Gunawan et al. analyze at single-cell resolution collective endocardial cell migration into the extracellular matrix and the cellular rearrangements forming leaflets during zebrafish heart valve formation. They show that focal adhesion activity driven by Integrin α5β1 and Talin1 are essential to drive cardiac valve morphogenesis in zebrafish.
Abstract
Elucidating the morphogenetic events that shape vertebrate heart valves, complex structures that prevent retrograde blood flow, is critical to understanding valvular development and aberrations. Here, we used the zebrafish atrioventricular (AV) valve to investigate these events in real time and at single-cell resolution. We report the initial events of collective migration of AV endocardial cells (ECs) into the extracellular matrix (ECM), and their subsequent rearrangements to form the leaflets. We functionally characterize integrin-based focal adhesions (FAs), critical mediators of cell–ECM interactions, during valve morphogenesis. Using transgenes to block FA signaling specifically in AV ECs as well as loss-of-function approaches, we show that FA signaling mediated by Integrin α5β1 and Talin1 promotes AV EC migration and overall shaping of the valve leaflets. Altogether, our investigation reveals the critical processes driving cardiac valve morphogenesis in vivo and establishes the zebrafish AV valve as a vertebrate model to study FA-regulated tissue morphogenesis.
Introduction
Throughout development, tissues undergo dynamic morphogenetic rearrangements to form complex structures. Active cellular processes, including migration, proliferation, and cell-shape changes, occur at the individual and collective levels, resulting in distinct structures within organs. In the heart, a specialized endocardial region at the atrioventricular canal (AVC) undergoes extensive remodelling to form the valves, which are essential to prevent retrograde blood flow (Butcher and Markwald, 2007; Lagendijk et al., 2010; Hinton and Yutzey, 2011; Calkoen et al., 2016; Steed et al., 2016a,b). In chicks and mammals, the valves arising as endocardial cells (ECs) respond to signals from the myocardium and undergo an epithelial-to-mesenchymal transition (EMT) to invade the ECM, which separates ECs and the myocardium (Butcher and Markwald, 2007; Joziasse et al., 2008; Calkoen et al., 2016). Aberrations in these early processes lead to faulty valves, which constitute a significant proportion of human congenital heart defects, and suboptimal circulation. Several studies have implicated an underlying genetic basis for these defects, highlighting the need to identify regulators of valve morphogenesis (Øyen et al., 2009; LaHaye et al., 2016).
Significant limitations in using chick and mouse embryos as model organisms to study heart valve development remain. For example, since studying valve development in live mouse embryos is not possible, results are derived from fixed and sectioned tissues. Valve imaging at single-cell resolution is difficult to achieve in vivo, and many studies use AVC cells explanted on collagen gels to study valve-forming EC behaviors. However, a recent study using explanted lineage-traced atrioventricular (AV) cells reveals that mouse AV tissues (from the 24-somite stage onward) contain a significant amount of epicardial cells, which behave similarly to ECs and undergo EMT (Criem and Zwijsen, 2018). Thus, the molecular mechanisms that have been shown to play a role during valve explant EMT ex vivo may not be required in vivo to promote AV EC EMT.
Zebrafish embryonic heart valves constitute ideal in vivo models that circumvent these limitations. Zebrafish embryos develop externally and are optically clear, allowing visualization of hearts at single-cell resolution. Several signaling pathways, including Notch (Timmerman et al., 2004; Luna-Zurita et al., 2010; Wang et al., 2013), Wnt (Hurlstone et al., 2003; Wang et al., 2013), and bone morphogenic protein (Sugi et al., 2004; Luna-Zurita et al., 2010; Wang et al., 2013), play crucial roles during valvulogenesis in zebrafish and higher vertebrates, suggesting a conservation in the regulation of this process. Studies have shown that AV ECs in zebrafish embryonic hearts undergo cell shape changes that allow their convergence (Boselli et al., 2017) and cellular rearrangements that lead to the formation of a multilayered tissue at the AVC (Beis et al., 2005; Scherz et al., 2008; Pestel et al., 2016; Steed et al., 2016b). However, the morphogenetic processes that occur after this multilayering to form the leaflets are not well characterized. Importantly, most studies examining molecular drivers of zebrafish valvulogenesis have not addressed the cellular processes they regulate or whether they have cell-autonomous roles in the AV ECs and their derivatives. Indeed, defects in contractility or cardiac looping have been reported to impair overall cardiac development (Bartman et al., 2004; Auman et al., 2007; Kalogirou et al., 2014), making it difficult to determine valve-specific requirements of many factors essential for earlier cardiogenic events.
The importance of the cardiac ECM in promoting valve development is well documented in a number of species. Mutations in genes encoding factors involved in ECM composition, including Fibronectin (Fn; Steed et al., 2016b), the proteoglycan Versican (Yamamura et al., 1997; Mjaatvedt et al., 1998; Hatano et al., 2012), or the enzymes that drive hyaluronan production (Camenisch et al., 2000, 2002; Walsh and Stainier, 2001; Lagendijk et al., 2011), severely impair AV EC invasion. Although much evidence has indicated that the ECM acts not only as a passive acellular substrate but also as an active influencer of cellular behavior (van Helvert et al., 2018), the mechanisms by which the ECM signals to valve ECs are poorly elucidated. Only two signaling pathways have been found to act downstream of the ECM (specifically hyaluronan) in valve ECs: the ErbB2/ErbB3 (Camenisch et al., 2002) and VEGF pathways (Rodgers et al., 2006). However, the mechanisms by which hyaluronan regulates these pathways are unclear, as it is not known to directly bind the ErbB2/ErbB3 or VEGF receptors.
Among the many membrane receptors that can mediate communication between the ECM and migrating cells, the integrin-mediated focal adhesion (FA) is one of the most important (Ciobanasu et al., 2013; Sun et al., 2016). Assembly of the FA complex is initiated as integrin heterodimers, composed of α and β subunits, form when bound to their ECM ligands. 24 vertebrate integrin receptors are known, each with its own specific expression patterns, cellular functions, and ECM ligands (Barczyk et al., 2010). Integrin activation induces recruitment of the adaptor molecule Talin and subsequently downstream actin organizers, including the mechanosensitive protein Vinculin and the GTPase-interacting protein Apbb1ip (Ciobanasu et al., 2013; Sun et al., 2016). These stepwise recruitment events result in the formation of stable membrane protrusions that adhere to the ECM in migrating cells. FAs also respond to physical tension, as mechanical forces induce the active conformation of FA-associated factors, including Integrins, Talin, and Vinculin (van Helvert et al., 2018).
Much of our current knowledge about the cellular processes that FAs regulate is derived from in vitro studies. Developmentally, FA-associated factors have been implicated in cardiogenesis, including Integrin α5 (Itgα5) in cardiac neural crest and outflow tract development (Mittal et al., 2010, 2013; Liang et al., 2014; Turner et al., 2015), as well as Itgβ1 (Shai et al., 2002; Carlson et al., 2008), Talin1 and Talin2 (Wu et al., 2015; Manso et al., 2017), and Vinculin (Xu et al., 1998; Zemljic-Harpf et al., 2007; Cheng et al., 2016) in endocardial and cardiomyocyte development and stability. However, their role in the development of heart valves, which interact extensively with the ECM and are exposed to the highest amount of shear stress, has not been investigated.
Our study aims to deepen our understanding of the cellular processes involved in establishing valve leaflets and the molecular mechanisms by which the ECM influences valvulogenesis. Using membrane-targeted EGFP to label cells, we tracked the developing valve-forming AV ECs at single-cell resolution. Our results show that protrusive activities observed at early stages of valvulogenesis precede the relocalization of AV ECs into the ECM, which appears to be at least partially driven by collective EC migration. These ECs do not appear to undergo a complete EMT at this stage. This event is followed by elongation of the folded tissue into the cardiac lumen to establish the valve leaflets. To identify regulators of AV EC invasion into the ECM, we focused on FA members that exert effects on actin reorganization and uncovered a function for FAs in promoting valve leaflet establishment. Loss-of-function analyses of different FA members reveal a requirement for the receptor Itgα5β1and its adaptor molecule, Talin1. These results indicate that the initial establishment of the primordial valve structure depends on Itgα5β1 and Tln1 signaling.
Results
Dynamic morphogenetic events shape functional valve leaflets
To better visualize the cellular processes involved in establishing the cardiac valves, we used the transgenic BAC line Tg(nfatc1:GAL4) (Pestel et al., 2016). This driver line recapitulates the expression pattern of the gene nfatc1, which encodes a transcription factor whose homologue in mouse has been shown to regulate valvulogenesis (Lange and Yutzey, 2006; Wu et al., 2011). After the heart has undergone looping, at ∼48 h postfertilization (hpf), and onward, expression of the reporter line appeared mostly restricted to AV ECs.
We analyzed the morphogenesis of AV ECs in the inner and outer curvatures, which respectively correspond to the superior and inferior AV ECs (Beis et al., 2005). To outline AV ECs at single-cell resolution, we drove the expression of Tg(UAS:EGFP-CAAX), which expresses membrane-targeted EGFP, with Tg(nfatc1:GAL4). Until ∼48 hpf, the ECs constituted a single layer of cells at both the superior and inferior sides of the AVC (Fig. 1, A–A″, single-layered stage). In the superior AVC (Fig. 1, A″–F″), from as early as 50 hpf, ECs from the ventricular side extended long protrusions into the ECM (Fig. 1, B and B′, initial migration). The protruding cells then appeared to invade the ECM in a ventricle-to-atrium direction (Fig. 1, C and C′, collective EC migration). The leading cells remained attached to other AV ECs that also appeared to move into the ECM. These results suggest that the AV ECs rearrange at least partially through collective migration. At 75 hpf, the ECs appeared to establish a folded, multilayered tissue at the AVC and no longer exhibited protrusive activity (Fig. 1, D and D′, folded structure). At later stages, the EC layers proceeded to elongate into the cardiac lumen, forming a leaflet that was apparent at 100 and 120 hpf (Fig. 1, E–F, leaflet elongation). This process appeared to be driven by EC shape changes, as the cells appeared more flattened and less cuboidal at these elongation phases (Fig. 1, E′ and F′) compared with the earlier stages (Fig. 1 D′).
