Abstract
Objectives:
Research on the genetic basis of tinnitus is still in its first steps. A group of scientists dedicated to tinnitus genetics within European Tinnitus Network (TINNET) network recognize that further progress requires multicenter collaborative efforts for defining contributing genes. The purpose of the present work is to provide instructions regarding collection, processing, storage, and shipment of samples intended for genetic studies in auditory research.
Design:
One part of the recommendations has a general character; another part is of particular importance for auditory healthcare practitioners such as otolaryngology physicians, audiologists, and general practitioners.
Results:
We provide a set of instructions and various options for obtaining samples. We give advice regarding sample processing, storage, and shipment and define the minimal and essential clinical information that should accompany the samples collected for genetic processing.
Conclusions:
These recommendations offer a basis to standardize and optimize collaborations between geneticists and healthcare practitioners specialized in tinnitus and hearing disorders.
Keywords: Auditory disorders, Biobanks, DNA, Genetics, Genomics, Hearing loss, RNA, Tinnitus
BACKGROUND
Subjective tinnitus is thought to be triggered by maladaptive plasticity in the auditory pathway that occurs across varying degrees of hearing loss (Shore et al. 2016). Tinnitus may emerge under a variety of conditions, but most investigators conceptualize it as the result of the auditory pathway employing compensatory adjustments that increase neural gain and promote central disinhibition in response to a hearing disorder that may or may not be accompanied by a change in threshold (Schaette & McAlpine 2011).
Recent evidence obtained from concordance studies in twins supports moderate to high heritability of bilateral tinnitus among men, as well as women less than 40 years old (Maas et al. 2017). In contrast, unilateral tinnitus is found to be more related to environmental factors (Maas et al. 2017). Although additional concordance studies in independent twin cohorts need to be conducted to confirm these novel findings, this observation supports the notion that genetic factors are important in the generation and persistence of tinnitus, and different biobanking initiatives have started to conduct genomic studies in tinnitus patients (Cederroth et al. 2017).
Genome-wide association studies (GWAS) have identified >24,000 single-nucleotide variants (SNVs) associated with common diseases or traits (MacArthur et al. 2017). Although the functional significance of many reliable SNV associations is unknown, bioinformatics integration of phenotype and genomic data has facilitated the understanding of disease mechanisms to establish targets for novel therapies. Examples of such approach include a GWAS conducted in major depression involving the HOMER1 gene, which mutant mice displayed evidence of depression and altered cortical-limbic circuitry (Rietschel et al. 2010); a collaborative effort of the International Multiple Sclerosis Genetics Consortium to demonstrate a primary role for adaptive immunity (International Multiple Sclerosis Genetics Consortium 2011) or the identification of missense SNVs in the MST1 gene in Crohn disease leading to reduced macrophage signaling (Pal & Moult 2015).
Two critical factors appear to contribute to the development and persistence of tinnitus: first, a hearing disorder, and second, the response of the cognitive and emotional neural networks to the tinnitus percept. It is rather well established that the interaction between these two components leads to the end-phenotype of tinnitus (Noreña 2015). While tinnitus in general is perceived by approximately 15% of the population, nearly one tenth of affected individuals report it as bothersome. We believe that the design of clinical trials to find an effective treatment for tinnitus should involve deep phenotyping (Hall 2017). Deep phenotyping requires the acquisition of structured clinical information that facilitates the classification of patients into tinnitus subgroups (Lopez-Escamez et al. 2016) and is one of the fundamental concepts of genetics research and precision medicine (Müller et al. 2016).
These recommendations are intended for health practitioners involved in auditory and tinnitus research. Our intention, which originated during networking in the European consortium TINNET, is to facilitate the formation of a collaborative international consortium and to develop a stratified collection and storage of human samples with corresponding medical data.
Increasing Genetic Knowledge About Hearing Disorders and Psychological Comorbidities Facilitates Understanding of the Molecular Basis of Tinnitus
Basic knowledge about tinnitus genetics is being acquired rather slowly (Vona et al. 2017), in particular when compared with other auditory disorders such as hearing loss. As of yet, a large number of genetic mutations leading to hearing loss have been identified, and their number continues to grow (Korver et al. 2017). Many of these mutations are responsible for congenital hearing loss and can affect genes that are associated with autosomal recessive or dominant forms of hearing loss. For instance, many well-known mutations have been reported in genes encoding autosomal recessive hearing loss such as GJB2, GJB3, and GJB6, encoding gap junction proteins; MYO3A, MYO7A, and MYO15A, encoding molecular motor proteins; OTOF encoding inner ear-specific synaptic protein otoferlin, or OTOG encoding a key protein of the extracellular matrix in the inner ear; as well as genes associated with autosomal dominant hearing loss such as KCNQ4 or MYH14. For a complete list of genes, please see the study by Korver et al. (2017) or the following regularly updated website (http://hereditaryhearingloss.org). Not surprisingly, the epigenetic control of gene expression appears equally important for proper auditory function. A recent study demonstrated the contribution of gene methylation, histone modification, and regulatory micro-RNAs to the development and functioning of the inner ear (Layman & Zuo 2014).
