Menin and glucagon-like peptide 1 (GLP-1) pathways play central yet opposing roles in regulating β cell function, with menin suppressing and GLP-1 promoting β cell function. Xing et al show that GLP-1 induces PKA-mediated phosphorylation of menin. Phosphorylation of menin triggers its interaction with nuclear actin to relieve menin’s transcriptional repression and increase insulin production.
Abstract
Both menin and glucagon-like peptide 1 (GLP-1) pathways play central yet opposing role in regulating β cell function, with menin suppressing, and GLP-1 promoting, β cell function. However, little is known as to whether or how GLP-1 pathway represses menin function. Here, we show that GLP-1 signaling–activated protein kinase A (PKA) directly phosphorylates menin at the serine 487 residue, relieving menin-mediated suppression of insulin expression and cell proliferation. Mechanistically, Ser487-phosphorylated menin gains increased binding affinity to nuclear actin/myosin IIa proteins and gets sequestrated from the Ins1 promoter. This event leads to reduced binding of repressive epigenetic histone modifiers suppressor variegation 3–9 homologue protein 1 (SUV39H1) and histone deacetylases 1 (HDAC1) at the locus and subsequently increased Ins1 gene transcription. Ser487 phosphorylation of menin also increases expression of proproliferative cyclin D2 and β cell proliferation. Our results have uncovered a previously unappreciated physiological link in which GLP-1 signaling suppresses menin function through phosphorylation-triggered and actin/myosin cytoskeletal protein–mediated derepression of gene transcription.
Introduction
Menin, encoded by the MEN1 gene (Men1 in mice), which is mutated in human multiple neoplasia type 1 (MEN1) syndrome, is mainly a nuclear protein (Chandrasekharappa et al., 1997; Thakker, 2010). Based on functional and x-ray crystallography studies, menin acts as a scaffold protein by interacting with various epigenetic regulators (Karnik et al., 2005; Murai et al., 2011; Huang et al., 2012). Menin represses gene transcription by interacting with epigenetic regulators, including histone deacetylases (HDACs; Agarwal et al., 1999; Gobl et al., 1999; Kim et al., 2003) or histone H3K9 methyltransferase-like suppressor variegation 3–9 homologue protein 1 (SUV39H1; Feng et al., 2017). Our previous studies have shown that menin is a prodiabetic factor, as ablation of the Men1 gene reverses preexisting hyperglycemia in diabetes and prevents development of diabetes in streptozotocin-treated mice (Yang et al., 2010a,b). Moreover, ectopic expression of menin in cultured β cells leads to reduction of insulin expression (Sayo et al., 2002). Numerous attempts have been made to understand whether posttranslational modifications influence menin function in regulating β cells, and multiple phosphorylation sites have been reported in menin protein (MacConaill et al., 2006; Francis et al., 2011). However, none of these phosphorylation sites has been shown crucial for regulating menin function.
Glucagon-like peptide 1 (GLP-1) is a cleaved peptide processed from a precursor encoded by the glucagon gene in intestinal L cells. GLP-1 binds to its cell surface receptors, generating second-messenger cAMP and thus activating protein kinase A (PKA) and cAMP-regulated guanine nucleotide exchange factor II (Epac2; Drucker and Rosen, 2011). GLP-1 has pleiotropic functions, including upregulation of β cell proliferation and insulin secretion (Stoffers et al., 2000; Buteau et al., 2003; De León et al., 2006; Yusta et al., 2006), acting as a major player in regulating islet function and a key target of therapy for type 2 diabetes.
While it is well documented that both the menin and GLP-1 pathways play a central yet opposite role in regulating β cell function and islet mass, little is known as to whether GLP-1 signaling has any influence on menin. In current studies, we investigated the interplay between these two pathways in regulating insulin expression, and the underlying mechanism in this process was also elucidated.
Results
GLP-1 signaling induces phosphorylation of menin at the Ser487 residue through PKA
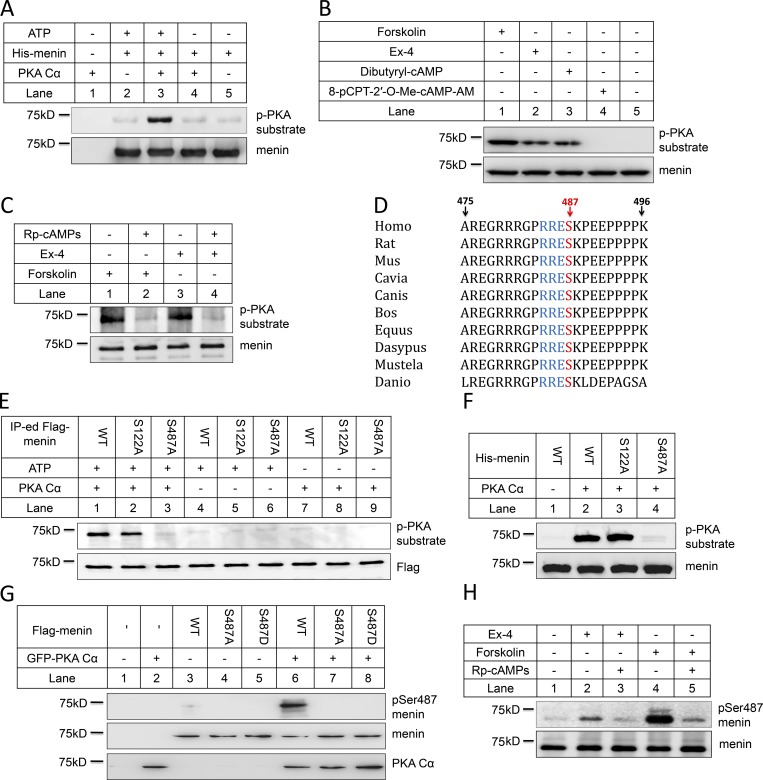
As both GLP-1 and menin are crucial regulators of the β cell function and GLP-1 signaling increases PKA activity, we determined whether PKA interacted with menin and thus affected its function. We expressed PKA (PKA Cα) and menin in HEK293 cells, followed by coimmunoprecipitation (coIP). The results indicated that ectopically expressed menin bound to PKA Cα (Fig. S1 A). In vitro pull-down assay using purified menin and PKA Cα showed that menin and PKA directly interacted with each other (Fig. S1 B). Consistently, interaction between endogenous menin and PKA Cα was also confirmed in mouse embryonic fibroblasts (MEFs; Fig. S1 C) and INS-1 cells (Fig. S1 D). These findings prompted us to examine whether PKA Cα could directly phosphorylate menin. We therefore used purified PKA Cα and full-length recombinant menin proteins to perform in vitro kinase assay. Proteins in various reactions were immunoblotted with a well-characterized phospho-(Ser/Thr) PKA substrate-specific antibody, which was designed to detect peptides and proteins containing a phospho-(Ser/Thr) residue. Indeed, our results showed that PKA Cα directly phosphorylated menin in vitro (Fig. 1 A). Given the well-established notion that GLP-1 signals through cAMP and eventually activates PKA, we investigated whether GLP-1 signaling enhanced menin phosphorylation inside β cells. Serum-starved INS-1 cells were treated with Exendin-4 (Ex-4), a potent GLP-1 analogue. Menin was immunoprecipitated and detected with the phospho-(Ser/Thr) PKA substrate-specific antibody. The results showed that Ex-4 treatment induced menin phosphorylation as a substrate of PKA (Fig. 1 B). Consistently, forskolin, a potent adenylate cyclase activator, also substantially increased menin phosphorylation (Fig. 1 B). To determine the possibility that this phosphorylation might be induced by the Epac2 pathway, which is also activated by cAMP, INS-1 cells were treated with either dibutyryl-cAMP, an analogue of cAMP that stimulates PKA, or 8-pCPT-2′-O-Me-cAMP-AM, a highly membrane-permeable analogue of cAMP in INS-1 cells that is specific for Epac2, but not PKA, activation (Chepurny et al., 2009). Treatment with dibutyryl-cAMP, but not 8-pCPT-2′-O-Me-cAMP-AM, led to menin phosphorylation (Fig. 1 B). Furthermore, Rp-cAMPS, a cell-permeable inhibitor of PKA, abolished forskolin- and Ex-4–induced menin phosphorylation (Fig. 1 C). PKA-mediated menin phosphorylation was also confirmed using treatment with H-89 (Fig. S2 A), another membrane-permeable inhibitor of PKA kinase activity (Schwede et al., 2015).
Figure 1.
Ex-4 induces phosphorylation of menin at the Ser487 residue through PKA. (A) Purified recombinant His-menin proteins were incubated with active PKA Cα in the presence of ATP. After SDS-PAGE, the phosphorylation was tested with anti-phospho-(Ser/Thr) PKA substrate antibody. Anti-menin antibody showed that equal amount of menin was used for the kinase assay. (B) After overnight starvation, INS-1 cells were treated with 10 nM Ex-4, 10 µM forskolin, 100 µM dibutyryl-cAMP, or 10 µM 8-pCPT-2′-O-Me-cAMP-AM for 30 min and then lysed for immunoprecipitation (IP) with anti-menin antibody. Proteins retained on sepharose were blotted with the indicated antibodies. (C) 2 h before forskolin or Ex-4 treatment, 2 µM Rp-cAMPS was added to the starvation medium, and the same experiment described in B was conducted. (D) The Ser487 residue of menin is highly conserved among species. (E) HEK293 cells were transfected with full-length Flag-menin WT, S122A, or S487A mutant, followed by IP using anti-Flag antibody. Proteins retained on sepharose were incubated with active PKA Cα for the kinase assay and then blotted with anti-phospho-(Ser/Thr) PKA substrate antibody. (F) Purified His-menin proteins (WT, S122A, or S487A mutant) were incubated with active PKA Cα in the presence of ATP. After SDS-PAGE, the phosphorylation was tested with anti-phospho-(Ser/Thr) PKA substrate antibody. (G) HEK293 cells cotransfected with Flag-menin (WT or S487A or S487D mutant) and GFP-PKA Cα were lysed and subjected to Western blot using the indicated antibodies. (H) INS-1 cells, treated with 10 nM Ex-4, 10 µM forskolin for 30 min after overnight starvation, were lysed for IP with anti-menin antibody, followed by Western blot with the indicated antibodies. 2 µM Rp-cAMPS was added to the starvation medium 2 h before treatment, if applicable.
PKA phosphorylates proteins that have the motif R/K-R/K-X-S/T exposed on the surface. Sequence analysis of human menin highlights two potential phosphorylation sites: KKVS122 and RRES487. Among them, the Ser487 residue is highly conserved (Fig. 1 D). To determine the exact site phosphorylated by PKA in menin, we generated S122A menin and S487A mutants (which mimic the unphosphorylated form) in constructs expressing Flag-tagged or His-tagged menin. Flag-tagged menin were transfected into HEK293 cells and immunoprecipitated for in vitro kinase assay. Although PKA Cα still induced menin phosphorylation when Ser122 was mutated to alanine, menin phosphorylation was completely abolished in S487A menin mutant (Fig. 1 E). Kinase assay using purified His-menin proteins also showed that S487A, but not S122A mutant, blocked PKA-induced menin phosphorylation (Fig. 1 F), demonstrating that Ser487 is the sole in vitro PKA phosphorylation site.
