Abstract
INTRODUCTION:
The tumor microenvironment within the breast is rich in adipose elements. The interaction between adipose cells and breast cancer is poorly understood, particularly as it pertains to patients with genetic susceptibility to breast cancer. Our study focuses on the phenotype of human adipose-derived stem cells (ASCs) with the BRCA1 mutation and the effect they may have on breast cancer cell behavior.
METHODS:
CRISPR/Cas9 was used to generate de novo BRCA1-knockdown human ASCs (BRCA1-KD). The effect of the BRCA1 knockdown on the ASC phenotype was compared to wild type ASCs and patient derived breast ASCs with known BRCA 1 mutations. Interactions between ASCs and the MDA-MB-231 breast cancer cell line were evaluated.
RESULTS:
BRCA1-KD ASCs stimulated MDA-MB-231 proliferation (1.4-fold increase on day4, P=0.0074) and invasion (2.3-fold increase on day2. P=0.0171) compared to wild type ASCs. Immunofluorescence staining revealed higher levels of phosphorylated ATM activation in BRCA1-KD ASCs (72.9±5.32% vs 42.9±4.97%. P=0.0147), indicating higher levels of DNA damage. Beta-galactosidase staining demonstrated a significantly higher level of senescence in BRCA1-KD ASCs compared to wild type ASCs (7.9±0.25% vs 0.17±0.17%, P<0.0001). Using quantitative ELISA to evaluate conditioned media, we found significantly higher levels of IL-8 in BRCA1-KD ASCs (2.57±0.32-fold, P=0.0049).
CONCLUSION:
We have shown here for the first time that the BRCA1 mutation affects the ASC phenotype. Moreover, CRISPR/Cas9 generated BRCA1-KD ASCs stimulate a more aggressive behavior in breast cancer cells than wild type ASCs. This appears to be related to increased inflammatory cytokine production via a DNA damage-mediated cell senescence pathway.
INTRODUCTION
With the rapid advancement of genomic programs in cancer care, management of patients with breast cancer-associated gene mutations is becoming more common. Of these, BRCA tumor suppressor gene mutations are the best characterized. Patients with BRCA1 mutations have a 50–65% lifetime risk of developing breast cancer with a preponderance of an aggressive subtype known as triple negative breast cancer (TNBC) (1). Research has shown that breast epithelial cells with the BRCA1 mutation can demonstrate accelerated breast carcinogenesis (2). Recent advances have demonstrated that hereditary mutations in breast stroma coincide significantly with genomic alterations in breast epithelial cells (3, 4). However, the possibility that inherited mutations may also impact breast stromal cells, thus influencing cancer progression, has not been tested.
It has been previously established that cells harboring BRCA mutations are sensitive to DNA-damage (5). DNA damage can lead to cellular senescence and a senescence-associated secretory phenotype (SASP), defined by their secretion of inflammatory cytokines, including IL-6 and IL-8 (6). Cytokines associated with SASP can drive aggressive cancer behaviors (7, 8) and may play a role in TNBC progression.
Targeting active pathways within the tumor microenvironment may be especially beneficial for TNBC. Our study focuses on the phenotype of human adipose-derived stem cells (ASCs) with the BRCA1 mutation and the resulting effects on breast cancer cells. We hypothesize that the BRCA1 mutation in ASCs leads to a functional abnormality of these cells. Furthermore, we hypothesize that the mutation in the BRCA1 protein, a key player in DNA damage response pathway (8), induces cell senescence in ASCs and promotes the secretion of inflammatory cytokines, which act in a paracrine manner to promote tumor cell growth/progression.
MATERIALS AND METHODS
Adipose-derived Stem Cells (ASCs)
One commercial ASC line (Lonza PT-5006) was obtained for CRISPR-mediated BRCA1 knockdown. ASCs were maintained in ASC growth media (Lonza PT-3273 & PT-4503). Cell media was changed every 3 days, and 0.5% Typsin were used to passage the cells every 6 days according to published protocol (9). Maintenance of adipogenesis of the ASCs was confirmed by culturing 1×105 ASCs in adipogenic induction media (Lonza PT-3004) for 14 days. Cells were fixed with 4% paraformaldehyde and stained with Oil Red O (Sigma O0625).
Human Primary Breast Adipose-derived Stem Cells (brASCs)
To confirm findings from our CRISPR-mediated BRCA1-knockdown ASC model, we also used human brASCs. Human breast adipose tissues were obtained at the time of mastectomy in accordance with our IRB approved protocol. Five female patient ASCs were used for this study. Patient age, gender, BMI, BRCA1 mutation status, breast cancer type, and receptor status were obtained using online electronic records at Duke University Health System. Breast adipose tissue was digested with collagenase (Sigma C0130) in a 3:1 ratio with agitation for 60 minutes. The stromal vascular fraction was isolated with centrifugation and a cellular pellet was generated in the standard fashion. The pellet was then incubated with erythrocyte lysis buffer (154 mM NH4Cl, 10mM KHCO3, 1mM EDTA) for 10 minutes, filtered, and centrifuged. Adipose-derived stem cells were then plated and expanded or stored for future experiments as per our standard protocol.