While there has been a substantial advance in the genetics of hearing loss, knowledge about the genetic basis of psychological disorders also progressed thanks to joint clinical efforts of large consortiums (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014; Marshall et al. 2017). For instance, genetic mutations leading to changes in calcium channel signaling were commonly found in autism, attention deficit hyperactivity disorder, bipolar disorder, major depression, and schizophrenia (Cross-Disorder Group of the Psychiatric Genomics Consortium 2013). Troublesome tinnitus is often accompanied by psychological comorbidities such as depressive symptoms, anxiety, and most importantly increased stress perception. An interesting genetic study of tinnitus could explore the relationship between promoter polymorphisms of a serotonin transporter and stress that may influence the development of depressive symptoms (Karg et al. 2011). Although an association was found between allelic variants of the SLC6A4 gene encoding the serotonin transporter promoter and the psychological distress caused by tinnitus, these findings have not yet been replicated in an independent cohort (Deniz et al. 2010).
Because anxiety and depression are frequently observed associated with tinnitus (Trevis et al. 2017) and differences in personality including neuroticism are associated with several noncoding variants (Lo et al. 2017), it is necessary to design genomic studies controlling these comorbidities (such as hearing loss or anxiety) to obtain comparable groups.
Collaborative Multicenter Genetic Projects
Human biobanks and genetic research databases (HBGRD) allow the sharing of human biological material and information derived from its analysis. In 2009, the Organization for Economic Cooperation and Development (OECD) Council adopted a recommendation on HBGRD to foster scientific research (http://www.oecd.org/sti/biotech/44054609.pdf). The recommendation provides guidelines for the establishment, management, governance, access, and discontinuation of HBGRDs. In general, it facilitate access to data and materials for biomedical advancement while ensuring that research is conducted in a fully ethical way that supports human dignity and fundamental human rights.
To describe the genetic architecture of tinnitus and to understand the complex interactions between hearing loss and tinnitus development, a large multicenter effort is necessary (Cederroth et al. 2017). Learning from other complex disorders, one can estimate that tens of thousands of subjects would be required to reach satisfactory statistical power for a GWAS. For instance, in a study where calcium channel signaling was determined as a common denominator of multiple psychiatric conditions, over 60,000 participants were enrolled by a large consortium consisting of 800 investigators from 38 countries (Cross-Disorder Group of the Psychiatric Genomics Consortium 2013). This prompted researchers to call for the involvement of international professional healthcare providers for tinnitus patients in such a project (Cederroth et al. 2017). The recent European Union-funded tinnitus networks (TINNET1, European School for Interdisciplinary Tinnitus Research (ESIT)2 and Tinnitus-Action (TIN-ACT)3) would address the project’s challenges, as these are comprised members with primary interests in tinnitus from several European countries.
Large, multicenter biobanking platforms are necessary to perform omics-studies targeting tinnitus (genomics, epigenomics, transcriptomics, and metabolomics). Various scientific associations have put together their own guidelines about DNA and tissue banking, and the Council for International Organizations of Medical Sciences (CIOMS), recognizing the importance of biobanks for epidemiological research, revised its guidelines in 2005 to integrate relevant issues from the biobank debate. The United Nations Educational, Scientific and Cultural Organization (UNESCO, Paris, France) adopted the International Declaration on Human Genetic Data in October 2003, and France, Germany, Canada, and Switzerland have all issued their own guidelines for biobanks or genetic databases (Elger & Caplan 2006).
The main aims of this publication are to provide biobanking recommendations for clinicians and tinnitus researchers. We describe the main steps for collecting, processing, and storing biological samples for genetic studies on tinnitus and hearing loss. Such recommendations are a cornerstone in the initiation of large-scale studies.
DESIGN OF GENOMIC STUDIES WITH BIOLOGICAL SAMPLES
The design and execution of genomic studies using samples obtained from human subjects must follow certain steps, which are presented in Figure 1 and discussed in detail below.
Fig. 1.
