Abstract
Background:
Organophosphates (OP) are widely used insecticides that acutely inhibit acetylcholinesterase enzyme activity. There is great interest in improving the understanding of molecular mechanisms related to chronic OP exposure induced toxicity. We aim to elucidate epigenetic changes associated with OP exposure, using untargeted analysis of genome-wide DNA methylation data.
Methods:
In a population-based case control study of Parkinson's disease (PD), we assessed ambient OP exposure via residential and workplace proximity to commercial applications. We investigated associations between OP exposure and genome-wide DNA methylation (Illumina 450 k) in 580 blood samples (342 PD patients, 238 controls) and 259 saliva samples (128 patients, 131 controls). To identify differential methylation related to OP exposure, we controlled for age, sex, European ancestry, and PD status; in addition, we stratified by disease status.
Results:
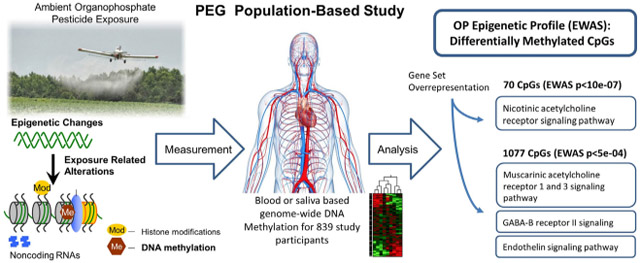
We identified 70 genome-wide significant CpGs, including cg01600516 in ALOX12 (cor = 0.27, p = 1.73E–11) and two CpGs in HLA genes, cg01655658 (cor = −0.24, p = 2.80E–09) in HLA-L (pseudogene) and cg15680603 (cor = 0.20, p = 7.94E–07) in HLA-DPA1. Among the 70 CpGs located in 41 genes, 14 were also differentially methylated in saliva samples. The most overrepresented pathway was the nicotinic acetylcholine receptor signaling pathway (fold enrichment = 15.63, p = 1.01E–03, FDR = 1.64E–01). Expanding to a larger number of genes (CpG p < 5E–04, FDR < 2.25E–01; 1077 CpGs, 662 genes), the most enriched pathway shifted to the muscarinic acetylcholine receptor 1 and 3 signaling pathway (p-value = 5.36E–04, FDR = 4.73E–02). When we stratified by PD status, results were similar. Of the 70 significant CpGs, 63 were detected among both patients and controls and 7 were only associated with OP exposure among patients.
Conclusions:
This study finds chronic low-level OP exposure is associated with differential DNA methylation in blood and saliva, both in elderly population controls and PD patients. Our study results suggest that long-term sub-acute OP exposure influences methylation in genes enriched for muscarinic and nicotinic acetylcholine receptor pathways.
Keywords: DNA methylation, Organophosphates, EWAS, Acetylcholine receptor pathways
GRAPHICAL ABSTRACT
1. Introduction
Pesticides represent a broad range of chemicals used for crop protection, agricultural food production, and disease control, which are designed to impact living systems. Organophosphates (OP) are widely used insecticides that acutely inhibit acetylcholinesterase enzyme activity, inducing neurotoxicity and cell death (Terry, 2012). Observational and experimental studies have shown detrimental health effects in humans from acute acetylcholinesterase inhibition toxicity and possibly neurodegeneration from long-term low level chronic OP exposure, such as seen in Parkinson's disease (PD) (Lukaszewicz-Hussain, 2010; Paul et al., 2016b). There is great interest in improving our understanding of molecular mechanisms related to OP and other pesticide induced chronic toxicity. A recent tool available to help us address the sequela of chronic exposures, which may not elicit acute toxicity, are epigenetic changes derived from methylation arrays. We can then assess whether methylation patterns may influence the effects of exposure on disease, potentially providing useful biomarkers of chronic low-level exposure.
To date, a variety of environmental toxicants, including pesticides, have been shown to influence DNA methylation. For example, exposure to persistent organic pollutants, endocrine disruptors, and metals have been shown to modulate DNA methylation (Baccarelli and Bollati, 2009; Ruiz-Hernandez et al., 2015). In fact, three commonly used OP pesticides induced DNA methylation changes in 712 genes in a cancer cell line in vitro (Zhang et al., 2012a). However, there is a lack of population-based epigenome-wide investigations in exposed humans such that knowledge of the effects of OP pesticides on DNA methylation is currently limited.
In a study of PD we conducted in the agriculturally intense Central Valley of California, we previously identified 82 differentially methylated CpG sites in blood comparing PD patients and population-based controls in an epigenome-wide association study (EWAS) (Chuang et al., 2017) and observed differences in epigenetic age and cell composition (Horvath and Ritz, 2015). In this rural community, pesticide exposure is ubiquitous and residential proximity to high levels of agricultural OP use is strongly associated with an increased risk of PD (Paul et al., 2016b). In the present study, we aim to elucidate epigenetic changes associated with OP exposure, using untargeted analysis of genome-wide DNA methylation data (Illumina 450 k), among both PD patients and population-controls.
2. Methods
All procedures described were approved by the University of California at Los Angeles (UCLA) Human Subjects Committee and informed consent was obtained from all participants.
2.1. Study population
This study used data from 839 participants enrolled in the Parkinson's Environment and Genes Study (PEG), designed to investigate causes of Parkinson's disease (PD) in agricultural regions of the California central valley. We analyzed blood samples from 580 participants (342 PD patients and 238 controls) enrolled during wave 1, known as PEG1, conducted from 2000 to 2007. We further analyzed saliva samples from another 259 participants (128 PD patients and 131 controls) enrolled during wave 2 (PEG2, 2010–2015). PEG1 patients were identified by neurologists, large medical groups, or public service announcements. In PEG2, PD patients were identified through the pilot California PD Registry, which was initiated in 2008 in the same CA counties (Kern, Fresno, and Tulare). Population controls were identified using Medicare lists and residential tax assessor records. Controls were randomly selected from clusters of five neighboring households, which study staff approached in person at their homes. One eligible household member was allowed to enroll.
PEG PD patients were enrolled within three years of PD diagnosis, confirmed as having idiopathic PD through in person exams by UCLA movement disorder specialists, and lived in Kern, Tulare, or Fresno counties. Controls enrolled from the same three counties were required to be over the age of 35, having lived within one of the counties for at least 5 years prior to enrollment, and not have a Parkinsonism diagnosis. Further details about participant recruitment and participation have been previously described (Costello et al., 2009; Paul et al., 2016a; Wang et al., 2011).
Trained interviewers recorded detailed information on demographic and risk factors for all patients and controls, and recorded detailed information on lifetime occupational and residential histories. Ancestry was self-reported and confirmed using ancestry informative genetic markers.
2.2. Organophosphate exposure
We estimated ambient OP pesticide exposure from residential and occupational proximity to commercial agricultural pesticide applications using geographic information systems (GIS) based model (Cockburn et al., 2011). This method links California state mandated pesticide use reports (CA-PUR), mandated for all commercial application in CA since 1974 (CDPR, 2013), land use surveys providing locations of specific crops (CDWR, 2013), and the geocoded residential and occupational addresses for each participant. For each pesticide reported to the CA-PUR, we summed the pounds applied per year and per acre within a 500-m buffer of each residential and occupational address of our participants. This method has been validated with biomarker studies (Ritz and Costello, 2006).
For each participant, we calculated a yearly average exposure for each chemical over the study period (from 1974 to year of blood draw), by summing the year-specific pounds of chemical applied and dividing by the total number of years in the relevant time period. There was residential or occupational proximity in our population to the application of 36 different chemicals classified as OPs, based on information from California Department of Pesticide Regulation (CDPR) and the pesticide action network (PAN) pesticide database (Kegley et al., 2014). We dichotomized the study period yearly average exposure to each of the 36 individual OP chemicals based on the median level in the exposed healthy controls. Exposures at both residential and occupational addresses were included, and each participant could have been exposed at both locations, only one, or neither. We then summed the number of OP chemicals that each participant was exposed to above the median, creating an OP count with a range of 0 to 46, to estimate OP exposure.
