By blocking an important signaling pathway (called NOTCH) and interfering with expression of two tumor suppressor genes in cells derived from human embryonic stem cells, Chen et al. have developed a model for studying small cell lung cancers.
Abstract
Cancer models based on cells derived from human embryonic stem cells (hESCs) may reveal why certain constellations of genetic changes drive carcinogenesis in specialized lineages. Here we demonstrate that inhibition of NOTCH signaling induces up to 10% of lung progenitor cells to form pulmonary neuroendocrine cells (PNECs), putative precursors to small cell lung cancers (SCLCs), and we can increase PNECs by reducing levels of retinoblastoma (RB) proteins with inhibitory RNA. Reducing levels of TP53 protein or expressing mutant KRAS or EGFR genes did not induce or expand PNECs, but tumors resembling early-stage SCLC grew in immunodeficient mice after subcutaneous injection of PNEC-containing cultures in which expression of both RB and TP53 was blocked. Single-cell RNA profiles of PNECs are heterogeneous; when RB levels are reduced, the profiles resemble those from early-stage SCLC; and when both RB and TP53 levels are reduced, the transcriptome is enriched with cell cycle–specific RNAs. Our findings suggest that genetic manipulation of hESC-derived pulmonary cells will enable studies of this recalcitrant cancer.
Introduction
Cancers presumed to arise from different cell lineages display characteristic genotypes, but cells of origin are generally uncertain, and the relationships between lineage-specific attributes and genotypic differences of tumors are not understood (Garraway and Lander, 2013; Weinstein et al., 2013). One of the main obstacles to greater knowledge about these relationships is the need for tractable systems that allow molecular changes observed in mature cancer cells to be evaluated for their contribution to hallmarks of neoplasia as they occur during the development of specific cell lineages.
Small cell lung cancer (SCLC), the most aggressive type of lung cancer, characterized by a poor prognosis, the rapid development of resistance to treatment, and nearly universal loss of function of tumor suppressor genes tumor protein P53 (TP53) and retinoblastoma (RB), is a relatively common human cancer in need of improved model systems (Peifer et al., 2012; George et al., 2015; Pietanza et al., 2015; Semenova et al., 2015; Gazdar et al., 2017). Studies of mouse models of SCLC indicate that the target cells for malignant transformation are likely to be pulmonary neuroendocrine cells (PNECs; Linnoila, 2006; Sutherland et al., 2011; Song et al., 2012), consistent with the morphology of the cancer cells and expression of neuroendocrine markers. Despite these recent advances, fundamental features of SCLC, especially its initiation, progression, and eventual resistance to therapy, are not understood in relation to its observed genotypes.
To study these problems, we have sought ways to assess functional changes that occur after specific genes are altered in human pulmonary cells at defined stages of tissue development. Recent advances in the induction, cultivation, and directed differentiation of human embryonic stem cells (hESCs) provide opportunities to study carcinogenesis in many human cell types derived from a variety of lineages (Funato et al., 2014), including cancers such as SCLC, in which rapid onset and progression limit the availability of clinical samples, especially from early-stage disease (Pietanza et al., 2015). In this report, we demonstrate that lung progenitor (LP) cells derived from hESCs can be induced to form PNECs by inhibition of NOTCH signaling, that the proportion of PNECs can be specifically increased by inhibition of expression of the RB tumor suppressor gene, and that subsequent interference with the P53 tumor suppressor gene allows xenografted cells to form early-stage tumors resembling SCLC.
Results
Generation of PNECs from cultured hESCs
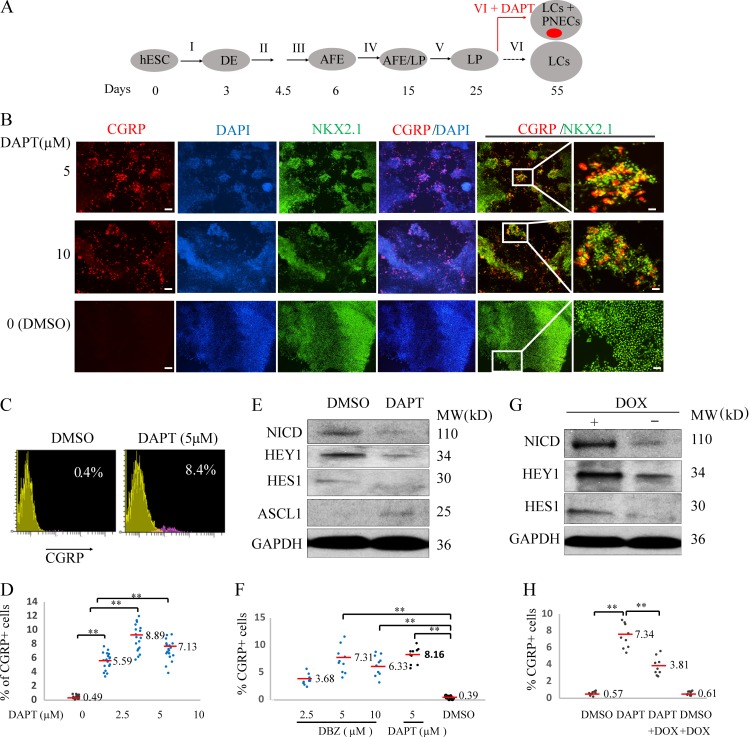
Methods have recently been described for generating most, but not all, of the cell types observed in adult lung tissues by using growth factors and chemicals to alter signaling pathways sequentially in cells derived from hESCs over several weeks (Fig. 1 A). Using a protocol developed by Huang et al. (2014, 2015), we have confirmed that by day 3, ∼90% of hESCs (the RUES2 and ES02 lines) differentiate into definitive endoderm (DE), triple positive for the markers KIT, EPCAM, and CXCR4 (Fig. S1, A and B); anterior foregut endoderm by day 6; increasing numbers of LPs, SOX2+, NKX2.1+, and FOXA2+ between days 15 and 25 (Fig. S1, C and D; and Fig. S2, A and B); and then a variety of airway and lung epithelial cells (basal progenitor cells, ciliated cells, goblet cells, club cells, and alveolar type 1 and type 2 cells [AT1 and AT2]; Warburton et al., 1998; Treutlein et al., 2014) by day 55 (Fig. S1, E–G). However, this protocol and others produce few, if any, PNECs (<0.5%; Fig. 1, B and C; and Fig. S1 G).
Figure 1.
Generating PNECs through directed differentiation of hESCs and suppression of NOTCH. (A) Schematic of the protocol used to generate PNECs by stepwise differentiation of hESCs to form DE by day 3, anterior foregut endoderm (AFE) by day 6, and increasing numbers of LPs from days 15 to 25, using the differentiation mixtures I–V (defined in Materials and methods section; Fig. 3 and Fig. S1). LPs were further differentiated in mixture VI from days 25 to 55 into the major types of LCs found in mature human lung parenchyma and airway epithelium (Warburton et al., 1998; Treutlein et al., 2014). Addition of DAPT to mixture VI induced formation of PNECs (red dot), as described in the text. (B) Detection of putative PNECs by IHC after treatment with DAPT. ESCs from the RUES2 line were differentiated according to the protocol in A to day 55 then stained to detect CGRP, NKX2.1, or both, with the indicated antisera; nuclei were detected by staining with DAPI. Scale bars, 100 µm (left) and 20 µm (right). (C and D) Percentages of CGRP+ cells were determined at day 55 by FACS and displayed as flow cytometry data (red, CGRP+; yellow, CGRP–) and a scatter graph (D). (E and F) Confirmation of mechanism of action of DAPT as inhibitor of γ-secretase cleavage of NOTCH. (E) DAPT (5 μM) treatment from day 25 to 55 decreased the level of the NICD and protein products of the NOTCH target genes, HES1 and HEY1, while increasing levels of ASCL1 in day 55 LCs, as detected by Western blot. (F) LPs treated with another γ-secretase inhibitor, DBZ, from day 25 to 55, also form CGRP+ cells at frequencies similar to those observed with DAPT (D). (G and H) Constitutive expression of NICD prevents the appearance of CGRP+ cells cotreated with DAPT. RUES2 cells carrying a DOX-inducible NICD were differentiated to form LPs and then treated with DOX, DAPT, or both for 30 d. The induction of NICD by DOX and expression of HES1 and HEY1 with Western blot are demonstrated in G; H shows by FACS that DOX (to induce expression of NICD) inhibits DAPT-mediated appearance of CGRP+ cells. **, P < 0.01; *, P < 0.05 by one-way ANOVA test or (for C) by Student’s t test. Horizontal red lines denote average values; number of biological repeats (n) = 18 for D, n = 10 for F, and n = 9 for H.
