Rowe and colleagues show that hematopoietic cells with MLL translocations undergoing leukemogenesis in neonatal mice yield mixed-lineage early B-lymphoid/myeloid leukemia while identical cells in adults generate myeloid leukemia, demonstrating that the age of the hematopoietic microenvironment impacts lineage.
Abstract
Leukemia phenotypes vary with age of onset. Delineating mechanisms of age specificity in leukemia could improve disease models and uncover new therapeutic approaches. Here, we used heterochronic transplantation of leukemia driven by MLL/KMT2A translocations to investigate the contribution of the age of the hematopoietic microenvironment to age-specific leukemia phenotypes. When driven by MLL-AF9, leukemia cells in the adult microenvironment sustained a myeloid phenotype, whereas the neonatal microenvironment supported genesis of mixed early B cell/myeloid leukemia. In MLL-ENL leukemia, the neonatal microenvironment potentiated B-lymphoid differentiation compared with the adult. Ccl5 elaborated from adult marrow stroma inhibited B-lymphoid differentiation of leukemia cells, illuminating a mechanism of age-specific lineage commitment. Our study illustrates the contribution of the developmental stage of the hematopoietic microenvironment in defining the age specificity of leukemia.
Introduction
The clinical and pathological features of leukemia, as well as its response to therapy, vary markedly with the age of onset. Among acute leukemias, B-cell acute lymphoblastic leukemia (B-ALL) is most prevalent in children, while acute myeloid leukemia (AML) prevails in older adults. B-ALL of infancy, occurring at <1 yr of age, is a unique entity. Infant B-ALL often shows biphenotypic or mixed-lineage B-lymphoid/myeloid differentiation and is frequently triggered by chromosomal translocations involving the MLL gene (Pieters et al., 2007). Compared with B-ALL of later childhood, infant B-ALL is associated with poor outcome and requires more intensive treatment with a higher risk of short- and long-term toxicities (Pieters et al., 2007). Despite these striking age-dependent leukemia phenotypes, the mechanisms by which age impacts the pathobiology of leukemia are largely uninvestigated.
Given the potency of MLL translocations in transforming normal hematopoietic stem and progenitor cells (HSPCs), many mouse models of MLL-driven leukemia exist (Chen et al., 2006; Krivtsov et al., 2008; Barabé et al., 2017; Milne, 2017). However, to date, these models have not fully recapitulated all aspects of human infant B-ALL. Although the MLL-AF9 translocation causes AML or B-ALL in humans, in mice, it almost invariably drives AML when introduced into mouse HSPCs (Meyer et al., 2013; Milne, 2017). However, in human cells, the lineage fate of MLL-AF9–transformed human CD34+ HSPCs is sensitive to experimental manipulation of the cytokine milieu when xenografted into adult mice (Barabé et al., 2007; Wei et al., 2008; Lin et al., 2016). Despite these observations, the significance of the endogenous neonatal hematopoietic microenvironment, particularly a very young microenvironment, to defining leukemia phenotypes during leukemogenesis in infants has not been explored.
Prenatally, the mammalian hematopoietic system follows a strict schedule of maturation whereby the predominant site of hematopoiesis relocates from the liver to the bone marrow (BM) during late gestation. Later, during postnatal aging, HSPCs shift from lymphoid toward myeloid lineage bias in both humans and mice (Rebel et al., 1996; Pang et al., 2011), which interestingly parallels the shift in predominance from B-ALL to AML that occurs during maturation from infancy to adulthood in humans. In line with this, previous studies comparing leukemogenesis in HSPCs from murine midgestation fetal liver (FL) to those from adult BM have suggested that the developmental age of the leukemic cells of origin may affect disease latency, immunophenotype, and molecular dependencies (Chen et al., 2011; Man et al., 2016). The developmental maturation of the hematopoietic system could therefore have relevance to the B-lymphoid to myeloid lineage switching that can occur in human infant leukemia at relapse (Rossi et al., 2012). As such, we hypothesized that the normal developmental maturation of the hematopoietic microenvironment might underpin the age specificity of leukemia pathobiology. Therefore, we aimed to define the relative contributions of both the age of the leukemic cell of origin and the hematopoietic microenvironment in specification of lineage bias in leukemia. We find that the neonatal hematopoietic microenvironment supported B-lymphoid differentiation of MLL-rearranged HSPCs during leukemogenesis, whereas identical cells generated nearly pure myeloid leukemia in adults. Aiming to define age-specific microenvironmental factors regulating leukemia lineage phenotypes, we implicate Ccl5 derived from the adult BM stroma in promoting myeloid differentiation of leukemia. These studies uncover a role for the hematopoietic microenvironment in specifying the lineage of MLL-rearranged leukemia and provide insight into the discordance between adult mouse models of MLL-rearranged leukemia and human infant disease driven by the same translocations.
Results and discussion
We first sought to experimentally evaluate the impact of the cell of origin on leukemic phenotypes. To evaluate cells as a function of age, we isolated Lineage– c-kit+ Sca-1+ CD127– (127-LSK) HSPCs from either murine E14.5 FL or 6–8-wk-old young adult BM by FACS (Fig. S1 A), transduced these cells with a retrovirus encoding the MLL-AF9 oncogene, and engrafted these cells into congenic sublethally irradiated 8-wk-old adult recipients. We initially chose the MLL-AF9 translocation because this has been reported to invariably induce myeloid leukemia in mice but which can also cause B-ALL in humans (Meyer et al., 2013; Milne, 2017), and so we aimed to elicit B-lymphoid differentiation in this mouse model using heterochronic transplantation without transgenic manipulation of the microenvironment. We found that leukemia from either cell source manifested as myelomonocytic AML with identical latency and leukemia-initiating cell (LIC) content as measured by in vivo limiting dilution secondary transplantation (Fig. S1, B–H).
