The role of guanylate-binding proteins in host defense.
Abstract
Guanylate-binding proteins (GBPs) have recently emerged as central orchestrators of immunity to infection, inflammation, and neoplastic diseases. Within numerous host cell types, these IFN-induced GTPases assemble into large nanomachines that execute distinct host defense activities against a wide variety of microbial pathogens. In addition, GBPs customize inflammasome responses to bacterial infection and sepsis, where they act as critical rheostats to amplify innate immunity and regulate tissue damage. Similar functions are becoming evident for metabolic inflammatory syndromes and cancer, further underscoring the importance of GBPs within infectious as well as altered homeostatic settings. A better understanding of the basic biology of these IFN-induced GTPases could thus benefit clinical approaches to a wide spectrum of important human diseases.
Introduction
No cell is an island (Weiss, 1971). Indeed, the vertebrate immune system—with its reliance on intercellular communication and functional interdependence—epitomizes this principle. Yet recent work with single-cell transcriptomics, proteomics, and mathematical modeling has revealed that most nucleated cells also possess a deeply interconnected responsivity of their own (Regev et al., 2017; Stubbington et al., 2017). This network often galvanizes hundreds of host defense proteins to guard diverse tissues or portals of entry, and operates on a nanoscopic rather than a macroscopic scale (MacMicking, 2012).
The ability of individual cells to marshal such broad-based defense programs is known as cell-autonomous immunity (Beutler et al., 2006; Randow et al., 2013). In addition to providing a barrier to infection, cell-autonomous immunity integrates homeostatic signals to ensure the maintenance of key housekeeping functions and genome integrity. Why is such integration necessary? Because as organismal lifespan increases, recurring threats to cellular homeostasis accumulate for the host (López-Otín et al., 2013). Here, repeated encounters with microbial pathogens or nonmicrobial allergens, toxins, venoms, and other xenobiotics represent external threats (Palm et al., 2012; Randow et al., 2013), whereas intrinsic challenges arise from endogenous retroelements that promote oncogenesis or from metabolic disturbances that elicit inflammatory sequelae (Kassiotis and Stoye, 2016; O’Neill et al., 2016; Hotamisligil, 2017). Each of these challenges is closely monitored by the innate and adaptive immune systems to restrict infection or cellular transformation and to mobilize repair mechanisms that help refurbish damaged tissue.
Among the most effective containment strategies used by longer-lived vertebrates are those elicited by the IFN family of cytokines. IFNs drive complex homeostatic defense networks comprising several hundred IFN-stimulated genes (ISGs; MacMicking, 2012; Schneider et al., 2014). Some ISGs exhibit direct microbicidal activity while others influence neoplastic progression through their antiproliferative or immune-editing effects (MacMicking, 2012; Schneider et al., 2014; Parker et al., 2016; Nirschl et al., 2017). Recent work has identified a family of IFN-inducible GTPases—the 65–73 kD guanylate-binding proteins (GBPs)—as a major nexus of this defensive repertoire (Kim et al., 2012, 2016). GBPs confer protectant functions against phylogenetically diverse pathogens and cooperate with the core inflammasome machinery to orchestrate pyrogenic cytokine production and pyroptosis. These activities impact localized microbial encounters as well as systemic derangements, for example during Gram-negative sepsis (Finethy et al., 2017). They also extend to microbiota-driven inflammatory diseases and cancer.
This review examines the newly emerging roles played by GBPs in protective immunity with an emphasis on cell-autonomous defense and inflammasome-driven responses during infection, inflammation, and cancer. Where appropriate, GBP activities in humans will be highlighted to underscore the clinical and therapeutic implications of these increasingly important immune proteins.
GBPs in humans and other vertebrates
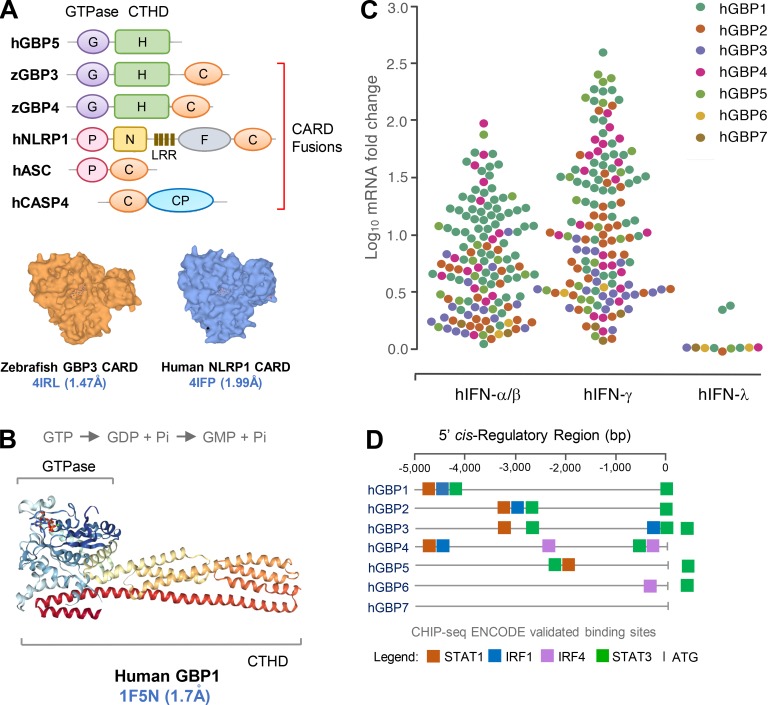
GBPs arose early in the eukaryote lineage, as shown by Hidden Markov Modeling that revealed 132 intact GBP genes across 32 taxa (Shenoy et al., 2012). Bona fide orthologues exist not only in vertebrates and cephalochordates but also protists, amoebae, plants, and algae (Fig. 1). Hence the birth of GBPs predates IFN signaling, with their primordial defense functions being refashioned in early gnathostomes (jawed vertebrates) to respond to cytokine stimulation (Kim et al., 2012; Gaudet et al., 2016; Secombes and Zou, 2017). Despite differences in inducibility, a consistent feature across most species is the expansion of GBPs into chromosomally clustered multigene families that include as many as 16 paralogues in Paramecium tetraurelia (Li et al., 2009; Shenoy et al., 2012; Fig. 1).
Figure 1.
GBPs are ancient and widely distributed in eukaryotes. A comprehensive in silico scan of 91 taxa from Shenoy et al. (2012) plus newer deposits yielded this circular dendrogram with emphasis on human GBP-related orthologues. Sequences were aligned with the E-INS-i algorithm (MAFFT v7) and taxa with incomplete data removed via MaxAlign version 1.1. Right: The phylogenetic tree was constructed using a modified version of the unweighted pair group method with arithmetic mean clustering as implemented by MAFFT. Left: A TimeTree diagram (http://www.timetree.org) depicts the ancient origin of GBPs in lineages leading to Homo sapiens.
In humans, seven GBP genes and one pseudogene reside in a single cluster on chromosome 1q22.2, with close orthologues present in most anthropomorphic primates (Olszewski et al., 2006; Shenoy et al., 2007, 2012; Li et al., 2009; Fig. 2). The evolution of primate GBPs is likely to yield important insights into human-tropic infections including HIV-1 and HIV-2 that originated via multiple transmissions of simian immunodeficiency viruses across species barriers (Sharp and Hahn, 2011). HIV-1 was thought to be introduced into humans by zoonotic spread from Western gorillas and chimpanzees, whereas HIV-2 arrived via Old World monkeys including sooty mangabeys and drills that probably lack a human GBP5 orthologue capable of restricting HIV-2 infectivity (Krapp et al., 2016; Palesch et al., 2018; Fig. 2). Thus, GBP5 might have helped limit HIV-2 pathogenicity within the human population. Rapidly evolving nonsynonymous codon usage in GBP5 and HIV-2 should highlight the basis for this host–pathogen arms race and reinforce the evolutionary impact of GBPs for pathogen-specific defense among different primate species. A similar arms race likely occurred in Neanderthal and Denisovan genomes, in which the enrichment of GBP4 and GBP7 alleles suggests that early hominid orthologues were also positively selected via host–pathogen conflicts (Vernot et al., 2016; Enard and Petrov, 2018).
Figure 2.
Composition of GBP families among closely related primates. Overlapping genomic contigs for primate GBP cluster assembly reveal differences for GBP5 in selected Great Apes versus Old World monkeys. Alignment of the GBP clusters were extracted from the Ensembl database with mRNA transcripts validated for the encoded genes in multiple repositories. Blue regions depict contig regions. Red bars cover individual GBP loci.
In genetically tractable vertebrate models such as mice and zebrafish, the presence of familial GBP clusters also dominates, in this case 11 and 8 Gbp genes, respectively (Degrandi et al., 2007; Shenoy et al., 2007, 2012; Kresse et al., 2008; Kim et al., 2016). Gbp KO mice were initially reported 7 yr ago (Kim et al., 2011), and engineered deletions in the murine chromosomal 3H1 cluster that harbors Gbp1 (Gbp2a), Gbp2, Gbp3, Gbp5, and Gbp7 have continued to aid our understanding of innate immunity and host defense by these proteins (Kim et al., 2016). For the 5E5 cluster that contains Gbp6, Gbp8, Gbp9, Gbp10, and Gbp11, chromosomal deletions have yet to be reported, although loss-of-function siRNA approaches indicate an important role for Gbp6 and Gbp10 in antibacterial immunity (Kim et al., 2011).
Genomic examination of Danio rerio has likewise unearthed links between GBPs and the core inflammasome machinery (Shenoy et al., 2012; Kim et al., 2016; Tyrkalska et al., 2016). Some zebrafish GBP genes (zGBP3, zGBP4) encode caspase activation and recruitment domains (CARDs) like those found within the human inflammasome-associated proteins, ASC (apoptosis-associated speck-like protein containing a CARD), NLRP1, and caspase-4 (Shenoy et al., 2012; Kim et al., 2016; Meunier and Broz, 2017; Fig. 3 A). Physical and functional interactions between inflammasome proteins and GBPs are seen in zebrafish neutrophils and human macrophages (Shenoy et al., 2012; Tyrkalska et al., 2016). In the protochordate, Branchiostomata floridae, Gbp genes are fused with death effector domains related to human caspase-8 and FADD (Fas-associated protein with death domain; Shenoy et al., 2012); these orthologues presage roles for mammalian GBPs in inflammasome-dependent and -independent cell death programs (Meunier et al., 2014; Pilla et al., 2014; Finethy et al., 2015; Man et al., 2015; Ingram et al., 2018). Thus, by comparing sequence and interdomain similarities across multiple genomes, new clues as to how GBPs may operate during infectious and inflammatory disease in humans have emerged.
Figure 3.
Human GBP domain structure and response to IFNs. (A) Top: Some GBPs share orthology with core inflammasome components as revealed by domain accretion in other taxa. Zebrafish GBP3 and GBP4 harbor GTPase (G) and C-terminal helical domains (H) fused to CARDs (C). The latter are orthologous to CARD fusions in human NLRP1, ASC, and caspase-4. Bottom: Similar CARD domain surface structures from zebrafish GBP3 (PDB 4IRL) and human NLRP1 (4IFP; https://www.ebi.ac.uk/pdbe). (B) Crystal structure of human GBP1 bound to the GTP analogue GMPPNP (RCSB Protein Data Bank accession no. 1F5N) reveals a bidomain architecture: N-terminal GTPase and C-terminal helical domains. Two-step GTP hydrolysis shown above. (C) GBP mRNA expression levels robustly induced with IFNs from multiple microarray and RNA sequencing studies deposited in the Interferome database and Bolen et al. (2014). (D) Position of IFN-dependent binding sites for transactivation validated in CHIP-seq ENCODE data from human cells induced with IFN-α or -β (IRF1), IFN-γ (STAT1), or untreated (STAT3, IRF4), or treated with OHTAM/ethanol (STAT3; https://genome.ucsc.edu). CP, cysteine protease domains; F, function to find; P, pyrin; N, NACHT domain; LRR, leucine-rich repeat; CHIP-seq, chromatin immunoprecipitation sequencing.
Biochemical and structural features of the GBPs
The proteins encoded by these GBP loci share similarities with dynamin-like GTPases that undergo guanosine nucleotide-driven self-assembly to form large homotypic complexes (Praefcke and McMahon, 2004; Ferguson and De Camilli, 2012). In this respect, the GBPs resemble other IFN-induced GTPases including the 21–48 kD immunity-related GTPases (IRGs), 72–82 kD myxoma resistance proteins, and ∼200–285 kD very large inducible GTPases (Kim et al., 2012). Unlike these latter groups, which bind guanosine-5′-triphosphate (GTP) to produce guanosine-5′-diphosphate (GDP), GBPs bind both GTP and GDP with equimolar affinity to also produce guanosine-5′-monophosphate (GMP; Cheng et al., 1991; Praefcke et al., 1999). The physiological importance of this nucleotide preference is currently unknown, but GBP mutants unable to catalyze the production of GMP could be informative in understanding their unique host defense profiles versus other IFN-induced GTPases.
Once nucleotide-bound, GBPs exhibit high intrinsic rates of GTPase and GDPase activity (catalytic rate constant, ∼80–150/min) that occurs in a structurally conserved two-step reaction (Schwemmle and Staeheli, 1994; Ghosh et al., 2006; Fig. 3 B). This high rate of catalysis is due to an internal GTPase-activating protein region which obviates the need for external GTPase-activating proteins to accelerate substrate hydrolysis for higher-order assembly (Praefcke et al., 1999, 2004; Kunzelmann et al., 2006; Abdullah et al., 2009; Kim et al., 2011; Kravets et al., 2012). Such unusual substrate binding can be traced to an atypical TLRD- or TVRD-containing G4 site that makes contact with the purine ring of guanosine nucleotides in the GBP catalytic domain; this differs from the canonical (N/T)(K/Q)xD G4 motif found in other dynamin-like proteins and smaller H-Ras GTPases (Cheng et al., 1991; MacMicking, 2004; Shenoy et al., 2007).
