Mutations in the essential retinal protein retinoschisin (RS1) cause a form of macular degeneration. Heymann et al. use cryo-EM to show that RS1 assembles into branched networks that may play a stabilizing role in maintaining the integrity of the retina.
Abstract
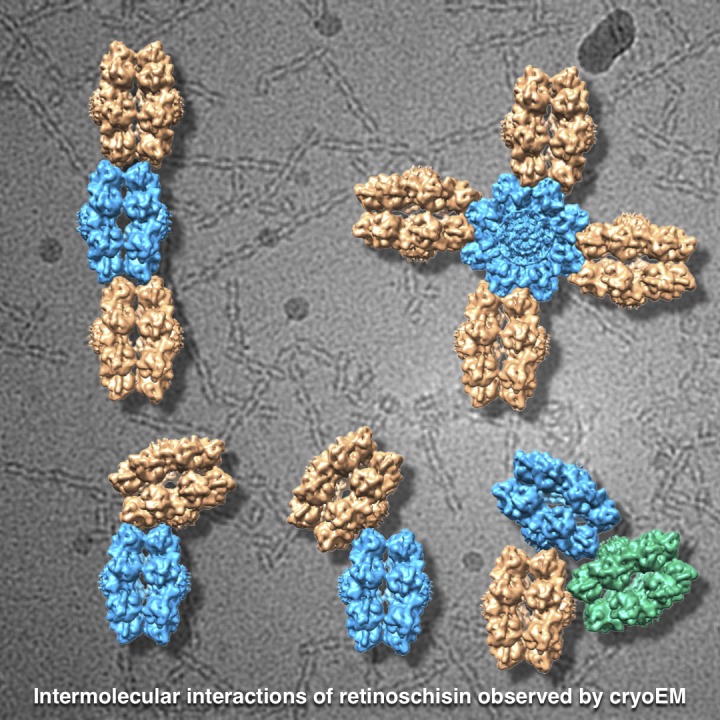
Mutations in the retinal protein retinoschisin (RS1) cause progressive loss of vision in young males, a form of macular degeneration called X-linked retinoschisis (XLRS). We previously solved the structure of RS1, a 16-mer composed of paired back-to-back octameric rings. Here, we show by cryo–electron microscopy that RS1 16-mers can assemble into extensive branched networks. We classified the different configurations, finding four types of interaction between the RS1 molecules. The predominant configuration is a linear strand with a wavy appearance. Three less frequent types constitute the branch points of the network. In all cases, the “spikes” around the periphery of the double rings are involved in these interactions. In the linear strand, a loop (usually referred to as spike 1) occurs on both sides of the interface between neighboring molecules. Mutations in this loop suppress secretion, indicating the possibility of intracellular higher-order assembly. These observations suggest that branched networks of RS1 may play a stabilizing role in maintaining the integrity of the retina.
Graphical Abstract
Introduction
Loss-of-function mutations in retinoschisin (RS1) cause a form of macular degeneration in young males called X-linked retinoschisis (XLRS; Weber et al., 2002). The hallmark of the disease is a separation (schisis) of the inner retinal layers and formation of macular microcysts, leading to progressive decline in vision. Functional RS1 is secreted as a covalently linked octameric ring by photoreceptors and bipolar cells (Molday, 2007). It is most prominently located on the cellular surface of photoreceptor inner segments (Vijayasarathy et al., 2007). Based on the clinical pathology and morphological consequences of XLRS, RS1 is implicated in cell–cell adhesion (Weber et al., 2002). RS1 is also thought to regulate fluid balance in the retina (Molday et al., 2012). Ongoing studies aim at reversing the deficiency for RS1 through gene replacement therapy (Zeng et al., 2004; Park et al., 2009; Bush et al., 2015).
Although Sauer et al. (1997) cloned the gene for RS1 20 yr ago, the structure was solved only recently (Bush et al., 2016; Ramsay et al., 2016; Tolun et al., 2016). These studies revealed that RS1 is actually secreted as a double octameric ring. Many of the disease-causing mutations in RS1 map to the interfaces between subunits (Tolun et al., 2016), indicating that any impedance of its assembly precludes secretion and leads to loss of function (also proposed by Wang et al., 2006). However, a number of additional disease-causing mutants still assemble into octameric rings and are secreted (Wang et al., 2006). These mutations occur in peripheral regions of the molecule, where they may be involved in essential interactions with other components in the intercellular space (Table 1).
Table 1. Disease-causing mutations in the spikes of retinoschisin.
The RS1 molecule is composed of 16 subunits arranged in back-to-back octameric rings (Fig. 1 A; Tolun et al., 2016). Each subunit is divided into an N-terminal RS1 domain of ∼40 residues, and a discoidin domain of ∼160 residues (Kiedzierska et al., 2007). All the subunits in an octameric ring are covalently linked through disulfide bonds between residues 59 and 223 (yellow parts in Fig. 1 A). Of particular note are the exposed surfaces of this molecule that likely interact with other entities in the intercellular space: the spikes (the peripheral loops), the tops of the discoidin domains, and the top of the central density contributed by the RS1 domains.
Figure 1.
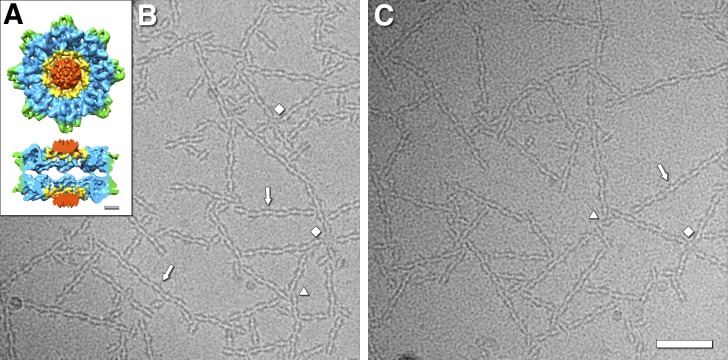
Cryo-electron micrographs of filamentous networks of RS1 molecules. (A) Architecture of the retinoschisin double rings, showing the N-terminal RS1 domains (red), the ring containing the disulfide bonds between subunits (yellow), the discoidin domain cores (blue), and the spikes (green; Tolun et al., 2016). Scale bar, 20 Å. (B and C) Two examples of micrographs of RS1 molecules forming branched networks of strands (arrows). The diamonds indicate four-way connections with a central RS1 molecule hub. The triangles indicate three-way connections. Scale bar, 500 Å.
As an adhesive molecule, RS1 can potentially interact with several other entities in the interphotoreceptor space. Foremost and simplest is that it should dock to the lipid bilayer, through either an ionic (Vijayasarathy et al., 2007; Kotova et al., 2010) or a hydrophobic (Fraternali et al., 2003) mechanism. Another is that it should bind to carbohydrate moieties on proteins or lipids, based on its retention on selected sugar-agarose beads (Dyka et al., 2008). Several proteins have been suggested as interacting partners, such as the sodium-potassium ATPase (NaKATPase; Molday et al., 2007; Friedrich et al., 2011; Plössl et al., 2017), L-type voltage-gated calcium channels (Shi et al., 2009), β2 laminin, and αB-crystallin (Steiner-Champliaud et al., 2006). RS1 is possibly a promiscuous binder, interacting with many counterparts between the photoreceptors, including the abundant NaKATPase.
Molday et al. (2012) suggested an alternative function: The fluid-filled cysts in the XLRS retina could result from a failure in regulating fluid balance. This may involve the NaKATPase through signaling cascades (Friedrich et al., 2011; Plössl et al., 2017). Decreasing the volume of these cysts using carbonic anhydrase inhibitors (such as dorzolamide) improves retinal function, countering the effects of RS1 deficiency (Walia et al., 2009; Verbakel et al., 2016). While this treatment does alleviate the symptoms of XLRS, it may not directly relate to the function of RS1 (Walia et al., 2009). Indeed, Zhour et al. (2012) did not find any improvement in a mouse model for XLRS when treated with dorzolamide.
Our initial aim in this study was to further probe the structure of RS1 by cryo-EM. What we found was that RS1 forms branched networks of strands. Analysis of the main strands and branching indicates different types of interaction between RS1 molecules. These interactions are primarily through the peripheral spikes of the molecule, suggesting how it could span the intercellular space and provide an adhesive function in the retina. Reconstruction of the linear interaction placed the spike 1 loop on both sides of the interface between molecules. Mutations in this loop suppress secretion, consistent with intracellular formation of higher-order assemblies.
