Abstract
Macrolides are a large group of natural products that display broad and potent biological activities and are biosynthesized by type I polyketide synthases (PKSs) and associated enzymatic machinery. There is an urgent need to access macrolides and unnatural macrolide derivatives for drug discovery, drug manufacture, and probe development. Typically, efforts to engineer the biosynthesis of macrolides and macrolide analogues in various microbial hosts are hampered by the complexity of macrolide biosynthetic pathways and our limited ability to rationally reprogram type I PKSs and post-PKS machinery. High-throughput approaches based on synthetic biology and directed evolution could overcome this problem by testing the function of large libraries of variants. Yet, methods that can identify mutant enzymes, pathways, and strains that produce the desired macrolide target are not generally available. Here we show that the promiscuous macrolide sensing transcription factor MphR is a powerful platform for engineering variants with tailored properties. We identified variants that displayed improved sensitivity towards erythromycin, tailored the inducer specificity, and significantly improved sensitivity to macrolides that were very poor inducers of the wild-type MphR biosensor. Designer macrolide biosensors should find broad utility and enable applications related to high-throughput synthetic biology and directed evolution of macrolide biosynthesis.
INTRODUCTION
Macrolides are a group of diverse natural products that display broad and potent biological activities, including antibacterial, anticancer, antifungal, antiprotozoal, and immuno-modulating activities.1, 2 Access to large quantities of macrolides and analogs thereof is critical for the discovery of new biological activities, optimization of pharmacological properties, and probe discovery and development.3, 4 Biosynthetic approaches to macrolide production offer enormous potential and numerous benefits compared to traditional chemical approaches. The scaffolds of macrolides are constructed by type I polyketide synthases (PKSs). These are large multifunctional protein complexes organized in a modular fashion.5 Each module is responsible for the selection and installation of a ketide unit into the polyketide, while a terminal cyclase is responsible for regiospecific macrocyclization and/or hydrolysis of the polyketide chain. The number, identity, order of modules, and specificity of the cyclase describes the structure of the corresponding polyketide. These scaffolds are often further elaborated by tailoring enzymes to afford the mature, biologically active natural product.
Accordingly, these systems offer the potential for the synthesis of large quantities of polyketides via microbial fermentation and combinatorial biosynthesis of analogues by mixing and matching modules and tailoring enzymes. However, the sheer size, mechanistic diversity,6 and poor understanding of how specificity and catalysis are controlled by type I PKSs and their associated post-PKS enzymatic machinery render rational design of new pathways difficult.5, 7, 8 For example, many hybrid PKSs designed to produce polyketide analogues fail or are less active than wild-type machinery.9, 10 Moreover, tailoring enzymes with the requisite regio-selectivity or substrate specificity/promiscuity are often unavailable to achieve late-stage functional modification of a given target macrolide. In addition, biosynthetic pathways often require expression in heterologous hosts to overcome limitations often associated with the intractable genetic systems, unknown regulatory systems, and poorly understood molecular biology of natural product producing microbes.11 Critically though, whereas DNA synthesis and assembly technologies can access large numbers of refactored and engineered gene clusters to address this need, methods to report their ability to produce the desired natural product in similarly high throughput are not always available. Consequently, the full synthetic potential of type I PKSs and associated biosynthetic machinery has yet to be realized. However, synthetic biology and directed evolution offer an opportunity to overcome these challenges by testing the functions of large libraries of variants. Successes in this area frequently leverage chromophores to facilitate screening12–15 or employ analytical methods that are only of moderate throughput.9, 16 Yet, the ability of synthetic biology and directed evolution approaches to be applied to macrolides is extremely limited because there are no generally applicable high-throughput tools available for screening steps of macrolide biosynthesis. For instance, macrolides are not chromophores or fluorophores and do not offer a spectrophotometric change upon tailoring that could be monitored. Moreover, post-PKS tailoring typically does not provide a sufficiently distinctive phenotype that can be leveraged. Mass spectrometry is suitable for screening small libraries of variants when the requisite instrumentation and expertise is available. Regardless, the ability of high-throughput mass spectrometry to quantify macrolides in complex mixtures and to distinguish congeners is unproven.17–20
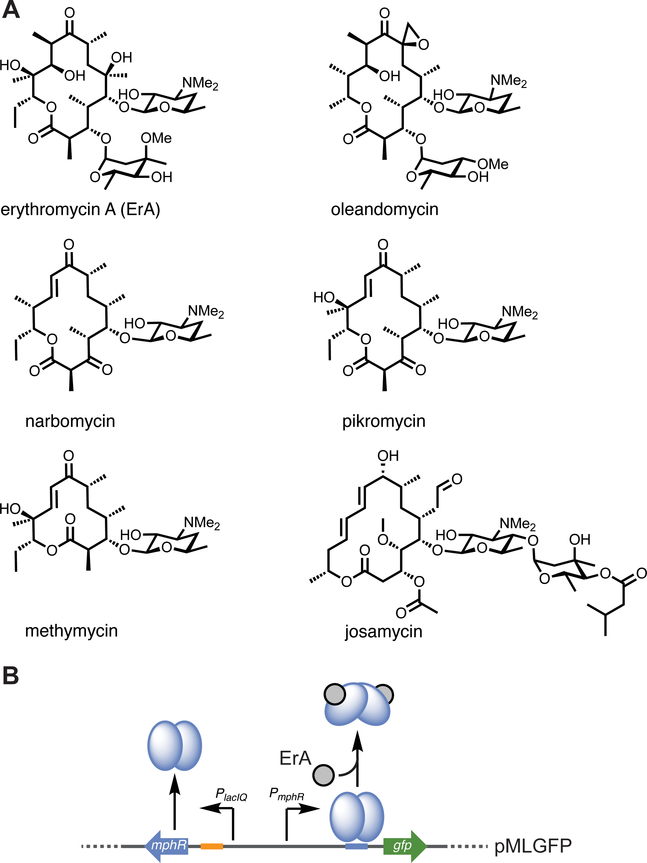
Regulatory proteins such as transcription factors have proven highly effective devices for sensitive and specific detection of small molecules.21, 22 These proteins bind a chemical effector which triggers an allosteric response that controls the transcription of one or more genes.23 Transcription factors have been described for sensing various small molecules and have enabled high-throughput screening to improve product titers, including dicarboxylic acids,24, 25 alcohols,24 phenylpropanoids,26 lactone,27 and the aromatic products of type II PKSs.28 To date though, engineered transcription factors have not been reported for the complex products of type I PKSs. MphR is a repressor protein that belongs to the TetR family of regulators,23 a group of transcription factors that are often associated with the ability to bind products of antibiotic biosynthetic pathways.29–31 Indeed, MphR controls the transcription of a gene cassette32, 33 responsible for resistance to macrolide antibiotics via phosphorylation of the desosamine 2’-hydroxy group.34 Interestingly, MphR is de-repressed by several naturally produced and semi-synthetic macrolide antibiotics, including erythromycin (ErA), josamycin, oleandomycin, narbomycin, methymycin and pikromycin, albeit with widely varied efficiencies (Figure 1A).35
Figure 1.
Inducer scope and function of the MphR biosensor. (A) Examples of naturally produced macrolides that are MphR inducers. (B) Scheme illustrating the artificial MphR regulatory system. In the presence of a macrolide activator, MphR undergoes a conformational change and leaves the cognate operator sequence, allowing transcription of the downstream reporter, GFP. For clarity, phosphorylation of ErA via MphA (MphA housed in pJZ12) is omitted from the figure.
In protein engineering, promiscuous enzymes often prove to be useful scaffolds for creating new functions by directed evolution.14, 36 We reasoned that the remarkable promiscuity of MphR could provide a platform for creating tailored MphR variants for applications related to macrolide synthetic biology and directed evolution beyond those offered by the wild-type biosensor. For example, MphR could be engineered to (1) recognize a wide variety of macrolides beyond the established inducers of MphR, (2) distinguish the desired mature natural product from biosynthetic intermediates or off-pathway byproducts, (3) recognize non-natural semi-synthetic derivatives or late-stage enzymatically functionalized analogues that enable directed evolution of novel enzymatic machinery for macrolide diversification, and (4) tailor the sensitivity, linear range of detection, and dynamic range for specific applications. Herein, we set out to explore the potential for leveraging the effector promiscuity of MphR as a platform to engineer its inducer sensitivity, specificity and selectivity to afford designer genetically encoded biosensors. The tailored MphR biosensors described here enable the application of high-throughput engineering strategies to be applied to solving long-standing problems in macrolide biosynthesis and diversification.
RESULTS AND DISCUSSION
Characterization of a prototype two-plasmid macrolide detection system in E. coli
A previously described two-plasmid system37 leverages the MphR macrolide resistance cassette to control the expression of GFP in response to erythromycin A (ErA) (Figure 1B). The plasmid pJZ12 is under control of the p15A origin (10 copies per cell) and carries a macrolide phosphotransferase (MphA), while pMLGFP houses the reporter module (PMphR-gfp) and biosensor (MphR) with a pBR322 origin of replication (15–20 copies per cell) (Figure 1B, and Supplementary Figure S1). Notably, the copy number of transcription factor based biosensor and reporter plasmids often dictates the performance of the resulting system. For example, previously described K1/2 values for MphR with ErA were 10 μM with a low (SC101 origin, 2–5 copies per cell) and 97 μM with a high copy (pUC origin, 100–500 copies per cell) number plasmid.38 Subsequently, in order to characterize the properties of the prototype sensor system, the ErA dose-response of GFP expression controlled by wild-type MphR in pMLGFP was determined and the data fitted to the Hill equation. Under these conditions, the K1/2 for ErA was 2.18 μM. Although this is several-fold more sensitive than previously reported MphR systems,38, 39 this concentration of macrolide antibiotic is still an order of magnitude higher that those typically produced by heterologous expression.40, 41 Thus, wild-type MphR might not be optimal for guiding high-throughput optimization of ErA biosynthesis in heterologous hosts. Based on these results, we set out to explore the ability of directed evolution to improve the sensitivity of MphR towards ErA.