Studies of mouse development have suggested that inhibition of signaling via NOTCH receptors might influence cells to adopt a neuroendocrine fate (Ito et al., 2000; Linnoila, 2006; Shan et al., 2007; Morimoto et al., 2012). In addition, inactivation of NOTCH genes is found in ∼25% of SCLCs (George et al., 2015). Based on these reports, we exposed LPs between day 25 and 55 to N-[(3,5-difluorophenyl)acetyl]-L-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester (DAPT; Geling et al., 2002)—a known inhibitor of γ-secretase, the protease that normally cleaves NOTCH receptors to yield a transcriptionally active, mobile, intracytoplasmic domain of NOTCH (NICD; Schroeter et al., 1998)—to ask whether the loss of NOTCH signaling might promote the production of PNECs. (We define PNECs here as cells containing a general lung-specific marker, the transcription factor NKX2.1, and expressing one or more genes encoding well-recognized neuroendocrine markers, especially the cell surface–associated protein that includes the calcitonin gene-related peptide [CGRP; Song et al., 2012] or the nuclear transcription factor ASCL1 [Borges et al., 1997; Borromeo et al., 2016].)
After exposure to 5–10 μM DAPT for 30 d, a substantial number of differentiating LP cells (∼8.9 ± 1.9% in RUES2 cells and 5.6 ± 1.4% in ES02 cells, versus 0.5 ± 0.20% or 0.4 ± 0.2% in control cultures) adopt properties of PNECs, as measured by counting CGRP+ cells with FACS and confirmed qualitatively by immunofluorescence of cells in monolayer cultures with antibodies against NXK2.1 and CGRP (Fig. 1, B–D; and Fig. S2, C–E).
DAPT induces PNECs by blocking cleavage of NOTCH
We took several approaches to confirm the mechanism by which DAPT induced PNECs. First, we measured the abundance of NICD in extracts from cells treated with DAPT and observed the expected loss of the γ-secretase cleavage product (Fig. 1 E). To confirm the reduction in NOTCH-mediated signaling, we measured the readout from two NOTCH target genes (Iso et al., 2003), HES1 and HEY1, and found a marked decrease in levels of transcripts from HEY1 and a slight but significant decrease of RNA from HES1 (Fig. S2 F). We also tested another known inhibitor of γ-secretase, dibenzazepine (DBZ; Milano et al., 2004), and produced percentages of PNECs at day 55 similar to those observed with DAPT (Fig. 1 F). Finally, we reversed the effects of DAPT by providing NICD, the normal product of γ-secretase–mediated cleavage of NOTCH: expression of a tetracycline-inducible transgene encoding NICD from days 25 to 55 in differentiated RUES2 cells decreased the appearance of CGRP+ cells in cultures concurrently exposed to DAPT (Fig. 1, G and H). We also showed that expression of HES1 and HEY1, which was down-regulated in DAPT-treated lung cells (LCs; Fig. 1 E and Fig. S2 F), was increased by induction of NICD (Fig. 1 G). In addition, DAPT induced expression of ASCL1 after the RUES2 line was differentiated to form LCs (Fig. 1 E).
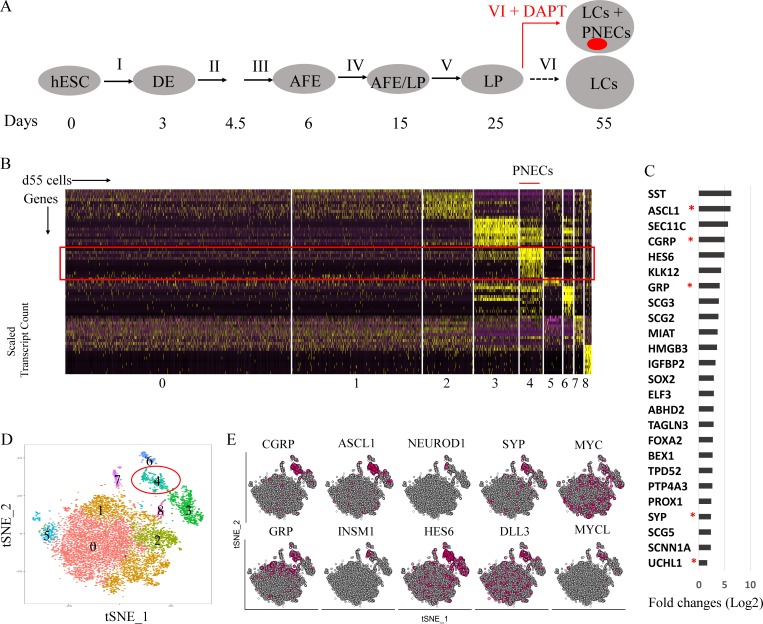
Single-cell transcriptional profiling of induced PNECs
To further characterize the presumptive PNECs generated by inhibition of NOTCH signaling, we used high-throughput single-cell RNA sequencing (scRNA-seq) applied to DAPT-treated and untreated cells at day 55 (Fig. 2, A–E; and Fig. S1, F and G; see Materials and methods section). Clustering of the single-cell profiles revealed one cluster enriched with cells expressing genes that encode CGRP or ASCL1, thus identifying the presumed PNEC cells. In total, the presumed PNEC cells constituted 7.72% of the 9,824 high-quality cells (pooled from two biological replicates). Analysis of differential gene expression in the various clusters revealed that cells in cluster 4 exhibit relatively high numbers of transcripts from a set of genes, including CGRP, ASCL1, GRP, SYP, and UCHL1, that encode canonical PNEC markers (Linnoila, 2006; Song et al., 2012; Fig. 2, C and E). Fig. 2 E displays the expression of those and other relevant genes, including MYC, MYCL, and NEUROD1 (Mollaoglu et al., 2017), characteristically expressed in neuroendocrine cells or SCLC. In comparison, we detected CGRP or ASCL1 RNA in only 1.3% percent of cells, and rarely together, from control cultures not treated with DAPT (Fig. S1 G).
Figure 2.
Single-cell RNA profiling of RUES2-derived LCs in which NOTCH signaling was blocked by DAPT. (A) Schematic of the protocol used to generate PNECs by stepwise differentiation of hESCs and DAPT treatment from day 25 to 55. AFE, anterior foregut endoderm. (B–E) Single-cell RNA profiling of day 55 LCs derived from RUES2 cells treated with DAPT (5 µM) from day 25 to 55. (B) Heat map representing scaled expression of the most differentially expressed genes specific to different cell clusters. Rows represent genes, and columns represent cells. (C) Putative PNEC markers differentially expressed in the PNEC-like cell cluster number 4 in B. Bars indicate log fold change versus non-PNEC cells. Asterisks indicate canonical PNEC markers. (D) Reduced-dimensionality tSNE map colored by cluster assignment (Materials and methods section). (E) Individual cells positive for PNEC markers and other genes associated with neuroendocrine differentiation are denoted by red dots. Number of biological repeats (n) = 2.
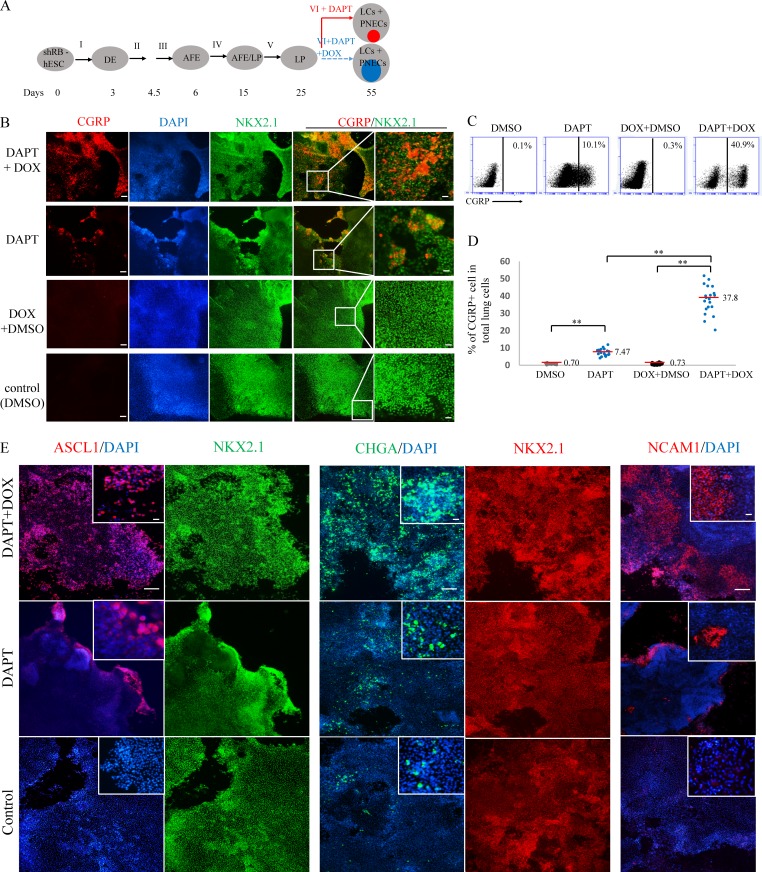
Reduced expression of RB enlarges the proportion of PNECs
Since a central objective of this work is to assess the influence of known lung cancer genes on the behavior of cells in the lung lineage, we next examined the consequences of expressing or simulating known oncogenic mutations in hESC cultures undergoing differentiation, with or without inhibition of NOTCH signaling (Figs. 3 and 4). To that end, we equipped the RUES2 hESC line with doxycycline (DOX)-inducible transgenes encoding shRNAs that inhibit production of RNA from either of the two tumor suppressor genes most commonly inactivated by mutations in SCLC, the RB or TP53 genes (Fig. S3, A and B; and Fig. 4 A). We also introduced into parallel cultures inducible transgenes encoding oncogenes commonly encountered in lung adenocarcinomas, mutant epidermal growth factor receptor (EGFR) or mutant KRAS (Cancer Genome Atlas Research Network, 2014; Fig. 4 B).
