This study reveals a previously unappreciated interplay between Rab5 GTPases and TORC2 function in yeast. TORC2 signaling stimulates the Rab5-specific guanine nucleotide exchange factor Muk1, and, in turn, activated Rab5 GTPases, especially Vps21, act as positive effectors to stimulate TORC2 activity.
Abstract
Target of rapamycin complex-2 (TORC2), a conserved protein kinase complex, is an indispensable regulator of plasma membrane homeostasis. In budding yeast (Saccharomyces cerevisiae), the essential downstream effector of TORC2 is protein kinase Ypk1 and its paralog Ypk2. Muk1, a Rab5-specific guanine nucleotide exchange factor (GEF), was identified in our prior global screen for candidate Ypk1 targets. We confirm here that Muk1 is a substrate of Ypk1 and demonstrate that Ypk1-mediated phosphorylation stimulates Muk1 function in vivo. Strikingly, yeast lacking its two Rab5 GEFs (Muk1 and Vps9) or its three Rab5 paralogs (Vps21/Ypt51, Ypt52, and Ypt53) or overexpressing Msb3, a Rab5-directed GTPase-activating protein, all exhibit pronounced reduction in TORC2-mediated phosphorylation and activation of Ypk1. Vps21 coimmunoprecipitates with TORC2, and immuno-enriched TORC2 is less active in vitro in the absence of Rab5 GTPases. Thus, TORC2-dependent and Ypk1-mediated activation of Muk1 provides a control circuit for positive (self-reinforcing) up-regulation to sustain TORC2-Ypk1 signaling.
Introduction
Studies in Saccharomyces cerevisiae (Loewith et al., 2002; Wedaman et al., 2003) were the first to show the presence of two evolutionarily conserved multicomponent protein kinase complexes—Target of rapamycin complex-1 (TORC1) and TORC2—in which a TOR polypeptide is the catalytic subunit, as now found in all eukaryotes (Eltschinger and Loewith, 2016; González and Hall, 2017; Saxton and Sabatini, 2017; Tatebe and Shiozaki, 2017). Whereas the role of TORC1 in both yeast and animal cells is in sensing at the lysosome/vacuole whether supplies of nutrients are sufficient to support protein synthesis and other anabolic processes required for cell growth (Jewell and Guan, 2013; Kim et al., 2013; Lim and Zoncu, 2016; Powis and De Virgilio, 2016), TORC2, located at the cell cortex in both yeast and animal cells, promotes the biosynthesis of glycerolipids and sphingolipids and controls other pathways required for plasma membrane (PM) biogenesis and homeostasis (Gaubitz et al., 2016; Guri et al., 2017; Roelants et al., 2017a, 2018).
The fact that basal signaling emanating from TORC2 is essential for cell viability also was first shown in yeast (Kunz et al., 1993; Helliwell et al., 1994). Subsequent work has demonstrated that TORC2 activity is responsive to various stresses and insults that can perturb the integrity of the cell envelope. Certain challenges (sphingolipid depletion, hypotonic conditions, heat shock, and elevated exogenous acetic acid) markedly stimulate TORC2 function (Roelants et al., 2011; Berchtold et al., 2012; Sun et al., 2012; Guerreiro et al., 2016), whereas others (hypertonic conditions and cell wall damage) markedly reduce TORC2 activity (Lee et al., 2012; Muir et al., 2015; Leskoske et al., 2018). The primary downstream effector of yeast TORC2 is the AGC family protein kinase Ypk1 (mammalian orthologue is SGK1; Casamayor et al., 1999) and its paralog Ypk2 because it has been shown that constitutively active alleles of either Ypk1 or Ypk2 rescue the inviability of tor2 mutations and other mutations (avo3) that result in deficient TORC2 function (Kamada et al., 2005; Aronova et al., 2008; Roelants et al., 2011; Leskoske et al., 2017). TORC2 activates Ypk1 function by phosphorylating multiple sites within its C-terminal regulatory tail (Leskoske et al., 2017). To understand what physiological processes are under TORC2-Ypk1 control, we conducted both genome-wide genetic (Roelants et al., 2002) and global biochemical (Muir et al., 2014) screens to identify candidate substrates and regulators of TORC2-Ypk1 signaling. Validation and characterization of many of these candidates has shed considerable light on those sectors of cellular physiology that are regulated by the TORC2-Ypk1 axis (Roelants et al., 2010, 2011, 2017b, 2018; Lee et al., 2012; Muir et al., 2014, 2015; Alvaro et al., 2016).
Among the presumptive, but as yet uncharacterized, targets of Ypk1 that we identified is Muk1 (Muir et al., 2014). Muk1 and its paralog Vps9 are guanine nucleotide exchange factors (GEFs) that catalyze GDP-GTP exchange in the Rab5 class of GTPases in S. cerevisiae, namely, Vps21/Ypt51 and its paralogs Ypt52 and Ypt53 (Burd et al., 1996; Hama et al., 1999; Paulsel et al., 2013; Bean et al., 2015). As in other eukaryotes, the function of Rab5 GTPases was thought to be confined to promoting vesicle formation, trafficking, and sorting in the early stages of the endocytic pathway (Singer-Krüger et al., 1994, 1995; Cabrera and Ungermann, 2013), and disruption of Rab5 function in yeast leads to accumulation of cargo in endosomes and disruption of their delivery to the vacuole (Horazdovsky et al., 1994; Gerrard et al., 2000). Muk1 and Vps9 have overlapping roles as Rab5 GEFs because a muk1Δ vps9Δ double mutant has a more severe phenotype than either single mutant and, tellingly, exhibits a phenotype closely resembling a vps21Δ ypt52Δ ypt53Δ triple mutant (Cabrera and Ungermann, 2013; Paulsel et al., 2013). Although Muk1 and Vps9 both act on Rab5 GTPases, the presence of two distinct proteins suggests that they may be differentially regulated.
In this study, we sought, first, to determine whether Muk1 is indeed an authentic and physiologically relevant target of Ypk1 and, if so, the consequences of its Ypk1-mediated phosphorylation. In the process, and as documented here, we discovered quite unexpectedly that Rab5 function is necessary to support maximal TORC2 function, revealing a previously unappreciated new connection between the vesicle trafficking machinery and the control of PM homeostasis by TORC2-Ypk1 signaling.
Results
Muk1 is a substrate of protein kinase Ypk1
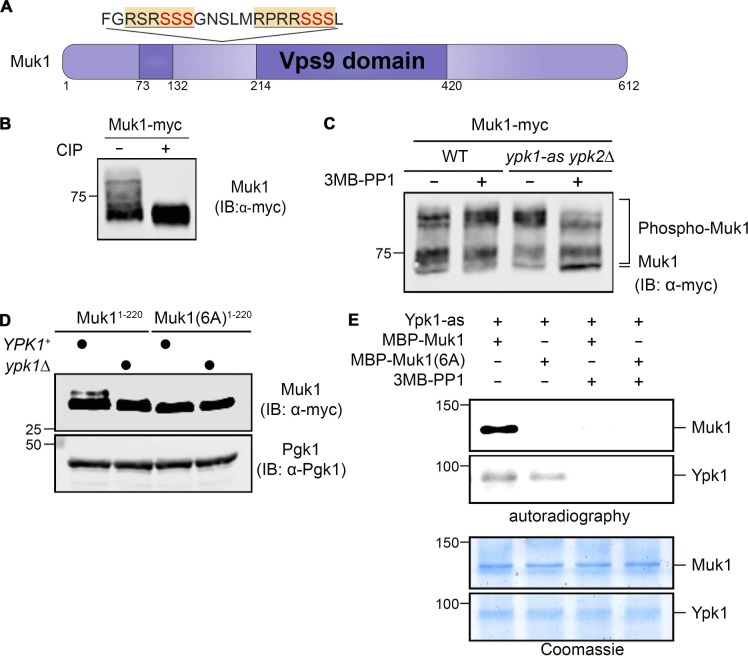
An ∼260-residue segment of Vps9 (451 residues) is necessary and sufficient for its Rab5 GEF activity (Carney et al., 2006; Barr and Lambright, 2010; Bean et al., 2015). Compared with Vps9 itself (Gough et al., 2001), the catalytic (Vps9 homology) domain of Muk1 (612 residues) is split by an 83-residue insert that contains, in tandem, two matches to the consensus phospho-acceptor site motif of Ypk1 (RxRxxS; 168RSRSSSG174 and 179RPRRSSS185; Mok et al., 2010; Muir et al., 2014; Fig. 1 A). These same sites are completely conserved among S. cerevisiae and all its sensu stricto relatives, as well as in more divergent yeast species (e.g., Candida glabrata, Kluyveromyces lactis, and Ashbya gossypii; see https://portals.broadinstitute.org/cgi-bin/regev/orthogroups/show_orthogroup.cgi?orf=YPL070W), and phosphorylation at these same sites in vivo has been detected in several global phosphoproteomic analyses of S. cerevisiae (Albuquerque et al., 2008; Holt et al., 2009; Swaney et al., 2013).
Figure 1.
Muk1 is phosphorylated by Ypk1 in vivo and in vitro. (A) Schematic depiction of Muk1. Dark purple, split catalytic (Vps9 homology) domain; yellow and underlined, consensus Ypk1 phospho-acceptor site; red, phosphorylated residues. (B) WT cells (BY4741) expressing Muk1-myc from the GAL promoter on a multi-copy (2 µm DNA) vector (pMLT22) were grown to mid-exponential phase, harvested, and lysed, and equivalent samples of the resulting extract protein were incubated in the absence (−) or presence (+) of CIP and, after treatment, resolved by SDS-PAGE in an 8% acrylamide gel and analyzed by immunoblotting (IB), all as described in Materials and methods. (C) WT (BY4741) or otherwise isogenic ypk1-as ypk2Δ (yAM123-A) cells expressing Muk1-myc as in B were grown to mid-exponential phase, treated with vehicle (DMSO) or 3MB-PP1 (10 µM final concentration) in the same solvent for 90 min, harvested, lysed, and examined as in B, except SDS-PAGE was conducted using a 7% acrylamide gel. (D) WT (BY4741) or otherwise isogenic ypk1Δ (JTY6142) cells expressing from the same vector as in B either Muk1(1–220; pMLT56) or Muk1(1–220 6A; pMLT57) were grown to mid-exponential phase, treated with AbA (1.8 µM final concentration) for 2 h, harvested, and lysed, and the resulting extracts were analyzed as in B, except SDS-PAGE was conducted using a 13.5% acrylamide gel. Pgk1 served as a loading control. (E) Equivalent amounts (∼0.5 µg) of MBP-Muk1-(His)6 or MBP-Muk16A-(His)6, expressed in and purified from E. coli, were incubated with [γ-32P]ATP and Ypk1-as purified from S. cerevisiae in the absence (–) or presence (+) of 3MB-PP1, as described in Materials and methods, and the resulting products were analyzed by both autoradiography (top) and staining with Coomassie Blue dye (bottom).
We confirmed that Muk1 is a phosphoprotein, first by showing that an epitope-tagged derivative (Muk1-myc) migrates even in standard SDS-PAGE as a series of multiple, slower-mobility isoforms that are collapsed to a faster-mobility species by treatment with calf intestinal phosphatase (CIP; Fig. 1 B). To determine whether any isoforms were attributable to phosphorylation by Ypk1, we expressed Muk1-myc in either WT or otherwise isogenic cells lacking Ypk2 and carrying an allele of Ypk1, Ypk1(L424A) (hereafter Ypk1-as), sensitive to acute inhibition by the adenine analogue 1-(tert-butyl)-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine (3MB-PP1; Lopez et al., 2014; Muir et al., 2014). We found that only in the analogue-sensitive cells and only in the presence of inhibitor were the slowest-mobility species greatly reduced and the most hypophosphorylated species markedly increased (Fig. 1 C), showing that Muk1 is phosphorylated in a Ypk1-dependent manner in vivo. We have shown previously that in other Ypk1 substrates containing Ser residues juxtaposed (immediately upstream, immediately downstream, or both) to the primary phospho-acceptor Ser, such residues can also be phosphorylated by Ypk1 both in vivo and in vitro (Roelants et al., 2011; Muir et al., 2014). Hence, to test whether Ypk1-mediated phosphorylation of Muk1 was occurring at the expected sites, we examined the mobility pattern of WT Muk1 tagged with a 3xFLAG epitope or a derivative in which all six potential Ser residues at the Ypk1 consensus motifs were mutated to Ala. For this purpose, we used Phos-tag SDS-PAGE, a technique that enhances retardation of a protein depending on the extent of its phosphorylation (Kinoshita et al., 2009). We found that lack of the six Ser residues resulted in a marked reduction in the slowest-mobility isoforms and a pronounced increase in the fastest-mobility (hypophosphorylated) isoform (Fig. S1), indicating that these sites are phosphorylated in vivo.