Figure 1.
Morphogenetic processes driving development of the superior and inferior leaflets of the AV valve. (A–F) AV ECs marked by Tg(nfatc1:GAL4); Tg(UAS:EGFP-CAAX) expression. (A′–F′) Superior AV ECs. (A″–F″) Inferior AV ECs. (A) At 48 hpf, single-layered ECs were found in the superior and inferior parts of the AVC. (B) At 55 hpf, leading ECs form protrusions in the superior AVC (B′), whereas inferior AV ECs in the majority of embryos examined (25/44) remained single layered (B″). (C) At 60 hpf, collective migration of superior AV ECs into the ECM in a ventricle-to-atrium direction (C′). Inferior AV ECs extended protrusions into the ECM but remain single layered (C″). (D) At 75 hpf, establishment of a folded multilayered structure in the superior part of the AVC (D′). Variability was observed in inferior leaflet formation, as inferior AV ECs appeared as a folded tissue in most larvae (D″; 18/27) and remained single layered in the others (9/27). (E and F) At 100 and 120 hpf, there was elongation of AV EC layers into the vascular lumen in both the superior (E′–F′) and inferior (E″–F″) parts of the AVC. Overviews are single planes (A–F); magnified images of AV ECs are 5-µm-thick maximum projections of five confocal planes (A′–F″). A, atrium; V, ventricle. In models A–F: blue, AV ECs; red, nonvalve ECs; brown, myocardium; yellow, vascular lumen; purple, ECM. Scale bars: (A–F) 20 µm; (A′–F″) 10 µm.
In the inferior AVC, we found that the AV ECs extended protrusions at a later time point (∼60 hpf) and appeared to undergo a shorter migration compared with the superior AV ECs, traversing a distance of one cell length (Fig. 1 D″). At 75 hpf, the inferior AV EC tissue in most larvae (67%) had adopted a folded structure (Fig. 1 D″). Whereas the EC rearrangement in the superior AVC appeared consistent, a proportion of ECs in the inferior AVC remained single layered at 60 (57%) and 75 (33%) hpf, which resulted in the appearance of an asymmetrical AV valve. Our findings that the inferior leaflet developed later than the superior leaflet are consistent with an earlier report (Beis et al., 2005), although we noted a higher percentage of larvae exhibiting multilayered inferior AVC tissues. All inferior AV EC tissue eventually became multilayered, and formation of inferior AV leaflets was apparent at 100 and 120 hpf (Fig. 1, E″–F″). Altogether, our results indicate that AV ECs in both the superior and inferior parts of the AVC undergo collective migration to establish a folded tissue, which forms the primordial structure for the later AV leaflets.
Enrichment of FA factor expression at the time of AV EC migration
Our results show that zebrafish valvulogenesis involves EC migration into the ECM, as it does in mammals. We hypothesized that factors transmitting ECM signals play critical roles in valvulogenesis. Using a microarray analysis of 52-hpf hearts (GEO accession number GSE124849), we identified potential candidates by comparing their expression values to those genes enriched in the AVC: notch1b (Beis et al., 2005), has2 (Lagendijk et al., 2011), and klf2a (Vermot et al., 2009). Interestingly, we found high expression levels of FA-associated factor genes crucial for actin regulation (Fig. S1 A).
Among the zebrafish genes encoding α (19 annotated) and β (13 annotated) subunits of Integrin, itga5 and itgb1b had the highest expression levels. Dimerization and activation of these subunits promote cellular motility (Collo and Pepper, 1999; Kiwanuka et al., 2013; Schiller et al., 2013). To validate these findings, we examined the expression of itga5 and itgb1b using in situ hybridization, together with immunostaining for the heart-enriched MF20 antigen (a sarcomeric myosin heavy chain). In 36-hpf hearts, we observed itga5 and itgb1b expression in the ventricles (Fig. S1, B and C). At the time of AV EC migration (56 hpf), both genes appeared to be noticeably expressed in the AVC (Fig. S1, G and H). Fluorescent in situ hybridization showed that within the heart, itga5 and itgb1b expression was highest in AV ECs (Fig. S1, L and M).
We then focused on Integrin-recruited cytoplasmic adaptors that regulate actin organization: Talin1, Vinculin, and Apbb1ip. Tln1 and Apbb1ip are encoded by one gene each, whereas Vinculin is encoded by two genes, vcla and vclb. The microarray data show that tln1 and apbb1ip are expressed at high levels in embryonic hearts (Fig. S1 A), but vcla is expressed at low levels (data not shown); no probe for vclb was included in the array. A recent report showed that vclb is the predominant vcl gene expressed in the embryonic heart (Cheng et al., 2016). Our in situ hybridization results showed that tln1, apbb1ip, and vclb were expressed in 36- and 56-hpf embryonic hearts (Fig. S1, D–F and I–K). Interestingly, the expression pattern of tln1, vclb, and appb1ip closely resembled that of itga5 and itgb1b, with noticeable expression in the AVC in 56-hpf hearts (Fig. S1, I–K).
FA factors are concentrated at the leading edge of migrating ECs
Considering the transcript enrichment in the AVC, we examined in more detail the localization of FA proteins during AV EC migration. We focused on the superior AV ECs, as their migration was more pronounced and had more consistent timing compared with the inferior ones. Itgα5 and β1 appeared enriched at the leading protrusions of the migrating AV ECs (Fig. 2, A–B). Talin1 expression appeared more ubiquitous in heart cells. However, whereas it appeared uniformly diffuse throughout the cytoplasm in most cells, it seemed concentrated near the membrane of the leading AV ECs (Fig. 2 C). Enrichment near the leading edge of the migrating AV ECs was found with Vinculin localization (Fig. 2 D). We also checked for phosphorylated Paxillin (p-Pax; Tyr-118), which is indicative of activated Fas, as Paxillin becomes phosphorylated upon Integrin engagement to the ECM (Burridge et al., 1992). p-Pax localization appeared enriched close to the leading edge of the migrating AV ECs (Fig. 2 E), further confirming the presence of activated FAs.
Figure 2.
Localization of factors involved in cytoskeletal organization in migrating AV ECs. (A–H) Immunohistochemistry of 58-hpf Tg(nfatc1:GAL4); Tg(UAS:EGFP-CAAX) (A–F) or Tg(nfatc1:GAL4); Tg(UAS:EGFP) (G and H) embryos. Green, GFP; cyan, DAPI; magenta (A–H) and white (A′–H′), specific factors of interest. (A–F) Localization of FA-associated factors. (A and B) Itgβ1 (A) and Itgα5 (B) appeared enriched in discrete puncta at the migrating cell protrusions. (C–E) Cytoplasmic FA adaptor molecules Talin1 (C), Vinculin (D), and p-Pax (E) appeared strongly localized in the leading edge of migrating cells. (F) The Itgα5β1 ligand Fn appeared distributed around the endocardial and myocardial cells at the AVC. (G and H) The tight junction–associated protein ZO-1 and adherens junction protein VE-cadherin were localized between the leading and follower AV ECs but appeared undetectable in the EC protrusions. Dashed red lines outline the leading edge of the migrating cells at a distance of 1–2 µm. Scale bars: (A–F) 20 µm; (A′–F′ and G–H′) 10 µm.
Itgα5β1 binds selectively to the ECM ligand Fn (Sun et al., 2005; Huveneers et al., 2008), whose dysfunction in zebrafish causes valvulogenesis defects (Steed et al., 2016b). We observed that Fn was localized around both endocardial and myocardial cells, including those in the AVC, at the time of AV EC migration (Fig. 2 F), consistent with the potential role of Fn as a guidance cue to induce or direct the migration of AV ECs.
As cell–cell adhesion molecules are known to play crucial roles in promoting cell movement (Theveneau and Mayor, 2012), we examined the localization of the tight junction–associated protein ZO-1 and the adherens junction molecule VE-cadherin. In contrast to the FA signals, ZO-1 and vascular endothelial (VE)–cadherin were localized to cell–cell boundaries and not at the protrusions of the migrating ECs (Fig. 2, G–H). These results further indicate that the cells migrate collectively and suggest that protrusion formation likely depends on FAs, but not junctional proteins.
Blocking FA signaling through dominant-negative (DN) Vinculin impairs valve morphogenesis
We proceeded to analyze the functional consequences of blocking FA signaling and generated an upstream activating sequence (UAS)–driven DN Vinculin transgene, Tg(UAS:DN-Vcl-EGFP), which expresses a form of Vinculin containing only its head domain (Fig. 3 A). In vitro, this protein blocks interactions between Talin and Talin-interacting proteins, thereby preventing association between FAs and actin (Cohen et al., 2005; Humphries et al., 2007).
Figure 3.
Blocking FA signaling through DN Vinculin overexpression severely affects AV EC migration and valve tissue establishment. (A) Schematics of the UAS-driven DN Vinculin transgene, expressing the head domain of Vinculin fused to GFP, Tg(UAS:DN-Vcl-EGFP). (A–C′) 60 hpf: AV EC migration stage. (B) In WT embryos, AV ECs extended protrusions and formed a multilayered tissue. (C) In embryos overexpressing DN Vinculin, AV ECs remained single layered and extended microvilli-like protrusions. (D) Categorization of migration stages of superior AV ECs in WT (n = 12) and DN Vinculin–overexpressing (n = 21) embryos. (E) Number of protruding ECs in superior and inferior AVC was significantly reduced when DN Vinculin was overexpressed (averages of 2.25 and 1.32 cells per embryo in WT and DN Vinculin, respectively; P = 3.1e−3, unpaired Student’s t test). WT, n = 47 embryos; DN Vinculin, n = 34 embryos. Error bars represent standard deviation; asterisks indicate statistical significance. (F–G′) 72 hpf: establishment of prevalvular structure. (F) WT AV ECs completed migration and established a folded multilayered prevalvular structure. (G) DN Vinculin–overexpressing AV ECs appeared disorganized and did not clearly establish a multilayered tissue. (H–I′) 96 hpf: establishment of functional AV valve leaflets. (H) In WT larvae, both superior and inferior leaflets formed and extensively elongated into the vascular lumen. (I) In DN Vinculin–overexpressing larvae, AV endocardial tissue remained single layered, without established leaflet structures. Scale bars: (B and C, and F–I) 20 µm; (B′ and C′ and E′–H′) 10 µm.
