Tfr cells regulate Tfh-mediated antibody responses. Hou et al. demonstrate that FoxP3 and Ezh2 control the Tfr transcriptional program, and loss of this program results in dysfunctional ex-Tfr cells.
Abstract
Follicular regulatory T (Tfr) cells are a regulatory T cell subset that controls antibody production by inhibiting T follicular helper (Tfh)–mediated help to B cells. Tfh and Tfr cells possess opposing functions suggesting unique programming. Here we elucidated the transcriptional program controlling Tfr suppressive function. We found that Tfr cells have a program for suppressive function fine-tuned by tissue microenvironment. The transcription factor FoxP3 and chromatin-modifying enzyme EZH2 are essential for this transcriptional program but regulate the program in distinct ways. FoxP3 modifies the Tfh program to induce a Tfr-like functional state, demonstrating that Tfr cells coopt the Tfh program for suppression. Importantly, we identified a Tfr cell population that loses the Tfr program to become “ex-Tfr” cells with altered functionality. These dysfunctional ex-Tfr cells may have roles in modulating pathogenic antibody responses. Taken together, our studies reveal mechanisms controlling the Tfr transcriptional program and how failure of these mechanisms leads to dysfunctional Tfr cells.
Introduction
Regulatory T (T reg) cells are a subset of CD4+ T lymphocytes that inhibit effector T cells and inflammation (Josefowicz et al., 2012). T reg cells are largely defined by the forkhead box transcription factor FoxP3, which acts as a master regulator for T reg cell differentiation and function (Ziegler, 2006; Hill et al., 2007; Josefowicz et al., 2012). Loss of FoxP3 leads to multiorgan autoimmune disease in mice and immune dysregulation, polyendocrinopathy, enteropathy, and X-linked syndrome in humans (Bennett et al., 2001; Brunkow et al., 2001). FoxP3 can bind to a large number of proteins and interact with thousands of genomic sites, which posits FoxP3 as a complex node of T reg cell regulation (Hill et al., 2007; Marson et al., 2007; Zheng et al., 2007; Rudra et al., 2012; Samstein et al., 2012).
FoxP3 can act as a transcriptional repressor in T reg cells by binding to specific genes and recruiting the polycomb repressor complex 2 (PRC2) through direct interactions with the enzymatic protein of the PRC2 complex, enhancer of zeste homologue 2 (Ezh2; Arvey et al., 2014). PRC2 recruitment results in altered chromatin accessibility of specific genes targeted by FoxP3, which elicits part of the T reg cell transcriptional program. Loss of Ezh2 in T reg cells results in defective T reg cell expansion and suppressive function (DuPage et al., 2015; Yang et al., 2015). However, FoxP3 can also act as a transcriptional activator. Recent studies have suggested that FoxP3 binds to active enhancers, and formation of a complex with either RELA-KAT5-EP300 or EZH2-IKZF3-YY1 can dictate target gene regulation (Samstein et al., 2012; Kwon et al., 2017). Interestingly, FoxP3 in complex with EZH2-IKZF3-YY1 may result in transcriptional activation or repression. Therefore, the role of FoxP3 in regulating transcriptional programs depends on specific enhancer regions as well as molecular complexes bound by FoxP3.
T reg cell transcriptional programs also may be distinct in different anatomical locations. Within particular tissue microenvironments, T reg cells can express the same transcription factors as the cells that they suppress, suggesting that T reg cells may coopt some of the transcriptional program of effector cells as part of their suppression machinery (Chaudhry et al., 2009; Zheng et al., 2009; Cipolletta et al., 2012).
Follicular regulatory T (Tfr) cells are a subset of effector T reg cells that express the chemokine receptor CXCR5, gain access to the B cell follicle, and have specialized roles in inhibiting T follicular helper (Tfh)–mediated B cell responses (Sage and Sharpe, 2015b, 2016). The vast majority of Tfr cells differentiate from natural T reg cell precursors in lymphoid organs, although a small number of Tfr cells may originate from induced T reg cells under very limited circumstances (Chung et al., 2011; Linterman et al., 2011; Wollenberg et al., 2011; Sage et al., 2013; Maceiras et al., 2017). Tfr cells require similar cues as Tfh cells for differentiation, including the requirements for dendritic cells and B cells, inducible T cell costimulator (ICOS) and CD28 signals, and the transcription factor Bcl6 (Linterman et al., 2011; Sage et al., 2014a). Tfr and Tfh cell differentiation is also similarly restrained by inhibitory receptors such as PD-1 and CTLA-4 (Sage et al., 2013, 2014b; Wing et al., 2014). Despite similar differentiation cues, Tfh and Tfr cells have opposing functions: Tfh cells stimulate, whereas Tfr cells inhibit B cell responses. Tfr cells inhibit IL-21 and IL-4 production in Tfh cells and suppress class switch recombination, antibody secretion, and somatic hypermutation in B cells through altering metabolism (Sage et al., 2014a, 2016). Tfr cells appear to be specialized in their ability to suppress B cell responses because conventional T reg cells are unable to suppress B cells to the same degree in vivo or in vitro, although some studies have suggested they may suppress similarly in vitro (Sage et al., 2013, 2014b, 2016). The transcriptional machinery facilitating maintenance of the Tfr cell phenotype and suppressive function are still unknown.
Here we investigated the Tfr cell transcriptional program and how it relates to Tfh and T reg cell transcriptional programs. We determined that Tfr cells have a transcriptional program that is a hybrid of a Tfh and T reg cell program and that the Tfr program can be fine-tuned by the tissue microenvironment. We also discovered that a population of Tfr cells loses FoxP3 expression to become ex-Tfr cells, which exhibit an aberrant Tfr cell transcriptional program and deficits in suppressive capacity. Expression of FoxP3 in Tfh cells was sufficient to convert Tfh cells to Tfr-like cells that can suppress B cell responses. In addition, we determined that Ezh2, like FoxP3, is essential for maintenance of the Tfr cell transcriptional program and optimal Tfr cell suppressive function. These results point to FoxP3 and Ezh2 as key regulators of the Tfr transcriptional program and identify mechanisms leading to the generation of dysfunctional ex-Tfr cells.
Results
The Tfr cell transcriptional program has features of Tfh and T reg transcriptional programs and is fine-tuned by tissue microenvironment
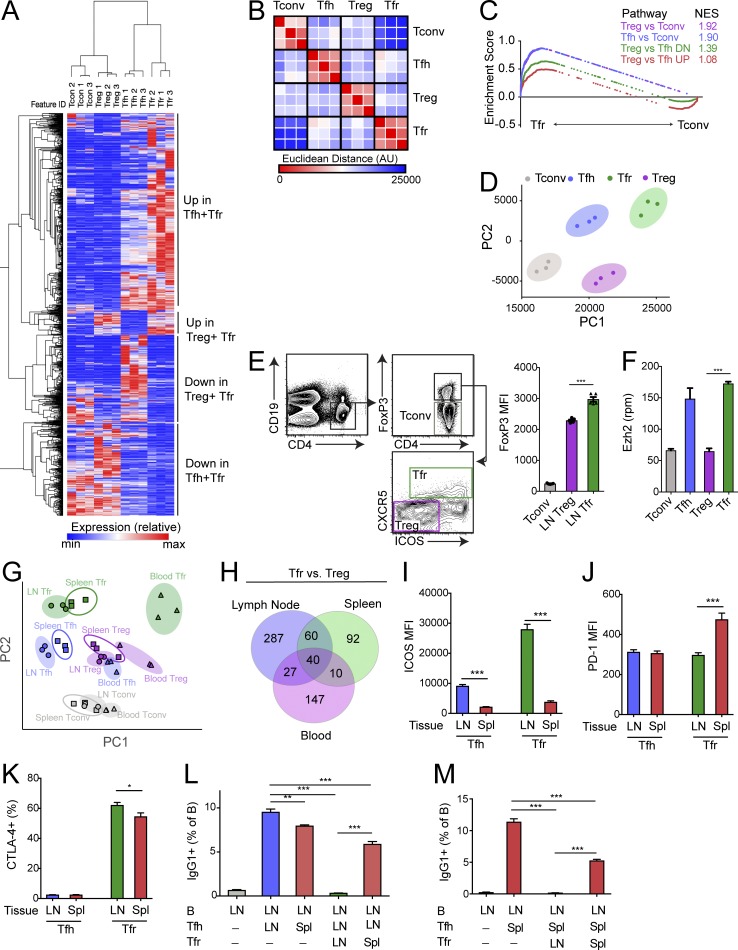
To delineate the relationships among follicular CD4+ T cell populations, we first compared the transcriptional programs of CD4+ T cell subsets residing inside and outside of B cell follicles during an immune response. To do this, we immunized FoxP3IRES-GFP mice with NP-OVA (emulsified in CFA) subcutaneously, and isolated draining lymph nodes 7 d later. We sorted conventional CD4+ T cells (referred to as “T conv” and sorted as CD4+ICOS−CXCR5−CD19−FoxP3−), T reg cells (sorted as CD4+ICOS−CXCR5−CD19−FoxP3+), Tfh cells (sorted as CD4+ICOS+CXCR5+CD19−FoxP3−), and Tfr cells (sorted as CD4+ICOS+CXCR5+CD19−FoxP3+) and subjected these cells to RNA sequencing (RNA-seq) transcriptional analysis (Fig. S1, A and B). Tfr cells had 945 differentially expressed (P < 0.01) genes compared with T reg cells and 480 differentially expressed genes (P < 0.01) compared with Tfh cells. When we compared all of these genes, we identified a subset of differentially expressed genes that were more highly expressed in Tfr and Tfh cells compared with T reg and T conv cells, and a smaller subset that was more highly expressed in Tfr and T reg cells compared with Tfh and T conv cells (Fig. 1 A).
Figure 1.
The Tfr cell transcriptional program is a combination of a Tfh and T reg cell transcriptional program. (A) Hierarchical clustering of genes differentially expressed (P < 0.05) between Tfr and T reg (945 genes) or Tfr and Tfh (480 genes) cells from RNA-seq data. (B) Similarity matrix showing Euclidean distance between Tfr, Tfh, T conv, and T reg cells using RNA-seq data from A. AU, arbitrary units. (C) GSEA of indicated gene sets (generated using differentially expressed genes from RNA-seq data as in A for Tfr versus T conv RNA-seq data). NES, normalized enrichment score. (D) PCA of T conv, Tfh, Tfr, and T reg cells using RNA-seq data from A. PC1, principal component 1; PC2, principal component 2. (E) Expression of FoxP3 in Tfr cells. Gating strategy to identify Tfr and T reg cells by flow cytometry (left). Quantification of FoxP3 expression in T conv, T reg, and Tfr cells (right). (F) Quantification of Ezh2 expression in T conv, Tfh, T reg, and Tfr cells taken from RNA-seq data as in A. (G) PCA of T conv, Tfh, Tfr, and T reg cells from lymph node, spleen, or blood using RNA-seq data. (H) Venn diagram of genes differentially expressed (P < 0.01) between Tfr and T reg from lymph node, spleen, and blood. (I–K) Expression of ICOS (I), PD-1 (J), and CTLA-4 (K) on Tfh and Tfr cells from lymph node and spleen. (L) Suppression assay in which lymph node B cells were cultured with lymph node or splenic Tfh and/or Tfr cells for 6 d. IgG1+ class-switched B cells were quantified. (M) Suppression assay in which lymph node B cells were cultured with splenic Tfh and lymph node or splenic Tfr cells for 6 d. IgG1+ class-switched B cells were quantified. All error bars indicate standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Student’s t test. Data are from individual experiments including 10 mice per replicate (A–G), from individual experiments of five mice per group and representative of three independent experiments (F and I–K), or from an individual experiment with triplicate wells and representative of three independent experiments (L and M).
