Transcriptome and proteome profiling in isogenic CRISPR/Cas9 engineered human colon organoids reveals qualitatively distinct Wnt responses in APC-deficient adenoma and normal stem/progenitor cells. The data represent a comprehensive resource for molecular markers that are linked with colon cancer prognosis.
Abstract
Constitutive Wnt activation upon loss of Adenoma polyposis coli (APC) acts as main driver of colorectal cancer (CRC). Targeting Wnt signaling has proven difficult because the pathway is crucial for homeostasis and stem cell renewal. To distinguish oncogenic from physiological Wnt activity, we have performed transcriptome and proteome profiling in isogenic human colon organoids. Culture in the presence or absence of exogenous ligand allowed us to discriminate receptor-mediated signaling from the effects of CRISPR/Cas9-induced APC loss. We could catalog two nonoverlapping molecular signatures that were stable at distinct levels of stimulation. Newly identified markers for normal stem/progenitor cells and adenomas were validated by immunohistochemistry and flow cytometry. We found that oncogenic Wnt signals are associated with good prognosis in tumors of the consensus molecular subtype 2 (CMS2). In contrast, receptor-mediated signaling was linked to CMS4 tumors and poor prognosis. Together, our data represent a valuable resource for biomarkers that allow more precise stratification of Wnt responses in CRC.
Graphical Abstract
Introduction
The gastrointestinal epithelium depends on precise regulation of the Wnt signaling pathway to coordinate stem cell maintenance, proliferation, and cell lineage differentiation (Clevers et al., 2014). During homeostasis, the stem cell niche confines Wnt activity to the crypt compartment by limiting the availability of Wnt ligands (Sato et al., 2011b; Farin et al., 2012; Valenta et al., 2016), R-spondin coactivators, and BMP antagonists (Kabiri et al., 2014; Aoki et al., 2016; Stzepourginski et al., 2017). Binding of Wnt to Frizzled receptors and LRP5/6 coreceptors results in β-catenin (CTNNB1) stabilization, nuclear import, and transcriptional activation of Wnt target genes. In the absence of Wnt ligands, the cytoplasmic destruction complex that contains Adenoma polyposis coli (APC), AXIN1/2, the serine/threonine kinases GSK3B, and CSNK1A1 mediates CTNNB1 phosphorylation, ubiquitinylation, and proteasomal degradation (Stamos and Weis, 2013). Several mutations have been identified that result in ligand-independent CTNNB1 stabilization in cancers (Zhan et al., 2017). In colorectal cancer (CRC), truncating APC mutations are most frequent and can be found in ∼80% of all patients (Cancer Genome Atlas Network, 2012). Homozygous APC loss causes adenoma-like growth, which is considered as a precursor lesion that can further progress into carcinoma by acquisition of additional driver mutations (Vogelstein et al., 1988).
Molecular characterization has led to the identification of a conserved transcriptional Wnt signature that is shared between cultured cell lines (van de Wetering et al., 2002; Van der Flier et al., 2007) and intestinal stem cells in mouse (Muñoz et al., 2012) and human (Jung et al., 2011). Wnt-responsive genes such as LGR5, EPHB2, TNFRSF19, and PTK7 have subsequently been identified as specific markers of actively cycling gastrointestinal stem cells (Barker et al., 2007; Jung et al., 2011, 2015; Stange et al., 2013). Interestingly, mouse Apc mutant adenomas (Sansom et al., 2007), as well as human CRC (Vermeulen et al., 2010; Merlos-Suárez et al., 2011) are also characterized by induction of a Wnt/Stem cell signature, emphasizing the progenitor status of normal crypts and tumors. The presence of functional stem cells has been described in mouse adenomas (Schepers et al., 2012; Kozar et al., 2013) and in xenotransplanted CRC cells (Cortina et al., 2017; Shimokawa et al., 2017), indicating a hierarchical organization of tumors despite constitutive Wnt activation.
Pronounced transcriptional Wnt activity has been associated with a tumor subtype with favorable prognosis (de Sousa E Melo et al., 2011; Guinney et al., 2015). Recent experiments, however, have shown that progressed CRC cells remain addicted to Wnt activity (Dow et al., 2015; O’Rourke et al., 2017), providing a rationale for therapeutic targeting. While pharmacological strategies are available to interfere with upstream pathway mutations (Gurney et al., 2012; Koo et al., 2015; Storm et al., 2016), only limited options exist for the majority of tumors that are driven by APC mutations (Novellasdemunt et al., 2015). In preclinical models, global interference with Wnt signaling resulted in gastrointestinal toxicity (Lau et al., 2013; Kabiri et al., 2014), emphasizing a demand for strategies that do not interfere with homeostatic signaling. APC mutant cells undergo extensive pathway rewiring (Billmann et al., 2018), which could create new vulnerabilities. Specific dependence of mouse adenomas has been described on Stat3 (Phesse et al., 2014), mTORC1 (Faller et al., 2015), Yap/Taz (Azzolin et al., 2014), Rac1 (Myant et al., 2013), or the ER stress regulator Grp78 (van Lidth de Jeude et al., 2017).
Despite these promising examples, a systematic characterization of normal and oncogenic Wnt has not been performed yet. Here we have set out to catalog the physiological and oncogenic Wnt responses in primary human colon epithelial cells on the transcriptome and proteome level. We take advantage of the organoid culture model that allows expansion of normal and tumor gastrointestinal epithelia (Sato et al., 2011a) and genetic engineering of oncogenic mutations by CRISPR/Cas9 technology (Schwank et al., 2013; Drost et al., 2015; Matano et al., 2015). By subjecting normal and APC mutant isogenic organoid lines to Wnt-stimulation, we aimed to generate an expression resource for stratification of extrinsic and intrinsic Wnt responses.
Results
Differential analysis of Wnt-receptor– and APC-KO–induced signaling in human colon organoids
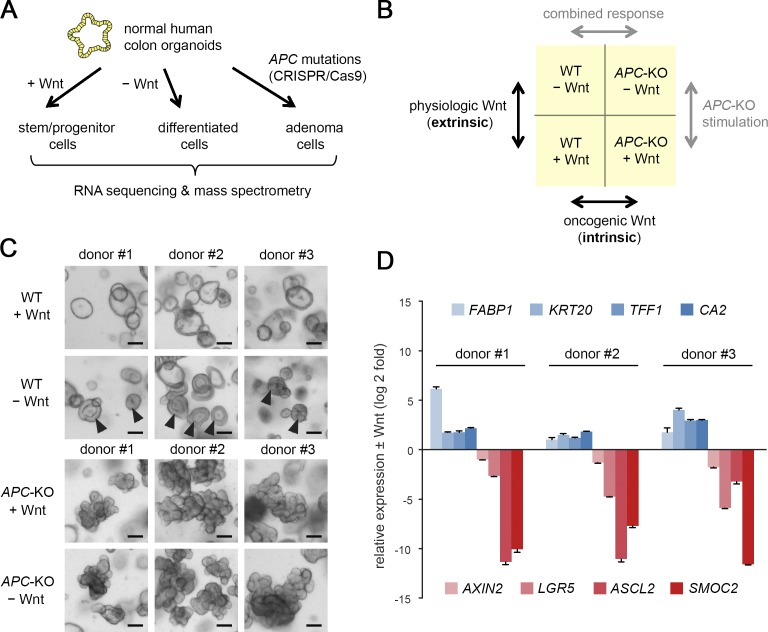
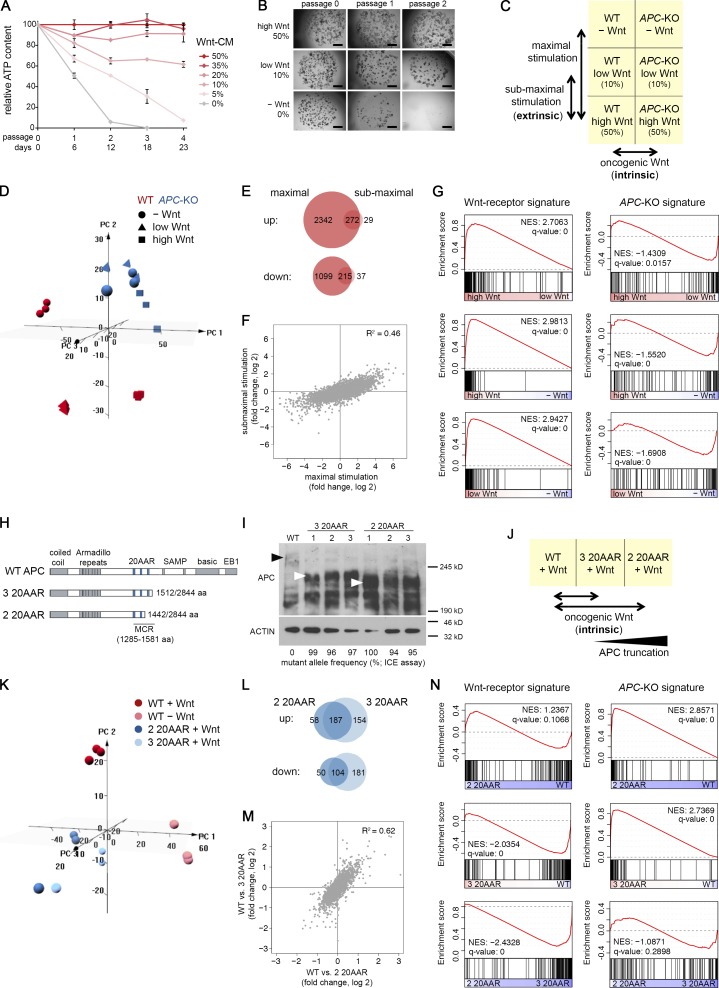
To distinguish receptor-induced from constitutive Wnt pathway activation, we have introduced truncating APC mutations within the mutation cluster region by the CRISPR/Cas9 technology in normal human colon organoids (Fig. 1 A). The cells were derived from nonpathological mucosa of three separate subjects to account for differences in gender, age, and location (Fig. S1 A). Growth independence from Wnt/R-spondin served as a stringent selection criterion for successful targeting of APC, the resulting organoids (APC-KO) were clonally expanded, and the introduced mutations were confirmed by Sanger sequencing and Western blot (WB) analysis (Fig. S1, B and C). By culturing WT and APC-KO organoids in Wnt/R-spondin–containing and deprived medium, we defined four biological conditions for differential analysis (Fig. 1 B): receptor-mediated, “extrinsic” stimulation was measured in WT cells ± Wnt/R-spondin. To address the “intrinsic” effect of APC-KO, both genotypes were compared in the presence of Wnt/R-spondin to normalize for the physiological stimulation. Comparison of APC-KO ± Wnt/R-spondin allowed us to study signal responsiveness in presence of constitutive activation, and comparison of both genotypes in the absence of Wnt/R-spondin represents the combination of extrinsic and intrinsic stimulation. 2 d after Wnt/R-spondin withdrawal, all WT lines showed a compact morphology indicative of cellular differentiation (Fig. 1 C). By quantitative RT-PCR (qRT-PCR), we observed robust induction of differentiation markers (FABP1, KRT20, TFF1, and CA2) and reduction of Wnt/Stem cell markers (AXIN2, LGR5, ASCL2, and SMOC2; Fig. 1 D), confirming responsiveness of our models.
