Croft et al. demonstrate the use of recombinant adeno-associated viruses and organotypic brain slice cultures to study the central nervous system. This strategy can be used to model Alzheimer’s and Parkinson’s disease inclusion pathologies and determine mechanisms underlying neurodegeneration and therapeutic targets.
Abstract
It has been challenging to produce ex vivo models of the inclusion pathologies that are hallmark pathologies of many neurodegenerative diseases. Using three-dimensional mouse brain slice cultures (BSCs), we have developed a paradigm that rapidly and robustly recapitulates mature neurofibrillary inclusion and Lewy body formation found in Alzheimer’s and Parkinson’s disease, respectively. This was achieved by transducing the BSCs with recombinant adeno-associated viruses (rAAVs) that express α-synuclein or variants of tau. Notably, the tauopathy BSC model enables screening of small molecule therapeutics and tracking of neurodegeneration. More generally, the rAAV BSC “toolkit” enables efficient transduction and transgene expression from neurons, microglia, astrocytes, and oligodendrocytes, alone or in combination, with transgene expression lasting for many months. These rAAV-based BSC models provide a cost-effective and facile alternative to in vivo studies, and in the future can become a widely adopted methodology to explore physiological and pathological mechanisms related to brain function and dysfunction.
Introduction
Genetic, pathological, and experimental modeling data all provide strong evidence that numerous neurodegenerative diseases are proteinopathies triggered by the accumulation of proteins within the brain (Forman et al., 2004; Golde et al., 2013a). Although there is reasonable consensus that protein aggregation is tightly associated with neurodegeneration, there is limited understanding regarding (1) how protein aggregation impacts neurodegeneration, (2) what events trigger protein aggregation in the absence of mutations or overexpression, and (3) whether therapeutically targeting this aggregation leads to disease modification.
Decades of research into neurodegenerative proteinopathies using in vivo and in vitro models have linked mutations and overexpression of these aggregation-prone proteins to the development of inclusions (Forman et al., 2004; Rademakers et al., 2004; Golde et al., 2013a; Goedert et al., 2017). Despite this intensive body of work in the field, mechanistic insights and therapeutic development have been limited by a lack of facile in vitro models that fully recapitulate proteinopathies found in humans. Exciting observations and preclinical development have mostly been conducted in vivo in mammalian models. Specifically, in the case of tau pathology, such as that observed in Alzheimer’s disease (AD), robust neurofibrillary tangle (NFT) development and pathology are only observed in transgenic rodent models (Lewis et al., 2000; Allen et al., 2002; Buée et al., 2010; Noble et al., 2010). These models restrict throughput and are expensive to maintain and age. Phenotypic variability in transgenic tau mice has been reported (Woerman et al., 2017), with gender differences and other confounding variables often cited (Noble et al., 2010; Jankowsky and Zheng, 2017), thereby hindering both preclinical therapeutic studies and studies probing mechanisms regulating tau pathology and tau-induced neurodegeneration. Nonmammalian models have been useful in enabling behavioral screening and the study of tau phosphorylation, but no evidence of true tau inclusion pathology has been observed (Jackson et al., 2002; Kraemer et al., 2006; Brandt et al., 2009). Primary neuronal cultures or neuronally differentiated human induced pluripotent stem cell cultures have been used in efforts to create a reliable culture system to recapitulate inclusion pathology reflective of that observed in AD or Parkinson’s disease (PD; Choi et al., 2014; Sposito et al., 2015). However, none have reproducibly and robustly shown mature neurofibrillary pathologies resembling those in human tauopathies or Lewy body (LB) pathology reminiscent of those found in PD. Further, these systems are not comprised of all the central nervous system (CNS) cell types, which may play a role in disease (Choi et al., 2014; Sposito et al., 2015). Indeed, in AD, where a genetic role of microglia has emerged recently (Guerreiro et al., 2013; Tejera and Heneka, 2016; Sims et al., 2017), an accessible system that enables the study of all the neuronal and nonneuronal cell types and their interactions in an environment where anatomical planes of connectivity are maintained would be highly useful.
On this basis, we explored the feasibility of combining over a decade of experience in our laboratories optimizing CNS delivery of recombinant adeno-associated viruses (rAAVs) with a three-dimensional intact brain slice culture (BSC) system to see if we could develop more robust ex vivo models of AD and PD inclusion pathologies. These three-dimensional BSCs are functionally and physiologically relevant (Beach et al., 1982; Bahr, 1995; De Simoni et al., 2003), can be derived from brain areas involved in the human disease, and are comprised of neuronal and nonneuronal cell types. In addition, BSCs can often predict in vivo findings such as acute treatment of BSCs with small molecule compounds, recapitulating data obtained in in vivo studies (Croft et al., 2017a); other similarities and differences between in vivo models and BSCs are reviewed extensively by others (Sundstrom et al., 2005; Humpel, 2015). We find that rAAVs can be used to efficiently target neurons, astrocytes, microglia, and oligodendrocytes in this system and that expression is sustained long-term in culture. In addition, various brain cell types can be individually or simultaneously targeted in the same cultures, allowing the study of both cell autonomous and nonautonomous effects.
By using rAAVs to express unique combinations of disease-associated microtubule-associated protein tau (MAPT) mutations in cis (Strang et al., 2018), we show that BSCs develop an abundance of mature neurofibrillary inclusions (>1,000 inclusions per BSC) over a period of 7–28 d in vitro (DIV). These inclusions are Thioflavin S–positive, comprised of highly phosphorylated, sarkosyl-insoluble tau, and contain a multitude of tau filaments upon examination by electron microscopy (EM). BSCs transduced with mutant pro-aggregant tau show increased cell death at 60 DIV. We show that these tau-transduced BSCs can be used to screen for compounds that alter tau pathology, bypassing the need for blood–brain barrier penetrant compound, showing the potential of this system to streamline preclinical investigations of tau candidates and rapidly identify the most appropriate pathways to target. We also demonstrate robust inclusion pathology in BSCs transduced with WT and mutant human α-synuclein, providing a second example of a neurodegenerative proteinopathy that can be modeled in this system. Our data exemplify an efficient ex vivo platform to study disease mechanisms associated with pathogenic inclusion formation, as well as a platform to screen for compounds targeting protein aggregation and inclusion formation in relevant CNS cell types.
Results
rAAV vectors drive long-term and cell type–specific transgene expression in BSCs
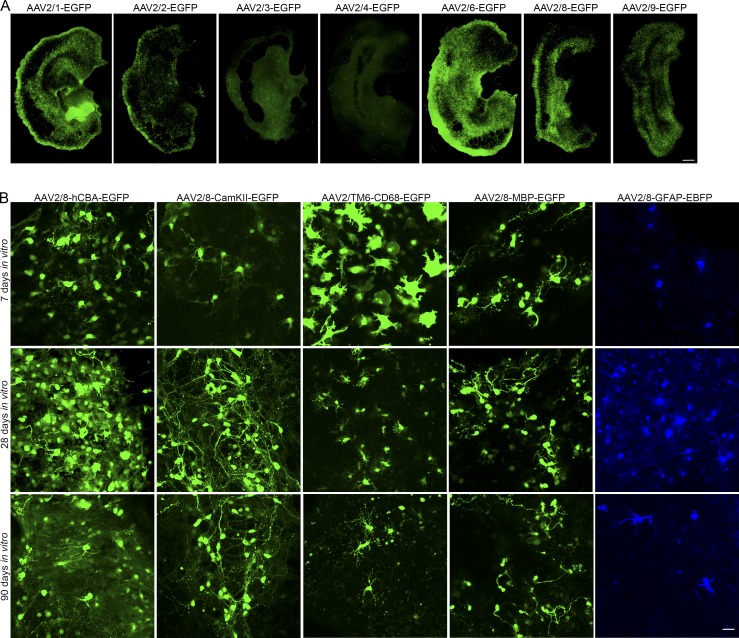
To identify appropriate capsid serotypes to transduce BSCs, we evaluated transduction of rAAV2 vectors expressing enhanced GFP (EGFP) pseudotyped with a number of rAAV capsids. BSCs were prepared and transduced at 0 DIV with rAAVs packaged in capsid serotype 1–9 expressing an EGFP transgene under control of the hybrid cytomegalovirus enhancer/chicken β-actin promoter (hCBA). BSCs were maintained in culture until 28 DIV, when slices were fixed and EGFP fluorescence was assessed. BSCs were efficiently transduced by rAAV2 vectors pseudotyped with capsids 1, 2, 6, 8, and 9, inefficiently transduced by capsids 3 and 4, and poorly transduced by capsids 5 and 7 (Fig. 1 A and Fig. S1).
Figure 1.
Neurons, astrocytes, oligodendrocytes, and microglia in BSCs can be targeted using different rAAV promoter-capsid combinations. (A) Organotypic BSCs were prepared and transduced at 0 DIV with rAAVs (1–2 × 1010 VGs per well) packaged in capsid serotype 2/1, 2/2, 2/3, 2/4, 2/6, 2/8, and 2/9 to express an EGFP transgene driven by the hCBA promoter. BSCs were maintained in culture until 28 DIV, and then slices were fixed, and fluorescence of EGFP throughout the BSC was imaged. Bar, 100 µm; n = 9 slices from n = 3 wells. (B) BSCs were prepared and transduced at 0 DIV with rAAVs (1–2 × 1010 VGs per well) with the hCBA promoter to express EGFP packaged in rAAV2/8, a CamKII promoter to express EGFP packaged in rAAV2/8, a CD68 promoter to express EGFP packaged in rAAV2/6 with three mutations—Y731F/Y705F/T492V (TM6)—an MBP promoter to express EGFP packaged in rAAV2/8, and a GFAP promoter to express EBFP packaged in rAAV2/8. BSCs were maintained in culture until 7, 28, and 90 DIV. Slices were then fixed, and fluorescence of EGFP/EBFP of CNS cells at these time points in the BSCs was imaged. Bar, 25 µm.
To extend these findings, we evaluated the effectiveness of targeting expression to specific brain cell populations using rAAV2 vectors containing neuronal and nonneuronal promoters pseudotyped with various capsids and expressing various fluorescent proteins (Fig. 1 B). rAAV2/8 with a hCBA promoter drove EGFP expression in neuronal and nonneuronal cells with rapid and robust expression observed from 7 to 90 DIV. rAAV2/6 with a CamKII promoter restricted expression to neurons, as predicted by in vivo neuronal targeting using the CamKII promoter with rAAVs (Lin et al., 2017). rAAV2/6 with three mutations—Y731F/Y705F/T492V (abbreviated as TM6)—and a cluster of differentiation 68 (CD68) promoter (Rosario et al., 2016) selectively targeted EGFP transgene expression to microglia (Fig. 1 B), with expression again detected from 7 to 90 DIV. To enable accurate quantification of the transduction of microglia, we also prepared BSCs from the CX3CR1-EGFP transgenic mouse line, which enabled visualization of EGFP in microglia (Jung et al., 2000) and transduced these with rAAV2/TM6-CD68-mCherry. We see 40% colocalization between mCherry and EGFP (Fig. S2 B). Counterstaining by immunohistochemistry with a marker for CD68 confirms colocalization between the EGFP and microglial staining (Fig. S2 E). Of particular note, at 7 DIV, microglia are in an amoeboid state, similar to other in vitro cultures of microglia (Stansley et al., 2012). This activated state is likely due to the initial trauma following slice generation; however, microglia appear to return to a resting state morphology beyond 7 DIV (Lossi et al., 2009). rAAV2/8 with a myelin basic protein (MBP) promoter drove EGFP transgene expression in cells of the oligodendrocyte lineage (Fig. 1 B); counterstaining with a marker for MBP confirms colocalization between the EGFP and an oligodendrocyte marker (Fig. S2 C). rAAV2/8 with a glial fibrillary acidic protein (GFAP) promoter drove expression of an enhanced blue fluorescent protein (EBFP) transgene to astrocytes in this culture system (Fig. 1 B), and counterstaining with a marker for GFAP confirms colocalization between the EBFP and the astrocyte marker (Fig. S2 D). Notably, there is no overlap in transgene expression when neuronal, astrocytic, and microglial promoters are used to drive fluorescent protein expression (Fig. S2 A; in this case, the MAP-2 promoter is used as an alternative to CamKII promoter for neuronal expression). These data show that neurons, microglia, oligodendrocytes, and astrocytes can be targeted by rAAVs in BSCs, with expression evident from 7 to ≥90 DIV. These findings are summarized in Table 1.