2.3. DNA extraction and illumina methylation array
DNA was extracted from peripheral whole blood and saliva. We profiled DNA samples using the Illumina Infinium 450 k platform (486 k CpGs). The array covers 99% of RefSeq genes, with an average of 17 CpG sites per gene region distributed across the promoter, 5′UTR, first exon, gene body, and 3′UTR. It covers 96% of CpG islands, with additional coverage in island shores and the regions flanking them (Illumina Infinium HumanMethylation450 BeadChip data sheet). The raw methylation data (beta values) were preprocessed using the background normalization method from the Genome Studio software. Sex concordance was confirmed.
2.4. Statistical methods
To examine differences in demographics, cell composition, and overall mean methylation by OP exposure, we used chi-square or Pearson correlations. All analysis was done with R (WGCNA, missMethyl, ggplot2, qqman, IlluminaHumanMethylation450kanno.ilmn12.hg19, org.Hs.eg.db, minfi, and limma packages).
We first performed an epigenome-wide association study (EWAS) to assess the relationship between OP exposure and methylation. We regressed individual CpG methylation levels on potential confounding variables (age, sex, PD, and European ancestry) and formed residuals (Horvath et al., 2016; van Eijk et al., 2012). For association testing, we then calculated correlation coefficients, implementing biweight midcorrelation calculation, between the adjusted methylation levels (residuals) at each CpG and OP exposure, and p-values based on Student’s t-test, using the standard screening for numeric trait (standardScreeningNumericTrait command of WGNCA in R (Langfelder and Horvath, 2008)). To adjust for multiple testing (~486 k CpGs on the Illumina array), we considered a strict Bonferroni correction (p-value < 0.05/500,000 = 10E–08) for genome-wide significance, and p-value < 10E–07 for suggestive associations (FDR < 7E–03). We conducted analysis in the full population, and further stratified by PD status. As secondary analysis, we also controlled for cell count. Abundance measures of cell types were estimated with the Houseman algorithm in the minfi R package and the epigenetic clock software (Horvath, 2013; Houseman et al., 2012; Jaffe and Irizarry, 2014).
We performed genome-wide analysis on the PEG1 blood-based DNA methylation markers and then analyzed the CpGs with p-value < 10E–07 in the PEG2 saliva-based DNA methylation.
To better understand the biologic function of our epigenetic profile (identified through EWAS), we linked each gene with a significant CpG to its' biologic function and performed gene set overrepresentation analysis. This analysis was used to identify which genetic pathways and functions are overrepresented among the genes with differentially methylated CpGs. We used the PANTHER gene ontology classification system (Mi et al., 2017; Mi et al., 2013). The PANTHER (protein annotation through evolutionary relationship) classification system (http://www.pantherdb.org/, accessed March 2018) is a comprehensive system that combines gene function, ontology, and pathways. PANTHER classifies genes according to their function both into families and subfamilies, annotated with ontology terms (Gene Ontology (GO) and PANTHER protein class), and as assigned to expert-curated PANTHER pathways (Mi and Thomas, 2009).
We created two gene lists, one for genes with at least one significantly associated CpG at p < 10E–07 and a second list with a less stringent p < 5E–04. Gene lists are compared with the human gene database (21,042 genes) to test for overrepresentation of pathways, molecular functions, biological processes, and cellular components, using a Fisher's exact test with FDR correction for multiple comparisons.
3. Results
Demographic characteristics of the study population by PD status are detailed in Table 1. In PEG1, patients were slightly older than controls (70.2 (SD = 9.8) vs 67.5 (SD = 12.8)), and had a higher proportion of European ancestry (87% of patients vs 81% of controls). Among the PEG2 population, the patients and controls were of similar age and proportions of sex/European ancestry. In both populations, the patients were more highly exposed to OPs as reported previously (Paul et al., 2016b; Ritz et al., 2016).
Table 1.
Population exposure and characteristics of PEG study population with DNA methylation.
| Population characteristics Mean ± SD/n (%) |
PEG1 blood DNA methylation |
PEG2 saliva DNA methylation |
||||
|---|---|---|---|---|---|---|
| Full PEG1 population (n = 580) |
PEG1 PD patients (n = 342) |
PEG1 controls (n = 238) |
Full PEG2 population (n = 259) |
PEG2 PD patients (n = 128) |
PEG2 controls (n = 131) |
|
| Age at blood draw (y) | 69.1 ±11.4 | 70.2 ± 9.8 | 67.5 ± 12.8a | 68.8 ± 9.7 | 69.3 ± 10.0 | 68.4 ± 9.5 |
| Male | 323 (56) | 197 (58) | 126 (53) | 146 (56) | 72 (56) | 74 (56) |
| European ancestry | 483 (83) | 276 (81) | 207 (87)a | 166 (64) | 82 (64) | 84 (64) |
| OP count | 8.2 ± 9.3 | 9.1 ± 9.8 | 7.0 ± 8.3b | 10.2 ± 10.2 | 12.3 ± 11.1 | 8.5 ± 9.2b |
| Mean methylation | 0.29 ± 0.01 | 0.29 ± 0.01 | 0.29 ± 0.01 | 0.32 ± 0.01 | 0.32 ± 0.01 | 0.32 ± 0.01 |
Abbreviations: PEG = Parkinson's Environmentand Gene study; OP = organophosphate.
PD patients vs controls (Wilcox or ChiSq) p-value < 0.05.
p-Value < 0.01.
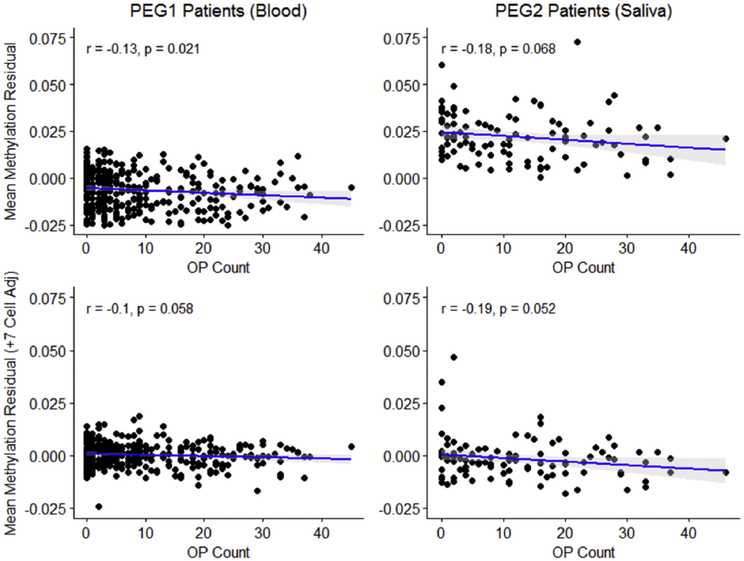
Global mean methylation levels (averaged genome-wide methylation levels across the 450 k CpG sites) from both tissues (blood and saliva DNA) were not different between patients and controls (Table 1). In both tissue samples, PD patients exhibited lower levels of global mean methylation with higher OP exposure (Pearson correlation r = −0.13, p = 0.021 and r = −0.18, p = 0.068; Fig. 1), after controlling for age, sex, and European ancestry. This pattern remained after additionally controlling for cell composition (r = −0.10, p = 0.058 and r = −0.19, p = 0.052; Fig. 1). OP exposure was not associated with global mean methylation levels among the controls.
Fig. 1.
Mean methylation residual across OP count among PEG PD patients (PEG1: blood-based, n = 342; PEG2: saliva-based, n = 128). Row one residual controlling for age, sex, European ancestry; row two residual additionally controlling for cell composition (7 cell counts). We show a Pearson correlation coefficient (r) and p-value.