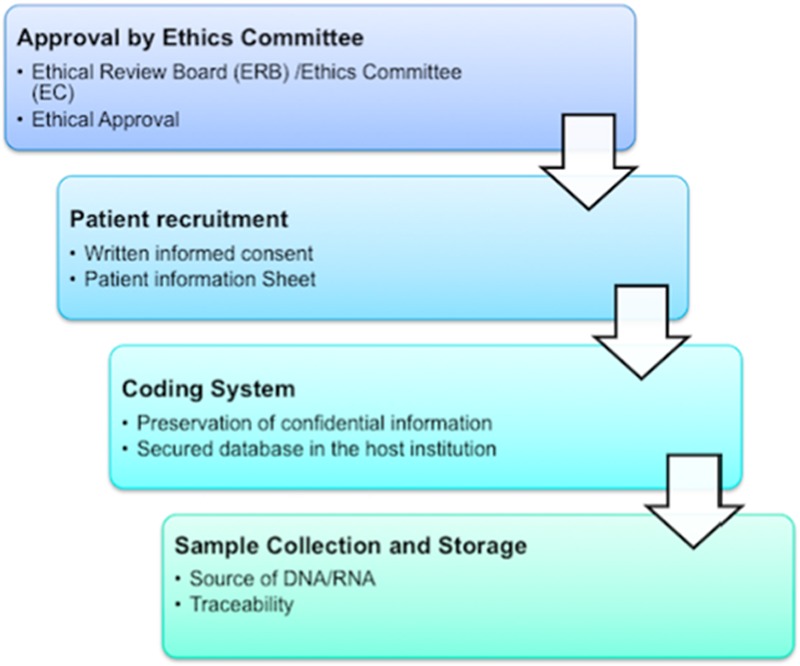
Flowchart for conducting genomic studies in patients with hearing loss or tinnitus.
Ethical Approval
All research involving human subjects should be conducted in accordance with three basic ethical principles, namely, respect toward individuals, beneficence, and justice. It is generally agreed that these principles, which have equal moral force, guide the conscientious preparation of proposals for scientific studies. Research with human subjects should be carried in accordance with a protocol that clearly states: (i) the aim of the research; the reasons for performing research in humans; (ii) the nature and degree of any known risks to the subjects; (iii) the sources from which it is proposed to recruit subjects and how; and (iv) the means for ensuring that any subjects participation is voluntary and that consent will be collected in written form (CIOMS, 2016)4. As mentioned before, all proposals for conducting research involving human subjects must be submitted for review of their ethical acceptability to at least one ethical review board (ERB). Any direct financial or other material benefits that may derive from the research should be contingent on the outcome of ERB review. The investigator must obtain their approval or clearance before undertaking the research. Considering the public interest in the use of human biological samples and data in biomedical research, the policies state a number of duties, which the institutions responsible for the collection and storage of samples have to respect, namely: (i) to properly conserve the samples; (ii) to manage the samples and the derived genetic data, (iii) and to control their use and distribution (Shekelle et al. 2013). All these aspects should be consistent with the new European General Data Protection Regulation (http://www.eugdpr.org).
Written Informed Consent
An informed consent form is a document signed by the participant after a dialogue between patients/donors and health care practitioners/researchers concerning the consequences, harms, benefits, risks, and expected results from the planned study (Shekelle et al. 2013). Such a document is required to confirm the individual’s consent that his/her samples and data will be collected, stored in a biobank to facilitate the analysis, and possibly shared with consortium partners, according to a specific material transfer agreement. The consent form is the first guarantee of patient’s data confidentiality because the results obtained from research studies might potentially have an impact not only on the participants themselves but also on the families, communities, and populations to which they belong.
The informed consent form consists of two parts: the information sheet and the consent certificate5. The information sheet explains to prospective participants the nature of the research project, why they were chosen as candidates for the research, what risks and benefits exist, what options are associated with the research, and what rights they have as research subjects. The consent form must include the expression of the free and informed authorization for using their own biological samples and additional authorization for the collection of derived data for research purpose. The consent form should also include the name of the institution that will receive biological material or data for analysis, as well as information specifying that the participant can retract from the study at any time, with or without the removal of samples or data from the biobank or the database.
The WHO recommends that consent forms should be written using language at the level of a local 6th/8th grade student, with short sentences and paragraphs, simple terms and concepts, and technical information explained in nontechnical ways6. The consent form consequently contains accurate and precise information that is easy to follow, presented in a clear layout, with white-space borders, and fonts that are easy to read.
Sensitive issues about how much information a participant should receive, how to collect consent in a way that is respectful for a local culture, how to collect the consent of minors, or whether community or group consent is required in some circumstances should be considered. Not every detail needs to be discussed in the consent form, but all details needed for a participant to make a decision about participation in the study must be provided; investigators understand that ERBs must assist in the forms’ development and approval.
Genetic databases are often presented as key biomedical resources that enable researchers to perform a diverse range of research projects. As a consequence, participants must be informed that their samples could possibly be used in the future for other research purposes that are presently unforeseen. European policies adopt different views on how “specific” the informed consent should be, ranging from recommending a broad consent to a consent specifying all future uses. One possibility is to offer to contact the participant when a sample is intended to be used for a purpose other than what was initially agreed on in the consent form. In any case, we recommend adding the specific information about how will the sample be pseudoanonymized by a biobank.
