GWAS. A signal between two GWAS is considered to be the same if the P value of an index variant of one GWAS conditioned on an index variant of the other GWAS is ≤ 10-5 or a P value difference between conditional and unconditional analysis of less than 2 orders of magnitude.
To investigate statistical independence between index variants from each GWAS and previously reported variants, we performed approximate conditional analysis, as implemented by COJO in GCTA7, of each index variant conditional on all previously reported variants within 1Mb region, each one at a time. The index variant was considered independent from a previously reported variant if it had a conditional P ≤ 10-5 or a P value difference between conditional and unconditional analysis of less than 2 orders of magnitude. Variants were classified as known (denoting either a previously reported variant, or a variant for which the association signal disappears after conditioning on the lead variant of a previously reported locus) or newly identified (denoting a variant which is conditionally independent of previously reported loci).
Fine-mapping
We constructed regions for fine-mapping by taking a window of 1Mb either side of each index variant. Within each region, we performed an approximate stepwise model selection procedure, as implemented by COJO in GCTA7, using the meta-analysis summary statistics and LD calculations based on the UK Biobank cohort to determine the number of independent signals. We consider conditionally distinct signals those where the stepwise regression association reaches genome-wide significance (P<3.0x10-8). We then perform single-SNP approximate association analyses conditional on the set of SNPs identified by the model selection procedure, again using COJO, and we calculate Wakefield’s asymptotic Bayes’ factors8 (ABF). In particular, when there is a single causal variant in the region, ABF is based on the marginal summary statistics of the meta-analysis. When there are multiple causal variants in the region, for each signal we calculate a set of ABF using the conditional summary statistics of the meta-analysis conditioned on all other signals. For each signal, we then calculate posterior probabilities of each variant being causal and a 95% credible set, which contains the minimum set of variants that jointly have at least 95% probability of including the causal variant. As this number can be large, we focus on the variants in the 95% credible set that have posterior probability of causality (PPC) over 3% and also on any variants in the 95% credible set with moderate or high consequence (irrespective of their PPC).
Genetic correlation analysis
To better understand the degree to which genetic architecture is shared across osteoarthritis and other complex traits, LD score regression5 was performed as implemented in the LDHub pipeline6 (URLs). We calculated the genome-wide genetic correlation between each of the osteoarthritis definitions and all available 832 human traits and diseases (accessed 15-18 June 2018). Of these, 597 traits were available within the UK Biobank resource. In each analysis, all variants in the major histocompatibility complex (MHC) region on chromosome 6 (26–34 MB) were removed and only variants with rsIDs were included in the analyses, yielding 1203892 - 1204029 variants overlapping with LDHub. We used the Benjamini-Hochberg false discovery rate and the effective number of independent traits tested for multiple testing correction. The level of significance was set at FDR-corrected P<0.05.
Mendelian randomization
We performed Mendelian randomization analyses using the MR-Base platform44. We tested the bidirectional causal associations of each of the four osteoarthritis datasets with 991 exposures/outcomes in MR-Base. Statistical significance was considered at P<6.3x10-6. To follow up on pain associations, we performed analyses of knee and hip pain as an outcome after excluding all individuals self-reporting or hospital-diagnosed with osteoarthritis in UK Biobank. All instruments were aggressively clumped prior to analysis (LD r2<0.001) and inverse variance-weighted (IVW), Median-weighted, and MR-Egger analyses were performed for multi-variant instruments, and Wald ratio estimators were used to assess causality for single variant instruments.
Transcriptome-wide association
We used a gene-based approach, MetaXcan45, to test for associations between the osteoarthritis traits and predicted expression levels in 48 human tissues from GTEx V746. MetaXcan leverages a set of reference individuals for whom both gene expression and genetic variation have been measured to impute the cis-genetic component of expression into a much larger set using the elastic net model. It then correlates the imputed gene expression to the trait of interest and performs a transcriptome-wide association study to identify significant expression-trait associations. We used a conservative Bonferroni correction to account for the gene-tissue pairs (20,000 genes across 48 tissues), leading to a significance threshold of 5.20x10-8. To reduce the effect of LD confounding on the MetaXcan results, when different causal SNPs are affecting expression levels and the phenotypic trait in a GWAS, we estimated the probability of colocalization of each GWAS and expression quantitative trait locus (eQTL) signal in each significant MetaXcan result using Coloc47 (Supplementary Note, Supplementary Figure8, Supplementary Tables 8 and 9).