To determine if Tfr cells more closely resembled Tfh or T reg cells transcriptionally, we performed a similarity matrix analysis using Euclidean distance as a measure of dissimilarity. Tfr cells had a similar Euclidean distance from Tfh and T reg cells, which was shorter than from T conv cells (Fig. 1 B). To determine similarity in more detail, we performed gene set enrichment analysis (GSEA) comparing Tfr cells to T conv cells using specific gene sets. Tfr cells had a high enrichment score for gene sets for T reg cell identity (T reg versus T conv) and Tfh cell identity (Tfh versus T conv; Fig. 1 C).
When we analyzed a gene set comprised of genes differentially expressed in T reg versus Tfh cells, we found that Tfr cells had higher enrichment in Tfh genes, suggesting that Tfr cells more closely resembled Tfh cells than T reg cells, which we confirmed using principal component analysis (PCA; Fig. 1 D). FoxP3, the master T reg cell transcription factor, is thought to function by recruiting distinct complexes, one of which is Ezh2 (Kwon et al., 2017). Therefore, we compared levels of FoxP3 and Ezh2 in T conv, Tfh, T reg, and Tfr cells. Tfr cells had significantly higher expression of FoxP3 compared with CXCR5−ICOS− T reg cells (Fig. 1 E). Similarly, Tfr cells had higher Ezh2 expression compared with CXCR5−ICOS− T reg cells; however, Tfr cells and Tfh cells had comparable expression of Ezh2 (Fig. 1 F). At the protein level, Tfr cells had higher expression of Ezh2 than ICOS+CXCR5− T regs which had intermediate expression compared with ICOS−CXCR5− T regs (Fig. S1 C).
We previously have found that Tfr cells from the blood have distinct functions compared with lymph node Tfr cells, suggesting that Tfr cells from different organs may have different transcriptional programs and/or functionality (Sage et al., 2014a). In addition, in mice and humans, blood Tfr cells arise independently of interactions with B cells, suggesting distinct differentiation cues (Sage et al., 2014a; Fonseca et al., 2017). To test whether Tfr cells from different anatomical locations have distinct transcriptional profiles, we performed RNA-seq transcriptional analysis on T conv, T reg, Tfh, and Tfr cells from spleens (of mice immunized with NP-OVA intraperitoneally) or blood (of mice immunized with NP-OVA either intraperitoneally or subcutaneously) and compared these populations to the RNA-seq performed in Fig. 1 A. Using PCA, we found that Tfr cells separated from each other based on anatomical location (Fig. 1 G). When we analyzed the differentially expressed genes between Tfr cells and T regs in each tissue, we found that only 40 genes were differentially expressed in all tissues (Fig. 1 H). These data suggest that the Tfr program may be fine-tuned based on anatomical location. Lymph node Tfr cells expressed higher levels of ICOS and CTLA-4 but lower amounts of PD-1 compared with splenic Tfr cells (Fig. 1, I–K). Changes in the Tfr phenotype from different tissues were not simply due to different activation states, because expression of the cell cycle marker Ki67 was similar in lymph node and spleen (Fig. S1 D).
Since PD-1 signaling inhibits Tfr cell suppressive function and CTLA-4 signaling is essential for Tfr cell suppressive function, we next determined how anatomical location impacted the suppressive capacity of Tfr cells. We performed suppression assays in which we cultured lymph node B cells with lymph node or splenic Tfh cells, and either lymph node or splenic Tfr cells for 6 d in the presence of anti-CD3/IgM, similarly to what we have published previously (Sage and Sharpe, 2015a). Lymph node Tfr cells suppressed class switching to a greater degree compared with splenic Tfr cells (Fig. 1 L). In addition, splenic Tfh cells were less potent at stimulating B cell class switching compared with lymph node Tfh cells. The diminished capacity of splenic Tfr cells to inhibit class switching was not due to tissue imprinting of Tfh or B cells because splenic Tfr cells also were less potent at suppressing splenic Tfh cells and B cells (Fig. 1 M and data not shown). Taken together, these data indicate that Tfr cells have a transcriptional program with features of both Tfh and T reg cells and that this program and Tfr cell suppressive function can be altered by tissue microenvironment.
Instability of FoxP3 in Tfr cells results in loss of suppressive function
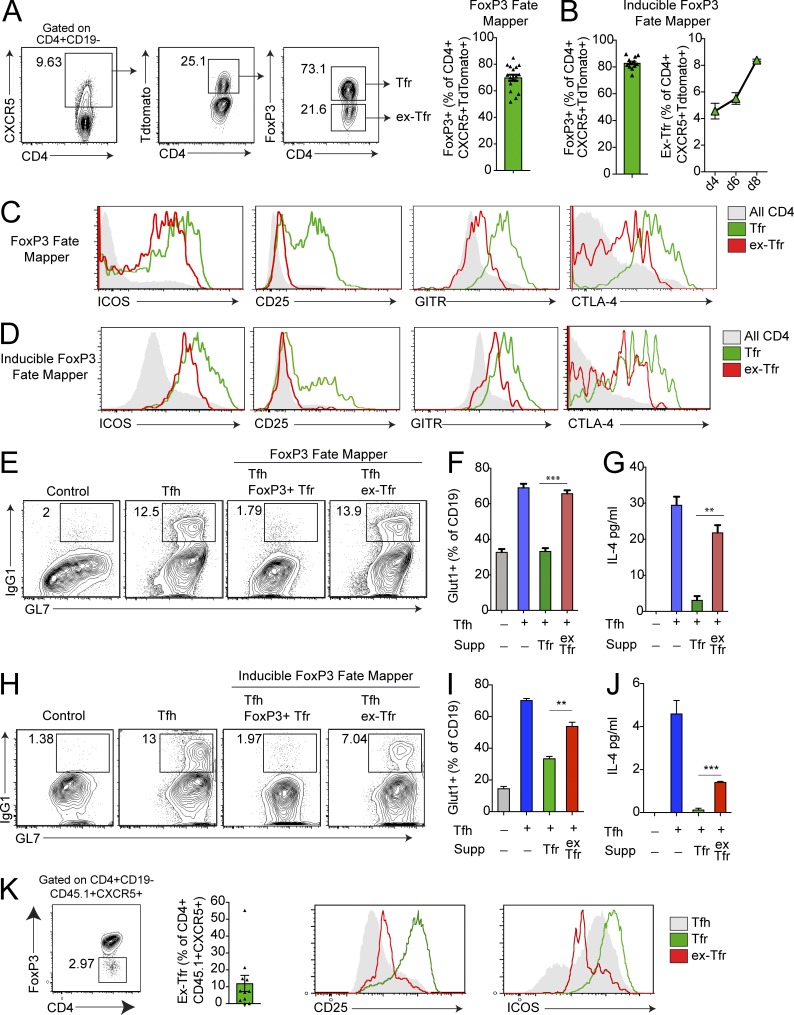
Since the Tfr cell transcriptional program has features of a Tfh and T reg program, we next determined how T reg cell transcription factors such as FoxP3 contribute to the identity and function of Tfr cells. First, we investigated whether FoxP3 was stably expressed in Tfr cells by crossing mice with a FoxP3-driven Cre recombinase allele with mice expressing a lox-stop-lox-TdTomato allele in the Rosa26 locus to generate FoxP3CreYFP Rosa26Lox-Stop-Lox-TdTomato mice that act both as a reporter for current FoxP3 expression as well as a “fate mapper” for cells that have expressed FoxP3 in the past. We immunized these mice with NP-OVA and 7 d later assessed the frequency of FoxP3+ and TdTomato+ follicular populations. Of the ∼25% of CXCR5+CD4+ cells that expressed TdTomato, ∼70% still expressed FoxP3, suggesting that ∼30% of the population of CXCR5+CD4+ cells had down-regulated FoxP3 (Fig. 2 A). We termed the in vivo generated CXCR5+CD4+TdTomato+FoxP3low population “ex-Tfr” cells. This result was not due to a “leaky” lox-stop-lox-TdTomato allele because mice were prescreened for leakiness and CD19+ B cells consistently expressed very low levels of TdTomato (data not shown). We confirmed these data using inducible FoxP3 fate mapper mice (FoxP3ERT2-Cre Rosa26Lox-Stop-Lox-TdTomato), which were injected with tamoxifen at the time of immunization to activate fate mapping. Since tamoxifen is required to induce Cre expression and TdTomato expression, this approach identifies cells that down-regulate FoxP3 specifically during the immunization period. In the inducible model, ∼80% of CD4+CXCR5+TdTomato+ cells retained FoxP3 (referred to as Tfr cells), whereas ∼20% had lost FoxP3 (referred to as ex-Tfr cells; Fig. 2 B). Temporal studies indicated that the frequency of ex-Tfr cells increased after day 4 of immunization (Fig. 2 B). In vivo generated ex-Tfr cells from both the FoxP3 fate mapper and inducible FoxP3 fate mapper models consistently expressed lower amounts of ICOS, CD25, GITR, and CTLA-4 compared with Tfr cells that retained high expression of FoxP3 (Fig. 2, C and D).
Figure 2.