Figure 1.
Differential profiling of receptor-induced and constitutive Wnt signaling. (A) Experimental approach for molecular profiling of CRISPR/Cas9 engineered human colon organoids. (B) Strategy for differential analysis of receptor-induced (extrinsic) and oncogene-induced (intrinsic) Wnt signaling. (C) Morphology of normal and APC-KO human colon organoids in control medium or 2 d after Wnt/R-spondin withdrawal. Arrowheads show differentiation of normal organoids. Scale bars are 200 µm. (D) qRT-PCR analysis of differentiation markers (blue) and Wnt/stem cell markers (red) in normal organoid lines 2 d after withdrawal of Wnt/R-spondin. Mean normalized expression (± SD in three technical replicates) is shown for all three organoid lines, and expression was measured twice independently. See also Fig. S1.
Normal and oncogenic Wnt induce distinct transcriptional responses
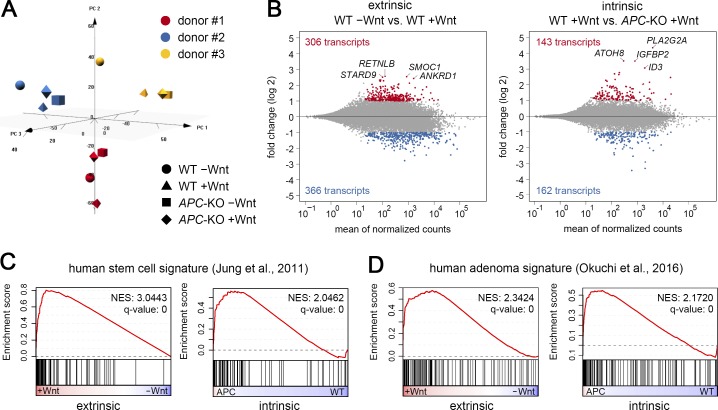
Next, we performed RNA sequencing to record the transcriptome-wide changes of the isogenic organoid lines. Principal component analysis (PCA; Fig. 2 A) showed that the variability of gene expression between donor lines largely exceeded the biological effects. However, this variability was small compared with the major transcriptional changes observed between normal organoids and paired CRC-derived organoids (Fig. S2, A and B). Thus, while a comparison of tumor and normal samples usually results in two discrete expression clusters (van de Wetering et al., 2015; Cristobal et al., 2017), the individual variability between normal organoids exceeds the effects induced by a single oncogenic hit such as APC-KO. To normalize for this donor-dependent variability, we performed paired differential analysis (Fig. 2 B). This allowed us to extract 306 and 143 transcripts that were significantly up-regulated among all lines (log twofold change >1; adjusted P value <0.05) after Wnt-receptor stimulation (extrinsic) and APC-KO (intrinsic), respectively. We found few transcriptomic changes when APC-KO cells were treated ± Wnt/R-spondin, while simultaneous modulation of medium and genotype (combined response) caused a more pronounced biological response (833 up-regulated transcripts; Fig. S2 C). We conclude that differential analysis of isogenic organoids allows sensitive detection of single gene/pathway responses despite strong individual variation of gene expression.
Figure 2.
Transcriptomic changes after extrinsic and intrinsic Wnt modulation. (A) PCA shows that donor line–specific differences are the dominant source of gene expression variation. The 3,000 most variant genes were included for the analysis. (B) Differential gene expression analysis. Mean log twofold changes in n = 3 colon organoid lines (paired analysis). Significantly up- and down-regulated genes (±1 log twofold change; P adjust < 0.05) are marked in red and blue, respectively. (C and D) GSEA using previously reported human signatures for stem cells (C) and adenomas (D). Each signature was studied in the extrinsic and intrinsic Wnt response, and NESs and q values are shown. See also Fig. S2.
To intersect our data with previous studies of gastrointestinal Wnt/Adenoma signaling, we performed gene set enrichment analysis (GSEA). Interestingly, both of our datasets showed strong enrichment of the human colon EPHB2 stem cell signature (Jung et al., 2011; Fig. 2 C) and of genes induced in human adenomas (Okuchi et al., 2016; Fig. 2 D), an observation that was confirmed using mouse-derived stem cell and adenoma signatures (Sansom et al., 2007; Muñoz et al., 2012; Fig. S2, D and E). While this overlap confirms our data and demonstrates conserved responses in mammals, it also suggests that the available signatures cannot specifically distinguish physiological from constitutive pathway activation. This is most likely due to the reported similarities between normal and cancer stem cells (Vermeulen et al., 2010; Merlos-Suárez et al., 2011; Schepers et al., 2012).
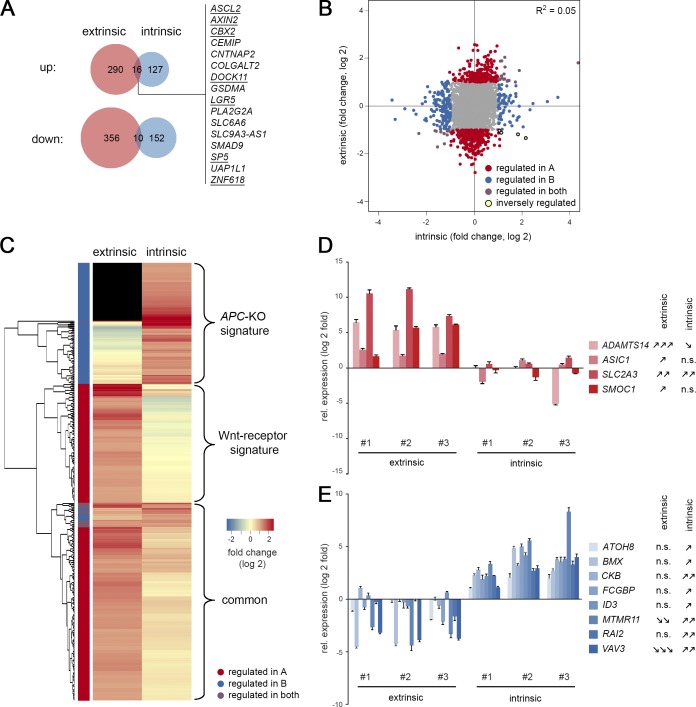
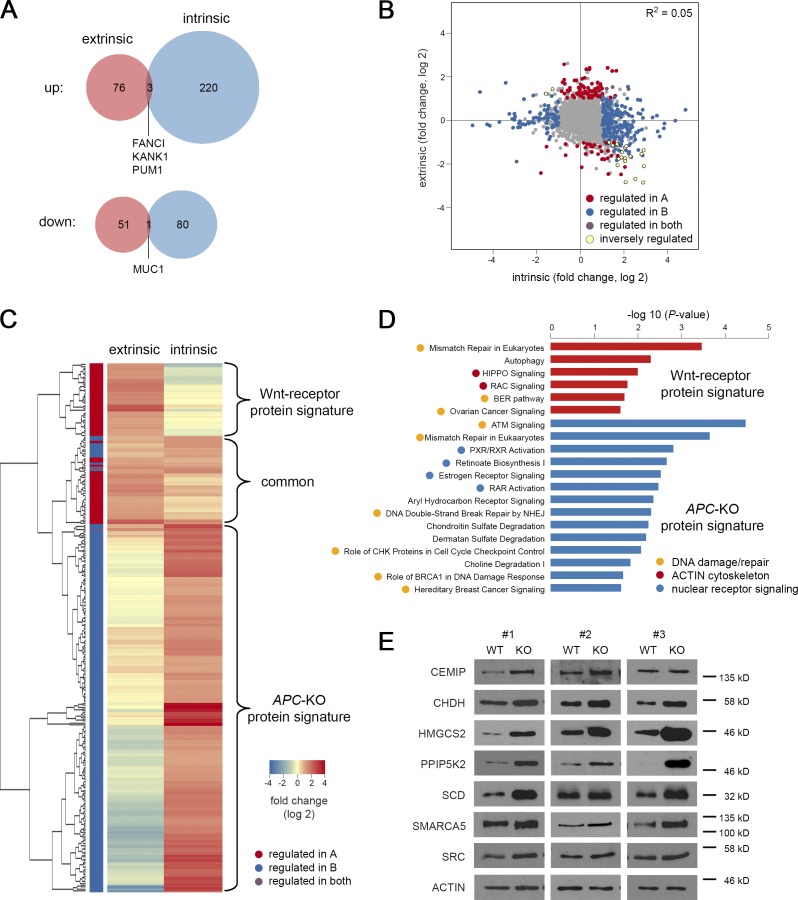
Interestingly, we only observed limited overlap between genes that were significantly changed after Wnt-receptor stimulation and APC loss, some of which contained well-characterized Wnt/stem cell markers such as ASCL2, AXIN2, LGR5, and SP5 (Fig. 3 A). However, on the global scale, no correlation was found between both responses (R2 = 0.05; Fig. 3 B). By hierarchical clustering, we could visualize two nonoverlapping classes of induced transcripts (Fig. 3 C and Table S1, A and B): a Wnt-receptor signature (146 genes) and an APC-KO signature (112 genes) that are expressed in a mutually exclusive manner. A third class of genes showed up-regulation in both situations, however, generally weaker in the intrinsic response, indicating that Wnt/R-spondin stimulation may not have fully saturated canonical signaling in WT organoids. By qRT-PCR, we independently validated specific markers of Wnt-receptor signaling (Fig. 3 D; ADAMTS14, ASIC1, SLC2A3, and SMOC1) and APC-KO cells (ATOH8, BMX, CKB, FCGBP, ID3, MTMR11, RAI2, and VAV3). Our profiling strategy thus allowed us to identify distinct Wnt responses in normal and adenoma cells.
