Zhao and Zhang summarize recent advances in our molecular understanding of the maturation of nascent autophagosomes into degradative autolysosomes in multicellular organisms.
Abstract
Macroautophagy involves the sequestration of cytoplasmic contents in a double-membrane autophagosome and their delivery to lysosomes for degradation. In multicellular organisms, nascent autophagosomes fuse with vesicles originating from endolysosomal compartments before forming degradative autolysosomes, a process known as autophagosome maturation. ATG8 family members, tethering factors, Rab GTPases, and SNARE proteins act coordinately to mediate fusion of autophagosomes with endolysosomal vesicles. The machinery mediating autophagosome maturation is under spatiotemporal control and provides regulatory nodes to integrate nutrient availability with autophagy activity. Dysfunction of autophagosome maturation is associated with various human diseases, including neurodegenerative diseases, Vici syndrome, cancer, and lysosomal storage disorders. Understanding the molecular mechanisms underlying autophagosome maturation will provide new insights into the pathogenesis and treatment of these diseases.
Introduction
Macroautophagy (hereafter autophagy) refers to the engulfment of cytoplasmic contents, such as a portion of cytosol, mitochondria, or the ER, in a double-membrane autophagosome and their delivery to the vacuole (in yeast) or lysosomes (in multicellular organisms) for degradation. Formation of the autophagosome involves multiple membrane remodeling processes, including initiation and nucleation of a cup-shaped isolation membrane (IM; also known as the phagophore) and its subsequent expansion and closure (Nakatogawa et al., 2009; Lamb et al., 2013; Feng et al., 2014). By degrading and recycling sequestrated materials, autophagy acts as a survival mechanism in response to a variety of stress conditions. Autophagy also removes damaged organelles and/or misfolded proteins in a highly selective manner to maintain cellular homeostasis. Selective autophagy requires adaptor molecules that link the cargo to the autophagic machinery (Stolz et al., 2014; Anding and Baehrecke, 2017).
Our basic understanding of the molecular mechanism of autophagosome formation mainly comes from the study of a set of autophagy-related (ATG) genes identified from yeast genetic screens (Nakatogawa et al., 2009; Lamb et al., 2013; Feng et al., 2014; Zhao and Zhang, 2018). ATG proteins form different complexes and act coordinately for autophagosome biogenesis. In brief, the ATG1 complex (composed of ATG1, ATG13, and ATG17) and the class III phosphatidylinositol 3-phosphate (PI(3)P) kinase complex (composed of VPS34, VPS15, ATG6/Beclin1, and ATG14) are required for initiation and nucleation of IMs. Vesicles carrying the integral membrane protein ATG9, known as ATG9 vesicles, may provide the membrane source for initiation of IMs. ATG9 is later retrieved from autophagosomal membranes, a process controlled by the ATG2/ATG18 complex. The two ubiquitin-like conjugation systems, including ATG7 (E1 enzyme)/ATG3 (E2 enzyme)/Atg8 family members (ubiquitin-like molecules) and ATG7/ATG10 (E2 enzyme)/ATG12 (ubiquitin-like molecule), function in autophagosomal membrane expansion and closure (Nakatogawa et al., 2009; Lamb et al., 2013; Feng et al., 2014).
The autophagy pathway in multicellular organisms is much more complex, containing steps that are not present in yeast (Lamb et al., 2013; Zhang and Baehrecke, 2015; Zhao and Zhang, 2018). For example, in yeast, autophagosomes are generated at a single site, called the preautophagosomal formation site, and directly fuse with the vacuole. In multicellular organisms, autophagosomes are formed simultaneously at different sites, and nascent autophagosomes fuse with vesicles originating from the endolysosomal compartments before eventually forming degradative autolysosomes. Lysosomes are regenerated after the sequestrated materials are degraded. In this review, we will discuss recent advances in understanding the mechanism underlying the conversion of nascent autophagosomes into functional autolysosomes.
It is useful to clarify a few terms describing the steps after autophagosome formation. Autophagosome maturation refers to the process involving the progression of nascent autophagosomes to degradative autolysosomes. It is often misused to refer to progression from IMs to autophagosomes. The term amphisome refers to structures containing both autophagic and endocytic materials, while the term autolysosome indicates structures containing autophagic and lysosome-specific contents such as hydrolases (Eskelinen, 2005). Although LAMP1 is routinely used as a marker for lysosomes, LAMP1-labeled compartments are actually heterogeneous in nature (Cheng et al., 2018). Lysosomal membrane proteins such as LAMP1 and LAMP2 are transported from the trans-Golgi network by vesicles, and they can accumulate in vesicles of different compartments if the trafficking process is impaired. Therefore, assays monitoring acidification and lysosomal hydrolases are more accurate than LAMP1/2-labeling methods to distinguish autolysosomes from autophagosomes/amphisomes. The term autophagic vacuole (AV) refers collectively to autophagic structures after autophagosome formation, including autophagosomes, amphisomes, and autolysosomes.
Autophagosomes are formed at the ER
In multicellular organisms, the ER functions as a platform for the recruitment of ATG proteins for autophagosome formation. Upon autophagy induction, the ULK1/FIP200 complex (the mammalian Atg1 complex) is targeted to the ER and recruits the PI(3)P kinase complex for generation of PI(3)P (Itakura and Mizushima, 2010; Zhao and Zhang, 2018). The specialized PI(3)P-enriched subdomains of the ER, named omegasomes, provide platforms for IM nucleation and expansion (Fig. 1; Axe et al., 2008). The possible membrane sources for the initial nucleation of IMs include COPII vesicles, ATG9 vesicles, and membranes originating from omegasome structures (Lamb et al., 2013; Karanasios et al., 2016). Plasma membrane, mitochondria, ER–Golgi intermediate compartment, COPII vesicles, and ATG9 vesicles have all been suggested to contribute membrane sources for IM expansion (Hailey et al., 2010; Ravikumar et al., 2010; Orsi et al., 2012; Ge et al., 2013; Graef et al., 2013; Puri et al., 2013; Zhao and Zhang, 2018). The IM is closely apposed with the ER and is also linked to the ER by tubular membrane extensions, collectively referred to as ER–IM contacts (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009), at which lipid transfer between the ER and IMs may occur (Prinz, 2014; Phillips and Voeltz, 2016; Zhang and Hu, 2016). Several factors have been identified that mediate the formation of dynamic ER–IM contacts. The two ER-resident tail-anchored VAP proteins (VAPA and VAPB) interact with FIP200 and ULK1 through their conserved FFAT motifs (two phenylalanines in an acidic tract), contributing to the recruitment of ULK1 and also stabilization of the ULK1/FIP200 complex at the autophagosome formation sites on the ER (Zhao et al., 2018). The interaction between FIP200 and the WD40-repeat containing PI(3)P-binding protein WIPI2 (one of four mammalian ATG18 homologues) also participates in mediating the ER–IM contact (Zhao et al., 2017, 2018). Yeast ATG2 tethers the ER to the leading edges of the growing IMs by simultaneously binding to the ER and also acting cooperatively with Atg18 to bind ATG9 at the IM extremities (Gómez-Sánchez et al., 2018; Kotani et al., 2018). Whether mammalian ATG2 also participates in ER–IM contact formation has yet to be determined. The ER-transmembrane protein EPG-3/VMP1 modulates disassembly of ER–IM contacts. VMP1 activates the ER Ca2+ channel SERCA to disassociate IMs from the ER, probably through perturbation of local Ca2+ concentration (Zhao et al., 2017). VMP1 depletion causes stable association of IMs with the ER, and the IMs then fail to proceed into closed autophagosomes (Zhao et al., 2017).
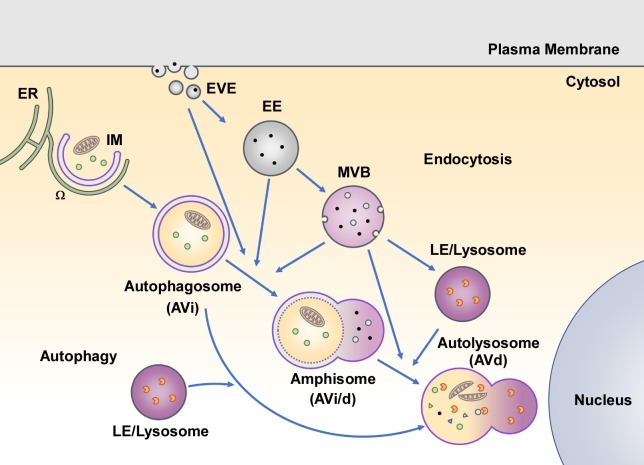
Figure 1.
Overview of the autophagy pathway. Autophagosomes are generated at PI(3)P-enriched subdomains of the ER, called omegasomes (Ω). A cup-shaped autophagosomal precursor structure, the IM, forms in close association with the omegasome. Upon closure of the IM, cytoplasmic contents are enclosed in double-membrane autophagosomes, also known as AVi. Autophagosomes undergo a series of fusion processes with various endolysosomal compartments, including the earliest vesicular endocytic vesicles (EVE), early endosomes (EE), MVBs, and LEs/lysosomes to form amphisomes, also known as AVi/d. Amphisomes finally mature into functional autolysosomes, also called AVd.
