Abstract
Robertson reflects on a new study showing how ensemble-biased metadynamics can be used to interpret BetP protein dynamics data.
Secondary active transporters are exquisite biological machines that finely tune the chemical composition of cells. They can extract a substrate that may be at a low concentration in the surrounding environment, such as a nutrient or amino acid, and sequester it into the cell against its concentration gradient. For example, under hyperosmotic conditions, the BetP transporter will move betaine or another biologically inert osmolyte into the cell to provide protection against osmotic stress (Krämer and Morbach, 2004). This transport process is uphill, but BetP makes it possible by cotransporting Na+, which is in excess in the extracellular environment. The binding of both Na+ and betaine unlocks the access pathway to the other side of the membrane, via a change in conformational dynamics, leading to downhill movement of Na+ into the cell along with the desired substrate. Although different states in the transport cycle have been identified, associating these states to dynamical changes has been less tractable. In this issue of the Journal of General Physiology, Leone et al. reveal the potential of a new method to interpret such protein dynamics.
The mechanism underlying coupled transport remains poorly understood, but pieces of the puzzle have gradually been added over time. In the case of BetP, there are at least 11 states in the reaction cycle, each depicted by different conformations (Perez et al., 2014). Remarkable efforts in x-ray crystallography have led to the determination of high-resolution structures of BetP in eight of these distinct states. This includes the inward-open (Koshy et al., 2013) and outward-open (Perez et al., 2014) structures that map the two endpoints of the alternating access mechanism. Yet, while these static pictures provide essential information about the molecular structures, they do not explain how the transport reaction occurs. For that, we also need to understand the protein dynamics and how different binding conditions change the balance of energetically accessible conformations (Liao et al., 2016). Fortunately, there are experimental methods that can provide direct information about the dynamics of membrane transporters. Fluorescence methods, such as Förster Resonance Energy Transfer (FRET) and pulsed electron–electron double resonance (PELDOR)/double electron–electron resonance (DEER), can provide information about the changes in distance between specific positions under different transport conditions. These types of experiments have been useful in following shifts in the conformational dynamics of transporters that undergo large distance changes, such as the elevator-like motions of the glutamate transporter GltPh (Akyuz et al., 2015).
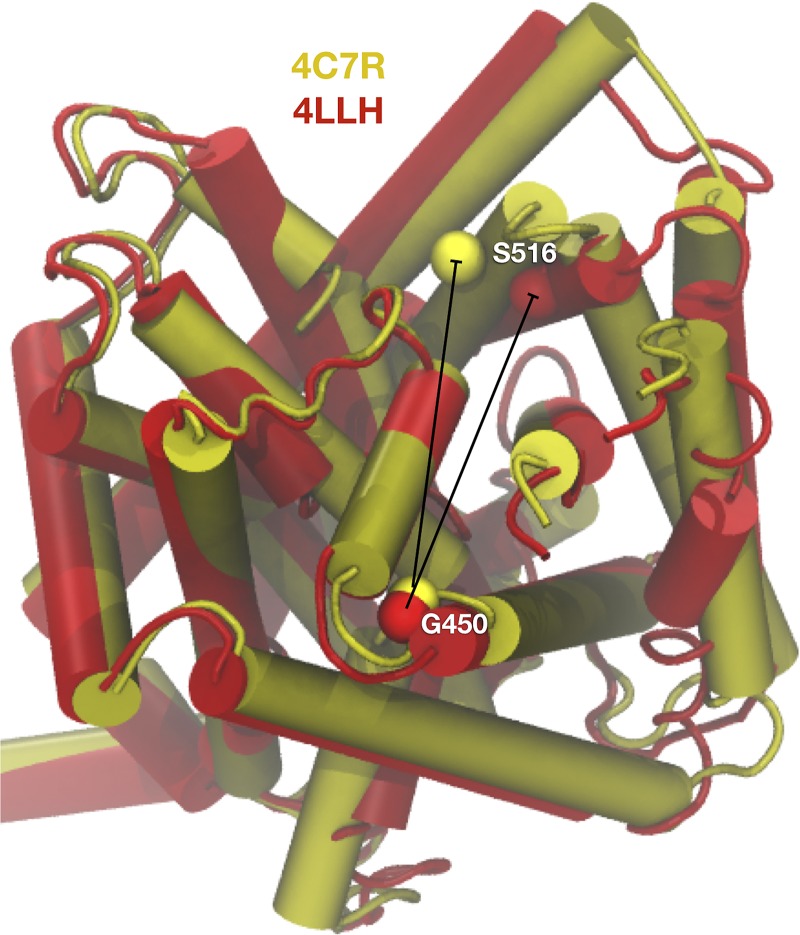