Figure 3.
Inhibited expression of the RB tumor suppressor gene augments induction of PNECs by DAPT. (A) Schematic of PNEC production from hESC cells carrying a tetracycline-inducible transgene that produces shRNA targeting RB1 (shRB). The format is similar to Fig. 1 A, except that mixture VI is supplemented with DAPT (5 µM) with or without DOX. Internal colored circles at day 55 indicate PNECs induced by DAPT (red) or by DAPT and DOX (blue). AFE, anterior foregut endoderm. (B–D) Increased numbers of putative PNECs detected by costaining for CGRP and NKX2.1 (B) or by FACS sorting with anti-human CGRP antibody (C and D) as described in the legend to Fig. 1 and the Materials and methods section. **, P < 0.01 by one-way ANOVA test. In D, horizontal red lines denote average values; biological repeats (n) = 20. Scale bars, 100 µm (left) and 20 µm (right). (E) Expression of shRNA-RB increases the percentages of ASCL1+, CHGA+, or NCAM+ cells. The indicated markers were detected by immunostaining as in B. Scale bars, 100 µm and 20 µm (in small window).
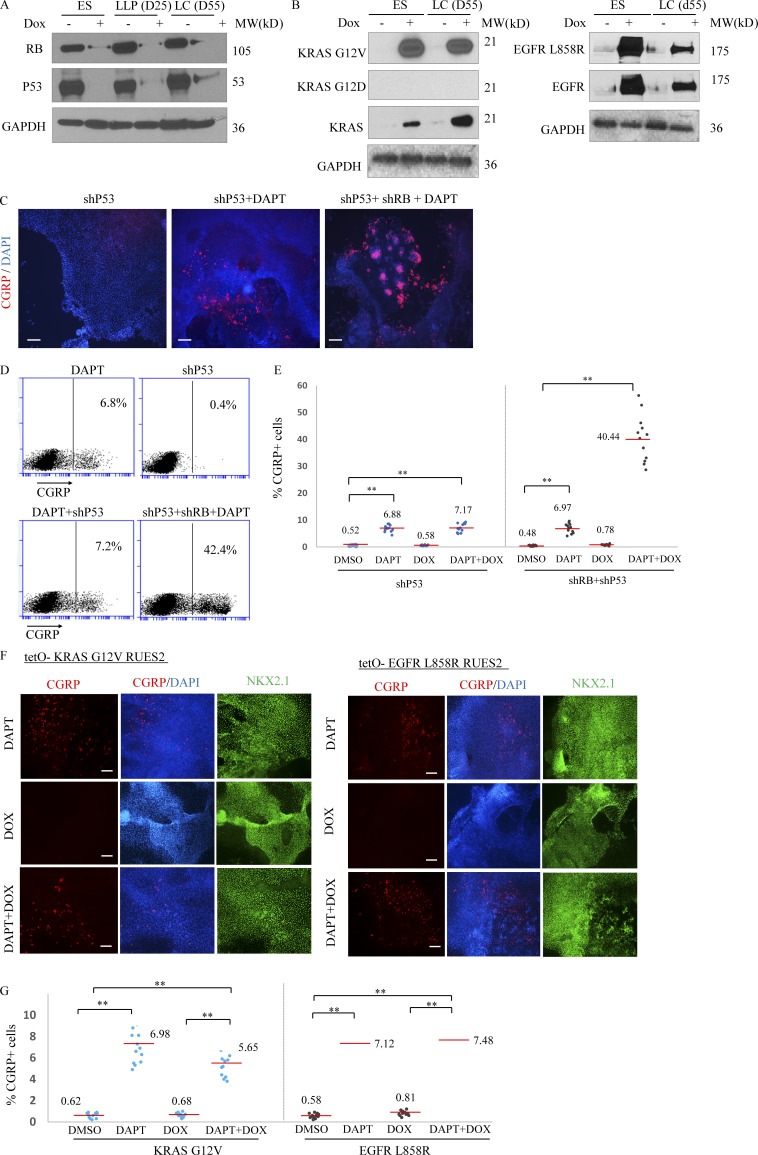
Figure 4.
Inhibition of the p53 tumor suppressor gene or expression of two common lung cancer oncogenes do not augment production of PNECs in the presence and absence of DAPT. RUES2 cells carrying DOX-inducible transgenes that encode shRNAs targeting P53 or RB or mutant alleles of KRAS (G12V) or EGFR (L858R) were used as in the experimental protocol shown in Fig. 2 A to measure the effects of DOX induction of the indicated transgenes from day 25 to 55 on production of PNECs. (A) DOX-dependent expression of oncogenes or shRNAs in RUES2 cells. Western blots show RB and P53 tumor suppressor proteins after DOX induction of shRNA cassettes. (B) Production of proteins encoded by DOX-regulated transgenes encoding KRAS-G12V (left) and EGFR-L858R (right). Proteins were detected by the indicated antibodies (Materials and methods section). (C–G) Decreased P53 or production of mutant KRAS or EGFR proteins fail to increase the percentage of PNECs. The indicated RUES2 cell lines were tested for the appearance of PNECs at day 55 by immunofluorescence staining for CGRP (C and F) and by FACS (D, E, and G). See text for interpretation. Scale bars, 100 µm (C) and 200 µm (F). **, P < 0.01 by one-way ANOVA test. Horizontal red lines denote average values. Biological repeats (n) = 12 for E and 13 for G.
Induction of RB-specific shRNA in RUES2 cells differentiating to form LCs between days 25 and 50 markedly reduced the amount of RB protein (Fig. S3 A) but not the amounts of the closely related proteins p107 and p130 (Fig. S3 B). Reduced expression of the RB gene was associated with a significantly increased number of CGRP+ NKX2.1+ cells (putative PNECs) from 7.5 ± 2.0% to 37.8 ± 8.2% (Fig. 3, A–D), as measured by FACS, but only when cells were also exposed to DAPT to inhibit processing of NOTCH (Fig. 2 D). Similarly, the proportions of cells expressing the PNEC transcription factor ASCL1 and the associated markers NCAM1 and CHGA were also significantly increased (Fig. 3 E).
In contrast, induction of TP53-specific shRNA during the same interval had no effect on the number of CGRP+ cells, with or without DAPT and with or without induction of RB-shRNA (Fig. 4, C–E). Similarly, induction of mutant forms of EGFR and KRAS proteins between days 25 and 55 (Fig. 4 B) failed to increase the number of CGRP+ cells grown in the absence or presence of DAPT (Fig. 4, F and G). These findings indicate that loss of TP53 or production of mutant EGFR or KRAS proteins does not induce or affect the abundance of PNECs.
The effects of reducing RB levels on PNEC transcriptomes
To examine the transcriptional phenotypes of cultures of differentiated (day 55) RUES2 cells in which both NOTCH and RB signaling were inhibited, we turned again to scRNA-seq. Cell clustering and differential expression analyses, similar to those used in Fig. 1, indicate the presence of multiple cell populations in our cultures, including an expanded PNEC-like cell compartment (11.7%), expressing markers similar to those observed in PNEC-like cells at day 55 with normal levels of RB protein (Fig. S3, C–G). (We attribute the relatively modest increase in PNEC-like cells—as judged by scRNA-seq, compared with the increase measured by FACS—to differences in the sensitivity of detection methods that measure RNA levels as opposed to cell surface proteins.)
Transcriptomes of PNECs with reduced RB levels resemble transcriptomes from early SCLC
To ask whether inhibition of NOTCH signaling, coupled with a reduction of RB protein, produces a transcriptional program that resembles the program in human SCLC, we compared the scRNA-seq profiles of PNECs and non-PNECs from day 55 RUES2 cells, with normal or reduced levels of RB gene expression, to the published RNA profiles from 29 early-stage (stage Ia or Ib) human SCLCs (Peifer et al., 2012; George et al., 2015; Fig. S3 H). This analysis confirmed that PNEC expression profiles more closely resemble SCLC profiles than do the profiles from non-PNECs. In addition, the similarity to SCLC profiles is greater (P < 2.2e-16 by two-sided Kolmogorov–Smirnov test) in PNECs in which RB levels were reduced than in PNECs in which RB levels were not perturbed.
