Abstract
MicroRNA (miRNA) expression is frequently deregulated in human disease, in contrast, disease-associated miRNA mutations are understudied. We developed Annotative Database of miRNA Elements, ADmiRE that combines multiple existing and new biological annotations to aid prioritization of causal miRNA variation. We annotated 10,206 mature (3,257 within seed region) miRNA variants from multiple large sequencing datasets including gnomAD (15,496 genomes; 123,136 exomes). The pattern of miRNA variation closely resembles protein-coding exonic regions, with no difference between intragenic and intergenic miRNAs (p=0.56), and high confidence miRNAs demonstrate higher sequence constraint (p<0.001). Conservation analysis across 100 vertebrates identified 765 highly conserved miRNAs that also have limited genetic variation in gnomAD. We applied ADmiRE to the TCGA PanCancerAtlas WES datasets from over 10,000 individuals across 33 adult cancers and annotated 1,267 germline (rare in gnomAD) and 1,492 somatic miRNA variants. Several miRNA families with deregulated gene expression in cancer have low levels of both somatic and germline variants, e.g. let-7, miR-10. In addition to known somatic miR-142 mutations in hematologic cancers, we describe novel somatic miR-21 mutations in esophageal cancers impacting downstream miRNA targets. Through the development of ADmiRE, we present a framework for annotation and prioritization of miRNA variation in disease datasets.
Keywords: microRNA, variant annotation, conservation, cancer, genomics
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs, 18–25 nucleotides in length, that regulate the expression of greater than 60% of genes by complementarily binding target messenger RNAs (mRNAs) (Bartel, 2009; Friedman, Farh, Burge, & Bartel, 2009). Dysregulation of miRNAs leads to altered expression of their downstream target genes as seen in a wide variety of human diseases such as cancer, cardiovascular and developmental diseases (Lee & Dutta, 2009; Lu et al., 2008; Lujambio & Lowe, 2012; Spizzo, Nicoloso, Croce, & Calin, 2009). Primary miRNA transcripts are sequentially processed by DROSHA and DICER1 into precursor and mature miRNAs, respectively (Friedman et al., 2009). Ablation of the miRNA repertoire in Dicer1−/− mutant zebrafish and mice leads to lethality in model organisms and heterozygous loss leads to cancer predisposition in humans (De Kock et al., 2014; Harfe, 2005; Kumar et al., 2009; Lin & Gregory, 2015). Loss of individual miRNAs also leads to disease-related phenotypes in model systems as shown by the knockout mouse model of miR-155 alters mammalian differentiation processes (Thai et al., 2007) while overexpression of miR-21 leads to oncogenic phenotypes in mice (Medina, Nolde, & Slack, 2010). In addition to transcriptional regulation, several mechanisms for the dysregulation of miRNAs have been proposed such as single nucleotide variation (SNV), genomic amplifications or deletions, and transcriptional regulation (Chan, Prado, & Weidhaas, 2011). Mature miRNAs complementarily bind the recognition site in target genes, and the highly conserved seed domains of mature miRNAs are vital for this function. As a result, mutations in mature miRNAs can drastically change their targeting ability (Hill, Jabbari, Matyunina, & McDonald, 2014; Roden et al., 2017; Zeng & Cullen, 2003). Seed or mature miRNA variants are relatively rare and some of them have been shown to have highly deleterious effects. Three out of the four known Mendelian disorders shown to be caused by mutations in miRNAs (OMIM (McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, 2018)) are due to seed domain mutations: miR-96 in hearing loss (Mencía et al., 2009), miR-184 in familial keratoconus (Hughes et al., 2011; Iliff, Riazuddin, & Gottsch, 2012), and miR-204 in inherited retinal dystrophy (Conte et al., 2015). In comparison, miRNA processing machinery recognizes several well-defined structural and primary sequence features such as CNNC, basal-UG, apical UGU/UGUG, and GHG motifs that enhance processing of primary miRNA transcripts (Auyeung, Ulitsky, McGeary, & Bartel, 2013; Bartel, 2018; Fang & Bartel, 2015; Nguyen et al., 2015; Roden et al., 2017). Expectedly, SNVs affecting these motifs can interfere with miRNA processing and affect the expression of mature miRNA (Fang & Bartel, 2015; Roden et al., 2017). Germline or somatic mutations in miRNA transcripts have been found in patients with several cancers (Ryan, Robles, & Harris, 2010; Tuna, Machado, & Calin, 2016) such as breast (W. Li et al., 2009; Shen, Ambrosone, & Zhao, 2009; Shen, DiCioccio, Odunsi, Lele, & Zhao, 2010), leukemia (Calin et al., 2005; Calin & Croce, 2006; Kotani et al., 2010) and pancreatic (Zhu, Gao, Qian, & Miao, 2009) cancers. Most of these variants ascribed to cancer predisposition or somatic mechanisms are primary or precursor miRNA transcript variants and alter the expression levels of mature miRNAs by interfering with the miRNA processing. Despite the potential significance of miRNA SNVs in disease, there have been limited reports of miRNA sequence variation across human diseases and population datasets.
An early study describing miRNA variation in human reference population identified a total of 527 miRNA variants by analyzing 720 miRNA sequences in the 1000 Genomes Project, Phase I dataset (Carbonell et al., 2012). Other databases such as miRNASNP and polymiRTS report approximately 500 SNVs in miRNA regions using the dbSNP137 database (Bhattacharya, Ziebarth, & Cui, 2014; Gong et al., 2012). However, there has been a significant increase in the number of recognized miRNA genes (miRBase v21: 1,881 precursor, 2,815 mature miRNAs) and in the availability of large whole genome sequencing (WGS) and whole exome sequencing (WES) datasets (Kozomara & Griffiths-Jones, 2011) since those earlier studies.
Annotation of variants within protein-coding genes utilize several criteria including but not limited to, predicted protein truncations, change in the amino acid code, evolutionary conservation and/or on sequence constraint metrics such as those developed using large human population datasets of protein-coding sequences like ExAC (Lek et al., 2016; Samocha et al., 2014). However, a similar framework for interpretation of miRNA variation is not readily available. Many commonly used variant annotation tools such as Variant Effect Predictor (VEP) (McLaren et al., 2016), ANNOVAR (K. Wang et al., 2010) or SnpEff (Cingolani et al., 2012) prioritize the effect of sequence changes in protein-coding regions over the changes in miRNAs (Lek et al., 2016). For example, one scheme for prioritizing variant deleteriousness ranks miRNA annotation at the 32nd position out of 44 annotations (Cingolani, Cunningham, McLaren, & Wang, 2018). Currently, the only dedicated miRNA variant annotation tool, miRVaS (Cammaerts, Strazisar, Dierckx, Del Favero, & De Rijk, 2015), does not include sequence constraint metrics or highly-relevant biological and functional annotations such as expression, downstream targets and disease associations. Thus, there is a need to determine the landscape of miRNA variation through the analysis of large human population datasets, determine evolutionary conservation and then combine this data with existing annotations to provide a comprehensive miRNA annotation focused tool.
In this study, we describe a novel miRNA variation module entitled Annotative Database for miRNA Elements, ADmiRE (https://github.com/nroak/ADmiRE), to annotate human miRNA variants. ADmiRE can be easily integrated into established variant annotation workflows. The ADmiRE annotations include existing biological annotations for downstream targets and upstream transcription factors combined with new data described herein from the analyses of miRNA sequence constraint derived from publicly large available datasets, sequence conservation across 100 vertebrates and per basepair annotations of the domain and conserved motifs. In particular, we analyzed gnomAD containing 123,136 WES and 15,496 WGS samples to define domain level and miRNA level sequence constraints for miRNA variation, similar to previous work done on protein-coding genes (Lek et al., 2016). Finally, we applied ADmiRE annotations to The Cancer Genome Atlas (TCGA) PanCancerAtlas adult cancer dataset comprising of WES data from over 10,000 samples across 33 cancer types to provide the patterns of germline and somatic miRNA variation in cancer. In sum, development, and deployment of a dedicated human miRNA variant annotation tool, ADmiRE, which includes variant frequency and conservation information in combination with existing miRBase annotations can facilitate the prioritization and identification of biologically relevant miRNA variation in control and disease datasets.
