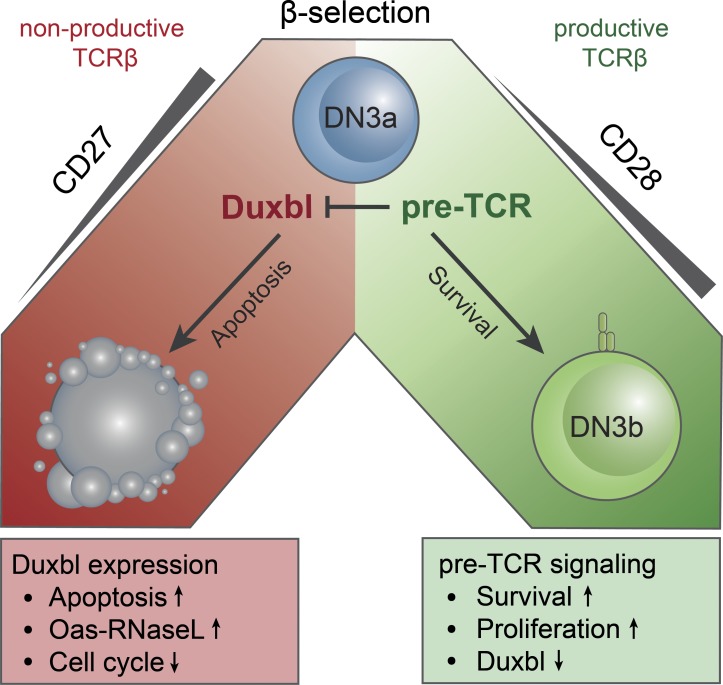
During β-selection, T cells without productive TCRβ rearrangements are eliminated. Klein et al. show that the transcription factor Duxbl regulates this process by inducing apoptosis through activation of the Oas/RNaseL pathway. Successful TCRβ rearrangement rescues cells by pre-TCR–mediated Duxbl suppression.
Abstract
T cell development is critically dependent on successful rearrangement of antigen-receptor chains. At the β-selection checkpoint, only cells with a functional rearrangement continue in development. However, how nonselected T cells proceed in their dead-end fate is not clear. We identified low CD27 expression to mark pre-T cells that have failed to rearrange their β-chain. Expression profiling and single-cell transcriptome clustering identified a developmental trajectory through β-selection and revealed specific expression of the transcription factor Duxbl at a stage of high recombination activity before β-selection. Conditional transgenic expression of Duxbl resulted in a developmental block at the DN3-to-DN4 transition due to reduced proliferation and enhanced apoptosis, whereas RNA silencing of Duxbl led to a decrease in apoptosis. Transcriptome analysis linked Duxbl to elevated expression of the apoptosis-inducing Oas/RNaseL pathway. RNaseL deficiency or sustained Bcl2 expression led to a partial rescue of cells in Duxbl transgenic mice. These findings identify Duxbl as a regulator of β-selection by inducing apoptosis in cells with a nonfunctional rearrangement.
Graphical Abstract
Introduction
T cell development occurs in the thymus and is initiated by a bone marrow–derived multipotent progenitor named thymus settling progenitor (Zlotoff and Bhandoola, 2011). The identity of the exact cell type that migrates to the thymus is still under debate, since several possible candidates have been described (Rodewald et al., 1994; Kondo et al., 1997; Krueger and von Boehmer, 2007; Serwold et al., 2009; Saran et al., 2010). There is consensus that this progenitor retains the ability to give rise to several lineages including B cells, natural killer cells, dendritic cells, and other myeloid lineages (Balciunaite et al., 2005b; Ceredig et al., 2007; Bell and Bhandoola, 2008; Wada et al., 2008; Luis et al., 2016). Final commitment to the T cell pathway is achieved upon Notch1 engagement (Radtke et al., 1999; Balciunaite et al., 2005a; Sambandam et al., 2005).
Thymic T cell development is a stepwise process that involves several successive stages, which are phenotypically distinguished by the expression of various cell surface markers. The most immature populations are characterized by the absence of CD4 and CD8 and are therefore named double-negative (DN) cells (Ceredig and Rolink, 2002). The DN population can be further subdivided based on the expression pattern of CD25, CD44, and CD117 (Godfrey et al., 1992, 1993; Massa et al., 2006). High-level expression of CD44 and CD117 and the absence of CD25 mark DN1 cells, which retain the potential to give rise to different lineages. At the next stage, DN2, progenitors are additionally characterized by expression of CD25. Upon progression to the DN3 stage, which displays lower CD44 and CD117 expression, final commitment to the T cell lineage takes place (Yui and Rothenberg, 2014). Down-regulation of CD25 marks the onset of the DN4 stage that is negative for all three surface markers (Godfrey et al., 1994). After the DN4 stage, CD4 as well as CD8 become up-regulated, and therefore cells are named double-positive (DP) cells. Finally, CD4 or CD8 single-positive cells expressing a functional TCR will undergo positive and negative selection, thereby completing their maturation in the thymus (Germain, 2002).
T cell development can also be subdivided into developmentally distinct stages by the use of the rearrangement status of the β- and the α-chain of the TCR. β-Chain rearrangement starts at the DN2 and is completed at the DN3 stage (Capone et al., 1998), whereas rearrangement of the α-chain takes place at the DP stage (Livák et al., 1999). An essential checkpoint for this process, called β-selection, selects cells with a productive rearrangement of their β-chain to continue in their development, whereas cells with a nonfunctional rearrangement will undergo apoptosis (Dudley et al., 1994). Pairing of productively rearranged β-chains with the pre-T cell receptor α (pTα) chain and the CD3 molecules results in the expression of the pre-TCR (Saint-Ruf et al., 1994), which induces survival and a massive proliferative expansion of these cells (Kreslavsky et al., 2012) by autonomous signaling (Saint-Ruf et al., 1994; Jacobs et al., 1996; Irving et al., 1998). The requirement for pre-TCR signaling during this checkpoint is manifested in the arrest of T cell development in mice with defects in the pTα chain (Fehling et al., 1995), the CD3 signaling components (Malissen et al., 1995), or the genes responsible for the recombination of the β-chain (Shinkai et al., 1992). Additionally, Notch1 signaling and engagement of the chemokine receptor Cxcr4 by its ligand Cxcl12 were shown to be essential for a successful passage through this selection point, since they are crucial for the survival as well as the proliferation of the cells (Ciofani et al., 2004; Ciofani and Zúñiga-Pflücker, 2005; Maillard et al., 2006; Trampont et al., 2010; Tussiwand et al., 2011). The up-regulation of several costimulatory surface molecules, such as CD27, CD28, and CD71 (Brekelmans et al., 1994; Gravestein et al., 1996; Williams et al., 2005), can also be used to subdivide the DN3 population into cells expressing an intracellular β-chain (DN3b) and cells that do not yet do so (DN3a; Taghon et al., 2006). The fact that VDJ-recombination can result in a nonproductive rearrangement, together with the inability of some β-chains to build a functional pre-TCR, leads to the elimination of the vast majority of cells during β-selection. The exact mechanism and regulation of this process remain unresolved.
In this study, we identify CD27 down-regulation at the DN3 stage to be a marker for cells that have failed rearrangement of the β-chain and will therefore undergo apoptosis. Gene expression analysis revealed high expression of the transcription factor Duxbl within this population. We show that conditional transgenic overexpression of Duxbl resulted in a block in T cell development at the DN3-to-DN4 transition due to reduced proliferation and increased apoptosis, whereas in vitro Duxbl knockdown decreased apoptosis, instead. Our results identify Duxbl as an important regulator of the elimination of cells with a nonfunctional rearrangement at the β-selection checkpoint.
Results
CD27 down-regulation marks cells failing β-chain rearrangement
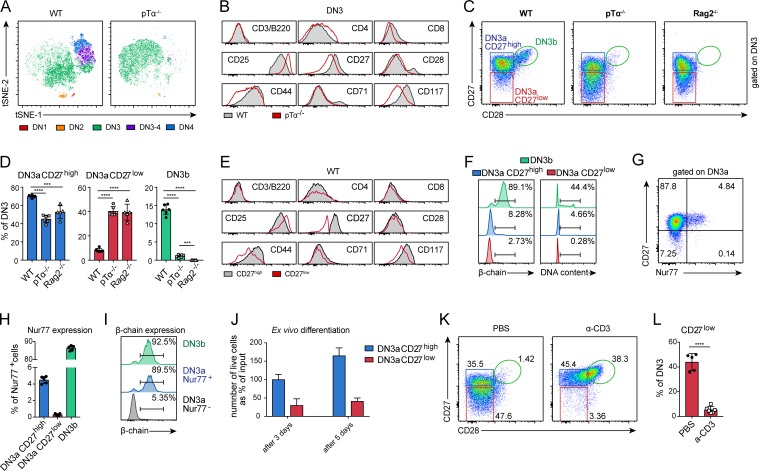
In mice with a defective pTα chain, developing thymocytes are arrested at the DN3 stage and undergo apoptosis due to their inability to form the pre-TCR (Fehling et al., 1995). Comparison of the DN compartments of WT versus pTα−/− mice by tSNE (t-distributed stochastic neighbor embedding) confirmed the absence of DN3-4 and DN4 cells in pTα−/− mice, but also a clear phenotypical change of the DN3 compartment, which showed lower CD27, CD44, and CD117 and higher CD25 expression (Fig. 1, A and B; and Fig. S1). While in pTα−/− mice, most cells undergo apoptosis due to the complete absence of pre-TCR signaling, in WT mice, only a fraction of the DN3 population is eliminated. We therefore sought to phenotypically identify this fraction in the WT DN3 population. CD27 down-regulation is also observed in WT mice, whereas high CD27 and CD28 expression marks DN3b cells, which have successfully rearranged their β-chain (Fig. 1 C). In pTα−/− and Rag2−/− mice, this CD27low population is significantly increased, whereas the DN3b population is barely detectable or completely absent (Fig. 1, C and D). Comparison of WT DN3a CD27low with CD27high cells displays similar differences with regard to lower CD44 and CD117 and increased CD25 expression, as observed in the WT versus pTα−/− comparison (Fig. 1 E).
Figure 1.
CD27 down-regulation marks cells failing β-selection. (A) tSNE-based comparison of pTα−/− and WT DN cells using the surface markers CD3/B220, CD4, CD8, CD25, CD27, CD28, CD44, CD71, and CD117. DN stages were identified by FACS as shown in Fig. S1. (B) Histograms of markers used in A of WT and pTα−/− DN3 cells. (A and B) Two independent experiments were performed, with representative data from one experiment shown. (C) Representative FACS plots for the separation of DN3 cells in pTα−/−, Rag2−/−, and WT mice using CD27 and CD28 expression. Three independent experiments were performed, with representative data from one experiment shown. (D) Frequencies of DN3a CD27high (left), DN3a CD27low (middle), and DN3b (right) cells as percentage of DN3 in WT (n = 6), pTα−/− (n = 6), and Rag2−/− (n = 5) mice. Data were collected from three independent experiments. (E) Histograms of markers used in A of WT DN3a CD27high and DN3a CD27low cells. Three independent experiments were performed, with representative data from one experiment shown. (F) Representative histograms showing intracellular β-chain expression (left) or DNA content (right) of WT DN3a CD27low, DN3a CD27high, and DN3b cells. Three independent experiments were performed, with representative data from one experiment shown. (G) Representative FACS plot of Nur77 expression in CD27high and CD27low DN3a cells in Nur77GFP mice. Three independent experiments were performed, with representative data from one experiment shown. (H) Frequencies of Nur77+ cells within the DN3a CD27high or CD27low and DN3b population (n = 5). Data were collected from three independent experiments. (I) Representative histograms showing intracellular β-chain expression in Nur77− or Nur77+ DN3a and DN3b cells. Two independent experiments were performed, with representative data from one experiment shown. (J) Numbers of sorted WT DN3a CD27high and DN3a CD27low cells as percentage of input after 3 and 5 d of culture in wells coated with Dll4 and supplemented with Cxcl12. Two independent experiments were performed, with representative data from one experiment shown. (K) Representative FACS plots of CD27 and CD28 expression in pTα−/− DN3 cells 1 d after intraperitoneal injection of 100 µg anti-CD3ε antibodies or PBS. Three independent experiments were performed, with representative data from one experiment shown. (L) Frequency of DN3a CD27low cells as percentage of DN3 in pTα−/− mice 1 d after intraperitoneal injection of 100 µg anti-CD3ε antibodies (n = 6) or PBS (n = 5). Data were collected from three independent experiments. Gate numbers in FACS plots and histograms indicate frequencies of parent gate. Statistical analysis was done with two-tailed unpaired Student’s t test. ***, P < 0.001; ****, P < 0.0001. Error bars indicate standard deviation.