To more readily visualize Ypk1-specific phosphorylation of Muk1, we expressed a smaller fragment (residues 1–220) of WT Muk1-myc or an otherwise identical fragment in which the six potential Ser residues at the Ypk1 consensus motifs were mutated to Ala. We also took advantage of the fact that TORC2-dependent activation of Ypk1 is stimulated in response to inhibition of sphingolipid biosynthesis by antibiotics such as myriocin (Myr; which inactivates l-Ser:palmitoyl-CoA acyltransferase; Miyake et al., 1995) or aureobasidin A (AbA; which inactivates inositolphosphate-ceramide synthase; Sugimoto et al., 2004), as we and others have shown (Roelants et al., 2011; Berchtold et al., 2012; Khakhina et al., 2015). Indeed, when cells were treated with AbA, we found that the Muk1(1–220) fragment displayed a readily detectable slower-mobility isoform that was eliminated by the 6A mutations or by absence of just Ypk1 itself (Fig. 1 D), because under normal growth conditions very little Ypk2 is expressed (Gasch et al., 2000; Roelants et al., 2002). Moreover, as expected, this species appeared when cells were treated with AbA, but not in its absence (Fig. S1 B). Finally, purified recombinant Muk1, but not Muk1(6A), was robustly phosphorylated in vitro by purified Ypk1-as in the absence of 3-MB-PP1, but not in its presence (Fig. 1 E). Thus, Muk1 is a bona fide Ypk1 substrate.
Muk1 function is stimulated by Ypk1-mediated phosphorylation
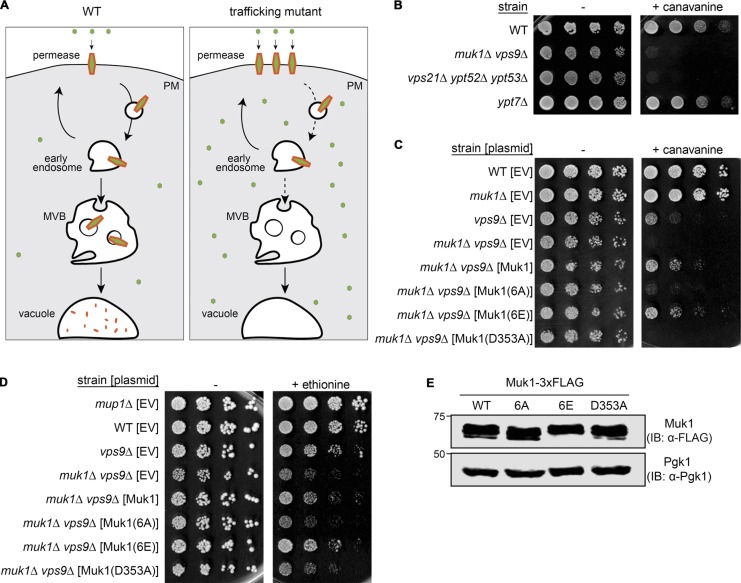
The three yeast Rab5 isoforms (Vps21/Ypt51, Ypt52, and Ypt53) are involved in the early stages of the endocytic pathway, which is responsible for clearing integral PM membrane proteins, including amino acid permeases (Singer-Krüger et al., 1994; Cabrera and Ungermann, 2013; Paulsel et al., 2013). Hence, defects in that process will increase the steady-state level of such permeases, enhancing uptake of cognate amino acid analogues, thus conferring on the cell greater sensitivity to the toxic effect of such compounds (Grenson et al., 1966; Lin et al., 2008; Fig. 2 A). Indeed, we found that compared with otherwise isogenic WT cells, cells lacking both Rab5 GEFs or all three Rab5 isoforms were markedly more sensitive to the growth-inhibitory action of canavanine, an Arg analogue taken up by the Arg permease (Can1), whereas cells lacking Ypt7, the sole yeast orthologue of Rab7, which acts later in the endocytic pathway (Schimmöller and Riezman, 1993; Rink et al., 2005), were not (Fig. 2 B), indicating that the observed defect was specific for Rab5.
Figure 2.
Ypk1-dependent phosphorylation of Muk1 promotes endocytosis of permeases. (A) Schematic depiction of how inefficient internalization of amino acid permeases that reside in the PM results in increased sensitivity to the growth-inhibitory effect of a toxic amino acid analogue (small green circle). (B) Samples of exponentially growing cultures of otherwise isogenic strains of the indicated genotype were plated in fivefold serial dilutions on growth medium lacking Arg in the absence (–) or presence (+) of canavanine (5.7 µM final concentration). (C) Strains of the indicated genotype carrying an empty vector (EV; pRS315) or expressing from the same vector either Muk1-3xFLAG (pMLT83) or the indicated Muk1 variants Muk1(6A)-3xFLAG (pMLT84), Muk1(6E)-3xFLAG (pMLT92), or catalytically inactive Muk1(D353A)-3xFLAG (pMLT85), were grown and plated as in B. (D) Strains of the indicated genotype, as in C, were plated in fivefold serial dilutions on growth medium lacking Met in the absence (−) or presence (+) of ethionine (10 µM final concentration). (E) To confirm equivalent levels of expression, WT cells (BY4741) expressing the indicated Muk1-3xFLAG constructs, as in C and D, were grown to mid-exponential phase, harvested, and lysed, and proteins in the resulting extracts were treated with CIP, as in Materials and methods, resolved by SDS-PAGE on an 8% acrylamide gel, and analyzed by immunoblotting (IB).
This phenotype provided a means to assess the effect of Ypk1-mediated phosphorylation on Muk1 function. Indeed, we found that a muk1Δ vps9Δ double mutant conferred a greater degree of canavanine sensitivity than a vps9Δ mutation alone and that reexpression of Muk1 from its endogenous promoter on a CEN plasmid reproducibly restored readily detectable canavanine resistance to the muk1Δ vps9Δ cells (Fig. 2 C). Moreover, a site-directed Muk1 mutant (D353A) that substituted a residue in its Vps9 homology domain known to be essential for catalytic function (Bean et al., 2015) was unable to confer canavanine resistance to the muk1Δ vps9Δ cells, demonstrating that the GEF function of Muk1 is required (Fig. 2 C). Most tellingly, Muk1(6A), which cannot be phosphorylated on any of its Ypk1 sites, was also unable to confer canavanine resistance to muk1Δ vps9Δ cells, whereas the phosphomimetic mutant Muk1(6E), which simulates full Ypk1-dependent phosphorylation at these same sites, was at least as potent as WT Muk1 at restoring canavanine resistance to the muk1Δ vps9Δ cells (Fig. 2 C). We found exactly the same trends when we examined the sensitivity of the same set of strains to the growth-inhibitory action of ethionine, a Met analogue taken up by the Met permease (Mup1; Isnard et al., 1996; Guiney et al., 2016; Fig. 2 D). In addition, we found exactly the same trends monitoring rescue of an independent phenotype—namely, the poor growth of muk1Δ vps9Δ cells at an elevated temperature (37°C; Paulsel et al., 2013; Shideler et al., 2015; Fig. S1 C). Furthermore, we confirmed that the difference in behavior between Muk1(6A) and Muk1(6E) could not be attributed to any difference in their relative level of expression, compared with each other or WT Muk1 (Fig. 2 E). Taken together, these findings indicated that Ypk1-mediated phosphorylation of Muk1 is required for the function of this Rab5 GEF.
Rab5 GTPases are necessary for efficient TORC2-mediated Ypk1 activation
Based on the precedent that TORC1 function requires its interaction with both the Rheb (Avruch et al., 2009; Yang et al., 2017) and Rag (Sancak and Sabatini, 2009; Nicastro et al., 2017) classes of small GTPases, and on the fact that Ryh1, a Schizosaccharomyces pombe GTPase that most closely resembles human Rab6 and its S. cerevisiae orthologue (Ypt6), was reported to stimulate TORC2 in fission yeast (Tatebe et al., 2010), we examined whether Rab5 function influenced TORC2 activity by monitoring its phosphorylation and activation of Ypk1.
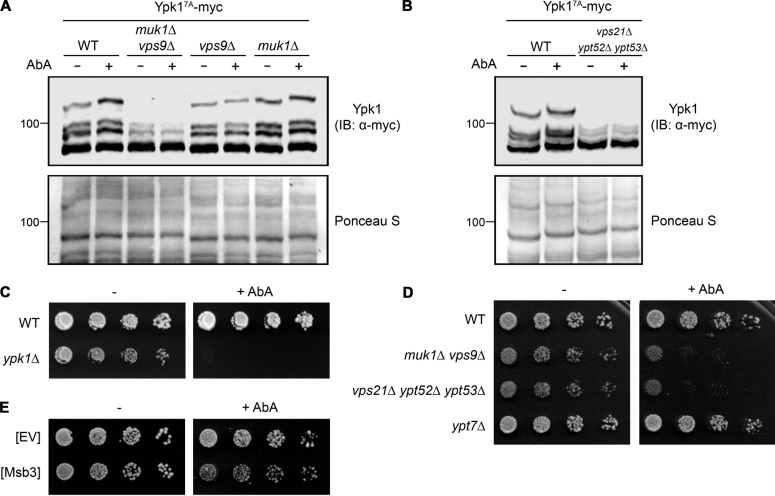
For that purpose, we used, first, a version of Ypk1 (Ypk17A) that contains only TORC2-dependent phosphorylation sites, which when examined by Phos-tag SDS-PAGE allows for convenient and reliable detection of the isoforms produced by its TORC2-mediated phosphorylation, as we have extensively documented elsewhere (Muir et al., 2015; Leskoske et al., 2017). Hence, we analyzed the phosphorylation pattern of the Ypk17A reporter in cells before and after treatment with AbA. TORC2-dependent phosphorylation of Ypk17A could be readily visualized in WT cells as a series of isoforms well resolved by Phos-tag SDS-PAGE, and inhibition of complex sphingolipid biosynthesis with AbA stimulated the appearance of the slowest-mobility (most highly phosphorylated) isoform (Fig. 3 A, left), in accord with our prior observations with Myr (Muir et al., 2015; Leskoske et al., 2017). Strikingly, however, in the absence of both Rab5 GEFs, basal and AbA-induced TORC2 phosphorylation of Ypk17A were both markedly reduced, and loss of Vps9 (the major Rab5 GEF) alone had a stronger effect than loss of Muk1 (Fig. 3 A). Likewise, compared with WT control cells, basal and AbA-induced TORC2 phosphorylation of Ypk17A was dramatically decreased in cells lacking all three Rab5 paralogs (Fig. 3 B). Thus, in cells that cannot generate active Rab5, TORC2-mediated phosphorylation of Ypk1 was very inefficient.
Figure 3.