CHOICE OF MATERIAL TO BE USED FOR GENETIC RESEARCH
Although DNA or RNA can be obtained from any biological sample, several factors must be considered in clinical practice. Blood and saliva samples are the most common source of DNA and RNA. A standardized operating procedure and quality controls must be implemented for any type of biological sample. Traceability from a coded sample to the biobank and finally to the genomic facility is essential. The source of DNA/RNA should be the same throughout the study to maintain sample homogeneity and reliability of the results. Gene expression (mRNA) or epigenetic markers are cell-specific and transcriptome or epigenetic studies in DNA obtained from saliva or blood samples are inappropriate to investigate patients with hearing loss or tinnitus; thus, different sources of DNA or RNA could induce a bias. Appropriate sample labeling is an important initial step that ensures a blind code with reference to the date of collection, location, and clinical information. Labeling must be done using appropriate materials, for example, permanent and ethanol-resistant markers.
Blood Samples
Blood is routinely used in research as a source of DNA or RNA. The most common way is to draw the blood using ethylenediaminetetraacetic acid (EDTA) Vacutainer system (or similar) and keep the samples refrigerated at +4 °C for up to 24 hr or in a freezer at −80 °C for longer preservation. However, when long-term viability of samples is necessary (for instance, for purified lymphocytes or other blood cells), storage in liquid nitrogen can be required.
The advantage of using blood for genetics studies is the small amount needed to extract DNA (3 to 5 mL are sufficient). Blood can also be collected on paper cards (Whatman Flinders Technology Associates [FTA] cards) allowing its maintenance at room temperature for at least 80 days (Dobbs et al. 2002) or up to 5 years, according to the manufacturer; however, the DNA yield is lower compared with using peripheral blood.
Buccal Swab
A buccal swab is among the fastest methods of collecting biological samples from individuals. The greatest advantage in swab collection is that it generally does not require medically trained personnel; however, the amount of DNA obtained may be not enough to conduct genomic studies. The first step for an effective buccal swab sample collection is to ensure that the subject’s mouth is thoroughly rinsed. Participants are advised to restrain from eating or drinking for 2 hr before the collection of buccal cells. Different factors, such as individual’s diet, oral hygiene, oral flora, and smoking habits, influence the quality of extracted DNA/RNA. It is also important not to touch the teeth, the lips, or any surface before placing the swab in the transport tube. Buccal swab samples can be stored for up to 2 weeks at 4 °C before processing for DNA extraction. Foam-tipped, cotton-tipped, or flocked swabs are common types of swab collectors. Foam-tipped swabs can range in size from small to large, being selected according to the need and purpose, in order to minimize the subject’s discomfort or collect high amounts of cells.
Saliva
Saliva is another source of a high-quality DNA and can be used for genetic studies. Up to 74% of DNA obtained from saliva samples originates from the white blood cells (Thiede et al. 2000)7. The quality and quantity of DNA extracted from a saliva sample must meet the same standards as those established for blood samples, and DNA obtained from saliva has been extensively used for exome-sequencing studies (Zhu et al. 2015). DNA extraction kits for saliva samples that ensure the highest integrity and prevent rapid DNA degradation by local enzymes and bacteria should be considered. Several commercially available products have been identified in multiple studies as ensuring the highest integrity of extracted salivary DNA8.
Other Types of Samples
Although other biological material could be used for obtaining DNA/RNA, such as those used for the identification of individuals in forensic studies (e.g., FTA cards, hair, scats, feces, or bone fragments), we do not recommend these materials for genetic studies. Although samples of the inner ear or brain tissues would be very interesting for gene expression studies, these are extremely rare and for ethical reasons difficult to obtain during surgical procedures or postmortem.
COLLECTION OF BIOLOGICAL SAMPLES
Collecting Samples to Analyze DNA
DNA is a two-stranded long molecule containing our unique genetic code. Although DNA has a double-helix shape, which makes it more resistant to degradation than single-stranded RNA, it is important to treat the samples carefully and process them in a timely manner. As mentioned above, quality DNA samples for genetic studies can be obtained from whole blood, saliva, or buccal swabs. It is our recommendation to use samples that are as fresh as possible, meaning that the sample should be processed within 24 hr after collection. If for any reason that would not be possible (due to shipment or other issues), whole blood and saliva should be immediately stored at +4 °C, at which the sample will be stable up to 7 days. For a long-term storage, saliva can be frozen at −20 °C for up to 4 months—after that time, samples should be stored at −80 °C. Of note, whole blood EDTA tubes should not be frozen if stored for the intended purpose of genomic DNA isolation because freezing EDTA tubes reduces the DNA quality and quantity.
Collecting Samples to Measure Gene Expression (RNA)
Samples that are processed for gene expression analyses need to be treated with extreme caution, as RNA decay occurs much faster than that of DNA. RNases are enzymes that destroy RNA, and these are present in bodily fluids, including saliva, tears, and sweat. This is the reason why touching the samples with bare hands or breathing in the direction of the sample may degrade the RNA in seconds. This is also why one should protect the samples by wearing powder-free nitrile gloves and a mouth mask. Fortunately, the scientific market offers special reagents that stabilize RNA. These reagents include RNAlater and Allprotect Tissue Reagent. The samples (blood and buccal swab) should be placed in the RNase-free tube containing one of the reagents, closed, and stored submerged in a protective solution for up to 6 months at 2 to 8 °C (fridge) or up to 7 days between +15 °C and +25 °C.