The final closure of IMs into autophagosomes appears to be completed by membrane abscission mediated by the endosomal sorting complex required for transport (ESCRT; Yu and Melia, 2017; Takahashi et al., 2018) and is accompanied by a change in morphology from elliptical expanding IMs to spherical sealed autophagosomes (Tsuboyama et al., 2016). This process is impaired in cells deficient in the ESCRT machinery and also the ATG8 conjugation system (Tsuboyama et al., 2016; Takahashi et al., 2018). Upon closure, nascent autophagosomes dissociate from the ER and undergo maturation.
Autophagosome maturation involves multiple entry sites for endocytic vesicles
AVs can be classified into three types in EM analysis based on their morphology and enzymatic characteristics. Nascent autophagosomes, termed AV-initial (AVi), consist of double-membrane structures containing unaltered cytoplasmic constituents. AV-intermediates (AVi/ds) represent amphisomes with a single limiting membrane that shows no or early signs of content degradation, while AV-degradative (AVd) structures are autolysosomes containing components at various stages of degradation (Eskelinen, 2005; Fig. 1). A series of elegant EM analyses using different endosome labeling methods were performed in the 1990s to determine the stage of the autophagic and endocytic pathway when the fusion occurs (Dunn, 1990; Tooze et al., 1990; Yokota et al., 1995). Dunn (1990) revealed that maturation of AVs into degradative vacuoles in rat liver cells occurs in a stepwise fashion by sequential acquisition of lysosome membrane proteins and lysosomal hydrolases and acidification of the lumen. These steps appear to involve fusion with different stages of endolysosomal compartments (Dunn, 1990). Tooze et al. (1990) showed that the endocytic tracer HRP (a marker of fluid phase endocytosis) is first detected in AVi/ds (amphisomes) in pig exocrine pancreatic cells, indicating the convergence of early endosomes and AVis. Using immunogold cytochemistry and morphometric analysis, Liou et al. (1997) also found that in rat liver cells, AVis coalesce with vesicular endosomes, including small vesicles of the size of the earliest endocytic vesicles, and multivesicular bodies (MVBs). AVi is preferred over AVi/d and AVd as the partner for fusion with endocytic structures (Liou et al., 1997). Thus, fusion of autophagosomes with endosomes and lysosomes does not occur in a random order or in a strict sequential order, but in a multistage fashion (Fig. 1). The composition and spatiotemporal distribution of endolysosomal compartments, which vary in different cell types and even under different growth conditions, may result in different preferential fusion combinations.
Fusion of AVs with functional early endosomes and MVBs is required for autophagy. Impairment of early endosome function by depleting COPI subunits (β′, β, or α) inhibits autophagosome maturation, resulting in accumulation of AVis and AVi/ds (Razi et al., 2009). The accumulated AVs may have fused with very early endocytic compartments (before the appearance of the early endosomal protein EEA1), but are not acidic and lack lysosomal proteases (Razi et al., 2009). Autophagosome maturation is also impaired when the formation of functional MVBs is disrupted by impairing the activities of ESCRT, which is required for the invagination of the endosomal membrane and the formation of intraluminal vesicles (Filimonenko et al., 2007; Lee et al., 2007). Both autophagosomes and amphisomes accumulate in ESCRT-depleted cells, depending on the cell type and cell context (Filimonenko et al., 2007; Lee et al., 2007; Rusten et al., 2007). ESCRT proteins may be involved in the recruitment of factors essential for fusion, such as Rab7, to modulate autophagosome maturation (Urwin et al., 2010).
Live-cell imaging revealed that in addition to complete fusion, LC3-labeled autophagosomes/amphisomes can undergo kiss-and-run fusion with late endosomes (LEs)/lysosomes. In this process, a portion of the vesicle contents is delivered, but the two vesicles ultimately stay separate (Jahreiss et al., 2008). The mechanisms underlying this multievent delivery remain unknown.
SNARE complexes mediate autophagosome–LE/lysosome fusion
Membrane fusion is driven by the formation of four-helix bundles from one of each of the Qa, Qb, Qc, and R SNARE proteins that are localized in opposing membranes (Jahn and Scheller, 2006). Assembly of the trans-SNAREs is spatiotemporally controlled by the actions of Rab GTPase and tethering factors (Fig. 2 A).
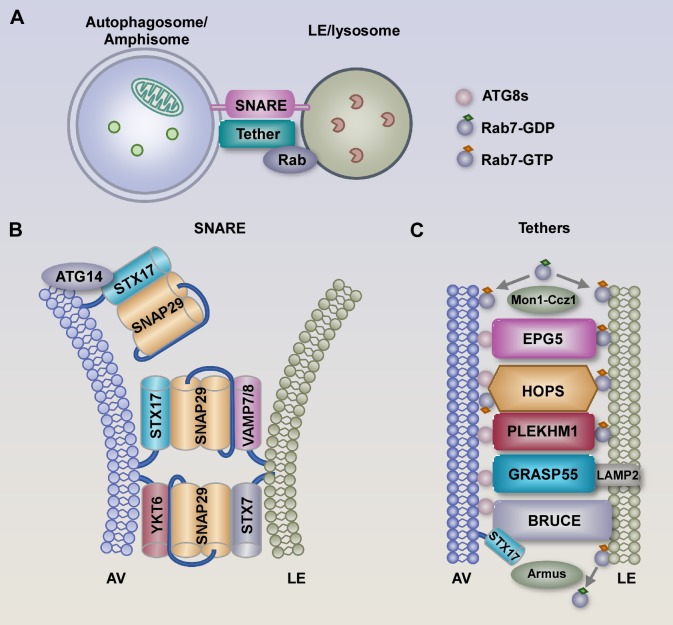
Figure 2.
SNAREs, tethers, and Rab proteins act in concert to mediate autophagosome–lysosome fusion. (A) Fusion of autophagosomes with LEs/lysosomes requires the concerted actions of SNARE proteins, Rab GTPases, and tethering factors. Amphisomes, which are single and membrane bound, also undergo fusion with LEs/lysosomes. (B) Two sets of cognate SNARE complexes (the autophagosomal STX17 [Qa]-SNAP29[Qbc]-endolysosomal VAMP7/8 [R] complex and the lysosomal STX7 [Qa]-SNAP29-endolysosomal YKT6 [R] complex) function in parallel with each other to mediate autophagosome–LE/lysosome fusion. ATG14 interacts with STX17 and promotes and stabilizes the assembly of the STX17 and SNAP29 complex. (C) Tethering proteins, including EPG5, HOPS, and PLEKHM1, simultaneously bind to ATG8s on autophagosomes and Rab7 on LEs/lysosomes to tether the autophagosomes/amphisomes with LEs/lysosomes for fusion. GRASP55 binds to Atg8s and LAMP2 on lysosomes. Lysosomal-localized BRUCE tethers autophagosomes through interaction with both ATG8s and STX17. HOPS has also been shown to target to autophagosomes via the Mon1-Ccz1-Rab7 module to promote autophagosome maturation. Rab7 is cycled between inactive GDP forms and active GTP forms by the Mon1-Ccz1 complex and the GAP protein Armus, respectively, during autophagosome maturation.
The STX17-SNAP29-VAMP8 complex and the YKT6-SNAP29-STX7 complex
Two cognate SNARE complexes act additively to mediate fusion of autophagosomes with LEs/lysosomes (Fig. 2 B). One complex consists of the autophagosomal-localized STX17 (Qa), SNAP29 (Qbc), and endolysosomal-localized VAMP8 (mammalian cells)/VAMP7 (Drosophila melanogaster and Caenorhabditis elegans; Itakura et al., 2012). Another complex is composed of the autophagosomal R-SNARE YKT6, SNAP29, and lysosomal-localized Qa SNARE STX7 (Matsui et al., 2018). Depletion of these SNARE proteins causes accumulation of autophagosomes in different systems (Sato et al., 2011; Itakura et al., 2012; Takáts et al., 2013; Matsui et al., 2018). YKT6 also forms a complex with STX17 and SNAP29 (Matsui et al., 2018), which could be involved in fusion of autophagosomes with amphisomes/autolysosomes or homotypic fusion of autophagosomes and/or autolysosomes, depending on their respective localization. Multiple sets of SNARE complexes may promote the fusion efficiency or mediate fusion of autophagosomes with different populations of LEs/lysosomes containing differential levels of VAMP7/8 and STX7. Disassembly of the cis-SNARE complex on postfusion membranes, a process catalyzed by NSF and αSNAP, is essential for completion of the autophagosome–LE/lysosome fusion process (Ishihara et al., 2001; Abada et al., 2017).
When and how are the STX17 and YKT6 proteins translocated to autophagic structures? STX17 and YKT6 are absent in IMs and are independently targeted to autophagosomes (Matsui et al., 2018). The recruitment of STX17 is accompanied by autophagosome closure. STX17 also labels elliptical autophagosome-like structures in cells deficient in ESCRT-III components and the ATG8 conjugation system, suggesting that STX17 can be recruited before the completion of autophagosome closure (Tsuboyama et al., 2016; Takahashi et al., 2018). The mechanisms mediating the targeting of STX17 and YKT6 to autophagosomes are distinct from their targeting to other membrane compartments. STX17 is a hairpin-tailed membrane-localized protein (Itakura et al., 2012). The charged residues at the C-terminal flanking region are important for targeting of STX17 to the ER and mitochondria, but are dispensable for its localization to the autophagosome, while the glycine zipper-like motif in the trans-membrane domains is essential for integration of STX17 into the autophagosomal membrane but not for its targeting to the ER and mitochondria (Itakura et al., 2012). In YKT6, the C-terminal cysteine residues for palmitoylation and farnesylation are essential for its Golgi localization but dispensable for its targeting to autophagosomes (Matsui et al., 2018). The N-terminal longin domain is required and sufficient for the autophagosomal localization of YKT6 (Matsui et al., 2018). Efficient translocation of STX17 to autophagosomes is facilitated by the small guanosine triphosphatase IRGM in human cells (Kumar et al., 2018). IRGM interacts directly with STX17 and also with ATG8s via a noncanonical LIR region (Kumar et al., 2018). The motifs involved in targeting STX17 and YKT6 to autophagosomes need to be released for subsequent assembly of trans-SNARE complexes. The LIR motif within the SNARE domain of STX17 binds to ATG8s (Kumar et al., 2018). The longin domain in YKT6 binds to the intramolecular SNARE domain to inhibit SNARE complex assembly. However, ATG8s are not essential for STX17 and YKT6 recruitment. In cells depleted of all six ATG8s, or lacking the Atg8 conjugation system, STX17 and YKT6 still target to autophagosomes (Tsuboyama et al., 2016). SNAP29 contains neither a trans-membrane domain nor a membrane-anchoring lipidation site, and thus it is recruited by interacting with STX17 or STX7.
