EHF transcriptionally inhibits the expressions of TGFβ1 and GM-CSF to decrease T reg cell and MDSC accumulation, making it a promising biomarker to evaluate the immune microenvironment in PDAC. EHF overexpression may improve the efficacy of checkpoint immunotherapy in PDAC.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly immune-suppressive tumor with a low response rate to single checkpoint blockade therapy. ETS homologous factor (EHF) is a tumor suppressor in PDAC. Here, we report a novel function of EHF in pancreatic cancer immune microenvironment editing and efficacy prediction for anti-PD1 therapy. Our findings support that the deficiency of tumoral EHF induced the accumulation of regulatory T (T reg) cells and myeloid-derived suppressor cells (MDSCs) and a decrease in the number of tumor-infiltrating CD8+ T cells. Mechanistically, EHF deficiency induced the conversion and expansion of T reg cells and MDSCs through inhibiting tumor TGFβ1 and GM-CSF secretion. EHF suppressed the transcription of TGFB1 and CSF2 by directly binding to their promoters. Mice bearing EHF overexpression tumors exhibited significantly better response to anti-PD1 therapy than those with control tumors. Our findings delineate the immunosuppressive mechanism of EHF deficiency in PDAC and highlight that EHF overexpression may improve PDAC checkpoint immunotherapy.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal tumor. Despite recent advances in combination chemotherapy regimens, the prognosis remains poor (Siegel et al., 2018; Teng et al., 2018). Checkpoint blockade is a pillar of cancer therapy for several tumor types including melanoma, lung cancer, renal cancer, and bladder cancer; however, its efficacy in PDAC remains poor when used as a single agent (Iorio et al., 2018; Michl and Krug, 2018).
Current research to date has identified that the efficacy of a single PD1/PD-L1 blockade may be limited due to tumor cell–extrinsic factors, including poor CD8+ T cell infiltration, accumulation of regulatory T (T reg) cells and myeloid-derived suppressor cells (MDSCs), and the up-regulation of other inhibitory immune checkpoints (Teng et al., 2015; Beatty et al., 2017). Increasing evidence suggests that PDAC secretes a series of immune-modulating factors, which induce an immune-suppressive microenvironment composed of T reg cells, MDSCs, and tumor-associated macrophages (TAMs). These factors result in an immunosuppressive environment resistant to PD1/PD-L1 blockade therapy (Bayne et al., 2012; Cox and Olive, 2012; Stromnes et al., 2014; Farren et al., 2016; Park et al., 2016; Principe et al., 2016; Kenkel et al., 2017; Lin and Lin, 2017; Pergamo and Miller, 2017; Pickup et al., 2017; Piro et al., 2017; Seo and Pillarisetty, 2017; Zhang et al., 2017). However, some advances still have been achieved in anti-PD1/PD-L1 treatment (Patnaik et al., 2015; Le et al., 2016; Feng et al., 2017). Careful assessment of the patient’s tumor immune microenvironment is critical to identify the suitable immunotherapeutic option. The identification of a molecular index predictive for immunotherapy efficacies will greatly help in the selection of tumor immunotherapy for patients.
EHF (ETS [E26 transformation-specific] homologous factor/epithelium-specific ETS factor family member 3 [ESE3]) is a member of the ETS superfamily. EHF was reported to be highly expressed in normal human pancreas and prostate tissues (Feldman et al., 2003). In prostate cancer, the expression of EHF was lower than in normal tissue; moreover, EHF loss leads to mesenchymal and stem-like features (Albino et al., 2012, 2016a,b; Longoni et al., 2013). Our previous investigation identified that EHF inhibits PDAC epithelial–mesenchymal transition and metastasis by transcriptionally up-regulating E-cadherin (Zhao et al., 2017). However, the function of tumoral EHF in tumor immune modulation has never been studied. Here, we report a novel function of EHF in tumor immune microenvironment editing. Our results indicate that EHF expression level could be used as a predictor for the efficacy of anti-PD1 therapy.
Results
The association between tumoral EHF expression and immune profiles in human PDAC tissue
We noticed that EHF overexpression in PANC02 cells significantly inhibited tumor growth in syngeneic C57BL/6 mice, but not in immune-deficient BALB/c nude mice (Fig. S1). We hypothesized that the differential effects on tumor growth in the two models might be due to effects of EHF on tumoral immune microenvironment. To examine these effects, we investigated the correlation between tumoral EHF and the infiltration of T reg cells, MDSCs, and CD8+ T cells in archived tissues from a retrospective cohort of 96 consecutive PDAC patients. For the retrospective cohort, tissue immunofluorescence (IF) of FOXP3/CD33/CD8, EHF, and DAPI were performed in three sets of tissues (Fig. 1, A–C). The average counts of tumor-infiltrating T reg cells, MDSCs, and CD8+ T cells per high-power field (HPF; 200×) were 19.97 ± 9.0 (range, 0–52), 12.49 ± 6.07 (range, 0–32), and 17.92 ± 17.14 (range, 0–100). Our results indicated that high T reg infiltration and MDSC infiltration significantly correlated with decreased overall survival (OS; P = 0.028 and 0.024, for T reg cells and MDSCs, respectively) and relapse-free survival (RFS; P = 0.016 for MDSCs). On the other hand, high CD8+ T cells correlated with increased OS (P = 0.039; Table 1 and Fig. S2, A–C).
Figure 1.
Tumoral EHF associates with the immune profile in human PDAC tissue. (A–C) IF staining (left) of EHF expression and the accumulation of FOXP3+ T reg cells (A), CD33+ MDSCs (B), and CD8+ T cells (C) in tumor tissues. An example from the 96 cases is shown. The arrows indicated tumor-infiltrating Foxp3+ T reg cells, CD33+ myeloid cells, and CD8+ T cells. Bars, 200 µm. Nonpaired Student’s t test was used as statistical analysis; n = 96, ***, P < 0.001. (D and E) Kaplan–Meier OS (D) and RFS (E) for different levels of EHF based on the log-rank statistic test (P < 0.001). (F–I) Single-cell suspensions were prepared from 45 cases of fresh PDAC tissues and stained with specific antibodies against human T reg cells (CD4+, CD25+, and FOXP3+), MDSCs (HLA-DR−, CD33+, and CD11b+), and CD8+ T cells (CD8+). Representative IHC staining of EHF is shown. Bars, 200 µm. Representative dot plots or histograms of T reg cells (gated on CD4+ T cells; G, left), MDSCs (gated on HLA-DR− cells; H, left), and CD8+ T cells (I, left). Spearman correlation analyses between EHF IHC score and the proportions of tumor-infiltrating T reg cells (G, right), MDSCs (H, right), and CD8+ T cells (I, right); n = 45.
Table 1. Univariate and multivariate Cox proportional hazards analysis of clinicopathological factors for OS and RFS.
| Variables | OS | RFS | ||
|---|---|---|---|---|
| HR (95.0% CI) | P | HR (95.0% CI) | P | |
| Univariate analysis | ||||
| Age | 0.932 (0.570–1.524) | 0.780 | 0.805 (0.508–1.275) | 0.355 |
| Sex | 1.398 (0.855–2.287) | 0.182 | 1.335 (0.842–2.114) | 0.219 |
| Histological grade | 1.772 (1.058–2.969) | 0.030a | 1.621 (1.001–2.627) | 0.05 |
| Tumor size | 1.133 (0.688–1.868) | 0.624 | 1.058 (0.664–1.684) | 0.813 |
| pTNM stage | 2.174 (1.153–4.100) | 0.016a | 2.462 (1.363–4.447) | 0.003a |
| LN metastasis | 1.417 (0.768–2.612) | 0.265 | 1.440 (0.816–2.543) | 0.209 |
| EHF expression | 0.257 (0.134–0.492) | 0.000a | 0.293 (0.162–0.532) | 0.000a |
| CD8 | 0.561 (0.324–0.972) | 0.039a | 0.676 (0.411–1.114) | 0.124 |
| T reg cell | 1.742 (1.061–2.862) | 0.028a | 1.520 (0.956–2.417) | 0.077 |
| MDSC | 1.906 (1.088–3.339) | 0.024a | 1.897 (1.126–3.195) | 0.016a |
| Multivariate analysis | ||||
| pTNM stage | 1.977 (1.041–3.754) | 0.037a | 2.344 (1.292–4.255) | 0.005a |
| EHF expression | 0.267 (0.139–0.514) | 0.000a | 0.338 (0.184–0.623) | 0.001a |
Data were based on tissue IF assay. Multivariate Cox proportional hazards analysis used backward selection model. HR, hazard ratio; CI, confidence interval.
Statistically significant (P < 0.05).
To examine the correlation between EHF and immune cell infiltrations, PDAC samples were divided into two groups (high EHF and low EHF) according to EHF-positive cell count/HPF. The count of EHF-positive points per field ranged from 0 to 251, mean ± SD 50.89 ± 65.04. EHF-positive cells/HPF >50.89 was considered the high-EHF group; EHF-positive cells/HPF <50.89 was considered the low-EHF group. The frequencies of T reg cells (Fig. 1 A) and MDSCs (Fig. 1 B) in the high-EHF group were significantly decreased compared with the low-EHF group (T reg count/HPF: low-EHF group, 22.33 ± 8.46; high-EHF group, 13.81 ± 7.43, P < 0.001; MDSC count/HPF: low-EHF group, 13.84 ± 5.75; high-EHF group, 9.04 ± 5.56, P < 0.001). Moreover, higher numbers of CD8+ tumor-infiltrating lymphocytes (TILs) were observed in the high-EHF group (low-EHF group, 13.14 ± 11.69; high-EHF group, 30.11 ± 22.39, P < 0.001; Fig. 1 C). The median OS for the low-EHF group was 14.53 mo, which was significantly shorter than that in the high-EHF group (45.00 mo, P < 0.001; Fig. 1 D). The median RFS in the low-EHF group was also significantly lower (low-EHF group, 8.00 mo; high-EHF group, 17.03 mo, P < 0.001; Fig. 1 E). χ2 analysis between EHF and clinicopathological factors in PDAC demonstrated that high EHF expression was significantly correlated with lower histological grade (r = −0.258, P = 0.012), lower pathological tumor node metastasis (pTNM) stage (r = −0.210, P = 0.04), decreased accumulation of T reg cells (r = −0.314, P = 0.002) and MDSCs (r = −0.228, P = 0.025), and an increased infiltration of CD8+ T cells (r = 0.377, P < 0.001; Table 2). Univariate and multivariate analysis identified EHF (high/low) and pTNM as independent prognostic factors of PDAC patients (Table 1).
Table 2. Correlation of EHF expression to clinicopathological features in PDAC.
| Parameters | EHF (n) | χ2 | P | Spearman r | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (yr) | |||||
| <60 | 35 | 12 | 0.306 | 0.580 | 0.056 |
| ≥60 | 34 | 15 | |||
| Sex | |||||
| Male | 36 | 16 | 0.392 | 0.531 | −0.064 |
| Female | 33 | 11 | |||
| Histological grade | |||||
| G1, G2 | 40 | 23 | 6.371 | 0.012a | −0.258 |
| G3 | 29 | 4 | |||
| pTNM stage | |||||
| IA, IB | 14 | 11 | 4.214 | 0.040a | −0.210 |
| IIA, IIB | 55 | 16 | |||
| CD8 | |||||
| ≤20/HP | 53 | 10 | 13.609 | 0.000a | 0.377 |
| >20/HP | 16 | 17 | |||
| T reg cell | |||||
| ≤20/HP | 27 | 20 | 9.483 | 0.002a | −0.314 |
| >20/HP | 42 | 7 | |||
| MDSC | |||||
| ≤10/HP | 17 | 13 | 4.993 | 0.025a | −0.228 |
| >10/HP | 52 | 14 | |||
Data were based on tissue IF assay. Statistical data on EHF expression in relation to clinic-histopathologic features for surgical PDAC specimens. P values were calculated using the χ2 test.
Statistically significant (P < 0.05).
EHF intensity was also evaluated by software (Fig. S1 D). Cases were divided into low-EHF and high-EHF groups using the mean values as the cutoff. Statistical analyses are shown in Fig. S2 (E–H) and Tables S1 and S2. EHF scores evaluated by software correlated well with those evaluated by pathologists (Spearman r = 0.919, P < 0.001; Fig. S2 I).
Immunohistochemical (IHC) staining of FOXP3, CD33, CD8, and EHF in four consecutive sets (96 cases) of human PDAC tissues were also performed. The results based on IHC assay were consistent with the IF findings (Fig. S2, J–L; and Tables S3 and S4). EHF IF score correlated well with EHF IHC score (r = 0.872, P < 0.001; Fig. S2 M).