However, there are many other transporters, for instance members of the LeuT-fold family, where conformational changes are far more conservative. In these cases, dynamics data cannot be clearly assigned to any particular structure and thus it can be difficult to interpret the data. Leone et al., show that this is a problem for the BetP transporter (Leone et al., 2019). They carry out site-directed spin labeling at two residue positions, G450C and S516C, which span the region of the protein where the largest conformational changes are observed between the inward-open (PDB ID 4C7R) and outward-open (PDB ID 4LLH) states. Importantly, despite the significant structural changes in this area, the average distances between the Cα atoms are similar in both states (29–30 Å; Fig. 1). In contrast, when measuring the experimental distance distribution of the spin-label nitroxides using PELDOR, large changes were observed. In the presence of saturating Na+, the intrasubunit distances are larger than the crystal structures indicate, with a peak close to 44 Å. However, when betaine is added in addition to Na+, the distribution shifts to reveal two populations with peaks at 25 and 42 Å, indicating a significant change in dynamics. The peak at the shorter distance agrees with the Cα distances observed in either crystal structure, yet the larger distance observed in both Na+ and Na+/betaine conditions must reflect extended conformations of the probes. To consolidate this broad distance range, the authors accounted for probe flexibility by modeling possible spin-label nitroxide positions using a rotamer library. While this expanded the distance range of the spin labels, the overall positions of the probes are still highly overlapping when modeled onto both crystal structures. Therefore, there is insufficient information in the static crystal structures to assign a particular state to the observed probe distances described in the experimental data.
Figure 1.
The problem of conformational ambiguity of BetP distance data. The inward-facing (4C7R, yellow) and outward-facing (4LLH, red) conformations of the Na+/betaine cotransporter BetP show small structural changes of 2–3 Å RMSD. Distances of Cα atoms between G450 and S516 are 28.7 ± 0.1 Å and 29.7 ± 0.8 Å respectively, calculated and averaged over the three subunits in the crystal structures. Structural alignment, RMSD, and distance calculations were performed using the analysis program VMD (Humphrey et al., 1996).
It is apparent from this analysis that conformational dynamics of the system are important to consider. One possibility is that the dynamics of the protein are different in the different conformations. For example, the helices that have conjugated probes attached may have increased flexibility in one particular state or in the absence of betaine. In addition, the dynamics of the spin-label must be a contributing factor, as it is possible that there are preferential rotamers in the different structural states that will change the weighting of the distribution. In complex situations such as this, MD simulations can be performed to sample local protein conformations and the configurational space of the spin-labels. However, the results that come out of calculated MD trajectories are only meaningful if sufficient sampling of the system has been performed to ensure that the representative weighting is observed. One has to make sure that the spin labels do not get trapped in local energy wells and that there is sufficient sampling of the protein conformation. Unfortunately, this bystander approach of conventional MD is generally impractical as it is time consuming, and oftentimes impossible to assess whether the sampling has converged locally instead of globally.