Previously, high expression of CD27 was linked to intracellular β-chain expression at the DN3 stage (Taghon et al., 2006). To assess whether CD27 down-regulation is associated with failure at the β-selection checkpoint, intracellular β-chain expression as well as the cell cycle status of the cells were examined. The DN3a CD27low population exhibited decreased intracellular β-chain expression as well as almost complete absence of cycling cells (Fig. 1 F). In addition, analysis of Nur77GFP mice, reporting activation of TCR or pre-TCR signaling, revealed a signal exclusive to the DN3a CD27high and not the CD27low population, whereas the vast majority of DN3b cells were positive (Fig. 1, G and H). Costaining for intracellular β-chain expression further revealed that Nur77 expression marks DN3a cells that started to express a β-chain (Fig. 1 I). Cultivation of ex vivo–isolated DN3 cells in wells coated with Delta-like 4 (Dll4) and supplemented with Cxcl12 results in a progression of cells with a functional rearrangement to the DP stage, which is accompanied by a proliferative burst (Tussiwand et al., 2011). This progression is vastly reduced in the DN3a CD27low compared with the CD27high population (Fig. 1 J). Injection of anti-CD3ε antibody in pTα−/− mice, which provides a signal that compensates for the absent pre-TCR signaling (Fehling et al., 1997), resulted in an almost complete disappearance of the CD27low population and in an up-regulation of CD28 in the CD27high cells, showing their induced progression to the DN4 stage (Fig. 1, K and L).
Thus, CD27 down-regulation is associated with an unsuccessful β-chain rearrangement and/or pre-TCR formation. On the other hand, high CD27 expression is associated with pre-TCR signaling and successful passage through the β-selection checkpoint.
Specific expression of the transcription factor Duxbl during β-selection
RNA sequencing (RNA-seq) of DN3a CD27high, DN3a CD27low, and DN3b cells was performed to further investigate the mechanism of β-selection. All three populations were clearly separated from each other on the first axes of a principal component analysis (PCA). However, there was a closer relationship between DN3a CD27high and CD27low samples, whereas the DN3b samples were more distant (Fig. S2).
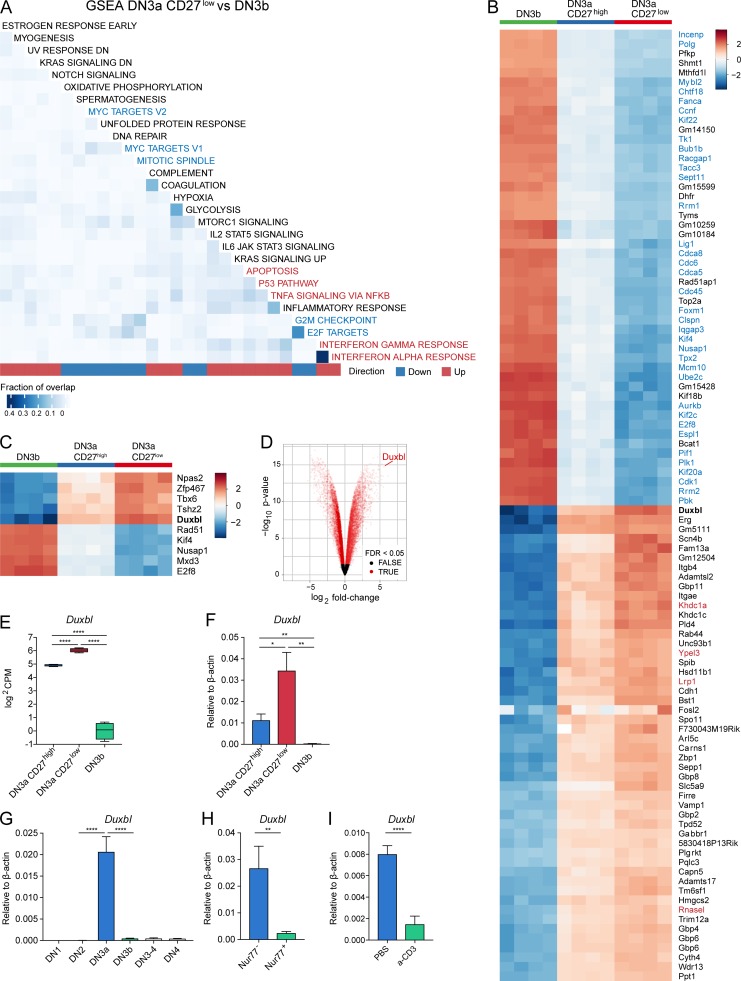
Gene set enrichment analysis for MsigDB hallmark signatures comparing DN3a CD27low and DN3b cells revealed a higher expression of genes involved in apoptosis-related pathways in DN3a CD27low cells, with a significant enrichment of the tumor necrosis factor α (TNFA) signaling, interferon-α and -γ responses, p53, and apoptosis pathways. On the other hand, hallmark signatures that were significantly decreased in the CD27low population were related to cell cycle and DNA replication, such as the Myc and E2F targets, mTORC1 signaling, mitotic spindle, and genes involved at the G2-to-M checkpoint in the cell cycle (Fig. 2 A). This enrichment of genes involved in cell cycle and proliferation is visible in the top 50 genes down-regulated in the DN3a CD27low compared with the DN3b population (Fig. 2 B). Therefore, global gene expression analysis is in agreement with the previous findings of cell cycle arrest and the developmental blockade of cells that will undergo apoptosis in the DN3a CD27low population, whereas DN3b cells, after receiving pre-TCR signaling, undergo proliferative expansion. The DN3a CD27high population seems to be located in between these two populations, as indicated by the gene expression levels shown in Fig. 2 B and the hierarchical clustering shown in Fig. S2 A.
Figure 2.
Transcriptome comparison reveals specific up-regulation of the transcription factor Duxbl in DN3a CD27low cells. (A–C) Bulk RNA-seq of DN3a CD27high, DN3a CD27low, and DN3b cells, performed as described in Materials and methods. (A) Heat map illustrating the results of the gene set enrichment analysis (GSEA) of DN3a CD27low versus DN3b cells using gene sets from the hallmark collection of the Molecular Signature Database. Only sets containing >10 genes and with a false discovery rate (FDR) <5% are illustrated. The fraction of genes overlapping across gene sets is indicated by the color intensity on the heat map. Down-regulated hallmark signatures related to cell cycle/division and DNA replication are highlighted with blue font, and up-regulated ones related to apoptosis pathways in red font. (B) Heat map displaying the centered gene expression levels of the top 50 significantly overexpressed genes and top 50 significantly underexpressed genes in DN3a CD27low compared with DN3b cells. Genes are clustered using hierarchical clustering, but the dendrogram is not displayed. Duxbl is marked in bold font. The color gradient illustrates the normalized log2CPM values centered across all samples for each gene. (C) Heat map displaying the centered gene expression levels of the top 10 transcription factors (i.e., annotated to Gene Ontology category GO:0003677) with the highest absolute fold change among differentially expressed genes (FDR <0.05) between DN3a CD27low and DN3b cells. Duxbl is marked in bold font. The color gradient illustrates the normalized log2CPM values centered across all samples for each gene. (D) Volcano plot of differentially expressed genes between DN3a CD27low and DN3b cells. Genes with an FDR <0.05 are marked in red, and genes with an FDR >0.05 in black. (E and F) Normalized log2CPM obtained from the bulk RNA-seq (E) and relative expression obtained by qPCR (F) of Duxbl in DN3a CD27high, DN3a CD27low, and DN3b cells. (G) Relative expression of Duxbl in DN1, DN2, DN3a, DN3b, DN3-4, and DN4 cells. (H) Relative expression of Duxbl in Nur77− and Nur77+ DN3a CD27high cells isolated from Nur77GFP mice. (I) Relative expression of Duxbl in DN3 cells isolated from pTα−/− mice 1 d after intraperitoneal injection of 100 µg anti-CD3ε antibodies (n = 7) or PBS (n = 5). As housekeeping gene, β-actin was used. Data were collected from four (G and H) or three (F and I) independent experiments. DN stages were identified by FACS as shown in Fig. S1. Statistical analysis was done with two-tailed unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Error bars indicate standard deviation.
The homeobox transcription factor Duxbl (double homeobox B-like gene) was found to be the DNA binding protein with the highest fold change between the DN3a CD27low and DN3b populations (Fig. 2, B–D). This factor was of specific interest since its deregulated expression was shown to result in an impaired DN thymocyte development in an in vitro setting (Kawazu et al., 2007). While Duxbl was expressed in the DN3a CD27high population almost twofold less compared with the DN3a CD27low population, it was expressed at very low levels in DN3b cells (Fig. 2 E). Quantitative PCR (qPCR) analysis confirmed these data and additionally highlighted that Duxbl is very specifically up-regulated at the DN3a stage during T cell development and immediately down-regulated at the DN3b stage after cells have passed β-selection (Fig. 2 F-G). This prompted us to test whether pre-TCR signaling is involved in the down-regulation of Duxbl expression. To this end, we quantified Duxbl expression in Nur77− as well as Nur77+ DN3a CD27high cells in Nur77GFP mice. Our analysis revealed diminished Duxbl expression in Nur77+ cells (Fig. 2 H). Furthermore, Duxbl expression was also decreased in DN3 cells from pTα−/− mice that were injected with anti-CD3ε antibodies compared with the PBS-injected controls (Fig. 2 I). The prominent expression of Duxbl at the stage where β-selection occurs, as well as its induced down-regulation by pre-TCR signaling, suggest a potential role of this transcription factor in the selection process.
Single-cell transcriptome clustering of DN cells elucidates Duxbl expression before β-selection
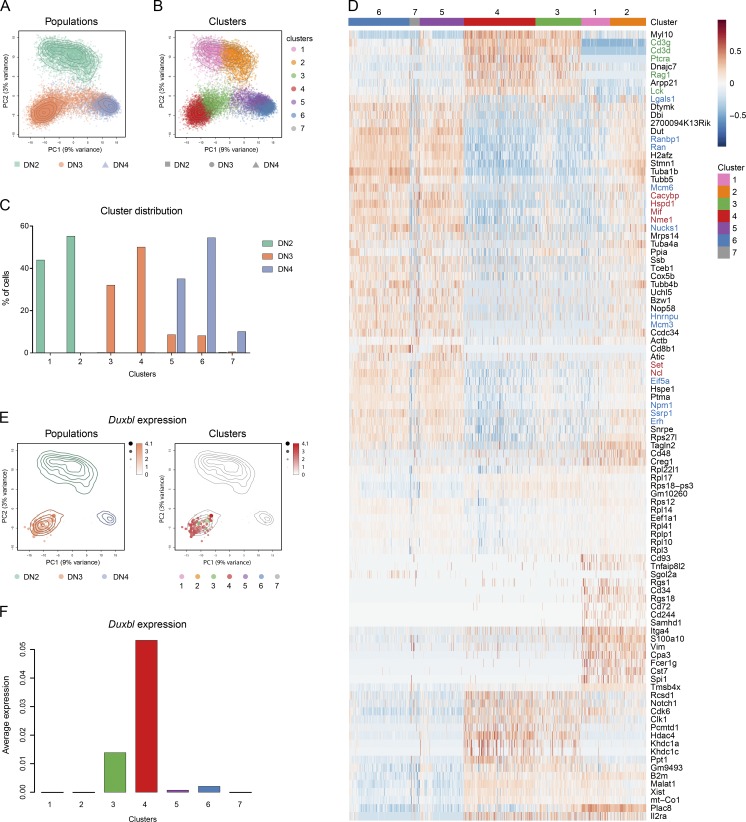
To further characterize the β-selection checkpoint, single-cell RNA-seq of WT DN2, DN3, and DN4 cells was performed. In total, after quality filtering, we obtained 3,952 DN2, 8,675 DN3, and 5,501 DN4 cells, with an average of 1,988 genes per cell detected. PCA clearly separated the individual cells of the DN2 population from the other populations on the second principal component (Fig. 3 A). While most of the DN3 cells separated from the DN4 population on the first principal component, some merged into the DN4 population. Clustering analysis resulted in seven clusters, with two clusters composed of DN2 cells, two clusters composed of DN3 cells, two clusters composed of both DN3 and DN4 cells, and one cluster composed of DN4 cells (Fig. 3, B and C; and Fig. S3 A).
