Lee et al. show that methionine triggers Ppz phosphatase-dependent dephosphorylation of the E3 ubiquitin ligase adaptor Art1. Art1 dephosphorylation promotes its interaction with the methionine transporter Mup1, as well as subsequent endocytosis and degradation of Mup1 in the vacuole.
Abstract
Regulation of plasma membrane (PM) protein abundance by selective endocytosis is critical for cellular adaptation to stress or changing nutrient availability. One example involves rapid endocytic turnover of Mup1, a yeast methionine transporter, in response to increased methionine availability. Here, we report that methionine triggers rapid translocation of the ubiquitin ligase adaptor Art1 to the PM and dephosphorylation of Art1 at specific threonine residues. This methionine-induced dephosphorylation of Art1 is mediated by Ppz phosphatases, and analysis of phosphomimetic and phosphorylation-defective variants of Art1 indicates that these events toggle Art1 recognition of Mup1 at the PM. Importantly, we find that Ppz phosphatases are dispensable for Art1 PM translocation, but are required for Art1 interaction with Mup1. Based on our findings, we propose that methionine influx triggers Art1 translocation to the PM, followed by Ppz-mediated dephosphorylation which promotes cargo recognition at the PM.
Introduction
Endocytosis is a dynamic process that requires the complex and ordered assembly of at least 60 different proteins to capture vesicle cargo, sculpt and bend membranes, assemble coat complexes, and ultimately mediate vesicle scission (Goh et al., 2010; Weinberg and Drubin, 2012; Schmid, 2017). As if the regulation of this extraordinary biophysical event were not complicated enough, there is the added task of specifying the cargo contents of endocytic vesicles, a sorting process that requires selection of specific plasma membrane (PM) proteins to target for internalization among the PM proteome. This selection and sorting process is critical since many aspects of cell identity and physiology within an organism depend on cell surface functionalities, such as receptor signaling and attenuation (Goh and Sorkin, 2013; Di Fiore and von Zastrow, 2014), nutrient and ion uptake (Rotin and Kumar, 2009; Rizzo and Staub, 2015), and protein quality control (Okiyoneda et al., 2011; MacGurn, 2014).
From yeast to mammalian cells, ubiquitylation of integral membrane proteins at the PM triggers capture by ubiquitin-binding elements in endocytic sorting machinery and sorting by the ESCRT pathway into intraluminal vesicles on the limiting membrane of endosomes (Henne et al., 2011; MacGurn et al., 2012). Thus, ubiquitylation is a critical decision point in the selection of endocytic cargo, and as such, E3 ubiquitin ligases and deubiquitylases are key determinants of PM protein stability. Nedd4 family E3 ubiquitin ligases are conserved across eukaryotic evolution and play a major role in endocytic down-regulation by mediating cargo ubiquitylation. One example involves the epithelial Na+ channel (ENaC), a complex of three transmembrane subunits expressed on the apical surface of kidney epithelial cells that mediate sodium reabsorption and thus control blood plasma sodium levels (Fakitsas et al., 2007; Rotin and Kumar, 2009; Ronzaud and Staub, 2014). Nedd4L ubiquitylates ENaC, triggering endocytosis, and mutations that disrupt the ENaC–Nedd4L interaction stabilize ENaC and result in a form of hereditary hypertension called Liddle Syndrome (Ronzaud and Staub, 2014). Physiological regulation of ENaC turnover is mediated by phosphoinhibition of Nedd4L, which stabilizes ENaC at the PM and increases sodium reabsorption (Debonneville et al., 2001; Ronzaud and Staub, 2014). This example illustrates how coordination of phosphorylation and ubiquitylation pathways contribute to regulation of PM remodeling processes.
Regulation of endocytosis by Nedd4 family E3 ubiquitin ligases is conserved across eukaryotic evolution. In yeast, most endocytic events are regulated by Rsp5, the lone Nedd4 family member encoded in the yeast genome. As is characteristic of all Nedd4 family members, Rsp5 contains a C-terminal HECT E3 ubiquitin ligase domain, an N-terminal C2 domain, and tandem WW domains in the middle of the protein (three in the case of Rsp5) that function as a network scaffold. Substrate targeting for Rsp5 is largely mediated by a network of adaptor proteins which contain PY motifs (PPxY) that bind with high affinity to the WW domains of Rsp5 (Léon et al., 2008; Lin et al., 2008; Nikko and Pelham, 2009a; O’Donnell et al., 2013). Many Rsp5 adaptors also contain arrestin fold domains which are thought to mediate substrate selection. These arrestin-related Rsp5 adaptors (called ARTs) are often required for endocytosis of specific cargo (Lin et al., 2008; Hatakeyama et al., 2010; Becuwe et al., 2012), although much redundancy has been reported within the network (Nikko et al., 2008; O’Donnell et al., 2013; Zhao et al., 2013). One important aspect of ART protein function is cargo selection, yet we understand little about how different ARTs govern the specificity of Rsp5 substrate targeting. Recent studies have shed light on cargo recognition mechanisms by defining sorting signals specifically recognized by ARTs (Guiney et al., 2016) and by characterizing substrate-induced conformational changes in transporters that are critical for ART-mediated ubiquitylation (Gournas et al., 2017). Ultimately, understanding the structural features that mediate recognition of specific cargo by ARTs will be critical to understanding how the ART-Rsp5 network manages the PM proteome.
An equally important property of ART family adaptors is their ability to target Rsp5 to specific cargo in a highly regulated and context-dependent manner. For example, Art1 is required for the methionine-induced ubiquitylation and endocytosis of the methionine transporter Mup1 (Lin et al., 2008) and the arginine-induced ubiquitylation and endocytosis of the arginine transporter Can1 (Gournas et al., 2017). Strikingly, activation of Art1 results in rapid translocation to the PM (Lin et al., 2008), while phosphorylation of N-terminal residues of Art1 by the Npr1 kinase inhibit this translocation (MacGurn et al., 2011). Importantly, these studies suggest that phosphatase activities are required for the activation of Art1, but to date, no Art1 phosphatases have been reported. Other ART family adaptors are reported to influence carbon metabolism by regulating the trafficking of hexose transporters (Becuwe et al., 2012; O’Donnell et al., 2015; Hovsepian et al., 2017). For example, the glucose-induced ubiquitylation and endocytosis of the lactate transporter Jen1 requires Art4/Rod1. In this case, glucose stimulation triggers dephosphorylation of Art4/Rod1 by the Glc7 PP1 family protein phosphatase (Becuwe et al., 2012), which correlates with translocation of Art4/Rod1 to the TGN, where it participates in post-endocytic regulation of Jen1 trafficking (Becuwe and Léon, 2014). Importantly, Art4/Rod1 was also shown to function in the ubiquitylation and endocytic trafficking of Ste2 (α-factor pheromone receptor, a G protein–coupled receptor) in a manner that requires dephosphorylation by calcineurin (protein phosphatase type 2B; Alvaro et al., 2014). Other examples of phosphoregulation in the Rsp5 adaptor network have been reported, including calcineurin-dependent dephosphorylation/activation of Aly1/Art6 (O’Donnell et al., 2013) and Npr1-dependent phosphoinhibition of Bul proteins (Merhi and André, 2012); but in most cases, little is known about the mechanistic basis of regulation by specific phosphorylation events. Ultimately, understanding how ART proteins coordinate specific ubiquitylation and endocytic trafficking outcomes at the PM will require quantitative characterization of specific ART phosphorylation events combined with establishing mechanisms of regulatory action.
Given our previous findings that hyperphosphorylation at specific residues inhibited Art1 function in endocytosis (MacGurn et al., 2011), we hypothesized that phosphatase activities may contribute to activation of Art1. Consistent with this hypothesis, we observed specific dephosphorylation events on Art1 that occur in response to methionine stimulation, a trigger for Art1-mediated turnover of Mup1. In our effort to identify the responsible phosphatase activity, we found that Ppz phosphatases regulate endocytic trafficking in a cargo-specific manner and that this mode of regulation is distinct from their reported role in ubiquitin homeostasis (Lee et al., 2017). Furthermore, we find that Ppz phosphatases localize to the PM and that the endocytic-trafficking defects observed in ppz mutant cells are attributable to elevated phosphorylation of Art1 at specific threonine residues. Importantly, Ppz phosphatases are required to detect Art1–Mup1 interactions, suggesting that Ppz phosphatases contribute to an Art1 activation event that promotes cargo recognition. Collectively, the results reported here indicate that dephosphorylation of Art1 by Ppz phosphatases is an important activation event that promotes cargo recognition at the PM.