CRISPR/Cas9 Sequence Cloning to generate BRCA1 knockdown in human ASCs (BRCA1-KD ASCs)
BRCA1 gene knockouts in human adipose derived stem cells were performed according to published methods using lentiCRISPRv2 vector system (10, 11). BRCA1 gene target was first selected using the online tool (12, 13): SgRNA1 sequence: TGGTCACACTTTGTGGAGAC; SgRNA2 sequence: GGTTTCTGTAGCCCATACTT. Then DNA oligos (Genewiz, Morrisville, NC) were digested and dephosphorylated with restriction enzyme Esp3I (Thermo Fisher ER0451). The plasmids were purified using QIAquick Gel Extraction Kit (Qiagen 28704) and eluted in EB buffer (10mM Tris-Cl, pH 8.5). Then the purified plasmids were annealed using T4 Polynucleotide Kinase (NEB M0201S). Annealed oligos were diluted 1:200 and cloned into the lentiCRISPRv2 backbone. The plasmids were then transformed into competent bacteria. Transfer plasmids (lentiCRISPR plasmids, luciferase plasmids) were co-transfected into HEK293T cells with packaging plasmids pMD2.G (AddGene 12259) and psPAX2 (AddGene 12260). Virus yield was collected and added to target cells. Successfully infected cells were selected using 1ug/mL Puromycin (ThermoFisher A1113802). CRISPR negative controls were generated using non-targeting gRNAs that do not recognize any sequence in the human genome.
Tumor Cell Proliferation Assays
MDA-MB-231 cells (triple negative breast cancer cells) were labelled with luciferase gene using lentiviruses. To do this, 1× 105 ASCs and 1E4 MDA-MB-231 cells were seeded together in 12-well plates containing 5% FBS media. The luciferase absorbance was measured every 48hrs over 8 days. Tumor cell growth rate at various time points was calculated by comparing the luciferase levels to day 0. Growth rates were compared using t-tests.
Western Blot Analysis
Cells were collected and lysed using lysis buffer. 20–30ug of lysates were loaded onto 10% SDS-PAGE gel with molecular markers and ran at 120V for 1–2hrs. Proteins were transferred to PVDF membranes. Then the membranes were stained with primary and secondary antibodies. Experiments were performed according to published protocol (14). BRCA1 protein expression was analyzed with western blotting using BRCA1 (Bethyl A300–000A) and anti-rabbit IgG antibodies (abcam ab6721). GADPH protein expressions were used as controls using GADPH (abcam ab8245) and anti-mouse antibodies (abcam ab6785). Images were acquired using darkroom development, and results were quantified using ImageJ (LOCI, Madison, Wisconsin, 11).
Transwell Assays
Basement membrane matrix was created using Cultrex BME solution and buffer (Trevigen 3455–096-02&03) and used to coat 0.8 um Millicell cell culture inserts (Millipore PICM01250). 2×104 ASCs were plated onto each of the 24-well plate with 600uL 10%FBS media, and 1×103 MDA-MB-231 cells were plated into each insert in the presence of 100 μL of serum-free media. Details can be found in published protocol (15). After 48 hours, tumor cell invasion to the lower side of the chamber insert was determined following Crystal Violet staining for 2hrs. All tumor cells with positive staining were counted in 10 high power fields per sample and compared between BRCA1-knockdown ASCs and controls using t tests.
Beta-galactosidase Enzyme Assays
Senescence beta-galactosidase staining kit (Cellsignal 9860) was used according to manufacture protocol after plating 1×105 ASCs in 6-well plates. Cells were first fixed with fixative buffer (Cellsignal 11674), and then stained with staining buffer (Cellsignal 11675) at 37 degrees. After 24hrs of staining, ASCs were imaged under 10x magnification, and 10 images were recorded per well. ASCs with positive beta-galactosidase staining and total cell numbers were counted. The ratio of positive beta-galactosidase ASCs was compared using t tests.
ELISA
Conditioned media was collected from ASCs after 48hr incubation in 10%FBS media. Inflammatory cytokine multi-analyte ELISArray kit (Qiagen MEH-004A) was used according to protocol to measure cytokine levels in the conditioned media. T-tests were used to compare the cytokine levels in different groups of ASCs.
Immunofluorescence Staining
Immunofluorescence staining was performed for phosphorylated ATM (pATM), an established marker for DNA double strand breaks. 1×105 ASCs were formalin fixed onto 3cm plates with glass bottoms, these plates were incubated in pATM (Rockland pS1981) and BRCA1 (Benthyl A301–377A) primary antibodies for 12 hours. After blocking in buffers, fluorochrome-conjugated secondary antibodies (ab150074, ab150117) were applied. Experiments were performed according to published protocol (16). Images were taken on the Zeiss Z1 Observer inverted fluorescence microscope (Carl Zeiss Microscopy, LLC, NY). 15 high power field (20x magnification, 20–35 cells per image) images were taken per sample, every 3 images were counted as a replicate. Cells positive for BRCA1 and/or pATM foci were identified. The ratio of pATM-positive cells in BRCA1-positive and BRCA1-negative cells were compared using t tests.