At the primary sequence level, human and mouse GBPs share 40–98% amino acid identity (Shenoy et al., 2007; Kim et al., 2011). A bidomain architecture comprising an N-terminal catalytic GTPase domain and extended C-terminal helical domain harboring a series of amphipathic α-helices has emerged from crystallization studies and is common to human and mouse GBPs (Prakash et al., 2000; Fig. 3 B). Both halves contribute to formation of the holoenzyme, as C-terminal truncations or GTPase mutants each prevent cooperative self-assembly (Shenoy et al., 2012; Vöpel et al., 2014; Ince et al., 2017). This cooperative self-assembly can in some cases lead to extreme supramolecular structures, containing as many as 6,000 monomeric units, being deposited on pathogen-associated membranes (Kravets et al., 2016). Such large nanomachines constitute sensory platforms which alert the host to infection and induce antimicrobial pathways, including oxidative and inflammasome components, that contribute to direct pathogen control (Kim et al., 2012, 2016). These nanomachines could also act as mechanoenzymes capable of directly disrupting the microbial cell wall or vacuole during pathogen elimination (Man et al., 2015; Meunier et al., 2015; Kravets et al., 2016).
Physical engagement of GBPs with microbial surfaces, pathogen-containing vacuoles, and endolysosomal membranes relies on posttranslational modifications within the C-terminal domain. Human and mouse GBP1, GBP2, and GBP5, for example, each possess a CaaX box at the C terminus. 15-Carbon farnesyl or 20-carbon geranylgeranyl hydrophobic groups are covalently added via the cysteine residue in this CaaX motif to facilitate membrane binding. For human GBP1, recent evidence suggests that farnesylation not only helps anchor this protein but also serves as a nucleating template to deposit more GBP1 on the membrane surface (Shydlovskyi et al., 2017). Partial geranylgeranylation of mouse Gbp1 may serve a similar purpose (Stickney and Buss, 2000). Mutations in or nearby the CaaX motif as well as C-terminal deletions both liberate membrane-bound GBP complexes; they also severely impair cell-autonomous immunity and inflammasome activation (Kim et al., 2011; Kravets et al., 2012; Shenoy et al., 2012; Finethy et al., 2017; Piro et al., 2017; Santos et al., 2018). Thus, membrane targeting and lipid binding are centrally important for immune-related GBP functions. To date, other posttranslational switches involving phosphorylation, acetylation, and succinylation have not been reported, although online databases (https://www.phosphosite.org) indicate that serine/threonine, tyrosine, and lysine residues are naturally modified in both human and mouse GBPs.
Mobilizing GBPs in cells and tissues
GBPs are among the most abundant ISGs expressed in humans, and they show enormous plasticity in their responsiveness to physiological stimuli. Human GBP1 and GBP5 expression, for example, can be inducibly increased by nearly three orders of magnitude, reaching up to 300,000 molecules per cell within 24 h of IFN-γ exposure (Cheng et al., 1983, 1985; Fig. 3 C).
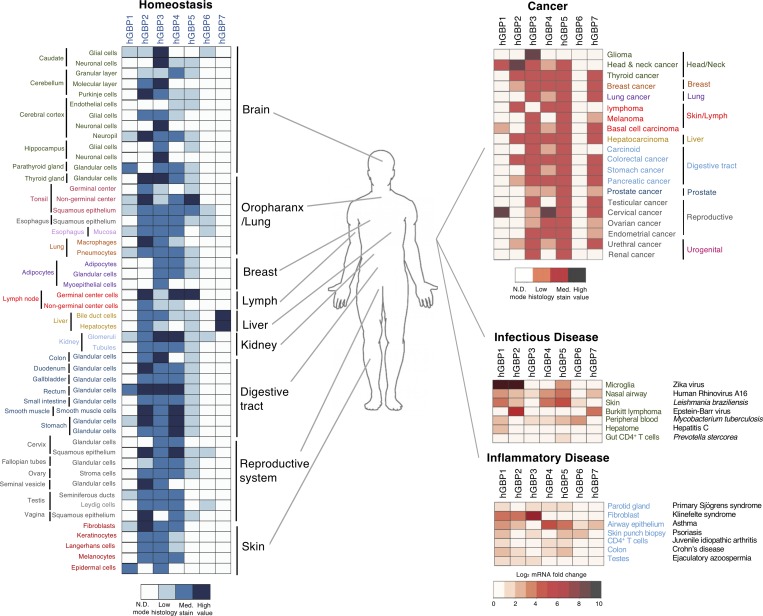
In the absence of acute activating signals, GBPs are typically expressed at low-to-medium basal or tonic levels in immune cells as well as in the stroma of the lung, liver, kidney, digestive tract, brain, and skin (Mostafavi et al., 2016; Fig. 4). Two notable exceptions are human GBP6 and GBP7, which are constitutively expressed only in the oropharyngeal tract and liver, respectively (Fig. 4). Human GBP4 is likewise present in nonactivated pluripotent embryonic stem cells, but expression is lost upon differentiation (Wu et al., 2018). Indeed, all seven human GBPs are detectable at low levels by single-cell RNA sequencing throughout preimplantation (<2–11 reads per kilobase million), with peaks for GBP1 in epiblasts, GBP2 in oocysts, and GBP5 in morula (Yan et al., 2013). These constitutive examples may reflect stage-, cell-, and tissue-specific epigenetic control. In most other settings, however, GBP expression requires robust immune-receptor signaling, including in adult pluripotent lineages such as neural stem cells or neocortical progenitors like radial glia (Llorens-Bobadilla et al., 2015; Thomsen et al., 2016). Similar findings emerge in murine systems. Gbps are expressed in numerous parenchymal and stromal cells as well as at least 11 different immune lineages that span phagocytes and innate-like lymphocytes including NKT, B1-, and CD8αα T cells (Yamagata et al., 2006; Mostafavi et al., 2016). The role of GBPs in the lymphoid lineages has yet to be delineated.
Figure 4.
Diverse GBP expression under homeostatic and disease conditions. Left: Homeostatic expression of different GBP proteins varies between human tissues. Heat maps generated from data deposited in the Human Protein Atlas consortium (https://www.proteinatlas.org/). mRNA data are shown for GBP7 due to a lack of antibody validation for this GBP. Right: GBP protein expression in human tissues during different disease states from the Human Protein Atlas consortium. Scale values shown in each case below heat maps. N.D., not detectable.
IFN-γ and IFN-α/β were the first immune signals described for GBP induction (Cheng et al., 1983; Decker et al., 1989). They remain the most potent stimuli when compared with equimolar concentrations of other cytokines or TLR agonists in humans and mice (Cheng et al., 1985; Boehm et al., 1998; Degrandi et al., 2007; Kim et al., 2011). Multiple gamma-activated sites (GASs) and IFN-stimulated response elements exist within the 5′ cis-regulatory regions of human GBP genes that engage STAT and IFN regulatory factor (IRF) complexes, respectively (Fig. 3 D). Pioneering work by the Darnell group showed that GAS elements were occupied by STAT1 homodimers following IFN-γ treatment, for example, in the human GBP1 promoter (Darnell et al., 1994). De novo synthesis of IRF1 is also obligate for the transcription of several GBPs that are classified as secondary response genes because of their IRF1 dependency and cycloheximide sensitivity (Boehm et al., 1998). Because of this dependency, GBP1 and GBP2 mRNAs typically accumulate ∼4–24 h after IFN exposure. IFN-stimulated response elements also bind heterotrimeric STAT-1/STAT-2/IRF-9 complexes (termed IFN-stimulated gene factor 3; Decker et al., 1991) downstream of IFN-α/β and IFN-λs1-3; thus, some GBPs can respond to type III IFN as well, for example GBP1 in human keratinocytes (Alase et al., 2015) and multiple GBPs in IFN-λ–treated hepatocytes (Bolen et al., 2014; Fig. 3 C).
The presence of NFKB1 binding sites for Rel-related dimers in some GBP promoters extends the inductive stimuli beyond IFNs. Proinflammatory cytokines like IL-1α, IL-1β, or TNF-α each induce human GBP1 expression within isolated and inflamed tissue endothelium, albeit at much lower levels than IFNs; this induction is opposed by vascular endothelial growth factor and basic fibroblast growth factor through unknown mechanisms (Guenzi et al., 2001; Lubeseder-Martellato et al., 2002; Naschberger et al., 2004; Tripal et al., 2007). Human GBP1 likewise responds to IL-1β and TNF-α in colonic epithelial cells, whereas both cytokines robustly elicit several mouse Gbps in murine fibroblasts (Nguyen et al., 2002; Britzen-Laurent et al., 2013). Besides inflammatory triggers, epidermal growth factor receptor–dependent Src and p38 MAPK signaling can also directly induce human GBP1 in glioblastoma targets (Li et al., 2011). Thus, several cytokines or growth factors mobilize GBPs across different host cell lineages. These findings highlight potential servo-regulatory functions for GBPs outside of the traditional immune system and extend their protective coverage to essentially every major organ system for cell-autonomous defense against infection.
Such widespread responsivity is reinforced in natura by curated expression profiles of 84 cell types and tissues from healthy individuals as well as patients with clinically confirmed diseases (Fig. 4). Here, human GBP levels are often dramatically heightened by bacterial, viral, or protozoan infection and are markedly altered during auto-inflammation and cancer. Further regulation is conferred by microRNAs and long intergenic noncoding RNAs. Human GBP2 is a directly down-regulated target of miR-433 in leukemia (Lin et al., 2013), and several mouse GBPs including Gbp2, Gbp3, Gbp4, Gbp5, Gbp6, Gbp8, Gbp9, and Gbp10 are heavily suppressed by long intergenic noncoding RNA–erythroid prosurvival (also known as Ttc39aos1) in LPS-stimulated murine macrophages (Atianand et al., 2016). How epigenetic regulation of GBP expression impacts their immune activities is still unknown, but it is clear that multiple levels of control govern their responsiveness in specific cell types and tissues.
GBPs in antibacterial defense
Following induction, GBPs confer cell-autonomous immunity against wide variety of microbial pathogens, especially intracellular bacteria that remain a serious medical problem across the globe. Shigella and Salmonella species, for example, are major contributors to the 1.3 million diarrheal deaths that occur annually (GBD Diarrhoeal Diseases Collaborators, 2017), and Mycobacterium tuberculosis accounts for 1.3 million deaths on its own, killing more people than any other pathogen and making it one of top 10 most lethal diseases from any cause (World Health Organization, 2018). Clearly, a better understanding of how native immunity combats these bacterial species is a high priority for public health agencies worldwide.
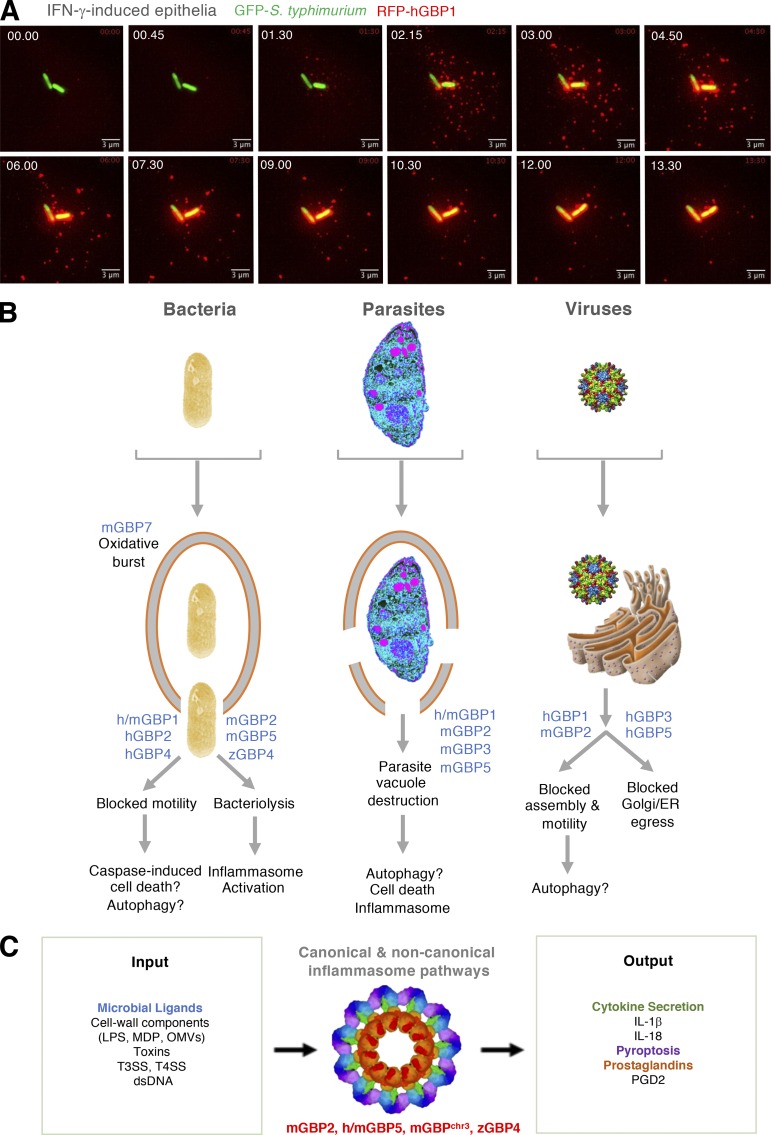
Antibacterial defense was the first function tested across a complete Gbp family, and this activity was the first defect was reported in genetically engineered Gbp1-deficient mice (Kim et al., 2011; see Table 1). In this study, Gram-positive Listeria monocytogenes, a cause of lethal food-borne illness in humans, and Mycobacterium bovis BCG, responsible for disseminated mycobacteriosis in IFN-γ receptor–deficient patients (Zhang et al., 2008), were examined. Both bacteria are sensitive to IFN-γ–mediated killing, and each serve as an example of how different bacterial lifestyles are adopted within host cells. L. monocytogenes escapes its vacuole shortly after uptake and replicates in the host cell cytosol, whereas M. bovis BCG lacks a chromosomal region encoding part of the bacterial type VII secretion (T7SS) apparatus needed for escape; hence it remains trapped inside a phagocytic compartment unless the vacuole becomes damaged (Kim et al., 2012; MacMicking, 2012). In IFN-γ–activated mouse macrophages, both bacteria were targeted by GBPs irrespective of their different lifestyles, and translocation of these immune GTPases to the site of microbial replication was obligate for killing, as shown via amino acid substitutions in GBP mutants that interfere with pathogen targeting (Kim et al., 2011; Fig. 5 A). Such findings fit with the discovery made at The Rockefeller University nearly a decade earlier that IFN-induced GTPases can directly target the bacterial niche, for example, against M. tuberculosis (MacMicking et al., 2003). This discovery led to a model in which IFN-induced GTPases were posited to recognize microbial and/or modified-self components for their recruitment to the “marked” pathogen (MacMicking, 2004, 2005). It now serves as a central tenet in the field.