Enhancing the sensitivity of MphR to erythromycin via multi-site saturation mutagenesis and random mutagenesis
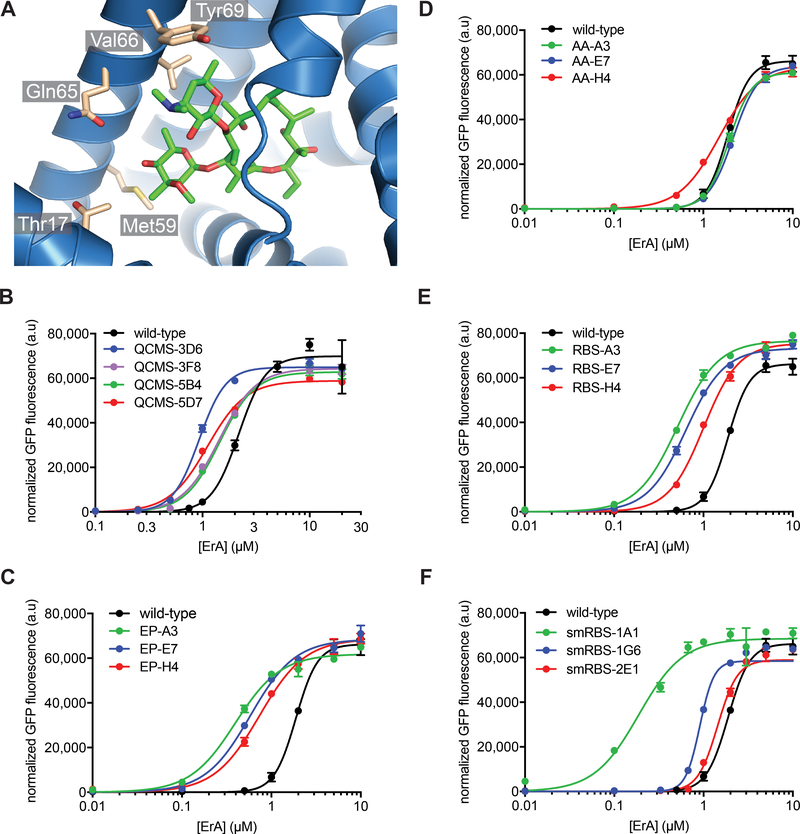
We sought to determine if the sensitivity of the wild-type MphR transcription factor with ErA could be improved via mutagenesis and screening. To this end, multi-site saturation mutagenesis and error-prone PCR (epPCR) of the MphR gene was performed, using plasmid pMLGFP as the template. The focused mutagenesis libraries were guided by the MphR crystal structure (PDB: 3FRQ).39 Five residues (Thr17, Met59, Gln65, Val66, and Tyr69) are in proximity to the ErA sugars and are predicted to drive macrolide recognition (Figure 2A). One library, designed QCMS3, included multi-site saturation mutagenesis of three residues (Thr17, Met59, and Tyr69), while the library QCMS5 was constructed by multi-site saturation mutagenesis of all five residues. A final library was generated via error-prone PCR. To simultaneously explore the potential effect of mutagenesis of both the MphR-coding sequence and the upstream non-coding region, the entire MphR transcriptional unit, including the PlacIQ promoter and ribosome binding site (RBS), were amplified by epPCR. Next, each mutant library was transformed into E. coli TOP10 cells that harbored plasmid pJZ12 and was subjected to an initial round of negative sorting in the absence of ErA via fluorescence activated cell sorting (FACS) to eliminate variants that were constitutively expressing GFP or activated by endogenous ligands. The negatively-sorted mutants were then cultured on agar plates, individual colonies were grown in 96-well microplates, and then aliquots of the microplate cultures were stored. Each remaining culture was then screened in the absence of ErA and in the presence of 0.8 μM ErA. Comparison of the normalized GFP fluorescence in each case provides the induction factor and a measure of the ErA sensitivity.
Figure 2.
Characterization of MphR biosensor variants with increased sensitivity to ErA. (A) Ligand binding site of MphR (monomer A, PDB: 3FRQ) with residues surrounding the sugars of ErA (green sticks) that were selected for multi-site saturation mutagenesis shown as sticks and labelled. (B) Dose-response curves of the wild-type MphR biosensor strain and selected QCMS biosensor variants. (C) Dose-response curves of the wild-type MphR biosensor strain and sensitivity clones containing RBS and amino acid mutations. (D) Dose-response curves of the wild-type MphR biosensor strain and variants with amino acid mutations and the wild-type RBS. (E) Dose-response curves of the wild-type MphR biosensor strain and variants with variant RBS sequences and wild-type amino acid sequence. (F) Dose-response curves of the wild-type MphR biosensor strain and selected variants from RBS saturation mutagenesis libraries. Error-bars represent the standard error of the mean (n=3) and are only visible when larger than the data point symbol.
After application of this sequential negative-positive screening procedure, several mutants from the QCMS libraries that displayed improved sensitivity with ErA compared to the wild-type MphR were identified. The GFP fluorescence of each biosensor strain in the presence of various concentrations of ErA was determined. The biosensor performance characteristics were extracted by fitting the dose response data to the Hill equation (Figure 2B and Table 1). Notably, Thr17 is mutated in four of the five variants that were analyzed, and the most sensitive variant, T17R, does not possess mutations at the other positions that were targeted for mutagenesis (Met59, Gln65, Val66, and Tyr69). As judged by the K1/2, T17R (QCMS-3D6) is at least two-fold more sensitive with ErA than wild-type MphR. In addition, mutants T17A/M59S (QCMS-3F8), T17G/Q65M (QCMS-5B4), and T17A/M59E (QCMS-5D7) were ~1.4-fold more sensitive with ErA compared to the wild-type MphR (Figure 2B and Table 1).
Table 1.
Performance features of MphR biosensor strains with improved ErA sensitivity.
| Biosensor | RBS sequencea | Amino acid mutations | Ligand | K1/2 (μM)b | Dynamic rangec | nd |
|---|---|---|---|---|---|---|
| Wild-type | AGAAGGT | - | ErA | 1.97 ± 0.03 | 65,000 | 3.8 ± 0.3 |
| QCMS-3D6 | AGAAGGT | T17R | ErA | 0.92 ± 0.03 | 67,000 | 3.5 ± 0.2 |
| QCMS-3F8 | AGAAGGT | T17A/M59S | ErA | 1.43 ± 0.03 | 64,000 | 2.5 ± 0.1 |
| QCMS-5B4 | AGAAGGT | Q65M | ErA | 1.45 ± 0.03 | 63,000 | 2.6 ± 0.1 |
| QCMS-5D7 | AGAAGGT | T17A/M59E | ErA | 1.11 ± 0.02 | 60,000 | 2.4 ± 0.1 |
| EP-A3 | GGAAGGT | G76C | ErA | 0.39 ± 0.02 | 64,000 | 2.0 ± 0.1 |
| EP-E7 | AGATGGT | V90I | ErA | 0.58 ± 0.02 | 70,000 | 2.5 ± 0.1 |
| EP-H4 | AGAAGGC | R24H/K35N | ErA | 0.74 ± 0.03 | 68,000 | 2.4 ± 0.1 |
| AA-A3 | AGAAGGT | G76C | ErA | 1.94 ± 0.02 | 61,000 | 3.4 ± 0.1 |
| AA-E7 | AGAAGGT | V90I | ErA | 2.17 ± 0.3 | 63,800 | 3.1 ± 0.1 |
| AA-H4 | AGAAGGT | R24H/K35N | ErA | 1.88 ± 0.03 | 63,700 | 3.6 ± 0.3 |
| RBS-A3 | GGAAGGT | - | ErA | 0.52 ± 0.02 | 78,000 | 1.9 ± 0.1 |
| RBS-E7 | AGATGGT | - | ErA | 0.64 ± 0.02 | 74,000 | 2.0 ± 0.1 |
| RBS-H4 | AGAAGGC | - | ErA | 1.00 ± 0.02 | 75,000 | 2.2 ± 0.1 |
| smRBS-1A1 | TTCAGGT | - | ErA | 0.19 ± 0.02 | 70,000 | 1.7 ± 0.3 |
| smRBS-1G6 | CTGAGGT | - | ErA | 0.91 ± 0.04 | 64,000 | 5.5 ± 1.2 |
| smRBS-2E1 | AAAGGTT | - | ErA | 1.44 ± 0.08 | 64,000 | 3.9 ± 0.5 |
Mutated nucleotides are underlined
Concentration of ligand at half maximum normalized GFP fluorescence
GFPmax – GFPmin
Hill coefficient, a measure of cooperativity within the MphR biosensor. Values >1 indicate positive cooperativity.