Statistical Analysis
Statistical analysis was conducted using GraphPad Prism 7.0 for MAC OS X, GraphPad Software, La Jolla California USA, www.graphpad.com. T-tests were performed for all experiments, and significance level was set at 0.05. Because the sample sizes were small (N<5), effect sizes were calculated for all samples to confirm the power of the analysis (17). Literature has shown that t-tests are acceptable for small sample sizes when the effect size is large (18) (effect size > 0.8) (19).
RESULTS
CRISPR/Cas9-mediated Knockdown of BRCA1 in Human ASCs
Using CRISPR/Cas9-mediated system, we generated human adipose stem cell (ASC) with stable BRCA1 knockdown models BRCA1-KD ASC. We demonstrated 79.1% knockdown expression of the BRCA1 gene in our BRCA1-KD ASCs, compared to CRISPR controls with empty vectors (Figure 1A). The ability of the ASCs to undergo adipogenesis was confirmed with Oil Red O staining (see Figure, Supplemental Digital Content 1, which demonstrates mature adipocytes differentiated from ASCs used in this study. The Oil Red O staining of differentiated adipocytes from ASCs used in the study at adipogenic induction day 14, INSERT HYPER LINK).
Figure 1.
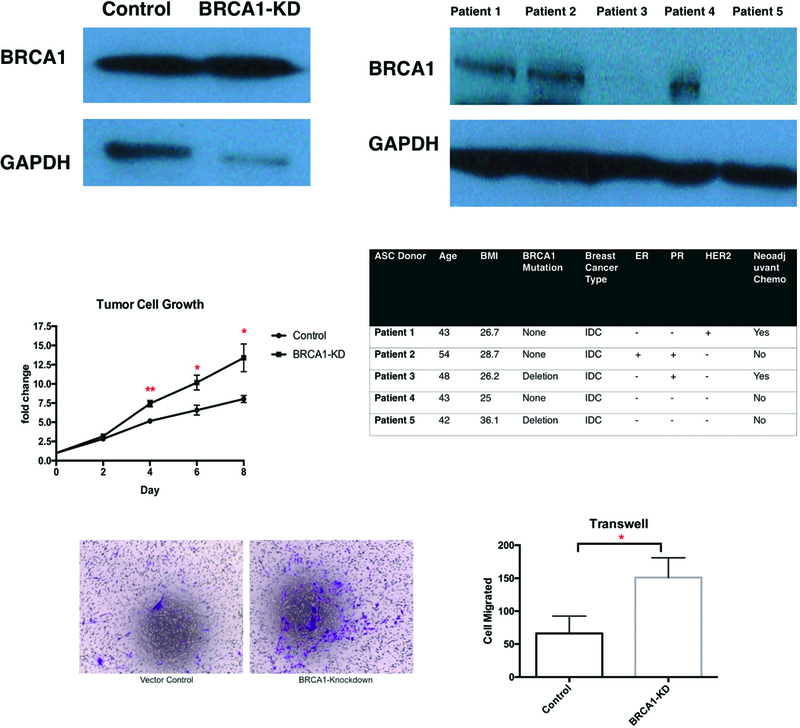
BRCA1-KD ASC promoted proliferation and invasion of breast tumor cells. A. WB of BRCA1 expression in CRISPR/Cas 9-mediated BRCA1-KD ASCs and primary brASC. B. WB of BRCA1 expression in patient brASCs. The table below shows the patient demographics, cancer details, and BRCA1 mutation status for these patients. C. Tumor growth rate when MDA-MB-231 cells were co-cultured with ASCs (N=3). D&E. Representative images and quantification of tumor cell invasion when ASCs were used as feeders in Transwell assays (N=3).
Decreased BRCA1 Expression in Primary brASCs
Western blot confirmed decreased expression of BRCA1 in brASCs in patients with deletion mutations (Figure 1B). Patient demographics, cancer details, and mutation status of the 5 patient donors were detailed below the Western blot.