We overexpressed the DN Vinculin transgene in AV ECs using Tg(nfatc1:GAL4) to determine the requirements of FAs during valvulogenesis and minimize earlier cardiogenesis defects. We assessed the efficacy of the DN Vinculin transgene by examining the p-Pax signal in AV ECs at 60 hpf. In all WT embryos, we observed a higher p-Pax signal at the cortex than in the cytoplasm of leading AV ECs (average of 1.8-fold enrichment; Fig. S3, A and C). Strikingly, p-Pax cortical enrichment, as well as the p-Pax signal throughout the cell, was significantly reduced in DN Vinculin–overexpressing leader ECs (Fig. S3, B and C), indicating a reduction in the activation of factors downstream of FAs. In contrast, no alteration in the expression or localization of ZO-1 was observed in DN Vinculin–overexpressing AV ECs (Fig. S3, D and E), suggesting that DN Vinculin does not alter adhesion between AV ECs.
We next assessed the behaviors of the DN Vinculin–overexpressing ECs at the AV EC migration stage, focusing again on the superior AV ECs. We performed a careful categorization of the EC rearrangement phases (Fig. S2). At 60 hpf, most (83%) WT embryos exhibited multilayered valve ECs, with leading ECs extending prominent protrusions in 50% of the embryos examined (Fig. 3, B and D). Strikingly, the majority (77%) of embryos with DN Vinculin–overexpressing AV ECs exhibited a single-layered arrangement, suggesting an impairment in their ability to migrate (Fig. 3, C and D). Several cells extended protrusions that more closely resembled microvilli than those in WT embryos (Fig. 3 C). A significant reduction in the number of AV ECs that extended protrusions was also observed upon DN Vinculin overexpression (Fig. 3 E; averages of 2.3 ECs per WT embryo versus 1.3 EC per DN Vinculin embryo). Overall heart morphology appeared comparable between WT and DN Vinculin–overexpressing embryos, indicating that the AV EC migration impairment is not secondary to earlier cardiogenesis defects. At 75 hpf, whereas the WT AV ECs adopted the folded multilayered structure (Fig. 3 F), most DN Vinculin–overexpressing AV ECs appeared single layered or adopted a disorganized multilayered arrangement (Fig. 3 G).
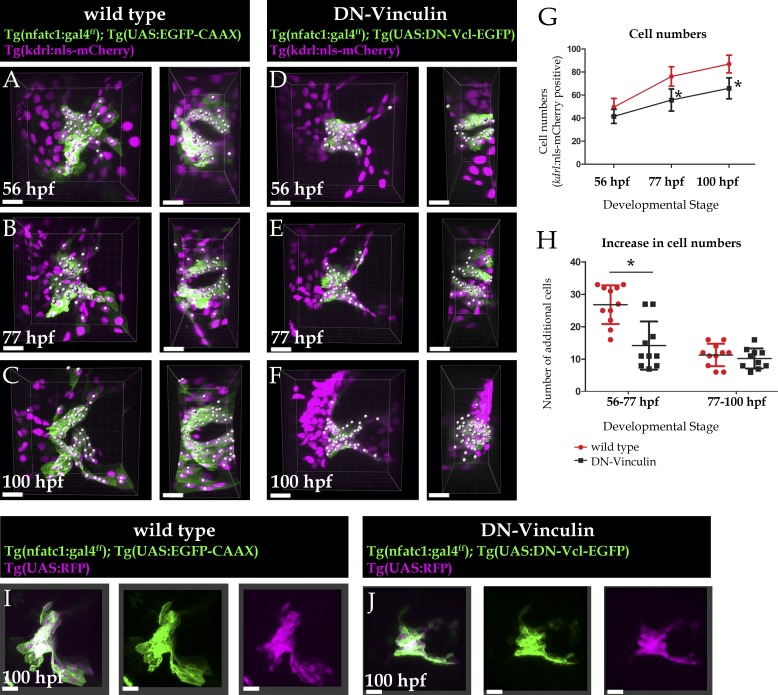
We aimed to observe these morphogenetic events in real time in WT and DN Vinculin–overexpressing AV ECs. Using light sheet microscopy, we performed hourly imaging of developing hearts from 50 to 75 hpf. We imaged Tg(nfatc1:GAL4); Tg(UAS:EGFP-CAAX) or Tg(nfatc1:GAL4); Tg(UAS:DN-Vcl-EGFP) animals together with Tg(kdrl:nls-mCherry) expression, which marks EC nuclei. Over a time window of 15 h, we observed that the hearts developed normally, including looping and chamber ballooning. In WT hearts, we indeed observed that AV ECs extended long protrusions into the ECM at ∼57 hpf and moved into the ECM collectively (Fig. 4, A–C; and Video 1). By 76 hpf, the AV ECs no longer extended protrusions and clearly established a multilayered tissue (Fig. 4, D–F; and Video 1). In contrast, we did not observe protrusions being extended by DN Vinculin–overexpressing ECs (Fig. 4, G–I; and Video 2). From 72 hpf onward, DN Vinculin–overexpressing ECs appeared disorganized and were abnormally multilayered (Fig. 4, J–L). The AVC also appeared collapsed in DN Vinculin–overexpressing animals, suggesting that AV EC rearrangements may be needed to maintain the AV lumen.
Figure 4.
Real-time imaging shows WT AV ECs migrating and establishing a multilayered tissue, whereas DN Vinculin–overexpressing ECs failed to extend protrusions. (A–L) Maximum projections of the entire heart. (A–F) At the AVC, WT ECs originated as a single layer of cells (A), extended long protrusions, moved into the overlying ECM (B–D), and established a multilayered tissue (E and F; no more protrusions were observed at these stages). n = 4 animals. (G–L) DN Vinculin–overexpressing ECs did not extend prominent protrusions and remained single layered until 60 hpf (G–I). By 72 hpf, they appeared disorganized and did not adopt the folded tissue arrangement, and the AVC appeared collapsed (J–L). n = 2 animals. Scale bars: 20 µm.
We also examined the effects of DN Vinculin overexpression at later stages. By 96 hpf, AV valve leaflet structures were apparent in WT larvae (Fig. 3 H), but not in any larvae overexpressing DN Vinculin (Fig. 3 I). 3D reconstruction analysis also showed that the AVCs in DN Vinculin–overexpressing larvae appeared much narrower than those in WT (Fig. 5, C and F), consistent with our observations with the light sheet imaging. Altogether, our data indicate that the initial EC migratory phase is crucial to establish the leaflet structure and that it depends on FA activation.
Figure 5.
Overexpression of DN Vinculin leads to significantly fewer nfatc1:GAL4-expressing ECs in the AVC. (A–F) 3D images of heart valves in WT (A–C) and DN Vinculin–overexpressing (D–F) animals. Shown are sagittal views (left) and atrium-to-ventricle views (right). (G) Number of AV ECs, marked by nfatc1:GAL4; UAS:EGFP and kdrl:nls-mCherry expression, in WT and DN Vinculin–overexpressing animals imaged over a 48-h time window (P = 4.48e−5 at 77 hpf, P = 1.6e−5 at 105 hpf). Error bars represent standard deviation; asterisks indicate statistical significance. (H) Increase in the number of AV ECs between 56 and 77 hpf and 77 and 105 hpf in WT and DN Vinculin–overexpressing animals. WT, n = 12; DN Vinculin, n = 10; P = 2.24e−5. An unpaired Student’s t test was used for G and H. (I and J) 3D images of 100-hpf heart valves. Scale bars: 10 µm.
In addition to affecting tissue structure, we observed that DN Vinculin overexpression modulated the number of AV ECs. Combining Tg(kdrl:nls-mCherry) with Tg(nfatc1:GAL4); Tg(UAS:EGFP-CAAX) or Tg(nfatc1:GAL4); Tg(UAS:DN-Vcl-EGFP), we imaged the same animals within 24-h intervals and quantified the number of nfatc1:GAL4-expressing ECs. In WT animals, we observed an average increase of 26.75 ± 5.7 cells between 56 and 77 hpf and 10.8 ± 3.6 AV ECs between 77 and 100 hpf (49.5 ± 7.6 cells at 56 hpf, 76.2 ± 8.4 cells at 77 hpf, and 86.9 ± 7.7 cells at 100 hpf; Fig. 5, A–C, G, and H). This observation indicates that the increase in AV EC numbers occurs early during valve morphogenesis. Interestingly, when DN Vinculin was overexpressed in AV ECs, the numbers of nfatc1:GAL4 expressing ECs at the earlier time point (56 to 77 hpf) failed to increase to the same extent as in WT animals, with an average increase of 14.2 ± 7.4 AV ECs (41.5 ± 6.1 to 55.7 ± 9.6 cells from 56 to 77 hpf; Fig. 5, D, E, G, and H). At the later time point, the increase in AV EC number was comparable between WT and DN Vinculin, which exhibited an average increase of 10.2 ± 3.1 AV ECs (55.7 ± 9.6 to 65.9 ± 9.1 cells from 77 to 100 hpf; Fig. 5, E–H).
The differences in cell numbers were not due to mosaic expression of the DN Vinculin transgene, as crossing the WT and DN Vinculin transgenes to an independent UAS-driven fluorophore (Tg(UAS:RFP) showed complete coexpression of RFP and GFP in both genotypes (Fig. 5, I and J). Proliferation assays using 24-h ethynyl deoxyuridine (EdU) incorporation also showed that the DN Vinculin–overexpressing ECs appeared to proliferate at a rate similar to WT at 80 and 102 hpf (Fig. S3, G–I). Interestingly, compared with WT AV ECs, the DN Vinculin–overexpressing ECs appeared to adopt an abnormally rounded morphology and sort out of the endothelial tissue (Fig. S3, J and K). We hypothesize that the DN Vinculin–overexpressing AV ECs may extrude from the endothelial tissue into the vascular lumen, thus accounting for the lower cell numbers observed in DN Vinculin–overexpressing AVCs.