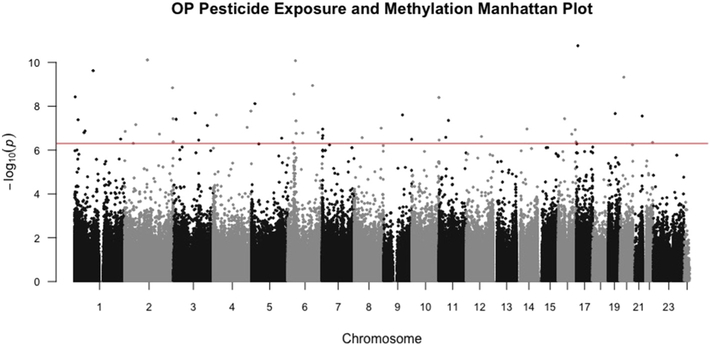
In our primary analysis, we related GIS derived OP exposure to each of the genome-wide DNA methylation markers (Illumina 450 k) in the whole population, adjusting for age, sex, PD, and European ancestry (Table 2). We also stratified by PD status to identify differentially methylated CpGs in each group separately and show these results in Table 2 as well. We identified 27 CpGs associated with exposure at a Bonferroni adjusted significance level in the whole population (p < 10E–08, FDR < 1.78E–03; Table 2; Manhattan Plot Fig. 2). This included our top hit cg01600516 in the ALOX12 gene (Chr 17, bp = 6,904,263), where methylation was positively correlated with exposure, r = 0.27 (p = 1.73E–11). This was seen among both patients (r = 0.29, p = 4.11E–0.8) and controls (r = 0.21, p = 1.04E–03), though results did not reach the same level of statistical significance. The GO molecular function for ALOX12 is oxidoreductase activity, with a biologic function in immune system processes and lipid metabolic processes (Supplemental Table S2). The other top hits include cg15177604 in POLR1B (r = −0.27, p = 7.70E–11), cg03655023 (intergenic, r = 0.27, p = 8.3E–11), cg01081438 in BRDT (r = −0.26, p = 4.74E–10), and cg24859648 in BANF2 (r = −0.26, p = 5.70E–10).
Table 2.
List of OP-associated CpGs from blood based DNA methylation.
| CpG | Chr | Position (bp) |
Probe SNPs | Probe SNPs 10 |
Gene | Gene region | Full PEG1 population (n = 579) |
PEG1 PD patients (n =342) |
PEG1 controls (n =237) |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cor | p value | cor | p value | cor | p value | |||||||
| cg01600516 | 17 | 6,904,263 | rs11571335 | ALOX12 | Body | 0.27 | 1.73E–11 | 0.29 | 4.11E–08 | 0.21 | 1.04E–03 | |
| cg15177604 | 2 | 113,300,283 | rs35724716 | rs2241804 | POLR1B | Body | −0.27 | 7.70E–11 | −0.23 | 2.39E–05 | −0.30 | 2.81E–06 |
| cg03655023 | 6 | 37,555,406 | rs2776900 | 0.27 | 8.33E–11 | 0.25 | 3.96E–06 | 0.26 | 5.22E–05 | |||
| cg01081438 | 1 | 92,417,998 | rs11165885 | BRDT | 5′UTR | −0.26 | 2.38E–10 | −0.30 | 1.51E–08 | −0.13 | 4.21E–02 | |
| cg24859648 | 20 | 17,680,544 | rs2277868 | BANF2 | TSS200; Body | −0.26 | 4.74E–10 | −0.22 | 5.70E–05 | −0.18 | 5.29E–03 | |
| cg11927033 | 6 | 122,793,073 | rs3752576 | PKIB; SERINC1 | 1st exon; 5′UTR; TSS200 | 0.25 | 1.14E–09 | 0.22 | 3.12E–05 | 0.25 | 1.29E–04 | |
| cg14271023 | 2 | 238,410,067 | MLPH | Body | 0.25 | 1.44E–09 | 0.22 | 4.11E–05 | 0.28 | 9.69E–06 | ||
| cg01655658 | 6 | 30,227,583 | rs9391807 | HLA-L | Body | −0.24 | 2.80E–09 | −0.21 | 1.00E–04 | −0.27 | 2.85E–05 | |
| cg02368820 | 1 | 3,052,501 | rs4648363 | PRDM16 | Body | 0.24 | 3.74E–09 | 0.25 | 2.63E–06 | 0.13 | 4.85E–02 | |
| cg15083522 | 10 | 134,188,873 | LRRC27 | 3′UTR | −0.24 | 4.00E–09 | −0.27 | 5.45E–07 | −0.17 | 9.60E–03 | ||
| cg15845365 | 5 | 16,785,399 | MYO10 | Body | −0.24 | 7.64E–09 | −0.22 | 4.16E–05 | −0.21 | 1.05E–03 | ||
| cg19952704 | 4 | 187,751,549 | −0.23 | 1.67E–08 | −0.21 | 1.13E–04 | −0.21 | 1.07E–03 | ||||
| cg03329597 | 3 | 108,125,523 | MYH15 | Body | 0.23 | 2.05E–08 | 0.18 | 7.21E–04 | −0.01 | 9.07E–01 | ||
| cg22996768 | 19 | 33,719,749 | −0.23 | 2.17E–08 | −0.20 | 2.19E–04 | −0.20 | 1.76E–03 | ||||
| cg02106072 | 9 | 94,182,501 | NFIL3 | 5′UTR | −0.23 | 2.50E–08 | −0.22 | 3.08E–05 | −0.22 | 5.14E–04 | ||
| cg17858192 | 4 | 16,077,807 | PROM1 | TSS200; 5′UTR | −0.23 | 2.51E–08 | −0.28 | 2.09E–07 | −0.13 | 5.35E–02 | ||
| cg08514194 | 21 | 46,075,092 | rs2838613 | KRTAP12-4; C21orf29 | TSS1500; body | 0.23 | 2.81E–08 | 0.16 | 2.96E–03 | 0.26 | 5.57E–05 | |
| cg02043600 | 16 | 31,021,933 | STX1B | TSS200 | −0.23 | 3.70E–08 | −0.20 | 1.37E–04 | −0.27 | 2.49E–05 | ||
| cg02059823 | 2 | 240,872,433 | rs73101773 | 0.23 | 3.70E–08 | 0.22 | 3.31E–05 | 0.32 | 7.07E–07 | |||
| cg26845082 | 3 | 13,555,664 | −0.23 | 3.93E–08 | −0.25 | 4.44E–06 | −0.11 | 1.06E–01 | ||||
| cg15600437 | 1 | 17,309,539 | MFAP2 | TSS1500 | −0.23 | 4.13E–08 | −0.22 | 5.49E–05 | −0.05 | 4.72E–01 | ||
| cg12856521 | 11 | 46,389,249 | rs34470543 | DGKZ | Body | −0.23 | 4.41E–08 | −0.15 | 4.18E–03 | −0.24 | 1.74E–04 | |
| cg13604933 | 6 | 40,145,993 | −0.22 | 4.62E–08 | −0.19 | 5.28E–04 | −0.09 | 1.86E–01 | ||||
| cg19384241 | 2 | 55,393,977 | rs11396095 | rs11382356 | −0.22 | 6.85E–08 | −0.13 | 2.01E–02 | −0.13 | 5.07E–02 | ||
| cg10590338 | 3 | 168,707,389 | −0.22 | 7.51E–08 | −0.23 | 1.71E–05 | −0.22 | 8.46E–04 | ||||
| cg10815671 | 4 | 168,824,531 | −0.22 | 9.15E–08 | −0.19 | 5.59E–04 | −0.23 | 3.66E–04 | ||||
| cg14044167 | 8 | 134,271,505 | NDRG1 | Body | 0.22 | 9.91E–08 | 0.16 | 3.21E–03 | 0.27 | 2.69E–05 | ||
| cg01926858 | 14 | 53,619,558 | DDHD1 | 1st exon | 0.22 | 1.09E–07 | 0.20 | 2.83E–04 | 0.19 | 2.69E–03 | ||