Table 1. Immune phenotypes in GBP KO mice and disease-associated SNPs/gene alterations in humans.
| GBP deficiency | Disease or immune challenge | Phenotype | Reference |
|---|---|---|---|
| GBP KO mice | |||
| Gbp1−/− | L. monocytogenes | Susceptible to orogastric infection | Kim et al. (2011) |
| M. bovis BCG | Susceptible to i.v. infection | Kim et al. (2011) | |
| T. gondii | Susceptible to s.c. infection | Selleck et al. (2013) | |
| Gbp2−/− | T. gondii | Susceptible to i.p. infection | Degrandi et al. (2013) |
| L. monocytogenes | Resistant to i.p. infection | Degrandi et al. (2013) | |
| F. novicida | Susceptible to s.c. infection and reduced serum IL-18 | Meunier et al. (2015) | |
| OMV i.p. challenge | Resistant to endotoxemia after poly I:C priming; reduced serum IL-1β plus IL-18 | Finethy et al. (2017); Santos et al. (2018) | |
| Gbp5−/− | L. monocytogenes | Susceptible to orogastric infection and insensitive to the caspase-1 inhibitor z-YVAD-FMK | Shenoy et al. (2012) |
| LPS i.p. challenge | Reduced serum IL-1β plus IL-18 and reduced active caspase-1 in splenic macrophages | Shenoy et al. (2012) | |
| MDP i.p. challenge | Impaired peritonitis and reduced active caspase-1 in peritoneal neutrophils | Shenoy et al. (2012) | |
| Alum i.p. challenge | Normal peritonitis | Shenoy et al. (2012) | |
| MSU i.p. challenge | Normal peritonitis | Shenoy et al. (2012) | |
| Gbpchr3−/− | T. gondii | Susceptible to i.p. infection | Yamamoto et al. (2012) |
| L. monocytogenes | Resistant to i.p. infection | Yamamoto et al. (2012) | |
| F. novicida | Susceptible to s.c. infection and reduced serum IL-18 | Meunier et al. (2015); Man et al. (2016); Wallet et al. (2017) | |
| L. pneumophila | Susceptible to cytosolic ΔsdhA Lpn administered oropharangeally | Liu et al. (2018) | |
| Norovirus | Susceptible to peroral infection in combination with Ifnar1 deficiency | Biering et al. (2017) | |
| OMV i.p. challenge | Resistant to endotoxemia after poly I:C priming; reduced serum IL-1β plus IL-18 | Finethy et al. (2017); Santos et al. (2018) | |
| LPS i.p. challenge | Resistant to endotoxemia after poly I:C priming; reduced serum IL-1β plus IL-18 | Santos et al. (2018) | |
| Human GBP SNPs/alterations | |||
| GBP2 | Hepatitis B virus | IFN treatment responsiveness vs. nonresponsiveness | King et al. (2002) |
| (41544 A/G)a | |||
| (41556 G/C)a | |||
| GBP3 (rs7911) | Fibromyalgia | Protective role in pathophysiology of FM | Parisien et al. (2017) |
| GBP5 (spliced GBP5ta-97 AA truncation) | Cutaneous T cell lymphoma | High isoform expression in tumor tissues and cell lines | Fellenberg et al. (2004) |
| GBP7 (rs1803632) | Malaria | Promoter mutations associated with higher disease incidence | Apinjoh et al. (2014) |
rs, dbSNP database number; MDP, muramyl dipeptide; MSU, monosodium ureate; FM, fibromyalgia.
JSNP, Japanese Single Nucleotide Polymorphisms database.
Figure 5.
Antimicrobial activities of GBPs. (A) Targeting of human GBP1 to cytosolic S. typhimurium via live epifluorescent imaging in IFN-γ–activated human HeLa epithelium. Bar, 3 µm. Courtesy of A. Maminska. (B) Specific cell-autonomous responses of GBPs to different pathogen classes and the downstream consequences of their antimicrobial actions in humans (h), mice (m), and zebrafish (z). (C) Involvement of different GBPs from humans, mice, and zebrafish in canonical and noncanonical inflammasome activation elicited by microbial ligands as input signals. Cytokine and prostaglandin secretion plus pyroptosis are identified outputs. Inflammasome cryoelectron tomography structure (3JBL) from https://www.ebi.ac.uk/pdbe. MDP, muramyl dipeptide.
Subsequent work on IFN-induced GBPs over the past 5 yr has extended the targeting paradigm to other intracellular bacteria that are acquired via contaminated food or water, human-to-human transmission, or zoonotic exposure. These include serovars of Salmonella typhimurium, Francisella novicida, Legionella pneumophila, Chlamydia trachomatis, Yersinia pseudotuberculosis, and Brucella abortus (Haldar et al., 2013, 2015; Meunier et al., 2014, 2015; Man et al., 2015; Feeley et al., 2017; Lindenberg et al., 2017; Zwack et al., 2017; Costa Franco et al., 2018; Liu et al., 2018; Santos et al., 2018; Fig. 5 A). GBP recruitment to these bacteria in human or mouse cells often coincides with reduced microbial viability. It can also lead to decreased or altered fluorescence in bacterial strains expressing GFP or RFP transgenes (Li et al., 2017; Liu et al., 2018) and loss of LPS staining in Gram-negative bacteria, as reported for S. typhimurium and F. novicida when they become decorated with endogenous Gbp2 or Gbp5 in mouse macrophages (Meunier et al., 2014, 2015; Man et al., 2015). Notably, these effects were abolished in Ifnar1−/−, Stat1−/−, or Irf1−/− cells, underscoring the importance of IFN signaling to elicit GBP expression.
Two major questions arise from GBP recruitment to intracellular bacteria (Kim et al., 2012; Bradfield, 2016). First, what are the structures recognized by these immune GTPases to target the pathogen niche? Second, how do GBPs control infection once they reach this destination? Attempts to answer the first question using bacterial cell surface mutants have shown that the lipid A moiety of Gram-negative LPS is recognized by Gbp5 in mouse macrophages, and that the LPS O antigen is detected by human GBP1 in lung epithelia infected by Shigella or Burkholderia (Piro et al., 2017; Santos et al., 2018). In neither case, however, was direct LPS binding by GBPs shown. Moreover, GBPs also target Gram-positive L. monocytogenes, actinobacteria such as M. bovis, protozoan parasites like Toxoplasma gondii, and the replication complexes of HIV-1 and murine norovirus (Degrandi et al., 2007; Kim et al., 2011; Virreira Winter et al., 2011; Bradfield, 2016; Krapp et al., 2016; Biering et al., 2017), indicating recognition of structures other than LPS.
Among the likely culprits are intraluminal ligands originating from the endolysosomal system (Bradfield, 2016). All GBP-restricted pathogens identified so far either escape from, dwell within, or intersect this system of cargo vesicles and multivesicular body intermediates at some point during their life cycle (Kim et al., 2012; Bradfield, 2016). In addition, pharmacologic agents that invoke sterile endosomal damage in the absence of infection also solicit GBPs to the site of membrane injury (Bradfield, 2016; Feeley et al., 2017). Thus, signals emanating from disrupted organelles probably serve as a proxy of infection, with bacterial pore-forming toxins or type III (T3SS) or IV (T4SS) secretion systems triggering the initial damage to help release these ligands in the context of bacterial infection (Bradfield, 2016; Feeley et al., 2017; Zwack et al., 2017; Liu et al., 2018). An ability to recognize “modified self” was also reported for the IRGs that bind phosphatidylinositides generated directly on the bacterial phagosome by host lipid kinases in IFN-γ–activated macrophages (Tiwari et al., 2009; Kim et al., 2012). For the GBPs, however, “altered-self” signals arise from inside rather than outside the vacuole and become available for detection only once pathogens escape into the cytosol (Bradfield, 2016).
Following their recruitment to bacteria, GBPs exert distinct antibacterial mechanisms depending on the protein partners they deploy (Fig. 5 B). Such partners often reflect the antimicrobial machinery expressed in immune versus nonimmune cells (Gaudet et al., 2016). For example, in IFN-γ–activated mononuclear phagocytes, certain GBPs interact with the phagocyte oxidase complex (phox; NADPH oxidase) as well as proteins involved in autolysosomal killing of bacteria and inflammasome activation (Kim et al., 2011; Shenoy et al., 2012; Haldar et al., 2015; Man et al., 2016; Feeley et al., 2017). Gbp7 binds endogenous gp91phox and p22phox, which comprise the native cytochrome b558 heterodimeric membrane (Nox2) complex, via its C-terminal tail, whereas it engages the cytosolic p67phox subunit via its GTPase domain. In this way, Gbp7 bridges the Nox2 holoenzyme for assembly on listerial and mycobacterial phagosomes. This finding may add mechanistic insight into the discovery made 35 yr ago that IFN-γ represents the coveted “macrophage-activating factor” responsible for priming the respiratory burst against intracellular pathogens (Nathan et al., 1983; MacMicking, 2009).
Besides oxidant defense, Gbp7 also binds the cysteine protease Atg4b for potential closure of autolysosomes around bacteria. Mouse Gbp1 and Gbp2 similarly interact with the autophagy-related proteins p62 and galectin-3, respectively, for recruitment to membranes ruptured by Listeria and Legionella in IFN-γ–activated macrophages (Kim et al., 2011; Haldar et al., 2015; Feeley et al., 2017; Fig. 5 B). A comparable interaction may help recruit Gbp2 to ruptured membranes in S. typhimurium–infected macrophages; the latter observation led to the idea that GBPs might directly lyse bacteria-containing vacuoles to release intraluminal ligands for inflammasome activation (Meunier et al., 2014). However, subsequent work suggests that this release probably occurs after bacteria have already escaped into the cytosol and that the IFN-induced 47-kD IRG family member Irgb10 lyses the bacterial membrane rather than the vacuolar membrane (Man et al., 2015, 2016; Meunier et al., 2015; Liu et al., 2018). Thus, several Gbps mobilize membrane-associated defense activities against Gram-positive, Gram-negative, and actinobacteria within immune phagocytes. Such activities often require GTPase activity and C-terminal isoprenylation (Kim et al., 2011). They also operate in vivo where these immune GTPases are expressed within circulating monocytes as well as mouse and human target organs during natural infection and after vaccination (Degrandi et al., 2007; Marquis et al., 2011; Ottenhoff et al., 2012; Soudja et al., 2014; Zak et al., 2016; Fig. 4). Importantly, Gbp1−/−, Gbp2−/−, Gbp5−/−, and chromosome 3H1 cluster–deficient (Gbpchr3−/−) mice are often highly susceptible to these same bacterial infections (see Table 1).
Additional GBP-related mechanisms protect nonimmune cells that lack some of the armamentarium found in phagocytes (Gaudet et al., 2016). Human GBP1, for example, binds F- and G-actin (Ostler et al., 2014) to potentially interfere with actin-based motility of Shigella flexneri and possibly Burkholderia thailandensis within IFN-γ–treated fibroblasts (Piro et al., 2017; Wandel et al., 2017; Fig. 5 B). GBP1 enlists a C-terminal RRR motif that acts as a catalyst for hierarchical recruitment of additional GBPs including GBP2, GBP3, and GBP4 to block bacterial dissemination to neighboring cells (Bradfield, 2016; Piro et al., 2017; Wandel et al., 2017). The importance of GBPs in this process is underscored by the discovery of a Shigella E3 ubiquitin ligase, IpaH9.8, which ubiquitinates several GBP family members for subsequent degradation by the host proteasome (Li et al., 2017; Piro et al., 2017; Wandel et al., 2017). This allows Shigella to escape restriction and highlights the selective pressure placed on intracellular pathogens by this class of defense proteins (MacMicking, 2017). A similar strategy applies to the human-tropic bacterial pathogen, C. trachomatis, which evades GBP targeting in IFN-γ–activated human epithelia but is detected by multiple Gbps within murine embryonic or NIH3T3 fibroblasts (Haldar et al., 2015, 2016; Lindenberg et al., 2017). Thus C. trachomatis has also invented methods for GBP avoidance within its natural human host.
GBP immunity to parasites
While intracellular bacteria remain the most studied group of pathogens that are restricted by GBPs, apicomplexan parasites such as T. gondii provide valuable information on the host defense activities conferred by these immune GTPases. Toxoplasmosis remains an important global health problem with nearly a third of the world’s population seropositive for the parasite (Centers for Disease Control and Prevention, 2018). Severe and occasionally lethal infections ensue in patients with AIDS and other immune-compromised individuals because CD4+ T cell–driven immunity is critical for controlling T. gondii growth. In this context, IFN-γ mobilizes toxoplasmacidal activity in both immune and nonimmune cells (Yap and Sher, 1999; Hunter and Sibley, 2012). Nitric oxide synthase, indoleamine-2,3-dioxygenase, and autophagy-related pathways are all important IFN-γ–induced effectors in multiple cell lineages of humans and mice (MacMicking, 2012; Selleck et al., 2015). However, additional mechanisms have recently been added to this list, including GBPs (Fig. 5 B).
Work by the Pfeffer group initially helped highlight a role for GBPs in response to T. gondii. Examination of the murine Gbp family revealed that many members—including Gbp1, Gbp2, Gbp3, Gbp6, Gbp7, and Gbp9—were targeted to the parasitophorous vacuole (PV) in embryonic fibroblasts and macrophages (Degrandi et al., 2007). The PV derives from the plasma membrane but typically excludes host proteins involved in later lysosomal fusion (Coppens and Romano, 2018); hence recognition of early altered-self signals probably elicits cooperative interactions among different GBPs, as subsequently reported by several groups (Virreira Winter et al., 2011; Yamamoto et al., 2012; Kravets et al., 2016). Elegant Förster resonance energy transfer and coimmunoprecipitation studies showed that murine Gbp1 directly binds Gbp2 and Gbp5, and that Gbp2 also engages Gbp3 within preformed complexes that are then recruited to the PV membrane (Virreira Winter et al., 2011; Kravets et al., 2016). Here, several thousand homo- and hetero-oligomeric Gbp complexes form densely packed polyvalent structures which precede loss of PV integrity, followed by direct binding of Gbp2 to the underlying parasite plasma membrane (Kravets et al., 2016; Fig. 5 B). Whether PV disruption results from GBP mechanoenzyme activity is unknown, although Gbp2 mutagenesis experiments indicate that GTP hydrolysis, multimerization, and isoprenylation are all involved (Kravets et al., 2012, 2016; Degrandi et al., 2013). In addition, p62 and the E3 ligases TRAF6 and TRIM21 may help ubiquitinate the PV, enabling its recognition as a damaged organelle and potentially aiding GBP targeting to this altered-self structure in mouse fibroblasts (Selleck et al., 2013; Finethy et al., 2015; Foltz et al., 2017; Sasai et al., 2017). p62, NDP52, LC3B, and galectin-8 perform similar functions in IFN-γ–activated human HeLa cells (Selleck et al., 2015).