To further confirm the effects of EHF on tumor immune cell infiltrations, a second cohort of 45 cases of fresh PDAC tissues were collected prospectively and cut into two parts. One part was used to quantify the proportions of CD8+ T cells (CD8+), T reg cells (CD4+, CD25+, and FOXP3+), and MDSCs (HLA-DR−, CD33+, and CD11b+) using flow cytometry; the other part was prepared to detect EHF expression by IHC (Fig. 1 F). As shown in Fig. 1 (G–I), tumoral EHF score (0–9) negatively correlated with the proportion of tumor-infiltrating T reg cells (r = −0.597, P < 0.001) and MDSCs (r = −0.529, P < 0.001) and positively correlated with the percentage of tumor-infiltrating CD8+ T cells (r = 0.570, P < 0.001) in the prospective cohort, confirming our findings from the archived PDAC tissues. Taken together, our data imply a role for EHF in immune editing of the PDAC microenvironment.
Tumoral EHF reduces MDSCs, T reg cells, and PD1+Tim3+CD8+ T cells while restoring IFNγ+CD8+ T cells in vivo
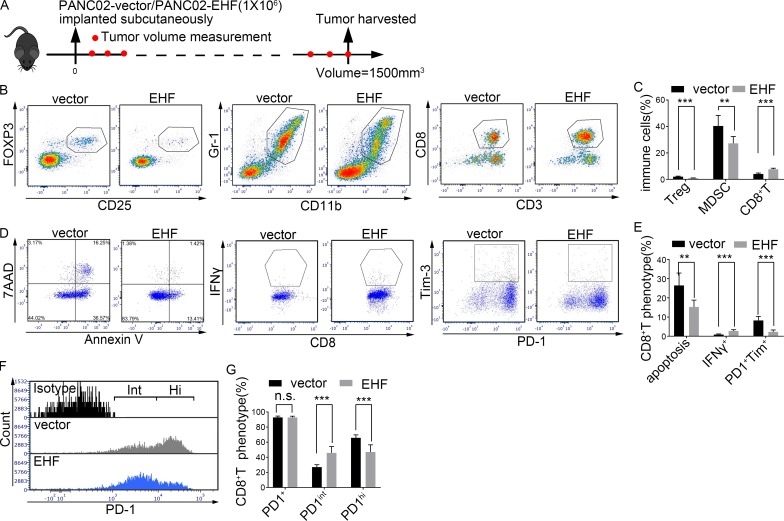
To determine whether EHF plays a causal role in tumor infiltration by immune cells, murine PANC02-vector and PANC02-EHF were subcutaneously implanted into immunocompetent C57BL/6 mice (Fig. S3 A). The mice were sacrificed when tumor volumes reached 1,500 mm3 (Fig. 2 A). Tumor-infiltrating immune cell content was analyzed by flow cytometry.
Figure 2.
Tumor-specific EHF reduces MDSCs, T reg cells, and PD1+Tim3+CD8+ T cells while restoring IFNγ+CD8+ T cells in vivo. (A) Immunocompetent C57BL/6 mice were subcutaneously inoculated with PANC02-vector or PANC02-EHF. Mice were sacrificed when tumor volumes reached a threshold of 1,500 mm3. (B and C) Representative dot plots (B) and statistical analysis (C) of the frequency of tumor-infiltrating T reg cells (left), MDSCs (middle), and CD3+CD8+ T cells (right). (D and E) Representative dot plots (D) and statistical analysis (E) of the frequency of tumor-infiltrating CD8+ T cell apoptosis (left), CD8+IFNγ+ T cells (middle), and CD8+ T cell exhaustion (PD1+Tim3+CD8+ T; right). CD8+ T cells were gated, and the percentage calculations used CD8+ T cell count as the denominator. (F) Representative histogram of PD1. CD8+ T cells were gated. (G) Statistical analysis of the frequency of PD1+CD8+ T (left), PD1intCD8+ T (middle), and PD1hiCD8+ T (right) cells. CD8+ T cell count was used as the denominator. The in vivo mouse experiments (A–G) were repeated three times independently, using seven mice per experimental group. Representative data are shown. Data are presented as mean ± SD. Nonpaired Student’s t test was used as statistical analysis. **, P < 0.01; ***, P < 0.001; n.s., not significant.
The proportions of CD45+ cells in PANC02-vector and PANC02-EHF tumors were comparable (P = 0.85; Fig. S3 B). The relative frequencies of T reg cells, MDSCs, TAMs, and CD8+ T cells in CD45+ leukocytes were evaluated. A notable decrease in T reg cells (vector, 2.02 ± 0.41%; EHF, 0.91 ± 0.18%, P < 0.0001) and MDSCs (vector, 40.3 ± 8.6%; EHF, 27.27 ± 5.17%, P = 0.0035) and an increased infiltration of CD8+ T cells (vector, 4.04 ± 0.75%; EHF, 7.62 ± 0.81%, P < 0.0001; Fig. 2, B and C) were observed in PANC02-EHF tumors. There was no significant difference in the percentage of TAMs (P = 0.16; Fig. S3 C). PANC02-EHF tumor tissues exhibited decrease in CD8+ T cell apoptosis rates (vector, 26.5 ± 6.47%; EHF, 15.29 ± 3.56%, P = 0.0017), higher percentages of IFNγ+CD8+ T cells (vector, 0.88 ± 0.33%; EHF, 2.76 ± 0.64%, P < 0.0001), and lower percentages of PD1+Tim3+CD8+ T cells (vector, 8.25 ± 2.11%; EHF, 2.33 ± 0.95%, P < 0.0001) than those from PANC02-vector control (Fig. 2, D and E). Moreover, CD8+ T cells from PANC02-EHF tumors displayed decreased proportions of PD1hiCD8+ T cells (vector, 8.25 ± 2.11%; EHF, 2.33 ± 0.95%, P = 0.0004), which were hyperexhausted and could not be reversed by anti-PD1 therapy (Fig. S3, D–F), and increased proportions of PD1intCD8+ T cells (vector, 26.96 ± 3.24%; EHF, 45.86 ± 8.44%, P = 0.0001), which were responsive to anti-PD1 therapy (Fig. S3, D–F), compared with PANC02-vector tumors (Fig. 2, F and G). The difference in total PD1+CD8+ T cell (PD1hiCD8+ T cells plus PD1intCD8+ T cells) percentages between the two groups was not significant (Fig. 2, F and G).
Results from an additional mouse model using KPC-vector and KPC-EHF cell lines are shown in Fig. S3 (G–R). Tumor volumes were significantly lower in the KPC-EHF group. Mice in the KPC-EHF group survived significantly longer than those in the KPC-vector group. A notable decrease in T reg cells and MDSCs and an increased infiltration of CD8+ T cells were observed in KPC-EHF tumors. KPC-EHF tumor tissues exhibited decreased CD8+ T cell apoptosis rates, higher percentages of IFN-γ+CD8+ T cells, and lower percentages of exhausted CD8+ T cells than those from KPC-vector tumor tissues, confirming our results from PANC02 models.
Tumoral EHF deficiency induced T reg cell conversion, expansion, and function in vitro
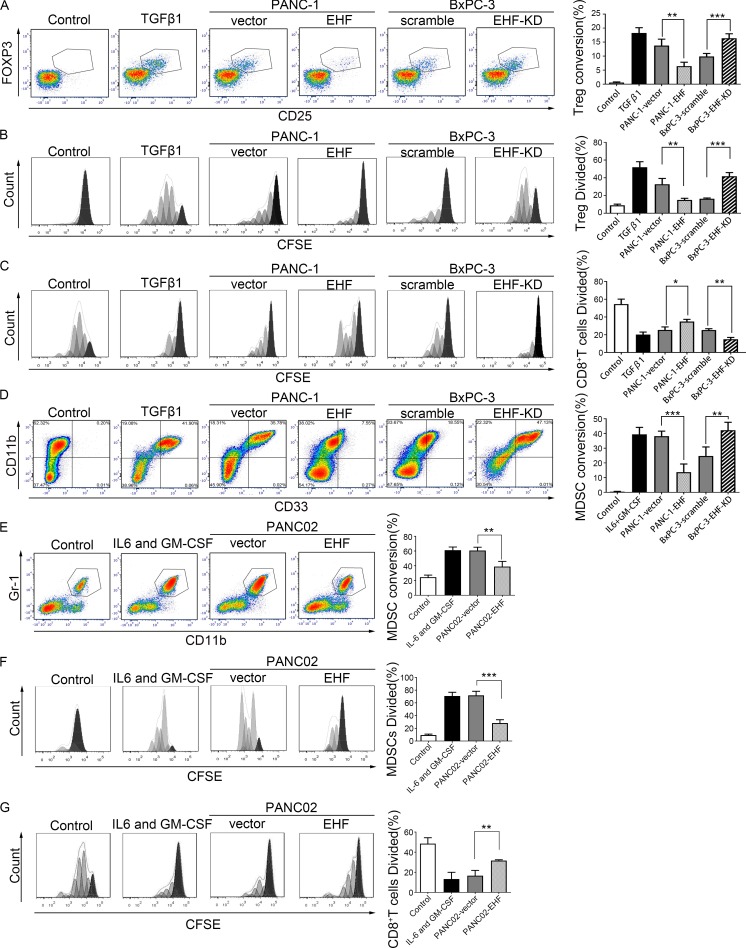
To determine whether tumoral EHF expression regulated the generation of T reg cells, coculture experiments with and without transwell chambers were performed with immune cells and four human pancreatic cancer cell lines. Results for PANC-1-vector/PANC-1-EHF and BxPC-3-scramble/BxPC-3-EHF-KD are shown in Fig. 3 (A–C). Results for AsPC-1-vector/AsPC-1-EHF and CFPAC-1-scramble/CFPAC-1-EHF-KD are shown in Fig. S4 (A–C).
Figure 3.
Tumor cells expressing low levels of EHF induce the conversion, expansion, and function in vitro of T reg cells and MDSCs. (A) Representative plots of T reg cell conversion from CD4+CD25− T cell induced by PANC-1-EHF cells relative to PANC-1-vector and T reg cell conversion induced by BxPC-3-EHF-KD cells relative to BxPC-3-scramble. TGFβ1 was used as positive control. RPMI 1640 was used as negative control. CD25 and FOXP3 expression was determined by flow cytometry after 3 d of coculture (left). Percentage of T reg cell conversion from CD4+CD25− T cells. CD4 was gated (right). (B) Representative histogram of T reg cell proliferation. Flow cytometry was performed after 5 d of coculture by gating on live cells to determine the percentage of T reg cells that diluted CFSE (left). Statistical analysis of the percentage of T reg cell division (right). (C) T reg cell suppression assay compared the percentage of CD8+ T cells division cocultured with PANC-1-EHF (BxPC-3-EHF-KD)–generated T reg cells and PANC-1-vector (BxPC-3-scramble)–generated T reg cells. Statistical analyses of the responder T cells division (right). (D) Human PBMCs were cultured in vitro under different conditions (negative control: RPMI 1640; positive control: IL6 and GM-CSF; experimental group: PANC-1-vector, PANC-1-EHF, BxPC-3-scramble, and BxPC-3-EHF-KD) and analyzed on day 6 to determine the MDSC marker expression by flow cytometry. Representative plots show the surface CD11b+ and CD33+ costaining of MDSCs. HLA-DR− cells were gated (left). The frequency of CD11b+CD33+ cells among HLA-DR− cells was quantified in the bar graph (right). (E) Mouse BMDCs were cultured in vitro under different conditions (negative control: RPMI 1640; positive control: IL6 and GM-CSF; experimental group: PANC02-EHF and PANC02-vector) and analyzed on day 6 to determine the MDSC marker expression by flow cytometry. Representative plots show the surface CD45+CD11b+Gr-1+ costaining of MDSCs (left). The frequencies of CD11b+Gr-1+ cells among CD45+ cells were quantified in the bar graph (right). (F) Representative histogram of MDSC expansion. IL-6 and GM-CSF were used as positive controls. Flow cytometry was performed after 3 d of coculture by gating on live cells to determine the percentage of MDSCs that diluted CFSE (left). Statistical analysis of the percentage of MDSCs division (right). (G) MDSCs induced by PANC02-vector or PANC02-EHF cells were isolated to evaluate their suppressive activities in vitro. Histograms were gated on CFSE+ cells to determine the percentage of CD8+ T cells that diluted CFSE (left). The percentage of division of responder T cells is summarized and quantified in the bar graph (right). The coculture experiments (A–G) were repeated five times independently. Representative data are shown. Data are presented as mean ± SD. Paired Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
For in vitro T reg cell conversion, isolated human CD4+CD25− T cells were cocultured with PDAC-vector or PDAC-EHF/PDAC-EHF-KD cells using transwell chambers. 3 d later, CD25 and Foxp3 expression by CD4+ T cells was determined by flow cytometry. As shown in Fig. 3 A, tumor EHF overexpression strongly inhibited the conversion of CD4+CD25− T cells to T reg cells (PANC-1-vector, 13.76 ± 2.30%; PANC-1-EHF, 6.43 ± 1.39%, P = 0.0012). Conversely, tumor EHF knockdown significantly enhanced T reg cell conversion (BxPC-3-scramble, 9.89 ± 1.07%; BxPC-3-EHF-KD, 16.36 ± 1.61%, P = 0.0002).