Methods
MicroRNA Annotation Resources
We obtained the precursor stem-loop (1,881) and mature (2,815) miRNA sequences (assembly GRCh38) for all human miRNAs from miRBase v21 (Kozomara & Griffiths-Jones, 2011). As most variation datasets are based on reference genome build GRCh37, we used UCSC LiftOver tool to convert miRBase genomic coordinates to this reference resulting in the final list of 1,878 precursors and 2,574 mature miRNAs (Kuhn, Haussler, & James Kent, 2013).
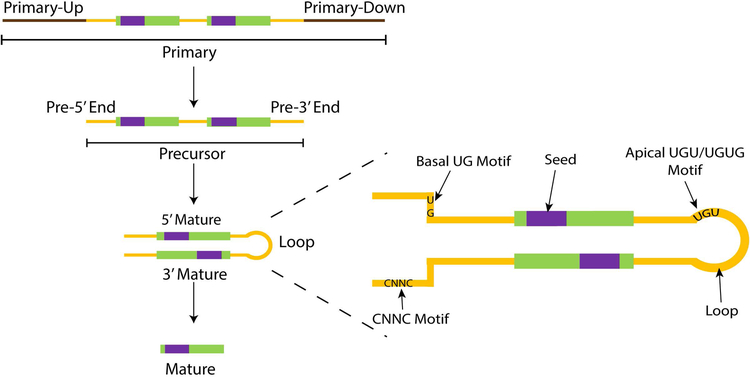
We defined the miRNA sequence domains (Figure 1) as primary (top, defined by the 100bp sequence flanking the precursor -- Primary-Up and Primary-Down (in brown), 5’ and 3’ precursor arms (yellow, “End”), 5’ and 3’ mature (green), seed region (purple, 2–8bp of mature miRNA), and precursor loop (yellow, between mature miRNA).
Figure 1. Schematic of miRNA sequence domains.
miRNA Sequence domains as annotated in ADmiRE pipeline. Precursor (yellow) and mature (green) domains as defined by miRBase, primary (brown) domains defined as 100bp flanking regions to precursor domains, and seed (purple) domains defined as 2–8bp of mature domains. For the precursor hairpin transcript, sequence motifs namely, seed, CNNC, basal UG, apical UGU/UGUG, and loop motifs are shown.
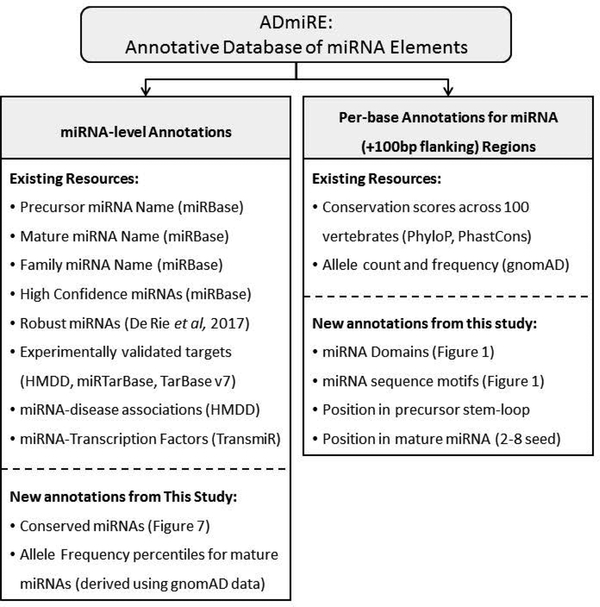
We utilized the previously curated list of miRNAs that pass the following criteria for miRNA gene annotation from high-throughput sequencing pipelines across different tissues, defined as ‘high confidence’ miRNAs by miRBase v21, (i) At least 10 reads must map with no mismatches to each of the two possible mature microRNAs derived from the hairpin precursor, (ii) The most abundant reads from each arm of the precursor must pair in the mature microRNA duplex with 0–4 nt overhang at their 3′ ends, (iii) At least 50% of reads mapping to each arm of the hairpin precursor must have the same 5′ end, (iv) The predicted hairpin structure must have a folding free energy of <−0.2 kcal/mol/nt, and (v) At least 60% of the bases in the mature sequences must be paired in the predicted hairpin structure (Kozomara & Griffiths-Jones, 2014). Furthermore, we also utilize a more recent set of miRNAs that satisfy at least four out of these five criteria, defined as FANTOM5 ‘robust’ set of miRNAs resulting in two sets of well-characterized miRNAs: a) 295 ‘high confidence’ miRNAs from miRBase v21 (Kozomara & Griffiths-Jones, 2014), and b) 795 ‘robust’ miRNAs from FANTOM5 project (Rie et al., 2017). Furthermore, we incorporated sequence motif information for precursor miRNAs such as CNNC, basalUG, and apical UGU/UGUG motifs (Roden et al., 2017). Lastly, annotations from the following databases were incorporated for functionally validated miRNA-target genes, literature-curated transcription factors, and human disease associations: Human microRNA Disease Database (HMDD) (Y. Li et al., 2014), PhenomiR (Ruepp et al., 2010), miRTarBase (Chou et al., 2016), TransmiR (J. Wang, Lu, Qiu, & Cui, 2010), and TarBase 7.0 (Vlachos et al., 2015). Additionally, measures of miRNA sequence variation and conservation described in this study were subsequently added (Supp. Table S1). These biologically relevant set of annotations were compiled into a tab-separated file containing each of these annotations for every base of primary miRNA transcript across all miRNAs called Annotative Database of miRNA Elements, ADmiRE (see Availability) (Figure 2). ADmiRE file and a Perl-script are made available on GitHub which can be utilized to annotate a user-supplied variant file for miRNA variant annotations (see section: Data Access).
Figure 2. Description of Annotative Database of miRNA Elements (ADmiRE).
List of ADmiRE annotations categorized as miRNA-level features that describe miRNA gene annotations compiled from miRBase, from analyses computed in this study, or derived from additional external databases (left box). Additionally, every base within precursor miRNA stem-loop and 100bp flanking region is annotated for its domain and sequence motif information (per Figure 1), PhyloP and PhastCons conservation scores, and gnomAD allele frequency information (right box).
Genomic Datasets
High-quality single nucleotide variants (SNVs) and small insertions and deletions (InDels) were downloaded from the following publicly available whole genome (WGS) and whole exome sequencing (WES) datasets a) genome aggregation database (gnomAD) consisting of 15,496 WGS and 123,136 WES samples (date accessed 2/28/2017) (Lek et al., 2016), b) annotated variants across 60,706 WES samples in the tab-separated format from the Exome Aggregation Dataset (ExAC; date accessed 6/23/2017) (Lek et al., 2016), c) UK10K project (date accessed 2/21/2017) (Walter et al., 2015), d) 1000 Genomes project phase 3 across 2,504 low-coverage WGS samples (date accessed 1/3/2016) (Auton et al., 2015), and e) National Heart, Lung and Blood Institute’s (NHLBI) Trans-Omics for Precision Medicine (TOPMed) dataset accessed from University of Michigan’s Bravo browser, consisting of variants from 62,784 WGS samples (date accessed 1/11/2018).
We obtained comprehensive germline variant calls (10,389 individuals) (Huang et al., 2018) and somatic mutation calls (10,000 individuals) (Ellrott et al., 2018) across 33 cancer types from TCGA PanCancerAtlas samples (mc3.v0.2.8.PUBLIC.maf). The data consisted of whole exome sequencing, miRNAseq, and RNAseq sequencing samples of 10,389 individuals across 33 cancer types.
Cross-species miRNA conservation heatmap
We retrieved per base conservation scores for all miRNAs using UCSC table browser utility for the following cross-species comparisons: Algorithms used were phyloP and phastCons across 100 vertebrates, 46 vertebrates, 46 placental mammals and 46 primates (Pollard, Hubisz, Rosenbloom, & Siepel, 2010; Siepel et al., 2005; Yang, 1995). For each conservation scoring system, phyloP and phastCons, scores were averaged across each mature miRNA. Each of these scoring systems generates a genome-wide conservation score distribution, phyloP (−13 to 10) and phastCons (0–1). Thus, we used a Z-score normalization for generating a heatmap by implementing ‘aheatmap’ function in the NMF package in R for hierarchical clustering and heatmap generation (Gaujoux & Seoighe, 2010).