Nearly all GBP convergence on PV membranes occurs with the widely distributed type II and III T. gondii strains that are less virulent, whereas virulent type I parasites interfere with sequestration by GBPs, as seen earlier for Shigella and Chlamydia (Degrandi et al., 2007, 2013; Virreira Winter et al., 2011; Kravets et al., 2012; Yamamoto et al., 2012; Selleck et al., 2013). Secreted parasitic rhoptry proteins (ROP) 16, ROP18, and ROP54 and dense granule antigen 15 (GRA15) are implicated in this process, possibly via direct GBP phosphorylation or indirect STAT3 and STAT6 activation since several of these type I parasite proteins constitute bone fide kinases (Virreira Winter et al., 2011; Hunter and Sibley, 2012). Additional parasite effectors including TgIST interfere with STAT1 activation, thereby impacting IFN-γ–induced GBP expression (Gay et al., 2016; Olias et al., 2016). Thus host–parasite coevolution reinforces how GBP suppression leads to evasive action by the virulent type I strain. For this reason, most challenge studies in Gbp KO mice have used the less virulent type II strain, against which Gbp1−/−, Gbp2−/−, and Gbpchr3−/− mice have all proved susceptible (see Table 1).
Are similar defense profiles evident in humans? Preliminary studies using siRNA or chromosomal CRISPR-Cas9 silencing suggest that GBPs can exhibit robust anti-toxoplasma activity in specific human cell types. For example, human GBP1 restricts T. gondii growth in IFN-γ–activated primary mesenchymal stem cells and transformed lung epithelia, with PVs clearly targeted in the former but not the latter cell type (Ohshima et al., 2014; Johnston et al., 2016; Qin et al., 2017). In contrast, human HAP1 cells derived from a myelogenous leukemia cell line do not require GBPs to control T. gondii despite the fact that these GTPases target PVs during infection (Ohshima et al., 2014). This suggests additional IFN-γ–induced factors are needed to disrupt PVs in HAP1 cells, which lack orthologues of the membranolytic IRGs found in mice (Martens et al., 2005; Kim et al., 2012). In human foreskin fibroblasts, halting T. gondii replication via host cell death occurs independently of GBP1 or GBP2 as well as caspases, receptor-interacting protein kinases, autophagy, or purinergic receptor activation (Niedelman et al., 2013). Whether other GBPs beside GBP1 and GBP2 participate in host cell death awaits testing. Clearly, understanding which human cell lineages enlist GBP-mediated immunity to restrict the broad cellular tropism of T. gondii will be a fruitful area of future investigation.
The role of GBPs in defense against other apicomplexan parasites, including Plasmodium species responsible for human malaria, remains functionally untested. However, a genetic association study unveiled a single nucleotide polymorphism (SNP; rs1803632) in the human GBP7 promoter that correlated with elevated parasitemia, malaria-related anemia, and hyperpyrexia in Cameroon populations (Apinjoh et al., 2014; Table 1). In mouse models of cerebral malaria, a number of family members including Gbp3, Gbp4, Gbp6, Gbp7, Gbp9, and Gbp11 are up-regulated in the liver of animals infected with P. berghei ANKA sporozoites, whereas Gbp2 and Gbp3 were highly expressed in the infected brain (Berghout et al., 2013; Liehl et al., 2014). Similar up-regulation was seen in otherwise resistant mice challenged with the kinetoplastid parasite Leishmania major, with increased expression of Gbp1 and Gbp5 in the skin, inguinal lymph nodes, spleen, and liver (Sohrabi et al., 2018). Moreover, Gbp1 colocalized with L. major–infected sites. Human GBPs are likewise up-regulated in the skin of patients infected with L. braziliensis (Fig. 4). A role for these large GTPases in immunity to cutaneous leishmaniasis therefore seems likely. In contrast, GBPs fail to target Trypanosoma cruzi to control parasite growth (Virreira Winter et al., 2011). The basis for these different GBP targeting profiles among kinetoplastids is currently unknown. Recognition of specific protozoan structures, subcellular location of individual GBPs, or cell type may all conceivably influence this parasite defense profile in humans as well as in other mammalian hosts.
Antiviral effects of GBPs
Viruses continue to plague the human population, with HIV-1, rotaviruses, chronic hepatitis viruses, and respiratory pathogens such as influenza A among the leading causes of death (GBD Diarrhoeal Diseases Collaborators, 2017; World Health Organization, 2018). Some of these viruses are reported targets of GBP-mediated immunity; however, the breadth of antiviral activity is still being delineated (Fig. 5 B). Early experiments implicated human GBPs in the control of several RNA viruses. GBP1 was found to restrict vesicular stomatitis, encephalomyocarditis, and hepatitis C viruses in IFN-γ– or IFN-α–treated HeLa cells and GBP1 overexpressing Huh 7 hepatocytes (Anderson et al., 1999; Itsui et al., 2009). GBP1, GBP3, and a truncated splice isoform of GBP3 that lacks part of the C-terminal domain (GBP3ΔC) also restrict influenza A replication in human lung epithelia (Nordmann et al., 2012). Notably, influenza inhibition was dependent on GTP binding but not hydrolysis, enabling antiviral activity to proceed in the face of viral antagonists that interfere with direct catalytic activity, such as the NS5B protein of hepatitis C virus (Itsui et al., 2009). Lastly, human nonsynonymous polymorphisms in GBP2 blunt responsiveness to IFN treatment during hepatitis B virus infection, implying that GBP2 activity is protective in these patients (King et al., 2002; Table 1).
More recent work on important RNA viruses responsible for major human epidemics has extended the reach of GBPs to HIV-1. Silencing GBP5 in IFN-γ–treated human monocyte-derived macrophages made them more permissive to HIV-1 infection as shown by increased production of progeny virions and expression levels of mature gp120 (Krapp et al., 2016). Human GBP5 hinders processing and incorporation of the viral envelope glycoprotein, presumably at the Golgi since the viral envelope is assembled at this site and anti-HIV activity is abolished in GBP5 mutants defective for isoprenylation, which is required for Golgi anchorage. Furthermore, macrophage GBP5 expression in HIV patients correlated inversely with infectious virus yields, suggesting that GBP-mediated protective mechanisms operate in natura (Krapp et al., 2016). These results fit with the evolutionary profiles of primate GBPs discussed earlier in relation to their antiretroviral potential and reinforce the idea that certain GBP family members may interfere with viruses that transit specific compartments, in this case GBP5 located near the Golgi. Other RNA viruses restricted by GBPs include noroviruses, which cause highly contagious gastroenteritis in people (Biering et al., 2017). Noroviruses propagate on membraneous replication complexes that become marked by the autophagy-related LC3 conjugation system resulting in recruitment of Gbp2, along with other GBPs, and disruption of the replication complexes (Biering et al., 2017). Such protection also manifests in vivo; compound Ifnar1−/− and Gbpchr3−/− mice show diminished survival in murine models of noroviral pathogenesis (Table 1).
DNA viruses such as Karposi’s sarcoma–associated herpesvirus (KSHV) have recently been added to the list of those targeted by GBPs. Here, human GBP1 was found to disrupt the formation of actin filaments needed to translocate KSHV particles into the nucleus (Zou et al., 2017). GTPase activity and dimerization were needed for these anti-KSHV effects. Notably, KSHV encodes a replication and transcriptional activator, RTA, that induces proteasomal degradation of GBP1 as part of an escape mechanism (Zou et al., 2017). This mechanism echoes earlier tactics deployed by S. flexneri (Li et al., 2017; Piro et al., 2017; Wandel et al., 2017). Hence immune evasion strategies are shared in certain bacterial and viral species, again highlighting the selective pressure that GBPs place on structurally diverse pathogens.
GBPs in inflammasome activation
GBP-driven destruction of the bacterial, protozoal, or viral niche can liberate products that then trigger innate immunity to potentially protect the host. These products include LPS and double-stranded DNA that activate inflammasomes, which in turn produce pyrogenic cytokines (IL-1β, IL-18) and induce pyroptosis in many cell types, including macrophages, monocytes, splenic and plasmacytoid dendritic cells, T cells, B cells, neutrophils, keratinocytes, and inflamed endothelium (Broz and Dixit, 2016; Kim et al., 2016). To date, the role of GBPs in activation and assembly of the inflammasome complex has been established in immunologically primed mouse macrophages and human monocytes (Kim et al., 2016; Fig. 5 C). Such activities are compromised in Gbp-deficient mice (Table 1).
Important early clues pointing to a relationship between GBPs and the inflammasome complex arose from genome-wide phylogenetic analyses (Shenoy et al., 2012). Hidden Markov Modeling retrieved 594 GBP-related sequences from 91 taxa including orthologues in highly divergent species that harbored CARDs. These CARDs resembled those within the core human inflammasome proteins NLRP1, ASC, and caspase-4 (Shenoy et al., 2012; Kim et al., 2016; Fig. 3 A). Similarities extend to the atomic level where crystallization of zebrafish IFN-induced guanylate-binding protein 1 (zGBP3) revealed a six-helix bundle almost identical to that of human NLRP1 and ASC (Jin et al., 2013). By “mining the fossil record,” ancestral links between vertebrate GBPs and the core inflammasome machinery involved in both the canonical and noncanonical pathways were therefore established.
Such evolutionary links operate at both molecular and organismal levels. Loss-of-function experiments initially identified human GBP5 and its mouse orthologue as facilitators of NLRP3 inflammasome activation in response to microbial triggers and intact bacteria (Rupper and Cardelli, 2008; Shenoy et al., 2012). IL-β secretion and pyroptosis were diminished in GBP5-deficient human monocytes and in Gbp5 siRNA-treated mouse macrophages from C57BL/6J, C57BL/6N, or Balb/c mice (Rupper and Cardelli, 2008; Shenoy et al., 2012); the latter experiment ruled out potential strain differences in Gbp1 expression conferred by the chr.3H1 cluster (Staeheli et al., 1984). Notably, inflammasome defects were most evident in cells primed by IFN-γ compared with those primed by LPS or lipid A (Rupper and Cardelli, 2008; Shenoy et al., 2012; Meunier et al., 2014; Pilla et al., 2014; Man et al., 2015; Marty-Roix et al., 2016). Such differences may reflect higher GBP expression levels or inflammasome activation after IFN-γ induction, which appears to be a common feature of all GBPs tested thus far (Shenoy et al., 2012; Meunier et al., 2014; Pilla et al., 2014; Marty-Roix et al., 2016; Finethy et al., 2017; Wallet et al., 2017). Human GBP5 was shown to bind the NLRP3 pyrin domain and accelerate inflammasome complex formation via its own tetrameric assembly, as revealed using GBP5 oligomerization mutants (Shenoy et al., 2012). By analogy, IFN-γ–induced zGBP4 likewise directly binds ASC via its CARD domain, facilitating assembly of a macromolecular complex that includes caspase-1 in response to S. typhimurium infection of zebrafish embryos (Tyrkalska et al., 2016; Fig. 5 C). Hence GBP oligomerization can help drive assembly of complexes with inflammasome proteins across distant vertebrate species; this may extend to inflammatory caspases like human caspase-4, which shares CARD sequence similarities with zebrafish GBPs (Shenoy et al., 2012; Fig. 3 A). Such assembly could conceivably take place at cytosolic preassembly sites or after recruitment of these partners onto bacteria themselves (Kim et al., 2016).
In fact, Gbp5 and Gbp2 directly target F. novicida for disruption, resulting in release of double-stranded DNA (dsDNA) in mouse macrophages and subsequent activation of the canonical AIM2 inflammasome (Man et al., 2015; Meunier et al., 2015; Wallet et al., 2017). Similar results emerge with L. pneumophila; Gbp-mediated release of Legionella dsDNA stimulated downstream cyclic GMP-AMP synthase–mediated IFN-β production and AIM2 inflammasome-induced pyroptosis. Here, autocrine IFN-β was sufficient for tonic Gbp expression (Liu et al., 2018). Gbp5 may also target outer membrane vesicles (OMVs) from Gram-negative bacteria to activate the noncanonical inflammasome in IFN-β–primed human monocytes, as well as the type III strain of T. gondii, to elicit IL-1β release in naive mouse macrophages (Matta et al., 2018; Santos et al., 2018). Thus, targeting GBPs to bacteria and protozoa often releases ligands to elicit canonical or noncanonical inflammasome pathways (Fig. 5 C), thereby linking cell-autonomous defenses with innate immune signaling further downstream.
These GBP-dependent inflammasome activities manifest in vivo (Table 1). Bacterial infections that require caspase-1 or caspase-11 for protection also involve GBPs. For example, Gbp2−/−, Gbp5−/−, or Gbpchr3−/− mice exhibit heightened vulnerability to Gram-positive L. monocytogenes and to a panel of Gram-negative pathogens, due in part to reduced IL-1β and IL-18 expression (Shenoy et al., 2012; Meunier et al., 2015; Man et al., 2016; Wallet et al., 2017; Liu et al., 2018). S. typhimurium, F. novicida, and L. pneumophila mutants (ΔSdhA) that engage the inflammasome within the cytosol also have defects in pyroptosis that may prevent clearance of pathogen-infected cells (Meunier et al., 2015; Man et al., 2016; Wallet et al., 2017; Liu et al., 2018). Gbp4-deficient zebrafish likewise fail to control flagellin-expressing S. typhimurium infection due to reduced caspase-1 activation and prostaglandin (PG) D2 synthesis (Tyrkalska et al., 2016). Flagellin-dependent PG release was initially shown for the murine Naip5-Nlrc4 inflammasome complex (von Moltke et al., 2012), but it appears that jawed fish can also enlist PGs as bona fide downstream inflammasome mediators in addition to IL-1β and IL-18 (Fig. 5 C). Whether comparable PG-dependent mechanisms exist in humans has yet to be identified.