HOW TO PROCESS THE SAMPLES?
For both DNA and RNA extraction, it must be emphasized that specialized equipment will be necessary: micropipettes, centrifuges, and DNase-free and RNase-free pipette tips and tubes.
In the absence of such equipment, the unprocessed samples should be directly shipped to the biobank or genomic center for their processing.
Isolation of DNA
When isolating DNA, it is important to take the source of tissue into account. We list the available kits for DNA extraction in Table 1. Although there are many options, the QIAamp DNA Mini Kit (Qiagen) is widely used as it provides silica-membrane–based nucleic acid purification from swabs and whole blood. High and consistent yields can be achieved when following the manufacturer’s protocol. If saliva is the starting material for genomic DNA extraction, many kits are specialized and have their own collection tubes to maintain and stabilize the DNA for longer periods of time, for example, the Oragene 2DNA kit (DNA Genotek), which allows saliva specimens to be stored for years at room temperature without DNA degradation.
TABLE 1.
Commercially available DNA and RNA extraction kits
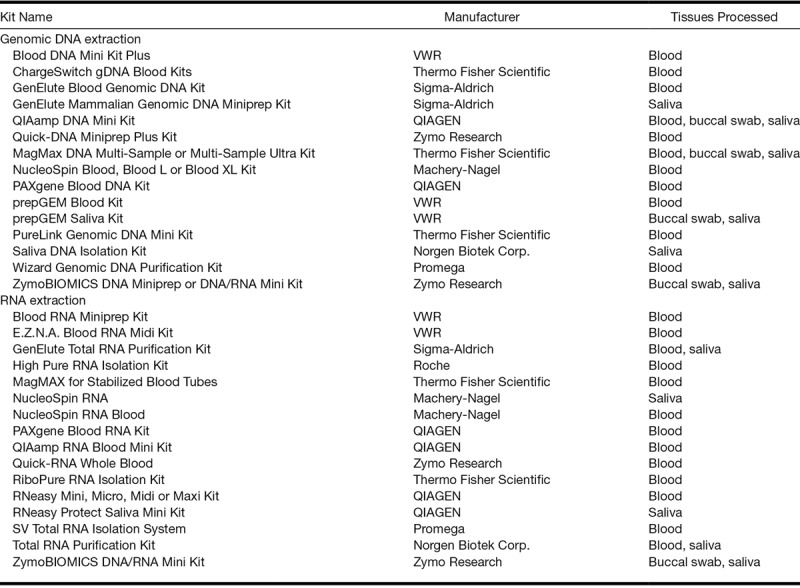
Isolation of RNA
RNA may show signs of significant degradation in as little as 30 min after collection (Sheldon et al. 2011). If the sample intended for RNA processing is placed and stored in one of the two solutions mentioned earlier (RNAlater or Allprotect Tissue Reagent, both developed by Qiagen) then the best RNA isolation method will be using RNeasy Mini or Micro Kit (Qiagen) following the protocol strictly provided by the manufacturer (protocol B). If, however, RNA is to be isolated from fresh samples, then a method of choice could be used.
Quality Controls for DNA and RNA
Sample quality is critical in genomic experiments, and it should always be checked before starting a genomics protocol. A quality control can be performed using the Agilent Bioanalyzer system for both DNA and RNA. Quantification of DNA/RNA can also be performed using the NanoDrop or Qubit assays. Quality control of genomic DNA could be done using Bioanalyzer or a 1% agarose gel. High-quality genomic DNA should appear as a major band of 10 to 20 kb on the gel.
For DNA, we recommend the Qubit fluorometric quantitation. One can also check the OD260/280 ratio of DNA or RNA using a NanoDrop. Pure DNA that is uncontaminated by proteins has an A260/280 ratio of 1.8, whereas pure RNA has an A260/280 ratio of 2.0. It is also of benefit to measure absorption at 230 nm, as high A230 values indicate contamination with salts or organic solvents.
Total RNA samples for RNAseq or microarray should be free of proteins, DNA, phenol, ethanol, and salt. RNA integrity must be checked by running RNA samples on an Agilent Bioanalyzer. If there is significant degradation (when RNA Integrity Number, or RIN, is below 7.0), we recommend obtaining new RNA samples of satisfactory quality.
STORAGE OF DNA AND RNA SAMPLES
The storage of biological samples is a key challenge that begins during sample collection, as sample integrity is a factor of higher relevance. Ideally, biological samples should be processed immediately after collection, but in practice, the majority of them will be stored. Biological samples must be stored in fully qualified, temperature-controlled storage. Any area in which biological samples are stored must remain within a controlled temperature range (e.g.: −196 °C; −80 °C; −20 °C; 2.0 to 8.0 °C; 15.0 to 25.0 °C), depending on biological samples’ requirements that range from controlled room temperature to cold storage, ultra-low-temperature storage and vapor-phase liquid nitrogen storage. Sample temperatures must be monitored, and the sample storage facility must be supported by multiple backup systems and an inventory tracking system.