Analysis of PanCancerAtlas Dataset: ≥10,000 Adult Cancers
We used BEDTools (Quinlan, 2014) intersect tool to subset the whole exome germline variant calls to the miRNA regions that corresponded to the mature and precursor miRNA sequences (miRBase v21). We annotated these germline variants using ADmiRE and filtered for rare germline miRNA variation using ExAC-nonTCGA subset (excluding 7,601 germline TCGA samples from the original ExAC dataset) with allele frequency threshold of <0.1%. Additionally, we retained variants with ‘PASS’ filter, variant allele fraction >30%, and variants with ExAC sub-population allele count of <10. Somatic mutation calls were also restricted to the mature miRNA regions, annotated using ADmiRE, and filtered for PASS-variants with variant allele fractions of >20%. Only the precursor miRNAs with >6X read-depth across at least 75% TCGA samples were retained for the analysis. We additionally filtered out any variants found in the gnomAD dataset that were reported as non-PASS variants using the online gnomAD browser.
Statistical Analysis
All statistical analyses described in the study were performed using R statistical software (R-3.4.3). To control for largely variable target size across different genomic and miRNA regions, we used the distribution of allele frequency (AF) as a variable. We found that the AF distribution did not follow a normal distribution and thus utilized non-parametric tests for statistical analyses. We used both Mann-Whitney test and Kruskal-Wallis tests, with a Bonferroni adjusted p-value (as implemented in R in functions pairwise.wilcox.test and dunn.test, respectively) for computing the significance of differences between the AF distribution of the following comparisons: a) the protein-coding exonic, intronic, and intergenic variation, b) the mature, precursor, and primary miRNA domain variation, c) variation within ‘high confidence’ or ‘robust’ miRNAs and the remainder of miRNAs. To determine the variability of each miRNA with respect to all other miRNAs, we calculated mean allele frequency for each mature miRNA, followed by computing a mean AF percentile for all the miRNAs using the empirical cumulative distribution function (ecdf), as implemented in R. We calculated whether ‘high confidence’ or ‘robust’ miRNAs are enriched within the cluster of conserved miRNAs using contingency table chi-square test. To test whether gnomAD AF distribution correlates with evolutionary conservation, we calculated Spearman’s correlation coefficient between gnomAD AF and PhyloP or PhastCons conservation scores using function ‘rcorr’ as implemented in R package Hmisc v4.1. Expression quantiles for each of the 20,501 genes and 1,072 miRNAs were computed using the ecdf function in R for each cancer type using RNAseq and miRNAseq expression data, respectively. To compare the expression quantiles of candidate genes between mutated and non-mutated samples, we used non-parametric Wilcoxon signed rank test as implemented in R function wilcox.test.
Results
Genomic Distribution of miRNAs and Coverage Analysis for WES and WGS datasets
Genomic locations of 1,020 miRNAs (1,020/1,878, 54%) overlap with RefSeq gene regions (RefSeq v78) and the remaining 858 miRNAs are intergenic in nature. We analyzed publicly available target-interval files for five commonly used whole exome target capture methods, Roche SeqCap EZ HGSC VCRome 2.1, Roche SeqCap EZ Exome v3, Agilent SureSelect v6, and Illumina TruSeq Exome for the inclusion of miRNAs in their target capture design. On average, 55% of all miRNAs listed in miRBase v21 (>90% precursor bases in capture bed file) were targeted either directly, as per the miRBase release at the time of capture design, or indirectly as a result of miRNAs overlapping the protein-coding exons. Exome capture of miRNA displayed substantial range with Agilent SureSelect v6 targeting over 78% miRNAs and Roche VCRome2.1 targeting only ~30% of miRNAs (using miRBase v16 but may have been improved since).
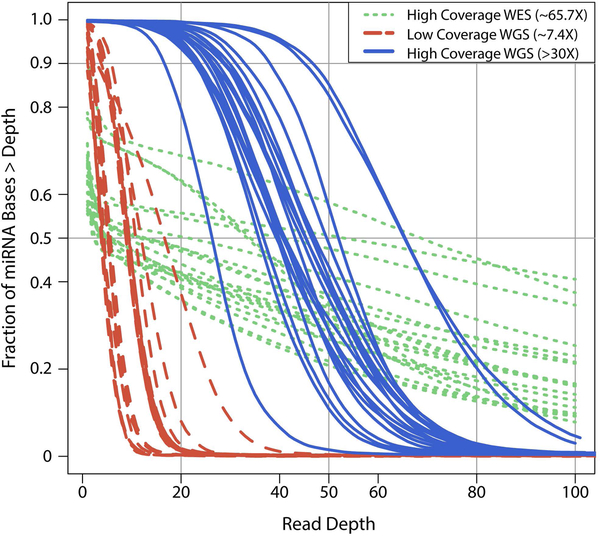
However, the inclusion of miRNAs on targeted capture does not fully represent the final coverage of miRNAs from the sequencing data. Our analysis of gnomAD whole exome sequencing dataset (n=123,136), which contains aggregated data across different WES capture methods, showed that only 774 precursor miRNAs (41%) and 1224 mature miRNAs (48%) have a read depth >10X across at least 75% of samples. In contrast, for gnomAD whole genome dataset (n=15,496), 1,804 (96%) precursor and 2,534 (99%) mature miRNAs passed the above coverage criteria. Out of the 39 miRNAs that fail the above-mentioned coverage criteria, 25 miRNAs were sequenced at 1–10X median read-depth across the majority of samples while the remainder have median read depth of less than one (Supp. Table S2). Only the miRNAs passing 10X coverage threshold were used in all the analyses described below. To further characterize miRNA coverage, we analyzed 20 samples from 1000 genomes project that have been sequenced by 3 longitudinal sequencing methods: high coverage WES (mean read depth =65X), high coverage WGS (mean read depth >30X), and low coverage WGS (mean read depth =7.4X) across four different sequencing centers (Figure 3). Overall, although high coverage WGS (mean read depth >30X) is optimal for capturing all known miRNAs (~99%), more commonly used WES methods capture ~50% of all miRNAs at a read depth of >10X.
Figure 3. Extent of capture and coverage of miRNA regions across different sequencing platforms.
Cumulative coverage distribution of the sequenced bases within miRNA regions across high-coverage WES (>65.7X), high-coverage WGS (>30X), and low-coverage WGS (~7.4X) sequencing platforms across 4 different sequencing centers for 20 samples from 1000 Genomes project. Each point on y-axis corresponds to the fraction of miRNA bases covered at the corresponding read depth on the x-axis.
ADmiRE Comprehensively Annotates miRNA Variation
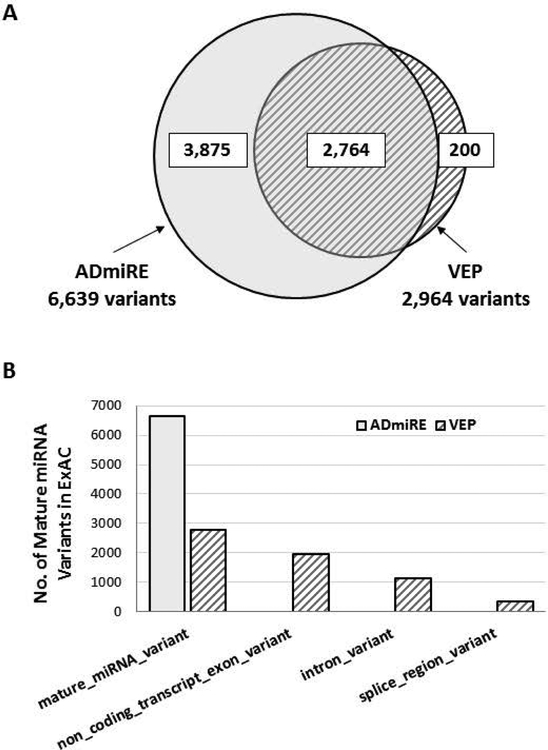
To assess variant annotation across miRNA regions, we used the set of pre-annotated variants from the ExAC dataset (n=60,706, 10 million variants) which had been annotated using the standard parameters in Variant Effect Predictor (VEP) tool prioritized for the most damaging variant consequences (Lek et al., 2016). Reannotation of this same set of variants with ADmiRE allows us to compare the sensitivity of both tools to annotate variants found within mature miRNA gene regions for their potential impact on miRNAs. Across the VEP annotated variants in the entire ExAC dataset, 2,764 variants were annotated as ‘mature miRNA’. In comparison, annotation of this variant set using ADmiRE annotations, which prioritizes miRNA variation, identified a total of 6,639 mature miRNA variants. (Figure 4 A). Thus, ADmiRE annotates an additional 3,875 variants as occurring within mature miRNA in comparison to VEP, which instead annotated these variants as ‘non_coding_transcript_exon_variant’ (50%) which is a generic category describing many non-coding RNAs, and ‘intron_variant’ (29%) (Figure 4 B). The 177 of 200 variants annotated as “miRNA” by VEP but not ADmiRE did not correspond to miRNA regions when validated using UCSC Genome Browser.