Rab5 GTPases and their GTP loading are necessary for efficient phosphorylation and activation of Ypk1 by TORC2. (A) WT (BY4741) or otherwise isogenic yMLT9 (muk1Δ vps9Δ), yMLT3 (vps9Δ), or yMLT5 (muk1Δ) cells, all expressing Ypk17A-myc from its native promoter on a CEN plasmid (pFR252), were grown to mid-exponential phase and treated with ethanol or an equivalent volume of the same solvent containing AbA (1.8 µM final concentration). After incubation for 2 h, the cells were harvested, and extracts were prepared, resolved by Phos-tag SDS-PAGE, transferred to a nitrocellulose filter, stained with Ponceau S, and then analyzed by immunoblotting (IB), as in Materials and methods. (B) As in A, except WT (BY4741) and yMLT42 (vps21Δ ypt52Δ ypt53Δ) cells were compared. (C) Cultures of WT (BY4741) and JTY6142 (ypk1Δ) cells were plated in fivefold serial dilutions on growth medium in the absence (–) or presence (+) of AbA (40 nM final concentration). (D) As in C, for WT (BY4741), yMLT9 (muk1Δ vps9Δ), yMLT42 (vps21Δ ypt52Δ ypt53Δ), and yMLT42 (ypt7Δ) cells. (E) As in C, for WT (BY4741) cells transformed with either empty multicopy vector (EV; pRS426) or the same vector expressing from its native promoter Msb3-FLAG (pMLT111).
We and others have demonstrated previously that phosphorylation by TORC2-activated Ypk1 of multiple targets up-regulates flux through the sphingolipid biosynthetic pathway and is required for the cell to survive inhibition of ceramide synthesis caused by the presence of Myr (Roelants et al., 2011; Berchtold et al., 2012; Muir et al., 2014; Leskoske et al., 2017). Similarly, as expected, we also found that Ypk1 function is required for the cell to survive the challenge of the depletion of complex sphingolipids caused by AbA treatment (Fig. 3 C). Thus, the degree of resistance that cells display to agents like Myr and AbA provides a convenient phenotypic readout for the efficiency of Ypk1 activation by TORC2. By this criterion and in agreement with our biochemical analysis (Fig. 3 A and B), cells lacking both Rab5 GEFs and cells lacking all three Rab5 paralogs were much more sensitive to the growth-inhibitory action of AbA than otherwise isogenic WT control cells (Fig. 3 D), whereas cells devoid of Ypt7 (as an additional control for specificity) were not. Thus, cells that cannot generate active Rab5 exhibit physiological behavior consistent with inefficient TORC2-mediated phosphorylation of Ypk1. As a further test of this conclusion, we examined cells overexpressing Msb3, one of two apparent Rab5-specific GAPs encoded in the S. cerevisiae genome. Although Msb3 has a paralog (Msb4), only Msb3 had demonstrable Rab5 GAP activity in vivo and in vitro in two independent studies (Lachmann et al., 2012; Nickerson et al., 2012). We reasoned, therefore, that overexpressed Msb3 should lower the steady-state pool of activated (GTP-bound) Rab5 in the cell. Indeed, we found that such cells grew significantly less robustly on AbA than the control cells harboring the empty vector (Fig. 3 E). Collectively, these data indicate that Rab5 in its GTP-bound state is necessary to support TORC2 phosphorylation and activation of Ypk1.
Active Rab5 stimulates TORC2 phosphorylation of Ypk1
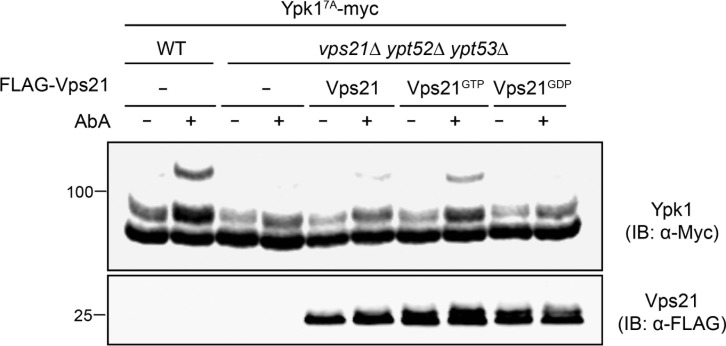
As a further test of the conclusion that activated Rab5 promotes TORC2 phosphorylation of Ypk1, we examined the effect of reexpressing in the Rab5-deficient vps21Δ ypt52Δ ypt53Δ triple mutant WT Vps21, Vps21(Q66L) (a GTP hydrolysis-defective mutant [so-called “GTP-locked” variant]), or Vps21(S21L) (a mutant that preferentially binds GDP [so-called “GDP-locked” variant]; Tall et al., 1999; Lo et al., 2011; Plemel et al., 2011). We focused on Vps21 because it is constitutively expressed and the most abundant yeast Rab5 isoform (Schmidt et al., 2017), has been shown to play the major role in endocytic vesicle trafficking (Horazdovsky et al., 1994), and when absent, causes more pronounced phenotypes compared with loss of the other two Rab5 GTPases (Singer-Krüger et al., 1994; Nickerson et al., 2012; Nakatsukasa et al., 2014). We found that, when present as the sole Rab5, WT Vps21 was able to rescue AbA-induced TORC2-phosphorylation of Ypk17A (Fig. 4). This rescue was reproducibly more efficacious in cells expressing GTP-locked Vps21 but undetectable in cells expressing GDP-locked Vps21 (Fig. 4), consistent with the need for a Rab5 GTPase to be in its GTP-bound state to promote TORC2 phosphorylation of Ypk1. The weaker rescue conferred by WT Vps21 in the vps21Δ ypt52Δ ypt53Δ mutant cells may be attributable to its reduced expression relative to GTP-locked Vps21. Alternatively, the high level of GAL promoter-driven expression could overwhelm the capacity of the Rab5 GEFs, thereby lowering the fraction of total WT Vps21 in the GTP-bound state. By contrast, once loaded, GTP-locked Vps21 persists in its active state. In any event, because GTP-locked Vps21 cannot cycle and is defective in endocytic trafficking (Tall et al., 1999; Lachmann et al., 2012), its ability to stimulate TORC2 phosphorylation of Ypk1 appears to be a direct effect of Vps21-GTP and not some secondary consequence of its roles in vesicle trafficking per se. Nonetheless, we explored whether the reduction in TORC2 phosphorylation of Ypk1 in cells deficient in GTP-Rab5 might be attributable to such indirect effects.
Figure 4.
GTP-bound Vps21 stimulates TORC2 phosphorylation of Ypk1. Equivalent samples of cultures of WT (BY4741) or yMLT42 (vps21Δ ypt52Δ ypt53) cells expressing Ypk17A-myc from its native promoter on a CEN plasmid (pFR252), and carrying either empty vector (−; pRS426) or coexpressing FLAG-Vps21 (pMLT101), FLAG-Vps21Q66L (pMLT103), or FLAG-Vps21S21L (pMLT102) from the GAL promoter on the same vector, were grown to mid-exponential phase in 2% raffinose-0.2% sucrose medium, shifted for 1 h to galactose medium (2% final concentration), then treated for 2 h with ethanol (–) or an equivalent volume of the same solvent containing AbA (1.8 µM final concentration; +), harvested, lysed, resolved by Phos-tag SDS-PAGE, and analyzed by immunoblotting (IB).
Lack of GTP-Rab5 does not cause mislocalization of either TORC2 or Ypk1
Deletion of the C-terminal PH domain in Avo1, an essential subunit of TORC2, reportedly rendered cells inviable, but they could be rescued by expression of an Avo1 construct in which a CaaX box was substituted for its PH domain (Berchtold and Walther, 2009), suggesting that association with the PM is crucial for TORC2 function. Given that Rab5 GTPases are important in endocytic vesicle internalization, and also have been implicated in recycling of endosomes back to the PM (Jovic et al., 2010; Feyder et al., 2015; MacDonald and Piper, 2016), it seemed possible that cells deficient in GTP-bound Rab5 might have decreased TORC2 activity simply owing to altered TORC2 localization. However, when we analyzed the subcellular distribution of TORC2, monitored by fluorescence microscopy in cells marked with Avo3-GFP, another essential TORC2 subunit very tightly bound to the catalytic subunit Tor2 (Wullschleger et al., 2005; Karuppasamy et al., 2017; see also Fig. S4 A), we saw no difference in PM localization of TORC2 between WT and muk1Δ vps9Δ mutant cells (Fig. S2 A). Thus, TORC2 does not appear to be mislocalized in the absence of active Rab5.
Alternatively, it seemed possible that the decrease in TORC2 phosphorylation of Ypk1 observed in cells deficient in GTP-bound Rab5 might be due to diminished accessibility of Ypk1 at the PM. For its proper folding and function, Ypk1 is obligatorily phosphorylated on T504 in its activation loop by Pkh1 (and Pkh2; Roelants et al., 2002, 2004), which are protein kinases very tightly bound to PM structures called eisosomes (Walther et al., 2007; Douglas and Konopka, 2014). However, we saw no difference in Ypk1 activation loop phosphorylation between WT and muk1Δ vps9Δ mutant cells (Fig. S2 B), suggesting that there is no impairment in Ypk1 availability at the PM. Similarly, substantial data support the conclusion that interaction with the so-called CRIM element in the Avo1 subunit of TORC2 is responsible for substrate presentation to the active site in the Tor2 catalytic subunit of TORC2 (Liao and Chen, 2012; Tatebe et al., 2017; Yao et al., 2017), and we saw no significant difference in Ypk1 association with Avo1 between WT and muk1Δ vps9Δ mutant cells in GST pull-down experiments (Fig. S2 C). Thus, lack of efficient phosphorylation by TORC2 in cells deficient in GTP-Rab5 cannot be explained by the inability of Ypk1 to encounter TORC2 at the PM. As a further test, we expressed PHSlm1-Ypk1-3xHA, which others have demonstrated is efficiently targeted to the PM via its PtdIns4,5P2-binding PH domain and serves as an effective substrate of TORC2 (Niles et al., 2012). We found, however, that TORC2-dependent phosphorylation of the C-terminal regulatory tail in PHSlm1-Ypk1-3xHA was still poor in cells deficient in GTP-Rab5 (Fig. S2 D). Therefore, we explored whether there might be a more direct, physical interaction between Vps21 and TORC2, or between Vps21 and Ypk1.
Vps21 associates with Tor2 in TORC2
As one means to test whether Vps21 and TORC2 are able to physically associate, we examined whether they were able to coimmunoprecipitate. For this purpose, we first expressed either FLAG-Vps21 or FLAG-mNeonGreen (mNG; Shaner et al., 2013) in cells coexpressing a 3xHA epitope-tagged version of the Tor2 catalytic subunit of TORC2, either from a CEN plasmid in tor1Δ tor2Δ cells (Fig. S3 A) or integrated at the TOR2 locus on chromosome XI (Fig. S3 B). We chose mNG as an initial control because mNG (237 residues) is quite comparable in size to Vps21 (210 residues), as well as quite similar in overall surface charge (pI = 5.7 for mNG versus pI = 5.0 for Vps21]. In multiple trials, when extracts of the cells were immunoprecipitated with anti-FLAG antibodies, we reproducibly found readily detectable coimmunoprecipitation of 3xHA-tagged Tor2 from the cells expressing FLAG-Vps21, but not from the cells expressing FLAG-mNG (Fig. S3, A and B). We conducted similar immunoprecipitation experiments using cells expressing either FLAG-Vps21 or FLAG-mNG and coexpressing Ypk1-myc, because phosphorylation and activation of certain other AGC family protein kinases requires association of their regulatory domains with other classes of small GTPases (Pearce et al., 2010; Leroux et al., 2018). However, only an equivalent level of nonspecific background interaction was observed for both FLAG-Vps21 and FLAG-mNG with Ypk1-myc (Fig. S3 C). Thus, these initial experiments indicated that Vps21 associates with TORC2, not Ypk1.