Figure 3.
Single-cell RNA-seq elucidates a developmental trajectory through β-selection. Single-cell RNA-seq of DN2, DN3, and DN4 cells, performed as described in the Materials and methods. (A and B) PCA based on the 500 most variable genes across all cells. The colors represent cells from the different populations (A) or the different clusters (B). Contour lines indicate the density of the DN2, DN3, and DN4 cells in the PCA space. (C) Relative frequency of DN2, DN3, and DN4 cells within the different clusters. (D) Heat map displaying the centered and scaled expression across the 18,128 cells of the top cluster-specific genes (resulting from the union of the top 23 differentially expressed genes from each pairwise comparison between clusters). The color gradient illustrates the normalized log-count values centered and scaled across all samples for each gene. Colors at the top indicate the membership of cells to the different clusters. Font colors indicate genes involved in recombination and pre-TCR (green), cell cycle/division and DNA replication (blue), and negative regulation of apoptosis (red). (E) Expression of Duxbl shown in the PCA space. Size and color intensity of the dots indicate relative expression level of Duxbl in each cell, and the colors correspond to the respective population (left) or cluster (right). (F) Bar plot showing the average normalized log-counts of Duxbl across all cells from each cluster.
Progression from the DN2 to the DN4 stage is accompanied by specific developmental gene expression changes. We used well-defined expression patterns of surface markers, genes involved in recombination, and transcriptional regulators of T cell development to establish a developmental trajectory of the obtained clusters (Yui and Rothenberg, 2014). As genes specific for the DN2 stage, Cd117, Cd44, Hoxa9, Lmo2, Mef2c, Gfi1b, Lyl1, Spi1, Bcl11a, Mycn, and Hhex were used. Il2ra, Il7r, Erg, Notch1, Hes1, and Runx1 were used as genes expressed in both DN2 and DN3 cells. DN3 cells in particular were defined by high expression of CD3γ, CD3δ, CD3ε, Rag1, Rag2, Ptcra, Tcf12, Ets, Ahr, Notch3, and SpiB. From the DN3 stage onward, Tcf12, Ets1, Bcl11b, Ets2, Lef1, Thy1, and Themis are highly expressed, whereas Cd4, Cd8, Rorc, and Id3 are increased once the cells have reached DN4 stage. The heat map shown in Fig. S3 B illustrates the expression patterns of these marker genes throughout the developmental trajectory. Although these markers are mainly studied at the protein expression level, their expression at the transcriptome level was overall in very good agreement with the expectation. As expected, clusters containing DN2 cells display high expression of the marker genes sets that are early expressed during development. On the other hand, DN3-specific genes split the four clusters containing DN3 cells into two fractions, since they are highly expressed in the clusters exclusively consisting of DN3 cells (clusters 3 and 4), whereas the clusters that map with DN4 cells (5 and 6) show reduced expression of these genes. The fact that many of these DN3-specific genes relate to recombination and pre-TCR signaling suggests a separation of DN3 cells into pre– (clusters 3 and 4) and post– (clusters 5 and 6) β-selection. Expression of DN4-specific genes in clusters 5 and 6 further supports this hypothesis. This is confirmed by isolating from the data the top genes that are the best at discriminating the seven different clusters: recombination- and pre-TCR signaling–related genes are highly expressed in clusters 3 and 4, while their expression is lower in clusters 5 and 6 (Fig. 3 D, genes in green). Furthermore, clusters 5 and 6 showed increased expression of genes associated with cell cycle and proliferation (in blue), which are only induced after passage of the β-selection checkpoint, and up-regulation of genes involved in the negative regulation of apoptotic processes (in red).
Confirming our previous results, Duxbl expression was almost exclusively detected in the clusters consisting of DN3 cells, with maximal expression in cluster 4 (Fig. 3, E and F), whose cells have an expression profile that is the farthest from cells that have passed β-selection (clusters 5 and 6; Figs. 3 B and S3 A). Thus, cluster 4 most likely represents cells that are undergoing recombination, including the cells that have failed productive rearrangement and are going to die. The highly specific expression of Duxbl within this cluster further suggests a potential role for this transcription factor during the β-selection process.
Conditional Duxbl transgenic mice display perturbed T cell development
To further investigate the role of Duxbl during T cell development and specifically during β-selection, we generated conditional transgenic mice that overexpress Duxbl and eGFP after removal of an mCherry stop-cassette by cre-mediated recombination (hereafter Duxblind; Fig. S4 A). To achieve specific expression at the DN3a stage during T cell development, these mice were crossed to mice expressing the cre recombinase under the control of the pTα promoter (hereafter DuxblindxpTαCre; Luche et al., 2013). The stage-specific loss of mCherry and gain of eGFP expression in these mice confirmed specificity of the system (Fig. S4, B–D).
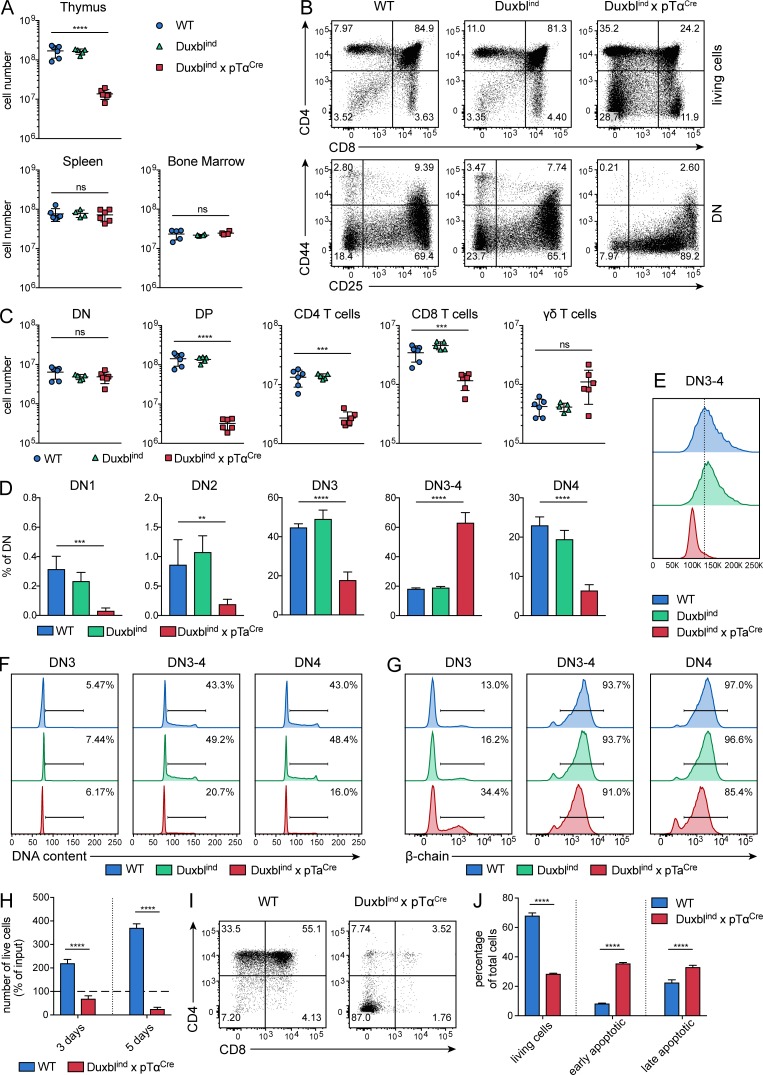
Thymic cellularity of DuxblindxpTαCre mice was reduced 10-fold compared with WT and Duxblind control mice. In contrast, the cellularity of the spleen and the bone marrow was not significantly changed (Fig. 4 A). Staining with CD4 and CD8 revealed an increase in DN percentage with simultaneous decrease in DP (Fig. 4 B). As a result, DN numbers were not increased, whereas DP cell numbers were reduced ∼45-fold. Mature CD4 and CD8 T cell compartments were reduced approximately fivefold and approximately threefold, respectively (Fig. 4 C). The γδ-T cell compartment was not significantly altered, indicating that Duxbl specifically acts during αβ-T cell development.
Figure 4.
Blocked T cell development in DuxblindxpTαCre mice. (A) Total numbers of live, nucleated cells in the thymus, spleen, and bone marrow of WT (n = 6), Duxblind (n = 5), and DuxblindxpTαCre (n = 6) mice. Data were collected from three independent experiments. (B) Representative FACS plots of CD4 and CD8 expression in total live cells (top) and CD44 and CD25 expression in DN cells (bottom) of WT, Duxblind, and DuxblindxpTαCre mice. DN cells were gated as CD4, CD8, CD3, and B220 negative. Three independent experiments were performed, with representative data from one experiment shown. (C) Numbers of DN, DP, CD4 T cells, CD8 T cells, and γδ-T cells in WT (n = 6), Duxblind (n = 5), and DuxblindxpTαCre (n = 6) mice. (D) Frequencies of DN1, DN2, DN3, DN3-4, and DN4 cells as percentage of DN cells in WT (n = 6), Duxblind (n = 5), and DuxblindxpTαCre (n = 6) mice. (C and D) Data were collected from three independent experiments. (E) Representative histogram showing the size (FSC-A) of DN3-4 cells in WT, Duxblind, and DuxblindxpTαCre mice. Three independent experiments were performed, with representative data from one experiment shown. (F and G) Histograms showing the DNA content (F) and intracellular β-chain expression (G) of DN3, DN3-4, and DN4 cells in WT, Duxblind, and DuxblindxpTαCre mice. Four independent experiments were performed, with representative data from one experiment shown. (H) Numbers of sorted DN3 cells from WT and DuxblindxpTαCre mice as percentage of input after 3 and 5 d of culture in wells coated with Dll 4 and supplemented with Cxcl12. (I) Representative FACS plots of sorted DN3 cells from WT and DuxblindxpTαCre mice showing CD4 and CD8 expression after 5 d of culture. (J) Frequencies of living, early apoptotic, and late apoptotic cells after 3 d of culture of sorted DN3 cells from WT and DuxblindxpTαCre mice. Living cells were defined as annexinV− 7AAD−, early apoptotic cells as annexinV+ 7AAD−, and late apoptotic cells as annexinV+ 7AAD+. (H–J) Three independent experiments were performed, with representative data from one experiment shown. Gate numbers in FACS plots and histograms indicate frequencies of parent gate. DN stages were identified by FACS as shown in Fig. S1. Statistical analysis was done with two-tailed unpaired Student’s t test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. Error bars indicate standard deviation.