Colocalization analysis
To assess whether the genome-wide significant osteoarthritis signals colocalise with eQTL signals, and therefore potentially share a causal molecular mechanism, we employed the Coloc method47, which uses asymptotic Bayes factors with summary statistics and regional LD structure to estimate five posterior probabilities: no association with either GWAS or eQTL (PP0), association with GWAS only (PP1), association with eQTL only (PP2), association with GWAS and eQTL but two independent SNPs (PP3), and association with GWAS and eQTL having one shared SNP (PP4). A large posterior probability for PP4 indicates support for a single variant affecting both GWAS and eQTL studies. For each of the GWAS signals, we defined a 100kb region either side of the index variant, and tested for colocalization within the entire cis-region of any overlapping eQTLs (transcription start and end position of an eQTL gene plus and/or minus 1Mb, as defined by GTEx) in 48 human tissues from GTEx V746. A PP4 over or equal to 80% was considered as evidence for colocalization (Supplementary Note, Supplementary Table 7).
Most colocalization methods, such as Coloc, rely on the availability of genome-wide eQTL results, which are not always readily available. For eQTL datasets with no publically available full summary statistics, we used an alternative approach that estimates the probability of colocalization using published top eQTL signals. First, we estimated the credible sets for the eQTLs using the Probabilistic Identification of Causal SNPs (PICS) method48 for each index SNP for each gene from 27 eQTL studies (Supplementary Table 6). PICS is a fine-mapping algorithm that assumes one causal signal tagged by a single index SNP per locus. For neutral SNPs (SNPs whose association signals are due to LD with the causal SNP), the strength of association scales linearly with the r2 relationship/distance to the index SNP. Under this assumption, PICS can estimate the posterior probability of a given SNP being causal using LD information from the 1000 Genomes database. Second, we generated PICs credible sets for osteoarthritis GWAS index SNPs. We then performed a colocalization analysis of the osteoarthritis GWAS and eQTL PICs credible sets using an adapted Coloc method49. Given that PICs calculates the posterior probabilities for each SNP in the credible set, we bypassed the need for calculating the Bayes Factors using Wakefield’s approximate Bayes Factor method which is reliant on full summary statistics. Colocalizations with a posterior probability greater than 0.8 were considered positive. This method was benchmarked on other GWAS datasets, and we found the false positive rate to be no higher than the standard Coloc package.
We observe evidence of colocalization in at least one tissue for 50 out of our 64 loci using any of the 3 methods (MetaXcan, Coloc, Piccolo), 41 of which are at newly associated osteoarthritis signals (Supplementary Table 7). MetaXcan alone identified 119 genes, Coloc 113 and Piccolo 58, while the overlap of all 3 methods implicate 20 genes (TGFA, ILF3, CSK, CYP1A1, ULK3, CHMP1A, TSKU, SUPT3H, GNL3, NT5DC2, LMX1B, SMAD3, MLXIP, COLGALT2, FAM89B, UQCC1, NFAT5, ALDH1A2, FAM53A, FGFR3; Supplementary Figure 7).
Heritability estimation
To investigate the narrow sense heritability for the four osteoarthritis disease definitions, we ran LDscore42, which uses summary statistics at common-frequency variants genome-wide (independent of P value thresholds) and LD estimates between variants while accounting for sample overlap. To calculate the population prevalence in the UK (65 million people), we consulted Arthritis Research UK figures: 8.75 million people have symptomatic osteoarthritis, while 2.46 and 4.11 million people have osteoarthritis of the hip and the knee, respectively. We assumed that 2.46+4.11 million people have osteoarthritis of the hip and/or the knee. We estimated the phenotypic variance explained by the 99previously and newly reported variants that reached genome-wide significance in the meta-analysis between UK Biobank and arcOGEN, as a function of allele frequency (Figure 2; Supplementary Table 17). The phenotypic variance explained by a variant is) ln(OR)2 × 2 × EAF × (1 − EAF), where ln(OR) is the natural logarithm of the OR of the variant in the meta-analysis and EAF is its weighted effect allele frequency across UK Biobank and arcOGEN. Variants associated with hip osteoarthritis tend to have larger effect size estimates and hence explain more of the phenotypic variability (Figure 2; Supplementary Table 17). The hip osteoarthritis dataset is the smallest in both the UK Biobank and arcOGEN cohorts (18% and 59% fewer cases compared to knee osteoarthritis and osteoarthritis at any joint in UK Biobank, respectively).