FoxP3 instability in Tfr cells results in loss of suppressive function. (A) Identification of Tfr cells that down-regulate the transcription factor FoxP3. Gating strategy of ex-Tfr cells by flow cytometry from Foxp3Cre Rosa26Lox-stop-Lox-TdTomato (FoxP3 fate mapper) mice immunized with NP-OVA 7 d previously (left). Quantification of FoxP3+ cells as a frequency of CD4+CXCR5+TdTomato+ cells (right). (B) Quantification of FoxP3+ cells as a frequency of CD4+CXCR5+TdTomato+ cells from Foxp3ERT2-Cre Rosa26Lox-stop-Lox-TdTomato (inducible FoxP3 fate mapper) mice immunized with NP-OVA and injected with tamoxifen 7 d previously. (C) Expression of ICOS, CD25, GITR, and CTLA-4 on all CD4+ T cells, Tfr cells, or ex-Tfr cells from FoxP3 fate mapper mice as in A. (D) Expression of ICOS, CD25, GITR, and CTLA-4 on all CD4+ T cells, Tfr cells, or ex-Tfr cells from inducible FoxP3 fate mapper mice as in B. (E) Ex-Tfr cells fail to suppress Tfh-mediated B cell responses. In vitro suppression assay in which B and Tfh cells from WT mice were cultured with or without Tfr or ex-Tfr cells sorted from FoxP3 fate mapper mice as in A in the presence of anti-CD3/IgM for 6 d. IgG1+GL7+ class-switched B cells are gated. Plots are pregated on CD19+IA+CD4− cells. (F) Quantification of Glut1 expression on B cells from suppression assays (Supp) as in E. (G) Quantification of IL-4 levels in culture supernatants from suppression assays as in E. (H) In vitro suppression assay in which B and Tfh cells from WT mice were cultured with or without Tfr or ex-Tfr cells sorted from inducible FoxP3 fate mapper mice as in B in the presence of anti-CD3/IgM for 6 d. IgG1+GL7+ class-switched B cells are gated. Plots are pregated on CD19+IA+CD4− cells. (I) Quantification of Glut1 expression on B cells from suppression assays as in H. (J) Quantification of IL-4 levels in culture supernatants from suppression assays as in H. (K) Analysis of ex-Tfr cells after adoptive transfer of CD45.1+ Tfr cells (along with CD45.2+ Tfh cells) to Tcra−/− recipients which were immunized with NP-OVA and harvested 6 d later. Identification of ex-Tfr cells (left) and expression of CD25 and ICOS (right) are shown. All error bars indicate standard error. **, P < 0.01; ***, P < 0.001 using Student’s t test. Data are from four combined experiments (A) or three combined experiments (B), are representative of three independent experiments (C and D), are from individual experiments of triplicate wells and are representative of three independent experiments (E–J), or are combined data from four independent experiments (K).
We next asked whether in vivo generated ex-Tfr cells have a similar capacity to suppress B cell responses as FoxP3-expressing Tfr cells. We immunized fate mapper (Foxp3CreYFP Rosa26Lox-Stop-Lox-TdTomato) mice with NP-OVA and 7 d later sorted Tfr cells (sorted as CD4+CXCR5+FoxP3+TdTomato+) or in vivo generated ex-Tfr cells (sorted as CD4+CXCR5+FoxP3lowTdTomato+) and cultured them with CD19+ B cells and Tfh cells (sorted as CD4+CXCR5+FoxP3−) from FoxP3CreGFP mice that were immunized with NP-OVA 7 d previously, plus anti-CD3 and anti-IgM for 6 d. Tfr cells completely inhibited Tfh cell–mediated class switch to IgG1 as previously published (Fig. 2 E; Sage et al., 2014b, 2016; Sage and Sharpe, 2015a). In contrast, in vivo generated ex-Tfr cells could not suppress Tfh cell–mediated class switch recombination to IgG1 in B cells (Fig. 2 E). Because Tfr cells potently suppress metabolic pathways such as glycolysis in B cells during suppression (Sage et al., 2016), we also measured levels of the glucose transporter Glut1 in B cells. Ex-Tfr cells suppressed Glut1 expression less well than Tfr cells, suggesting that ex-Tfr cells are unable to suppress glycolysis in B cells (Fig. 2 F). Moreover, ex-Tfr cells were less effective at suppressing IL-4 production compared with Tfr cells, suggesting that ex-Tfr cells are not able to suppress cytokine production in Tfh cells (Fig. 2 G). Likewise, in the inducible FoxP3 fate mapper model, ex-Tfr cells were less potent at suppressing IgG1 class switching in B cells, Glut1 expression in B cells, and IL-4 levels in the culture supernatant, suggesting that ex-Tfr cells have defects in B cell suppression (Fig. 2, H–J). To confirm that ex-Tfr cells arise from loss of FoxP3 expression in Tfr cells (and are not ex–T reg cells that up-regulate CXCR5), we adoptively transferred CD45.1+ Tfr cells (from FoxP3IRESGFP mice) along with CD45.2+ Tfh cells to TCRα−/− mice that were immunized with NP-OVA. After 6 d, we assessed the frequency of CD45.1+CD4+CXCR5+CD19− cells that lost FoxP3. We found a small, but substantial, population of ex-Tfr cells that had attenuated levels of ICOS and CD25 compared with FoxP3-sufficient Tfr cells (Fig. 2 K). Taken together, these data suggest that a small population of Tfr cells can down-regulate (or lose) FoxP3, resulting in diminished suppressive function.
Ex-Tfr cells lose the Tfr transcriptional program
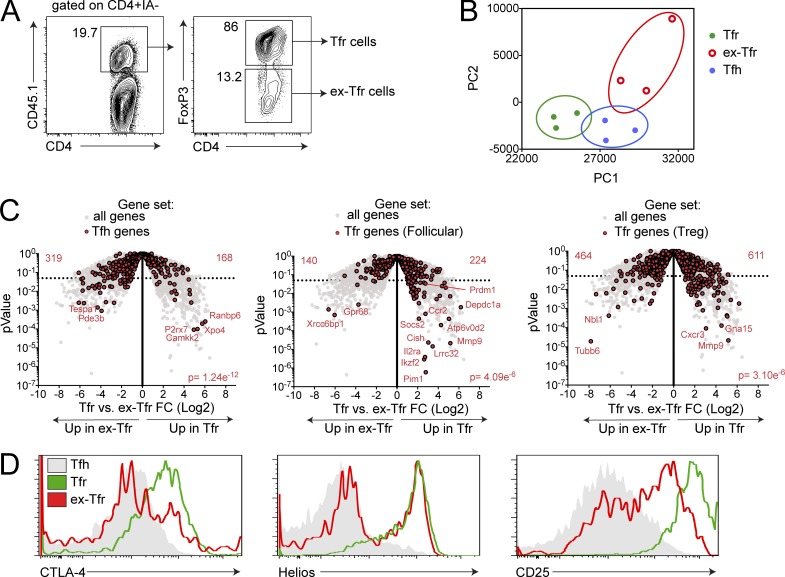
Next we determined how the Tfr transcriptional program is altered in the dysfunctional ex-Tfr cells. To investigate this issue, we required a system in which we could ensure that ex-Tfr cells were in fact Tfr cells at the time of FoxP3 down-regulation (and not an ex–T reg that up-regulates CXCR5 expression) and in which FoxP3 loss occurs proximal to the time of analysis. Our in vitro suppression assay fulfilled these criteria. We immunized CD45.1+ FoxP3GFP mice with NP-OVA and 7 d later sorted Tfr cells (sorted as CD4+CXCR5+FoxP3+) and cultured these CD45.1 Tfr cells with CD19+ B and Tfh (sorted as CD4+CXCR5+FoxP3−) cells sorted from immunized CD45.2 mice, along with anti-CD3 and anti-IgM. After 3 d, we harvested the cultures and sorted Tfr cells that still maintained FoxP3 (sorted as CD45.1+FoxP3+) and ex-Tfr cells that had down-regulated FoxP3 expression in vitro (sorted as CD45.1+FoxP3−; Fig. 3 A). We characterized these CD45.1 Tfr and ex-Tfr cells by RNA-seq transcriptional profiling and compared them with CD45.2 Tfh cells from B, Tfh, and Tfr cell cultures as controls. PCA of these populations revealed that in vitro generated ex-Tfr cells separated from both Tfr and Tfh cells, but phenotypically resided more closely to Tfh cells in PCA space (Fig. 3 B). Of the 499 genes differentially expressed (P < 0.01) in Tfr versus ex-Tfr cells, only 72 genes overlapped with the 241 genes differentially expressed (P < 0.01) between Tfr versus Tfh cells (Fig. S2, A and B). A comparison of Tfh genes (Tfh versus Tfr, from analysis in Fig. 1) in Tfr versus ex-Tfr cells revealed that ex-Tfr cells expressed more Tfh genes compared with Tfr cells, suggesting that ex-Tfr cells are more Tfh-like than Tfr cells (Fig. 3 C). Next, we determined if ex-Tfr cells lost Tfr signature genes. To do this, we derived two Tfr signature gene sets, one that contains Tfr genes compared with a follicular signature (by comparing Tfr versus Tfh cells) and a second that contains Tfr genes as they compare to a T reg signature (by comparing Tfr versus T reg). Using these two different Tfr gene sets is necessary, since Tfr cells possess a transcriptional program that contains both Tfh and T reg genes. A comparison of Tfr genes derived from follicular phenotype in Tfr versus ex-Tfr showed that Tfr cells expressed more Tfr genes (including Prdm1/Blimp1) compared with ex-Tfr cells. When we compared Tfr genes as they relate to the T reg phenotype in Tfr versus ex-Tfr, we found that Tfr cells expressed more Tfr genes than ex-Tfr cells. Lastly, we compared protein expression of molecules that were differentially expressed from in vivo ex-Tfr cells (compared with Tfr cells) including CTLA-4, as well as CD25 and Helios, which were hits from our in vitro RNA-seq analysis, and found reduced expression levels in ex-Tfr cells compared with Tfr cells (Fig. 3 D). Taken together, these data indicate that in vitro generated ex-Tfr cells lose the Tfr transcriptional program and have a transcriptional signature more similar to Tfh cells.
Figure 3.
Ex-Tfr cells lose the Tfr cell transcriptional program. (A) Representative gating strategy to identify in vitro generated ex-Tfr cells. CD45.2+ Tfh and CD45.1+ Tfr cells were cultured for 4 d in the presence of anti-CD3/IgM before analysis. (B) Ex-Tfr cells are phenotypically separate from Tfr cells. PCA analysis of Tfr, ex-Tfr, and Tfh cells using RNA-seq data as in A. (C) Ex-Tfr cells down-regulate the Tfr cell–specific transcriptional program and up-regulate Tfh genes. Volcano plots showing Tfr versus ex-Tfr RNA-seq data for all genes, Tfh genes (left), Tfr genes (follicular; derived from Tfr versus Tfh analysis; middle), and Tfr genes (T reg; derived from Tfr versus T reg analysis; right; gene sets were generated from RNA-seq in Fig. 1 A). P value indicates χ2 test. (D) Expression of CTLA-4, Helios, and CD25 on in vitro generated ex-Tfr, Tfr, and Tfh cells. Cultures were performed as in A. Data are combined data from three independent experiments (A–C) or are from an individual experiment that is representative of three individual experiments (D).