Three improved clones were also chosen for detailed analysis from the epPCR library, including “EP-A3” which was nearly five times more sensitive with ErA than the wild-type MphR, as judged by the K1/2 (Figure 2C and Table 1). Interestingly, all three top hits from the epPCR library included mutations to the ribosome binding site (RBS) of MphR in addition to amino acid mutations within the MphR coding region. To determine whether the RBS and/or amino acid mutations contribute to improved sensitivity in each case, site-directed mutagenesis was used to generate variants that either carried the amino acid mutations from each original clone (AA-A3, AA-E7, and AA-H4) or the RBS mutations alone (RBS-A3, RBS-E7, and RBS-H4). Dose response curves of the six new variants with ErA were determined, revealing that the RBS mutations alone were sufficient to afford ErA sensitivity to nearly the same levels of the original variants (Figure 2D/E and Table 1). The amino acid mutations alone (Figure 2D) resulted in MphR variants with dose response curves that were indistinguishable from that of the wild-type MphR. Taken together, these results indicate the ErA sensitivity of variants EP-A3, EP-E7, and EP-H4 is a direct consequence of mutations to the RBS of MphR.
Random Mutagenesis of the RBS of MphR
Given that random mutagenesis throughout the entire MphR coding and non-coding region provided hits with improved ErA sensitivity due to RBS mutations, we hypothesized that since the epPCR library was not exhaustively screened, further improvements could be identified by focusing random mutations to the RBS and screening a small portion of the corresponding library. Accordingly, a saturation mutagenesis library of the MphR RBS was constructed and a small number of library members were used to determine the normalized GFP fluorescence at various concentrations of ErA (~200 unique colonies for each concentration). Most of these library members did not support GFP expression at any concentration of ErA that was tested (Supplementary Figure S2). For example, at 2.0 μM ErA, <20% of the library members resulted in detectable GFP expression. However, even at the lowest concentration of ErA that was tested (0.2 μM), which is two-fold lower than the K1/2 of the most sensitive MphR variant from the previous libraries (EP-A3), several variants were identified that were up to 10-fold more strongly induced with ErA than the wild-type MphR. The top three strains were further characterized by dose-response curve analysis (Figure 2F). The most sensitive clone, smRBS-1A1, was fully ten-fold more sensitive to ErA than the wild-type MphR, and two-fold more sensitive than previously described EP-A3, as judged by the K1/2’s (Table 1). Interestingly, the predicted strength42, 43 of the RBS sequences of the RBS mutants did not correlate with the sensitivity (K1/2) of each variant (R2=0.27, data not shown). The Hill coefficient of each RBS mutant was >1, indicating positive cooperativity, as expected, while that of smRBS-1A1 was the lowest (1.7 ± 0.3) among those analyzed. The benefit of a lower Hill coefficient is a broader linear range of detection afforded by smRBS-1A1 compared to the other variants (Figure 2F). Gratifyingly, the dynamic range of each RBS mutant biosensor was close (within 15%) to that of the wild-type MphR. Ultimately, by screening less than 200 MphR variants, out of a theoretical library consisting of 4092 variants, the sensitivity of MphR towards its native inducer, ErA, was improved 10-fold. Thus, RBS engineering is an efficient strategy to improve the sensitivity of MphR.
Narrowing the Macrolide Inducer Specificity of MphR
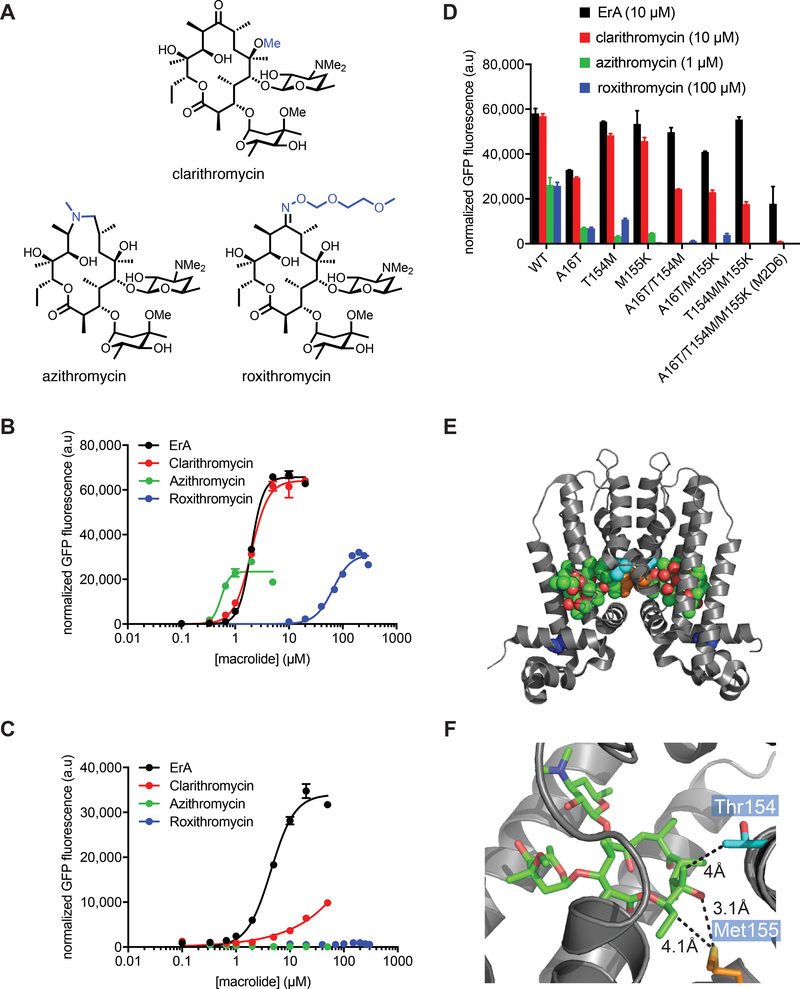
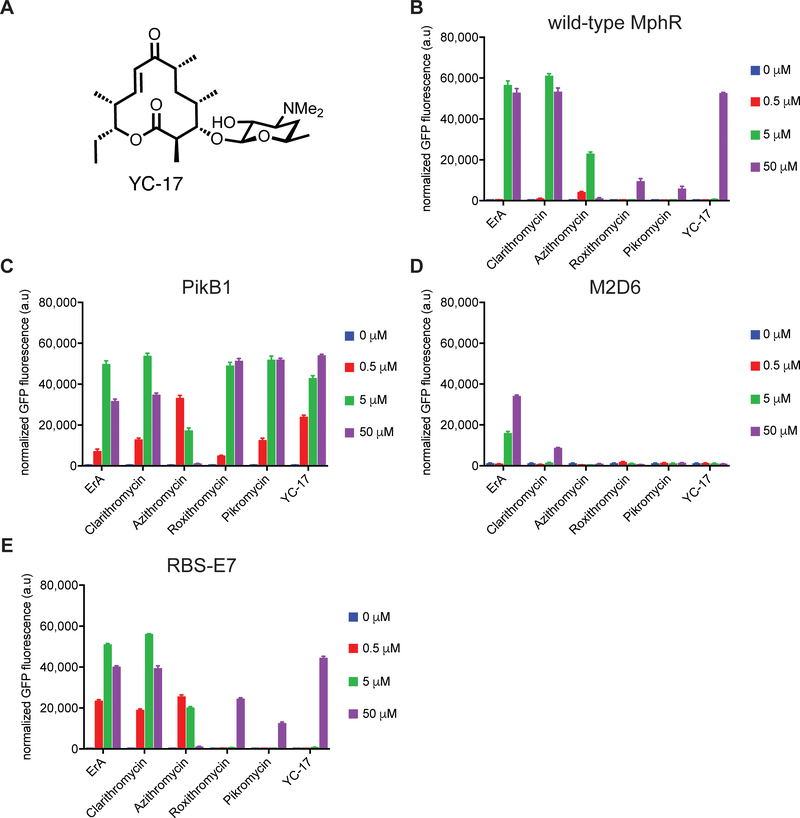
Wild-type MphR is activated by a variety of 12-, 14-, 15-, and 16-membered macrolides, including the semi-synthetic analogues clarithromycin, azithromycin, and roxithromycin (Figure 3A).35 A dose-response analysis of wild-type MphR with ErA, clarithromycin, azithromycin, and roxithromycin reveals a broad inducer specificity (Figure 3B). The K1/2’s for clarithromycin, azithromycin, and roxithromycin are 1.4-, 0.3-, and 34-fold that of ErA, respectively (Table 2). Thus, while roxithromycin is detected much more poorly than ErA, wild-type MphR does not discriminate between clarithromycin and ErA, and azithromycin is a stronger inducer than ErA with respect to the K1/2, but also displays a several-fold poorer dynamic range than ErA. To utilize MphR as a biosensor, its specificity likely requires tuning towards certain ligands to render it more selective. For example, future MphR-based applications might include determining the concentration of a target macrolide in a complex macrolide mixture, in which case the biosensor should be induced by a narrow range of ligands to avoid false positives. Similarly, to guide ultra high-throughput engineering of macrolide producing microbial strains, a biosensor should detect the final product of a macrolide biosynthetic pathway (e.g. ErA). In this case, MphR should not be induced by intermediates along the biosynthetic route to the final product (e.g. erythromycins B, C, and D).44, 45 We therefore turned our attention to exploring whether directed evolution could be used to narrow the ligand specificity of MphR. Thus, we set out to explore whether MphR variants could be identified that were induced by ErA but not clarithromycin, azithromycin, and roxithromycin. Given that clarithromycin only differs from ErA by the presence of a single O-methyl group at C6 and it is a strong inducer of MphR, shifting specificity away from this ligand along with azithromycin and roxithromycin was considered a particularly challenging goal and a robust test of our molecular engineering approach.