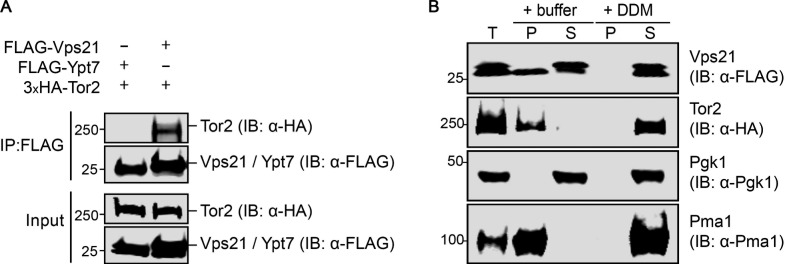
As a critical control for specificity, we repeated these same experiments using both FLAG-Vps21 and FLAG-Ypt7, and found again that only Vps21 exhibited a readily detectable association with Tor2 (Fig. 5 A), indicating that the observed interaction does not occur with an unrelated small GTPase. Because both Vps21 and Ypt7 are membrane-anchored proteins, this specificity made it highly unlikely that Vps21–TORC2 interaction resulted from the fact that both Vps21 and TORC2 are membrane associated. To rule out this possibility, we directly documented that the level of detergent present in the cell lysis and immunoprecipitation buffers, as described in Materials and methods, was sufficient to fully solubilize both proteins (Fig. 5 B). Finally, we found that both WT and GTP-locked Vps21 were efficacious in pulling down Tor2, but we also observed detectable association with GDP-locked Vps21 (Fig. S3 D), similar to prior reports showing that both GTP-locked Rheb and GDP-locked Rheb bound to mTORC1 (Long et al., 2005). Collectively, these results demonstrate that Vps21 makes specific contact with TORC2.
Figure 5.
Vps21 physically interacts with TORC2. (A) Strain yMLT78 expressing 3xHA-Tor2 from its endogenous locus was transformed with plasmids expressing either FLAG-Ypt7 (pMLT110) or FLAG-Vps21 (pMLT101). The resulting transformants were grown to mid-exponential phase, and expression of the FLAG-tagged proteins was induced with 2% galactose for 4 h. The induced cells were harvested and lysed, and FLAG-tagged proteins were immuno-isolated from the extracts using resin coated with anti-FLAG antibodies as described in Materials and methods. Samples of the bound proteins were resolved by SDS-PAGE on a 4–20% gradient gel analyzed by immunoblotting (IB) with anti-HA and anti-FLAG antibodies. (B) Strain yMLT78 expressing 3xHA-Tor2 from its endogenous locus was transformed with a plasmid expressing FLAG-Vps21 (pMLT101). The resulting transformants were cultured and harvested as in A. Cells were lysed without detergent. Samples were split and treated with buffer containing 0.67% DDM or no detergent to solubilize proteins for 30 min before clarifying lysates by centrifugation, as described in Materials and methods. IP, immunoprecipitation; T, total lysate; P, pellet fraction; S, supernatant fraction. Protein extracts were resolved and analyzed as in A, with the indicated antibodies.
Rab5 GTPase stimulates TORC2 activity
Given the interaction between Vps21 and Tor2 that we detected by coimmunoprecipitation, we explored the possibility that this association, whether stable or transient, is able to stimulate the catalytic function of TORC2, analogous to the stimulation of mTORC1 activity by Rheb in mammalian cells (Avruch et al., 2009; Yang et al., 2017). In vegetatively growing and unstressed cells, Ypt53 expression is repressed (Schmidt et al., 2017). Therefore, to obtain TORC2 from cells depleted of Rab5 GTPases, we used immuno-isolation with anti-FLAG antibodies from extracts of vps21Δ ypt52Δ mutant cells expressing Avo3-3xFLAG, which we were able to demonstrate also successfully enriched for the Tor2 catalytic subunit of TORC2, which was marked with a 3xHA tag (Fig. S4 A). We used the identical approach to prepare TORC2 from otherwise isogenic WT cells (Fig. S4 A). Regardless of the strain background, our procedure using FLAG-tagged Avo3 for immuno-isolation recovered TORC2 complexes that contain equivalent amounts of HA-tagged Tor2. As the substrate, we used a purified recombinant protein in which the 78 C-terminal residues of Ypk1 (680 residues), which contain all of its TORC2 phosphorylation sites (Leskoske et al., 2017), was fused to the C terminus of maltose-binding protein (MBP; 370 residues; Duplay et al., 1984), hereafter referred to as MBP-Ypk1CT.
We first incubated TORC2 from WT cells with MBP-Ypk1CT and [γ-32P]ATP and found robust incorporation, as expected. We found that this incorporation was almost completely eliminated when we used the mutant substrate MBP-Ypk1CT(6A), in which all six of the TORC2-specific phospho-acceptor residues were mutated to Ala, confirming that the observed phosphorylation was occurring at the previously defined TORC2-specific sites in Ypk1 (Fig. S4 B). TORC2 itself, and not some other protein kinase present in the preparation, was responsible for the observed phosphorylation because incubation with a compound (QL-IX-55) specifically designed and demonstrated by others to selectively inhibit yeast TORC2 (Liu et al., 2012) markedly reduced the incorporation into MBP-Ypk1CT (Fig. S4 C). Likewise, when we prepared TORC2 in the same way from cells expressing a Tor2 allele (Tor2(L2178A)) designed to be specifically sensitive to an orthogonal inhibitor (NVP-BEZ235; Kliegman et al., 2013), incorporation into MBP-Ypk1CT was almost completely eliminated (Fig. S4 D). These controls established that the observed phosphorylation of MBP-Ypk1CT was occurring in a strictly TORC2-dependent manner. We further established that, under our assay conditions, incorporation was roughly linear with time of incubation (Fig. S4, E and F).
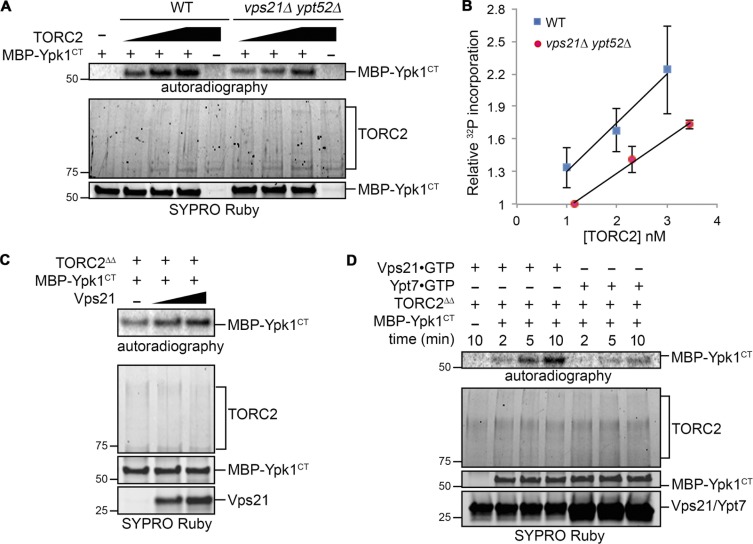
Having thoroughly validated the specificity of this in vitro reaction, we then compared TORC2 isolated from WT cells to TORC2 isolated from the vps21Δ ypt52Δ cells and found, in multiple trials, that the specific activity of the WT TORC2 was reproducibly higher (Fig. 6, A and B). Moreover, addition of GTP-loaded Vps21 to the TORC2 isolated from vps21Δ ypt52Δ mutant cells reproducibly stimulated incorporation into MBP-Ypk1CT (Fig. 6 C), whereas under the exact same assay conditions, GTP-loaded Ypt7 did not (Fig. 6 D). The most parsimonious model to explain these in vitro results is that GTP-bound Rab5 associates with Tor2 and causes a conformational change in TORC2 that allows it to access the C-terminal end of Ypk1 more efficiently, fully consistent with all of the in vivo evidence presented in this study that active (GTP-bound) Rab5 GTPases promote TORC2 activity toward its substrate Ypk1 (Fig. 7).
Figure 6.
Vps21 stimulates TORC2 activity on Ypk1. (A) Equivalent amounts of TORC2 immunopurified from WT (BY4741) or yMLT85 (vps21Δ ypt52Δ) strains (see Fig. S4 A) were added in increasing amounts and incubated with [γ-32P]ATP and MBP-Ypk1CT. The reaction products were resolved by SDS-PAGE and analyzed by autoradiography and staining with SYPRO Ruby, as described in Materials and methods. Results of one representative experiment are shown. (B) The results of multiple experiments (n = 3) as in A are plotted as the mean 32P incorporation into MBP-Ypk1CT versus the amount of TORC2 complex present. Error bars, SEM. 32P incorporation was measured by quantifying the corresponding autoradiograms using ImageJ and estimating the amount of TORC2 present from the staining intensity of the SYPRO Ruby-stained gel and accompanying standards (not depicted). The values were then normalized to the mean 32P incorporation into MBP-Ypk1CT catalyzed by the lowest amount of TORC2 from the vps21Δ ypt52Δ cells, which was set at [1]. (C) TORC2 immunopurified from yMLT85 (vps21Δ ypt52), denoted TORC2ΔΔ, was incubated with [γ-32P]ATP and MBP-Ypk1CT in the absence or presence of Vps21-GTPγS (0.2 µg and 0.4 µg). (D) Equivalent amounts of TORC2 immunopurified from yMLT85 (vps21Δ ypt52Δ) as in A, denoted TORC2ΔΔ, were incubated with [γ-32P]ATP, MBP-Ypk1CT, and either Vps21-GTPγS (2 µg) or Ypt7-GTPγS (4 µg), as indicated. Reactions were terminated at the indicated time points.
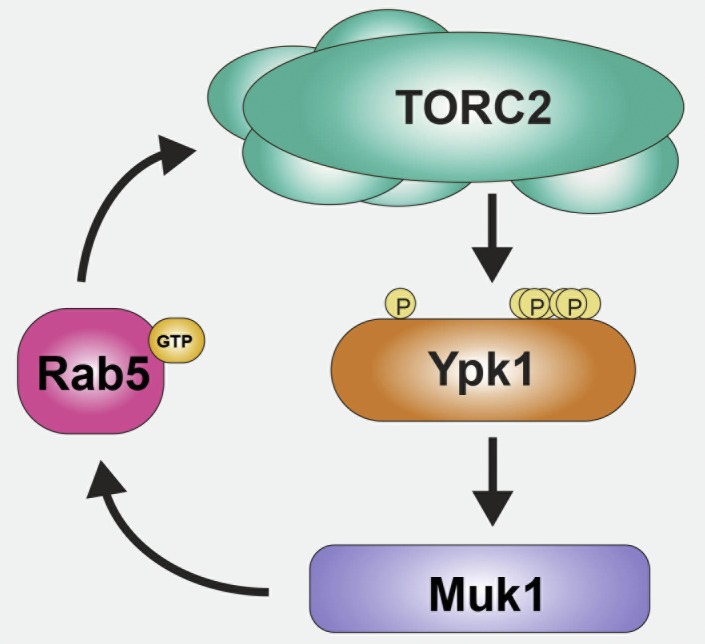
Figure 7.
Muk1-dependent Rab5-mediated mechanism for sustained activation of TORC2-Ypk1 signaling. Basal Ypk1 (orange) activity requires phosphorylation of T504 in its activation loop (indicated by the left-most P) and TORC2-mediated phosphorylation at S644 near its C terminus. However, under many stress conditions, TORC2 (aqua) up-regulates Ypk1 by phosphorylating T662 and additional multiple sites at its C-terminal end (indicated by the cluster of Ps to the right). As documented in this study, activated Ypk1 phosphorylates the Rab5-specific GEF Muk1 (purple), which in turn is activated by its Ypk1-mediated phosphorylation. Activated Muk1 will generate more GTP-bound Rab5 (pink), and as we also have demonstrated here, activated Rab5 promotes the ability of TORC2 to phosphorylate Ypk1. Based on the observed coimmunoprecipitation of the Rab5 Vps21 with the Tor2 catalytic subunit of TORC2, the stimulation of TORC by Rab5 may be direct.