Deletion of FoxP3 is sufficient for an ex-Tfr phenotype
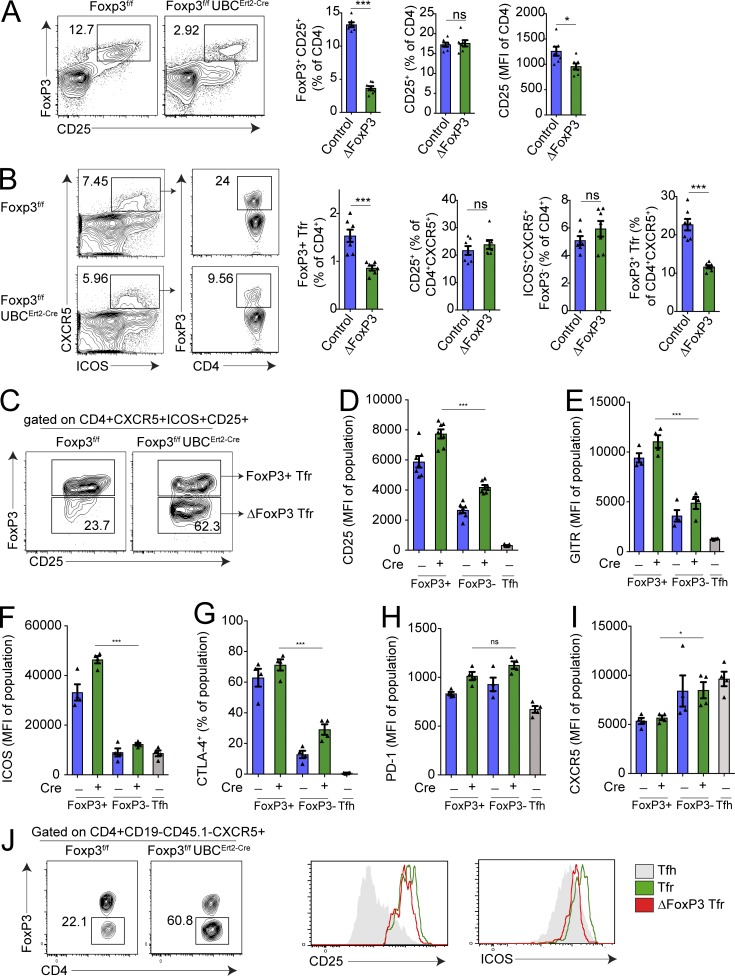
One possible explanation for the ex-Tfr phenotype may be that FoxP3 controls the Tfr program by directly promoting Tfr genes, by repressing Tfh genes, or both. Alternatively, loss of FoxP3 in unstable Tfr cells may occur simultaneously with loss of additional molecules that may directly regulate Tfr cells. To discriminate among these possibilities, we next determined how the Tfr cell phenotype changes with selective deletion of FoxP3 using a FoxP3f/f UBCERT2-Cre strain in which FoxP3 can be inducibly deleted by administration of tamoxifen. We immunized these mice or control FoxP3f/f mice with NP-OVA subcutaneously, administered tamoxifen starting on day 0, and harvested draining lymph nodes on day 7. Tamoxifen administration resulted in a profound, but not complete, deletion of FoxP3 expression in CD4+ T cells (Fig. 4 A). Importantly, deletion of FoxP3 with this protocol did not alter the frequency of total CD4+ cells expressing the high-affinity IL-2 receptor, CD25; the frequency of CD25-positive CD4+ cells was similar in mice that had deleted FoxP3 and those that had not (Fig. 4 A). However, CD25 levels were attenuated on T regs that had deleted FoxP3.
Figure 4.
Deletion of FoxP3 is sufficient for an ex-Tfr phenotype. (A) Quantification of inducible FoxP3 deletion. Foxp3f/fUBCErt2-Cre mice (ΔFoxP3) or Foxp3f/f control mice were immunized with NP-OVA and given tamoxifen daily starting on day 2. Gating strategy to identify FoxP3+CD25+ T regs (left; plots are pregated on CD4+CD19− cells). Quantification of CD25+ CD4+ (middle) or CD25 MFI of CD4+ cells (right). (B) Quantification of FoxP3 deletion in Tfr cells. Flow cytometric plots showing the Tfr frequency of all CD4+CXCR5+ cells in mice as in A (left). Quantification of FoxP3+Tfr cells, CD4+ICOS+CXCR5+CD25+ cells, CD4+ICOS+CXCR5+FoxP3− cells, and FoxP3+Tfr frequency of follicular cells in mice as in A (right) are shown. (C) Gating strategy to identify Tfr cells in which FoxP3 was deleted. Plots are pregated on CD4+CXCR5+ICOS+CD25+ cells. (D–I) Analysis of CD25 (D) and GITR (E), ICOS (F), CTLA-4 (G), PD-1 (H), and CXCR5 (I) expression on FoxP3+ or FoxP3− Tfr cells (gated as in C) in mice as in A. Tfh (CD4+ICOS+CXCR5+FoxP3−CD19−) cells from control mice are included. MFI, mean fluorescence intensity. (J) Analysis of FoxP3 deletion on Tfr cells after transfer. Tfr cells (gated as CD4+ICOS+CXCR5+GITR+CD19−) from immunized control or Foxp3f/fUBCErt2-Cre mice were transferred to Tcra−/− mice along with CD45.1+ Tfh cells which were immunized with NP-OVA. 6 d later, ex-Tfr/ΔFoxP3 cells were identified (left), and CD25 and ICOS levels were assessed on FoxP3-expressing versus deleted Tfr cell populations (right). All error bars indicate standard error. *, P < 0.05; ***, P < 0.001 using Student’s t test. ns, not significant. Data are from individual experiments and are representative of three (A–I) or two (J) independent experiments.
The frequency of FoxP3+ Tfr cells (gated as CD4+CXCR5+ICOS+FoxP3+ cells) was reduced by ∼50% of total CD4+ T cells in immunized FoxP3f/f UBCERT2-Cre mice compared with controls (Fig. 4 B). However, the frequency of total CD4+CXCR5+ICOS+CD25+ cells was unchanged with FoxP3 deletion, suggesting that deletion of FoxP3 does not result in diminished Tfr cell differentiation or increases in cell death. The frequency of total CD4+CXCR5+ICOS+FoxP3− cells (containing Tfh and FoxP3-deleted Tfr cells) was slightly increased, likely due to FoxP3-deleted Tfr cells appearing in this gating strategy, but this did not reach statistical significance. The frequency of FoxP3+ Tfr cells of all follicular CD4+ cells (otherwise known as the Tfr ratio) was attenuated by >50%, suggesting deletion of FoxP3 in ∼50% of Tfr cells. The percentage of germinal center (GC) B cells and NP-specific antibody 14 d after NP OVA immunization was unchanged between Cre+ and Cre− mice, suggesting that the Tfr cells that maintained FoxP3 in the Cre+ mice were able to control B cell responses (Fig. S3 A).
To determine the role of FoxP3 in Tfr cells, we took advantage of the suboptimal deletion strategy to delete FoxP3 on ∼50% of Tfr cells so that we could compare Tfr cells in which FoxP3 was deleted or not in the same microenvironment, which was devoid of autoimmunity. We immunized Foxp3f/f or Foxp3f/f UBCErt2-Cre mice with NP-OVA and 7 d later harvested draining lymph nodes, gated FoxP3-expressing Tfr cells as CD4+CXCR5+ICOS+CD25+FoxP3+ and FoxP3-deleted Tfr cells as CD4+CXCR5+ICOS+CD25+FoxP3− cells, and compared expression of proteins that are more highly expressed on Tfr cells than Tfh cells to determine if FoxP3 is required for the ex-Tfr phenotype (Fig. 4 C). We additionally gated on CD4+CXCR5+ICOS+FoxP3−CD25− Tfh cells from Foxp3f/f mice as controls. Although recent reports have suggested that some Tfr cells do not express CD25 (Botta et al., 2017; Wing et al., 2017), the majority of Tfr cells in skin draining lymph nodes after immunization with NP-OVA expressed CD25 (Fig. S3 B). Moreover, most (but not all) CD25-expressing cells were derived from cells that expressed FoxP3 previously, with the remaining cells being CD25+ Tfh cells (Fig. S3 C). FoxP3-deleted Tfr cells expressed less GITR and CD25 compared with FoxP3-expressing Tfr cells, but moderately higher GITR and CD25 compared with Tfh cells (Fig. 4, D and E). The expression of the essential follicular costimulatory molecule ICOS was severely diminished on FoxP3-deleted Tfr cells compared with FoxP3-expressing Tfr cells, but similar to Tfh cells (Fig. 4 F). Moreover, CTLA-4 expression was markedly attenuated on FoxP3-deleted Tfr cells compared with FoxP3-expressing Tfr cells, consistent with previous studies showing that FoxP3 regulates expression of CTLA-4 (Fig. 4 G; Sage et al., 2014b; Wing et al., 2014). Expression of PD-1 and CXCR5, however, was not reduced in FoxP3-deleted versus FoxP3-expressing Tfr cells (Fig. 4, H and I). Similar results were found if we used GITR as an alternative gating strategy to CD25 (Fig. S3 D).
To further demonstrate that deletion of FoxP3 leads to an ex-Tfr phenotype in vivo, we adoptively transferred CD4+ICOS+CXCR5+GITR+CD19− Tfr cells from either control (Foxp3f/f) or Foxp3f/f UBCErt2-Cre mice along with CD45.1+ Tfh cells into Tcrα−/− recipients that were subsequently immunized with NP-OVA and given tamoxifen treatment to delete FoxP3. After 6 d, we assessed CD25 and ICOS expression on FoxP3-deleted Tfr cells and found that FoxP3-deleted Tfr cells had lower expression of CD25 and ICOS (Fig. 4 J). These data indicate that FoxP3 is responsible for optimal expression of proteins that phenotypically separate Tfh and Tfr cells including GITR, ICOS, and CTLA-4 and suggest that the phenotype of ex-Tfr cells is likely related to specific loss of the transcription factor FoxP3 in these cells.
We next asked whether loss of FoxP3 resulted in loss of the Tfr transcriptional program and/or a shift to a Tfh transcriptional program. We immunized Foxp3f/f or Foxp3f/f UBCErt2-Cre mice with NP-OVA, induced deletion with tamoxifen, and 7 d later harvested draining lymph nodes and sorted Tfr cells (by gating on CD4+CXCR5+ICOS+GITR+CD25+ cells) for RNA-seq transcriptional analysis. It is important to note that without a FoxP3 reporter allele, this gating strategy isolates Tfr cells from Foxp3f/f UBCErt2-Cre mice in which ∼50% have FoxP3 deleted. In addition to Tfr cells, we analyzed FoxP3-expressing or FoxP3-deleted T reg (sorted as CD4+CXCR5−ICOS−GITR+CD25+) and T conv cells (sorted as CD4+CXCR5−ICOS−CD25−). Using a similarity matrix that calculates Euclidean distance, we found that Tfr cells from Foxp3f/f UBCErt2-Cre (Cre+) mice were more similar to Tfh cells than Foxp3f/f (Cre−) Tfr cells were to Tfh cells, suggesting that deletion of FoxP3 in Tfr cells results in a Tfh-like state transcriptionally (Fig. S3, E–G). Taken together, these data indicate that specific deletion of FoxP3 in Tfr cells results in an ex-Tfr surface phenotype.