VAMP7/8 are targeted to lysosomes via endocytic internalization and trafficking. The endolysosomal trafficking of VAMP7/8 is regulated by Rab21 and its guanine nucleotide exchange factor (GEF) protein MTMR13 (Jean et al., 2015). Rab21 interacts with VAMP7/8, and the interaction is enhanced in response to starvation. In Rab21- or MTMR13-depleted cells, VAMP8 accumulates at early endosomes and leads to an increased number of autophagosomes (Jean et al., 2015).
To maintain membrane identity and also to drive new rounds of fusion events, post-fusion SNAREs traffic back to their steady-state locations. Dissociation of STX17 from autolysosomes coincides with the collapse of the autophagosomal inner membrane and is independent of luminal acidification (Tsuboyama et al., 2016). The molecular mechanisms sensing the integrity of the inner membrane and retrieval of SNAREs from autolysosomes have yet to be determined.
Tether proteins determine the fusion specificity of autophagosomes with LEs/lysosomes
Tethers enhance vesicle fusion specificity and efficiency. Tethering factors capture intracellular trafficking vesicles and bring them closer with targeting membranes, promoting and/or stabilizing the assembly of SNARE complexes (Cai et al., 2007; Yu and Hughson, 2010). Tethers are recruited to specific membranes via their coordinated binding to Rab proteins, phospholipids, and SNAREs (Cai et al., 2007; Yu and Hughson, 2010). Multiple tethering factors have been identified that localize on autophagosomes and/or LEs/lysosomes to promote autophagosome maturation (Fig. 2 C).
EPG5
The essential autophagy gene EPG5 was identified from genetic screens in C. elegans (Tian et al., 2010). EPG5 is localized on LEs/lysosomes under normal growth conditions and targets amphisomes/autolysosomes upon autophagy induction (Wang et al., 2016). EGP5 is a Rab7 effector, and Rab7 mediates its membrane targeting. EPG5 recognizes autophagosomes by interacting with LC3 and also with assembled STX17-SNAP29. EPG5 stabilizes and facilitates the assembly of trans-SNARE proteins to facilitate fusion between autophagosomes and LEs/lysosomes (Wang et al., 2016). EPG5 depletion causes formation of nondegradative autolysosomes (Zhao et al., 2013a; Wang et al., 2016). The membrane tethering and fusion-promoting activity of EPG5 has also been demonstrated in STX17-SNAP29-VAMP7–mediated fusion of reconstituted proteoliposomes (Wang et al., 2016).
ATG14
ATG14, a component of the VPS34 PI(3)P kinase complex, is also localized on sealed autophagosomes. ATG14 binds to the SNARE core domain of STX17 and stabilizes the STX17-SNAP29 complex to promote fusion of autophagosomes to endolysosomes (Diao et al., 2015). In reconstituted proteoliposome systems, ATG14 contains tethering activity and also promotes membrane fusion mediated by the STX17-SNAP29-VAMP8 complex (Diao et al., 2015).
Homotypic fusion and protein sorting (HOPS) complex
The multisubunit HOPS complex is involved in multiple fusion processes with LEs/lysosomes (Yu and Hughson, 2010). HOPS is recruited to the membrane by binding to Rab7 and phospholipids such as PI(3)P (Stroupe et al., 2006). HOPS facilitates membrane fusion by promoting the assembly of the trans-SNARE complex, and also by functioning as an SM protein to promote SNARE-mediated fusion (Cai et al., 2007; Yu and Hughson, 2010). The HOPS complex is recruited to autophagosomes via its interaction with STX17 (Jiang et al., 2014; Takáts et al., 2014) or by the Mon1-Ccz1-Rab7 module (Hegedűs et al., 2016; Gao et al., 2018b) to promote autophagosome maturation.
PLEKHM1
PLEKHM1 acts as a Rab7 effector and is localized on LEs/lysosomes (Tabata et al., 2010). PLEKHM1 binds to ATG8 family proteins (see below), preferentially to GABARAPs, for autophagosome capture (McEwan et al., 2015; Nguyen et al., 2016). PLEKHM1 recruits the HOPS complex to LEs/lysosomes and hence promotes the assembly of the SNARE complex (McEwan et al., 2015). The function of PLEKHM1 in autophagosome maturation appears to be cell type specific. Depletion of PLEKHM1 causes no autophagy defect in A549 lung adenocarcinoma cells (Tabata et al., 2010) or in C. elegans (Wang et al., 2016).
Baculovirus IAP repeat-containing ubiquitin-conjugating enzyme (BRUCE)
BRUCE is localized on LEs/lysosomes and promotes autolysosome formation independent of its ubiquitin-conjugating activity (Ebner et al., 2018). BRUCE interacts with ATG8s, preferentially with GABARAP and GABARAPL1, via noncanonical LIR-containing regions, and also interacts with STX17 and SNAP29 (Ebner et al., 2018). Depletion of BRUCE leads to a defect in autophagosome–lysosome fusion (Ebner et al., 2018).
GRASP55
The Golgi stacking protein GRASP55 is O-GlcNAc modified and locates in medial and trans-Golgi cisternae. Upon de-O-GlcNAcylation under glucose starvation, GRASP55 is targeted to autophagosomes and LEs/lysosomes (Zhang et al., 2018). GRASP55 interacts via a LIR motif with Atg8 members, preferentially with LC3B, and simultaneously binds to LAMP2 on LEs/lysosomes to promote fusion (Zhang et al., 2018). Depletion of GRASP55 inhibits autophagosome maturation, while expression of an O-GlcNAcylation–defective GRASP55 mutant accelerates autophagic flux (Zhang et al., 2018).
Multiple tethering factors may act coordinately to enhance the efficiency and specificity of fusion, and mediate the fusion of autophagosomes with different populations of LEs/lysosomes. Different tethering factors may be differentially preferred by distinct SNARE complexes. Furthermore, different tethers may have distinct functions in mediating autophagosome–lysosome and amphisome–lysosome fusion in different cell types, tissues, and stress conditions, resulting in accumulation of AVs at different stages in a cell context–dependent manner. Tethers and SNARE proteins governing the fusion of autophagosomes with early endocytic vesicles remain to be determined.
ATG8 family members promote autophagosome maturation
ATG8 family members play multiple roles in autophagy. The single ATG8 in yeast is essential for IM expansion (Nakatogawa et al., 2009; Feng et al., 2014). LGG-1 and LGG-2 in C. elegans, which belong to the GABARAP and LC3 families, respectively, act differentially in autophagy. LGG-1 depletion blocks autophagosome formation, while loss of LGG-2 causes the formation of smaller autophagosomes (Wu et al., 2015). Mammalian cells contain six ATG8 homologues, in the LC3 subfamily (LC3A, LC3B, and LC3C) and the GABARAP subfamily (GABARAP, GABARAPL1, and GABARAPL2). The LC3 subfamily promotes elongation of IMs, while the GABARAP subfamily acts at the step of autophagosome closure (Weidberg et al., 2010). In cells depleted of all six ATG8s, sealed autophagosomes can be formed, but the process is severely delayed and the autophagosomes are smaller (Nguyen et al., 2016).
ATG8s, predominantly the GABARAP subfamily, play important roles in autophagosome maturation (Nguyen et al., 2016; Pontano Vaites et al., 2017). Autophagosome–lysosome fusion is severely impaired by depletion of the three GABARAP members, and is almost completely blocked by depletion of all six ATG8s (Nguyen et al., 2016; Pontano Vaites et al., 2017). Tethering factors, including EPG5, PLEKHM1, BRUCE, and GRASP55, interact with ATG8s for initial capture of autophagosomes. ATG8s also target HOPS to the autophagosome via direct interaction or indirectly by recruiting the Mon1-Ccz1-Rab7 module (Manil-Ségalen et al., 2014; Gao et al., 2018b). GABARAPs also target palmitoylated PI4KIIα to autophagosomes for generation of PI(4)P, which is critical for autophagosome–lysosome fusion (Wang et al., 2015). LC3 and GABARAP possess membrane tethering and homotypic fusion activity (Nakatogawa et al., 2007; Weidberg et al., 2011; Wu et al., 2015), which may directly contribute to fusion of autophagic structures. In contrast to cells depleted of all ATG8s, cells deficient in the ATG8 conjugation systems (e.g., ATG3, ATG5, and ATG7 KO cells) can still support the fusion of STX17-positive autophagosome-like structures with lysosomes (Tsuboyama et al., 2016). Thus, lipidation of ATG8s may not be absolutely required for their role in autophagosome maturation. The ATG8 conjugation system is also important for efficient degradation of the inner autophagosomal membrane (Tsuboyama et al., 2016).