The separation of the DN stages using CD25, CD44, and CD117 expression revealed a reduction in DN1, DN2, DN3, and DN4 fractions, whereas the DN3-4 stage, comprising cells with intermediate levels of CD25, was increased in DuxblindxpTαCre mice (Fig. 4, B and D). This clearly indicates a block in the development of T cells at the transition from the DN3 to the DN4 stage. Cells accumulating at the DN3-4 stage were much smaller in size compared with the ones in WT or Duxblind mice (Fig. 4 E). Cell cycle analysis of DN3, DN3-4, and DN4 cells from all mouse strains revealed a decreased percentage of cycling cells in DuxblindxpTαCre mice (Figs. 4 F and S5 A). On the other hand, the subsequent selection of cells with successful rearrangement of the β-chain seemed not to be affected by Duxbl, since the majority of cells stained positive for intracellular β-chain at the DN3-4 stage in all genotypes (Figs. 4 G and S5 B). Ultimately, the disturbed T cell development resulted in reduced peripheral CD4 and CD8 T cell compartments in DuxblindxpTαCre mice (Fig. S5 C). On the contrary, γδ-T cells were increased in the periphery (Fig. S5 C).
To test the abnormalities in the progression to the DP stage in more detail, we sorted WT and DuxblindxpTαCre DN3 cells and cultivated them for up to 5 d in wells coated with Dll4 and medium supplemented with Cxcl12. As shown in Fig. 4 (H and I), WT cells expanded approximately twofold after 3 d and ∼3.5-fold after 5 d and expressed CD4 and CD8 on their surface. This progression was almost completely blocked in cells derived from DuxblindxpTαCre mice, demonstrated by the reduction in cell numbers over time and the absence of CD4 and CD8 expression after 5 d, thus recapitulating the situation observed in vivo. For analysis of apoptosis, annexin V and 7AAD staining was performed after 3 d of culture (Fig. S5 D). A strong decrease in the number of living cells in accordance with an increase in the number of early and late apoptotic cells in cultures containing DN3 cells from DuxblindxpTαCre mice was observed (Fig. 4 J).
Taken together, these data indicate that conditional overexpression of Duxbl specifically affects the β-selection checkpoint by blocking the developmental progression of cells that should normally continue to their next stage, as shown by the tremendous reduction in DP cells. Induction of apoptosis and cell cycle arrest seem to cause the observed block, indicating that Duxbl is involved in the elimination of cells that fail β-selection.
Silencing of Duxbl results in decreased apoptosis at the DN3 stage
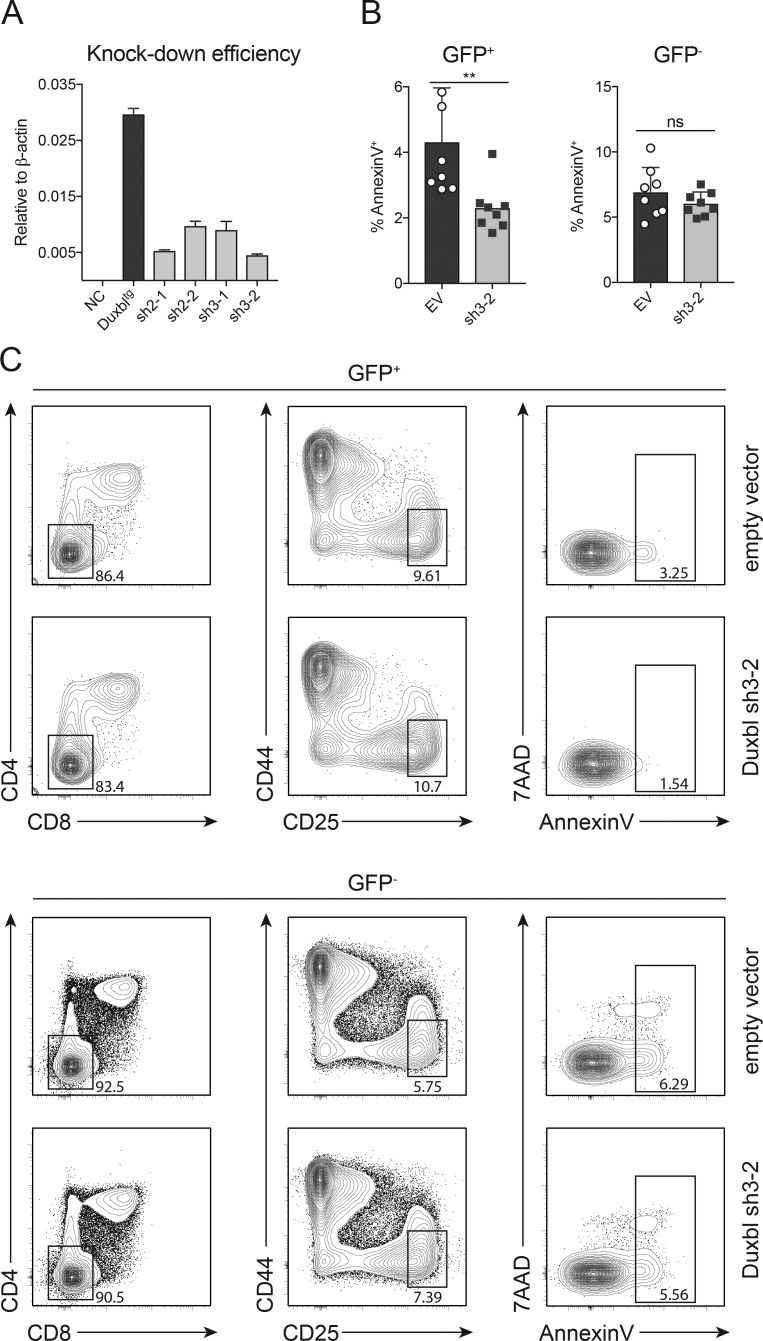
We designed four different shRNA vectors for retroviral knockdown of Duxbl expression via RNA interference. Additionally, transduced cells were marked by GFP expression. Efficiency of the different shRNAs was tested on a cell line that overexpressed Duxbl due to transposon-mediated integration of a construct containing the Duxbl cDNA under control of a constitutive active promoter. Transduction of this cell line with the different shRNAs and subsequent sorting of the GFP+ fraction revealed reduced mRNA expression of Duxbl as determined by qPCR analysis (Fig. 5 A). The shRNA with the highest knockdown efficiency (sh3-2) was selected for further experiments.
Figure 5.
Decreased apoptosis in DN3 cells after Duxbl knockdown. (A) Knockdown efficiency of four different shRNAs illustrated by relative expression of Duxbl in the 40E1 cell line overexpressing Duxbl with or without additional transduction of the described shRNAs. As housekeeping gene, β-actin was used. Two independent experiments were performed, with representative data from one experiment shown. (B) Frequencies of annexinV+ cells within GFP+ (left) or GFP− (right) cells transduced with an empty vector control or shRNA 3–2 4 d after removal of IL-7 from the fetal liver OP9-Dll1 cultures. Data were collected from three independent experiments. (C) Representative FACS plots showing CD4 and CD8 expression (left), CD44 and CD25 expression within DN (middle), and annexinV and 7AAD levels within DN3 cells (right) 4 d after removal of IL-7 from the fetal liver OP9-Dll1 cultures. Plots were gated either on GFP+ or GFP− cells. Three independent experiments were performed, with representative data from one experiment shown. Gate numbers in FACS plots indicate frequencies of parent gate. Statistical analysis was done with two-tailed unpaired Student’s t test. **, P < 0.01. Error bars indicate standard deviation.
For the establishment of an in vitro simulation of T cell development, fetal liver cells were cultured on OP9-Dll1 stromal cells. After several days of culture, the majority of cells adopted a DN2 phenotype, and only after removal of IL-7 was a transition through β-selection initiated, which resulted in the generation of DP cells (data not shown). Since pre-TCR signaling results in CD25 down-regulation, we specifically assessed the potential effect of Duxbl knockdown on CD44−CD25high cells, the majority of which represent cells before β-selection and in which Duxbl is expressed. Knockdown of Duxbl in this system diminished apoptosis at the DN3 stage specifically in the GFP+ fraction, whereas untransduced GFP− cells showed no significant change compared with the empty vector control (Fig. 5, B and C). Thus, in accordance with apoptosis induction after Duxbl overexpression (Fig. 4), silencing of Duxbl expression results in reduced apoptosis in DN3 cells, further supporting its role in mediating the elimination of DN3 cells that failed β-selection.
Activation of apoptosis in DuxblindxpTαCre mice
To reveal the mechanism involved in the Duxbl-mediated apoptosis induction in T cell development, we investigated gene expression changes in DuxblindxpTαCre DN3-4 cells by RNA-seq of these cells in parallel with their WT counterparts. Comparison of these two populations with the previous RNA-seq data revealed a close similarity between WT DN3-4 cells and DN3b cells (Fig. 6 A), which is expected, since at the level of surface protein expression they differ only slightly in their CD25 expression. Interestingly, after batch-effect correction, DN3-4 cells derived from DuxblindxpTαCre mice exhibited a gene expression profile most similar to WT DN3a CD27low cells, even though they have a successfully rearranged β-chain. This further suggests that continuous Duxbl expression prevents developmental progression and instead induces a transcriptomic phenotype resembling the cells that fail β-selection.
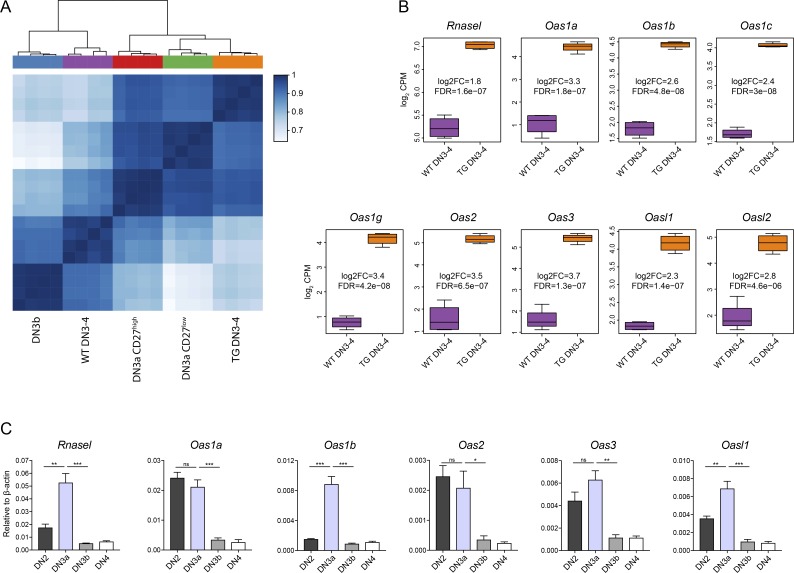
Figure 6.
Gene expression analysis identifies up-regulation of the Oas/RNaseL system in Duxbl transgenic DN3-4 cells. (A and B) Bulk RNA-seq of WT and DuxblindxpTαCre DN3-4 cells, performed as described in the Materials and methods. (A) Heat map and hierarchical clustering based on the matrix of Spearman correlation across the samples of WT and TG (derived from DuxblindxpTαCre mice) DN3-4 cells together with WT DN3a CD27high, DN3a CD27low, and DN3b cells (using the 25% most variable genes). Data were corrected for batch effects across datasets. (B) Boxplots showing the normalized log2CPM of Rnasel, Oas1a, Oas1b, Oas1c, Oas1g, Oas2, Oas3, Oasl1, and Oasl2 in WT and TG (derived from DuxblindxpTαCre mice) DN3-4 cells. Log2 FC and false discovery rate (FDR) are indicated for each gene. (C) Relative expression of Rnasel, Oas1a, Oas1b, Oas2, Oas3, and Oasl1 in WT DN2, DN3a, DN3b, and DN4 cells. As housekeeping gene, β-actin was used. Data were collected from four independent experiments. Statistical analysis was done with two-tailed unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ns, not significant. Error bars indicate standard deviation (B) or standard error of the mean (C).