Figure 4. Annotation of mature miRNA variation from ExAC dataset by ADmiRE and VEP tools.
A. The overlap between number of mature miRNA variants annotated by ADmiRE and VEP for the pre-annotated variants from ExAC dataset B. Variant consequence prediction categories from VEP annotations (hatched) for mature miRNA variation as annotated by ADmiRE (solid).
We further compared other commonly used variant annotation tools, ANNOVAR and snpEFF, to ADmiRE for their ability to annotate miRNA variation from the above set of ExAC variants. None of the ANNOVAR annotation consequence included mature miRNA annotations, so we used -regionanno wgRNA option in ANNOVAR. There were a total of 10,615 precursor miRNA variants (63%) as compared to 16,657 precursor miRNA variants annotated by ADmiRE. Similarly, snpEFF annotations for mature miRNA variants are annotated as snpEFF gene biotype ‘miRNA’ for non_coding_transcript_exon category. We identified a total of 1,725 mature miRNA variants (26% of those by ADmiRE). Overall, ADmiRE accurately annotates substantially more mature miRNA variants from WES datasets compared to many other variant annotation platforms.
To improve annotation and prioritization of miRNA variation we provide a bed-formatted ADmiRE file that can be incorporated as a custom database to improve VEP’s annotation of miRNA variants (--custom flag, see Availability). We have also submitted ADmiRE to ANNOVAR to be included under ‘User-contributed datasets’ for easy integration.
To comprehensively identify and annotate human miRNA sequence variation, we performed ADmiRE annotations of the gnomAD dataset, which contains 15,496 WGS samples and more than twice the number of WES samples (n= 123,136) compared to the ExAC dataset. ADmiRE annotations identified 3,257 seed, 10,206 mature, 14,699 precursor, and 52,705 primary miRNA variants across 1,878 miRNAs (Table 1). In addition to the mature miRNA domains, certain sequence motifs in the precursor stem-loop region are known to affect miRNA processing efficiency (Roden et al., 2017). We annotated 997 variants within CNNC motif, 79 in basalUG, and 67 variants in apical UGU/UGUG motifs (Table 1). An additional 3,857 variants were in the stem-loop of the precursor miRNA transcript. This collection of 70,007 miRNA variants largely surpasses previously described miRNA sequence variation as reported in miRNASNP (Gong et al., 2012) (2,257 precursor, 706 mature variants) and Cui et al (Bhattacharya & Cui, 2015) (611 seed variants) datasets. In addition to the far more complete list of annotated miRNA variants for the gnomAD dataset, we also provide miRNA variation data from the ExAC, 1000 Genomes, UK10K, and NHLBI’s TOPMed datasets in the Supplementary Data, although some of these datasets are contained within the gnomAD dataset.
Table 1.
Number of unique miRNA variants identified using ADmiRE from the gnomAD dataset
| miRNA Domain | gnomAD WGS n= 15,496 |
gnomAD WES n= 123,125 |
gnomAD Total |
|---|---|---|---|
| miRNA Functional Domains | |||
| Total | 40,581 | 45,172 | 76,007 |
| Mature (Seed) | 4,626 (1,499) | 7,039 (2,224) | 10,206 (3,257) |
| Precursor | 6,918 | 9,462 | 14,428 |
| Primary | 29,231 | 28,671 | 51,542 |
| Precursor miRNA Sequence Motifs | |||
| Total | 2,196 | 3,492 | 5,000 |
| basalUG | 40 | 56 | 79 |
| UGU/UGUG | 16 | 58 | 67 |
| CNNC | 438 | 681 | 997 |
| Loop | 1,702 | 2,697 | 3,857 |
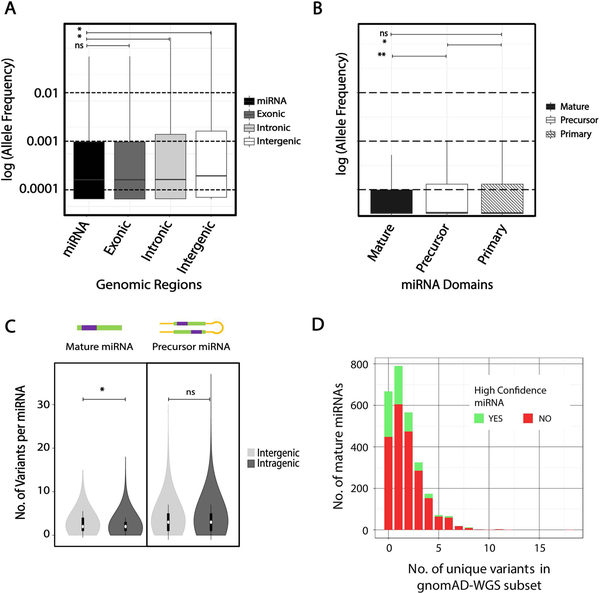
A previous study that analyzed miRNA variation from ~1000 individuals (1000 Genomes project phase I) had noted that level of variability for miRNAs is comparable to coding genes (Carbonell et al., 2012). We compared the allele frequency distribution from the 1878 precursor miRNA variants in the much larger gnomAD dataset to protein-coding exonic, intronic, and intergenic regions. We find the allele frequency distribution of miRNA variants resembles that of the protein-coding exonic variants and is significantly depleted compared to that of intronic and intergenic variants (Mann-Whitney adjusted p-value <0.001) (Figure 5 A). Not surprisingly, functionally important miRNA domains, mature and seed, have a significantly lower allele frequency distribution compared to the precursor domains (p<0.001) and the flanking 100bp primary domains (p<0.01) across the entire gnomAD dataset (Figure 5 B). We then compared the impact of the genomic context of miRNAs, i.e. intragenic or intergenic location on variation. We found no significant difference in the allele frequency (AF) distribution of miRNA variants across precursor domains (p=0.56, t-test) (Figure 5 C). The majority of mature miRNAs contain 0–2 variants in the entire gnomAD WGS dataset, with a few outlier miRNAs accumulating relatively high sequence variation (Figure 5 D).
Figure 5. Patterns of miRNA variation in gnomAD WGS (n=15,496) dataset.