Table 1. Summary of targeting neurons, microglia, oligodendrocytes, and astrocytes using rAAVs in BSCs.
| BSC cell type | Promoter | Capsid |
|---|---|---|
| Non-specific | hCBA | 2/1, 2/2, 2/6, 2/8, 2/9 |
| Microglia | CD68 | 2/6 with three mutations, Y731F/Y705F/T492V (TM6) |
| Neurons | CamKII, MAP-2 | 2/6, 2/8 |
| Oligodendrocytes | MBP | 2/8 |
| Astrocytes | GFAP | 2/8 |
In some studies, we have shown that GFAP promoter–driven expression of BFP in astrocytes and CamKII promoter–driven expression of EGFP in neurons within the rAAV-transduced BSCs can last for at least 9 mo (Fig. S3).
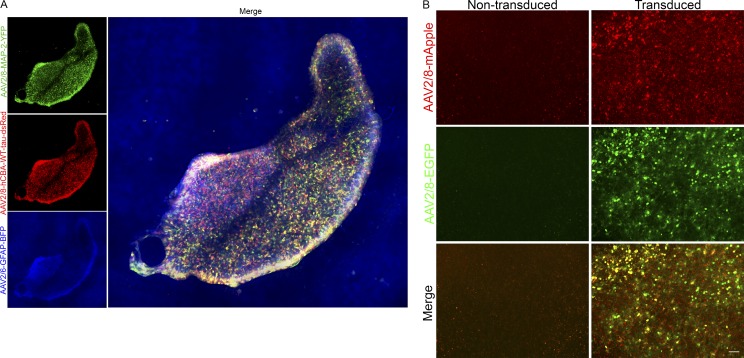
We also investigated whether we could deliver multiple rAAVs to the same BSCs. As shown in Fig. S2 A, rAAV2/TM6-CD68-mCherry, rAAV2/8-MAP-2-YFP, and rAAV2/8-GFAP-BFP applied to BSCs at 0 DIV and fluorescence imaged at 14 DIV show no overlap. This indicates that cell-autonomous and -nonautonomous effects can be studied using different promoter and capsid serotype combinations to deliver genes of interest to the same BSCs. To apply this to a context to understand the biology of human MAPT in these BSCs, we examined transduction of BSCs at 0 DIV with rAAV2/8-GFAP-BFP, rAAV2/8-MAP-2-YFP, and rAAV2/8-hCBA-WT-tau-dsRed and imaged them at 28 DIV (Fig. 2 A). These images confirm that cells expressing both dsRed and the cells expressing YFP and dsRed enable the study of human tau in astrocytes and neurons, respectively, in a single BSC. In addition, rAAV2/8-hCBA-EGFP and rAAV2/8-hCBA-mApple expressed in the same slice cultures show excellent colocalization (Fig. 2 B), enabling the study of two genes in the same cells in this system.
Figure 2.
Multiple CNS cell types can be targeted simultaneously in BSCs or cotransduced using rAAV vectors. (A) Organotypic BSCs were prepared and transduced at 0 DIV with three rAAVs (1–2 × 1010 VGs per well): a MAP-2 promoter to express YFP packaged in rAAV2/8 (green), a hCBA promoter to express human WT tau packaged in rAAV2/8 (red), and a GFAP promoter to express EBFP packaged in rAAV2/8 (blue), fixed at 28 DIV. Then fluorescence throughout the entire BSC was imaged. Bar, 200 µm. (B) Organotypic BSCs were prepared and transduced at 0 DIV with two rAAVs: a hCBA promoter to express EGFP packaged in rAAV2/8 (green), and a hCBA promoter to express mApple (red), fixed at 14 DIV. Then fluorescence throughout the entire BSC was imaged to identify colocalization. Bar, 50 µm.
Progressive and robust neurofibrillary inclusion pathology driven by rAAVs expressing MAPT in BSCs
Tau inclusions are a pathological hallmark of the tauopathies including AD, progressive supranuclear palsy, corticobasal degeneration, Pick’s disease, and frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17). The tauopathies are symptomatically and pathologically heterogeneous (Wang and Mandelkow, 2016; Guo et al., 2017). Tau inclusions are comprised of numerous forms of aggregated, post-translationally modified tau including highly phosphorylated, cleaved, and acetylated species (Guo et al., 2017). Previous BSC models prepared from tau transgenic mice have enabled insight into mechanisms of tau phosphorylation, kinase activation, and physiological and pathological tau release (Duff et al., 2002; Messing et al., 2013; Croft et al., 2017a,b), but robust neurofibrillary inclusions have not been reported.
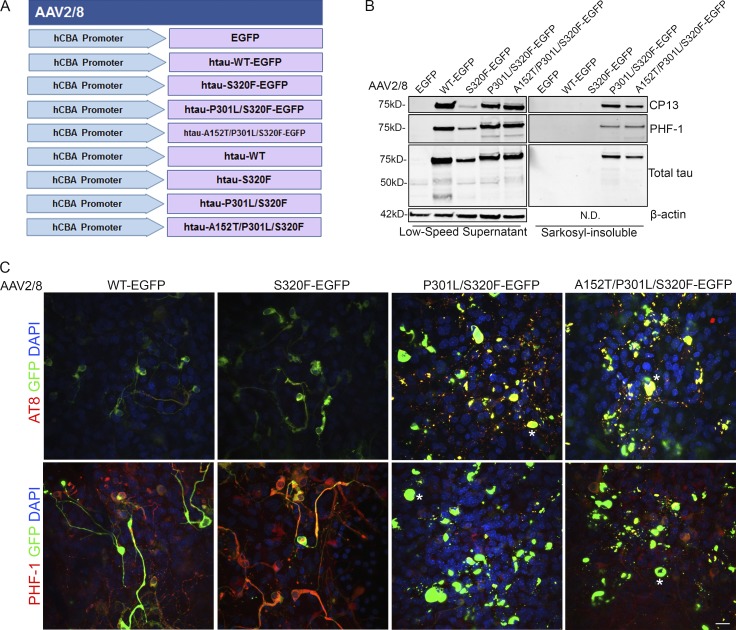
We recently identified a FTDP-17–associated mutation—S320F (Rosso et al., 2002)—that is prone to aggregate in an HEK293T cell assay (Strang et al., 2018). In combination with other disease-associated MAPT variants, one can generate increasingly more pro-aggregant constructs (Strang et al., 2018). We used rAAV to express these and other 0N4R MAPT constructs (Fig. 3 A) in BSCs in an attempt to elicit neurofibrillary pathology in this model. Initially, BSCs were transduced with rAAV2/8-EGFP, rAAV2/8-WT-htau-EGFP, rAAV2/8-S320F-htau-EGFP, rAAV2/8-P301L/S320F-htau-EGFP, or rAAV2/8-A152T/P301L/S320F-htau-EGFP fusion constructs. Low-speed supernatant (soluble) and sarkosyl-insoluble fractions were prepared (Li et al., 2015) from the BSC lysates and run on immunoblots. The low-speed supernatant fraction shows overexpression of total tau detected by the total tau antibody 3026 (Strang et al., 2017) and phosphorylated soluble tau at Ser202 (CP13) and Ser396/404 (PHF-1) induced by all htau-EGFP constructs (Fig. 3 B). Additionally, BSCs expressing tau with the unique combinations of MAPT mutations, P301L/S320F or A152T/P301L/A152T, developed an abundance of sarkosyl-insoluble, hyperphosphorylated tau (Fig. 3 B), a finding not observed in BSCs transduced with EGFP, WT, or S320F tau. Immunohistochemistry of tau phosphorylated at Ser202/Thr205 (AT8) and at Ser396/404 (PHF-1) was performed in order to assess the inclusion pathology (Fig. 3 C). BSCs from nontransgenic mice have previously been shown to develop some soluble phosphorylated tau in culture (Croft et al., 2017b), and this can also be observed in BSCs transduced with EGFP. Both WT and S320F tau cause the accumulation of phosphorylated tau, which is distributed throughout the axons and somatodendritic compartments of neurons. The P301L/S320F and P301L/S320F/A152T constructs, however, drive the accumulation of phosphorylated tau to the somatodendritic compartment, a finding resembling that described for tau inclusions in humans (Götz et al., 1995; Uchihara et al., 2001).
Figure 3.
rAAV tau-EGFP fusion protein transduced BSCs develop highly phosphorylated and sarkosyl-insoluble tau inclusions. (A) Schematic diagram of rAAV human mutant and WT tau constructs under a hCBA promoter used to transduce organotypic BSCs. Slice cultures were prepared and transduced with rAAV2/8-EGFP, rAAV2/8-WT-htau-EGFP, rAAV2/8-S320F-htau-EGFP, rAAV2/8-P301L/S320F-htau-EGFP, or rAAV2/8-A152T/P301L/S320F-htau-EGFP (1–2 × 1010 VGs per well) at 0 DIV and then maintained in culture until 28 DIV. (B) Transduced slices were sequentially extracted in order to prepare sarkosyl-insoluble tau fractions, and then lysates were immunoblotted for tau phosphorylated at Ser202 (CP13), tau phosphorylated at Ser396/404 (PHF-1), total tau (3026), and β-actin as a loading control. Representative Western blots of tau in the low speed supernatant and sarkosyl-insoluble fraction are shown. n = 9. (C) Transduced slice cultures were fixed, immunostained for tau phosphorylated at Ser396/Ser404 (PHF-1) and tau phosphorylated at Ser202/Thr205 (AT8), and confocal imaged to identify the location of phosphorylated tau in these sections. Asterisks mark examples of cells showing somatodendritic accumulation of phosphorylated tau. Bar, 25 µm. n = 3.