Figure 3.
Distinct transcriptomic signatures induced by normal and oncogenic Wnt signaling. (A) Venn diagrams show limited overlap between significantly changed genes (± 1 log twofold change; P adjust < 0.05) after Wnt-receptor stimulation (extrinsic) and APC loss-of-function (intrinsic). Genes that are part of the mouse intestinal stem cell signature are underlined. (B) Global correlation shows independence of intrinsic and extrinsic responses. (C) Unsupervised clustering identifies specific APC-KO and Wnt-receptor signatures. Note that a number of adenoma genes are not expressed in WT cells (black). (D and E) qRT-PCR validation of identified marker genes. Genes induced after Wnt-receptor stimulation (D) and APC-KO–induced genes (E) are shown as mean normalized expression (± SD in three technical replicates). Significant responses in all three organoid lines were determined by Student’s t test and labeled as follows: one arrow, P < 0.05; two arrows, P < 0.01; three arrows, P < 0.001; n.s., not significant. Expression was measured twice independently. See also Table S1, A and B.
Next, we tested if CRC samples that are driven by alternative Wnt pathway mutations may differentially express the identified signatures. RNF43 has been identified as a tumor suppressor in microsatellite instable CRC (Giannakis et al., 2014) that causes Wnt-ligand dependent pathway activation (Hao et al., 2012; Koo et al., 2012). We used public TCGA (The Cancer Genome Atlas) data for differential expression analysis between colon cancers that are deficient in either RNF43 or APC. Here, we could confirm that RNF43 mutation is associated with the consensus molecular subtype 1 (CMS1), indicative of microsatellite instable CRC, whereas APC mutation is linked to the canonical CMS2 subtype as described before (Fig. S2 F; Guinney et al., 2015). In parallel, we could observe reciprocal and highly significant enrichment of our Wnt-receptor and APC-KO signatures (Fig. S2 G), supporting that also in tumors distinct responses are induced by upstream and downstream activation.
Transcriptomic signatures are preserved at different levels of intrinsic and extrinsic stimulation
The divergent responses could result from quantitative differences in Wnt signaling between normal and APC-KO cells or involve cell differentiation in the WT. To address these questions, we performed titration experiments and first determined a Wnt concentration that causes mild reduction of growth of WT organoids rather than complete growth arrest and differentiation (Fig. 4, A and B). RNA sequencing was then performed in the absence of Wnt or in the presence of low and high concentration in WT and APC-KO cells (Fig. 4 C). Global analysis showed that Wnt modulation had a strong effect on WT but not on APC-KO cells (Fig. 4 D), consistent with our previous results (Fig. S2 C). By differential gene expression analysis, we found that maximal and submaximal stimulation induced largely overlapping responses in WT cells (Fig. 4, E and F; R2 = 0.46). Induction of the Wnt-receptor signature but not of the APC-KO signature was observed at distinct levels of stimulation (Fig. 4 G) and is thus independent from the extent of differentiation. Because the experiments were performed in the presence of a constant R-spondin concentration, R-spondin in this system does not influence the specificity of response but rather acts as a facilitator of Wnt-signaling, as suggested before (Hao et al., 2012; Koo et al., 2012).
Figure 4.
Wnt responses after modulation of the extrinsic (A–E) and intrinsic (H–N) signaling level. (A) Growth of normal colon organoids after titration of Wnt-conditioned medium. In each passage, the mean ATP level was measured (± SD; n = 3 technical replicates; donor #3). Splitting factor was 1:5. Experiment was repeated twice with similar results. (B) Morphological images of organoids after culture at different Wnt concentrations. Scale bar is 1 mm. (C) Strategy to study maximal and submaximal Wnt stimulation. (D) PCA shows dose-dependent changes in WT cells and separate clustering from APC-KO cells (n = 3 technical replicates each). (E and F) Venn diagrams of regulated transcripts (± 1 log twofold change; P adjust < 0.05; E) and global correlation of transcriptomic responses after maximal and submaximal Wnt stimulation (F). (G) GSEA shows incremental induction of the Wnt-receptor signature at distinct levels of receptor stimulation in WT cells. (H) Schematic representation of the APC protein and truncated variants containing three or two 20-aa repeat regions (20AAR, blue). The mutation cluster region (MCR) is indicated. (I) WB analysis of APC (and ACTIN for normalization) in whole-cell lysates of WT and CRISPR/Cas9 induced clonal lines (donor #3). Black and white arrowheads show WT and truncated proteins, respectively. High frequency of a single mutant allele and absence of WT allele was measured by ICE assay. WB and ICE analyses were repeated twice independently. (J) Strategy to study the influence of different APC truncations. (K) PCA shows separate clustering of normal and APC-KO organoids (n = 3 each). Stimulation was performed as in Fig. 1. (L and M) Similar intrinsic response by APC variants with two and three 20AARs. Venn diagrams of regulated transcripts (L; ± 0.5 log twofold change; P adjust < 0.05) and global correlation of changes (M). (N) GSEA shows similar induction of the APC-KO signature by both allelic variants.
Next, we tested if distinct APC mutations influence the output of our intrinsic response. For this purpose, we generated further guide RNAs (gRNAs) targeting the mutation cluster region of the APC locus inducing C-terminal truncations that have been associated with distinct tumor locations and capacity of CTNNB1 down-regulation (Rosin-Arbesfeld et al., 2003; Christie et al., 2013; see Table S2 A). APC alleles with zero or one remaining 20-aa repeat regions (20AARs) could not be efficiently generated, indicating that short variants may compromise growth. Besides the APC variant with two 20AARs (studied above), we could generate variants with three remaining 20AARs. As technical replicates, three hemizygous clonal lines each were derived from donor #3 and confirmed by Sanger sequencing (interference of CRISPR edits [ICE] assay; Hsiau et al., 2018) and WB analysis (Fig. 4 I). RNA sequencing showed global similarity among the APC-KO organoids but pronounced differences from normal organoids that were largely shared between both allelic variants (Fig. 4, J–M; R2 = 0.62). Most importantly, the APC-KO signature remained strongly enriched independent from the level of truncation (Fig. 4 N). In contrast, the Wnt-receptor signature was unaffected in presence of two 20AARs or was even down-regulated indicating weaker activity of the three 20AAR allele.
Specific proteomic responses induced by physiological Wnt signaling and APC-KO
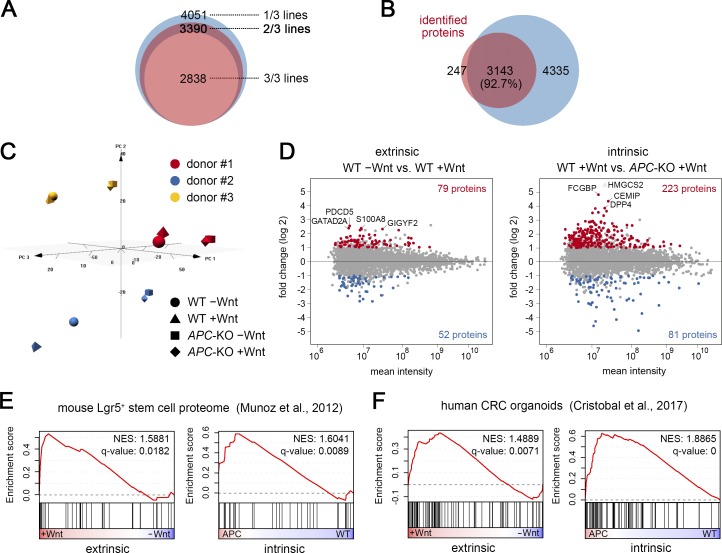
To investigate the changes on the proteomic level, we performed a label-free quantitative mass spectrometry (MS) approach. Protein lysates were prepared from all three donors after stimulation as above (WT and two 20AAR; see Fig. 1) and in total 4,051 proteins could be identified by at least two independent peptides. For subsequent analyses, we focused on the 3,390 proteins that were detected in at least two of three organoid lines (Fig. 5 A), a large majority of which overlapped with the described characterization of normal and CRC organoids (Fig. 5 B; Cristobal et al., 2017). Global data inspection again revealed a dominant line-specific variation (Figs. 5 C and S3 A). We performed pairwise differential analysis with a cutoff of P < 0.25 to filter for proteins that show common regulation between the lines. For extrinsic and intrinsic stimulation, we identified 79 and 223 proteins, respectively, that were greater than onefold (log 2) induced (Fig. 5 D). 14 and 308 proteins were induced after APC-KO and combined stimulation (Fig. S3 B). GSEA revealed a highly significant enrichment of protein signatures identified in CRC organoids (Cristobal et al., 2017) and in mouse Lgr5+ intestinal stem cells (Muñoz et al., 2012; Fig. 5, E and F). However, similar to the transcriptome, the previous signatures could not discriminate between intrinsic and extrinsic activation. We observed weak correlation between transcriptomic and proteomic changes (Fig. S3 C), indicating a strong impact of posttranscriptional regulation that has been noted before in human colon samples (Zhang et al., 2014; Cristobal et al., 2017). Gene ontology categories showed stronger correlation, indicating that the biological responses are preserved between RNA and protein (Fig. S3 D).
Figure 5.
Proteomic changes after extrinsic and intrinsic Wnt modulation. (A) Number of identified proteins (represented by at least two peptides) in one, two, or three organoid lines. (B) Venn diagram shows overlap of identified proteins (present in at least two of three lines) and the data from Cristobal et al. (2017). (C) PCA of proteomic data. Note that the three lines cluster separately. (D) Pairwise differential expression analysis. Up- and down-regulated proteins (± 1 log twofold change; P < 0.25) are marked in red and blue. (E and F) GSEA shows that previous proteome signatures in mouse Lgr5+ stem cells (E) and human CRC organoids (F) cannot discriminate between extrinsic and extrinsic Wnt responses. See also Fig. S3.