Results
Methionine stimulation triggers dephosphorylation of Art1
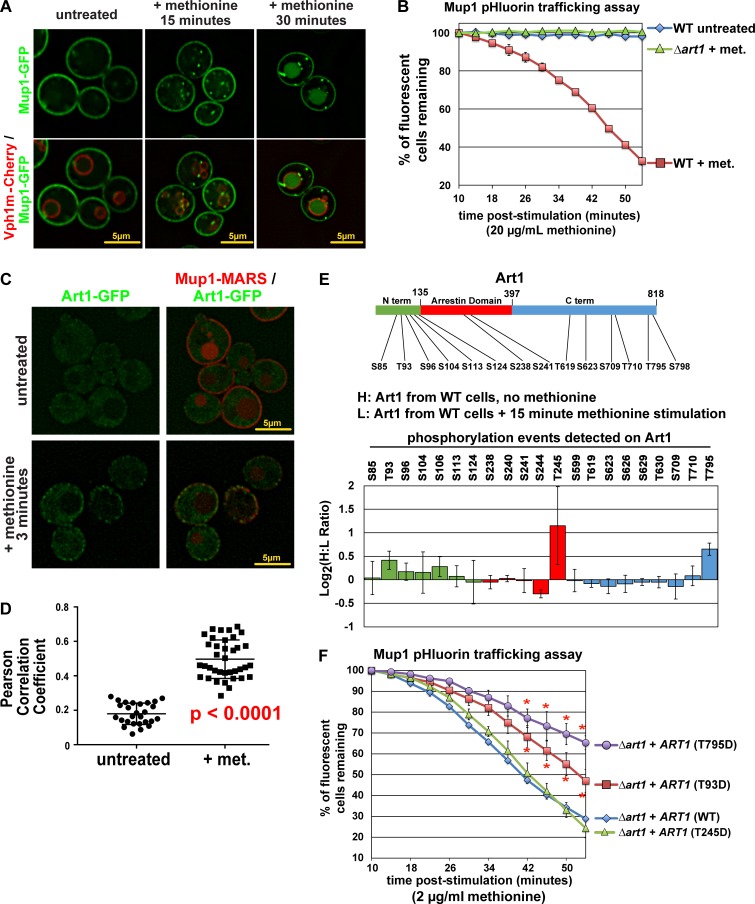
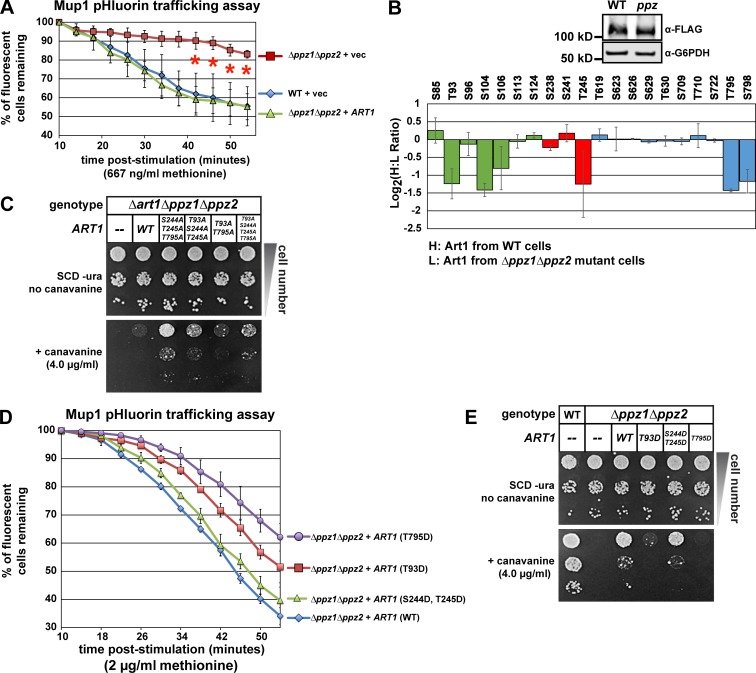
Yeast cells grown in the absence of methionine stably express the high affinity methionine transporter Mup1 at the cell surface, and acute addition of methionine triggers endocytosis and vacuolar degradation of Mup1 (Lin et al., 2008; Guiney et al., 2016). The response of Mup1-GFP to methionine is rapid, with delivery to endosomes observed within 15 min and vacuolar delivery observed within 30 min of stimulation (Fig. 1 A, with the limiting membrane of the vacuole marked by mCherry-tagged Vph1). This response can be analyzed by monitoring the fluorescence signal of Mup1-pHluorin fusion proteins, which is lost when Mup1 is delivered to the acidic environment of the vacuolar lumen (Prosser et al., 2010; Lee et al., 2017; Fig. S1 A). Consistent with previous reports (Lin et al., 2008; MacDonald et al., 2015), methionine-induced endocytic trafficking of Mup1-pHluorin was found to be dependent on the Rsp5 E3 ubiquitin ligase adaptor Art1 (Fig. 1 B). Strikingly, we found that Art1 translocates to the PM within 3 min of methionine stimulation (Fig. 1, C and D), indicating the acute cellular response to methionine involves translocation of Art1. To further probe this Art1 response to methionine stimulation, we performed stable isotope labeling with amino acids in culture (SILAC) mass spectrometry analysis to quantify how phosphorylation events on Art1 change in response to methionine stimulation (Fig. S1 B). This analysis revealed that within 15 min of methionine stimulation, Art1 undergoes dephosphorylation at Thr245 and Thr795 and to a lesser extent Thr93 (Fig. 1 E). Importantly, we found that phosphomimetic (Asp) mutations at Thr795 and Thr93 resulted in partial Art1 loss of function, while phosphomimetic mutation at Thr245 had no effect on Art1 function (Fig. 1 F). Phosphorylation-resistant Art1 variants exhibited slight but significant increases in Art1 activity in response to methionine stimulation (Fig. S1, C and D). Collectively, these results indicate that phosphorylation of Art1 at Thr93 and Thr795 inhibit Art1-mediated endocytosis of Mup1 and that methionine-triggered dephosphorylation at these sites promotes Art1 activation.
Figure 1.
Methionine stimulation induces dephosphorylation of Art1. (A) Yeast cells expressing Mup1-GFP (green) and Vph1-mCherry (red; localized to the limiting membrane of the vacuole) were grown to mid-log phase and imaged by fluorescence deconvolution microscopy (untreated) or stimulated with methionine for the indicated amount of time and then imaged. (B) Yeast cells (WT or Δart1) expressing Mup1-pHluorin were cultured to mid-log phase, and flow cytometry was used to measure pHluorin-positive cells at the indicated time points following methionine stimulation (+ met.) or an untreated control culture. Sample data of the Mup1-pHluorin–trafficking analysis by microscopy and flow cytometry are provided in Fig. S1 A. Error bars indicate standard deviation from multiple biological replicate experiments (n = 3). (C) Yeast cells expressing Art1-GFP (green) and Mup1-MARS (red) were grown to mid-log phase and imaged by fluorescence deconvolution microscopy (untreated) or stimulated with methionine for 3 min and then imaged. (D) Pearson’s correlation coefficients were measured for yeast cells (n > 25 cells each for untreated or methionine stimulation conditions) expressing Art1-GFP and Mup1-MARS-RFP (as shown in C). Error bars indicate standard deviation for the set, and significance was determined by an unpaired t test. (E) SILAC-based quantification of phosphorylation events was performed (according to the scheme shown in Fig. S1 B) using endogenous Art1-FLAG as bait. A schematic representation of Art1 (top) is shown with domains color-coded to indicate location of phosphopeptides detected and quantified in this analysis (bottom graph). Normalized H:L ratios were measured and averaged over multiple biological replicates (n = 3). Error bars indicate standard deviation of measurements on peptides from three experiments (n = 3), except for S104 and S113, which were only resolved and quantified in two replicate experiments. (F) Complementation analysis of Δart1 mutant yeast cells using the Mup1-pHluorin–trafficking assay. For all Mup1-pHluorin–trafficking assays plots represents average of multiple biological replicate experiments (n ≥ 4), and error bars indicate standard deviation. For the final five time points of each time course, statistical significance was calculated using an unpaired t test (P < 0.005).
Ppz phosphatases regulate Methionine-triggered Mup1 endocytosis in yeast
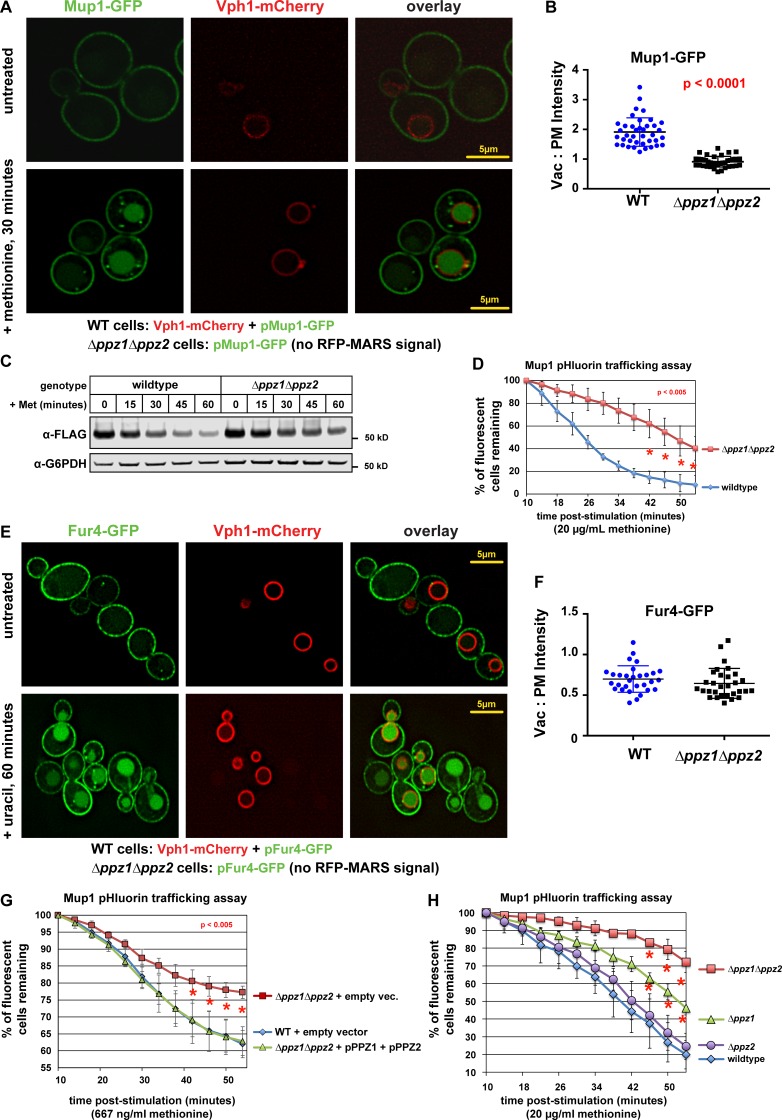
Based on our findings that Art1 undergoes methionine-induced dephosphorylation, we hypothesized that a corresponding phosphatase activity plays a critical role in the activation of Art1-mediated Mup1 endocytosis in response to methionine stimulation. Although phosphatases have been reported to regulate other Rsp5 adaptors (Becuwe et al., 2012; O’Donnell et al., 2013), no phosphatases have been implicated in the methionine response or in the regulation of Art1. Given their reported role in the regulation of nutrient transporter function (Ruiz et al., 2004; Yenush et al., 2005) and ubiquitin homeostasis (Lee et al., 2017), we decided to test if Ppz1 and Ppz2, a pair of highly similar (57% identical) phosphatases, regulate Art1-mediated Mup1 endocytosis. We found that yeast cells lacking Ppz1 and Ppz2 (Δppz1Δppz2 double mutant cells or ppz mutants) exhibited defects in trafficking of Mup1-GFP to the vacuole (Fig. 2, A and B), a decreased rate of Mup1-FLAG degradation (Fig. 2 C), and delayed quenching of Mup1–pHluorin signal (Fig. 2 D) in response to methionine stimulation. Notably, ppz mutants exhibited canavanine hypersensitivity (Fig. S2, A and B) and a corresponding defect in endocytic trafficking of the arginine transporter Can1 (Fig. S2, C and D), which is also regulated by Art1. However, Ppz phosphatases were dispensable for uracil-triggered endocytic trafficking of the uracil transporter Fur4 (Fig. 2, E and F), indicating that Ppz phosphatases are not generally required for endocytosis. Furthermore, Ppz phosphatases were dispensable for the endocytosis and turnover of Mup1 that occurs in late stationary phase (Fig. S2, E and F), an Art1-independent mode of Mup1 trafficking that requires the Cos family of tetraspan proteins (MacDonald et al., 2015). Additional genetic analysis of ppz mutants revealed that expression of both Ppz1 and Ppz2 fully complements the Mup1-trafficking defect (Fig. 2 G), and loss of Ppz1, but not Ppz2, is sufficient to confer a Mup1-trafficking defect (Fig. 2 H). These results indicate that Ppz1 and Ppz2 phosphatases exhibit some redundancy of function, although loss of Ppz1 results in a significant (albeit less severe) Mup1 endocytic-trafficking defect. We also found that expression of both Ppz1 and Ppz2 in WT cells or overexpression of either Ppz1-FLAG or Ppz2-FLAG accelerated the trafficking of Mup1-pHluorin (Fig. S2, G and H). Collectively, our results indicate that Ppz phosphatases function in methionine-dependent endocytosis and vacuolar trafficking of Mup1, but are not generally required for endocytic trafficking.