Rabs, GEFs, and GTPase-activating proteins (GAPs) regulate autophagosome maturation
Rab GTPases specify membrane identity and regulate various processes of intracellular vesicle trafficking, including budding, uncoating, transport, and fusion, by recruiting effector proteins (Stenmark, 2009; Mizuno-Yamasaki et al., 2012). Rab proteins cycle between GTP-bound active forms and GDP-bound inactive forms. The recruitment, activation, and function of Rab proteins at a specific membrane compartment is controlled by the concerted actions of GEFs and GAPs (Mizuno-Yamasaki et al., 2012). GEFs target Rabs to a specific membrane and also catalyze exchange of GDP to GTP, which triggers recruitment of downstream effectors by Rabs (Stenmark, 2009; Mizuno-Yamasaki et al., 2012). GAPs terminate Rab function in trafficking by accelerating conversion from the GTP- to the GDP-bound form (Stenmark, 2009).
Rab7 is located on LEs/lysosomes (Gutierrez et al., 2004; Jäger et al., 2004; Szatmári and Sass, 2014). In Rab7-knockdown cells, fusion of autophagosomes with early endosomes and MVBs still occurs, but fusion with lysosomes is blocked (Gutierrez et al., 2004; Jäger et al., 2004; Eskelinen, 2005). Rab7 on LEs/lysosomes recruits tethering factors, including EPG5, PLEKHM1, and HOPS, to promote the assembly of trans-SNARE complexes for fusion (Fig. 2 C). Rab7 also functions on autophagosomes to promote maturation (Hegedűs et al., 2016; Gao et al., 2018b). The Mon1-Ccz1 complex, which acts as the Rab7 GEF and stabilizes its localization (Nordmann et al., 2010), is targeted to autophagosomes via direct interaction with ATG8, which in turn recruits Rab7 and HOPS to promote autophagosome fusion with LEs/lysosomes (Hegedűs et al., 2016; Pontano Vaites et al., 2017; Gao et al., 2018b).
The membrane trafficking function of Rab7 is under tight spatiotemporal control. Rab7 inactivation triggers its release and facilitates completion of fusion. TBC/Rab GAP Armus (TBC1D2A), which is recruited to autophagosomes via interaction with LC3, promotes Rab7 cycling and thus facilitates autophagosome maturation (Carroll et al., 2013). Armus depletion impairs Rab7 inactivation and delays autophagic flux (Carroll et al., 2013).
Rab2, previously known to regulate vesicle trafficking at the ER and Golgi, is also involved in autophagosome maturation. Rab2 localizes to autophagosomes and recruits the HOPS complex to promote autophagosome maturation (Fujita et al., 2017; Lőrincz et al., 2017). Rab2 depletion causes accumulation of autophagosomes in fly cells and amphisomes or autolysosomes in MEF cells (Fujita et al., 2017; Lőrincz et al., 2017).
Vps34 PI(3)P kinase complexes modulate autophagosome maturation
Two Vps34 PI(3)P kinase complexes have been identified that act at different steps of the autophagy pathway. Both of them contain Beclin 1, hVps34, and hVps15, while UVRAG and Atg14L bind to Beclin 1 in a mutually exclusive manner (Matsunaga et al., 2009). The Atg14L-containing Vps34 complex generates PI(3)P, which is essential for autophagosome formation (Feng et al., 2014). Temporal turnover of PI(3)P from end-stage IMs and/or nascent autophagosomes triggers disassociation of PI(3)P-binding proteins and other Atg proteins and is a prerequisite for fusion of autophagosomes with endolysosomal compartments. Loss of function of the myotubularin PI(3)P phosphatase Ymr1 results in persistent association of Atg proteins with autophagosomes that fail to fuse with the vacuole in yeast (Cebollero et al., 2012). Loss of function of the C. elegans myotubularin family PI(3)P phosphatase MTM-3 causes persistent association of ATG-18 and impairs autophagosome maturation (Wu et al., 2014). PI(3)P, however, is also required for autophagosome maturation. PI(3)P on autophagosomes facilitates the recruitment of Mon1/Ccz1 or directly stabilizes Rab7 (Hegedűs et al., 2016; Bas et al., 2018; Gao et al., 2018b). Therefore, PI(3)P levels on autophagic structures must be deliberately controlled to ensure autophagosome maturation.
The UVRAG-containing PI(3)P kinase complex promotes autophagosome maturation (Matsunaga et al., 2009). UVRAG is localized on early endosomal compartments under normal growth conditions (Liang et al., 2008). Upon autophagy induction, UVRAG efficiently recruits the HOPS complex to AVs, thus stimulating Rab7 activity and autophagosome maturation (Liang et al., 2008). UVRAG also activates the kinase function of the hVps34 complex (Sun et al., 2010). Targeting of the UVRAG-Beclin1-Vps34 complex to autophagosomes is mediated by protein associated with UVRAG as autophagy enhancer (Pacer), which is recruited to autophagosomes by its interaction with STX17 and phosphoinositides (Cheng et al., 2017). The role of the UVRAG-containing hVps34 complex in autophagosome maturation is negatively regulated by Rubicon (Matsunaga et al., 2009; Zhong et al., 2009). Rubicon is highly enriched on early endosomes and directly interacts with Rab7, preferentially with GTP-bound Rab7 (Sun et al., 2010; Tabata et al., 2010). Rubicon interacts with Rab7 and UVRAG in a mutually exclusive manner (Sun et al., 2010). Rubicon may regulate autophagosome maturation by sequestration of UVRAG, thus preventing it from stimulating Vps34 kinase activity and binding HOPS (Liang et al., 2010; Sun et al., 2010). The role of UVRAG in autophagosome maturation has been challenged by several recent studies. UVRAG is not present in the HOPS complex that mediates autophagosome maturation (Jiang et al., 2014). The fusion of autophagosomes with lysosomes is largely unaffected by depletion of UVRAG in Drosophila and also in mammalian cells (Itakura et al., 2008; Jiang et al., 2014; Takáts et al., 2014). The role of UVRAG in autophagosome maturation may depend on specific cell contexts, which differ in endolysosomal compartments, and also on experimental conditions, such as UVRAG overexpression.
The location of PI(3)P, Rabs, and tethering factors for autophagosome maturation
PI(3)P, Rabs, and tethering factors act coordinately to promote the assembly of the trans-SNARE complex. The sites where these different components function during autophagosome maturation have not been fully elucidated. Some components act at multiple steps of the autophagy pathway, and their late function during autophagosome maturation may be masked by their role in earlier steps of autophagosome formation. For example, Atg14 and PI(3)P act at both autophagosome formation and maturation (Feng et al., 2014; Diao et al., 2015; Bas et al., 2018). The location at which these factors act and the stage of the AVs that accumulate after their depletion are mainly determined by their colocalization with LC3, STX17, and markers for LEs/lysosomes. LC3 and STX17 label nascent autophagosomes, amphisomes, and also autolysosomes (early-stage autolysosomes for STX17; Tsuboyama et al., 2016). More comprehensive analysis using a combination of assays, such as immuno-EM analysis, reporters for different vesicles, and tracers that are delivered to different stages of endocytic vesicles, is lacking for most of the factors involved in autophagosome maturation.
In vitro reconstitution of the fusion of autophagosomes with endosomes/lysosomes is useful to identify the essential factors and also to define the location of these factors during autophagosome maturation. Using isolated yeast vacuoles, autophagosomes, and cytosolic material derived from wild-type and mutants depleted of a specific autophagy gene, it was determined that Atg14 and Vps34 must be present in the cytosolic fraction for fusion to occur (Bas et al., 2018). Ypt7 (yeast Rab7 homologue) acts on both autophagosomes and vacuoles to enhance fusion efficiency (Bas et al., 2018). Consistent with this, purified autophagosomes carry Ypt7 and Mon1-Ccz1 on their surface (Gao et al., 2018a). Autophagosome-localized PI(3)P, generated by cytosolic Atg14/Vps34, may help to recruit and stabilize the Mon1-CCz1-Ypt7 complex on autophagosomes (Bas et al., 2018; Gao et al., 2018a). HOPS function is required on vacuoles but not on autophagosomes in the in vitro reconstitution assay (Bas et al., 2018), although previous studies showed that HOPS also acts on autophagosomes to promote fusion (Bas et al., 2018; Gao et al., 2018a). The in vitro reconstitution assay also revealed that autophagosome–vacuole fusion requires the R-SNARE Ykt6 on the autophagosome and Vam3 (Qa), Vti1 (Qb), and Vam7 (Qc) on the vacuole (Bas et al., 2018; Gao et al., 2018a).
Lysosomal properties modulate fusion with autophagosomes and amphisomes
Lysosomes are the final destination for degradation of autophagosome-enclosed materials and also serve as a platform for mTORC1 activation to regulate autophagy induction. The acidic environment of lysosomes is not essential for autophagosome–lysosome fusion (Mauvezin et al., 2015; Tsuboyama et al., 2016). However, the lysosomal membrane protein LAMP2 promotes fusion of lysosomes with autophagosomes/amphisomes (Tanaka et al., 2000; Eskelinen, 2005). LAMP2 has been shown to interact with GRASP55 to facilitate the fusion process (Zhang et al., 2018).