Other forms of systemic inflammation incited by microbial products are controlled by GBPs. Inflammasome-related defects in Gbp2−/−, Gbp5−/−, and Gbpchr3−/− mice challenged with LPS or exposed to Gram-negative OMVs include impaired CD11b+ macrophage caspase-1 activity, diminished IL-1β and IL-18 production, and delayed mortality during sepsis (Shenoy et al., 2012; Finethy et al., 2017; Santos et al., 2018; Table 1). In muramyl dipeptide–induced peritonitis models, Gbp5−/− mice likewise exhibit neutrophil caspase-1 defects that block IL-18–induced polymorphonuclear neutrophil recruitment (Shenoy et al., 2012). Thus, GBPs impact inflammatory responses to localized infection and systemic microbial products in part through activation and assemblage of different inflammasome complexes.
It will be interesting to discern whether manipulation of such activities can impact Gram-negative sepsis in humans, a condition that is responsible for 250,000 annual deaths in the United States alone following surgical, burn, or infectious trauma (Hotchkiss et al., 2013). While blocking the initial cytokine-mediated hyper-inflammatory phase of sepsis improves survival, patients often undergo a protracted immunosuppressive phase that renders them vulnerable to both primary and secondary infections (Hotchkiss et al., 2013). Rescuing “deactivated” monocytes using IFN-γ in individuals with sepsis has been shown to improve antimicrobial defense (Döcke et al., 1997). Whether specific GBPs can also be pharmacologically targeted to mobilize IFN-γ–driven host defense while simultaneously avoiding inflammasome-mediated tissue injury is an important and exciting clinical question going forward.
GBPs in metabolic inflammatory diseases and microbiota-driven colitis
Other inflammatory conditions besides sepsis enlist nucleotide binding–leucine-rich repeat (NLR)–mediated cytokine production or eicosanoid synthesis. Metabolic inflammatory syndromes such as nonalcoholic fatty acid liver diseases and obesity as well as with inflammatory bowel diseases (IBDs) are subject to control by NLRP3 and NLRP6 inflammasomes and their product, IL-18, in response to dysregulated microbiota (Zaki et al., 2010; Henao-Mejia et al., 2012). Atherogenesis may similarly be further worsened by NLRP3 inflammasome activation in response to excess cholesterol crystals accumulating within fatty plaques (Duewell et al., 2010). GBPs have been implicated in both of these processes.
The gastrointestinal tract functions not only as a place of nutrient uptake but also as a barrier to infection. When innate and acquired immunity is not adequate in maintaining control of commensal flora and food-borne pathogens, these gut infections can lead to several forms of IBD, including Crohn’s disease (CD) and ulcerative colitis (UC; Abraham and Cho, 2009). Several reports point to a potential role for the GBPs in human IBD and animal models of colitis. The Gbp1 locus (also termed Gbp2a) resides in a quantitative trait locus susceptibility region for spontaneous enterocolitis in Il10−/− CdcsC3H/HeJBir mice (de Buhr et al., 2006), and human GBP1 localizes to tight junctions of intestinal epithelial cells in patients with both CD and UC; in this context, GBP1 has been posited to regulate commensal Escherichia coli–induced apoptosis and barrier integrity to control gastrointestinal inflammation (Schnoor et al., 2009). GBP5 stimulates the inflammasome-related activity of NLRP3 and may be involved in the assembly of NOD2 (Shenoy et al., 2012); mutations in both NLRP3 and NOD2 have been linked to susceptibility to CD in humans (Abraham and Cho, 2009; Villani et al., 2009). In this regard, several GBPs are also up-regulated in the colon during CD and in murine models of microbial colonization of the developing intestine (Mirpuri et al., 2010; Fig. 4). Hence GBPs could act as regulators of microbiota-driven inflammation due to their involvement in epithelial cell defense and inflammasome responses that are needed for gastrointestinal immunity or dysbiosis. Experiments in Gbp-deficient mice should help establish direct causal links with IBD progression or suppression as well as shifts in microbial consortia of the gut.
An association of GBPs with atherogenesis has recently emerged from murine models of hypercholesterolemia (Goo et al., 2016). ApoE−/− mice fed a high-fat Western diet exhibited increased Gbp2, Gbp3, Gbp5, Gbp6, and Gbp7 expression within lesional foam cells (Goo et al., 2016). In ApoE−/− macrophages, ex vivo exposure to oxidized low-density lipoproteins elicited high Gbp3 and Gbp6 expression, suggesting that this metabolic stimulus may trigger Gbp induction at a cell-intrinsic level. Whether such expression impacts the development of atherogenic lesions and how different inflammasome subtypes operate during atherogenesis remain untested. This also applies to several other inflammatory and autoimmune disorders such as asthma, psoriasis, arthritis, and Sjögren’s syndrome in which heightened GBP expression has been described in human patient samples (Fig. 4). Some of these IFN-associated diseases are amenable to JAK inhibitors (Moodley et al., 2016; Schwartz et al., 2016). Whether GBPs are involved in the mechanism of action of JAK inhibitors is unknown; however, this is poised to be an active area of clinical investigation in the future.
GBPs in oncogenic surveillance and cancer
Cancer immune-editing models show IFN-γ–driven surveillance is essential for eliminating tumors and maintaining residual malignant cells in a dormant state (Alspach et al., 2018). IFN-γ can also be coopted, however, by certain tumors to escape anti-cancer responses by up-regulating PD-L1 expression (Benci et al., 2016; Overacre-Delgoffe et al., 2017). Both pro-and anti-tumor roles are therefore likely to result from IFN-γ–dependent gene induction depending on the cancer cell type, clinical context, and the specific ISGs enlisted (Parker et al., 2016).
GBP family members are among the IFN-γ–dependent genes most highly induced in patients with different cancers at various stages of disease or remission (Fig. 4). Increased tissue expression of human GBP1, GBP2, GBP4, and GBP5 reveals favorable prognostic outcomes with certain tumors, for example, breast, and colorectal cancer, but highly unfavorable associations for others, such as renal cancers (Godoy et al., 2014). The basis for this disparity is currently unknown, although poor prognosis in renal cancer may be independent of IFN signaling since both type I and II IFNs exhibit survival benefits for patients with kidney cancer (Otto et al., 1988; Parker et al., 2016).
One explanation for these disparate outcomes involves distinct cellular mechanisms used by different GBP family members. For example, GBP1 binds F-actin to regulate cytoskeletal dynamics (Ostler et al., 2014); these interactions may influence cancer cell migration or cell division via effects on mitotic spindle assembly. It could also offer some insight into how GBP1 confers paclitaxel resistance in tumors such as ovarian cancer (Duan et al., 2006; Tipton et al., 2016; Wadi et al., 2016). Paclitaxel stabilizes the microtubule polymer and protects it from disassembly, whereas GBP1 is thought to prevent cytoskeletal polymerization (Wandel et al., 2017). Incorporation of GBP1 into the cytoskeletal network facilitates binding of pro-survival kinases such as PIM1 and NEK6; such interactions may initiate a resistance pathway to paclitaxel (Andreoli et al., 2014; Donato et al., 2015).
Human GBP1 has also been reported to suppress matrix metalloproteinase 1 expression for glioblastoma and endothelial cell invasiveness, the former in response to EGFR-dependent signaling (Guenzi et al., 2001; Li et al., 2011). Subsequent silencing of GBP1 led to metastatic invasion in nu/nu mouse models of glioblastoma multiforme (Lan et al., 2016). Thus, GBP1 has onco-protective effects beyond direct actin remodeling. Other cytoskeletal-independent activities include interactions between the GBP1 C-terminal domain and Hippo signaling transcription factor TEA domain protein that interferes with expression of YAP1–TEA domain protein target genes needed for cellular hyper-proliferation (Unterer et al., 2018). This may provide some basis for the ability of GBP1 to act as a tumor suppressor in colorectal cancers which arise from dysregulated Hippo signaling (Britzen-Laurent et al., 2013; Hong et al., 2016).
A similar function has been ascribed to human GBP2, which serves as a p53 target gene elicited together with IRF1 in esophageal squamous cell carcinomas (Guimarães et al., 2009; Britzen-Laurent et al., 2013). Whether these three proteins form an inhibitory complex is unknown. Gbp2 has also been posited to block Drp1-mediated mitochondrial fission needed for breast cancer invasion (Zhang et al., 2017). Clearly more work is needed to establish the molecular basis for how individual GBPs facilitate tumor suppression or inhibition as part of their protective activities. This may extend to cancers having an infectious origin, which currently comprise an estimated 20% of all cases (Casper and Fitzmaurice, 2016) and where the antimicrobial effects of GBPs could be highly influential. Last, truncated human GBP5 isoforms lacking the C-terminal 97 amino acids (including the isoprenylation motif) have been associated with some cutaneous T cell lymphomas (Fellenberg et al., 2004; Table 1). Here, truncated GBP5 could act as a dominant-negative variant by interfering with full-length GBP5 and its partners; indeed, large-scale human studies should begin to unveil cancer-associated GBP alleles and inhibitory splice isoforms if these proteins participate in innate immune surveillance and tumor cell killing. Each is a fertile topic for further scientific and clinical exploration.
Conclusions
Janeway’s influential treatise “Approaching the asymptote?” rekindled interest in the innate immune system as a platform for mobilizing acquired cellular resistance (Janeway, 1989). Yet like all asymptotes, it remains out of reach. Major gaps in our understanding still exist in terms of how individual cells bring infection and inflammation under control, whether through direct sterilization or indirect tissue tolerance. Indeed, novel aspects of host immune defense continue to emerge in all nucleated cells—not only those of hematopoietic origin—with the GBPs occupying a central position in this newly discovered arsenal (Kim et al., 2012, 2016).
With a combined 18 members in humans and mice, the GBPs draw parallels with the 24 TLRs jointly encoded by these two species, but with a different biological purpose and design. Most GBPs seem to act as integrators and even effectors of cell-autonomous immunity once they recognize their microbial targets, unlike TLRs, which constitute classical immune sensors. Importantly, GBPs serve as major protective hubs both inside and outside of the immune system. This ubiquitous profile is reinforced by recent studies in Gbp-deficient mice that reveal susceptibility to intracellular pathogens exhibiting a wide range of cellular tropisms (Table 1). Whether GBP involvement is equally pervasive in different cancer and inflammatory models awaits inquiry. Clearly much remains to be discovered on this tour d’horizon of GBP biology (MacMicking, 2017), especially within the human population where targeted therapeutics of these immune GTPases could eventually serve as novel modifiers of disease (Kim et al., 2016).
Acknowledgments
We thank Patricia Nargi for help with manuscript editing and references.
Work in the MacMicking laboratory is supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (R01AI068041-12, R01AI108834-05). J.D. MacMicking is an investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Author contributions: K. Tretina helped conceive, write, and edit the manuscript and generated figures; E-S. Park scrutinized the relevant literature and helped edit the text; A. Maminska provided unpublished data and helped edit the text; J.D. MacMicking conceived, wrote, and edited the manuscript and helped generate figures.
References
- Abdullah N., Srinivasan B., Modiano N., Cresswell P., and Sau A.K.. 2009. Role of individual domains and identification of internal gap in human guanylate binding protein-1. J. Mol. Biol. 386:690–703. 10.1016/j.jmb.2008.12.060 [DOI] [PubMed] [Google Scholar]
- Abraham C., and Cho J.H.. 2009. Inflammatory bowel disease. N. Engl. J. Med. 361:2066–2078. 10.1056/NEJMra0804647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alase A.A., El-Sherbiny Y.M., Vital E.M., Tobin D.J., Turner N.A., and Wittmann M.. 2015. IFNλ Stimulates MxA Production in Human Dermal Fibroblasts via a MAPK-Dependent STAT1-Independent Mechanism. J. Invest. Dermatol. 135:2935–2943. 10.1038/jid.2015.317 [DOI] [PubMed] [Google Scholar]
- Alspach E., Lussier D.M., and Schreiber R.D.. 2018. Interferon γ and Its Important Roles in Promoting and Inhibiting Spontaneous and Therapeutic Cancer Immunity. Cold Spring Harb. Perspect. Biol.:a028480 10.1101/cshperspect.a028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.L., Carton J.M., Lou J., Xing L., and Rubin B.Y.. 1999. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 256:8–14. 10.1006/viro.1999.9614 [DOI] [PubMed] [Google Scholar]
- Andreoli M., Persico M., Kumar A., Orteca N., Kumar V., Pepe A., Mahalingam S., Alegria A.E., Petrella L., Sevciunaite L., et al. . 2014. Identification of the first inhibitor of the GBP1:PIM1 interaction. Implications for the development of a new class of anticancer agents against paclitaxel resistant cancer cells. J. Med. Chem. 57:7916–7932. 10.1021/jm5009902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apinjoh T.O., Anchang-Kimbi J.K., Njua-Yafi C., Ngwai A.N., Mugri R.N., Clark T.G., Rockett K.A., Kwiatkowski D.P., and Achidi E.A.. MalariaGEN Consortium . 2014. Association of candidate gene polymorphisms and TGF-beta/IL-10 levels with malaria in three regions of Cameroon: a case-control study. Malar. J. 13:236 10.1186/1475-2875-13-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J., et al. . 2016. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 165:1672–1685. 10.1016/j.cell.2016.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benci J.L., Xu B., Qiu Y., Wu T.J., Dada H., Twyman-Saint Victor C., Cucolo L., Lee D.S.M., Pauken K.E., Huang A.C., et al. . 2016. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. 167:1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghout J., Langlais D., Radovanovic I., Tam M., MacMicking J.D., Stevenson M.M., and Gros P.. 2013. Irf8-regulated genomic responses drive pathological inflammation during cerebral malaria. PLoS Pathog. 9:e1003491 10.1371/journal.ppat.1003491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B., Jiang Z., Georgel P., Crozat K., Croker B., Rutschmann S., Du X., and Hoebe K.. 2006. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 24:353–389. 10.1146/annurev.immunol.24.021605.090552 [DOI] [PubMed] [Google Scholar]
- Biering S.B., Choi J., Halstrom R.A., Brown H.M., Beatty W.L., Lee S., McCune B.T., Dominici E., Williams L.E., Orchard R.C., et al. . 2017. Viral Replication Complexes Are Targeted by LC3-Guided Interferon-Inducible GTPases. Cell Host Microbe. 22:74–85.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm U., Guethlein L., Klamp T., Ozbek K., Schaub A., Fütterer A., Pfeffer K., and Howard J.C.. 1998. Two families of GTPases dominate the complex cellular response to IFN-gamma. J. Immunol. 161:6715–6723. [PubMed] [Google Scholar]
- Bolen C.R., Ding S., Robek M.D., and Kleinstein S.H.. 2014. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 59:1262–1272. 10.1002/hep.26657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfield C.J.2016. Sulfated DAMPs mobilize human GBPs for cell-autonomous immunity against bacterial pathogens. PhD thesis. Yale University, New Haven, CT. 276 pp. [Google Scholar]
- Britzen-Laurent N., Lipnik K., Ocker M., Naschberger E., Schellerer V.S., Croner R.S., Vieth M., Waldner M., Steinberg P., Hohenadl C., and Stürzl M.. 2013. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis. 34:153–162. 10.1093/carcin/bgs310 [DOI] [PubMed] [Google Scholar]
- Broz P., and Dixit V.M.. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16:407–420. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Casper C., and Fitzmaurice C.. 2016. Infection-related cancers: prioritising an important and eliminable contributor to the global cancer burden. Lancet Glob. Health. 4:e580–e581. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 2018. Parasites – Toxoplasmosis (Toxoplasma infection). Available at: https://www.cdc.gov/parasites/toxoplasmosis/index.html (accessed December 14, 2018).