| cg21598190 | 7 | 2,099,404 | rs6953670 | MAD1L1 | Body | 0.22 | 1.09E–07 | 0.19 | 4.20E–04 | 0.23 | 4.27E–04 | |
| cg06052372 | 16 | 83,967,808 | rs5016957 | −0.22 | 1.17E–07 | −0.18 | 6.87E–04 | −0.22 | 7.69E–04 | |||
| cg10701801 | 1 | 52,082,593 | OSBPL9 | TSS200 | −0.22 | 1.34E–07 | −0.24 | 6.19E–06 | −0.17 | 7.31E–03 | ||
| cg19274180 | 2 | 2,726,437 | rs75478670 | 0.22 | 1.40E–07 | 0.17 | 2.12E–03 | 0.23 | 3.36E–04 | |||
| cg09374123 | 6 | 150,379,008 | −0.22 | 1.57E–07 | −0.12 | 2.30E–02 | −0.31 | 1.55E–06 | ||||
| cg03327352 | 1 | 46,979,222 | rs4660355 | DMBX1 | 3′UTR | −0.22 | 1.58E–07 | −0.20 | 2.17E–04 | −0.17 | 7.69E–03 | |
| cg08145292 | 6 | 73,867,933 | rs6453649 | rs6453647 | KCNQ5 | Body | −0.22 | 1.66E–07 | −0.23 | 1.67E–05 | −0.17 | 9.19E–03 |
| cg03644281 | 6 | 41,068,752 | NFYA; LOC221442 | 3′UTR; TSS200 | 0.22 | 1.72E–07 | 0.22 | 4.38E–05 | 0.19 | 2.57E–03 | ||
| cg13226272 | 2 | 173,189,711 | rs7577264 | −0.21 | 1.86E–07 | −0.21 | 1.15E–04 | −0.21 | 1.02E–03 | |||
| cg05477582 | 16 | 66,600,478 | CMTM1 | 1st exon | 0.21 | 1.87E–07 | 0.19 | 4.99E–04 | 0.17 | 9.36E–03 | ||
| cg23627948 | 7 | 2,760,692 | 0.21 | 2.16E–07 | 0.19 | 5.00E–04 | 0.20 | 2.02E–03 | ||||
| cg23601664 | 12 | 75,780,994 | 0.21 | 2.39E–07 | 0.23 | 1.83E–05 | 0.19 | 3.77E–03 | ||||
| cg11008123 | 11 | 33,097,335 | LOC283267 | TSS1500 | 0.21 | 2.60E–07 | 0.17 | 1.25E–03 | −0.03 | 6.86E–01 | ||
| cg17527589 | 8 | 39,380,435 | ADAM3A | Body | 0.21 | 2.70E–07 | 0.19 | 3.95E–04 | 0.21 | 1.41E–03 | ||
| cg14388237 | 5 | 150,051,778 | MYOZ3 | Body | −0.21 | 2.85E–07 | −0.21 | 8.73E–05 | −0.18 | 5.22E–03 | ||
| cg27468880 | 7 | 965,995 | rs55956927 | ADAP1 | Body | 0.21 | 2.89E–07 | 0.23 | 2.07E–05 | 0.23 | 3.14E–04 | |
| cg08354527 | 1 | 229,252,042 | rs2125454 | −0.21 | 3.12E–07 | −0.19 | 3.04E–04 | −0.20 | 2.56E–03 | |||
| cg00540295 | 9 | 139,616,839 | rs11145847 | rs3739939 | FAM69B | Body | 0.21 | 3.21E–07 | 0.15 | 4.96E–03 | 0.22 | 4.93E–04 |
| cg11445109 | 10 | 135,343,248 | CYP2E1 | Body | 0.21 | 3.47E–07 | 0.18 | 8.65E–04 | 0.16 | 1.27E–02 | ||
| cg15145296 | 3 | 125,709,740 | 0.21 | 3.49E–07 | 0.21 | 1.27E–04 | 0.17 | 6.98E–03 | ||||
| cg06193597 | 2 | 241,896,910 | rs10173220 | −0.21 | 4.22E–07 | −0.20 | 2.63E–04 | −0.19 | 3.14E–03 | |||
| cg06896988 | 16 | 87,952,610 | CA5A | Body | 0.21 | 4.25E–07 | 0.17 | 1.46E–03 | 0.20 | 2.47E–03 | ||
| cg05995465 | 2 | 240,313,602 | rs6147245 | HDAC4 | 5′UTR | 0.21 | 4.28E–07 | 0.15 | 7.06E–03 | 0.21 | 1.19E–03 | |
| cg18514595 | 22 | 49,579,968 | 0.21 | 4.41E–07 | 0.23 | 1.16E–05 | 0.13 | 3.86E–02 | ||||
| cg18433519 | 6 | 24,646,494 | rs6456625 | KIAA0319 | TSS1500; TSS200 | 0.21 | 4.48E–07 | 0.16 | 2.44E–03 | 0.10 | 1.15E–01 | |
| cg14883135 | 2 | 43,051,175 | rs12997857 | 0.21 | 4.88E–07 | 0.21 | 6.88E–05 | 0.12 | 7.07E–02 | |||
| cg23676314 | 17 | 1,835,482 | rs4790841 | 0.21 | 5.13E–07 | 0.14 | 1.15E–02 | 0.25 | 1.14E–04 | |||
| cg06653140 | 5 | 36,157,329 | SKP2 | Body | −0.21 | 5.18E–07 | −0.21 | 1.20E–04 | −0.24 | 1.62E–04 | ||
| cg06108461 | 20 | 60,628,389 | rs34023475 | TAF4 | Body | −0.21 | 5.76E–07 | −0.18 | 9.84E–04 | −0.22 | 8.12E–04 | |
| cg13589109 | 7 | 36,124,843 | 0.21 | 5.76E–07 | 0.19 | 2.97E–04 | 0.27 | 3.38E–05 | ||||
| cg08508337 | 8 | 144,660,607 | NAPRT1 | TSS200 | −0.21 | 6.19E–07 | −0.23 | 1.89E–05 | −0.16 | 1.11E–02 | ||
| cg04497611 | 17 | 80,860,250 | TBCD | Body | −0.20 | 7.25E–07 | −0.13 | 1.45E–02 | −0.11 | 9.91E–02 | ||
| cg11035303 | 3 | 43,465,503 | ANO10 | Body | −0.20 | 7.28E–07 | −0.26 | 9.46E–07 | −0.26 | 5.82E–05 | ||
| cg26889118 | 15 | 49,342,629 | −0.20 | 7.64E–07 | −0.15 | 4.99E–03 | −0.22 | 5.09E–04 | ||||
| cg24927769 | 7 | 147,732,939 | rs73168563 | MIR548F3; CNTNAP2 | Body | −0.20 | 7.76E–07 | −0.20 | 1.88E–04 | −0.19 | 4.11E–03 | |
| cg18705301 | 15 | 41,695,430 | rs56283962 | NDUFAF1 | TSS1500 | −0.20 | 7.76E–07 | −0.21 | 1.00E–04 | −0.16 | 1.21E–02 | |
| cg15680603 | 6 | 33,037,413 | rs1126542 | rs35171346 | HLA-DPA1 | Body | 0.20 | 7.94E–07 | 0.16 | 3.21E–03 | 0.24 | 2.58E–04 |
| cg15138543 | 4 | 987,391 | rs3806756 | IDUA; SLC26A1 | Body; TSS1500; TSS200 | −0.20 | 8.12E–07 | −0.24 | 9.70E–06 | −0.15 | 1.98E–02 | |
| cg18576044 | 14 | 76,015,669 | rs11626877 | −0.20 | 8.46E–07 | −0.11 | 3.48E–02 | −0.30 | 2.70E–06 | |||
| cg04461143 | 3 | 31,494,586 | −0.20 | 9.77E–07 | 0.00 | 9.29E–01 | −0.11 | 8.76E–02 | ||||
| cg27012203 | 1 | 11,865,920 | CLCN6; MTHFR; | TSS1500; 5′UTR | 0.20 | 9.89E–07 | 0.17 | 1.81E–03 | 0.21 | 1.18E–03 | ||
| cg23779604 | 7 | 2,760,784 | 0.20 | 9.93E–07 | 0.18 | 1.09E–03 | 0.17 | 7.75E–03 | ||||
Chr chromosome, bp base pair, TSS transcription start site, TSS1500 within 1500 bps of a TSS, TSS200 within 200 bps of a TSS, UTR untranslated region, SNPs listing dbSNP entries within a probe, SNPs_10 listing dbSNP entries within 10bpof the CpG site.