Consistent with our transcriptomic analysis, we observed two largely nonoverlapping responses after Wnt-receptor stimulation and APC-KO (Fig. 6, A and B). Hierarchical clustering of up-regulated proteins identified mutually exclusive responses (Fig. 6 C) and we defined two protein signatures by excluding proteins that were <0.25-fold (log 2) induced in the respective other condition. We obtained a 38-protein signature for Wnt-receptor signaling and 167 proteins upon APC loss (Table S1, C and D). Gene ontology analysis revealed significant association of the Wnt-receptor signature with processes such as autophagy and small GTPase signaling (“RAC signaling,” “HIPPO signaling”; Figs. 6 D and S3 E). In contrast, terms related to “nuclear receptor signaling” and “chondroitin/dermatan sulfate degradation” were found in the APC-KO signature (Figs. 6 D and S3 F). The most strongly enriched terms were associated with DNA damage and repair such as “mismatch repair” that were shared in both signatures, indicating that both common and divergent biological programs act downstream of intrinsic and extrinsic Wnt activation. To validate the mass spectrometric data, we performed WB analysis using organoid lysates and could confirm that the proteins CEMIP, CHDH, HMGCS2, PPIPK2, SCD, SMARCA5, and SRC were indeed consistently induced upon APC loss (Fig. 6 E).
Figure 6.
Specific protein signatures for normal and oncogenic Wnt signaling. (A) Venn diagrams show limited overlap of proteins up- or down-regulated after Wnt-receptor (extrinsic) stimulation and APC loss-of-function (intrinsic). Proteins were filtered for log twofold changes ± 1 and P < 0.25. (B) Global correlation shows independence of intrinsic and extrinsic responses. (C) Unsupervised clustering of up-regulated proteins marks distinct Wnt-receptor and APC-KO signatures. (D) Ingenuity pathway analysis. Significantly enriched gene ontology terms for Wnt-receptor and APC-KO protein signature are shown (red and blue bars) that were further grouped into biological categories. (E) WB validation of proteins. Lysates from normal and APC-KO organoids (n = 3 isogenic pairs) cultured in the presence of Wnt/R-spondin were probed. ACTIN was used for normalization. See also Fig. S3 and Table S1, C and D.
New biomarkers for human adenoma cells and normal colonic stem/progenitor cells
Next, we sought to determine the representation of our protein signatures within human tissues. We took advantage of the web-based Human Protein Atlas that contains well-annotated immunohistochemistry data for normal and tumor tissues (Uhlén et al., 2015). For the Wnt-receptor signature, we hypothesized a gradient-like expression in colonic crypts that is typical for Wnt/Stem cell markers (Jung et al., 2015). After excluding markers with absent or ubiquitous expression, we could identify a specific staining pattern for 11 of 38 proteins (Table S3 A), of which nine were crypt specific. From our APC-KO signature, we could identify a specific staining pattern for 36 of 167 proteins (Table S3 B). Here, we found 50% with pronounced expression in tumors and normal crypts, and a further 36% that were induced in a tumor-specific manner, confirming the in vivo relevance of our data.
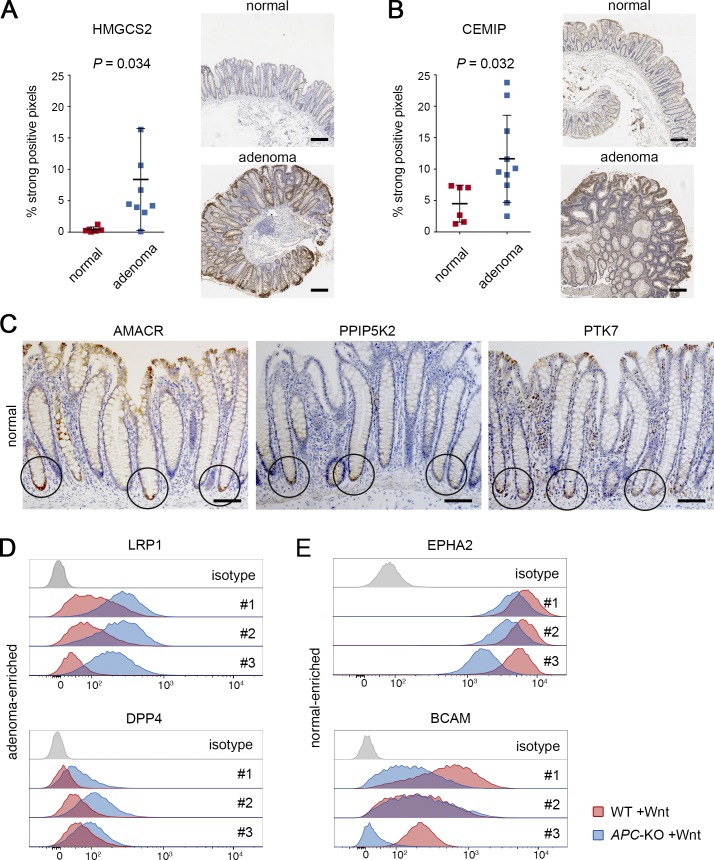
Because the CRC tissues contained in the Human Proteome Atlas likely have acquired numerous additional genetic hits, we collected material from early adenomas for further validation of our APC-KO protein signature. We included endoscopic biopsies of tubular adenomas (<1 cm, stage 1, n = 9–10) and found significant enrichment of HMGCS2 (Fig. 7 A) and CEMIP (Fig. 7 B) identifying these proteins as new biomarkers for human colon adenomas. Interestingly, we noted that PPIP5K2 and AMACR (a protein induced upon extrinsic and intrinsic stimulation) displayed highly specific expression at the bottom of normal crypts in a domain similar to the known stem cell marker PTK7 (Fig. 7 C).
Figure 7.
Validation of protein biomarkers for adenomas and normal stem cells. (A and B) Adenoma-specific expression of HMGCS2 (A) and CEMIP (B). Staining was performed on n = 6 normal tissues and n = 9–10 adenomas each. Expression was scored as percentage of strong pixels per total area, and mean percentage ± SD and P values (Student’s t test) are shown. Representative histological images of normal colon and adenoma tissue are shown next to each graph. Scale bars are 250 µm. (C) Immunodetection of AMACR, PPIP5K2, and the stem cell marker PTK7. Circles indicate increased expression at the bottom of colonic crypts. Scale bars are 100 µm. (D) FACS analysis shows increased surface expression of LRP1 and DPP4 in APC-KO cells (blue) compared with normal cells (red). Histogram plots in three isogenic organoid pairs that were cultured in Wnt/R-spondin containing medium. (E) Surface expression EPHA2 and BCAM shows reduced expression in APC-KO cells compared with normal cells. Experiments as in D. Stainings in A–E were independently reproduced at least twice. See also Fig. S4.
In addition, we distinguished a number of surface molecules in our protein signatures that could represent powerful new tools for identification and purification of tumor cells. To explore this possibility, we performed FACS analysis using Wnt-stimulated WT and APC-KO organoids and found that LRP1 and DPP4 showed consistent enrichment in all three APC-KO lines (Fig. 7 D). Of note, these markers could discriminate normal from APC-KO cells better than previously reported stem cell markers EPHB2 or PTK7 in this setting (Fig. S4 A). Furthermore, we identified the cell surface proteins EPHA2 and BCAM as specifically down-regulated in APC-KO cells (Fig. 7 E), which allows costaining strategies to improve separation of normal and adenoma cells by flow cytometry (Fig. S4 B). Together, these results show that in vitro profiling of isogenic organoids allows identification of highly specific markers for normal and adenoma cells that are conserved in human tissues.
Opposing prognostic value of Wnt-receptor and oncogenic signatures in CRC patients
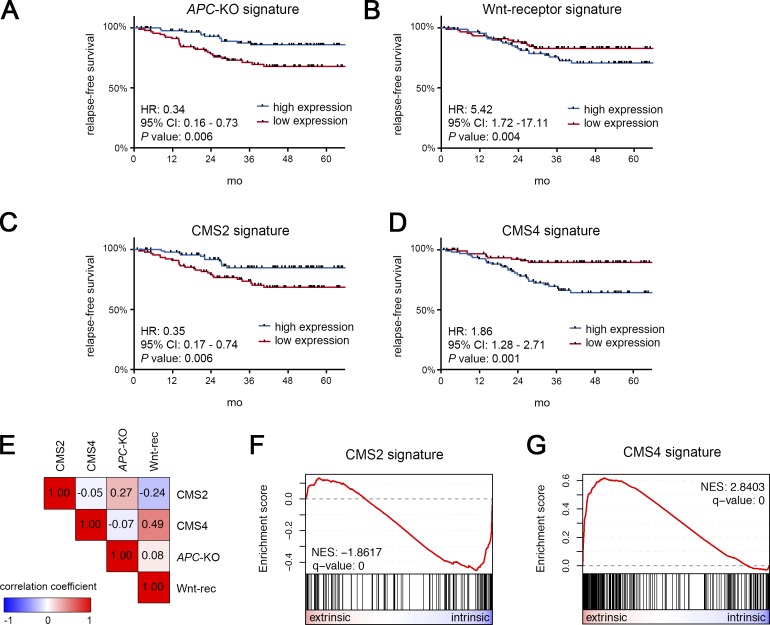
Subsequently, we sought to test if our organoid-derived signatures are associated with distinct clinical outcome in CRC. Previous transcriptome-based CRC classification has revealed active Wnt signaling in CMS2 tumors that have favorable prognosis (Guinney et al., 2015). To further differentiate this Wnt response in CRC, we performed Kaplan–Meier analysis after bifurcation according to the combined expression of the top 16 transcripts of each of our signatures (Table S1, A and B). In an age-, gender-, and stage-adjusted cohort (GSE14333; 187 cases), we found that tumors characterized by high expression of the APC-KO signature indeed displayed significantly prolonged relapse-free survival (Fig. 8 A). In contrast, high expression of the Wnt-receptor signature predicted poor outcome (Fig. 8 B), an association that was further validated in an independent cohort (GSE39582; 265 cases) for both relapse-free and overall survival (Fig. S5, A–D). We then studied published signatures for CMS2 and CMS4 tumors (Linnekamp et al., 2018) and found a similar, mutually exclusive association (Fig. 8, C and D; and Fig. S5, E–H). To determine the overlap between these expression-based classifications, we correlated the assignment of tumors to high and low expression groups. This analysis revealed that APC-KO and Wnt-receptor signatures indeed mark two sets of patients that correlate with CMS2 and CMS4 tumors (Fig. 8 E). This notion was further supported by GSEA of CMS2 and CMS4 gene signatures that displayed highly significant enrichment for the intrinsic and extrinsic responses, respectively (Fig. 8, F and G). Together, our results reveal that oncogenic and physiological Wnt responses represent two independent and opposing prognostic determinants in CRC.
Figure 8.