Figure 2.
Ppz phosphatases are required for Mup1 endocytic trafficking in response to methionine. (A) WT (expressing Vph1-mCherry) or Δppz1Δppz2 mutant (lacking mCherry expression) yeast cells (SEY6210 strain background) expressing Mup1-GFP were cultured to mid-log phase, mixed, stimulated by addition of methionine for 30 min, and imaged by fluorescence deconvolution microscopy. (B) Mup1-GFP signal intensity was measured at the PM and the vacuole lumen, and a Vac:PM ratio was calculated for many individual yeast cells (n ≥ 40). Statistical significance was calculated using a two-way ANOVA test (P values indicated in red). (C) A methionine stimulation time course was performed on WT and ppz mutant yeast cells expressing Mup1-FLAG. At the indicated time points after stimulation, cell lysates were analyzed by SDS-PAGE and immunoblot. (D) Flow cytometry analysis of yeast cells expressing Mup1-pHluorin grown in the absence of methionine and then stimulated by addition of methionine. Fluorescence of pHluorin is pH sensitive and lost during endocytic trafficking when the cargo encounters an acidic environment. Plot represents average of multiple biological replicate experiments (n ≥ 3), and error bars indicate standard deviation. Statistical significance was calculated using a Student’s t test (P < 0.005). (E) WT (expressing Vph1-mCherry) or Δppz1Δppz2 mutant (lacking mCherry expression) yeast cells (SEY6210 strain background) expressing Fur4-GFP were cultured to mid-log phase, mixed, stimulated by addition of methionine for 30 min, and imaged by fluorescence deconvolution microscopy. (F) Fur4-GFP signal intensity was measured at the PM and the vacuole lumen, and a Vac:PM ratio was calculated for many individual yeast cells (n ≥ 30). Statistical significance was calculated using a two-way ANOVA test, but no statistically significant difference between WT and Δppz1Δppz2 mutant cells was observed. (G and H) Flow cytometry analysis of yeast cells expressing Mup1-pHluorin grown in the absence of methionine and then stimulated by addition of methionine. Fluorescence of pHluorin is pH sensitive and lost during endocytic trafficking when the cargo encounters an acidic environment. Plot represents average of multiple biological replicate experiments (n ≥ 3), and error bars indicate standard deviation. Statistical significance was calculated using a Student’s t test (P < 0.005). Vac, vacuole lumen.
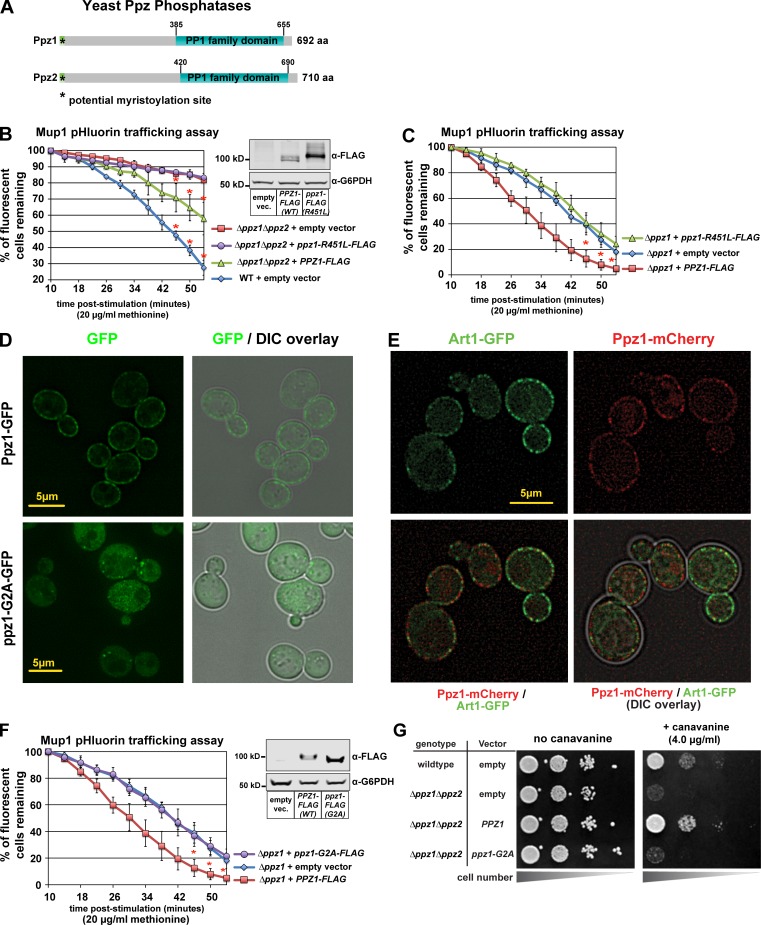
We decided to explore and characterize the structural features of Ppz phosphatases (Fig. 3 A) that function in methionine-induced Mup1 endocytic trafficking in yeast. Importantly, we found that expression of WT Ppz1-FLAG, but not a catalytic dead variant (R451L; both under control of the native PPZ1 promoter), complemented Mup1-pHluorin–trafficking defects in Δppz1Δppz2 and Δppz1 mutant cells (Fig. 3, B and C). Similarly, Ppz1 catalytic activity was required to complement the canavanine-hypersensitivity phenotype of Δppz1Δppz2 double mutant cells (Fig. S3, A and B). These findings demonstrate that Ppz phosphatase activity functions in the regulation of methionine-induced Mup1 endocytosis.
Figure 3.
Ppz catalytic activity and PM localization are required for Mup1 endocytosis. (A) Schematic representation of functional features known in Ppz1 and Ppz2 phosphatases. (B) Complementation analysis of Δppz1Δppz2 mutant yeast cells using the Mup1-pHluorin–trafficking assay. Error bars indicate standard deviation from multiple biological replicate experiments (n = 3). Expression of WT and catalytic dead (R451L) Ppz1-FLAG was confirmed by immunoblot (inset). (C) Complementation analysis of Δppz1 mutant yeast cells using the Mup1-pHluorin–trafficking assay. Error bars indicate standard deviation from multiple biological replicate experiments (n = 4). (D) Yeast cells expression WT Ppz1-GFP or a ppz1-G2A-GFP mutant were analyzed by fluorescence microscopy. (E) WT (strain SEY6210) yeast cells expressing Art1-GFP and Ppz1-mCherry were stimulated with methionine for 3 min and imaged using fluorescence deconvolution microscopy. (F) Complementation analysis of Δppz1 mutant yeast cells using the Mup1-pHluorin–trafficking assay. Immunoblot (inset) shows expression of Ppz-FLAG from indicated vectors. For all Mup1-pHluorin–trafficking assays, plots represents average of multiple biological replicate experiments (n ≥ 3), and error bars indicate standard deviation. For the final three time points of each time course, statistical significance was calculated using a Student’s t test (P < 0.005). (G) WT or Δppz1Δppz2 mutant yeast cells (SEY6210 strain background) harboring the indicated Ppz1 expression plasmids were plated on minimal media in the presence of canavanine (toxic arginine analogue).
In addition to their C-terminal PP1 family phosphatase domain, Ppz phosphatases also contain an N-terminal domain that is largely uncharacterized (Fig. 3 A). Previous studies have reported evidence that Ppz1 is N-myristoylated (Clotet et al., 1996), and Ppz2 is also predicted to be N-myristoylated. Furthermore, these prior studies found that deleting N-terminal portions of Ppz1 (Δ1–344 or Δ241–318) or preventing myristoylation with a G2A mutation resulted in failure to complement the LiCl-resistance phenotype of Δppz1Δppz2 double mutant cells (Clotet et al., 1996). Given these prior observations, we hypothesized that N-terminal myristoylation of Ppz1 might be important for its localization and its role in endocytic trafficking. To test this, we first analyzed subcellular localization of WT Ppz1-GFP and a G2A mutant in yeast cells using fluorescence deconvolution microscopy. We found that WT Ppz1-GFP exhibited PM and diffuse cytosolic localization, while the G2A mutant failed to localize to the PM (Fig. 3 D).
Importantly, we found that WT Ppz1-GFP does not localize to known PM structures, including sites of endocytosis (as marked by Abp1-mCherry [Fig. S3 C] and Ede1-mCherry [Fig. S3 D]) or eisosomes (as marked by Pil1-mCherry [Fig. S3 E]). Additionally, we found that the PM localization patterns of Art1-GFP and Ppz1-mCherry were largely distinct, although some punctae were found to contain both Art1-GFP and Ppz1-mCherry (Fig. 3 E).
Having confirmed that N-myristoylation of Ppz1 is critical for its localization to the PM, we next tested if the mislocalized Ppz1-G2A variant is functional. Strikingly, expression of ppz1-G2A was increased relative to WT Ppz1, yet failed to complement the Mup1-pHluorin–trafficking defect in Δppz1 mutant cells (Fig. 3 F) or the canavanine-hypersensitivity phenotype of Δppz1Δppz2 double mutant cells (Fig. 3 G). These results indicate that N-myristoylation and PM localization of Ppz1 are required for its role in the regulation of methionine-triggered Mup1 endocytosis in yeast.