To further characterize the effects of the reduction of RB protein on the transcriptional profile of the PNEC cells, we performed a differential expression analysis that compared PNECs appearing after DAPT with PNECs appearing after DAPT combined with reduced expression of RB (Fig. S4 A). Subsequent gene function enrichment analysis (Chen et al., 2009) indicated that the most differentially expressed genes following reduction of RB gene expression are involved in regulation of nerve development, apoptosis, TP53 signal transduction, and other processes (Fig. S4 B). When investigating the heterogeneity within the DAPT-induced PNEC cell cluster, we detected three subpopulations (Fig. S4 C) with substantially different expression profiles (Fig. S4 C). A similar analysis of PNECs from DAPT-treated cultures in which RB levels were reduced also revealed three subpopulations but with transcriptional profiles different from those in cultures in which RB expression was not altered (Fig. S4 C). Some of these differentially expressed genes, including HES1, SST, SCG3, STMN2, ELAVL3, and IGFBP4, as well as NEUROD1, have previously been reported to be responsible for the heterogeneity of pulmonary neuroendocrine tumors and implicated in the initiation and progression of human SCLC (Borromeo et al., 2016; Lim et al., 2017).
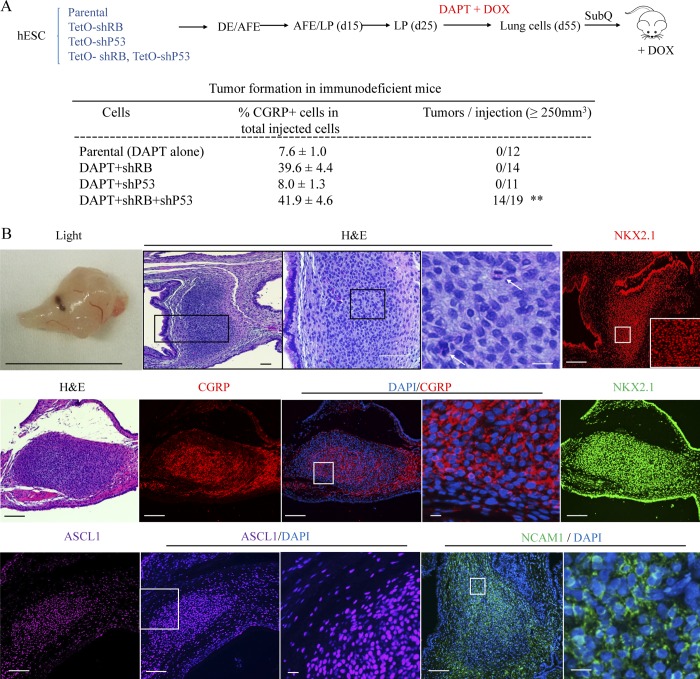
Reduction of both P53 and RB in DAPT-induced PNECs allows xenografts to form tumors
To assess the ability of cells in differentiated RUES2 cultures to form tumors, we performed subcutaneous injections of day 55 cells treated in various ways into immune-deficient NSG mice (NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ; Shultz et al., 2005). The four tested cell populations all contained presumptive PNECs (measured as CGRP+ cells by FACS), ranging from ∼10 to ∼40% of the cultured cells. No growths >250 mm3 in volume were observed within 7 wk after injection of parental cells exposed to DAPT alone or cells also carrying the TP53-shRNA or the RB-shRNA expression cassettes and treated with DOX (Fig. 5 A). In contrast, cells with both shRNA cassettes and treated with DAPT and DOX formed visible tumors at 14 of 19 injection sites within 6–7 wk. In general, these tumors were ∼1 cm in diameter, formed of compact, darkly staining cells, with mitotic figures observed in some of them, morphologically resembling SCLC in mice and humans (Fig. 5 B), and not locally invasive. The origin of the tumor cells was confirmed by detection of GFP encoded by a component of the RB-shRNA cassette in nuclei of RUES2 cells (Fig. S5, A and B); in addition, the nuclei were stained with antisera specific for human antigens (Fig. S5 C). A PNEC-like phenotype was documented using immunohistochemistry (IHC) to display the neuroendocrine biomarkers CGRP, NCAM1, and ASCL1, as well as the lung marker NKX2.1 (Fig. 5 B). In addition, we observed that some cells within the xenograft tumors produced C-MYC and MYCL (Mollaoglu et al., 2017) but not NEUROD1, another neuroendocrine transcription factor (Fig. S5, F–H).
Figure 5.
Characterization of xenografts formed with hESC-derived LCs. (A) Schematic representation of tumorigenesis experiment. The indicated transgenic and control lines of RUES2 hESCs were differentiated and grown in DAPT and DOX from day 25 to 55. At day 55, total LCs were injected subcutaneously into NSG mice that then received DOX in their feed as described in the Materials and methods. Xenografts grew into visible tumors after 6–7 wk only from cells containing transgenes encoding shRNAs for both RB and P53. **, P < 0.01 by Fisher’s test. AFE, anterior foregut endoderm. (B) Characterization of xenografts. Top row: Left segment, a representative tumor by light microscopy; middle three segments, H&E staining of that tumor at different magnifications (white arrows designate the cells with mitotic figures; right segment, staining for NKX2.1. Scale bars from left to right: 1 cm, 200 µm, 100 µm, 10 µm, and 200 µm. Middle bottom row: Additional samples stained with H&E, DAPI, or antibodies specific for the indicated marker proteins. Scale bars, 200 µm (long); 10 µm (short). Biological repeats (n) = 3.
We excluded the possibility that the observed tumors were mischaracterized teratomas known to be formed in mice injected with hESCs. When we produced teratomas in NSG mice with undifferentiated RUES2 cells, they contained embryonic tissue markers (Liu et al., 2010), such as α-fetoprotein, Nanog, Oct4, and SSEA4 (Fig. S5 D); exhibited morphological features different from tumors formed with our differentiated cultures in which NOTCH, RB, and P53 pathways were disrupted (Fig. S5 A); and did not contain PNEC markers CGRP, ASCL1, and NKX2.1 (Fig. S5 E). However, since the neuroendocrine tumors in Fig. 5 arose from injections of heterogeneous cultures of RUES2-derived cells, we cannot completely exclude the highly unlikely possibility that the tumors originated from non-PNEC cells in the cultures (Huang et al., 2018).
The effects on PNEC transcriptomes of reduced expression of both RB and P53
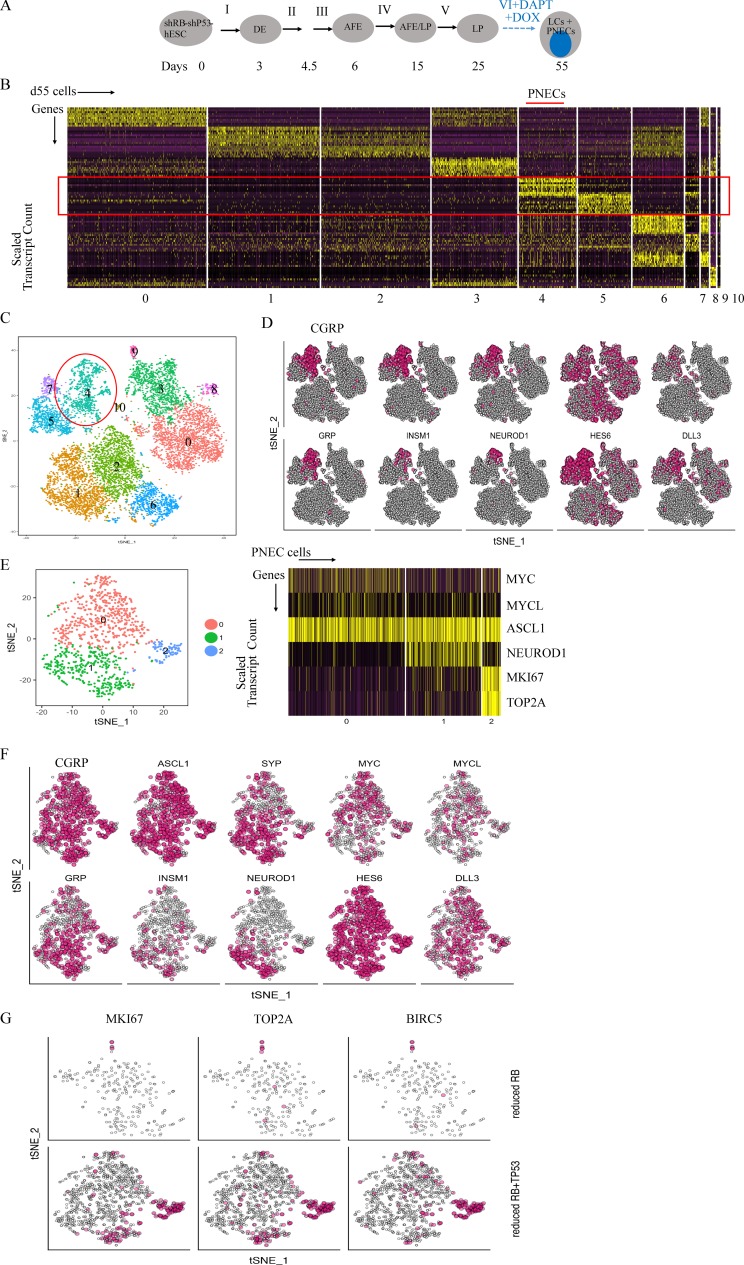
Since induction of TP53-specific shRNA, as well as RB-specific shRNA, was required to produce tumor-forming cells in DAPT-treated cells derived from RUES2 cells, we used scRNA-seq to seek evidence that might associate the tumorigenic phenotype with changes in PNEC transcriptomes. As indicated earlier (Fig. 4, C–E), inhibition of expression of TP53 does not affect the proportion of PNECs in these cultures; as expected, scRNA-seq revealed that the PNEC-like compartment of cultures in which both RB and TP53 were inhibited was similar in size (10.8%) to that of cultures in which only RB was inhibited (Fig. S3, C–G). Cell clustering and analysis of differentially expressed genes in these unfractionated cultures indicate the presence of multiple cell populations (Fig. 6, A–D), similar to cultures in which P53 was not reduced (Fig. 4, C–G).