As the antibody against pan-phospho R/K-R/K-X-S/T site also recognizes many other PKA-phosphorylated proteins, we generated pSer487-menin monoclonal antibody by immunizing mice with the pSer487-containing peptide. To evaluate the specificity of the pSer487 menin antibody, HEK293 cells were transfected with Flag-menin and GFP-PKA Cα, and we found that the antibody only detected phosphorylated menin from the cells cotransfected with WT menin and PKA Cα (Fig. 1 G). Samples from in vitro kinase assay, performed with purified PKA Cα and recombinant His-menin proteins, were also detected with pSer487-menin antibody, showing that the antibody specifically recognized His-menin phosphorylated by PKA Cα in the presence of ATP (Fig. S2 B). To determine whether physiologically or pharmacologically induced menin phosphorylation could also be recognized by our pSer487-menin antibody, we treated INS-1 cells with Ex-4, forskolin, dibutyryl-cAMP, or 8-pCPT-2′-O-Me-cAMP-AM and found that this antibody detected menin phosphorylation induced by Ex-4, forskolin, or dibutyryl-cAMP, but not 8-pCPT-2′-O-Me-cAMP-AM (Figs. 1 H and S2 C). As expected, the phosphorylation was abolished by Rp-cAMPS (Fig. 1 H).
It was previously reported that the Ser/Thr kinase inhibitor Ro-31-8220 blocked the action of Ex-4 to stimulate insulin gene promoter activity via the cAMP response element in INS-1 cells (Chepurny et al., 2002), we therefore investigated whether this PKA-independent mechanism of Ex-4 action was also involved in menin phosphorylation by testing Ro-31-8220 at various concentrations in Ex-4–treated INS-1 cells. The results indicated that Ro-31-8220 could not block Ex-4–induced menin phosphorylation at lower concentrations (0.1 µM, 0.5 µM, or 1.0 µM), but it decreased menin phosphorylation by ∼39% at 10 µM (Fig. S2, D and E). Since Ro-31-8220 can also inhibit PKA activity (IC50 = 900 nM; Davis et al., 1992), we reasoned that the decreased menin phosphorylation was due to the inhibition of PKA activity by Ro-31-8220 at the high concentration. Taken together, our data suggested that PKA primarily mediated the action of Ex-4 in menin phosphorylation.
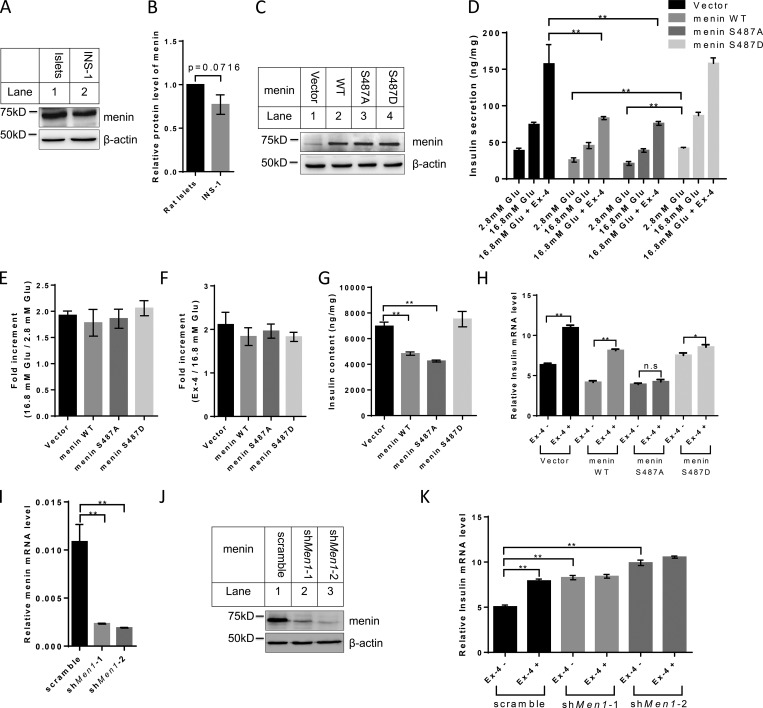
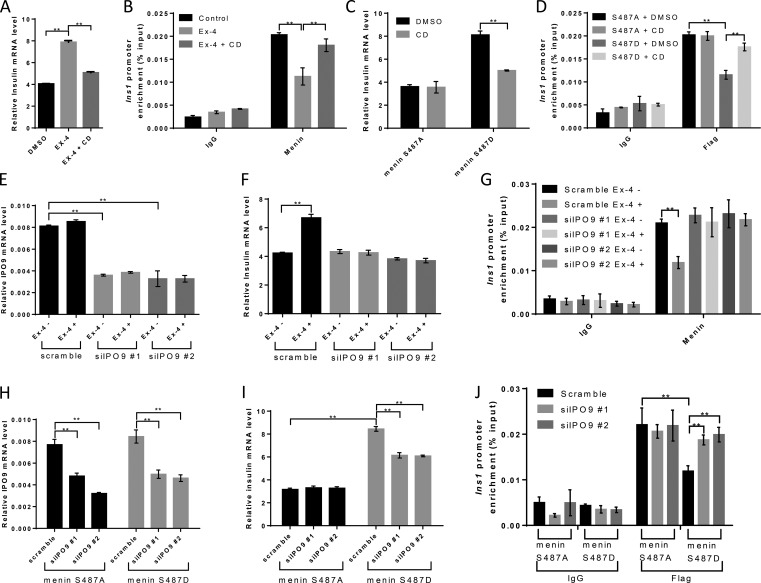
Ser487 phosphorylation of menin influences the synthesis of insulin, but not the fold induction of insulin secretion by Ex-4, by regulating insulin gene transcription
We sought to determine whether mutation of the phosphorylation acceptor site Ser487 to aspartate (D), a residue that mimics negatively charged phospho-group, or alanine (A), which is resistant to phosphorylation, has any effect on menin function. To this end, we stably transduced INS-1 cells, which expressed an amount of menin comparable to that in primary rat islets (Fig. 2, A and B), with empty vector–, WT-, S487A-, or S487D menin–expressing retroviruses, and the expression level of menin was confirmed by Western blot (Fig. 2 C). These various types of cells were treated for a short time with Ex-4, and supernatants of the culture were collected. We found that ectopic expression of WT or S487A menin reduced the amount of secreted insulin in the supernatants, while S487D mutant failed to do so (Fig. 2 D). It is worth noting that Ex-4 potentiated glucose stimulated insulin secretion similarly in terms of the fold induction in all four cell lines (Fig. 2, E and F), suggesting that Ser487 phosphorylation of menin regulated the amount of insulin content rather than the secretory capacity of β cells. Indeed, insulin content from the total cell lysates was reduced in INS-1 cells expressing WT or S487A menin as compared with that of vector- or S487D menin–expressing cells (Fig. 2 G).
Figure 2.
Ser487 phosphorylation of menin affects Ex-4–induced insulin secretion by regulating its transcription. (A) Cell lysate from rat islets and INS-1 cells was examined for menin expression by Western blot. (B) Band density analysis was executed with ImageJ. Menin protein levels were quantified and normalized to β-actin (n = 3). (C) INS-1 cells transduced with control retroviral vector or retroviruses expressing menin WT and Ser487 mutants were examined for menin expression by Western blot. (D–F) Insulin secretion was stimulated by incubation of INS-1 cells with or without 10 nM Ex-4 at 16.8 mM glucose for 30 min and then determined using an insulin-ultrasensitive ELISA kit. Values were normalized to total protein (D), and fold increment was calculated (E and F; n = 3). (G) Whole-cell protein of INS-1 cells was extracted and insulin content was determined using an insulin-ultrasensitive ELISA kit. Insulin levels were normalized to total cellular protein content (n = 3). (H) After overnight starvation, indicated INS-1 cell lines were treated with or without 10 nM Ex-4 for 12 h, followed by quantitative real-time PCR analysis of insulin mRNA level (n = 3). Data were normalized to the level of GAPDH. n.s, no significance. (I and J) Menin mRNA (I) and protein (J) levels were determined in menin knockdown INS-1 cells (n = 3). (K) After overnight starvation, menin-knockdown INS-1 cells were treated with 10 nM Ex-4 for 12 h, followed by quantitative real-time PCR analysis of insulin mRNA level (n = 3). Statistics in B and I: two-tailed t tests. Statistics in D–H and K: one-way ANOVA with least significant difference (LSD) post hoc test. *, P < 0.05; **, P < 0.01. Error bars indicate SD.
We then performed RNA-sequencing analysis and found that the expression of Ins1 gene was 3.7 times higher in S487D menin cells than in S487A menin cells. Insulin mRNA level was further confirmed with quantitative real-time PCR in all four cell lines using primers designed to Ins1 gene (Fig. 2 H). Long-term treatment with Ex-4 showed that vector or WT menin-transduced INS-1 cells were sensitive to Ex-4 in terms of induction of insulin transcription (mRNA level). On the other hand, Ex-4 treatment only slightly increased the insulin mRNA level in S487D menin cells while having no effect in S487A menin cells (Fig. 2 H). Since transfected INS-1 cells expressing recombinant menin might work as a nonphysiological context, we stably knocked down menin in INS-1 cells (Fig. 2, I and J) and found that knockdown of menin increased insulin expression, which was consistent with previous report that menin inhibited insulin transcription in rat insulinoma cells (Sayo et al., 2002). However, the knockdown cells no longer responded to Ex-4–induced insulin transcription (Fig. 2 K), suggesting that endogenous menin mediated the action of Ex-4.