DN Vinculin overexpression severely impairs valve function
We further examined the correlation between the observed morphogenetic defects and valve function in DN Vinculin–overexpressing larvae. Interestingly, in 77-hpf WT hearts, the superior leaflet is able to close the AVC and prevent retrograde blood flow, even without the inferior leaflet (Video 3 and Fig. 6, A and A′). The hanging superior leaflet strikes against the single-layered inferior AVC and closes the lumen for ∼25 ms per contraction. At 98 hpf, the contact between the superior and growing inferior AV leaflets becomes more pronounced (Video 4 and Fig. 6, C and C′). In contrast, DN Vinculin–overexpressing AV ECs in superior and inferior AVCs collided with each other briefly and did not remain closed at 77 hpf (Video 5 and Fig. 6 B) or 98 hpf (Video 6 and Fig. 6 D). Failure of the leaflets to close the AV lumen led to severe retrograde flow (Videos 5 and 6). Together, these results indicate that inhibiting FA signaling in valve ECs through DN Vinculin overexpression severely impairs valve function.
Figure 6.
DN Vinculin overexpression impairs valve function. (A–D) Still images of beating hearts from WT (A and C) and DN Vinculin–overexpressing (B and D) larvae. Images were taken from videos that comprised one ventricular contraction (a duration of ∼40 ms). Larvae at 77 (A and B) and 100 (C and D) hpf are shown. WT valve leaflets closed the AVC for ∼30 ms per contraction, whereas AVC leaflets in DN Vinculin–overexpressing larvae did not close efficiently and did not prevent retrograde blood flow. Scale bars: 20 µm.
DN Itgβ1 overexpression impairs AV EC migration in a cell-autonomous manner
We further tested the requirements of FA activation in the migrating ECs by overexpressing DN Itgβ1, a truncated form of Itgβ1 that lacks the extracellular domain and is predicted to prevent interactions between endogenous Itgβ1 and intracellular FA adaptors (LaFlamme et al., 1994; Lukashev et al., 1994; Retta et al., 1998; Relvas et al., 2001; Tanentzapf et al., 2006). Recently, Iida et al. (2018) have shown that expressing this UAS-driven DN Itgβ1 (Tg(UAS:mCherry-itgb1DN) in endothelial cells leads to defective cerebral and trunk vascular development as is observed in itgb1b mutants.
We drove the overexpression of DN Itgβ1 as well as a control construct that possesses the same domains without the Itgβ1 intracellular domain (Fig. 7 A; Iida et al., 2018) specifically in AV ECs. Using the Tg(nfatc1:GAL4) driver, we found mosaic overexpression for both control and DN Itgβ1 in 75% and 56% of the total AV ECs, respectively (Fig. 7, B, C, E, F, and H). Close examination at the time of AV EC migration reveals that the control transgene was expressed in 100% of the leading AV ECs (Fig. 7, B, C, and I). Strikingly, DN Itgβ1 overexpression was only observed in 24% of the leading AV ECs, with the remaining 73% appearing to be follower, nonmigrating cells (Fig. 7, E, F, and I). Overexpression of DN Itgβ1 also significantly reduced the number of AV ECs that extended protrusions (Fig. 7 J). Interestingly, overexpression of DN Itgβ1 also led to cell rounding and extrusion out of the endocardial layer (Fig. 7, D and G), similar to our observations of DN Vinculin–overexpressing ECs. Our data indicate that DN Itgβ1 impaired the migratory capabilities of AV ECs and further strengthen our hypothesis that FA activation promotes AV EC migration in a cell-autonomous manner.
Figure 7.
DN Itgβ1 overexpression affects the protrusive activity of AV ECs. (A) Domain organization of the control Tg(UAS:mCherry) and the DN Itgβ1 Tg(UAS:mCherry-itgb1DN transgenes. SP, signal peptide; TM, transmembrane; ICD, intracellular domain. (B–G) Representative images and schematics of the control (B–D) and DN Itgβ1–overexpressing (E–G) AV ECs at 56 and 75 hpf. (H and I) Proportions of cells expressing mCherry-tagged control or DN Itgβ1 in total AV ECs (H; control, n = 242 cells/12 embryos; DN Itgβ1, n = 361 cells/26 embryos) or leader AV ECs (I; control, n = 31 cells/12 embryos; DN Itgβ1, n = 36 cells/26 embryos). Whereas 100% of leader ECs expressed the control transgene, only 24% expressed DN Itgβ1 (P = 2.03e−9). (J) Number of leader ECs in control (n = 12 embryos) and DN Itgβ1–overexpressing (n = 26 embryos) animals. The number of leader ECs in the superior and inferior AVC was significantly reduced when DN Itgβ1 was overexpressed (averages of 2.58 and 1.38 cells per embryo in control and DN Itgβ1, respectively; P = 4.5e−4). An unpaired Student’s t test was used for I and J. Error bars represent standard deviation; asterisks indicate statistical significance. Scale bars: 20 µm.
Loss of the receptor Itgα5β1b or the adaptor molecule Tln1 delays AV EC migration
We also examined the loss-of-function effects of several FA factors enriched in the AVC to identify specific ones necessary during valvulogenesis. Zebrafish itga5 mutants exhibit defects in somite formation (Jülich et al., 2005; Koshida et al., 2005) and retinal lens fiber formation (Hayes et al., 2012), but the role of itga5 in cardiovascular development has not been reported. We used the predicted null allele itga5kt451 (Koshida et al., 2005). Interestingly, until ∼80 hpf, most itga5 mutant hearts appeared relatively unaffected (Fig. 8, A–C and E–G).
Figure 8.
Loss of Itgα5 leads to delays in AV EC migration. (A–G) Phalloidin staining of itga5 WT, heterozygous, and mutant hearts at 58 (A–D) and 80 (E–H) hpf (magenta in overviews [left]; white in magnified AV areas [right]); green, ZO-1; cyan, DAPI. Representative images of hearts and superior AVCs of itga5 WT (A and E), heterozygous (B and F), and mutant (C and G) animals at 58 (A–C) and 80 (E–G) hpf. Categorization of migration stages at 58 hpf (D: +/+, n = 9; +/−, n = 13; −/−, n = 10) and formation of valvular structures at 80 hpf (H: +/+, n = 4; +/−, n = 17; −/−, n = 11). itga5 mutants exhibited delayed AV EC migration at 58 hpf and delayed establishment of valve structure at 80 hpf. Scale bars: (A–C and E–G, left) 20 µm; (right) 10 µm.
To analyze the morphology of itga5 mutants AV ECs, we stained embryos with the F-actin marker phalloidin and DAPI. Interestingly, at 60 hpf, we observed a higher proportion of AV ECs in itga5 homozygous (70%) and heterozygous (46%) embryos that did not exhibit the multilayering seen in WT (13%; Fig. 8, A–D). A higher proportion of itga5 homozygous embryos (40% compared with 24% in WT) also did not appear to extend noticeable protrusions. At 80 hpf, the folded AV EC tissue structure seen in WT and heterozygous larvae was not apparent in many itga5 mutants (64%; Fig. 8, E–H), suggesting that the ECs had not completed migration. We further examined the rate of retrograde blood flow in itga5 mutants at 80 hpf and found that almost half (43%) exhibited retrograde flow, a significantly higher proportion compared with WT (14%) and heterozygous (8%) larvae (Video 5). These results suggest that loss of Itgα5 leads to a delay in AV EC migration, which subsequently impacts valve function.
In contrast to itga5 mutants, tln1 and itgb1b mutants exhibit severe cardiovascular malformations at a much earlier stage (Wu et al., 2015; Iida et al., 2018). As complete loss of these genes severely affected embryonic development, we used morpholinos to induce partial loss of function and examined valves in embryos that did not exhibit early cardiac malformations. When very low doses of itgb1b (0.75 ng) and tln1 (0.5 ng) morpholinos were injected, we observed a noticeable delay in AV EC migration at 60 hpf (Fig. S4, A–C). At a stage when all WT AV ECs have undergone migration to be multilayered, high percentages of itgb1b (77%) and tln1 (47%) morphants remained single layered (Fig. S4 D). The AV EC shapes appeared abnormally rounded, and only 35% of itgb1b morphants and 42% of tln1 morphants exhibited AV ECs with protrusions compared with 82% of WT. These results suggest that the presence of Itgβ1b and Tln1 is, like that of Itgα5, crucial for the migration and rearrangement of embryonic AV ECs.
Mutations in the actin-regulating factor genes vclb and apbb1ip do not strongly affect AV valve morphogenesis
We further hypothesized that Talin-associated factors that regulate actin, particularly the mechanosensitive factor Vinculin, are required for AV EC migration. We generated a vclb allele carrying a 16-bp deletion (vclbbns247; Fig. S5 A), predicted to produce a truncated protein lacking its actin-binding domain. These mutants did not survive to adulthood, a finding consistent with other recent studies of vclb function (Cheng et al., 2016; Han et al., 2017; unpublished data). However, we found that vclb mutants did not exhibit strong delays in AV EC migration and valve formation (Fig. S5, C and G).
As DN Vinculin overexpression and tln1 mutations generated stronger defects than vclb mutations, other Talin-interacting factors might compensate for defects in Vinculin. Our microarray data indicate that apbb1ip is one of the most highly expressed FA genes in the embryonic heart (Fig. S1, A and K). In cell culture, the dynamics of Vinculin and Apbb1ip binding to Talin mediate FA formation in membrane protrusions (Goult et al., 2013; Lee et al., 2013). To analyze apbb1ip function, we generated an allele carrying a 5-bp deletion, which is predicted to cause a truncated protein lacking its domains that interact with actin organizers (apbb1ipbns275; Fig. S5 B). The apbb1ip mutants survived to adulthood without gross morphological abnormalities or valve defects (Fig. S5, D and H). We then examined vclb; apbb1ip double-homozygous mutants at 77 hpf and observed a subset of the animals with pericardial edema, a phenotype that increased in frequency after heat stress at 33°C for 24 h starting at 53 hpf (data not shown). However, initial valve morphogenesis did not appear to be strongly affected in these double mutants, as the AV endocardial tissue appeared properly folded at 77 hpf (Fig. S5, E and I). These results suggest that mutations in vclb and apbb1ip do not strongly perturb AV EC migration, potentially revealing different degrees of requirement for FA factors during cardiac valvulogenesis.