The role of Ppz1 in ubiquitin homeostasis is distinct from its role in endocytosis
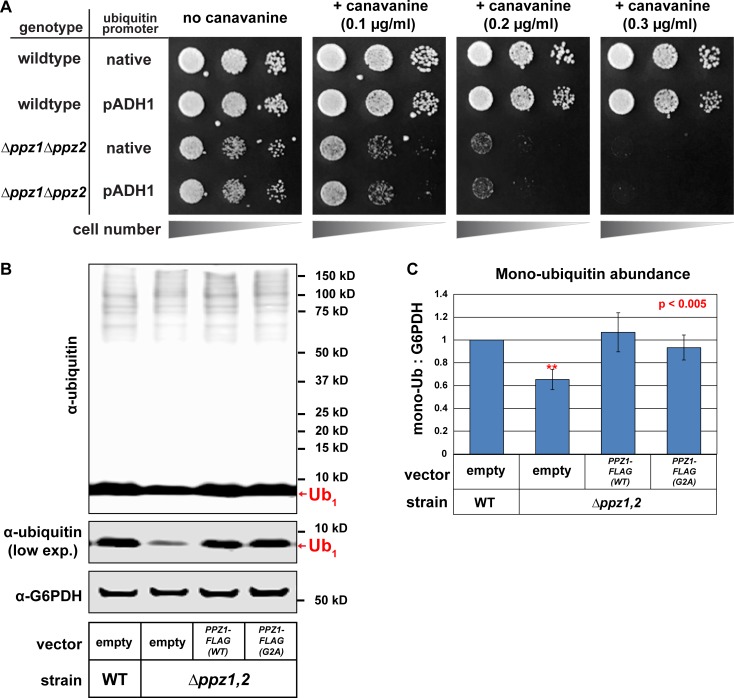
Recently, we reported that Ppz phosphatases play a critical role in the management of ubiquitin homeostasis in yeast. Specifically, we found that ppz mutant cells exhibit a ubiquitin deficiency that is partially attributable to increased levels of pSer57 phospho-ubiquitin (Lee et al., 2017). Since ubiquitin deficiency is known to cause defects in endocytic trafficking (Swaminathan et al., 1999; Amerik et al., 2000), we hypothesized that ubiquitin deficiency might underlie the endocytic- trafficking defect observed in ppz mutant cells (Lee et al., 2017). Importantly, we previously reported that several phenotypes observed in ppz mutant cells are suppressed by restoring ubiquitin levels, suggesting that these phenotypes are indirect effects of ubiquitin deficiency. In contrast to these other ppz mutant phenotypes, we found that ubiquitin overexpression does not suppress the canavanine hypersensitivity of Δppz1Δppz2 cells (Fig. 4 A). Unexpectedly, we also found that the Ppz1-G2A mutant, which was unable to complement the endocytic-trafficking defects observed in ppz mutant cells (Fig. 3, F and G), fully complemented the ubiquitin deficiency phenotype reported for ppz mutant cells (Fig. 4, B and C), indicating that PM localization of Ppz1 is not required for its role in the regulation of ubiquitin homeostasis. These data suggest that (1) Ppz phosphatases have distinct and separable functions in cargo-specific regulation of endocytic trafficking and ubiquitin homeostasis and (2) PM localization of Ppz1 is required for its endocytic function but dispensable for its role in ubiquitin homeostasis.
Figure 4.
Ppz function in ubiquitin homeostasis is separable from its endocytic function. (A) WT or Δppz1Δppz2 mutant yeast cells (SUB280 strain background) harboring the indicated ubiquitin expression plasmids were plated on minimal media in the presence of canavanine (toxic arginine analogue). (B) The indicated yeast cells (SEY6210 background) containing either empty vector or PPZ1-FLAG vectors were analyzed for total cellular ubiquitin levels by immunoblot analysis. (C) Quantification of ubiquitin expression yeast cells (SEY6210 background) expressing the indicated Ppz expression plasmids. Error bars indicate standard deviation from multiple biological replicate experiments (n = 4). Statistical significance was calculated using a Student’s t test (P < 0.005).
Ppz phosphatases regulate Art1 activity via dephosphorylation
Based on these findings, we hypothesized that Ppz phosphatases might operate on the ART-Rsp5 adaptor network, which is known to regulate the endocytosis of many cargo, including Mup1 and Can1 (Lin et al., 2008; Léon and Haguenauer-Tsapis, 2009; Hatakeyama et al., 2010; Nikko and Pelham, 2009; Nikko et al., 2008; O’Donnell et al., 2010). Indeed, ART family proteins are known to be regulated by phosphorylation (MacGurn et al., 2011; Becuwe et al., 2012), making them potential targets for signaling relays to regulate endocytosis of specific transporters. Since the endocytosis of both Can1 and Mup1 requires Art1 (Lin et al., 2008), and Art1 is subjected to extensive regulation by phosphorylation (MacGurn et al., 2011), we explored the possibility that Ppz phosphatases operate on Art1. One kinase known to phosphorylate and inhibit Art1 function is Npr1, and loss of Npr1 leads to hyperactivation of Art1 and a canavanine-resistance phenotype (MacGurn et al., 2011). Importantly, the canavanine-resistance phenotype of Δnpr1 mutant cells required Ppz phosphatases, as Δppz1Δppz2Δnpr1 triple mutant cells exhibited canavanine hypersensitivity similar to Δppz1Δppz2 mutant cells (Fig. S4 A). Furthermore, while Npr1-mediated phosphorylation of Art1 is readily detectible by SDS-PAGE mobility shifts (MacGurn et al., 2011), we found that loss of Ppz phosphatases had no effect on Art1 mobility on SDS-PAGE (Fig. S4 B). These results indicate that Ppz phosphatases do not antagonize Npr1 kinase activity toward Art1. However, we also found that increased expression of Art1 suppresses both the Mup1-pHluorin–trafficking defect and the canavanine-hypersensitivity phenotype of ppz mutant cells (Fig. 5 A and Fig. S4 C), indicating that the trafficking phenotype in ppz mutant cells may be due to a defect in Art1 function or regulation. Therefore, we performed SILAC-based quantitative proteomic analysis (Fig. S1 B) to determine if phosphorylation of endogenous Art1 is altered in a ppz mutant background. We found that loss of Ppz phosphatases did not affect Art1 interaction with Rsp5 (Fig. S4 D) or overall Art1 abundance (Fig. 5 B, inset blot; Fig S4, B and D). However, we resolved several Art1 phosphorylation events that were significantly elevated (approximately twofold) in ppz mutant cells (Fig. 5 B), including Thr93, Thr245, and Thr795, which were previously found to be dephosphorylated in response to methionine (Fig. 1 E). Notably, Thr93 and Thr245 were detected in previous studies, but were found to be unaffected by either Npr1 overexpression or deletion (MacGurn et al., 2011), while the other Ppz-regulated events (Ser104 and Thr795) were not detected in previous studies. These results suggest that Ppz phosphatases regulate Art1 phosphorylation at sites not targeted by the Npr1 kinase.
Figure 5.
Ppz phosphatases regulate the phosphorylation and activity of Art1. (A) Mup1-pHluorin–trafficking assays were performed to characterize endocytic trafficking in the indicated yeast strains. ART1 is expressed from its native promoter on a pRS416 centromere plasmid and contains a C-terminal 3× FLAG fusion. Error bars indicate standard deviation from multiple biological replicate experiments (n = 3). (B) SILAC-based quantification of phosphorylation events was performed (according to the scheme shown in Fig. S1 A) using endogenous Art1-FLAG as bait. Color coding of regions of Art1 corresponds to the schematic representation of Art1 (Fig. 1 E). Normalized H:L ratios were measured and averaged over multiple biological replicates (n = 3). Error bars indicate standard deviation of measurements on peptides from three experiments (n = 3), except for S85, S106, S722, and S798, which were only resolved and quantified in two replicate experiments. (C) The indicated yeast strains (SEY6210 background) expressing empty vector or the indicated Art1 expression plasmid were plated on minimal media (SCD-ura) containing the indicated concentration of canavanine. (D) Mup1-pHluorin–trafficking assays were performed to characterize endocytic trafficking in the indicated yeast strains with Art1 variants expressed from a pRS416 centromere plasmid (as in Fig. 4 A and Fig. S1 A). Error bars indicate standard deviation from multiple biological replicate experiments (n = 3). (E) The indicated yeast strains (SEY6210 background) expressing empty vector or the indicated Art1 expression plasmid were plated on minimal media (SCD-ura) containing the indicated concentration of canavanine.
We hypothesized that increased phosphorylation of Art1 at Thr93, Thr245, and Thr795 (or some combination of these residues) contributes to the endocytic defect observed for Mup1 and Can1 trafficking in ppz mutant cells. Importantly, we found that expression of phosphorylation-resistant (Ala substitution) variants of Art1 generally increased the rate of Mup1 endocytic trafficking (Fig. S4 E) and suppressed the canavanine-hypersensitivity phenotype of ppz mutants (Fig. 5 C). Furthermore, T795D and T93D (but not T245D) phosphomimetic mutations prevented the ability of Art1 to suppress Mup1-trafficking defects (Fig. 5 D) and canavanine-hypersensitivity (Fig. 5 E) observed in ppz mutants. Collectively, these results indicate that Ppz-mediated dephosphorylation of Art1 at Thr93 and Thr795 plays an important role in the activation of Art1 in response to methionine stimulation.
Regulation of the Rsp5 network by Ppz phosphatases
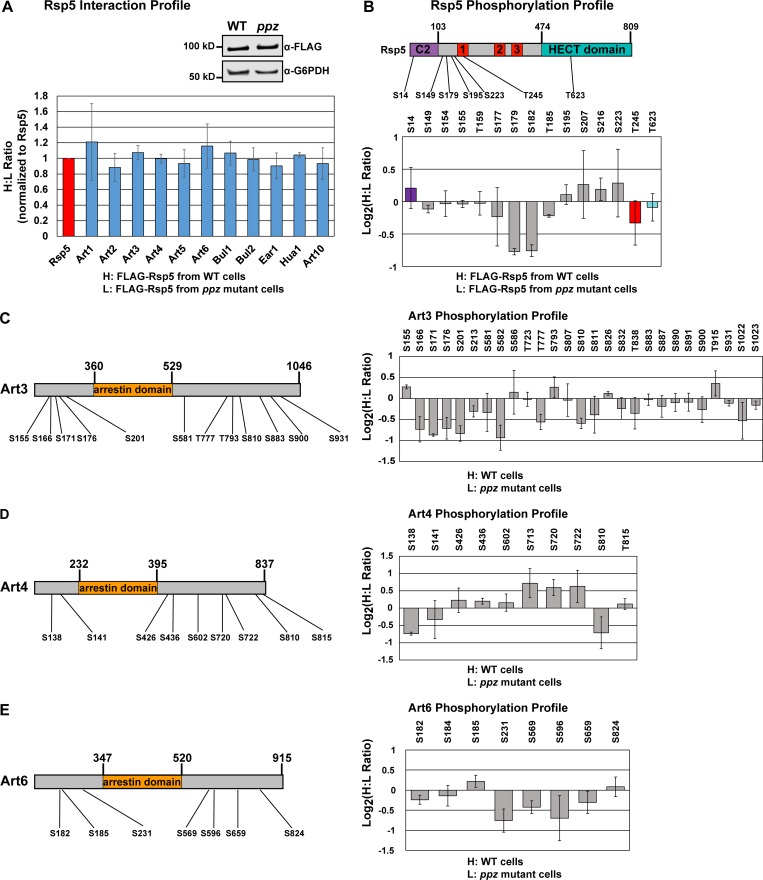
Our finding that Ppz phosphatases regulate phosphorylation and activity of Art1 led us to consider the possibility that Ppz phosphatases may broadly impact Rsp5 function via phosphoregulation of other ART family adaptors. Using FLAG-Rsp5 as bait, we performed SILAC-based quantitation of (1) the Rsp5 interaction network and (2) phosphorylation events in the Rsp5 interaction network. This analysis revealed that loss of Ppz phosphatases did not significantly impact Rsp5 interactions with adaptors (Fig. 6 A) or other known interactors (Fig. S4 F). In contrast, SILAC-based quantitative phosphoprofiling revealed some phosphorylation events within the Rsp5 interaction network that were elevated in the absence of Ppz phosphatases, although most were unaffected (Fig. 6, B–E; and Fig. S4, G–J). Specifically, phosphorylation events on N-terminal residues of Rsp5 (Fig. 6 B; Ser179 and Ser182) and N-terminal residues of Art3 (Fig. 6 C; Ser166, Ser171, and Ser201) were elevated in ppz mutants, while phosphorylation events detected on other adaptors, including Art2, Art4, Art6, Bul1, and Bul2, were less affected by the absence of Ppz phosphatases (Fig. 6, D–E; and Fig. S4, G–I). It is noteworthy that for several known Rsp5 adaptors, only a handful of phosphorylation events were resolved (Fig. S4 J). Indeed, using FLAG-Rsp5 as bait yielded fewer quantified phosphopeptides for Art1, compared with using Art1-FLAG as bait, but confirmed elevated phosphorylation of Thr93 and Thr795 in ppz mutant cells (Fig. S4 J). Additionally, this analysis resolved some phosphorylation events on Npr1, although these were not affected by loss of Ppz phosphatases (Fig. S5 A). Overall, this analysis indicates that Ppz phosphatases antagonize specific phosphorylation events on Art1, and possibly Art3 and Rsp5, but do not broadly impact phosphorylation of other Rsp5 adaptors or interactions within the Rsp5 network.