Results
RS1 molecules self-associate into networks
We purified RS1 as in Tolun et al., 2016, but omitted dialysis and Mono Q chromatography. In particular, the dialysis against 150 mM salt buffer leads to extensive aggregation, rendering the specimen poorly suitable for cryo-EM. We resolved this problem by using the high-salt elution buffer from the cobalt-agarose column (500 mM sodium chloride and 200 mM imidazole), which prevents extensive aggregation, and yielded cryo-EM grids with an acceptable distribution of specimen (Fig. 1, B and C). Under these conditions, we found that RS1 assembles into networks of strands (Fig. 1, B and C). The predominant configuration is a wavy strand (arrows in Fig. 1, B and C), while there is also an abundance of branching interactions. These include simple T-like junctions, three-way joints (triangles in Fig. 1, B and C), four-way intersections (diamonds in Fig. 1, B and C), and larger formations.
Because the cobalt-agarose elution buffer is not at physiological ionic strength, we wanted to better define how RS1 behaves under different conditions. From our accumulated experience, we think that the imidazole in our purification buffer serves as a chelator of a cation that interacts with retinoschisin. Previous studies have shown that EDTA is required to release retinoschisin from retinal membranes (Vijayasarathy et al., 2007). We therefore dialyzed RS1 against buffers with calcium or EDTA, and with high or low salt, in comparison with imidazole-salt buffer. We performed light scattering (optical density) as a crude test of aggregation (Fig. S1), followed by microscopy on the same samples (Fig. S2). In this experiment, the controls in imidazole–high-salt buffer (Fig. S2, A and B) yielded only short strands of RS1 compared with those in Fig. 1 (B and C). We found that whether we get such long strands is variable from sample to sample but is highly reproducible for a given sample. We therefore performed all the analyses shown in Figs. S1 and S2 at the same time. Calcium induced massive aggregation (blue and black curves in Fig. S1), precluding interpretation by microscopy. EDTA at high and low salt concentrations appears to favor some dissociation of RS1 into single rings (arrowheads in Fig. S2, C and D) with more extensive dissociation at high salt. However, some strands of RS1 molecules are apparent in the low-salt EDTA buffer, comparable to those in the controls (Fig. S2, A and B). The behavior of RS1 is therefore complicated but definitely a function of the buffer conditions. However, it has the capability to form strands in both high- and low-salt buffers.
We were also interested in the interaction of RS1 with galactose, because Dyka et al. (2008) showed that RS1 binds to galactose-agarose. We observed a mild enhancement of strand formation in some of our specimens (Fig. S3). At this time, we cannot make a definitive statement about the effect of galactose on RS1 behavior.
2D classification of images yields four types of configurations
To explore the interactions between the RS1 molecules, we generated a data set consisting of short linear segments or junctions, each containing two to four molecules. We subjected them to 2D classification, followed by averaging within each class to enhance the resulting images (Fig. S4 A). The resolution of a class average depends on the number of contributing subimages: here, that of the most abundant class was ∼5 Å (Fig. S4 B). Such a high resolution indicates that—within this class at least—the subimages averaged are consistent, and consequently the interaction conveyed in the class average is specific.
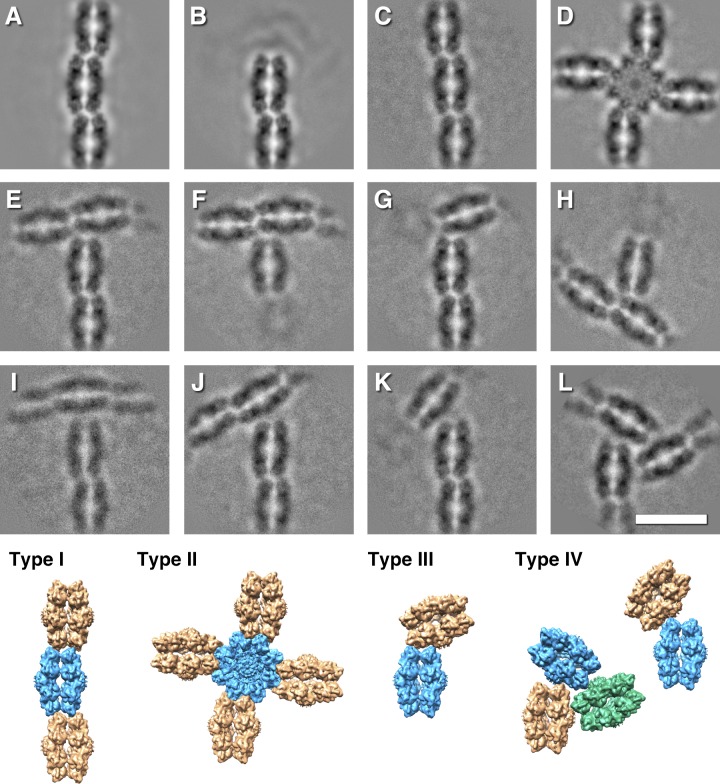
The major configurations are the wavy strand (58%; Fig. 2 A) and strand ends (24%; Fig. 2 B). A stepped strand (Fig. 2 C) is much less abundant. All of these involve the spikes from both molecules at the periphery of the RS1 octameric rings, and we refer to them as type I interactions. Another class of spike–spike interaction is the four-way or type II, where the central molecule is rotated by 90° with respect to the four attached molecules (Fig. 2 D). Here, the interacting spike partners differ from those involved in the linear strand segments.
Figure 2.
Selected class averages, illustrating the different interactions between molecules of RS1 and models illustrating the configurations. (A–C) The most common interaction (type I) between the spikes of molecules with alternating offset (58%; A), ends (24%; B), and stepped offset (1.4%; C). (D) A fourfold symmetric spoke-wheel arrangement with 90° interactions between the spikes (type II: 1.5%). (E–H) Interactions where spikes of one molecule are bound to one side of the top of the ring of another molecule (type III, 8%). (I–L) Interactions where spikes of one molecule are bound close to the center of the top of the ring of another molecule with various angles (type IV, 7%). (The image in L was threefold symmetrized). Several of these class averages are included in the larger and more diverse set shown in Fig. S4. Scale bar, 150 Å.
In the remainder, the spikes of one molecule interact with the top of a ring from another molecule (Fig. 2, E–L). We distinguish between those interacting with the periphery of the ring (type III; Fig. 2, E–H) and those closer to the center of the ring (type IV; Fig. 2, I–L). The latter likely involves the RS1 domains of one of the molecules. The abundances of the two types of interaction are similar (7–8%), suggesting that they contribute equally to the branching of the network. However, the three-way joint in Fig. 2 L occurs in only ∼1% of the extracted images.
The main strand configuration (type I)
The most prevalent configuration is the linear strand of RS1 molecules (type I; Fig. 2 A). Curiously, the central planes through the rings from one molecule to the next do not line up but are offset in an alternating fashion, giving a slightly zigzag or wavy appearance. The lengths of the strands are highly variable, with an average of approximately seven molecules long (∼850 Å). The repeat along the strand is ∼122 Å, and the lateral offset is ∼13 Å. This constitutes a 1D unit cell consistent with space group P2221 and repeat length of 244 Å (Fig. S5 C). The two rings in each molecule also look somewhat different, with one ring slightly smaller than the other (∼120 Å vs. ∼125 Å). We aligned all the particles against projections from a 3D reference map of RS1, obtaining a consistent orientation for each double-ring molecule in the alternating configuration to within a small uncertainty (±3°). This allowed us to place 3D maps in the correct relationship with each other (Fig. 2, type I).
The stepped offset configuration (Fig. 2 C) is rarer than the alternating configuration, although the repeat and offset are similar (∼121 Å and ∼14 Å, respectively) and is therefore included in the type I class. One octameric ring appears offset with respect to the other ring in the direction of the strand. This varying diameter of the rings in both the alternating and stepped configurations is consistent with either a small radial displacement of the subunits or a remodeling of the spikes at the interface between neighboring RS1 molecules.