For T reg cell proliferation, isolated CD4+CD25+CD127dim T reg cells were cocultured with different tumor cells using transwell chambers. CFSE assays showed that tumoral EHF overexpression strongly inhibited T reg cell proliferation (PANC-1-vector, 32.74 ± 6.56%; PANC-1-EHF, 15.04 ± 1.86%, P = 0.0064), whereas tumoral EHF knockdown significantly increased T reg cell proliferation (BxPC-3-scramble, 16.44 ± 0.76%; BxPC-3-EHF-KD, 41.76 ± 4.13%, P = 0.0001; Fig. 3 B). Direct contact coculture studies obtained similar results as the indirect contact coculture (data not shown).
T reg cell–suppression assays were conducted to determine T reg cell function. The proliferation of targeted CD8+ T cells was measured by CFSE dilution after 5 d of coculture with educated T reg cells. T reg cells induced by EHF-low tumors had greater immune-suppressive effects than cells induced by EHF-high tumor (PANC-1-vector, 25.42 ± 3.39%; PANC-1-EHF, 34.90 ± 2.44%, P = 0.014; BxPC-3-scramble, 25.30 ± 1.56%; BxPC-3-EHF-KD, 14.9 ± 2.09%, P = 0.0018; Fig. 3 C).
Tumoral EHF deficiency induced MDSCs conversion, expansion, and function in vitro
To determine whether tumoral EHF expression regulated the generation of MDSCs, coculture experiments with and without transwell chambers were performed using immune cells and pancreatic cancer cell lines. Results for PANC-1-vector/PANC-1-EHF, BxPC-3-scramble/BxPC-3-EHF-KD, and PANC02-vector/PANC02-EHF are shown in Fig. 3 (D–G). Results for AsPC-1-vector/AsPC-1-EHF and CFPAC-1-scramble/CFPAC-1-EHF-KD are shown in Fig. S4 D.
For MDSC conversion analysis, human peripheral blood mononuclear cells (PBMCs) were cocultured with PDAC at 5:1 using the transwell system. After 6 d of coculture, cells in the lower chamber were collected and stained for markers consistent with the MDSC phenotype. PDACs promoted the differentiation of PBMCs to HLA-DR−CD11b+CD33+ cells. As shown in Fig. 3 D, MDSC conversion was partially reversed when EHF was overexpressed (PANC-1-vector, 38.18 ± 3.44%; PANC-1-EHF, 13.73 ± 5.52%, P = 0.0003). By contrast, PDACs with EHF knockdown significantly increased the proportion of MDSC conversion (BxPC-3-scramble, 24.73 ± 6.13%; BxPC-3-EHF-KD, 42.23 ± 5.37%, P = 0.0086). Bone marrow–derived cells (BMDCs) from C57BL/6 mice were isolated and cocultured with PANC02-EHF or PANC02-vector cells. 6 d later, MDSC proportion was determined. PANC02-vector cells induced a significantly larger proportion of MDSCs than PANC02-EHF cells (PANC02-vector, 60.68 ± 4.50%; PANC02-EHF, 38.88 ± 7.10%, P = 0.0095; Fig. 3 E).
For MDSC expansion, CFSE-labeled purified murine MDSCs were cocultured with PANC02-vector or PANC02-EHF using transwell chambers. CFSE assay showed that tumoral EHF overexpression strongly inhibited MDSC expansion (PANC02-vector, 71.87 ± 6.45%; PANC02-EHF, 28.32 ± 5.24%, P = 0.0002; Fig. 3 F). Direct contact coculture studies yielded similar results as the indirect contact coculture (data not shown).
MDSC-suppression assays were conducted by coculturing with CFSE-labeled CD8+ T cells. MDSCs induced by PANC02-vector cells had greater immune-suppressive effects toward CD8+ T cell proliferation than cells induced by PANC02-EHF cells (CD8+ T divided %: PANC02-vector, 16.60 ± 2.36%; PANC02-EHF, 31.68 ± 0.36%, P = 0.0028; Fig. 3 G).
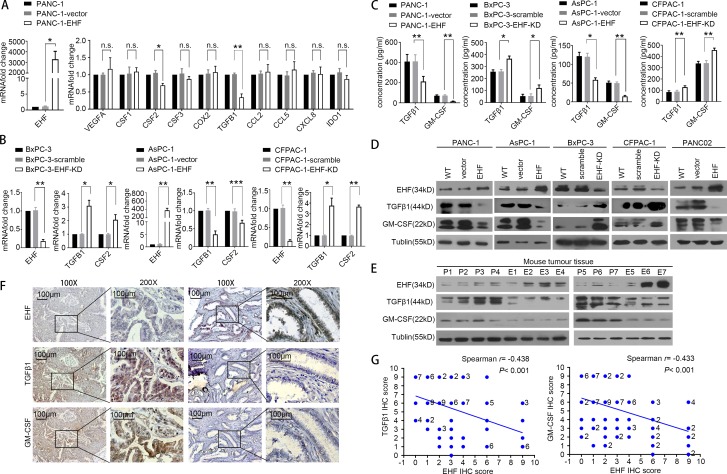
EHF loss is associated with overexpression of the potent immune-suppressive factors TGFβ1 and GM-CSF
As the differences in immune cell education between PDACs-vector/PDACs-scramble and PDACs-EHF/PDACs-EHF-KD cells were not dependent on cell–cell contact, we focused on changes in the secretory proteins that can induce T reg cell and MDSC accumulation. The changes in VEGFA, CSF1, CSF2 (GM-CSF), CSF3, COX2, TGFB1 (TGFβ1), CCL2, CCL5, CXCL8, and IDO1 mRNA levels were analyzed using quantitative PCR (qPCR) in PANC-1-vector and PANC-1-EHF cell lines. Among them, TGFβ1 and GM-CSF were the most significantly down-regulated cytokines (Fig. 4 A). The TGFB1 (TGFβ1) and CSF2 (GM-CSF) mRNAs were negatively regulated by EHF, which was further confirmed in three other cell lines (Fig. 4 B). Western blot and ELISA demonstrated that TGFβ1 and GM-CSF were decreased in the EHF-overexpressing cell lines and increased in the EHF-knockdown cell lines (Fig. 4, C and D). The down-regulation of TGFβ1 and GM-CSF by EHF was also confirmed using mouse subcutaneous tumor tissues (Fig. 4 E). In addition, EHF expression negatively correlated with TGFβ1 and GM-CSF in consecutive PDAC tumor tissues (EHF, TGFβ1: r = −0.438, P < 0.001; EHF, GM-CSF: r = −0.433, P < 0.001; Fig. 4 G).
Figure 4.
EHF negatively regulates TGFβ1 and GM-CSF expression in PDAC. (A) Changes in gene expression profiling upon EHF expression. qPCR was performed to detect the transcriptional change in the secreted factors that can induce T reg cell and MDSC accumulation. RNA was purified from PANC-1, PANC-1-vector, and PANC-1-EHF tumors. Relative expression is shown as fold change relative to GAPDH. (B) qPCR on EHF, TGFB1, and CSF2 was performed in the following cell lines: BxPC3, BxPC3-scramble, and BxPC3-EHF-KD; AsPC1, AsPC1-vector, and AsPC1-EHF; and CFPAC-1, CFPAC-1-scramble, and CFPAC-1-EHF-KD. (C) ELISA of TGFβ1 and GM-CSF in indicated cell lines. qPCR and ELISA experiments (A–C) were repeated three times independently. Paired Student’s t test was used as statistical analysis. *, P < 0.01; **, P < 0.01; ***, P < 0.001; n.s., not significant. (D) Western blot analysis of EHF, TGFβ1, and GM-CSF in indicated cell lines. Experiments were repeated three times independently. Representative data are shown. (E) Expression of EHF, TGFβ1, and GM-CSF in harvested subcutaneous mouse tumor tissues. P, PANC02-vector (n = 7); E, PANC02-EHF (n = 7). Experiments were repeated three times independently. Representative data are shown. (F) Representative IHC images of EHF, TGFβ1, and GM-CSF expression using human PDAC tissue (n = 96). Bars, 100 µm. (G) Spearman rank correlation analysis was used to evaluate the correlation of tumor-specific EHF and TGFβ1/GM-CSF expression (n = 96). The number at the right side of the plots represents the case number; plots without number at the right side represent only one case. All experimental data verified in three independent experiments. Data are presented as mean ± SD.
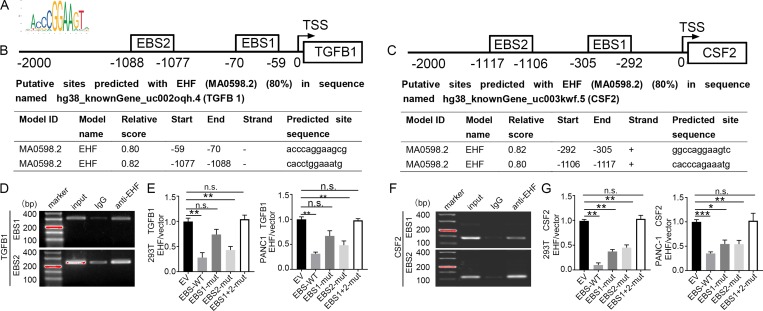
EHF transcriptionally represses TGFβ1 and GM-CSF
Since EHF is a transcription factor, we surveyed the TGFβ1 (TGFB1) and GM-CSF (CSF2) gene promoter regions for potential EHF binding sites (EBSs; Fig. 5 A). Computational analysis showed two high-confidence EBSs corresponding to the promoter regions of TGFB1 and CSF2 in the JASPAR database (Fig. 5, B and C; Khan et al., 2018). To evaluate whether EHF directly binds to the promoters of TGFB1 and CSF2, a chromatin immunoprecipitation (ChIP) assay was performed using the PANC-1 cell line. In chromatin fractions pulled down by the anti-EHF antibody, we detected EBS1 and EBS2 in both the TGFB1 promoter (Fig. 5 D) and the CSF2 promoter (Fig. 5 F) by PCR, indicating the binding of EHF to the promoter of these two genes.
Figure 5.
EHF directly down-regulates the expression of TGFβ1 and GM-CSF in PDACs by binding to the EBS in their gene promoters. (A) EHF-scanned motif logo. (B and C) Predicted ETS binding sites (EBS) in the human TGFB1 (B) and CSF2 (C) promoters. Position relative to the transcription start site of the gene, sequence, and corresponding scores. (D and F) Binding of EHF to the promoters of TGFB1 (D) and CSF2 (F) in PANC-1-EHF determined by ChIP. Experiments were repeated three times independently. Representative data are shown. (E and G) The HEK293 (left) and PANC-1 cells (right) were transfected with either vector control or pCDH-EHF in conjunction with the luciferase reporter pGL3-empty vector, WT pGL3-TGFB1/CSF2-promoter, or pGL3-TGFB1/CSF2-promoter with EBS1 mutation (EBS1-mut), EBS2 mutation (EBS2-mut), or EBS1+2 mutation (EBS1+2-mut) vector. Results are expressed as fold induction relative to that of the corresponding cells transfected with the control vector after normalization of firefly luciferase activity according to Renilla luciferase activity. Experiments were repeated three times independently. Data are presented as mean ± SD. Paired Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s. not significant.
To further determine whether binding of EHF to the TGFB1/CSF2 promoter suppressed gene transcription, we constructed four TGFB1 and four CSF2 luciferase promoter vectors: pGL3-TGFB1/CSF2-EBS-WT, pGL3-TGFB1/CSF2-EBS1-mutation, pGL3-TGFB1/CSF2-EBS2-mutation, and pGL3-TGFB1/CSF2-EBS1+2-mutation (Table S4). Then, we transfected them with or without an EHF expression vector (pCDH-EHF) into HEK293 and PANC-1 cells. pGL3-empty vector was used as the control. Luciferase analysis demonstrated that EHF overexpression significantly decreased TGFB1 promoter activity at the WT promoter vector (P < 0.01) and pGL3-TGFB1-EBS2-mutation in HEK293 and PANC-1 cells (P < 0.01) but not in pGL3-TGFB1-EBS1-mutation and pGL3-TGFB1-EBS1+2-mutation (Fig. 5 E). EHF overexpression significantly decreased CSF2 promoter activity at the WT promoter vector (P < 0.01), pGL3-CSF2-EBS1-mutation (P < 0.05), and pGL3-CSF2-EBS2-mutation (P < 0.01) but not at the pGL3-CSF2-EBS1+2-mutation in HEK293 and PANC-1 cells (Fig. 5 G). Collectively, these data indicated that EHF directly suppressed TGFB1 (mainly through EBS1) and CSF2 (mainly through both EBS1 and EBS2) transcription in PDAC by binding to their promoters.
Tumor EHF deficiency induces immune suppression through TGFβ1 and GM-CSF
We then evaluated whether EHF deficiency induced immune suppression through TGFβ1 and GM-CSF. In vivo and in vitro experiments were performed using TGFβ1 neutralization, GM-CSF neutralization, and combined GM-CSF and TGFβ1 neutralization.