FoxP3 is sufficient to convert Tfh cells to Tfr-like cells
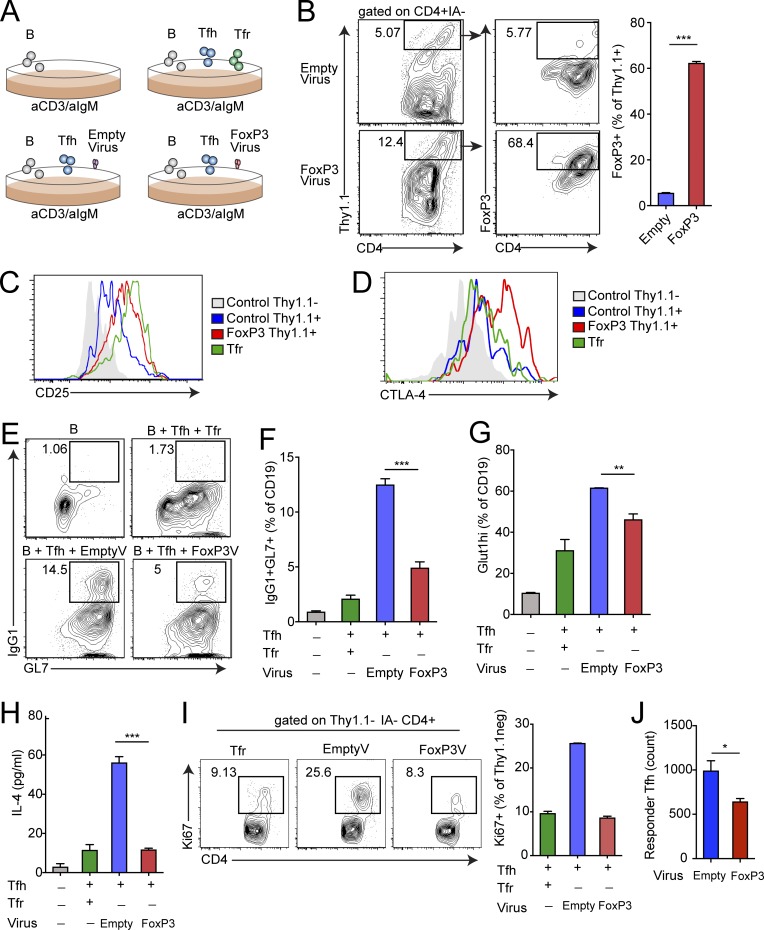
Because FoxP3 can regulate the Tfr cell transcriptional program but is not sufficient for Tfr cell functionality (since conventional T reg cells cannot suppress similarly to Tfr cells), we hypothesized that FoxP3 may be modulating Tfh genes to induce a Tfr transcriptional program. To determine if FoxP3 is sufficient to convert a Tfh cell to a Tfr-like cell, we forced FoxP3 expression in Tfh cells. To do this, we cultured B and Tfh cells (sorted from Foxp3IRES-GFP mice immunized with NP-OVA) with a retrovirus carrying the Thy1.1 transduction reporter with or without WT FoxP3. After culture overnight, the virus was washed away and anti-CD3/IgM was added to stimulate B cell responses, and the culture was analyzed 3 d later (Fig. 5 A). There was a small population of Thy1.1 highly expressing cells in cultures transduced with the Thy1.1 transduction reporter with or without WT FoxP3 (Fig. 5 B). When cells were gated on the Thy1.1 highly expressing cells, FoxP3 expression was found on Tfh cells from cultures containing the FoxP3-encoding virus, but not the control virus. Moreover, FoxP3-transduced Tfh cells expressed FoxP3 similarly to Tfr cells (Fig. S4 A). FoxP3 was not induced in B cells, demonstrating that the virus elicits FoxP3 expression in Tfh cells specifically (Fig. S4 B and data not shown). Next, we determined if FoxP3 expression in Tfh cells can result in a Tfr-like phenotype by examining expression of surface receptors that are typically highly expressed in Tfr cells, but not Tfh cells. We gated on Thy1.1low and Thy1.1hi Tfh cells as well as Tfr controls from cultures and stained for CD25 and CTLA-4. Transduction with the FoxP3 virus resulted in much higher CD25 expression compared with control virus, with CD25 levels almost reaching those of Tfr cells (Fig. 5 C). Moreover, FoxP3 transduction resulted in higher expression of CTLA-4, which is necessary for full Tfr cell suppressive capacity (Fig. 5 D; Sage et al., 2014b; Wing et al., 2014). FoxP3 transduction did not alter expression of Bcl6 or CXCR5 (Fig. S4 C).
Figure 5.
FoxP3 is sufficient to convert a Tfh program to a Tfr program. (A) Schematic of in vitro assay to express FoxP3 in Tfh cells. Control Thy1.1 retrovirus or FoxP3-encoding Thy1.1 retrovirus was added to cultures (as described in Materials and methods) along with anti-CD3/IgM for 4 d. (B) Representative gating of Thy1.1+ (transduced) and FoxP3-expressing Tfh cells from cultures as in A (left) and quantification of FoxP3+ Tfh cells reported as a percentage of Thy1.1+ transduced cells (right). Cells are pregated on CD4+IA− cells. (C) Expression of CD25 on Thy1.1+ Tfh cells from experiments as in A. (D) CTLA-4 expression on Thy1.1+ Tfh cells from experiments as in A. (E and F) FoxP3 expression in Tfh cells results in suppression of B cell responses. Analysis of IgG1+GL7+ class-switched B cells from cultures as in A. Representative gating (E) and quantification (F) are shown. (G) Glut1 expression in B cells from cultures as in A. (H) IL-4 levels in culture supernatants as in A. (I) Tfr-like FoxP3+ Tfh cells suppress nontransduced Tfh cells. Ki67 expression in Thy1.1− Tfh cells from cultures as in A. Representative gating (left) and quantification (right) are shown. (J) Relative count of responder CD45.1+ Tfh cells which were added to cultures as in A 2 d after removal of virus and cultured for an additional 3 d. All error bars indicate standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Student’s t test. Data represent triplicate wells from an individual experiment and are representative of three (B–H) or two (J) independent experiments.
Next we determined if B cell responses were altered in cultures in which FoxP3 was expressed in Tfh cells. Control retrovirus treatment resulted in a substantial induction of class-switched B cells. When FoxP3 was transduced in Tfh cells, there was an enormous decrease in the frequency of IgG1 class-switched B cells, suggesting that B cell responses were profoundly attenuated (Fig. 5, E and F). The reduction in class-switched B cells in cultures in which FoxP3 was expressed in Tfh cells was not due to presence of virus in the cultures, because titrating the amount of control virus did not alter the frequency of class-switched B cells (Fig. S4 D). Moreover, the suppressive capacity of FoxP3-transduced Tfh cells was not due to higher ratios of suppressive cells than Tfr cells, because the total number of FoxP3-expressing Tfh cells was less than Tfr cells in typical suppression wells (Fig. S4 E). Glut1 was more highly attenuated on B cells when FoxP3 was expressed in Tfh cells, suggesting diminished glycolytic potential (Fig. 5 G). Moreover, the Tfh cytokine IL-4 (which is suppressed in Tfh cells by Tfr cells) was also attenuated in the presence of FoxP3-expressing Tfh cells to the same extent as Tfr cell suppression (Fig. 5 H). To determine if the FoxP3-expressing Tfh cells became Tfr-like cells with suppressive capacity (and not just dysfunctional Tfh cells), we compared the levels of Ki67 in Thy1.1neg Tfh cells (which were not transduced with FoxP3) and Thy1.1 FoxP3− Tfh cells. Nontransduced FoxP3− Tfh cells in the FoxP3 retrovirus culture had consistently less Ki67 compared with cultures with control Thy1.1 retrovirus (Fig. 5 I). Moreover, when we added freshly isolated responder CD45.1+ Tfh cells, we found that these cells expanded less in wells containing FoxP3-transduced Tfh cells, suggesting active suppression (Fig. 5 J). These data indicate that FoxP3 can coopt Tfh programs to induce a Tfr-like phenotype and suppressive state.
Ezh2 is required for suppressive function and transcriptional program of Tfr cells
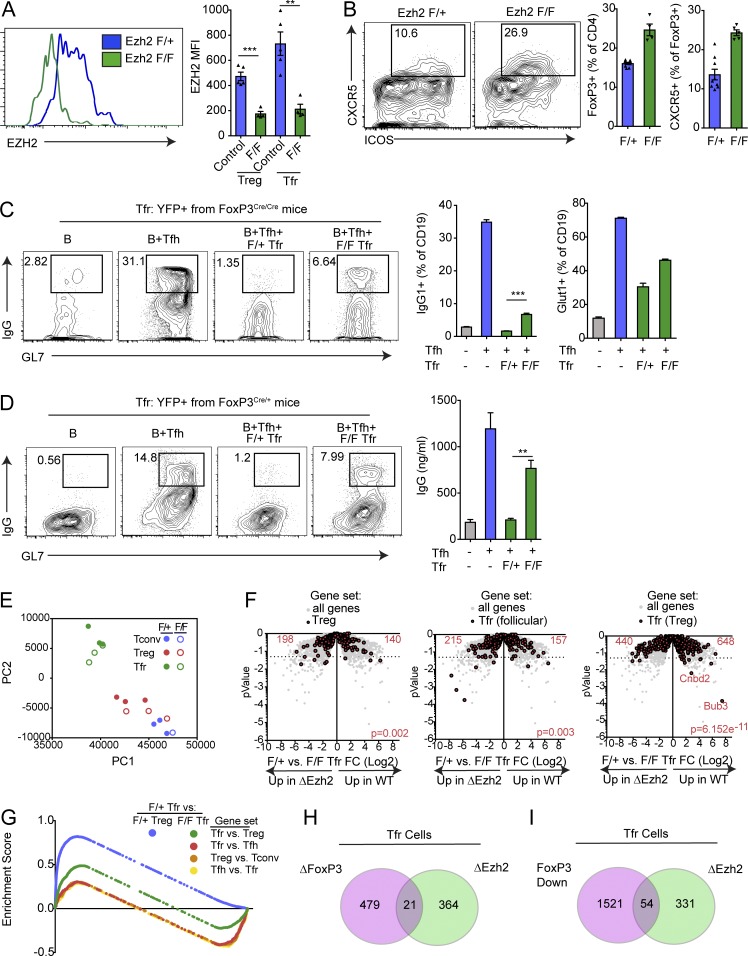
Recent work has suggested that one potential mechanism by which FoxP3 can regulate transcriptional identity is by directly binding to Ezh2 in the polycomb repressor complex, resulting in targeted alterations in gene expression (Arvey et al., 2014; Kwon et al., 2017). However, Ezh2 may have non-FoxP3 related roles in modulating gene transcription, since Ezh2 has roles in non–T reg cells. Since Ezh2 has been implicated as a potential mechanism by which FoxP3 can change programs, we next asked whether Ezh2 played a role in Tfr function. To test this, we bred Ezh2-floxed (Ezh2f/f) mice with FoxP3-Cre (Foxp3CreYFP) mice to generate mice that will delete Ezh2 in T reg cell subsets. We first assessed whether Ezh2 was deleted in T reg cells and Tfr cells by immunizing Ezh2f/f Foxp3Cre or Ezh2f/+Foxp3Cre control mice with NP-OVA and 7 d later staining for Ezh2 expression. There was a profound reduction of Ezh2 expression in T reg cells and Tfr cells from Ezh2f/f Foxp3Cre but not control mice, suggesting that Ezh2 was deleted in both subsets (Fig. 6 A). As previously reported, loss of Ezh2 on T reg cells resulted in autoimmunity manifested as smaller size, hair loss, and increased cellularity of lymphoid organs (DuPage et al., 2015; data not shown). To determine if Ezh2 affects Tfr cell development, we assessed the frequency of total T reg cells and Tfr cells in control versus Ezh2 deleted mice. The frequencies of total T reg cells and Tfr cells (calculated as a percentage of total T reg cells) were increased (Fig. 6 B). Tfr cells in Ezh2 T reg–deficient mice did not have a defect in migration, because the B cell zone and GCs in these mice had infiltrating FoxP3+ cells (Fig. S5 A). Despite the increased frequency of Tfr cells in Ezh2-deficient mice and the ability of these cells to migrate to B cell follicles, we found an increased frequency of GC B cells (Fig. S5 B). These data suggest that Ezh2-deficient Tfr cells (or other T reg populations) may not be able to fully suppress B cell responses.