The major difficulty in collecting biological material for genetic purposes is keeping the DNA from degrading. The main degrading agents of DNA in most samples are enzymes with DNase activity, present in the sample or produced by growing microorganisms; therefore, a procedure for enzyme inactivation must be used. Table 2 summarizes the effect of temperature on DNA storage. Several options exist for storing samples according to the regulations and resources of local facilities. We recommend preparing stock and aliquots with diluted samples to prevent repeated freeze–thaw cycles. Several possibilities exist for storing samples according to the local facilities.
TABLE 2.
Recommendations for DNA and RNA storage
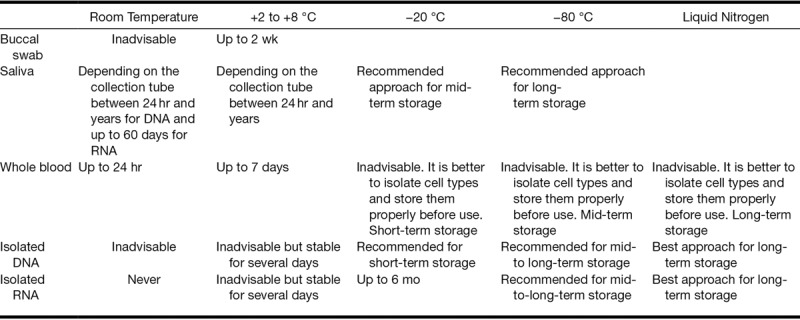
Storing at Room Temperature
We recommend storing the samples at room temperature (even those in protective solutions) for as briefly as possible. The reasons are multiple, but the primary reason is that room temperature may vary widely between 16 and 35 °C, depending on time of year and availability of air conditioning. We recommend the following storage temperatures to create as uniform processing conditions as possible.
Storing at +2 to +8 °C
This is the most common type of storage, and it provides an almost constant temperature that is available in all healthcare units. When using AllProtect, DNA samples are stabilized for an extended storage period of up to 6 months without damaging the samples. RNA samples should not be stored at +2 to +8 °C.
Storing at −20 °C
Storage at −20 °C is recommended for purified DNA or purified RNA (when in ethanol). Aqueous RNA samples cannot be stored for more than 3 months.
Storing at −80 °C
Storage at −80 °C is the most advisable approach for long-term storage of all purified nucleic acids, but this is likely the least available freezer in clinical settings.
SAMPLE SHIPMENT
The international nature of collaboration requires streamlined shipping logistics to reduce specimen quality issues. Sample collection and shipping protocols are important to minimize potentially detrimental biomolecular artefacts that are the result of delayed sample processing. If samples are not processed and shipped according to best-practice standard operating procedures, downstream applications of collected samples could be impacted (Hatzis et al. 2011).
Shipment of human samples from clinical centers to biobanks must comply with federal packaging (country-specific) and labeling regulations. The package should include only the coded biological specimen. Packages must be clearly marked with sender and recipient addresses. Utilizing express or overnight courier services is highly recommended if possible. Packaging should follow the best practice guidelines of the courier9.
Biological samples intended for extraction and analysis of genomic DNA should be shipped either at room temperature or chilled and expedite shipped to the biobank. Blood or saliva samples for RNA extraction and analysis should be collected in tubes containing additive RNA-stabilizing chemicals (0.5 mL of EDTA blood with 1.3 mL RNAlater or PAXgene blood vacutainers; Weber et al. 2010). These tubes should be shipped on dry ice for overnight delivery. This procedure has demonstrated higher RNA yields and integrity and is recommended for implementation into biobanking protocols where RNA isolation will follow.
As a final best-practice standard operating procedure for sample collection and shipment (unless the biobank is located locally with a guarantee for same-day pick-up and delivery), it is highly recommended to plan collection and shipment of samples to avoid extended delays in transit due to weekends or holidays. Unplanned or anticipated delays can drastically influence sample quality.
CLINICAL INFORMATION
As tinnitus is a symptom of multiple conditions involving numerous audiological and psychological comorbidities, we believe that an effective genetic study could only emerge from a detailed phenotyping strategy allowing the classification and selection of individuals with a homogeneous tinnitus phenotype (Lopez-Escamez et al. 2016). The Human Phenotype Ontology (HPO) Project provides a standardized vocabulary of phenotypic abnormalities encountered in human diseases (Köhler et al. 2017). Each term in the HPO describes a phenotypic abnormality, such as hearing loss (HPO:0000365) or tinnitus (HPO:0000360). However, the heterogeneity in tinnitus phenotype is not considered, with the exception of pulsatile tinnitus (HPO: 00008629), and further additional terms in hearing and tinnitus should be included, such as those that distinguish unilateral from bilateral tinnitus. Moreover, psychological comorbidities (anxiety and depression) and mental health status should be included in clinical questionnaires to obtain a comprehensive phenotype, as these conditions display a genetic contribution.