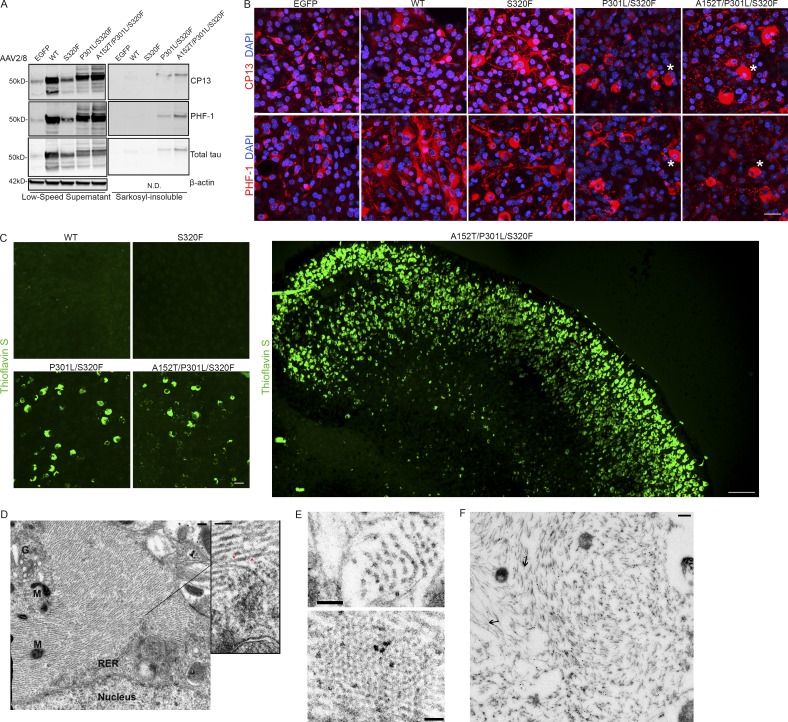
To further examine tauopathy formation in tau-transduced BSCs and the nature of inclusion pathology, untagged tau constructs, rAAV2/8-WT-htau, rAAV2/8-S320F-htau, rAAV2/8-P301L/S320F-htau, or rAAV2/8-A152T/P301L/S320F-htau (Fig. 3 A), were delivered to BSCs at 0 DIV, and BSCs were analyzed at 28 DIV. The low-speed supernatant fraction confirms overexpression of total tau (3026) and phosphorylated soluble tau at Ser202 (CP13) and Ser396/404 (PHF-1) in all BSCs transduced with untagged tau constructs (Fig. 4 A). BSCs expressing untagged P301L/S320F and A152T/P301L/S320F forms of tau accumulate appreciable amounts of sarkosyl-insoluble hyperphosphorylated tau (Fig. 4 A). The intraneuronal distribution of phosphorylated tau was examined using immunofluorescence. Small amounts of tau phosphorylated at Ser202 (CP13) and Ser396/404 (PHF-1) can be seen in the EGFP control (Fig. 4 B). Both WT and S320F tau-transduced BSCs show increased tau phosphorylated at Ser202 (CP13) and Ser396/404 (PHF-1) throughout the axons and somatodendritic compartments of neurons (Fig. 4 B). In contrast, P301L/S320F and P301L/S320F/A152T tau constructs cause the accumulation of phosphorylated tau within the somatodendritic compartment and form inclusion pathology resembling human tau inclusions (Fig. 4 B). These inclusions in BSCs expressing P301L/S320F and A152T/P301L/S320F tau were Thioflavin S–positive, similar to those seen in human AD (Ksiezak-Reding et al., 1987; Fig. 4 C), whereas Thioflavin S staining was absent from BSCs expressing WT or S320F tau at 28 DIV. Thioflavin S–positive inclusions in the A152T/P301L/S320F tau-transduced BSCs can be seen across the entire slice, with thousands of Thioflavin S–positive inclusions detectable (Fig. 4 C). rAAV-mediated transduction of tau containing two of the most commonly used FTDP-17 MAPT mutations to model disease—P301L and P301S (Hutton et al., 1998; Bugiani et al., 1999)—like S320F tau did not show Thioflavin S staining at 28 DIV (Fig. S4). To examine the ultrastructural nature of tau accumulation in A152T/P301L/S320F tau-transduced BSCs, we performed electron-microscopic ultrastructure analyses on BSCs that were transduced at 0 DIV and maintained until 28 DIV. The tau inclusions in these BSCs were comprised predominantly of a mass of orderly straight tau filaments, some with constrictions (Fig. 4, D and E) as in human AD (Grundke-Iqbal et al., 1986; Crowther, 1991; Murray et al., 2014) and were CP13-positive by Immuno-EM (Fig. 4 F). Based on this aggregate data, we conclude that the tau inclusions in these BSCs resemble mature neurofibrillary pathology.
Figure 4.
rAAV tau-transduced organotypic BSCs develop mature neurofibrillary inclusion pathology. Organotypic BSCs were prepared and transduced with rAAV2/8-EGFP, rAAV2/8-WT-htau, rAAV2/8-S320F-htau, rAAV2/8-P301L/S320F-htau, or rAAV2/8-A152T/P301L/S320F-htau under a hCBA promoter (1–2 × 1010 VGs per well) at 0 DIV and then maintained in culture until 28 DIV. (A) BSCs were sequentially extracted in order to prepare sarkosyl-insoluble tau fractions, and then lysates were immunoblotted for tau phosphorylated at Ser202 (CP13), tau phosphorylated at Ser396/404 (PHF-1), total tau (3026), and β-actin as a loading control. Representative Western blots of tau in the low-speed supernatant and sarkosyl-insoluble fraction are shown. n = 9. (B) Transduced slice cultures were fixed at 28 DIV, immunostained for CP13 and PHF-1, and imaged to identify the location of phosphorylated tau in these sections. Asterisks mark examples of cells showing somatodendritic accumulation of phosphorylated tau. Bar, 25 µm. n = 3. (C) Transduced slice cultures were fixed, stained with 0.0125% Thioflavin S, and confocal imaged to identify any β-sheet structures in these sections. Bar, 25 µm. n = 9. A large section spanning an entire BSC transduced with rAAV2/8-A152T/P301L/S320F-htau is also shown. Bar, 100 µm. (D) Transduced slices were fixed and examined by EM for the presence of filamentous inclusions. Nucleus, mitochondria (M), Golgi (G), and rough endoplasmic reticulum (RER) are marked. Enlarged section shows bundles of straight tau filaments with some constrictions (red arrows). Bars, 0.2 µm (left); 100 nm (right, enlargement). (E) Cross sections show numerous bundles of tau filaments. Bars, 50 nm (top); 100 nm (bottom). (F) Immunogold labeling with antibody to tau phosphorylated at Ser202 (CP13) confirms the presence of phosphorylated tau throughout the inclusions (arrows show examples of immunogold-labeled CP13 on tau filaments). Bar, 0.2 µm.
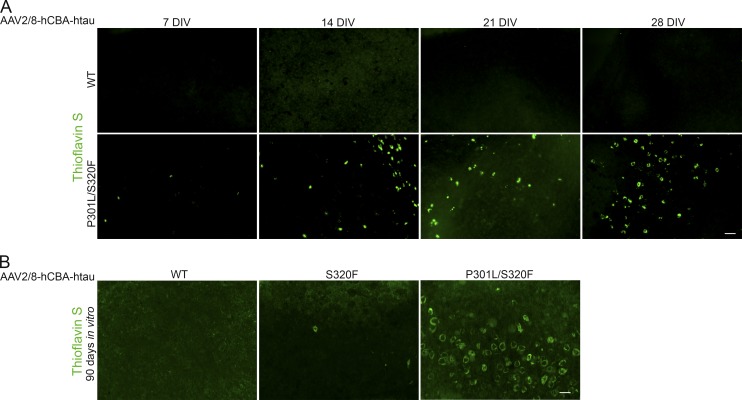
Initially, we had assessed tau pathology in tau-transduced BSCs at 28 DIV. To assess the timeline of pathology progression in P301L/S320F tau transduced BSCs, BSCs were transduced at 0 DIV and inclusion pathology assessed at 7, 14, 21, and 28 DIV (Fig. 5 A). A small number of Thioflavin S–positive cells are observed in P301L/S320F tau-transduced BSCs at 7 DIV, and there is a steady increase until 28 DIV. At 28 DIV for WT and S320F tau-transduced BSCs, neither sarkosyl-insoluble tau nor Thioflavin S–positive cells were detectable, despite evidence for high levels of tau expression and increased soluble phosphophorylated tau. However, at 90 DIV, rare Thioflavin S–positive inclusions can be observed in S320F tau-transduced BSCs (Fig. 5 B), suggesting a progressive development of tau inclusion pathology in this BSC model relating directly to the intrinsic aggregatablity of the expressed tau protein (Strang et al., 2018). We would speculate that expression of P301L or P301S may also drive rare inclusions upon extended culture periods but was not assessed here.
Figure 5.
Progressive development of NFTs from 7 to 28 DIV in rAAV tau-transduced BSCs. (A) Organotypic BSCs were prepared and transduced with rAAV2/8-WT-htau or rAAV2/8-P301L/S320F-htau (1–2 × 1010 VGs per well) at 0 DIV and then maintained in culture until 7, 14, 21, or 28 DIV. Transduced slice cultures were fixed, stained with 0.0125% Thioflavin S, and confocal imaged to identify any β-sheet structures in these sections. Bar, 100 µm. n = 3. (B) BSCs were prepared and transduced with rAAV2/8-WT-htau, rAAV2/8-S320F-htau, or rAAV2/8-P301L/S320F-htau at 0 DIV and then maintained in culture until 90 DIV. Transduced slice cultures were fixed, stained with 0.0125% Thioflavin S, and confocal imaged to identify any β-sheet structures in these sections. Bar, 50 µm. n = 3.
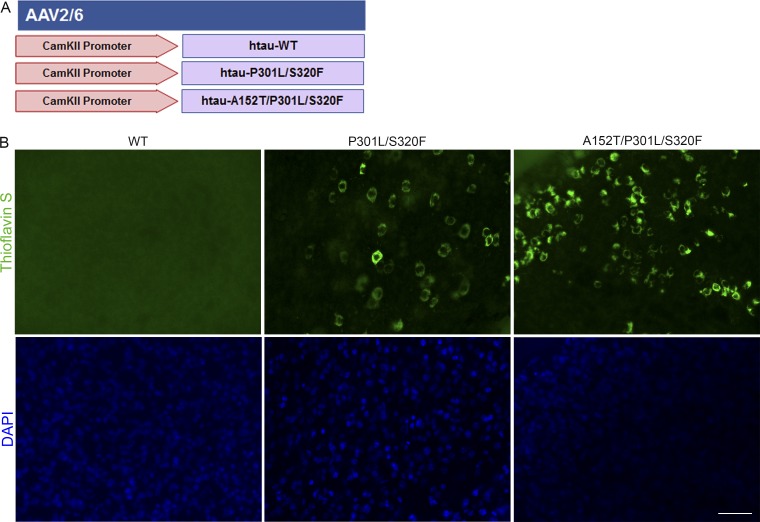
We also explored whether tau expression targeted to neurons would drive pathology in this model. BSCs were transduced with WT and mutant rAAV tau constructs driven by a CamKII promoter (Fig. 6 A). Numerous Thioflavin S–positive inclusions can be observed with CamKII driven expression of mutant tau in these BSCs (Fig. 6 B), albeit to a lesser extent than hCBA-driven mutant tau, allowing the observation and understanding of tau inclusions selectively driven to neurons in BSCs.
Figure 6.
Neuronal driven expression of tau to BSCs drives Thioflavin S–positive tau inclusions. (A) Schematic diagram of rAAV human mutant and WT tau constructs used to transduce organotypic BSCs to neurons only with a CamKII promoter. (B) Slice cultures were prepared and transduced with rAAV2/8-CamKII-WT-htau, rAAV2/8-CamKII-P301L/S320F-htau, or rAAV2/8-CamKII-A152T/P301L/S320F-htau (1–2 × 1010 VGs per well) at 0 DIV and then maintained in culture until 28 DIV. Transduced slice cultures were fixed, stained with 0.0125% Thioflavin S, and imaged to identify any β-sheet structures in these sections. DAPI is also shown to mark cell nuclei. Bar, 50 µm. n = 3.