Figure 6.
Ezh2 is required for Tfr suppressive function and transcriptional program. (A) Conditional deletion of Ezh2 on T reg cells. Ezh2f/fFoxP3Cre/Cre or control (Ezh2f/+FoxP3Cre/Cre) mice were immunized with NP-OVA, and 7 d later, T reg and Tfr cells were analyzed for expression of Ezh2 by flow cytometry. Representative gating (left) and quantification (right) are shown. MFI, mean fluorescence intensity. (B) Loss of Ezh2 results in increased Tfr cell percentages. Mice as in A were analyzed for Tfr cells. Representative gating (left) and quantification (middle, right) are shown. (C) Ezh2-deficient Tfr cells are less suppressive. In vitro suppression assay in which B and Tfh cells were cultured alone or along with Ezh2-sufficient or -deficient cells sorted as in B. IgG1+GL7+ class-switched B cells gating (left), IgG1+GL7+ class switched B cell quantification (middle), and quantification of Glut1 expression on B cells (right) are shown. (D) Ezh2 deficiency leads to a cell-intrinsic loss of Tfr cell suppressive function. Tfr cells from Ezh2f/fFoxP3Cre/+ or Ezh2f/+FoxP3Cre/+ mice (sorted based on Cre-YFP) were used for suppression assays as in C. Representative flow cytometry of IgG1+GL7+ class-switched B cells (left) and IgG secretion (right) are shown. (E) Loss of Ezh2 results in loss of the Tfr cell transcriptional program. PCA of RNA-seq transcriptional data from Ezh2-deficient (Ezh2f/fFoxP3Cre) or Ezh2-sufficient (Ezh2f/+FoxP3Cre) Tfr and T reg cells sorted as in B. (F) Volcano plots showing control or Ezh2-deficient Tfr cell RNA-seq data with total T reg genes (T reg versus T conv; left), Tfr (follicular) genes (Tfr versus Tfh; middle), and Tfr (T reg) genes (Tfr versus T reg; right; from Fig. 1; in red) compared with all genes (gray). P value was calculated using a χ2 test. (G) Enrichment score traces from GSEA analysis comparing Ezh2 F/+ Tfr cells versus F/+ T reg or F/F Tfr cells using indicated gene sets (generated from data in Fig. 1). (H and I) Venn diagram illustrating the overlap of genes differentially expressed in Ezh2-deficient Tfr cells versus control Tfr cells (ΔEzh2) compared with either genes differentially expressed in FoxP3-deleted Tfr cells compared with control Tfr cells (ΔFoxP3) or genes differentially expressed in in vitro generated ex-Tfr versus Tfr cells (FoxP3 Down). All error bars indicate standard error. **, P < 0.01; ***, P < 0.001 using Student’s t test. Data are from individual experiments and are representative of three independent experiments (A and B), are from individual experiments of triplicate wells and are representative of three independent experiments (C and D), or are combined data from three independent experiments (E and F).
To determine if loss of Ezh2 alters suppressive function of Tfr cells, we assessed Tfr cell suppressive capacity using an in vitro suppression assay. We immunized Ezh2f/f Foxp3Cre/Cre or Ezh2f/+Foxp3Cre/Cre control mice with NP-OVA; 7 d later sorted B, Tfh, and Tfr cells; and cultured B and Tfh cells (from control mice) with or without Ezh2-deficient or control Tfr cells, plus anti-CD3/IgM for 6 d. Control Tfr cells potently suppressed class switching of B cells to IgG1, while Ezh2-deficient Tfr cells suppressed class switching to a lesser degree (Fig. 6 C). Ezh2-deficient Tfr cells also inhibited Glut1 expression on B cells less than control Tfr cells. Ezh2-deficient conventional T reg cells (gated as ICOS−CXCR5−FoxP3+ cells) did not have altered suppressive capacity; however, total conventional T reg suppressive capacity was quite low, as we have described previously (Fig. S5 C).
Since Ezh2f/f Foxp3Cre/Cre mice have baseline inflammation due to loss of Ezh2 on all T regs, we further investigated whether Ezh2 deficiency exerted cell-intrinsic changes in Tfr cells by taking advantage of random X-chromosome inactivation. We immunized Ezh2f/f Foxp3Cre/+ or Ezh2f/+Foxp3Cre/+ control mice with NP-OVA and 7 d later sorted Tfr cells to add into the in vitro suppression assay. Because of X-linked inactivation, both experimental and control mice have at least 50% normal T regs, which prevents potential autoimmunity. Ezh2-deficient Tfr cells were less potent at suppressing class switch recombination to IgG1 and antibody secretion compared with control Tfr cells (Fig. 6 D). These data suggest that Ezh2 has a cell-intrinsic role in Tfr cells to ensure optimal suppressive capacity.
Since Ezh2 alters Tfr cell suppressive capacity, we determined whether Ezh2 alters the Tfr cell transcriptional program. We sorted T conv, T reg, and Tfr cells from Ezh2f/f Foxp3Cre/Cre or Ezh2f/+Foxp3Cre/Cre mice that were immunized with NP-OVA 7 d previously and performed RNA-seq transcriptional analysis. Ezh2-deficient and control Tfr cells had 78 differentially expressed (P < 0.01) genes, including the transcript for Ezh2 (Fig. S5 D and data not shown). PCA revealed that Ezh2-deficient and control Tfr cells occupied a similar space in PC1 and PC2, which was separate from T reg and T conv cells (Fig. 6 E). To determine if loss of Ezh2 alters T reg, Tfh, or Tfr genes in Tfr cells, we visualized sets of genes identified from data in Fig. 1 in control versus Ezh2-deleted Tfr cells. T reg genes and Tfr (follicle) genes (generated by comparing Tfr versus Tfh) were not lost upon deletion of Ezh2 in Tfr cells (Fig. 6, F and G). Tfr (T reg) genes (generated by comparing Tfr versus T reg) as a group were expressed less in Ezh2-deleted Tfr cells, suggesting that Ezh2 controls the follicular Tfr program (and not simply a T reg program in Tfr cells). However, only a few of these genes reached statistical significance individually. To determine the frequency of genes that are coregulated by Ezh2 and FoxP3, we compared the differentially expressed genes in Ezh2-deficient Tfr cells and compared them to genes differentially expressed in Tfr cells that down-regulated FoxP3 (in vitro generated ex-Tfr cells; Fig. 3) or had FoxP3 deleted (FoxP3 floxed; Fig. S3). We found that only a subset of genes differentially expressed in Ezh2-deficient Tfr cells overlapped with FoxP3 down-regulated or FoxP3 deleted, suggesting that Ezh2 and FoxP3 have nonoverlapping roles in modulating the Tfr transcriptional program (Fig. 6, F, H, and I).
Discussion
In this study, we assessed how Tfr cells are transcriptionally programmed to suppress B cell responses. We determined that Tfr cells have a transcriptional program that can be fine-tuned by the tissue microenvironment. We also identified important, but nonoverlapping, roles for FoxP3 and Ezh2 in transcriptional control of Tfr cell identity and function. FoxP3 is necessary for Tfr cell identity and suppressive function, and some Tfr cells can down-regulate FoxP3 to become dysfunctional ex-Tfr cells. Ezh2 also is essential for Tfr cell identity and suppressive function. However, FoxP3 and Ezh2 appear to control the Tfr program in distinct ways, because loss of FoxP3 or Ezh2 resulted in distinct alterations to the Tfr cell transcriptional program. Together, these data demonstrate that alterations in the Tfr transcriptional program can lead to dysfunctional ex-Tfr cells. Further work is needed to determine whether dysfunctional ex-Tfr cells contribute to pathogenic antibody responses.
FoxP3 has a complex role in controlling T reg cell differentiation and function. FoxP3 is thought to act predominantly as a transcriptional repressor through direct binding and recruitment of Ezh2 to specific loci, resulting in chromatin modification and diminished gene accessibility (Arvey et al., 2014). However, newer data have suggested that this model is oversimplified and that FoxP3 predominantly binds to enhancer regions and then potentiates or represses activity through associations with RelA or Ezh2, respectively (Kwon et al., 2017). Interestingly, loss of FoxP3 through natural down-regulation (or through deletion) resulted in alterations in T reg genes and Tfr genes, suggesting that FoxP3 is maintaining a T reg cell state as well as a distinct Tfr state. Based on our data showing that FoxP3 can convert a Tfh cell to a functional Tfr cell, we hypothesize that FoxP3 is binding to enhancer regions that are uniquely accessible in Tfh and Tfr (but not in conventional) T reg cells. These data are reminiscent of adipose tissue T regs in which FoxP3 expression and the adipose transcription factor Pparγ were required to obtain an adipose tissue–like T reg cell signature (Cipolletta et al., 2012), or where T reg cells require the transcription factor IRF4 to specifically inhibit Th2 responses (Zheng et al., 2009). In this way, FoxP3 can coopt transcription factors of the cells they suppress in order to have specialized functional roles. It is currently unclear which transcription factors in addition to FoxP3 are required to form a Tfr-like cell capable of suppressing B cell responses. Canonical Tfh transcription factors such as Bcl6 or Ascl2 may be involved, because these transcription factors control the Tfh transcriptional program.