A. Distribution of allele frequency of all variants in log-scale across 4 different genomic regions; precursor miRNA (miRBase v21), protein-coding exonic (GENCODEv19), intronic (GENCODEv19), and intergenic (remaining fraction of the hg19 reference, slanted line fill). B. Allele frequency distribution of variants (log-scale) in mature, precursor, and primary domains of miRNAs, C. Allele frequency distribution of variants across mature and precursor miRNA domains compared between miRNAs in intragenic and intergenic regions (within RefSeq genes). D. Number of miRNAs across each bin of number of unique mature domain variants. *- Mann-Whitney adjusted p-value <0.001, ns- not significant
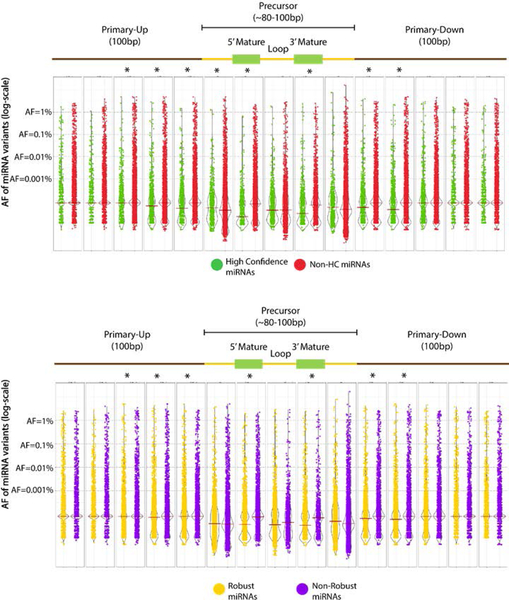
We analyzed variation in two sets of curated miRNAs, ‘high-confidence’ miRNAs from miRBase and ‘robust’ miRNAs from FANTOM5 studies, which both use miRNA expression data across several datasets to exclude false positive miRNAs (Kozomara & Griffiths-Jones, 2014; Rie et al., 2017) (see Methods). For both high confidence and robust (Figure 6) miRNAs, the allele frequency distribution is significantly lower across mature miRNAs and 20–40 bases flanking the precursor transcripts compared to the remainder of miRNAs. Whereas the high confidence and FANTOM5 robust miRNAs that pass the high confidence criteria of expression tend to harbor more rare variation (gnomAD AF < 0.01; Mann-Whitney test p-value <0.001 for both datasets).
Figure 6. Distribution of miRNA variation in gnomAD across miRNA domains.
Allele frequency (AF) distribution (log-scale) for all miRNA variants in gnomAD (WGS and WES) across primary miRNA transcript domains, normalized by domain length, median shown. Allele frequency distribution of all variants within each of the miRNA domains is shown, grouped by A. high confidence miRNAs (green), and B. robust miRNAs (yellow). *- Mann Whitney adjusted p-value <0.001
Notably, we found that 667 mature miRNAs (of the 2534 mature miRNAs that passed the 10X coverage threshold) did not harbor any germline variants across gnomAD-WGS subset (n=15,496). We next analyzed the 375 of these 667 miRNAs with adequate coverage in the gnomAD-WES subset, which contains ~8X as many samples (n=123,136). This analysis identifies 60 miRNAs with no variants and 298 miRNAs with less than 13 variant alleles per miRNA across the gnomAD reference dataset. Although this relative lack of variation may indicate constraint, the conclusions are limited by the short length of mature miRNAs, lack of larger WGS datasets, and a lack of comparable well-defined deleteriousness mechanisms for miRNAs to carry out any further statistical analyses to define a constraint “score”. As an alternative, to provide a measure of miRNA sequence variation using the gnomAD reference dataset, we computed variant allele frequency percentiles across all miRNAs and added this annotation to ADmiRE. The allele frequency percentile gives a reference for the variability of each mature miRNA with respect to all other miRNAs. There are 557 miRNAs within the lower quartile (25th percentile) of allele frequency in gnomAD including 334 miRNAs without any variants in gnomAD WGS. Many top published miRNA families (reported by web-tool mirPub) that play critical roles in development or disease states (generally defined by studies of miRNA expression) are in this set of miRNAs depleted for variation, including most miRNAs from hsa-let-7, hsa-mir-21, hsa-mir-155, hsa-mir-15, hsa-mir-221, hsa-mir-145, and hsa-mir-29 families, and of the miR-17–92 cluster (Vergoulis et al., 2015).
Analysis of miRNA sequence conservation across 100 vertebrates
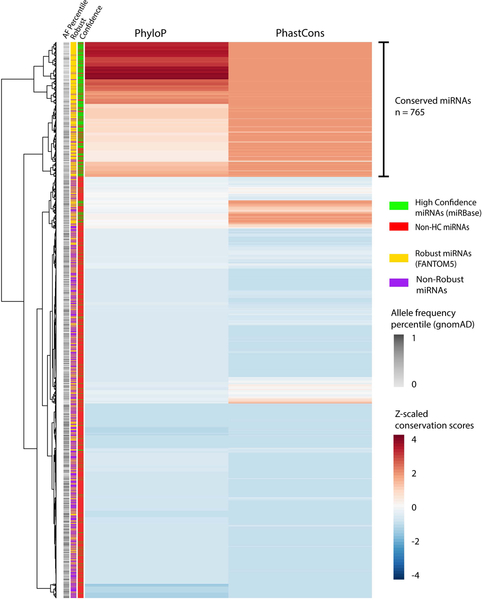
Analysis of evolutionary sequence conservation is a powerful approach to identify functionally important coding and regulatory non-coding genomic regions (Pennacchio & Rubin, 2001; Siepel et al., 2005). We examined human miRNAs using conservation scores from phyloP and phastCons algorithms across 100 vertebrates (Pollard et al., 2010; Siepel et al., 2005; Siepel, Pollard, & Haussler, 2006). Hierarchical clustering of z-normalized conservation scores across 2,571 mature miRNAs identified 765 (30%) highly conserved miRNAs (Figure 7). We also identified a similar proportion of conserved precursor miRNA transcripts (n=434/1,878, 23%) (Supp. Figure S1). Similar to the analysis of sequence variation (Figure 5 C), we observed no significant effect of the genomic context of miRNAs (intragenic and intergenic) on the conservation of miRNAs (Mann-Whitney U test, p-value = 0.23) (Supp. Figure S2). In comparison, gnomAD allele frequency distribution is negatively correlated with PhyloP (Spearman correlation r=−0.29) and PhastCons (Spearman correlation r=−0.26) conservation scores across all mature miRNAs.
Figure 7. Hierarchical clustering of phyloP and phastCons 100way vertebrate conservation scores across mature miRNAs.
PhyloP and phastCons scores are centered and z-score adjusted across each mature miRNA (n=2,571). Each miRNA is annotated for gnomAD AF percentile (grayscale), ‘high confidence’ miRNA (green) or otherwise (red), and ‘robust’ miRNA (yellow) or otherwise (purple).
We also observed significant enrichment of the high-confidence and robust miRNA subsets (these subsets annotated based on robust miRNA expression data) within the cluster of conserved miRNAs (Chi-square p<0.001); with 77% of conserved miRNAs annotated as robust miRNAs (Supp. Figure S3). The miRNAs depleted for variation in the gnomAD dataset (bottom 25th AF percentile) are also enriched within the clusters of conserved miRNAs (Chi-square p<0.05) and 90% of these are robust and/or high confidence. In contrast, ~50% of miRNAs annotated as robust are poorly conserved and also vary in the degree of variation, suggesting that further study is needed to assess their function.
Overall, our analysis of miRNA variation, sequence context, and evolutionary conservation provides critical information for the study of miRNA variation in population and disease datasets. We incorporated into the ADmiRE database the allele frequency percentiles from gnomAD dataset and conservation analysis for each mature miRNA. We next demonstrate the application of ADmiRE annotations to the TCGA PanCancerAtlas dataset to identify and prioritize causal miRNA variation.
Discovery of miRNA variation across 33 cancer types
We analyzed The Cancer Genome Atlas (TCGA) PanCancerAtlas dataset containing whole exome sequencing data from over 10,000 individuals (both germline and tumor samples) across 33 cancer types (Huang et al., 2018). We assessed germline and somatic miRNA variation (see Methods) in 1,317 mature (47%) and 844 precursor (44%) miRNAs that pass the coverage threshold (>6X read depth across >75% of samples) for this WES dataset resulting in a total miRNA target region of 70kb. Among the mature miRNA variants, 1,267 were rare germline variants (as defined by ExAC nonTCGA AF <0.1%) found in approximately 11% of all PanCancerAtlas samples and 1,492 were somatic mutations in 8% of all samples (Table 2). There were an additional 3,771 rare germline and somatic variants when including the entire precursor miRNAs of which 1,165 variants targeted sequence motifs such as CNNC (223 variants), basal UG (16 variants), apical UGU (18 variants) motifs that are important for pre-miRNA processing (Table 2, Supp. Figure S4). The complete list of rare germline and somatic variation across mature and precursor miRNAs in the TCGA PanCancerAtlas dataset is available in the Supplementary Data (Tables 3–6).