Robust neurofibrillary pathology by transduction of MAPT drives neurodegeneration in BSCs
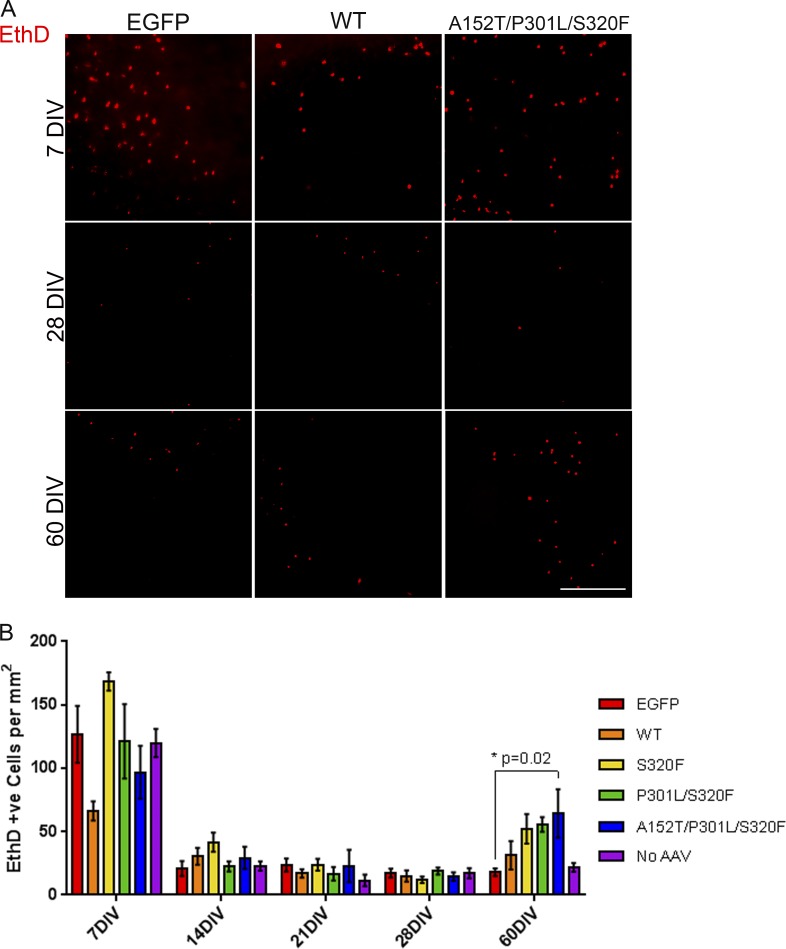
As burden of tau inclusion pathology correlates with neuronal loss in AD (Gómez-Isla et al., 1997; Augustinack et al., 2002), we examined any changes in cell death associated with tau transduction in BSCs. At 7, 14, 21, 28, and 60 DIV, slice cultures were incubated with ethidium homodimer-1 (EthD-1), and uptake by dead cells was imaged (Fig. 7 A) and quantified (Fig. 7 B). At 7 DIV, EthD-1 uptake was increased in transduced and nontransduced BSCs as previously reported (Lossi et al., 2009). This initial cell death reduced to a background level by 14 DIV, but by 60 DIV, cell death is significantly increased in BSCs transduced with A152T/P301L/S320F tau (P = 0.02) coincident with abundant tau inclusions. This suggests that at this time point, tau-induced neurodegeneration begins to be recapitulated in this model. In BSCs, the increased cell death is associated with an increased propensity of tau aggregation and with additive MAPT mutations. It is also important to note that at all of the time points in culture examined, there is no difference in cell death between the EGFP-transduced control and BSCs that have not been transduced.
Figure 7.
Increased cell death follows mature neurofibrillary inclusion formations in tau-transduced BSCs. (A) Representative images of EthD-1 uptake (red fluorescence) in BSCs transduced with rAAV2/8-EGFP, rAAV2/8-WT-htau, or rAAV2/8-A152T/P301L/S320F-htau (1–2 × 1010 VGs per well) and imaged at 7, 28, and 60 DIV. Bar, 400 µm. (B) Bar chart shows numbers of EthD-1–positive (red) cells per mm2 in rAAV2/8-EGFP, rAAV2/8-WT-htau, rAAV2/8-S320F-htau, rAAV2/8-P301L/S320F-htau, rAAV2/8-A152T/P301L/S320F-htau, or no rAAV-transduced BSCs at 7, 14, 21, 28, and 60 DIV. Data are mean ± SEM. *, P < 0.05. n = 6–12.
Compounds that reduce tau aggregation can be screened in tau-transduced BSCs
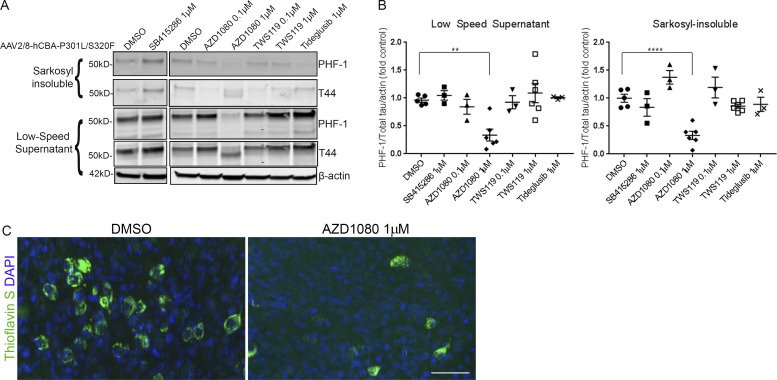
Tau is a well-established target in AD and other tauopathies (Golde et al., 2013b; Holtzman et al., 2016; Khanna et al., 2016). Tau therapeutic candidates have been chiefly screened in vivo in transgenic tau mouse models, but this is expensive and laborious, and requires large numbers of aged mice in order to assess effects on tau aggregation and burden. BSC models prepared from tau transgenic mice have previously been used to assess acute effects of compounds on tau phosphorylation (Croft et al., 2017a), but no chronic treatment studies have been reported. To assess whether compounds can be administered in a chronic manner in BSCs to modulate tau aggregation, four GSK-3β inhibitors (SB415286, AZD1080, TWS119, and Tideglusib) were applied at 0.1 µM or 1 µM from 14 to 28 DIV in P301L/S320F tau-transduced BSCs. GSK-3β phosphorylates tau and has been implicated as a therapeutic target in tauopathies (Hanger et al., 1992; Leroy et al., 2007; Terwel et al., 2008). At 28 DIV, one of these four GSK3β inhibitors, 1 µM AZD1080 (Georgievska et al., 2013), showed a significant reduction in PHF-1–positive phosphorylated soluble (P = 0.0031) and sarkosyl-insoluble (P = 0.0001) tau in BSCs (Fig. 8, A and B) and a reduction in Thioflavin S–positive inclusions (Fig. 8 C). These data highlight the utility of these tauopathy BSCs for cost-effective preclinical efficacy studies.
Figure 8.
Tauopathy BSCs can be used for testing tau-targeting therapeutics. (A) Representative Western blots of lysates from organotypic BSCs transduced at DIV 0 with rAAV2/8-P301L/S320F-htau (1–2 × 1010 VGs per well) and then treated from 14 to 28 DIV with 0.1 µM or 1 µM GSK-3β inhibitors. Half maximal inhibitory concentrations (IC50s) for GSK-3β for each compound are listed in parentheses as follows: SB415286 (78 nM), AZD1080 (324 nM), TWS119 (30 nM), and Tideglusib (60 nM). Blots were probed with antibodies against total tau (both nonphosphorylated and phosphorylated; 3026) and PHF-1 (phospho-Ser396/404). Blots were also probed with an antibody against β-actin as a loading control. (B) Bar charts show amounts of phospho-tau as a proportion of total tau in low speed supernatant and sarkosyl-insoluble samples after treatment with 0.1 µM or 1 µM GSK-3β inhibitors or DMSO (control). n = 3–6. **, P < 0.01; ****, P < 0.0001. Data are mean ± SEM and are shown as fold change from DMSO (control). (C) Transduced and treated slice cultures were fixed, stained with 0.0125% Thioflavin S, and imaged to identify any β-sheet structures in these sections. DAPI is also shown to mark cell nuclei. Bar, 50 µm. n = 6.
Robust LB pathology can also be driven by rAAVs expressing α-synuclein in BSCs
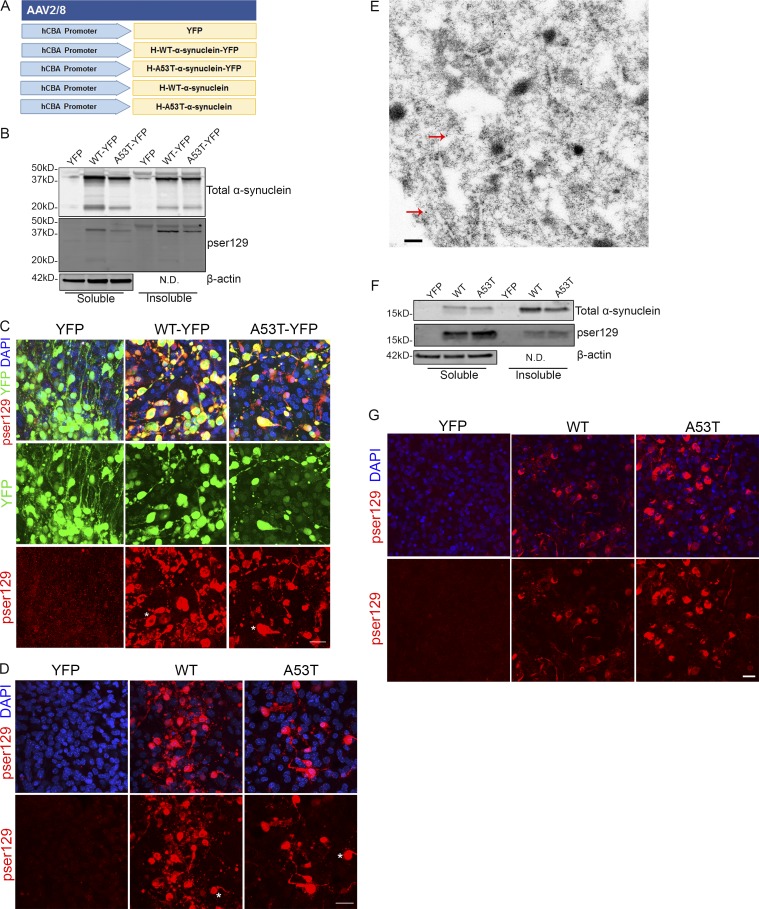
PD is the most common motor neurodegenerative disease (Poewe et al., 2017). PD is characterized by the presence of α-synuclein inclusions, which are found in PD and other α-synucleinopathies including LB dementia and multiple system atrophy (Kim et al., 2014; Dehay et al., 2015). These inclusions are comprised of α-synuclein phosphorylated at Ser129, and this is predominantly used as a marker of α-synuclein inclusions (Fujiwara et al., 2002; Anderson et al., 2006). To assess whether we could also drive α-synucleinopathy and the formation of LBs in BSCs, we developed both YFP-fused and untagged WT and A53T SNCA rAAV constructs (Fig. 9 A). BSCs were transduced with WT or A53T human α-synuclein tagged with YFP at 0 DIV, and then BSCs were lysed, and insoluble and soluble α-synuclein was extracted from the BSCs at 28 DIV, as previously described (Sacino et al., 2013). These fractions were then immunoblotted for total and pSer129 α-synuclein (Fig. 9 B). Insoluble and pSer129 α-synuclein accumulated in BSCs transduced with both WT and A53T α-synuclein. BSCs were also fixed at 28 DIV, and immunohistochemistry for pSer129 α-synuclein was performed to assess the distribution of α-synuclein in these transduced BSCs (Fig. 9 C). The α-synuclein inclusions in both WT- and A53T-transduced BSCs show LB-like inclusions with some Lewy neurites, recapitulating what is seen in human α-synucleinopathies (Kim et al., 2014). The presence of these pSer129 α-synuclein inclusions was also confirmed by immunohistochemistry in BSCs transduced with untagged WT and A53T α-synuclein (Fig. 9 D), and LBs and Lewy neurites like those found in human α-synucleinopathies (Spillantini et al., 1998) are observed in these BSCs. Furthermore, when examining rAAV2/8-WT–α-synuclein transduced BSCs by immunogold EM, labeling with an antibody to non-amyloid-β component of plaques (NACP)/α-synuclein confirms the presence of α-synuclein fibrils by 28 DIV (Fig. 9 E). In these experiments, α-synuclein inclusion pathology was induced using rAAVs from a microscale production of rAAV, avoiding costly purification of the virus (see Materials and methods). To confirm this was not an artifact of using secreted “microscale” rAAVs, BSCs were prepared and transduced with purified rAAVs to express YFP, WT human α-synuclein, or A53T human α-synuclein at 0 DIV and maintained until 28 DIV. BSCs were lysed, and insoluble and soluble α-synuclein was extracted from the BSCs at 28 DIV as performed for “microscale” rAAV-transduced BSCs. These fractions were then immunoblotted and probed for total and pSer129 α-synuclein (Fig. 9 F). Insoluble and pSer129 α-synuclein accumulated in BSCs transduced with purified rAAVs to express WT and A53T α-synuclein. BSCs were also fixed at 28 DIV, and immunohistochemistry for pSer129 α-synuclein was performed to assess the distribution of α-synuclein in these transduced BSCs (Fig. 9 G). BSCs transduced with WT or A53T purified or microscale rAAVs show LB-like inclusions and also highlight that the more cost-effective microscale rAAV production method can even further increase the accessibility of this system to model proteinopathies.