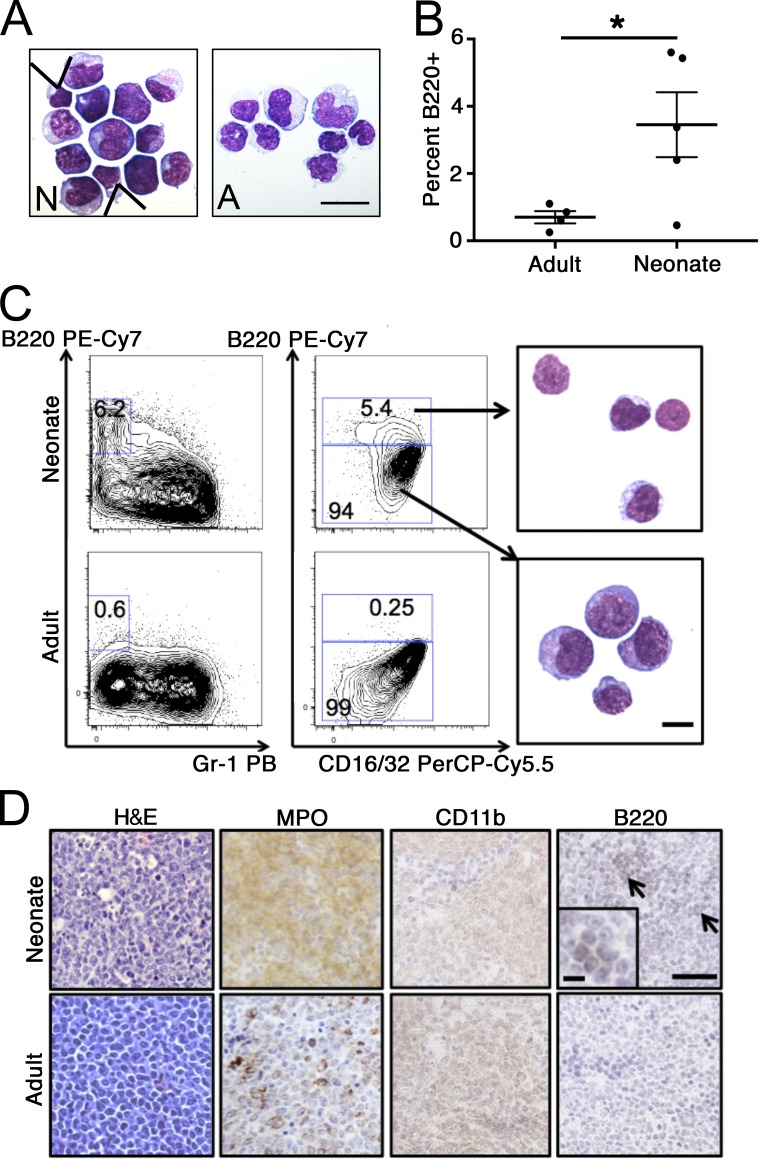
We next asked if the developmental stage of the microenvironment impacts leukemia differentiation. We transplanted MLL-AF9–transduced adult BM 127-LSK cells into congenic adult (8-wk-old) or neonatal (postnatal [P] day 0–1) recipients and performed necropsy when mice became moribund. Leukemia onset occurred between 68 and 109 d in adults (mean, 79 d; n = 7) and between 76 and 101 d in neonatal recipients (mean, 86 d; n = 9; P = 0.2 by Student’s t test compared with adults). Morphological analysis revealed the expected myelomonocytic AML in adult recipients (Fig. 1 A). However, leukemia in neonatal recipients contained a small population of agranular cells that appeared to have undergone lymphoid differentiation, interspersed with myelomonocytic cells (Fig. 1 A). Flow cytometry analysis of neonatal MLL-AF9 leukemia identified a small proportion of cells expressing the B-cell marker B220/CD45R in some leukemias, with coexpression of the myeloid progenitor marker CD16/32 (Fig. 1, B and C). Purified B220+ leukemic cells were morphologically small, with scant cytoplasm, while B220− cells appeared myelomonocytic (Fig. 1 C). At necropsy, neonatal recipients showed effacement of splenic architecture due to infiltration by leukemia-expressing myeloperoxidase, CD11b, as well as focal B220 staining, which was not present in adult tissue (Fig. 1 D). These results suggested that transformation of HSPCs by MLL-AF9 in the neonatal microenvironment elicits leukemic B-lymphoid differentiation in a proportion of leukemia cells.
Figure 1.
Leukemogenesis in adults and neonates. (A) Representative morphology of leukemic BM of mice engrafted with MLL-AF9–transformed 127-LSKs transplanted at P0 (N) or 8 wk of age (scale bar, 20 µm; arrowheads indicate cells with lymphoid morphology). (B) B220+ cells were quantified by flow cytometry in BM from mice engrafted at the indicated ages (n = 5 neonatal and 4 congenic adults; by Student’s t test; results are mean ± SEM compiled from two independent transplantation experiments; *, P = 0.04). (C) Flow cytometry analysis of leukemias arising from the indicated recipients. Representative morphology of sorted B220+ (top) and B220− (bottom) neonatal leukemia cells is shown (scale bar, 10 µm; samples from animals analyzed in B; numbers on plots indicate percentage of cells in each gate). (D) Representative photomicrographs of tissue stained with H&E or for myeloperoxidase (MPO), CD11b, or B220 (with inset showing B220+ focus; arrows indicate foci of B220 staining; scale bars, 100 µm [10 µm in the inset]; samples from animals analyzed in B).
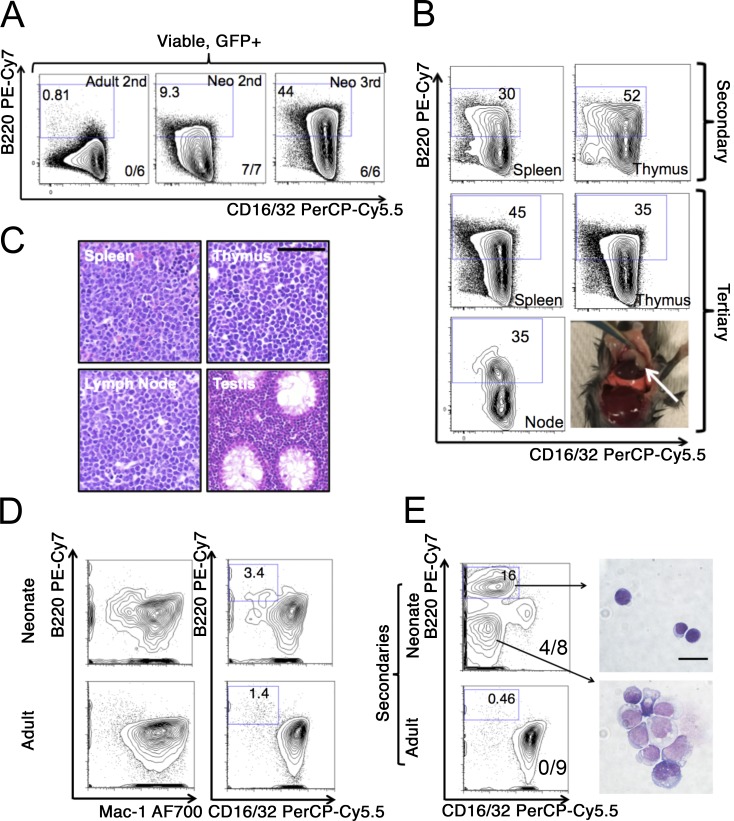
To further investigate this observation, we used serial transplantation to shorten leukemia latency (Puram et al., 2016), as mice engrafted as neonates with MLL-AF9–expressing HSPCs reach maturity by the time of leukemia onset, which could result in diminishment of the B220+ population as the microenvironment matures. By limiting dilution analysis, neonatal leukemia was more efficient than adult leukemia at engrafting secondary neonatal recipients, while both adult- and neonatal-derived LICs engrafted secondary adult recipients similarly (Fig. S1, I–K). Cells from primary neonatal grafts induced leukemia in secondary neonates with a shortened latency of 20–51 d (mean, 26 d; n = 21; P = 0.001 by Student’s t test versus primary neonatal recipients). Serial transplantation of neonatal-derived leukemia through neonatal recipients resulted in expansion of the B220+ component, with mixed-lineage leukemia (defined here as a minimum proportion of 5% B220+ cells) in seven out of seven transplanted secondary neonatal recipients, whereas serial transplantation of adult leukemia maintained AML with no mixed-lineage leukemic mice observed (P = 0.0003 by χ2 test compared with neonatal secondaries; Figs. 2 A and S2 A). We observed maintenance of mixed-lineage leukemia with expansion of the B220+ component in tertiary neonatal recipients (Figs. 2 A and S2 A). Infiltration of the thymus, spleen, lymph nodes, and testes with leukemic blasts occurred in secondary and tertiary neonatal recipients of neonatal MLL-AF9 leukemia (Fig. 2 B). Analysis of B cell differentiation in MLL-AF9 leukemia showed that B220+ cells were CD24-low CD43+ CD19− sIgM− and did not undergo Igh rearrangement, consistent with early pre-/pro-B differentiation (Fig. S2, B and C). Moreover, serially transplanted neonatal leukemia expressed the lymphoid-primed multipotent progenitor marker Flk2, which is indicative of an early lymphoid progenitor state and not expressed in adult leukemia (Fig. S2, D and E; Pietras et al., 2015). Interestingly, we found that serial transplantation of adult myeloid leukemia through the neonate could elicit slight B-lymphoid differentiation in some mice (Fig. S2 A).
Figure 2.