Discussion
Using the zebrafish model, we were able to investigate at higher resolution the morphogenetic processes that lead to functional heart valves. After cardiac looping, AV ECs in both the inner and outer heart curvatures undergo cellular rearrangements. Based on the morphology of the ECs, particularly the protrusions extending into the ECM (Fig. 1, B and C; Fig. 4, B and C; and Video 1), as well as the localization of FA-associated factors (Fig. 2, A–E; and Fig. S3 A) and F-actin (Fig. 8 A), we propose that these rearrangements are at least partially driven by collective cell migration. However, other mechanisms, including reorganization of the neighboring ECs and/or proliferation of the AV ECs, may contribute to the “pushing” of the AV ECs into the ECM and their subsequent multilayering. As observed by the maintenance of membrane borders between AV ECs (Fig. 1, A–C) and the localization of ZO-1 and VE-cadherin to these borders (Fig. 2, G and H; and Fig. S3 A) at this stage, the cells do not appear to completely transition to a mesenchymal state. The cellular rearrangements result in a folded multilayered tissue. This initial step appears to differ between zebrafish and mammals, as mammalian AV ECs have been reported to become mesenchymal as they invade the ECM (Butcher and Markwald, 2007; Hinton and Yutzey, 2011). However, it has been reported that the lack of the adhesion molecule Cadherin-11 impairs the migratory abilities of mouse AV ECs, which is attributed to defects in communications between ECs (Bowen et al., 2015).
After the folded AV endocardial structure is established, tissue elongation leads to the formation of leaflets, which prevent retrograde blood flow. Our work shows that formation of the superior valve leaflet precedes that of the inferior leaflet and that the superior leaflet alone is sufficient to prevent retrograde blood flow at early larval stages. We hypothesize that development of the inferior leaflet is necessary to ensure unidirectional flow at later stages as the heart grows and pumps more blood.
Several questions regarding the processes driving AV valve morphogenesis remain. First, how the leading AV ECs are specified is unclear. As they originate from the ventricular side of the AVC, it is possible that ECs from this region experience a greater shear stress due to retrograde flow. Indeed, reduction of oscillatory flow (Vermot et al., 2009) or abrogation of flow through inhibiting cardiac contractility (Beis et al., 2005; Vermot et al., 2009) results in impaired valve morphogenesis, implicating flow as an important regulator of this process. Second, the mechanisms behind the different developmental timing between the superior and inferior leaflets (e.g., whether these differences are due to differential activation of signaling pathways or different physical environments) remain to be investigated. Third, the processes by which the valve tissue elongates to form leaflets need to be determined and could involve cell proliferation, changes in EC shapes, and/or infiltration of other cells.
Our study investigates the importance of FA activities in regulating cardiac valve morphogenesis. Our expression analyses show an up-regulation of FA factors in AV ECs at the time of EC migration. Using the DN Vinculin and DN Itgβ1 transgenes, we discovered a cell-autonomous requirement of the FA in AV ECs to promote their migration. Through genetic loss-of-function studies, we discovered a specific requirement for the Itgα5β1 receptor and the adaptor Tln1 during AV EC migration. Of course, it remains possible that other Integrins have important roles during valve development. Indeed, itga5 mutations lead to a delay in, but not complete abrogation of, AV EC migration. In several systems, including during mouse cardiac neural crest cell development (Turner et al., 2015) and zebrafish gastrulation (Jülich et al., 2005), Itgα5 and αV can partially compensate for each other. However, we found itgaV to be expressed at nearly undetectable levels in the developing heart (data not shown), which is why we did not pursue it further.
Our study is the first to address the potential in vivo consequences of loss of Vinculin-b and Apbb1ip. Although mutants in these genes exhibited other phenotypes, including pericardial edema, AV EC migration appeared largely unaffected. This lack of effects of Vinculin-b and Apbb1ip deficiency on AV EC migration may be due to the ability of Tln1, a large 270-kD adaptor, to recruit molecules that compensate for the loss of both proteins, including FA kinase (Chen et al., 1995) and Paxillin (Zacharchenko et al., 2016). It is also possible that the mutant alleles we generated for vclb and apbb1ip are not null alleles and/or that vcla can compensate for the loss of vclb (Rossi et al., 2015).
Based on our data, we propose the following model: The ECM ligand Fn is secreted by cardiomyocytes and ECs at the time of AV EC migration. The leading AV ECs respond to Fn through activation of Itgα5β1, which recruits Talin1 and other downstream factors to confer migratory properties to the leader ECs. Subsequently, AV ECs migrate and establish a multilayered tissue, followed by tissue elongation of the EC layers to form leaflets.
Given the importance of the ECM in valvulogenesis in other vertebrates, our study presents the possibility that the functional relevance of FAs in heart valve development is conserved all the way to humans. Indeed, a link between hereditary mitral valve defects and mutations in the FA-associated factor gene TENSIN-1 has been uncovered (Dina et al., 2015). Interestingly, clinical geneticists at the National Institutes of Health have recently discovered mutations in FA-associated genes investigated in this paper in patients with congenital valve defects (Kruszka, P., and M. Muenke, personal communication). These findings further highlight the evolutionary relevance of FAs in heart valve development, and present the potential for using mutations in these genes as diagnostic tools for congenital heart valve defects in humans.
Materials and methods
Zebrafish husbandry
Zebrafish husbandry was performed in accordance with institutional (Max Planck Society) and national (German) ethical and animal welfare regulations. Embryos were grown at 28°C and staged at 75% epiboly for synchronization. Previously described lines used in this study include Tg(nfatc1:GAL4)mu246 (Pestel et al., 2016), Tg(UAS:EGFP-CAAX)m1230 (Fernandes et al., 2012), Tg(UAS:EGFP)nkuasgfp1a (Asakawa et al., 2008), Tg(UAS:RFP)nkuasrfp1a (Asakawa et al., 2008), Tg(kdrl:nls-mCherry)iS5 (Wang et al., 2010), Tg(UAS:mCherry-itgb1DNko112) (Iida et al., 2018), Tg(UAS:mCherryko113) (Iida et al., 2018), and itga5kt451 (Koshida et al., 2005). Heat stress experiments with apbb1ipbns275; vclbbns247 mutants were performed for 24 h (53–77 hpf) at 33°C.
In situ hybridization
The following primers were used to generate DNA templates for RNA probes: itga5 forward, 5′-TGTCCGTGAATGACACAGACC-3′; reverse, 5′-TAATACGACTCACTATAGGGCCCTGAGTCCTCCAGCATTCC-3′; itgb1b forward, 5′-CAGATCACCCAGATTCAGCCTCAGC-3′; reverse, 5′-TAATACGACTCACTATAGGGGAGGACGACCTCCACCTCC-3′; tln1 forward, 5′-ATGGTACGGGGGCTGGAG-3′; reverse, 5′-TAATACGACTCACTATAGGGCAGCTGATTAATGCTGTCTGTCACCGCGC-3′; vclb forward, 5′-TGGCTGAGATGTCTCGTCTG-3′; reverse, 5′-TAATACGACTCACTATAGGGGTTTGGACACCCCTGCTTTA-3′; apbb1ip forward, 5′-AAAATCAAGCTCGCTTTGGA-3′; reverse, 5′-TAATACGACTCACTATAGGGGAGCCTGATGCTTTCCTCAC-3′. Digoxigenin (DIG)-labeled probes were synthesized in vitro, using a MegaScript T7 Transcription Kit (Thermo Fisher Scientific).
Whole-mount in situ hybridization was performed on 36- and 56-hpf AB embryos. Embryos were fixed in 4% paraformaldehyde overnight at 4°C and then washed with RNase-free PBS/0.1% Tween for 1 h. They were permeabilized in serial methanol dilutions for 5 min each and then incubated overnight at −20°C. They were then rehydrated in PBS/Tween for 1 h, incubated in 1 µg/ml Proteinase K for 30 min, and fixed again in 4% paraformaldehyde for 20 min. After washing with PBS/Tween for 1 h, the embryos were incubated in hybridization buffer (Hyb buffer; 50% formamide, 5× SSC, 0.1% Tween-20, 50 µg/ml heparin, and 500 µg/ml tRNA) at 65°C for ≥2 h. They were then incubated in DIG-labeled RNA probe (1 µg/ml in Hyb buffer) at 65°C overnight. We then performed serial dilutions with Hyb buffer/2× SSC at 65°C, then 0.2× SSC/PBS/Tween at room temperature. We then incubated the embryos for 3–4 h in blocking buffer (1× PBS/Tween, 2% sheep serum, and 2 mg/ml BSA), and anti-DIG antibody (1:5,000) at 4°C overnight. After washing in PBS/Tween for 1 h, the embryos were incubated in alkaline Tris buffer (100 mM Tris-CL, pH 9.5, 100 mM NaCl, and 0.1% Tween-20) for 15 min, and BM Purple staining solution (Roche) until the desired staining intensity. MF20 primary antibody (mouse monoclonal, 1:500 dilution; eBioscience), which labels cardiomyocytes was added after BM Purple treatment for overnight staining at 4°C, followed by PBS/Tween washes and secondary antibody treatment (anti-mouse Alexa Fluor 488; Thermo Fisher Scientific) for 2 h at room temperature.
For fluorescence in situ hybridization, after overnight incubations in RNA probes and anti-DIG-peroxidase, embryos were incubated in anti-tyramide-Alexa Fluor 568 (Thermo Fisher Scientific) in TSA buffer (100 mM borate, pH 8.5, 0.1% Tween-20, 2% dextran sulfate, 0.003% H2O2, and 450 µg/ml 4-iodophenol) for 30 min at room temperature.
Immunostaining
Embryos were collected and fixed in 4% paraformaldehyde. After removing the fixative with PBS/0.1% Tween washes, yolks were removed using forceps and then washed with PBS/1% BSA/1%DMSO/0.5% Triton X-100 (PBDT) and blocked with PBDT/10% goat serum before incubating in primary antibody at 4°C overnight. The embryos were washed in PBDT and incubated in secondary antibody for 2 h at room temperature and then incubated with 2 µg/ml DAPI for 10 min and washed with PBS/0.1% Tween.