Figure 6.
Ppz phosphatases regulate specific phosphorylation events within the Rsp5 adaptor network. (A) SILAC-based quantitation of the FLAG-Rsp5 interaction network in WT (H) and ppz mutant (L) cells. Experiment was performed as shown in Fig. S1 A using FLAG-Rsp5 as the bait. Normalized H:L ratios were measured and averaged over multiple biological replicates (n = 3). Error bars indicate standard deviation of measurements on peptides from at least two experiments. Immunoblot (inset) shows recovery (1%) of FLAG-Rsp5 as bait in these samples. Additional data for this experiment are shown in Fig. S4. (B) SILAC-based quantification was performed on Rsp5 phosphopeptides enriched by immobilized metal affinity chromatography. A schematic (top) illustrates the phosphorylation events detected and quantified relative to known domain organization. Color coding of domain organization applies to the graph depicting Rsp5 phosphopeptide quantification (bottom panel). (C–E) SILAC-based quantification of phosphorylation events resolved and quantified in the Rsp5 adaptor network, including Art3/Aly2 (C), Art4/Rod1 (D), and Art6/Aly1 (E). For all panels in Figure 6, normalized H:L ratios were measured and averaged over multiple biological replicates (n = 3). Error bars indicate standard deviation of measurements on peptides from at least two experiments (n ≥ 2). Schematic representations of each adaptor are shown at the left, and phosphopeptide quantification is shown on the right. Additional phosphoprofiling data from these experiments are shown in Fig. S4.
Ppz phosphatases promote Art1 interaction with Mup1
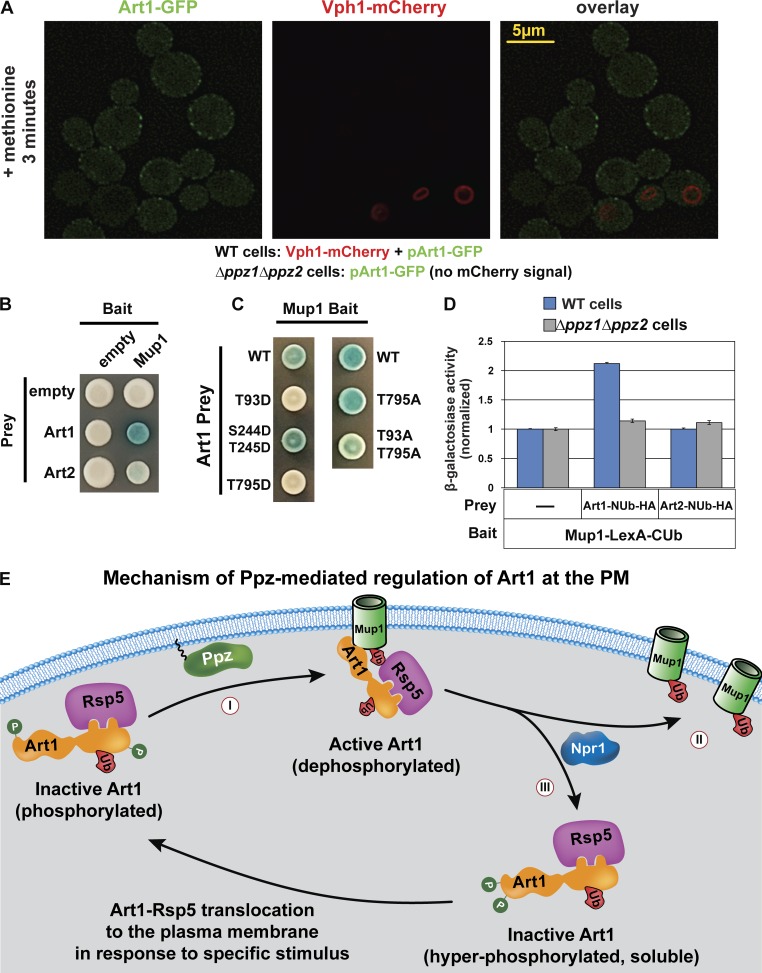
Based on our findings that Ppz phosphatases promote Art1-mediated Mup1 endocytosis and mediate dephosphorylation of Art1, we hypothesized that Ppz phosphatases promote the PM translocation of Art1 in response to methionine stimulation (Fig. 1, C and D). To test this, we characterized Art1 localization in ppz mutant cells, but we observed no PM translocation defect in response to methionine stimulation (Fig. 7 A) or in Δnpr1 mutant cells, where Art1 aberrantly localizes to the PM in the absence of stimulus (MacGurn et al., 2011; Fig. S5 B). Thus, we concluded that Ppz-mediated dephosphorylation is not required for Art1 translocation to the PM. Furthermore, loss of Ppz phosphatases did not affect Npr1 phosphorylation (Fig. S5, A and C), indicating that they do not regulate trafficking indirectly via regulation of Npr1. Since Ppz phosphatases localize to the PM (Fig. 3 D), we next considered the possibility that Ppz phosphatases operate downstream of Art1 PM translocation to regulate interaction with cargo. To probe Art1 interaction with cargo, we used a split-ubiquitin two-hybrid system, using Art1 as prey and various cargo as bait. The split-ubiquitin two-hybrid system detected the interaction between Art1 and Mup1, but not other cargo (Fig. 7 B and Fig. S5, D and E). Importantly, T93D or T795D phosphomimetic mutations resulted in decreased Art1 interaction with Mup1 (Fig. 7 C and Fig. S5 F) indicating that phosphorylation at these positions interferes with cargo interaction. Furthermore, the Art1–Mup1 interaction was decreased in the absence of Ppz phosphatases (Fig. 7 D and Fig. S5 E), indicating that Ppz phosphatases promote the Art1–Mup1 interaction. Collectively, our results indicate that Ppz-mediated dephosphorylation contributes to Art1 activation by promoting cargo interaction following translocation to the PM (Fig. 7 E).
Figure 7.
Ppz phosphatases promote Art1 interaction with Mup1 at the PM. (A) WT yeast cells (expressing Vph1-mCherry) and Δppz1Δppz2 yeast cells (lacking mCherry expression) were grown to mid-log phase, mixed, stimulated with methionine for 3 min, and imaged by fluorescence deconvolution microscopy. (B and C) A split-ubiquitin two-hybrid system was used to probe for Art1-cargo interactions. Different cargo (Mup1, Can1, and Lyp1) were expressed as bait constructs fused to LexA and CUb. Art1 was expressed as a prey construct fused to NUb-HA. (D) β-Galactosidase activity assays were performed to quantify the Art1-Mup1 interaction in WT and Δppz1Δppz2 yeast cells. Error bars indicate standard deviation from multiple biological replicate experiments (n = 7). Expression of bait and prey constructs is shown in Fig. S5 (E and F). (E) An integrated model summarizing current understanding of Art1 phosphoinhibition at the PM. Ppz phosphatases promote dephosphorylation of Art1 at Thr245, Thr795, and Thr93 positions, which contributes to Art1 activation at the PM (I). Specifically, Ppz phosphatase activity promotes the interaction of Art1 with Mup1 at the PM. Activated Art1 targets Rsp5 activity to Mup1, which is ubiquitylated (II) and subsequently sorted for endocytosis. At the PM, Npr1 kinase antagonizes Art1 activity by phosphorylating its N terminus, resulting in Art1 departure from the PM (III); thus, both Ppz phosphatases and Npr1 kinase participate in a complex phosphorylation/dephosphorylation cycle that controls Art1 activity and cargo ubiquitylation.
Discussion
We have uncovered and characterized a novel feature of the yeast methionine response that involves Ppz phosphatase-mediated dephosphorylation of Art1 to trigger endocytosis of the methionine transporter Mup1. Specifically, we report that (1) Art1 undergoes rapid Ppz-mediated dephosphorylation at specific threonine residues in response to methionine stimulation, (2) these specific threonine residues are critical regulatory sites for Art1 function, and (3) Ppz phosphatases promote Art1 interaction with the methionine transporter, Mup1. Although our data indicate that Ppz-mediated dephosphorylation of Art1 is part of the cellular response to methionine, it occurs downstream of Art1 translocation to the PM. Prior studies identified N-terminal residues of Art1 that are subject to phosphorylation by the Npr1 kinase and are important regulators of Art1 localization to the PM (MacGurn et al., 2011). However, it is noteworthy that the phosphorylation events regulated by Ppz phosphatases, including Thr93, Thr245, and Thr795, were not previously found to be affected by loss or gain of Npr1 function (MacGurn et al., 2011). Thus, the data indicate that Ppz phosphatases regulate Npr1-independent phosphorylation events that occur at the N and C terminus of Art1. Overall, these findings highlight complex phosphoregulation as a mechanism for fine tuning activation of Art1 and cargo recognition at the PM (Fig. 7 E).