Figure 9.
rAAV α-synuclein-transduced organotypic BSCs develop insoluble α-synuclein inclusions reminiscent of LBs. (A) Schematic diagram of rAAV human mutant and WT α-synuclein constructs used to transduce organotypic BSCs. BSCs were prepared and transduced with rAAV2/8-YFP, rAAV2/8-h-WT-α-synuclein-YFP, rAAV2/8-h-A53T-α-synuclein-YFP, rAAV2/8-h-WT-α-synuclein, or rAAV2/8-h-A53T-α-synuclein prepared using a microscale method of rAAV preparation at 0 DIV and then maintained in culture until 28 DIV. (B) Transduced slices were sequentially extracted in order to prepare soluble and insoluble fractions, and then lysates were immunoblotted for total α-synuclein (SNL-4), phospho-Ser129 α-synuclein (81A), and β-actin as a loading control. Representative Western blots of insoluble and soluble fractions are shown. n = 9. (C and D) Transduced slice cultures were fixed, immunostained for phospho-Ser129 α-synuclein (EP1536Y), and confocal imaged to identify the location of phosphorylated α-synuclein in these sections. DAPI is also shown to mark cell nuclei. Asterisks mark examples of cells showing accumulation of phosphorylated α-synuclein and a LB-like appearance. Bar, 25 µm. n = 3. (E) rAAV2/8-h-WT-α-synuclein transduced slices were fixed and examined by immuno-EM for the presence of filamentous inclusions. Immunogold labeling with antibody to NACP98 confirms the presence of α-synuclein in inclusions (red arrows show examples of immunogold-labeled NACP98 on filaments). Bar, 0.2 µm. BSCs were also prepared and transduced with rAAV2/8-YFP, rAAV2/8-h-WT-α-synuclein, or rAAV2/8-h-A53T-α-synuclein prepared using the traditional purified method of rAAV preparation at 0 DIV and then maintained in culture until 28 DIV. (F) Transduced slices were sequentially extracted in order to prepare soluble and insoluble fractions, and then lysates were immunoblotted for total α-synuclein (SNL-4), phospho-Ser129 α-synuclein (81A), and β-actin as a loading control. Representative Western blots of insoluble and soluble fractions are shown. n = 3. (G) Transduced slice cultures were fixed, immunostained for phospho-Ser129 α-synuclein (EP1536Y), and confocal imaged to identify the location of phosphorylated α-synuclein in these sections. DAPI is also shown to mark cell nuclei. Bar, 20 µm. N.D., not done.
Discussion
Determining mechanisms underpinning proteinopathies including tauopathies and synucleinopathies and developing therapeutics for these diseases have posed an experimental challenge on the basis that ex vivo models recapitulating robust and bona fide inclusion pathology and any associated neurotoxicity do not exist. Here we have experimentally addressed this unmet need and developed models of both tauopathy and α-synucleinopathy. BSCs transduced with rAAV vectors encoding mutant tau progressively develop mature neurofibrillary tau pathology. Over a thousand tau inclusions can reproducibly develop in a single BSC, and the tau inclusion pathologies are characterized by highly phosphorylated, filamentous, sarkosyl-insoluble tau with a β-sheet structure; in other words, they are bona fide mature neurofibrillary tau inclusions by all standard measures (Hanger and Wray, 2010; Murray et al., 2011; Guo et al., 2017). BSCs transduced with a single MAPT mutation do develop sparse inclusions by 3 mo, thus providing a panel of tools to analyze varying degrees of tau pathology with differing temporal progression due to the intrinsic aggregatability of the encoded tau protein. Further, restricting tau expression to neurons using a CamKII promoter provides proof of concept that we can drive tau pathology in specific cell types. Coupled with data showing our ability to target expression using various promoter capsid combinations, such data portend the ability to probe how inclusion pathology alters cellular function in each of the major CNS cell types and also examine cell-autonomous and noncell autonomous mechanisms of action. Of note, by 3 mo in vitro, BSCs transduced with WT MAPT do not show inclusion pathology but do develop high levels of phosphorylated tau. As with in vivo models expressing WT human tau (Götz et al., 1995; Brion et al., 1999; Andorfer et al., 2005), development of inclusion pathology may require further time in culture in this system or may require other triggers to transition to its aggregated state, all of which remain open avenues to explore in this system.
Despite intensive study, a detailed mechanistic understanding of how neurofibrillary tau inclusion formation contributes to cellular dysfunction and cell death remains enigmatic, partly due to the absence of suitable models to study. In AD, the burden of tau inclusion pathology correlates with neuronal loss (Gómez-Isla et al., 1997; Augustinack et al., 2002). However, NFTs appear to exist in living neurons for decades in the human brain (Buée et al., 2010), and NFT-bearing neurons remain functional in mouse models for considerable periods of time (Kuchibhotla et al., 2014). In the BSC tauopathy model, we do not see increased cell death at the time during which massive neurofibrillary inclusions are developing. However, we note increased cell death 1 mo later that appears to be associated with the inherent aggregability of the transduced tau.
One of the challenges in the tau field is that a large number of mutations have been linked to human tauopathies, but only a much smaller number of these variants have been investigated with respect to their effects on tau pathology in relevant model systems. As highlighted by our recent study (Strang et al., 2018), it appears that these tau mutants have different rates of intrinsic aggregation as well as marked differences in their ability to aggregate following seeding. The rAAV-transduced BSCs will be an excellent tool to evaluate aggregate formation of additional tau variants in relevant CNS cell types as well as effects on cellular function. Although the simplistic explanation is that A152T in cis with other mutants makes a more pro-aggregant tau, we cannot firmly conclude that this is the reason that the A152T/P301L/S320F tau shows the most abundant pathology and toxicity. Expression of the A152T variant in model systems has previously been linked to a variety of effects in addition to increased NFT formation, including neurotoxicity, enhanced neuronal excitotoxicity, neuroinflammation, altered tau phosphorylation, and tau missorting (Decker et al., 2016; Maeda et al., 2016; Pir et al., 2016; Sydow et al., 2016).
Despite intense interest in tau as a therapeutic target for nearly two decades, only a small number of tau-targeting therapeutics have advanced to clinical trials, again due to the slow pipeline with an absence of suitable in vitro models that currently exist (Golde et al., 2013b; Golde, 2014). Few small molecules targeting tau remain in the clinical development pipeline, while the majority of clinical programs are now using active and passive immunotherapies to target tau. Our data evaluating select GSK-3β inhibitors show that BSCs can be used as a preclinical tool to gain confidence in both target engagement and mechanism of action of a tau-targeting therapeutic. Indeed, these studies yielded rather provocative results showing that only one of several GSK-3β inhibitors reduced tau aggregation in a model that recapitulates the three-dimensional architecture of the CNS. Effects on inclusion pathology can be monitored in the BSCs, and effects on tau-induced neurodegeneration could be evaluated. Although a large-scale screen of thousands of agents would not likely be practical in this system, unless highly automated, targeted screens of tens to hundreds of agents are certainly feasible.
Similarly, we extend our findings to show robust inclusion pathology in slices transduced with WT and mutant α-synuclein, providing a second example of a proteinopathy that can be modeled by using rAAVs and BSCs. By 28 DIV, BSCs transduced with WT and mutant α-synuclein develop inclusions that contain phosphorylated, insoluble α-synuclein which morphologically recapitulate LBs and show some Lewy neurites like those found in human α-synucleinopathies (Spillantini et al., 1998; Kim et al., 2014). Again, these provide a rapid in vitro model to study α-synucleinopathies and mechanistically understand or therapeutically target this inclusion pathology. Of importance, this LB pathology was elicited in BSCs transduced from microscale production of the rAAV vectors. More generally, microscale production enables rAAV vectors to be rapidly and cost-effectively prepared at scales suitable for transduction of multiple BSCs at minimal cost, and also enables efficient generation of multiple vectors without the need for a large-scale standard vector preparation. Notably, many proteins linked to CNS proteinopathies fit within the packing limits of rAAV vectors, and in many cases, rAAV-mediated expression and subsequent inclusion pathology in the brain has been documented (Verbeeck et al., 2012; Ceballos-Diaz et al., 2015). Thus, it is highly likely that rAAV-mediated expression of proteins linked to other neurodegenerative disorders within BSCs can be developed.
The versatility of rAAV-mediated transgene expression in the BSC system enables targeting of select CNS cell types, either individually or at the same time, and highlights its applicability not only for study of neurodegenerative diseases but also for the study of the physiology of the CNS and other brain diseases. Various rAAV capsid serotypes can be used to effectively transduce BSCs, and these capsids in combination with several neuronal and nonneuronal cell type promoters enable the selective targeting of the major CNS cell types in slice cultures. In all cases, expression is maintained for ≥3 mo in culture, and we have noted neuronal expression even at 9 mo in vitro. Of importance, we also observe that microglia in this system show morphologies of quiescent in vivo–like adult microglia after 1 mo in vitro, enabling the study of microglia in physiology and disease in this system.
Overall, this work highlights this rAAV and BSC “toolkit” as a platform to study neurodegenerative disease mechanisms associated with pathogenic inclusion formation in a rapid, scalable manner. Given the sustained expression and flexibility of this system with the major CNS cell types in an anatomically connected milieu, their roles in physiology and pathology can be studied. This toolkit can also be leveraged to screen for compounds targeting protein aggregation and inclusion formation while remaining time-, resource-, and cost-effective.
Materials and methods
rAAV preparation
rAAV 2/1, 2/2, 2/3, 2/4, 2/5, 2/6, 2/7, 2/8, and 2/9 expressing EGFP; rAAV2/1 expressing 0N4R human tau, htau-WT, htau-S320F, htau-P301L, htau-P301S, and htau-P301L/S320F; and rAAV2/8 expressing htau-WT, htau-S320F, htau-P301L/S320F, htau-A152T/P301L/S320F, htau-WT-EGFP, htau-EGFP, htau-P301L/S320F-EGFP, htau-A152T/P301L/S320F-EGFP, YFP, WT-hα-synuclein, and A53T-hα-synuclein, under the control of the hCBA promoter, were generated as described previously (Chakrabarty et al., 2010; Ceballos-Diaz et al., 2015; Rosario et al., 2016). rAAV2/6 with three mutations—Y731F/Y705F/T492V (abbreviated as TM6; Rosario et al., 2016)—expressing EGFP or mCherry driven by a CD68 promoter, rAAV2/6 expressing EGFP, htau-WT, htau-P301L/S320F, or htau-A152T/P301L/S320F driven by a CamKII promoter (Lin et al., 2017) or YFP driven by a MAP-2 promoter, rAAV2/8 expressing EGFP driven by an MBP promoter, or EBFP driven by a GFAP promoter were also generated as previously described (Chakrabarty et al., 2010; Ceballos-Diaz et al., 2015; Rosario et al., 2016). rAAVs were applied to BSCs by adding rAAVs into the culture medium on the first day of culture (0 DIV) at 1–2 × 1010 viral genomes (VGs) per well containing three BSCs.
Microscale preparation of rAAV
rAAV 2/8 expressing YFP, WT-hα-synuclein, A53T-hα-synuclein, WT-hα-synuclein-YFP, and A53T-hα-synuclein-YFP under the control of the hCBA promoter was prepared as microscale rAAVs. HEK293T cells (ATCC) were maintained as recommended in DMEM (Lonza Group) with 10% (vol/vol) FBS (Thermo Fisher Scientific), 2 mM l-glutamine, and 1% (vol/vol) penicillin/streptomycin (Thermo Fisher Scientific). HEK293T cells were plated in 2 ml media in 6-well plates 24 h before transfection of transgene plasmid with rAAV2/8 (pDP8.ape; Plasmid Factory GmbH & Co. KG) using Polyethylenimine Linear (Polysciences) at 60–80% confluency. The microscale virus was harvested 72 h after transfection by centrifuging the media at 500 g for 5 min and collecting the supernatant. Media containing secreted rAAVs were then applied to BSCs by rAAVs into the culture medium on the first day of culture (0 DIV) at 1–2 × 1010 VGs per well containing three BSCs.