The 90° branching configuration (type II)
The type II interaction in Fig. 2 D has a 90° rotation from that of the main strand. With four peripheral molecules surrounding the central molecule, all spikes of the latter are involved (Fig. 2, type II). The distance between the center and a peripheral molecule is ∼116 Å, less than the distance between main strand molecules (∼122 Å). The central molecule appears distorted with a more fourfold symmetric nature than eightfold (Fig. 2 D). The spike tips of the four subunits in the corners are separated by ∼136 Å, while the intervening spikes are only ∼120 Å apart. The rings of the peripheral molecules also appear different in size. One ring of the attached molecule appears larger (∼125 Å) and presses one subunit of the central molecule inwards, while the other ring appears smaller (∼118 Å) and interacting between the spikes. The next attached molecule (second from the central molecule) then follows the alternating offset pattern of the main strand (see class average 26 in Fig. S4 A). These details further support the ability of the subunits within a ring to shift radially.
The lateral branching configurations (types III and IV)
Most branch points feature interactions between the spikes of one molecule and the top of another (Fig. 2, E–L, types III and IV). The angle between the main strand and lateral branch varies considerably. To quantify this, we measured the angle between the planes through the octamer rings. The angles for the type IV branches vary more (37–90°) than those of the type III branches (63–90°), indicating greater freedom of movement. The triplet interaction (Fig. 2 L) is a special case of the type IV configuration. It is sufficiently rigid to allow symmetrization, indicating strong constraints on the relative orientations of the RS1 molecules.
Interactions of the spikes in the main strand configuration
We reconstructed the “unit cell” of the main strand configuration (Fig. S5 C) in three dimensions to understand which parts of the RS1 spikes are involved. To overcome the preferred orientations of RS1 in the first data set (e.g., Fig. 1, B and C), we imaged the specimen with a 30° tilt (Fig. S5, A and B), producing oblique views suitable for 3D reconstruction (statistics given in Table 2 and the Fourier shell correlation [FSC] curve in Fig. S5 D). Fig. 3 A shows the two molecules in the unit cell.
Table 2. Image processing report.
| Parameter | Data set 1 | Data set 2 |
|---|---|---|
| Micrographs | ||
| Camera | K2, superresolution | K2, superresolution |
| Magnification | 48077 | 48077 |
| Number | 218 | 1136 |
| Tilt (°) | 0 | 30 |
| Frames per micrograph | 54 | 50 |
| Frame rate (/s) | 4 | 4 |
| Dose per frame (e−/pixel) | 1.08a | 0.76a |
| Accumulated dose (e−/Å2) | 51 | 33 |
| Frame alignment | bseries | bseries |
| CTF | ||
| Defocus range (µm) | 1.04–2.92 | 1.06–3.23 |
| Correction | Phase flip | Phase flip |
| Particles | ||
| Picked | 39536 | 26702 |
| Used in final 2D averages/3D reconstruction | 19701 (50%) | 18646 (70%) |
| Alignment | ||
| Initial reference map | PBD: 3JD6 | PBD: 3JD6 |
| Low-pass filter limit (Å) | 35 | 20 |
| Projection-matching algorithm | Cross-correlation | Cross-correlation |
| Number of iterations | 5 | 4 |
| Reconstruction | ||
| Classes | 70 | 1 |
| Size | 330 × 330 × 1 | 224 × 224 × 224 |
| Pixel size (Å) | 1.04 | 1.04 |
| Symmetry | C1 | C2 |
| Resolution limit (Å) | 2.1 | 3.0 |
| Resolution estimate (Å, FSC0.143) | 5–25 | 10.0 |
| Masked | No | Yes |
| Sharpened | No | Yesb |
| Filtered | No | Low-pass to 10 Å |
The micrographs were taken in superresolution mode and subsequently binned for further processing (final size: 3838 × 3710).
Amplitudes weighted to match PDB accession no. 3JD6.
Figure 3.
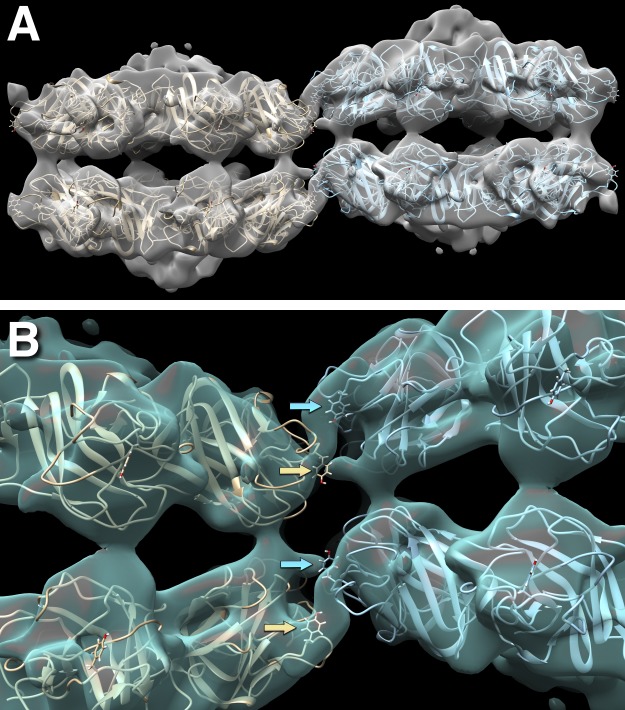
Reconstruction of the type I alternating configuration unit cell at ∼10-Å resolution (EMDB accession no. 7907) with two docked 16-mer molecules (PDB accession no. 3JD6). (A) Side view of the two 16-mer molecules in the unit cell, with the outside rings (top right and bottom left) somewhat more curved than the inside rings (top left and bottom right). (B) Close-up view of the interaction between neighboring molecules, showing the Y93 side chains as stick models (arrows). Y93 is part of an apparently flexible loop, P56EQYVGWY93SSWTA98, that fits into the connecting density between the molecules and is thus thought to form the main interaction. In this loop, mutations in Y89, G91, W92, and Y93 are associated with XLRS.
Four subunits from one RS1 molecule interact with the corresponding four subunits in the neighboring molecule (Fig. 3 B). Because of symmetry, there is only one unique interface formed by the spikes from two opposing subunits. Fig. 3 B shows the residue, Y93, that occurs prominently on both sides of the interface. This loop (spike 1) is interesting because several mutations in it have been linked to XLRS (Table 1). The structure fitted into the map is the one calculated for free RS1 molecules (PDB accession no. 3JD6; Tolun et al., 2016), and may not show the correct conformation of the loop in the assembled strand. We did not consider it reasonable to attempt modeling of the loops at the limited resolution (10 Å) of the map. The density connection shown in Fig. 3 B indicates that the conformations of these loops from the two sides probably differ.
Mutants in spike 1 are poorly secreted
There is evidence that the spike 1 loop is important in XLRS (Table 1). However, most of the mutations are to cysteine (except W96R), which may lead to aberrant cross-linking and a disrupted fold. We therefore constructed three mutants, G91V, W92S, and Y93A, and as control, the well-studied mutant, R141H (Park et al., 2000; Wang et al., 2006; Dyka and Molday, 2007; Dyka et al., 2008; Ramsay et al., 2016; Plössl et al., 2018). All of these mutants are comparably expressed in HEK293 and ARPE-19 cells, but the spike 1 mutants are secreted poorly (W92S and Y93A) or not at all (G91V), compared with WT RS1 or the R141H mutant (Fig. 4). As expected, assembly of the double rings is not affected in the small amounts of W92S and Y93A that are secreted. Plössl et al. (2018) reported that W96R is also poorly secreted. These observations are consistent with the importance of spike 1 in the higher-order intermolecular interactions of RS1.
Figure 4.