Effects of BRCA1 Mutation on Proliferation and Invasion of Breast Cancer Cells
We tested the effect of the BRCA1 knockdown on ASC stimulation of breast cancer cell progression. Proliferation of MDA-MB-231 cells in vitro was significantly faster when co-cultured with BRCA1-KD ASCs (Figure 1C, 7.43-fold vs 5.16-fold increase at day 4 comparing to day0, P=0.0074; 10.17-fold vs 6.57-fold at day6, P=0.036; 13.39-fold vs 8.02-fold at day8, P=0.045, N=3, effect size Cohen’s d = 0.96). We found that BRCA-KD ASC also significantly increased breast cancer cell invasion. This is demonstrated using Transwell assays. Serving as feeders, BRCA1-KD ASCs induced significantly higher level of tumor cell invasion in MDA-MB-231 cells using Transwell assays (Figure 1D&1E, 151±17.39 vs 66±13.17 cells migrated. P=0.0171, N=3, effect size Cohen’s d = 5.62). We also found that brASCs from patients with BRCA1 deletion mutation also increased breast cancer cell invasion in the Transwell assay experiment (see Figure 2, Supplemental Digital Content 2, which demonstrates higher tumor cell invasion brASCs from BRCA1 mutation carriers were plated in the bottom chamber, INSERT HYPER LINK). Therefore, our data suggests that BRCA1 mutation in ASCs results in a secretome which promotes in vitro breast cancer cell proliferation and invasion.
Figure 2.
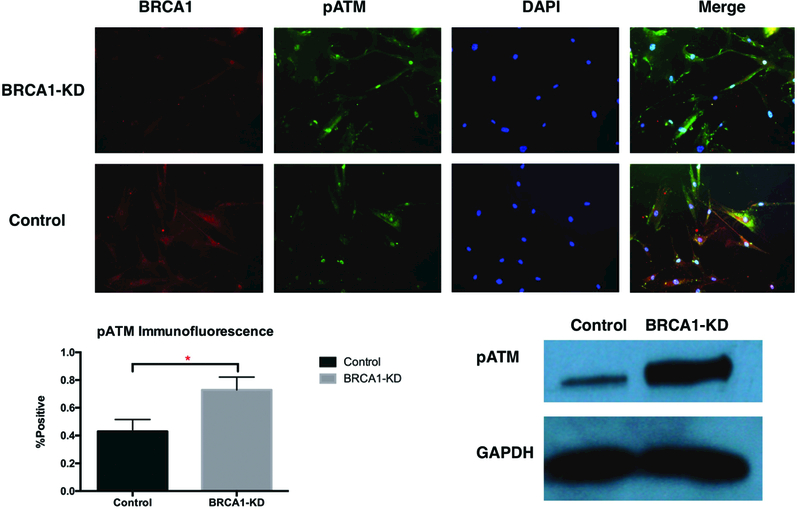
DNA Damage in BRCA1-Deficient ASCs. A&B. Representative immunofluorescence images and quantification of phosphorylated ATM. C. WB of pATM expression (N=3, * indicates P<0.05).
Increased Spontaneous DNA Damage Cell and ATM Activation in BRCA1-Deficient ASCs
We then aimed to elucidate the mechanism by which ASCs with the BRCA1 mutation stimulate breast cancer cells. The BRCA1 gene is well-known to play a role in DNA damage repair. Since the BRCA1 protein is required for mediating ATM-dependent DNA damage repair, and ATM activation induces cellular senescence, we hypothesized that senescence-associated secretory phenotype was part of the underlying mechanism that drove BRCA1-deficient ASCs to secrete inflammatory factors that induce breast tumor aggression. To test this, we performed immunofluorescence staining and western blot to quantify ATM activation in BRCA1-knockdown ASCs. Immunofluorescence staining revealed higher levels of phosphorylated ATM in BRCA1-knockdown cells (Figure 2A&2B, 72.9±5.32% vs 42.9±4.97%. P=0.0147, N=3, effect size Cohen’s d=5.83). Western blot also demonstrated higher expression of phosphorylated ATM in BRCA1-knockdown ASCs (Figure 2C). This confirms higher level of spontaneous DNA damage and ATM activation in BRCA1-deficient ASCs.
Senescence-Associated Secretory Phenotype and Its Role in Breast Cancer Microenvironment
We assessed cell senescence in different ASC groups with beta-galactosidase assay and western blotting. The beta-galactosidase assay demonstrated a higher degree of cell senescence in both BRCA1-knockdowns ASCs and brASCs with the BRCA1 mutation (Figure 3A&3B, 7.9±0.25% vs 0.17±0.17%, P<0.0001 in BRCA1-KD, N=3, effect size Cohen’s d=36.2; 41.7±2.30% vs 18.4±2.34%, P<0.0001 in patient brASCs, N=3, effect size Cohen’s d=10.0). Western blots also showed increased expression of p21, an established intermediate in cell senescence pathway (20) (Figure 3C). In this senescent state, we hypothesized that BRCA1-deficient ASCs secrete inflammatory cytokines. To confirm this, we used ELISA assays on supernatant of BRCA1-knockdown and control ASCs. First, our Screening ELISA assays demonstrated the most significant differences in cytokine production between the two ASC groups in IL-6 and IL-8 (see Figure, Supplemental Digital Content 3, which demonstrated the most significant differences in IL-6 and IL-8 production among 12 different cytokines, INSERT HYPER LINK). We found higher level of IL-6 and significantly higher level of IL-8 inflammatory cytokine in BRCA1-knockdown ASCs (Figure 4A, 2.57±0.32-fold change, P=0.0049, N=4, effect size Cohen’s d=4.97). We then demonstrated the effects of IL-6 and IL-8 inflammatory cytokines on invasion of breast tumor cells using Transwell assays. We found a significantly higher degree of breast tumor cell invasion when exposed to exogenous IL-6 and/or IL-8(Figure 4B&C, P=0.03 for IL-6, P=0.01 for IL-8, P=0.002 for IL-6&IL-8, N=3, effect size Cohen’s d=4.39).