Serial transplantation of adult and neonatal leukemia. (A) Leukemia was transplanted from primary adult recipients into secondary adult recipients or serially from primary neonatal recipients into secondary and then tertiary, neonatal recipients. The number of transplanted recipients with mixed-lineage leukemia (defined as a minimum of 5% B220+ cells) out of the total number of recipients is shown in the bottom right of each panel. The baseline B220+ fraction of the primary neonatal leukemia used for secondary transplants was 5.4%, and representative flow cytometry plots from at least two independent transplantations at each passage are shown; numbers on plots indicate percentage of cells in each gate. (B) Representative flow cytometry analysis of leukemia involving the indicated organs following serial transplantation through neonatal recipients. In the gross photograph, a mediastinal tumor in situ is indicated; numbers on plots indicate percentage of cells in each gate. (C) Representative H&E-stained tissues from a tertiary neonatal transplant are shown (scale bar, 100 µm). (D) Mouse adult 127-LSK cells were transduced with a retrovirus for MLL-ENL and transplanted into either adult or recipient mice. At the onset of leukemia, BM was analyzed by flow cytometry with the indicated markers, with representative plots presented; numbers on plots indicate percentage of cells in each gate. (E) Leukemic BM from adult or neonatal primary recipients was transplanted into adult or neonatal secondary recipients, and leukemic BM was analyzed by flow cytometry with the indicated markers. From neonatal secondary transplants, B220+ and B220− cells were sorted by FACS and morphology examined following May–Grünwald–Giemsa staining (scale bar, 20 µm). Five out of five adult secondary transplants (22–33 d) and eight out of eight neonatal secondary transplants (23–26 d) developed leukemia. Proportions of mixed-lineage leukemias are indicated on flow cytometry plots, with results representative of two independent transplantations; numbers on gates indicate the percentage of cells in that gate.
Adult recipients of MLL-ENL–transformed murine HSPCs have been reported to develop a mixed-lineage B-lymphoid/myeloid leukemia (Zeisig et al., 2003). To examine the effects of the neonatal microenvironment on leukemia driven by a second MLL translocation, we transduced normal mouse adult HSPCs with MLL-ENL and engrafted these cells in adult or neonatal recipients. In primary recipients, we observed incomplete leukemia penetrance. Leukemia developed in three out of six neonates transplanted (latency, 67–102 d) and five out of eight adults (latency, 68–85 d), but we observed no obvious differences in immunophenotype, with coexpression of myeloid markers and a low B220 signal without mixed-lineage leukemia observed in any primary recipients (Fig. 2 D). However, upon secondary transplantation of primary neonatal leukemia into secondary neonatal recipients, we observed the development of mixed-lineage leukemia with the emergence of a population of B220+ cells with lymphoid morphology that were not present in secondary adult recipients, which maintained a phenotype similar to primary adults (mixed-lineage leukemia in four out of eight secondary neonatal recipients compared with zero out of nine secondary adult recipients; P = 0.02 by χ2 test; Fig. 2 E). There was complete penetrance of leukemia in secondary transplants (neonatal latency, 23–26 d; adult latency, 22–33 d). These MLL-ENL–driven leukemias underwent Igh recombination as reported previously (Fig. S2 C; Zeisig et al., 2003). Mixed-lineage MLL-ENL neonatal leukemia contained cells expressing CD19 that did not coexpress mature myeloid markers, indicating a more mature B-cell phenotype in this mixed-lineage leukemia compared with MLL-AF9 leukemia (Fig. S2, G–I; Zeisig et al., 2003). These data demonstrate that leukemogenesis in an infant murine microenvironment augments pro–B-lymphoid differentiation of MLL-ENL leukemia.
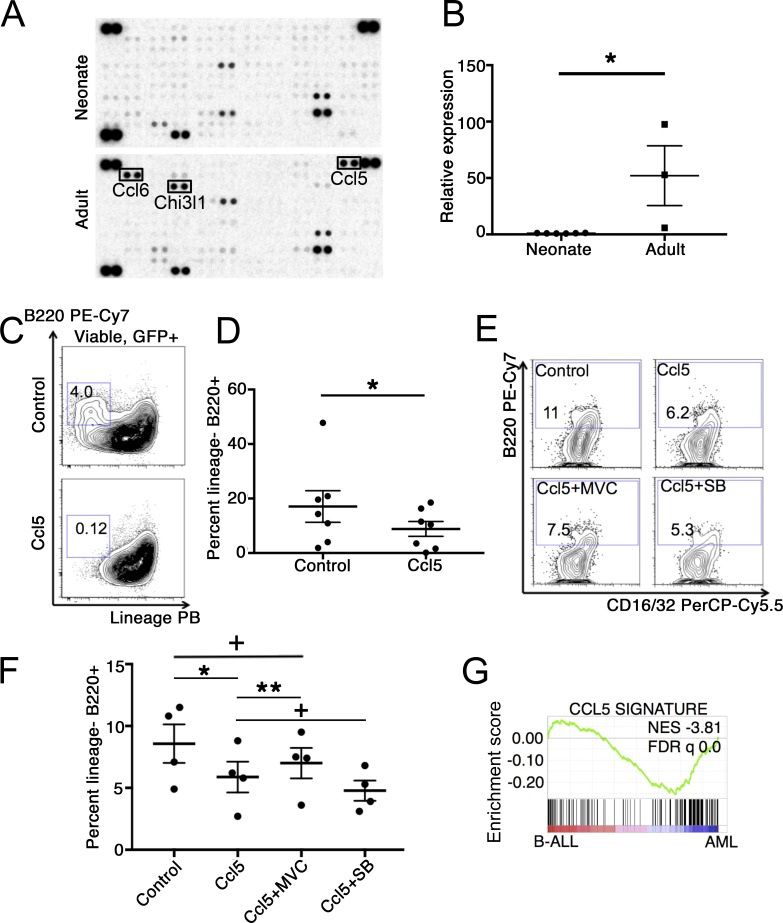
We next sought to identify the niche-derived factors responsible for age-specific effects on lineage. To this end, we isolated whole adult and neonatal BM stromal cells by enzymatic digestion. We found that neonatal and adult BM differed in their stromal cell compositions, in line with developmental changes in BM endothelial cells described previously (Fig. S3, A–C; Langen et al., 2017). We used a cytokine array to identify differentially expressed soluble factors in the conditioned medium released from the adherent fraction of adult or neonatal BM stroma after 48 h of culture. We found that adult BM stromal cells secreted markedly higher amounts of Rantes/Ccl5, Ccl6, and Chi3l1 compared with neonatal stroma (Fig. 3 A), a result that we confirmed by quantitative PCR (Fig. 3 B).
Figure 3.
Role of Ccl5 in specification of leukemia lineage. (A) Supernatant from BM stromal cells plated 48 h previously from pooled adult (6–8 wk old) or neonatal (P0) mice were used to probe a mouse cytokine array. (B) mRNA expression of Ccl5 was examined in neonatal or adult BM stroma from individual mice relative to adult BM stroma. Expression was calculated relative to the average of neonatal values, and results were compared by Student’s t test; mean ± SEM is presented, n = 6 neonatal and 3 adult samples isolated over three independent experiments; *, P = 0.02. (C and D) Neonatal MLL-AF9 leukemia cells were cultured under pro-myeloid (SCF, IL-3, and IL-6) conditions with or without 100 ng/ml recombinant mouse Ccl5 for 10 d, and lineage− (Gr-1−/Mac-1−) B220+ B-lymphoid cells were quantified by flow cytometry (n = 7 individual neonatal leukemia samples tested over seven independent experiments; analyzed pairwise by two-tailed Wilcoxon signed rank test; mean ± SEM is presented; *, P = 0.02). Numbers on plots indicate percentage of cells in each gate. (E and F) Neonatal primary transplanted leukemia cells were cultured under myeloid conditions with the indicated molecules for 8 d, after which time the cultures were analyzed by flow cytometry (MVC, maraviroc; SB, SB297006); results are four biological replicates for each condition (i.e., four independent neonatal leukemia donors) analyzed over three independent experiments, analyzed by paired Student’s t test; mean ± SEM is presented; *, P = 0.03; **, P = 0.01; +, not significant. Numbers on plots indicate percentage of cells in each gate. (G) Neonatal leukemia cells were cultured with or without Ccl5 for 48 h, at which point RNA was isolated for next-generation sequencing. Differentially expressed genes in Ccl5-treated cells relative control were identified at a P cutoff of 0.05 to establish a signature, which was used for GSEA against a preranked list of differentially expressed genes in human MLL-AF9–driven B-ALL versus AML. FDR, false discovery rate; NES, normalized enrichment score.