Figure 3.
Characterization of MphR biosensor variants with engineered narrow macrolide specificity. (A) Structures of ErA and semisynthetic derivatives. Structural differences compared to ErA are highlighted blue. (B) Wild-type MphR dose-response curves. (C) MphR variant M2D6 dose-response curves. (D) Activities of individual and combination mutations from M2D6 using 10 μM ErA, 10 μM clarithromycin, 1 μM azithromycin, and 100 μM roxithromycin. Error-bars represent the standard error of the mean (n=3). (E) The crystal structure of MphR (PDB: 3FRQ) complexed with ErA (green space filled) showing locations of amino acid sidechains (space filled) mutated in M2D6. Blue, Ala16; cyan, Thr54; orange, Met55. (F) Ligand binding site of MphR showing locations of Thr54, Met55, and ErA (green sticks).
Table 2.
Performance features of the wild-type MphR, ErA-selective biosensor strain M2D6, and PikB1.
| Biosensor | Amino acid mutations | Ligand | K1/2 (μM)a | Dynamic Rangeb | nc | Selectivityd |
|---|---|---|---|---|---|---|
| Wild-type | - | ErA | 1.97 ± 0.03 | 65,000 | 3.83 ± 0.3 | 1 |
| - | clarithromycin | 1.98 ± 0.06 | 64,000 | 2.71 ± 0.2 | 0.99 | |
| - | azithromycin | 0.55 ± 0.04 | 28,000 | 5.31 ± 1.6 | 3.58 | |
| - | roxithromycin | 65.7 ± 3.5 | 32,000 | 2.60 ± 0.3 | 0.03 | |
| M2D6 | M2D6 A16T/T154M/M155K | ErA | 4.64 ± 0.2 | 39,000 | 2.27 ± 0.2 | 1.00 |
| clarithromycin | 21.5 ± 2 | 7,000 | 2.20 ± 0.1 | 0.22 | ||
| azithromycin | N.D.e | N.D.e | N.D.e | N.D.e | ||
| roxithromycin | N.D.e | N.D.e | N.D.e | N.D.e | ||
| Wild-type | - | pikromycin | 97± 2 | 34,000 | 2.5 ± 0.1 | N.Af |
| PikB1 | S106F | pikromycin | 0.79 ± 0.02 | 62,000 | 1.80 ± 0.1 | N.Af |
Concentration of ligand at half maximum normalized GFP fluorescence
GFPmax – GFPmin
Hill coefficient, a measure of cooperativity within the MphR biosensor. Values >1 indicate positive cooperativity.
Ratio of K1/2(ErA) / K1/2(ligand)
Not Detected
Not Applicable
An epPCR MphR library was sorted by FACS in the presence of clarithromycin, azithromycin, and roxithromycin, and a portion of the least fluorescent cells (~7% of the total population) was collected. This population of MphR variants was incubated on agar plates and several hundred individual clones were then cultured in microplates and screened against ErA, clarithromycin, azithromycin and roxithromycin. Several clones were identified that supported high GFP fluorescence in the presence of ErA, but low GFP fluorescence in the presence of clarithromycin, azithromycin and roxithromycin, indicating that these clones might be more specific to ErA. One particularly promising clone, M2D6, was chosen for further analysis, and a dose-response curve was determined with each macrolide. In stark contrast to wild-type MphR, GFP induction of M2D6 was not detected with azithromycin and roxithromycin over a four-order of magnitude range of macrolide concentration (Figure 3C). The K1/2 value of M2D6 for clarithromycin was 10-fold higher than the wild-type MphR (Table 2). Together, these data describes a variant MphR with dramatically shifted specificity towards ErA.
Sequencing of the gene encoding M2D6 revealed three amino acid changes compared to the wild-type MphR sequence, A16T, T154M, and M155K. To determine which of these mutations contribute to the dramatic specificity shift of the engineered MphR variant, all six single and double mutant combinations were constructed by site-directed mutagenesis. Subsequently, the induction of each variant was analyzed at a fixed concentration of macrolide by measuring the GFP fluorescence in the presence and absence of each macrolide (Figure 3D). Notably, five of the single and double mutant combinations are induced by all four macrolides, in a similar fashion to the wild-type MphR. The double mutant T154M/M155K is not induced by azithromycin and roxithromycin at the concentrations tested, but displays significant induction by clarithromycin. The remaining mutation, A16T, only impacts specificity when in combination with the other two mutations. These data indicates that all three amino acid changes in M2D6 are necessary to achieve the full specificity shift towards ErA. According to the MphR crystal structure,39 Thr154 and Met155 (from the other subunit in the homodimer) are located at the rear of the ligand binding cavity close to the dimerization motif (Figure 3E) and are within ~4Å of the C12-C13 substituents of ErA (Figure 3F). Ala16 on the other hand, is located within the DNA-binding domain of MphR (Figure 3E). The proximity of Thr154 and Met155 to the ligand binding site of MphR, coupled with the proximity of Ala16 to the DNA binding site of MphR, could explain why all three are ultimately required to drive the narrow inducer specificity of M2D6. However, these exact amino acid mutations are difficult to precisely rationalize a posteriori.
Improving the Sensitivity of MphR Towards Macrolides that are Poor Inducers of Wild-Type MphR
Although the known inducer specificity of wild-type MphR is quite broad, some macrolides are only poorly detected, and many others do not activate MphR at all.35 Given that MphR, to the best of our knowledge, is the only experimentally confirmed macrolide-specific transcription factor known, we set out to improve the sensitivity of MphR towards macrolides that are very poor inducers of the wild-type biosensor strain. Pikromycin is a 14-membered macrolide natural product produced by Streptomyces venezuelae ATCC 15439.46 Compared to ErA, the structure of pikromycin lacks the cladinose sugar at C3 of ErA, lacks a hydroxyl group at C6, and includes a double bond at C9-C10 in place of a methyl and hydroxyl, respectively (Figure 1A). Several other macrolides are also produced by S. venezuelae including methymycin, neomethymycin, novamethymycin, neopikromycin, and novapikromycin.46 The ability to utilize a biosensor-based platform for screening the production of macrolides from S. venezuelae is currently hindered by the poor activation of the wild-type biosensor by pikromycin and related compounds.35
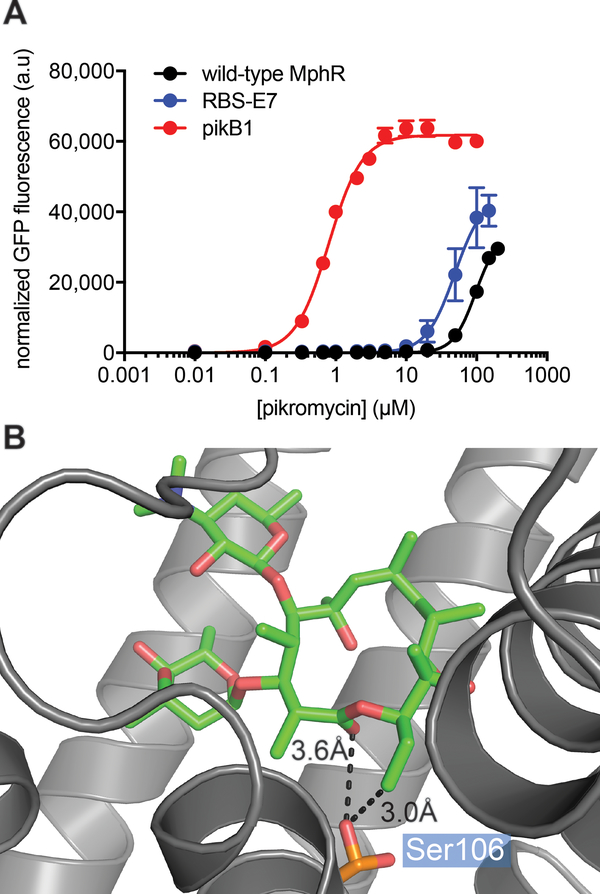
Given the success of our previous epPCR libraries to yield biosensors with altered inducer specificities, a previously negatively sorted (in the absence of ligand) epPCR MphR library was screened in the presence of pikromycin. One specific clone, designated pikB1, was chosen for further analysis. Dose-response curve analysis revealed that the wild-type MphR was ~50-fold less sensitive with pikromycin than with ErA, as judged by the K1/2’s. In contrast, the mutant biosensor pikB1 is 123-fold more sensitive to pikromycin than the wild-type, as judged by the K1/2’s (Figure 4A and Table 2). Sequencing analysis revealed that a single amino acid change, S106F, was responsible for the improvement in sensitivity. The structure of pikromycin differs from that of ErA most notably at the C3 position, where a sugar is attached in ErA but not pikromycin. Interestingly, according to the crystal structure of MphR bound with ErA, Ser106 is located far from this site, and is in proximity to the ester and C12-ethyl side chain of ErA (Figure 4B). This highlights the ability of random mutations to arrive at molecular solutions that are difficult to predict a priori. To determine whether this amino acid change is a broad sensitivity enhancer, or if it is specific to pikromycin, the inducer specificity of MphR pikB1 along with that of the RBS-E7, M2D6, and wild-type biosensors was determined with ErA, clarithromycin, azithromycin, roxithromycin, pikromycin, and YC-17 (a 12-membered polyketide produced by S. venezuelae, Figure 5A)46. Notably, even though the ability of YC-17 to induce wild-type MphR at high concentration (50 μM) is similar to that of ErA (Figure 5B), YC-17 does not support detectable GFP fluorescence at any of the lower concentrations that were tested. In contrast, PikB1 is induced by YC-17 at every concentration tested (Figure 5C). The sensitivity of PikB1 was also significantly improved towards roxithromycin, compared to the wild-type MphR biosensor strain, even though the structure of roxithromycin is most similar to that of ErA. The sensitivity of PikB1 towards ErA, clarithromycin, and azithromycin were only improved compared to the wild-type biosensor at 0.5 μM macrolide. Thus, while S106F does impart improved sensitivity towards every macrolide tested, the ligand used in screening the MphR library, pikromycin, displays the largest improvement. Interestingly, M2D6 was not induced by pikromycin or YC-17 (Figure 5D), further emphasizing the narrow inducer specificity of this evolved MphR variant. In addition, the ErA sensitivity mutant RBS-E7 displayed some improvements in sensitivity towards clarithromycin, azithromycin, but specificity against roxithromycin, pikromycin, and YC-17 was similar to that of the wild-type MphR (Figure 5E). Cumulatively, these results highlight the facile ability of random mutations to improve the sensitivity of MphR towards otherwise very poor inducers, including 12- and 14-membered macrolides.