Our data demonstrate that FoxP3 is not only essential for differentiation of Tfr cells, but also required to maintain the Tfr cell program. FoxP3 expression was unstable and could be lost in a subset of Tfr cells in vitro and in vivo. Ex-Tfr cells had lower expression of CTLA-4 and CD25 compared with Tfr cells, but higher expression compared with Tfh cells. In addition, ex-Tfr cells had a transcriptional program distinct from both Tfr and Tfh cells. Therefore, ex-Tfr cells seem to lose a proportion of the Tfr program, but never fully become Tfh cells. Although the precise frequency of ex-Tfr cells is still unclear, these data have several important implications. First, ex-Tfr cells are likely contained in Tfh gating strategies, which may explain variability in the association of Tfh cell percentages and/or ratios with B cell responses. Second, ex-Tfr cells may have special modulatory roles in the B cell follicle or GC reaction, which may or may not overlap with Tfh and Tfr cell functions. For instance, it is possible that ex-Tfr cells may create a state of reduced suppression due to lower, but still positive, levels of CTLA-4. Alternatively, ex-Tfr cells may compete with Tfr cells for survival or costimulatory signals, thereby preventing Tfr cell suppression. Since ex-Tfr cells lack suppressive capacity and may have other functionalities, they may contribute to pathogenic antibody responses in some settings such as autoimmunity. It is challenging to identify ex-Tfr cells in humans. However, CD25+Bcl6lowBlimp-1low FoxP3− Tfh cells are found in human tonsils, which match ex-Tfr cells, at least phenotypically (Li and Pauza, 2015).
Our data also identified Ezh2 as essential for full suppressive capacity of Tfr cells. Ezh2 has important roles in epigenetic control of lymphocyte development (Raaphorst et al., 2001; Su et al., 2003; Mandal et al., 2011), T helper cytokine production (Tumes et al., 2013), and T reg cell stability (DuPage et al., 2015; Yang et al., 2015). T reg cells require a specific epigenetic landscape for activity (Ohkura et al., 2012). We found that Ezh2 was essential for full suppressive capacity of Tfr cells, but seemed to maintain the Tfr program in distinct ways compared with FoxP3, since the overlap in differentially expressed genes was fairly minor. Therefore, the role of Ezh2 in controlling the Tfr cell program is likely multifaceted, with Ezh2 activity having both FoxP3-dependent and -independent functions. This result makes sense considering that Ezh2 has profound roles in controlling gene expression in cell types which do not express FoxP3. Similarly, FoxP3 likely has both Ezh2-dependent and -independent functions, as would be predicted by the recent finding that FoxP3 can bind to a number of different complexes, only one of which utilizes Ezh2 for gene regulation (Kwon et al., 2017). Based on these findings, we hypothesize that the transcriptional program allowing Tfr cell identity relies on multiple nodes of regulation that cooperate to allow full Tfr cell suppressive function. Further studies are needed to determine how Ezh2 affects the Tfh transcriptional program compared with the Tfr transcriptional program, since Tfh cells also express high levels of Ezh2, and Tfh cell differentiation can be regulated by histone demethylases (Cook et al., 2015). These data will be useful to determine if inhibiting Ezh2 can result in diminished Tfr cell functionality and might be a useful target for boosting antibody responses to vaccines.
Our comparison of Tfr cells from different tissues demonstrated that the Tfr cell transcriptional program and functionality can be altered by the local tissue microenvironment. In murine models, Tfh and Tfr cells in the circulation can act as memory cells, and it has been suggested that the reduced suppression of Tfr cells in the blood and increased stimulatory capacity of Tfh cells in the blood facilitate strong antibody production during memory responses (Sage et al., 2014a). Moreover, numerous groups have shown that Tfh cells in human circulation are distinct from Tfh cells found in lymphoid organs (Rasheed et al., 2006; Crotty, 2011; Morita et al., 2011). Our data demonstrate that Tfh and Tfr cells from distinct lymphoid tissue such as lymph node and spleen also can have distinct functions. These findings may help resolve differences in the literature between studies using splenic Tfr cells and lymph node Tfr cells. Further work is needed to determine the cause of changes to the Tfr program in different tissues; however, distinct dendritic cell subsets or cytokine milieu are likely causes.
In summary, we have demonstrated that the Tfr cell transcriptional program requires FoxP3 and Ezh2 for maintenance, and loss of either FoxP3 or Ezh2 results in diminished suppression of B cell responses. Our studies showed that FoxP3 and Ezh2 exert multifaceted and partially overlapping functions to control Tfr cells. FoxP3 can coopt the Tfh program and turn a Tfh cell into a functional suppressive Tfr-like cell. These findings elucidate mechanisms that control the Tfr transcriptional program and suppressive function and suggest potential targets to modulate in Tfr cells to enhance vaccine responses or ameliorate autoimmune diseases.
Materials and methods
Mice
Foxp3IRES-GFP, Foxp3CreYFP, Rosa26Lox-Stop-Lox-TdTomato, Ptprca (CD45.1), and Ezh2 floxed mice were purchased from The Jackson Laboratory. Foxp3 floxed mice were from the Rudensky laboratory (Williams and Rudensky, 2007). All mice were between 6 and 8 wk of age at the time of experiments and were housed in a specific pathogen–free facility. Each individual experiment contained one sex of mice, but replicates were performed with males or females. All mice were used according to the Harvard Medical School and Brigham and Women's Hospital Standing Committees on Animals and National Institutes of Health Guidelines.
Immunizations
Mice were immunized with 100 µg NP-OVA (Biosearch Technologies) emulsified in H37RA CFA subcutaneously in the mouse flanks or intraperitoneally as previously described (Sage et al., 2013; Sage and Sharpe, 2015a). Mice were sacrificed 7 d later, and inguinal lymph nodes or spleen were harvested.
Antibodies
The following antibodies were used for surface staining: anti-CD4 (RM4-5), anti-ICOS (15F9), anti-CD19 (6D5), anti-CD25 (PC61), anti-CXCR5 biotin (2G8), GL7 (GL-7), anti-PD-1 (RMP1-30), anti-GITR (DTA-1), anti-Thy1.1 (OX-7), anti-CD45.1 (A20), and anti-IA (M5/114.15.2). For intracellular staining, samples were fixed with the FoxP3 Fix/Perm buffer set according to the manufacturer’s instructions (eBioscience). Samples were then intracellularly stained with anti-IgG1 (A85-1), anti-FoxP3 (FJK-16S), anti-Ki67 (B56), anti-Glut1 (polyclonal; Abcam), anti-Helios (22F6), or CTLA-4 (UC10-4B9).
Sorting
Single-cell suspensions were diluted in PBS supplemented with 1% FBS containing 1 mM EDTA. Tfh and Tfr cells were isolated by first enriching CD4+ cells with positive selection (Miltenyi Biotec). CD4+-enriched cells were then stained and sorted as follows: Tfh (CD4+ICOS+CXCR5+FoxP3−CD19−), Tfr (CD4+ICOS+CXCR5+FoxP3+CD19−), T conv (CD4+ICOS−CXCR5−FoxP3−CD19−), and T reg (CD4+ICOS−CXCR5−FoxP3+CD19−). B cells were isolated from the flow-through of CD4+ selection, which was then positively selected using CD19 beads (Miltenyi Biotec) to >98% purity.
Suppression assays
In vitro suppression assays were performed as described previously (Sage et al., 2014b, 2016; Sage and Sharpe, 2015a). Briefly, 5 × 104 B cells, 3 × 104 Tfh cells, and/or 1.5 × 104 Tfr cells were plated in 96-well plates along with 2 µg/ml anti-CD3 (BioXcell) and 5 µg/ml anti-IgM (Jackson ImmunoResearch). Cultures were harvested after 4–6 d as described. For analysis, B cells were gated as CD19+IA+CD4− cells, Tfh cells were gated as CD4+IA−CD19−FoxP3− cells, and Tfr cells were gated as CD4+IA−CD19−FoxP3+ cells. For FoxP3 stability assays, Tfr cells were gated as CD45.1+CD4+IA−CD19− cells.
Retroviral transduction
WT FoxP3-encoding and empty retroviruses were a gift from the Mathis/Benoist laboratory (Harvard Medical School, Boston, MA; Kwon et al., 2017). For transducing FoxP3 in Tfh cells, cultures were plated as above and cultured for 6 h. After 6 h, cultures were spin-infected with virus in the presence of 8 µg/ml polybrene and 50 U/ml IL-2 at 805 g for 2 h and subsequently incubated at 37°c for 8–10 h. Culture supernatants were then replaced with media containing anti-CD3/IgM as above and cultured for an additional 3 d before analysis.
RNA-seq
Samples were sorted as described above. Each replicate indicates a biological replicate that was prepared using different sets of mice on different experimental days. RNA-seq library preparations were performed as previously described (Sage et al., 2016; Kadoki et al., 2017). Briefly, RNA was isolated using MyOne Silane Dynabeads (Thermo Fisher Scientific). RNA was fragmented and barcoded using 8-bp barcodes in conjunction with standard Illumina adaptors. Primers were removed using Agencourt AMPure XP bead cleanup (Beckman Coulter/Agencourt), and samples were amplified with 14 PCR cycles. Libraries were gel purified and quantified using a Qubit high-sensitivity DNA kit (Invitrogen), and library quality was confirmed using Tapestation high-sensitivity DNA tapes (Agilent Technologies). RNA-seq reactions were sequenced on an Illumina NextSeq sequencer (Illumina) according to the manufacturer’s instructions, sequencing 50-bp single-end reads. Analysis was performed using the CLC Genomics Workbench version 8.0.1 RNA-seq analysis software package (Qiagen). Briefly, reads were aligned (mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.8, similarity fraction = 0.8) to the mouse genome, and differential expression analysis was performed (total count filter cutoff = 5.0). Results were normalized to reads per million. Morpheus (Broad Institute) was used to generate heat maps and similarity matrices.
GSEA
For GSEA analysis, RNA-seq data were converted to human nomenclature and compared with GSEA mSigDatabases including Hallmarks and C3 and C5 collections using standard settings (Broad Institute). For enrichment plots of specific gene sets, pathways were analyzed along with 20 randomized gene sets to ensure specificity.
ELISA
ELISA measurements of total IgG from culture supernatants were performed as described previously (Sage and Sharpe, 2015a).
Confocal microscopy
Confocal microscopy was performed as previously described (Sage et al., 2013). Briefly, lymph nodes were embedded in OCT (Tissue-Tek), and 10-µm sections were cut and stained for indicated antibodies. Sections were imaged on an Olympus FV3000 confocal microscope. Linear contrast enhancement was performed using ImageJ (National Institutes of Health).
Statistics
Most statistical tests were performed using Prism 6.0 (GraphPad) using Student’s two-tailed unpaired t test or one-way ANOVA with Tukey’s correction as specified assuming Gaussian distribution. Statistics for RNA-seq were performed using CLC Genomics Workbench (Qiagen). Statistics for gene set enrichment were performed in GSEA (Broad Institute). Statistics for Volcano plots were performed in Excel using a χ2 test.