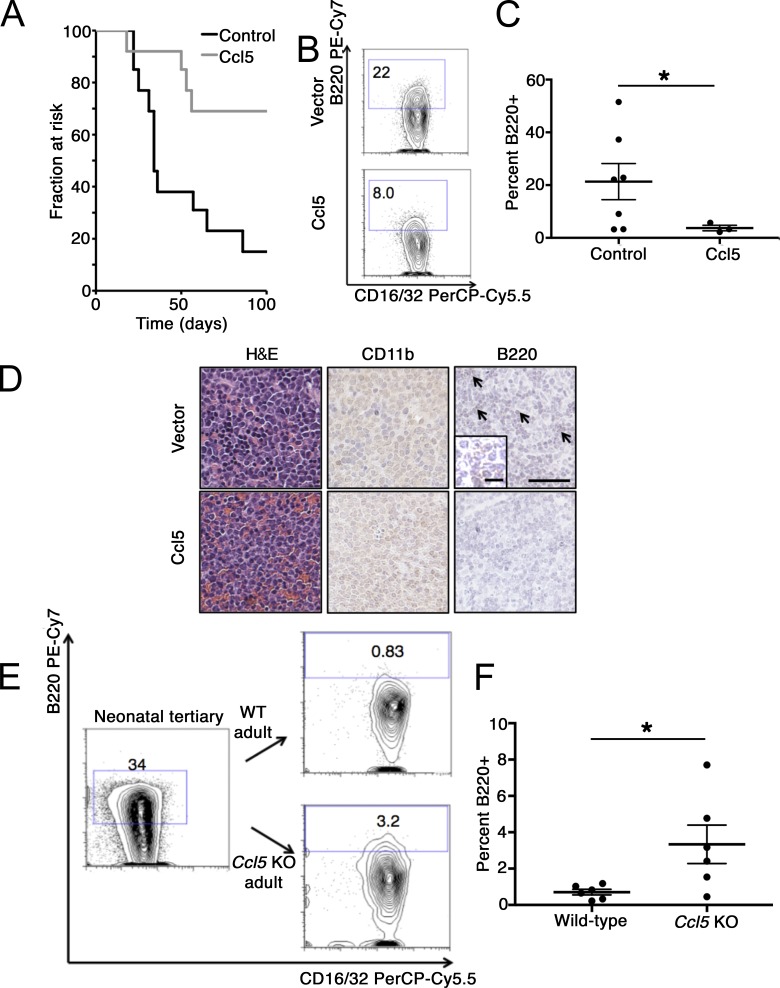
Since Ccl5 was previously reported to influence age-specific lineage bias in normal hematopoietic stem cells (Ergen et al., 2012), we examined the effects of Ccl5 on neonatal MLL-AF9 leukemia. Neonatal leukemia cells express the Ccl5 receptors Ccr3 and Ccr5 (Fig. S3 D). Furthermore, culture of neonatal leukemia cells with Ccl5 for 7–10 d led to a depletion of the B220+ Gr-1−/Mac-1− B-lymphoid population (Fig. 3, C and D). Exposure of purified populations of B220+ Gr-1−/Mac-1− cells or B220− cells to Ccl5 did not affect proliferation or clonogenicity (Fig. S3, E–H). By using a selective inhibitor of Ccr5 (maraviroc), we found that Ccr5 inhibition blunted the effect of Ccl5 on B-lymphoid differentiation, while culture with an inhibitor of Ccr3 (SB297006) in the presence of Ccl5 did not affect differentiation of neonatal leukemia cells in culture (Fig. 3, E and F). RNA sequencing (RNA-seq) of neonatal leukemia cells treated with or without Ccl5 showed that a Ccl5 signature was highly enriched in human MLL-AF9–driven AML, consistent with elicitation of myeloid programs by Ccl5 exposure (Fig. 3 G). Next, we overexpressed Ccl5 in mixed-lineage neonatal MLL-AF9 leukemia cells and engrafted these cells into neonatal recipients. This approach substituted for the more cumbersome strategy of engineering ectopic expression specifically in the recipient stroma. We unexpectedly found that overexpression of Ccl5 signaling significantly delayed both the onset and incidence of leukemia (Fig. 4 A), with a trend toward a smaller proportion of B220+ B-lymphoid cells in the resulting leukemia compared with cells expressing a control vector (Fig. 4, B–D). Moreover, transplantation of mixed-lineage neonatal MLL-AF9 leukemia into Ccl5−/− adult mice resulted in preservation of B220 expression in the resulting leukemia relative to Ccl5+/+ recipients (Fig. 4, E and F). Finally, although we found that endothelial cells express Ccl5 at the highest level compared with other types of stromal cells, they likely contribute only a small amount to the total adult BM microenvironmental Ccl5, as these cells are significantly outnumbered by other populations expressing detectable Ccl5 in adult BM, such as osteoblastic cells (Fig. S3, B and I). These data indicate that microenvironmental Ccl5 restrains lymphoid differentiation of MLL-AF9 leukemia and that ectopic Ccl5 signaling can impair mixed leukemogenesis in the neonatal hematopoietic microenvironment.
Figure 4.
Ccl5 regulates lineage of MLL-AF9 leukemia in vivo. (A) Neonatal leukemia cells were transduced with a lentivirus bearing a full-length Ccl5 cDNA and engrafted into sublethally irradiated neonatal recipients. When recipients became moribund, mice were euthanized, and survival data are presented. Results are combined from two independent transplantation experiments. n = 13 mice each in control and Ccl5 group. z = 2.75, P = 0.0006 by log-rank test. (B and C) Neonatal leukemia cells expressing either ectopic Ccl5 or transduced with empty vector were engrafted into neonatal recipients, and the percentage of lineage− (Gr-1−/Mac-1−) B220+ cells in the resultant leukemia was quantified. Results are aggregated over two independently transplanted cohorts analyzed by Student’s t test (mean ± SEM is presented; n = 7 control and 3 Ccl5 mice obtained for analysis; *, P = 0.14). Numbers on plots indicate the percentage of cells in each gate. (D) Representative histology including H&E staining (left) immunohistochemistry for CD11b and B220 (arrows indicate B220+ foci, shown at higher magnification in inset; scale bars, 100 µm (10 µm in the inset). (E) Serially transplanted neonatal mixed-lineage leukemia cells (either tertiary or secondary transplant in two independent transplant experiments) were engrafted into 6–8-wk-old Ccl5 KO mice or age-matched, congenic recipients. Representative flow cytometry profile of the input leukemia is shown, as well as flow cytometry profiles of the resultant leukemia in the indicated recipients. Numbers on plots indicate the percentage of cells in each gate. (F) Quantification of the percentage of B220+ leukemia cells in the indicated recipients. Results are compiled from two independent transplantation experiments with different leukemias used for engraftment in each experiment (analysis by Student’s t test; results are presented as mean ± SEM; n = 6 mice obtained for analysis for both the wild-type and Ccl5 KO conditions; *, P = 0.03).