Primary antibodies (used at 1:500 dilution) were Itgα5 (rabbit, STJ97284; Antibodyplus), Itgβ1 (rabbit, STJ93731; Antibodyplus), Talin1 (rabbit, STJ97271; Antibodyplus), Vinculin (rabbit, V4139; Sigma-Aldrich), p-Pax (rabbit, NPB2-24459; Novus Biologicals), Fn (rabbit, F3648; Sigma-Aldrich), ZO-1 (mouse, 33-9100; Thermo Fisher Scientific), GFP (chicken, ab13970; Abcam), and mCherry (ab125096; Abcam). To mark F-actin, phalloidin Alexa Fluor 568 (A12380; Thermo Fisher Scientific) was used at 1:100 dilution. Secondary antibodies (used at 1:500 concentration) were goat anti-rabbit Alexa Fluor 568 and goat-anti mouse Alexa Fluor 488 (Thermo Fisher Scientific).
Generation of the transgenic line
To generate the DN Vinculin transgene, the first 774 bp of the vcla coding sequence was amplified by PCR using the following primers: forward, 5′-ATGCCAGTTTTCCATACCAAGAC-3′; reverse, 5′-CAGGCGTCCTCGTCCC-3′. The amplicon was cloned into a pT2-UAS vector upstream of a five-glycine linker and GFP. The plasmid was then injected into AB embryos at the one-cell stage (50 pg/embryo) together with Tol2 mRNA (25 pg/embryo) to establish the Tg(UAS:dn-vclaa1-258-GFPbns206) line.
Generation and genotyping of mutants
apbb1p and vclb mutants were generated using CRISPR/Cas9 technology. DNA sequences used to transcribe the gRNAs were designed into an oligonucleotide containing a T7 promoter sequence (Gagnon et al., 2014). gRNA sequences were designed using the CRISPOR program (http://crispor.tefor.net/) and CRISPR Design (http://crispr.mit.edu). gRNAs were transcribed in vitro using MegaShortScript T7 Transcription Kit (Thermo Fisher Scientific). cas9 mRNA was transcribed in vitro using a MegaScript T3 Transcription Kit (Thermo Fisher Scientific) using pT3TS-nCas9n as template. Both gRNAs and cas9 RNA were purified with RNA Clean and Concentrator Kit (Zymo Research). gRNAs (∼25 pg/embryo) and cas9 mRNA (∼300 pg/embryo) were coinjected at the one-cell stage to generate mutant fish.
The gRNA target sequence within the apbb1ip gene is 5′-GTGGCGATCACTGCAAA-3′. To genotype the mutants (apbb1ipbns275), the following primers were used to amplify a 180-bp amplicon through PCR: forward, 5′-CTTGATGGCTGACCTTGTGGC-3′; reverse, 5′-AGCTGCGATGGAGTTGGAGGAAG-3′. The PCR product was incubated with DdeI (NEB), which digests only the apbb1ip WT PCR product into 70-bp and 110-bp fragments.
The gRNA target sequence within the vclb gene is 5′-AGATGGAGCAGGCCGGCCGC-3′. To genotype the mutants (vclbbns247), high-resolution melting was used, with the following primers to amplify a 70-bp band: forward, 5′-CGTTCATCTGGCCGGAAA-3′; reverse, 5′-CATCTCTCTCACCGACGC-3′. WT vclb HRM products had a melting temperature of 84°C, whereas vclbΔ16 HRM products had a melting temperature of 77°C.
The tln1hi3093 mutants were genotyped as previously described (Wu et al., 2015). To genotype itga5kt451 mutants, the following primers were used to amplify a 420-bp band: forward, 5′-CTTTACTCCAGATGTAGCGCTGACC-3′; reverse, 5′-CATTGGTAATAAAGACATAATAAAATAAAATTACATC-3′. The PCR product was incubated with BglI (NEB), which digests only the itga5kt451 mutant PCR product into 210-bp bands.
Morpholinos
Morpholino oligomers were designed to target the ATG regions of tln1 (5′-TCAGGAGCAGCCTTACGTCCATCTT-3′) and itgb1b (5′-TCAGGAGCAGCCTTACGTCCATCTT-3′) by Gene Tools. tln1 and itgb1b ATG morpholinos were injected into the yolk at the one-cell stage at 0.5 and 0.75 ng per embryo, respectively.
EdU assays
Zebrafish were incubated in 0.5 mM EdU in egg water/0.5% DMSO (four animals/500 µl in 24-well plates) for 24 h (from 56 to 80 hpf and from 78 to 102 hpf). They were fixed in 4% paraformaldehyde and incubated in Click-iT EdU Alexa Fluor 647 Kit (Thermo Fisher Scientific) for 30 min. They were then processed for immunostaining with anti-GFP, anti-mCherry, and DAPI using the procedure described above.
Microscopy imaging
For confocal imaging, embryos were mounted in 1% low-melting agarose/egg water with 0.2% or 0.01% Tricaine to image stopped or beating hearts, respectively. Images of stopped hearts were acquired using a Zeiss LSM700 Axio Imager, LSM800 Axio Observer, LSM800 Axio Examiner, or LSM880 Axio Examiner confocal microscope with a W Plan-Apochromat 20×/1.0 or W Plan-Apochromat 40×/1.0 dipping lens (LSM700, LSM800 Axio Examiner, and LSM880), or LD C-Apochromat 40×/1.1 water-immersion lens (LSM800 Axio Observer; Zeiss). Videos of beating hearts were acquired at 100 frames per second with the LD C-Apochromat 40×/1.1 water immersion lens (Zeiss Cell Observer spinning disk microscope with a CSU-X1 scanner unit [Yokogawa] and Hamamatsu ORCAflash4.0 sCMOS cameras). During imaging, embryos were kept in 28°C chambers. All images were acquired using the Zeiss ZEN program.
For light sheet imaging, embryos were mounted in 1% low-melting agarose/egg water without Tricaine and imaged using a W Plan-APO 20×/1.0 dipping lens (Zeiss Lightsheet Z.1) using a s-CMOS Camera PCO.edge. Immediately before imaging, the embryos were incubated in 0.1% Tricaine to stop the heart. After z-stacks were acquired, the Tricaine water was replaced with no-Tricaine water to allow cardiac contraction to resume. Images were acquired every hour using the Zeiss ZEN program. Image realignment and stitching were done with Imaris (Bitplane) and ImageJ (National Institutes of Health).
For whole-mount in situ hybridization, embryos were mounted on 1% low-melting agarose/egg water and imaged using a Nikon SMZ25 stereomicroscope with a 2×/0.3 objective.
Quantification of cell numbers and fluorescence intensity
For cell counting, nuclei were marked by Tg(kdrl:nls-mCherry) expression and quantified using Imaris (Bitplane). For fluorescence intensity quantification of p-Pax, we selected a confocal section with the brightest detectable signal of p-Pax in each cell. We then drew a 13-µm line from the center of the nucleus through the leading edge and quantified the fluorescence in ImageJ (National Institutes of Health). We took the average of the fluorescence intensity from five points (0.1 µm apart) at the cortex and the cytoplasm and quantified the cortical/cytoplasmic fluorescence signal ratio.
Online supplemental material
Fig. S1 shows RNA expression levels and expression patterns of FA-associated genes, as detected by microarrays (GEO accession number GSE124849) and in situ hybridization, respectively. Fig. S2 shows representative images of AV ECs at different phases of initial AV EC rearrangements. Fig. S3 shows the effects of DN Vinculin overexpression on expression levels and patterns of p-Pax and ZO-1, cellular proliferation, and cellular morphology. Fig. S4 shows the effects of itgb1b and tln1 morpholino-mediated knockdown on AV EC rearrangements at 60 hpf. Fig. S5 shows AV EC morphology and endocardial tissue rearrangements in apbb1ip and vclb single and double mutants at 60 and 75 hpf. Videos 1 and 2 show AV EC morphogenesis obtained from hourly imaging of WT and DN Vinculin–overexpressing animals from 50 to 75 hpf, respectively. Videos 3 and 4 show AV valve functioning in beating hearts to prevent retrograde blood flow in WT at 77 and 98 hpf, respectively. Videos 5 and 6 show severe dysfunction of the AV valve, resulting in retrograde blood flow, in DN Vinculin–overexpressing animals at 77 and 98 hpf, respectively. Video 7 shows representative videos of itga5+/+ and itga5−/− beating hearts at 78 hpf, with retrograde blood flow evident in the itga5−/− AVC.
Supplementary Material
Acknowledgments
We thank Hans-Martin Maischein for expert technical assistance; Anabela Bensimon-Brito, Michelle Collins, and Yu Hsuan Carol Yang for critical reading and comments on the manuscript; and Jeff Gross (University of Pittsburgh, Pittsburg, PA) for sharing the itga5kt451 allele.
This work was supported by a European Molecular Biology Organization fellowship (ALTF 437-2016 to F. Gunawan) and the Max Planck Society (D.Y.R. Stainier).
The authors declare no competing financial interests.
Author contributions: F. Gunawan and D.Y.R. Stainier developed the concept. F. Gunawan, A. Gentile, R. Fukuda, A.T. Tsedeke, R. Ramadass, A. Iida, and V. Jiménez-Amilburu performed the investigation. F. Gunawan wrote the original draft. F. Gunawan, A. Gentile, R. Fukuda, A.T. Tsedeke, V. Jiménez-Amilburu, R. Ramadass, and D.Y.R. Stainier reviewed and edited the draft. F. Gunawan and A. Gentile produced the visualizations. F. Gunawan, A. Sehara-Fujisawa, and D.Y.R. Stainier acquired funding. A. Sehara-Fujisawa, and D.Y.R. Stainier provided resources. F. Gunawan and D.Y.R. Stainier supervised the work.