Human CD4+CD25− T cells were cultured in the T reg cell–induction system; purified CFSE-labeled T reg cells were cultured in the T reg cell proliferation model as previously described. No significant differences were observed in T reg cell induction rate (PANC1-vector+blockage, 2.53 ± 0.75%; PANC1-EHF+blockage, 2.43 ± 0.97%, P = 0.58; Fig. 6 A, left) and T reg divided proportion (PANC1-vector+blockage, 11.24 ± 1.68%; PANC1-EHF+blockage, 12.08 ± 2.42%, P = 0.59; Fig. 6 A, right) between PANC1-vector and PANC1-EHF cells after TGFβ1 neutralization. Similarly, when mouse BMDCs were cocultured with tumor cells in the MDSC induction model, mouse isolated MDSCs were cocultured with tumor cells in the MDSC expansion model. No significant differences were observed in MDSC conversion rate (PANC02-vector+blockage, 21.76 ± 3.04%; PANC02-EHF+blockage, 23.1 ± 2.19%, P = 0.12; Fig. 6 B, left) and MDSC divided proportion (PANC02-vector+blockage, 21.28 ± 3.50%; PANC02-EHF+blockage, 23.44 ± 5.56%, P = 0.52; Fig. 6 B, right) between PANC02-vector and PANC02-EHF group after GM-CSF neutralization.
Figure 6.
Tumor EHF deficiency induces immune suppression through TGFβ1 and GM-CSF. (A) Statistical analysis of CD4+CD25− T cells to T reg cell conversion (left) and T reg cell proliferation (right) induced by PANC-1-EHF cells relative to PANC-1-vector with or without TGFβ1 neutralization. CD4+CD25− T cells cultured in RPMI 1640 were used as negative controls. (B) Statistical analysis of mouse BMDC to MDSC conversion and MDSC expansion induced by PANC02-vector cells relative to PANC02-EHF with or without GM-CSF neutralization. BMDCs cultured in RPMI 1640 were used as negative controls. Experiments were repeated three times independently. Paired Student’s t test was used for statistical analysis. (C) PANC02-vector or PANC02-EHF was injected subcutaneously in a Matrigel plug containing neutralizing anti–GM-CSF mAb and anti-TGFβ1 mAb (vs. isotype IgGs as control) into the flanks of C57BL/6 mice. After 8 d, TGFβ1 and GM-CSF antibodies or isotype IgGs were intratumorally injected at 20 µg/mouse two times a week. (D and E) Representative dot plots of the proportion of tumor infiltration by T reg cells (D, left) and MDSCs (E, left) in the PANC02-vector and PANC02-EHF groups with or without combined TGFβ1 and GM-CSF depletion. Statistical analysis of the proportion of tumor-infiltrating T reg cells (D, right) and MDSCs (E, right) in the PANC02-vector and PANC02-EHF groups with or without combined TGFβ1 and GM-CSF depletion. The mouse experiments were repeated three times independently, using seven mice per experimental group. Representative data are shown. Nonpaired Student’s t test was used for statistical analysis. (F) Schematic of the roles of EHF in tumor immune modulation. EHF decreased the tumor-infiltrating T reg cells and MDSCs by transcriptionally suppressing the expression of TGFβ1 and GM-CSF. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
To determine whether EHF modulated the immune microenvironment through TGFβ1 and GM-CSF in vivo, mouse models for TGFβ1 and GM-CSF neutralization within the local tumor microenvironment were established as shown in Fig. 6 C. On the 28th day after tumor cells were implanted, all mice were sacrificed for tumor harvesting and flow cytometry. No significant differences were observed in the proportion of tumor-infiltrating T reg cells (PANC02-vector+blockage, 0.60 ± 0.095%; PANC02-EHF+blockage, 0.59 ± 5.560.19%, P = 0.82; Fig. 6 D) and MDSCs (PANC02-vector+blockage, 11.83 ± 2.30%; PANC02-EHF+blockage, 10.22 ± 1.72%, P = 0.16; Fig. 6 E) after TGFβ1 plus GM-CSF neutralization.
Furthermore, T reg cell/MDSC conversion and expansion blocking experiments were conducted using cell lines with genetic knockdown of TGF-B1 and CSF2 in vitro (Fig. S5). In vivo blocking experiments were conducted using murine PANC02-vector and PANC02-EHF cell lines that genetically knock down both TGF-B1 and CSF2. Related scramble cell lines were used as control. The consequences of EHF loss on the tumor microenvironment can be reversed by genetically knocking down TGF-B1 and CSF2 (Fig. S5). Taken together, our data support that EHF loss in PDAC relieves the transcriptional suppression of TGFβ1 and GM-CSF, which in turn promotes the conversion and expansion of T reg cells and MDSCs, respectively (Fig. 6 F).
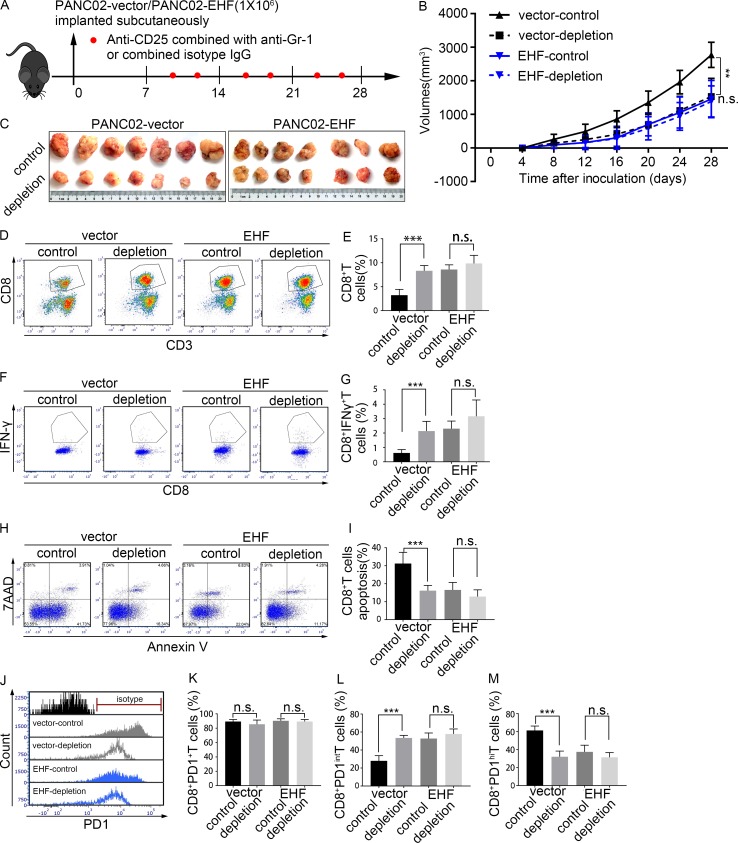
EHF-low tumors benefit from T reg cell and MDSC depletion therapy
An in vivo study was conducted to evaluate the treatment efficacy of T reg cell and MDSC depletion in PANC02-vector and PANC02-EHF tumors (Fig. 7 A). Tumor growth was significantly reduced following T reg cell and MDSC depletion in PANC02-vector tumors (P = 0.001; Fig. 7, B and C). When the tumors were harvested, tumor volumes of the PANC02-vector-control group and PANC02-vector-depletion group were 2,774.00 ± 374.17 and 1,533.32 ± 541.75 mm3 (P < 0.001), respectively. No significant difference was observed in the tumor volume between the PANC02-vector-depletion group and the PANC02-EHF-control group (P = 0.197). Moreover, the efficacy of T reg cell and MDSC depletion was not significant in PANC02-EHF tumor (P = 0.738; Fig. 7, B and C). With T reg cell and MDSC depletion, the proportions of CD8+ T cells (Fig. 7, D and E), CD8+IFNγ+ T cells (Fig. 7, F and G), and PD1intCD8+ T cells (Fig. 7, J and L) were significantly increased, and the proportions of apoptotic CD8+ T cells (Fig. 7, H and I) and PD1hiCD8+ T cells (Fig. 7, J and M) were significantly decreased in the PANC02-vector tumor (P < 0.001). The percentage of total PD1+CD8+ T cells (PD1hiCD8+ T cells plus PD1intCD8+ T cells; Fig. 7, J and K) was unchanged after depletion. However, in PANC02-EHF tumor, no significant differences were observed in the percentage of CD8+IFNγ+ T cells (Fig. 7, H and I), PD1intCD8+ T cells (Fig. 7, J and L), PD1hiCD8+ T cells (Fig. 7, J and M), total PD1+CD8+ T cells (Fig. 7, J and K), or apoptotic CD8+ T cells (Fig. 7, F and G) after T reg cell and MDSC depletion. No significant difference was observed between the PANC02-EHF-control and PANC02-vector depletion. Elevated tumoral EHF could achieve an effect similar to T reg cell and MDSC depletion therapy.
Figure 7.
EHF-low tumors can benefit from T reg cell and MDSC depletion treatment. (A) Anti-mouse CD25 antibody combined with anti-mouse Gr-1 antibody (or combined isotype control) was injected intraperitoneally two times a week, 7 d after tumor inoculation (red plots represent the time points of drug administration). T reg cell and MDSCs depletion therapy efficacy was evaluated in PANC02-vector and PANC02-EHF tumors, respectively. Tumor volumes were measured every 4 d using calipers. (B) Tumor growth curves of the four groups. Repeated measure two-way ANOVA (time × tumor volume) and post hoc analysis were used to test mouse tumor growth between groups. (C) Effects of T reg cell and MDSC depletion therapy on tumor growth in the PANC02-vector and PANC02-EHF groups. Tumors at the end point of the experiment are also shown (left, PANC02-vector; right, PANC02-EHF). (D and E) Representative plots (D) and statistical analyses (E) of tumor-infiltrating CD8+ T cells in tumors that received T reg cell and MDSCs depletion therapy or isotype IgG (left, vector group; right, EHF group). (F and G) Representative plots (F) and statistical analyses (G) of tumor-infiltrating IFNγ+CD8+ T cells. (H and I) Representative plots (H) and statistical analyses (I) of tumor-infiltrating CD8+ T cell apoptosis. (J) Histogram of tumor infiltration by PD1+CD8+ T cells, PD1intCD8+ T cells, and PD1hiCD8+ T cells in the four groups. (K–M) Percentage of PD1+CD8+ T cells, PD1intCD8+ T cells, and PD1hiCD8+ T cells in tumors that received T reg cell and MDSC depletion therapy or isotype IgG. Data are presented as mean ± SD. Nonpaired Student’s t test was used for statistical analysis; **, P < 0.01, ***, P < 0.001; n.s., not significant. All mouse experiments were repeated three times independently, using seven mice per experimental group. Representative data are shown.
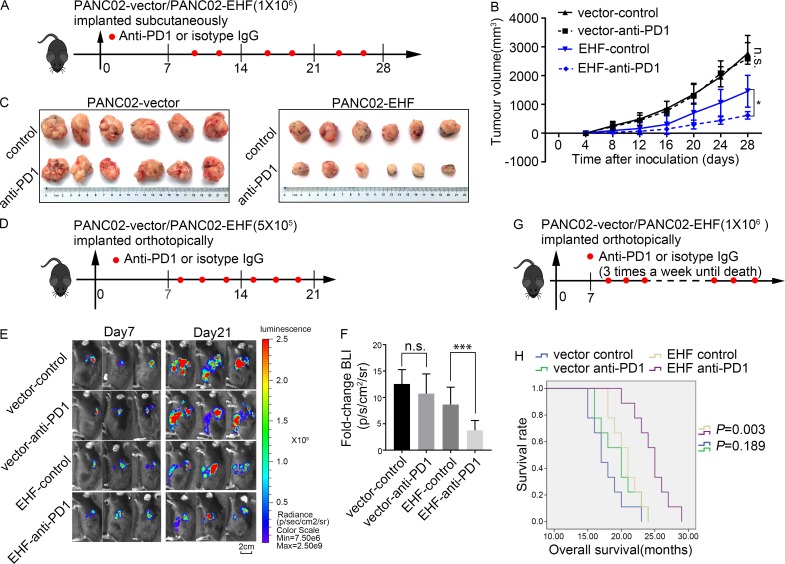
Potential role for EHF as a marker for single anti-PD1 therapy
As tumors with high EHF expression exhibit relatively high infiltration by CD8+ T cells and decreased accumulation of T reg cells and MDSCs, we used a subcutaneous (Fig. 8, A–C) and orthotopic (Fig. 8, D–G) mouse tumor model to compare the efficacy of anti-PD1 therapy in PANC02-vector and PANC02-EHF tumors.
Figure 8.
