Abstract
A half century of research and more than 75,000 publications have cemented the foundational role of integrins as major conduits of communication between the extracellular environment and the interior of cells. By serving as cell-surface receptors for numerous extracellular ligands, integrin α and β heterodimers are integrally involved in the physiological and pathophysiological responses of virtually all cell types. Excessive or suppressed ligand binding to integrins are implicated in many diseases, including cancer, inflammation and cardiovascular diseases.
From the earliest studies of integrins on blood cells, it became evident that their capacity to serve as functional receptors must be tightly controlled. Integrins on platelets and leukocytes must rapidly transit from a resting state where they display minimal ligand binding activity to an activated state where they can engage ligands with high affinity and/or avidity. Hence, the molecules and mechanisms governing integrin activation became and have remained major themes of research. While many modulators, both activators and inhibitors, have been described, two intracellular proteins, talins and kindlins, are now known to play essential roles in integrin activation. Talins and kindlins bind directly to similar but non-overlapping motifs in the short cytoplasmic tails of integrin β subunits. This prospective focuses on the kindlins; it provides a brief summary of our present understanding of their structure and function and then predicts some future directions where research on the kindlin family is likely to proceed.
The Kindlins.
There are three kindlins in mammals. Kindlin-1 is found primarily in ectodermal cells, kindlin-2 is broadly distributed and found in cardiomyocytes, endothelial cells and fibroblasts, and kindlin-3 is restricted primarily to hematopoietic cells. However, several cell types express more than one kindlin. Each kindlin is a FERM domain-containing protein, so named for a motif found in protein 4.1, ezrin, radixin and monesin, which contain F1, F2 and F3 subdomains. In kindlins, F1 is preceded by an N-terminal F0 subdomain (Fig.1), which is not infrequent among FERM domain proteins. Distinguishing kindlins from other FERM domain proteins is the transection of F2 by a PH domain. PH domains are often lipid binding, and lipid binding to kindlins helps to target them to membranes, a localization important in integrin activation. The three kindlins family members are similar to each other (53% amino acid sequence identity between kindlin-2 and kindlin-3). In comparing amino acid sequences of the kindlins, variable regions are interspersed among conserved regions; presumably the common regions mediate shared functions among the kindlins and the variable regions impart functions unique to the individual kindlins1.
Figure.
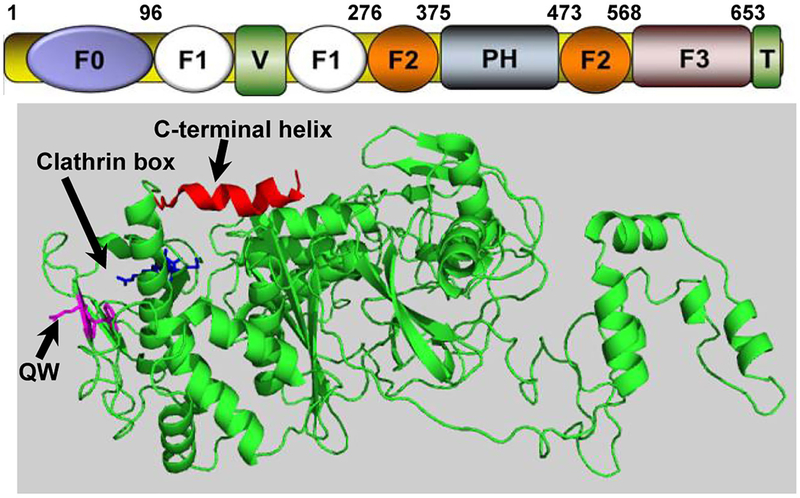
Subdomain organization of Kindlin-2. (Top) Subdomains of Kindlin-2 and their corresponding sequence numbers. (Bottom) Crystal structure of Kindlin-2 (PDB code 5xpy) with subdomains highlighted. Note that F2-N is the N-terminal portion of F2 prior to PH domain since the structure lacks PH domain and C-terminal F2 part. Certain regions that are under investigation are highlighted by arrows including C-terminal helix that bridges F1 and F3; QW that interacts with integrin; and clathrin box that potentially binds to clathrin.