Organotypic BSC
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Florida. Organotypic BSCs were prepared from postnatal day 8–9 B6/C3H mice (Envigo) or CX3CR1-GFP transgenic mice (Jung et al., 2000; 005582; The Jackson Laboratory) as previously described (Croft et al., 2017a). In brief, pups were cryoanesthetized and decapitated, and the brains removed and dissected to retain the cortex, hippocampus, and connecting regions in each hemi-brain in sterile filtered ice-cold dissection buffer (HBSS, calcium, magnesium, no phenol red [Thermo Fisher Scientific], 2 mM ascorbic acid [Sigma-Aldrich], 39.4 µM ATP [Sigma-Aldrich], and 1% (vol/vol) penicillin/streptomycin [Thermo Fisher Scientific]). The hemi-brain was placed on filter paper, and 350-µm coronal slices were cut using a McIllwain tissue chopper (Mickle Laboratory Engineering Co. Ltd.). Slices were collected and plated randomly to contain three slices per semi-porous membrane insert (0.4 µm pore diameter; Millipore; Thermo Fisher Scientific) in 6-well sterile culture plates. Slices from different parts of the brain are therefore distributed randomly across wells of each plate. Slices were maintained at 37°C and 5% CO2 in sterile-filtered culture medium (basal medium eagle [Thermo Fisher Scientific], 26.6 mM Hepes [pH 7.1; Thermo Fisher Scientific], 511 µM ascorbic acid, 1% [vol/vol] GlutaMAX [Thermo Fisher Scientific], 0.033% [vol/vol] insulin [Sigma-Aldrich], 1% [vol/vol] penicillin/streptomycin [Thermo Fisher Scientific], and 25% [vol/vol] heat-inactivated horse serum [Sigma-Aldrich]). Culture medium was changed every 3–4 d. rAAVs were applied directly into the culture medium on the first day of culture (0 DIV) at 1–2 × 1010 genome particles of rAAV per well containing three BSCs.
Treatment of organotypic slice cultures
Slice cultures were treated with GSK-3β inhibitors from the DiscoveryProbe Kinase Inhibitor Library (ApeXBio) reconstituted in DMSO (Sigma-Aldrich). GSK-3β inhibitors were added at 14, 18, 21, and 25 DIV during media changes to a final concentration of 0.1 or 1 µM of GSK-3β inhibitors in a final concentration of 0.1% DMSO in the media. 0.1% DMSO was used as the control.
Sarkosyl extraction of BSC lysates
Slice cultures for assessment of insoluble tau from three wells (nine slices in total) were harvested into ice-cold PBS and presented as n = 1. Slices were washed in PBS and then pelleted, and sarkosyl extractions were performed as previously described (Li et al., 2015). BSCs were homogenized in Tris-buffered saline (TBS; 50 mM Tris, 274 mM NaCl, and 5 mM KCl, pH 8) containing a protease/phosphatase cocktail (Roche) and centrifuged at 27,000 g for 20 min at 4°C. The supernatant (S1) was kept and run on immunoblots as the low-speed supernatant. The pellet (P1) was homogenized in Tris-sucrose buffer (10 mM Tris, pH 7.4, 0.8 M NaCl, and 10% sucrose) and similarly centrifuged. The supernatant (S2) was incubated in 1% sarkosyl for 60 min at 37°C and was centrifuged at 150,000 g for 30 min at 4°C. The pellet (P3) was resuspended in Tris-EDTA buffer (10 mM Tris, pH 8, and 1 mM EDTA) and run on immunoblots as the sarkosyl-insoluble fraction.
Triton X-100 extraction of BSC lysates
Slice cultures for assessment of insoluble α-synuclein from three wells (nine slices in total) were harvested into ice-cold PBS and presented as n = 1. Slice cultures were washed and pelleted, and extraction was performed as previously described for primary neuroglia (Sacino et al., 2013). Slices were lysed in 1% Triton X-100 TBS (50 mM Tris and 150 mM NaCl, pH 7.4) with protease and phosphatase inhibitors. Lysates were then centrifuged at 100,000 g for 20 min at 4°C. Supernatants were removed (Triton-soluble fraction), and the pellet was washed with the TBS buffer and centrifuged at 100,000 g for 20 min at 4°C. The pellet was then resuspended in 2% SDS, sonicated, and heated to 100°C for 10 min (Triton-insoluble fraction). Protein concentrations were determined by bicinchoninic acid assay (Thermo Fisher Scientific) before equal amounts of protein were resolved by immunoblotting.
Antibodies
The following antibodies were used for immunoblotting and/or immunohistochemistry. A rabbit polyclonal antibody 3026 raised against full-length recombinant 0N3R human tau (Strang et al., 2017) and AT8 (Thermo Fisher Scientific) mouse monoclonal antibody to tau phosphorylated at Ser202 and Thr 205 (Goedert et al., 1995) were used. The following mouse monoclonal tau antibodies were provided by Peter Davies (Feinstein Institute for Medical Research, Manhasset, NY): CP13 (tau phosphorylated at Ser202) and PHF-1 (tau phosphorylated at Ser396 and Ser404; Jicha et al., 1997). SNL-4, a rabbit polyclonal antibody raised against amino acid sequence 2–12 of human α-synuclein (Giasson et al., 2000); 81A, a mouse monoclonal antibody specific for α-synuclein phosphorylated at Ser129; and a rabbit monoclonal antibody specific for α-synuclein phosphorylated at Ser129 (EP1536Y; Abcam) were used. Rabbit polyclonal antibodies to GSK-3β phosphorylated at Ser9 (Cell Signaling Technology) and total GSK-3β (Cell Signaling Technology), mouse monoclonal antibodies to β-actin (Sigma-Aldrich), MAP-2 (Millipore), and GFAP (Cell Signaling Technology) and rat monoclonal antibodies to MBP (Bio-Rad Laboratories) and to CD68 (BioLegend) were also used.
SDS-PAGE and immunoblotting
5–20 µg protein was separated on 4–12% or 10% (wt/vol) SDS-PAGE gels (Bio-Rad) and electrophoretically transferred to polyvinylidene difluoride membranes, as described previously (Li et al., 2015). Membranes were blocked in 0.5% casein for 1 h and then incubated with primary antibodies overnight at 4°C, washed three times with TBS before incubation with fluorophore-conjugated Alexa Fluor 680 anti-mouse IgG (Thermo Fisher Scientific) or IRDye 800 goat anti-rabbit IgG (Li-Cor Biosciences) secondary antibodies, and washed three times with TBS. Protein bands were detected and quantified using the multiplex Li-Cor Odyssey Infrared Imaging system (Li-Cor Biosciences).
Immunohistochemistry
Organotypic BSCs were washed in PBS and then fixed on their inserts in 4% paraformaldehyde for 1 h and stained according to Gogolla et al. (2006). Individual slice cultures (n = 1) were cut out from their membranes after fixation and then treated as free-floating sections for the following steps. Slice cultures were permeabilized for 18 h in 0.5% Triton X-100 at 4°C and then blocked in 20% BSA (Sigma-Aldrich) for 4 h at room temperature (RT). Slice cultures were then incubated with appropriate primary antibodies overnight at 4°C in 5% BSA, washed, and then incubated with fluorophore-coupled secondary antibodies for 4 h at RT. Slice cultures were washed a final time before mounting on slides with Fluoromount-G with DAPI (Southern Biotech) and then imaged.
Thioflavin S staining
Organotypic BSCs were washed in PBS, fixed on their inserts with 4% paraformaldehyde for 1 h, and then stained with 0.0125% Thioflavin S as described in Strang et al. (2018). In brief, a stock solution of 1% Thioflavin S (Sigma-Aldrich) in 50% EtOH in PBS was filtered through a 0.2-µm filter. Individual slices (n = 1) were cut from their membranes and autofluorescence reagent (Millipore) was applied for 5 min, and then slices were washed in 40% ethanol (EtOH). BSCs were incubated with 0.0125% Thioflavin S in 50% EtOH in PBS for 3 min in the dark, followed by 50% EtOH and PBS washes. BSCs were then mounted on slides with Fluoromount-G with DAPI (Southern Biotech) and then imaged to identify any amyloidogenic β-sheet structures in these sections.
Imaging of slice cultures
Images of slice cultures were captured using a Nikon A1RMPsi-STORM4.0 multiphoton/super resolution imaging system (Nikon Instruments) on the laser-scanning confocal mode or using a Keyence BZ-X700 all-in-one fluorescence microscope (Keyence Corp. of America) using the optical sectioning mode. Z-stacks were captured over 20 µm at recommended step-sizes and projected onto a full focus image using the BZ-analyzer or Nikon Elements software.
EthD-1 dead cell uptake assay
Cytotoxicity was assessed by measuring EthD-1 uptake. EthD-1 was prepared according to the manufacturer’s instructions (LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells; Thermo Fisher Scientific). Culture medium was aspirated from slice cultures, and wells were washed in prewarmed PBS. Slice cultures were then incubated with EthD-1 in PBS for 15 min and then imaged. Three images of each slice were captured blindly from predetermined regions using a 10× objective lens, positive cells were counted using the analyze particles feature on ImageJ (Version 1.51k; National Institutes of Health), and the mean number of positive cells calculated from the three images captured.
EM
BSCs were fixed on the culture insert with 2.5% glutaraldehyde-2% paraformaldehyde in PBS at 4°C overnight and washed in PBS. EM was performed as previously described (Lin et al., 2003). Slices were scraped into PBS and centrifuged for 1 min at 16,000 g to form tissue pellets and were transferred to glass vials and fixed in aqueous osmium tetroxide (2%) for 1 h at RT, followed by 2% uranyl acetate in 50% EtOH for 30 min at RT. Tissues were then dehydrated in a series of EtOHs and propylene oxide, infiltrated and embedded in epon 812 (Polysciences). Thin sections were stained with uranyl acetate and lead citrate, examined with a Philips 208s electron microscope fitted with a Gatan 831 Orius digital camera. Digital images were processed with Adobe Photoshop software.
Post-embedding immunogold EM
BSCs were fixed in 4% paraformaldehyde in PBS at 4°C overnight, and collected into pellets as for regular EM. Pellets were transferred to glass vials dehydrated in 50, 70, 80, and 90% EtOH for 10 min at RT, infiltrated, and embedded in LR White resin (medium grade; Polysciences). They were polymerized in a vacuum oven at 50°C for 2 d. Thin sections were collected on Formvar-coated grids and were incubated with antibody CP13 or NACP-98 (Klein et al., 2005) overnight at 4°C, followed by goat anti-mouse IgG conjugated with 18 nm colloidal gold particles (Jackson ImmunoResearch Laboratories) for 30 min at RT. The sections were stained and examined as described for Regular EM.
Statistical analysis
Data were analyzed using either Student’s two-tailed unpaired t test or one-way analysis of variance followed by Dunnett’s post-hoc tests (Graphpad Prism 7.0 Software). Differences were considered statistically significant when P < 0.05. For slice cultures used for preparation of insoluble fractions and immunoblotting, three wells containing three slice cultures each (nine total) were pooled together, and n refers to each group of pooled slices. For immunohistochemistry and Thioflavin S staining, n refers to each individual slice. For EthD-1 uptake assays, n refers to each individual slice, which was imaged three times and positive cell numbers averaged across the three images.
Online supplemental material
Fig. S1 shows the inefficient transduction by rAAV2/5 and 2/7. Fig. S2 shows transduction of multiple CNS cell types in the same BSC and counterstaining of cell markers to show cell-specific transduction in BSCs. Fig. S3 shows long-term expression of rAAVs in neurons and astrocytes. Fig. S4 shows an absence of Thioflavin S inclusions by 28 DIV with transduction of WT, S320F, P301L, or P301S tau.