The Wnt-receptor signature is associated with poor prognosis and overlaps with CMS4 tumors. (A–D) Prognostic value of the APC-KO signature (A), the Wnt-receptor signature (B), and signatures for CMS2 (C) and CMS4 (D) tumors. Kaplan–Meier plots for relapse-free survival. For each signature, the expression cohort (GSE14333; 187 CRC cases; adjusted for age, gender, and stage) was divided into high expression (blue) and low expression (red) groups. Hazard ratios (HRs), 95% confidence intervals (CIs), and P values (log-rank test, multivariate analysis) are shown. (E) Pearson correlation of the individual tumor assignment to high/low expression groups in A–D. Note that CMS2/APC-KO signature and CMS4/Wnt-receptor signature mark two distinct groups of patients. (F and G) GSEA shows that CMS2 (F) and CMS4 genes (G) are strongly associated with the intrinsic and extrinsic Wnt responses in organoids, respectively. See also Fig. S5 and Table S5, A and B.
Discussion
Canonical Wnt signals govern homeostasis and tumorigenesis in the gut. Here, we report the first comprehensive profiling that stratifies between Wnt responses in normal human colon and adenoma cells. The usage of isogenic organoid pairs has allowed us to normalize for donor-specific heterogeneity and to identify novel tumor and putative stem cell markers that were validated in human tissues. The described strategy permits to better differentiate the signaling activities in human CRC and link them to distinct tumor and prognostic subtypes.
Organoids provide an accessible model to dissect signals induced by the extrinsic culture environment or intrinsic activation after genetic manipulation. By comparing normal and APC-KO cells in the presence of Wnt-containing medium, we could normalize the physiological stimulation to obtain an oncogene-specific signature. Importantly, we report that constitutive signaling induces distinct responses compared with receptor-mediated activation. Because APC is lost in the majority of CRC cases, proteins identified and validated in this study could represent a set of sensitive new biomarkers (Table S4). Among this list, several proteins have enzymatic activity and could serve as therapeutic targets, which should be addressed in further studies. We suggest that a similar profiling strategy in organoids could allow identification of tumor-specific traits for other oncogenic deregulations that commonly hijack signaling pathways critical for normal cell physiology (e.g., KRAS, PI3K, NOTCH).
Another important finding is the considerable heterogeneity between normal organoids from different donors that was observed both on the transcriptome and proteome level. Previous studies have shown separate clustering of normal/adenoma and carcinoma organoids (Matano et al., 2015; van de Wetering et al., 2015; Cristobal et al., 2017). However, this dissimilarity might lead to an underestimation of the actual variation among normal organoids that largely exceeded the effect of Wnt stimulation or APC loss. Genetic differences such as expression quantitative trait loci might account for this individual variability (Kilpinen et al., 2017). In addition, preserved expression of gut region-specific genes might be involved (Middendorp et al., 2014), although our analysis suggests to a minor extent (Fig. S2 A). Irrespective of the source of this individual variation, we conclude that bulk comparisons of tumor and normal samples have limited power to identify single gene/mutation effects. Paired analysis of engineered organoids can circumvent this problem and provide a sensitive approach to extract pathway or mutational signatures that are valid across a heterogeneous population.
We report two mutually exclusive Wnt signatures in normal and APC-KO cells, a distinction that was not feasible using previous signatures derived from either stem cells or primary adenomas. The most likely explanation for this lack of definition is the close association between Wnt signaling, stemness and tumorigenesis: mouse adenomatous crypts show increased β-catenin expression and consequently expand their stem cell compartment (Schepers et al., 2012). A tumor-normal comparison, thus, always includes direct and indirect effects. Likewise, differential comparison of stem cells and daughter cells detects alterations in other pathways (e.g., NOTCH), as well as the induction of differentiation genes. We aimed to normalize these confounding effects by extrinsic stimulation in order to extract the tumor-specific signaling component. The identified signatures were preserved after variation of the level of extrinsic and intrinsic stimulation (Fig. 4), indicating that the divergent responses are caused by qualitative rather than quantitative changes in Wnt signaling. In support, our signatures were differentially enriched in RNF43 and APC mutant CRC (Fig. S2, G and H), which are driven by upstream and downstream pathway activity. Alternatively, ligand-dependent activation may not activate the pathway to the same extent as truncation of APC. Remarkably, we could observe only limited gene expression changes in APC-KO cells after Wnt/R-spondin stimulation (Fig. S2 C), indicating that canonical responses are saturated and that noncanonical transcriptional Wnt responses are negligible in our experimental system. In addition, the multiple layers of negative feedback regulation (e.g., by AXIN2, NKD1, RNF43, DKKs) could cause the limited response in APC-KO cells. On the proteomic level, the changes in APC-KO cells were more prominent (Fig. S3 B), indicating that posttranslational regulation, e.g., protein stabilization by Wnt-STOP signaling, could be involved (Acebron et al., 2014) that is independent of nuclear CTNNB1 function. Differential phospho-proteomic analyses could provide further mechanistic insights in future. In our analyses, we have only found a few genes that were shared between intrinsic and extrinsic responses. Interestingly, these genes were enriched for known stem cell markers such as LGR5 and ASCL2 (Fig. 3 A). As an in vivo correlate, we have identified adenoma-induced proteins AMACR and PPIP5K2 that also showed highly crypt bottom-specific expression in the normal colon. We conclude that stem cell markers are either characterized by a particular dose sensitivity to Wnt, or dependent on additional signals that are induced in a secondary manner both upon Wnt stimulation and in adenomas.
CRC subtypes with high Wnt-activity have been previously associated with a favorable prognosis (de Sousa E Melo et al., 2011; De Sousa E Melo et al., 2013; Guinney et al., 2015). It is plausible that this indicates resemblance to benign adenomas that are driven by APC loss and that more malignant carcinomas gradually lose Wnt profiles. In contrast, other reports have associated intestinal stem cell profiles with high risk of relapse (Merlos-Suárez et al., 2011; Marisa et al., 2013; Sadanandam et al., 2013), where presence of Wnt-active tumor stem cells could reflect increased invasiveness (Brabletz et al., 2001) and therapy resistance. These two opposing concepts emphasize a need to better understand the biology that underlies the consensus CRC subtypes. We postulate that rather than Wnt activity per se, stratification of specific downstream responses is more informative on the tumor status. Association of our APC-KO signature with good prognosis and the canonical subtype (CMS2) is fully consistent with the dominant role of APC mutations in these cases (Guinney et al., 2015). However, we found that increased receptor-mediated signaling is linked to poor prognosis in CMS4 tumors that are also rich in mesenchymal stroma. Whether this activity represents cell-autonomous expression in tumor cells, e.g., induced by the microenvironment (Vermeulen et al., 2010), or activation in stromal cells remains to be addressed. It appears unlikely that this expression is caused by RNF43 deficiency, which is mainly found in microsatellite instable tumors (CMS1; Giannakis et al., 2014), or the more rare R-spondin translocations that also remain dependent on external Wnt signals (Storm et al., 2016). Yet, our findings may help to reconcile how the “stem-like” tumor subtype (described by Sadanandam et al., 2013) can combine malignancy with expression of specific Wnt targets. Future experiments should reveal if interference with receptor-mediated Wnt activity might allow the modulation of invasiveness.
Materials and methods
Patient samples
Normal human colon organoid lines were derived from nonpathological mucosa that was collected after prior written informed consent either during preemptive colonoscopy or from tumor-adjacent normal colon after tumor resection (Fig. S1 A). Additional organoid pairs from CRC and adjacent normal tissues were obtained from resected primary tumor tissues with written informed consent (Fig. S2 A). Tumor samples and patient data used in this study were provided by the University Cancer Center Frankfurt (UCT). Written informed consent was obtained from all patients, and the study was approved by the institutional review boards of the UCT and the Ethical Committee at the University Hospital Frankfurt (project-number: SGI-06-2015). Adenoma sections were collected from endoscopically resected benign colorectal polyps (T1/Dukes stage A). Only tubular adenomas and tubulovillous adenomas were included that were derived from nonhereditary cases and familial adenomatous polyposis patients. Hyperplasia and sessile (serrated) adenomas, as well as hereditary nonpolyposis CRC cases, were excluded. Normal control sections were prepared from pathologically normal tumor-adjacent mucosa (as above).
Organoid culture and transgenesis
Organoid cultures were established and maintained as described previously (Sato et al., 2011a). Human normal organoids were cultured in complete medium (advanced DMEM/F12 supplemented with 10 mM Hepes, 1× Glutamax, 1× penicillin/streptomycin, 2% B27, 1 mM nicotinamide, 12.5 mM N-acetylcysteine, 500 nM A83-01 [R&D Systems], 10 µM SB202190 [Sigma-Aldrich], 50% Wnt3a conditioned media [CM], 20% R-spondin 1 CM, 10% Noggin CM, 50 ng/ml human EGF [Peprotech], and 100 µg/ml Primocin [InvivoGen]). 10 µM of Y-27632 was added to the medium for the first 3 d after seeding. CM was prepared as described (Farin et al., 2012), and control CM was prepared in parallel from parental L-cells and Hek293T cells. The normal phenotype of all WT organoids was confirmed by growth arrest in medium lacking Wnt3a. APC-KO and CRC organoids were cultured in medium lacking Wnt3a and R-spondin. For stimulations, the organoids were first seeded in regular medium for 3 d before Wnt/R-spondin was either added (+Wnt) or left out (–Wnt) for an additional 2 d, before RNA/protein collection. For normalization, the –Wnt medium contained 50% L-cell control CM and 20% Hek293T CM. For Wnt-titration experiments, cells were maintained continuously in complete medium containing 0%, 5%, 10%, 20%, 35%, or 50% of Wnt3a CM, and L-cell control CM was added for normalization. Growth was quantified at the end of each passage using the CellTiter-Glo assay (Promega) as per manufacturer’s instructions, and RNA was collected at day 6 (passage 0) after seeding.
For CRISPR/Cas9 engineering, organoid lines were stably transduced with lentiCRISPR v2, a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA (Addgene plasmid 52961; Sanjana et al., 2014). Lentivirus production and transfection was performed as described (Koo et al., 2011). Cas9 expression was selected by addition of 0.5–1 µg/ml puromycin to the culture medium. APC loss-of-function mutations were introduced by transient transfection of the plasmid gRNA_GFP-T2, a gift from George Church, Harvard Medical School, Boston, MA (Addgene plasmid 41820; Mali et al., 2013) that contained the single-guide RNA sequences (Table S2 A). Transfection and functional selection for growth independence on Wnt/R-spondin was performed as described previously (Schwank et al., 2013). Clonal lines were expanded and confirmed by Sanger sequencing of multiple genomic amplicons (Fig. S1 B) or by ICE assay (Hsiau et al., 2018). Genotyping primers for the APC locus were one 20AAR, 5′-GCCACAGATATTCCTTCATCACAGA-3′ and 5′-TGGCAATCGAACGACTCTC-3′; two 20AAR, 5′-GTTCGATTGCCAGCTCCG-3′ and 5′-TCATTTTCCTGAACTGGAGGC-3′; and three 20AAR, 5′-CAAGCTGCAGTAAATGCTGCA-3′ and 5′-TGATGACTTTGTTGGCATGGC-3′.