Potential roles for EHF as a marker for anti-PD1 therapy. (A) PANC02-vector or PANC02-EHF was subcutaneously inoculated at day 0. Tumor volume was measured every 4 d using calipers. Anti-PD1 antibody or isotype IgG was injected intraperitoneally two times a week (red plots represent time points of drug administration). Anti-PD1 therapy efficacy was evaluated in PANC02-vector and PANC02-EHF tumors. (B and C) Effects of anti-PD1 therapy on tumor growth in the PANC02-vector and PANC02-EHF groups. (B) Tumor growth curves of the four groups. Repeated measure two-way ANOVA (time × tumor volume) and post hoc analysis were used to test mouse tumor growth between groups. *, P < 0.05; n.s., not significant. n = 6 per group. (C) Tumors at the endpoint of the experiment are shown (left, PANC02-vector; right, PANC02-EHF). (D) C57BL/6 mice were orthotopically injected with 5 × 105 luciferase-expressing PANC02-vector cells or luciferase-expressing PANC02-EHF cells in Matrigel. Anti-PD1 antibody or isotype IgG was injected intraperitoneally three times a week (red plots represent time points of drug administration). Tumor growth was assessed by BLI at day 7 before treatment and at day 21 after six treatments. (E) Three representative bioluminescent images of the four groups on days 7 and 21 after tumor implantation. (F) Statistical analysis of the fold change of BLI after treatment (BLI on day 21 to BLI on day 7). ***, P < 0.001 by nonpaired Student’s t test (n = 9 per group). (G) C57BL/6 mice were orthotopically injected with 1 × 106 luciferase-expressing PANC02-vector cells or luciferase-expressing PANC02-EHF cells in Matrigel. Anti-PD1 antibody or isotype IgG was injected intraperitoneally three times a week until death (red plots represent time points of drug administration). (H) Kaplan–Meier survival curves with log-rank test for significance between isotype control and anti-PD1 therapy (PANC02-vector control vs. PANC02-vector anti-PD1, P = 0.189; PANC02-EHF control vs. PANC02-EHF anti-PD1, P = 0.003). All mouse experiments were repeated three times independently, using six mice per experimental group in subcutaneous tumor model and nine mice per experimental group in orthotopic tumor model. Representative data are shown. Data are presented as mean ± SD.
In the subcutaneous C57BL/6 tumor mouse model, the tumor’s growth was significantly inhibited by anti-PD1 therapy in PANC02-EHF tumor (P = 0.017; Fig. 8 B). When tumors were harvested, the tumor volumes of PANC02-EHF-control and PANC02-EHF-anti-PD1 were 1,457.39 ± 617.21 and 554.64 ± 129.94 (P = 0.0021). However, no significant difference in tumor volume was observed between PANC02-vector-control and PANC02-vector-anti-PD1 (P = 0.784; Fig. 8, B and C).
In the orthotopic C57BL/6 tumor mouse model, tumors were randomly divided into treatment and control groups on the day 7 after inoculation. After receiving anti-PD1 treatment or the isotype control six times, tumor growth was significantly inhibited by anti-PD1 therapy in the PANC02-EHF group. The bioluminescent imaging (BLI) on day 21 normalized to day 7 was 3.76 ± 1.87 in the PANC02-EHF-anti-PD1 group, which was significantly lower than in the PANC02-EHF-control group (8.66 ± 3.29, P = 0.013). The normalized BLI was not significantly different in the PANC02-vector-control group (12.56 ± 2.72) and the PANC02-vector-anti-PD1 group 10.72 ± 3.73 (P = 0.25; Fig. 8, E and F). In the orthotopic tumor model, the mice in the PANC02-EHF anti-PD1 group survived significantly longer than those in the PANC02-EHF-control group (P = 0.003). However, no improvement in survival was observed in PANC02-vector tumor after receiving anti-PD1 treatment (Fig. 8, G and H). Our data indicate that restoration of EHF expression in PDAC may sensitize these tumors to anti-PD1 treatment, and that EHF expression levels in PDAC could be an indicator for anti-PD1 therapy efficacy.
Discussion
Antibodies targeting the PD1/PD-L1 axis have been proven to be an effective immuno-oncology strategy (Foley et al., 2016; Steuer and Ramalingam, 2018); however, its efficacy in PDAC remains uncertain (Feng et al., 2017). The main reason for the resistance of anti-PD1/PD-L1 therapy includes tumor cell–intrinsic and –extrinsic factors (Foley et al., 2016; Feng et al., 2017; Sakellariou-Thompson et al., 2017). Tumor cell–intrinsic factors include the activation of the MAPK and WNT/β-catenin signaling pathways, the loss of PTEN, IFNγ signaling pathways, and tumor mutation burden. Tumor cell–extrinsic mechanisms include hyperinfiltration of T reg cells, MDSCs, and T cells with high expression of inhibitory immune checkpoint molecules, such as mucin-domain containing protein 3 (TIM-3), lymphocyte activation gene 3 (LAG-3), and V-domain Ig suppressor of T cell activation (VISTA; Johnston et al., 2015; Maleki Vareki et al., 2017; Taylor et al., 2017; Steuer and Ramalingam, 2018).
PDAC was characterized by prominent stroma with abundant immunosuppressive cells, including T reg cells, MDSCs, and TAMs, and low numbers of tumor-infiltrating CD8+ T cells, all of which correlated with compromised immune surveillance, poor prognosis, and low response to single anti-PD1/PD-L1 treatment (Sideras et al., 2014; Chang et al., 2016; Melstrom et al., 2017). Patients who did not benefit from the single regimen may benefit from combined therapy of anti-PD1/PD-L1 treatment and radiotherapy (Wang et al., 2017), MDSC depletion (Steele et al., 2016), T reg cell depletion (Taylor et al., 2017), TAM depletion (Borgoni et al., 2017), anti-BAG3 (Iorio et al., 2018), and anti-CD4 (Ueha et al., 2015). To identify a predictive marker for responders to anti-PD1/PD-L1 therapy or combined immunotherapy in preclinical studies, the precise selection of the immunotherapy strategy is crucial (Steuer and Ramalingam, 2018; Teng et al., 2018).
EHF is a member of the highly diverse ETS superfamily, consisting of both transcription activators and repressors that mediate growth factor signaling and regulate gene expression (Fossum et al., 2017). Previous studies have demonstrated that EHF inhibits tumor invasion, metastasis, and mesenchymal phenotype (Albino et al., 2012, 2016a,b; Dallavalle et al., 2016; Zhao et al., 2017). Our group first demonstrated EHF as a tumor-suppressing transcription factor that directly inhibits PDAC metastasis by up-regulating E-cadherin. In addition, our current research further identified EHF deficiency as the primary molecular event that induced T reg cell and MDSC accumulation and caused a decrease in CD8+ T cell infiltration.
In the C57BL/6 tumor model, the PDAC-EHF group (PANC02-EHF group/KPC-EHF group) has decreased numbers of tumor-infiltrating T reg cells and MDSCs and an increased number of functional CD8+ T cells. Results from mouse models were consistent with the observations of paraffin sections from tumor tissues from a 96-case retrospective cohort and fresh tissues collected from a 45-case prospective cohort. In vitro coculture studies and in vivo experiments verified that tumoral EHF deficiency induced the conversion, expansion, and function of T reg cells and MDSCs, all of which significantly changed the makeup of the tumor-infiltrating immune cell content. As a direct-contact coculture study and an indirect coculture study using transwells yielded the same results, we compared the secreted cytokines between PANC-1-vector and PANC-1-EHF cells, which are critical in T reg cell and MDSC accumulation, by using qPCR. The downstream effects of EHF on TGFβ1 (TGFB1) and GM-CSF (CSF2) were the most significant among the cytokines that induce T reg cell and MDSC accumulation; this significant effect was further confirmed at the protein and mRNA levels in four stably transfected PDAC cell lines. We found several putative EHF binding motifs in the promoter regions of CSF2 and TGFB1. ChIP and dual-luciferase assays revealed that EHF directly bound to the promoter regions of TGFB1 and CSF2 to suppress transcription. Our study was the first to identify EHF deficiency as the primary molecular event that triggered T reg cell and MDSC accumulation in PDAC by directly regulating TGFβ1 and GM-CSF expression. TGF-β1 is an indispensable cytokine in fostering T reg cell accumulation, whereas T reg cells significantly suppressed CD8+ T cell function (Wörmann et al., 2014). GM-CSF is a key cytokine in the induction of MDSC accumulation and is often overexpressed by PDAC cells. In turn, MDSCs suppressed the function of CD8+ T cells, preventing them from recognizing and clearing transformed pancreatic ductal cells (Stromnes et al., 2014; Paschall et al., 2015; Sahin et al., 2017). In our study, we found that TGFβ1 and GM-CSF were the main mechanisms for the function of EHF in tumor immune modulation. However, some other studies that focused on tumoral EHF may also hold important clues to its immune modulation functions. In prostate cancer, EHF deficiency induces stem-like properties by directly suppressing IL6 secretion (Albino et al., 2016b). IL6 is a critical cytokine that can induce MDSC conversion and activation. In primary human bronchial epithelial cells, EHF depletion altered the expression of cytokines important for neutrophil migration (Fossum et al., 2017). When TGFβ1 and GM-CSF were neutralized or genetically knocked down, the effect of EHF deficiency could be fully reversed. EHF overexpression achieved a similar effect of TGFβ1 and GM-CSF knockdown.
We used the combination of anti-CD25 and anti-Gr1 to deplete T reg cells and MDSCs. In PANC02-vector tumors, the tumor volumes were significantly inhibited by T reg cell plus MDSC depletion. In addition to changes in tumor volume, we also observed significant elevation in the proportion of tumor-infiltrating CD8+ T cells and a decrease in the apoptosis rate of CD8+ T cells. With depletion, the total percentage of PD1+CD8+ T cells remained unchanged; however, the PD1hiCD8+ T cell percentage was significantly decreased, and the PD1intCD8+ T cell percentage was significantly elevated. The proportion of PD1intCD8+ T cells was a determinant of anti-PD1 therapy efficacy; however, PD1hiCD8+ T cells had a hyperexhausted phenotype that could not be reversed by anti-PD1 therapy (Ngiow et al., 2015). No significant difference was observed between PANC02-EHF-control and PANC02-vector depletion. As a result, we propose that elevated tumoral EHF may achieve a similar effect as T reg cell and MDSC depletion treatments. Drugs elevating tumoral EHF may also decrease both tumor-infiltrating T reg cells and MDSCs. Moreover, the proportion of tumor-infiltrating T reg cells and MDSCs in the PANC02-EHF group is already low. Thus, no significant efficacy of T reg cell plus MDSC depletion therapy was observed in PANC02-EHF tumor. EHF could be used as a marker predicting decreased tumor infiltration by T reg cells, MDSCs, and hyperexhausted CD8+ T cells but with an increasing accumulation of PD1intCD8+ T cells. Treatment efficacy of anti-PD1 therapy was evaluated in PANC02-EHF tumors and PANC02-vector tumors using an orthotopic and subcutaneous tumor model of pancreatic cancer in immunocompetent C57BL/6 mice. The PANC02-EHF tumor-bearing mice significantly benefited from anti-PD1 therapy; however, no significant difference was observed between PANC02-vector tumors that received anti-PD1 and those that received isotype IgG. EHF is a promising biomarker to identify patients who are suitable for single anti-PD1 treatment in our preclinical study.
Various combination regimens have been tested in preclinical studies in order to treat anti-PD1/PD-L1 resistance, and preliminary therapeutic effects have been achieved. For tumors enriched with T reg cells, T reg cell depletion combined with immune checkpoint inhibition can enhance the immune response, impair tumor growth, and prolong survival (Ueha et al., 2015; Taylor et al., 2017). Similarly, when tumor trafficking of MDSCs was inhibited by anti-CXCR2 therapy, anti-PD1 treatment could induce significant anti-tumor effects (Highfill et al., 2014; Chesney et al., 2017; Najjar et al., 2017). For tumor infiltration with T reg cells, MDSCs, or other immune suppression cells, a single anti-PD1/PD-L1 therapy may not be a wise choice. The subgroup with overexpressed EHF has relatively elevated numbers of PD1intCD8+ T cells but decreased T reg cells and MDSCs, thereby indicating a relatively good response to single anti-PD1 therapy. For a tumor with EHF deficiency, a combined regimen of T reg cell plus MDSC depletion and anti-PD1 therapy could be considered. The identification of the function of tumoral EHF could potentially lead the way for anti-PD1/PD-L1 therapy to enter the era of precision medicine (Steuer and Ramalingam, 2018; Teng et al., 2018).
There are some limitations in our study. In the tissue IF experiments of the 96-case retrospective cohort, we used only single markers for T reg cells (FOXP3) and MDSCs (CD33). To make up for the shortcomings of the single markers, we conducted a 45-case prospective cohort and tested the proportion of tumor-infiltrating immune cells by multiparameter flow cytometry. A large prospective cohort using multiparameter evaluation methods is still needed. Experiments using patient-derived xenograft tumor models and clinical trials are still needed to verify our findings.
Our study is the first to identify that EHF transcriptionally inhibited the expression of TGFβ1 and GM-CSF, which are the two critical suppressive cytokines involved in the induction of T reg cells and accumulation of MDSCs. Tumoral EHF can be used as a promising biomarker to evaluate the immune microenvironment status of PDAC and to screen for patient response to anti-PD1 therapy.