Discussion
This study was initiated to determine whether we could extend our understanding of TORC2-Ypk1 signaling by establishing whether a candidate substrate for Ypk1 that emerged in a global screen (Muir et al., 2014) was authentic and physiologically relevant. As we have documented here, the Rab5-specific GEF Muk1 is indeed a target of Ypk1. Even though Muk1 is clearly phosphorylated by other protein kinases, which may influence other aspects of Muk1 function, our genetic evidence demonstrates that Ypk1-mediated phosphorylation is required for the GEF activity of Muk1 in vivo. S. cerevisiae contains just two functional Rab5-specific GEFs that possess the Vps9 homology domain, Vps9 itself and Muk1 (Paulsel et al., 2013; Bean et al., 2015). The human genome encodes nine proteins with readily discernible Vps9 homology domains (Carney et al., 2006; Barr and Lambright, 2010); however, none of them is the obvious orthologue of either Vps9 or Muk1. It has been reported that SGK1, the mammalian homologue of Ypk1 (Casamayor et al., 1999), facilitates PM recycling of KCNQ1 channels in a Rab5-dependent manner, but whether that behavior was dependent on SGK1-mediated phosphorylation of any Rab5 GEF was not explored (Seebohm et al., 2008). Likewise, it has been reported that SGK1 function promotes association of a Rho- and Rac-specific GEF (ARHGEF2/GEF-H1) with a component (Sec5/EXOC2) of the mammalian exocyst complex, but again whether that behavior was dependent on SGK1-mediated phosphorylation of the GEF was not determined (Wang et al., 2015).
Given TORC2-Ypk1–mediated activation of a Rab5-specific GEF, we then explored what the role of that pathway might be. It is now well established that TORC1 localization and activity are influenced by small GTPases of the Rag and Rheb classes, respectively. The Rag GTPases tether TORC1 at the surface of the lysosome/vacuole through interaction with the RAPTOR (yeast counterpart Kog1/Las24) subunit (Sancak and Sabatini, 2009; Nicastro et al., 2017). By contrast, recent structural work (Yang et al., 2017) has shown that TORC2 activation by Rheb (Avruch et al., 2009) is mediated by its direct interaction with a pocket constituted by the M- and N-HEAT repeats and FAT domain of mTOR. Whether small GTPases modulate TORC2 in any similar fashion has been more elusive. It has been reported that mammalian Rab35 affects TORC2 signaling to the AGC kinase AKT, but only indirectly through the effect of Rab35 on intracellular trafficking of PtdIns 3-kinase (Wheeler et al., 2015). The closest S. cerevisiae orthologue of mammalian Rab35 is Ypt1, which clearly functions in the ER-to-Golgi step of the yeast secretory pathway (Novick, 2016), not at the PM. Evidence for a more direct involvement of a small GTPase in TORC2 regulation was provided for fission yeast Ryh1 (Tatebe et al., 2010). However, the closest S. cerevisiae orthologue of Ryh1 is Ypt6 (mammalian counterpart Rab6), which functions in cis- to trans-Golgi vesicle trafficking (Suda et al., 2013), not at the PM. We were quite surprised, therefore, when we observed that deficiencies of active GTP-bound Rab5—due to absence of the Vps9 and Muk1 GEFs, to absence of all three Rab5 isoforms (Vps21, Ypt52, and Ypt53), or to elevation of the Rab5-specific GAP Msb3—all lowered both basal and stress-induced TORC2-mediated phosphorylation of Ypk1, as assessed biochemically, genetically, or both. Thus, in S. cerevisiae, Rab5 GTPases are an important positive effector of TORC2 function.
Yeast TORC2 components are located at the cell cortex in close apposition to the PM but are highly dynamic (Berchtold and Walther, 2009; Leskoske et al., 2018). Muk1 and its target Vps21 have both been localized to endosomes (Cabrera and Ungermann, 2013; Paulsel et al., 2013; Bean et al., 2015; Shideler et al., 2015). Our findings that Muk1 is a substrate of the TORC2-Ypk1 kinase cascade and that Vps21 is required for full TORC2 activity raise the possibilities that the observed dynamic behavior of yeast TORC2 reflects, first, that a fraction of the population of active TORC2 is on endosomes and, second, that a fraction of the GTP-bound Vps21 is at the PM. Similarly, by monitoring a fluorescent mSIN1 reporter (S. cerevisiae counterpart Avo1), it was observed in mammalian cells that active mTORC2 could be found at both the PM and on endosomes (Ebner et al., 2017).
On the other hand, despite the fact that Rab5 function is involved in endosome internalization, trafficking, and recycling, we could find no impairment in TORC2 localization in cells deficient in active Rab5. So, unlike Rag GTPases, Rab5 does not seem to have a role in tethering TORC2 to the PM per se. Rather, our data, especially the ability of Vps21 to coimmunoprecipitate with Tor2, even in the presence of detergent, and the ability of GTP-loaded Vps21 to stimulate TORC2-mediated phosphorylation of Ypk1 in vitro, indicate a more direct role for GTP-bound Rab5 in TORC2 function, as suggested for Ryh1 in fission yeast (Tatebe et al., 2010). Even though S. pombe Ryh1 has greater overall relatedness to S. cerevisiae Ypt6 (63% identity; 76% similarity) than to S. cerevisiae Vps21 (37% identity; 61% similarity), it is the case that mammalian Rab5B, Vps21, and Ryh1 all share with each other greater resemblance in their Switch I, Switch II, and hypervariable loop regions than they do with other classes of Ras superfamily GTPases (Fig. S5). In the case of Rheb, it is residues mainly in its Switch I and Switch II regions that make the main contacts with the M- and N-heat repeat regions and FAT domain in mTOR (Yang et al., 2017). Thus, it is possible that Rab5 binds to the Tor2 subunit of yeast TORC2 in the same binding pocket as that occupied by Rheb in the mTOR subunit in mTORC1. In both yeast and mammals, aside from Lst8, the ancillary subunits in TORC2 are all completely different from those in TORC1 (Eltschinger and Loewith, 2016; Tatebe and Shiozaki, 2017), and interaction of these other components with the catalytic subunit likely modifies the geometry of the binding pocket, so that it accommodates the right GTPase effector and excludes spurious ones. Furthermore, yeast and other fungi encode two highly related, but not identical, TOR isoforms; Tor2 is the primary catalytic subunit in TORC2, whereas Tor1 is the primary catalytic subunit in TORC1 (Helliwell et al., 1994; Roelants et al., 2017a). Hence, subtle sequence differences between Tor2 and Tor1 in their M- and N-heat repeat regions and FAT domain (Hill et al., 2018) may further fine tune the pocket in Tor2 to be selective for Rab5.
Regardless of the detailed molecular mechanism by which Rab5 stimulates the function of TORC2 (Fig. 7), Muk1 is activated in a TORC2-dependent and Ypk1-mediated manner, presumably generating more GTP-bound Rab5. Given that active Rab5 promotes TORC2 function, this control circuit provides self-reinforcing positive feedback that would lead to sustained TORC2-Ypk1 signaling, once it has been initiated. In this same regard, using the split-YFP (Venus) system to tag Vps21 and Msb3 for bimolecular fluorescence complementation, which traps protein–protein complexes (Sung and Huh, 2007; Kerppola, 2009), a substantial fraction of this Rab5-GAP complex was found at the PM, not on endosomes (Lachmann et al., 2012), indicating that Msb3 may be responsible for deactivation of the PM-associated pool of GTP-bound Vps21, thus providing a mechanism for down-regulation of Rab5-mediated stimulation of TORC2.
Materials and methods
Strains and plasmids
S. cerevisiae strains (Table 1) were constructed using standard genetic methods (Amberg et al., 2005). For integrations into the genome, proper insertion was confirmed by PCR amplification and sequencing. Plasmids (Table 2) were constructed using standard molecular biology techniques (Green and Sambrook, 2012) and verified by sequencing.
Table 1. S. cerevisiae strains used in this study.
| Strain | Genotype | Source/reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| yAM123A | BY4741 Ypk1(L424A)::URA3 ypk2Δ::KanMX4 | Muir et al., 2014 |
| JTY6142 | BY4741 ypk1Δ::KanMX4 | Research Genetics |
| yMLT42 | BY4741 vps21Δ::NatMX ypt52Δ::KanMX ypt53Δ::HphMX | This study |
| yMLT60 | BY4741 ypt7Δ::KanMX | This study |
| yMLT9 | BY4741 vps9Δ::HphMX muk1Δ::NatMX | This study |
| yMLT3 | BY4741 vps9Δ::HphMX | This study |
| yMLT5 | BY4741 muk1Δ::NatMX | This study |
| yMLT78 | BY4741 3xHA-TOR2 | This study |
| yMLT86 | BY4741 mup1Δ::HisMX MET15 | This study |
| yMLT87 | BY4741 MET15 | This study |
| yAEA343 | BY4741 vps9Δ::HphMX muk1Δ::NatMX MET15 | This study |
| yAEA344 | BY4741 vps9Δ::HphMX MET15 | This study |
| yMLT64 | BY4741 AVO3-GFP::KAN | This study |
| yMLT66 | BY4741 AVO3-GFP::KAN vps9Δ::HphMX muk1Δ::NatMX | This study |
| y2470 | BY4741 tor1::HIS3 tor2::LEU2 [pRS314(CEN4 TRP1)-3xHA-TOR2] | Jiang and Broach, 1999 |
| yMLT85 | BY4741 AVO3-3xFLAG::KanMX 3xHA-TOR2 vps21Δ::HphMX ypt52Δ::NatMX | This study |
| yMLT69 | BY4741 AVO1-6xHA::HIS3MX6 | This study |
| yKL7 | BY4741 AVO3-3xFLAG::KanMX TOR2(L2178A)::Hyg | K. Leskoske, this laboratory |
| yMLT70 | BY4741 AVO1-6xHA::HIS3MX6 vps9Δ::HphMX muk1Δ::NatMX | This study |
Table 2. Plasmids used in this study.
| Plasmid | Description | Source/reference |
|---|---|---|
| BG1805 | 2 µm, URA3, GAL1prom, C-terminal tandem affinity (TAP) tag vector | Open Biosystems |
| pMAL-c5X | Bacterial expression vector for production of MBP (MalE) fusion proteins | New England BioLabs |
| pESC-Leu | 2 µm, GAL1/10prom, LEU2 | Agilent |
| YCpLG | CEN, LEU2, GAL1/10prom vector | Bardwell et al., 1998 |
| pRS425 | 2 µm, LEU2 vector | Sikorski and Hieter, 1989 |
| pRS426 | 2 µm, URA3 vector | Sikorski and Hieter, 1989 |
| pRS315 | CEN, LEU2 vector | Sikorski and Hieter, 1989 |
| pET30a | Bacterial expression vector | Novagen |
| pMLT35 | pMAL-c5X MBP-MUK1-(His)6 | This study |
| pMLT74 | pMAL-c5X MBP-muk1(S171A S172A S173A S183A S184A S185A)-(His)6 | This study |
| pMLT22 | pESC-LEU GAL1prom-MUK1-Myc | This study |
| pMLT56 | pESC-LEU GAL1prom-muk1(1-220)-Myc | This study |
| pMLT57 | pESC-LEU GAL1prom-muk1(1-220; S171A S172A S173A S183A S184A S185A)-Myc | This study |
| pAX50 | BG1805 YPK1(L424A)-TAP | Muir et al., 2014 |
| pFR252 | pRS315 ypk1(S51A T57A S71A T504A S644A S653A T662A)-Myc | Muir et al., 2015 |
| pAM20 | pRS315 Ypk1-myc | Roelants et al., 2011 |
| pMLT101 | pRS426 GAL1prom-FLAG-VPS21 | This study |
| pMLT102 | pRS426 GAL1prom-FLAG-vps21(S21L) | This study |
| pMLT103 | pRS426 GAL1prom-FLAG-vps21(Q66L) | This study |
| pMLT110 | pRS426 GAL1prom-FLAG-YPT7 | This study |
| pMLT114 | pRS426 GAL1prom-FLAG-mNG | This study |
| pMLT111 | pRS426 MSB3-FLAG | This study |
| pMLT83 | pRS315 MUK1-3xFLAG | This study |
| pMLT84 | pRS315 muk1(S171A S172A S173A S183A S184A S185A)-3xFLAG | This study |
| pMLT85 | pRS315 muk1(D353A)-3xFLAG | This study |
| pMLT92 | pRS315 MUK1(S171E S172E S173E S183E S184E S185E)-3xFLAG | This study |
| pMLT132 | pET30a ypk1(D242 D470A)-(His)6 | This study |
| pMLT135 | pMAL-c5X MBP-Ypk1(603-680) [MBP-Ypk1CT] | This study |
| pMLT138 | pMAL-c5X MBP-Ypk1(603-680; S644 S653A T662A S671A S672A S678) [MBP-Ypk1CT (6A)] | This study |
| pMLT88 | pET30a VPS21-(His)6 | This study |
| pMLT141 | pET30a YPT7-(His)6 | This study |
| pPL495 | pRS315 PHSlm1-YPK1-3xHA | Niles et al., 2012 |
| pMLT115 | pRS425 GAL1prom-GST | This study |
| pML116 | pRS425 GAL1prom-GST-Ypk1 | This study |
Cell culture conditions
Yeast strains were propagated in either standard rich medium (YP) containing 2% dextrose/glucose or a defined synthetic medium (SC; Sherman, 2002) with the same carbon source and supplemented with appropriate nutrients to maintain selection for plasmids. For galactose induction of gene expression, cells were pregrown in SC medium containing 2% raffinose-0.2% sucrose and induced by addition of galactose (2% final concentration) for 3 h. To inhibit complex sphingolipid synthesis, a stock of AbA (630466; Takara Bio USA) dissolved in ethanol was added at the indicated final concentrations, and an equivalent volume of the same solvent was added to control cultures. Stocks of the amino acid analogues l-canavanine (C1625; Sigma-Aldrich) and l-ethionine (E1260; Sigma-Aldrich) dissolved in water were added to plates of SD-containing agar medium at the indicated concentrations. For inhibition of analogue-sensitive Ypk1(L424A), a stock of the adenine analogue 3MB-PP1 (17860; Cayman Chemical) dissolved in DMSO was added at the indicated final concentration, and an equivalent volume of the same solvent was added to control cultures.