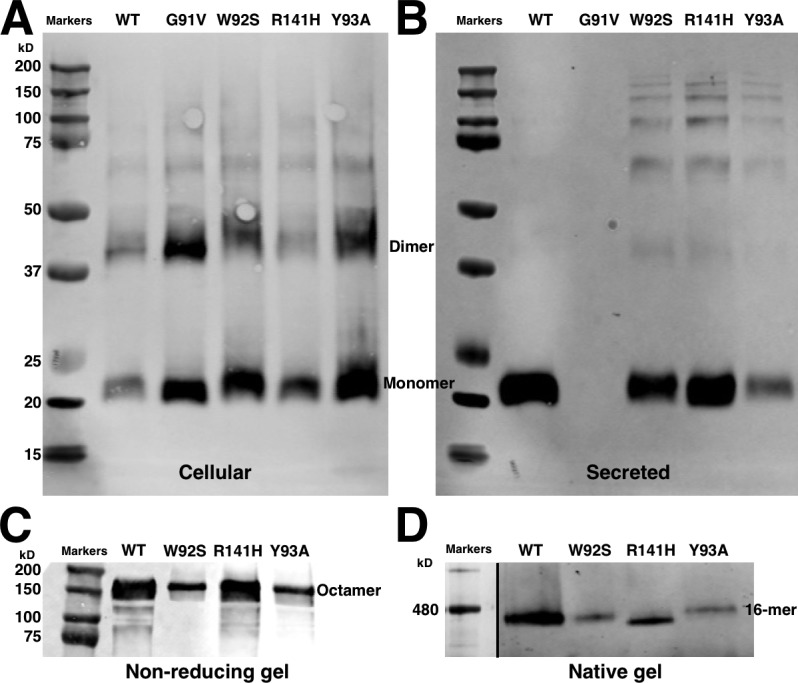
Expression and secretion from HEK-293 cells of WT and mutant retinoschisin. (A) Reducing SDS-PAGE of the cells. The spike 1 mutants (G91V, W92S, and Y93A) are better retained in the cells than the WT and spike 3 mutant (R141H). (B) Reducing SDS-PAGE of the medium. Compared with WT and R141H, W92S and Y93A are poorly secreted, and G91V is secreted not at all. (C) Nonreducing SDS-PAGE of the medium. All secreted RS1 form disulfide cross-linked octamers. (D) Blue-native gel of the medium. All secreted RS1 form double octamers.
Discussion
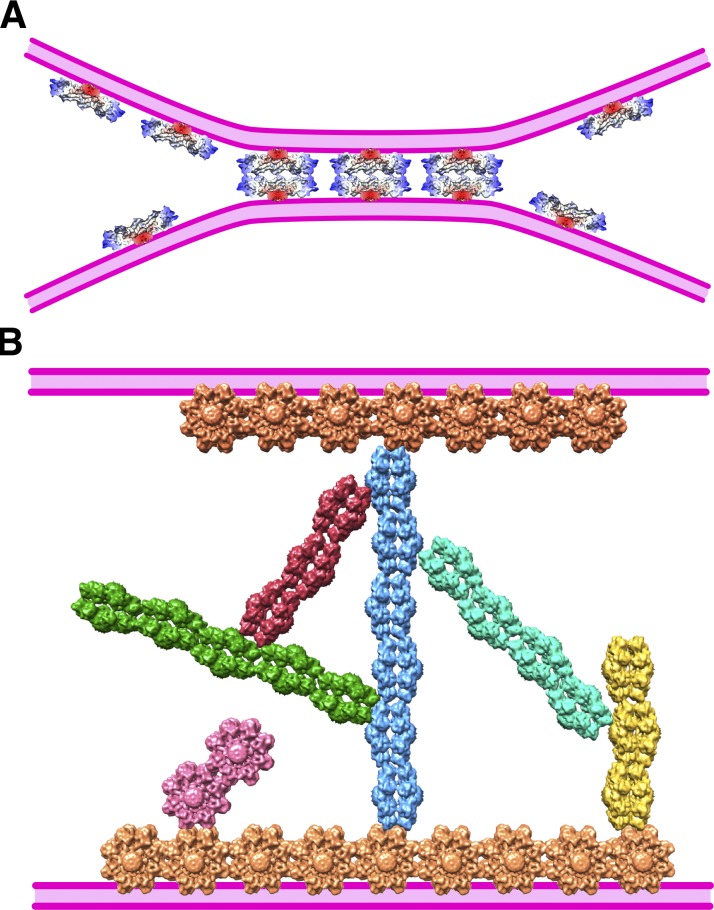
Deficiencies in RS1 have a debilitating effect on the structural integrity of the retina (Weber et al., 2002; Bush et al., 2015). While other explanations of this property are possible, an impairment of adhesive function (Weber et al., 2002) seems the most likely. However, the fact that RS1 is a soluble molecule is potentially problematic because known cell–cell adhesion systems such as cadherins (Al-Amoudi et al., 2011), connexins (Sosinsky and Nicholson, 2005), and claudins (Haseloff et al., 2015) tend to have obvious membrane-embedded moieties. On the other hand, all these junctions are built from paired back-to-back oligomers, a property shared by RS1. A second issue is that a single RS1 molecule (ring diameter of ∼140 Å and thickness of ∼80 Å) could only adhere to opposing membranes where the intercellular space is very narrow (Fig. 5 A). The closest membrane separation between cells ranges from 20 to 30 Å in gap junctions (Sosinsky and Nicholson, 2005) to 300 to 500 Å in desmosomes (Al-Amoudi et al., 2007, 2011), and much larger in nonjunctional regions. The reach of RS1 could be extended by binding to other components, such as the lipid bilayer, membrane-embedded proteins, or components of the intercellular matrix. Here, we describe higher-order, branched assemblies of RS1 that could traverse wider intercellular spaces (Fig. 5 B).
Figure 5.
Models of cell–cell junctions mediated by RS1. (A) The model from Tolun et al. (2016) depends on a close apposition of the cellular membranes (purple), with a separation of equivalent to the ∼80-Å-thick RS1 molecule. (B) The ability of RS1 to self-assemble into branched networks suggests that it can span a wider intercellular space. In this model, the hydrophobic spikes intercalate into the membrane, with branched strands extending toward the opposing membrane.
The RS1 spikes are able to form multiple interactions
We cataloged the higher-order interactions between RS1 molecules into four classes (Fig. 2). The consistent feature is the spikes of one molecule interacting with either the spikes of another (types I and II) or with other parts, including the RS1 domain (types III and IV). The models in Fig. 2 (bottom) for the four types of configuration show similar side view orientations for all the RS1 molecules, with the exception of the central molecule for type II. This orientation is such that the spikes of four subunits (oriented toward the viewer in Fig. 3) interact with the air–water interface, with lateral interactions also involving four subunits for configurations I–III. This provides a rich set of configurations available to form an intercellular scaffold (Fig. 5 B).
In configuration IV (Fig. 2), only spikes of two subunits are interacting with the RS1 domain of another molecule. The angular variability in the latter may also indicate a more flexible connection. This type of branching may not be essential because, to date, no disease-related mutations have been found in the RS1 domain. We also note that any stacking interactions (i.e., expanding the stacking of the two rings) are conspicuously absent. Had stacking occurred, it would have meant a greater involvement of the RS1 domains that protrude from the rings.
Spike 1 is important for higher-order assembly
The 3D map (Fig. 3) of the most abundant conformation (type I) indicates a prominent role for spike 1 in the intermolecular interaction. Because one RS1 molecule is shifted relative to its neighbor (giving rise to the wavy appearance of the strand of molecules), the interaction between the loops is not symmetric and it likely adopts different conformations. Mutagenesis of several residues in this loop results in partial or complete abolishment of secretion (Fig. 4). The W92S, Y93A, and W96R (Plössl et al., 2018) mutations represent a decrease in residue size that could affect the strength of interaction. The G91V mutant is not secreted at all, suggesting a more severe effect on the spike 1 conformation. This is in contrast to the R141H mutant (spike 3) that is well secreted (Fig. 4; Plössl et al., 2018), but still causes disease (Table 1). Essential steps in producing a functional RS1 are synthesis followed by oligomerization and secretion (Wang et al., 2006). Any mutations interfering with forming the octamer also prevent secretion and are associated with disease (Tolun et al., 2016). Our findings indicate that higher-order assembly may already occur intracellularly, and any deficiency in it leads to decreased secretion.
The physiological relevance of higher-order RS1 assemblies
We observed the network of RS1 molecules in high salt and confined to a thin layer of solvent (just before plunge freezing) during preparation for microscopy. Both these features require examination relative to its production in cells and occurrence in the intercellular space of the retina.