Figure 6.
Single-cell RNA profiling of RUES2-derived LCs in which NOTCH signaling was blocked by DAPT and RB and P53 protein levels were reduced by shRNA. (A) Schematic of PNEC production from RUES2 cells carrying DOX-inducible transgenes encoding shRNAs against RB and TP53 after exposure to DAPT and DOX (as in Fig. 2 A). AFE, anterior foregut endoderm. (B) Single-cell RNA profiling of day 55 LCs treated with DAPT (5 µM) and DOX from day 25 to 55. Scaled expression of the most differentially expressed genes (horizontal rows) specific to different clusters of cells (vertical columns). (C) PNEC-like cells are principally in cluster 4, but additional cells expressing some PNEC markers are found in other clusters. tSNE map colored by cluster assignment as in Fig. 2 D. (D) Individual cells positive for RNA encoding the indicated PNEC markers and for other proteins associated with neuroendocrine differentiation are denoted with red dots as in Fig. 2 E. (E) Left: A tSNE map of reclustering of the PNEC-enriched cluster only (cluster 4 in C); Right: A scaled expression of the most differentially expressed key genes (horizontal rows) specific to cells in three PNEC subclusters called 0, 1, and 2 on the left (vertical columns). (F) Individual cells positive for PNEC markers and other genes associated with neuroendocrine differentiation are denoted by red dots. (G) Individual cells positive for cell cycle–related genes are denoted by red dots in PNECs in which RB levels were reduced (top row) or both RB and TP53 levels were reduced (bottom row). Biological repeats (n) = 2.
In the analysis of cells in which P53 levels were reduced, however, we identified three well-defined clusters, one differing from the others because its cells show increased expression of genes associated with active cell cycling (Fig. 6, E–G). This is consistent with the idea that different proliferation rates exist within the PNEC compartment, suggesting that reduction of P53 levels, coupled with low levels of RB protein, enables a subset of PNECs to enter a proliferative mode, a phenomenon we did not observe in cells with normal levels of P53, regardless of RB status (Fig. S4 C). We also found that this subset of presumptively proliferating PNECs highly expresses genes associated with inhibition of apoptosis, as well as cell cycle genes, including BIRC5, TOP2A, MKI67, CDK1, CDKN3, and CDC20. This indicates that the main effects of reducing P53 levels in PNECs are likely to be proliferative and antiapoptotic (Fig. 6, E–G). These effects might account for the appearance of tumor-forming potential in xenografted mice.
A significant increase in the expression of RNA encoding the neuroendocrine transcription factor, NEUROD1, which has been associated with advanced stages of SCLC, was also observed in cultures in which P53 protein levels were reduced (Osborne et al., 2013a,b; Borromeo et al., 2016; Fig. 6, E and F). The percentages of NEUROD1 RNA-positive cells in PNEC clusters produced by NOTCH inhibition alone, by NOTCH inhibition with reduced RB protein, and by NOTCH inhibition with reduction of both RB and P53 are 6.61 ± 0.52%, 3.02 ± 0.75%, and 15.75 ± 0.50%, respectively (mean ± SD, following repeated subsampling to account for sequencing depth). In addition, the expression of genes associated with neuroendocrine differentiation (such as CGRP, ASCL1, and NEUROD1; Fig. S4 D) and genes encoding canonical markers for other LC types (Fig. S4 E) was compared at day 55, using scRNA profiles of LCs treated from day 25 to 55 with DMSO (Parental), DAPT (DAPT only), and DAPT and DOX to induce shRNA targeting RB (RB reduced) or to induce shRNAs targeting both RB and P53 (RB and P53 reduced). Whereas some markers (e.g., NKX2.1, SOX2, and SOX9) are clearly detected in particular subpopulations of cells, other markers were not detected often enough to allow clear determination of cell lineage. In particular, the mRNA for transcription factor POU2F3, associated with tuft cells (Huang et al., 2018; Zhang et al., 2018), was not detected in our scRNA data; however, the depth of sequencing was not sufficient to distinguish between the limited power to detect the transcripts or the absence of this cell type.
Discussion
In this report, we describe the development of a human cell-based model for the initiation of early-stage tumors resembling SCLC. To do this, we discovered a means to induce PNEC-like cells from cultured hESCs after differentiation into LPs by inhibition of NOTCH signaling; expanded the proportion of PNEC cells by blocking the expression of the RB tumor suppressor gene; and conferred a tumorigenic phenotype on those cells by also blocking the expression of TP53, another tumor suppressor gene commonly inactivated in SCLC. We also used scRNA-seq to characterize gene expression in hESC-derived PNECs, revealing heterogeneity in cells with normal or reduced levels of RB protein. These studies revealed the resemblance of transcriptional profiles in PNECs in which expression of RB was reduced to those reported for early-stage human SCLC, further supporting the idea that our manipulations of hESCs in culture are generating cells on a pathway to full-fledged SCLC phenotypes. We also found that the transcriptomes of PNECs in which levels of both RB and P53 proteins were reduced include enhanced expression of genes associated with cell proliferation and inhibition of apoptosis. These changes may explain the acquisition of tumor-forming potential when both tumor suppressor genes have been inhibited, as commonly occurs in human SCLC. They also indicate that it may be possible to assign specific hallmarks of the cancer phenotype to each of the two most common genotypic changes observed in SCLCs, inactivating mutations of the RB and TP53 genes. Additional studies will be required to establish this idea more firmly.
Since the SCLC-like tumors grown subcutaneously in immunodeficient mice appear to have low tumor potency (slow-growing and noninvasive), it is likely that this system will enable studies of tumor progression. In addition, it should be possible to examine cells at different stages of tumor development for susceptibility and resistance to therapeutic strategies, in a manner similar to that used in an earlier study, in which a rare form of glioma was derived from hESCs by genetic manipulation of neural precursors (Funato et al., 2014).
We recognize that tumorigenesis in an intact organism is a complex process that involves interactions between the cells from which cancer arises and the several types of normal cells: stromal, epithelial, vascular, immunological, and other blood cells. These other cell types, and the adjacencies and paracrine effects that might influence tumor initiation and progression are, of course, missing in the hESC-derived culture system we have described here. Nevertheless, this system has several advantages: it is based on the use of human rather than nonhuman cells, and it allows the characterization of substantial numbers of cells subjected to synchronized changes in genotype, gene expression, or hormonal effects.
Although we have begun our evaluation of the ability of manipulated LCs to form tumors from xenografts at an ectopic site (subcutaneous tissue), it may be possible to study the cells at more appropriate locations, including lung tissue, and we are presently exploring that possibility. Other models, based on the use of genetically altered mice, offer an opportunity to study the generation of cancers like SCLC in a setting in which tissue architecture has not been perturbed, and we view the two approaches as complementary. However, we believe that the use of developmentally controlled human cells to probe the relationship between cancer genotypes and stages of lineage development to be an especially promising feature of the work we have described here.
Notably, we have generated early-stage SCLC-like growths only after inducing neuroendocrine cell types and reducing expression of tumor suppressor genes implicated in human SCLC. Thus, it seems likely that our differentiation protocol yields cell types, like LPs and PNECs, that are vulnerable to specific types of oncogenic lesions, such as inactivation of certain tumor suppressor genes. Further, such cells do not appear to be susceptible to the transforming effects of other genes implicated in human cancers, including oncogenes, such as mutated EGFR and KRAS, known to be drivers of other types of human lung cancer (e.g., adenocarcinomas). Understanding the determinants of this apparent specificity may help account for the widely recognized correlations between cancer genotypes and lineage phenotypes in the major forms of lung cancers and many other forms of human neoplasia (Garraway and Lander, 2013; Weinstein et al., 2013).