The lipid composition of the lysosomal membrane also regulates the fusion capability of lysosomes. Reduction or elevation of the lysosomal PI(3,5)P2 level impairs autophagosome/amphisome–lysosome fusion (de Lartigue et al., 2009; Ferguson et al., 2009). PI(3,5)P2 is synthesized by the PI(3)P 5-kinase Fab1 (PIKfyve) and degraded by FIG4 (the PI(3,5)P2 5-phosphatase)/VAC14 (the scaffold protein; Gary et al., 1998, 2002). Fab1 is required for the maturation of amphisomes to autolysosomes (Rusten et al., 2007). Mice deficient in Fig4 and Vac14 have a severely reduced cellular concentration of PI(3,5)Ps and also exhibit a defect in formation and recycling of autolysosomes (Ferguson et al., 2009). The lysosomal-localized inositol polyphosphate-5-phosphatase E (INPP5E) down-regulates lysosomal PI(3,5)P2, which stabilizes actin filaments on the lysosome surface that are essential for lysosome-autophagosome fusion (Hasegawa et al., 2016). Autophagosomes accumulate in INPP5E-depleted cells (Hasegawa et al., 2016). Levels of lysosomal cholesterol also modulate autophagic flux. The myosin MYO1C facilitates recycling of cholesterol-enriched lipid rafts from intracellular compartments to the cell surface. Aberrant accumulation and distribution of cholesterol in the endolysosomal membrane in MYO1C-depleted cells or in lysosomal storage disorder cells are linked to reduced fusion of lysosomes with AVs (Fraldi et al., 2010; Brandstaetter et al., 2014). Cholesterol-enriched endolysosomal membranes sequestrate assembled SNARE complexes, including Syntaxin 7 and VAMP7 and thus impair their sorting and recycling (Fraldi et al., 2010). Other aspects of LEs/lysosomes influenced by a change in cholesterol levels, such as membrane rigidity and fluidity, may also impact autophagosome–lysosome fusion.
Lysosome positioning regulates fusion with autophagosomes and amphisomes
Lysosomes are most concentrated in the perinuclear region in nonpolarized cells and their position influences their function (reviewed in Pu et al., 2016). Autophagosomes are formed throughout the cytoplasm (Jahreiss et al., 2008). Fusion is modulated by bidirectional movement of autophagosomes and lysosomes. Autophagosomes in the cell periphery move centripetally to fuse with juxtanuclear lysosomes or with lysosomes that undergo centrifugal transport. The centripetal movement (also known as minus end transport) of autophagosomes and lysosomes is driven by the motor protein dynein, while their centrifugal movement (plus end transport) is mediated by kinesin motors (Jahreiss et al., 2008; Pu et al., 2016). Lysosome distribution is responsive to nutrient availability and influences mTORC1 activity. Starvation induces perinuclear clustering of lysosomes and inhibits mTORC1, while relocalization of lysosomes to the periphery by overexpression of kinesin activates mTORC1 (Korolchuk et al., 2011). Movement of lysosomes, autophagosomes, and amphisomes affects their fusion frequency and also influences recruitment of tethering factors. The Rab7 effector FYCO1, which binds to LC3 and PI(3)P, mediates plus end–directed AV transport (Pankiv et al., 2010). Depletion of FYCO1 leads to the perinuclear accumulation of clustered AVs. Transport and positioning of lysosomes and AVs are also regulated by the cholesterol-sensing Rab7 effector ORP1L (Rocha et al., 2009; Wijdeven et al., 2016). ORP1L is localized on LEs/lysosomes, amphisomes, and autolysosomes and adopts different conformational states specified by the cholesterol level on these vesicles (Rocha et al., 2009; Wijdeven et al., 2016). Under low cholesterol conditions, ORP1L forms a complex with VAPA on the ER, which removes the p150Glued subunits of the dynein-dynactin motor from the Rab7-RILP complex and subsequently suppresses the centripetal movement of these vesicles (Rocha et al., 2009; Wijdeven et al., 2016). Formation of the ORPIL-VAPA ER complex also inhibits the Rab7-RILP–mediated recruitment of PLEKHM1 and the HOPS complex, which impairs autophagosome maturation (Wijdeven et al., 2016; Fig. 3).
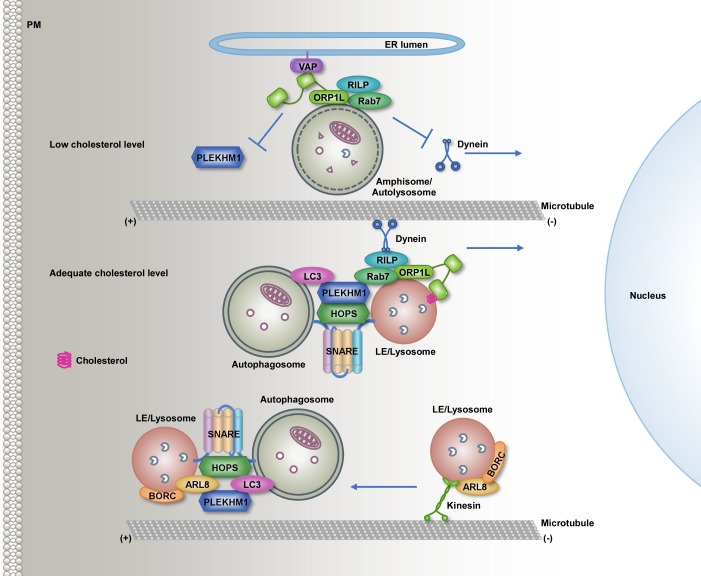
Figure 3.
Lysosome positioning modulates autophagosome–lysosome fusion efficiency. Center: Amphisome- and lysosome-localized ORP1L senses cholesterol levels. Under adequate cholesterol conditions, ORP1L binds to cholesterol on the membrane through its ORD domain, which further binds to Rab7/RILP for subsequent recruitment of PLEKHM1 and HOPS to facilitate autophagosome–endolysosome fusion. ORP1L also promotes the centripetal movement of amphisomes/autolysosomes through RILP-mediated recruitment of the motor protein dynein. Top: Under low-cholesterol conditions, the interaction of ORP1L with the ER protein VAPA tethers AVs to the ER, which inhibits dynein-mediated centripetal transport and also impairs autophagosome–lysosome fusion by reducing the recruitment of tether proteins. Bottom: The BORC complex increases the ARL8-dependent interaction with kinesins, leading to the centrifugal movement of lysosomes to the cell periphery for efficient fusion with autophagosomes. BORC also facilitates the fusion process by recruiting HOPS and promoting assembly of the STX17-SNAP29-VAMP8 SNAREs. PM, plasma membrane.
Lysosome dispersal increases the fusion of lysosomes with peripheral autophagosomes, which is also required for autophagic flux (Fig. 3). The multisubunit BORC complex interacts with ARL8 on lysosomes to promote ARL8-dependent association to kinesins, resulting in lysosome movement toward the cell periphery. Depletion of BORC causes the juxtanuclear accumulation of lysosomes without affecting mTORC1 activity and reduces the frequency of encounter of peripheral autophagosomes with lysosomes (Jia et al., 2017). BORC also promotes the ARL8-mediated recruitment of the HOPS complex and subsequently facilitates the assembly of STX17-SNAP29-VAMP8 for autophagosome–lysosome fusion (Jia et al., 2017). ARL8b also directly binds to PLEKHM1 (Marwaha et al., 2017). Therefore, the movement of autophagosomes and lysosomes affects the frequency of encounter and also the recruitment of fusion factors.
Nutrient status integrates into the autophagosome maturation machinery
Nutrient status is sensed by distinct signals that ultimately impinge on the Atg1 complex and the Vps34 complex to regulate autophagy induction. Starvation also promotes autophagosome maturation by enhancing the fusion process and lysosome biogenesis (Fig. 4). Levels of UDP-GlcNAc, the precursor for post-translational O-GlcNAcylation, are highly responsive to the availability of glucose, fatty acids, uridine, and glutamine (Slawson et al., 2010). Some components of the autophagosome fusion machinery are O-GlcNAc modified. SNAP29 is O-GlcNAcylated at multiple sites (Ser2, Ser61, Thr130, and Ser153 in mammals and Ser70, Ser134, Thr143, and Ser249 in C. elegans), and levels of O-GlcNAcylated SNAP29 are reduced by nutrient starvation (Guo et al., 2014). O-GlcNAc modification attenuates the interaction of SNAP29 with STX17 and VAMP8/7. Loss of function of OGT-1 (O-GlcNAc transferase) or mutation of the O-GlcNAc sites in SNAP29 promotes formation of the SNAP29-containing SNARE complex and enhances autophagic flux (Guo et al., 2014). De-O-GlcNAcylation of GRASP55 elicits its translocation to autophagosomes and LEs/lysosomes to promote fusion (Zhang et al., 2018). The interaction of UVRAG with Rubicon is enhanced by mTORC1-mediated phosphorylation of UVRAG at Ser498 (Kim et al., 2015). Dephosphorylation of UVRAG triggers the release of Rubicon, which then positively regulates Vps34 kinase activity and also interacts with the HOPS complex (Kim et al., 2015). Therefore, post-translational modifications such as O-GlcNAcylation and phosphorylation serve as a mechanism for integrating nutrient availability with autophagosome maturation.