To better understand the transcriptomic differences between neonatal mixed-lineage or adult myeloid MLL-AF9 leukemia, we examined the lineage− c-kit+ CD16/32+ CD34+ leukemic granulocyte-monocyte progenitor (LGMP) compartment, which is enriched for functional LICs (Krivtsov et al., 2013). We found that neonatal mixed-lineage LGMPs variably expressed B220 and the lymphoid-primed progenitor marker Flk2 (Pietras et al., 2015), which were not expressed on adult myeloid LGMPs, suggestive of B-lymphoid priming at the LIC level (Fig. 5 A). We sorted Flk2+ neonatal LGMPs and Flk2− adult LGMPs and performed RNA-seq analysis. By gene set enrichment analysis (GSEA), the Flk2+ neonatal LGMPs were enriched in profiles consistent with normal human B cells, while myeloid leukemia cells were enriched for a normal myeloid cell profile, indicative of priming of ultimate lineage fate (Hutcheson et al., 2008; Figs. 5 B and S3 J). Moreover, we found that neonatal LGMPs showed a signature of a primitive HSPC state, while adult LGMPs expressed a transcriptome of more mature blood cells (Fig. 5 B; Jaatinen et al., 2006). To examine the transcriptomes of uniform populations of leukemic blasts, we transduced murine 127-LSK cells with MLL-AF9 and cultured the cells under pro-lymphoid (stem cell factor [SCF], Flt3, and IL-7) or pro-myeloid (SCF, IL-3, and IL-6) conditions during transformation. Leukemic cells cultured under lymphoid conditions were uniformly B220+ with a dedifferentiated morphology, while cells cultured under myeloid conditions were uniformly B220− and appeared as early granulated myeloblasts (Fig. 5 C). RNA-seq analysis revealed enrichment of profiles of B-lymphoid cells as well as human MLL-AF9 B-ALL in cells transformed under lymphoid conditions, while cells under myeloid conditions expressed a myeloid program as well as that of human MLL-AF9–driven AML (Barabé et al., 2017). Together, these data indicate that compared with murine adult MLL-AF9 leukemia cells, which manifest myeloid identity, murine neonatal leukemia cells possess transcriptional programs consistent with early B-cell lineage commitment and a primitive HSPC state, consistent with a multipotent B-lymphoid/myeloid identity.
Figure 5.
Gene expression analysis of murine lymphoid and myeloid leukemia. (A) Serially transplanted neonatal or adult MLL-AF9 leukemia was analyzed by flow cytometry with the indicated antibodies to examine markers of early lymphoid commitment in the LGMP (lineage− c-kit+ CD16/32+ CD34+) population. Numbers on plots indicate percentage of cells in each gate. (B) RNA was isolated from either neonatal (Flk2+ B220−) or adult (Flk2− B220−) LGMPs and analyzed by RNA-seq in triplicate (and LGMPs isolated from three independent leukemias as biological triplicates). GSEA was performed to compare preranked lists of differentially expressed genes between neonatal and adult LGMPs to signatures of normal HSPCs relative to mature cells and B-lymphoid cells relative to myeloid cells. Normalized enrichment scores (NES) and false discovery rate (FDR) q-values (FDR q) are reported. (C) Adult mouse BM 127-LSK cells were isolated by FACS and transduced with MLL-AF9. 48 h following transduction, GFP+ cells were isolated by FACS and cultured in the presence of either pro-lymphoid (SCF, FLT3L, and IL-7) or pro-myeloid (SCF, IL-3, and IL-6) cytokines. Cells were passaged at least three times to select for immortalized cells and analyzed by flow cytometry. Cells were also examined morphologically following May–Grünwald–Giemsa staining (scale bar, 10 µm). (D) RNA-seq was performed on leukemia cells cultured either under pro-lymphoid or pro-myeloid conditions (three biological replicates from two independent transformations). A list of differentially expressed genes with log2 fold changes comparing lymphoid to myeloid leukemia cells was generated (samples sequenced in biological triplicates), and GSEA was used to compare the datasets of transcripts either enriched (top left) or depleted (top right) in normal B cells relative to myeloid cells or enriched (bottom left) or depleted (bottom right) in human MLL-AF9–driven B-ALL compared with AML (Hutcheson et al., 2008; Barabé et al., 2017). NESs and FDR q-values are reported for each analysis.
Here, we used heterochronic transplantation to vary the age of the hematopoietic microenvironment while holding constant the leukemic cell of origin, an approach that has, to our knowledge, not been reported previously in the study of leukemia. Given the inherent lineage plasticity that is manifested under experimental conditions and in patients, we hypothesized that MLL-rearranged leukemia would be responsive to the changes in the composition of the hematopoietic niche that occur with age. Indeed we found that the developmental stage of the hematopoietic microenvironment in which leukemogenesis occurs impacts leukemia lineage when the leukemogenic mutation and source of the cell of origin are held constant. In MLL-AF9–transformed HSPCs, the neonatal hematopoietic microenvironment supports early pre–/pro–B-lymphoid progenitor differentiation, with the proportion of B-lymphoid differentiated cells correlating with the age of onset of leukemia in serial transplantation. In contrast, transformation of the identical HSPCs in the adult microenvironment promotes myeloid differentiation. By switching the B220+ neonatal leukemia to the adult microenvironment, B220+ expression diminishes (see Fig. 4 E), while the neonatal microenvironment can elicit modest B220 expression from adult leukemia (see Fig. S2 A), indicating that the microenvironment can partially reset epigenetically specified lineage fates. Leukemia driven by MLL-ENL undergoes a B-lymphoid expansion when serially transplanted in neonates, an effect not seen in adults. These two translocations generally produce either pure myeloid (MLL-AF9) or mixed myeloid/B-lymphoid (MLL-ENL) leukemia in mice but can cause either B-ALL or AML in humans (Meyer et al., 2013; Milne, 2017). Our finding that the young hematopoietic niche can elicit B-lymphoid differentiation in these mouse models suggests that the age of the microenvironment at the time of HSPC transformation may contribute to defining leukemia lineage in humans and explains the discordance of mouse and human systems, since mouse model systems of MLL-rearranged leukemia have been reported using adult mice in the vast majority of cases. Furthermore, our findings may explain the observation that a B-lymphoid-to-myeloid lineage switch can occur when infant B-ALL relapses after treatment later in life in an older microenvironment (Rossi et al., 2012).
While it is known that experimental variation of the cytokine microenvironment elicits lineage plasticity of MLL-rearranged leukemia in culture and in vivo using genetically modified mice (Barabé et al., 2007; Wei et al., 2008), we wish to draw a clear distinction between these prior reports and our results. Here, we extend this observation to the clinical relevance of human infant leukemia by tying the lineage state of MLL-rearranged leukemia to the temporal state of the hematopoietic microenvironment by transplanting the identical leukemic cells of origin into isogenic recipients differing only in age. Although infant-like MLL-driven mixed-lineage B-lymphoid/myeloid leukemia can be induced in adult mice by ectopic expression of certain MLL fusion oncogenes seen rarely in humans (So et al., 2003), we show that this lineage phenotype could be elicited using the otherwise myeloid-biased MLL-AF9 and MLL-ENL in the neonatal hematopoietic microenvironment. The data presented here posit that the young hematopoietic niche contributes to the hybrid B-lymphoid/myeloid differentiation characteristic of human infant B-ALL and suggest that modulation of factors that mediate crosstalk between LICs and the hematopoietic microenvironment, such as Ccl5/Ccr5, may provide a novel therapeutic approach to direct differentiation toward a single lineage that may be more amenable to therapy. Although we implicate Ccr5/Ccl5 in age-specific regulation of leukemia lineage in our system, based on our finding that transplantation of neonatal leukemia into Ccl5−/− recipients incompletely preserves a mixed-lineage phenotype, it is reasonable to conclude that other factors that change with aging, including changes in stromal cell composition, extracellular matrix content, vascularization, and hormonal signaling, collaborate to define the lineage bias of LICs during leukemogenesis at various ages (Langen et al., 2017; Maryanovich et al., 2018). Current culture-based systems are limited in their capacity to mechanistically account for these factors, as are in vivo transgenic studies, which are biased toward implication of soluble factors. A more complete understanding of developmental maturation within the hematopoietic microenvironment and its impact on leukemia is needed. Nonetheless, our findings demonstrate that normal development and aging of the blood-forming system, which is regulated in part by changes in the hematopoietic microenvironment over time (Rossi et al., 2005; Ergen et al., 2012), directly impact the pathobiology of age-specific hematologic disease.