Materials and methods
Generation of LCs, including PNECs, from hESCs
Protocols for maintenance of hESCs and generation of LCs were slightly modified from previous studies (Huang et al., 2014, 2015). Two hESC lines, RUES2 (National Institutes of Health approval no. NIHhESC-09-0013, registration no. 0013; passage 7–10) and ES02 (HES2; National Institutes of Health registry, WiCell Research Institute, Inc.; passage 3–7), were cultured on irradiated mouse embryonic fibroblasts (Global Stem; cat. no. GSC-6001G) at a density of 20,000–25,000 cells per cm2 in a medium of DMEM/F12, 20% knockout serum replacement (Life Technologies), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), and 20 ng/ml FGF-2 (R&D Systems), and medium was changed daily. hESC cultures were maintained in an undifferentiated state at 37°C in a 5% CO2/air environment until stem cells reached ∼90% confluence.
hESC differentiation into endoderm was performed in serum-free differentiation (SFD) media of DMEM/F12 (3:1; Life Technologies) supplemented with N2 (Life Technologies), B27 (Life Technologies), ascorbic acid (50 µg/ml; Sigma), Glutamax (2 mM; Life Technologies), monothioglycerol (0.4 µM; Sigma), and 0.05% BSA (Life Technologies) at 37°C in a 5% CO2/5% O2/95% N2 environment. hESCs were treated with Accutase (Stemcell Technologies) and plated onto low attachment 6-well plates (Corning Inc.), resuspended in endoderm induction media containing Y-27632 (10 µM; R&D Systems), human BMP4 (0.5 ng/ml; R&D Systems), human basic fibroblast growth factor (bFGF; 2.5 ng/ml; R&D Systems); and human activin A (100 ng/ml; R&D Systems) for 72–84 h dependent on the formation rates of endoderm cells. On day 3 or 3.5, the embryoid bodies were dissociated into single cells using 0.05% trypsin/0.02% EDTA and plated onto fibronectin (Sigma)-coated, 24-well tissue culture plates (∼100,000–150,000 cells per well). For induction of anterior foregut endoderm, the endoderm cells were cultured in SFD medium supplemented with 1.5 µM dorsomorphin dihydrochloride (R&D Systems) and 10 µM SB431542 (R&D Systems) for 36–48 h, and then switched to 36–48 h of 10 µM SB431542 and 1 µM IWP2 (R&D Systems) treatment.
For induction of early-stage LP cells (day 6–15), the resulting anterior foregut endoderm was treated with CHIR99021 (3 µM; R&D Systems), human FGF10 (10 ng/ml; R&D Systems), human FGF7 (10 ng/ml; R&D Systems), human BMP4 (10 ng/ml; R&D Systems), and all-trans retinoic acid (ATRA; 50–60 nM; Sigma) in SFD medium for 8–10 d. The day 10–15 cultures were maintained in a 5% CO2/air environment. On days 15 and 16, the lung progenitor cells were replated after brief 1-min trypsinization onto fibronectin-coated plates in the presence of SFD containing either a combination of five factors (CHIR99021, 3 µM; human FGF10, 10 ng/ml; human FGF7, 10 ng/ml; human BMP4, 10 ng/ml; and ATRA, 50 nM), or three factors (CHIR99021, 3 µM; human FGF10, 10 ng/ml; human FGF7, 10 ng/ml) for day 14–16. Day 16–25 cultures of late-stage LP cells were maintained in SFD media containing CHIR99021 (3 µM), human FGF10 (10 ng/ml), and human FGF7 (10 ng/ml) in a 5% CO2/air environment.
For differentiation of mature LCs (day 25–55), cultures were replated after brief trypsinization onto 3.3% matrigel coated 24-well plates in SFD media containing maturation components containing 3 µM CHIR99021, 10 ng/ml human FGF10, 10 ng/ml human FGF7, and DCI (50 nM dexamethasone, 0.1 mM 8-bromo-cAMP [Sigma], and 0.1 mM IBMX [3,7-dihydro-1-methyl-3-(2-methylpropyl)-1H-purine-2,6-dione; Sigma]). DAPT or DBZ (5–10 µM; Sigma) was added to the maturation media for induction of PNECs.
In Figs. 1 A, 2 A, 6 A, S1 A, S2 A, and S3 C, I–VI denote the mixtures of factors shown in Table 1.
Table 1. Growth factors used for the differentiation of hESCs to lung lineage cells.
| I | II | III | IV | V | VI |
|---|---|---|---|---|---|
| Activin A, Y-27632, BMP4, bFGF | DSM, SB | IWP2, SB | CHIR, BMP4, FGF10, KGF, RA | CHIR, FGF10, KGF | CHIR, FGF10, KGF, DCI |
Single-cell sequencing and transcriptomics
Single-cell capture, reverse transcription, cell lysis, and library preparation were performed using the Single Cell 3′ version 2 kit and chip according to the manufacturer’s protocol (10x Genomics). Single-cell suspensions were generated by dissociating the cultured RUES2 cells with 0.05% trypsin/0.02% EDTA for 10–15 min, followed with passing through 40 µM strainer. The single-cell suspension was achieved through sorting the dissociated cells in flow cytometry singlets. Cell count was adjusted to 6,000–9,000 cells to achieve an estimated capture of 4,000–5,000 cells. Six input wells were used.
Sequencing was performed on a HiSeq 2500 (Illumina; paired-end protocol with 26 base pairs for read 1 and 98 for read 2). Alignment of the raw reads to the human reference genome (hg19), removing duplicated transcripts using the unique molecular identifiers (UMIs), and assignment to single cells was performed using CellRanger (10x Genomics).
Single-cell analyses, including quality filtering, clustering, differential gene expression, and reduced-dimensionality visualization, were performed using the Seurat package as described in the package tutorial (version 2.1.0; Satija et al., 2015). Briefly, cells to be included in the analysis were required to have ≥2,000 and ≤40,000 UMIs. In addition, cells were excluded if >7.5% of the determined RNA sequences mapped to mitochondrial genes. In total, 8,716 cells from the sample without DAPT treatment, 9,824 cells from the DAPT samples, 4,148 cells from the samples treated with DAPT and with DOX to induce RB-shRNA, and 11,361 from the samples treated with DAPT and with DOX to induce RB and P53-shRNA passed these filters for quality. Following the package suggestions, a linear model was used to mitigate the variation stemming from the number of detected unique molecules per cell. Principal component analysis was performed on a subset of the gene expression matrix using 1,500–2,000 genes with the most variable expression level. Using the top 50 principal components, clustering and t-distributed stochastic neighbor embedding analysis (tSNE) visualization were performed. Clustering resolution, which affects the number of clusters, was set at 0.6, 0.4, 0.6, and 0.4 for the four samples, respectively (for more information, see package tutorial at http://satijalab.org/seurat/). Differential gene expression across clusters was performed using the Wilcoxon rank sum test implemented in the Seurat package.
Correlation with SCLC patient samples
Each of the single-cell data sets from DAPT-treated, RUES2-derived LCs and from cells treated with DAPT and with DOX to induce shRNA-RB were divided to two groups classified as PNECs and non-PNECs. To correct for the effects of UMI/cell, the single-cell transcriptomes from either PNECS or non-PNECs in each treatment group (the samples treated with DAPT only or samples treated with DAPT and DOX) were subsampled to form transcriptomes with an equal number of UMIs and correlated (Spearman’s correlation) with the previously published bulk transcriptomes of 29 early-stage (stage Ia or Ib) SCLC patients (George et al., 2015). The 29 correlation values for each cell were averaged, and the distributions of the mean correlations were compared using the Kolmogorov–Smirnov test. Subsampling to 2,000, 4,000, or 10,000 UMIs resulted in a similar pattern.
Gene list enrichment analysis
Genes that showed at least 2.5-fold changes in expression in comparisons of PNECs derived by DAPT treatment alone and PNECs derived by DAPT treatment plus reduction of RB RNA were used for ToppGene analysis (Chen et al., 2009; https://toppgene.cchmc.org/).
Intra-PNEC heterogeneity
Following the clustering procedure described above, the PNEC single-cell transcriptomes from DAPT-treated cells, DAPT-treated cells in which RB RNA was reduced, and DAPT-treated cells in which RB and P53 RNA was reduced were assigned separately to subclusters, using up to 14 top principal components and a clustering resolution of 0.6, 0.6, and 0.1. Each of the two data sets was assigned to three clusters, and a differential expression analysis was performed as described above. The percentage of PNECs positive for NEUROD1 was calculated following repetitive subsampling (50 repeats) of the transcriptomes to 7,500 UMIs per cell to account for sequencing depth.
Lentivirus transduction of hESCs
The lentiviral vectors expressing tetracycline-inducible (TetO) shRNAs against human RB constructed in the pSLIK vector system were obtained as a gift from Julien Sage’s laboratory at Stanford University (Conklin et al., 2012). The lentiviral vector expressing tetracycline-inducible shRNA against human P53 was purchased from Gentarget, Inc. (cat. no. LVP-343-RB-PBS). The lentiviral vectors expressing TetO KRAS G12V or EGFR L858R were constructed in the pInducer vector system by the Varmus laboratory (Unni et al., 2015). The lentiviral vector with a tetracycline-inducible cassette encoding the human NOTCH1-intracytoplasmic domain (NICD) in TetO-FUW vector system was obtained from Addgene (plasmid no. 61540). shRNA target sequences are as follows: RB 1, 5′-GGACATGTGAACTTATATA-3′; RB 2, 5′-GAACGATTATCCATTCAAA-3′; and p53, 5′-CACCATCCACTACAACTACAT-3′.