References
- Asakawa K., Suster M.L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., and Kawakami K.. 2008. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA. 105:1255–1260. 10.1073/pnas.0704963105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman H.J., Coleman H., Riley H.E., Olale F., Tsai H.J., and Yelon D.. 2007. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 5:e53 10.1371/journal.pbio.0050053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczyk M., Carracedo S., and Gullberg D.. 2010. Integrins. Cell Tissue Res. 339:269–280. 10.1007/s00441-009-0834-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman T., Walsh E.C., Wen K.K., McKane M., Ren J., Alexander J., Rubenstein P.A., and Stainier D.Y.. 2004. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2:E129 10.1371/journal.pbio.0020129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis D., Bartman T., Jin S.W., Scott I.C., D’Amico L.A., Ober E.A., Verkade H., Frantsve J., Field H.A., Wehman A., et al. 2005. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 132:4193–4204. 10.1242/dev.01970 [DOI] [PubMed] [Google Scholar]
- Boselli F., Steed E., Freund J.B., and Vermot J.. 2017. Anisotropic shear stress patterns predict the orientation of convergent tissue movements in the embryonic heart. Development. 144:4322–4327. 10.1242/dev.152124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen C.J., Zhou J., Sung D.C., and Butcher J.T.. 2015. Cadherin-11 coordinates cellular migration and extracellular matrix remodeling during aortic valve maturation. Dev. Biol. 407:145–157. 10.1016/j.ydbio.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., Turner C.E., and Romer L.H.. 1992. Tyrosine phosphorylation of paxillin and pp125FAK accompanies cell adhesion to extracellular matrix: a role in cytoskeletal assembly. J. Cell Biol. 119:893–903. 10.1083/jcb.119.4.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher J.T., and Markwald R.R.. 2007. Valvulogenesis: the moving target. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1489–1503. 10.1098/rstb.2007.2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkoen E.E., Hazekamp M.G., Blom N.A., Elders B.B., Gittenberger-de Groot A.C., Haak M.C., Bartelings M.M., Roest A.A., and Jongbloed M.R.. 2016. Atrioventricular septal defect: From embryonic development to long-term follow-up. Int. J. Cardiol. 202:784–795. 10.1016/j.ijcard.2015.09.081 [DOI] [PubMed] [Google Scholar]
- Camenisch T.D., Spicer A.P., Brehm-Gibson T., Biesterfeldt J., Augustine M.L., Calabro A. Jr., Kubalak S., Klewer S.E., and McDonald J.A.. 2000. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J. Clin. Invest. 106:349–360. 10.1172/JCI10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch T.D., Schroeder J.A., Bradley J., Klewer S.E., and McDonald J.A.. 2002. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat. Med. 8:850–855. 10.1038/nm742 [DOI] [PubMed] [Google Scholar]
- Carlson T.R., Hu H., Braren R., Kim Y.H., and Wang R.A.. 2008. Cell-autonomous requirement for beta1 integrin in endothelial cell adhesion, migration and survival during angiogenesis in mice. Development. 135:2193–2202. 10.1242/dev.016378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.C., Appeddu P.A., Parsons J.T., Hildebrand J.D., Schaller M.D., and Guan J.L.. 1995. Interaction of focal adhesion kinase with cytoskeletal protein talin. J. Biol. Chem. 270:16995–16999. 10.1074/jbc.270.28.16995 [DOI] [PubMed] [Google Scholar]
- Cheng F., Miao L., Wu Q., Gong X., Xiong J., and Zhang J.. 2016. Vinculin b deficiency causes epicardial hyperplasia and coronary vessel disorganization in zebrafish. Development. 143:3522–3531. 10.1242/dev.132936 [DOI] [PubMed] [Google Scholar]
- Ciobanasu C., Faivre B., and Le Clainche C.. 2013. Integrating actin dynamics, mechanotransduction and integrin activation: the multiple functions of actin binding proteins in focal adhesions. Eur. J. Cell Biol. 92:339–348. 10.1016/j.ejcb.2013.10.009 [DOI] [PubMed] [Google Scholar]
- Cohen D.M., Chen H., Johnson R.P., Choudhury B., and Craig S.W.. 2005. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 280:17109–17117. 10.1074/jbc.M414704200 [DOI] [PubMed] [Google Scholar]
- Collo G., and Pepper M.S.. 1999. Endothelial cell integrin alpha5beta1 expression is modulated by cytokines and during migration in vitro. J. Cell Sci. 112:569–578. [DOI] [PubMed] [Google Scholar]
- Criem N., and Zwijsen A.. 2018. The epicardium obscures interpretations on endothelial-to-mesenchymal transition in the mouse atrioventricular canal explant assay. Sci. Rep. 8:4722 10.1038/s41598-018-22971-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C., Bouatia-Naji N., Tucker N., Delling F.N., Toomer K., Durst R., Perrocheau M., Fernandez-Friera L., Solis J., Le Tourneau T., et al. Leducq Transatlantic MITRAL Network . 2015. Genetic association analyses highlight biological pathways underlying mitral valve prolapse. Nat. Genet. 47:1206–1211. 10.1038/ng.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A.M., Fero K., Arrenberg A.B., Bergeron S.A., Driever W., and Burgess H.A.. 2012. Deep brain photoreceptors control light-seeking behavior in zebrafish larvae. Curr. Biol. 22:2042–2047. 10.1016/j.cub.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., and Schier A.F.. 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 9:e98186 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult B.T., Zacharchenko T., Bate N., Tsang R., Hey F., Gingras A.R., Elliott P.R., Roberts G.C., Ballestrem C., Critchley D.R., and Barsukov I.L.. 2013. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J. Biol. Chem. 288:8238–8249. 10.1074/jbc.M112.438119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.K.L., van der Krogt G.N.M., and de Rooij J.. 2017. Zygotic vinculin is not essential for embryonic development in zebrafish. PLoS One. 12:e0182278 10.1371/journal.pone.0182278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano S., Kimata K., Hiraiwa N., Kusakabe M., Isogai Z., Adachi E., Shinomura T., and Watanabe H.. 2012. Versican/PG-M is essential for ventricular septal formation subsequent to cardiac atrioventricular cushion development. Glycobiology. 22:1268–1277. 10.1093/glycob/cws095 [DOI] [PubMed] [Google Scholar]
- Hayes J.M., Hartsock A., Clark B.S., Napier H.R., Link B.A., and Gross J.M.. 2012. Integrin α5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish. Mol. Biol. Cell. 23:4725–4738. 10.1091/mbc.e12-09-0672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton R.B., and Yutzey K.E.. 2011. Heart valve structure and function in development and disease. Annu. Rev. Physiol. 73:29–46. 10.1146/annurev-physiol-012110-142145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J.D., Wang P., Streuli C., Geiger B., Humphries M.J., and Ballestrem C.. 2007. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179:1043–1057. 10.1083/jcb.200703036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlstone A.F.L., Haramis A.P., Wienholds E., Begthel H., Korving J., Van Eeden F., Cuppen E., Zivkovic D., Plasterk R.H., and Clevers H.. 2003. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature. 425:633–637. 10.1038/nature02028 [DOI] [PubMed] [Google Scholar]
- Huveneers S., Truong H., Fässler R., Sonnenberg A., and Danen E.H.. 2008. Binding of soluble fibronectin to integrin alpha5 beta1 - link to focal adhesion redistribution and contractile shape. J. Cell Sci. 121:2452–2462. 10.1242/jcs.033001 [DOI] [PubMed] [Google Scholar]
- Iida A., Wang Z., Hirata H., and Sehara-Fujisawa A.. 2018. Integrin β1 activity is required for cardiovascular formation in zebrafish. Genes Cells. 23:938–951. 10.1111/gtc.12641 [DOI] [PubMed] [Google Scholar]
- Joziasse I.C., van de Smagt J.J., Smith K., Bakkers J., Sieswerda G.J., Mulder B.J., and Doevendans P.A.. 2008. Genes in congenital heart disease: atrioventricular valve formation. Basic Res. Cardiol. 103:216–227. 10.1007/s00395-008-0713-4 [DOI] [PubMed] [Google Scholar]
- Jülich D., Geisler R., and Holley S.. 2005. Integrinalpha5 and delta/notch signaling have complementary spatiotemporal requirements during zebrafish somitogenesis. Dev. Cell. 8, 575–586. [DOI] [PubMed] [Google Scholar]
- Kalogirou S., Malissovas N., Moro E., Argenton F., Stainier D.Y., and Beis D.. 2014. Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc. Res. 104:49–60. 10.1093/cvr/cvu186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwanuka E., Andersson L., Caterson E.J., Junker J.P., Gerdin B., and Eriksson E.. 2013. CCN2 promotes keratinocyte adhesion and migration via integrin α5β1. Exp. Cell Res. 319:2938–2946. 10.1016/j.yexcr.2013.08.021 [DOI] [PubMed] [Google Scholar]
- Koshida S., Kishimoto Y., Ustumi H., Shimizu T., Furutani-Seiki M., Kondoh H., and Takada S.. 2005. Integrinalpha5-dependent fibronectin accumulation for maintenance of somite boundaries in zebrafish embryos. Dev. Cell. 8:587–598. 10.1016/j.devcel.2005.03.006 [DOI] [PubMed] [Google Scholar]
- LaFlamme S.E., Thomas L.A., Yamada S.S., and Yamada K.M.. 1994. Single subunit chimeric integrins as mimics and inhibitors of endogenous integrin functions in receptor localization, cell spreading and migration, and matrix assembly. J. Cell Biol. 126:1287–1298. 10.1083/jcb.126.5.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagendijk A.K., Smith K.A., and Bakkers J.. 2010. Genetics of Congenital Heart Defects: A Candidate Gene Approach. Trends Cardiovasc. Med. 20:124–128. [DOI] [PubMed] [Google Scholar]
- Lagendijk A.K., Goumans M.J., Burkhard S.B., and Bakkers J.. 2011. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ. Res. 109:649–657. 10.1161/CIRCRESAHA.111.247635 [DOI] [PubMed] [Google Scholar]