Overall, our findings highlight the importance of leveraging paradigms from normal development and aging to improve understanding of the pathogenesis of age-specific disease. Our work reinforces the notion that an important tool in age-specific disease modeling is heterochronic transplantation (Cho et al., 2017). Future models of human age-specific blood diseases should consider cell-extrinsic microenvironmental factors and their impact on disease phenotypes. This approach has broad applicability to the study and modeling of blood disorders of childhood, which we anticipate will aid the development of novel therapeutic approaches for patients.
Materials and methods
Isolation and transformation of hematopoietic cells
All studies were performed with approval of the Institutional Animal Care and Use Committee at Boston Children’s Hospital. Male, adult (6–8-wk-old) C57BL/6J mice were purchased from The Jackson Laboratory (stock number 000664). Mice were euthanized by carbon dioxide asphyxiation and femurs and tibiae isolated. BM was isolated and red blood cells lysed using Red Blood Cell Lysis Buffer (Sigma). BM mononuclear cells were lineage depleted using a mouse Lineage Cell Depletion Kit (Miltenyi Biotech) with magnetic separation. Following lineage depletion, cells were stained with lineage cocktail (Pacific Blue conjugated; details below) and antibodies against CD127 (PE conjugated), c-kit (APC-Cy7 conjugated), and Sca-1 (PE-Cy7 conjugated). 127-LSK cells were sorted on a FACS-Aria cytometer (BD Biosciences) under sterile conditions.
Purified 127-LSK cells were infected with retrovirus bearing the human MLL-AF9 cDNA (derived from pMig-MLL-AF9-EGFP, or pMSCV-neo-MLL-ENL; gifts of Scott Armstrong, Dana-Farber Cancer Institute, Boston, MA) or a control empty vector with GFP alone in some experiments. Retrovirus was packaged using 293-Tx cells (ATCC), and unmanipulated supernatant was used to infect 127-LSK cells. Cells were plated in retronectin-coated 96-well plates (50,000 cells per well) in the presence of retroviral supernatant as well as 3 µg/ml polybrene (Sigma) and 100 ng/ml SCF, 50 ng/ml IL-3, and 10 ng/ml IL-6 (all from R&D Systems). The plates were spun at 2,500 rpm at room temperature for 1 h followed by transfer to 37°/5% carbon dioxide. Cells were infected for 48 h, followed by a second round of FACS to isolate transduced GFP+ cells. Following this second sorting, cells were either directly transplanted into conditioned neonatal (P0–P1) or adult (8-wk-old) recipients or transformed in culture.
To overexpress Ccl5 in leukemia cells, we obtained a full-length Ccl5 cDNA cloned into a lentiviral vector from Genecopoiea. High-titer lentivirus was produced in HEK-293 cells and concentrated by ultracentrifugation, and leukemia cells were transduced for 48 h before transplantation into sublethally irradiated neonatal recipients. Flow cytometry analysis was performed on the GFP+ Cherry+ (transduced) fraction of the resultant leukemia. In these studies, neonatal recipients were randomized to receive either vector- or Ccl5-transduced leukemia cells.
Transplantation and limiting dilution assays
For primary transplantation into adult recipients, we used strains isogenic to the engrafted cells, either female C57BL6/J (stock 000664; The Jackson Laboratory) or B6.SJL-Ptprca Pepcb/BoyJ (stock 002014; The Jackson Laboratory). 2,000–4,000 GFP+ 127-LSK cells were injected into adult recipients via the tail vein 1–4 h following a sublethal dose of total body radiation of 675 rad. For primary transplantation into neonatal recipients, timed pregnancies were used to isolate C57BL/6J neonatal mice on the planned date of transplant. On the day of transplant, neonatal mice received a sublethal dose of 350 rad of radiation (Arora et al., 2014), and a dose of 100–200 GFP+ 127-LSK cells were injected via the facial vein. After transplantation, all mice were maintained on antibiotic prophylaxis and in autoclaved cages with sterile food. In all experiments, mice were monitored daily for evidence of morbidity, such as hunched posture, decreased activity, lethargy, or emaciation, and were euthanized by carbon dioxide asphyxiation at this time. Complete blood counts were not routinely monitored before the onset of clinical signs of illness. Occasionally, leukemic mice died before showing signs of morbidity, and by the time the carcass was found, the viability of the BM cells was typically lost. In these cases, these mice were excluded from downstream phenotypic analysis of leukemia cells but included in survival analyses. The onset of detectable morbidity or death was defined as the endpoint for every leukemic transplant recipient.
For secondary transplantation, C57BL/6J recipients were used following an identical conditioning and transplantation procedure as primary transplants, varying only cell dose as indicated in the text. Limiting dilution analysis was performed using Extreme Limiting Dilution Analysis (Hu and Smyth, 2009; http://bioinf.wehi.edu.au/software/elda/). Obviously, comparison of adult and neonatal transplant recipients cannot use littermates, and so highly inbred congenic C57BL/6J mice were used in all experiments.
For transplantation into Ccl5 KO mice, the same transplantation protocol was used for these mice as well as age-matched congenic nonlittermate adult recipients in parallel. Mice were 6–8 wk of age. Ccl5 KO mice were obtained from The Jackson Laboratory (stock 5090; Makino et al., 2002).
Animals were housed in a specific pathogen–free facility with standard light–dark cycles. After irradiation and transplantation, mice were maintained with autoclaved food and water, with water supplemented with Sulfatrim. Health was monitored on a daily basis. Studies adhered to the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
Statistical analysis
The statistical methodology used and sample sizes are described in the individual figure legends. t tests were two tailed unless otherwise stated. Results are presented as mean ± SEM unless otherwise stated. A significance level cutoff of 0.05 was used unless otherwise stated. Statistical analysis was performed using GraphPad Prism, Microsoft Excel, and R.
Antibodies and flow cytometry
For flow cytometry, the following mouse antibodies were used in the lineage cocktail: Gr-1 eFluor 450 (clone RB6-865), B220 eFluor 450 (RA3-6B2), CD3 eFluor 450 (17A2), Ter119 eFluor 450 (TER-119); other antibodies used were Flk-2 PE (A2F10), c-kit APC-eFluor 780 (ACK2), CD16/32 PerCp-Cy5.5 (93), c-kit APC (2B8), B220 PECy7 (RA3-6B2), sIgM eFluor 660 (II/41), and CD19 PE (eBioD3) from eBioscience; CD34 Alexa Fluor 700 (RAM34) from BD Biosciences; Sca-1 PECy7 (D7), CD43 PECy5 (1B11), CD24 Alexa Fluor 700 M1/69), CCR1 APC (S15040E), CCR3 PE (J073E5), and CCR5 PE (HM-CCR5) from BioLegend. Antibodies used in FACS sorting of BM stromal cell populations were CD31 FITC (390), CD51 PE (RMV-7), Sca-1 PE-Cy7 (D7; BioLegend), and Mac-1/CD11b Alexa Fluor 700 (M1/70; BD Biosciences). Flow cytometry analysis was performed on an LSR Fortessa (BD Biosciences) and sorting performed on a FACS-Aria (BD Biosciences).