Figure 3.
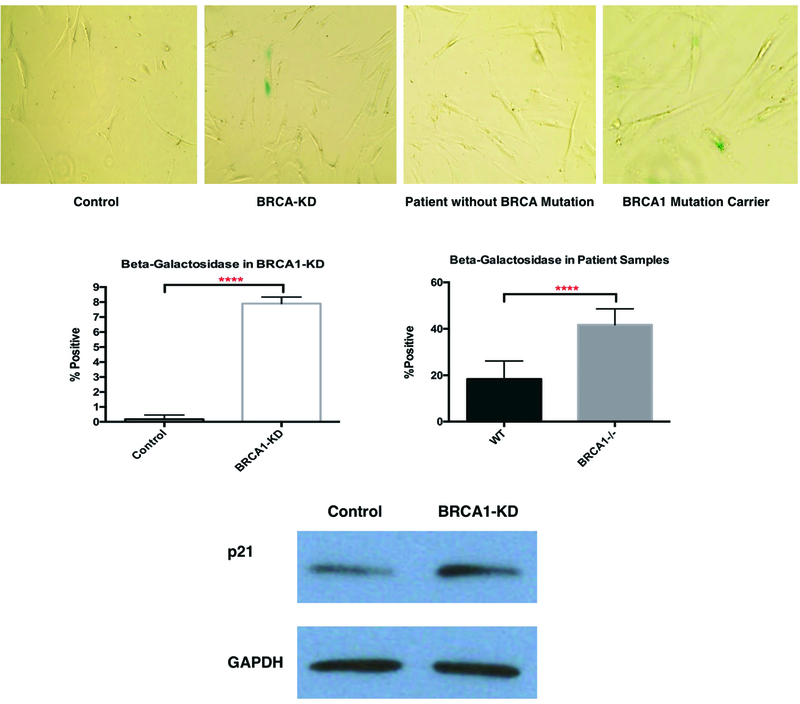
Cell Senescence in BRCA1-deficient ASCs. A&B. Representative images and quantifications of beta-galactosidase assays in both BRCA1-KD ASCs and brASCs (N=3, **** indicates P<0.0001). C. WB of p21 expression.
Figure 4.
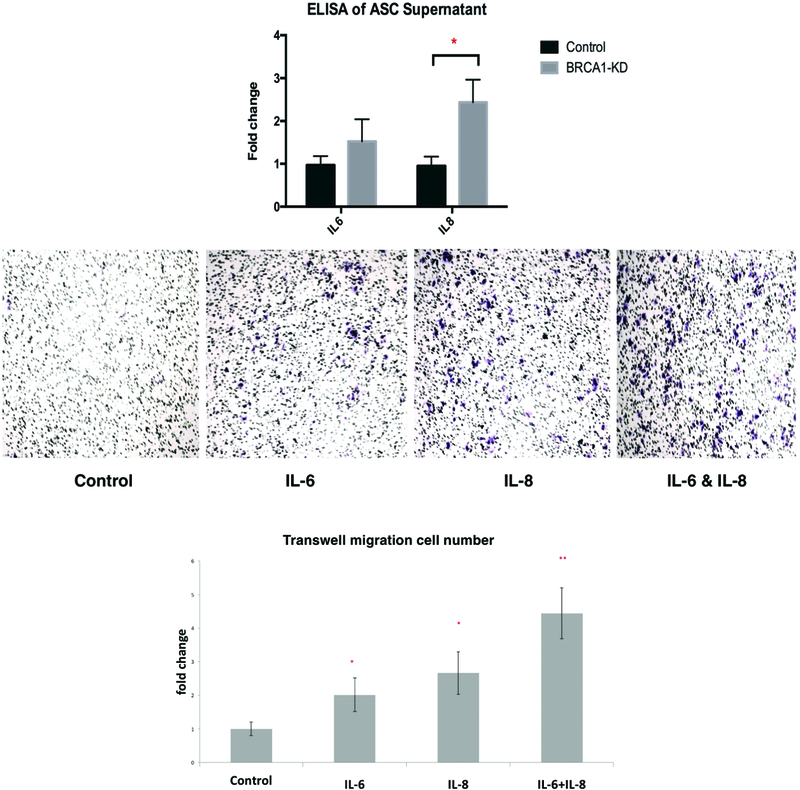
IL-8 in BRCA1-KD ASC cell supernatant that promotes cell invasion. A. Quantification of ASC supernatant using ELISA (N=4). B&C. Representative images and quantifications of cell invasion using IL6 and IL8 in transwell assays comparing to control (N=3, * indicates P<0.05, ** indicates P<0.01).