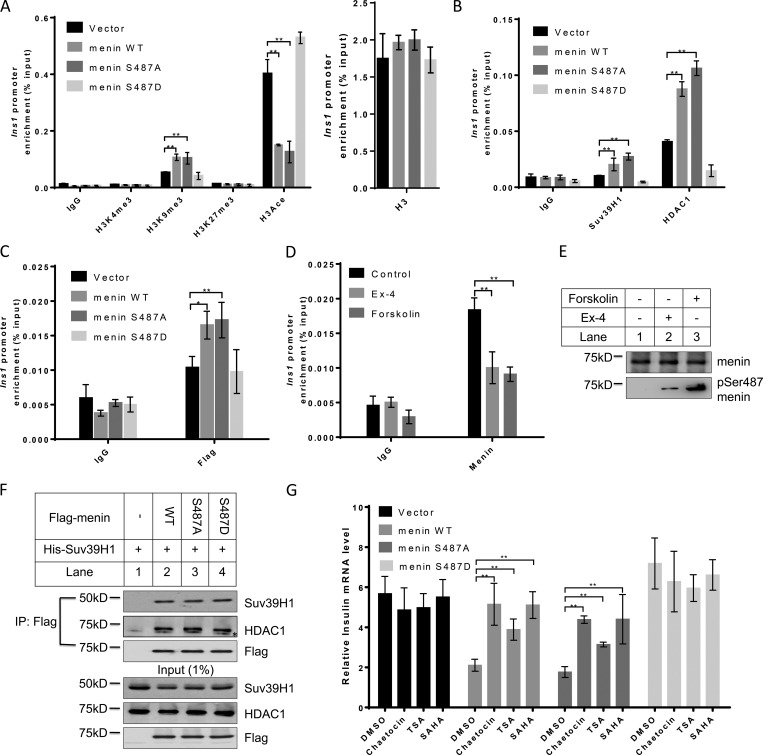
Ser487 phosphorylation of menin epigenetically promotes insulin transcription by attenuating SUV39H1- and HDAC1-mediated repression
We next investigated how menin phosphorylation might affect Ins1 transcription. As menin interacts with various epigenetic regulators that catalyze histone modifications, we examined the trimethylation levels of H3K4, H3K9, and H3K27, as well as acetylation level of histone H3 at the Ins1 promoter, the four established histone marks involving transcriptional regulation. Using a chromatin immunoprecipitation (ChIP) assay, we found that the level of H3K9 trimethylation, a transcription repression–associated histone marker, was significantly increased at the Ins1 locus in menin WT- or S487A-overexpressing cells as compared with that in vector- or S487D menin–overexpressing cells; however, H3K4 and H3K27 trimethylation levels showed no substantial changes. We also detected a profoundly decreased level of histone H3 acetylation in menin WT- or S487A-overexpressing cells (Fig. 3 A). Together, these data indicated that menin phosphorylation at Ser487 promoted Ins1 transcription, at least partially, through H3K9 trimethylation and H3 acetylation.
Figure 3.
Ser487 phosphorylation of menin epigenetically promotes insulin transcription by attenuating SUV39H1- and HDAC1-mediated repression. (A and B) ChIP assays were performed using anti-H3K4me3, H3K9me3, H3K27me3, H3 acetylation (A), Suv39H1 or HDAC1 (B) antibodies in INS-1 cells expressing vector, menin WT, or S487A or S487D mutant. IgG served as a negative control (n = 3). (C) ChIP assays for detecting menin at the Ins1 promoter, using anti-Flag antibody, in INS-1 cells expressing vector, menin WT, or S487A or S487D mutant. IgG served as a negative control (n = 3). (D and E) After overnight starvation, INS-1 cells were treated with 10 µM forskolin or 10 nM Ex-4 for 30 min, and then ChIP assays were performed to detect menin at the Ins1 promoter. IgG served as a negative control (n = 4; D). During the ChIP assay, just before the protein–DNA complexes were eluted, part of the agarose was taken out from each group at equal amount for Western blot using the indicated antibodies (E). (F) HEK293 cells were cotransfected with Flag-menin and His-SUV39H1, followed by IP and Western blot with the indicated antibodies. Asterisk (*) indicates the nonspecific binding. (G) Indicated INS-1 cell lines were treated 20 nM Chaetocin, 0.5 µM trichostatin A (TSA), or 1 µM SAHA for 24 h, followed by quantitative real-time PCR analysis of insulin mRNA levels (n = 5). Data were normalized to the level of GAPDH. All statistics: one-way ANOVA with LSD post hoc test. *, P < 0.05; **, P < 0.01. Error bars indicate SD.
We then explored whether SUV39H1 and HADC1 bound to the Ins1 promoter, and if so, whether S487A menin or S487D mutant affected their association with the promoter, as menin interacted with these factors in certain non-β cells (Yang et al., 2013). As compared with vector control cells, INS-1 cells expressing WT menin or S487A mutant showed increased binding of SUV39H1 and HDAC1 to the Ins1 locus. Notably, phospho-mimicking S487D mutant led to substantial reduction of these transcription repressors at the locus (Fig. 3 B), consistent with the epigenetic histone marker changes at the Ins1 locus shown in Fig. 3 A.
We reasoned that S487D mutant might fail to reach the Ins1 locus or/and fail to recruit epigenetic repressors to the locus. ChIP results showed that, unlike WT menin and S487A mutant, which bound to the Ins1 locus, S487D mutant was not detectible at the locus (Fig. 3 C). Furthermore, treatment of INS-1 cells with Ex-4 or forskolin, which induced Ser487 menin phosphorylation, also led to significant reduction of menin binding to the Ins1 locus (Fig. 3, D and E). Meanwhile, coIP conducted in HEK293 cells overexpressing Flag-menin and His-SUV39H1 indicated that Ser487 phosphorylation of menin did not affect its interaction with SUV39H1 and HDAC1 (Fig. 3 F). INS-1 cells were then treated with the SUV39H1 inhibitor Chaetocin or the pan-HDAC inhibitors trichostatin A and suberoylanilidehydroxamic acid (SAHA). The results showed that the three inhibitors each had little impact on Ins1 mRNA level in the control cells (Fig. 3 G). As indicated earlier (Fig. 2 H), ectopic expression of WT menin or S487A mutant led to marked reduction of Ins1 mRNA levels, and this effect was repressed by each of the three inhibitors. Consistently, in S487D menin mutant–expressing cells, the Ins1 mRNA level was not increased by treatment with any of the three inhibitors (Fig. 3 G). These results indicated that Ser487 phosphorylation of menin epigenetically promoted insulin transcription through SUV39H1 and HDAC1.
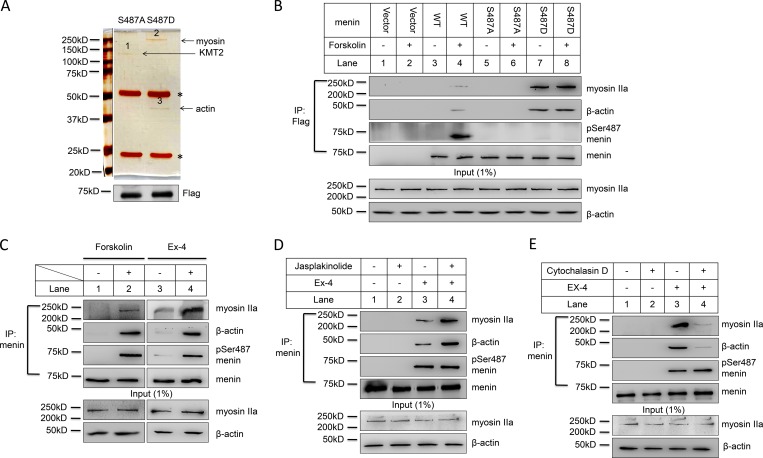
Menin Ser487 phosphorylation leads to enhanced binding to nuclear actin/myosin proteins
How does phosphorylation of menin at Ser487 lead to reduced binding to the loci of its target genes? According to our results, Ser487 phosphorylated menin did not substantially translocate out to the cytoplasm in MEF or forskolin-treated INS-1 cells (Fig. S3, A and B). We hypothesized that phosphorylated menin might bind to certain partner proteins and thus be sequestered from its target genes. To test our hypothesis, we performed coIP using anti-Flag antibody in nuclear extract from menin-null MEF cells complemented with S487A or S487D menin mutant. Based on silver staining, three bands that differentially associated with either S487A or S487D mutant were identified (Fig. 4 A, upper panel). Sequencing analysis of these bands using mass spectrometry technique showed that band 1, associating with S487A mutant, was KMT2, a protein known to bind menin (Hughes et al., 2004), while band 2 and 3 only associated with S487D mutant and contained myosin IIa and β actin, respectively, both of which are cytoskeletal proteins. The immunoprecipitated menin was not visible in the silver staining gel but could be detected by Western blot (Fig. 4 A, lower panel). It is possible that although silver staining is a highly sensitive method for staining certain proteins in polyacrylamide gels, it is not as sensitive as Western blot.
Figure 4.
Menin Ser487 phosphorylation leads to enhanced binding to nuclear actin/myosin proteins. (A) Nuclear extract from menin-null MEF cells complemented with Flag–S487A menin or S487D menin mutant was isolated for IP using anti-Flag antibody. Proteins were separated by SDS-PAGE and visualized with silver staining. Asterisk (*) indicates IgG heavy chain and light chain (upper panel). The immunoprecipitated Flag-menin proteins were detected by Western blot (lower panel). (B) Menin-null MEF cells complemented with vector, Flag-menin WT, or S487A or S487D mutant were treated with or without 10 µM forskolin for 1 h after overnight starvation. Nuclear extract was isolated for coIP using anti-Flag antibody. Proteins were separated by SDS-PAGE, followed by Western blot with the indicated antibodies. (C) INS-1 cells were starved overnight and then treated with 10 µM forskolin or 10 nM Ex-4 for 1 h. Nuclear extract was isolated for coIP using anti-menin antibody. Proteins were separated by SDS-PAGE, followed by Western blot with the indicated antibodies. (D and E) After overnight starvation, INS-1 cells were treated with 10 nM Ex-4 in the absence or presence of 1 µM jasplakinolide (D) or 20 µM cytochalasin D (E) for 1 h. Nuclear extract was isolated for coIP using anti-menin antibody. Proteins were separated by SDS-PAGE, followed by Western blot with the indicated antibodies.
To further confirm that S487D mutant, but not WT menin or S487A mutant, differentially binds these cytoskeletal proteins, we performed coIP experiment using anti-Flag antibody with the control or menin WT, S487A or S487D expressing MEF cells, followed by Western blot to detect myosin IIa and β actin. The results showed that indeed only S487D, but not WT or S487A, bound these cytoskeletal proteins. Meanwhile, treatment of the cells with forskolin, which potently induced menin phosphorylation at Ser487, led to increased binding of myosin IIa and β actin to WT menin but had no impact on cells expressing S487A or S487D mutant (Fig. 4 B). We also treated INS-1 cells with forskolin or Ex-4 to detect interaction between phosphorylated menin and myosin IIa or β actin. The results showed that forskolin or Ex-4 could induce interaction of menin with myosin IIa and β actin in β cells (Fig. 4 C).
While it is well known that myosin interacts with actin to control muscle contraction, nonmuscle myosin IIa also interacts with F-actin to influence motility in nonmuscle cells (Doller et al., 2013). Moreover, nuclear actin is also involved in regulating gene transcription via influencing nuclear protein shuttling between the nucleus and cytoplasm (Zheng et al., 2009). To determine whether monomeric globular actin (G-actin) or polymerized F-actin prefers to bind phosphorylated menin, we treated INS-1 cells with Ex-4, jasplakinolide (an F-actin polymerizing and stabilizing agent), or their combination and found that jasplakinolide substantially enhanced pSer487 menin binding to actin and myosin IIa (Fig. 4 D). In contrast, cytochalasin D, an F-actin–disrupting agent (Fig. 4 E), or a more selective G-actin–binding and sequestering agent, latrunculin B (Fig. S3 C), almost completely blocked actin and myosin IIa’s interaction with pSer487 menin. Together, this series of experiments suggested that Ser487 phosphorylated menin had enhanced affinity with the nuclear cytoskeletal proteins, which could lead to, or at least participate in, the sequestration of phosphorylated menin away from loci of the target genes such as Ins1.