Figure 4.
Characterization of a MphR biosensor optimized towards pikromycin. (A) Dose-response analysis of wild-type MphR and PikB1 with the inducer pikromycin. Error-bars represent the standard error of the mean (n=3) and are only visible when larger than the data point symbol. (B) The crystal structure of MphR (PDB: 3FRQ) complexed with ErA (green sticks). Ser106 is shown as orange sticks.
Figure 5.
Macrolide specificity of wild-type MphR and variant biosensor strains. (A) Structure of YC-17. Macrolide specificity of (B) wild-type MphR, (C) PikB1, (D) M2D6, and (E) RBS-E7. Error-bars represent the standard deviation of the mean (n=3).
Validation of a Biosensor Microplate Screen and Growth Selection
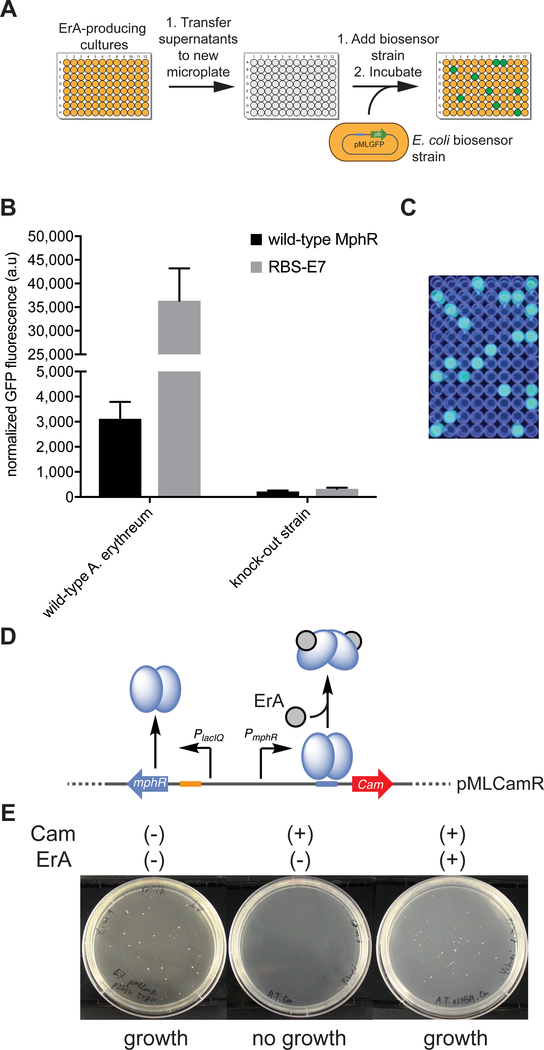
Leveraging genetically-encoded biosensors to guide high-throughput engineering of macrolide biosynthesis is an attractive approach to overcome our limited understanding of how to rationally redesign complex biosynthetic pathways in producing microbes or heterologous hosts. In many cases, a suitable phenotypic screen is not available to specifically detect the target natural product, biosynthetic intermediate, or semi-synthetic derivative. To demonstrate the utility of engineered MphR to detect macrolide production in situ, we set out to couple ErA production in an ErA-producing strain to our whole cell biosensor strain in microplates. In principle, this approach is highly versatile and modular: it can be coupled to any producing microbe of interest, is relatively high-throughput, and the sensitivity, dynamic range, and ligand scope of detection can be controlled by the choice of MphR variant housed in the biosensor strain. The soil-dwelling microbe Aeromicrobium erythreum47, 48 was chosen as the test strain to showcase this screening approach. In contrast to the well-known ErA producer Saccharopolyspora erythraea, A. erythreum does not undergo a multistage life-cycle and produces ErA from the start of culturing.48, 49 Moreover, A. erythreum produces ErA as the sole major product,48 whereas other producing strains also accumulate the intermediates erythromycins B and C. The A. erythreum genome has been fully sequenced50 and the ErA biosynthetic cluster fully annotated.48 Accordingly, application of plasmid-based and genomic engineering methods to manipulate macrolide biosynthesis in A. erythreum is an attractive option. To simulate the parallel biosensor-guided screening of ErA production in multiple samples, and to validate the ability of MphR biosensors to discriminate between producing and non-producing strains, single colonies of A. erythreum and a knock-out strain51 that does not produce ErA were used to inoculate individual wells of a microplate. Following incubation, the culture supernatants were mixed with the E. coli sensor strain harboring either the wild-type MphR or the MphR variant RBS-E7, incubated, and the GFP fluorescence measured (Figure 6A). The difference in fluorescence between the wild-type producing strain vs. the knock-out strain was ~90-fold with the engineered biosensor, compared to ~18-fold with the wild-type MphR biosensor strain (Figure 6B). As expected, the biosensor strain clearly identified every well that corresponded to the producing strain (Figure 6C). These data is consistent with the improved ErA sensitivity of RBS-E7 compared to the wild-type MphR and demonstrates that the dynamic range of ErA detection with the engineered MphR variant is far greater than that of the wild-type biosensor strain. Moreover, our results validates the ability of MphR to detect ErA production in culture supernatants without purification or concentration.
Figure 6.
MphR-enabled high-throughput screening and growth selection. (A) General workflow for MphR-based biosensor screening of macrolide producing microbial strains in liquid culture. (B) Fluorescent readout of ErA production in culture supernatants of A. erythreum wild-type and knock-out strain using wild-type MphR and variant RBS-E7. Error-bars represent the standard deviation of the mean (n=3). (C) Photograph of a MphR biosensor strain microplate under UV light showing clear distinction of wells containing supernatants from wild-type A. erythreum or the knock-out strain. (D) Configuration of the MphR biosensor as a growth selection system. Cam, chloramphenicol resistance gene. (E) E. coli pMLCamR biosensor strain (MphR variant RBS-E7) grows in the presence of ErA and chloramphenicol (Cam), but not when ErA is absent.
Next, we set out to demonstrate the ability of the MphR biosensor regulatory system to function as a growth selection. The gfp reporter gene in pMLGFP was replaced with the chloramphenicol (Cam) resistance gene, Cam, generating the plasmid pMLCamR (Figure 6D). Subsequently, E. coli cells that harbor pMLCamR and the accessory plasmid pJZ12 should only grow in the presence of Cam when ErA is provided in the culture media or produced by the host strain. As expected, after incubating the MphR biosensor strain on agar plates, colonies were observed only when ErA was provided in the presence of Cam (Figure 6E). Thus, coupling MphR to the regulation of antibiotic resistance markers enables a simple growth selection that could be leveraged to select ErA-producing microbial strains from large libraries of variants.
Discussion
In general, transcription factor based biosensors offer tremendous potential as tools to guide high-throughput engineering of biosynthetic pathways.21, 22, 25 However, in many cases transcription factors with the requisite inducer specificity or performance characteristics are not available. Here we demonstrate for the first time, that the inherent macrolide inducer promiscuity35 of the MphR repressor protein provides a highly flexible platform for creating new inducer specificities and sensitivities. In this study, the sensitivity of MphR towards its likely native ligand, ErA, was improved as much as 10-fold, compared to the wild-type MphR, without significant loss of dynamic range. The largest improvements in sensitivity came from a simple RBS engineering strategy, whereby only a relatively small portion of the MphR RBS library was screened. Our data and other recent examples52 suggests that other transcription factor based biosensors could be engineered using a similar strategy.