Morphological analysis
Mouse BM, spleen, thymic, or nodal tissue was dissociated into a single-cell suspension. Cells were spun onto Cytoslides (Thermo Scientific) and stained with May-Grünwald and Giemsa stains (Sigma). In some experiments, cells were purified by FACS before spinning onto slides and staining. Cell morphology was examined for stage of differentiation arrest and lymphoid or myeloid morphology blindly. For examination of tissue sections, fresh tissue was fixed in 4% paraformaldehyde overnight and dehydrated with 70% ethanol before embedding and staining with H&E by standard techniques.
Drug treatment
Maraviroc was obtained from Sigma and used at 2 μM. SB-297006 was obtained from Tocris and used at 1 μM.
Isolation of BM stroma
BM stromal cell populations were isolated by enzymatic digestion and sorted according to published protocols (Suire et al., 2012; Schepers et al., 2013). Cells were either analyzed by flow cytometry or sorted by FACS immediately following enzymatic digestion or plated on fibronectin-coated plates and cultured in minimal essential medium with 10% fetal calf serum. CD11b was excluded from the hematopoietic cell lineage cocktail in these experiments to quantify macrophage content.
Quantitative PCR
The following quantitative PCR primers were used: mouse Ccl5: forward, 5′-TGCTCCAATCTTGCAGTCGT-3′; reverse, 5′-GCAAGCAATGACAGGGAAGC-3′; and mouse β-actin primers: forward, 5′-CAGAAGGAGATTACTGCTCTGGCT-3′; reverse, 5′-TACTCCTGCTTGCTGATCCAC-3′.
Cytokine analysis
BM stromal cells were isolated from adult (6–8 wk) or neonatal (P0–P1) C57BL6/J mice by enzymatic digestion with collagenase and dispase and plated on fibronectin. Cells were cultured in minimal essential medium with 10% fetal bovine serum and plated at identical density. 48 h after plating, the culture supernatant was collected and used to probe the mouse XL cytokine array (R&D Systems) per the manufacturer’s guidelines.
Histology and immunohistochemistry
For tissue analysis, tissue was fixed with buffered formalin or 4% paraformaldehyde, embedded, sectioned, and stained with H&E according to standard techniques. Immunostaining was performed using the following antibodies at 1:50 dilutions: anti-myeloperoxidase (ab9535; Abcam), anti-B220 (ab64100; Abcam), and anti-CD11b (ab133357; Abcam). Slides were dewaxed with xylene and rehydrated through a series of washes with decreasing percentages of ethanol. Antigen retrieval was performed in 10 mM sodium citrate buffer, pH 6.0, by placement in decloaking chamber for 30 min at 95°C. Immunohistochemistry was performed with Elite ABC kit and DAB substrate (Vector Laboratories) according to the manufacturer’s protocol.
IgH recombination assay
The germline mouse Igh locus was detected as described previously (Schlissel et al., 1991). D-J recombination was detected using the DFS, DQ52, and JH4A primers as described previously (Ehlich et al., 1994).
RNA-seq and data analysis
Total RNA was isolated using Trizol (Invitrogen) per the manufacturer’s protocol. Purified RNA was subjected to polyA selection using the NEBNext Polyadenylated mRNA Magnetic Isolation Module and subsequently used for library preparation with the NEBNext Ultra RNA Library Prep Kit (both from New England Biolabs). Libraries were analyzed on a Tapestation (Agilent) for quality control and quantified using the Qubit dsDNA HS Assay (Invitrogen), and equimolar pools were sequenced on HiSeq 2500 using 75-bp single-end protocols.
For raw data processing, fastq files containing single-end RNA-seq reads were aligned with Tophat 2.0.12 (Kim et al., 2013) against the UCSC mm10 reference genome using Bowtie 2.2.4 with default settings (Langmead and Salzberg, 2012). Gene level counts were obtained using the subRead featureCounts program (v1.5.1) using the parameter “--primary” and gene models from the UCSC mm10 Illumina iGenomes annotation package (Liao et al., 2014). Read counts were normalized using size factors as available by the DESeq2 package (Love et al., 2014). Unsupervised hierarchical clustering of pairwise sample-to-sample Spearman’s rank correlation coefficients was performed using Euclidean distance as the distance metric and ward.D as the linkage method. Heat maps were plotted using the pheatmap R package.
For comparing mouse and human leukemia, we used published RNA-seq data from human CD34 cells transformed with MLL-AF9 and engrafted into immunodeficient mice, where both AML and ALL are generated in vivo (Barabé et al., 2017). These human leukemia datasets were downloaded from the Gene Expression Omnibus (accession no. GSE71691).
Differential expression of mouse RNA-seq data was performed using DESeq2 with standard parameters. Differential expression of human RNA-seq data (fragments per kilobase per million values downloaded from the Gene Expression Omnibus, accession no. GSE71691) was performed by computing the ratio between the mean expression of each gene in AML samples compared with ALL samples. P values were computed using an unpaired t test.
For GSEA, we generated lists of differentially expressed genes and used a preranked list of relative transcript expression comparing myeloid versus lymphoid mouse leukemia cells to the top 100 enriched or depleted transcripts in human AML compared with ALL. GSEA was performed as described using the Molecular Signatures Database (Mootha et al., 2003; Subramanian et al., 2005). Gene ontology analysis was performed using PANTHER classification (http://www.geneontology.org; Mi et al., 2013).
RNA-seq data were deposited in the Gene Expression Omnibus under the SuperSeries accession no. GSE105087, including the datasets GSE105085, GSE105086, and GSE113974.
Online supplemental material
Fig. S1 shows the scheme for sorting 127-LSK cells, characterization of leukemias from FL and BM sources, and serial transplantation with limiting dilution data. Fig. S2 shows quantification of serial transplantation through neonates and adults and further characterization of MLL-AF9 and MLL-ENL adult and neonatal leukemias. Fig. S3 shows analysis of adult and neonatal stromal populations, Ccl5 effects on leukemia cell subpopulations, and RNA sequencing data of B cell–specific transcripts.
Supplementary Material
Acknowledgments
We thank Drs. David Scadden, Youmna Kfoury, Scott Armstrong, and Haihua Chu for helpful conversations.
This work was supported by the National Heart, Lung, and Blood Institute Progenitor Cell Translation Consortium (grant U01HL134812), the National Institute of Diabetes and Digestive and Kidney Diseases (grant R24DK092760 to G.Q. Daley and grant K08DK114527 to R.G. Rowe), Alex’s Lemonade Stand Foundation for Childhood Cancer and the Doris Duke Medical Charitable Foundation (G.Q. Daley), and a St. Baldrick’s Foundation Fellowship and a Pedals for Pediatrics grant to R.G. Rowe.
The authors declare no competing financial interests.
Author contributions: R.G. Rowe designed the research, performed experiments, analyzed data, and wrote the paper. E. Lummertz da Rocha and M.D. Fleming analyzed data. P. Sousa, P. Missios, D.K. Jha, M. Morse, W. Marion, A. Yermalovich, J. Barragan, and R. Mathieu performed experiments and analyzed data. T.E. North and G.Q. Daley designed the research, analyzed data, and wrote the paper.