These are CpGs with p values < 10E–7 for blood DNA methylation analyses adjusting for PD (full population only), age, sex, and European ancestry.
Fig. 2.
Manhattan plot showing significance of association between methylation levels of CpGs and OP exposure prior to blood draw.
Another 43 CpGs were associated with OP exposure at a less stringent significance level (p < 10E–07, FDR < 6.88E–03; Table 2). It should be noted that when stratifying by PD status, a number of CpGs had similar correlation coefficients as seen in the whole population, but did not reach formal Bonferroni correction withstanding significance (0.05/70 = p < 7.1E–04), we attribute this mostly to the reduction in sample size when stratifying. After controlling for cell composition, 56 CpGs remained significant (Supplemental Table S3). Among the 70 CpGs we identified, seven seemed to be specific to PD patients only, as associations were not detected among controls (Table 2).
When examining these 70 CpGs (blood based OP EWAS p < 10E–07) in the saliva samples, 14 CpGs were also found to be associated with OP exposure (p < 0.05), though the direction of association was not consistent for three CpGs (Table 3). Two CpGs were significantly associated with OP exposure in both the blood and saliva samples at a strict Bonferroni level (p < 0.05/70 = 7E–04). These two CpGs were cg19952704 (intergenic; chr 4, bp = 187,751,549) which was hypo-methylated (blood: r = −0.23, p = 1.67E-08; saliva: r = −0.24, p = 2.10E–04) and cg05995465 in the 5′UTR region of HDAC4, which was hyper-methylated (blood: r = 0.21, p = 4.28E–07; saliva: r = 0.22, p = 7.02E–04). HDAC4 is involved in chromatin organization and negative regulation of apoptotic process (Supplemental Table S2). HLA-DPA1 was also suggestively associated with OP exposure among both tissue sources (blood: r = 0.20, p = 7.94E–07; saliva: r = 0.16, p = 1.42E–02).
Table 3.
List of OP-associated CpGs from blood based DNA methylation and their association in saliva based DNA methylation replication samples.
| CpG | Chr | Position (bp) | Probe SNPs | Probe SNPs 10 | Gene | Gene region | PEG1 - blood DNA (n = 579) |
PEG2 - saliva DNA (n = 231) |
||
|---|---|---|---|---|---|---|---|---|---|---|
| cor | p value | cor | p value | |||||||
| cg19952704 | 4 | 187,751,549 | −0.23 | 1.67E−08 | −0.24 | 2.10E−04 | ||||
| cg05995465 | 2 | 240,313,602 | rs6147245 | HDAC4 | 5′UTR | 0.21 | 4.28E−07 | 0.22 | 7.02E−04 | |
| cg06193597 | 2 | 241,896,910 | rs10173220 | −0.21 | 4.22E−07 | −0.22 | 8.75E−04 | |||
| cg08354527 | 1 | 229,252,042 | rs2125454 | −0.21 | 3.12E−07 | −0.22 | 9.42E−04 | |||
| cg21598190 | 7 | 2,099,404 | rs6953670 | MAD1L1 | Body | 0.22 | 1.09E−07 | 0.21 | 1.44E−03 | |
| cg12856521 | 11 | 46,389,249 | rs34470543 | DGKZ | Body | −0.23 | 4.41E−08 | −0.21 | 1.57E−03 | |
| cg22996768 | 19 | 33,719,749 | −0.23 | 2.17E−08 | 0.20 | 1.79E−03 | ||||
| cg02059823 | 2 | 240,872,433 | rs73101773 | 0.23 | 3.70E−08 | 0.19 | 3.98E−03 | |||
| cg15680603 | 6 | 33,037,413 | rs1126542 | rs35171346 | HLA-DPA1 | Body | 0.20 | 7.94E−07 | 0.16 | 1.42E−02 |
| cg11927033 | 6 | 122,793,073 | rs3752576 | PKIB; SERINC1 | 1st exon; 5′UTR; TSS200 | 0.25 | 1.14E−09 | 0.16 | 1.49E−02 | |
| cg15145296 | 3 | 125,709,740 | 0.21 | 3.49E−07 | 0.16 | 1.84E−02 | ||||
| cg14044167 | 8 | 134,271,505 | NDRG1 | Body | 0.22 | 9.91E−08 | −0.15 | 1.86E−02 | ||
| cg17858192 | 4 | 16,077,807 | PROM1 | TSS200; TSS200; 5′UTR | −0.23 | 2.51E−08 | 0.14 | 3.04E−02 | ||
| cg15138543 | 4 | 987,391 | rs3806756 | IDUA; SLC26A1 | Body; TSS1500; TSS200 | −0.20 | 8.12E−07 | −0.13 | 5.15E−02 | |
Chr Chromosome, bp base pair, TSS transcription start site, TSS1500 within 1500 bps of a TSS, TSS200 within 200 bps of a TSS, UTR untranslated region, SNPs listing dbSNP entries within a probe, SNPs_10 listing dbSNP entries within 10 bp of the CpG site.
These are CpGs with p values < 0.05 from saliva DNA methylation replication analyses; adjusting for PD, age, sex, and European ancestry.
Blood-based DNA population (PEG1) and saliva-based DNA population (PEG2) are comprised of different participants.
The GO and biologic function information about each of the identified genes (intragenic CpGs, 41 genes), based on the PANTHER Classification System (Mi et al., 2017; Mi et al., 2013), can be found in Supplemental Table S2. These CpGs are located in genes related to cellular defense (HLA-DPA1) and immune system processes (KIAA0319) among other functions.
Employing gene set overrepresentation analysis, comparing the list of OP related CpG genes (70 CpGs p < 10E–07; 41 genes) to all genes in the human database (21,042 genes), we identified only one PANTHER pathway with near significant overrepresentation after multiple testing correction (FDR = 1.64E–01), the nicotinic acetylcholine receptor signaling pathway. This pathway had a fold enrichment of 15.63 (Fisher's exact p-value = 1.01E–03, FDR = 1.64E–01; Table 4). This is noteworthy, though not unexpected, as OP pesticides are designed to directly inhibit acetylcholinesterase enzyme activity.
Table 4.
PANTHER analysis of gene overrepresentation among identified OP-related CpG genes.
| Annotation data set | Term | Fold enrichment | Raw p-value | FDR |
|---|---|---|---|---|
| Top 70 CpGs (EWAS p < 10E–07), 41 genes | ||||
| PANTHER Pathways | Nicotinic acetylcholine receptor signaling pathway (P00044) | 15.24 | 1.08E–03 | 1.77E–01 |
| Top 1077 CpGs (EWAS p < 5e–04), 662 genes | ||||
| PANTHER Pathways | Muscarinic acetylcholine receptor 1 and 3 signaling pathway (P00042) | 3.90 | 5.36E–04 | 4.37E–02 |
| GABA-B receptor II signaling (P05731) | 4.38 | 2.01E–03 | 8.20E–02 | |
| Endothelin signaling pathway (P00019) | 3.08 | 1.67E–03 | 9.06E–02 | |
| Muscarinic acetylcholine receptor 2 and 4 signaling pathway (P00043) | 3.02 | 7.64E–03 | 2.07E–01 | |
| Nicotinic acetylcholine receptor signaling pathway (P00044) | 2.36 | 1.43E–02 | 3.32E–01 | |
| PANTHER GO-Slim Molecular Function | Pyrophosphatase activity: catalytic activity (GO:0003824) | 1.30 | 2.75E–05 | 2.64E–03 |
| Pyrophosphatase activity (GO:0016462) | 1.80 | 1.43E–04 | 9.14E–03 | |
| Microtubule binding (GO:0008017) | 2.87 | 1.20E–03 | 4.62E–02 | |
| Transporter activity (GO:0005215) | 1.54 | 1.15E–03 | 5.53E–02 | |
| PANTHER GO-Slim Biological Process | Cellular process (GO:0009987) | 1.15 | 3.37E–04 | 2.74E–02 |
| Phosphate–containing compound metabolic process (GO:0006796) | 1.46 | 2.99E–04 | 3.64E–02 | |
| PANTHER GO-Slim Cellular Component | Intracellular: Cell part (GO:0044464) | 1.22 | 1.59E–04 | 5.08E–03 |
| Intracellular (GO:0005622) | 1.21 | 6.64E–04 | 1.42E–02 | |
| Postsynaptic membrane (GO:0045211) | 4.76 | 1.33E–03 | 2.13E–02 | |
We examined overrepresentation in PANTHER pathways, molecular function, biological process, and cellular components (compared to the all the genes in the database, n = 21,042). Genes lists are from top CpGs with genes mapped (excludes intergenic CpGs and CpGs mapped to multiple genes, see Supplemental text for gene lists).