The macrolide inducer specificity of MphR has been previously reported to be quite broad.35 In our study, the ligand promiscuity was quantitatively described with a panel of three naturally occurring macrolides and three semi-synthetic derivatives. According to this analysis, while all six macrolides are inducers of MphR, the strength of induction varies widely across the series. For example, roxithromycin and pikromycin induce MphR ~10-fold more poorly than ErA, even at concentrations as high as 50 μM. While these differences in the ability of MphR to detect these macrolides might in part be dependent on the uptake of each effector by the host strain, we set out to determine whether engineering the transcription factor directly could impact the detection capabilities of the biosensor strain. Remarkably, a single round of random mutagenesis and screening was sufficient to yield an MphR variant with a narrow inducer specificity. By first negatively screening the library of random variants in the presence of a mixture of clarithromycin and azithromycin, and then positively screening in the presence of ErA, the variant M2D6 was obtained that was almost completely selective for ErA. This is a considerable achievement given that the structure of clarithromycin differs from ErA by just one methyl group, while at the same time, azithromycin and roxithromycin are structurally quite different from ErA. Notably, while two amino acid mutations in M2D6 are located in the ligand binding domain of MphR, and the third is located in the DNA binding domain, all three mutations were shown to be required for the full ErA selectivity of M2D6. Moreover, MphR functions as a homodimer, and in M2D6, the mutation M155K likely impacts ligand binding in the other subunit, given its location in the crystal structure.39 In general, mutations to MphR could therefore impact dimerization of the two MphR subunits, DNA binding, ligand binding in both subunits, or allosteric signal transduction. Consequently, the mutations found in M2D6 (and other variants) would have been difficult to rationally predict. We were also able to improve the sensitivity of MphR towards pikromycin 120-fold via random mutagenesis and screening. This is notable because pikromycin, along with several other macrolides, is an exceedingly poor inducer of the wild-type MphR. Thus, directed evolution of MphR offers the possibility of improving the detection capabilities of MphR to include a wide range of cell-permeable macrolides.
To showcase the potential utility of an MphR-based biosensor strain, ErA producing strains of A. erythreum were rapidly identified from a non-producing knock-out strain in microplates. The ability to screen macrolide production in microplates by simply mixing the biosensor strain with supernatants or extracts from producing trains is potentially highly versatile and modular. MphR biosensors may be applied in a strain agnostic manner to screen liquid cultures of a variety of microorganisms for production of macrolides. Furthermore, because this approach relies directly on the chemical structure of a given macrolide and not its biological activity, this strategy can be applied to any macrolide that is an inducer of the wild-type or engineered MphR, including unnatural macrolides that might be valuable scaffolds for semi-synthesis and drug discovery.
Designer MphR variants with tailored macrolide inducer specificities or selectivities should find utility in many applications related to macrolide synthetic biology and metabolic engineering. For example, the sensitivity of MphR could be fine-tuned to match a specific screening regime expected for a given macrolide. This could enable MphR variants as high-throughput tools to guide high-throughput gene cluster refactoring, directed evolution of chimeric or hybrid biosynthetic pathways, cell-free macrolide biosynthesis, and metagenomic searches for macrolide producing or modifying genes. Furthermore, we envision that the ability of engineered MphR to discriminate between closely related macrolide derivatives (e.g. ErA vs. clarithromycin) might be leveraged in the design of high-throughput screens and selections to identify mutant enzymes that catalyze the regioselective modification of macrolides. In addition, the ability to tune selectivity towards a narrow range of macrolide inducers should prove useful to construct MphR variants that specifically recognize the end-products of macrolide biosynthetic pathways, or that recognize target precursors en route to the final product. Finally, the ability to significantly improve the sensitivity of MphR towards macrolides that are otherwise very poor inducers of the wild-type biosensor should dramatically expand the scope and utility to include a broad range of macrolides beyond those described here.
Curiously, the structure of MphR revealed a chloride anion and a lysine side-chain in the ligand binding pocket in proximity to the 2’-phosphorylation site of ErA, suggesting that the products of self-resistance might also be effectors.39 However, previous studies have not yet focused on determining the role that the kinase MphA plays in macrolide detection by MphR, and it is currently not clear whether MphR absolutely depends on macrolide phosphorylation for induction. Subsequently, we cannot rule out the possibility that the mutations described here have impacted the ability of MphR to discriminate between phosphorylated and non-phosphorylated macrolide derivatives. Along with corresponding structural studies, this is the subject of future work in our laboratories.
The MphR repressor protein has been demonstrated to have a remarkable plasticity for engineering new sensitivities and selectivity. By tuning these characteristics, the MphR biosensor system may be tailored for many applications including environmental testing, metagenomics, synthetic biology, and metabolic engineering, in a way that is completely independent of the macrolide biological activity.
MATERIALS AND METHODS
Strains, Media, and Chemicals
The strains and plasmids used in this study are listed in Table S1. E. coli strains 10G (Lucigen) and TOP10 (Invitrogen) were used for cloning and biosensor expression, respectively. Aeromicrobium erythreum NRRL B3381 and the corresponding knock-out strain were a kind gift from Prof. Eric Miller, NC State University. The plasmid pMLGFP was used as a template for MphR mutagenesis and to express the reporter gene in the biosensor strains. Bacteria were grown in Luria Broth supplemented with ampicillin and tetracycline as appropriate. ErA, clarithromycin, azithromycin, and roxithromycin were from Sigma-Aldrich and pikromycin was from Abcam. YC-17 was a kind gift from Prof. David Sherman, University of Michigan. Each macrolide was prepared in dimethyl sulfoxide (DMSO) to a stock concentration of 50 mM, 5 mM, 500 μM, or 50 μM. All other chemicals were purchased from Sigma-Aldrich unless stated otherwise. PCR products were extracted with a Bio Basic Gel Extraction Kit. Restriction enzymes were purchased from New England Biolabs. Plasmids were isolated using a plasmid miniprep kit from Bio Basic.
Error-prone PCR of MphR
Error-prone PCR was performed using a Genemorph II kit (Agilent) in a total volume of 25 μL using 0.5 ng, 5.0 ng, or 50 ng of pMLGFP as template, 0.8 mM dNTPs, 1.25% (v/v) DMSO, 0.5 μL Mutazyme II, and primers 1-EP-for and 2-EP-rev (Supplemental Table S1). Thermocycling conditions were those recommended by the manufacturer instructions. The PCR product was gel purified and double digested using XbaI and EcoNI following standard protocols. The double digested product was ligated at 16 °C for 18 h into similarly treated pMLGFP. The ligation mixture was electroporated into E. cloni 10G electrocompetent cells and grown overnight according to the manufacturer instructions. An aliquot of the overnight culture was plated onto LB agar plates supplemented with 100 μg/mL ampicillin. Randomly selected colonies were used to prepare plasmids for DNA sequencing, revealing on average 4 nucleotide mutations per MphR gene. The rest of the original overnight culture was grown as an overnight liquid culture in LB supplemented with 100 μg/mL ampicillin and a glycerol stock was prepared of the mutant library. The epPCR plasmid library was purified and stored.
Multi-Site Saturation Mutagenesis of MphR
Plasmid pMLGFP was used as template for multi-site saturation mutagenesis using the QuickChange Multi Site-Directed Mutagenesis Kit (Agilent). Briefly, 200 ng of pMLGFP, 1 mM of dNTPs, 3.5 μL of buffer, 1 μL of DNA polymerase, and primer 3–3QCMS, 4–3QCMS, 5–3QCMS (3 site) and 6–5QCMS, 7–5QCMS, 8–5QCMS, or 9–5QCMS (5 site) (see Supplemental Table 1) in a total volume of 25 μL were used in a PCR reaction following the manufacturer instructions. The reaction products were digested with DpnI for 1 h at 37 °C and electroporated into E. cloni 10G electrocompetent cells and incubated overnight according to the manufacturer’s instructions. An aliquot of the overnight culture was plated onto LB agar plates supplemented with 100 μg/mL ampicillin. DNA sequencing of purified plasmids from randomly selected colonies confirmed that the targeted residues were mutated. The rest of the original overnight culture was grown as an overnight liquid culture in LB supplemented with 100 μg/mL ampicillin and a glycerol stock was prepared of the mutant library. The plasmid library was purified and stored.
Site-directed Mutagenesis of MphR
Site-directed mutagenesis for construction of RBS-A3, RBS-E7, and RBS-H4 was carried out using QuikChange Mutagenesis (Agilent) in a total volume of 25 μL containing 200 ng of pMLGFP, 200 μM dNTPs, 5 μL buffer, 0.25 μL Phusion Hot Start Polymerase (Thermo Scientific), and the primers 10-RBSA3-for and 11-RBSA3-rev (RBS-A3), 12-RBSE7-for and 13-RBSE7-rev (RBS-E7), 14-RBSH4-for and 15-RBSH4-rev (RBS-H4) and 16-WTRBS-for and 17-WTRBS-rev (WT RBS) (see Supplemental Table S1). For construction of AA-A3, AA-E7, AA-H4, the plasmid from the corresponding error-prone-PCR library was used (EP-A3, EP-E7, and EP-H4, respectively) along with the primers 16-WTRBS-for and 17-WTRBS-rev. For construction of the single and double mutants of M2D6, the template pMLGFP was used along with a suitable combination of primers (entries 20–25, see Supplemental Table S1). The double mutant T154M/M155K was constructed via “round the horn” mutagenesis53 using primers 22-M155K-for 21-T154M-rev and the template pMLGFP. Standard Phusion thermocycling protocols were used with the site-directed mutagenesis protocol as described by the manufacturer instructions. The reaction product was digested with DpnI for 1 h at 37 °C and electroporated into E. cloni 10G electrocompetent cells and incubated overnight according to the manufacturer’s instructions. An aliquot of the overnight culture was plated onto LB agar plates supplemented with 100 μg/mL ampicillin. The DNA sequences of plasmids from individual transformants confirmed that the correct mutation was obtained and that there were no spurious mutations. The sequence of variant RBS-H4 included two nucleotide mutations upstream of the MphR RBS. Purified plasmids were transformed into TOP10 competent cells with plasmid pJZ12 and grown overnight in LB media supplemented with 3.4 μg/mL tetracycline and 67 μg/mL ampicillin. Glycerol stocks were prepared.