RNA collection and RT-PCR
Total RNA was extracted using Macherey-Nagel NucleoSpin RNA kit according to the manufacturer’s instructions. For cDNA synthesis, random hexamers and M-MLV Reverse transcription enzyme (Promega) were used. Relative gene expression was measured using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) on a StepOne Real-Time PCR instrument (Applied Biosystems) and calculated using the ΔΔCT method by normalization to HPRT expression. Data were analyzed by Student’s t test. A list of used primers is shown in Table S2 B.
RNA sequencing and data analysis
RNA sequencing was performed using the TruSeq RNA 50 cycle kit and run on a Hiseq 2000 instrument (Illumina). Phred +33 Quality score encoding was used. 40,000,000 total reads per sample were measured. Quality of the data was monitored using FastQC Analysis. We performed quality score filtering, poly-A trimming, artifact removal, removal of N containing reads, and clearing of rRNA contamination using a pipeline provided by the HUSAR platform, German Cancer Research Center. Genomic mapping to human genome 38 was performed using TopHat2 (version 2.0.14). The number of reads per gene was determined using HTSeq count and the gencode annotations (release 24). Overlaps were handled as union. Further analysis and preparation of graphs was performed in R (versions 3.3.3 and 3.4.3) using RStudio (versions 1.0.136 and 1.1.423). We performed differential expression analysis using the DESeq2 tool (versions 1.12.4 and 1.18.1), and paired analysis was performed except in Fig. 4, where technical replicates were compared by unpaired analysis. Differential gene expression was considered significant if log twofold change was ≥1 or ≤−1 and the adjusted P value was <0.05. Data were annotated with HGNC symbols using biomaRt (version 2.28.0). For Wnt-receptor and APC-KO signatures (Table S1, A and B), only transcripts were included that were changed <0.2 log twofold in the respective other comparison. Hierarchical clustering was performed with the hclust function of the stats package (version 3.3.3) in R using Spearman correlation and average as agglomeration setting. The heatmap.2 function from gplots (version 3.0.1) was used for heat map visualization. For pairwise correlation matrices, the R dist function from the stats package (version 3.3.3) was used for distance matrix computation, and results were visualized with the help of the pheatmap package (version 1.0.8). PCAs were computed from all IDs if not specified elsewise with the prcomp function, and the pca3d package (version 0.10) was used for visualization.
Protein collection and WB analysis
Organoids were collected in cold medium and mechanically separated from the Matrigel by pipetting several times. The organoids were centrifuged at 1,200 rpm for 5 min and resuspended in cold medium, and this process was repeated at least twice to dissolve the Matrigel. Cells were lysed using modified RIPA lysis buffer (150 mM NaCl, 50 mM Tris HCl, pH 7.6, 5 mM NaF, 5 mM β-glycerophosphate, 1 mM EDTA, 1 mM Na-orthovanadate, 1% Na-deoxycholate, and 1% NP-40) containing complete protease and phosphatase inhibitors (both from Roche). Protein content was quantified using standard Bradford assay. For WB analysis, 15 µg of protein was loaded per lane and transferred to a nitrocellulose membrane and probed with the antibodies described in Table S2 C.
Liquid chomatography–MS measurements
Protein samples were processed as described by Shevchenko et al. (1996). The peptides were resuspended in sample loading buffer (2% acetonitrile and 0.1% TFA) and then fractionated and analyzed by an online UltiMate 3000 RSLCnano HPLC system (Thermo Fisher Scientific) coupled online to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). First, the peptides were desalted on a reverse phase C18 precolumn (5 × 0.3 mm diameter). After 3 min, the precolumn was switched online with the analytical column (30 cm long, 75 µm inner diameter) prepared in-house using ReproSil-Pur C18 AQ 1.9 µm reversed phase resin (Dr. Maisch GmbH). The peptides were separated with a linear gradient of 5–35% buffer (80% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl/min (with back pressure of 500 bars) over 90-min gradient time. The precolumn and column temperature were set to 50°C during the chromatography. The MS data were acquired by scanning the precursors in a mass range from 350 to 1,600 m/z at a resolution of 70,000 at m/z 200. The top 20 precursor ions were chosen for MS2 by using data-dependent acquisition mode at a resolution of 17,500 at m/z 200 with maximum ion injection time of 50 ms.
MaxQuant search
The MS raw files were processed by MaxQuant (Cox and Mann, 2008; version 1.5.2.8), and MS/MS spectra were searched against UniProt human database (155,990 entries) via the Andromeda search. Mass tolerance after recalibration of precursor mass and fragment ion mass were set as 6 and 20 ppm; trypsin was selected as protease. Allowed variable modifications included protein deamidation (N) and oxidation (M). Cysteine carbamidomethylation was defined as a fixed modification. Minimal peptide length was set to 7 aa with the maximum of two enzymatic missed-cleavages. Minimum quantification radio of 2 was chosen. The false discovery rate was set to 1% for both peptide and protein identifications. MaxLFQ was chosen as algorithm for quantification.
After MaxQuant analysis, data were analyzed using Perseus software (version 1.5.5.3; MPI for Biochemistry). Reverse hits and hits identified only by side and potential contaminants were removed, and label-free quantification values from MaxQuant were log 2 transformed. Proteins were included in the analysis if they were found in at least two of three patients and if more than one peptide was found in at least one patient. Imputation of remaining missing values was performed from a normal distribution (width 0.3, down shift 1.8). We used a two-sample t test with a permutation-based false discovery rate of 0.05 to identify proteins with a significantly different abundance between the different comparisons. We performed paired differential analysis in Perseus and considered all proteins as regulated if the log twofold changes were ≥1 or ≤−1 and the P value was ≤0.25. Data were annotated with gene symbols using the annotation function included in Perseus. R (version 3.4.0) and RStudio (version 1.0.143) were used for data visualization. For Wnt-receptor and APC-KO signatures (Table S1, C and D) only proteins were included that were changed <0.25 log twofold in the respective other comparison. Hierarchical clustering, pairwise correlation, and PCA were performed in R as described above. Identified proteins were compared with the quantified proteome data from Cristobal et al. (2017) (refer to supplemental material therein). For protein RNA comparison, the transcript IDs were converted in Perseus and graphs were plotted in R.
Enrichment analysis and functional annotation
GSEA was performed using the preranked tool (version 2.2.3; Broad Institute; Subramanian et al., 2005). Data were visualized using the replotGSEA function from the Rtoolbox package. We generated ranked list files for extrinsic (WT –Wnt vs. WT +Wnt) and intrinsic (WT +Wnt vs. APC-KO +Wnt) regulation for the RNA sequencing and the proteome data. Additionally, we created a differential ranked list file from the intrinsic versus extrinsic RNA sequencing data by subtraction of both log twofold change values. RNA sequencing data were analyzed using the following gene sets. For the EPHB2 human intestinal stem signature, we used the supplementary table in Jung et al. (2011) and filtered all genes with an average fold change of >2 in the category “all samples high vs. med” and which were marked as “TRUE” in the high versus med statistics. Duplicated gene names were removed, resulting in a set of 108 genes. For mouse Lgr5 intestinal stem cells, we used the mRNA signature from Muñoz et al. (2012) (supplemental material therein), which contains 384 genes. The human adenoma signature was derived using a publicly available dataset (Gene Expression Omnibus [GEO] accession no. GSE80981; Okuchi et al., 2016), and differential expression of control and adenoma genes was performed in GEO2R (National Center for Biotechnology Information) using the accession nos. GSM2139717, GSM2139718, and GSM2139719 (control) and GSM2139723, GSM2139723, and GSM2139725 (adenomas). The list was filtered for genes with a log twofold change of >1, resulting in a 225-gene signature. The mouse adenoma signature was published previously (Sansom et al., 2007; also refer to MSigDB signature: SANSOM_APC_TARGETS_UP, containing 126 genes). For CMS gene sets, we classified the publicly available gene expression dataset GSE39582, which was also used in the original publication from Guinney et al. (2015). Frozen robust multiarray analysis–normalized data were downloaded from the Colorectal Cancer Subtyping Consortium’s Synapse platform. Gene symbols were converted to Entrez IDs using the R package biomaRt and classified by random forest analysis provided by the R package CMS classifier (Guinney et al., 2015). We identified 30 high-confidence cases for each subtype by filtering for a posterior probability value of >50% in one CMS and <20% in the other three CMSs (Table S5 A). For differential analysis, the identified cases were compared with all cases of the three other subtypes using the GEO2R tool. Genes with significantly induced expression (log twofold change >1 and adjusted P < 0.05; 341 for CMS1, 143 for CMS2, 245 for CMS3, and 681 for CMS4; see Table S5 B) were used to generate .grp files. For differential expression analysis of APC- and RNF43-deficient tumors, the mutation status was accessed from public data using cBioPortal for cancer genomics (v.1.18.0). The datasets Colorectal Adenocarcinoma (TCGA, Provisional; coadread_tcga) and Colon Adenocarcinoma (TCGA, PanCancer Atlas; coad_tcga_pan_cancer_atlas_2018) were used. Only colonic samples were included, and all 29 available samples with RNF43 mutations (and no APC mutations) and 30 randomly chosen samples with APC mutations (and no RNF43 or ZNRF3 mutations) were used (see Table S5 C). HT-Seq–derived read count files were downloaded from the NIH GDC data portal, and differential analysis was performed using the DESeq2 tool as described above.
GSEA for the proteome data were performed using the following signatures: 200 proteins with log twofold change >1 from Muñoz et al. (2012) (refer to supplemental material therein) and all 199 significant proteins in stem cells reported by Cristobal et al. (2017) (refer to supplemental material therein).