Pathway analysis
We performed gene-set analyses for each of the osteoarthritis phenotypes separately, using MAGMA v1.0650. We mapped variants to 19,427 protein-coding genes (NCBI 37.3), including a 10kb window on either side of the gene. We then computed gene P values based on individual variant association P values. We used the ‘snp-wise=mean’ model, which calculates the mean of the χ2-statistic amongst the single variant P values in each gene, and applied default MAGMA QC steps. Genotype data of 10,000 individuals (subset of self-reported plus hospital-diagnosed osteoarthritis at any site analysis), were used to calculate LD (as measured by r2). We carried out a one-sided competitive gene-set analysis for each phenotype, implemented as a linear regression model on a gene data matrix created internally from the gene-based results. Briefly, this converts the gene-based P values to Z-scores, and tests if the mean association with the phenotype of genes in the gene set is greater than that of all other genes. We used Kyoto Encyclopedia of Genes and Reactome (accessed through MSigDB113 (version 5.2) on 23 January 2017). We also downloaded Gene Ontology (GO) biological process and molecular function gene annotations from Ensembl (version 87). We used annotations with the following evidence codes: a) Inferred from Mutant Phenotype (IMP); b) Inferred from Physical Interaction (IPI); c) Inferred from Direct Assay (IDA); d) Inferred from Expression Pattern (IEP); and e) Traceable Author Statement (TAS). KEGG/Reactome and GO annotations were analysed separately and only pathways that contained between 20 and 200 genes were included (594 for KEGG/Reactome, 619 for GO). We used MAGMA’s built-in permutation method (k=10,000 permutations) to produce corrected competitive P values with a family-wise error rate (FWER) of 5%. We then further adjusted these corrected competitive P values for the effective number of independent traits tested (1.6046).
We also performed gene set enrichment analysis by using DEPICT (URLs) and PASCAL (URLs). DEPICT version 1 rel194 was downloaded from GitHub (URLs) on 14/06/2018. We run DEPICT separately in each of the four osteoarthritis definitions for the variants with a meta-analysis P<1x10-5. Briefly, DEPICT first clumped the variants with P<1x10-5 using 500 kb flanking regions as physical distance threshold and an r2>0.1 with PLINK51 to obtain lists of independent SNPs, resulting in 864 clumps. Variants within the major histocompatibility complex region on chromosome 6 were excluded. DEPICT analyses were conducted using the default settings: 50 repetitions to compute FDR and 500 permutations based on 500 null GWAS to compute P values adjusted for gene length. All 14,461 available reconstituted gene sets were used representing a wide spectrum of biological and mouse phenotypic annotations. We also used the method implemented in PASCAL to perform gene set enrichment analysis which accounts for LD structure in the genome and particularly of highly correlated chromosomal regions containing multiple genes that can negatively impact the results of the analysis. In this approach, variants were first mapped to genes, including a 10kb window on either side of the gene. We then computed gene scores by aggregating the single-marker association values with the LD structure. Finally, the scores of genes that belong to the same pathways (i.e. gene sets) were used to compute pathway scores and determine the statistical significance of the association between the pathway and each of the osteoarthritis phenotypes. Here we used exactly the same pathways of the MAGMA analysis. The gene and the pathway scores were performed by using the sum gene score and the chi-squared approach respectively, as implemented in PASCAL. All pathway P values obtained by either software were adjusted for multiple testing correction by using FDR and the effective number of independent traits. The level of significance was set at FDR-corrected P<0.05.
Monogenic enrichment analysis
We compiled a systematic list of genes causing bone phenotypes in humans by scanning the STOPGAP database 52, which uses OMIM (URLs) and Orphanet (URLs) to define genes underlying monogenic/Mendelian diseases. We selected all genes causing monogenic diseases and annotated with MeSH terms (Medical Subject Headings) related to bone, cartilage or joint disease, including: “bone disease, developmental”, “osteochondrodysplasias”, “osteogenesis imperfecta”, “osteoporosis”, “osteopetrosis”, “arthritis, juvenile” and “arthrogryposis”. Other bone-, cartilage or joint related mesh terms linked to less than 10 genes in the STOPGAP database were excluded from the analysis. Additionally, we selected a list of well-validated genes underlying syndromic or non-syndromic forms of early onset osteoarthritis (EO-OA) from a review by Aury-Landas et al.53. For enrichment analysis, genes residing within 500kb of each index variant identified in our GWAS were considered as osteoarthritis loci, and the rest of the genes in the genome associated to any mesh term in STOPGAP were considered non-osteoarthritis loci. We built a 2x2 table by counting the number of genes annotated to each of the above-mentioned MeSH terms among osteoarthritis and non-osteoarthritis loci. We assessed evidence for enrichment using a Fisher’s exact test.