- LaHaye S., Corsmeier D., Basu M., Bowman J.L., Fitzgerald-Butt S., Zender G., Bosse K., McBride K.L., White P., and Garg V.. 2016. Utilization of Whole Exome Sequencing to Identify Causative Mutations in Familial Congenital Heart Disease. Circ Cardiovasc Genet. 9:320–329. 10.1161/CIRCGENETICS.115.001324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A.W., and Yutzey K.E.. 2006. NFATc1 expression in the developing heart valves is responsive to the RANKL pathway and is required for endocardial expression of cathepsin K. Dev. Biol. 292:407–417. 10.1016/j.ydbio.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Lee H.-S., Anekal P., Lim C.J., Liu C.C., and Ginsberg M.H.. 2013. Two modes of integrin activation form a binary molecular switch in adhesion maturation. Mol. Biol. Cell. 24:1354–1362. 10.1091/mbc.e12-09-0695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Wang X., Mittal A., Dhiman S., Hou S.Y., Degenhardt K., and Astrof S.. 2014. Mesodermal expression of integrin α5β1 regulates neural crest development and cardiovascular morphogenesis. Dev. Biol. 395:232–244. 10.1016/j.ydbio.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev M.E., Sheppard D., and Pytela R.. 1994. Disruption of integrin function and induction of tyrosine phosphorylation by the autonomously expressed β 1 integrin cytoplasmic domain. J. Biol. Chem. 269:18311–18314. [PubMed] [Google Scholar]
- Luna-Zurita L., Prados B., Grego-Bessa J., Luxán G., del Monte G., Benguría A., Adams R.H., Pérez-Pomares J.M., and de la Pompa J.L.. 2010. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J. Clin. Invest. 120:3493–3507. 10.1172/JCI42666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manso A.M., Okada H., Sakamoto F.M., Moreno E., Monkley S.J., Li R., Critchley D.R., and Ross R.S.. 2017. Loss of mouse cardiomyocyte talin-1 and talin-2 leads to β-1 integrin reduction, costameric instability, and dilated cardiomyopathy. Proc. Natl. Acad. Sci. USA. 114:E6250–E6259. 10.1073/pnas.1701416114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A., Pulina M., Hou S.Y., and Astrof S.. 2010. Fibronectin and integrin alpha 5 play essential roles in the development of the cardiac neural crest. Mech. Dev. 127:472–484. 10.1016/j.mod.2010.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A., Pulina M., Hou S.Y., and Astrof S.. 2013. Fibronectin and integrin alpha 5 play requisite roles in cardiac morphogenesis. Dev. Biol. 381:73–82. 10.1016/j.ydbio.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt C.H., Yamamura H., Capehart A.A., Turner D., and Markwald R.R.. 1998. The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev. Biol. 202:56–66. 10.1006/dbio.1998.9001 [DOI] [PubMed] [Google Scholar]
- Øyen N., Poulsen G., Boyd H.A., Wohlfahrt J., Jensen P.K., and Melbye M.. 2009. Recurrence of congenital heart defects in families. Circulation. 120:295–301. 10.1161/CIRCULATIONAHA.109.857987 [DOI] [PubMed] [Google Scholar]
- Pestel J., Ramadass R., Gauvrit S., Helker C., Herzog W., and Stainier D.Y.. 2016. Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development. 143:2217–2227. 10.1242/dev.133272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relvas J.B., Setzu A., Baron W., Buttery P.C., LaFlamme S.E., Franklin R.J., and ffrench-Constant C.. 2001. Expression of dominant-negative and chimeric subunits reveals an essential role for β1 integrin during myelination. Curr. Biol. 11:1039–1043. 10.1016/S0960-9822(01)00292-5 [DOI] [PubMed] [Google Scholar]
- Retta S.F., Balzac F., Ferraris P., Belkin A.M., Fässler R., Humphries M.J., De Leo G., Silengo L., and Tarone G.. 1998. beta1-integrin cytoplasmic subdomains involved in dominant negative function. Mol. Biol. Cell. 9:715–731. 10.1091/mbc.9.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers L.S., Lalani S., Hardy K.M., Xiang X., Broka D., Antin P.B., and Camenisch T.D.. 2006. Depolymerized hyaluronan induces vascular endothelial growth factor, a negative regulator of developmental epithelial-to-mesenchymal transformation. Circ. Res. 99:583–589. 10.1161/01.RES.0000242561.95978.43 [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M., and Stainier D.Y.. 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 524:230–233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Scherz P.J., Huisken J., Sahai-Hernandez P., and Stainier D.Y.. 2008. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 135:1179–1187. 10.1242/dev.010694 [DOI] [PubMed] [Google Scholar]
- Schiller H.B., Hermann M.R., Polleux J., Vignaud T., Zanivan S., Friedel C.C., Sun Z., Raducanu A., Gottschalk K.E., Théry M., et al. 2013. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15:625–636. 10.1038/ncb2747 [DOI] [PubMed] [Google Scholar]
- Shai S.Y., Harpf A.E., Babbitt C.J., Jordan M.C., Fishbein M.C., Chen J., Omura M., Leil T.A., Becker K.D., Jiang M., et al. 2002. Cardiac myocyte-specific excision of the β1 integrin gene results in myocardial fibrosis and cardiac failure. Circ. Res. 90:458–464. 10.1161/hh0402.105790 [DOI] [PubMed] [Google Scholar]
- Steed E., Boselli F., and Vermot J.. 2016a Hemodynamics driven cardiac valve morphogenesis. Biochim. Biophys. Acta. 1863:1760–1766. 10.1016/j.bbamcr.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Steed E., Faggianelli N., Roth S., Ramspacher C., Concordet J.P., and Vermot J.. 2016b klf2a couples mechanotransduction and zebrafish valve morphogenesis through fibronectin synthesis. Nat. Commun. 7:11646 10.1038/ncomms11646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugi Y., Yamamura H., Okagawa H., and Markwald R.R.. 2004. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev. Biol. 269:505–518. 10.1016/j.ydbio.2004.01.045 [DOI] [PubMed] [Google Scholar]
- Sun Z., Martinez-Lemus L.A., Trache A., Trzeciakowski J.P., Davis G.E., Pohl U., and Meininger G.A.. 2005. Mechanical properties of the interaction between fibronectin and alpha5beta1-integrin on vascular smooth muscle cells studied using atomic force microscopy. Am. J. Physiol. Heart Circ. Physiol. 289:H2526–H2535. 10.1152/ajpheart.00658.2004 [DOI] [PubMed] [Google Scholar]
- Sun Z., Guo S.S., and Fässler R.. 2016. Integrin-mediated mechanotransduction. J. Cell Biol. 215:445–456. 10.1083/jcb.201609037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanentzapf G., Martin-Bermudo M.D., Hicks M.S., and Brown N.H.. 2006. Multiple factors contribute to integrin-talin interactions in vivo. J. Cell Sci. 119:1632–1644. 10.1242/jcs.02859 [DOI] [PubMed] [Google Scholar]
- Theveneau E., and Mayor R.. 2012. Cadherins in collective cell migration of mesenchymal cells. Curr. Opin. Cell Biol. 24:677–684. 10.1016/j.ceb.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman L.A., Grego-Bessa J., Raya A., Bertrán E., Pérez-Pomares J.M., Díez J., Aranda S., Palomo S., McCormick F., Izpisúa-Belmonte J.C., and de la Pompa J.L.. 2004. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18:99–115. 10.1101/gad.276304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C.J., Badu-Nkansah K., Crowley D., van der Flier A., and Hynes R.O.. 2015. α5 and αv integrins cooperate to regulate vascular smooth muscle and neural crest functions in vivo. Development. 142:797–808. 10.1242/dev.117572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helvert S., Storm C., and Friedl P.. 2018. Mechanoreciprocity in cell migration. Nat. Cell Biol. 20:8–20. 10.1038/s41556-017-0012-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermot J., Forouhar A.S., Liebling M., Wu D., Plummer D., Gharib M., and Fraser S.E.. 2009. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 7:e1000246 10.1371/journal.pbio.1000246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.C., and Stainier D.Y.R.. 2001. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 293:1670–1673. 10.1126/science.293.5535.1670 [DOI] [PubMed] [Google Scholar]
- Wang Y., Kaiser M.S., Larson J.D., Nasevicius A., Clark K.J., Wadman S.A., Roberg-Perez S.E., Ekker S.C., Hackett P.B., McGrail M., and Essner J.J.. 2010. Moesin1 and Ve-cadherin are required in endothelial cells during in vivo tubulogenesis. Development. 137:3119–3128. 10.1242/dev.048785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu B., Chamberlain A.A., Lui W., Koirala P., Susztak K., Klein D., Taylor V., and Zhou B.. 2013. Endocardial to myocardial notch-wnt-bmp axis regulates early heart valve development. PLoS One. 8:e60244 10.1371/journal.pone.0060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Wang Y., Lui W., Langworthy M., Tompkins K.L., Hatzopoulos A.K., Baldwin H.S., and Zhou B.. 2011. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circ. Res. 109:183–192. 10.1161/CIRCRESAHA.111.245035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Zhang J., Koh W., Yu Q., Zhu X., Amsterdam A., Davis G.E., Arnaout M.A., and Xiong J.W.. 2015. Talin1 is required for cardiac Z-disk stabilization and endothelial integrity in zebrafish. FASEB J. 29:4989–5005. 10.1096/fj.15-273409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Baribault H., and Adamson E.D.. 1998. Vinculin knockout results in heart and brain defects during embryonic development. Development. 125:327–337. 10.1371/journal.pone.0011530 [DOI] [PubMed] [Google Scholar]
- Yamamura H., Zhang M., Markwald R.R., and Mjaatvedt C.H.. 1997. A heart segmental defect in the anterior-posterior axis of a transgenic mutant mouse. Dev. Biol. 186:58–72. 10.1006/dbio.1997.8559 [DOI] [PubMed] [Google Scholar]
- Zacharchenko T., Qian X., Goult B.T., Jethwa D., Almeida T.B., Ballestrem C., Critchley D.R., Lowy D.R., and Barsukov I.L.. 2016. LD Motif Recognition by Talin: Structure of the Talin-DLC1 Complex. Structure. 24:1130–1141. 10.1016/j.str.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemljic-Harpf A.E., Miller J.C., Henderson S.A., Wright A.T., Manso A.M., Elsherif L., Dalton N.D., Thor A.K., Perkins G.A., McCulloch A.D., and Ross R.S.. 2007. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol. Cell. Biol. 27:7522–7537. 10.1128/MCB.00728-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.