For global pairwise correlation of the GSEA and protein/RNA expression, we first performed enrichment analysis using the KEGG and REACTOME gene sets from the curated gene sets (C2) of the Molecular Signatures Database (MSigDB database v6.1; Broad Institute). All 307 normalized enrichment score (NES) values and 3,247 RNA/protein fold changes were included that were identified in both protein and RNA data, and the values were analyzed by Pearson correlation (average linkage) using the Morpheus tool (Broad Institute). Ingenuity Pathway Analysis software (version 01-07; Qiagen; default settings) was applied for gene ontology analysis of the protein signatures, and a P value of <0.05 was considered significant.
Immunohistochemistry
Formalin fixed tissues were stored in 70% ethanol or directly embedded in paraffin following standard procedures. 3-µm sections were generated, and immunostaining was performed on a Leica Bond-Max, using the Bond Polymer Refine Detection System (Leica). The antibodies and staining conditions are described in Table S4 C. Following embedding, the sections were documented on a ScanScopeCS2 scanner equipped with a 20× objective (Olympus UPLSAPO; numerical aperture 0.75). Using Aperio eSlide Manager software (v12.3; Leica), scanned sections were analyzed, submucosal areas were excluded, and the number of positive pixels was quantified in the mucosal compartment using the pixel count algorithm. From the software output, the expression was scored as percentage of strong positive pixels per total pixels (including negative and positive pixels). Data were analyzed by Student’s t test.
FACS analysis
Organoids were collected in cold medium, centrifuged at 1,200 rpm for 5 min, and dissociated by incubation for 5 min with Accutase (Life Technologies; A1110501) at 37°C. Single cells were filtered through a 0.45-µm filter, centrifuged at 1,500 rpm for 5 min, and resuspended in FACS buffer (PBS, 2% FBS, and 2 mM EDTA). 1 million cells per 300 µl were incubated with antibody or isotype control for 20 min protected from light on ice. NucBlue Fixed cell (Invitrogen) was added for live cell gating. Flow cytometry was performed on BD Fortessa, and the results were analyzed using FlowJo v10 software. The list of antibodies and respective isotype controls is shown in Table S2 C.
Kaplan–Meier survival analysis
The PROGgeneV2 tool (Goswami and Nakshatri, 2014) was used to analyze combined expression in the precomputed GEO datasets GSE14333 and GSE39582. The cohorts were adjusted for all available covariates and bifurcated based on median expression. The hazard ratios, 95% confidence intervals, and statistical analysis using log-rank test were retrieved from the program. The 16 most up-regulated transcripts were used for the Wnt-receptor and APC-KO signatures, respectively. Validity of the signatures was confirmed by variation of the input gene sizes to 8, 12, or 20 genes, which yielded similar results. For CMS2 and CMS4, the previously described signatures of 25 genes each were used (Linnekamp et al., 2018; refer to supplement therein). To link case assignment to high and low expression groups for all four signatures, Pearson correlation by average linkage was performed using the Morpheus tool (Broad Institute).
Data availability
The following datasets from the GEO (National Center for Biotechnology Information) were used in this study: accession nos. GSE14333 (Jorissen et al., 2009), GSE39582 (Marisa et al., 2013), and GSE80981 (Okuchi et al., 2016). Gene expression datasets generated in this study are accessible under accession nos. GSE125472 and GSE125578. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD012650.
Online supplemental material
Fig. S1 provides information on the CRISPR/Cas9 engineered organoids, including Sanger sequencing and WB analysis. Fig. S2 shows detailed characterization of the transcriptomic data and comparison to colon cancer samples. Fig. S3 shows detailed characterization of the proteomic data, including correlation of RNA and protein changes and gene ontology analysis. Fig. S4 shows analysis of EPHB2, PTK7, LRP1/EPHA2, and DPP4/EPHA2 surface expression in normal and APC-KO organoids. Fig. S5 shows relapse-free survival and overall survival for APC-KO and Wnt-receptor signatures in expression cohort GSE39582. Table S1 lists the protein- and RNA-based Wnt signatures. Table S2 provides all gRNAs, qPCR primers, and antibodies used. Table S3 lists the tissue expression of protein signatures in the Human Protein Atlas. Table S4 summarizes literature information of all validated proteins, and Table S5 provides information on the CMS classification and tumor subtype-specific RNA signatures.
Supplementary Material
Acknowledgments
We thank Petra Dinse for help with histology; Agnes Hotz-Wagenblatt and Ingo Ebersberger for bioinformatic support; Chirayu Goswami for help with Kaplan–Meier analysis and support with the ProgGENEV2 tool; and Uwe Plessmann and Björn Häupl for MS analyses. We thank the High Throughput Sequencing Unit of the Genomics & Proteomics Core Facility, German Cancer Research Center, for providing excellent services. The UCT biobank is acknowledged for support with collection of patient tissues, and Bon-Kyoung Koo and Florian Greten for critical comments on the manuscript. Cell lines for preparation of conditioned media were a kind gift from Hans Clevers (Hubrecht Institute, Utrecht, Netherlands) and Calvin Kuo (Stanford University, Stanford, CA). We thank Andreas Bosio, David Agorku, and Olaf Hardt from Miltenyi Biotec for kind support with FACS antibodies and Angela Brieger for sharing additional antibodies.
H.F. Farin has been supported by German Cancer Consortium junior researcher group funding and by the LOEWE Center Frankfurt Cancer Institute funded by the Hessen State Ministry for Higher Education, Research and the Arts (III L 5 - 519/03/03.001 - (0015)).
The authors declare no competing financial interests.
Author contributions: B.E. Michels, M.H. Mosa, B.M. Grebbin, and D. Yepes performed the experiments and analyzed the results. Bioinformatic analysis was done by B.E. Michels. D. Yepes and T. Oellerich performed mass spectrometric analysis. H. Urlaub supervised MS analyses. T. Darvishi provided technical support. H.F. Farin conceived the study and supervised the experiments. H.F. Farin, B.E. Michels, and M.H. Mosa drafted the manuscript. J. Hausmann, S. Zeuzem, and H.M. Kvasnicka provided biomaterial. H.F. Farin obtained funding. All authors discussed the results and commented on the manuscript.
References
- Acebron S.P., Karaulanov E., Berger B.S., Huang Y.-L., and Niehrs C.. 2014. Mitotic wnt signaling promotes protein stabilization and regulates cell size. Mol. Cell. 54:663–674. 10.1016/j.molcel.2014.04.014 [DOI] [PubMed] [Google Scholar]
- Aoki R., Shoshkes-Carmel M., Gao N., Shin S., May C.L., Golson M.L., Zahm A.M., Ray M., Wiser C.L., Wright C.V.E., and Kaestner K.H.. 2016. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell. Mol. Gastroenterol. Hepatol. 2:175–188. 10.1016/j.jcmgh.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V., et al. 2014. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 158:157–170. 10.1016/j.cell.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., and Clevers H.. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Billmann M., Chaudhary V., ElMaghraby M.F., Fischer B., and Boutros M.. 2018. Widespread rewiring of genetic networks upon cancer signaling pathway activation. Cell Syst. 6:52–64.e4. 10.1016/j.cels.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T., Jung A., Reu S., Porzner M., Hlubek F., Kunz-Schughart L.A., Knuechel R., and Kirchner T.. 2001. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA. 98:10356–10361. 10.1073/pnas.171610498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487:330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie M., Jorissen R.N., Mouradov D., Sakthianandeswaren A., Li S., Day F., Tsui C., Lipton L., Desai J., Jones I.T., et al. 2013. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/β-catenin signalling thresholds for tumourigenesis. Oncogene. 32:4675–4682. 10.1038/onc.2012.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Loh K.M., and Nusse R.. 2014. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 346:1248012 10.1126/science.1248012 [DOI] [PubMed] [Google Scholar]
- Cortina C., Turon G., Stork D., Hernando-Momblona X., Sevillano M., Aguilera M., Tosi S., Merlos-Suárez A., Stephan-Otto Attolini C., Sancho E., and Batlle E.. 2017. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol. Med. 9:869–879. 10.15252/emmm.201707550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cristobal A., van den Toorn H.W.P., van de Wetering M., Clevers H., Heck A.J.R., and Mohammed S.. 2017. Personalized Proteome Profiles of Healthy and Tumor Human Colon Organoids Reveal Both Individual Diversity and Basic Features of Colorectal Cancer. Cell Reports. 18:263–274. 10.1016/j.celrep.2016.12.016 [DOI] [PubMed] [Google Scholar]
- de Sousa E Melo F., Colak S., Buikhuisen J., Koster J., Cameron K., de Jong J.H., Tuynman J.B., Prasetyanti P.R., Fessler E., van den Bergh S.P., et al. 2011. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 9:476–485. 10.1016/j.stem.2011.10.008 [DOI] [PubMed] [Google Scholar]
- De Sousa E Melo F., Wang X., Jansen M., Fessler E., Trinh A., de Rooij L.P., de Jong J.H., de Boer O.J., van Leersum R., Bijlsma M.F., et al. 2013. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 19:614–618. 10.1038/nm.3174 [DOI] [PubMed] [Google Scholar]
- Dow L.E., O’Rourke K.P., Simon J., Tschaharganeh D.F., van Es J.H., Clevers H., and Lowe S.W.. 2015. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell. 161:1539–1552. 10.1016/j.cell.2015.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., van Jaarsveld R.H., Ponsioen B., Zimberlin C., van Boxtel R., Buijs A., Sachs N., Overmeer R.M., Offerhaus G.J., Begthel H., et al. 2015. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 521:43–47. 10.1038/nature14415 [DOI] [PubMed] [Google Scholar]
- Faller W.J., Jackson T.J., Knight J.R., Ridgway R.A., Jamieson T., Karim S.A., Jones C., Radulescu S., Huels D.J., Myant K.B., et al. 2015. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 517:497–500. 10.1038/nature13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin H.F., Van Es J.H., and Clevers H.. 2012. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 143:1518–1529.e7. 10.1053/j.gastro.2012.08.031 [DOI] [PubMed] [Google Scholar]
- Giannakis M., Hodis E., Jasmine Mu X., Yamauchi M., Rosenbluh J., Cibulskis K., Saksena G., Lawrence M.S., Qian Z.R., Nishihara R., et al. 2014. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 46:1264–1266. 10.1038/ng.3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami C.P., and Nakshatri H.. 2014. PROGgeneV2: enhancements on the existing database. BMC Cancer. 14:970 10.1186/1471-2407-14-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J., Dienstmann R., Wang X., de Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. 2015. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21:1350–1356. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney A., Axelrod F., Bond C.J., Cain J., Chartier C., Donigan L., Fischer M., Chaudhari A., Ji M., Kapoun A.M., et al. 2012. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc. Natl. Acad. Sci. USA. 109:11717–11722. 10.1073/pnas.1120068109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H.-X., Xie Y., Zhang Y., Charlat O., Oster E., Avello M., Lei H., Mickanin C., Liu D., Ruffner H., et al. 2012. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 485:195–200. 10.1038/nature11019 [DOI] [PubMed] [Google Scholar]