Taken together, we established that BRCA1-deficienct ASCs were associated with persistently high levels of stress-induced DNA damage. This accumulation of DNA damage lead to more persistently active ATM complex, which in turns increased cellular senescence. In the senescence state, BRCA1- deficient ASCs secreted increased number of inflammatory cytokines, which promoted breast tumor proliferation and invasion (Figure 5). Our results confirmed that human ASCs with BRCA1 mutation created a secretome that lead to more aggressive tumor phenotypes. This was mediated through the DNA damage cell senescence pathway.
Figure 5.
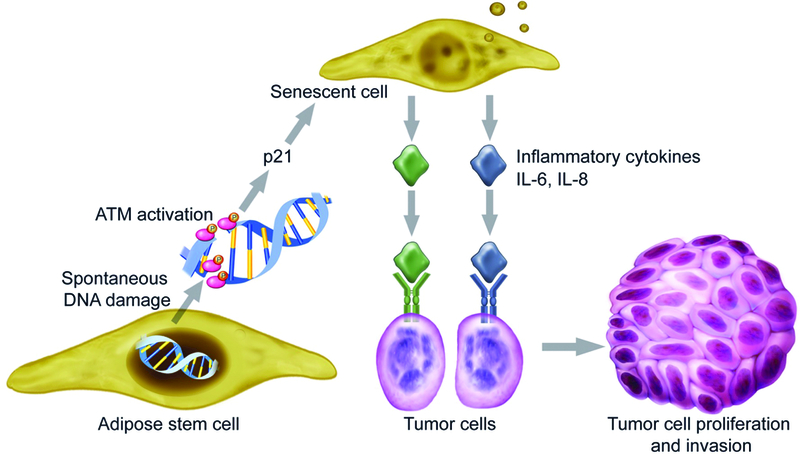
The mechanism of how BRCA1-deficient ASCs promote breast cancer progression. BRCA1-deficienct ASCs are unable to repair both spontaneous and stress-induced DNA damage. This accumulation of DNA damage leads to more persistently active ATM complex, which activates p21, and induces cellular senescence. In the senescence state, BRCA1- deficient ASCs secrete increased number of inflammatory cytokines, which promotes breast tumor proliferation and invasion.
DISCUSSION
Adipose tissue makes up a large proportion of the breast gland and consists of mature adipocytes, adipose-derived stem cells, stromal supporting cells and inflammatory cells. Studies have supported that cancer-associated adipocytes (CAAs) play important roles in breast cancer progression and carcinogenesis (21). This study confirmed our hypothesis that the BRCA1 mutation in ASCs results in a phenotype that can promote breast cancer growth and invasion via increased production of IL6 and IL8, mediated by the DNA-damage-senescence pathway.
One important implication for this study relates to patients with the BRCA1 mutation, in that the aggressive nature of these tumors may be partially related to alterations within the surrounding adipose tissue itself. Studies have shown that ASCs may contribute to tumor cell proliferation and invasion, particularly in biologically aggressive breast cancers, such as triple negative or basal-type (22). Our study showed the increased IL-6, IL-8 production by BRCA1-defecient ASCs promotes breast cancer progression in vitro. This observation is consistent with previous studies that showed cancer-associated adipose cells secrete cytokines, including IL-6 and IL-8, which promote cancer growth in breast and prostate cancers. (23–31). Our study explored this concept within the context of the BRCA1 mutation and describes a mechanism by which ASCs may alter the tumor microenvironment in a previously unrecognized manner. Another study highlighted the role of BRCA1 in modulation of estrogen biosynthesis in ASCs, contributing to tumor suppressor function (32). Results from others and this study further confirmed the importance of BRCA1 mutation in breast tumor microenvironment on breast cancer progression (33, 34).
Cellular senescence, originally thought to be a tumor-suppressive mechanism, induces secretion of inflammatory cytokines that in fact promote tumor progression. Studies have found that IL-6 and IL-8 are two of the most prominent cytokines of SASP, which could be induced by persistent DNA-damage signaling mediated by ATM activation (35). Our findings of elevated levels of IL-6 and IL-8 in cell supernatant, as well as the higher percentage of phosphorylated ATM in BRCA1-deficient ASCs are consistent with the SASP pathway. This mechanism explains how BRCA1 mutation affects ASCs in a way that may lead to breast cancer progression. More importantly, this pathway can be targeted for more effective anticancer therapy, especially in the setting of high risk breast cancer. Our future directions include investigating the effects of inhibiting SASP in vitro and in vivo through inhibiting its inflammatory cytokines IL-6 and IL-8.
Risk of breast cancer in BRCA1 and BRCA2 mutation carriers can be modified by changes in body weight (36) and there is some evidence linking BRCA1 mutations with abnormal fat physiology, metabolism, and homeostasis (37, 38). Additionally, CAAs undergo delipidation and de-differentiation (39) which has been shown to occur in human obesity (40, 41). Thus, our investigation of the breast cancer tumor microenvironment helps provide insights into the effects of obesity on breast cancer development.