Protein purification
Expression from the corresponding plasmids (Table 2) of MBP-Muk1-(His)6, MBP-Muk1(6A)-(His)6, Vps21-(His)6, and Ypt7-(His)6 in the Escherichia coli derivative LOBSTR-BL21(DE3) (Andersen et al., 2013) was induced by addition of IPTG (0.75 mM final concentration). After incubation of the cultures at 16°C for 16 h, the cells were harvested by centrifugation and lysed by sonication, and the corresponding proteins were purified by immobilized metal ion affinity chromatography using Ni2+-charged, nitrilotriacetate-derivatized agarose beads (Qiagen) following standard procedures. After elution with 200 mM imidazole, pooled eluates were dialyzed into PBS containing 2 mM DTT. For Vps21-(His)6, the dialyzed protein was further purified by ion exchange chromatography on a MonoQ 10/100GL column (Sigma-Aldrich) using an AKTA Fast Protein Liquid Chromatography system (GE Healthcare). To load purified Vps21-(His)6 or purified Ypt7-(His)6 with the desired guanine nucleotide, samples were incubated for 10 min in 5 mM EDTA to strip any bound Mg2+-nucleotide complexes, then incubated for 30 min with 2 mM GTPγS (Roche Diagnostics), followed by addition of MgCl2 (20 mM final concentration) and incubation for an additional 30 min. MBP-Ypk1(603–680) [denoted MBP-Ypk1CT] and MBP-Ypk1CT(6A) were purified by affinity chromatography on amylose-agarose (New England Biolabs), following the manufacturer’s protocol. Analogue-sensitive Ypk1-as [Ypk1(L424A)-TAP] was purified from yeast by affinity purification using IgG-agarose and elution by cleavage with Pre-Scission protease (GE Healthcare), as described in detail previously (Muir et al., 2014).
Ypk1 protein kinase assays
To monitor the protein kinase activity of Ypk1, samples of purified Ypk1-as (0.15 µg) were mixed with, typically, 1 µg of the purified substrate of interest in kinase assay buffer (125 mM potassium acetate, 12 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 1% glycerol, 0.2 mg/ml BSA, 12.5 mM β-glycerol phosphate, 1 mM NaVO4, and 50 mM Hepes, pH 7.4, containing cOmplete EDTA-free protease inhibitor cocktail [1 mini-tablet per 10 ml; Roche]) in the absence or presence of 3MB-PP1 (10 µM final concentration), and reaction was initiated by addition of ATP (10 µM final concentration) containing 2 µCi [γ-32P]ATP. After incubation at 30°C for 30 min, reactions were terminated by the addition of an equal volume of 6× SDS gel sample buffer followed by boiling for 5 min, and the resulting products were resolved by SDS-PAGE and analyzed by both protein staining (0.05% Coomassie Blue R-250, 10% acetic acid, and 25% isopropanol) and autoradiography of the same fixed and stained gel performed using a Phosphorimager screen (Molecular Dynamics) and a Typhoon imaging system (GE Healthcare).
Immuno-isolation of TORC2 and other FLAG-tagged proteins
For immunoprecipitation of FLAG-tagged proteins expressed in yeast, including TORC2 tagged with Avo3-3xFLAG, frozen cell pellets were resuspended in an equal volume of ice-cold lysis buffer (200 mM KCl, 10 mM magnesium acetate, 10 mM β-glycerol-phosphate, 10 mM NaF, 10% glycerol, 0.1 mM EDTA, and 50 mM Hepes, pH 7.4, containing 2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride and cOmplete EDTA-free protease inhibitor cocktail [1 mini-tablet per 10 ml; Roche]), mixed with an equivalent volume of chilled glass beads, and lysed by vigorous vortexing for 10 min at 4°C. The resulting lysate was separated from the beads by draining after puncturing the conical tip of the plastic centrifuge tube with a 25-gauge hypodermic needle. The recovered liquid was diluted with 0.5 volume of lysis buffer containing 2% n-dodecyl β-d-maltoside (DDM; D4641; Sigma-Aldrich) and incubated with gentle agitation on a rollerdrum for 20 min at 4°C. The detergent-treated extract was clarified by centrifugation at maximum rpm in a microfuge (Model 5424; Eppendorf), and the resulting supernatant fraction was withdrawn and mixed with 15 µl of a slurry of magnetic affinity resin decorated with anti-FLAG M2 antibodies (M8823; Sigma-Aldrich) that had been preequilibrated in lysis buffer containing 0.5% DDM. After incubation with resin for 2 h with gentle agitation on a rollerdrum at 4°C, the resin was removed from the bulk solution and moved to the side of the tube using a magnet tube rack (Dynal; Thermo Fisher Scientific) and then resuspended in and washed six times with 1 ml lysis buffer containing 0.5% DDM. For TORC2 purification, the remaining bound protein was eluted with 3xFLAG peptide (Sigma-Aldrich) in 1× kinase assay buffer, and samples of the resulting eluate were used immediately thereafter in in vitro kinase assays. For other FLAG-tagged proteins, the remaining bound protein was eluted with urea-SDS gel sample buffer (6 M urea, 6% SDS, 25% glycerol, 0.1% bromophenol blue, and 150 mM Tris-HCl, pH 6.8).
TORC2 protein kinase assays
To monitor the protein kinase activity of TORC2, samples of TORC2 (each derived by immuno-isolation from the equivalent of 50 µg clarified lysate, as described above) were incubated with purified MBP-Ypk1CT or MBP-Ypk1CT(6A), in kinase assay buffer in the absence or presence of the indicated amounts of GTPγS-loaded Vps21 or Ypt7, and reactions were initiated by the addition of ATP (10 µM final concentration) containing 2 µCi [γ-32P]ATP. Reactions were incubated at 30°C for 30 min unless otherwise noted, and the products were analyzed in the same manner as for the Ypk1 protein kinase assays, except SYPRO Ruby (Thermo Fisher Scientific) was used for protein staining. For specific inhibition of WT TORC2, the enzyme was incubated with QL-IX-55 (gift of Nathanael Gray, Harvard Medical School, Cambridge, MA) dissolved in DMSO or with an equivalent volume of the same solvent at 30°C for 5 min before addition of the other reaction components. For specific inhibition of the TORC2 containing Tor2(L2178A), the enzyme was incubated with NVP-BEZ235 (Cayman Chemical) dissolved in dimethyl formamide or with an equivalent volume of the same solvent.
Preparation of cell extracts and immunoblotting
Whole-cell lysates of yeast were prepared by alkaline lysis with 1.85 M NaOH and 7.4% 2-mercaptoethanol followed by TCA precipitation, as described in detail previously (Westfall et al., 2008). For samples treated with CIP, the TCA precipitates of whole-cell lysates were washed with ice-cold acetone, resuspended in 100 µl CIP resuspension buffer (126 mM sorbitol, 42 mM NaCl, 10.5 mM MgCl2, 0.24 mM EDTA, 2% 2-mercaptoethanol, and 180 mM Tris base), diluted with 900 µl of 50 mM Tris-HCl, pH 8.0, and incubated with 45 units of CIP (New England Biolabs) for 2 h at 30°C. Reactions were terminated by addition of TCA (25% final concentration), and the precipitated protein was resuspended in a buffer containing 0.1 M Tris base and 5% SDS before addition of an appropriate volume of 5× SDS sample buffer.
In most cases, protein samples were separated by standard SDS-PAGE in gels containing the appropriate concentration of acrylamide to resolve the species of interest, transferred electrophoretically to nitrocellulose filter paper, and analyzed by immunoblotting. To resolve the spectrum of phospho-isoforms of particular phosphorylated proteins (such as Muk1 and Ypk1), resuspended whole-cell lysates were separated using Phos-tag SDS-PAGE in 8% acrylamide gels containing 35 µM Phos-Tag reagent (Fujifilm Wako Chemicals USA), according to the manufacturer’s instructions, and, after transfer to nitrocellulose, analyzed by immunoblotting.
Primary antibodies used in this study (at the dilution indicated) were mouse anti-FLAG (1:5,000, F1804; Sigma-Aldrich); rabbit anti-DYKDDDDK (1:2,500, 2368; Cell Signaling Technologies); mouse anti-c-myc mAb 9E10 (1:100; Monoclonal Antibody Facility, Cancer Research Laboratory, University of California, Berkeley, Berkeley, CA); mouse anti-HA 6E2 (1:5,000, 2367; Cell Signaling Technologies); rabbit anti-GST (1:10,000, SC-459; Santa Cruz Biotechnology); rabbit anti-Pgk1 (1:200,000; Baum et al., 1978); rabbit anti-SGK1(pT256) (1:1,000, SC-16744; Santa Cruz Biotechnology); mouse anti-Pma1 (1:5,000, ab4645; Abcam); and rabbit anti-Ypk1(pT662) (1:20,000; gift from Ted Powers, University of California, Davis, Davis, CA). Infrared dye–labeled secondary antibodies were CF770-derivatized goat anti–mouse IgG (1:20,000, 20077; Biotium) and CF680-derivatized goat anti–rabbit IgG (1:20,000, 20067; Biotium). The resulting filter-bound immune complexes were visualized using an infrared imaging system (Model Odyssey CLx; LI-COR Biosciences).
GST pull-down analysis
Lysates were prepared in the same manner as for immunoprecipitation of FLAG-tagged proteins (above). The resulting extracts were mixed with a slurry (10 µl) of glutathione-Sepharose 4B beads (GE Healthcare) preequilibrated in lysis buffer containing 0.5% DDM. After incubation for 2 h at 4°C, the beads were separated from the protein solution and washed 6× with 1 ml lysis buffer, and the bound proteins were eluted with urea-SDS gel sample buffer and analyzed.