Among the most differentially expressed genes between WT DN3-4 cells and DN3-4 cells derived from DuxblindxpTαCre mice, the Oas/RNaseL pathway was clearly overexpressed in transgenic samples (Fig. 6 B). This system is important in interferon responses and was shown to be mainly involved in antiviral immunity by inducing apoptosis in infected cells (Silverman, 2007). RNaseL deficiency also results in an increased resistance of thymocytes to apoptosis, suggesting further involvement of this pathway in thymic development (Zhou et al., 1997). qPCR analysis of the expression of different Oas genes, as well as of the Rnasel gene, during T cell development revealed that some, such as Rnasel and Oas1b, are specifically up-regulated at the DN3a stage (Fig. 6 C). Even more striking was the silencing of all Oas/RNaseL genes in DN3b cells after β-selection passage (Fig. 6 C). This specific expression, together with the up-regulation in DuxblindxpTαCre DN3-4 cells, indicates that this pathway is involved in the β-selection checkpoint, potentially acting downstream of Duxbl, and thus mediating the T cell developmental block observed in the DuxblindxpTαCre mice.
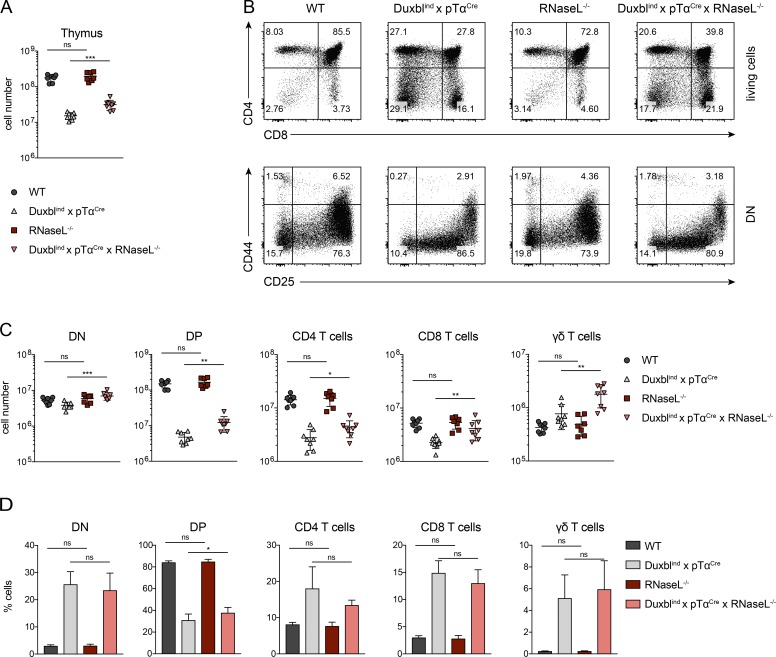
To test whether RNaseL is involved in the Duxbl-mediated T cell developmental block, we generated RNaseL-defective mice (Fig. S4) and crossed them to the DuxblindxpTαCre line. Thymic cellularities of DuxblindxpTαCrexRNaseL−/− mice were significantly increased compared with their DuxblindxpTαCre counterparts, due to higher numbers of DN, DP, CD4, and CD8 T cells, as well as γδ-T cells (Fig. 7, A–C). The DP stage was particularly affected, since this population was also increased in percentage, confirming that the T cell developmental block was at least partly suppressed (Fig. 7, B and D). The observed up-regulation of the Oas/RNaseL system by transgenic Duxbl expression is therefore in part responsible for the disturbed developmental progression of DN3 cells in DuxblindxpTαCre mice. Of note, since DuxblindxpTαCrexRNaseL−/− mice retain an altered T cell development compared with WT mice (Fig. 7 B), other mechanisms must be additionally involved.
Figure 7.
Disruption of the Oas/RNaseL system in DuxblindxpTαCre mice extenuates the perturbed T cell development. (A) Total number of live, nucleated cells in the thymus of WT (n = 8), DuxblindxpTαCre (n = 8), RNaseL−/− (n = 7), and DuxblindxpTαCrexRNaseL−/− (n = 8) mice. Data were collected from four independent experiments. (B) Representative FACS plots of CD4 and CD8 expression in total live cells (top) and CD44 and CD25 expression in DN cells (bottom) of WT, DuxblindxpTαCre, RNaseL−/−, and DuxblindxpTαCrexRNaseL−/− mice. DN cells were gated as CD4, CD8, CD3, and B220 negative. Four independent experiments were performed, with representative data from one experiment shown. (C) Numbers of DN, DP, CD4 T cells, CD8 T cells, and γδ-T cells in WT (n = 8), DuxblindxpTαCre (n = 8), RNaseL−/− (n = 7), and DuxblindxpTαCrexRNaseL−/− (n = 8) mice. (D) Frequencies of DN, DP, CD4 T cells, CD8 T cells, and γδ-T cells in WT (n = 8), DuxblindxpTαCre (n = 8), RNaseL−/− (n = 7), and DuxblindxpTαCrexRNaseL−/− (n = 8) mice. (C and D) Data were collected from four independent experiments. Gate numbers in FACS plots indicate frequencies of parent gate. DN stages were identified by FACS as shown in Fig. S1. Statistical analysis was done with two-tailed unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. Error bars indicate standard deviation.
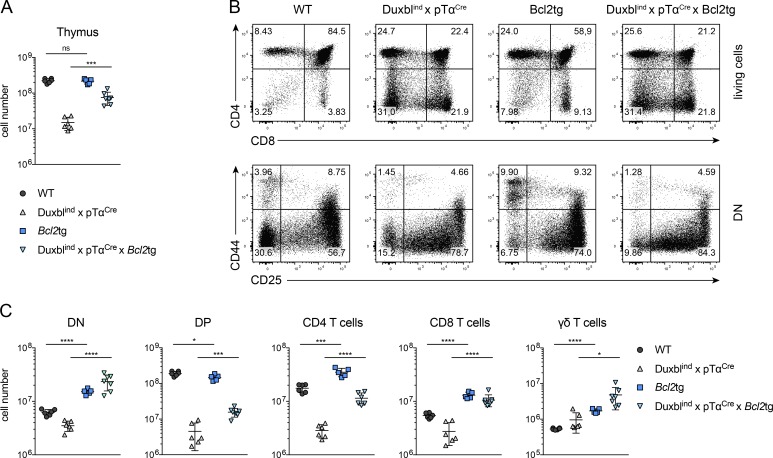
We further investigated the connection of Duxbl to apoptosis by crossing the DuxblindxpTαCre to Bcl2tg mice, to examine whether an additional survival signal would extenuate the block in T cell development, as observed in the case of IL-7 receptor–deficient mice (Akashi et al., 1997). Indeed, the thymic cellularity of DuxblindxpTαCrexBcl2tg DN, CD4, and CD8 and γδ-T cells, but especially of the DP (∼3.5-fold change), were all significantly increased compared with DuxblindxpTαCre mice. The number of DP cells did not reach WT levels, and phenotypically, the block showing an increased DN and decreased DP compartment was still apparent (Fig. 8, A–F), indicating that the development is not completely rescued by additional expression of Bcl2 alone. However, the partial rescue of DP cells in DuxblindxpTαCrexBcl2tg mice provides further evidence for a role of Duxbl in mediating the elimination of nonselected DN3 cells by inducing apoptosis.
Figure 8.
Transgenic Bcl2 expression restores T cell populations in DuxblindxpTαCre mice. (A) Total number of live, nucleated cells in the thymus of WT (n = 6), DuxblindxpTαCre (n = 6), Bcl2tg (n = 5), and DuxblindxpTαCrexBcl2tg (n = 7) mice. Data were collected from four independent experiments. (B) Representative FACS plots of CD4 and CD8 expression in total live cells (top) and CD44 and CD25 expression in DN cells (bottom) of WT, DuxblindxpTαCre, Bcl2tg, and DuxblindxpTαCrexBcl2tg mice. DN cells were gated as CD4, CD8, CD3, and B220 negative. Four independent experiments were performed, with representative data from one experiment shown. (C) Numbers of DN, DP, CD4 T cells, CD8 T cells, and γδ-T cells in WT (n = 6), DuxblindxpTαCre (n = 6), Bcl2tg (n = 5), and DuxblindxpTαCrexBcl2tg (n = 7) mice. Data were collected from four independent experiments. Gate numbers in FACS plots indicate frequencies of parent gate. DN stages were identified by FACS as shown in Fig. S1. Statistical analysis was done with two-tailed unpaired Student’s t test. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Error bars indicate standard deviation.
Discussion
Thymic selection of αβ-T cells requires a series of events in which TCR engagement follows defined rules. The majority of cells will undergo apoptosis during β-selection, and the underlying mechanisms of this process have not been completely resolved. In the present study, CD27 down-regulation within the DN3 population was identified as a marker for cells that have failed to rearrange their β-chain and therefore are eliminated. Transcriptome analysis at both population and single-cell levels revealed high expression of the transcription factor Duxbl exclusively in a cluster of cells that have not yet passed β-selection and display high rearrangement activity. Conditional transgenic expression of Duxbl led to a disrupted DN3-to-DN4 transition due to suppressed proliferation and induction of apoptosis, with the developmentally arrested cells highly resembling WT DN3a CD27low Duxblhigh cells. Silencing of its expression, on the other hand, resulted in decreased apoptosis. We identified up-regulation of the apoptosis-inducing Oas/RNaseL pathway as a potential mechanism by which Duxbl facilitates the elimination of nonselected cells at the β-selection checkpoint.
CD27 expression was previously described to separate intracellular β-chain positive from negative cells, since it gets slightly up-regulated in DN3b cells (Taghon et al., 2006). However, the observation of CD27 down-regulation in DN3a cells extends the current resolution of the developmental staging during T cell development, as it marks cells that failed successful rearrangement and that will therefore undergo apoptosis (Figs. 1 and 2). The DN3a CD27high population, on the other hand, seems to be one stage before the rescue-versus-apoptosis decision, as indicated by its gene expression profile, which clusters in between CD27low and DN3b cells (Fig. 2). This suggests that DN3a CD27high cells are still in the process of β-selection and are not yet assigned to a specific fate.
The expression of Duxbl in DN3a cells, which mostly occurs in the CD27low population, indicates a role during β-selection (Fig. 2, C and D). An association, whose molecular mechanisms were not defined, was suggested by in vitro studies in which expression of Duxbl affected the development of the DN stages (Kawazu et al., 2007). Our findings provide in vivo proof for this role of Duxbl and further identify this transcription factor as an initiator of developmental arrest and apoptosis in DN3a cells that fail β-selection. It is currently unknown what signals initiate Duxbl expression at the DN3a stage, and two scenarios can be envisaged. In the first case, Duxbl could be induced specifically in cells with a nonfunctional rearrangement. In the second case, it could be induced in all cells that enter the DN3a stage, thus acting as a timer for successful rearrangement. Duxbl expression would increase over time in cells failing β-chain rearrangement, leading to apoptosis induction. On the contrary, cells with successful rearrangement would receive a pre-TCR signal that silences Duxbl expression and leads to their rescue and developmental progression (Fig. 2, F and G).
Our studies also showed a profound heterogeneity within the DN3 population. Indeed, single-cell RNA-seq of DN2, DN3, and DN4 cells unveiled four different clusters in DN3 thymocytes, of which two were shared with DN4 cells (Fig. 3, A–C; and Fig. S3 A). These findings showed partitioning of the DN3 population into cells that have not yet undergone β-selection (clusters 3 and 4) and cells that already passed this checkpoint (clusters 5 and 6). Consequently, the latter cells are closer to the gene expression profile of DN4 cells. Interestingly, Duxbl expression was mainly detected in cluster 4, which is transcriptionally farthest from the DN4 clusters (Fig. 3, E and F). Considering the high expression of genes involved in β-chain recombination and the pre-TCR in this cluster, it probably consists of cells continuing the process of rearrangement, including some that have failed rearrangement.