No molecular function, biological process, or cellular component terms neared significance (after FDR correction) for the top 70 CpGs, so these categories are not shown.
PANTHER Overrepresentation Test (released 2017-12-05); PANTHER version 13.1 released 2018-02-03; Fisher's exact with FDR multiple test correction (Mi et al., 2017; Mi et al., 2013), accessed March 2018.
Among the top 1077 CpGs (p < 5E–04, FDR < 2.25E–01), mapped to 662 genes (excluding intergenic CpGs and those which mapped to multiple genes), the only significantly overrepresented PANTHER pathway was the muscarinic acetylcholine receptor 1 and 3 signaling pathway, with a fold enrichment of 3.90 (Fisher's exact p-value = 5.36E–04, FDR = 4.73E–02; Table 4). Notably, the muscarinic acetylcholine receptor 2 and 4 signaling pathway (fold enrichment = 3.02, p-value = 7.64E–03, FDR = 2.07E–01) and nicotinic acetylcholine receptor signaling pathway (fold enrichment = 2.36, p-value = 1.43E–02, FDR = 3.32E–01), were also among the most significant pathways. Though given the high FDR for these two pathways chance cannot be ruled out. However, acetylcholine signaling is what OP pesticides target. The muscarinic acetylcholine receptor 1 and 3 signaling pathway was also the top pathway when focusing on the top CpGs after controlling for blood cell composition (fold enrichment = 3.57, p-value = 3.69E–03, FDR = 3.01E–02; Supplemental Table S4). Other top pathways after controlling for cell composition include Heterotrimeric G-protein signaling pathways (P00026, P00027), Ionotropic glutamate receptor pathway (P00037), and FAS signaling pathway (P00020), Supplemental Table S4.
To ensure that we were picking up an OP exposure signal and not a PD signal, we checked pathways for the top CpGs found in controls only (n = 259 blood-based samples; p < 5E–04, 250 CpGs in 181 genes). Both acetylcholine receptor pathways were among the top pathways (top 5 and 6 pathways), though neither reached significance (nicotinic acetylcholine receptor signaling pathway (fold enrichment = 2.63, p = 1.11E–01) and muscarinic acetylcholine receptor 1 and 3 signaling pathway (fold enrichment = 2.90, p = 1.59E–01)). It should be noted the sample size in controls is less than half of the total population with blood samples (n = 580).
A number of other molecular functions, biological processes, and cellular components were also enriched in the group of 662 genes. These include pyrophosphatase related catalytic activity (p-value = 2.75E–05, FDR = 2.64–03) and postsynaptic membrane components (p-value = 1.33E–03, FDR = 2.13E–02), both overrepresented in the set of differentially methylated genes (Table 4).
The gene lists used for overrepresentation analysis and the list of differentially methylated genes in the acetylcholine pathways can be found in the supplemental text.
4. Discussion
Our study presents population-based human evidence that chronic low-level OP exposure differentially affects DNA methylation levels in leukocytes and buccal cell epithelia in both elderly population controls and also in PD patients. Our large population size (n = 839) allowed us to use an untargeted methylation approach (genome-wide Illumina 450 k array) to build an epigenetic profile of differentially methylated CpGs associated with this exposure. Our objective was to identify differences related to OP exposure, not Parkinson's disease (PD). To this end, in blood samples, we identified correlations between our agricultural pesticide use based measure of chronic OP exposure and methylation levels at 70 CpG sites. Interestingly, these CpGs were in a gene sets enriched for muscarinic and nicotinic acetylcholine (ACh) receptor pathways, though perhaps not surprising given the main toxic action of OPs is related to inhibition of acetylcholinesterase with consequences of increasing ACh levels. When we assessed methylation at these 70 CpGs in a separate population using saliva samples, fourteen of these same sites were also differentially methylated in this source.
In the following, we will discuss our top methylation and gene overrepresentation results. However, it should be noted, signals for single CpGs (or sets of CpGs) should be interpreted carefully as biologic function is highly dynamic across multiple molecular layers (genome, methylome, transcriptome, etc.). Our EWAS only captured a snapshot of associations at a single molecular layer, and investigation across multiple molecular layers may be necessary.
The CpG most significantly correlated with OP exposure was cg01600516 (cor = 0.27, p = 1.73E–11) in the ALOX12 gene. ALOX12 (Arachidonate 12-lipoxygenase, 12S-type) belongs to a family of lipid peroxidizing enzymes, which oxidize polyunsaturated fatty acids (Kuhn et al., 2015). These enzymes are key players in cellular redox homeostasis. Many studies have shown that oxidative stress occurs after both acute and chronic OP exposure (Lukaszewicz-Hussain, 2010). Based on current understanding, ALOX12 is primarily expressed in thrombocytes and skin (Kuhn et al., 2015), which is notable given we detected this association with blood-based DNA. Other notable hits include two CpGs in HLA genes, cg01655658 (cor = −0.24, p = 2.80E–09) in HLA-L (pseudogene) and cg15680603 (cor = 0.20, p = 7.94E–07) in HLA-DPA1. What is even more compelling for cg15680603, this CpG was also differentially methylated in our separate sample population of saliva-based DNA (cor = 0.16, p = 1.42E–02). This may highlight the involvement of cellular defenses encouraged by chronic OP exposure.
Additionally, we saw differential methylation of CpGs in genes directly involved in gene transcription, including in cg15177604 (cor = −0.27, p = 7.70E–11) in POLR1B (DNA-directed RNA polymerase I subunit RPA2), which is involved in catalyzing the transcription of DNA to RNA, and cg05995465 (cor = 0.21, p = 4.28E–07) in HDAC4 (Histone deacetylase 4). Deacetylation of histones is involved in transcriptional repression. This CpG was also significantly associated in the saliva samples of our study participants (cor = 0.22, p = 7.02E–04).
To date, we have not found any previous population-based reports of genome-wide DNA methylation and OP pesticides specifically. However, a study recently published on occupational pesticide exposure and DNA methylation (450 k) in a Dutch population-based cohort (n = 1656) also reported differential methylation related to any occupational pesticide exposure at the most recent job held (van der Plaat et al., 2018). This cohort was considerably younger (median = 46 years) with a much lower prevalence of pesticide exposure (89% “No exposure”, 7% “Low exposure”, 4% “High exposure”). Although none of the same CpGs were mentioned by this group, they also detected differential DNA methylation in HLA genes and in LYPD6 (Ly6/PLAUR domain-containing protein 6), which interacts with the nicotinic acetylcholine receptors (van der Plaat et al., 2018). Another study examined genome-wide DNA methylation after exposing the human hematopoietic K562 cell line with the OP diazinon in-vitro (Zhang et al., 2012b). Interestingly, of the 1069 CpGs associated with exposure in this study, a top CpG was in HDAC3, which is in the same histone deacetylase superfamily as HDAC4 we found to be differentially methylated in blood- and saliva-based analyses in our population. Other genes detected in this cell line and in our population are NFYA (Nuclear Transcription Factor Y Subunit Alpha), CLCN6 (Chloride Voltage-Gated Channel 6), and C21orf29/TSPEAR (Thrombospondin Type Laminin G Domain and EAR Repeats) (Zhang et al., 2012b).