- Cheng Y.S., Colonno R.J., and Yin F.H.. 1983. Interferon induction of fibroblast proteins with guanylate binding activity. J. Biol. Chem. 258:7746–7750. [PubMed] [Google Scholar]
- Cheng Y.S., Becker-Manley M.F., Chow T.P., and Horan D.C.. 1985. Affinity purification of an interferon-induced human guanylate-binding protein and its characterization. J. Biol. Chem. 260:15834–15839. [PubMed] [Google Scholar]
- Cheng Y.S., Patterson C.E., and Staeheli P.. 1991. Interferon-induced guanylate-binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Mol. Cell. Biol. 11:4717–4725. 10.1128/MCB.11.9.4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppens I., and Romano J.D.. 2018. Hostile intruder: Toxoplasma holds host organelles captive. PLoS Pathog. 14:e1006893 10.1371/journal.ppat.1006893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Franco M.M., Marim F., Guimarães E.S., Assis N.R.G., Cerqueira D.M., Alves-Silva J., Harms J., Splitter G., Smith J., Kanneganti T.D., et al. . 2018. Brucella abortus Triggers a cGAS-Independent STING Pathway To Induce Host Protection That Involves Guanylate-Binding Proteins and Inflammasome Activation. J. Immunol. 200:607–622. 10.4049/jimmunol.1700725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E. Jr., Kerr I.M., and Stark G.R.. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 264:1415–1421. 10.1126/science.8197455 [DOI] [PubMed] [Google Scholar]
- de Buhr M.F., Mähler M., Geffers R., Hansen W., Westendorf A.M., Lauber J., Buer J., Schlegelberger B., Hedrich H.J., and Bleich A.. 2006. Cd14, Gbp1, and Pla2g2a: three major candidate genes for experimental IBD identified by combining QTL and microarray analyses. Physiol. Genomics. 25:426–434. 10.1152/physiolgenomics.00022.2005 [DOI] [PubMed] [Google Scholar]
- Decker T., Lew D.J., Cheng Y.S., Levy D.E., and Darnell J.E. Jr. 1989. Interactions of alpha- and gamma-interferon in the transcriptional regulation of the gene encoding a guanylate-binding protein. EMBO J. 8:2009–2014. 10.1002/j.1460-2075.1989.tb03608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T., Lew D.J., and Darnell J.E. Jr. 1991. Two distinct alpha-interferon-dependent signal transduction pathways may contribute to activation of transcription of the guanylate-binding protein gene. Mol. Cell. Biol. 11:5147–5153. 10.1128/MCB.11.10.5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrandi D., Konermann C., Beuter-Gunia C., Kresse A., Würthner J., Kurig S., Beer S., and Pfeffer K.. 2007. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 179:7729–7740. 10.4049/jimmunol.179.11.7729 [DOI] [PubMed] [Google Scholar]
- Degrandi D., Kravets E., Konermann C., Beuter-Gunia C., Klümpers V., Lahme S., Wischmann E., Mausberg A.K., Beer-Hammer S., and Pfeffer K.. 2013. Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc. Natl. Acad. Sci. USA. 110:294–299. 10.1073/pnas.1205635110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döcke W.D., Randow F., Syrbe U., Krausch D., Asadullah K., Reinke P., Volk H.D., and Kox W.. 1997. Monocyte deactivation in septic patients: restoration by IFN-gamma treatment. Nat. Med. 3:678–681. 10.1038/nm0697-678 [DOI] [PubMed] [Google Scholar]
- Donato M.D., Fanelli M., Mariani M., Raspaglio G., Pandya D., He S., Fiedler P., Petrillo M., Scambia G., and Ferlini C.. 2015. Nek6 and Hif-1α cooperate with the cytoskeletal gateway of drug resistance to drive outcome in serous ovarian cancer. Am. J. Cancer Res. 5:1862–1877. [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Foster R., Brakora K.A., Yusuf R.Z., and Seiden M.V.. 2006. GBP1 overexpression is associated with a paclitaxel resistance phenotype. Cancer Chemother. Pharmacol. 57:25–33. 10.1007/s00280-005-0026-3 [DOI] [PubMed] [Google Scholar]
- Duewell P., Kono H., Rayner K.J., Sirois C.M., Vladimer G., Bauernfeind F.G., Abela G.S., Franchi L., Nuñez G., Schnurr M., et al. . 2010. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 464:1357–1361. 10.1038/nature08938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D., and Petrov D.A.. 2018. Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell. 175:360–371.e13. 10.1016/j.cell.2018.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley E.M., Pilla-Moffett D.M., Zwack E.E., Piro A.S., Finethy R., Kolb J.P., Martinez J., Brodsky I.E., and Coers J.. 2017. Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc. Natl. Acad. Sci. USA. 114:E1698–E1706. 10.1073/pnas.1615771114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg F., Hartmann T.B., Dummer R., Usener D., Schadendorf D., and Eichmüller S.. 2004. GBP-5 splicing variants: New guanylate-binding proteins with tumor-associated expression and antigenicity. J. Invest. Dermatol. 122:1510–1517. 10.1111/j.0022-202X.2004.22613.x [DOI] [PubMed] [Google Scholar]
- Ferguson S.M., and De Camilli P.. 2012. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13:75–88. 10.1038/nrm3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R., Jorgensen I., Haldar A.K., de Zoete M.R., Strowig T., Flavell R.A., Yamamoto M., Nagarajan U.M., Miao E.A., and Coers J.. 2015. Guanylate binding proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect. Immun. 83:4740–4749. 10.1128/IAI.00856-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R., Luoma S., Orench-Rivera N., Feeley E.M., Haldar A.K., Yamamoto M., Kanneganti T.D., Kuehn M.J., and Coers J.. 2017. Inflammasome Activation by Bacterial Outer Membrane Vesicles Requires Guanylate Binding Proteins. MBio. 8:e01188-17 10.1128/mBio.01188-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz C., Napolitano A., Khan R., Clough B., Hirst E.M., and Frickel E.M.. 2017. TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP-positive parasite vacuoles. Sci. Rep. 7:5209 10.1038/s41598-017-05487-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R.G., Bradfield C.J., and MacMicking J.D.. 2016. Evolution of Cell-Autonomous Effector Mechanisms in Macrophages versus Non-Immune Cells. Microbiol. Spectr. 4 10.1128/microbiolspec.MCHD-0050-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay G., Braun L., Brenier-Pinchart M.P., Vollaire J., Josserand V., Bertini R.L., Varesano A., Touquet B., De Bock P.J., Coute Y., et al. . 2016. Toxoplasma gondii TgIST co-opts host chromatin repressors dampening STAT1-dependent gene regulation and IFN-γ-mediated host defenses. J. Exp. Med. 213:1779–1798. 10.1084/jem.20160340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD Diarrhoeal Diseases Collaborators 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 17:909–948. 10.1016/S1473-3099(17)30276-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Praefcke G.J., Renault L., Wittinghofer A., and Herrmann C.. 2006. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 440:101–104. 10.1038/nature04510 [DOI] [PubMed] [Google Scholar]
- Godoy P., Cadenas C., Hellwig B., Marchan R., Stewart J., Reif R., Lohr M., Gehrmann M., Rahnenführer J., Schmidt M., and Hengstler J.G.. 2014. Interferon-inducible guanylate binding protein (GBP2) is associated with better prognosis in breast cancer and indicates an efficient T cell response. Breast Cancer. 21:491–499. 10.1007/s12282-012-0404-8 [DOI] [PubMed] [Google Scholar]
- Goo Y.H., Son S.H., Yechoor V.K., and Paul A.. 2016. Transcriptional Profiling of Foam Cells Reveals Induction of Guanylate-Binding Proteins Following Western Diet Acceleration of Atherosclerosis in the Absence of Global Changes in Inflammation. J. Am. Heart Assoc. 5:e002663 10.1161/JAHA.115.002663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenzi E., Töpolt K., Cornali E., Lubeseder-Martellato C., Jörg A., Matzen K., Zietz C., Kremmer E., Nappi F., Schwemmle M., et al. . 2001. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 20:5568–5577. 10.1093/emboj/20.20.5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães D.P., Oliveira I.M., de Moraes E., Paiva G.R., Souza D.M., Barnas C., Olmedo D.B., Pinto C.E., Faria P.A., De Moura Gallo C.V., et al. . 2009. Interferon-inducible guanylate binding protein (GBP)-2: a novel p53-regulated tumor marker in esophageal squamous cell carcinomas. Int. J. Cancer. 124:272–279. 10.1002/ijc.23944 [DOI] [PubMed] [Google Scholar]
- Haldar A.K., Saka H.A., Piro A.S., Dunn J.D., Henry S.C., Taylor G.A., Frickel E.M., Valdivia R.H., and Coers J.. 2013. IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog. 9:e1003414 10.1371/journal.ppat.1003414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar A.K., Foltz C., Finethy R., Piro A.S., Feeley E.M., Pilla-Moffett D.M., Komatsu M., Frickel E.M., and Coers J.. 2015. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc. Natl. Acad. Sci. USA. 112:E5628–E5637. 10.1073/pnas.1515966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar A.K., Piro A.S., Finethy R., Espenschied S.T., Brown H.E., Giebel A.M., Frickel E.M., Nelson D.E., and Coers J.. 2016. Chlamydia trachomatis Is Resistant to Inclusion Ubiquitination and Associated Host Defense in Gamma Interferon-Primed Human Epithelial Cells. MBio. 7:e01417-16 10.1128/mBio.01417-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W.Z., Strowig T., Thaiss C.A., Kau A.L., Eisenbarth S.C., Jurczak M.J., et al. . 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 482:179–185. 10.1038/nature10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong A.W., Meng Z., and Guan K.L.. 2016. The Hippo pathway in intestinal regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 13:324–337. 10.1038/nrgastro.2016.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S. 2017. Inflammation, metaflammation and immunometabolic disorders. Nature. 542:177–185. 10.1038/nature21363 [DOI] [PubMed] [Google Scholar]
- Hotchkiss R.S., Monneret G., and Payen D.. 2013. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 13:862–874. 10.1038/nri3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., and Sibley L.D.. 2012. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat. Rev. Microbiol. 10:766–778. 10.1038/nrmicro2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince S., Kutsch M., Shydlovskyi S., and Herrmann C.. 2017. The human guanylate-binding proteins hGBP-1 and hGBP-5 cycle between monomers and dimers only. FEBS J. 284:2284–2301. 10.1111/febs.14126 [DOI] [PubMed] [Google Scholar]
- Ingram J.P., Tursi S., Zhang T., Guo W., Yin C., A Wynosky-Dolfi M., van der Heijden J., Cai K.Q., Yamamoto M., Finlay B.B., et al. . 2018. A Nonpyroptotic IFN-γ-Triggered Cell Death Mechanism in Nonphagocytic Cells Promotes Salmonella Clearance In Vivo. J. Immunol. 200:3626–3634. 10.4049/jimmunol.1701386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsui Y., Sakamoto N., Kakinuma S., Nakagawa M., Sekine-Osajima Y., Tasaka-Fujita M., Nishimura-Sakurai Y., Suda G., Karakama Y., Mishima K., et al. . 2009. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology. 50:1727–1737. 10.1002/hep.23195 [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Jr 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54:1–13. 10.1101/SQB.1989.054.01.003 [DOI] [PubMed] [Google Scholar]
- Jin T., Huang M., Smith P., Jiang J., and Xiao T.S.. 2013. Structure of the caspase-recruitment domain from a zebrafish guanylate-binding protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69:855–860. 10.1107/S1744309113015558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A.C., Piro A., Clough B., Siew M., Virreira Winter S., Coers J., and Frickel E.M.. 2016. Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii. Cell. Microbiol. 18:1056–1064. 10.1111/cmi.12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis G., and Stoye J.P.. 2016. Immune responses to endogenous retroelements: taking the bad with the good. Nat. Rev. Immunol. 16:207–219. 10.1038/nri.2016.27 [DOI] [PubMed] [Google Scholar]
- Kim B.H., Shenoy A.R., Kumar P., Das R., Tiwari S., and MacMicking J.D.. 2011. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 332:717–721. 10.1126/science.1201711 [DOI] [PubMed] [Google Scholar]
- Kim B.H., Shenoy A.R., Kumar P., Bradfield C.J., and MacMicking J.D.. 2012. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 12:432–444. 10.1016/j.chom.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.H., Chee J.D., Bradfield C.J., Park E.S., Kumar P., and MacMicking J.D.. 2016. Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nat. Immunol. 17:481–489. 10.1038/ni.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.K., Yeh S.H., Lin M.W., Liu C.J., Lai M.Y., Kao J.H., Chen D.S., and Chen P.J.. 2002. Genetic polymorphisms in interferon pathway and response to interferon treatment in hepatitis B patients: A pilot study. Hepatology. 36:1416–1424. 10.1002/hep.1840360618 [DOI] [PubMed] [Google Scholar]
- Krapp C., Hotter D., Gawanbacht A., McLaren P.J., Kluge S.F., Stürzel C.M., Mack K., Reith E., Engelhart S., Ciuffi A., et al. . 2016. Guanylate Binding Protein (GBP) 5 Is an Interferon-Inducible Inhibitor of HIV-1 Infectivity. Cell Host Microbe. 19:504–514. 10.1016/j.chom.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Kravets E., Degrandi D., Weidtkamp-Peters S., Ries B., Konermann C., Felekyan S., Dargazanli J.M., Praefcke G.J., Seidel C.A., Schmitt L., et al. . 2012. The GTPase activity of murine guanylate-binding protein 2 (mGBP2) controls the intracellular localization and recruitment to the parasitophorous vacuole of Toxoplasma gondii. J. Biol. Chem. 287:27452–27466. 10.1074/jbc.M112.379636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravets E., Degrandi D., Ma Q., Peulen T.O., Klümpers V., Felekyan S., Kühnemuth R., Weidtkamp-Peters S., Seidel C.A., and Pfeffer K.. 2016. Guanylate binding proteins directly attack Toxoplasma gondii via supramolecular complexes. eLife. 5:e11479 10.7554/eLife.11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse A., Konermann C., Degrandi D., Beuter-Gunia C., Wuerthner J., Pfeffer K., and Beer S.. 2008. Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC Genomics. 9:158 10.1186/1471-2164-9-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzelmann S., Praefcke G.J., and Herrmann C.. 2006. Transient kinetic investigation of GTP hydrolysis catalyzed by interferon-gamma-induced hGBP1 (human guanylate binding protein 1). J. Biol. Chem. 281:28627–28635. 10.1074/jbc.M604911200 [DOI] [PubMed] [Google Scholar]