Our study may also be applicable to BRCA1 mutation carrier patients undergoing fat transfer procedures. This finding raises the question of safety of fat grafting in patients with BRCA1 mutation. Since our data demonstrate that BRCA1-mutated ASCs stimulate breast cancer cells, a cautious approach to fat grafting in patients with confirmed BRCA1 genetic mutation and residual breast cancer risk is reasonable until more clinical data is available.
CONCLUSION
We demonstrate that BRCA1 mutation in tumor microenvironment promotes breast cancer progression. This effect is mediated by increased secretion of inflammatory cytokines in BRCA1-deficient ASCs via DNA-damage-senescence pathway. Our study not only helps identify targets for more effective anticancer therapies, but also provides insights into how obesity might contribute to breast cancer.
Supplementary Material
which shows the Oil Red O staining of differentiated adipocytes from ASCs used in the study at adipogenic induction day 14, INSERT HYPER LINK.
which shows Representative images and quantification of tumor cell invasion when brASCs were used as feeders in Transwell assays (N=3), INSERT HYPER LINK.
which shows Screening ELISA in ASC supernatant in BRCA1-KD ASCs and Control ASCs, INSERT HYPER LINK.
ACKNOWLEDGEMENT
This study is supported in part by grants ES024015, CA208852, CA216876 from US National Institutes of Health (to C-Y. Li), and Eugene A. Stead Research Scholarship from Duke University School of Medicine (to R Zhao).
Footnotes
Financial Disclosure: None of the authors has a financial interest in any of the products, devices, or drugs mentioned in this manuscript
REFERENCES
- 1.Okuma HS, Yonemori K BRCA Gene Mutations and Poly(ADP-Ribose) Polymerase Inhibitors in Triple-Negative Breast Cancer. Adv Exp Med Biol 2017;1026:271–286. [DOI] [PubMed] [Google Scholar]
- 2.Konishi H, Mohseni M, Tamaki A, et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc Natl Acad Sci U S A 2011;108:17773–17778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eng C, Leone G, Orloff MS, Ostrowski MC Genomic alterations in tumor stroma. Cancer Res 2009;69:6759–6764. [DOI] [PubMed] [Google Scholar]
- 4.Weber F, Shen L, Fukino K, et al. Total-genome analysis of BRCA1/2-related invasive carcinomas of the breast identifies tumor stroma as potential landscaper for neoplastic initiation. Am J Hum Genet 2006;78:961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aleskandarany M, Caracappa D, Nolan CC, et al. DNA damage response markers are differentially expressed in BRCA-mutated breast cancers. Breast Cancer Res Treat 2015;150:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppe JP, Desprez PY, Krtolica A, Campisi J The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 2010;5:99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan NJ, Sasser AK, Axel AE, et al. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 2009;28:2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J, Lu LY, Yu X The role of BRCA1 in DNA damage response. Protein Cell 2010;1:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JM, Gu Y, Pan CJ, Yin LR Isolation, culture and identification of human adipose-derived stem cells. Exp Ther Med 2017;13:1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 2016;44:W272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 2014;42:W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu ZQ, Mahmood T, Yang PC Western blot: technique, theory and trouble shooting. N Am J Med Sci 2014;6:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV In vitro cell migration and invasion assays. J Vis Exp 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchisio PC, Trusolino L Immunofluorescence of cultured cells. Methods Mol Biol 1999;96:85–92. [DOI] [PubMed] [Google Scholar]
- 17.Faul F, Erdfelder E, Lang AG, Buchner A G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007;39:175–191. [DOI] [PubMed] [Google Scholar]
- 18.Winter J. C. F. d. Using the Student’s t-test with extremely small sample sizes. Practical Assessment, Research & Evaluation, Vol. 18, 2013. [Google Scholar]
- 19.Sullivan GM, Feinn R Using Effect Size-or Why the P Value Is Not Enough. J Grad Med Educ 2012;4:279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Li J, Sejas DP, Pang Q The ATM/p53/p21 pathway influences cell fate decision between apoptosis and senescence in reoxygenated hematopoietic progenitor cells. J Biol Chem 2005;280:19635–19640. [DOI] [PubMed] [Google Scholar]
- 21.Mao Y, Keller ET, Garfield DH, Shen K, Wang J Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev 2013;32:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, Sachs PC, Wang X, et al. Mesenchymal stem cells in mammary adipose tissue stimulate progression of breast cancer resembling the basal-type. Cancer Biol Ther 2012;13:782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilroy GE, Foster SJ, Wu X, et al. Cytokine profile of human adipose-derived stem cells: expression of angiogenic, hematopoietic, and pro-inflammatory factors. J Cell Physiol 2007;212:702–709. [DOI] [PubMed] [Google Scholar]