Fluorescence microscopy
Yeast cells expressing Avo3-GFP (excitation λmax 489 nm; emission λmax 508) were grown to mid-exponential phase in synthetic medium lacking Trp, mounted onto 1% agarose pads, and viewed immediately at 23°C using a confocal microscope (Model LSM 710; Zeiss) equipped with a 100× oil-immersion objective (1.40 NA), excited with an argon laser at 488 nm at 2.3% power (100 mW), and emission monitored using a bandpass filter (495–550-nm window). Images were acquired with an iXon3 EM-CCD camera (Andor) using Metamorph software (Molecular Devices) and processed using ImageJ (Collins, 2007).
Online supplemental material
Fig. S1 shows that phosphorylation of full-length Muk1 occurs at its Ypk1 consensus phospho-acceptor site motifs and is stimulated by treatment of the cells with AbA; and, using a growth assay for temperature sensitivity, that Ypk1 phosphorylation is necessary for Muk1 function. Fig. S2 shows that, in the absence of the Rab5 GEFs, localization of TORC2 is unaffected, phosphorylation of Ypk1 by Pkh1 is unimpaired, Ypk1 association with the Avo1 subunit of TORC2 is unimpaired, and forced PM association of Ypk1 cannot restore TORC2-dependent phosphorylation. Fig. S3 shows that Vps21 coimmunoprecipitates with Tor2, but not Ypk1, and the effect of guanine nucleotide on Vps21-Tor2 association. Fig. S4 shows that phosphorylation of MBP-Ypk1CT occurs on its TORC2-specific sites, is blocked by specific inhibitors of TORC2, and is stimulated by Vps21-GTPγS. Fig. S5 shows a sequence alignment of the yeast Rab5 isoforms to other classes of small GTPases from both yeast and human cells.
Supplementary Material
Acknowledgments
We thank Nathanael Gray for the generous gift of QL-IX-55, Alex Merz (University of Washington, Seattle, WA) for Vps21 mutants, Kirsten Young for technical assistance, Dr. Anita Emmerstorfer-Augustin for help with strain construction, the staff of the University of California Berkeley DNA Sequencing Facility for sequence analysis, Dr. Françoise M. Roelants for her constant guidance, Ben Wharton for proofreading, and all other members of the Thorner Lab for their constructive criticism and enthusiastic support.
This work was supported by National Institutes of Health Predoctoral Traineeship GM07232, by a Summer Research Grant from the University of California Berkeley Graduate Division, and by a Cancer Research Coordinating Committee Fellowship from University of California Systemwide (all to M.N. Locke) and by National Institutes of Health, National Institute of General Medical Sciences Research Grant R01GM21841 (to J. Thorner).
The authors declare no competing financial interests.
Author contributions: M.N. Locke designed, executed, and analyzed experiments and drafted the manuscript. J. Thorner designed and analyzed experiments and revised the manuscript.
References
- Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., and Zhou H.. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 7:1389–1396. 10.1074/mcp.M700468-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro C.G., Aindow A., and Thorner J.. 2016. Differential phosphorylation provides a switch to control how α-arrestin Rod1 down-regulates mating pheromone response in Saccharomyces cerevisiae. Genetics. 203:299–317. 10.1534/genetics.115.186122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D.C., Burke D.J., and Strathern J.N.. 2005. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Andersen K.R., Leksa N.C., and Schwartz T.U.. 2013. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins. 81:1857–1861. 10.1002/prot.24364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Aronov P.A., Fontes K., Ramos K., Hammock B.D., and Powers T.. 2008. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 7:148–158. 10.1016/j.cmet.2007.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J., Long X., Lin Y., Ortiz-Vega S., Rapley J., Papageorgiou A., Oshiro N., and Kikkawa U.. 2009. Activation of mTORC1 in two steps: Rheb-GTP activation of catalytic function and increased binding of substrates to raptor. Biochem. Soc. Trans. 37:223–226. 10.1042/BST0370223 [DOI] [PubMed] [Google Scholar]
- Bardwell L., Cook J.G., Zhu-Shimoni J.X., Voora D., and Thorner J.. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA. 95:15400–15405. 10.1073/pnas.95.26.15400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F., and Lambright D.G.. 2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22:461–470. 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Thorner J., and Honig L.. 1978. Identification of tubulin from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 75:4962–4966. 10.1073/pnas.75.10.4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean B.D., Davey M., Snider J., Jessulat M., Deineko V., Tinney M., Stagljar I., Babu M., and Conibear E.. 2015. Rab5-family guanine nucleotide exchange factors bind retromer and promote its recruitment to endosomes. Mol. Biol. Cell. 26:1119–1128. 10.1091/mbc.E14-08-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D., and Walther T.C.. 2009. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell. 20:1565–1575. 10.1091/mbc.e08-10-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D., Piccolis M., Chiaruttini N., Riezman I., Riezman H., Roux A., Walther T.C., and Loewith R.. 2012. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14:542–547. 10.1038/ncb2480 [DOI] [PubMed] [Google Scholar]
- Burd C.G., Mustol P.A., Schu P.V., and Emr S.D.. 1996. A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol. Cell. Biol. 16:2369–2377. 10.1128/MCB.16.5.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M., and Ungermann C.. 2013. Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J. Biol. Chem. 288:28704–28712. 10.1074/jbc.M113.488213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney D.S., Davies B.A., and Horazdovsky B.F.. 2006. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16:27–35. 10.1016/j.tcb.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Casamayor A., Torrance P.D., Kobayashi T., Thorner J., and Alessi D.R.. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9:186–197. 10.1016/S0960-9822(99)80088-8 [DOI] [PubMed] [Google Scholar]
- Collins T.J. 2007. ImageJ for microscopy. Biotechniques. 43(Suppl):25–30. 10.2144/000112517 [DOI] [PubMed] [Google Scholar]
- Douglas L.M., and Konopka J.B.. 2014. Fungal membrane organization: the eisosome concept. Annu. Rev. Microbiol. 68:377–393. 10.1146/annurev-micro-091313-103507 [DOI] [PubMed] [Google Scholar]
- Duplay P., Bedouelle H., Fowler A., Zabin I., Saurin W., and Hofnung M.. 1984. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J. Biol. Chem. 259:10606–10613. [PubMed] [Google Scholar]
- Ebner M., Sinkovics B., Szczygieł M., Ribeiro D.W., and Yudushkin I.. 2017. Localization of mTORC2 activity inside cells. J. Cell Biol. 216:343–353. 10.1083/jcb.201610060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltschinger S., and Loewith R.. 2016. TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 26:148–159. 10.1016/j.tcb.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Feyder S., De Craene J.-O., Bär S., Bertazzi D.L., and Friant S.. 2015. Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int. J. Mol. Sci. 16:1509–1525. 10.3390/ijms16011509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., and Brown P.O.. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11:4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubitz C., Prouteau M., Kusmider B., and Loewith R.. 2016. TORC2 structure and function. Trends Biochem. Sci. 41:532–545. 10.1016/j.tibs.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Gerrard S.R., Bryant N.J., and Stevens T.H.. 2000. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol. Biol. Cell. 11:613–626. 10.1091/mbc.11.2.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., and Hall M.N.. 2017. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36:397–408. 10.15252/embj.201696010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J., Karplus K., Hughey R., and Chothia C.. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313:903–919. 10.1006/jmbi.2001.5080 [DOI] [PubMed] [Google Scholar]
- Green M.R., and Sambrook J.. 2012. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Grenson M., Mousset M., Wiame J.M., and Bechet J.. 1966. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim. Biophys. Acta. 127:325–338. 10.1016/0304-4165(66)90387-4 [DOI] [PubMed] [Google Scholar]
- Guerreiro J.F., Muir A., Ramachandran S., Thorner J., and Sá-Correia I.. 2016. Sphingolipid biosynthesis upregulation by TOR complex 2-Ypk1 signaling during yeast adaptive response to acetic acid stress. Biochem. J. 473:4311–4325. 10.1042/BCJ20160565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney E.L., Klecker T., and Emr S.D.. 2016. Identification of the endocytic sorting signal recognized by the Art1-Rsp5 ubiquitin ligase complex. Mol. Biol. Cell. 27:4043–4054. 10.1091/mbc.e16-08-0570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guri Y., Colombi M., Dazert E., Hindupur S.K., Roszik J., Moes S., Jenoe P., Heim M.H., Riezman I., Riezman H., and Hall M.N.. 2017. mTORC2 promotes tumorigenesis via lipid synthesis. Cancer Cell. 32:807–823.e12. 10.1016/j.ccell.2017.11.011 [DOI] [PubMed] [Google Scholar]
- Hama H., Tall G.G., and Horazdovsky B.F.. 1999. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. 274:15284–15291. 10.1074/jbc.274.21.15284 [DOI] [PubMed] [Google Scholar]
- Helliwell S.B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., and Hall M.N.. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell. 5:105–118. 10.1091/mbc.5.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Niles B., Cuyegkeng A., and Powers T.. 2018. Redesigning TOR kinase to explore the structural basis for TORC1 and TORC2 assembly. Biomolecules. 8:E36 10.3390/biom8020036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., and Morgan D.O.. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 325:1682–1686. 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky B.F., Busch G.R., and Emr S.D.. 1994. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 13:1297–1309. 10.1002/j.1460-2075.1994.tb06382.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnard A.D., Thomas D., and Surdin-Kerjan Y.. 1996. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J. Mol. Biol. 262:473–484. 10.1006/jmbi.1996.0529 [DOI] [PubMed] [Google Scholar]
- Jewell J.L., and Guan K.L.. 2013. Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 38:233–242. 10.1016/j.tibs.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., and Broach J.R.. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782–2792. 10.1093/emboj/18.10.2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic M., Sharma M., Rahajeng J., and Caplan S.. 2010. The early endosome: a busy sorting station for proteins at the crossroads. Histol. Histopathol. 25:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Fujioka Y., Suzuki N.N., Inagaki F., Wullschleger S., Loewith R., Hall M.N., and Ohsumi Y.. 2005. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25:7239–7248. 10.1128/MCB.25.16.7239-7248.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppasamy M., Kusmider B., Oliveira T.M., Gaubitz C., Prouteau M., Loewith R., and Schaffitzel C.. 2017. Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nat. Commun. 8:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. 2009. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem. Soc. Rev. 38:2876–2886. 10.1039/b909638h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhina S., Johnson S.S., Manoharlal R., Russo S.B., Blugeon C., Lemoine S., Sunshine A.B., Dunham M.J., Cowart L.A., Devaux F., and Moye-Rowley W.S.. 2015. Control of plasma membrane permeability by ABC transporters. Eukaryot. Cell. 14:442–453. 10.1128/EC.00021-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.G., Buel G.R., and Blenis J.. 2013. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol. Cells. 35:463–473. 10.1007/s10059-013-0138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., and Koike T.. 2009. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4:1513–1521. 10.1038/nprot.2009.154 [DOI] [PubMed] [Google Scholar]
- Kliegman J.I., Fiedler D., Ryan C.J., Xu Y.F., Su X.Y., Thomas D., Caccese M.C., Cheng A., Shales M., Rabinowitz J.D., et al. 2013. Chemical genetics of rapamycin-insensitive TORC2 in S. cerevisiae. Cell Reports. 5:1725–1736. 10.1016/j.celrep.2013.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N.R., and Hall M.N.. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 73:585–596. 10.1016/0092-8674(93)90144-F [DOI] [PubMed] [Google Scholar]
- Lachmann J., Barr F.A., and Ungermann C.. 2012. The Msb3/Gyp3 GAP controls the activity of the Rab GTPases Vps21 and Ypt7 at endosomes and vacuoles. Mol. Biol. Cell. 23:2516–2526. 10.1091/mbc.e11-12-1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Jeschke G.R., Roelants F.M., Thorner J., and Turk B.E.. 2012. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell. Biol. 32:4705–4717. 10.1128/MCB.00897-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux A.E., Schulze J.O., and Biondi R.M.. 2018. AGC kinases, mechanisms of regulation ýand innovative drug development. Semin. Cancer Biol. 48:1–17. 10.1016/j.semcancer.2017.05.011 [DOI] [PubMed] [Google Scholar]