We experienced severe problems of aggregation of RS1 when dialyzed from the high-salt elution buffer used for purification. We therefore decided to simply image RS1 in this buffer, discovering that it forms this branched network. We continued trying to establish conditions for its formation with limited supplies of the protein (Fig. S1). We found that calcium leads to massive aggregation, while a chelator such as imidazole or EDTA prevents it. It is obvious that the intermolecular interactions of RS1 are highly dependent on its environment. Unfortunately, understanding the actual physiological environment is complicated by the crowded nature of both intracellular compartments and intercellular space. At the same time, the interactions (particularly type I and II; Fig. 2) are very specific and rigid, suggesting this to be an inherent capability of the molecule and therefore physiologically relevant.
The second aspect of our experiment was its disposition in a thin layer of water, with extensive air–water interfaces. These are considered hydrophobic–hydrophilic interfaces in nature (Trigg et al., 2013; Campioni et al., 2014) and a potential mimic for membrane interfaces. Our micrographs clearly show that RS1 mostly orients such that the spikes of four subunits in each 16-mer are located toward the hydrophobic air–water interface (Fig. 2). Fraternali et al. (2003) suggested from modeling results that RS1 binds to membranes through a hydrophobic patch in the spikes (involving the residues Y89, V90, W92, Y93, F108, and I144). Vijayasarathy et al. (2007) showed that mutating Y89 and W92 to cysteine decreases binding to phosphatidylserine (acknowledging that phosphatidylserine mostly occurs on the inner leaflet; Leventis and Grinstein, 2010). Furthermore, Plössl et al. (2018) found altered membrane interaction for several spike mutants. Together, these observations indicate that the RS1 spikes are probably involved in membrane association.
RS1 may form an intercellular 3D scaffold
We show that RS1 has the potential to self-assemble into a branched network. Protein filaments of various kinds are widely encountered, and they fulfill numerous functions; however, despite their diversity, branching is unusual. The prime example is cortical actin, in which the networks are built up from daughter filaments anchored on Arp2/3 complexes emanating laterally from mother filaments (Pollard and Borisy, 2003; Rouiller et al., 2008). This architecture confers stability to the peripheral cytoskeleton and directs motile forces, coupled to the actin polymerization engine (Theriot, 2000). Similarly, the factor SSNA1 facilitates remodeling of microtubules into branched forms (Basnet et al., 2018). Numerous biophysical studies have investigated how the mechanical properties of a network—in particular, enhanced stability—may be affected by branching (e.g., Pujol et al., 2012; Bieling et al., 2016). There is, as yet, no evidence that RS1 polymerizes into networks in situ, but there is a perceived need to stabilize the boundary regions between retinal cells in order to prevent adverse effects such as those encountered in XLRS. This requirement, viewed in light of the rarity of branching, makes it tempting to speculate that RS1 networks may contribute to the required stability.
Conclusion
Under appropriate conditions (e.g., high salt), RS1 is able to assemble into branched networks. From our 2D averaged images, it follows that the interactions between the RS1 molecules have some specificity, but they also show some flexibility. The one peripheral spike involved in the linear strand interaction (residues 56–98) is also the location where several disease-causing mutations occur. Decreased secretion of such mutants suggests that self-assembly of RS1 already occurs intracellularly. A branched network of RS1 would be capable of bridging the space between adjacent cell membranes and thereby facilitate the adhesive properties attributed to this molecule.
Materials and methods
RS1 cloning, expression, and purification
Cloning of RS1 sequences
Regions from a pTOPO-RS1 vector (Zeng et al., 2004) were amplified, using standard PCR to generate mature RS1 in tandem with a hexahistine tag (aa 24–224-6xHis). The sense (Topo-RS1-F) and antisense (Topo-RS1-R) primers listed in Table S1 also included sequences for Gateway recombination sites and were obtained from Eurofins MWG Operon. PCR was performed using Phusion polymerase (New England Biolabs) under standard conditions with a 30-s/kB extension time for five cycles. After that time, 200 nM of the adapter primer (see Table S1) was added, and amplification was continued for an additional 15 cycles to introduce the honeybee melittin (HBM) leader sequence and Gateway sites. PCR products were cleaned using the QIAQuick PCR purification kit (Qiagen). The final PCR product contains RS1 cDNA with an amino terminal insect HBM secretion signal sequence on the 5′ end and appropriate Gateway recombination sites on both ends. The cleaned PCR products were recombined into pDonr253 (Gateway Donor vector modified from pDonr201; Thermo Fisher Scientific) vector using the Gateway BP recombination reaction according to the manufacturer’s protocols. The subsequent Entry clones (11109-E01 HBM-RS1aa24-224-6xHis;) were sequence verified throughout the entire cloned region.
Subcloning of RS1 sequences
The Entry clone was subcloned by Gateway LR recombination into pDest-8 vector (Thermo Fisher Scientific) to create clone 11109-X01 (RS1-6xHis) for insect cell expression. The expression clones were then transformed into Escherichia coli DH10Bac (Thermo Fisher Scientific) and plated on selective media containing gentamycin, kanamycin, tetracycline, IPTG, and X-gal as per the manufacturer’s protocols. White colonies were selected from these plates, and bacmid DNA was generated by alkaline lysis plasmid preparation and verified by PCR amplification across the bacmid junctions.
Baculovirus production and insect cell expression
Bacmid DNAs were transfected into 1 × 107 Sf9 cells using polyethyleneimine (Thermo Fisher Scientific), and baculovirus supernatants were harvested after incubation at 27°C for 72 h. 1 ml supernatant was transferred to 50 ml High Five cells (107 cells/ml; Thermo Fisher Scientific) and grown at 21°C for 72 h before harvest.
Protein purification
RS1-6xHis and mutant RS1-6xHis secreted into the culture medium were purified by cobalt-agarose affinity chromatography (HisPur Cobalt Resin; Thermo Fisher Scientific). All steps were performed at 4°C. The culture medium was extensively dialyzed against equilibration buffer (20 mM sodium phosphate, pH 7.4, 0.5 M NaCl, and protease inhibitor cocktail) and centrifuged at 10,000 g to remove the cellular debris. Approximately 200 ml dialyzed medium was loaded onto a 10-ml cobalt-agarose column. After a wash step of 10 vol with buffer A (20 mM sodium phosphate, pH 7.4, 0.5 M NaCl, and 20 mM imidazole), RS1-6xHis was eluted with buffer B (20 mM sodium phosphate, pH 7.4, 0.5 M NaCl, and 200 mM imidazole). The fractions were analyzed on 10% SDS-PAGE, and fractions enriched in RS1-6xHis were pooled. For experiments with galactose, we added galactose to a final concentration of 10 mM.
We assessed the aggregation behavior of purified RS1-6xHis by light scattering (optical density) using an HP G1103A (Hewlett Packard) spectrophotometer after dialysis in various buffers, as given in Fig. S1. The optical density was recorded from 200 to 1,000 nm of each sample in a 100-µl quartz cuvette.
RS1 spike mutants
Construction
Human RS1 cDNA in frame with 6xHis tag was subcloned into the pCMV Tag4 vector (Takara Bio USA) using the NotI–XhoI restriction sites. This mammalian expression vector encodes the cytomegalovirus promoter for constitutive expression of the cloned DNA in a wide variety of mammalian cell lines and a C-terminal Flag tag. We silenced the flag epitope and introduced a His tag as an alternative for purification. The sense (RS1-F) and antisense (RS1-R) primers used in the experiment are listed in Table S1. We also introduced a Kozak consensus sequence upstream of the ATG start codon to provide optimal expression of the RS1-His fusion protein. We used the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) to introduce the following mutations: G91V, W92S, Y93A, and R141H. The primers used for mutagenesis and their sequences are listed in Table S1 (Integrated DNA Technologies). All constructs were sequenced to verify the presence of the desired mutations and absence of random mutations. WT and mutant plasmids were isolated from bacterial cell cultures using Maxiprep plasmid purification kit (Qiagen Sciences).