- Lan Q., Wang A., Cheng Y., Mukasa A., Ma J., Hong L., Yu S., Sun L., Huang Q., Purow B., and Li M.. 2016. Guanylate binding protein-1 mediates EGFRvIII and promotes glioblastoma growth in vivo but not in vitro. Oncotarget. 7:9680–9691. 10.18632/oncotarget.7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhang J., Sun Y., Wang H., and Wang Y.. 2009. The evolutionarily dynamic IFN-inducible GTPase proteins play conserved immune functions in vertebrates and cephalochordates. Mol. Biol. Evol. 26:1619–1630. 10.1093/molbev/msp074 [DOI] [PubMed] [Google Scholar]
- Li M., Mukasa A., Inda M.M., Zhang J., Chin L., Cavenee W., and Furnari F.. 2011. Guanylate binding protein 1 is a novel effector of EGFR-driven invasion in glioblastoma. J. Exp. Med. 208:2657–2673. 10.1084/jem.20111102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Jiang W., Yu Q., Liu W., Zhou P., Li J., Xu J., Xu B., Wang F., and Shao F.. 2017. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature. 551:378–383. [DOI] [PubMed] [Google Scholar]
- Liehl P., Zuzarte-Luís V., Chan J., Zillinger T., Baptista F., Carapau D., Konert M., Hanson K.K., Carret C., Lassnig C., et al. . 2014. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med. 20:47–53. 10.1038/nm.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Rice K.L., Buzzai M., Hexner E., Costa F.F., Kilpivaara O., Mullally A., Soares M.B., Ebert B.L., Levine R., and Licht J.D.. 2013. miR-433 is aberrantly expressed in myeloproliferative neoplasms and suppresses hematopoietic cell growth and differentiation. Leukemia. 27:344–352. 10.1038/leu.2012.224 [DOI] [PubMed] [Google Scholar]
- Lindenberg V., Mölleken K., Kravets E., Stallmann S., Hegemann J.H., Degrandi D., and Pfeffer K.. 2017. Broad recruitment of mGBP family members to Chlamydia trachomatis inclusions. PLoS One. 12:e0185273 10.1371/journal.pone.0185273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.C., Sarhan J., Panda A., Muendlein H.I., Ilyukha V., Coers J., Yamamoto M., Isberg R.R., and Poltorak A.. 2018. Constitutive Interferon Maintains GBP Expression Required for Release of Bacterial Components Upstream of Pyroptosis and Anti-DNA Responses. Cell Reports. 24:155–168.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., and Martin-Villalba A.. 2015. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 17:329–340. 10.1016/j.stem.2015.07.002 [DOI] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., and Kroemer G.. 2013. The hallmarks of aging. Cell. 153:1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeseder-Martellato C., Guenzi E., Jörg A., Töpolt K., Naschberger E., Kremmer E., Zietz C., Tschachler E., Hutzler P., Schwemmle M., et al. . 2002. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am. J. Pathol. 161:1749–1759. 10.1016/S0002-9440(10)64452-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J.D. 2004. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25:601–609. 10.1016/j.it.2004.08.010 [DOI] [PubMed] [Google Scholar]
- MacMicking J.D. 2005. Immune control of phagosomal bacteria by p47 GTPases. Curr. Opin. Microbiol. 8:74–82. 10.1016/j.mib.2004.12.012 [DOI] [PubMed] [Google Scholar]
- MacMicking J.D. 2009. Recognizing macrophage activation and host defense. Cell Host Microbe. 5:405–407. 10.1016/j.chom.2009.05.006 [DOI] [PubMed] [Google Scholar]
- MacMicking J.D. 2012. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12:367–382. 10.1038/nri3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J.D. 2017. Microbiology: Bacteria disarm host-defence proteins. Nature. 551:303–305. 10.1038/nature24157 [DOI] [PubMed] [Google Scholar]
- MacMicking J.D., Taylor G.A., and McKinney J.D.. 2003. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 302:654–659. 10.1126/science.1088063 [DOI] [PubMed] [Google Scholar]
- Man S.M., Karki R., Malireddi R.K., Neale G., Vogel P., Yamamoto M., Lamkanfi M., and Kanneganti T.D.. 2015. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16:467–475. 10.1038/ni.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., Sasai M., Place D.E., Kesavardhana S., Temirov J., Frase S., Zhu Q., Malireddi R.K.S., Kuriakose T., et al. . 2016. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell. 167:382–396.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis J.F., Kapoustina O., Langlais D., Ruddy R., Dufour C.R., Kim B.H., MacMicking J.D., Giguère V., and Gros P.. 2011. Interferon regulatory factor 8 regulates pathways for antigen presentation in myeloid cells and during tuberculosis. PLoS Genet. 7:e1002097 10.1371/journal.pgen.1002097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S., Parvanova I., Zerrahn J., Griffiths G., Schell G., Reichmann G., and Howard J.C.. 2005. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 1:e24 10.1371/journal.ppat.0010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty-Roix R., Vladimer G.I., Pouliot K., Weng D., Buglione-Corbett R., West K., MacMicking J.D., Chee J.D., Wang S., Lu S., and Lien E.. 2016. Identification of QS-21 as an inflammasome-activating molecular component of saponin adjuvants. J. Biol. Chem. 291:1123–1136. 10.1074/jbc.M115.683011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta S.K., Patten K., Wang Q., Kim B.H., MacMicking J.D., and Sibley L.D.. 2018. NADPH Oxidase and Guanylate Binding Protein 5 Restrict Survival of Avirulent Type III Strains of Toxoplasma gondii in Naive Macrophages. MBio. 9:e01393-18 10.1128/mBio.01393-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E., and Broz P.. 2017. Evolutionary Convergence and Divergence in NLR Function and Structure. Trends Immunol. 38:744–757. 10.1016/j.it.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Meunier E., Dick M.S., Dreier R.F., Schürmann N., Kenzelmann Broz D., Warming S., Roose-Girma M., Bumann D., Kayagaki N., Takeda K., et al. . 2014. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 509:366–370. 10.1038/nature13157 [DOI] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R.F., Costanzo S., Anton L., Rühl S., Dussurgey S., Dick M.S., Kistner A., Rigard M., et al. . 2015. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16:476–484. 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirpuri J., Brazil J.C., Berardinelli A.J., Nasr T.R., Cooper K., Schnoor M., Lin P.W., Parkos C.A., and Louis N.A.. 2010. Commensal Escherichia coli reduces epithelial apoptosis through IFN-alphaA-mediated induction of guanylate binding protein-1 in human and murine models of developing intestine. J. Immunol. 184:7186–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley D., Yoshida H., Mostafavi S., Asinovski N., Ortiz-Lopez A., Symanowicz P., Telliez J.B., Hegen M., Clark J.D., Mathis D., and Benoist C.. 2016. Network pharmacology of JAK inhibitors. Proc. Natl. Acad. Sci. USA. 113:9852–9857. 10.1073/pnas.1610253113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S., Yoshida H., Moodley D., LeBoité H., Rothamel K., Raj T., Ye C.J., Chevrier N., Zhang S.Y., Feng T., et al. Immunological Genome Project Consortium . 2016. Parsing the interferon transcriptional network and its disease associations. Cell. 164:564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naschberger E., Werner T., Vicente A.B., Guenzi E., Töpolt K., Leubert R., Lubeseder-Martellato C., Nelson P.J., and Stürzl M.. 2004. Nuclear factor-kappaB motif and interferon-alpha-stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem. J. 379:409–420. 10.1042/bj20031873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C.F., Murray H.W., Wiebe M.E., and Rubin B.Y.. 1983. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158:670–689. 10.1084/jem.158.3.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.T., Hu Y., Widney D.P., Mar R.A., and Smith J.B.. 2002. Murine GBP-5, a new member of the murine guanylate-binding protein family, is coordinately regulated with other GBPs in vivo and in vitro. J. Interferon Cytokine Res. 22:899–909. 10.1089/107999002760274926 [DOI] [PubMed] [Google Scholar]
- Niedelman W., Sprokholt J.K., Clough B., Frickel E.M., and Saeij J.P.. 2013. Cell death of gamma interferon-stimulated human fibroblasts upon Toxoplasma gondii infection induces early parasite egress and limits parasite replication. Infect. Immun. 81:4341–4349. 10.1128/IAI.00416-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirschl C.J., Suárez-Fariñas M., Izar B., Prakadan S., Dannenfelser R., Tirosh I., Liu Y., Zhu Q., Devi K.S.P., Carroll S.L., et al. . 2017. IFNγ-Dependent Tissue-Immune Homeostasis Is Co-opted in the Tumor Microenvironment. Cell. 170:127–141.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann A., Wixler L., Boergeling Y., Wixler V., and Ludwig S.. 2012. A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication. FASEB J. 26:1290–1300. 10.1096/fj.11-189886 [DOI] [PubMed] [Google Scholar]
- O’Neill L.A., Kishton R.J., and Rathmell J.. 2016. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16:553–565. 10.1038/nri.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima J., Lee Y., Sasai M., Saitoh T., Su Ma J., Kamiyama N., Matsuura Y., Pann-Ghill S., Hayashi M., Ebisu S., et al. . 2014. Role of mouse and human autophagy proteins in IFN-γ-induced cell-autonomous responses against Toxoplasma gondii. J. Immunol. 192:3328–3335. 10.4049/jimmunol.1302822 [DOI] [PubMed] [Google Scholar]
- Olias P., Etheridge R.D., Zhang Y., Holtzman M.J., and Sibley L.D.. 2016. Toxoplasma Effector Recruits the Mi-2/NuRD Complex to Repress STAT1 Transcription and Block IFN-γ-Dependent Gene Expression. Cell Host Microbe. 20:72–82. 10.1016/j.chom.2016.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski M.A., Gray J., and Vestal D.J.. 2006. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interferon Cytokine Res. 26:328–352. 10.1089/jir.2006.26.328 [DOI] [PubMed] [Google Scholar]
- Ostler N., Britzen-Laurent N., Liebl A., Naschberger E., Lochnit G., Ostler M., Forster F., Kunzelmann P., Ince S., Supper V., et al. . 2014. Gamma interferon-induced guanylate binding protein 1 is a novel actin cytoskeleton remodeling factor. Mol. Cell. Biol. 34:196–209. 10.1128/MCB.00664-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenhoff T.H., Dass R.H., Yang N., Zhang M.M., Wong H.E., Sahiratmadja E., Khor C.C., Alisjahbana B., van Crevel R., Marzuki S., et al. . 2012. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One. 7:e45839 10.1371/journal.pone.0045839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto U., Schneider A., Denkhaus H., and Conrad S.. 1988. [Treatment of metastatic kidney cancer with recombinant alpha-2 or gamma interferon. Results of 2 clinical phase II and III studies]. Onkologie. 11:185–191. [DOI] [PubMed] [Google Scholar]
- Overacre-Delgoffe A.E., Chikina M., Dadey R.E., Yano H., Brunazzi E.A., Shayan G., Horne W., Moskovitz J.M., Kolls J.K., Sander C., et al. . 2017. Interferon-γ Drives Treg Fragility to Promote Anti-tumor Immunity. Cell. 169:1130–1141.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palesch D., Bosinger S.E., Tharp G.K., Vanderford T.H., Paiardini M., Chahroudi A., Johnson Z.P., Kirchhoff F., Hahn B.H., Norgren R.B., et al. . 2018. Sooty mangabey genome sequence provides insight into AIDS resistance in a natural SIV host. Nature. 553:77–81. 10.1038/nature25140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm N.W., Rosenstein R.K., and Medzhitov R.. 2012. Allergic host defences. Nature. 484:465–472. 10.1038/nature11047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M., Khoury S., Chabot-Doré A.J., Sotocinal S.G., Slade G.D., Smith S.B., Fillingim R.B., Ohrbach R., Greenspan J.D., Maixner W., et al. . 2017. Effect of human genetic variability on gene expression in dorsal root ganglia and association with pain phenotypes. Cell Reports. 19:1940–1952. 10.1016/j.celrep.2017.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B.S., Rautela J., and Hertzog P.J.. 2016. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer. 16:131–144. 10.1038/nrc.2016.14 [DOI] [PubMed] [Google Scholar]
- Pilla D.M., Hagar J.A., Haldar A.K., Mason A.K., Degrandi D., Pfeffer K., Ernst R.K., Yamamoto M., Miao E.A., and Coers J.. 2014. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. USA. 111:6046–6051. 10.1073/pnas.1321700111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro A.S., Hernandez D., Luoma S., Feeley E.M., Finethy R., Yirga A., Frickel E.M., Lesser C.F., and Coers J.. 2017. Detection of Cytosolic Shigella flexneri via a C-Terminal Triple-Arginine Motif of GBP1 Inhibits Actin-Based Motility. MBio. 8:e01979-17 10.1128/mBio.01979-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G.J., and McMahon H.T.. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147. 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- Praefcke G.J., Geyer M., Schwemmle M., Robert Kalbitzer H., and Herrmann C.. 1999. Nucleotide-binding characteristics of human guanylate-binding protein 1 (hGBP1) and identification of the third GTP-binding motif. J. Mol. Biol. 292:321–332. 10.1006/jmbi.1999.3062 [DOI] [PubMed] [Google Scholar]