To generate the lentiviral particles, the above plasmids were transfected into HEK293T cells with the PC-Pack2 lentiviral packaging mix (Cellecta, Inc.) according to the manufacturer’s protocol. High-titer viral particles were used to transduce hESCs in serum-free conditions, and the antibiotic selection of transduced hESCs was performed without MEF feeder cells, using mTeSR1 stem cell media (Stemcell Technologies). The efficiency of RB or P53 knockdown and production of KRAS G12V, EGFR L858R, or NICD were verified by Western blotting after antibiotic selection using the following antibodies: anti-RB (Cell Signaling; clone 4H1, cat. no. 9309), anti-P107 (Santa Cruz; clone C-18, cat. no. Sc-318), anti-P-130 (Santa Cruz; clone C-20, cat. no. Sc-317), anti-P53 (Santa Cruz; clone DO-1, cat. no. Sc-126), anti-KRAS G12V (Cell Signaling; clone D2H12, cat. no. 14412), anti-KRAS G12D (Cell Signaling; clone D8H7, cat. no. 14429), anti-KRAS (Cell Signaling; clone D2C1, cat. no. 8955), anti-EGFR L858R (Cell Signaling; clone 43B2, cat. no. 3197), anti-EGFR (Cell Signaling; clone D38B1, cat. no. 4267), or anti-NICD (Cell Signaling; clone D3B8, cat. no. 4147).
Immunohistochemistry
Living cells in culture were directly fixed in 4% paraformaldehyde for 25 min, followed with 15 min permeabilization in 1% Triton X-100. Histology on tissues from mice was performed on paraffin-embedded or frozen sections from xenografted tumors and corresponding normal tissues as previously described (Chen et al., 2015). Tissues were either fixed overnight in 10% buffered formalin and transferred to 70% ethanol, followed by paraffin embedding, or snap frozen in O.C.T. (Fisher Scientific) and fixed in 10% buffered formalin, followed by paraffin embedding. For immunofluorescence, cells or tissue sections were immunostained with antibodies and counterstained with DAPI. Adjacent sections stained with H&E were used for comparison.
The antibodies used for immunostaining or Western blot experiments are as follows: anti-c-Kit (Invitrogen; cat. no. CD11705), anti-CXCR4 (BioLegend; clone 12G5, cat. no. 306506), anti-EpCAM (Invitrogen; clone G8.8, cat. no. 17-5791-80), anti-NKX2.1 (Seven Hills; cat. no. WRAB-1231), anti-NKX2.1 (Invitrogen; clone 8G7G3/1, cat. no. 18-0221), anti-SOX2 (Santa Cruz; clone Y-17, cat. no. sc-17320), anti-FOXA2 (Santa Cruz; clone M-20, cat. no. sc-6554), anti-SP-B (Seven Hills; cat. no. WRAB-48604), anti-SP-C (Seven Hills; cat. no. WRAB-76694), anti-pro-SP-C (Seven Hills; cat. no. WRAB-9337), anti-PDPN (Santa Cruz; clone FL-162, cat. no. sc-134482), anti-CC10 (Santa Cruz; clone T-18, cat. no. sc-9772), anti-P63 (BioLegend; cat. no. 619001), anti-CGRP (Sigma; clone CD8, cat. no. c9487), anti-ASCL1 (Sigma; clone 2D9, cat. no. SAB1403577), anti-NCAM1 (R&D Systems; cat. no. AF2408), anti-CHGA (Sigma; cat. no. HPA017369), anti-NOTCH1 or Cleaved NOTCH1 (Cell Signaling; clone D3B8, cat. no. 4147), anti-NOTCH1 or NICD (R&D Systems; clone 2D9, cat. no. AF3647), anti-NOTCH2 (Cell Signaling; clone 8a1, cat. no. 2420), anti-Ki67 (Cell Signaling; clone D2H10, cat. no. 9027), anti-human nuclei (EMD Millipore; cat. no. MAB1281), anti-OCT4 (Abcam; cat. no. ab19857), anti-Nanog (Abcam; cat. no. ab21624), anti-SSEA4 (Abcam; clone MC813, cat. no. ab16287), anti-β-Actin (Cell Signaling; clone D6A8, cat. no. 8457), and anti-GAPDH (Cell Signaling; clone 14C10, cat. no. 2118).
FACS
FACS with anti-c-Kit (Invitrogen; cat. no. CD11705), anti-CXCR4 (BioLegend; clone 12G5, cat. no. 306506), and anti-EpCAM (Invitrogen; clone G8.8, cat. no. 17-5791-80) was used to detect DE cells. Basically, cells were incubated with antibodies for 30 min at 4°C, followed by being washed and suspended in 0.1% BSA/PBS buffer. PE and APC filters were then used to detect cells double positive for Kit and CXCR4 or EpCAM and CXCR4 by signal intensity gating. FACS with anti-CGRP antibody (Abcam; clone 4902, cat. no. ab81887) was used to detect CGRP+ cells. Cells were first incubated with anti-human CGRP antibody for 30 min at room temperature followed with incubation of secondary antibody conjugated with R-PE for 30 min at room temperature. Then cells were washed and suspended in 0.1% BSA/PBS buffer. PE filter was then used to separate cells into CGRP+ and CGRP− subgroups by signal intensity gating. Negative controls stained with control IgG instead of primary antibodies were always performed with sample measurements. Flow cytometry machine of BD FACSAria II and software of Flowjo were mainly used to collect and analyze the flow cytometry data.
Xenograft formation
1–2 × 106 undifferentiated hESCs or hESC-derived LCs (at day 55 with or without 30 d of prior exposure to DAPT alone or to DAPT and DOX to reduce P53 or RB RNA or both RNAs) were subcutaneously injected into 6–8-wk-old NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ (NSG) mice (The Jackson Laboratory; Chen et al., 2012). DOX was added to mouse food beginning the day after injection; tumor incidence was monitored at least two to three times weekly. When mice became moribund or tumor size reached the Institutional Animal Care and Use Committee allowable burden, they were sacrificed immediately, necropsy was performed, and tumors were harvested for further histological or molecular study.
Statistical analyses
Sample sizes for all figures and tables were estimated based on our previous studies (Huang et al., 2014, 2015; Unni et al., 2015; Chen et al., 2016). For mouse experiments, animals were randomly assigned in each experimental group, and no animals were excluded from the analyses. For each set of experiments, samples were prepared for all experimental arms at the same time. All statistical tests are two sided. No adjustments were made for multiple comparisons. The relevant investigators (H.J. Chen and A. Poran) were blinded to experimental allocations among different experimental arms for all experiments. For all parametric statistical analyses, data were determined to be normally distributed by the D’Agostino–Pearson test. For all parametric and nonparametric tests, variances were similar between groups being compared. For comparison between experimental and control groups at a specific time point or tissue site in Figs. 1, 2, 3, 4, S2, S3, and S4, two-sided Student’s t tests, one-way or two-way ANOVA test, Fisher’s exact tests, and two-sided Kolmogorov–Smirnov test were used. All cells were purchased from ATCC or WiCell in the past 2 yr and were negative for mycoplasma. The hESC lines were regularly checked for chromosome abnormalities and maintained with normal chromosome numbers.
Study approval
All embryonic stem cell studies were approved by the Tri-Institutional Embryonic Stem Cell Research Oversight committee (Weill Cornell Medicine, Memorial Sloan-Kettering Cancer Center, and Rockefeller University). All animal protocols in this study were approved by the Institutional Animal Care and Use Committee of Weill Cornell Medicine.
Data availability and accession
Single-cell sequence data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRP136659).
Online supplemental material
Figs. S1 and S2 show differentiation of two hESC lines (RUES2 and ES02) to form endoderm and lung lineage cells. Fig. S1 also shows the scRNA profiles of day 55 LCs derived from hESCs not exposed to DAPT. Fig. S3 presents the generation of PNECs through directed differentiation of hESCs in which NOTCH signaling was blocked by DAPT and RB protein levels were reduced by shRNA. The cultures containing PNECs were further characterized by scRNA-seq, and the profiles were compared with the RNA profiles of early-stage SCLC. Fig. S4 compares scRNA profiles from cultured RUES2 cells in which RB and P53 levels were and were not reduced by induction of shRNA. Fig. S5 presents the further characterization of xenografts formed with hESC-derived LCs in which both RB and P53 proteins were reduced.
Supplementary Material
Acknowledgments
We thank Oksana Mashadova and Sukanya Goswami in the Varmus Laboratory for technical support and Eric Gardner, Dennis Fei, and John Ferrarone in the Varmus Laboratory; Yawen Chen in the Snoeck Laboratory; Rahul Satija (New York Genome Center); Viviane Tabar (Memorial Sloan-Kettering Cancer Center); and Shuibing Chen (Weill Cornell Medicine) for useful advice.