The main tinnitus variables should be defined in the protocol of the study and a core set of relevant questionnaires included (Müller et al. 2016), along with a number of recommended audiological procedures (Table 3). The clinical information to be collected must be easy to obtain and should be stored in a secure database with coded access according to national/European regulations on personal data protection.
TABLE 3.
Minimum clinical information required for phenotyping patients with tinnitus for genetic studies

SECURITY AND PROTECTION
A contingency plan should be established to access, eliminate, and share data with potential collaborators, keeping personal data secure and guaranteeing the right of each individual to withdraw or participate in future studies.
The disposal of DNA, RNA, or any human biological sample is governed by international regulation in biomedical research, and all samples should be handled according to ERB and national/European regulations on health information or HIPAA Privacy Rule in the United States (https://www.hhs.gov/hipaa/index.html).
Access to Data and Transfer to the Consortium
Data from biological databases cannot be accessed for nonresearch purposes by insurers, employers, law enforcement agencies, or other civil-law agencies (OECD, 2009).
Different access forms to data are possible, according to the main goal of the study. Essentially, two possibilities are accepted: database querying is performed by the staff who returns collected results to the researchers, or researchers directly access databases to query only certain aspects of the data held by the database. Access to genetic data must be consistent with the research protocols, the participant’s informed consent, and conform to the privacy and confidentiality policies established in the ERB TINNET consortium will facilitate access to anonymized genomic data according to EU regulation on data protection.
Secured Storage of Human Biological Samples and Data
The storage of biological material as a collection for future research (including genetics research) should be considered as an option, and in case of a positive decision, permission must be obtained from the ERB. However, the essential topic is that the written consent form has to address the issue of sample storage length and possibility for further research use (Wolf et al. 2008).
The storage of tissue samples and data either linked to the samples or derived from them needs to be clearly considered, with protection of data that contains information about the donor (demographic characteristics, type of disease, and outcomes associated with the sample and its treatment) a priority.
Consequently, all protected health information must be considered sensitive data and addressed to ensure patient confidentiality, avoiding the possibility of linking genetic data back to a patient’s identity. Some guidelines suggest a de-identification of samples such that a coded sample is relabeled with a unique second code, while maintaining a link between the two codes (i.e., double-coded); or anonymization of samples such that the link between the two codes of a double-coded sample is permanently deleted. While the process of anonymization provides a maximum security, it does not allow for returning results, sample withdrawal, clinical monitoring, or patient follow-up.
CONCLUDING REMARKS
We have provided detailed instructions for nongeneticist healthcare professionals who would like to be involved in genetics research in the field of hearing science. Collaborative multidisciplinary research with a deep phenotyping of tinnitus is an optimal way to identify well-defined targets for the development of effective therapies. TINNET consortium encourages all health professionals dealing with tinnitus patients who read this paper and became interested in the project to contact us directly (see authors information).
ACKNOWLEDGMENTS
The work was supported by an independent research program funded under the Biomedicine and Molecular Biosciences European Cooperation in Science and Technology (COST) Action framework (TINNET, BM1306) and from the European Union H2020 research and innovation program under the MSC-ITN 722046 (Schlee et al. 2017). A.J.S. received funding from German Tinnitus Foundation, Sonnenfeld Fundation, and Dürr Foundation. C.R.C. received funding from Karolinska Institutet, Tysta Skolan, Hörselforskningsfonden, Magnus Bergvalls, and Lars Hiertas Minne. J.A.L.-E. received funding from Meniere Society, UK, and the Luxembourg National Research Fund (INTER/Mobility/17/11772209).
The authors have no conflicts of interest to disclose.
Publication fees were partially supported by Polytechnic Institute of Setúbal, Portugal.