Among the 70 OP-related CpGs we identified with genome-wide significance, seven were picked up in PD patients only, i.e. associations were not seen among controls. These include cg03329597 in MYH15 (myosin, heavy chain 15) involved in muscle contraction, cg15600437 in MFAP2 (microfibrillar-associated protein 2) a component of the elastin-associated microfibrils, and cg18433519 in KIAA0319, which is involved in neuronal migration during development of the cerebral neocortex and may play a role in adhesion between migrating neurons. Other PD specific OP-related CpGs were intergenic or in genes of unknown function (LOC283267).
When we considered the OP associated gene set (genes with at least one significantly associated CpG), rather than individual CpG sites, the most overrepresented pathways were acetylcholine (ACh) receptor pathways. OP pesticides are specifically designed to inhibit acetylcholinesterase enzyme activity, resulting in an excess of ACh and thus cholinergic receptor overstimulation, acutely disrupting the function of the peripheral and central nervous system (Terry, 2012; van der Plaat et al., 2018). Concentrating only on genome-wide significant CpGs (p < 10E–07, FDR < 7E–03; 70 CpGs, 41 genes), the most enriched pathway of the gene set pointed to the nicotinic ACh receptors (p-value = 1.01E–03, FDR = 1.64E–01). However, when we expanded to include a larger number of CpGs (p < 5E–04, FDR < 2.25E–01; 1077 CpGs, 662 genes), the most enriched pathway shifted to the muscarinic ACh receptor 1 and 3 signaling pathway (p-value = 5.36E–04, FDR = 4.73E–02). ACh stimulates both nicotinic and muscarinic receptors and both receptors are found throughout the nervous systems. However, they are involved in quite different cholinergic responses. For example, N1 nicotinic receptors are found at neuromuscular junctions, with integral involvement in skeletal muscle movement. Muscarinic receptors are important in the parasympathetic nervous system. The M3 muscarinic receptors, for example, affect smooth muscle function, such as on blood vessels and vasodilation or constriction.
Although ACh primarily occurs and acts in the nervous system as a neurotransmitter, the presence of ACh in blood has been demonstrated (review: Kawashima et al., 2012) and expression of both muscarinic and nicotinic acetylcholine receptors has also been shown in various immune cells, such as lymphocytes (Kawashima and Fujii, 2000; Kawashima and Fujii, 2003; Kawashima and Fujii, 2004; Kawashima et al., 2012). It has been hypothesized that ACh released from parasympathetic cholinergic nerve terminals acts on muscarinic and nicotinic ACh receptors expressed on immune cells and their stimulation influences the immune system. Some evidence supporting this include studies which have found that in-vitro stimulation of muscarinic and nicotinic ACh receptors with agonists causes various functional and biochemical changes in immune cells (Kawashima and Fujii, 2000; Kawashima and Fujii, 2003; Kawashima and Fujii, 2004; Kawashima et al., 2012). ACh has been measured in blood and localized to T cells; furthermore, T cell activation enhances ACh synthesis, suggesting lymphocytic cholinergic activity is related to immunological activity (Kawashima et al., 2012). Muscarinic ACh receptor gene knockout mice have shown that M1/M5 muscarinic receptors are involved in up-regulating TNF-α, IFN-γ and IL-6 (Fujii et al., 2007b), while nicotinic ACh receptor gene knockout mice showed that α7 nicotinic ACh receptors down-regulate these pro-inflammatory cytokines (Fujii et al., 2007a).
These findings are very intriguing, as this suggests OP action on acetylcholinesterase enzyme activity may not only induce neurotoxicity through the known cholinergic pathways but may also act at the same time through cholinergic pathways in the immune system. Since we measured DNA methylation from blood and saliva, which also contains blood cells, the overrepresentation of methylated genes in ACh receptor pathway signals seems to be related to ACh activity in the blood on immune cells. The muscarinic ACh receptor 1 and 3 signaling pathway (p-value = 3.69E–04, FDR = 3.01E–02) was still the most overrepresented pathway when we looked at methylation after controlling for cell composition (Supplemental Table S4). Additionally, the gene set enrichment from CpGs identified in our control population only also indicated ACh pathways, supporting that we were not picking up a Parkinson's disease signal. However, given the strong association we have shown previously between OP exposure and Parkinson's disease risk (Paul et al., 2016b), these methylation results may suggest that we should not only consider the neurotoxicity of OP exposure but also its effects on immune system and the contributions of systemic immune regulation on PD and neurodegeneration.
Furthermore, general cell toxicity from OP exposures may also result from non-cholinergic mechanisms, including the induction of mitochondrial dysfunction and oxidative stress through the generation of reactive oxygen species (Soltaninejad and Abdollahi, 2009; Terry, 2012; Zaganas et al., 2013). This is suggested in our methylation analyses as well, with overrepresentation of genes related to other pathways including GABA-B receptor II signaling (p = 2.01E–03, FDR = 8.20E–02) which affects neurotransmitter release in both the peripheral and central nervous system. OP exposure thus may not only influence ACh activity, but also other neurotransmitters, including GABA. The endothelin signaling pathway was also overrepresented (p = 1.67E–03, FDR = 9.06E–02). This pathway has a role in potent stimulation of long-lasting and persistent vasoconstriction, but also in inflammation and oxidative stress. Other neurotransmission and immune related pathways were also overrepresented in the gene sets derived from epigenome-wide analysis after controlling for cell composition (Supplemental Table S4). These notably include heterotrimeric G-protein pathways, which are common signal transducers that mediate the effects of many xenobiotics but can also bind to muscarinic acetylcholine receptors. Two other pathways we identified are the ionotropic glutamate receptor pathway and the FAS signaling pathway. Glutamate is a neurotransmitter involved in learning and memory and also a key compound in cellular metabolism. The FAS signaling pathway is involved in the regulation of the immune system.
Although it may not come as a surprise that OP exposure influences molecular components in acetylcholine receptor pathways, it is noteworthy that our findings suggest that not only acute, but chronic, low-level ambient OP exposure, as modeled here based on residential and workplace proximity to commercial pesticide application, left a epigenetic fingerprint of exposure. This was detected in an untargeted, population-based analysis in two independent subsamples of both PD patients and controls with either blood samples or saliva samples. Our study population lives and works in an agriculturally active region of Central California, where a large amount of pesticides are applied commercially. A state mandated pesticide use reports registry (see Methods), allowed us to investigate chronic, low-level exposure that occurred over a 30+ year period. An advantage of this method is that it does not rely on participant recall of exposure. While we have previously validated our exposure assessment method with biomarkers for organochlorine pesticides, which have a long half live, and found it to correlate well with measures derived from our GIS-based PUR exposure system (Ritz and Costello, 2006), such validation for long-term low level OP exposure is not possible due to the short half-life of OPs in general. Thus, finding these highly plausible bio-signatures for long term OP exposure is very encouraging. With the limitations of our ambient pesticide exposure assessment, including currently unaccounted for factors, such as wind patterns at the time of application and geographic features that may influence pesticide drift, or actual participant location, any exposure misclassification would very likely be non-differential, biasing results toward the null.
This study is the largest and most comprehensive population-based DNA methylation study of OP exposure to date. We used strict significance criteria given methylation levels across the genome are often highly correlated to account for multiple testing (Bonferroni; p < 10E–8 and suggestive p < 10E-7, FDR < 7E–3). Additionally, our objective was to identify differential methylation related to OP exposure, not Parkinson's disease (PD), thus we controlled for PD in our primary analysis, and further, stratified all results by PD status. There are differential methylation patterns related to PD, which we have previously published on Chuang et al.(2017)However, there was no overlap in the CpGs (or genes) related to PD status and those we detected with OP exposure at genome-wide significance. This is expected as we controlled for PD status to remove the PD signals in our primary analyses of the whole population. Of the 70 identified CpGs, only 7 appear to be disease specific, meaning associations with OP exposures were only seen in PD patients (discussed above). Given the strong association between OP exposure and PD, future analysis will aim to elucidate the differential methylation patterns and pathways overrepresented among patients only and aim to suggest biologic mechanism associated with exposure and PD pathogenesis.