Table 2.
Number of unique miRNA variants identified using ADmiRE from over 10,000 individuals from PanCancerAtlas dataset
| miRNA Domain | Rare Germline (AF <0.1%) |
Somatic | Total |
|---|---|---|---|
| miRNA Functional Domains | |||
| Total | 2,991 | 3,539 | 6,530 |
| Mature (Seed) | 1,267 (395) | 1,492 (460) | 2,759 (855) |
| Precursor | 1,724 | 2,047 | 3,771 |
| Precursor miRNA Sequence Motifs | |||
| Total | 547 | 618 | 1,165 |
| basalUG | 9 | 7 | 16 |
| UGU/UGUG | 4 | 14 | 18 |
| CNNC | 116 | 107 | 223 |
| Loop | 418 | 490 | 908 |
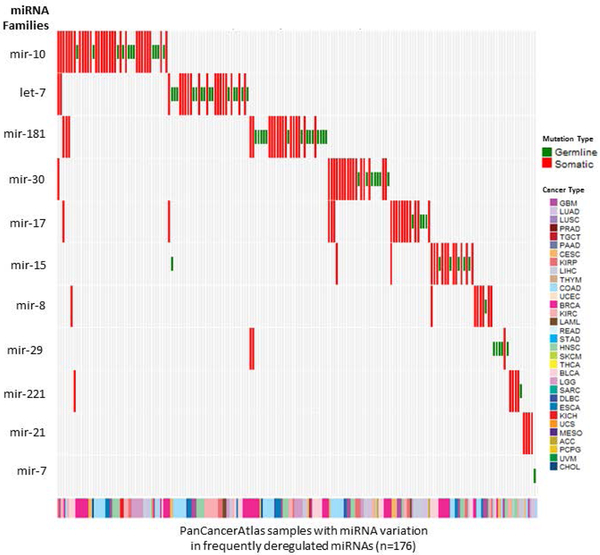
We first analyzed those variants found in miRNAs in the KEGG pathway ‘MicroRNAs in Cancer’ that catalogues differentially expressed miRNAs across 10 cancer types (Lee & Dutta, 2009). We found rare germline and/or somatic variation within the same miRNA-cancer pairs in 1.2% of samples. We further combined these miRNAs according to their miRNA families and found that a subset of miRNA families e.g. miR-10, let-7, miR-17, miR-30, and miR-15 harbor frequent somatic and rare germline variants in the above mentioned 10 cancer types as well as some other cancer types in the PanCancerAtlas dataset, (Figure 8). For the top 2 frequently deregulated miRNA families, mir-10 and let-7, the spread of variation in PanCancerAtlas dataset is distinct from that in the large population dataset of ExAC (nonTCGA subset). (Supp. Figures S6 and S7).
Figure 8. Somatic and rare germline variants in miRNA families that are frequently deregulated in adult cancers as per KEGG pathway, ‘miRNA in Cancer’.
PanCancerAtlas samples with somatic (red) or rare germline (green) mutations in most frequently deregulated miRNAs listed in KEGG pathway ‘miRNAs in Cancer’ grouped by miRNA families. The full name of each cancer type corresponding to the TCGA standard abbreviation is provided in the Abbreviations section.
We further assessed miRNA germline sequence variation in the entire dataset and within each of the 33 cancers. There were 1,267 rare germline variants spread among 631 miRNAs, however, variation within many miRNAs did not significantly cluster within one tumor type (Supplementary Data). To filter out miRNAs that accumulate rare germline variation in control human population and thus indicate low sequence constraint, we removed miRNAs with >50th AF percentile for variation in gnomAD. We further prioritized miRNAs with >1 variant in a cancer type, resulting in a set of 179 mature miRNAs containing 267 rare germline variants. For each miRNA, we quantified the number of samples with rare germline variation normalized by the total number of samples in the entire dataset and for each cancer type. Most of these miRNAs harbored only ~1–2 rare germline variants for each cancer type, at a similar frequency to that observed in the gnomAD dataset. Analysis of rare germline variation across the entire precursor miRNA transcripts yielded similar results. Overall, our analysis of rare germline miRNA variants in the TCGA PanCancerAtlas Dataset compared with gnomAD suggests that rare germline variation plays little to no role in predisposition to the most common histologic types of adult cancers, however, this does not preclude smaller effects of common variants, or miRNA variation in other tumors types or histopathologic subtypes.
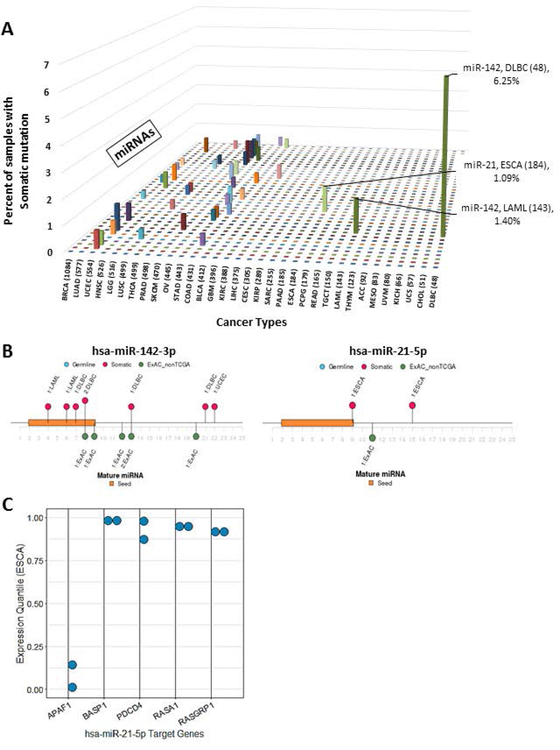
We then assessed the landscape of somatic miRNA mutation in the entire TCGA PanCancerAtlas dataset. Overall, the distribution per sample of somatic mutations resembles that previously described for protein-coding variants across respective adult cancer types, as samples with higher somatic mutation rate had more somatic miRNA variants (Supp. Figure S5A) (Kandoth et al., 2013). We identified the 10 most frequently mutated miRNAs within each cancer type (Figure 9 A). We found that the majority of miRNAs were mutated in <1% of samples in the respective cancer type. In addition, the cancer types known to have high mutation burdens such as lung squamous cell carcinoma (LUSC), skin cutaneous melanomas (SKCM), and endometrial (UCEC) cancers have multiple mutated miRNAs each in <1% of samples. The tumor types with at least 1% somatic mutation in a specific miRNA (Figure 9 A) included diffuse large B-cell lymphoma (DLBC) and acute myeloid leukemia (LAML) with miR-142 mutations, 6.2%, and 1.3% respectively. This miRNA was mutated among <1% samples (majority singleton variants) in all other cancer types. This result is consistent with the previously reported association of somatic mutations of miR-142 (miR142) mutations in hematologic malignancies, lymphoma (20% DLBC) and leukemia (2% LAML) (Kwanhian et al., 2012; Silva et al., 2013; Yao et al., 2016) (Figure 9 B). In addition, we found somatic mutations in miR-21 (miR21) among 1% (2/184) of esophageal cancer (ESCA) samples (Figure 9 B). Multiple prior studies have described miR-21 deregulation in esophageal cancer (Chu, Zhu, Lv, Zhou, & Huo, 2013; Kan & Meltzer, 2009; Smith, Watson, Michael, & Hussey, 2010; Song & Meltzer, 2012), however, to our knowledge somatic mutations in this miRNA have not been previously reported. Using the experimentally validated targets of miR-21–5p annotated in ADmiRE (miRTarBase (Chou et al., 2016)), we further assessed whether the two mature miR-21–5p variants, at +9 (C>T) and +15 (G>T) bp, alter target regulation using the RNAseq data provided by the TCGA project. Indeed, expression of 5 miR-21 target genes, APAF1, BASP1, PDCD4, RASA1, and RASGRP1, show significantly altered expression in mutated samples as compared to the rest of the samples in this cancer type (Wilcoxon test p<0.05) suggesting the miRNA variants impact miR-21 function (Figure 9 C). It is more challenging to analyze the importance of somatic variation in the cases of highly mutated cancer types such as UCEC, SKCM, and LUSC, with several miRNAs each having a few somatic mutations. Overall, somatic mutations in miRNAs appear to play an important role in a small number of the most common forms of adult cancers. In those samples, mutations may contribute to the loss of miRNA-mediated gene regulation.