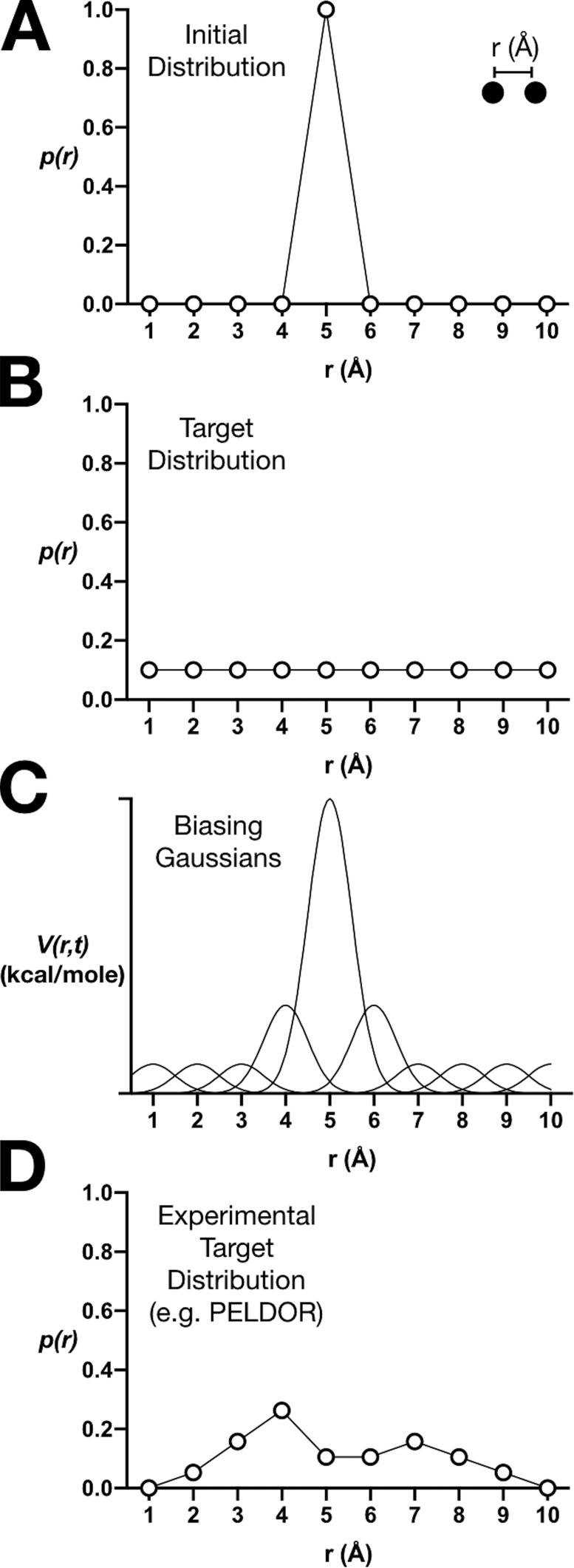
Instead, enhanced sampling methods, such as Metadynamics (MetaD), offer a way of carrying out MD simulations using tools that can help to address the problem of local trapping (Laio and Parrinello, 2002). Imagine that two atoms in a protein are positioned 5 Å apart in the crystal structure and that they are trapped in a local energy well so that conventional MD leads to limited sampling around this initial condition (Fig. 2 A). However, if our goal is to sample uniformly all distances between 1–10 Å (Fig. 2 B), external input will be necessary to encourage sampling outside of the local energy minimum. In MetaD, repulsive energy potentials in the form of Gaussian hills are gradually added to the energy function to prevent the simulation from getting stuck in its initial energy well (Fig. 2 C). As the cumulative sum of the Gaussians nullifies the potential well in the underlying energy landscape, the two atoms that were naturally confined near their 5-Å starting distance are increasingly less likely to stay put and at some point will begin to sample other distances. This process is repeated for every local energy well that might be encountered so that the resulting MD trajectory contains an ensemble of configurations where all distances have been uniformly sampled. Convergence of the sampling is directly monitored, and since the magnitudes of the biasing potentials are known, the results can be unbiased to reveal the underlying energy landscape of the system.
Figure 2.
Enhanced sampling by metadynamics and ensemble biasing. (A) A hypothetical initial distance distribution of two atoms in a crystal structure that are confined in a local energy well. (B) A target distribution for uniform sampling of all distances. (C) Metadynamics uses repulsive Gaussian energy potentials to bias the sampling to converge to the target distribution. (D) In EBMetaD, the target distribution is nonuniform and can be an experimental distance distribution, such as one measured using PELDOR.
Leone et al. (2019) demonstrate how to use a variation of the Metadynamics strategy, called ensemble-biased Metadynamics (EBMetaD; Marinelli and Faraldo-Gómez, 2015), to interrogate which BetP structure most likely corresponds to the experimental dynamics data. Ensemble biasing means that the target of the simulation is a nonuniform distribution, such as a distance distribution measured by PELDOR (Fig. 2 D). Using this approach, the authors performed simulations of the inward- and outward-open structures of BetP bound to either Na+ alone or to Na+ and betaine. Following a blinded computational experiment, they biased the sampling in each case to reproduce the two different experimental PELDOR distributions. The authors’ expectation was that the sampling would converge to the target distribution in all cases, as dictated by the EBMetaD method. However, their premise was also that the effort that was required to meet a target, i.e., the work, would differ. For instance, if the initial structure is already portrayed in the experimental distribution, little biasing would be required the reproduce the PELDOR data. On the other hand, if the input structure does not correspond to the experimental conditions of the PELDOR measurement, the work would be significantly larger.