References
- Arora N., Wenzel P.L., McKinney-Freeman S.L., Ross S.J., Kim P.G., Chou S.S., Yoshimoto M., Yoder M.C., and Daley G.Q.. 2014. Effect of developmental stage of HSC and recipient on transplant outcomes. Dev. Cell. 29:621–628. 10.1016/j.devcel.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabé F., Kennedy J.A., Hope K.J., and Dick J.E.. 2007. Modeling the initiation and progression of human acute leukemia in mice. Science. 316:600–604. 10.1126/science.1139851 [DOI] [PubMed] [Google Scholar]
- Barabé F., Gil L., Celton M., Bergeron A., Lamontagne V., Roques E., Lagace K., Forest A., Johnson R., Pecheux L., et al. 2017. Modeling human MLL-AF9 translocated acute myeloid leukemia from single donors reveals RET as a potential therapeutic target. Leukemia. Leukemia. 31:1166–1176. [DOI] [PubMed] [Google Scholar]
- Chen W., Li Q., Hudson W.A., Kumar A., Kirchhof N., and Kersey J.H.. 2006. A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood. 108:669–677. 10.1182/blood-2005-08-3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., O’Sullivan M.G., Hudson W., and Kersey J.. 2011. Modeling human infant MLL leukemia in mice: leukemia from fetal liver differs from that originating in postnatal marrow. Blood. 117:3474–3475. 10.1182/blood-2010-11-317529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G.S., Lee D.I., Tampakakis E., Murphy S., Andersen P., Uosaki H., Chelko S., Chakir K., Hong I., Seo K., et al. 2017. Neonatal Transplantation Confers Maturation of PSC-Derived Cardiomyocytes Conducive to Modeling Cardiomyopathy. Cell Reports. 18:571–582. 10.1016/j.celrep.2016.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlich A., Martin V., Müller W., and Rajewsky K.. 1994. Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 4:573–583. 10.1016/S0960-9822(00)00129-9 [DOI] [PubMed] [Google Scholar]
- Ergen A.V., Boles N.C., and Goodell M.A.. 2012. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood. 119:2500–2509. 10.1182/blood-2011-11-391730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Hutcheson J., Scatizzi J.C., Siddiqui A.M., Haines G.K. III, Wu T., Li Q.Z., Davis L.S., Mohan C., and Perlman H.. 2008. Combined deficiency of proapoptotic regulators Bim and Fas results in the early onset of systemic autoimmunity. Immunity. 28:206–217. 10.1016/j.immuni.2007.12.015 [DOI] [PubMed] [Google Scholar]
- Jaatinen T., Hemmoranta H., Hautaniemi S., Niemi J., Nicorici D., Laine J., Yli-Harja O., and Partanen J.. 2006. Global gene expression profile of human cord blood-derived CD133+ cells. Stem Cells. 24:631–641. 10.1634/stemcells.2005-0185 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov A.V., Feng Z., Lemieux M.E., Faber J., Vempati S., Sinha A.U., Xia X., Jesneck J., Bracken A.P., Silverman L.B., et al. 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 14:355–368. 10.1016/j.ccr.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov A.V., Figueroa M.E., Sinha A.U., Stubbs M.C., Feng Z., Valk P.J., Delwel R., Döhner K., Bullinger L., Kung A.L., et al. 2013. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 27:852–860. 10.1038/leu.2012.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen U.H., Pitulescu M.E., Kim J.M., Enriquez-Gasca R., Sivaraj K.K., Kusumbe A.P., Singh A., Di Russo J., Bixel M.G., Zhou B., et al. 2017. Cell-matrix signals specify bone endothelial cells during developmental osteogenesis. Nat. Cell Biol. 19:189–201. 10.1038/ncb3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin S., Luo R.T., Ptasinska A., Kerry J., Assi S.A., Wunderlich M., Imamura T., Kaberlein J.J., Rayes A., Althoff M.J., et al. 2016. Instructive Role of MLL-Fusion Proteins Revealed by a Model of t(4;11) Pro-B Acute Lymphoblastic Leukemia. Cancer Cell. 30:737–749. 10.1016/j.ccell.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y., Cook D.N., Smithies O., Hwang O.Y., Neilson E.G., Turka L.A., Sato H., Wells A.D., and Danoff T.M.. 2002. Impaired T cell function in RANTES-deficient mice. Clin. Immunol. 102:302–309. 10.1006/clim.2001.5178 [DOI] [PubMed] [Google Scholar]
- Man N., Sun X.J., Tan Y., García-Cao M., Liu F., Cheng G., Hatlen M., Xu H., Shah R., Chastain N., et al. 2016. Differential role of Id1 in MLL-AF9-driven leukemia based on cell of origin. Blood. 127:2322–2326. 10.1182/blood-2015-11-677708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanovich M., Zahalka A.H., Pierce H., Pinho S., Nakahara F., Asada N., Wei Q., Wang X., Ciero P., Xu J., et al. 2018. Adrenergic nerve degeneration in bone marrow drives aging of the hematopoietic stem cell niche. Nat. Med. 24:782–791. 10.1038/s41591-018-0030-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C., Hofmann J., Burmeister T., Gröger D., Park T.S., Emerenciano M., Pombo de Oliveira M., Renneville A., Villarese P., Macintyre E., et al. 2013. The MLL recombinome of acute leukemias in 2013. Leukemia. 27:2165–2176. 10.1038/leu.2013.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., and Thomas P.D.. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8:1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne T.A. 2017. Mouse models of MLL leukemia: recapitulating the human disease. Blood. 129:2217–2223. 10.1182/blood-2016-10-691428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Pang W.W., Price E.A., Sahoo D., Beerman I., Maloney W.J., Rossi D.J., Schrier S.L., and Weissman I.L.. 2011. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 108:20012–20017. 10.1073/pnas.1116110108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters R., Schrappe M., De Lorenzo P., Hann I., De Rossi G., Felice M., Hovi L., LeBlanc T., Szczepanski T., Ferster A., et al. 2007. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 370:240–250. 10.1016/S0140-6736(07)61126-X [DOI] [PubMed] [Google Scholar]
- Pietras E.M., Reynaud D., Kang Y.A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Göttgens B., and Passegué E.. 2015. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 17:35–46. 10.1016/j.stem.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram R.V., Kowalczyk M.S., de Boer C.G., Schneider R.K., Miller P.G., McConkey M., Tothova Z., Tejero H., Heckl D., Järås M., et al. 2016. Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell. 165:303–316. 10.1016/j.cell.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel V.I., Miller C.L., Eaves C.J., and Lansdorp P.M.. 1996. The repopulation potential of fetal liver hematopoietic stem cells in mice exceeds that of their liver adult bone marrow counterparts. Blood. 87:3500–3507. [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., and Weissman I.L.. 2005. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA. 102:9194–9199. 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J.G., Bernasconi A.R., Alonso C.N., Rubio P.L., Gallego M.S., Carrara C.A., Guitter M.R., Eberle S.E., Cocce M., Zubizarreta P.A., and Felice M.S.. 2012. Lineage switch in childhood acute leukemia: an unusual event with poor outcome. Am. J. Hematol. 87:890–897. 10.1002/ajh.23266 [DOI] [PubMed] [Google Scholar]
- Schepers K., Pietras E.M., Reynaud D., Flach J., Binnewies M., Garg T., Wagers A.J., Hsiao E.C., and Passegué E.. 2013. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 13:285–299. 10.1016/j.stem.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., and Baltimore D.. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173:711–720. 10.1084/jem.173.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C.W., Karsunky H., Passegué E., Cozzio A., Weissman I.L., and Cleary M.L.. 2003. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 3:161–171. 10.1016/S1535-6108(03)00019-9 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suire C., Brouard N., Hirschi K., and Simmons P.J.. 2012. Isolation of the stromal-vascular fraction of mouse bone marrow markedly enhances the yield of clonogenic stromal progenitors. Blood. 119:e86–e95. 10.1182/blood-2011-08-372334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Wunderlich M., Fox C., Alvarez S., Cigudosa J.C., Wilhelm J.S., Zheng Y., Cancelas J.A., Gu Y., Jansen M., et al. 2008. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 13:483–495. 10.1016/j.ccr.2008.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig B.B., García-Cuéllar M.P., Winkler T.H., and Slany R.K.. 2003. The oncoprotein MLL-ENL disturbs hematopoietic lineage determination and transforms a biphenotypic lymphoid/myeloid cell. Oncogene. 22:1629–1637. 10.1038/sj.onc.1206104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.