Nuclear actin may control insulin transcription by influencing menin binding at the Ins1 locus
To further test our hypothesis that the nuclear F-actin–menin interaction is crucial for the regulation of pSer487 menin-mediated insulin transcription, we treated INS-1 cells with cytochalasin D and found that cytochalasin D blocked Ex-4–induced insulin transcription (Fig. 5 A), concomitant with increased menin binding at the Ins1 locus, as shown by ChIP assay (Fig. 5 B). Cytochalasin D also repressed insulin transcription in S487D menin–overexpressing INS-1 cells while having no effect on S487A menin INS-1 cells (Fig. 5 C). Consistently, cytochalasin D treatment resulted in increased menin binding at the Ins1 locus in S487D menin cells, but not in S487A menin cells (Fig. 5 D), suggesting that depolymerized actin failed to move S487D menin out of the Ins1 locus. As reduced expression of the import factor importin 9 (IPO9) decreased F-actin in somatic cell nuclei (Belin et al., 2013), we knocked down IPO9 in INS-1 cells with siRNAs (Fig. 5 E) and found that with IPO9 knocked down, Ex-4 could not promote insulin transcription anymore (Fig. 5 F). Meanwhile, the binding of menin at the Ins1 locus stayed the same even with Ex-4 treatment (Fig. 5 G). IPO9 was also knocked down in S487A- or S487D menin–overexpressing INS-1 cells (Fig. 5 H). quantitative real-time PCR results showed that reduced expression of IPO9 decreased insulin expression in S487D INS-1 cells, but not in S487A INS-1 cells (Fig. 5 I). Simultaneously, IPO9 knockdown led to increased binding of menin at the Ins1 locus in S487D menin cells while having no effect on S487A menin cells (Fig. 5 J). To demonstrate whether cytochalasin D treatment or IPO9 knockdown actually disassembled the nuclear actin filaments in our cells, we first transfected INS-1 cells with pEGFP-C1 Utr230-EGFP-3XNLS (Utr230-EN) plasmid for detection of nuclear actin filaments (Belin et al., 2013). The distribution of nuclear Utr230-EN exhibited three patterns in INS-1 cells: (i) punctate (∼6%), (ii) clustered (∼29%), and (iii) diffuse (∼65%; Fig. S3 D). Based on previous report, the punctate and clustered types represent structures with F-actin while the diffuse type indicates the absence of F-actin (Belin et al., 2013). Both cytochalasin D treatment (Fig. S3 E) and IPO9 knockdown (Fig. S3 F) decreased the fraction of INS-1 cells with detectable nuclear F-actin structures. Similarly, we showed the existence of nuclear F-actin in MEF cells (Fig. S3 G), and cells treated with cytochalasin D (Fig. S3 H), or from which IPO9 had been depleted, revealed reduced nuclear F-actin structures (Fig. S3, I and J). These results suggested the possibility that blocking importation of nuclear actin reduced F-actin formation and thus decreased the removal of phosphorylated menin at the Ins1 promoter, leading to derepression of insulin transcription.
Figure 5.
Disassembling of nuclear F-actin relieves menin to repress insulin transcription. (A) After overnight starvation, INS-1 cells were treated with 10 nM Ex-4 with or without 0.05 µM cytochalasin D (CD) for 12 h, followed by quantitative real-time PCR analysis of insulin mRNA level (n = 3). Data were normalized to the level of GAPDH. (B) After overnight starvation, INS-1 cells were treated with 10 nM Ex-4 for 30 min, and then ChIP assays were performed to detect menin at the Ins1 promoter. 0.05 µM cytochalasin D was added to the starvation medium 12 h before treatment. IgG served as a negative control (n = 3). (C and D) INS-1 cells expressing S487A menin or S487D menin mutant were treated with or without 0.05 µM cytochalasin D for 12 h in serum-free medium, followed by quantitative real-time PCR analysis of insulin mRNA levels (n = 3). Data were normalized to the level of GAPDH (C). ChIP assays using Flag antibody were performed to detect menin at the ins1 promoter. IgG served as a negative control (n = 3; D). (E and F) INS-1 cells were transfected with IPO9 siRNAs for 48 h before serum was withdrawn from the culture medium. After overnight starvation, 10 nM Ex-4 was added to the starvation medium for 12 h, followed by quantitative real-time PCR analysis of IPO9 (E) and insulin (F) mRNA levels (n = 3). Data were normalized to the level of GAPDH. (G) SiIPO9 INS-1 cells were starved overnight before 30-min treatment with 10 nM Ex-4, and then ChIP assays were performed to detect menin at the Ins1 promoter. IgG served as a negative control (n = 3). (H–J) INS-1 cells expressing S487A menin or S487D menin mutant were transfected with IPO9 siRNAs for 72 h, followed by quantitative real-time PCR analysis of IPO9 (H) and insulin (I) mRNA levels (n = 3). Data were normalized to the level of GAPDH. ChIP assays using Flag antibody were performed to detect menin at the Ins1 promoter. IgG served as a negative control (n = 3). All statistics: one-way ANOVA with LSD post hoc test. **, P < 0.01. Error bars indicate SD.
As export factor exportin 6 (XPO6) was demonstrated to produce the opposite effect of IPO9, and depletion of XPO6 increased F-actin in the nucleus (Belin et al., 2013), we also knocked down XPO6 in INS-1 cells with siRNAs (Fig. S4, A and B). The result indicated that reduced expression of XPO6 led to moderate yet consistent increase in Ex-4 treatment–induced insulin mRNA as compared with that in the control (scramble) cells (Fig. S4 C). These data suggested that actin in the nucleus rendered INS-1 cells more sensitive to Ex-4–induced insulin transcription, from ∼1.5 times to ∼1.8 times (Fig. S4 D), fine-tuning the control of insulin expression.
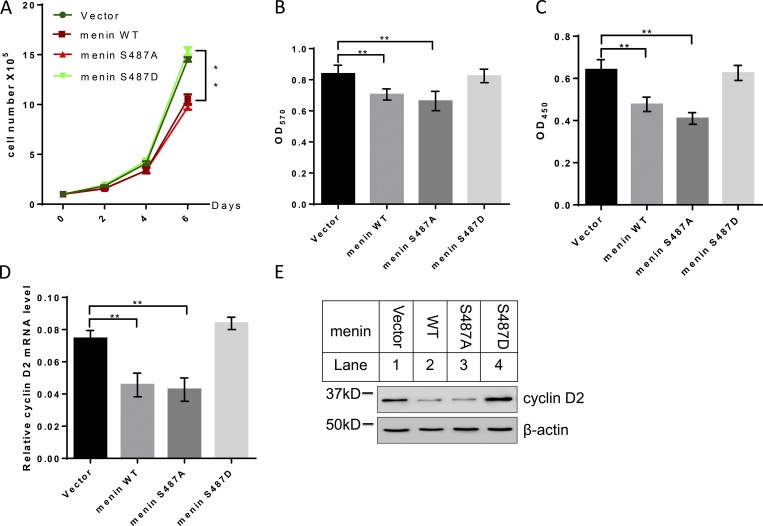
Ser487 phosphorylated menin loses its ability to suppress cell proliferation
As a putative tumor suppressor, menin has been shown to preferentially repress cell proliferation in endocrine tissues including pancreatic β cells (Crabtree et al., 2003). We therefore examined whether Ser487 phosphorylation of menin had any effect on β cell proliferation using cell counting (Fig. 6 A), MTT cell proliferation assay (Fig. 6 B), and BrdU assays (Fig. 6 C). While menin overexpression (WT or S487A) inhibited INS-1 cell growth, phospho-mimicking S487D menin mutant lost its ability to repress INS-1 cell proliferation. Based on our RNA-sequencing data from S487A- or S487D-expressing INS-1 cells, we found that cyclin D2, which has been demonstrated to be essential for postnatal pancreatic β-cell growth (Kushner et al., 2005), was upregulated in S487D-expressing cells. Consistently, quantitative real-time PCR and Western blot results showed that WT and S487A mutant menin repressed cyclin D2 expression in mRNA (Fig. 6 D) and protein levels (Fig. 6 E), but expression of S487D menin failed to do so.
Figure 6.
Ser487-phosphorylated menin loses its ability to repress cell proliferation. (A) INS-1 cells expressing vector, menin WT, S487A, or S487D mutant were examined for cell number with standard cytometry (n = 3). (B) MTT assay for assessing metabolic activity of INS-1 cells (n = 6). (C) INS-1 cells expressing vector, menin WT, S487A, or S487D mutant were examined for proliferation with a BrdU assay kit (n = 7). (D) Quantitative real-time PCR analysis of cyclin D2 mRNA level in INS-1cells expressing vector, menin WT, or S487A or S487D mutant (n = 3). Data were normalized to the level of GAPDH. (E) INS-1 cells harboring vector, menin WT, or S487A or S487D mutant were examined for cyclin D2 expression by Western blot. All statistics: one-way ANOVA with LSD post hoc test. **, P < 0.01. Error bars indicate SD.
Like pancreatic β cells, MEF cell proliferation is also suppressed by menin expression (Schnepp et al., 2006). As such, menin-null MEFs complemented with vector, Flag-menin WT, or S487A or S487D mutant (Fig. S5 A) were also monitored by cell counting (Fig. S5 B), MTT (Fig. S5 C), BrdU staining (Fig. S5 D), and colony-formation assays (Fig. S5, E and F). Consistent with the results from INS-1 cells, Ser487 phosphorylation of menin led to loss of its ability to repress MEF cell proliferation. As expected, cyclin D2 expression level was also lower in WT or S487A menin–expressing cells as compared with vector or S487D menin cells (Fig. S5, G and H). Taken together, these results suggested that Ser487 phosphorylation of menin regulated cell proliferation, at least partially, through cyclin D2.
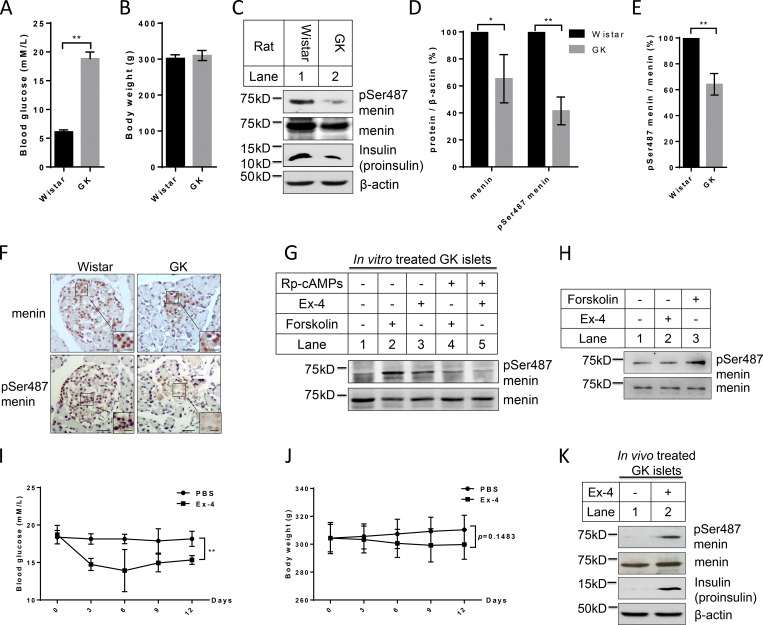
Ex-4 stimulates the decreased Ser487 phosphorylation of menin in islets of Goto–Kakizaki (GK) rats
The finding that Ser487 phosphorylation of menin promotes insulin expression encouraged us to investigate whether the menin phosphorylation is altered in islets of diabetic model. We therefore isolated islets from Wistar rats and spontaneous diabetic GK rats for Western blot (Fig. 7, A and B). As expected, the insulin level, detected as proinsulin, was decreased in GK islets, and total menin was also slightly lower in GK islets (Fig. 7 C), possibly reflecting the autosuppressing of menin to compensate the reduced β cell mass and function. Notably, Ser487 phosphorylated menin was much more reduced in islets from diabetic GK rats than in those from Wistar rats (Fig. 7, D and E).