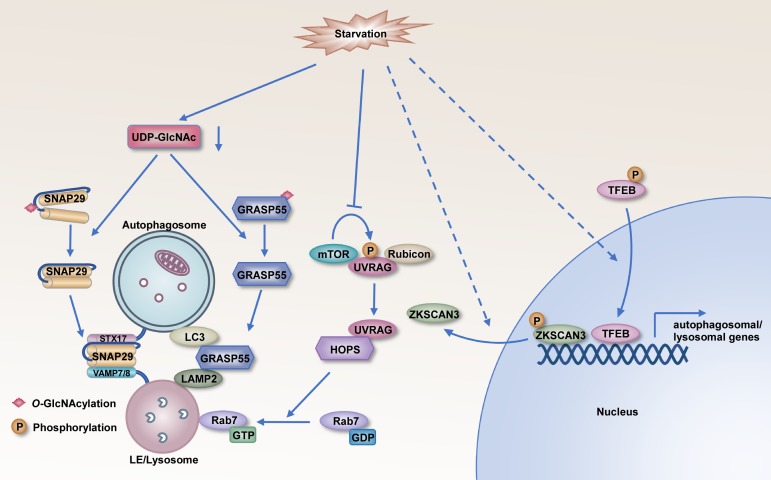
Figure 4.
Nutrient status regulates autophagosome maturation through multiple mechanisms. Starvation decreases O-GlcNAcylation of SNAP29 and promotes its interaction with STX17 and VAMP8 to form trans-SNARE complexes for autophagosome–LE/lysosome fusion. Starvation-induced de-O-GlcNAcylation of GRASP55 causes its translocation to autophagosomes. De-O-GlcNAcylated GRASP55 simultaneously binds to LC3 and LAMP2 to facilitate autophagosome maturation. Inhibition of mTORC1 activity by starvation leads to dephosphorylation of UVRAG, which releases Rubicon and subsequently recruits HOPS to promote autophagosome fusion. The transcription of a network of genes involved in autophagosome–lysosome fusion is activated by the transcription factor TFEB and inhibited by the transcriptional repressor ZKSCAN3. Starvation triggers dephosphorylation of TFEB, resulting in its nuclear translocation. The translocation of ZKSCAN3 out of the nucleus is also promoted by nutrient deficiency.
Starvation and stress also promote nuclear localization and activity of the bHLH-leucine zipper transcription factors TFEB and/or TFE3, which activate a network of genes involved in autophagosome and lysosome biogenesis and also autophagosome–lysosome fusion (Raben and Puertollano, 2016). Phosphorylation controls the cytoplasmic-nuclear shuttling of TFEB/TFE3; the nonphosphorylated proteins translocate into the nucleus. Distinct nutrient stimuli are transduced via different signaling pathways to mediate phosphorylation of TFEB/TFE3. Phosphorylation of TFEB at Ser 142 via extracellular signal-regulated kinase 2 retains TFEB in the cytosol (Settembre et al., 2011). mTORC1, which senses amino acid status and other stresses, phosphorylates TFEB at Ser211 and TFE3 at Ser321, creating binding sites for 14-3-3 proteins that mediate cytosolic retention (Settembre et al., 2012; Raben and Puertollano, 2016). Under starvation conditions, TFEB/TFE3 are dephosphorylated by calcineurin, a phosphatase activated by starvation-triggered lysosomal calcium release (Medina et al., 2015). PKC also controls nuclear translocation and activation of TFEB by regulating GSK3β activity (Li et al., 2016). GSK3β phosphorylates TFEB at Ser 134 and Ser 138 (Li et al., 2016).
The zinc-finger family DNA-binding protein ZKSCAN3 acts in opposition to TFEB/TFE3 to repress transcription of a set of genes involved in lysosome biogenesis/function and autophagosome–lysosome fusion (Chauhan et al., 2013). Subcellular localization of ZKSCAN3 is modulated by nutrient availability and stress (Chauhan et al., 2013). PKC activates JNK and p38 MAPK to phosphorylate ZKSCAN3 at Thr 153, which triggers ZKSCAN3 translocation out of the nucleus (Li et al., 2016).
Autophagosome maturation and neurodegenerative diseases
In neurons, distinct from other cell types, autophagosome formation and maturation exhibit spatiotemporal features. Autophagosomes are preferentially generated at the distal tip of the axon, and then undergo retrograde trafficking through the axon to the soma, accompanied by stepwise maturation into autolysosomes (Maday et al., 2012). Autophagosome formation in neurons requires the same set of autophagy proteins, although the membrane source for expansion of IMs may be distinct (Maday and Holzbaur, 2014). The STX17-containing SNARE complex, Rab7, EPG5, and the HOPS complex are involved in autophagosome maturation (Bains et al., 2011; Peng et al., 2012; Takáts et al., 2013; Zhao et al., 2013a; Cheng et al., 2015; Zhen and Li, 2015). Neurodegenerative diseases, such as Huntington’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (ALS), show impaired formation and function of autolysosomes, resulting in gradual accumulation of AVs in affected neurons and eventual cell death (Nixon et al., 2008). Mice with neural-specific depletion of genes essential for autophagosome formation, including Atg5, Atg7, Fip200, and Ei24, exhibit massive neuronal death (Hara et al., 2006; Komatsu et al., 2006; Liang et al., 2010; Zhao et al., 2012). However, these knockout mice fail to recapitulate the features of neurodegenerative diseases, including accumulation of AVs and damage of only a certain population of neurons (Hardy and Gwinn-Hardy, 1998).
Loss of function of EPG5 causes accumulation of nondegradative AVs (Zhao et al., 2013a). Epg5 KO mice develop age-dependent selective neuronal damage and exhibit characteristics of ALS (Zhao et al., 2013a). The pyramidal neurons in the fifth layer of the cerebral cortices and motor neurons in the anterior horn of the spinal cord are gradually lost in Epg5 KO mice, resulting in muscle atrophy and muscle denervation (Zhao et al., 2013a). Epg5 KO mice also display features of retinitis pigmentosa (Miao et al., 2016). Depletion of EPG5 impairs autophagy in various types of neurons; the mechanism underlying damage of only a specific population of neurons has yet to be determined (Zhao et al., 2013a). Recent human genetic studies revealed that recessive EPG5 mutations are causatively linked with the multisystem disorder Vici syndrome, characterized by neurodegenerative features as displayed in Epg5 KO mice, and also a defective immune response (Cullup et al., 2013; Zhao et al., 2013b). Mutations in the ESCRT-III complex subunit CHMP2B are associated with frontotemporal dementia and ALS (Skibinski et al., 2005; Parkinson et al., 2006). Different factors may act differentially in autophagosome maturation in distinct types of neurons, so that their loss of function causes autophagy defects of variable severity. Accumulation of AVs may interfere with endocytic trafficking, thus contributing to selective damage of a certain population of neurons.
Conclusion
Numerous factors involved in fusion of autophagosomes with LEs/lysosomes have been identified, including Rabs, tethers, and SNARE proteins; however, many fundamental questions remain to be answered. What is the molecular machinery that mediates the fusion of the earliest endocytic vesicles with autophagosomes? How do different tethering factors act coordinately in autophagosome maturation? Do different tethering factors mediate fusion of autophagosomes with distinct populations of endocytic vesicles? Is the homotypic fusion of autophagosomes/amphisomes/autolysosomes mediated by the same set of SNARE proteins as heterotypic fusion? How is the fusion of autophagosomes with other vesicles, such as recycling endosomes and secretory vesicles, prevented? In EPG5-deficient cells, autophagosomes undergo nonspecific fusion with other vesicles (Zhao et al., 2013a). How does the accumulation of AVs affect other endocytic trafficking and recycling processes? Understanding the mechanisms underlying autophagosome maturation will provide more insights into the pathogenesis of relevant human diseases as well as providing more therapeutic clues for treatment.
Acknowledgments
We are grateful to Dr. Isabel Hanson for editing the manuscript.
Work in the authors’ laboratory was supported by the National Natural Science Foundation of China (grants 31630048, 31421002, and 31561143001), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB19000000), the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (grant QYZDY-SSW-SMC006), and the Orphan Disease Center’s Million Dollar Bike Ride pilot grant program (MDBR-18-104-BPAN).
The authors declare no competing financial interests.
Author contributions: Y.G. Zhao and H. Zhang wrote and edited the manuscript.