We previously showed that Ppz phosphatases are involved in the management of ubiquitin homeostasis (Lee et al., 2017), suggesting the ubiquitin deficiency exhibited in ppz mutants might indirectly contribute to endocytic trafficking defects resulting in the accumulation of specific nutrient transporters at the cell surface. Unexpectedly, we found that endocytic defects in ppz mutant cells cannot be suppressed by ubiquitin supplementation and that the mislocalized Ppz1-G2A variant can fully suppress the ubiquitin deficiency of ppz mutants without rescuing endocytic-trafficking defects. These results establish a separation of function between Ppz-mediated management of ubiquitin homeostasis and Ppz-mediated regulation of endocytosis.
Regulation of ARTs for fine tuning of PM protein composition
By investigating the role of Ppz phosphatases in endocytic trafficking, we have uncovered a novel mechanism for the regulation of Art1, an Rsp5 adaptor required for endocytosis of specific cargo, including Can1, Mup1, and Lyp1 (Lin et al., 2008). ART proteins have been shown to target Rsp5 activity to specific cargo at the PM, but many important questions remain regarding the regulation of ART activity and mechanisms of substrate targeting. For example, it is still unclear how different ART family adaptors recognize cargo and how these specific recognition events are regulated. Although previous studies have detected interactions between ARTs and cargo (Lin et al., 2008; Hatakeyama et al., 2010; O’Donnell et al., 2010; Alvaro et al., 2014; Llopis-Torregrosa et al., 2016), these interactions have been difficult to quantify and are generally considered to be transient. Furthermore, the regulatory steps and requirements for cargo engagement at the PM are not well understood. Here, leveraging a split-ubiquitin two-hybrid system, we were able to investigate the Art1–Mup1 interaction and found that this interaction is promoted by the presence of Ppz phosphatases (Fig. 7 D) and disrupted by specific phosphomimetic mutations of Art1 (T93D and T795D; Fig. 7 C). To our knowledge, this is the first example of a regulatory activity at the PM which controls ART-cargo recognition. Importantly, our data indicate that Ppz phosphatases are critical for the Art1-mediated endocytosis of both Mup1 and Can1, two known targets of Art1 that are regulated by distinct stimuli. Thus, we hypothesize that Ppz-mediated dephosphorylation of Art1 contributes to its activation at the PM, rather than the specificity of cargo recognition. We speculate that similar phosphoregulatory switches may operate at the PM to promote cargo recognition by other ART family adaptors in response to specific stimuli.
Multiple lines of evidence suggests that Art1 is subject to complex phosphoregulation by multiple kinases and phosphatases, including the Npr1 kinase (MacGurn et al., 2011) and Ppz phosphatases. Ppz phosphatases appear to primarily antagonize phosphorylation events (Thr93, Thr245, and Thr795) distinct from those mediated by Npr1 (Ser79, Ser82, Ser96, and Ser113; MacGurn et al., 2011). Importantly, the data presented here do not establish that Art1 is a direct substrate of Ppz phosphatases, and it is possible that loss of Ppz function indirectly impacts Art1 phosphorylation. We considered the possibility that Ppz phosphatases indirectly regulate Art1 phosphorylation by operating on the Npr1 kinase, which is itself subject to extensive phosphoregulation (MacGurn et al., 2011). However, our data do not suggest a role for Ppz phosphatases in the regulation of Npr1. Additionally, there are many phosphorylated residues detected on Art1 which remain uncharacterized, particularly at the C terminus, which has no characterized domain structure, but does include two PY motifs that bind to the WW domains of Rsp5. Overall, these findings indicate that Art1 functions like a microprocessor, receiving multiple phosphorylation inputs from distinct signaling relays and executing specific cargo ubiquitylation outputs at the PM to regulate endocytosis.
Using Rsp5 as bait we were able to generate quantitative phosphorylation profiles for most of the known Rsp5 adaptors (Fig. 6 and Fig. S4, G–J). This analysis reveals many previously uncharacterized phosphorylation events in this network, and it is noteworthy that most of the phosphopeptides detected in these experiments lie outside predicted arrestin fold domains (Fig. 6, C–E; and Fig. S4 G). Importantly, most of the phosphorylation events detected remain unchanged in ppz mutants, indicating that Ppz phosphatases do not operate broadly within the Rsp5 network and may be relatively restricted to Art1. One exception appears to be Art3/Aly2, which exhibits elevated phosphorylation in ppz mutants, particularly at N-terminal residues including Ser171, Ser176, and Ser201. It is notable that we previously reported Npr1-dependent phosphorylation of Ser155 on Art3/Aly2 (MacGurn et al., 2011). Thus, as is the case with Art1, Ppz-mediated regulation appears to be orthogonal to Npr1-mediated phosphorylation of Art3/Aly2. It will be important to determine how these different phosphorylation events affect the function of Art3/Aly2, which has previously been shown to mediate the ubiquitylation and endocytosis of the Asp/Glu transporter Dip5 (Hatakeyama et al., 2010). In general, the extensive phosphorylation of the Rsp5 network indicates a significant capacity to integrate input from multiple signaling systems in the cell, hinting at the complexity associated with the management of PM protein homeostasis. Adding to the complexity are previous studies reporting regulation of specific ubiquitylation events that are activating (Lin et al., 2008) or inhibitory (Ho et al., 2017), suggesting the potential for even more complex integration of signals. We speculate that mammalian orthologues of the ARTs, the arrestin domain–containing proteins, are likely subject to similar modes of regulation by phosphorylation and ubiquitylation and may similarly function like microprocessors.
Interestingly, loss of Ppz phosphatases did not impact interactions within the Rsp5 network but it did impact phosphorylation of Rsp5 itself, specifically residues Ser179 and Ser182. These residues lie in the region between the C2 domain and WW1, along with several other N-terminal phosphorylated residues that appear unaffected in ppz mutant cells. The functional significance of Rsp5 phosphorylation at Ser179 and Ser182 remains to be determined. However, in mammalian cells, phosphorylation of the Rsp5 homologue Nedd4L promotes interaction with 14-3-3 proteins, which effectively inhibits activity toward channels at the PM (Ichimura et al., 2005; Nagaki et al., 2006). It is tempting to speculate that similar modes of Rsp5 phosphoregulation may occur in yeast, although previous studies have reported phosphorylation-dependent interactions between yeast 14-3-3 proteins and the Rsp5 adaptors Bul1 and Bul2 (Merhi and André, 2012). Additional studies will be required to determine if phosphorylation of Rsp5 affects interaction with 14-3-3 proteins or Bul1/Bul2 adaptors.
Ppz phosphatases: regulators of endocytic trafficking and ubiquitin homeostasis
Ppz phosphatases have long been known to regulate salt tolerance and ion homeostasis (Posas et al., 1993; Yenush et al., 2002; Ruiz et al., 2004), and here we report a role for Ppz phosphatases in the regulation of endocytosis. The observation that the endocytic trafficking function of Ppz phosphatases is separable from their function in ubiquitin homeostasis is notable and raises important questions about cellular strategies for coupling endocytic trafficking and ubiquitin metabolism. Our finding that PM localization is critical for endocytic trafficking but dispensable for ubiquitin homeostasis suggests a compartmentalization of function that may be related to cycling of Ppz phosphatases on and off the PM. We propose a major cytosolic function for soluble Ppz phosphatases relates to ubiquitin homeostasis, while PM-localized Ppz phosphatases are poised to operate on Art1. It is noteworthy that Art1 and other Rsp5 adaptors have been reported to be regulated by ubiquitylation (Lin et al., 2008; Ho et al., 2017), so the ubiquitin deficiency reported for ppz mutant cells could indirectly impact the function of Rsp5 adaptors. Although our analysis of Art1 did not detect altered ubiquitylation in ppz mutant cells (Fig. S4 B), we cannot exclude the possibility that ubiquitin deficiency may affect the function of other Rsp5 adaptors, thus contributing to defects in other endocytic processes.
Our analysis revealed that loss of myristoylation results in relocalization of Ppz1-GFP from the PM to the cytosol and cytosolic punctae (Fig. 3 D). Although not characterized in this study, we speculate that these Ppz1-enriched punctae may function in the regulation of ubiquitin homeostasis. Although N-myristoylation of Ppz1 seems critical for its localization to the PM, we cannot exclude the possibility that other determinants may contribute to PM binding. Indeed, the N terminus of Ppz phosphatases has been shown to be important for Ppz function (Clotet et al., 1996) and is itself subject to phosphorylation. It is possible that the N termini of Ppz phosphatases are also subject to phosphoregulation and, like Art1 and Npr1, integrate signals from different relays to coordinate operations that affect endocytosis and ubiquitin homeostasis.
Ppz phosphatases have been reported to operate in complex with several cofactors, although the precise regulatory function of these cofactors is still unclear. Two factors that inhibit Ppz phosphatases, Hal3 and Vhs3, also function in complex with Cab3 in the biosynthesis of coenzyme A (Abrie et al., 2015). This raises the intriguing possibility that coenzyme A biosynthesis is somehow coupled to the regulation of endocytic trafficking and/or ubiquitin homeostasis via Ppz phosphatases. Ppz phosphatases have also been reported to interact with Ypi1, Sds22, and Glc8 (Venturi et al., 2000; Ceulemans et al., 2002; García-Gimeno et al., 2003), all known cofactors of yeast PP1 protein phosphatase Glc7. However, the fact that these cofactors may regulate both Glc7 and Ppz phosphatases makes it difficult to separate their Glc7-mediated functions from their Ppz-mediated functions in vivo, and doing so may require isolation and characterization of Ypi1, Sds22, and Glc8 variants that interact exclusively with either Glc7 or Ppz phosphatases. These interactions suggest that Ppz phosphatases, like Glc7, may function in complex with multiple regulatory factors. Characterizing these interactions and their role in Ppz phosphatase function will be critical for improved characterization of specific Ppz activities and substrate interactions.
It is tempting to speculate that the role of Ppz phosphatases in salt tolerance and ion homeostasis is due to defects in endocytic trafficking. Indeed, it was previously proposed that Ppz phosphatases may regulate the trafficking of the Trk1-Trk2 potassium transporters (Ruiz et al., 2004; Yenush et al., 2005). However, we previously reported that ubiquitin supplementation can suppress many of the phenotypes associated with salt tolerance, including resistance to lithium and sensitivity to potassium (Lee et al., 2017). Evidence presented here indicates that the ubiquitin deficiency is distinct and separable from the endocytic defects of ppz mutant cells, leading us to conclude that the salt tolerance phenotypes are linked to ubiquitin deficiency, but not endocytic-trafficking defects. Indeed, previous studies have reported that potassium sensitivity phenotypes in ppz mutant cells are not solely attributable to Trk1-Trk2 transporters (Ruiz et al., 2004) and that salt tolerance in ppz mutant cells may be due to increased expression of the Ena1 Na+ ATPase (Ruiz et al., 2003). Ultimately, dissecting the regulation and effector mechanisms of Ppz phosphatases, and specifically those that converge on the ART-Rsp5 network, is likely to yield new insights into how eukaryotic cells coordinate the regulation of endocytic trafficking and ubiquitin metabolism in a manner critical for the execution of complex biological processes, including ion homeostasis and salt tolerance.