Figure 7.
Ex-4 stimulates the decreased Ser487 phosphorylation of menin in islets of GK rats. (A and B) Blood glucose (A) and body weight (B) of Wistar and GK rats that were used for islet isolation (n = 5). (C) Islets isolated from diabetic GK and control Wistar rats were lysed for Western blot using the indicated antibodies. (D) Band density analysis was executed with ImageJ. Menin and pSer487 menin protein levels were quantified and normalized to β-actin (n = 3). (E) Comparison of pSer487-menin/menin ratio between GK and Wistar islets (n = 3). (F) Immunohistochemistry was performed in paraffin-embedded sections of Wistar and GK islets using anti-menin or anti-pSer487 menin antibodies (scale bars, 25 µm). Insets show higher magnification of the selected areas; scale bars, 10 µm. (G) GK islets were treated with 10 µM forskolin or 10 nM Ex-4 for 30 min with or without 2 µM Rp-cAMPS and then lysed for Western blot using the indicated antibodies. (H) Wistar islets were treated with 10 µM forskolin or 10 nM Ex-4 for 30 min and then lysed for Western blot using the indicated antibodies. (I and J) Subcutaneous injection of Ex-4 (8 µg/kg/day) or PBS was conducted in age-, body weight–, and blood glucose–matched GK rats daily. Blood glucose (I) and body weight (J) of the rats were monitored every 3 d (n = 5). (K) 2 wk after subcutaneous injection of Ex-4 (8 µg/kg/day) or PBS, GK rats were sacrificed for islets isolation, followed by Western blot using the indicated antibodies. All statistics: two-tailed t tests. *, P < 0.05; **, P < 0.01. Error bars indicate SD.
We also performed immunohistochemistry in paraffin-embedded sections of Wistar and GK islets with anti-menin or anti-pSer487 menin antibody, showing that Ser487 phosphorylated menin was hardly detectable in GK islets while total menin only slightly reduced (Fig. 7 F). Even though GK islets had a low level of Ser487 phosphorylation, forskolin or Ex-4 treatment substantially boosted this phosphorylation, and as expected, the induced Ser487 phosphorylation was much repressed with the PKA inhibitor Rp-cAMPS (Fig. 7 G). As for Wistar islets, forskolin could still induce menin phosphorylation, but Ex-4 could not, probably because the menin phosphorylation background is already high in Wistar islets, so that forskolin, which has much stronger efficiency in activating PKA than Ex-4, was able to increase menin phosphorylation in the islets (Fig. 7 H). We then subcutaneously injected GK rats with Ex-4 or control PBS daily, and blood glucose level (Fig. 7 I) and body weight (Fig. 7 J) of the rats were monitored every 3 d. 2 wk later, the rats were sacrificed and islets were isolated for Western blot. Our results indicated that Ex-4 injections increased menin phosphorylation as well as insulin production but did not change the amount of total menin (Fig. 7 K). Together, these data demonstrated that Ex-4 also physiologically induced menin phosphorylation at Ser487 in vivo.
Discussion
The GLP-1 signaling pathway is crucial for positively regulating β cell function and islet mass (Nadkarni et al., 2014). On the other hand, menin is the only genetically proven human gene whose inhibition leads to enhanced β cell proliferation and increased islet mass (Marx et al., 1999; Crabtree et al., 2001; Bertolino et al., 2003; Schnepp et al., 2006; Thakker, 2010; Matkar et al., 2013). Although our recent research shows that the small-molecule menin inhibitor MI-2-2 can increase β cell function (Muhammad et al., 2017), a challenge has been that little is known about how menin is physiologically and mechanistically linked to the GLP-1 pathway in regulating β cells and whether this process is dysregulated in diabetes. In this regard, we have demonstrated that GLP-1 signaling–activated PKA directly phosphorylates menin at Ser487 to suppress menin function. These findings lead to a new concept that the genetically proven islet regulator menin is directly connected to the key GLP-1 pathway, revealing a novel molecular target of GLP-1 signaling in its promotion of β cell function.
While thus far menin has been found to be phosphorylated at several sites, including Ser487 (MacConaill et al., 2006; Francis et al., 2011), to our knowledge, no functional changes in phosphorylation or protein kinases responsible for phosphorylation have been definitively identified. In this aspect, we have, for the first time, identified that Ser487 menin phosphorylation induced by Ex-4/PKA is a key regulatory event that abolishes menin-mediated suppression of insulin expression by releasing HDAC1 and SUV39H1-related epigenetic suppression. Nevertheless, the current studies do not rule out the possibility that other positive or negative regulators are also crucial for regulating Ins1 genes in concert with menin. One key that allowed us to discover this phosphorylation-regulated step was mutating Ser487 to the phospho-mimicking aspartate residue, as expression of this S487D mutant leads to menin loss of function by suppressing Ins1 transcription. In contrast, the S487A mutant that many studies have adopted works largely as a WT or even super-WT and thus failed to show this loss of function. The second key was that we generated a specific and highly sensitive pSer487 menin antibody that allowed us to examine acute changes in GLP-1 signaling–induced phosphorylation and accompanying functional changes. Sundaresan et al. (2017) recently reported that gastrin or forskolin could induce nuclear export and proteasome degradation of menin in neuroendocrine cells, but in our system, menin did not translocate out to the cytoplasm in forskolin-treated INS-1 cells. Even though Ser487 locates in the nuclear localization sequence 1 (479–497 aa) domain of menin, phosphorylation at this site did not show any effect on its nuclear–cytoplasmic shuttling. Overall, the current studies suggest an elaborate mechanism whereby GLP-1 signaling–induced menin phosphorylation suppresses menin activity in regulating insulin expression and cell proliferation.
Nuclear G-actin regulates transcription factor serum response factor–mediated transcription by promoting nuclear export of its cofactor, megakaryocyte acute leukemia (Miralles et al., 2003; Vartiainen et al., 2007; Baarlink et al., 2013), indicating the crucial role of nuclear actin in regulating gene transcription. Our work clearly shows that GLP-1 signaling leads to menin phosphorylation and up-regulation of insulin transcription by de-repressing the insulin promoter, likely through nuclear F-actin–mediated removal of the pSer487 menin from the promoter. However, further work remains to precisely demonstrate how GLP-1 signaling controls nuclear F-actin formation and how F-actin interacts with pSer487 menin and removes it from the insulin promoter. It is noteworthy that majority of Utr230-EN–expressing cells (∼65% for INS-1 and ∼50% for MEFs) exhibit diffuse localization of the signals throughout the nucleoplasm, an indicator of free actin. We consider this conceivable because actin is dynamic between the free form and the polymerized form (F-actin), and rapid conversion between the two forms is important for actin to perform diverse tasks within various microenvironments of the cell, including the nucleus (Dopie et al., 2012). Consistently, subcellular distribution of Utr230-EN in HeLa cells also indicates that most of cells display the diffuse pattern (Belin et al., 2015). Utr230-EN–transfected U2OS cells show that large blobs (clustered structures) only account for a small portion of the stained structures (Belin et al., 2013); however, the probe reveals more large blobs in INS-1 cells, exceeding the number of punctate structures (Fig. S3, E and F). This is likely due to the variation of the staining patterns in different types of cells. Consistent with this, MEF cells display more punctate structures when transfected with precisely the same nuclear F-actin probe (Fig. S3, H and I). Although Utr230-EN–transfected MEF cells appear normal, the probe is toxic to INS-1 cells and causes severe cell death ∼36 h after transfection. Fortunately, expression of the Utr230-EN probe for less than 24 h is tolerable to INS-1 cells. As such, we limited the probe-expressing time to 12 h before cytochalasin D treatment or expressed the probe by a second round of transfection after IPO9 knockdown to keep the expression time of the Utr230-EN probe within 24 h in INS-1 cells.
It is interesting to note that Ser487 menin phosphorylation was substantially reduced in the islets of diabetic GK rats. It is conceivable that in diabetic rats, the GLP-1 signaling pathway is weakened or less sensitive, leading to reduced menin phosphorylation and enhanced menin activity in suppressing insulin production. On the other hand, it is also possible that the diabetes-induced stress of β cells causes less accessibility of menin to PKA-mediated phosphorylation. While the detailed scenarios remain to be further investigated, our current studies have demonstrated the key role of Ex-4–induced and PKA-mediated menin phosphorylation at Ser487 in β cells.
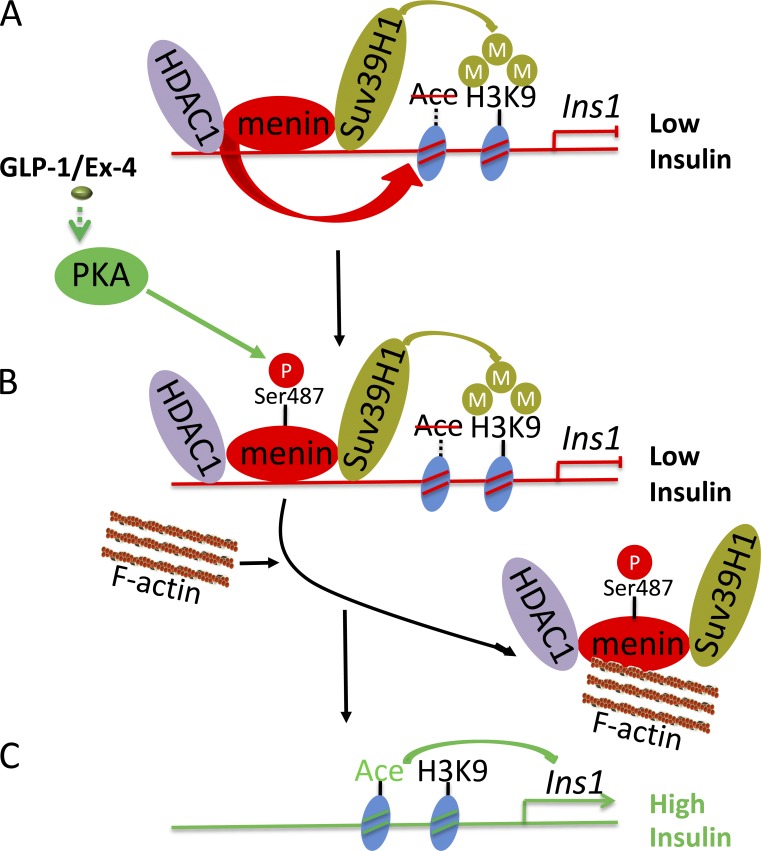
Our findings are consistent with a model of the interplay between the menin pathway and GLP-1 signaling pathway. Menin normally associates with SUV39H1 and HDAC1 to maintain the repressive histone markers at the insulin promoter to keep a subdued level of insulin expression (Fig. 8 A). When β cells receive signal from Ex-4/GLP-1, menin is phosphorylated at Ser487 and subsequently sequestrated to nuclear F-actin (Fig. 8 B) and thus loses its function to maintain the repressive epigenetic regulators at the locus, resulting in increased insulin expression (Fig. 8 C).