Fluorescence Activated Cell Sorting (FACS) of Biosensor Libraries
The QCMS and error-prone pMLGFP plasmid libraries were transformed into TOP10 cells with plasmid pJZ12 and grown overnight in LB media supplemented with 3.4 μg/mL tetracycline and 67 μg/mL ampicillin. The culture was diluted 10-fold in fresh LB media and sorted using a Beckman Coulter MoFlo cell sorter equipped with a 100 μM nozzle operating at 22 psi using a 488 nm excitation laser and measuring emission at 525 nm. A portion of the collected population was plated on LB agar supplemented with 3.4 μg/mL tetracycline and 67 μg/mL ampicillin and the remaining population was grown overnight in LB media with 3.4 μg/mL tetracycline and 67 μg/mL ampicillin and glycerol stocks were prepared. For identifying MphR variants with improved ErA sensitivity, negative sorting was done using whole cell biosensor cells with no inducing ligand. For identifying MphR variants that were selective against semi-synthetic macrolides, negative sorting was done in the presence of 5 μM clarithromycin and 5 μM azithromycin.
General Procedure for Microplate Screening of Libraries of MphR Variants
Individual colonies from biosensor mutant libraries were picked from LB agar plates and used to inoculate 500 μL of LB media containing 3.4 μg/mL tetracycline and 67 μg/mL ampicillin in wells of a deep-well 96-well microplate for 5 hours at 37 °C and with shaking at 350 rpm. Then, 10 μL of each culture was added to 490 μL of LB media containing 3.4 μg/mL tetracycline, 67 μg/mL ampicillin and 1 μM ErA (for ErA sensitivity mutants) or either 5 μM ErA, 5 μM clarithromycin, or 5 μM azithromycin (for identification of variants with ErA selectivity). Plates were grown overnight at 37 °C and with shaking at 350 rpm. Cultures were centrifuged at 3,000 rpm for 5 min and the cell pellet was re-suspended in 1 mL of phosphate-buffered saline (PBS). Then, 100 μL of the cell suspension was used for analyzing optical density at 600 nm and fluorescence (ex 395 nm/ em 509 nm). The fluorescence intensity was divided by the OD600 to yield a normalized GFP fluorescence value. All experiments were conducted in triplicate unless stated otherwise.
Dose-Response Analysis of Wild-Type and Variant MphR
Individual colonies of the wild-type biosensor or variant MphR biosensor strains that also included pJZ12 were picked from LB agar plates and used to inoculate 500 μL of LB media containing 3.4 μg/mL tetracycline and 67 μg/mL ampicillin in wells of a deep-well 96-well microplate for 5 hours at 37 °C and with shaking at 350 rpm. Then, 10 μL of each culture was added to 490 μL of LB media containing 3.4 μg/mL tetracycline, 67 μg/mL ampicillin and either ErA, clarithromycin, roxithromycin, azithromycin, or pikromycin at concentrations between 0 and 100 μM macrolide. Plates were incubated overnight at 37 °C and with shaking at 350 rpm. The cultures were centrifuged 3,000 rpm for 5 min and the cell pellet was washed once with 1 mL PBS and then re-suspended in 1 mL of PBS. Next, 100 μL of the cell suspension was used for analyzing optical density at 600 nm and fluorescence (ex 395 nm/em 509 nm). The fluorescence intensity was divided by the OD600 to yield a normalized GFP fluorescence value. All experiments were conducted in triplicate.
The data was fit to the Hill equation to derive the biosensor performance parameters:
where GFP0 is normalized GFP expression in the absence of inducer, GFPmax is the maximum observed normalized GFP expression, [I] is the inducer concentration, K1/2 is the inducer concentration resulting in half-maximal induction, and n is the Hill coefficient describing biosensor cooperativity.
Aeromicrobium erythreum Culturing and Screening with the Whole-Cell Biosensor Strain
A. erythreum colonies of either the wild-type or knock-out strain51 were picked from an LB agar plate and used to inoculate overnight cultures in LB media. The overnight culture was diluted 100-fold in LB media and 500 μL of the wild-type or knock-out strain was each added to each well of a 96-well microplate. Cultures were incubated at 33 °C with shaking at 250 rpm for two days and were then centrifuged at 4,700 rpm for 10 min. Then, 50 μL of each A. erythreum culture supernatant was added to 475 μL of a culture of E. coli TOP10/pJZ12 that harbored plasmids containing wild-type MphR or RBS-E7 that had been cultured for 5 h. The whole cell biosensor strains were incubated overnight at 37 °C with shaking at 350 rpm, centrifuged at 3,000 rpm for 5 min and each cell pellet re-suspended in 1 mL of phosphate-buffered saline (PBS). Next, 100 μL of the cell suspension was used for analyzing optical density at 600 nm and GFP fluorescence (ex 395 nm/ em 509 nm). The fluorescence intensity was divided by the OD600 to yield a normalized GFP fluorescence value.
Growth Selection
To enable growth selection, the gfp gene in pMLGFP-RBS-E7 was replaced with the chloramphenicol resistance gene (Cam). Briefly, Cam was amplified from the vector pACYDuet using the primers 28-Cam-for and 29-Cam-rev (Supplemental Table S1). Thermocycling conditions were those recommended by the manufacturer. The PCR product was gel purified and double digested using SpeI and HindIII following standard protocols. The double digested product was ligated at 16 °C for 18 h into similarly treated pMLGFP-RBS-E7, generating pMLCamR. The ligation mixture was electroporated into E. cloni 10G electrocompetent cells according to the manufacturer instructions, grown overnight, and the DNA sequence of plasmid prepared from a single colony was obtained to confirm the sequence identity. Next, pMLCamR was transformed into E. coli TOP10/pJZ12 cells and plated onto LB agar plates that were supplemented with various combinations of chloramphenicol (12.5 μg/mL), ErA (0.37 μg/mL (i.e. 0.5 μM)), tetracycline (3.4 μg/mL), and ampicillin (67 μg/mL).
Supplementary Material
Acknowledgments
This study was supported in part by National Institutes of Health grant GM104258 (G.J.W.) and an NC State Chancellors Innovation Award (G.J.W.).
Footnotes
Competing Financial Interests
G.J.W., C.M.K., and Y.L. have filed a patent application (PCT/US17/31962) covering designer MphR variants and associated applications.
Supporting Information
Supplementary Table S1
Supplementary Figures S1-S2
Supplemental Methods including: Saturation mutagenesis of MphR RBS, general procedure for microplate screening libraries of MphR saturation variants.