Materials and methods
Yeast strains and growth conditions
Yeast strains used in this study are reported in Table S1, and plasmids used are reported in Table S2. The SEY6210 strain background (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9) was used for most experiments. BY4741 and SUB280 (MATa lys2-801 leu2-3,112 ura3-52 his3-Δ200 trp 1–1 ubi1-Δ1::TRP1 ubi2-Δ2::ura3 ubi3-Δub-2 ubi4-Δ2::LEU2 [pUB39 Ub, LYS2] [pUB100, HIS3]; generously supplied by D. Finley and M. Boselli [Harvard Medical School, Boston, MA]; Spence et al., 1995) yeast strain backgrounds were used where indicated. Unless otherwise indicated, yeast strains were cultured in liquid and solid synthetic complete (SC) media containing dextrose (2%) as a carbon source. To assess canavanine sensitivity, yeast strains were cultured in liquid media (SC + dextrose) to saturation and 10-fold serial dilutions (starting with a culture adjusted to a density of 1 OD600/ml) were plated onto the indicated media using a 48-pin replicator. Higher concentrations (4.0 µg/ml) of canavanine were used to assess resistance, while lower concentrations (0.1 µg/ml, 0.2 µg/ml, and 0.3 µg/ml) were used to assess hypersensitivity. Sensitivity to thialysine (toxic lysine analogue; 2.5 µg/ml on SC + dextrose solid media) was tested in the BY4741 strain background (since SEY6210 is a lysine auxotroph).
Fluorescence microscopy
Yeast cells expressing fluorescent fusion proteins were cultured at 26°C to mid-log phase in synthetic media and then imaged in synthetic media at 26°C. All fluorescence microscopy was performed using a DeltaVision Elite Imaging system with a customized Olympus IX-71 inverted microscope, an Olympus 100× oil objective (1.4 NA), a DV Elite sCMOS camera, and temperature-controlled environmental chamber (GE Healthcare). All images were collected (as z stacks ≥3 µm), deconvolved, and analyzed using SoftWorx software (GE Healthcare). For a given cargo (Mup1-GFP, Can1-GFP, or Fur4-GFP), GFP signal intensity at the PM and at the vacuole (labeled Vph1-mCherry, which marks the limiting membrane of the vacuole) was measured, and a ratio was computed over a population of cells (n > 30). Two-way ANOVA tests were performed to assess significance. For Pearson correlation coefficient measurements in cells expressing Mup1-Mars and Art1-GFP, individual cells (n > 25 per condition) were measured in SoftWorx, and the data were analyzed using unpaired t test analysis in Prism (GraphPad).
Analysis of protein expression in yeast
Yeast lysates were prepared by growing cultures according to the indicated conditions, pelleting 5 OD600 equivalents of cells and precipitating by addition of 10% TCA. Precipitates were washed in acetone, aspirated, dried under vacuum in a speed-vac, resuspended in lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 1 mM EDTA, and 1% SDS), and mechanically disrupted with acid-washed glass beads. Protein sample buffer (150 mM Tris, pH 6.8, 6 M Urea, 6% SDS, 10% β-mercaptoethanol, and 20% Glycerol) was added, and extracts were analyzed by SDS-PAGE and immunoblotting. Quantitative fluorescence imaging of immunoblots was performed using an Odyssey infrared imaging system (LI-COR Biosciences). Antibodies used in this study include α-FLAG (M2; mouse monoclonal; Sigma), α-HA (12CA5; mouse monoclonal; Sigma); α-G6PDH (rabbit polyclonal; Sigma), α-ubiquitin (MAB1510; mouse monoclonal; Millipore), and α-LexA (rabbit polyclonal; Millipore).
SILAC-based quantitative proteomic analysis
Quantitative mass spectrometry analysis by SILAC was performed on yeast strains auxotrophic for lysine and arginine that express the indicated bait, fused to a 3× FLAG tag at the chromosomal locus. Cells were grown to mid-log phase (OD600 = 0.7; ∼700 OD equivalents) in SC medium containing either heavy or light isotopes (lysine and arginine). Lysate was generated by bead beating cell pellets in lysis buffer (50 mM Tris-HCl, pH 7.5, 0.2% NP-40, and 150 mM NaCl with protease and phosphatase inhibitors). Affinity purification was performed by binding to anti-FLAG M2 affinity gel (Sigma) for 2 h at 4°C, washing beads three times with lysis buffer, and elution by boiling beads for 5 min in elution solution (50 mM Tris, pH 8.0, and 1% SDS), followed by collection of the sample through a tip-column, sample reduction (10 mM DTT; 5 min at 95°C), and alkylation (20 mM iodoacetamide; room temperature). Samples were digested with 1 µg of trypsin overnight at 37°C and purified over using immobilized metal affinity chromatography (Albuquerque et al., 2008; MacGurn et al., 2011). Purified peptides were dried, reconstituted in 0.1% trifluoroacetic acid, and analyzed by liquid chromatography tandem mass spectrometry using a Q Exactive mass spectrometer (Thermo). Database search and heavy:light isotope ratio quantitation was performed using MaxQuant software (version 1.5.3.30).
Mup1-pHluorin–trafficking assays
Mup1-pHluorin–trafficking assays were performed by stimulating cells with either a high concentration of methionine (2 µg/ml) or a low concentration of methionine (667 ng/ml) to test for endocytic-trafficking loss-of-function or gain-of-function phenotypes, respectively. Indicated yeast strains expressing Mup1-pHluorin were grown to mid-log phase in synthetic media and distributed in 250 µl aliquots in 96-well plates. Following stimulation with the indicated concentrations of methionine, pHluorin signal was measured by flow cytometry (Guava; Millipore). Over 10,000 cells per time point were detected and analyzed per condition based on gating in the FITC channel, which detects signal from pHluorin. Flow cytometry data were analyzed using GuavaSoft 2.6 software with InCyte analyzer (Millipore).
Split-ubiquitin membrane yeast two-hybrid analysis
To probe interactions between Art1 and cargo, we used a split-ubiquitin membrane yeast two-hybrid system as previously described (Iyer et al., 2005). Prey vectors were constructed using the pPRC3 backbone and bait vectors were constructed using the pRS415-Cub-PLV backbone. Bait and prey fusion protein expression was confirmed by immunoblotting for LexA and HA epitope tag, respectively. The pSH18-34 plasmid (which encodes the LacZ gene under the control of tandem LexA operators) was transformed into indicated strains to report for two-hybrid interactions. To detect β-galactosidase activity, cells were grown on X-Gal plates containing 40 µg/ml of X-Gal and 0.7 M potassium phosphate (pH 7.0). Since Δppz1Δppz2 cells are sensitive to high concentrations of potassium (Lee et al., 2017) and did not grow on X-Gal plates, we instead measured β-galactosidase activity in yeast lysates. In brief, cells expressing bait and prey constructs were cultured till mid-log phase (OD600 < ∼0.7), and 5 OD600 of cells were collected. Cells were resuspended in Z buffer containing 0.1 M sodium phosphate (pH 7.0) and lysed by vortexing in 0.1% SDS and chloroform. 4 mg/ml of ONPG (ortho-Nitrophenyl-β-galactoside) was added and incubated for 50 min. 1 M sodium carbonate was added and OD420 was measured. The activity units were calculated using the formula: 1,000 × OD420/volume of cells (ml) × incubation time × OD600.
Online supplemental material
Fig. S1 (related to Fig. 1) illustrates the Mup1-pHluorin–trafficking assay and the SILAC-MS methodology and provides additional data for characterization of Art1 mutants in the Mup1-pHluorin–trafficking assay. Fig. S2 (related to Fig. 2) provides additional data characterizing the endocytic-trafficking defects observed in ppz mutant cells. Fig. S3 (related to Fig. 3) includes additional complementation analysis of the ppz mutant canavanine-hypersensitivity phenotype and includes fluorescence microscopy imaging analysis of Ppz1-GFP. Fig. S4 (related to Fig. 4 and Fig. 5) provides additional genetic interaction analysis of PPZ1, PPZ2, NPR1, and ART1 and provides additional SILAC-MS phosphorylation profiling analysis related to the data presented in Fig. 5. Fig. S5 (related to Fig. 7) reports additional data and provides additional controls for the split-ubiquitin two-hybrid analysis. A list of yeast strains and plasmids used in this study are provided in Table S1 and Table S2, respectively. Raw data and statistical reporting for this study are provided in Table S3.
Supplementary Material
Acknowledgments
We are very grateful to K. Rose for helpful advice regarding analysis of quantitative proteomic data. We are very grateful to T. Graham, M. Baile, and E. Guiney for critical reading of the manuscript and E. MacGurn for assistance with graphic design. We are especially grateful to S. Emr for invaluable feedback and supreme mentorship.
J.M. Tumolo was supported by National Institutes of Health (NIH) training grant T32 CA119925. S. Lee was supported by NIH training grant T32 HL069765. This research was supported by NIH grant R00 GM101077 (to J.A. MacGurn) and NIH grant R01 GM118491 (to J.A. MacGurn).
The authors declare no competing financial interests.
Author contributions: All authors contributed to data collection and analysis and provided intellectual input. S. Lee and J. MacGurn designed the study and wrote the manuscript. H.C. Ho, J.M. Tumolo, and P.C. Hsu contributed to review and editing of the manuscript.