Materials and methods
Patients and tissue samples
Data from 96 patients who had received radical surgery R0 resection with histological diagnosis of PDAC at the Tianjin Medical University Cancer Institute and Hospital, China, from October 2010 to February 2013 were retrospectively collected in this study. Until the last follow-up date of January 30, 2015, 14 patients were lost to follow-up (14.58%). Clinicopathological data of the 96 consecutive PDAC patients, including age, sex, tumor size, regional lymph node status, TNM stage, and differentiation, were obtained. None of the patients had received neoadjuvant chemotherapy or radiotherapy before tissue samples were collected. Systemic gemcitabine-based chemotherapy was given to all the patients. From June 2016 to June 2017, 45 consecutive cases of fresh PDAC tissues were prospectively collected during operation. The tissue mass was cut into two parts: one was prepared into single-cell suspension for flow cytometry, and the other was used for IHC detection of EHF expression. The use of these specimens and patients’ information was approved by the Ethics Committee of the Tianjin Medical University Cancer Institute and Hospital.
Cell culture and transfection
Human PDAC cell lines PANC-1, AsPC-1, BxPC-3, and CFPAC-1 were obtained from the Type Culture Collection Committee of the Chinese Academy of Sciences. The murine PDAC cell line PANC02 was a gift from Prof. Shengyu Yang (Moffitt Cancer Center, Tampa, FL). The PDA cell line from KPC (LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre) mouse was a gift from Dr. Tingbo Liang (Department of Surgery, the Second Affiliated Hospital, Zhejiang University, China). Cell lines were authenticated through the short tandem repeat analysis method. The KPC cell line was validated by RT-PCR to assess for Cre-mediated recombination of the mutant Kras and Trp53 alleles. Primer sequences to detect recombined Kras and Trp53 loci are as follows: Kras, forward 5′-GTCTTTCCCCAGCACAGTGC-3′ and reverse 5′-CTCTTGCCTACGCCACCAGCTC-3′; and Trp53, forward 5′-AGCCTGCCTAGCTTCCTCAGG-3′ and reverse 5′-CTTGGAGACATAGCCACACTG-3′. Mycoplasma contamination was excluded in these cell lines. Cells were cultured at 37°C in a humidified atmosphere of 95% air and 5% CO2 with DMEM, RPMI-1640, or IMDM basic medium supplemented with 10% FBS as medium. KPC cell lines used for implantation studies were used at <10 passages.
Human EHF cDNA (NM_012153.5) was cloned into a pCDH plasmid expression vector (pCDH-EHF). pCDH vector was used as control (Zhao et al., 2017). For mouse Ehf stable expression cell lines, mouse Ehf cDNA (NM_007914.3) was cloned into a pLV plasmid expression vector (pLV-Ehf). pLV vector was used as control. Lentiviral infections were performed according to standard procedures. For the stable knockdown cell lines, shRNA sequences were designed by Sigma-Aldrich shRNA designer (Table S5). Three recommended sequences for EHF genes were synthesized and cloned into the pLVi-shRNA-bsd Vectors (Biosettia). Blasticidin (InvivoGen) was added for selection. For the TGFB1/Tgfb1 stable knockdown pancreatic cancer cell lines, three recommended sequences for TGFB1/Tgfb1 genes were synthesized and cloned into the pLV-RNAi-Zeocin (Biosettia). Zeocin (InvivoGen) was added for selection. To generate murine pancreatic cancer cell lines with stable knockdown of Csf2, three shRNAs targeting murine Csf2 were designed and cloned into pLKO.1-Hygro (Addgene 24150). Hygromycin was added for selection. Scramble sequences were transfected to use as control. Among the three shRNA sequences, the most efficient one was used for relevant assays.
IHC
IHC analysis of the PDAC tissue for EHF (Thermo Fisher Scientific; PA5-30716), FOXP3 (eBioscience; PCH101), CD33 (Abcam; ab199432), CD8 (ZSGB-BIO; ZA-0508), TGFβ1 (Abcam; ab92486), GM-CSF (Proteintech; 17762-1-AP) expression, TGFβ1 (Abcam; ab92486), and GM-CSF (Proteintech; 17762-1-AP) were performed using a DAB substrate kit (Maxin). The score was determined using the following criteria. The final staining scores were determined by multiplying the staining intensity scores by the staining extent scores. The final score ranged from 0 to 9. The staining intensity score was scored as 0, negative; 1, low; 2, medium; and 3, high. The staining extent score was scored as 0, 0% stained; 1, 1–25% stained; 2, 26–50% stained; and 3, 51–100% stained. Five random fields (100× magnification) were evaluated under a light microscope. IHC score was determined by two independent pathologists who were blinded to the patients’ clinical features and outcomes.
Tissue IF
Three sets of 96 sequential PDAC tissues were used for immunologic assessment of EHF and FOXP3/CD33/CD8 expression. Rabbit anti-human EHF in conjugation with goat anti-rabbit Alexa Fluor H555 (Invitrogen) and mouse anti-human FOXP3 (Abcam; ab22510)/mouse anti-human CD33 (Abcam; ab11032)/mouse anti-human CD8 (Abcam; ab199016) in conjugation with goat anti-mouse Alexa Fluor H488 (Invitrogen) were used.
Anti-fade DAPI (Southern Biotech) solution was employed to stain nuclei. Isotype controls were used for all assays. Slides were viewed with Leica microscopy. Tissue IF scores were determined by two independent pathologists who were blinded to the patients’ clinical features, outcomes, and IHC score of EHF. Five random fields (200× magnification) were evaluated under microscope. FOXP3+, CD33+, and CD8+ cells in tumor stroma were used to represent T reg cells, MDSCs, and CD8+ T cells. T reg cells, MDSCs, and CD8+ T cells were counted under a microscope. T reg cells >20/HPF, MDSCs >10/HPF, and CD8+ T cells >20/HPF were defined as T reg high, MDSC high, and CD8+ T cell high infiltration, respectively. EHF-positive cells/HPF was used to evaluate EHF expression. The count of EHF-positive points per field ranged from 0 to 251, mean ± SD 50.89 ± 65.04. EHF-positive cells/HPF >50.89 was considered the EHF-high group; EHF-positive cells/HPF <50.89 was considered the EHF-low group. Furthermore, mean EHF intensity was quantified using TissueQuest software 6.0 (TissueGnostics). Tumor areas were manually outlined to exclude stromal nuclei. DAPI was used to identify nuclei. EHF was then measured on a cell-nucleus–based mode. The mean EHF intensity ranged from 0.06 to 189.26, median 12.03, mean 39.74 ± 52.01. Mean EHF intensity >39.74 was considered the EHF-high group; EHF-positive cells/HPF <39.74 was considered the EHF-low group.
Flow cytometry
Harvested tumors were processed into the single-cell suspensions with 1 mg/ml collagenase, 2.5 U/ml hyaluronidase, and 0.1 mg/ml DNase. Human tumor-infiltrating CD8+ T cells (CD8+), T reg cells (CD4+, CD25+, and FOXP3+), MDSCs (HLA-DR−, CD33+, and CD11b+) were detected. Mouse tumor-infiltrating CD8+ T cells (CD3+ and CD8+), T reg cells (CD4+, CD25+, and FOXP3+), MDSCs (CD45+, CD11b+, and Gr-1+), TAM (CD45+CD11b+F4/80+), CD8+ T cell phenotype (IFNγ+, PD1+, and Tim3+), and CD8+ T cell apoptosis (annexin V+ and 7-AAD) were analyzed. For intracellular IFNγ staining, 1 × 106 cells were stimulated with PMA (5 ng/ml) and ionomycin (500 ng/ml; Sigma-Aldrich) in the presence of GolgiStop (BD Biosciences) for 4 h, followed by surface and intracellular staining. Isotype controls were used as negative controls. The data were analyzed using FCS Express 6. Related antibodies are listed in Table S6.
Sorting of TIL subsets
A total of 1 × 106 PANC02 murine tumor cells were subcutaneously injected into each mouse to establish tumors. 21 d later, tumors were harvested sterilely and filtered through a 200-µm cell strainer. TILs were enriched through mouse lymphocyte separation medium (Dakewe Biotech; DKW33-R0100) according to the production protocol. TILs were labeled with CD8-APC (BioLegend; 100712) and PD1-PE (BioLegend; 135206) antibodies for CD8+PD1int and CD8+PD1hi cell sorting by flow cytometry.
In vitro functional restoration assay
To assess the restoration of the cytokine-secreting capacity of CD8+PD1int and CD8+PD1hi TILs, IFNγ-ELISPOT assay was performed according to the production protocol with 200,000 CD8+ T cells per well. 1 µg/ml anti-CD3, 5 µg/ml anti-CD28, and 100 IU/ml IL-2 were added in the presence of blocking antibodies for PD1 or isotype control. After 36-h culture, plates were developed. IFNγ spot-forming units (SFUs) were counted by ELISPOT reader. IFNγ+ T cells were quantified by subtracting the mean IFNγ-SFUs in negative control wells from the mean SFUs in experimental wells and expressed as IFNγ-SFUs/106 cells. RPMI 1640 was used as the negative control.
To assess the restoration of the functional capacity of CD8+PD1int and CD8+PD1hi TILs, the isolated CD8+PD1int T cells and CD8+PD1hi T cells were labeled with Cell Trace Reagent CFSE. 2 × 105 CFSE-labeled CD8+PD1int T cells and CD8+PD1hi T cells were plated onto 24-well plates and stimulated with 1 µg/ml anti-CD3 and 5 µg/ml anti-CD28 in the presence of 10 µg/ml of blocking antibodies for PD1 or isotype control. IL-2 was added at a final concentration of 100 IU/ml. After 108 h of culture, the percentage of CD8+ T cell division was measured by assessing the CFSE dilution.
Human PBMC, CD8+ T cell, CD4+CD25− T cell, and CD4+CD25+CD127dim/– T reg cell isolation and coculture system
Human PBMCs were isolated from healthy donors by density gradient centrifugation over Ficoll Paque. CD8+ T cells, naive CD4+ T cells, and T reg cells were purified from PBMCs using CD8+ T Cell Isolation Kit (Miltenyi Biotec; 130-096-495), Naive CD4 T Cell Isolation Kit II (Miltenyi Biotec; 130-094-131), and CD4+CD25+CD127dim/– regulatory T cell isolation kit (Miltenyi Biotec; 130-094-775) under sterile conditions following the instructions of the manufacturer. Our analysis revealed a purity of >98% CD8+ T cells and CD4+CD25− T cells and a purity of >95% CD4+CD25+FOXP3+ T reg cells. Cell viability was checked by Trypan blue dye exclusion. Purified cells were cultured in the presence of 1 µg/µl anti-CD3, anti-CD28 (eBiosciences). 3 d after stimulation, cells were used for further experiments. In vitro T reg conversion assays, T reg proliferation assays, and MDSC conversion assays were conducted.
For the in vitro T reg conversion assay, 1 × 105 CD4+CD25−Foxp3− T cells were cocultured with 5 × 105 tumor cells for 72 h in 1640 complete medium. TGF-β1 (R&D Systems) was used as positive control. Additionally, in blocking experiments, TGF-β1 was blocked by using anti-human TGF-β1 antibody (10 ng/µl; R&D Systems; AF-246-NA) with proven neutralizing activity. For the in vitro T reg proliferation assay, isolated T reg were stained with Cell Trace CFSE (eBioscience; 65-0850-84). Then, labeled T reg cells were cocultured with tumor cells at 5:1 for 72 h. Proliferation was assessed by measuring CFSE dilution by flow cytometry. For tumor-induced MDSC conversion assays, PBMCs were derived from healthy donors and cocultured with PDAC at a 5:1 ratio. After 6-d coculture, cells were collected and stained for markers consistent with MDSC phenotype. Related antibodies are listed in Table S6.
Mouse BMDC, mouse CD8+ T cell, and MDSC isolation and coculture system
For mouse BMDCs, femurs were obtained from 4–6-wk-old C57BL/6 mice, and bone marrow was flushed aseptically with RPMI medium using a syringe fitted with a 27-gauge needle. Mouse CD8+ T cells were isolated from mouse spleen using a CD8+ isolation kit (Miltenyi Biotec; 130-104-075). MDSCs were isolated from spleens of tumor-bearing mice using a mouse MDSCs isolation kit (Miltenyi Biotec; 130-094-538). Cell purity was checked by flow cytometric analysis using anti-CD11b and Gr-1 antibodies (>95%), and cell viability was checked by Trypan blue dye exclusion. The resulting cells were cultured using RPMI medium supplemented with 10% FBS at 37°C, 5% CO2, and used for further tumor-induced MDSC conversion, MDSC expansion, and MDSC function assays.
For the mouse tumor-induced MDSC conversion assay, BMDCs were derived from healthy 4–6-wk-old C57BL/6 mice and cocultured with PANC02-vector/PANC02-EHF. After 6-d coculture, cells were collected and stained for CD45, CD11b, and Gr-1. For mouse MDSC expansion assays, isolated MDSCs were stained with CFSE. Then, labeled MDSCs were cocultured with PANC02-vector/PANC02-EHF at 5:1 for 72 h. Proliferation was assessed by measuring CFSE dilution by flow cytometry.