- Leskoske K.L., Roelants F.M., Marshall M.N.M., Hill J.M., and Thorner J.. 2017. The stress-sensing TORC2 complex activates yeast AGC-family protein kinase Ypk1 at multiple novel sites. Genetics. 207:179–195. 10.1534/genetics.117.1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskoske K.L., Roelants F.M., Emmerstorfer-Augustin A., Augustin C.M., Si E.P., Hill J.M., and Thorner J.. 2018. Phosphorylation by the stress-activated MAPK Slt2 down-regulates the yeast TOR complex 2. Genes Dev. 32:1576–1590. 10.1101/gad.318709.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.C., and Chen M.Y.. 2012. Target of rapamycin complex 2 signals to downstream effector yeast protein kinase 2 (Ypk2) through adheres-voraciously-to-target-of-rapamycin-2 protein 1 (Avo1) in Saccharomyces cerevisiae. J. Biol. Chem. 287:6089–6099. 10.1074/jbc.M111.303701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.Y., and Zoncu R.. 2016. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 214:653–664. 10.1083/jcb.201607005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., MacGurn J.A., Chu T., Stefan C.J., and Emr S.D.. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 135:714–725. 10.1016/j.cell.2008.09.025 [DOI] [PubMed] [Google Scholar]
- Liu Q., Ren T., Fresques T., Oppliger W., Niles B.J., Hur W., Sabatini D.M., Hall M.N., Powers T., and Gray N.S.. 2012. Selective ATP-competitive inhibitors of TOR suppress rapamycin-insensitive function of TORC2 in Saccharomyces cerevisiae. ACS Chem. Biol. 7:982–987. 10.1021/cb300058v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.Y., Brett C.L., Plemel R.L., Vignali M., Fields S., Gonen T., and Merz A.J.. 2011. Intrinsic tethering activity of endosomal Rab proteins. Nat. Struct. Mol. Biol. 19:40–47. 10.1038/nsmb.2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., and Hall M.N.. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 10:457–468. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz-Vega S., Yonezawa K., and Avruch J.. 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15:702–713. 10.1016/j.cub.2005.02.053 [DOI] [PubMed] [Google Scholar]
- Lopez M.S., Kliegman J.I., and Shokat K.M.. 2014. The logic and design of analog-sensitive kinases and their small molecule inhibitors. Methods Enzymol. 548:189–213. 10.1016/B978-0-12-397918-6.00008-2 [DOI] [PubMed] [Google Scholar]
- MacDonald C., and Piper R.C.. 2016. Cell surface recycling in yeast: mechanisms and machineries. Biochem. Soc. Trans. 44:474–478. 10.1042/BST20150263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y., Kozutsumi Y., Nakamura S., Fujita T., and Kawasaki T.. 1995. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem. Biophys. Res. Commun. 211:396–403. 10.1006/bbrc.1995.1827 [DOI] [PubMed] [Google Scholar]
- Mok J., Kim P.M., Lam H.Y., Piccirillo S., Zhou X., Jeschke G.R., Sheridan D.L., Parker S.A., Desai V., Jwa M., et al. 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3:ra12 10.1126/scisignal.2000482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A., Ramachandran S., Roelants F.M., Timmons G., and Thorner J.. 2014. TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLife. 3:e03779 10.7554/eLife.03779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A., Roelants F.M., Timmons G., Leskoske K.L., and Thorner J.. 2015. Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. eLife. 4:e09336 10.7554/eLife.09336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Kanada A., Matsuzaki M., Byrne S.D., Okumura F., and Kamura T.. 2014. The nutrient stress-induced small GTPase Rab5 contributes to the activation of vesicle trafficking and vacuolar activity. J. Biol. Chem. 289:20970–20978. 10.1074/jbc.M114.548297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro R., Sardu A., Panchaud N., and De Virgilio C.. 2017. The architecture of the Rag GTPase signaling network. Biomolecules. 7:E48 10.3390/biom7030048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D.P., Russell M.R., Lo S.Y., Chapin H.C., Milnes J., and Merz A.J.. 2012. Termination of isoform-selective Vps21/Rab5 signaling at endolysosomal organelles by Msb3/Gyp3. Traffic. 13:1411–1428. 10.1111/j.1600-0854.2012.01390.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles B.J., Mogri H., Hill A., Vlahakis A., and Powers T.. 2012. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. USA. 109:1536–1541. 10.1073/pnas.1117563109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P. 2016. Regulation of membrane traffic by Rab GEF and GAP cascades. Small GTPases. 7:252–256. 10.1080/21541248.2016.1213781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsel A.L., Merz A.J., and Nickerson D.P.. 2013. Vps9 family protein Muk1 is the second Rab5 guanosine nucleotide exchange factor in budding yeast. J. Biol. Chem. 288:18162–18171. 10.1074/jbc.M113.457069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce L.R., Komander D., and Alessi D.R.. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9–22. 10.1038/nrm2822 [DOI] [PubMed] [Google Scholar]
- Plemel R.L., Lobingier B.T., Brett C.L., Angers C.G., Nickerson D.P., Paulsel A., Sprague D., and Merz A.J.. 2011. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol. Biol. Cell. 22:1353–1363. 10.1091/mbc.e10-03-0260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis K., and De Virgilio C.. 2016. Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov. 2:15049 10.1038/celldisc.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Roelants F.M., Torrance P.D., Bezman N., and Thorner J.. 2002. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell. 13:3005–3028. 10.1091/mbc.e02-04-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Torrance P.D., and Thorner J.. 2004. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology. 150:3289–3304. 10.1099/mic.0.27286-0 [DOI] [PubMed] [Google Scholar]
- Roelants F.M., Baltz A.G., Trott A.E., Fereres S., and Thorner J.. 2010. A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. USA. 107:34–39. 10.1073/pnas.0912497106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Breslow D.K., Muir A., Weissman J.S., and Thorner J.. 2011. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 108:19222–19227. 10.1073/pnas.1116948108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Leskoske K.L., Martinez Marshall M.N., Locke M.N., and Thorner J.. 2017a The TORC2-dependent signaling network in the yeast Saccharomyces cerevisiae. Biomolecules. 7:E66 10.3390/biom7030066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Leskoske K.L., Pedersen R.T., Muir A., Liu J.M., Finnigan G.C., and Thorner J.. 2017b TOR Complex 2-regulated protein kinase Fpk1 stimulates endocytosis via inhibition of Ark1/Prk1-related protein kinase Akl1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 37:e00627-16 10.1128/MCB.00627-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Chauhan N., Muir A., Davis J.C., Menon A.K., Levine T.P., and Thorner J.. 2018. TOR complex 2-regulated protein kinase Ypk1 controls sterol distribution by inhibiting StARkin domain-containing proteins located at plasma membrane-endoplasmic reticulum contact sites. Mol. Biol. Cell. 29:2128–2136. 10.1091/mbc.E18-04-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., and Sabatini D.M.. 2009. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochem. Soc. Trans. 37:289–290. 10.1042/BST0370289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR signaling in growth, metabolism, and disease. Cell. 168:960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmöller F., and Riezman H.. 1993. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J. Cell Sci. 106:823–830. [DOI] [PubMed] [Google Scholar]
- Schmidt O., Weyer Y., Fink M.J., Müller M., Weys S., Bindreither M., and Teis D.. 2017. Regulation of Rab5 isoforms by transcriptional and post-transcriptional mechanisms in yeast. FEBS Lett. 591:2803–2815. 10.1002/1873-3468.12785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebohm G., Strutz-Seebohm N., Ureche O.N., Henrion U., Baltaev R., Mack A.F., Korniychuk G., Steinke K., Tapken D., Pfeufer A., et al. 2008. Long QT syndrome-associated mutations in KCNQ1 and KCNE1 subunits disrupt normal endosomal recycling of IKs channels. Circ. Res. 103:1451–1457. 10.1161/CIRCRESAHA.108.177360 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Lambert G.G., Chammas A., Ni Y., Cranfill P.J., Baird M.A., Sell B.R., Allen J.R., Day R.N., Israelsson M., et al. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 10:407–409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. 2002. Getting started with yeast. Methods Enzymol. 350:3–41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Shideler T., Nickerson D.P., Merz A.J., and Odorizzi G.. 2015. Ubiquitin binding by the CUE domain promotes endosomal localization of the Rab5 GEF Vps9. Mol. Biol. Cell. 26:1345–1356. 10.1091/mbc.E14-06-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Krüger B., Stenmark H., Düsterhöft A., Philippsen P., Yoo J.S., Gallwitz D., and Zerial M.. 1994. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125:283–298. 10.1083/jcb.125.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Krüger B., Stenmark H., and Zerial M.. 1995. Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J. Cell Sci. 108:3509–3521. [DOI] [PubMed] [Google Scholar]
- Suda Y., Kurokawa K., Hirata R., and Nakano A.. 2013. Rab GAP cascade regulates dynamics of Ypt6 in the Golgi traffic. Proc. Natl. Acad. Sci. USA. 110:18976–18981. 10.1073/pnas.1308627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y., Sakoh H., and Yamada K.. 2004. IPC synthase as a useful target for antifungal drugs. Curr. Drug Targets Infect. Disord. 4:311–322. 10.2174/1568005043340597 [DOI] [PubMed] [Google Scholar]
- Sun Y., Miao Y., Yamane Y., Zhang C., Shokat K.M., Takematsu H., Kozutsumi Y., and Drubin D.G.. 2012. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell. 23:2388–2398. 10.1091/mbc.e12-03-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.K., and Huh W.K.. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 24:767–775. 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Swaney D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N.J., and Villén J.. 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 10:676–682. 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall G.G., Hama H., DeWald D.B., and Horazdovsky B.F.. 1999. The phosphatidylinositol 3-phosphate binding protein Vac1p interacts with a Rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol. Biol. Cell. 10:1873–1889. 10.1091/mbc.10.6.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H., and Shiozaki K.. 2017. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules. 7:E77 10.3390/biom7040077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H., Morigasaki S., Murayama S., Zeng C.T., and Shiozaki K.. 2010. Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr. Biol. 20:1975–1982. 10.1016/j.cub.2010.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H., Murayama S., Yonekura T., Hatano T., Richter D., Furuya T., Kataoka S., Furuita K., Kojima C., and Shiozaki K.. 2017. Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. eLife. 6:e19594 10.7554/eLife.19594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Aguilar P.S., Fröhlich F., Chu F., Moreira K., Burlingame A.L., and Walter P.. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26:4946–4955. 10.1038/sj.emboj.7601933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.L., Yang C.H., Lee H.H., Kuo J.C., Hur S.S., Chien S., Lee O.K., Hung S.C., and Chang Z.F.. 2015. Dexamethasone-induced cellular tension requires a SGK1-stimulated Sec5-GEF-H1 interaction. J. Cell Sci. 128:3757–3768. 10.1242/jcs.169961 [DOI] [PubMed] [Google Scholar]
- Wedaman K.P., Reinke A., Anderson S., Yates J. III, McCaffery J.M., and Powers T.. 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:1204–1220. 10.1091/mbc.e02-09-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall P.J., Patterson J.C., Chen R.E., and Thorner J.. 2008. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc. Natl. Acad. Sci. USA. 105:12212–12217. 10.1073/pnas.0805797105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.B., Zoncu R., Root D.E., Sabatini D.M., and Sawyers C.L.. 2015. Identification of an oncogenic RAB protein. Science. 350:211–217. 10.1126/science.aaa4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Oppliger W., and Hall M.N.. 2005. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 280:30697–30704. 10.1074/jbc.M505553200 [DOI] [PubMed] [Google Scholar]
- Yang H., Jiang X., Li B., Yang H.J., Miller M., Yang A., Dhar A., and Pavletich N.P.. 2017. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 552:368–373. 10.1038/nature25023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C.A., Ortiz-Vega S., Sun Y.Y., Chien C.T., Chuang J.H., and Lin Y.. 2017. Association of mSin1 with mTORC2 Ras and Akt reveals a crucial domain on mSin1 involved in Akt phosphorylation. Oncotarget. 8:63392–63404. 10.18632/oncotarget.18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.