References
- Abada A., Levin-Zaidman S., Porat Z., Dadosh T., and Elazar Z.. 2017. SNARE priming is essential for maturation of autophagosomes but not for their formation. Proc. Natl. Acad. Sci. USA. 114:12749–12754. 10.1073/pnas.1705572114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding A.L., and Baehrecke E.H.. 2017. Cleaning House: Selective Autophagy of Organelles. Dev. Cell. 41:10–22. 10.1016/j.devcel.2017.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains M., Zaegel V., Mize-Berge J., and Heidenreich K.A.. 2011. IGF-I stimulates Rab7-RILP interaction during neuronal autophagy. Neurosci. Lett. 488:112–117. 10.1016/j.neulet.2010.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas L., Papinski D., Licheva M., Torggler R., Rohringer S., Schuschnig M., and Kraft C.. 2018. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J. Cell Biol. 217:3656–3669. 10.1083/jcb.201804028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstaetter H., Kishi-Itakura C., Tumbarello D.A., Manstein D.J., and Buss F.. 2014. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome-lysosome fusion. Autophagy. 10:2310–2323. 10.4161/15548627.2014.984272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Reinisch K., and Ferro-Novick S.. 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 12:671–682. 10.1016/j.devcel.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Carroll B., Mohd-Naim N., Maximiano F., Frasa M.A., McCormack J., Finelli M., Thoresen S.B., Perdios L., Daigaku R., Francis R.E., et al. 2013. The TBC/RabGAP Armus coordinates Rac1 and Rab7 functions during autophagy. Dev. Cell. 25:15–28. 10.1016/j.devcel.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E., van der Vaart A., Zhao M., Rieter E., Klionsky D.J., Helms J.B., and Reggiori F.. 2012. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 22:1545–1553. 10.1016/j.cub.2012.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S., Goodwin J.G., Chauhan S., Manyam G., Wang J., Kamat A.M., and Boyd D.D.. 2013. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell. 50:16–28. 10.1016/j.molcel.2013.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.T., Zhou B., Lin M.Y., Cai Q., and Sheng Z.H.. 2015. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 209:377–386. 10.1083/jcb.201412046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Ma X., Ding X., Li L., Jiang X., Shen Z., Chen S., Liu W., Gong W., and Sun Q.. 2017. Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17. Mol. Cell. 65:1029–1043.e5. 10.1016/j.molcel.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Cheng X.T., Xie Y.X., Zhou B., Huang N., Farfel-Becker T., and Sheng Z.H.. 2018. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 217:3127–3139. 10.1083/jcb.201711083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullup T., Kho A.L., Dionisi-Vici C., Brandmeier B., Smith F., Urry Z., Simpson M.A., Yau S., Bertini E., McClelland V., et al. 2013. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat. Genet. 45:83–87. 10.1038/ng.2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lartigue J., Polson H., Feldman M., Shokat K., Tooze S.A., Urbé S., and Clague M.J.. 2009. PIKfyve regulation of endosome-linked pathways. Traffic. 10:883–893. 10.1111/j.1600-0854.2009.00915.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., Zhou Q., Wilz L.M., Li J., Vivona S., et al. 2015. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 520:563–566. 10.1038/nature14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A., Jr 1990. Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J. Cell Biol. 110:1935–1945. 10.1083/jcb.110.6.1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner P., Poetsch I., Deszcz L., Hoffmann T., Zuber J., and Ikeda F.. 2018. The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat. Commun. 9:599 10.1038/s41467-018-02823-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E.L. 2005. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 1:1–10. 10.4161/auto.1.1.1270 [DOI] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C.J., Lenk G.M., and Meisler M.H.. 2009. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 18:4868–4878. 10.1093/hmg/ddp460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M.C., Isaacs A., Brech A., Stenmark H., and Simonsen A.. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500. 10.1083/jcb.200702115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraldi A., Annunziata F., Lombardi A., Kaiser H.J., Medina D.L., Spampanato C., Fedele A.O., Polishchuk R., Sorrentino N.C., Simons K., and Ballabio A.. 2010. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 29:3607–3620. 10.1038/emboj.2010.237 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fujita N., Huang W., Lin T.H., Groulx J.F., Jean S., Nguyen J., Kuchitsu Y., Koyama-Honda I., Mizushima N., Fukuda M., and Kiger A.A.. 2017. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife. 6:e23367 10.7554/eLife.23367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Langemeyer L., Kümmel D., Reggiori F., and Ungermann C.. 2018b Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. eLife. 7:e31145 10.7554/eLife.31145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Reggiori F., and Ungermann C.. 2018a A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J. Cell Biol. 217:3670–3682. 10.1083/jcb.201804039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary J.D., Wurmser A.E., Bonangelino C.J., Weisman L.S., and Emr S.D.. 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143:65–79. 10.1083/jcb.143.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary J.D., Sato T.K., Stefan C.J., Bonangelino C.J., Weisman L.S., and Emr S.D.. 2002. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol. Biol. Cell. 13:1238–1251. 10.1091/mbc.01-10-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Melville D., Zhang M., and Schekman R.. 2013. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2:e00947 10.7554/eLife.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sánchez R., Rose J., Guimarães R., Mari M., Papinski D., Rieter E., Geerts W.J., Hardenberg R., Kraft C., Ungermann C., and Reggiori F.. 2018. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 217:2743–2763. 10.1083/jcb.201710116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M., Friedman J.R., Graham C., Babu M., and Nunnari J.. 2013. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 24:2918–2931. 10.1091/mbc.e13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A.L., Zhang Z., et al. 2014. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16:1215–1226. 10.1038/ncb3066 [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Munafó D.B., Berón W., and Colombo M.I.. 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117:2687–2697. 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., and Lippincott-Schwartz J.. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667. 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hardy J., and Gwinn-Hardy K.. 1998. Genetic classification of primary neurodegenerative disease. Science. 282:1075–1079. 10.1126/science.282.5391.1075 [DOI] [PubMed] [Google Scholar]
- Hasegawa J., Iwamoto R., Otomo T., Nezu A., Hamasaki M., and Yoshimori T.. 2016. Autophagosome-lysosome fusion in neurons requires INPP5E, a protein associated with Joubert syndrome. EMBO J. 35:1853–1867. 10.15252/embj.201593148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., and Yamamoto A.. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437. 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Hegedűs K., Takáts S., Boda A., Jipa A., Nagy P., Varga K., Kovács A.L., and Juhász G.. 2016. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell. 27:3132–3142. 10.1091/mbc.e16-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., and Ohsumi Y.. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702. 10.1091/mbc.12.11.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., and Mizushima N.. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 6:764–776. 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., and Mizushima N.. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19:5360–5372. 10.1091/mbc.e08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., and Eskelinen E.L.. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837–4848. 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- Jahn R., and Scheller R.H.. 2006. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Jahreiss L., Menzies F.M., and Rubinsztein D.C.. 2008. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 9:574–587. 10.1111/j.1600-0854.2008.00701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S., Cox S., Nassari S., and Kiger A.A.. 2015. Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep. 16:297–311. 10.15252/embr.201439464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Guardia C.M., Pu J., Chen Y., and Bonifacino J.S.. 2017. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy. 13:1648–1663. 10.1080/15548627.2017.1343768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., and Mizushima N.. 2014. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 25:1327–1337. 10.1091/mbc.e13-08-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Walker S.A., Okkenhaug H., Manifava M., Hummel E., Zimmermann H., Ahmed Q., Domart M.C., Collinson L., and Ktistakis N.T.. 2016. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7:12420 10.1038/ncomms12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.M., Jung C.H., Seo M., Kim E.K., Park J.M., Bae S.S., and Kim D.H.. 2015. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol. Cell. 57:207–218. 10.1016/j.molcel.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., and Tanaka K.. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T., Kirisako H., Koizumi M., Ohsumi Y., and Nakatogawa H.. 2018. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA. 115:10363–10368. 10.1073/pnas.1806727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Jain A., Farzam F., Jia J., Gu Y., Choi S.W., Mudd M.H., Claude-Taupin A., Wester M.J., Lidke K.A., et al. 2018. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol. 217:997–1013. 10.1083/jcb.201708039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Lee J.A., Beigneux A., Ahmad S.T., Young S.G., and Gao F.B.. 2007. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17:1561–1567. 10.1016/j.cub.2007.07.029 [DOI] [PubMed] [Google Scholar]
- Li Y., Xu M., Ding X., Yan C., Song Z., Chen L., Huang X., Wang X., Jian Y., Tang G., et al. 2016. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 18:1065–1077. 10.1038/ncb3407 [DOI] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., and Jung J.U.. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10:776–787. 10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C.C., Wang C., Peng X., Gan B., and Guan J.L.. 2010. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 285:3499–3509. 10.1074/jbc.M109.072389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou W., Geuze H.J., Geelen M.J., and Slot J.W.. 1997. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol. 136:61–70. 10.1083/jcb.136.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P., Tóth S., Benkő P., Lakatos Z., Boda A., Glatz G., Zobel M., Bisi S., Hegedűs K., Takáts S., et al. 2017. Rab2 promotes autophagic and endocytic lysosomal degradation. J. Cell Biol. 216:1937–1947. 10.1083/jcb.201611027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., and Holzbaur E.L.. 2014. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev. Cell. 30:71–85. 10.1016/j.devcel.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Wallace K.E., and Holzbaur E.L.. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417. 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manil-Ségalen M., Lefebvre C., Jenzer C., Trichet M., Boulogne C., Satiat-Jeunemaitre B., and Legouis R.. 2014. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell. 28:43–55. 10.1016/j.devcel.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Marwaha R., Arya S.B., Jagga D., Kaur H., Tuli A., and Sharma M.. 2017. The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J. Cell Biol. 216:1051–1070. 10.1083/jcb.201607085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Jiang P., Nakano S., Sakamaki Y., Yamamoto H., and Mizushima N.. 2018. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 217:2633–2645. 