REFERENCES
- Cederroth C. R., Kähler A. K., Sullivan P. F., et al. Genetics of tinnitus: Time to biobank phantom sounds. Front Genet, 2017). 8, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. (Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet, 2013). 381, 1371–1s379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz M., Bayazit Y. A., Celenk F., et al. Significance of serotonin transporter gene polymorphism in tinnitus. Otol Neurotol, 2010). 31, 19–24. [DOI] [PubMed] [Google Scholar]
- Dobbs L. J., Madigan M. N., Carter A. B., et al. Use of FTA gene guard filter paper for the storage and transportation of tumor cells for molecular testing. Arch Pathol Lab Med, 2002). 126, 56–63. [DOI] [PubMed] [Google Scholar]
- Elger B. S., Caplan A. L. Consent and anonymization in research involving biobanks: Differing terms and norms present serious barriers to an international framework. EMBO Rep, 2006). 7, 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. A. Designing clinical trials for assessing the effectiveness of interventions for tinnitus. Trends Hear, 2017). 21, 2331216517736689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis C., Sun H., Yao H., et al. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. J Natl Cancer Inst, 2011). 103, 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S., Hellenthal G., Pirinen M., et al. ; International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium, Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature, 2011). 476, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg K., Burmeister M., Shedden K., et al. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: Evidence of genetic moderation. Arch Gen Psychiatry, 2011). 68, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S., Vasilevsky N. A., Engelstad M., et al. The human phenotype ontology in 2017. Nucleic Acids Res, 2017). 45, D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver A. M., Smith R. J., Van Camp G., et al. Congenital hearing loss. Nat Rev Dis Primers, 2017). 3, 16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman W. S., Zuo J. Epigenetic regulation in the inner ear and its potential roles in development, protection, and regeneration. Front Cell Neurosci, 2014). 8, 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. T., Hinds D. A., Tung J. Y., et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet, 2017). 49, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Escamez J. A., Bibas T., Cima R. F., et al. Genetics of tinnitus: An emerging area for molecular diagnosis and drug development. Front Neurosci, 2016). 10, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas I. L., Brüggemann P., Requena T., et al. Genetic susceptibility to bilateral tinnitus in a Swedish twin cohort. Genet Med, 2017). 19, 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J., Bowler E., Cerezo M., et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res, 2017). 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. R., Howrigan D. P., Merico D., et al. ; Psychosis Endophenotypes International Consortium; CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium. (Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet, 2017). 49, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Edvall N. K., Idrizbegovic E., et al. Validation of online versions of tinnitus questionnaires translated into Swedish. Front Aging Neurosci, 2016). 8, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman C. W., Jacobson G. P., Spitzer J. B. Development of the Tinnitus Handicap Inventory. Arch Otolaryngol Head Neck Surg, 1996). 122, 143–148. [DOI] [PubMed] [Google Scholar]
- Newman C. W., Sandridge S. A., Jacobson G. P. Psychometric adequacy of the Tinnitus Handicap Inventory (THI) for evaluating treatment outcome. J Am Acad Audiol, 1998). 9, 153–160. [PubMed] [Google Scholar]
- Noreña A. J. Revisiting the cochlear and central mechanisms of tinnitus and therapeutic approaches. Audiol Neurootol, 2015). 20(Suppl 1)53–59. [DOI] [PubMed] [Google Scholar]
- Pal L. R., Moult J. Genetic basis of common human disease: Insight into the role of missense SNPs from genome-wide association studies. J Mol Biol, 2015). 427, 2271–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M., Mattheisen M., Frank J., et al. Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry, 2010). 68, 578–585. [DOI] [PubMed] [Google Scholar]
- Schaette R., McAlpine D. Tinnitus with a normal audiogram: Physiological evidence for hidden hearing loss and computational model. J Neurosci, 2011). 31, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. (Biological insights from 108 schizophrenia-associated genetic loci. Nature, 2014). 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee W., Hall D. A., Canlon B., et al. Innovations in doctoral training and research on tinnitus: The European School on Interdisciplinary Tinnitus Research (ESIT) Perspective. Front Aging Neurosci, 2017). 9, 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekelle P. G., Wachter R. M., Pronovost P. J., et al. Making health care safer II: An updated critical analysis of the evidence for patient safety practices. Evid Rep Technol Assess (Full Rep), 2013). 1–945. [PMC free article] [PubMed] [Google Scholar]
- Sheldon E., Vo K. C., McIntire R. A., et al. Biobanking human endometrial tissue and blood specimens: Standard operating procedure and importance to reproductive biology research and diagnostic development. Fertil Steril, 2011). 95, 2120–2122, 2122.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore S. E., Roberts L. E., Langguth B. Maladaptive plasticity in tinnitus–Triggers, mechanisms and treatment. Nat Rev Neurol, 2016). 12, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C., Prange-Krex G., Freiberg-Richter J., et al. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant, 2000). 25, 575–577. [DOI] [PubMed] [Google Scholar]
- Trevis K. J., McLachlan N. M., Wilson S. J. A systematic review and meta-analysis of psychological functioning in chronic tinnitus. Clin Psychol Rev, 2018). 60, 62–86. [DOI] [PubMed] [Google Scholar]
- Vona B., Nanda I., Shehata-Dieler W., et al. Genetics of tinnitus: Still in its infancy. Front Neurosci, 2017). 11, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D. G., Casjens S., Rozynek P., et al. Assessment of mRNA and microRNA stabilization in peripheral human blood for multicenter studies and biobanks. Biomark Insights, 2010). 5, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S. M., Lawrenz F. P., Nelson C. A., et al. Managing incidental findings in human subjects research: Analysis and recommendations. J Law Med Ethics, 2008). 36, 219–248, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Hu Q., Shepherd L., et al. The impact of DNA input amount and DNA source on the performance of whole-exome sequencing in cancer epidemiology. Cancer Epidemiol Biomarkers Prev, 2015). 24, 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]