While reduced Duxbl expression leads to a decrease in apoptosis at the DN3 stage, the detrimental effect of its expression in cells undergoing β-selection is exhibited in mice conditionally overexpressing Duxbl. The increased DN and decreased DP stages in the thymus of these mice showed arrest of T cell development after the DN3 stage (Fig. 3). However, the block is not absolute, since mature CD4 and CD8 T cells can still be found in the thymus as well as in the periphery. DN3-4 cells of DuxblindxpTαCre mice stain positive for a functionally rearranged β-chain (Fig. 3 F). Thus, these cells receive a survival and proliferation signal from the pre-TCR and at the same time express Duxbl, which is inhibiting their cell cycle and induces apoptosis (Figs. 3 G and 4, A–C). In a WT situation, Duxbl expression is silenced after pre-TCR activation, but in our system, Duxbl expression remains also in cells that already received signals via the pre-TCR, which seems to prevent their further differentiation. We therefore envisage that Duxbl down-regulation upon pre-TCR signaling is an absolute requirement for further differentiation of the cells. In the few cells that have reached the DP stage in DuxblindxpTαCre mice, pre-TCR signaling must have overcome the negative signals of the continuous Duxbl expression. However, as indicated by the ex vivo differentiation, this seems to be rarely the case (Fig. 4, A–C). Another interesting finding was the impact of Duxbl specifically on αβ-T cell development, since there was no detrimental effect on the development of γδ-T cells in DuxblindxpTαCre mice. In contrast, γδ-T cells were even increased, similar to what was described for pTα−/− mice (Fehling et al., 1995).
The link of Duxbl to apoptosis and in particular to the activation of the Oas/RNaseL pathway was further strengthened by the crossing of the DuxblindxpTαCre to Bcl2tg and RNaseL−/− mice, respectively. Both approaches resulted in a significant increase in thymic cellularity (Figs. 7 and 8). Remarkably, the DP stage in particular was rescued in these mice, as manifested by a ∼3.5-fold increase in DuxblindxpTαCrexBcl2tg and ∼3.2-fold in DuxblindxpTαCreRNaseL−/− mice. However, neither of the two resulted in completely normalized T cell development. In the case of Bcl2 expression, this can be explained by the fact that only the apoptotic effects of Duxbl are antagonized, whereas the antiproliferative effects that we observed are not affected. In fact, Bcl2 overexpression itself has a negative effect on the proliferation of cells (O’Reilly et al., 1996), thus potentially compromising the full assessment of its anti-apoptotic effect. Similarly, activation of the Oas/RNaseL system seems to be just part of the mechanism by which Duxbl prevents developmental progression. Therefore, we can conclude that apoptosis and the Oas/RNaseL system are connected to the Duxbl-mediated effects in T cell development, and that additional pathways are potentially involved.
Knockdown experiments confirmed the link of Duxbl to apoptosis induction. As implied by the increase of apoptosis in Duxbl transgenic DN3 cells (Fig. 4), silencing resulted in a reduced number of apoptotic cells at the DN3 stage (Fig. 5, B and C). This further indicates that Duxbl mediates the elimination of cells that have failed β-selection, and therefore its decreased expression rescues these cells from cell death. Since our experimental setting leads only to a reduction in Duxbl levels and does not result in its complete absence, it can be envisaged that the remaining Duxbl is still able to efficiently eliminate a fraction of the cells, thus resulting in only a partial rescue.
The Duxbl gene is part of a triplicated region in the mouse genome, which spans >300 kb and contains three almost identical Duxbl copies, thus making the generation of a conditional knockout mouse particularly challenging. Each of the three identical Duxbl copies contains two homeodomains that are responsible for sequence-specific DNA binding. The amino acid sequences of these homeodomains exhibit the highest similarity to those of human DUX4 (Wu et al., 2010). The physiological role of DUX4 in humans has not been identified so far. However, misexpression of DUX4 in skeletal muscle cells is associated with facioscapulohumeral dystrophy, triggered by DUX4-induced apoptosis. A recent study also identified the RNaseL pathway to be involved in DUX4-mediated toxicity. Silencing of RNaseL resulted in increased survival following DUX4 transgene induction in a rhabdomyosarcoma cell line (Shadle et al., 2017). Thus, human DUX4 has characteristics resembling the herein described function of mouse Duxbl with regard to apoptosis and the link to the Oas/RNaseL pathway. In accordance with that, high expression of DUX4 was detected in the thymus, suggesting its actual function in this tissue (Das and Chadwick, 2016). Therefore, it is presumable that, as with Duxbl in mice, DUX4 might be involved in human T cell development.
Collectively, the findings described in the present study demonstrate that failed β-chain rearrangement coincides with low expression of CD27 in DN3 T cell progenitors. Detailed analysis of this CD27low population revealed the homeobox transcription factor Duxbl as a key regulator of β-selection by inducing apoptosis in cells that have failed to rearrange their β-chain.
Materials and methods
Mice
C57BL/6 WT controls, pTα−/− (Fehling et al., 1995), Nur77GFP (Moran et al., 2011), pTαCre (Luche et al., 2013), Bcl2tg (Domen et al., 1998), Rag2−/− (Shinkai et al., 1992), Duxblind, and RNaseL−/− mice were bred and maintained in our animal facility unit under specific pathogen–free conditions. All mice used were 5–7 wk old and were of the C57BL/6 strain or were bred to it for >10 generations (Bcl2tg). As controls, littermates were used in every experiment. All animal experiments were performed within institutional guidelines (authorization numbers 1886 and 1888 from cantonal veterinarian office, Canton Basel-Stadt).
Duxblind and RNaseL−/− mice were generated at the Center for Transgenic Models in Basel. Duxblind mice were produced by pronuclear DNA microinjection of C57BL/6 zygotes essentially as described in Palmiter et al. (1982). RNaseL−/− mice were generated using CRISPR/Cas9 technology. All Cas9 reagents were purchased from IDT. Briefly, ribonucleoproteins consisting of Cas9 protein (40 ng/µl), trans-activating CRISPR RNA (20 ng/µl), and CRISPR RNA (10 ng/µl each) targeting exon 1 of the Rnasel gene were microinjected into C57BL/6 zygotes essentially as described in Jacobi et al. (2017). Embryos surviving the DNA and Cas9 ribonucleoproteins microinjections were transferred into pseudopregnant females generated by mating with genetically vasectomized males (Haueter et al., 2010), and the offspring were allowed to develop to term.
Fig. S4 A illustrates the construct used to generate the Duxblind mice by random integration. Fig. S4 E describes the generation of RNaseL−/− mice. The following 20-mers sequences were used to produce the CRISPR RNAs that guided Cas9 for the induction of double-strand breaks in exon 1 of the Rnasel gene: 5′-AATGCCTGTGAAGACACCTG-3′ and 5′-GACAAAAGGCGATTGAAGCA-3′.
Flow cytometry and cell sorting
For analysis, cells were flushed from femurs of the two hind legs of mice, or single-cell suspensions of spleen, thymus, and lymph node (inguinal and axillary) cells were made. Stainings were performed in PBS containing 0.5% BSA and 5 mM EDTA. For intracellular β-chain staining, cells were fixed and permeabilized after cell-surface staining using a Fix/Perm buffer set (eBioscience) and subsequently stained with Bv421-conjugated anti-TCRβ (H57). The following antibodies were used for flow cytometry (from BD Biosciences, eBioscience, BioLegend, R&D Systems, or produced in-house): anti-CD3 (145-2C11), anti-B220 (RA3-6B2), anti-CD4 (GK1.5), anti-CD8 (53-6.7), anti-CD25 (PC61), anti-CD44 (IM7), anti-CD117 (2B8), anti-CD27 (LG.3A10), anti-CD28 (37.51), anti-TCRβ (H57-597), anti-TCRγ (GL3), anti-CD69 (H1.2F3), anti-MHCI (Y3), anti-CD62L (MEL-14), and anti-S1PR1 (713412). For flow cytometry, an LSRFortessa (BD Biosciences) was used, and data were analyzed using FlowJo Software (Treestar). MACS enrichment was performed using an autoMACS separator (Miltenyi) after cells were stained with anti-CD4/CD8-bio and anti-biotin microbeads (Miltenyi). For cell sorting, a FACSAria IIu (BD Biosciences) was used (>98% purity).
Cell cycle analysis
Cells were sorted with an Aria II FACS sorter (BD Biosciences). Sorted cells were resuspended in 1 ml of 70% ethanol and stored at 4°C overnight. Cells were then centrifuged at 2,000 rpm for 5 min at 4°C and subsequently resuspended in 250 µl RNaseA (0.5 mg/ml). After 15 min of incubation at 37°C, 250 µl pepsin (0.5 mg/ml) was added and incubated for 15 min at 37°C. Finally, nuclei were stained by adding 500 µl ethidium bromide solution and incubation for 10 min at room temperature before analysis by flow cytometry.
Retroviral production
For Duxbl knockdown, MSCV-LMP empty vector plus four different MSCV-LMP vectors containing Duxbl-directed shRNAs were generated using the following oligo templates: sh2-1, 5′-TGCTGTTGACAGTGAGCGAACTCTTCGTGTGTGGTTTGAATAGTGAAGCCACAGATGTATTCAAACCACACACGAAGAGTGTGCCTACTGCCTCGGA-3′; sh2-2, 5′-TGCTGTTGACAGTGAGCGCAGAATCCCAATCCTGATCTAGTAGTGAAGCCACAGATGTACTAGATCAGGATTGGGATTCTTTGCCTACTGCCTCGGA-3′; sh3-1, 5′-TGCTGTTGACAGTGAGCGCTTGCATGCTGTTCTGAAGAAATAGTGAAGCCACAGATGTATTTCTTCAGAACAGCATGCAAATGCCTACTGCCTCGGA-3′; and sh3-2, 5′-TGCTGTTGACAGTGAGCGAAGTTTGCATGCTGTTCTGAAGTAGTGAAGCCACAGATGTACTTCAGAACAGCATGCAAACTCTGCCTACTGCCTCGGA-3′.
A total of 5 × 106 Plat-E cells were plated 1 d before transfection in 10-cm dishes (Falcon) with 10 ml DMEM (Sigma) supplemented with 10% FBS, 100 U/ml penicillin (BioConcept), 100 mg/ml streptomycin (BioConcept), and 1 mM sodium pyruvate (Sigma). The next day, 20 µg plasmid and 80 µg polyethylenimine were mixed in 1 ml DMEM without supplements, incubated for 15 min at room temperature, and subsequently added dropwise to the Plat-E cells. 24 h after transfection, supernatant was replaced with fresh medium. Viral supernatants were collected 48 and 72 h after transfection and either used directly for transductions or stored at −80°C.
Cell culture
For culturing of sorted DN3 cells, 96-well Maxisorp Nunc-Immuno plates (Thermo Fisher Scientific) were used. Wells were precoated overnight or longer with 10 µg/ml anti-human IgG-Fc (clone Huf-5.4, generated in-house) in PBS (50 µl per well) at 4°C. Thereafter, wells were washed twice with IMDM supplemented with 5% FBS, 5 × 10−5 M β-mercaptoethanol, 1 mM glutamine, 0.03% wt/vol Primatone, 100 U/ml penicillin, and 100 mg/ml streptomycin and then coated with Dll4-Fc at 3 µg/ml in IMDM (50 µl per well) overnight at 4°C. Before addition of the cells, wells were washed again twice with IMDM. Sorted cells were adjusted to a concentration of 2 × 105 cells/ml with IMDM, supplemented additionally with 10 nM Cxcl12, and cultured for 3 or 5 d (200 µl per well).