T cell suppression assay
Isolated human or mouse CD8+ T cells were labeled with CFSE. The labeled CD8+ T cells were then plated onto 24-well plates and stimulated with 1 µg/ml anti-CD3 and 5 µg/ml anti-CD28. Purified suppressor cells (i.e., mouse bone marrow–derived MDSCs or human CD4+CD25+CD127dim T regs) were cultured in RPMI 1640 (10% FBS) with 2 × 105 CFSE-labeled CD8+ T cells at 1:5 at 37°C. The proportion of CD8+ T cell proliferation was measured by assessing dilution of the CFSE by flow cytometry after 72 h of coculture.
RT-PCR
Total RNA was extracted from cells using TRIzol and converted to cDNA using TaqMan reverse transcription reagents. qPCR was performed using SYBR Green master mix. The products of semiquantitative PCR were detected by agarose gel electrophoresis, and β-actin was used as loading control. Primers are listed in Table S7.
Western blotting
Target proteins were detected by Western blotting with the following primary antibodies: anti-EHF (Lifespan; LS-B11884, 1:1,000), anti-TGFβ1 (Abcam; ab92486, 1:2,000,), anti–GM-CSF (Proteintech; 17762-1-AP, 1:1,000), and anti–β-tubulin (Abmart; 1:5,000). Secondary antibodies were goat anti-rabbit or mouse antibody at 1:5,000 dilution (Abmart).
ELISA
PDAC cell lines (2 × 106 cells) were implanted in 6-well plates and cultured for 72 h, and the conditioned medium was collected after centrifugation at 700 g for 5 min at 4°C. TGFβ1 and GM-CSF protein were quantified using TGFβ1 and GM-CSF ELISA kit (R&D Systems) according to the manufacturer’s instructions. The same culture medium was used as control to adjust zero.
ChIP and luciferase analysis
ChIP assays were performed using a ChIP kit (Millipore), according to the manufacturer’s instructions. Briefly, EHF stably transduced PANC-1 cells were immunoprecipitated with anti-EHF antibody (Thermo Fisher Scientific; PA5-30716). The immunoprecipitated products were detected by RT-PCR. For luciferase analyses, PANC-1 cells transfected with pCDH-EHF or control vector (pCDH-vector) were transfected with the following vectors: pGL3-TGFB1-EBS (WT), pGL3-TGFB1-EBS1-mutation, pGL3-TGFB1-EBS2-mutation, pGL3-TGFB1-EBS1+2-mutation, pGL3-CSF2-EBS (WT), pGL3-CSF2-EBS1-mutation, pGL3-CSF2-EBS2-mutation, pGL3-CSF2-EBS1+2-mutation, and pGL3-empty vectors (pGL3.1 EV). 48 h later, cells were subjected to dual luciferase analyses. The results were expressed as fold induction relative to the cells transfected with the control vector after normalization to renilla activity. Primers are listed in Table S7. Sequences of all the vectors used for luciferase analysis are listed in Table S8.
Animals and tumor model
Female 4–6-wk-old BALB/c nude mice and C57BL/6 immunocompetent mice were purchased from Vital River Company. All mice were maintained in specific pathogen–free conditions, and animal experiment procedures were approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital, in compliance with the principles and procedures of the NIH Guide for the Care and Use of Laboratory Animals.
All groups were randomly divided. A total of 1 × 106 or 5 × 105 tumor cells were subcutaneously or orthotopically injected into each mouse to form tumors. For the subcutaneous tumor model, tumor growth was monitored using a caliper and calculated by the following formula: volume = 1/2 L1 × (L2)2, where L1 is the long axis and L2 is the short axis of the tumor. For the orthotopic tumor model, tumor growth was analyzed by BLI. The orthotopic model was established using a Matrigel plug containing PANC02-vector-luc (luciferase expressing), PANC02-EHF-luc, KPC-vector-luc, and KPC-EHF-luc cell lines.
In vivo TGFβ1 and GM-CSF neutralization within the local tumor microenvironment
PANC02-vector or PANC02-EHF cells in a Matrigel plug containing neutralizing anti–GM-CSF mAb and anti-TGFβ1 mAb (vs. isotype IgGs as a control) were injected subcutaneously into the flanks of C57BL/6 mice to allow for GM-CSF and TGFβ1 neutralization within the local tumor microenvironment. 8 d later, TGFβ1 and GM-CSF antibody or isotype IgG (20 µg/mouse) were intratumorally injected twice a week.
In vivo T reg plus MDSC depletion efficacy study
Efficacy of T reg plus MDSC depletion was evaluated in PANC02-vector and PANC02-EHF, respectively. Mice were randomly divided into PANC02-vector and PANC02-EHF groups. 1 × 106 PANC02-vector or PANC02-EHF cells were injected subcutaneously in the flank of C57BL/6 mice of indicated group. 7 d after tumor cell inoculation, mice in the PANC02-vector group were randomized into PANC02-vector-control group and PANC02-vector-depletion group; mice in the PANC02-EHF group were randomized into PANC02-EHF-control group and PANC02-EHF-depletion group. Anti-CD25 Abs (BioXcell; PC61) combined with anti-Gr1 Abs (BioLegend; 108414, RB6-8C5) or combined isotype IgG were intraperitoneal injected (200 µg per mouse twice a week). Tumors were harvested at day 28 after implantation.
In vivo anti-PD1 efficacy study
Mice were randomly divided into PANC02-vector and PANC02-EHF groups. 1 × 106 PANC02-vector (PANC02-vector-luc) or PANC02-EHF (PANC02-EHF-luc) cells were injected subcutaneously in the flank of C57BL/6 mice (or orthotopically at pancreas) of the indicated group. 7 d after tumor inoculation, mice in the PANC02-vector group were randomized into PANC02-vector-control group and PANC02-vector-anti-PD1 group; mice in the PANC02-EHF group were randomized into PANC02-EHF-control group and PANC02-EHF-anti-PD1 group. Subcutaneous and orthotopic tumor-bearing mice were treated by PD1 blockade antibody (GoInVivo; RMP1-14) or isotype IgG via intraperitoneal injection. For the subcutaneous tumor model, mice were treated with anti-PD1 or isotype IgG intraperitoneally twice a week at 200 µg/mouse. Tumor volume were measured every 4 d using a caliper. For the orthotopic tumor model, mice were treated with anti-PD1 or isotype IgG intraperitoneally three times each week at 200 µg/mouse. The tumor loads were evaluated at day 7 (before therapy) and at day 21 (after six treatments) by BLI. The OS of each mouse was recorded.
Statistical analyses
Statistical analyses were performed using SPSS software version 21.0. Each experiment was conducted independently at least three times. Data are presented as mean ± SD (square deviation). The variance between the groups was compared. Student’s t test was used to compare the mean values. Median survival was estimated using the Kaplan–Meier method, and the difference between survival curves was tested by the log-rank test. Spearman rank correlation analysis was used to determine the correlation between parameters. Repeated measure two-way ANOVA (time × tumor volume) and post hoc analyses were used to test mouse tumor growth between groups. P < 0.05 was considered significant. All experimental data were verified in at least three independent experiments. The shRNA sequences, antibodies, and primers used in this investigation are listed in Tables S5, S6, S7, and S8.
Online supplemental material
Fig. S1 shows the different effects of EHF on tumor growth in immune-competent and immune-deficient hosts. Fig. S2 shows the prognostic value of EHF. Data were based on EHF objective IF score and IHC assays. Fig. S3 shows results from an additional mouse model using KPC-vector and KPC-EHF cell lines. Fig. S4 shows the in vitro coculture results from other pancreatic cancer cell lines (AsPC-1 and CFPAC-1). Fig. S5 shows in vitro and in vivo blocking experiments using cell lines with genetic knockdown of TGF-B1 and CSF2. Table S1 shows the Cox proportional hazards analysis of clinicopathological factors for OS and RFS (data were based on IF assay; analyses were made using mean intensity of EHF evaluated by software). Table S2 shows the correlation of EHF expression to clinicopathological features (data were based on IF assay; analyses were made using mean intensity of EHF evaluated by software). Table S3 shows the Cox proportional hazards analysis of clinicopathological factors for OS and RFS (data based on IHC assay). Table S4 shows the correlation of EHF expression to clinicopathological features (data based on IHC assay). Table S5 shows shRNA sequences for stable knockdown cell lines. Table S6 shows antibodies for flow cytometry used in this study. Table S7 shows primers used for qRT-PCR and ChIP in this study. Table S8 shows sequence of the vectors for luciferase analysis.
Supplementary Material
Acknowledgments
We thank Prof. Tingbo Liang and Prof. Xueli Bai (Department of Surgery, the Second Affiliated Hospital, Zhejiang University, China) for providing the PDA cell line from KPC (LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre) mouse as a gift. We also thank Prof. Dongming Kuang (State Key Laboratory of Oncology in Southern China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, China) and Dr. Zhen Tao (Department of Radiotherapy, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin, China) for their technical assistance and helpful discussions.
This work was supported by the National Natural Science Foundation of China (81525021, 81672431, 81672435, 81720108028, 81772633, 81702426, 81702427, 81572618, 81802432, 81802433, 81871968, and 81871978), the National Key Clinical Specialist Construction Programs of China (2013-544), Key Program of Prevention and Treatment of Chronic Diseases of Tianjin (17ZXMFSY0010), Key Program of Public Health Bureau Foundation of Tianjin (15KG144), the Science and Technology Development Fund of Tianjin Education Commission for Higher Education (2017KJ198), and the Foundation of Tianjin Medical University (2016KYZQ17). The research in S. Yang’s laboratory is supported by the National Cancer Institute (R01 CA175741) and the Elsa U. Pardee Foundation.
The authors declare no competing financial interests.
Author contributions: J. Hao, H. Ren, and J. Liu conceived and designed the experiments; J. Liu, W. Jiang, and K. Zhao performed most of the experiments; H. Wang, T. Zhou, W. Bai, X. Wang, T. Qin, W. Yu, B. Yang, X. Li, and D. Fu performed some experiments. H. Ren, T. Zhao, C. Huang, and S. Gao provided patient samples and technical support. J. Hao, H. Ren, J. Liu, W. Jiang, W. Tan, and S. Yang analyzed and discussed the data. J. Liu and J. Hao wrote and completed the paper. J. Hao supervised the entire project.