Figure 8.
A model for GLP-1 signaling–induced and pSer487 menin-induced upregulation of insulin expression. (A) Menin normally associates with SUV39H1 and HDAC1 to maintain the repressive histone markers in the Ins1 gene to suppress its expression. (B and C) When cells receive signal from Ex-4/GLP-1, menin is phosphorylated at Ser487 and sequestrated to nuclear F-actin (B) and thus loses its function to maintain the repressive epigenetic regulators at the locus, resulting in increased insulin expression (C).
In summary, there are four highlights described in the current work: (1) Menin is demonstrated to be a novel molecular target of GLP-1 signaling in its promotion of β cell function. (2) Ex-4-induced and PKA-mediated menin phosphorylation at Ser487, identified as the first functional posttranslational modification of menin, plays a crucial role in derepressing insulin transcription, fine-tuning the regulation of insulin production. Dysregulation of this circuitry may contribute to the development of type 2 diabetes. (3) We uncover a previously unrecognized mechanism whereby phosphorylated menin leads to reduced binding of repressive epigenetic regulators SUV39H1 and HDAC1 to the Ins1 locus, increasing insulin expression by derepression at the locus. (4) Nuclear F-actin actively participates in the regulation of Ins1 gene expression. Further studies of Ex-4/PKA/menin phosphorylation signaling in regulating other target genes in mouse and human islets will be informative to fully appreciate the importance of this crucial pathway.
Materials and methods
Antibodies and chemicals
The antibodies and chemicals used in this study were as follows: rabbit polyclonal anti-menin (A300-115A, 1:5,000 dilution; Bethyl), mouse monoclonal anti-Flag (clone M2; F3165, 1:1,000 dilution; Sigma-Aldrich), rabbit polyclonal anti-PKA Cα (4782, 1:1,000 dilution; Cell Signaling Technology), rabbit monoclonal anti-phospho-PKA substrate (RRXS*/T*; 9624, 1:1,000 dilution; Cell Signaling Technology), rabbit monoclonal anti-GFP (2956S, 1:1,000 dilution; Cell Signaling Technology), mouse monoclonal anti-insulin (clone L6B10; 8138, 1:1,000 dilution; Cell Signaling Technology), rabbit monoclonal anti-GAPDH (2118, 1:1,000 dilution; Cell Signaling Technology), rabbit polyclonal anti-Myosin IIa (3403, 1:1,000 dilution; Cell Signaling Technology), rabbit polyclonal anti-histone H3 (trimethyl K4) antibody (ab8580; Abcam), rabbit polyclonal anti-histone H3 (trimethyl K9) antibody (ab8898; Abcam), mouse monoclonal anti-histone H3 (trimethyl K27) antibody (ab6002; Abcam), rabbit polyclonal anti-histone H3 (ab1791; Abcam), mouse monoclonal anti-Suv39H1 (ab12405, 1:1,000 dilution; Abcam), rabbit polyclonal anti-HDAC1 (ab7028, 1:1,000 dilution; Abcam), rabbit polyclonal anti-IgG (ab171870; Abcam), rabbit polyclonal anti-lamin A (ab26300, 1:2,000 dilution; Abcam), rabbit polyclonal anti-XPO6 (ab72333, 1:1,000 dilution; Abcam), rabbit polyclonal anti-acetyl-histone H3 (06–599; Millipore), mouse monoclonal anti-β actin (SC47778, 1:1,000 dilution; Santa Cruz), mouse monoclonal anti-pSer487 menin (1:1,000 dilution; this paper), goat anti-mouse IgG (H+L) HRP conjugate (170–6516, 1:5,000 dilution; Bio-Rad), Goat Anti-Rabbit IgG (H+L) HRP conjugate (170–6515, 1:5,000 dilution; Bio-Rad), Clean-Blot IP detection kit (HRP; 21232, 1:400 dilution; Thermo Scientific), Alexa Fluor 488 goat anti-mouse IgG (H+L; A11001, 1:1,000 dilution; Invitrogen), Ex-4 (1269105; Sigma-Aldrich), forskolin (F6886; Sigma-Aldrich), ATP disodium salt hydrate (FLAAS; Sigma-Aldrich), PKA catalytic subunit from bovine heart (P2645; Sigma-Aldrich), Chaetocin from Chaetomium minutum (C9492; Sigma-Aldrich), trichostatin A (T1952; Sigma-Aldrich), SAHA (SML0061; Sigma-Aldrich), 8-pCPT-2′-O-Me-cAMP (sc-257020; Santa Cruz), dibutyryl-cAMP (sc-201567; Santa Cruz), Rp-cAMPS (sc-24010; Santa Cruz), leptomycin B (9676; Cell Signaling Technology), jasplakinolide (ab141409; Abcam), cytochalasin D (ab143484; Abcam), and latrunculin B (ab144291; Abcam).
Cell culture and authentication
MEF and HEK293 cells were maintained in DMEM (GIBCO BRL Shanghai) supplemented with 10% FBS (Hyclone) and 1% penicillin-streptomycin. INS-1 cells (passages 62–80) were kindly provided by Yong Liu (Shanghai Institute for Nutritional Sciences of Chinese Academy of Sciences, Shanghai, China; Yu et al., 2012) and maintained in RPMI 1640 medium (GIBCO BRL Shanghai) supplemented with 10% FBS, 10 mM Hepes, 2 mM glutamine, 1 mM sodium pyruvate, 50 µM β-mercaptoethanol, and 1% penicillin-streptomycin. All cells were cultured in a 37°C, 5% CO2 incubator. Authentication of cell lines was performed by IDEXX Bioresearch. Genetic profiles were generated for the actual cell lines by using a panel of microsatellite markers for genotyping. MEF cells were confirmed to be of mouse origin, and no mammalian interspecies contamination was detected. HEK293 cells were confirmed to be of human origin, and no mammalian interspecies contamination was detected. INS-1 cells were confirmed to be of rat origin, and no mammalian interspecies contamination was detected. Recent short tandem repeats analysis was performed within 6 mo before the beginning or in the course of the experiments for all cell lines (Feng et al., 2017).
Animals
Wistar and GK rats (male, ∼14–16 wk) were purchased from Shanghai slack animal center and maintained on a 12-h light/dark cycle in the animal facility at Shenzhen University. All of the experimental procedures received approval from Animal Ethical and Welfare Committee of Medical College at Shenzhen University.
Quantitative real-time PCR
Total RNA was extracted from cultured cells using the RNAiso plus reagents (TaKaRa) according to the supplier’s instructions. Quantitative real-time PCR was conducted to assess insulin expression using an ABI prism 7500 Sequence Detection System (Applied Biosystems). The relative mRNA level was calculated by the ΔΔCt values calibrated with GAPDH using SYBR green dye from Qiagen for detection. Primer sequences are as follows: rat Ins1 forward: 5′-CACCTTTGTGGTCCTCACCT-3′; rat Ins1 reverse: 5′-CCAGTTGGTAGAGGGAGCAG-3′; rat or mouse CCND2 forward: 5′-TTACCTGGACCGTTTCTTGG-3′; rat or mouse CCND2 reverse: 5′-TGCTCAATGAAGTCGTGAGG-3′; rat GAPDH forward: 5′-GGCAAGTTCAACGGCACAGT-3′; rat GAPDH reverse: 5′-TGGTGAAGACGCCAGTAGACTC-3′; rat IPO9 forward: 5′-GCAAGGCCACAATGGACTAT-3′; rat IPO9 reverse: 5′-TGATCCAGATTCTCCCCAAG-3′; rat XPO6 forward: 5′-GCCAGAGCTTGGAGGTAGTG-3′; rat XPO6 reverse: 5′-TTCTCCGCTGTGATGTTCAG-3′; mouse IPO9 forward: 5′-GCACCGTTAAGAAGGCTCTG-3′; mouse IPO9 reverse: 5′-AGTCTTGAGCGGCAATCCTA-3′; mouse GAPDH forward: 5′-AACTTTGGCATTGTGGAAGG-3′; and mouse GAPDH reverse: 5′-ACACATTGGGGGTAGGAACA-3′.
Protein purification
Escherichia coli BL21 transformed with His-menin (WT, S122A, S487A, or S487D) was incubated at 25°C and then induced by 0.25 mM IPTG for 4 h. Cells were harvested and lysed with 1% Triton X-100 in PBS, 1.0 mM PMSF, 4 µg/ml bacterial protease inhibitor cocktail (Sigma), and 100 µg/ml lysozyme. Lysates were spun down, and the supernatant was added to Ni-NTA beads (Qiagen) overnight at 4°C. After washing with 1 mM imidazole in PBS, proteins were eluted from Ni-NTA beads with 150 mM imidazole in PBS.
In vitro PKA Cα phosphorylation assay
The in vitro kinase assay was undertaken as previously reported (Xing et al., 2013). Briefly, 2 µg purified recombinant menin proteins was incubated with 0.15 µg purified recombinant active PKA Cα protein in kinase buffer (35 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 0.5 mM EGTA, and 0.1 mM CaCl2) containing 100 µM ATP for 30 min at 30°C in a final volume of 40 µl. Reactions were terminated by addition of concentrated sample buffer.
Generation of pSer487 menin antibody
The sequences of the normal and pSer487 peptides from human menin were CRRGPRRESKPEEPP and CRRGPRRE{p}SKPEEPP. The Keyhole Limpet Hemocyanin–conjugated peptides were synthesized by Genescript and injected into BALB/c mice with adjuvant more than five times. The sera from the immunized mice were monitored with ELISA for binding to the control or phosphorylated peptide. When a positive sera result was detected, the mice were sacrificed and spleen cells were isolated to perform fusion with a myeloma cell line as previously described (Harlow et al., 1988). The supernatant of the fused hybridoma was used to screen for clones that bound the well coated with phosphorylated peptide, but not the control peptide using ELISA. The positive hybridomas were further cloned and amplified.