The Past
Evidence that kindlins are integrally involved in integrin regulation converged from several distinct lines of investigation: a) numerous in vitro studies showed that over-expression, knockdown or knockout of kindlins alters integrin activation2,3; b) in C. elegans, the absence of unc-112, the kindlin homolog, recapitulates the phenotype arising the absence of the homologues of integrin alpha and beta subunits and ILK4; c) human diseases have been associated with mutations/deficiencies of kindlin-1 (Kindler Syndrome) and kindlin-3 (LADIII) and reflect deficits in integrin activation in affected individuals5; d) the phenotypes of mice where the genes of kindlin-1 and kindlin-3 were inactivated closely mirror the human diseases; and d) reduction or knockout of individual kindlins leads to tissue-specific pathologies, consistent with deficits in integrin activation. Together, these observations establish the essential role of kindlin in functions that depend on integrin activation.
The Present
Initially, it was thought that engagement of integrin β-subunits with the head domain of talin was necessary and sufficient for integrin activation. Binding of talin does favor a conformational transition of integrin from a bent to an extended conformation which is more permissive to ligand engagement6. However, the phenotypes of mice and humans with deficiencies of each kindlin establish their importance in integrin activation. Furthermore, in examining the time dependent sequence of events in formation of integrin containing nascent focal adhesions in cells, kindlins were recruited prior to talin7, contrary to the envisioned sequence of talin first and then kindlins to complete the activation process. Two more recent observations have provided some clarity to the role of kindlins in activation. First, the roles of kindlins and talin in integrin activation are distinct: while talin was able to enhance binding of a monovalent ligand, kindlins could not achieve this, talin was necessary for affinity modulation, but kindlins were required for integrin clustering leading to avidity modulation8. Since integrins in some LADIII patients cannot support productive cellular responses, such as platelet activation and innate immunity, integrin clustering mediated by kindlins must be required to mount functional responses. Second, using cells in which the genes kindlin-2 and talin were inactivated by gene knockout technology, it was concluded that neither is sufficient and both are needed for optimal integrin activation9.
Over the past 5 years, novel insights into kindlin biology have emerged: they not only control integrin-dependent responses, but they also govern some cellular responses that are independent of their integrin binding. The distinction depends on the capacity of a kindlin bearing mutations of its integrin binding Gln-Trp (QW) sequence in its F3 subdomain to still mediate specific functional responses. These responses depend on the capacities of kindlins to function as adapter proteins. More than 20 kindlin binding partners have been identified do date10. As each new binding partner is identified, the repertoire of kindlins’ functions expands.
The Future.
From our current vantage point, several specific areas of future progress on the kindlins can be envisioned.
High Resolution Structures of Kindlins and its Multi-molecular Complexes.
A major advance in our understanding of kindlins was provided by the crystal structure of mutant kindlin-2 in complex with integrin cytoplasmic tails. As expected, QW within the F3 subdomain was engaged in the complex. Of particular note, a kindlin-2 dimer was detected and implicated in integrin activation11. This interesting finding will need to be corroborated with a full-length kindlin. Beyond the structure of kindlins per se, their complexes with other binding partners would advance the field significantly. Cryo-electron microscopy of molecules that assemble with kindlins will provide high resolution of the organization of focal adhesion complexes.
Dynamics of integrin activation.
Very ambiguous is the temporal sequence of events that occur during assembly of integrin activation. Approaches that trace the recruitment of binding partners during integrin inside-out and outside-in signaling across integrins is needed. High resolution microscopy offers the opportunity to trace not only the recruitment of molecules into forming focal complexes but also the timed exit of these molecules as they compete for overlapping binding sites on integrins.
How kindlins mediate integrin activation.