One of both the strengths and limitations of our study was using blood-based DNA methylation for our primary analysis. Blood is readily available and thus replication will be easier than for other tissues. Additionally, blood may in fact be the ideal tissue source to investigate OP-related disruption of certain biologic pathways. This includes immune system pathways as we've discussed in this manuscript. Other tissues or available biofluids, such as cerebrospinal fluid, could additionally inform further about neurotoxic mechanisms associated with exposure.
5. Conclusions
Our study presents some of the first population-based evidence associating chronic low-level OP exposure with differential DNA methylation in blood and saliva in both in elderly population controls and in PD patients. Our study results suggest that long-term sub-acute OP exposure influences methylation in genes enriched for muscarinic and nicotinic acetylcholine (ACh) receptor pathways.
Supplementary Material
HIGHLIGHTS.
Organophosphates (OP) are insecticides designed to inhibit acetylcholinesterase activity.
Chronic OP exposure may influence disease pathogenesis through epigenetic changes.
Long-term ambient OP exposure was associated with differential methylation at 70 CpG sites.
The methylated gene set was enriched for genes in acetylcholine receptor signaling pathways.
Pathways involving neurotransmission and inflammation were also found to be enriched.
Acknowledgments
Funding sources
This work was supported by the National Institute of Environmental Health Science (grant numbers F32-ES028087 (KP), 2R01-ES010544, U54ES012078); the American Parkinson’s disease Association; the Levine Foundation, and the Parkinson Alliance (JB).
Footnotes
Conflict of interest
The authors declare no competing or conflicts of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2018.07.143.
References
- Baccarelli A, Bollati V, 2009. Epigenetics and environmental chemicals. Curr. Opin Pediatr. 21 (2),243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDPR, 2013. California Department of Pesticide Regulation Pesticide Use Reporting. http://www.cdpr.ca.gov/docs/pur/purmain.htm.
- CDWR, 2013. California Department of Water Resources Land Use Surveys. http://www.water.ca.gov/landwateruse/lusrvymain.cfm.
- Chuang Y-H, Paul KC, Bronstein JM, Bordelon Y, Horvath S, Ritz B, 2017. Parkinson's disease is associated with DNA methylation levels in human blood and saliva. Genome Med. 9 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn M, Mills P, Zhang X, Zadnick J, Goldberg D, Ritz B, 2011. Prostate cancer and ambient pesticide exposure in agriculturally intensive areas in California. Am. J. Epidemiol. 173 (11), 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello S, Cockburn M, Bronstein J, Zhang X, Ritz B, 2009. Parkinson's disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am. J. Epidemiol. 169 (8), 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Plaat DA, deJong K, de Vries M, van Diemen CC, Nedeljković I, et al. , 2018. Occupational exposure to pesticides is associated with differential DNA methylation. Occup. Environ. Med. 75 (6), 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk KR, de Jong S, Boks MPM, Langeveld T, Colas F, et al. , 2012. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics 13, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii YX, Fujigaya H, Moriwaki Y, Misawa H, Kasahara T, et al. , 2007a. Enhanced serum antigen-specific IgG1 and proinflammatory cytokine production in nicotinic acetylcholine receptor α7 subunit gene knockout mice. J. Neuroimmunol. 189 (1–2), 69–74. [DOI] [PubMed] [Google Scholar]
- Fujii YX, Tashiro A, Arimoto K, Fujigaya H, Moriwaki Y, et al. , 2007b. Diminished antigen-specific IgG1 and interleukin-6 production and acetylcholinesterase expression in combined M1 and M5 muscarinic acetylcholine receptor knockout mice. J. Neuroimmunol. 188 (1), 80–85. [DOI] [PubMed] [Google Scholar]
- Horvath S, 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Ritz BR, 2015. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging (Albany NY) 7 (12), 1130–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, et al. , 2016. Huntington's disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY) 8 (7), 1485–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, et al. , 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA, 2014. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima K, Fujii T, 2000. Extraneuronal Cholinergic System in Lymphocytes. [DOI] [PubMed] [Google Scholar]
- Kawashima K, Fujii T, 2003. The Lymphocytic Cholinergic System and Its Contribution to the Regulation of Immune Activity. [DOI] [PubMed] [Google Scholar]
- Kawashima K, Fujii T, 2004. Expression of non-neuronal acetylcholine in lymphocytes and its contribution to the regulation of immune function. Front. Biosci. 9, 2063–2085. [DOI] [PubMed] [Google Scholar]
- Kawashima K, Fujii T, Moriwaki Y, Misawa H, 2012. Critical Roles of Acetylcholine and the Muscarinic and Nicotinic Acetylcholine Receptors in the Regulation of Immune Function. [DOI] [PubMed] [Google Scholar]
- Kegley SE, Hill BR, Orme S, Choi AH, 2014. PAN Pesticide Database, Pesticide Action Network, North America; (Oakland, CA, 2014). [Google Scholar]
- Kuhn H, Banthiya S, Van Leyen K, 2015. Mammalian Lipoxygenases and Their Biological Relevance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S, 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz-Hussain A, 2010. Role of oxidative stress in organophosphate insecticide toxicity - short review. Pestic. Biochem. Physiol. 98 (2), 145–150. [Google Scholar]
- Mi H, Thomas P, 2009. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 563, 123–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD, 2013. Large-scale gene function analysis with the panther classification system. Nat. Protoc. 8 (8), 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, et al. , 2017. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45 (D1), D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul KC, Rausch R, Creek MM, Sinsheimer JS, Bronstein JM, et al. , 2016a. APOE, MAPT, and COMT and Parkinson's disease susceptibility and cognitive symptom progression. J. Parkinsons. Dis. 6 (2), 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul KC, Sinsheimer JS, Rhodes SL, Cockburn M, Bronstein J, Ritz B, 2016b. Organophosphate pesticide exposures, nitric oxide synthase gene variants, and gene-pesticide interactions in a case-control study of Parkinson's disease, California (USA). Environ. Health Perspect. 124 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz B, Costello S, 2006. Geographic Model and Biomarker-derived Measures of Pesticide Exposure and Parkinson's Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz BR, Paul KC, Bronstein JM, 2016. Of pesticides and men: a California story of genes and environment in Parkinson's disease. Curr. Environ. Heal. Reports. 3 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Hernandez A, Kuo CC, Rentero-Garrido P, Tang WY, Redon J, et al. , 2015. Environmental chemicals and DNA methylation in adults: a systematic review of the epidemiologic evidence. Clin. Epigenetics 7 (1), 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltaninejad K, Abdollahi M, 2009. Current opinion on the science of organophosphate pesticides and toxic stress: a systematic review. Med. Sci. Monit. 15 (3), RA75–A90. [PubMed] [Google Scholar]
- Terry AV, 2012. Functional consequences of repeated organophosphate exposure: potential non-cholinergic mechanisms. Pharmacol. Ther. 134 (3), 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Costello S, Cockburn M, Zhang X, Bronstein J, Ritz B, 2011. Parkinson's disease risk from ambient exposure to pesticides. Eur. J. Epidemiol. 26 (7), 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaganas I, Kapetanaki S, Mastorodemos V, Kanavouras K, Colosio C, et al. , 2013. Linking pesticide exposure and dementia: what is the evidence? Toxicology 307, 3–11. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wallace AD, Du P, Kibbe WA, Jafari N, et al. , 2012a. DNA methylation alterations in response to pesticide exposure in vitro. Environ. Mol. Mutagen. 53 (7), 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wallace AD, Du P, Lin S, Baccarelli AA., et al. , 2012b. Genome-wide study of DNA methylation alterations in response to diazinon exposure in vitro. Environ. Toxicol. Pharmacol. 34 (3), 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.