- 24.De Girolamo L, Stanco D, Salvatori L, et al. Stemness and osteogenic and adipogenic potential are differently impaired in subcutaneous and visceral adipose derived stem cells (ASCs) isolated from obese donors. Int J Immunopathol Pharmacol 2013;26:11–21. [DOI] [PubMed] [Google Scholar]
- 25.Finley DS, Calvert VS, Inokuchi J, et al. Periprostatic adipose tissue as a modulator of prostate cancer aggressiveness. J Urol 2009;182:1621–1627. [DOI] [PubMed] [Google Scholar]
- 26.Yu JM, Jun ES, Bae YC, Jung JS Mesenchymal stem cells derived from human adipose tissues favor tumor cell growth in vivo. Stem Cells Dev 2008;17:463–473. [DOI] [PubMed] [Google Scholar]
- 27.Walter M, Liang S, Ghosh S, Hornsby PJ, Li R Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene 2009;28:2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welte G, Alt E, Devarajan E, Krishnappa S, Jotzu C, Song YH Interleukin-8 derived from local tissue-resident stromal cells promotes tumor cell invasion. Mol Carcinog 2012;51:861–868. [DOI] [PubMed] [Google Scholar]
- 29.Pinilla S, Alt E, Abdul Khalek FJ, et al. Tissue resident stem cells produce CCL5 under the influence of cancer cells and thereby promote breast cancer cell invasion. Cancer Lett 2009;284:80–85. [DOI] [PubMed] [Google Scholar]
- 30.Devarajan E, Song YH, Krishnappa S, Alt E Epithelial-mesenchymal transition in breast cancer lines is mediated through PDGF-D released by tissue-resident stem cells. Int J Cancer 2012;131:1023–1031. [DOI] [PubMed] [Google Scholar]
- 31.Muehlberg FL, Song YH, Krohn A, et al. Tissue-resident stem cells promote breast cancer growth and metastasis. Carcinogenesis 2009;30:589–597. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh S, Lu Y, Katz A, Hu Y, Li R Tumor suppressor BRCA1 inhibits a breast cancer-associated promoter of the aromatase gene (CYP19) in human adipose stromal cells. Am J Physiol Endocrinol Metab 2007;292:E246–252. [DOI] [PubMed] [Google Scholar]
- 33.Chodankar R, Kwang S, Sangiorgi F, et al. Cell-nonautonomous induction of ovarian and uterine serous cystadenomas in mice lacking a functional Brca1 in ovarian granulosa cells. Curr Biol 2005;15:561–565. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Ghosh S, Amleh A, et al. Modulation of aromatase expression by BRCA1: a possible link to tissue-specific tumor suppression. Oncogene 2005;24:8343–8348. [DOI] [PubMed] [Google Scholar]
- 35.Jarvis IW, Bergvall C, Bottai M, Westerholm R, Stenius U, Dreij K Persistent activation of DNA damage signaling in response to complex mixtures of PAHs in air particulate matter. Toxicol Appl Pharmacol 2013;266:408–418. [DOI] [PubMed] [Google Scholar]
- 36.Kotsopoulos J, Olopado OI, Ghadirian P, et al. Changes in body weight and the risk of breast cancer in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res 2005;7:R833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortega FJ, Moreno-Navarrete JM, Mayas D, et al. Breast cancer 1 (BrCa1) may be behind decreased lipogenesis in adipose tissue from obese subjects. PLoS One 2012;7:e33233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones LP, Buelto D, Tago E, Owusu-Boaitey KE Abnormal Mammary Adipose Tissue Environment of Brca1 Mutant Mice Show a Persistent Deposition of Highly Vascularized Multilocular Adipocytes. J Cancer Sci Ther 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andarawewa KL, Motrescu ER, Chenard MP, et al. Stromelysin-3 is a potent negative regulator of adipogenesis participating to cancer cell-adipocyte interaction/crosstalk at the tumor invasive front. Cancer Res 2005;65:10862–10871. [DOI] [PubMed] [Google Scholar]
- 40.McLaughlin T, Sherman A, Tsao P, et al. Enhanced proportion of small adipose cells in insulin-resistant vs insulin-sensitive obese individuals implicates impaired adipogenesis. Diabetologia 2007;50:1707–1715. [DOI] [PubMed] [Google Scholar]
- 41.Isakson P, Hammarstedt A, Gustafson B, Smith U Impaired preadipocyte differentiation in human abdominal obesity: role of Wnt, tumor necrosis factor-alpha, and inflammation. Diabetes 2009;58:1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
which shows the Oil Red O staining of differentiated adipocytes from ASCs used in the study at adipogenic induction day 14, INSERT HYPER LINK.
which shows Representative images and quantification of tumor cell invasion when brASCs were used as feeders in Transwell assays (N=3), INSERT HYPER LINK.
which shows Screening ELISA in ASC supernatant in BRCA1-KD ASCs and Control ASCs, INSERT HYPER LINK.