Supplementary Material
Acknowledgments
All work performed in this manuscript was in accordance with the Institutional Animal Care and Use Committee at the University of Florida and with the approval of local research ethics committees. We thank Peter Davies (Feinstein Institute for Medical Research, Manhasset, NY) for providing mouse monoclonal tau antibodies CP13 and PHF-1.
This work was supported by funding from the Stop Alzheimer’s Now and Marjorie Thomas Fund (to T.E. Golde), the National Institutes of Health (U01AG046139, R01AG018454, and P50AG047266 to T.E. Golde), the Florida Department of Health (7AZ25 to B.I. Giasson), and the BrightFocus Foundation (A2018149F to C.L. Croft). Access to the Nikon A1RMPsi-STORM4.0 multiphoton/super resolution imaging system was provided by a National Institutes of Health Shared Instrumentation Grant (1S10OD020026).
The authors declare no competing financial interests.
Author contributions: Study concept and design: C.L. Croft, P. Chakrabarty, Y. Levites, B.I. Giasson, and T.E. Golde. Acquisition of data: C.L. Croft, P.E. Cruz, C. Ceballos-Diaz, K.H. Strang, D.H. Ryu, B.M. Woody, W.-L. Lin, and M. Deture. Statistical analysis: C.L. Croft and T.E. Golde. Analysis and interpretation of the data: C.L. Croft, D.W. Dickson, P. Chakrabarty, Y. Levites, B.I. Giasson, and T.E. Golde. Drafting of the manuscript: C.L. Croft, E. Rodríguez-Lebrón, P. Chakrabarty, Y. Levites, B.I. Giasson, and T.E. Golde. All authors read and approved the final manuscript.
References
- Allen B., Ingram E., Takao M., Smith M.J., Jakes R., Virdee K., Yoshida H., Holzer M., Craxton M., Emson P.C., et al. 2002. Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J. Neurosci. 22:9340–9351. 10.1523/JNEUROSCI.22-21-09340.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.P., Walker D.E., Goldstein J.M., de Laat R., Banducci K., Caccavello R.J., Barbour R., Huang J., Kling K., Lee M., et al. 2006. Phosphorylation of Ser-129 is the dominant pathological modification of α-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 281:29739–29752. 10.1074/jbc.M600933200 [DOI] [PubMed] [Google Scholar]
- Andorfer C., Acker C.M., Kress Y., Hof P.R., Duff K., and Davies P.. 2005. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human tau isoforms. J. Neurosci. 25:5446–5454. 10.1523/JNEUROSCI.4637-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinack J.C., Schneider A., Mandelkow E.M., and Hyman B.T.. 2002. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol. 103:26–35. 10.1007/s004010100423 [DOI] [PubMed] [Google Scholar]
- Bahr B.A. 1995. Long-term hippocampal slices: a model system for investigating synaptic mechanisms and pathologic processes. J. Neurosci. Res. 42:294–305. 10.1002/jnr.490420303 [DOI] [PubMed] [Google Scholar]
- Beach R.L., Bathgate S.L., and Cotman C.W.. 1982. Identification of cell types in rat hippocampal slices maintained in organotypic cultures. Brain Res. 255:3–20. 10.1016/0165-3806(82)90071-2 [DOI] [PubMed] [Google Scholar]
- Brandt R., Gergou A., Wacker I., Fath T., and Hutter H.. 2009. A Caenorhabditis elegans model of tau hyperphosphorylation: induction of developmental defects by transgenic overexpression of Alzheimer’s disease-like modified tau. Neurobiol. Aging. 30:22–33. 10.1016/j.neurobiolaging.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Brion J.-P., Tremp G., and Octave J.-N.. 1999. Transgenic expression of the shortest human tau affects its compartmentalization and its phosphorylation as in the pretangle stage of Alzheimer’s disease. Am. J. Pathol. 154:255–270. 10.1016/S0002-9440(10)65272-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buée L., Troquier L., Burnouf S., Belarbi K., Van der Jeugd A., Ahmed T., Fernandez-Gomez F., Caillierez R., Grosjean M.E., Begard S., et al. 2010. From tau phosphorylation to tau aggregation: what about neuronal death? Biochem. Soc. Trans. 38:967–972. 10.1042/BST0380967 [DOI] [PubMed] [Google Scholar]
- Bugiani O., Murrell J.R., Giaccone G., Hasegawa M., Ghigo G., Tabaton M., Morbin M., Primavera A., Carella F., Solaro C., et al. 1999. Frontotemporal dementia and corticobasal degeneration in a family with a P301S mutation in tau. J. Neuropathol. Exp. Neurol. 58:667–677. 10.1097/00005072-199906000-00011 [DOI] [PubMed] [Google Scholar]
- Ceballos-Diaz C., Rosario A.M., Park H.-J., Chakrabarty P., Sacino A., Cruz P.E., Siemienski Z., Lara N., Moran C., Ravelo N., et al. 2015. Viral expression of ALS-linked ubiquilin-2 mutants causes inclusion pathology and behavioral deficits in mice. Mol. Neurodegener. 10:25 10.1186/s13024-015-0026-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P., Ceballos-Diaz C., Beccard A., Dickson D., Golde T.E., and Das P.. 2010. IFN-gamma promotes complement expression and attenuates amyloid plaque deposition in APP transgenic mice. J. Immunol. 184:5333–5343. 10.4049/jimmunol.0903382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D’Avanzo C., Chen H., Hooli B., Asselin C., Muffat J., et al. 2014. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 515:274–278. 10.1038/nature13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft C.L., Kurbatskaya K., Hanger D.P., and Noble W.. 2017a Inhibition of glycogen synthase kinase-3 by BTA-EG4 reduces tau abnormalities in an organotypic brain slice culture model of Alzheimer’s disease. Sci. Rep. 7:7434 10.1038/s41598-017-07906-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft C.L., Wade M.A., Kurbatskaya K., Mastrandreas P., Hughes M.M., Phillips E.C., Pooler A.M., Perkinton M.S., Hanger D.P., and Noble W.. 2017b Membrane association and release of wild-type and pathological tau from organotypic brain slice cultures. Cell Death Dis. 8:e2671 10.1038/cddis.2017.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R.A. 1991. Straight and paired helical filaments in Alzheimer disease have a common structural unit. Proc. Natl. Acad. Sci. USA. 88:2288–2292. 10.1073/pnas.88.6.2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker J.M., Krüger L., Sydow A., Dennissen F.J.A., Siskova Z., Mandelkow E., and Mandelkow E.M.. 2016. The Tau/A152T mutation, a risk factor for frontotemporal-spectrum disorders, leads to NR2B receptor-mediated excitotoxicity. EMBO Rep. 17:552–569. 10.15252/embr.201541439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B., Bourdenx M., Gorry P., Przedborski S., Vila M., Hunot S., Singleton A., Olanow C.W., Merchant K.M., Bezard E., et al. 2015. Targeting α-synuclein for treatment of Parkinson’s disease: mechanistic and therapeutic considerations. Lancet Neurol. 14:855–866. 10.1016/S1474-4422(15)00006-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simoni A., Griesinger C.B., and Edwards F.A.. 2003. Development of rat CA1 neurones in acute versus organotypic slices: role of experience in synaptic morphology and activity. J. Physiol. 550:135–147. 10.1113/jphysiol.2003.039099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff K., Noble W., Gaynor K., and Matsuoka Y.. 2002. Organotypic slice cultures from transgenic mice as disease model systems. J. Mol. Neurosci. 19:317–320. 10.1385/JMN:19:3:317 [DOI] [PubMed] [Google Scholar]
- Forman M.S., Trojanowski J.Q., and Lee V.M.Y.. 2004. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 10:1055–1063. 10.1038/nm1113 [DOI] [PubMed] [Google Scholar]
- Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M.S., Shen J., Takio K., and Iwatsubo T.. 2002. α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4:160–164. 10.1038/ncb748 [DOI] [PubMed] [Google Scholar]
- Georgievska B., Sandin J., Doherty J., Mörtberg A., Neelissen J., Andersson A., Gruber S., Nilsson Y., Schött P., Arvidsson P.I., et al. 2013. AZD1080, a novel GSK3 inhibitor, rescues synaptic plasticity deficits in rodent brain and exhibits peripheral target engagement in humans. J. Neurochem. 125:446–456. 10.1111/jnc.12203 [DOI] [PubMed] [Google Scholar]
- Giasson B.I., Jakes R., Goedert M., Duda J.E., Leight S., Trojanowski J.Q., and Lee V.M.. 2000. A panel of epitope-specific antibodies detects protein domains distributed throughout human alpha-synuclein in Lewy bodies of Parkinson’s disease. J. Neurosci. Res. 59:528–533. [DOI] [PubMed] [Google Scholar]
- Goedert M., Jakes R., and Vanmechelen E.. 1995. Monoclonal antibody AT8 recognises tau protein phosphorylated at both serine 202 and threonine 205. Neurosci. Lett. 189:167–169. 10.1016/0304-3940(95)11484-E [DOI] [PubMed] [Google Scholar]
- Goedert M., Jakes R., and Spillantini M.G.. 2017. The Synucleinopathies: Twenty Years On. J. Parkinsons Dis. 7(s1):S51–S69. 10.3233/JPD-179005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolla N., Galimberti I., DePaola V., and Caroni P.. 2006. Staining protocol for organotypic hippocampal slice cultures. Nat. Protoc. 1:2452–2456. 10.1038/nprot.2006.180 [DOI] [PubMed] [Google Scholar]
- Golde T.E. 2014. Open questions for Alzheimer’s disease immunotherapy. Alzheimers Res. Ther. 6:3 10.1186/alzrt233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde T.E., Borchelt D.R., Giasson B.I., and Lewis J.. 2013a Thinking laterally about neurodegenerative proteinopathies. J. Clin. Invest. 123:1847–1855. 10.1172/JCI66029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde T.E., Lewis J., and McFarland N.R.. 2013b Anti-tau antibodies: hitting the target. Neuron. 80:254–256. 10.1016/j.neuron.2013.10.009 [DOI] [PubMed] [Google Scholar]
- Gómez-Isla T., Hollister R., West H., Mui S., Growdon J.H., Petersen R.C., Parisi J.E., and Hyman B.T.. 1997. Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer’s disease. Ann. Neurol. 41:17–24. 10.1002/ana.410410106 [DOI] [PubMed] [Google Scholar]
- Götz J., Probst A., Spillantini M.G., Schäfer T., Jakes R., Bürki K., and Goedert M.. 1995. Somatodendritic localization and hyperphosphorylation of tau protein in transgenic mice expressing the longest human brain tau isoform. EMBO J. 14:1304–1313. 10.1002/j.1460-2075.1995.tb07116.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y.C., Zaidi M.S., and Wisniewski H.M.. 1986. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 261:6084–6089. [PubMed] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S.K., Younkin S., et al. Alzheimer Genetic Analysis Group . 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Noble W., and Hanger D.P.. 2017. Roles of tau protein in health and disease. Acta Neuropathol. 133:665–704. 10.1007/s00401-017-1707-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanger D.P., and Wray S.. 2010. Tau cleavage and tau aggregation in neurodegenerative disease. Biochem. Soc. Trans. 38:1016–1020. 10.1042/BST0381016 [DOI] [PubMed] [Google Scholar]
- Hanger D.P., Hughes K., Woodgett J.R., Brion J.-P., and Anderton B.H.. 1992. Glycogen synthase kinase-3 induces Alzheimer’s disease-like phosphorylation of tau: generation of paired helical filament epitopes and neuronal localisation of the kinase. Neurosci. Lett. 147:58–62. 10.1016/0304-3940(92)90774-2 [DOI] [PubMed] [Google Scholar]
- Holtzman D.M., Carrillo M.C., Hendrix J.A., Bain L.J., Catafau A.M., Gault L.M., Goedert M., Mandelkow E., Mandelkow E.-M., Miller D.S., et al. 2016. Tau: From research to clinical development. Alzheimers Dement. 12:1033–1039. 10.1016/j.jalz.2016.03.018 [DOI] [PubMed] [Google Scholar]