References
- [1].Weissman KJ, and Leadlay PF (2005) Combinatorial biosynthesis of reduced polyketides, Nat. Rev. Microbiol 3, 925–936. [DOI] [PubMed] [Google Scholar]
- [2].Omura S (2002) Macrolide Antibiotics, 2nd ed., Academic Press. [Google Scholar]
- [3].Wright PM, Seiple IB, and Myers AG (2014) The evolving role of chemical synthesis in antibacterial drug discovery, Angew. Chem. Int. Ed. Engl 53, 8840–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Seiple IB, Zhang Z, Jakubec P, Langlois-Mercier A, Wright PM, Hog DT, Yabu K, Allu SR, Fukuzaki T, Carlsen PN, Kitamura Y, Zhou X, Condakes ML, Szczypinski FT, Green WD, and Myers AG (2016) A platform for the discovery of new macrolide antibiotics, Nature 533, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ladner CC, and Williams GJ (2016) Harnessing natural product assembly lines: structure, promiscuity, and engineering, J Ind Microbiol Biotechnol 43, 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hertweck C (2015) Decoding and reprogramming complex polyketide assembly lines: prospects for synthetic biology, Trends Biochem. Sci 40, 189–199. [DOI] [PubMed] [Google Scholar]
- [7].Dunn BJ, and Khosla C (2013) Engineering the acyltransferase substrate specificity of assembly line polyketide synthases, J R Soc Interface 10, 20130297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Weissman KJ (2016) Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology, Nat. Prod. Rep 33, 203–230. [DOI] [PubMed] [Google Scholar]
- [9].Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, and Santi DV (2005) Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes, Nat Biotechnol 23, 1171–1176. [DOI] [PubMed] [Google Scholar]
- [10].Klaus M, Ostrowski MP, Austerjost J, Robbins T, Lowry B, Cane DE, and Khosla C (2016) Protein-protein interactions, not substrate recognition, dominate the turnover of chimeric assembly line polyketide synthases, J. Biol. Chem 291, 16404–16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Luo Y, Enghiad B, and Zhao H (2016) New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters, Nat. Prod. Rep 33, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, and Church GM (2009) Programming cells by multiplex genome engineering and accelerated evolution, Nature 460, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schmidt-Dannert C, Umeno D, and Arnold FH (2000) Molecular breeding of carotenoid biosynthetic pathways, Nat Biotechnol 18, 750–753. [DOI] [PubMed] [Google Scholar]
- [14].Williams GJ, Zhang C, and Thorson JS (2007) Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution, Nat Chem Biol 3, 657–662. [DOI] [PubMed] [Google Scholar]
- [15].Furubayashi M, Ikezumi M, Kajiwara J, Iwasaki M, Fujii A, Li L, Saito K, and Umeno D (2014) A high-throughput colorimetric screening assay for terpene synthase activity based on substrate consumption, PLoS One 9, e93317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Smanski MJ, Bhatia S, Zhao D, Park Y, L BAW, Giannoukos G, Ciulla D, Busby M, Calderon J, Nicol R, Gordon DB, Densmore D, and Voigt CA (2014) Functional optimization of gene clusters by combinatorial design and assembly, Nat Biotechnol 32, 1241–1249. [DOI] [PubMed] [Google Scholar]
- [17].Roddis M, Gates P, Roddis Y, and Staunton J (2002) Structural elucidation studies on 14- and 16-membered macrolide aglycones by accurate-mass electrospray sequential mass spectrometry, J. Am. Soc. Mass Spectrom 13, 862–874. [DOI] [PubMed] [Google Scholar]
- [18].Wills RH, Tosin M, and O’Connor PB (2012) Structural characterization of polyketides using high mass accuracy tandem mass spectrometry, Anal. Chem 84, 8863–8870. [DOI] [PubMed] [Google Scholar]
- [19].Crowe MC, Brodbelt JS, Goolsby BJ, and Hergenrother P (2002) Characterization of erythromycin analogs by collisional activated dissociation and infrared multiphoton dissociation in a quadrupole ion trap, J. Am. Soc. Mass Spectrom 13, 630–649. [DOI] [PubMed] [Google Scholar]
- [20].Ashley GW, and Carney JR (2004) API-mass spectrometry of polyketides. II. Fragmentation analysis of 6-deoxyerythronolide B analogs, J. Antibiot (Tokyo) 57, 579–589. [DOI] [PubMed] [Google Scholar]
- [21].Rogers JK, Taylor ND, and Church GM (2016) Biosensor-based engineering of biosynthetic pathways, Curr Opin Biotechnol 42, 84–91. [DOI] [PubMed] [Google Scholar]
- [22].Zhang J, Jensen MK, and Keasling JD (2015) Development of biosensors and their application in metabolic engineering, Current Opinion in Chemical Biology 28, 1–8. [DOI] [PubMed] [Google Scholar]
- [23].Cuthbertson L, and Nodwell JR (2013) The TetR family of regulators, Microbiol. Mol. Biol. Rev 77, 440–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dietrich JA, Shis DL, Alikhani A, and Keasling JD (2013) Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis, ACS Synth. Biol 2, 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Raman S, Rogers JK, Taylor ND, and Church GM (2014) Evolution-guided optimization of biosynthetic pathways, Proc Natl Acad Sci USA 111, 17803–17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].de los Santos EL, Meyerowitz JT, Mayo SL, and Murray RM (2016) Engineering Transcriptional Regulator Effector Specificity Using Computational Design and In Vitro Rapid Prototyping: Developing a Vanillin Sensor, ACS Synth. Biol 5, 287–295. [DOI] [PubMed] [Google Scholar]
- [27].Tang SY, Qian S, Akinterinwa O, Frei CS, Gredell JA, and Cirino PC (2013) Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter, J. Am. Chem. Soc 135, 10099–10103. [DOI] [PubMed] [Google Scholar]
- [28].Tahlan K, Ahn SK, Sing A, Bodnaruk TD, Willems AR, Davidson AR, and Nodwell JR (2007) Initiation of actinorhodin export in Streptomyces coelicolor, Mol. Microbiol 63, 951–961. [DOI] [PubMed] [Google Scholar]
- [29].Abbas A, Morrissey JP, Marquez PC, Sheehan MM, Delany IR, and O’Gara F (2002) Characterization of interactions between the transcriptional repressor PhlF and its binding site at the phlA promoter in Pseudomonas fluorescens F113, J. Bacteriol 184, 3008–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Le TB, Schumacher MA, Lawson DM, Brennan RG, and Buttner MJ (2011) The crystal structure of the TetR family transcriptional repressor SimR bound to DNA and the role of a flexible N-terminal extension in minor groove binding, Nucleic Acids Res. 39, 9433–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Namwat W, Lee CK, Kinoshita H, Yamada Y, and Nihira T (2001) Identification of the varR gene as a transcriptional regulator of virginiamycin S resistance in Streptomyces virginiae, J. Bacteriol 183, 2025–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Szczepanowski R, Krahn I, Bohn N, Puhler A, and Schluter A (2007) Novel macrolide resistance module carried by the IncP-1beta resistance plasmid pRSB111, isolated from a wastewater treatment plant, Antimicrob. Agents Chemother 51, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Szczepanowski R, Krahn I, Linke B, Goesmann A, Puhler A, and Schluter A (2004) Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system, Microbiology 150, 3613–3630. [DOI] [PubMed] [Google Scholar]
- [34].Noguchi N, Emura A, Matsuyama H, O’Hara K, Sasatsu M, and Kono M (1995) Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2’-phosphotransferase I in Escherichia coli, Antimicrob. Agents Chemother 39, 2359–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mohrle V, Stadler M, and Eberz G (2007) Biosensor-guided screening for macrolides, Anal. Bioanal. Chem 388, 1117–1125. [DOI] [PubMed] [Google Scholar]
- [36].Bloom JD, and Arnold FH (2009) In the light of directed evolution: pathways of adaptive protein evolution, Proc Natl Acad Sci USA 106 Suppl 1, 9995–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gardner L, Zou Y, Mara A, Cropp TA, and Deiters A (2011) Photochemical control of bacterial signal processing using a light-activated erythromycin, Mol. Biosyst 7, 2554–2557. [DOI] [PubMed] [Google Scholar]
- [38].Rogers JK, Guzman CD, Taylor ND, Raman S, Anderson K, and Church GM (2015) Synthetic biosensors for precise gene control and real-time monitoring of metabolites, Nucleic Acids Res. 43, 7648–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zheng J, Sagar V, Smolinsky A, Bourke C, LaRonde-LeBlanc N, and Cropp TA (2009) Structure and function of the macrolide biosensor protein, MphR(A), with and without erythromycin, J. Mol. Biol 387, 1250–1260. [DOI] [PubMed] [Google Scholar]
- [40].Zhang H, Wang Y, Wu J, Skalina K, and Pfeifer BA (2010) Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host, Chem. Biol 17, 1232–1240. [DOI] [PubMed] [Google Scholar]
- [41].Chemler JA, Tripathi A, Hansen DA, O’Neil-Johnson M, Williams RB, Starks C, Park SR, and Sherman DH (2015) Evolution of Efficient Modular Polyketide Synthases by Homologous Recombination, J. Am. Chem. Soc 137, 10603–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Espah Borujeni A, Channarasappa AS, and Salis HM (2014) Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites, Nucleic Acids Res. 42, 2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Salis HM, Mirsky EA, and Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression, Nat Biotechnol 27, 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kibwage IO, Hoogmartens J, Roets E, Vanderhaeghe H, Verbist L, Dubost M, Pascal C, Petitjean P, and Levol G (1985) Antibacterial activities of erythromycins A, B, C, and D and some of their derivatives, Antimicrob. Agents Chemother 28, 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Majer J, Martin JR, Egan RS, and Corcoran JW (1977) Antibiotic glycosides. 8. Erythromycin D, a new macrolide antibiotic, J. Am. Chem. Soc 99, 1620–1622. [DOI] [PubMed] [Google Scholar]
- [46].Kittendorf JD, and Sherman DH (2009) The methymycin/pikromycin pathway: a model for metabolic diversity in natural product biosynthesis, Bioorg Med Chem 17, 2137–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Miller ES, Woese CR, and Brenner S (1991) Description of the erythromycin-producing bacterium Arthrobacter sp. strain NRRL B-3381 as Aeromicrobium erythreum gen. nov., sp. nov, International journal of systematic bacteriology 41, 363–368. [DOI] [PubMed] [Google Scholar]
- [48].Brikun IA, Reeves AR, Cernota WH, Luu MB, and Weber JM (2004) The erythromycin biosynthetic gene cluster of Aeromicrobium erythreum, J Ind Microbiol Biotechnol 31, 335–344. [DOI] [PubMed] [Google Scholar]
- [49].Reeves AR, Cernota WH, Brikun IA, Wesley RK, and Weber JM (2004) Engineering precursor flow for increased erythromycin production in Aeromicrobium erythreum, Metab Eng 6, 300–312. [DOI] [PubMed] [Google Scholar]
- [50].Harrell EA, and Miller ES (2016) Genome Sequence of Aeromicrobium erythreum NRRL B-3381, an Erythromycin-Producing Bacterium of the Nocardioidaceae, Genome announcements 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Miller ES (1991) Cloning vectors, mutagenesis, and gene disruption (ermR) for the erythromycin-producing bacterium Aeromicrobium erythreum, Appl. Environ. Microbiol 57, 2758–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Siedler S, Khatri NK, Zsohar A, Kjaerbolling I, Vogt M, Hammar P, Nielsen CF, Marienhagen J, Sommer MOA, and Joensson HN (2017) Development of a Bacterial Biosensor for Rapid Screening of Yeast p-Coumaric Acid Production, ACS Synth. Biol [DOI] [PubMed] [Google Scholar]
- [53].OpenWetWare-contributors. (2010) Round-the-horn site-directed mutagenesis, In OpenWetWare 26 February 2015 ed., OpenWetWare.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.