Deposition of data
RNA sequencing data have been deposited in the Gene Expression Omnibus database (accession no. GSE124884).
Online supplemental material
Fig. S1 shows gating strategies and further RNA-seq analysis of Tfr cells. Fig. S2 shows additional RNA-seq analysis of in vitro generated ex-Tfr cells. Fig. S3 shows additional analysis of FoxP3-deleted Tfr cells. Fig. S4 shows additional controls for FoxP3 expression in Tfh cells. Fig. S5 shows additional analysis of Ezh2-deficient Tfr cells.
Supplementary Material
Acknowledgments
We would like to thank Ho-keun Kwon and Christophe Benoist (Harvard Medical School, Boston, MA) for reagents and help with retroviral transduction protocols.
This work was supported by the National Institutes of Health through grants R01AI40614 (A.H. Sharpe), P01AI56299 (A.H. Sharpe), R01 HL11879 (B.R. Blazar), and K22AI132937 (P.T. Sage) and the Evergrande Center for Immunological Diseases.
The authors declare no competing financial interests.
Author contributions: S. Hou and R.L. Clement performed experiments and analyzed data. A. Diallo provided bioinformatic support. B.R. Blazar contributed to experimental design and edited the manuscript. A.Y. Rudensky provided essential reagents and contributed to experimental design. A.H. Sharpe contributed to experimental design and helped write the manuscript. P.T. Sage designed and performed experiments, analyzed data and wrote the manuscript.
References
- Arvey A., van der Veeken J., Samstein R.M., Feng Y., Stamatoyannopoulos J.A., and Rudensky A.Y.. 2014. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 15:580–587. 10.1038/ni.2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., and Ochs H.D.. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27:20–21. 10.1038/83713 [DOI] [PubMed] [Google Scholar]
- Botta D., Fuller M.J., Marquez-Lago T.T., Bachus H., Bradley J.E., Weinmann A.S., Zajac A.J., Randall T.D., Lund F.E., León B., and Ballesteros-Tato A.. 2017. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat. Immunol. 18:1249–1260. 10.1038/ni.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow M.E., Jeffery E.W., Hjerrild K.A., Paeper B., Clark L.B., Yasayko S.A., Wilkinson J.E., Galas D., Ziegler S.F., and Ramsdell F.. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73. 10.1038/83784 [DOI] [PubMed] [Google Scholar]
- Chaudhry A., Rudra D., Treuting P., Samstein R.M., Liang Y., Kas A., and Rudensky A.Y.. 2009. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 326:986–991. 10.1126/science.1172702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., Tanaka S., Chu F., Nurieva R.I., Martinez G.J., Rawal S., Wang Y.H., Lim H., Reynolds J.M., Zhou X.H., et al. 2011. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 17:983–988. 10.1038/nm.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D., Feuerer M., Li A., Kamei N., Lee J., Shoelson S.E., Benoist C., and Mathis D.. 2012. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 486:549–553. 10.1038/nature11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook K.D., Shpargel K.B., Starmer J., Whitfield-Larry F., Conley B., Allard D.E., Rager J.E., Fry R.C., Davenport M.L., Magnuson T., et al. 2015. T Follicular Helper Cell-Dependent Clearance of a Persistent Virus Infection Requires T Cell Expression of the Histone Demethylase UTX. Immunity. 43:703–714. 10.1016/j.immuni.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- DuPage M., Chopra G., Quiros J., Rosenthal W.L., Morar M.M., Holohan D., Zhang R., Turka L., Marson A., and Bluestone J.A.. 2015. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity. 42:227–238. 10.1016/j.immuni.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca V.R., Agua-Doce A., Maceiras A.R., Pierson W., Ribeiro F., Romão V.C., Pires A.R., da Silva S.L., Fonseca J.E., Sousa A.E., et al. 2017. Human blood Tfr cells are indicators of ongoing humoral activity not fully licensed with suppressive function. Sci. Immunol. 2:eaan1487 10.1126/sciimmunol.aan1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.A., Feuerer M., Tash K., Haxhinasto S., Perez J., Melamed R., Mathis D., and Benoist C.. 2007. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 27:786–800. 10.1016/j.immuni.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., and Rudensky A.Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoki M., Patil A., Thaiss C.C., Brooks D.J., Pandey S., Deep D., Alvarez D., von Andrian U.H., Wagers A.J., Nakai K., et al. 2017. Organism-level analysis of vaccination reveals networks of protection across tissues. Cell. 171:398–413.e21. 10.1016/j.cell.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.K., Chen H.M., Mathis D., and Benoist C.. 2017. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 18:1238–1248. 10.1038/ni.3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Pauza C.D.. 2015. CD25(+) Bcl6(low) T follicular helper cells provide help to maturing B cells in germinal centers of human tonsil. Eur. J. Immunol. 45:298–308. 10.1002/eji.201444911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Pierson W., Lee S.K., Kallies A., Kawamoto S., Rayner T.F., Srivastava M., Divekar D.P., Beaton L., Hogan J.J., et al. 2011. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17:975–982. 10.1038/nm.2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceiras A.R., Almeida S.C.P., Mariotti-Ferrandiz E., Chaara W., Jebbawi F., Six A., Hori S., Klatzmann D., Faro J., and Graca L.. 2017. T follicular helper and T follicular regulatory cells have different TCR specificity. Nat. Commun. 8:15067 10.1038/ncomms15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Maienschein-Cline M., Bartom E.T., Hamel K.M., Kee B.L., Dinner A.R., and Clark M.R.. 2011. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12:1212–1220. 10.1038/ni.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Kretschmer K., Frampton G.M., Jacobsen E.S., Polansky J.K., MacIsaac K.D., Levine S.S., Fraenkel E., von Boehmer H., and Young R.A.. 2007. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 445:931–935. 10.1038/nature05478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R., Schmitt N., Bentebibel S.E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N., et al. 2011. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 34:108–121. 10.1016/j.immuni.2010.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N., Hamaguchi M., Morikawa H., Sugimura K., Tanaka A., Ito Y., Osaki M., Tanaka Y., Yamashita R., Nakano N., et al. 2012. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 37:785–799. 10.1016/j.immuni.2012.09.010 [DOI] [PubMed] [Google Scholar]
- Raaphorst F.M., Otte A.P., van Kemenade F.J., Blokzijl T., Fieret E., Hamer K.M., Satijn D.P., and Meijer C.J.. 2001. Distinct BMI-1 and EZH2 expression patterns in thymocytes and mature T cells suggest a role for Polycomb genes in human T cell differentiation. J. Immunol. 166:5925–5934. 10.4049/jimmunol.166.10.5925 [DOI] [PubMed] [Google Scholar]
- Rasheed A.U., Rahn H.P., Sallusto F., Lipp M., and Müller G.. 2006. Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur. J. Immunol. 36:1892–1903. 10.1002/eji.200636136 [DOI] [PubMed] [Google Scholar]
- Rudra D., deRoos P., Chaudhry A., Niec R.E., Arvey A., Samstein R.M., Leslie C., Shaffer S.A., Goodlett D.R., and Rudensky A.Y.. 2012. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 13:1010–1019. 10.1038/ni.2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage P.T., and Sharpe A.H.. 2015a In vitro assay to sensitively measure T(FR) suppressive capacity and T(FH) stimulation of B cell responses. Methods Mol. Biol. 1291:151–160. 10.1007/978-1-4939-2498-1_13 [DOI] [PubMed] [Google Scholar]
- Sage P.T., and Sharpe A.H.. 2015b T follicular regulatory cells in the regulation of B cell responses. Trends Immunol. 36:410–418. 10.1016/j.it.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage P.T., and Sharpe A.H.. 2016. T follicular regulatory cells. Immunol. Rev. 271:246–259. 10.1111/imr.12411 [DOI] [PubMed] [Google Scholar]
- Sage P.T., Francisco L.M., Carman C.V., and Sharpe A.H.. 2013. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat. Immunol. 14:152–161. 10.1038/ni.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage P.T., Alvarez D., Godec J., von Andrian U.H., and Sharpe A.H.. 2014a Circulating T follicular regulatory and helper cells have memory-like properties. J. Clin. Invest. 124:5191–5204. 10.1172/JCI76861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage P.T., Paterson A.M., Lovitch S.B., and Sharpe A.H.. 2014b The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity. 41:1026–1039. 10.1016/j.immuni.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage P.T., Ron-Harel N., Juneja V.R., Sen D.R., Maleri S., Sungnak W., Kuchroo V.K., Haining W.N., Chevrier N., Haigis M., and Sharpe A.H.. 2016. Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nat. Immunol. 17:1436–1446. 10.1038/ni.3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein R.M., Arvey A., Josefowicz S.Z., Peng X., Reynolds A., Sandstrom R., Neph S., Sabo P., Kim J.M., Liao W., et al. 2012. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 151:153–166. 10.1016/j.cell.2012.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., and Tarakhovsky A.. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124–131. 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- Tumes D.J., Onodera A., Suzuki A., Shinoda K., Endo Y., Iwamura C., Hosokawa H., Koseki H., Tokoyoda K., Suzuki Y., et al. 2013. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 39:819–832. 10.1016/j.immuni.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Williams L.M., and Rudensky A.Y.. 2007. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8:277–284. 10.1038/ni1437 [DOI] [PubMed] [Google Scholar]
- Wing J.B., Ise W., Kurosaki T., and Sakaguchi S.. 2014. Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity. 41:1013–1025. 10.1016/j.immuni.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Wing J.B., Kitagawa Y., Locci M., Hume H., Tay C., Morita T., Kidani Y., Matsuda K., Inoue T., Kurosaki T., et al. 2017. A distinct subpopulation of CD25- T-follicular regulatory cells localizes in the germinal centers. Proc. Natl. Acad. Sci. USA. 114:E6400–E6409. 10.1073/pnas.1705551114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg I., Agua-Doce A., Hernández A., Almeida C., Oliveira V.G., Faro J., and Graca L.. 2011. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J. Immunol. 187:4553–4560. 10.4049/jimmunol.1101328 [DOI] [PubMed] [Google Scholar]
- Yang X.P., Jiang K., Hirahara K., Vahedi G., Afzali B., Sciume G., Bonelli M., Sun H.W., Jankovic D., Kanno Y., et al. 2015. EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci. Rep. 5:10643 10.1038/srep10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S.Z., Kas A., Chu T.T., Gavin M.A., and Rudensky A.Y.. 2007. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 445:936–940. 10.1038/nature05563 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Chaudhry A., Kas A., deRoos P., Kim J.M., Chu T.T., Corcoran L., Treuting P., Klein U., and Rudensky A.Y.. 2009. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 458:351–356. 10.1038/nature07674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler S.F. 2006. FOXP3: of mice and men. Annu. Rev. Immunol. 24:209–226. 10.1146/annurev.immunol.24.021605.090547 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.