- Humpel C. 2015. Organotypic brain slice cultures: A review. Neuroscience. 305:86–98. 10.1016/j.neuroscience.2015.07.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M., Lendon C.L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., et al. 1998. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 393:702–705. 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- Jackson G.R., Wiedau-Pazos M., Sang T.-K., Wagle N., Brown C.A., Massachi S., and Geschwind D.H.. 2002. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 34:509–519. 10.1016/S0896-6273(02)00706-7 [DOI] [PubMed] [Google Scholar]
- Jankowsky J.L., and Zheng H.. 2017. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol. Neurodegener. 12:89 10.1186/s13024-017-0231-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jicha G.A., Lane E., Vincent I., Otvos L. Jr., Hoffmann R., and Davies P.. 1997. A conformation- and phosphorylation-dependent antibody recognizing the paired helical filaments of Alzheimer’s disease. J. Neurochem. 69:2087–2095. 10.1046/j.1471-4159.1997.69052087.x [DOI] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M.R., Kovalevich J., Lee V.M.Y., Trojanowski J.Q., and Brunden K.R.. 2016. Therapeutic strategies for the treatment of tauopathies: Hopes and challenges. Alzheimers Dement. 12:1051–1065. 10.1016/j.jalz.2016.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.S., Kågedal K., and Halliday G.M.. 2014. Alpha-synuclein biology in Lewy body diseases. Alzheimers Res. Ther. 6:73 10.1186/s13195-014-0073-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.L., Dayton R.D., Lin W.-L., and Dickson D.W.. 2005. Tau gene transfer, but not alpha-synuclein, induces both progressive dopamine neuron degeneration and rotational behavior in the rat. Neurobiol. Dis. 20:64–73. 10.1016/j.nbd.2005.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B.C., Burgess J.K., Chen J.H., Thomas J.H., and Schellenberg G.D.. 2006. Molecular pathways that influence human tau-induced pathology in Caenorhabditis elegans. Hum. Mol. Genet. 15:1483–1496. 10.1093/hmg/ddl067 [DOI] [PubMed] [Google Scholar]
- Ksiezak-Reding H., Dickson D.W., Davies P., and Yen S.H.. 1987. Recognition of tau epitopes by anti-neurofilament antibodies that bind to Alzheimer neurofibrillary tangles. Proc. Natl. Acad. Sci. USA. 84:3410–3414. 10.1073/pnas.84.10.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchibhotla K.V., Wegmann S., Kopeikina K.J., Hawkes J., Rudinskiy N., Andermann M.L., Spires-Jones T.L., Bacskai B.J., and Hyman B.T.. 2014. Neurofibrillary tangle-bearing neurons are functionally integrated in cortical circuits in vivo. Proc. Natl. Acad. Sci. USA. 111:510–514. 10.1073/pnas.1318807111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy K., Yilmaz Z., and Brion J.P.. 2007. Increased level of active GSK-3β in Alzheimer’s disease and accumulation in argyrophilic grains and in neurones at different stages of neurofibrillary degeneration. Neuropathol. Appl. Neurobiol. 33:43–55. 10.1111/j.1365-2990.2006.00795.x [DOI] [PubMed] [Google Scholar]
- Lewis J., McGowan E., Rockwood J., Melrose H., Nacharaju P., Van Slegtenhorst M., Gwinn-Hardy K., Paul Murphy M., Baker M., Yu X., et al. 2000. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 25:402–405. 10.1038/78078 [DOI] [PubMed] [Google Scholar]
- Li A., Ceballos-Diaz C., DiNunno N., Levites Y., Cruz P.E., Lewis J., Golde T.E., and Chakrabarty P.. 2015. IFN-γ promotes τ phosphorylation without affecting mature tangles. FASEB J. 29:4384–4398. 10.1096/fj.15-275834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.-L., Lewis J., Yen S.-H., Hutton M., and Dickson D.W.. 2003. Ultrastructural neuronal pathology in transgenic mice expressing mutant (P301L) human tau. J. Neurocytol. 32:1091–1105. 10.1023/B:NEUR.0000021904.61387.95 [DOI] [PubMed] [Google Scholar]
- Lin Y.-T., Chen C.-C., Huang C.-C., Nishimori K., and Hsu K.-S.. 2017. Oxytocin stimulates hippocampal neurogenesis via oxytocin receptor expressed in CA3 pyramidal neurons. Nat. Commun. 8:537 10.1038/s41467-017-00675-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi L., Alasia S., Salio C., and Merighi A.. 2009. Cell death and proliferation in acute slices and organotypic cultures of mammalian CNS. Prog. Neurobiol. 88:221–245. 10.1016/j.pneurobio.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Maeda S., Djukic B., Taneja P., Yu G.Q., Lo I., Davis A., Craft R., Guo W., Wang X., Kim D., et al. 2016. Expression of A152T human tau causes age-dependent neuronal dysfunction and loss in transgenic mice. EMBO Rep. 17:530–551. 10.15252/embr.201541438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing L., Decker J.M., Joseph M., Mandelkow E., and Mandelkow E.-M.. 2013. Cascade of tau toxicity in inducible hippocampal brain slices and prevention by aggregation inhibitors. Neurobiol. Aging. 34:1343–1354. 10.1016/j.neurobiolaging.2012.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.E., Graff-Radford N.R., Ross O.A., Petersen R.C., Duara R., and Dickson D.W.. 2011. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. Lancet Neurol. 10:785–796. 10.1016/S1474-4422(11)70156-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.E., Kouri N., Lin W.-L., Jack C.R. Jr., Dickson D.W., and Vemuri P.. 2014. Clinicopathologic assessment and imaging of tauopathies in neurodegenerative dementias. Alzheimers Res. Ther. 6:1 10.1186/alzrt231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble W., Hanger D.P., and Gallo J.M.. 2010. Transgenic mouse models of tauopathy in drug discovery. CNS Neurol. Disord. Drug Targets. 9:403–428. 10.2174/187152710791556131 [DOI] [PubMed] [Google Scholar]
- Pir G.J., Choudhary B., Mandelkow E., and Mandelkow E.-M.. 2016. Tau mutant A152T, a risk factor for FTD/PSP, induces neuronal dysfunction and reduced lifespan independently of aggregation in a C. elegans Tauopathy model. Mol. Neurodegener. 11:33 10.1186/s13024-016-0096-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poewe W., Seppi K., Tanner C.M., Halliday G.M., Brundin P., Volkmann J., Schrag A.-E., and Lang A.E.. 2017. Parkinson disease. Nat. Rev. Dis. Primers. 3:17013 10.1038/nrdp.2017.13 [DOI] [PubMed] [Google Scholar]
- Rademakers R., Cruts M., and van Broeckhoven C.. 2004. The role of tau (MAPT) in frontotemporal dementia and related tauopathies. Hum. Mutat. 24:277–295. 10.1002/humu.20086 [DOI] [PubMed] [Google Scholar]
- Rosario A.M., Cruz P.E., Ceballos-Diaz C., Strickland M.R., Siemienski Z., Pardo M., Schob K.-L., Li A., Aslanidi G.V., Srivastava A., et al. 2016. Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol. Ther. Methods Clin. Dev. 3:16026 10.1038/mtm.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso S.M., van Herpen E., Deelen W., Kamphorst W., Severijnen L.-A., Willemsen R., Ravid R., Niermeijer M.F., Dooijes D., Smith M.J., et al. 2002. A novel tau mutation, S320F, causes a tauopathy with inclusions similar to those in Pick’s disease. Ann. Neurol. 51:373–376. 10.1002/ana.10140 [DOI] [PubMed] [Google Scholar]
- Sacino A.N., Thomas M.A., Ceballos-Diaz C., Cruz P.E., Rosario A.M., Lewis J., Giasson B.I., and Golde T.E.. 2013. Conformational templating of α-synuclein aggregates in neuronal-glial cultures. Mol. Neurodegener. 8:17 10.1186/1750-1326-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R., van der Lee S.J., Naj A.C., Bellenguez C., Badarinarayan N., Jakobsdottir J., Kunkle B.W., Boland A., Raybould R., Bis J.C., et al. GERAD/PERADES, CHARGE, ADGC, EADI . 2017. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49:1373–1384. 10.1038/ng.3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini M.G., Crowther R.A., Jakes R., Hasegawa M., and Goedert M.. 1998. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA. 95:6469–6473. 10.1073/pnas.95.11.6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposito T., Preza E., Mahoney C.J., Setó-Salvia N., Ryan N.S., Morris H.R., Arber C., Devine M.J., Houlden H., Warner T.T., et al. 2015. Developmental regulation of tau splicing is disrupted in stem cell-derived neurons from frontotemporal dementia patients with the 10 + 16 splice-site mutation in MAPT. Hum. Mol. Genet. 24:5260–5269. 10.1093/hmg/ddv246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansley B., Post J., and Hensley K.. 2012. A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J. Neuroinflammation. 9:115 10.1186/1742-2094-9-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang K.H., Goodwin M.S., Riffe C., Moore B.D., Chakrabarty P., Levites Y., Golde T.E., and Giasson B.I.. 2017. Generation and characterization of new monoclonal antibodies targeting the PHF1 and AT8 epitopes on human tau. Acta Neuropathol. Commun. 5:58 10.1186/s40478-017-0458-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang K.H., Croft C.L., Sorrentino Z.A., Chakrabarty P., Golde T.E., and Giasson B.I.. 2018. Distinct differences in prion-like seeding and aggregation between Tau protein variants provide mechanistic insights into tauopathies. J. Biol. Chem. 293:2408–2421. 10.1074/jbc.M117.815357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom L., Morrison B. III, Bradley M., and Pringle A.. 2005. Organotypic cultures as tools for functional screening in the CNS. Drug Discov. Today. 10:993–1000. 10.1016/S1359-6446(05)03502-6 [DOI] [PubMed] [Google Scholar]
- Sydow A., Hochgräfe K., Könen S., Cadinu D., Matenia D., Petrova O., Joseph M., Dennissen F.J., and Mandelkow E.-M.. 2016. Age-dependent neuroinflammation and cognitive decline in a novel Ala152Thr-Tau transgenic mouse model of PSP and AD. Acta Neuropathol. Commun. 4:17 10.1186/s40478-016-0281-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera D., and Heneka M.T.. 2016. Microglia in Alzheimer’s disease: the good, the bad and the ugly. Curr. Alzheimer Res. 13:370–380. 10.2174/1567205013666151116125012 [DOI] [PubMed] [Google Scholar]
- Terwel D., Muyllaert D., Dewachter I., Borghgraef P., Croes S., Devijver H., and Van Leuven F.. 2008. Amyloid activates GSK-3β to aggravate neuronal tauopathy in bigenic mice. Am. J. Pathol. 172:786–798. 10.2353/ajpath.2008.070904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihara T., Nakamura A., Yamazaki M., and Mori O.. 2001. Evolution from pretangle neurons to neurofibrillary tangles monitored by thiazin red combined with Gallyas method and double immunofluorescence. Acta Neuropathol. 101:535–539. [DOI] [PubMed] [Google Scholar]
- Verbeeck C., Deng Q., Dejesus-Hernandez M., Taylor G., Ceballos-Diaz C., Kocerha J., Golde T., Das P., Rademakers R., Dickson D.W., and Kukar T.. 2012. Expression of Fused in sarcoma mutations in mice recapitulates the neuropathology of FUS proteinopathies and provides insight into disease pathogenesis. Mol. Neurodegener. 7:53 10.1186/1750-1326-7-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., and Mandelkow E.. 2016. Tau in physiology and pathology. Nat. Rev. Neurosci. 17:5–21. 10.1038/nrn.2015.1 [DOI] [PubMed] [Google Scholar]
- Woerman A.L., Patel S., Kazmi S.A., Oehler A., Freyman Y., Espiritu L., Cotter R., Castaneda J.A., Olson S.H., and Prusiner S.B.. 2017. Kinetics of human mutant tau prion formation in the brains of 2 transgenic mouse lines. JAMA Neurol. 74:1464–1472. 10.1001/jamaneurol.2017.2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.