Expression and analysis
We used human embryonic kidney (HEK) 293 cells (ATCC CRL-1573; Graham et al., 1977) as the host for the heterologous expression of RS1-His fusion protein (ATCC). We cultured the cells in T75 culture dishes in 10 ml EMEM supplemented with 10% FBS and transiently transfected them at 80% confluence with plasmid DNAs (30 µg) encoding either WT or mutant RS1 with 60 µl Lipofectamine 2000 reagent (Thermo Fisher Scientific). After 6 h, we replaced the medium containing Lipofectamine with EMEM+10% FBS. At 48 h after transfection, we harvested the cells, rinsed them with PBS, and lysed them in RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, and 1 mM EGTA supplemented with Halt protease inhibitor cocktail; Thermo Fisher Scientific). To detect RS1 protein, we analyzed whole-cell extracts (retained RS1) and media (secreted RS1) by immunoblotting using anti-RS1 antibody. We loaded sample volumes corresponding to 3% of total media or 3% of total cell lysates onto Novex 10% Bis-Tris gels (Thermo Fisher Scientific) in Laemmli sample buffer. The gels were electrophoresed under reducing (with β-mercaptoethanol; Sigma-Aldrich) or nonreducing (no β-mercaptoethanol) conditions to detect monomeric and octameric forms of RS1, respectively (Vijayasarathy et al., 2010). To resolve RS1 double octamers, we used Blue-Native PAGE (Tolun et al., 2016; Thermo Fisher Scientific) for which identical sample volumes were loaded on 3–12% Novex Bis-Tris gel in Native PAGE sample buffer with G-250 sample additive. Following electrophoresis, the gels were electrotransferred to polyvinylidene difluoride membrane (Immobilon P; Bio-Rad) and blocked for 1 h in blocking buffer (LI-COR Biosciences). We incubated the membrane with a rabbit polyclonal RS1 antibody (raised against the N terminus of retinoschisin amino acid residues 24–37; Zeng et al., 2004), diluted in 1% BSA in PBS containing 0.05% Tween 20, at 4°C overnight, followed by IRDye 800CW–conjugated goat anti–rabbit IgG (LI-COR). We scanned the blots on a LI-COR Odyssey Infrared Imaging System (model 9120; LI-COR) and analyzed them using Odyssey software. Data presented are representative of two independent experiments.
EM
We applied 3 µl of a preparation of retinoschisin (∼0.14 mg/ml) with galactose (10 mM final concentration) to 40-s glow-discharged 400 mesh C-flat electron microscopy grids and plunge froze it into liquid ethane in an EM GP instrument (Leica Microsystems). We imaged the grids in a 300-kV Titan Krios microscope (FEI) at the Howard Hughes Medical Institute Janelia Research Campus (Ashburn, VA), which was equipped with a Cs corrector and a post-GIF K2 Summit camera (Gatan) in superresolution mode. The micrographs were taken as series of dose-fractionated images (Table 1) and stored as compressed TIFF files without gain correction. Automated data collection was done at a calibrated magnification of 48,077× in SerialEM (Mastronarde, 2005). We first obtained a data set of zero-tilt micrographs that were subsequently used in 2D image classification. Because of the preferred orientation of RS1 on the microscope grids (edge-on views of the double rings), we also acquired a second set of micrographs with the specimen tilted to 30°.
Image processing
3D processing of 0°-tilt micrographs
We did all processing in Bsoft (Heymann, 2018; see Table 2 in this paper for details). The images were gain-corrected by multiplying with the gain reference (program bop) and binned twofold for a final pixel size of 1.04 Å (program bint). The frames were aligned to yield a shift-compensated average for each micrograph (program bseries). A power spectrum was calculated for each micrograph as the average over tiles of size 512 × 512 (program bctf). The contrast transfer function (CTF) was automatically fitted to the radial power spectrum (program bctf) and checked manually (program bshow). A particle template was generated in bshow from a small set of manually selected particles. This template was used to automatically pick particles in all the micrographs (program bpick). We manually eliminated a large number of bad hits (that were obviously different from the double-ring structure, including ice contamination and odd aggregated densities) from selected particle sets, arriving at the numbers picked as given in Table 2. The CTF was corrected during reconstruction, adjusting for tilted specimens when necessary. We assessed the quality of 3D reconstructions by comparing maps from independently processed subsets by FSC using a shaped mask appropriately low-pass filtered. The particles from the first data set were aligned initially to a 25-Å resolution limit, using the map from Tolun et al. (2016) (EMD-6425; http://www.emdatabank.org). Unfortunately, the 3D reconstruction with D8 symmetry yielded low-quality maps due to the overwhelming preference for one orientation. We therefore decided to explore the interactions between RS1 molecules by 2D classification.
2D classification from 0°-tilt micrographs
We started the 2D alignment using a reference of the 420 particle images extracted from the first micrograph from the first data set (program borient). We selected particles with high correlation, obtaining 420 2D class average images corresponding to the original reference images. We eliminated classes with <10 contributing images and then repeated the alignment. After the fifth iteration, 150 classes remained, representing ∼50% of the total number of boxes. There were several classes with the same appearance. We compared the classes by correlation and clustered them using affinity propagation (Frey and Dueck, 2007) with a high threshold, resulting in 70 final classes (program bmapdist). The resolutions for the 2D averages were calculated at an FSC cutoff of 0.3 using the method of Unser et al. (1987) as implemented in the program borient. We used these class averages as a basis for characterizing the interactions between RS1 molecules.
3D processing of 30°-tilt micrographs
We acquired the second data set at a fixed tilt angle of 30° to obtain more views of the RS1 molecule for 3D reconstruction. As with the first data set, we started the particle alignment with a resolution limit of 25 Å on two independent subsets of the second data set (program borient). After 20 iterations and increasing the resolution limit to 8 Å (final alignment with program brefine), we calculated a D8 symmetrized reconstruction with FSC0.143 = 6.8 Å (size 168 × 168 × 168; 21,364 particle images). Our main goal was to visualize the interactions between RS1 molecules. We therefore generated a “unit cell” reference with C2 symmetry of two RS1 molecules in a slightly larger volume (224 × 224 × 224) placed in positions inferred from the 2D averages from the first data set (Fig. S5 C). We then used the orientations from the previous runs to find the D8 symmetry-equivalent orientations that correspond to the C2 unit cell (program brefine). We further refined these orientations, yielding a final reconstruction with FSC0.143 = 10.0 Å (Table 2 and Fig. S5 D). The quality of this map is lower than the D8 map because of lower symmetry and larger volume.
Building 3D models based on 2D class averages
We matched a projection of the 3D RS1 map (EMDB 6425) to each occurrence in a class average (Fig. 2, A–L) to obtain accurate translations and rotations. We then constructed 3D maps with the RS1 molecules in the correct orientations (Fig. 2, bottom). The assumption is that all the molecules showing side views are interacting with the air–water interface and are therefore in the same plane (supported by the fact that most showed the same view as determined by alignment). The RS1 atomic model (PDB accession no. 3JD6) was then docked as a rigid body into the 3D map obtained from the 30°-tilted data set (Fig. 3). All 3D visualization was done using UCSF Chimera (Pettersen et al., 2004).
Data and software availability
The 3D reconstruction of two RS1 molecules in the main strand configuration (Table 2, last column) has been deposited in the EM Databank (http://www.emdatabank.org), accession no. EMD-7907. The Bsoft package (Heymann, 2018) is freely available at http://bsoft.ws.
Online supplemental material
In Fig. S1, RS1 dialyzed in various buffers and assessed by light scattering massively aggregates in the presence of calcium, but not with the chelators EDTA and imidazole. Fig. S2 shows cryo-electron micrographs of RS1 in buffers from Fig. S1 with chelators, indicating an ability to form higher-order assemblies, even at lower salt concentrations. We observed more single octamer rings in the presence of EDTA (C and D). Fig. S3 displays an example of cryo-electron micrographs with a higher prevalence of linear strands of RS1 in the presence of galactose. However, the effect is modest and varies from experiment to experiment, and we are uncertain whether galactose affects the formation of RS1 networks. Fig. S4 presents 2D class averages (A) of the RS1 strands and branches. These are of high quality, as indicated by a strong relationship between the resolution estimated from the spectral signal-to-noise ratio and the number of images contributing to an average (B). Fig. S5 shows cryo-electron micrographs of RS1 at tilted angles (A and B) to overcome the preferred side orientation of the strands observed in the original micrographs. The main strand conformation (type I) is rigid, allowing us to describe it as a linear or 1D crystal (C). We calculated a 3D map within this unit cell and obtained a reconstruction at a resolution of 10 Å (D). Table S1 lists primers used for cloning RS1 and generating mutants.
Supplementary Material
Acknowledgments
We wish to thank Drs. Zhiheng Yu and Chuan Hong for assistance at the HHMI Janelia CryoEM Facility and Dr. Dennis Winkler for acquiring supplemental micrographs.