- Hsiau T., Maures T., Waite K., Yang J., Kelso R., Holden K., and Stoner R.. 2018. Inference of CRISPR Edits from Sanger Trace Data. bioRxiv. 251082. [DOI] [PubMed] [Google Scholar]
- Jorissen R.N., Gibbs P., Christie M., Prakash S., Lipton L., Desai J., Kerr D., Aaltonen L.A., Arango D., Kruhøffer M., et al. 2009. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin. Cancer Res. 15:7642–7651. 10.1158/1078-0432.CCR-09-1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P., Sato T., Merlos-Suárez A., Barriga F.M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M.A., Sancho E., et al. 2011. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 17:1225–1227. 10.1038/nm.2470 [DOI] [PubMed] [Google Scholar]
- Jung P., Sommer C., Barriga F.M., Buczacki S.J., Hernando-Momblona X., Sevillano M., Duran-Frigola M., Aloy P., Selbach M., Winton D.J., and Batlle E.. 2015. Isolation of Human Colon Stem Cells Using Surface Expression of PTK7. Stem Cell Reports. 5:979–987. 10.1016/j.stemcr.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabiri Z., Greicius G., Madan B., Biechele S., Zhong Z., Zaribafzadeh H., Edison J., Aliyev J., Wu Y., Bunte R., et al. 2014. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 141:2206–2215. 10.1242/dev.104976 [DOI] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J., et al. 2017. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 546:370–375. 10.1038/nature22403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.-K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., and Clevers H.. 2011. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 9:81–83. 10.1038/nmeth.1802 [DOI] [PubMed] [Google Scholar]
- Koo B.-K., Spit M., Jordens I., Low T.Y., Stange D.E., van de Wetering M., van Es J.H., Mohammed S., Heck A.J.R., Maurice M.M., and Clevers H.. 2012. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 488:665–669. 10.1038/nature11308 [DOI] [PubMed] [Google Scholar]
- Koo B.-K., van Es J.H., van den Born M., and Clevers H.. 2015. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc. Natl. Acad. Sci. USA. 112:7548–7550. 10.1073/pnas.1508113112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar S., Morrissey E., Nicholson A.M., van der Heijden M., Zecchini H.I., Kemp R., Tavaré S., Vermeulen L., and Winton D.J.. 2013. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell. 13:626–633. 10.1016/j.stem.2013.08.001 [DOI] [PubMed] [Google Scholar]
- Lau T., Chan E., Callow M., Waaler J., Boggs J., Blake R.A., Magnuson S., Sambrone A., Schutten M., Firestein R., et al. 2013. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 73:3132–3144. 10.1158/0008-5472.CAN-12-4562 [DOI] [PubMed] [Google Scholar]
- Linnekamp J.F., Hooff S.R.V., Prasetyanti P.R., Kandimalla R., Buikhuisen J.Y., Fessler E., Ramesh P., Lee K.A.S.T., Bochove G.G.W., de Jong J.H., et al. 2018. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ. 25:616–633. 10.1038/s41418-017-0011-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisa L., de Reyniès A., Duval A., Selves J., Gaub M.P., Vescovo L., Etienne-Grimaldi M.-C., Schiappa R., Guenot D., Ayadi M., et al. 2013. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 10:e1001453 10.1371/journal.pmed.1001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., and Sato T.. 2015. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21:256–262. 10.1038/nm.3802 [DOI] [PubMed] [Google Scholar]
- Merlos-Suárez A., Barriga F.M., Jung P., Iglesias M., Céspedes M.V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P., et al. 2011. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 8:511–524. 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Middendorp S., Schneeberger K., Wiegerinck C.L., Mokry M., Akkerman R.D.L., van Wijngaarden S., Clevers H., and Nieuwenhuis E.E.S.. 2014. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. 32:1083–1091. 10.1002/stem.1655 [DOI] [PubMed] [Google Scholar]
- Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.-K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S., et al. 2012. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4′ cell markers. EMBO J. 31:3079–3091. 10.1038/emboj.2012.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant K.B., Cammareri P., McGhee E.J., Ridgway R.A., Huels D.J., Cordero J.B., Schwitalla S., Kalna G., Ogg E.-L., Athineos D., et al. 2013. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 12:761–773. 10.1016/j.stem.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novellasdemunt L., Antas P., and Li V.S.W.. 2015. Targeting Wnt signaling in colorectal cancer. A Review in the Theme: Cell Signaling: Proteins, Pathways and Mechanisms. Am. J. Physiol. Cell Physiol. 309:C511–C521. 10.1152/ajpcell.00117.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke K.P., Loizou E., Livshits G., Schatoff E.M., Baslan T., Manchado E., Simon J., Romesser P.B., Leach B., Han T., et al. 2017. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 35:577–582. 10.1038/nbt.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuchi Y., Imajo M., Mizuno R., Kamioka Y., Miyoshi H., Taketo M.M., Nagayama S., Sakai Y., and Matsuda M.. 2016. Identification of Aging-Associated Gene Expression Signatures That Precede Intestinal Tumorigenesis. PLoS One. 11:e0162300 10.1371/journal.pone.0162300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47(D1):D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phesse T.J., Buchert M., Stuart E., Flanagan D.J., Faux M., Afshar-Sterle S., Walker F., Zhang H.-H., Nowell C.J., Jorissen R., et al. 2014. Partial inhibition of gp130-Jak-Stat3 signaling prevents Wnt-β-catenin-mediated intestinal tumor growth and regeneration. Sci. Signal. 7:ra92 10.1126/scisignal.2005411 [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R., Cliffe A., Brabletz T., and Bienz M.. 2003. Nuclear export of the APC tumour suppressor controls β-catenin function in transcription. EMBO J. 22:1101–1113. 10.1093/emboj/cdg105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandam A., Lyssiotis C.A., Homicsko K., Collisson E.A., Gibb W.J., Wullschleger S., Ostos L.C.G., Lannon W.A., Grotzinger C., Del Rio M., et al. 2013. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 19:619–625. 10.1038/nm.3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom O.J., Meniel V.S., Muncan V., Phesse T.J., Wilkins J.A., Reed K.R., Vass J.K., Athineos D., Clevers H., and Clarke A.R.. 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature. 446:676–679. 10.1038/nature05674 [DOI] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G.J., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., and Clevers H.. 2011a Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141:1762–1772. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., and Clevers H.. 2011b Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 469:415–418. 10.1038/nature09637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A.G., Snippert H.J., Stange D.E., van den Born M., van Es J.H., van de Wetering M., and Clevers H.. 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 337:730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K., et al. 2013. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13:653–658. 10.1016/j.stem.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Jensen O.N., Podtelejnikov A.V., Neubauer G., Shevchenko A., Mortensen P., and Mann M.. 1996. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24:893–896. 10.1042/bst0240893 [DOI] [PubMed] [Google Scholar]
- Shimokawa M., Ohta Y., Nishikori S., Matano M., Takano A., Fujii M., Date S., Sugimoto S., Kanai T., and Sato T.. 2017. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature. 545:187–192. 10.1038/nature22081 [DOI] [PubMed] [Google Scholar]
- Stamos J.L., and Weis W.I.. 2013. The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5:a007898 10.1101/cshperspect.a007898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange D.E., Koo B.-K., Huch M., Sibbel G., Basak O., Lyubimova A., Kujala P., Bartfeld S., Koster J., Geahlen J.H., et al. 2013. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 155:357–368. 10.1016/j.cell.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm E.E., Durinck S., de Sousa e Melo F., Tremayne J., Kljavin N., Tan C., Ye X., Chiu C., Pham T., Hongo J.A., et al. 2016. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature. 529:97–100. 10.1038/nature16466 [DOI] [PubMed] [Google Scholar]
- Stzepourginski I., Nigro G., Jacob J.-M., Dulauroy S., Sansonetti P.J., Eberl G., and Peduto L.. 2017. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA. 114:E506–E513. 10.1073/pnas.1620059114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347:1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Valenta T., Degirmenci B., Moor A.E., Herr P., Zimmerli D., Moor M.B., Hausmann G., Cantù C., Aguet M., and Basler K.. 2016. Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Reports. 15:911–918. 10.1016/j.celrep.2016.03.088 [DOI] [PubMed] [Google Scholar]
- Van der Flier L.G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M.E., Suijkerbuijk S., Van de Wetering M., Marra G., and Clevers H.. 2007. The Intestinal Wnt/TCF Signature. Gastroenterology. 132:628–632. 10.1053/j.gastro.2006.08.039 [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A.P., et al. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 111:241–250. 10.1016/S0092-8674(02)01014-0 [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L., et al. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161:933–945. 10.1016/j.cell.2015.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lidth de Jeude J.F., Meijer B.J., Wielenga M.C.B., Spaan C.N., Baan B., Rosekrans S.L., Meisner S., Shen Y.H., Lee A.S., Paton J.C., et al. 2017. Induction of endoplasmic reticulum stress by deletion of Grp78 depletes Apc mutant intestinal epithelial stem cells. Oncogene. 36:3397–3405. 10.1038/onc.2016.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., De Sousa E Melo F., van der Heijden M., Cameron K., de Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H., et al. 2010. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12:468–476. 10.1038/ncb2048 [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M., Nakamura Y., White R., Smits A.M., and Bos J.L.. 1988. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319:525–532. 10.1056/NEJM198809013190901 [DOI] [PubMed] [Google Scholar]
- Zhan T., Rindtorff N., and Boutros M.. 2017. Wnt signaling in cancer. Oncogene. 36:1461–1473. 10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M.C., Zimmerman L.J., Shaddox K.F., Kim S., et al. NCI CPTAC . 2014. Proteogenomic characterization of human colon and rectal cancer. Nature. 513:382–387. 10.1038/nature13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following datasets from the GEO (National Center for Biotechnology Information) were used in this study: accession nos. GSE14333 (Jorissen et al., 2009), GSE39582 (Marisa et al., 2013), and GSE80981 (Okuchi et al., 2016). Gene expression datasets generated in this study are accessible under accession nos. GSE125472 and GSE125578. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD012650.