This work is supported by the US Department of Defense (award LC160136 to H. Varmus and O. Elemento); funds from the Meyer Cancer Center, Weill Cornell Medicine (to H. Varmus); an Arnold O. Beckman postdoctoral fellowship (to H.J. Chen); and a Weill Cornell graduate fellowship (to A. Poran).
The authors declare no competing financial interests.
Author contributions: H.J. Chen and A. Poran performed the experiments. All authors participated in the design and interpretation of some or all experiments. H. Varmus, H.J. Chen, and A. Poran wrote the manuscript, and all authors suggested editorial changes. H. Varmus conceived the study and recruited the collaborating partners.
References
- Borges M., Linnoila R.I., van de Velde H.J., Chen H., Nelkin B.D., Mabry M., Baylin S.B., and Ball D.W.. 1997. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature. 386:852–855. 10.1038/386852a0 [DOI] [PubMed] [Google Scholar]
- Borromeo M.D., Savage T.K., Kollipara R.K., He M., Augustyn A., Osborne J.K., Girard L., Minna J.D., Gazdar A.F., Cobb M.H., and Johnson J.E.. 2016. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Reports. 16:1259–1272. 10.1016/j.celrep.2016.06.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 511:543–550. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.J., Edwards R., Tucci S., Bu P., Milsom J., Lee S., Edelmann W., Gümüs Z.H., Shen X., and Lipkin S.. 2012. Chemokine 25-induced signaling suppresses colon cancer invasion and metastasis. J. Clin. Invest. 122:3184–3196. 10.1172/JCI62110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.J., Sun J., Huang Z., Hou H. Jr., Arcilla M., Rakhilin N., Joe D.J., Choi J., Gadamsetty P., Milsom J., et al. 2015. Comprehensive models of human primary and metastatic colorectal tumors in immunodeficient and immunocompetent mice by chemokine targeting. Nat. Biotechnol. 33:656–660. 10.1038/nbt.3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.J., Wei Z., Sun J., Bhattacharya A., Savage D.J., Serda R., Mackeyev Y., Curley S.A., Bu P., Wang L., et al. 2016. A recellularized human colon model identifies cancer driver genes. Nat. Biotechnol. 34:845–851. 10.1038/nbt.3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., and Jegga A.G.. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37(Web Server issue, Web Server):W305-11 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J.F., Baker J., and Sage J.. 2012. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 3:1244 10.1038/ncomms2254 [DOI] [PubMed] [Google Scholar]
- Funato K., Major T., Lewis P.W., Allis C.D., and Tabar V.. 2014. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 346:1529–1533. 10.1126/science.1253799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway L.A., and Lander E.S.. 2013. Lessons from the cancer genome. Cell. 153:17–37. 10.1016/j.cell.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Gazdar A.F., Bunn P.A., and Minna J.D.. 2017. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat. Rev. Cancer. 17:725–737. 10.1038/nrc.2017.87 [DOI] [PubMed] [Google Scholar]
- Geling A., Steiner H., Willem M., Bally-Cuif L., and Haass C.. 2002. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 3:688–694. 10.1093/embo-reports/kvf124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J., Lim J.S., Jang S.J., Cun Y., Ozretić L., Kong G., Leenders F., Lu X., Fernández-Cuesta L., Bosco G., et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature. 524:47–53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X., Islam M.N., O’Neill J., Hu Z., Yang Y.G., Chen Y.W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., and Snoeck H.W.. 2014. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32:84–91. 10.1038/nbt.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X., Green M.D., de Carvalho A.T., Mumau M., Chen Y.W., D’Souza S.L., and Snoeck H.W.. 2015. The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc. 10:413–425. 10.1038/nprot.2015.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.H., Klingbeil O., He X.Y., Wu X.S., Arun G., Lu B., Somerville T.D.D., Milazzo J.P., Wilkinson J.E., Demerdash O.E., et al. 2018. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 32:915–928. 10.1101/gad.314815.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T., Kedes L., and Hamamori Y.. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237–255. 10.1002/jcp.10208 [DOI] [PubMed] [Google Scholar]
- Ito T., Udaka N., Yazawa T., Okudela K., Hayashi H., Sudo T., Guillemot F., Kageyama R., and Kitamura H.. 2000. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 127:3913–3921. [DOI] [PubMed] [Google Scholar]
- Lim J.S., Ibaseta A., Fischer M.M., Cancilla B., O’Young G., Cristea S., Luca V.C., Yang D., Jahchan N.S., Hamard C., et al. 2017. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 545:360–364. 10.1038/nature22323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnoila R.I. 2006. Functional facets of the pulmonary neuroendocrine system. Lab. Invest. 86:425–444. 10.1038/labinvest.3700412 [DOI] [PubMed] [Google Scholar]
- Liu A., Cheng L., Du J., Peng Y., Allan R.W., Wei L., Li J., and Cao D.. 2010. Diagnostic utility of novel stem cell markers SALL4, OCT4, NANOG, SOX2, UTF1, and TCL1 in primary mediastinal germ cell tumors. Am. J. Surg. Pathol. 34:697–706. [DOI] [PubMed] [Google Scholar]
- Milano J., McKay J., Dagenais C., Foster-Brown L., Pognan F., Gadient R., Jacobs R.T., Zacco A., Greenberg B., and Ciaccio P.J.. 2004. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82:341–358. 10.1093/toxsci/kfh254 [DOI] [PubMed] [Google Scholar]
- Mollaoglu G., Guthrie M.R., Böhm S., Brägelmann J., Can I., Ballieu P.M., Marx A., George J., Heinen C., Chalishazar M.D., et al. 2017. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to Aurora kinase inhibition. Cancer Cell. 31:270–285. 10.1016/j.ccell.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M., Nishinakamura R., Saga Y., and Kopan R.. 2012. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development. 139:4365–4373. 10.1242/dev.083840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J.K., Larsen J.E., Gonzales J.X., Shames D.S., Sato M., Wistuba I.I., Girard L., Minna J.D., and Cobb M.H.. 2013a NeuroD1 regulation of migration accompanies the differential sensitivity of neuroendocrine carcinomas to TrkB inhibition. Oncogenesis. 2:e63 10.1038/oncsis.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J.K., Larsen J.E., Shields M.D., Gonzales J.X., Shames D.S., Sato M., Kulkarni A., Wistuba I.I., Girard L., Minna J.D., and Cobb M.H.. 2013b NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc. Natl. Acad. Sci. USA. 110:6524–6529. 10.1073/pnas.1303932110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Fernández-Cuesta L., Sos M.L., George J., Seidel D., Kasper L.H., Plenker D., Leenders F., Sun R., Zander T., et al. 2012. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 44:1104–1110. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietanza M.C., Byers L.A., Minna J.D., and Rudin C.M.. 2015. Small cell lung cancer: will recent progress lead to improved outcomes? Clin. Cancer Res. 21:2244–2255. 10.1158/1078-0432.CCR-14-2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., and Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33:495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter E.H., Kisslinger J.A., and Kopan R.. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 393:382–386. 10.1038/30756 [DOI] [PubMed] [Google Scholar]
- Semenova E.A., Nagel R., and Berns A.. 2015. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 29:1447–1462. 10.1101/gad.263145.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., Aster J.C., Sklar J., and Sunday M.E.. 2007. Notch-1 regulates pulmonary neuroendocrine cell differentiation in cell lines and in transgenic mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L500–L509. 10.1152/ajplung.00052.2006 [DOI] [PubMed] [Google Scholar]
- Shultz L.D., Lyons B.L., Burzenski L.M., Gott B., Chen X., Chaleff S., Kotb M., Gillies S.D., King M., Mangada J., et al. 2005. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174:6477–6489. 10.4049/jimmunol.174.10.6477 [DOI] [PubMed] [Google Scholar]
- Song H., Yao E., Lin C., Gacayan R., Chen M.H., and Chuang P.T.. 2012. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc. Natl. Acad. Sci. USA. 109:17531–17536. 10.1073/pnas.1207238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland K.D., Proost N., Brouns I., Adriaensen D., Song J.Y., and Berns A.. 2011. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 19:754–764. 10.1016/j.ccr.2011.04.019 [DOI] [PubMed] [Google Scholar]
- Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., and Quake S.R.. 2014. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 509:371–375. 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unni A.M., Lockwood W.W., Zejnullahu K., Lee-Lin S.Q., and Varmus H.. 2015. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. eLife. 4:e06907 10.7554/eLife.06907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D., Wuenschell C., Flores-Delgado G., and Anderson K.. 1998. Commitment and differentiation of lung cell lineages. Biochem. Cell Biol. 76:971–995. 10.1139/o98-104 [DOI] [PubMed] [Google Scholar]
- Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., and Stuart J.M.. Cancer Genome Atlas Research Network . 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45:1113–1120. 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Girard L., Zhang Y.A., Haruki T., Papari-Zareei M., Stastny V., Ghayee H.K., Pacak K., Oliver T.G., Minna J.D., and Gazdar A.F.. 2018. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl. Lung Cancer Res. 7:32–49. 10.21037/tlcr.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell sequence data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRP136659).