Transcriptomic and proteomic analyses
Patients and samples
We collected cartilage samples from 38 patients undergoing total joint replacement surgery: 12 knee osteoarthritis patients (cohort 1; 2 women, 10 men, age 50-88 years); knee osteoarthritis patients (cohort 2; 12 women 5 men, age 54-82 years); 9 hip osteoarthritis patients (cohort 3; 6 women, 3 men, age 44-84 years). We collected matched intact and degraded cartilage samples from each patient. Cartilage was separated from bone and chondrocytes were extracted from each sample. From each isolated chondrocyte sample, we extracted DNA, RNA and protein. All patients provided full written informed consent prior to participation. The human biological samples were sourced ethically and their research use was in accord with the terms of the informed consents under an IRB/EC approved protocol. All sample collection, DNA, RNA and protein analysis steps are described in detail in Steinberg et al54.
Proteomics and RNA sequencing
Proteomics analysis was performed on intact and degraded cartilage samples from 24 individuals (15 from cohort 2, 9 from cohort 3). We performed a gene expression analysis on samples from all 38 patients (Supplementary Note).
Animal model data
The presence of abnormal skeletal phenotypes in mice was evaluated for all genes within 500kb of an osteoarthritis index variant and extracted from Open Targets55. This platform integrates all abnormal phenotype annotations for mutations in mouse genes reported in the literature and curated at MGI (URLs). Given the list of genes located less than 1 Mb away of the 64 genome-wide significant signals for osteoarthritis, abnormal skeletal system phenotypes from mutant mice were extracted systematically for all mouse orthologs of the human genes using the programmatic interface of the Open Targets platform (Supplementary Table 21). For instance, mutant mice homozygous for a targeted mutation of Smad3 (the ortholog of human SMAD family member 3) developed degenerative joint disease by progressive loss of articular cartilage56. Additional manual PubMed searches were conducted on selected genes to obtain information regarding animal models specific for osteoarthritis (Supplementary Table 20).
Supplementary Material
Editorial Summary.
Genome-wide meta-analysis of UK Biobank and arcOGEN (77,052 cases and 378,169 controls) identifies 52 new osteoarthritis risk loci. Integrated eQTL colocalization, fine-mapping, and rare disease data identify putative effector genes for osteoarthritis.
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Numbers 26041 and 9979. This work was funded by the Wellcome Trust (206194). We are grateful to Roger Brooks, Andrew McCaskie, Jyoti Choudhary and Theodoros Roumeliotis for their contribution to the transcriptomic and proteomic data collection, and to Arthur Gilly for help with figures. The Human Research Tissue Bank is supported by the NIHR Cambridge Biomedical Research Centre. arcOGEN was funded by a special purpose grant from Arthritis Research UK (grant 18030). The UK Household Longitudinal Study was funded by grants from the Economic & Social Research Council (ES/H029745/1) and the Wellcome Trust (WT098051). UKHLS is led by the Institute for Social and Economic Research at the University of Essex. The survey was conducted by NatCen and the genome-wide scan data were analysed and deposited by the Wellcome Sanger Institute. Information on how to access the data can be found on the Understanding Society website https://www.understandingsociety.ac.uk/. PICCOLO was developed by Karsten Sieber and Karl Guo. GDS and TRG receive funding from the UK Medical Research Council (MC_UU_00011/1 and MC_UU_00011/4). The authors would like to acknowledge Open Targets for enabling the collaboration on this work.
Footnotes
Data Availability
All RNA sequencing data have been deposited to the European Genome/Phenome Archive (cohort 1: EGAD00001001331; cohort 2: EGAD00001003355; cohort 3: EGAD00001003354). Genotype data of the arcOGEN cases and UKHLS controls have been deposited at the European Genome-phenome Archive under study accession numbers EGAS00001001017 and EGAS00001001232, respectively.