- Praefcke G.J., Kloep S., Benscheid U., Lilie H., Prakash B., and Herrmann C.. 2004. Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J. Mol. Biol. 344:257–269. 10.1016/j.jmb.2004.09.026 [DOI] [PubMed] [Google Scholar]
- Prakash B., Praefcke G.J., Renault L., Wittinghofer A., and Herrmann C.. 2000. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 403:567–571. 10.1038/35000617 [DOI] [PubMed] [Google Scholar]
- Qin A., Lai D.H., Liu Q., Huang W., Wu Y.P., Chen X., Yan S., Xia H., Hide G., Lun Z.R., et al. . 2017. Guanylate-binding protein 1 (GBP1) contributes to the immunity of human mesenchymal stromal cells against Toxoplasma gondii. Proc. Natl. Acad. Sci. USA. 114:1365–1370. 10.1073/pnas.1619665114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F., MacMicking J.D., and James L.C.. 2013. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 340:701–706. 10.1126/science.1233028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., et al. Human Cell Atlas Meeting Participants . 2017. The Human Cell Atlas. eLife. 6:e27041 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupper A.C., and Cardelli J.A.. 2008. Induction of guanylate binding protein 5 by gamma interferon increases susceptibility to Salmonella enterica serovar Typhimurium-induced pyroptosis in RAW 264.7 cells. Infect. Immun. 76:2304–2315. 10.1128/IAI.01437-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.C., Dick M.S., Lagrange B., Degrandi D., Pfeffer K., Yamamoto M., Meunier E., Pelczar P., Henry T., and Broz P.. 2018. LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. 37:e98089 10.15252/embj.201798089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M., Sakaguchi N., Ma J.S., Nakamura S., Kawabata T., Bando H., Lee Y., Saitoh T., Akira S., Iwasaki A., et al. . 2017. Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat. Immunol. 18:899–910. 10.1038/ni.3767 [DOI] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., and Rice C.M.. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32:513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoor M., Betanzos A., Weber D.A., and Parkos C.A.. 2009. Guanylate-binding protein-1 is expressed at tight junctions of intestinal epithelial cells in response to interferon-gamma and regulates barrier function through effects on apoptosis. Mucosal Immunol. 2:33–42. 10.1038/mi.2008.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D.M., Bonelli M., Gadina M., and O’Shea J.J.. 2016. Type I/II cytokines, JAKs, and new strategies for treating autoimmune diseases. Nat. Rev. Rheumatol. 12:25–36. 10.1038/nrrheum.2015.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmle M., and Staeheli P.. 1994. The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem. 269:11299–11305. [PubMed] [Google Scholar]
- Secombes C.J., and Zou J.. 2017. Evolution of interferons and interferon receptors. Front. Immunol. 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck E.M., Fentress S.J., Beatty W.L., Degrandi D., Pfeffer K., Virgin H.W. IV, Macmicking J.D., and Sibley L.D.. 2013. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 9:e1003320 10.1371/journal.ppat.1003320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck E.M., Orchard R.C., Lassen K.G., Beatty W.L., Xavier R.J., Levine B., Virgin H.W., and Sibley L.D.. 2015. A Noncanonical Autophagy Pathway Restricts Toxoplasma gondii Growth in a Strain-Specific Manner in IFN-γ-Activated Human Cells. MBio. 6:e01157–e15. 10.1128/mBio.01157-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., and Hahn B.H.. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1:a006841 10.1101/cshperspect.a006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A.R., Kim B.H., Choi H.P., Matsuzawa T., Tiwari S., and MacMicking J.D.. 2007. Emerging themes in IFN-gamma-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology. 212:771–784. 10.1016/j.imbio.2007.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A.R., Wellington D.A., Kumar P., Kassa H., Booth C.J., Cresswell P., and MacMicking J.D.. 2012. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 336:481–485. 10.1126/science.1217141 [DOI] [PubMed] [Google Scholar]
- Shydlovskyi S., Zienert A.Y., Ince S., Dovengerds C., Hohendahl A., Dargazanli J.M., Blum A., Günther S.D., Kladt N., Stürzl M., et al. . 2017. Nucleotide-dependent farnesyl switch orchestrates polymerization and membrane binding of human guanylate-binding protein 1. Proc. Natl. Acad. Sci. USA. 114:E5559–E5568. 10.1073/pnas.1620959114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi Y., Volkova V., Kobets T., Havelková H., Krayem I., Slapničková M., Demant P., and Lipoldová M.. 2018. Genetic Regulation of Guanylate-Binding Proteins 2b and 5 during Leishmaniasis in Mice. Front. Immunol. 9:130 10.3389/fimmu.2018.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudja S.M., Chandrabos C., Yakob E., Veenstra M., Palliser D., and Lauvau G.. 2014. Memory-T-cell-derived interferon-γ instructs potent innate cell activation for protective immunity. Immunity. 40:974–988. 10.1016/j.immuni.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeheli P., Prochazka M., Steigmeier P.A., and Haller O.. 1984. Genetic control of interferon action: mouse strain distribution and inheritance of an induced protein with guanylate-binding property. Virology. 137:135–142. 10.1016/0042-6822(84)90016-3 [DOI] [PubMed] [Google Scholar]
- Stickney J.T., and Buss J.E.. 2000. Murine guanylate-binding protein: incomplete geranylgeranyl isoprenoid modification of an interferon-gamma-inducible guanosine triphosphate-binding protein. Mol. Biol. Cell. 11:2191–2200. 10.1091/mbc.11.7.2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington M.J.T., Rozenblatt-Rosen O., Regev A., and Teichmann S.A.. 2017. Single-cell transcriptomics to explore the immune system in health and disease. Science. 358:58–63. 10.1126/science.aan6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen E.R., Mich J.K., Yao Z., Hodge R.D., Doyle A.M., Jang S., Shehata S.I., Nelson A.M., Shapovalova N.V., Levi B.P., and Ramanathan S.. 2016. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods. 13:87–93. 10.1038/nmeth.3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton A.R., Nyabuto G.O., Trendel J.A., Mazur T.M., Wilson J.P., Wadi S., Justinger J.S., Moore G.L., Nguyen P.T., and Vestal D.J.. 2016. Guanylate-Binding Protein-1 protects ovarian cancer cell lines but not breast cancer cell lines from killing by paclitaxel. Biochem. Biophys. Res. Commun. 478:1617–1623. 10.1016/j.bbrc.2016.08.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Choi H.P., Matsuzawa T., Pypaert M., and MacMicking J.D.. 2009. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat. Immunol. 10:907–917. 10.1038/ni.1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripal P., Bauer M., Naschberger E., Mörtinger T., Hohenadl C., Cornali E., Thurau M., and Stürzl M.. 2007. Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 27:44–52. 10.1089/jir.2007.0086 [DOI] [PubMed] [Google Scholar]
- Tyrkalska S.D., Candel S., Angosto D., Gómez-Abellán V., Martín-Sánchez F., García-Moreno D., Zapata-Pérez R., Sánchez-Ferrer Á., Sepulcre M.P., Pelegrín P., and Mulero V.. 2016. Neutrophils mediate Salmonella Typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat. Commun. 7:12077 10.1038/ncomms12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterer B., Wiesmann V., Gunasekaran M., Sticht H., Tenkerian C., Behrens J., Leone M., Engel F.B., Britzen-Laurent N., Naschberger E., et al. . 2018. IFN-γ-response mediator GBP-1 represses human cell proliferation by inhibiting the Hippo signaling transcription factor TEAD. Biochem. J. 475:2955–2967. 10.1042/BCJ20180123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B., Tucci S., Kelso J., Schraiber J.G., Wolf A.B., Gittelman R.M., Dannemann M., Grote S., McCoy R.C., Norton H., et al. . 2016. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 352:235–239. 10.1126/science.aad9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A.C., Lemire M., Fortin G., Louis E., Silverberg M.S., Collette C., Baba N., Libioulle C., Belaiche J., Bitton A., et al. . 2009. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat. Genet. 41:71–76. 10.1038/ng.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virreira Winter S., Niedelman W., Jensen K.D., Rosowski E.E., Julien L., Spooner E., Caradonna K., Burleigh B.A., Saeij J.P., Ploegh H.L., and Frickel E.M.. 2011. Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS One. 6:e24434 10.1371/journal.pone.0024434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Trinidad N.J., Moayeri M., Kintzer A.F., Wang S.B., van Rooijen N., Brown C.R., Krantz B.A., Leppla S.H., Gronert K., and Vance R.E.. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 490:107–111. 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vöpel T., Hengstenberg C.S., Peulen T.O., Ajaj Y., Seidel C.A., Herrmann C., and Klare J.P.. 2014. Triphosphate induced dimerization of human guanylate binding protein 1 involves association of the C-terminal helices: a joint double electron-electron resonance and FRET study. Biochemistry. 53:4590–4600. 10.1021/bi500524u [DOI] [PubMed] [Google Scholar]
- Wadi S., Tipton A.R., Trendel J.A., Khuder S.A., and Vestal D.J.. 2016. hGBP-1 Expression Predicts Shorter Progression-Free Survival in Ovarian Cancers, While Contributing to Paclitaxel Resistance. J. Cancer Ther. 7:994–1007. 10.4236/jct.2016.713097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallet P., Benaoudia S., Mosnier A., Lagrange B., Martin A., Lindgren H., Golovliov I., Michal F., Basso P., Djebali S., et al. . 2017. IFN-γ extends the immune functions of Guanylate Binding Proteins to inflammasome-independent antibacterial activities during Francisella novicida infection. PLoS Pathog. 13:e1006630 10.1371/journal.ppat.1006630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandel M.P., Pathe C., Werner E.I., Ellison C.J., Boyle K.B., von der Malsburg A., Rohde J., and Randow F.. 2017. GBPs Inhibit Motility of Shigella flexneri but Are Targeted for Degradation by the Bacterial Ubiquitin Ligase IpaH9.8. Cell Host Microbe. 22:507–518.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P.A. 1971. A cell is not an island entire of itself. Perspect. Biol. Med. 14:182–205. 10.1353/pbm.1971.0006 [DOI] [PubMed] [Google Scholar]
- World Health Organization 2018. The top 10 causes of death. Available at: https://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed December 14, 2018).
- Wu X., Dao Thi V.L., Huang Y., Billerbeck E., Saha D., Hoffmann H.H., Wang Y., Silva L.A.V., Sarbanes S., Sun T., et al. . 2018. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell. 172:423–438.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T., Benoist C., and Mathis D.. 2006. A shared gene-expression signature in innate-like lymphocytes. Immunol. Rev. 210:52–66. 10.1111/j.0105-2896.2006.00371.x [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Okuyama M., Ma J.S., Kimura T., Kamiyama N., Saiga H., Ohshima J., Sasai M., Kayama H., Okamoto T., et al. . 2012. A cluster of interferon-γ-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 37:302–313. 10.1016/j.immuni.2012.06.009 [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., et al. . 2013. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20:1131–1139. 10.1038/nsmb.2660 [DOI] [PubMed] [Google Scholar]
- Yap G.S., and Sher A.. 1999. Effector cells of both nonhemopoietic and hemopoietic origin are required for interferon (IFN)-gamma- and tumor necrosis factor (TNF)-alpha-dependent host resistance to the intracellular pathogen, Toxoplasma gondii. J. Exp. Med. 189:1083–1092. 10.1084/jem.189.7.1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak D.E., Penn-Nicholson A., Scriba T.J., Thompson E., Suliman S., Amon L.M., Mahomed H., Erasmus M., Whatney W., Hussey G.D., et al. ACS and GC6-74 cohort study groups . 2016. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 387:2312–2322. 10.1016/S0140-6736(15)01316-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M.H., Boyd K.L., Vogel P., Kastan M.B., Lamkanfi M., and Kanneganti T.D.. 2010. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 32:379–391. 10.1016/j.immuni.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang Y., Wu W., Wang F., Liu X., Shui G., and Nie C.. 2017. Guanylate-binding protein 2 regulates Drp1-mediated mitochondrial fission to suppress breast cancer cell invasion. Cell Death Dis. 8:e3151 10.1038/cddis.2017.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Boisson-Dupuis S., Chapgier A., Yang K., Bustamante J., Puel A., Picard C., Abel L., Jouanguy E., and Casanova J.L.. 2008. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol. Rev. 226:29–40. 10.1111/j.1600-065X.2008.00698.x [DOI] [PubMed] [Google Scholar]
- Zou Z., Meng Z., Ma C., Liang D., Sun R., and Lan K.. 2017. Guanylate-Binding Protein 1 Inhibits Nuclear Delivery of Kaposi’s Sarcoma-Associated Herpesvirus Virions by Disrupting Formation of Actin Filament. J. Virol. 91:e00632-17 10.1128/JVI.00632-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack E.E., Feeley E.M., Burton A.R., Hu B., Yamamoto M., Kanneganti T.D., Bliska J.B., Coers J., and Brodsky I.E.. 2017. Guanylate Binding Proteins Regulate Inflammasome Activation in Response to Hyperinjected Yersinia Translocon Components. Infect. Immun. 85:e00778-16 10.1128/IAI.00778-16 [DOI] [PMC free article] [PubMed] [Google Scholar]