For the establishment of in vitro T cell differentiation, fetal liver cells were cultured on OP9-Dll1 stromal cells. In short, 3 × 104 OP9-Dll1 cells were seeded in 24-well dishes the day before addition of fetal liver cells with 500 µl IMDM supplemented with 10% FBS, 5 × 10−5 M β-mercaptoethanol, 1 mM glutamine, 0.03% wt/vol Primatone, 100 U/ml penicillin, and 100 mg/ml streptomycin. The next day, cells were irradiated with 2,000 rad. Fetal liver cells were taken from embryonic day 14.5 C57BL/6 embryos, and mononuclear cells were separated by centrifugation on a Ficoll-Paque PLUS (GE Healthcare) gradient. A total of 5 × 104 cells per 24-well dish were added to the irradiated OP9-Dll1 and cultured in 1 ml of the previously described medium plus 10% IL-7 supernatant. After 5 d, cells were harvested, and 106 cells were transduced with 1.5 ml of viral supernatants of either MSCV-LMP empty vector or the corresponding Duxbl shRNAs using 3 h of spinoculation with 10 µg/ml polybrene. Subsequently, 5 × 104 transduced cells were put back into culture onto freshly prepared irradiated OP9-Dll1 in the presence of 10% IL-7 for another 5 d. Finally, a total of 105 cells were passed again onto freshly prepared OP9-Dll1 and cultured for 4 more days in the absence of IL-7. Then cells were harvested and stained with the corresponding surface markers as described in Flow cytometry and cell sorting, as well as with annexin V (BD Biosciences) and 7AAD (BioLegend) in Annexin V Binding Buffer (BD Biosciences) following the manufacturer’s protocol.
qPCR
Total RNA was extracted using RNAqueous Micro Kit (Invitrogen) followed by cDNA synthesis using GoScript reverse transcription (Promega) according to the manufacturer’s protocol. qPCR was performed using SYBR green PCR Master Mix (Applied Biosystems), and samples were run on an Applied Biosystems StepOnePlus qPCR machine. The following primers were used for PCR amplification: β-actin, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′ and 5′-TAAAACGCAGCTCAGTAACAG-3′; Duxbl, 5′-AAGCAGAGTTTGCATGCTGTT-3′ and 5′-TTGTGAAGTGCGTTCTGCTC-3′; Rnasel, 5′-TTGATCAGAGCATCCGATGGATGGGAGAG-3′ and 5′-TTCTCCAGGAGAAAACAGGCAATGAATGAGGT-3′; Oas1a, 5′-ATTACCTCCTTCCCGACACC-3′ and 5′-AAGGAACACCACCAGGTCAG-3′; Oas1b, 5′-TCTGCTTTATGGGGCTTCGG-3′ and 5′-TCGACTCCCATACTCCCAGG-3′; Oas2, 5′-GGCCTGGTACAGCCTTGGAA-3′ and 5′-AAAGCCTTGGTCCTGCCACA-3′; Oas3, 5′-AAGCCGACACCCAACGTGTA-3′ and 5′-TTTCGGGGCTCAGTGAAGCA-3′; and Oasl1, 5′-GCATCGGCCGACGAAGCTGA-3′ and 5′-CCGTGGCGAGGCCTTCATCC-3′.
Bulk and single-cell RNA-seq
For bulk RNA-seq, total RNA was isolated from 100,000–200,000 cells with an Ambion RNAqueous Micro Kit (Invitrogen). RNA quality was assessed with a Fragment Analyzer (Advanced Analytical). 50 ng of RNA was used for library preparation (Illumina Truseq stranded kit), and sequencing was performed on an Illumina NexSeq 500 machine at the Genomics Facility Basel of ETH Zurich. Single-end 51-mers reads were obtained, and their quality was assessed with the FastQC tool (version 0.11.3). Reads were mapped to the mouse mm10 genome assembly with STAR (version 2.5.2a; Dobin et al., 2013) with default parameters, except reporting only one hit in the final alignment for multimappers (outSAMmultNmax = 1) and filtering reads without evidence in the spliced junction table (outFilterType = “BySJout”). All subsequent gene expression data analysis was performed using R software (version 3.4).
The qCount function of the Bioconductor package QuasR (version 1.18; Gaidatzis et al., 2015) was used to count the number of reads (5′ ends) overlapping with the exons of the Ensembl 84 annotation, assuming an exon union model. Read and alignment quality was evaluated using the qQCReport function of QuasR.
Differentially expressed genes were identified with the Generalized Linear Model framework in the edgeR package (version 3.20.9; Robinson et al., 2010). For the DN3a CD27high/DN3a CD27low/DN3b dataset, a “replicate” covariate was incorporated into the model to control for systematic differences across flow cytometry sorting runs. Genes with a false discovery rate <5% were considered differentially expressed. Gene set enrichment analysis was performed with the function camera (Wu and Smyth, 2012) from the edgeR package (using the default parameter value of 0.01 for the correlations of genes within gene sets) using gene sets from the hallmark collection (Liberzon et al., 2015) of the Molecular Signature Database (MSigDB v6.0; Subramanian et al., 2005). We considered only sets containing >10 genes, and gene sets with a false discovery rate <5% were considered significant. For the heat map combining the two RNA-seq datasets, data were corrected for batch effects for visualization purposes, using the function removeBatchEffect from the Bioconductor limma package (version 3.34.9).
For single-cell RNA-seq, DN2, DN3, and DN4 cells were sorted from six mice and counted. An estimate of 5,000 cells from the DN2 and DN4 populations were loaded on one well each, and 10,000 cells from the DN3 population on two wells of a 10× Genomics Chromium Single Cell Controller. Single-cell capture and cDNA and library preparation were performed with a Single Cell 3′ v2 Reagent Kit (10× Genomics) according to the manufacturer’s instructions. Sequencing was performed on an Illumina NexSeq 500 machine at the Genomics Facility Basel of ETH Zurich. Paired-end reads were obtained, and their quality was assessed with the FastQC tool (version 0.11.3). The length of the first read was 26 mers, composed of individual cell barcodes (16 nt) and molecular barcodes (unique molecular identifiers [UMIs]; 10 nt). The length of the second read, composed of the transcript sequence, was 58 mers. The samples in the different wells were identified using sample barcodes of 8 nt. Sequencing files were processed with Cell Ranger software (version 2.1.0, provided by 10× Genomics) to perform sample and cell demultiplexing, read alignment to the mouse mm10 genome assembly with STAR, and generate a read count table (using the reference transcriptome refdata-cellranger-mm10-1.2.0 based on Ensembl release 84, provided by 10× Genomics). Default settings and parameters were used, except for the version of STAR updated to 2.5.3a, and the STAR parameters outSAMmultNmax set to 1 and alignIntronMax set to 10,000. Samples were merged with the cellranger aggregate procedure without downsampling.
Further analysis was performed using the scran (1.6.9; Lun et al., 2016) and scater (1.6.3; McCarthy et al., 2017) Bioconductor packages on the UMI counts matrix, following mostly the steps illustrated in the simpleSingleCell Bioconductor (release 3.6) workflow.
Cells with log library sizes (or log total number of features detected) more than two median absolute deviations below the median log library size (or log total number of features detected) were filtered out. Cells with >10% of UMI counts attributed to the mitochondrial genes were removed. Cells with <20% of UMI counts attributed to the ribosomal proteins genes were removed. Cells with >1% of UMI counts attributed to the hemoglobin genes were removed. Low-abundance genes with average log2 counts per million (log2CPM) values <0.005 were filtered out.
Expression values of 11,448 genes for 18,128 cells were obtained after filtering. The raw UMI counts were normalized with the size factors estimated from pools of cells to avoid dominance of zeros in the matrix (Lun et al., 2016). A mean-dependent trend was fitted to the variances of the log expression values of endogenous genes to distinguish between genuine biological variability and technical noise, under the assumption that most genes are not differentially expressed across cells, and their variance is mainly technical (trendVar function of the scran package with loess trend and span of 0.4 to better fit the sparse data). The fitted technical noise was subtracted, and the residual “biological” component of the genes variance was used to denoise the PCA with the denoisePCA function of the scran package.
Clustering of cells into putative subpopulations was done on normalized log-counts values using hierarchical clustering on the Euclidean distances between cells (with Ward’s criterion to minimize the total variance within each cluster). The clusters of cells were identified by applying a dynamic tree cut, which resulted in seven putative subpopulations. Marker genes specific for each cluster were identified with the findMarkers function of the scran package, which fits a linear model to the expression values for each gene using the limma framework.
In all analyses, reads assigned to the Duxbl1 (ENSMUSG00000048502), Duxbl2 (ENSMUSG00000072675), and Duxbl3 (ENSMUSG00000072672) genes, 100% identical at the cDNA level, were all assigned to Duxbl1.
Data availability
The RNA-seq data generated are available at the Gene Expression Omnibus database under accession no. GSE118059.
Online supplemental material
Fig. S1 illustrates the gating strategy used to define the different DN stages of T cell development in WT and pTα−/− mice. Fig. S2 compares the transcriptomic sequencing data of DN3a CD27low, DN3a CD27high, and DN3b cells, showing a heat map of Spearman correlation and a PCA. Fig. S3 shows the single-cell RNA-seq–based clustering of DN2, DN3, and DN4 cells. Fig. S4 illustrates the generation of conditional Duxbl transgenic and RNaseL−/− mice. Fig. S5 shows cell cycle analysis, intracellular β-chain expression, apoptosis, and peripheral T cell numbers in DuxblindxpTαCre mice.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of our group leader, mentor, colleague, and friend Antonius Rolink.
We thank Drs. Rhodri Ceredig, Jan Andersson, and Gennaro De Libero for helpful comments and critical reading of the manuscript. We also thank Dr. Gleb Turchinovich for valuable advice on retroviral production and Katrin Hafen for help with Nur77GFP mice. Calculations were performed at sciCORE scientific computing center at University of Basel.
A. Rolink was holder of the chair in immunology endowed by L. Hoffmann-La Roche Ltd, Basel. This study was supported by the Swiss National Science Foundation (310030B_160330/1) and by the People Program (Marie Curie Actions) of the European Union’s Seventh Framework Program FP7/2007-2013 under Research Executive Agency grant agreement 315902.
The authors declare no competing financial interests.
Author contributions: F. Klein, M. Mitrovic, C. Engdahl, L. von Muenchow, P. Pelczar, and P. Tsapogas performed experiments; F. Klein, J. Roux, and L. Alberti-Servera analyzed data; H.J. Fehling and P. Pelczar provided mice and materials; F. Klein wrote the manuscript; F. Klein, P. Tsapogas, and A. Rolink designed the study.