Insulin secretion and content test
INS-1 cells were seeded at 2 × 105 in six-well plates. 2 d later, the cells were washed and preincubated for 1 h in Krebs–Ringer bicarbonate buffer, and then the medium was replaced with Krebs–Ringer bicarbonate buffer containing 16.8 mM glucose with or without 10 nM Ex-4 for an additional 30 min. Secreted and cellular insulin (extracted in NP-40 lysis buffer containing 50 mM Tris, pH 7.4, 250 mM NaCl, 5 mM EDTA, and 1% NP-40) was assessed with rat insulin ELISA kits (ALPCO) according to the manufacturer’s protocol.
RNA sequencing
RNA sequencing was conducted in RiboBio to analyze the cellular transcriptome. In brief, total RNA was extracted using the RNAiso plus reagents (TaKaRa). mRNAs were isolated from Total RNA and fragmented to ∼200 bp. Subsequently, the collected mRNAs were subjected to first- and second-strand cDNA synthesis followed by adaptor ligation and enrichment with a low cycle according to the instructions provided with the TruSeq RNA LT/HT Sample Prep Kit (Illumina). The purified library products were evaluated using the Agilent 2200 TapeStation and Qubit 2.0 (Life Technologies) and then diluted to 10 pM for cluster generation in situ on the HiSeq2500 single-end flow cell followed by sequencing (1 × 50 bp) on HiSeq2500.
Viral infection
Menin shRNAs for retroviral packaging were cotransfected with psi-2 helper plasmid into HEK293 cells using the calcium chloride precipitation method, and the resulting recombinant virus were used to transduce INS-1 cells, as previously reported (Yang et al., 2013; Feng et al., 2017). The target sequences for menin shRNA are as follows: rat Men1 shRNA-1: 5′-CTCGGATGTCATATGGAAC-3′; and rat Men1 shRNA-2: 5′-GCTGTATGACCTAGGACAT-3′.
ChIP assay
The ChIP assay was performed as previously described using a ChIP assay kit (Millipore) to study protein–DNA interactions and chromatin changes associated with gene expression (Chen et al., 2006). Briefly, 106–107 formaldehyde cross-linked cells were lysed and sonicated to obtain sheared DNA. Lysates were incubated with antigen-specific antibodies and agarose beads at 4°C overnight. Samples were eluted from the beads and reverse cross-linked at 65°C. Eluted DNA was quantified using quantitative PCR and normalized to percent input of genomic DNA. The following primers were used for ChIP assays: Ins1 ChIP forward: 5′-CGGGCTAGAACCACACATTT-3′; and Ins1 ChIP reverse: 5′-TTTGCTGCTAGCCTGGAGTT-3′.
IPO9 and XPO6 knockdown by transient transfection
The duplex siRNA oligonucleotides used for silencing the expression of IPO9 (NM_001107180: SASI_Rn02_00226300 as siIPO9#1, SASI_Rn02_00226302 as siIPO9#2), XPO6 (NM_001011935: SASI_Rn02_00199945 as siXPO6#1, SASI_Rn02_00199946 as siXPO6#2), and nonspecific scrambled control (5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Sigma-Aldrich. INS-1 cells were transfected with siRNA duplexes at 50 nM using Lipofectamine 2000 reagent (11668-019; Invitrogen), and cells were cultured for 48 h before further analysis.
Immunohistochemistry
Pancreases of Wistar and GK rats were fixed in 4% paraformaldehyde overnight at 4°C, dehydrated in an ascending ethanol series, routinely embedded in paraffin, and sectioned at 5 µm. After conventional deparaffing, hydration, and antigen retrieval, the sections were subjected to UltraVision Quanto Detection System HRP DAB Kit (Thermo Scientific) for visualization of menin or pSer487 menin according to the manufacturer’s protocol.
Microscopy
Images of paraffin section (immunohistochemistry) were acquired using the Nikon Eclipse Ci microscope equipped with the Nikon digital sight DS-U3 camera and NIS Elements imaging software 4.51 (Nikon) using NIKON 20×/0.4 objective at room temperature. Confocal images were acquired with a Leica TSP SP5II confocal microscope equipped with a Leica DMI6000 camera and LAS AF software using a Leica HCXPLFLUOTARL 40×/0.6 objective or a Carl Zeiss LSM710 confocal microscope equipped with LSM T-PTM and ZEN (2011) software using a 40×/1.30 objective at room temperature. Images were processed for contrast enhancement and background noise reduction using ImageJ (National Institutes of Health) or Zen 2012 (blue edition).
Islet isolation
Wistar or GK islet isolation was performed as previously described (de Haan et al., 2004). Collagenase P (Sigma-Aldrich) dissolved to a concentration of 1.0 mg/ml in Hank’s balanced salts solution was transfused into the pancreatic ducts via the common bile duct. The pancreas was chopped into 1-mm2 pieces and brought into a 15-ml tube for incubation at 37°C with 1.0 mg/ml collagenase. After a 10-min digestion, the tissue was washed and sedimented twice to remove collagenase and leaked exocrine-derived proteases. Islets were separated from exocrine tissue by handpicking under a stereomicroscope (Ying et al., 2012).
Identification of proteins interacting with pSer487 menin
Nuclear extract from menin-null MEFs complemented with Flag–S487A menin or S487D menin mutant was isolated as described previously (Liu and Fagotto, 2011) and then incubated with 100 µl Protein A agarose beads (Invitrogen) coupled with the anti-Flag antibody at 4°C for 4 h. The beads were thoroughly washed and eluted by addition of 1× sample buffer. The purified proteins were separated by SDS-PAGE, stained with a Pierce Silver Stain Kit (Thermo Scientific), and excised under a sterile hood. The peptide sequences from these proteins were identified by mass spectrometry (TripleTOF 5600+ System; SCIEX) at Wininnovate Bio-Tech.
Quantification and statistical analysis
Statistical analyses were performed with GraphPad Prism (version 6.0; GraphPad Software) for t tests or IBM SPSS Statistics (version 19) for ANOVA. The statistical significances between datasets were indicated as P values, and P < 0.05 was considered statistically significant.
Data availability
The accession number for the raw and analyzed data files for the RNA-sequencing analysis reported in this paper is GEO: GSE106653.
Online supplemental material
Fig. S1 shows that PKA interacts with menin. Fig. S2 shows that Ex-4/PKA phosphorylates menin at the Ser487 residue. Fig. S3 shows that nuclear-localized pSer487 menin binds to F-actin, which could be disassembled by cytochalasin D or IPO9 knockdown. Fig. S4 shows that reduced expression of XPO6 renders INS-1 cells more sensitive to Ex-4–induced insulin transcription. Fig. S5 shows that Ser487-phosphorylated menin loses its ability to suppress cell proliferation.
Supplementary Material
Acknowledgments
We thank Yong Liu for the INS-1 cells.
This work was supported by the National Natural Science Foundation of China (grants 81600596, 81600597, 81570733, and 81670708), Shenzhen Peacock Plan (grant KQTD20140630100746562), the Shenzhen Basic Research Project (grant JCYJ20160307155501522 to X. Kong), and the National Institutes of Health (grant R01DK097555).
The authors declare no competing financial interests.
Author contributions: Conceptualization, X. Hua, B. Xing, and X. Ma; Methodology, B. Xing; Investigation, B. Xing, with assistance from J. Ma, Z. Jiang, Z. Feng, S. Ling, K. Szigety, W. Su, L. Zhang, and X. Kong; Writing – Original Draft, X. Hua and B. Xing; Writing – Review & Editing, X. Hua, B. Xing, X. Ma, W. Su, Z. Jiang, and Z. Feng; Resources, L. Zhang, R. Jia, and Y. Sun; Supervision, X. Hua and X. Ma.
References
- Agarwal S.K., Guru S.C., Heppner C., Erdos M.R., Collins R.M., Park S.Y., Saggar S., Chandrasekharappa S.C., Collins F.S., Spiegel A.M., et al. 1999. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell. 96:143–152. 10.1016/S0092-8674(00)80967-8 [DOI] [PubMed] [Google Scholar]
- Baarlink C., Wang H., and Grosse R.. 2013. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 340:864–867. 10.1126/science.1235038 [DOI] [PubMed] [Google Scholar]
- Belin B.J., Cimini B.A., Blackburn E.H., and Mullins R.D.. 2013. Visualization of actin filaments and monomers in somatic cell nuclei. Mol. Biol. Cell. 24:982–994. 10.1091/mbc.e12-09-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin B.J., Lee T., and Mullins R.D.. 2015. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-½ that promotes efficient DNA repair. [corrected]. eLife. 4:e07735 10.7554/eLife.07735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino P., Tong W.M., Galendo D., Wang Z.Q., and Zhang C.X.. 2003. Heterozygous Men1 mutant mice develop a range of endocrine tumors mimicking multiple endocrine neoplasia type 1. Mol. Endocrinol. 17:1880–1892. 10.1210/me.2003-0154 [DOI] [PubMed] [Google Scholar]
- Buteau J., Foisy S., Joly E., and Prentki M.. 2003. Glucagon-like peptide 1 induces pancreatic beta-cell proliferation via transactivation of the epidermal growth factor receptor. Diabetes. 52:124–132. 10.2337/diabetes.52.1.124 [DOI] [PubMed] [Google Scholar]
- Chandrasekharappa S.C., Guru S.C., Manickam P., Olufemi S.E., Collins F.S., Emmert-Buck M.R., Debelenko L.V., Zhuang Z., Lubensky I.A., Liotta L.A., et al. 1997. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 276:404–407. 10.1126/science.276.5311.404 [DOI] [PubMed] [Google Scholar]
- Chen Y.X., Yan J., Keeshan K., Tubbs A.T., Wang H., Silva A., Brown E.J., Hess J.L., Pear W.S., and Hua X.. 2006. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc. Natl. Acad. Sci. USA. 103:1018–1023. 10.1073/pnas.0510347103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepurny O.G., Hussain M.A., and Holz G.G.. 2002. Exendin-4 as a stimulator of rat insulin I gene promoter activity via bZIP/CRE interactions sensitive to serine/threonine protein kinase inhibitor Ro 31-8220. Endocrinology. 143:2303–2313. 10.1210/endo.143.6.8870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepurny O.G., Leech C.A., Kelley G.G., Dzhura I., Dzhura E., Li X., Rindler M.J., Schwede F., Genieser H.G., and Holz G.G.. 2009. Enhanced Rap1 activation and insulin secretagogue properties of an acetoxymethyl ester of an Epac-selective cyclic AMP analog in rat INS-1 cells: studies with 8-pCPT-2′-O-Me-cAMP-AM. J. Biol. Chem. 284:10728–10736. 10.1074/jbc.M900166200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree J.S., Scacheri P.C., Ward J.M., Garrett-Beal L., Emmert-Buck M.R., Edgemon K.A., Lorang D., Libutti S.K., Chandrasekharappa S.C., Marx S.J., et al. 2001. A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc. Natl. Acad. Sci. USA. 98:1118–1123. 10.1073/pnas.98.3.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree J.S., Scacheri P.C., Ward J.M., McNally S.R., Swain G.P., Montagna C., Hager J.H., Hanahan D., Edlund H., Magnuson M.A., et al. 2003. Of mice and MEN1: Insulinomas in a conditional mouse knockout. Mol. Cell. Biol. 23:6075–6085. 10.1128/MCB.23.17.6075-6085.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis P.D., Elliott L.H., Harris W., Hill C.H., Hurst S.A., Keech E., Kumar M.K., Lawton G., Nixon J.S., and Wilkinson S.E.. 1992. Inhibitors of protein kinase C. 2. Substituted bisindolylmaleimides with improved potency and selectivity. J. Med. Chem. 35:994–1001. 10.1021/jm00084a004 [DOI] [PubMed] [Google Scholar]