URLs
LDHub, http://ldsc.broadinstitute.org/; OMIM, https://www.omim.org/; Orphanet, http://www.orpha.net/; HRC pre-imputation checking tool, http://www.well.ox.ac.uk/~wrayner/tools/#Checking; MGI, http://www.informatics.jax.org; Open targets, https://www.opentargets.org/; Understanding Society, https://www.understandingsociety.ac.uk/; DEPICT, www.broadinstitute.org/depict; PASCAL www2.unil.ch/cbg/index.php?title=Pascal; DEPICT version 1 rel194 GitHub https://github.com/perslab/depict; Chembl, (https://www.ebi.ac.uk/chembl/); Clinical trials.gov, (https://www.clinicaltrials.gov/)
Author Contributions
UK Biobank association analyses: IT, LYA, RS, TJ, JH, EZ, JEG, KH, MK
arcOGEN analyses: arcOGEN, LS
Mendelian randomization: VH, JZ, RS, TG, GDS
Functional genomics: JMW, JEG, LMC, JS, LS, SB, DS, EZeggini
Translation work: LMC, JEG, NB, EZeggini
Manuscript writing: IT, KH, LS, JEG, LMC, RS, EZeggini
Competing Interests
IT, JEG, TJ, LYA, JDH, NB, RS, LMC are employees of GlaxoSmithKline and may own company stock. TRG receives research funding from GlaxoSmithKline and Biogen. VH is funded by a research grant from GlaxoSmithKline.
References
- 1.Vos T, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2163–96. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiligsmann M, et al. Health economics in the field of osteoarthritis: an expert's consensus paper from the European Society for Clinical and Economic Aspects of Osteoporosis and Osteoarthritis (ESCEO) Semin Arthritis Rheum. 2013;43:303–13. doi: 10.1016/j.semarthrit.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Baker PN, et al. The effect of surgical factors on early patient-reported outcome measures (PROMS) following total knee replacement. J Bone Joint Surg Br. 2012;94:1058–66. doi: 10.1302/0301-620X.94B8.28786. [DOI] [PubMed] [Google Scholar]
- 4.Zengini E, et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet. 2018;50:549–558. doi: 10.1038/s41588-018-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 9.arcOGEN Consortium et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–23. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi Y, et al. Interleukin-11 as a stimulatory factor for bone formation prevents bone loss with advancing age in mice. J Biol Chem. 2002;277:49011–8. doi: 10.1074/jbc.M207804200. [DOI] [PubMed] [Google Scholar]
- 11.Sims NA, et al. Interleukin-11 receptor signaling is required for normal bone remodeling. J Bone Miner Res. 2005;20:1093–102. doi: 10.1359/JBMR.050209. [DOI] [PubMed] [Google Scholar]
- 12.Chou CH, et al. Insights into osteoarthritis progression revealed by analyses of both knee tibiofemoral compartments. Osteoarthritis Cartilage. 2015;23:571–80. doi: 10.1016/j.joca.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanktree MB, et al. Meta-analysis of Dense Genecentric Association Studies Reveals Common and Uncommon Variants Associated with Height. Am J Hum Genet. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmegnies D, et al. Characterization of a potent human interleukin-11 agonist. Biochem J. 2003;375:23–32. doi: 10.1042/BJ20030459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keynan S, Hooper NM, Felici A, Amicosante G, Turner AJ. The renal membrane dipeptidase (dehydropeptidase I) inhibitor, cilastatin, inhibits the bacterial metallo-beta-lactamase enzyme CphA. Antimicrob Agents Chemother. 1995;39:1629–31. doi: 10.1128/aac.39.7.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssens K, et al. Camurati-Engelmann disease: review of the clinical, radiological, and molecular data of 24 families and implications for diagnosis and treatment. J Med Genet. 2006;43:1–11. doi: 10.1136/jmg.2005.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuldashev AJ, et al. Orthopedic Manifestations of Type I Camurati-Engelmann Disease. Clin Orthop Surg. 2017;9:109–115. doi: 10.4055/cios.2017.9.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M, Chen G, Li YP. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4 doi: 10.1038/boneres.2016.9. 16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y, et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757–65. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, et al. Transforming Growth Factor beta1/Smad4 Signaling Affects Osteoclast Differentiation via Regulation of miR-155 Expression. Mol Cells. 2017;40:211–221. doi: 10.14348/molcells.2017.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou S. TGF-beta regulates beta-catenin signaling and osteoblast differentiation in human mesenchymal stem cells. J Cell Biochem. 2011;112:1651–60. doi: 10.1002/jcb.23079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou S, Eid K, Glowacki J. Cooperation between TGF-beta and Wnt pathways during chondrocyte and adipocyte differentiation of human marrow stromal cells. J Bone Miner Res. 2004;19:463–70. doi: 10.1359/JBMR.0301239. [DOI] [PubMed] [Google Scholar]