References
- Akashi K., Kondo M., von Freeden-Jeffry U., Murray R., and Weissman I.L.. 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 89:1033–1041. 10.1016/S0092-8674(00)80291-3 [DOI] [PubMed] [Google Scholar]
- Balciunaite G., Ceredig R., Fehling H.J., Zúñiga-Pflücker J.C., and Rolink A.G.. 2005a The role of Notch and IL-7 signaling in early thymocyte proliferation and differentiation. Eur. J. Immunol. 35:1292–1300. 10.1002/eji.200425822 [DOI] [PubMed] [Google Scholar]
- Balciunaite G., Ceredig R., and Rolink A.G.. 2005b The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 105:1930–1936. 10.1182/blood-2004-08-3087 [DOI] [PubMed] [Google Scholar]
- Bell J.J., and Bhandoola A.. 2008. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 452:764–767. 10.1038/nature06840 [DOI] [PubMed] [Google Scholar]
- Brekelmans P., van Soest P., Voerman J., Platenburg P.P., Leenen P.J., and van Ewijk W.. 1994. Transferrin receptor expression as a marker of immature cycling thymocytes in the mouse. Cell. Immunol. 159:331–339. 10.1006/cimm.1994.1319 [DOI] [PubMed] [Google Scholar]
- Capone M., Hockett R.D. Jr., and Zlotnik A.. 1998. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) Pro-T thymocytes. Proc. Natl. Acad. Sci. USA. 95:12522–12527. 10.1073/pnas.95.21.12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceredig R., and Rolink T.. 2002. A positive look at double-negative thymocytes. Nat. Rev. Immunol. 2:888–897. 10.1038/nri937 [DOI] [PubMed] [Google Scholar]
- Ceredig R., Bosco N., and Rolink A.G.. 2007. The B lineage potential of thymus settling progenitors is critically dependent on mouse age. Eur. J. Immunol. 37:830–837. 10.1002/eji.200636728 [DOI] [PubMed] [Google Scholar]
- Ciofani M., and Zúñiga-Pflücker J.C.. 2005. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6:881–888. 10.1038/ni1234 [DOI] [PubMed] [Google Scholar]
- Ciofani M., Schmitt T.M., Ciofani A., Michie A.M., Cuburu N., Aublin A., Maryanski J.L., and Zúñiga-Pflücker J.C.. 2004. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J. Immunol. 172:5230–5239. 10.4049/jimmunol.172.9.5230 [DOI] [PubMed] [Google Scholar]
- Das S., and Chadwick B.P.. 2016. Influence of Repressive Histone and DNA Methylation upon D4Z4 Transcription in Non-Myogenic Cells. PLoS One. 11:e0160022 10.1371/journal.pone.0160022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domen J., Gandy K.L., and Weissman I.L.. 1998. Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood. 91:2272–2282. [PubMed] [Google Scholar]
- Dudley E.C., Petrie H.T., Shah L.M., Owen M.J., and Hayday A.C.. 1994. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1:83–93. 10.1016/1074-7613(94)90102-3 [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Krotkova A., Saint-Ruf C., and von Boehmer H.. 1995. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 375:795–798. 10.1038/375795a0 [DOI] [PubMed] [Google Scholar]
- Fehling H.J., Iritani B.M., Krotkova A., Forbush K.A., Laplace C., Perlmutter R.M., and von Boehmer H.. 1997. Restoration of thymopoiesis in pT alpha-/- mice by anti-CD3epsilon antibody treatment or with transgenes encoding activated Lck or tailless pT alpha. Immunity. 6:703–714. 10.1016/S1074-7613(00)80446-X [DOI] [PubMed] [Google Scholar]
- Gaidatzis D., Lerch A., Hahne F., and Stadler M.B.. 2015. QuasR: quantification and annotation of short reads in R. Bioinformatics. 31:1130–1132. 10.1093/bioinformatics/btu781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2:309–322. 10.1038/nri798 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Zlotnik A., and Suda T.. 1992. Phenotypic and functional characterization of c-kit expression during intrathymic T cell development. J. Immunol. 149:2281–2285. [PubMed] [Google Scholar]
- Godfrey D.I., Kennedy J., Suda T., and Zlotnik A.. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150:4244–4252. [PubMed] [Google Scholar]
- Godfrey D.I., Kennedy J., Mombaerts P., Tonegawa S., and Zlotnik A.. 1994. Onset of TCR-beta gene rearrangement and role of TCR-beta expression during CD3-CD4-CD8- thymocyte differentiation. J. Immunol. 152:4783–4792. [PubMed] [Google Scholar]
- Gravestein L.A., van Ewijk W., Ossendorp F., and Borst J.. 1996. CD27 cooperates with the pre-T cell receptor in the regulation of murine T cell development. J. Exp. Med. 184:675–685. 10.1084/jem.184.2.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haueter S., Kawasumi M., Asner I., Brykczynska U., Cinelli P., Moisyadi S., Bürki K., Peters A.H., and Pelczar P.. 2010. Genetic vasectomy-overexpression of Prm1-EGFP fusion protein in elongating spermatids causes dominant male sterility in mice. Genesis. 48:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving B.A., Alt F.W., and Killeen N.. 1998. Thymocyte development in the absence of pre-T cell receptor extracellular immunoglobulin domains. Science. 280:905–908. 10.1126/science.280.5365.905 [DOI] [PubMed] [Google Scholar]
- Jacobi A.M., Rettig G.R., Turk R., Collingwood M.A., Zeiner S.A., Quadros R.M., Harms D.W., Bonthuis P.J., Gregg C., Ohtsuka M., et al. 2017. Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods. 121-122:16–28. 10.1016/j.ymeth.2017.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H., Iacomini J., van de Ven M., Tonegawa S., and Berns A.. 1996. Domains of the TCR beta-chain required for early thymocyte development. J. Exp. Med. 184:1833–1843. 10.1084/jem.184.5.1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu M., Yamamoto G., Yoshimi M., Yamamoto K., Asai T., Ichikawa M., Seo S., Nakagawa M., Chiba S., Kurokawa M., and Ogawa S.. 2007. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J. Immunol. 179:5335–5345. 10.4049/jimmunol.179.8.5335 [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., and Akashi K.. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Kreslavsky T., Gleimer M., Miyazaki M., Choi Y., Gagnon E., Murre C., Sicinski P., and von Boehmer H.. 2012. β-Selection-induced proliferation is required for αβ T cell differentiation. Immunity. 37:840–853. 10.1016/j.immuni.2012.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger A., and von Boehmer H.. 2007. Identification of a T lineage-committed progenitor in adult blood. Immunity. 26:105–116. 10.1016/j.immuni.2006.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., and Tamayo P.. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1:417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livák F., Tourigny M., Schatz D.G., and Petrie H.T.. 1999. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 162:2575–2580. [PubMed] [Google Scholar]
- Luche H., Nageswara Rao T., Kumar S., Tasdogan A., Beckel F., Blum C., Martins V.C., Rodewald H.R., and Fehling H.J.. 2013. In vivo fate mapping identifies pre-TCRα expression as an intra- and extrathymic, but not prethymic, marker of T lymphopoiesis. J. Exp. Med. 210:699–714. 10.1084/jem.20122609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis T.C., Luc S., Mizukami T., Boukarabila H., Thongjuea S., Woll P.S., Azzoni E., Giustacchini A., Lutteropp M., Bouriez-Jones T., et al. 2016. Initial seeding of the embryonic thymus by immune-restricted lympho-myeloid progenitors. Nat. Immunol. 17:1424–1435. 10.1038/ni.3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T., McCarthy D.J., and Marioni J.C.. 2016. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000 Res. 5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I., Tu L., Sambandam A., Yashiro-Ohtani Y., Millholland J., Keeshan K., Shestova O., Xu L., Bhandoola A., and Pear W.S.. 2006. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 203:2239–2245. 10.1084/jem.20061020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Gillet A., Ardouin L., Bouvier G., Trucy J., Ferrier P., Vivier E., and Malissen B.. 1995. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J. 14:4641–4653. 10.1002/j.1460-2075.1995.tb00146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa S., Balciunaite G., Ceredig R., and Rolink A.G.. 2006. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur. J. Immunol. 36:526–532. 10.1002/eji.200535760 [DOI] [PubMed] [Google Scholar]
- McCarthy D.J., Campbell K.R., Lun A.T., and Wills Q.F.. 2017. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly L.A., Huang D.C., and Strasser A.. 1996. The cell death inhibitor Bcl-2 and its homologues influence control of cell cycle entry. EMBO J. 15:6979–6990. 10.1002/j.1460-2075.1996.tb01090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R.D., Chen H.Y., and Brinster R.L.. 1982. Differential regulation of metallothionein-thymidine kinase fusion genes in transgenic mice and their offspring. Cell. 29:701–710. 10.1016/0092-8674(82)90186-6 [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., and Aguet M.. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. 10.1016/S1074-7613(00)80054-0 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald H.R., Kretzschmar K., Takeda S., Hohl C., and Dessing M.. 1994. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 13:4229–4240. 10.1002/j.1460-2075.1994.tb06743.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Ruf C., Ungewiss K., Groettrup M., Bruno L., Fehling H.J., and von Boehmer H.. 1994. Analysis and expression of a cloned pre-T cell receptor gene. Science. 266:1208–1212. 10.1126/science.7973703 [DOI] [PubMed] [Google Scholar]
- Sambandam A., Maillard I., Zediak V.P., Xu L., Gerstein R.M., Aster J.C., Pear W.S., and Bhandoola A.. 2005. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 6:663–670. 10.1038/ni1216 [DOI] [PubMed] [Google Scholar]
- Saran N., Łyszkiewicz M., Pommerencke J., Witzlau K., Vakilzadeh R., Ballmaier M., von Boehmer H., and Krueger A.. 2010. Multiple extrathymic precursors contribute to T-cell development with different kinetics. Blood. 115:1137–1144. 10.1182/blood-2009-07-230821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwold T., Ehrlich L.I., and Weissman I.L.. 2009. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 113:807–815. 10.1182/blood-2008-08-173682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadle S.C., Zhong J.W., Campbell A.E., Conerly M.L., Jagannathan S., Wong C.J., Morello T.D., van der Maarel S.M., and Tapscott S.J.. 2017. DUX4-induced dsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLoS Genet. 13:e1006658 10.1371/journal.pgen.1006658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867. 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Silverman R.H. 2007. A scientific journey through the 2-5A/RNase L system. Cytokine Growth Factor Rev. 18:381–388. 10.1016/j.cytogfr.2007.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon T., Yui M.A., Pant R., Diamond R.A., and Rothenberg E.V.. 2006. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 24:53–64. 10.1016/j.immuni.2005.11.012 [DOI] [PubMed] [Google Scholar]
- Trampont P.C., Tosello-Trampont A.C., Shen Y., Duley A.K., Sutherland A.E., Bender T.P., Littman D.R., and Ravichandran K.S.. 2010. CXCR4 acts as a costimulator during thymic beta-selection. Nat. Immunol. 11:162–170. 10.1038/ni.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R., Engdahl C., Gehre N., Bosco N., Ceredig R., and Rolink A.G.. 2011. The pre-TCR-dependent DN3 to DP transition requires Notch signaling, is improved by CXCL12 signaling and is inhibited by IL-7 signaling. Eur. J. Immunol. 41:3371–3380. 10.1002/eji.201141824 [DOI] [PubMed] [Google Scholar]
- Wada H., Masuda K., Satoh R., Kakugawa K., Ikawa T., Katsura Y., and Kawamoto H.. 2008. Adult T-cell progenitors retain myeloid potential. Nature. 452:768–772. 10.1038/nature06839 [DOI] [PubMed] [Google Scholar]
- Williams J.A., Hathcock K.S., Klug D., Harada Y., Choudhury B., Allison J.P., Abe R., and Hodes R.J.. 2005. Regulated costimulation in the thymus is critical for T cell development: dysregulated CD28 costimulation can bypass the pre-TCR checkpoint. J. Immunol. 175:4199–4207. 10.4049/jimmunol.175.7.4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., and Smyth G.K.. 2012. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40:e133 10.1093/nar/gks461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.L., Tsai M.S., Wong S.H., Hsieh-Li H.M., Tsai T.S., Chang W.T., Huang S.L., Chiu C.C., and Wang S.H.. 2010. Characterization of genomic structures and expression profiles of three tandem repeats of a mouse double homeobox gene: Duxbl. Dev. Dyn. 239:927–940. 10.1002/dvdy.22210 [DOI] [PubMed] [Google Scholar]
- Yui M.A., and Rothenberg E.V.. 2014. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 14:529–545. 10.1038/nri3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Paranjape J., Brown T.L., Nie H., Naik S., Dong B., Chang A., Trapp B., Fairchild R., Colmenares C., and Silverman R.H.. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355–6363. 10.1093/emboj/16.21.6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotoff D.A., and Bhandoola A.. 2011. Hematopoietic progenitor migration to the adult thymus. Ann. N. Y. Acad. Sci. 1217:122–138. 10.1111/j.1749-6632.2010.05881.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated are available at the Gene Expression Omnibus database under accession no. GSE118059.