References
- Abrie J.A., Molero C., Ariño J., and Strauss E.. 2015. Complex stability and dynamic subunit interchange modulates the disparate activities of the yeast moonlighting proteins Hal3 and Vhs3. Sci. Rep. 5:15774 10.1038/srep15774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., and Zhou H.. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 7:1389–1396. 10.1074/mcp.M700468-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro C.G., O’Donnell A.F., Prosser D.C., Augustine A.A., Goldman A., Brodsky J.L., Cyert M.S., Wendland B., and Thorner J.. 2014. Specific α-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol. Cell. Biol. 34:2660–2681. 10.1128/MCB.00230-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik A.Y., Nowak J., Swaminathan S., and Hochstrasser M.. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 11:3365–3380. 10.1091/mbc.11.10.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M., and Léon S.. 2014. Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and the ubiquitin ligase Rsp5. eLife. 3:e03307 10.7554/eLife.03307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M., Vieira N., Lara D., Gomes-Rezende J., Soares-Cunha C., Casal M., Haguenauer-Tsapis R., Vincent O., Paiva S., and Léon S.. 2012. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J. Cell Biol. 196:247–259. 10.1083/jcb.201109113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H., Vulsteke V., De Maeyer M., Tatchell K., Stalmans W., and Bollen M.. 2002. Binding of the concave surface of the Sds22 superhelix to the alpha 4/alpha 5/alpha 6-triangle of protein phosphatase-1. J. Biol. Chem. 277:47331–47337. 10.1074/jbc.M206838200 [DOI] [PubMed] [Google Scholar]
- Clotet J., Posas F., de Nadal E., and Ariño J.. 1996. The NH2-terminal extension of protein phosphatase PPZ1 has an essential functional role. J. Biol. Chem. 271:26349–26355. 10.1074/jbc.271.42.26349 [DOI] [PubMed] [Google Scholar]
- Debonneville C., Flores S.Y., Kamynina E., Plant P.J., Tauxe C., Thomas M.A., Münster C., Chraïbi A., Pratt J.H., Horisberger J.D., et al. 2001. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na(+) channel cell surface expression. EMBO J. 20:7052–7059. 10.1093/emboj/20.24.7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore P.P., and von Zastrow M.. 2014. Endocytosis, signaling, and beyond. Cold Spring Harb. Perspect. Biol. 6:a016865 10.1101/cshperspect.a016865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakitsas P., Adam G., Daidié D., van Bemmelen M.X., Fouladkou F., Patrignani A., Wagner U., Warth R., Camargo S.M., Staub O., and Verrey F.. 2007. Early aldosterone-induced gene product regulates the epithelial sodium channel by deubiquitylation. J. Am. Soc. Nephrol. 18:1084–1092. 10.1681/ASN.2006080902 [DOI] [PubMed] [Google Scholar]
- García-Gimeno M.A., Muñoz I., Ariño J., and Sanz P.. 2003. Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J. Biol. Chem. 278:47744–47752. 10.1074/jbc.M306157200 [DOI] [PubMed] [Google Scholar]
- Goh L.K., and Sorkin A.. 2013. Endocytosis of receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 5:a017459 10.1101/cshperspect.a017459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh L.K., Huang F., Kim W., Gygi S., and Sorkin A.. 2010. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 189:871–883. 10.1083/jcb.201001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournas C., Saliba E., Krammer E.M., Barthelemy C., Prévost M., and André B.. 2017. Transition of yeast Can1 transporter to the inward-facing state unveils an α-arrestin target sequence promoting its ubiquitylation and endocytosis. Mol. Biol. Cell. 28:2819–2832. 10.1091/mbc.e17-02-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney E.L., Klecker T., and Emr S.D.. 2016. Identification of the endocytic sorting signal recognized by the Art1-Rsp5 ubiquitin ligase complex. Mol. Biol. Cell. 27:4043–4054. 10.1091/mbc.e16-08-0570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama R., Kamiya M., Takahara T., and Maeda T.. 2010. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol. Cell. Biol. 30:5598–5607. 10.1128/MCB.00464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Ho H.C., MacGurn J.A., and Emr S.D.. 2017. Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs. Mol. Biol. Cell. 28:1271–1283. 10.1091/mbc.e17-01-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovsepian J., Defenouillère Q., Albanèse V., Váchová L., Garcia C., Palková Z., and Léon S.. 2017. Multilevel regulation of an α-arrestin by glucose depletion controls hexose transporter endocytosis. J. Cell Biol. 216:1811–1831. 10.1083/jcb.201610094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T., Yamamura H., Sasamoto K., Tominaga Y., Taoka M., Kakiuchi K., Shinkawa T., Takahashi N., Shimada S., and Isobe T.. 2005. 14-3-3 proteins modulate the expression of epithelial Na+ channels by phosphorylation-dependent interaction with Nedd4-2 ubiquitin ligase. J. Biol. Chem. 280:13187–13194. 10.1074/jbc.M412884200 [DOI] [PubMed] [Google Scholar]
- Iyer K., Bürkle L., Auerbach D., Thaminy S., Dinkel M., Engels K., and Stagljar I.. 2005. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci. STKE. 2005:pl3. [DOI] [PubMed] [Google Scholar]
- Lee S., Tumolo J.M., Ehlinger A.C., Jernigan K.K., Qualls-Histed S.J., Hsu P.C., McDonald W.H., Chazin W.J., and MacGurn J.A.. 2017. Ubiquitin turnover and endocytic trafficking in yeast are regulated by Ser57 phosphorylation of ubiquitin. eLife. 6:e29176 10.7554/eLife.29176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S., and Haguenauer-Tsapis R.. 2009. Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp. Cell Res. 315:1574–1583. 10.1016/j.yexcr.2008.11.014 [DOI] [PubMed] [Google Scholar]
- Léon S., Erpapazoglou Z., and Haguenauer-Tsapis R.. 2008. Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell. 19:2379–2388. 10.1091/mbc.e08-01-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., MacGurn J.A., Chu T., Stefan C.J., and Emr S.D.. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 135:714–725. 10.1016/j.cell.2008.09.025 [DOI] [PubMed] [Google Scholar]
- Llopis-Torregrosa V., Ferri-Blázquez A., Adam-Artigues A., Deffontaines E., van Heusden G.P., and Yenush L.. 2016. Regulation of the Yeast Hxt6 Hexose Transporter by the Rod1 α-Arrestin, the Snf1 Protein Kinase, and the Bmh2 14-3-3 Protein. J. Biol. Chem. 291:14973–14985. 10.1074/jbc.M116.733923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C., Payne J.A., Aboian M., Smith W., Katzmann D.J., and Piper R.C.. 2015. A family of tetraspans organizes cargo for sorting into multivesicular bodies. Dev. Cell. 33:328–342. 10.1016/j.devcel.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J.A. 2014. Garbage on, garbage off: new insights into plasma membrane protein quality control. Curr. Opin. Cell Biol. 29:92–98. 10.1016/j.ceb.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.C., Smolka M.B., and Emr S.D.. 2011. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 147:1104–1117. 10.1016/j.cell.2011.09.054 [DOI] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.C., and Emr S.D.. 2012. Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81:231–259. 10.1146/annurev-biochem-060210-093619 [DOI] [PubMed] [Google Scholar]
- Merhi A., and André B.. 2012. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol. Cell. Biol. 32:4510–4522. 10.1128/MCB.00463-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K., Yamamura H., Shimada S., Saito T., Hisanaga S., Taoka M., Isobe T., and Ichimura T.. 2006. 14-3-3 Mediates phosphorylation-dependent inhibition of the interaction between the ubiquitin E3 ligase Nedd4-2 and epithelial Na+ channels. Biochemistry. 45:6733–6740. 10.1021/bi052640q [DOI] [PubMed] [Google Scholar]
- Nikko E., and Pelham H.R.. 2009a Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 10:1856–1867. 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Sullivan J.A., and Pelham H.R.B.. 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9:1216–1221. 10.1038/embor.2008.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A.F., Apffel A., Gardner R.G., and Cyert M.S.. 2010. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell. 21:3552–3566. 10.1091/mbc.e10-07-0636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A.F., Huang L., Thorner J., and Cyert M.S.. 2013. A calcineurin-dependent switch controls the trafficking function of α-arrestin Aly1/Art6. J. Biol. Chem. 288:24063–24080. 10.1074/jbc.M113.478511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A.F., McCartney R.R., Chandrashekarappa D.G., Zhang B.B., Thorner J., and Schmidt M.C.. 2015. 2-Deoxyglucose impairs Saccharomyces cerevisiae growth by stimulating Snf1-regulated and α-arrestin-mediated trafficking of hexose transporters 1 and 3. Mol. Cell. Biol. 35:939–955. 10.1128/MCB.01183-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Apaja P.M., and Lukacs G.L.. 2011. Protein quality control at the plasma membrane. Curr. Opin. Cell Biol. 23:483–491. 10.1016/j.ceb.2011.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Casamayor A., and Ariño J.. 1993. The PPZ protein phosphatases are involved in the maintenance of osmotic stability of yeast cells. FEBS Lett. 318:282–286. 10.1016/0014-5793(93)80529-4 [DOI] [PubMed] [Google Scholar]
- Prosser D.C., Whitworth K., and Wendland B.. 2010. Quantitative analysis of endocytosis with cytoplasmic pHluorin chimeras. Traffic. 11:1141–1150. 10.1111/j.1600-0854.2010.01088.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F., and Staub O.. 2015. NEDD4-2 and salt-sensitive hypertension. Curr. Opin. Nephrol. Hypertens. 24:111–116. 10.1097/MNH.0000000000000097 [DOI] [PubMed] [Google Scholar]
- Ronzaud C., and Staub O.. 2014. Ubiquitylation and control of renal Na+ balance and blood pressure. Physiology (Bethesda). 29:16–26. [DOI] [PubMed] [Google Scholar]
- Rotin D., and Kumar S.. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10:398–409. 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- Ruiz A., Yenush L., and Ariño J.. 2003. Regulation of ENA1 Na(+)-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot. Cell. 2:937–948. 10.1128/EC.2.5.937-948.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., del Carmen Ruiz M., Sánchez-Garrido M.A., Ariño J., and Ramos J.. 2004. The Ppz protein phosphatases regulate Trk-independent potassium influx in yeast. FEBS Lett. 578:58–62. 10.1016/j.febslet.2004.10.069 [DOI] [PubMed] [Google Scholar]
- Schmid S.L. 2017. Reciprocal regulation of signaling and endocytosis: Implications for the evolving cancer cell. J. Cell Biol. 216:2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J., Sadis S., Haas A.L., and Finley D.. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15:1265–1273. 10.1128/MCB.15.3.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S., Amerik A.Y., and Hochstrasser M.. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell. 10:2583–2594. 10.1091/mbc.10.8.2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi G.M., Bloecher A., Williams-Hart T., and Tatchell K.. 2000. Genetic interactions between GLC7, PPZ1 and PPZ2 in saccharomyces cerevisiae. Genetics. 155:69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J., and Drubin D.G.. 2012. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22:1–13. 10.1016/j.tcb.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenush L., Mulet J.M., Ariño J., and Serrano R.. 2002. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J. 21:920–929. 10.1093/emboj/21.5.920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenush L., Merchan S., Holmes J., and Serrano R.. 2005. pH-Responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell. Biol. 25:8683–8692. 10.1128/MCB.25.19.8683-8692.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Macgurn J.A., Liu M., and Emr S.. 2013. The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. eLife. 2:e00459 10.7554/eLife.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.