The prevailing model for talin’s role in integrin activation is its dissociation/separation of the α and β cytoplasmic tails12, causing a change in the juxa-positioning of the transmembrane helices and ultimately transitions of the extracellular region from a bent to an extended conformation13. During this process, talin may also compete with filamin, an integrin inactivator14; and displacement of filamin may promote integrin activation. Talin must undergo an “unmasking” of its integrin binding site to induce these conformational changes in the integrin15. Kindlin’s role is to cluster integrins but are these the sole roles of these molecules in the activation process? It remains to be seen whether talin also inserts into the clustering process and whether kindlin can induce conformational changes with the integrin heterodimer that enhances integrin activation. Kindlins are presumed to be constitutively bound to integrins, but yet an activating event in regulating kindlin activity has not been systematically excluded.
It would be customary to end a commentary with a brief summary, but as we have tried to convey, the story of kindlins is still in development. There are many areas of kindlin biology in need of more in depth studies. The structural basis and dynamic involvement of kindlins in integrin activation and novel integrin-independent functions are obvious directions requiring further emphasis. Unanticipated interactions may further broaden the roles of kindlins in cell biology.
Acknowledgements.
This is a very brief commentary on kindlins. We are severely limited in length and number of references that can be cited and genuinely apologize to colleagues whose contributions could not be adequately acknowledge.
Sources of Funding. This work was supported by NIH grants R01 HL096062, PO1 HL073311, P01 HL076491.
Footnotes
Disclosures: The authors of no conflicts of interest to declare.
Reference List
- (1).Malinin NL, Plow EF, Byzova TV Kindlins in FERM adhesion. Blood 2010;115:4011–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ma YQ, Qin J, Wu C, Plow EF. Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J Cell Biol 2008; 181:439–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Montanez E, Ussar S, Schifferer M, Bosl M, Zent R, Moser M, Fassler R. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev 2008; 15;1325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG. The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J Cell Biol 2000;150:253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Malinin NL, Zhang L, Choi J, Ciocea A, Razorenova O, Ma Y-Q, Podrez EA, Tosi M, Lennon DP, Caplin AI, Shurin SB, Plow EF, Byzova TV. A point mutation in kindlin-3 ablates activation of three integrin subfamilies in humans. Nature Med 2009;15:313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol 2007;25:619–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bachir AI, Zareno J, Moissoglu K, Plow EF, Gratton E, Horwitz AR. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr Biol 2014;24:1845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ye F, Petrich BG, Anekal P, Lefort CT, Kasirer-Friede A, Shattil SJ, Ruppert R, Moser M, Fassler R, Ginsberg MH. The mechanism of kindlin-mediated activation of integrin alphaIIbbeta3. Curr Biol 2013; 18:2288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Theodosiou M, Widmaier M, Bottcher RT, Rognoni E, Veelders M, Bharadwaj M, Lambacher A, Austen K, Muller DJ, Zent R, Fassler R. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. Elife 2016;5:e10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Plow EF, Das M, Bialkowska K, Sossey-Alaoui K. Of Kindlins and Cancer. Discoveries (Craiova ) 2016;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Li H, Deng Y, Sun K, Yang H, Liu J, Wang M, Zhang Z, Lin J, Wu C, Wei Z, Yu C. Structural basis of kindlin-mediated integrin recognition and activation. Proc Natl Acad Sci U S A 2017;114:9349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, Qin J. A structural mechanism of integrin aIIbb3 “inside-out” activation as regulated by its cytoplasmic face. Cell 2002;110:587–97. [DOI] [PubMed] [Google Scholar]
- (13).Humphries JD, Chastney MR, Askari JA, Humphries MJ. Signal transduction via integrin adhesion complexes. Curr Opin Cell Biol 2018; 5;56:14–21. [DOI] [PubMed] [Google Scholar]
- (14).Liu J, Das M, Yang J, Ithychanda SS, Yakubenko VP, Plow EF, Qin J. Structural mechanism of integrin inactivation by filamin. Nat Struct Mol Biol 2015:383–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Goksoy E, Ma Y-Q, Wang X, Kong X, Perera D, Plow EF, Qin J. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol Cell 2008;31:124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]