10.1083/jcb.201712058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., et al. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11:385–396. 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- Mauvezin C., Nagy P., Juhász G., and Neufeld T.P.. 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6:7007 10.1038/ncomms8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., et al. 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., et al. 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17:288–299. 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G., Zhao Y.G., Zhao H., Ji C., Sun H., Chen Y., and Zhang H.. 2016. Mice deficient in the Vici syndrome gene Epg5 exhibit features of retinitis pigmentosa. Autophagy. 12:2263–2270. 10.1080/15548627.2016.1238554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E., Rivera-Molina F., and Novick P.. 2012. GTPase networks in membrane traffic. Annu. Rev. Biochem. 81:637–659. 10.1146/annurev-biochem-052810-093700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., and Ohsumi Y.. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 130:165–178. 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., and Ohsumi Y.. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Nguyen T.N., Padman B.S., Usher J., Oorschot V., Ramm G., and Lazarou M.. 2016. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 215:857–874. 10.1083/jcb.201607039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R.A., Yang D.S., and Lee J.H.. 2008. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 4:590–599. 10.4161/auto.6259 [DOI] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Orsi A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M., and Tooze S.A.. 2012. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 23:1860–1873. 10.1091/mbc.e11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Alemu E.A., Brech A., Bruun J.A., Lamark T., Overvatn A., Bjørkøy G., and Johansen T.. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188:253–269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson N., Ince P.G., Smith M.O., Highley R., Skibinski G., Andersen P.M., Morrison K.E., Pall H.S., Hardiman O., Collinge J., et al. FReJA Consortium . 2006. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology. 67:1074–1077. 10.1212/01.wnl.0000231510.89311.8b [DOI] [PubMed] [Google Scholar]
- Peng C., Ye J., Yan S., Kong S., Shen Y., Li C., Li Q., Zheng Y., Deng K., Xu T., and Tao W.. 2012. Ablation of vacuole protein sorting 18 (Vps18) gene leads to neurodegeneration and impaired neuronal migration by disrupting multiple vesicle transport pathways to lysosomes. J. Biol. Chem. 287:32861–32873. 10.1074/jbc.M112.384305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.J., and Voeltz G.K.. 2016. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17:69–82. 10.1038/nrm.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontano Vaites L., Paulo J.A., Huttlin E.L., and Harper J.W.. 2017. Systematic analysis of human cells lacking ATG8 proteins uncovers roles for GABARAPs and the CCZ1/MON1 regulator C18orf8/RMC1 in macro and selective autophagic flux. Mol. Cell. Biol. 38:MCB.00392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W.A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205:759–769. 10.1083/jcb.201401126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Guardia C.M., Keren-Kaplan T., and Bonifacino J.S.. 2016. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129:4329–4339. 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C., Renna M., Bento C.F., Moreau K., and Rubinsztein D.C.. 2013. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 154:1285–1299. 10.1016/j.cell.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N., and Puertollano R.. 2016. TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress. Annu. Rev. Cell Dev. Biol. 32:255–278. 10.1146/annurev-cellbio-111315-125407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., and Rubinsztein D.C.. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757. 10.1038/ncb2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M., Chan E.Y., and Tooze S.A.. 2009. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185:305–321. 10.1083/jcb.200810098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J.. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185:1209–1225. 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., and Stenmark H.. 2007. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17:1817–1825. 10.1016/j.cub.2007.09.032 [DOI] [PubMed] [Google Scholar]
- Sato M., Saegusa K., Sato K., Hara T., Harada A., and Sato K.. 2011. Caenorhabditis elegans SNAP-29 is required for organellar integrity of the endomembrane system and general exocytosis in intestinal epithelial cells. Mol. Biol. Cell. 22:2579–2587. 10.1091/mbc.e11-04-0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. 2005. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37:806–808. 10.1038/ng1609 [DOI] [PubMed] [Google Scholar]
- Slawson C., Copeland R.J., and Hart G.W.. 2010. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 35:547–555. 10.1016/j.tibs.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Stolz A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Stroupe C., Collins K.M., Fratti R.A., and Wickner W.. 2006. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 25:1579–1589. 10.1038/sj.emboj.7601051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Westphal W., Wong K.N., Tan I., and Zhong Q.. 2010. Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl. Acad. Sci. USA. 107:19338–19343. 10.1073/pnas.1010554107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmári Z., and Sass M.. 2014. The autophagic roles of Rab small GTPases and their upstream regulators: a review. Autophagy. 10:1154–1166. 10.4161/auto.29395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K., Matsunaga K., Sakane A., Sasaki T., Noda T., and Yoshimori T.. 2010. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol. Biol. Cell. 21:4162–4172. 10.1091/mbc.e10-06-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., He H., Tang Z., Hattori T., Liu Y., Young M.M., Serfass J.M., Chen L., Gebru M., Chen C., et al. 2018. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9:2855 10.1038/s41467-018-05254-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539. 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Pircs K., Nagy P., Varga Á., Kárpáti M., Hegedűs K., Kramer H., Kovács A.L., Sass M., and Juhász G.. 2014. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell. 25:1338–1354. 10.1091/mbc.e13-08-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Guhde G., Suter A., Eskelinen E.L., Hartmann D., Lüllmann-Rauch R., Janssen P.M., Blanz J., von Figura K., and Saftig P.. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 406:902–906. 10.1038/35022595 [DOI] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 141:1042–1055. 10.1016/j.cell.2010.04.034 [DOI] [PubMed] [Google Scholar]
- Tooze J., Hollinshead M., Ludwig T., Howell K., Hoflack B., and Kern H.. 1990. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J. Cell Biol. 111:329–345. 10.1083/jcb.111.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., and Mizushima N.. 2016. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 354:1036–1041. 10.1126/science.aaf6136 [DOI] [PubMed] [Google Scholar]
- Urwin H., Authier A., Nielsen J.E., Metcalf D., Powell C., Froud K., Malcolm D.S., Holm I., Johannsen P., Brown J., et al. FReJA Consortium . 2010. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet. 19:2228–2238. 10.1093/hmg/ddq100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sun H.Q., Zhu X., Zhang L., Albanesi J., Levine B., and Yin H.. 2015. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc. Natl. Acad. Sci. USA. 112:7015–7020. 10.1073/pnas.1507263112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Miao G., Xue X., Guo X., Yuan C., Wang Z., Zhang G., Chen Y., Feng D., Hu J., and Zhang H.. 2016. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol. Cell. 63:781–795. 10.1016/j.molcel.2016.08.021 [DOI] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., and Elazar Z.. 2010. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29:1792–1802. 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shpilka T., Shvets E., Abada A., Shimron F., and Elazar Z.. 2011. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell. 20:444–454. 10.1016/j.devcel.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Wijdeven R.H., Janssen H., Nahidiazar L., Janssen L., Jalink K., Berlin I., and Neefjes J.. 2016. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7:11808 10.1038/ncomms11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Watanabe Y., Guo X.Y., Qi X., Wang P., Zhao H.Y., Wang Z., Fujioka Y., Zhang H., Ren J.Q., et al. 2015. Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy. Mol. Cell. 60:914–929. 10.1016/j.molcel.2015.11.019 [DOI] [PubMed] [Google Scholar]
- Wu Y., Cheng S., Zhao H., Zou W., Yoshina S., Mitani S., Zhang H., and Wang X.. 2014. PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep. 15:973–981. 10.15252/embr.201438618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., and Eskelinen E.L.. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 5:1180–1185. 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Yokota S., Himeno M., and Kato K.. 1995. Formation of autophagosomes during degradation of excess peroxisomes induced by di-(2-ethylhexyl)-phthalate treatment. III. Fusion of early autophagosomes with lysosomal compartments. Eur. J. Cell Biol. 66:15–24. [PubMed] [Google Scholar]
- Yu I.M., and Hughson F.M.. 2010. Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26:137–156. 10.1146/annurev.cellbio.042308.113327 [DOI] [PubMed] [Google Scholar]
- Yu S., and Melia T.J.. 2017. The coordination of membrane fission and fusion at the end of autophagosome maturation. Curr. Opin. Cell Biol. 47:92–98. 10.1016/j.ceb.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., and Baehrecke E.H.. 2015. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol. 25:376–387. 10.1016/j.tcb.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., and Hu J.J.. 2016. Shaping the endoplasmic reticulum into a social network. Trends Cell Biol. 26:934–943. 10.1016/j.tcb.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang L., Lak B., Li J., Jokitalo E., and Wang Y.. 2018. GRASP55 Senses Glucose Deprivation through O-GlcNAcylation to Promote Autophagosome-Lysosome Fusion. Dev. Cell. 45:245–261.e6. 10.1016/j.devcel.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., and Zhang H.. 2018. Formation and maturation of autophagosomes in higher eukaryotes: a social network. Curr. Opin. Cell Biol. 53:29–36. 10.1016/j.ceb.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Zhao H., Zhao Y.G., Wang X., Xu L., Miao L., Feng D., Chen Q., Kovács A.L., Fan D., and Zhang H.. 2013a Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J. Cell Biol. 200:731–741. 10.1083/jcb.201211014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., Zhao H., Miao L., Wang L., Sun F., and Zhang H.. 2012. The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J. Biol. Chem. 287:42053–42063. 10.1074/jbc.M112.415968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., Zhao H., Sun H., and Zhang H.. 2013b Role of Epg5 in selective neurodegeneration and Vici syndrome. Autophagy. 9:1258–1262. 10.4161/auto.24856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. 2017. The ER-Localized Transmembrane Protein EPG-3/VMP1 Regulates SERCA Activity to Control ER-Isolation Membrane Contacts for Autophagosome Formation. Mol. Cell. 67:974–989.e6. 10.1016/j.molcel.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., Liu N., Miao G., Chen Y., Zhao H., and Zhang H.. 2018. The ER Contact Proteins VAPA/B Interact with Multiple Autophagy Proteins to Modulate Autophagosome Biogenesis. Curr. Biol. 28:1234–1245.e4. 10.1016/j.cub.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Zhen Y., and Li W.. 2015. Impairment of autophagosome-lysosome fusion in the buff mutant mice with the VPS33A(D251E) mutation. Autophagy. 11:1608–1622. 10.1080/15548627.2015.1072669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Wang Q.J., Li X., Yan Y., Backer J.M., Chait B.T., Heintz N., and Yue Z.. 2009. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11:468–476. 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]