- de Haan B.J., Faas M.M., Spijker H., van Willigen J.W., de Haan A., and de Vos P.. 2004. Factors influencing isolation of functional pancreatic rat islets. Pancreas. 29:e15–e22. 10.1097/00006676-200407000-00063 [DOI] [PubMed] [Google Scholar]
- De León D.D., Farzad C., Crutchlow M.F., Brestelli J., Tobias J., Kaestner K.H., and Stoffers D.A.. 2006. Identification of transcriptional targets during pancreatic growth after partial pancreatectomy and exendin-4 treatment. Physiol. Genomics. 24:133–143. 10.1152/physiolgenomics.00156.2005 [DOI] [PubMed] [Google Scholar]
- Doller A., Schulz S., Pfeilschifter J., and Eberhardt W.. 2013. RNA-dependent association with myosin IIA promotes F-actin-guided trafficking of the ELAV-like protein HuR to polysomes. Nucleic Acids Res. 41:9152–9167. 10.1093/nar/gkt663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J., Skarp K.P., Rajakylä E.K., Tanhuanpää K., and Vartiainen M.K.. 2012. Active maintenance of nuclear actin by importin 9 supports transcription. Proc. Natl. Acad. Sci. USA. 109:E544–E552. 10.1073/pnas.1118880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker D.J., and Rosen C.F.. 2011. Glucagon-like peptide-1 (GLP-1) receptor agonists, obesity and psoriasis: diabetes meets dermatology. Diabetologia. 54:2741–2744. 10.1007/s00125-011-2297-z [DOI] [PubMed] [Google Scholar]
- Feng Z., Wang L., Sun Y., Jiang Z., Domsic J., An C., Xing B., Tian J., Liu X., Metz D.C., et al. 2017. Menin and Daxx Interact to Suppress Neuroendocrine Tumors through Epigenetic Control of the Membrane Metallo-Endopeptidase. Cancer Res. 77:401–411. 10.1158/0008-5472.CAN-16-1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J., Lin W., Rozenblatt-Rosen O., and Meyerson M.. 2011. The menin tumor suppressor protein is phosphorylated in response to DNA damage. PLoS One. 6:e16119 10.1371/journal.pone.0016119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobl A.E., Berg M., Lopez-Egido J.R., Oberg K., Skogseid B., and Westin G.. 1999. Menin represses JunD-activated transcription by a histone deacetylase-dependent mechanism. Biochim. Biophys. Acta. 1447:51–56. 10.1016/S0167-4781(99)00132-3 [DOI] [PubMed] [Google Scholar]
- Harlow E., and Lane D.; eds. . 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: 726. [Google Scholar]
- Huang J., Gurung B., Wan B., Matkar S., Veniaminova N.A., Wan K., Merchant J.L., Hua X., and Lei M.. 2012. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 482:542–546. 10.1038/nature10806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.M., Rozenblatt-Rosen O., Milne T.A., Copeland T.D., Levine S.S., Lee J.C., Hayes D.N., Shanmugam K.S., Bhattacharjee A., Biondi C.A., et al. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell. 13:587–597. 10.1016/S1097-2765(04)00081-4 [DOI] [PubMed] [Google Scholar]
- Karnik S.K., Hughes C.M., Gu X., Rozenblatt-Rosen O., McLean G.W., Xiong Y., Meyerson M., and Kim S.K.. 2005. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c. Proc. Natl. Acad. Sci. USA. 102:14659–14664. 10.1073/pnas.0503484102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee J.E., Cho E.J., Liu J.O., and Youn H.D.. 2003. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 63:6135–6139. [PubMed] [Google Scholar]
- Kushner J.A., Ciemerych M.A., Sicinska E., Wartschow L.M., Teta M., Long S.Y., Sicinski P., and White M.F.. 2005. Cyclins D2 and D1 are essential for postnatal pancreatic beta-cell growth. Mol. Cell. Biol. 25:3752–3762. 10.1128/MCB.25.9.3752-3762.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., and Fagotto F.. 2011. A method to separate nuclear, cytosolic, and membrane-associated signaling molecules in cultured cells. Sci. Signal. 4:pl2 10.1126/scisignal.2002373 [DOI] [PubMed] [Google Scholar]
- MacConaill L.E., Hughes C.M., Rozenblatt-Rosen O., Nannepaga S., and Meyerson M.. 2006. Phosphorylation of the menin tumor suppressor protein on serine 543 and serine 583. Mol. Cancer Res. 4:793–801. 10.1158/1541-7786.MCR-06-0123 [DOI] [PubMed] [Google Scholar]
- Marx S.J., Agarwal S.K., Kester M.B., Heppner C., Kim Y.S., Skarulis M.C., James L.A., Goldsmith P.K., Saggar S.K., Park S.Y., et al. 1999. Multiple endocrine neoplasia type 1: clinical and genetic features of the hereditary endocrine neoplasias. Recent Prog. Horm. Res. 54:397–438. [PubMed] [Google Scholar]
- Matkar S., Thiel A., and Hua X.. 2013. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem. Sci. 38:394–402. 10.1016/j.tibs.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.I., and Treisman R.. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 113:329–342. 10.1016/S0092-8674(03)00278-2 [DOI] [PubMed] [Google Scholar]
- Muhammad A.B., Xing B., Liu C., Naji A., Ma X., Simmons R.A., and Hua X.. 2017. Menin and PRMT5 suppress GLP1 receptor transcript and PKA-mediated phosphorylation of FOXO1 and CREB. Am. J. Physiol. Endocrinol. Metab. 313:E148–E166. 10.1152/ajpendo.00241.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai M.J., Chruszcz M., Reddy G., Grembecka J., and Cierpicki T.. 2011. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J. Biol. Chem. 286:31742–31748. 10.1074/jbc.M111.258186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni P., Chepurny O.G., and Holz G.G.. 2014. Regulation of glucose homeostasis by GLP-1. Prog. Mol. Biol. Transl. Sci. 121:23–65. 10.1016/B978-0-12-800101-1.00002-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayo Y., Murao K., Imachi H., Cao W.M., Sato M., Dobashi H., Wong N.C., and Ishida T.. 2002. The multiple endocrine neoplasia type 1 gene product, menin, inhibits insulin production in rat insulinoma cells. Endocrinology. 143:2437–2440. 10.1210/endo.143.6.8950 [DOI] [PubMed] [Google Scholar]
- Schnepp R.W., Chen Y.X., Wang H., Cash T., Silva A., Diehl J.A., Brown E., and Hua X.. 2006. Mutation of tumor suppressor gene Men1 acutely enhances proliferation of pancreatic islet cells. Cancer Res. 66:5707–5715. 10.1158/0008-5472.CAN-05-4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede F., Chepurny O.G., Kaufholz M., Bertinetti D., Leech C.A., Cabrera O., Zhu Y., Mei F., Cheng X., Manning Fox J.E., et al. 2015. Rp-cAMPS Prodrugs Reveal the cAMP Dependence of First-Phase Glucose-Stimulated Insulin Secretion. Mol. Endocrinol. 29:988–1005. 10.1210/me.2014-1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers D.A., Kieffer T.J., Hussain M.A., Drucker D.J., Bonner-Weir S., Habener J.F., and Egan J.M.. 2000. Insulinotropic glucagon-like peptide 1 agonists stimulate expression of homeodomain protein IDX-1 and increase islet size in mouse pancreas. Diabetes. 49:741–748. 10.2337/diabetes.49.5.741 [DOI] [PubMed] [Google Scholar]
- Sundaresan S., Meininger C.A., Kang A.J., Photenhauer A.L., Hayes M.M., Sahoo N., Grembecka J., Cierpicki T., Ding L., Giordano T.J., et al. 2017. Gastrin Induces Nuclear Export and Proteasome Degradation of Menin in Enteric Glial Cells. Gastroenterology. 153:1555–1567.e15. 10.1053/j.gastro.2017.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker R.V. 2010. Multiple endocrine neoplasia type 1 (MEN1). Best Pract. Res. Clin. Endocrinol. Metab. 24:355–370. 10.1016/j.beem.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Vartiainen M.K., Guettler S., Larijani B., and Treisman R.. 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 316:1749–1752. 10.1126/science.1141084 [DOI] [PubMed] [Google Scholar]
- Xing B., Wang L., Guo D., Huang J., Espenel C., Kreitzer G., Zhang J.J., Guo L., and Huang X.Y.. 2013. Atypical protein kinase Cλ is critical for growth factor receptor-induced dorsal ruffle turnover and cell migration. J. Biol. Chem. 288:32827–32836. 10.1074/jbc.M113.489427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gurung B., Wu T., Wang H., Stoffers D.A., and Hua X.. 2010a Reversal of preexisting hyperglycemia in diabetic mice by acute deletion of the Men1 gene. Proc. Natl. Acad. Sci. USA. 107:20358–20363. 10.1073/pnas.1012257107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wang H., and Hua X.. 2010b Deletion of the Men1 gene prevents streptozotocin-induced hyperglycemia in mice. Exp. Diabetes Res. 2010:876701 10.1155/2010/876701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.J., Song T.Y., Park J., Lee J., Lim J., Jang H., Kim Y.N., Yang J.H., Song Y., Choi A., et al. 2013. Menin mediates epigenetic regulation via histone H3 lysine 9 methylation. Cell Death Dis. 4:e583 10.1038/cddis.2013.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y., Li L., Cao W., Yan D., Zeng Q., Kong X., Lu L., Yan M., Xu X., Qu J., et al. 2012. The microtubule associated protein syntabulin is required for glucose-stimulated and cAMP-potentiated insulin secretion. FEBS Lett. 586:3674–3680. 10.1016/j.febslet.2012.08.025 [DOI] [PubMed] [Google Scholar]
- Yu Y., Sun Y., He S., Yan C., Rui L., Li W., and Liu Y.. 2012. Neuronal Cbl controls biosynthesis of insulin-like peptides in Drosophila melanogaster. Mol. Cell. Biol. 32:3610–3623. 10.1128/MCB.00592-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusta B., Baggio L.L., Estall J.L., Koehler J.A., Holland D.P., Li H., Pipeleers D., Ling Z., and Drucker D.J.. 2006. GLP-1 receptor activation improves beta cell function and survival following induction of endoplasmic reticulum stress. Cell Metab. 4:391–406. 10.1016/j.cmet.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Zheng B., Han M., Bernier M., and Wen J.K.. 2009. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 276:2669–2685. 10.1111/j.1742-4658.2009.06986.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw and analyzed data files for the RNA-sequencing analysis reported in this paper is GEO: GSE106653.