Considering this valuable information hidden in each simulation, the scientists were able to ask a single specific question: Which structural state requires more work for the simulation to converge to the experimental data? They find that in the Na+-only condition, the inward-facing structure requires less work and is thus more likely to represent the experimental state. The authors discuss their surprise in this finding because the expectation for the Na+ bound condition is that the transporter would be primed in the outward-open state to capture betaine from the extracellular solution. However, this expectation does correspond with observations that BetP preferentially crystallizes in the inward-facing conformation in the presence of Na+. In addition, the authors find that both inward- and outward-facing structures require a considerable amount of work to reproduce the experimental distribution in the presence of Na+ and betaine, implying that neither structure represents these data.
It is important to note that the aim of these simulations was not to sample large-scale transitions between conformational states, but instead to sample the full dynamical range of the spin labels for a given state. Therefore, the authors studied the different conformational states using independent simulations of each of the BetP structures, and ranked the likelihood of one structure over the other. However, local conformational changes can occur during such simulations. For example, the authors observe structural changes of 1–2 Å RMSD compared with the initial structure in their simulations, whereas the RMSD between the inward and outward conformations is 2–3 Å. Nevertheless, the majority of the biasing applied during the simulations is toward sampling the spin label configurations. This is something that has been observed in other systems (Marinelli and Faraldo-Gómez, 2015) and demonstrates that it is essential to consider that different structures and experimental conditions might translate into changes in the dynamics of the probes.
This study demonstrates a useful way of applying the EBMetaD enhanced sampling method to interpret potentially ambiguous dynamics data, using the BetP Na+/betaine cotransporter as an example. The methods are clearly mapped out to show how this type of analysis could be performed for other transporters that are be expected to undergo small, nondiscriminating structural changes. For BetP, it marks a first step along a new pathway of analysis. This could involve the analysis of other high-resolution structural states, such as the occluded state, that may correspond to the Na+/betaine experimental results. Furthermore, it is expected that this approach will become even more useful with additional PELDOR/DEER spectra mapping out protein dynamics at different positions within the protein. Altogether, this method has significant potential in relating structural states to dynamics data when no obvious interpretation is available from the static structures.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01GM120260 and R21GM126476).
José D. Faraldo-Gómez served as editor.
References
- Akyuz N., Georgieva E.R., Zhou Z., Stolzenberg S., Cuendet M.A., Khelashvili G., Altman R.B., Terry D.S., Freed J.H., Weinstein H., et al. 2015. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature. 518:68–73. 10.1038/nature14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Koshy C., Schweikhard E.S., Gärtner R.M., Perez C., Yildiz O., and Ziegler C.. 2013. Structural evidence for functional lipid interactions in the betaine transporter BetP. EMBO J. 32:3096–3105. 10.1038/emboj.2013.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer R., and Morbach S.. 2004. BetP of Corynebacterium glutamicum, a transporter with three different functions: betaine transport, osmosensing, and osmoregulation. Biochim. Biophys. Acta. 1658:31–36. 10.1016/j.bbabio.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Laio A., and Parrinello M.. 2002. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA. 99:12562–12566. 10.1073/pnas.202427399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V., Waclawska I., Kossmann K., Koshy C., Sharma M., Prisner T.F., Ziegler C., Endeward B., and Forrest L.R.. 2019. Interpretation of spectroscopic data using molecular simulations for the secondary active transporter. J. Gen. Physiol. 10.1085/jgp.201812111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Marinelli F., Lee C., Huang Y., Faraldo-Gómez J.D., and Jiang Y.. 2016. Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger. Nat. Struct. Mol. Biol. 23:590–599. 10.1038/nsmb.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli F., and Faraldo-Gómez J.D.. 2015. Ensemble-Biased Metadynamics: A Molecular Simulation Method to Sample Experimental Distributions. Biophys. J. 108:2779–2782. 10.1016/j.bpj.2015.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C., Faust B., Mehdipour A.R., Francesconi K.A., Forrest L.R., and Ziegler C.. 2014. Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na+ coupling. Nat. Commun. 5:4231 10.1038/ncomms5231 [DOI] [PMC free article] [PubMed] [Google Scholar]