Figure 9. Somatic mutations in mature miRNAs across 33 cancers and spectrum of mutations in top candidate miRNAs.
A. Percent of samples with somatic mutations within each of the mature miRNAs ordered by number of samples sequenced for each cancer type (number of mature miRNA mutations normalized by the number of samples sequenced for respective cancer type). B. Distribution of somatic and rare germline variation along two candidate miRNAs, miR-142 and miR-21, that show accumulation of seed and mature domain variation as compared to ExAC (nonTCGA subset) dataset. C. Expression quantile of miR-21 target genes from mutated samples that are significantly different as compared to non-mutated samples of the same cancer type (ESCA) (Wilcoxon test p <0.05). The full name of each cancer type corresponding to the TCGA standard abbreviation is provided in the Abbreviations section.
Discussion
Through the development of the Annotative Database of miRNA Elements, ADmiRE, we present a dedicated miRNA variant annotation tool that substantially adds to our knowledge of variation in miRNA genes across human datasets. Not surprisingly, our analyses demonstrate that high coverage WGS comprehensively captures >99% of miRNAs at mean read-depth >30X. The remaining 1% of mature miRNAs (n=39) largely fall into two categories, there are 25 miRNAs with sequence data across the majority of samples at the median read depth of 1–10X. Knowledge of variation in these miRNAs will be better captured by deeper WGS. However, the remaining 14 miRNAs are completely missed by the current WGS sequencing platforms and consist of more recently discovered as well as non-conserved miRNAs. Moreover, given a large number of research and clinical WES efforts, we demonstrate that current exome capture platforms capture approximately 50% of miRNAs in miRBase v.21 and these capture designs could be further updated to improve evaluation of miRNA variation through WES analysis.
We compared the performance of existing variant annotation tools for miRNA variation in comparison to ADmiRE. Variant annotation is comprised of two different processes. First, accurate annotation of variants in the region of interest, miRNAs in this case. Second, the breadth of useful information provided to aid variant interpretation. In comparison to ADmiRE many of the existing variant annotation tools, e.g. Variant Effect Predictor (VEP), ANNOVAR, and snpEFF either fail to accurately annotate mature miRNA variants or deprioritize them over potentially benign protein-coding variation. For protein-coding regions, Many tools provide a variety of annotations such as deleteriousness predictions from algorithms focused on changes in protein function (Polyphen, Sift), conservation scores (phyloP, GERP), population frequency, and other information about protein-function or structure and disease associations. However, these same tools provide limited information for the interpretation of miRNA variation and often only annotate these variants for their potential role in the nearby protein-coding gene. In comparison, ADmiRE provides extensive biological and functional annotations for every basepair within miRNA genes by combining miRNA sequence information such as miRNA domains and sequence motifs evolutionary conservation with variation constraint in the human population (gnomAD allele frequency percentile), high confidence of expression, miRNA-disease associations, and experimentally validated target genes. The goal of developing this tool is to facilitate the detection and downstream prioritization of candidate variation within miRNA sequences from disease discovery and clinical pipelines as described here for the TCGA dataset.
Analysis of miRNA variation from the large human population sequencing dataset (gnomAD) also provided new insights into the landscape of miRNA variation. Nearly 50% of miRNAs are expressed from protein-coding exonic or intronic regions (within RefSeq v78 transcripts), and the remainder are in the intergenic regions. Despite this difference in the genomic environment, we found comparable variant AF distribution of intragenic and intergenic miRNA genes and similar conservation profiles of the mature miRNA sequence. For both intergenic and intragenic miRNAs, the pattern of sequence variation in miRNAs (SNP density and domain-level constraints) is similar to that reported for protein-coding exons with less variation in functionally important miRNA sequences. Most existing methods that predict the biological impact of protein-coding variants utilize domain level information (exons, introns, UTRs, etc.) or AF distribution (z-scores, pLI scores, etc.); however, such prior efforts for non-coding RNAs have been challenging (Worth, Gong, & Blundell, 2009). We corroborated an earlier study of a smaller set of 729 miRNAs from 1000 Genomes Phase I dataset (Carbonell et al., 2012) and found that seed and mature domain variation is rare as compared to nearby regions (Figure 5 B). Moreover, we report the largest catalog of seed and mature domain miRNA variation surpassing current datasets by many folds (Bhattacharya et al., 2014; Gong et al., 2012; Liu et al., 2012). Annotation of variants for their location within precursor miRNA sequence motifs such as CNNC, basal UG, apical UGU/UGUG, etc. reveals that variation within these regions is also extremely rare in gnomAD and should be prioritized if found in disease datasets. In addition to ExAC/gnomAD datasets, we also provide ADmiRE annotations of miRNA variation from analysis of the WGS samples from 1000 Genomes, UK10K, and National Heart, Lung and Blood Institute’s (NHLBI’s) Trans-Omics for Precision Medicine (TOPMed- dbSNP VCF provided on the bravo server) projects (Supplementary Data).
We further evaluated previously curated sets of miRNAs which utilize expression criteria from high-throughput sequencing data, namely, ‘high confidence’ miRNAs (Kozomara & Griffiths-Jones, 2014) (miRBase v21) and ‘robust’ miRNAs (Rie et al., 2017) (FANTOM5 project). Recently, it was highlighted that the miRNAs that have passed these stringent criteria should be prioritized in order to filter out false positive miRNA entries (Bartel, 2018). Consistent with this recommendation, we find that both high confidence and robust miRNA datasets are enriched within the highly conserved miRNAs. Furthermore, both miRNA groups harbor significantly different allele frequency distributions (shifted towards rarity) compared to the remainder miRNAs in the entire gnomAD dataset (Mann-Whitney test p<0.001).
MicroRNA-mediated silencing pathways are conserved across eukaryotic lineages and many miRNA loci are also conserved across evolution (Altuvia et al., 2005; Chapman & Carrington, 2007). As expected, evolutionarily conserved miRNAs also tend to harbor less variation in the human population as indicated by the negative correlation between conservation and gnomAD minor allele frequency across miRNAs. Interestingly, miRNA families with a larger number of members tend to be highly conserved across 100 vertebrates suggesting the expansion of functionally important miRNA families. We find approximately 66% miRNAs with low conservation across 100 vertebrates and suggest that they are part of the group of miRNAs which are rapidly gained and lost during evolution that are yet to undergo evolutionary selection in humans (Bartel, 2018). This hypothesis can be supported by the observation that these less conserved miRNAs tend to accumulate common variation in the human population as indicated by our analysis of the gnomAD dataset. (Figure 7).
Our goal in developing ADmiRE and providing analysis of the gnomAD dataset was to support future analyses of miRNA variation in disease datasets. We demonstrate the application of ADmiRE annotations to the PanCancerAtlas dataset of WES from over 10,000 individuals spanning 33 cancer types, substantially adding to the knowledge of miRNA variation in cancer previously cataloged in SomamiR DB and other studies (Bhattacharya, Ziebarth, & Cui, 2013; Tuna et al., 2016). Using data derived from our analysis of miRNAs in gnomAD we were able to filter out variants found in highly variable miRNAs as well as common variants within any miRNA from the analysis of germline miRNA variants in the PanCancerAtlas dataset. As a result, we did not find any miRNAs with variants in more than 1% of any of the TCGA tumor types suggesting that germline variation in those miRNAs effectively sequenced by WES analysis is unlikely to play a substantial role in cancer susceptibility in the tumors studied by TCGA. Additional analysis using rare variant burden tests was limited by the statistical power to analyze relatively small genomic region (~22bp) and the small number of samples in some cancer types (seven TCGA tumor types had ≤100 tumor samples analyzed). Further study is needed to determine the potential role of germline variants in miRNA genes in rare pathologic subtypes, other rare adult cancers, pediatric cancers and those miRNAs not effectively sequenced by WES. Genome-wide association approaches could also be explored by focusing on the common miRNAs variants (AF >5%) described here, as some of these common variants are predicted to alter miRNA target gene expression (Gong et al., 2012; Iuliano et al., 2013; Ryan et al., 2010).