- 23.Kim MK, et al. A Multicenter, Double-Blind, Phase III Clinical Trial to Evaluate the Efficacy and Safety of a Cell and Gene Therapy in Knee Osteoarthritis Patients. Hum Gene Ther Clin Dev. 2018;29:48–59. doi: 10.1089/humc.2017.249. [DOI] [PubMed] [Google Scholar]
- 24.Nuchel J, et al. TGFB1 is secreted through an unconventional pathway dependent on the autophagic machinery and cytoskeletal regulators. Autophagy. 2018;14:465–486. doi: 10.1080/15548627.2017.1422850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koli K, Ryynanen MJ, Keski-Oja J. Latent TGF-beta binding proteins (LTBPs)-1 and -3 coordinate proliferation and osteogenic differentiation of human mesenchymal stem cells. Bone. 2008;43:679–88. doi: 10.1016/j.bone.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Cheung KS, et al. MicroRNA-146a regulates human foetal femur derived skeletal stem cell differentiation by down-regulating SMAD2 and SMAD3. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098063. e98063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tardif G, et al. NFAT3 and TGF-beta/SMAD3 regulate the expression of miR-140 in osteoarthritis. Arthritis Res Ther. 2013;15:R197. doi: 10.1186/ar4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura R, Hata K, Nakamura E, Murakami T, Takahata Y. Transcriptional network systems in cartilage development and disease. Histochem Cell Biol. 2018;149:353–363. doi: 10.1007/s00418-017-1628-7. [DOI] [PubMed] [Google Scholar]
- 29.Kanaan RA, Kanaan LA. Transforming growth factor beta1, bone connection. Med Sci Monit. 2006;12:RA164–9. [PubMed] [Google Scholar]
- 30.Song J, et al. MicroRNA-488 regulates zinc transporter SLC39A8/ZIP8 during pathogenesis of osteoarthritis. J Biomed Sci. 2013;20:31. doi: 10.1186/1423-0127-20-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, et al. Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell. 2014;156:730–43. doi: 10.1016/j.cell.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang R, et al. A blood pressure-associated variant of the SLC39A8 gene influences cellular cadmium accumulation and toxicity. Hum Mol Genet. 2016;25:4117–4126. doi: 10.1093/hmg/ddw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D, et al. A Pleiotropic Missense Variant in SLC39A8 Is Associated With Crohn's Disease and Human Gut Microbiome Composition. Gastroenterology. 2016;151:724–32. doi: 10.1053/j.gastro.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panoutsopoulou K, et al. Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann Rheum Dis. 2011;70:864–7. doi: 10.1136/ard.2010.141473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evangelou E, et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann Rheum Dis. 2014;73:2130–6. doi: 10.1136/annrheumdis-2012-203114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prins BP, et al. Genome-wide analysis of health-related biomarkers in the UK Household Longitudinal Study reveals novel associations. Sci Rep. 2017;7 doi: 10.1038/s41598-017-10812-1. 11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–56. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbeira A, et al. MetaXcan: Summary Statistics Based Gene-Level Association Method Infers Accurate PrediXcan Results. Preprint at bioRxiv. 2016 doi: 10.1101/045260. [DOI] [Google Scholar]
- 46.Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank. 2015;13:307–8. doi: 10.1089/bio.2015.29031.hmm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo C, et al. The American Society of Human Genetics. San Diego: 2018. A little data goes a long way: Finding target genes across the GWAS Catalog by colocalizing GWAS and eQTL top hits. [Google Scholar]
- 50.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen J, Song K, Slater AJ, Ferrero E, Nelson MR. STOPGAP: a database for systematic target opportunity assessment by genetic association predictions. Bioinformatics. 2017;33:2784–2786. doi: 10.1093/bioinformatics/btx274. [DOI] [PubMed] [Google Scholar]
- 53.Aury-Landas J, Marcelli C, Leclercq S, Boumediene K, Bauge C. Genetic Determinism of Primary Early-Onset Osteoarthritis. Trends Mol Med. 2016;22:38–52. doi: 10.1016/j.molmed.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 54.Steinberg J, et al. Integrative epigenomics, transcriptomics and proteomics of patient chondrocytes reveal genes and pathways involved in osteoarthritis. Sci Rep. 2017;7 doi: 10.1038/s41598-017-09335-6. 8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koscielny G, et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 2017;45:D985–D994. doi: 10.1093/nar/gkw1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X, et al. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153:35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.