This work was supported by the Intramural Research Programs of the National Institutes of Health, National Eye Institute, the National Institute on Deafness and Other Communication Disorders, and the National Institute for Arthritis, Musculoskeletal and Skin Diseases, National Institutes of Health.
The authors declare no competing interests.
Author contributions: Conceptualization: J.B. Heymann, C. Vijayasarathy, P.A. Sieving, and A.C. Steven. Methodology: C. Vijayasarathy, J.B. Heymann, A.D. Dearborn, and R.K. Huang. Investigation: C. Vijayasarathy, A.D. Dearborn, and R.K. Huang. Software and formal analysis: J.B. Heymann. Writing, original draft: J.B. Heymann. Writing, review and editing: J.B. Heymann, C. Vijayasarathy, A.D. Dearborn, R.K. Huang, P.A. Sieving, and A.C. Steven.
References
- Al-Amoudi A., Díez D.C., Betts M.J., and Frangakis A.S.. 2007. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 450:832–837. 10.1038/nature05994 [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A., Castaño-Diez D., Devos D.P., Russell R.B., Johnson G.T., and Frangakis A.S.. 2011. The three-dimensional molecular structure of the desmosomal plaque. Proc. Natl. Acad. Sci. USA. 108:6480–6485. 10.1073/pnas.1019469108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnet N., Nedozralova H., Crevenna A.H., Bodakuntla S., Schlichthaerle T., Taschner M., Cardone G., Janke C., Jungmann R., Magiera M.M., et al. 2018. Direct induction of microtubule branching by microtubule nucleation factor SSNA1. Nat. Cell Biol. 20:1172–1180. 10.1038/s41556-018-0199-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Li T.D., Weichsel J., McGorty R., Jreij P., Huang B., Fletcher D.A., and Mullins R.D.. 2016. Force Feedback Controls Motor Activity and Mechanical Properties of Self-Assembling Branched Actin Networks. Cell. 164:115–127. 10.1016/j.cell.2015.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M., Setiaputra D., Yip C.K., and Molday R.S.. 2016. Cog-Wheel Octameric Structure of RS1, the Discoidin Domain Containing Retinal Protein Associated with X-Linked Retinoschisis. PLoS One. 11:e0147653 10.1371/journal.pone.0147653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush R.A., Wei L.L., and Sieving P.A.. 2015. Convergence of Human Genetics and Animal Studies: Gene Therapy for X-Linked Retinoschisis. Cold Spring Harb. Perspect. Med. 5:a017368 10.1101/cshperspect.a017368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campioni S., Carret G., Jordens S., Nicoud L., Mezzenga R., and Riek R.. 2014. The presence of an air-water interface affects formation and elongation of α-Synuclein fibrils. J. Am. Chem. Soc. 136:2866–2875. 10.1021/ja412105t [DOI] [PubMed] [Google Scholar]
- Dyka F.M., and Molday R.S.. 2007. Coexpression and interaction of wild-type and missense RS1 mutants associated with X-linked retinoschisis: its relevance to gene therapy. Invest. Ophthalmol. Vis. Sci. 48:2491–2497. 10.1167/iovs.06-1465 [DOI] [PubMed] [Google Scholar]
- Dyka F.M., Wu W.W., Pfeifer T.A., Molday L.L., Grigliatti T.A., and Molday R.S.. 2008. Characterization and purification of the discoidin domain-containing protein retinoschisin and its interaction with galactose. Biochemistry. 47:9098–9106. 10.1021/bi800938g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraternali F., Cavallo L., and Musco G.. 2003. Effects of pathological mutations on the stability of a conserved amino acid triad in retinoschisin. FEBS Lett. 544:21–26. 10.1016/S0014-5793(03)00433-2 [DOI] [PubMed] [Google Scholar]
- Frey B.J., and Dueck D.. 2007. Clustering by passing messages between data points. Science. 315:972–976. 10.1126/science.1136800 [DOI] [PubMed] [Google Scholar]
- Friedrich U., Stöhr H., Hilfinger D., Loenhardt T., Schachner M., Langmann T., and Weber B.H.. 2011. The Na/K-ATPase is obligatory for membrane anchorage of retinoschisin, the protein involved in the pathogenesis of X-linked juvenile retinoschisis. Hum. Mol. Genet. 20:1132–1142. 10.1093/hmg/ddq557 [DOI] [PubMed] [Google Scholar]
- Gehrig A., White K., Lorenz B., Andrassi M., Clemens S., and Weber B.H.. 1999. Assessment of RS1 in X-linked juvenile retinoschisis and sporadic senile retinoschisis. Clin. Genet. 55:461–465. 10.1034/j.1399-0004.1999.550611.x [DOI] [PubMed] [Google Scholar]
- Graham F.L., Smiley J., Russell W.C., and Nairn R.. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74. 10.1099/0022-1317-36-1-59 [DOI] [PubMed] [Google Scholar]
- Haseloff R.F., Dithmer S., Winkler L., Wolburg H., and Blasig I.E.. 2015. Transmembrane proteins of the tight junctions at the blood-brain barrier: structural and functional aspects. Semin. Cell Dev. Biol. 38:16–25. 10.1016/j.semcdb.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Omoto S., Takeuchi T., Kozaki K., Ueoka Y., and Kitahara K.. 2004. Four Japanese male patients with juvenile retinoschisis: only three have mutations in the RS1 gene. Am. J. Ophthalmol. 138:788–798. 10.1016/j.ajo.2004.06.031 [DOI] [PubMed] [Google Scholar]
- Heymann J.B. 2018. Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 27:159–171. 10.1002/pro.3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiriyanna K.T., Bingham E.L., Yashar B.M., Ayyagari R., Fishman G., Small K.W., Weinberg D.V., Weleber R.G., Lewis R.A., Andreasson S., et al. 1999. Novel mutations in XLRS1 causing retinoschisis, including first evidence of putative leader sequence change. Hum. Mutat. 14:423–427. [DOI] [PubMed] [Google Scholar]
- Hu Q.R., Huang L.Z., Chen X.L., Xia H.K., Li T.Q., and Li X.X.. 2017. Genetic analysis and clinical features of X-linked retinoschisis in Chinese patients. Sci. Rep. 7:44060 10.1038/srep44060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedzierska A., Smietana K., Czepczynska H., and Otlewski J.. 2007. Structural similarities and functional diversity of eukaryotic discoidin-like domains. Biochim. Biophys. Acta. 1774:1069–1078. 10.1016/j.bbapap.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Kotova S., Vijayasarathy C., Dimitriadis E.K., Ikonomou L., Jaffe H., and Sieving P.A.. 2010. Retinoschisin (RS1) interacts with negatively charged lipid bilayers in the presence of Ca2+: an atomic force microscopy study. Biochemistry. 49:7023–7032. 10.1021/bi1007029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis P.A., and Grinstein S.. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39:407–427. 10.1146/annurev.biophys.093008.131234 [DOI] [PubMed] [Google Scholar]
- Ma X., Li X., and Wang L.. 2008. Novel XLRS1 gene mutations cause X-linked juvenile retinoschisis in Chinese families. Jpn. J. Ophthalmol. 52:48–51. 10.1007/s10384-007-0488-4 [DOI] [PubMed] [Google Scholar]
- Mashima Y., Shinoda K., Ishida S., Ozawa Y., Kudoh J., Iwata T., Oguchi Y., and Shimizu N.. 1999. Identification of four novel mutations of the XLRS1 gene in Japanese patients with X-linked juvenile retinoschisis. Mutation in brief no. 234. Online. Hum. Mutat. 13:338 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Molday R.S. 2007. Focus on molecules: retinoschisin (RS1). Exp. Eye Res. 84:227–228. 10.1016/j.exer.2005.12.013 [DOI] [PubMed] [Google Scholar]
- Molday L.L., Wu W.W., and Molday R.S.. 2007. Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J. Biol. Chem. 282:32792–32801. 10.1074/jbc.M706321200 [DOI] [PubMed] [Google Scholar]