References
- Albino D., Longoni N., Curti L., Mello-Grand M., Pinton S., Civenni G., Thalmann G., D’Ambrosio G., Sarti M., Sessa F., et al. 2012. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res. 72:2889–2900. 10.1158/0008-5472.CAN-12-0212 [DOI] [PubMed] [Google Scholar]
- Albino D., Civenni G., Dallavalle C., Roos M., Jahns H., Curti L., Rossi S., Pinton S., D’Ambrosio G., Sessa F., et al. 2016a Activation of the Lin28/let-7 Axis by Loss of ESE3/EHF Promotes a Tumorigenic and Stem-like Phenotype in Prostate Cancer. Cancer Res. 76:3629–3643. 10.1158/0008-5472.CAN-15-2665 [DOI] [PubMed] [Google Scholar]
- Albino D., Civenni G., Rossi S., Mitra A., Catapano C.V., and Carbone G.M.. 2016b The ETS factor ESE3/EHF represses IL-6 preventing STAT3 activation and expansion of the prostate cancer stem-like compartment. Oncotarget. 7:76756–76768. 10.18632/oncotarget.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne L.J., Beatty G.L., Jhala N., Clark C.E., Rhim A.D., Stanger B.Z., and Vonderheide R.H.. 2012. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 21:822–835. 10.1016/j.ccr.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty G.L., Eghbali S., and Kim R.. 2017. Deploying Immunotherapy in Pancreatic Cancer: Defining Mechanisms of Response and Resistance. Am. Soc. Clin. Oncol. Educ. Book. 37:267–278. 10.14694/EDBK_175232 [DOI] [PubMed] [Google Scholar]
- Borgoni S., Iannello A., Cutrupi S., Allavena P., D’Incalci M., Novelli F., and Cappello P.. 2017. Depletion of tumor-associated macrophages switches the epigenetic profile of pancreatic cancer infiltrating T cells and restores their anti-tumor phenotype. OncoImmunology. 7:e1393596 10.1080/2162402X.2017.1393596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.H., Jiang Y., and Pillarisetty V.G.. 2016. Role of immune cells in pancreatic cancer from bench to clinical application: An updated review. Medicine (Baltimore). 95:e5541 10.1097/MD.0000000000005541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney J.A., Mitchell R.A., and Yaddanapudi K.. 2017. Myeloid-derived suppressor cells-a new therapeutic target to overcome resistance to cancer immunotherapy. J. Leukoc. Biol. 102:727–740. 10.1189/jlb.5VMR1116-458RRR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A.D., and Olive K.P.. 2012. Silencing the killers: paracrine immune suppression in pancreatic cancer. Cancer Cell. 21:715–716. 10.1016/j.ccr.2012.05.029 [DOI] [PubMed] [Google Scholar]
- Dallavalle C., Albino D., Civenni G., Merulla J., Ostano P., Mello-Grand M., Rossi S., Losa M., D’Ambrosio G., Sessa F., et al. 2016. MicroRNA-424 impairs ubiquitination to activate STAT3 and promote prostate tumor progression. J. Clin. Invest. 126:4585–4602. 10.1172/JCI86505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farren M.R., Mace T.A., Geyer S., Mikhail S., Wu C., Ciombor K., Tahiri S., Ahn D., Noonan A.M., Villalona-Calero M., et al. 2016. Systemic Immune Activity Predicts Overall Survival in Treatment-Naïve Patients with Metastatic Pancreatic Cancer. Clin. Cancer Res. 22:2565–2574. 10.1158/1078-0432.CCR-15-1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman R.J., Sementchenko V.I., and Watson D.K.. 2003. The epithelial-specific Ets factors occupy a unique position in defining epithelial proliferation, differentiation and carcinogenesis. Anticancer Res. 23 (3A):2125–2131. [PubMed] [Google Scholar]
- Feng M., Xiong G., Cao Z., Yang G., Zheng S., Song X., You L., Zheng L., Zhang T., and Zhao Y.. 2017. PD-1/PD-L1 and immunotherapy for pancreatic cancer. Cancer Lett. 407:57–65. 10.1016/j.canlet.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Foley K., Kim V., Jaffee E., and Zheng L.. 2016. Current progress in immunotherapy for pancreatic cancer. Cancer Lett. 381:244–251. 10.1016/j.canlet.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossum S.L., Mutolo M.J., Tugores A., Ghosh S., Randell S.H., Jones L.C., Leir S.H., and Harris A.. 2017. Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. J. Biol. Chem. 292:10938–10949. 10.1074/jbc.M117.775304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill S.L., Cui Y., Giles A.J., Smith J.P., Zhang H., Morse E., Kaplan R.N., and Mackall C.L.. 2014. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 6:237ra67 10.1126/scitranslmed.3007974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio V., Rosati A., D’Auria R., De Marco M., Marzullo L., Basile A., Festa M., Pascale M., Remondelli P., Capunzo M., et al. 2018. Combined effect of anti-BAG3 and anti-PD-1 treatment on macrophage infiltrate, CD8 (+) T cell number and tumour growth in pancreatic cancer. Gut. 67:780–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Yu X., and Grogan J.L.. 2015. The checkpoint inhibitor TIGIT limits antitumor and antiviral CD8+ T cell responses. OncoImmunology. 4:e1036214 10.1080/2162402X.2015.1036214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel J.A., Tseng W.W., Davidson M.G., Tolentino L.L., Choi O., Bhattacharya N., Seeley E.S., Winer D.A., Reticker-Flynn N.E., and Engleman E.G.. 2017. An immunosuppressive dendritic cell subset accumulates at secondary sites and promotes metastasis in pancreatic cancer. Cancer Res. 77:4158–4170. 10.1158/0008-5472.CAN-16-2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Chèneby J., Kulkarni S.R., Tan G., et al. 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46(D1):D260–D266. 10.1093/nar/gkx1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D.T., Uram J.N., Wang H., Kemberling H., Eyring A., Bartlett B.., Goldberg R.M., Crocenzi T.S., Fisher G.A., Lee J.J., et al. 2016. PD-1 blockade in mismatch repair deficient non-colorectal gastrointestinal cancers. J. Clin. Oncol. 34(4_suppl):195 10.1200/jco.2016.34.4_suppl.195 [DOI] [Google Scholar]
- Lin H.J., and Lin J.. 2017. Seed-in-Soil: Pancreatic Cancer Influenced by Tumor Microenvironment. Cancers (Basel). 9:93 10.3390/cancers9070093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longoni N., Kunderfranco P., Pellini S., Albino D., Mello-Grand M., Pinton S., D’Ambrosio G., Sarti M., Sessa F., Chiorino G., et al. 2013. Aberrant expression of the neuronal-specific protein DCDC2 promotes malignant phenotypes and is associated with prostate cancer progression. Oncogene. 32:2315–2324. 10.1038/onc.2012.245 [DOI] [PubMed] [Google Scholar]
- Maleki Vareki S., Garrigós C., and Duran I.. 2017. Biomarkers of response to PD-1/PD-L1 inhibition. Crit. Rev. Oncol. Hematol. 116:116–124. 10.1016/j.critrevonc.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Melstrom L.G., Salazar M.D., and Diamond D.J.. 2017. The pancreatic cancer microenvironment: A true double agent. J. Surg. Oncol. 116:7–15. 10.1002/jso.24643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl P., and Krug S.. 2018. Overcoming immune evasion in pancreatic cancer: the combination matters. Gut. 67:997–999. 10.1136/gutjnl-2017-315443 [DOI] [PubMed] [Google Scholar]
- Najjar Y.G., Rayman P., Jia X., Pavicic P.G. Jr., Rini B.I., Tannenbaum C., Ko J., Haywood S., Cohen P., Hamilton T., et al. 2017. Myeloid-Derived Suppressor Cell Subset Accumulation in Renal Cell Carcinoma Parenchyma Is Associated with Intratumoral Expression of IL1β, IL8, CXCL5, and Mip-1α. Clin. Cancer Res. 23:2346–2355. 10.1158/1078-0432.CCR-15-1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngiow S.F., Young A., Jacquelot N., Yamazaki T., Enot D., Zitvogel L., and Smyth M.J.. 2015. A Threshold Level of Intratumor CD8+ T-cell PD1 Expression Dictates Therapeutic Response to Anti-PD1. Cancer Res. 75:3800–3811. 10.1158/0008-5472.CAN-15-1082 [DOI] [PubMed] [Google Scholar]
- Park B.V., Freeman Z.T., Ghasemzadeh A., Chattergoon M.A., Rutebemberwa A., Steigner J., Winter M.E., Huynh T.V., Sebald S.M., Lee S.J., et al. 2016. TGFβ1-Mediated SMAD3 Enhances PD-1 Expression on Antigen-Specific T Cells in Cancer. Cancer Discov. 6:1366–1381. 10.1158/2159-8290.CD-15-1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschall A.V., Zhang R., Qi C.F., Bardhan K., Peng L., Lu G., Yang J., Merad M., McGaha T., Zhou G., et al. 2015. IFN regulatory factor 8 represses GM-CSF expression in T cells to affect myeloid cell lineage differentiation. J. Immunol. 194:2369–2379. 10.4049/jimmunol.1402412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A., Kang S.P., Rasco D., Papadopoulos K.P., Elassaiss-Schaap J., Beeram M., Drengler R., Chen C., Smith L., Espino G., et al. 2015. Phase I Study of Pembrolizumab (MK-3475; Anti-PD-1 Monoclonal Antibody) in Patients with Advanced Solid Tumors. Clin. Cancer Res. 21:4286–4293. 10.1158/1078-0432.CCR-14-2607 [DOI] [PubMed] [Google Scholar]
- Pergamo M., and Miller G.. 2017. Myeloid-derived suppressor cells and their role in pancreatic cancer. Cancer Gene Ther. 24:100–105. 10.1038/cgt.2016.65 [DOI] [PubMed] [Google Scholar]
- Pickup M.W., Owens P., Gorska A.E., Chytil A., Ye F., Shi C., Weaver V.M., Kalluri R., Moses H.L., and Novitskiy S.V.. 2017. Development of Aggressive Pancreatic Ductal Adenocarcinomas Depends on Granulocyte Colony Stimulating Factor Secretion in Carcinoma Cells. Cancer Immunol. Res. 5:718–729. 10.1158/2326-6066.CIR-16-0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro G., Simionato F., Carbone C., Frizziero M., Malleo G., Zanini S., Casolino R., Santoro R., Mina M.M., Zecchetto C., et al. 2017. A circulating TH2 cytokines profile predicts survival in patients with resectable pancreatic adenocarcinoma. OncoImmunology. 6:e1322242 10.1080/2162402X.2017.1322242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe D.R., DeCant B., Mascariñas E., Wayne E.A., Diaz A.M., Akagi N., Hwang R., Pasche B., Dawson D.W., Fang D., et al. 2016. TGFβ Signaling in the Pancreatic Tumor Microenvironment Promotes Fibrosis and Immune Evasion to Facilitate Tumorigenesis. Cancer Res. 76:2525–2539. 10.1158/0008-5472.CAN-15-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin I.H., Askan G., Hu Z.I., and O’Reilly E.M.. 2017. Immunotherapy in pancreatic ductal adenocarcinoma: an emerging entity? Ann. Oncol. 28:2950–2961. 10.1093/annonc/mdx503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakellariou-Thompson D., Forget M.A., Creasy C., Bernard V., Zhao L., Kim Y.U., Hurd M.W., Uraoka N., Parra E.R., Kang Y., et al. 2017. 4-1BB Agonist Focuses CD8+ Tumor-Infiltrating T-Cell Growth into a Distinct Repertoire Capable of Tumor Recognition in Pancreatic Cancer. Clin. Cancer Res. 23:7263–7275. 10.1158/1078-0432.CCR-17-0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y.D., and Pillarisetty V.G.. 2017. T-cell programming in pancreatic adenocarcinoma: a review. Cancer Gene Ther. 24:106–113. 10.1038/cgt.2016.66 [DOI] [PubMed] [Google Scholar]
- Sideras K., Braat H., Kwekkeboom J., van Eijck C.H., Peppelenbosch M.P., Sleijfer S., and Bruno M.. 2014. Role of the immune system in pancreatic cancer progression and immune modulating treatment strategies. Cancer Treat. Rev. 40:513–522. 10.1016/j.ctrv.2013.11.005 [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., and Jemal A.. 2018. Cancer statistics, 2018. CA Cancer J. Clin. 68:7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- Steele C.W., Karim S.A., Leach J.D.G., Bailey P., Upstill-Goddard R., Rishi L., Foth M., Bryson S., McDaid K., Wilson Z., et al. 2016. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell. 29:832–845. 10.1016/j.ccell.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer C.E., and Ramalingam S.S.. 2018. Tumor Mutation Burden: Leading Immunotherapy to the Era of Precision Medicine? J. Clin. Oncol. 36:631–632. 10.1200/JCO.2017.76.8770 [DOI] [PubMed] [Google Scholar]
- Stromnes I.M., Brockenbrough J.S., Izeradjene K., Carlson M.A., Cuevas C., Simmons R.M., Greenberg P.D., and Hingorani S.R.. 2014. Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut. 63:1769–1781. 10.1136/gutjnl-2013-306271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.A., Vick S.C., Iglesia M.D., Brickey W.J., Midkiff B.R., McKinnon K.P., Reisdorf S., Anders C.K., Carey L.A., Parker J.S., et al. 2017. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J. Clin. Invest. 127:3472–3483. 10.1172/JCI90499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F., Meng X., Kong L., and Yu J.. 2018. Progress and challenges of predictive biomarkers of anti PD-1/PD-L1 immunotherapy: A systematic review. Cancer Lett. 414:166–173. 10.1016/j.canlet.2017.11.014 [DOI] [PubMed] [Google Scholar]
- Teng M.W., Ngiow S.F., Ribas A., and Smyth M.J.. 2015. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res. 75:2139–2145. 10.1158/0008-5472.CAN-15-0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueha S., Yokochi S., Ishiwata Y., Ogiwara H., Chand K., Nakajima T., Hachiga K., Shichino S., Terashima Y., Toda E., et al. 2015. Robust Antitumor Effects of Combined Anti-CD4-Depleting Antibody and Anti-PD-1/PD-L1 Immune Checkpoint Antibody Treatment in Mice. Cancer Immunol. Res. 3:631–640. 10.1158/2326-6066.CIR-14-0190 [DOI] [PubMed] [Google Scholar]
- Wang X., Schoenhals J.E., Li A., Valdecanas D.R., Ye H., Zang F., Tang C., Tang M., Liu C.G., Liu X., et al. 2017. Suppression of Type I IFN Signaling in Tumors Mediates Resistance to Anti-PD-1 Treatment That Can Be Overcome by Radiotherapy. Cancer Res. 77:839–850. 10.1158/0008-5472.CAN-15-3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörmann S.M., Diakopoulos K.N., Lesina M., and Algül H.. 2014. The immune network in pancreatic cancer development and progression. Oncogene. 33:2956–2967. 10.1038/onc.2013.257 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Velez-Delgado A., Mathew E., Li D., Mendez F.M., Flannagan K., Rhim A.D., Simeone D.M., Beatty G.L., and Pasca di Magliano M.. 2017. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut. 66:124–136. 10.1136/gutjnl-2016-312078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Jiang W., Wang X., Wang H., Zheng C., Li Y., Sun Y., Huang C., Han Z.B., Yang S., et al. 2017. ESE3 inhibits pancreatic cancer metastasis by upregulating E-cadherin. Cancer Res. 77:874–885. 10.1158/0008-5472.CAN-16-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.