- Molday R.S., Kellner U., and Weber B.H.. 2012. X-linked juvenile retinoschisis: clinical diagnosis, genetic analysis, and molecular mechanisms. Prog. Retin. Eye Res. 31:195–212. 10.1016/j.preteyeres.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Ott S.H., Wang X., Appukuttan B., Patel R.J., Van Boemel G.B., and Stout J.T.. 2000. Clinical phenotype associated with the arg141 his mutation in the X-linked retinoschisis gene. Arch. Ophthalmol. 118:127–129. 10.1001/archopht.118.1.127 [DOI] [PubMed] [Google Scholar]
- Park T.K., Wu Z., Kjellstrom S., Zeng Y., Bush R.A., Sieving P.A., and Colosi P.. 2009. Intravitreal delivery of AAV8 retinoschisin results in cell type-specific gene expression and retinal rescue in the Rs1-KO mouse. Gene Ther. 16:916–926. 10.1038/gt.2009.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Plössl K., Royer M., Bernklau S., Tavraz N.N., Friedrich T., Wild J., Weber B.H.F., and Friedrich U.. 2017. Retinoschisin is linked to retinal Na/K-ATPase signaling and localization. Mol. Biol. Cell. 28:2178–2189. 10.1091/mbc.e17-01-0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plössl K., Schmid V., Straub K., Schmid C., Ammon M., Merkl R., Weber B.H.F., and Friedrich U.. 2018. Pathomechanism of mutated and secreted retinoschisin in X-linked juvenile retinoschisis. Exp. Eye Res. 177:23–34. 10.1016/j.exer.2018.07.021 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Pujol T., du Roure O., Fermigier M., and Heuvingh J.. 2012. Impact of branching on the elasticity of actin networks. Proc. Natl. Acad. Sci. USA. 109:10364–10369. 10.1073/pnas.1121238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay E.P., Collins R.F., Owens T.W., Siebert C.A., Jones R.P.O., Wang T., Roseman A.M., and Baldock C.. 2016. Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function. Hum. Mol. Genet. 25:5311–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiller I., Xu X.P., Amann K.J., Egile C., Nickell S., Nicastro D., Li R., Pollard T.D., Volkmann N., and Hanein D.. 2008. The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180:887–895. 10.1083/jcb.200709092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer C.G., Gehrig A., Warneke-Wittstock R., Marquardt A., Ewing C.C., Gibson A., Lorenz B., Jurklies B., and Weber B.H.. 1997. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat. Genet. 17:164–170. 10.1038/ng1097-164 [DOI] [PubMed] [Google Scholar]
- Shi L., Jian K., Ko M.L., Trump D., and Ko G.Y.. 2009. Retinoschisin, a new binding partner for L-type voltage-gated calcium channels in the retina. J. Biol. Chem. 284:3966–3975. 10.1074/jbc.M806333200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorczyk A., and Krawczyński M.R.. 2012. Four novel RS1 gene mutations in Polish patients with X-linked juvenile retinoschisis. Mol. Vis. 18:3004–3012. [PMC free article] [PubMed] [Google Scholar]
- Sosinsky G.E., and Nicholson B.J.. 2005. Structural organization of gap junction channels. Biochim. Biophys. Acta. 1711:99–125. 10.1016/j.bbamem.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Steiner-Champliaud M.F., Sahel J., and Hicks D.. 2006. Retinoschisin forms a multi-molecular complex with extracellular matrix and cytoplasmic proteins: interactions with beta2 laminin and alphaB-crystallin. Mol. Vis. 12:892–901. [PubMed] [Google Scholar]
- The Retinoschisis Consortium 1998. Functional implications of the spectrum of mutations found in 234 cases with X-linked juvenile retinoschisis. Hum. Mol. Genet. 7:1185–1192. 10.1093/hmg/7.7.1185 [DOI] [PubMed] [Google Scholar]
- Theriot J.A. 2000. The polymerization motor. Traffic. 1:19–28. 10.1034/j.1600-0854.2000.010104.x [DOI] [PubMed] [Google Scholar]
- Tolun G., Vijayasarathy C., Huang R., Zeng Y., Li Y., Steven A.C., Sieving P.A., and Heymann J.B.. 2016. Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina. Proc. Natl. Acad. Sci. USA. 113:5287–5292. 10.1073/pnas.1519048113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg B.J., Lee C.F., Vaux D.J., and Jean L.. 2013. The air-water interface determines the outcome of seeding during amyloidogenesis. Biochem. J. 456:67–80. 10.1042/BJ20130605 [DOI] [PubMed] [Google Scholar]
- Unser M., Trus B.L., and Steven A.C.. 1987. A new resolution criterion based on spectral signal-to-noise ratios. Ultramicroscopy. 23:39–51. 10.1016/0304-3991(87)90225-7 [DOI] [PubMed] [Google Scholar]
- Verbakel S.K., van de Ven J.P., Le Blanc L.M., Groenewoud J.M., de Jong E.K., Klevering B.J., and Hoyng C.B.. 2016. Carbonic Anhydrase Inhibitors for the Treatment of Cystic Macular Lesions in Children With X-Linked Juvenile Retinoschisis. Invest. Ophthalmol. Vis. Sci. 57:5143–5147. 10.1167/iovs.16-20078 [DOI] [PubMed] [Google Scholar]
- Vijayasarathy C., Takada Y., Zeng Y., Bush R.A., and Sieving P.A.. 2007. Retinoschisin is a peripheral membrane protein with affinity for anionic phospholipids and affected by divalent cations. Invest. Ophthalmol. Vis. Sci. 48:991–1000. 10.1167/iovs.06-0915 [DOI] [PubMed] [Google Scholar]
- Vijayasarathy C., Sui R., Zeng Y., Yang G., Xu F., Caruso R.C., Lewis R.A., Ziccardi L., and Sieving P.A.. 2010. Molecular mechanisms leading to null-protein product from retinoschisin (RS1) signal-sequence mutants in X-linked retinoschisis (XLRS) disease. Hum. Mutat. 31:1251–1260. 10.1002/humu.21350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia S., Fishman G.A., Molday R.S., Dyka F.M., Kumar N.M., Ehlinger M.A., and Stone E.M.. 2009. Relation of response to treatment with dorzolamide in X-linked retinoschisis to the mechanism of functional loss in retinoschisin. Am. J. Ophthalmol. 147:111–115.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Waters C.T., Rothman A.M., Jakins T.J., Römisch K., and Trump D.. 2002. Intracellular retention of mutant retinoschisin is the pathological mechanism underlying X-linked retinoschisis. Hum. Mol. Genet. 11:3097–3105. 10.1093/hmg/11.24.3097 [DOI] [PubMed] [Google Scholar]
- Wang T., Zhou A., Waters C.T., O’Connor E., Read R.J., and Trump D.. 2006. Molecular pathology of X linked retinoschisis: mutations interfere with retinoschisin secretion and oligomerisation. Br. J. Ophthalmol. 90:81–86. 10.1136/bjo.2005.078048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B.H., Schrewe H., Molday L.L., Gehrig A., White K.L., Seeliger M.W., Jaissle G.B., Friedburg C., Tamm E., and Molday R.S.. 2002. Inactivation of the murine X-linked juvenile retinoschisis gene, Rs1h, suggests a role of retinoschisin in retinal cell layer organization and synaptic structure. Proc. Natl. Acad. Sci. USA. 99:6222–6227. 10.1073/pnas.092528599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Takada Y., Kjellstrom S., Hiriyanna K., Tanikawa A., Wawrousek E., Smaoui N., Caruso R., Bush R.A., and Sieving P.A.. 2004. RS-1 Gene Delivery to an Adult Rs1h Knockout Mouse Model Restores ERG b-Wave with Reversal of the Electronegative Waveform of X-Linked Retinoschisis. Invest. Ophthalmol. Vis. Sci. 45:3279–3285. 10.1167/iovs.04-0576 [DOI] [PubMed] [Google Scholar]
- Zhour A., Bolz S., Grimm C., Willmann G., Schatz A., Weber B.H., Zrenner E., and Fischer M.D.. 2012. In vivo imaging reveals novel aspects of retinal disease progression in Rs1h(-/Y) mice but no therapeutic effect of carbonic anhydrase inhibition. Vet. Ophthalmol. 15(Suppl 2):123–133. 10.1111/j.1463-5224.2012.01039.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3D reconstruction of two RS1 molecules in the main strand configuration (Table 2, last column) has been deposited in the EM Databank (http://www.emdatabank.org), accession no. EMD-7907. The Bsoft package (Heymann, 2018) is freely available at http://bsoft.ws.