Analysis of somatic mutations in miRNAs demonstrated higher specificity with cancer type including the previously described association of miR-142 somatic mutations in hematologic malignancies such as lymphoma (DLBC) and leukemia (LAML). Our data also suggest a potentially novel association of somatic mutations in miR-21 in esophageal cancer with evidence that these rare somatic mutations affect the function of this miRNA as evidenced by altered regulation of multiple miR-21 target genes. Independently, we also analyzed miRNA variation in 155 frequently up- or down-regulated miRNAs in 9 cancer types as documented in the KEGG pathway, ‘miRNA in Cancer’ (ID: hsa05206) (Lee & Dutta, 2009). Strikingly, 132 (85%) of these miRNAs are highly conserved across 100 vertebrates and 86 miRNAs are in the lowest quartile for variation in the gnomAD dataset. This result further underscores the utility of miRNA sequence annotations computed in this study and included in ADmiRE to detect potential causal miRNA variation.
Overall, we show that through the development and application of a dedicated annotation tool for miRNA variation, we characterized miRNA variation in multiple human population datasets and identify disease-related miRNA variation. WES datasets only effectively identify variation in 50% of miRNA genes and the growth in WGS datasets will provide greater power to identify disease-associated variation. Integrating the measures of miRNA sequence variation and conservation developed in this study with other existing miRNA annotations through ADmiRE will facilitate the interpretation of miRNA variation from human sequencing datasets.
Supplementary Material
Acknowledgments
This work was supported by the Cancer Prevention and Research Institute of Texas [RP10189]; National Institutes of Health [NIH-R01-CA138836]; and National Human Genome Research Institute [5U01HG007436–03, U41HG009649–01] to SEP.
The authors would like to thank the Genome Aggregation Database (gnomAD) team and the groups that provided exome and genome variant data towards this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about. The authors would also like to thank Dr. Stephen Montgomery, Stanford University and Charles Lin, Baylor College of Medicine for their comments on the manuscript.
This work was supported by the Cancer Prevention and Research Institute of Texas [RP10189]; National Institutes of Health [NIH-R01-CA138836]; and the National Human Genome Research Institute [5U01HG007436–03, U41HG009649–01] to SEP.
Abbreviations
- ACC
Adrenocortical carcinoma
- BLCA
Bladder Urothelial Carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- DLBC
Lymphoid Neoplasm Diffuse Large B-cell Lymphoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HNSC
Head and Neck squamous cell carcinoma
- KICH
Kidney Chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LAML
Acute Myeloid Leukemia
- LGG
Brain Lower Grade Glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and Paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin Cutaneous Melanoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular Germ Cell Tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- UCEC
Uterine Corpus Endometrial Carcinoma
- UCS
Uterine Carcinosarcoma
- UVM
Uveal Melanoma
Footnotes
Availability
ADmiRE workflow for annotation of user-supplied variant files available in the GitHub repository: https://github.com/nroak/admire GitHub page contains a stand-alone program for annotation using ADmiRE or a database file (BED format) that can be used as a ‘--custom’ database to VEP annotation tool. ADmiRE is also available as a ‘User-contributed dataset’ on ANNOVAR website for ease of integration into existing workflows, ADmiRE has also been dockerized as an app on CAVATICA cloud based platform (https://cavatica.sbgenomics.com).
Supplementary Data
Supplementary Data are available online (Supplementary_Figures.pdf, Supplementary_Data.zip).
Conflict of Interest
SEP is a member of the Baylor Genetics laboratory scientific advisory panel.
References
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, … Margalit H (2005). Clustering and conservation patterns of human microRNAs. Nucleic Acids Research, 33(8), 2697–2706. 10.1093/nar/gki567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, … Abecasis GR (2015). A global reference for human genetic variation. Nature, 526(7571), 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung VCC, Ulitsky I, McGeary SEE, & Bartel DPP (2013). Beyond secondary structure: Primary-sequence determinants license Pri-miRNA hairpins for processing. Cell, 152(4), 844–858. 10.1016/j.cell.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009). MicroRNAs: target recognition and regulatory functions. Cell, 136(2), 215–33. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell, 173(1), 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, & Cui Y (2015). Knowledge-based analysis of functional impacts of mutations in microRNA seed regions. Journal of Biosciences, 40(4), 791–798. 10.1007/s12038-015-9560-2 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Ziebarth JD, & Cui Y (2013). SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Research, 41(Database issue), D977–82. 10.1093/nar/gks1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Ziebarth JD, & Cui Y (2014). PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Research, 42(D1), D86–D91. 10.1093/nar/gkt1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, & Croce CM (2006). Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Seminars in Oncology, 33(2), 167–73. 10.1053/j.seminoncol.2006.01.010 [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, … Sc M (2005). A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England Journal of Medicine, 353(17), 1793–801. 10.1056/NEJMoa050995 [DOI] [PubMed] [Google Scholar]
- Cammaerts S, Strazisar M, Dierckx J, Del Favero J, & De Rijk P (2015). miRVaS: a tool to predict the impact of genetic variants on miRNAs. Nucleic Acids Research, 44(3), gkv921 10.1093/nar/gkv921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell JJ, Alloza E, Arce P, Borrego S, Santoyo J, Ruiz-Ferrer M, … Dopazo JJJ (2012). A map of human microRNA variation uncovers unexpectedly high levels of variability. Genome Medicine, 4(8), 62 10.1186/gm363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E, Prado DE, & Weidhaas JB (2011). Cancer microRNAs: from subtype profiling to predictors of response to therapy. Trends in Molecular Medicine, 17(5), 235–43. 10.1016/j.molmed.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, & Carrington JC (2007). Specialization and evolution of endogenous small RNA pathways. Nature Reviews. Genetics, 8(11), 884–896. 10.1038/nrg2179 [DOI] [PubMed] [Google Scholar]
- Chou C-HH, Chang N-WW, Shrestha S, Hsu S.-D.Da, Lin Y-LL, Lee W-HH, … Huang H-TH-DDH-TH-DHT (2016). miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Research, 44(D1), D239–D247. 10.1093/nar/gkv1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Zhu H, Lv L, Zhou Y, & Huo J (2013). MiRNA s in oesophageal squamous cancer. The Netherlands Journal of Medicine, 71(2), 69–75. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/23462054 [PubMed] [Google Scholar]
- Cingolani P, Cunningham F, McLaren W, & Wang K (2018). Variant annotations in VCF format. Retrieved from http://snpeff.sourceforge.net/VCFannotationformat_v1.0.pdf
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, … Ruden DM (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6(2), 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte I, Hadfield KD, Barbato S, Carrella S, Pizzo M, Bhat RS, … Black GCM (2015). MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proceedings of the National Academy of Sciences of the United States of America, 112(25), E3236–45. 10.1073/pnas.1401464112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kock L, Sabbaghian N, Plourde F, Srivastava A, Weber E, Bouron-Dal Soglio D, … Foulkes WD (2014). Pituitary blastoma: A pathognomonic feature of germ-line DICER1 mutations. Acta Neuropathologica, 128, 111–122. 10.1007/s00401-014-1285-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, … Mariamidze A (2018). Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Systems, 6(3), 271–281.e7. 10.1016/J.CELS.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, & Bartel DP (2015). The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Molecular Cell, 60(1), 131–145. 10.1016/j.molcel.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB, & Bartel DP (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Research, 19(1), 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, & Seoighe C (2010). A flexible R package for nonnegative matrix factorization. BMC Bioinformatics, 11(1), 367 10.1186/1471-2105-11-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Tong Y, Zhang H-M, Wang K, Hu T, Shan G, … Guo A-Y (2012). Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Human Mutation, 33(1), 254–63. 10.1002/humu.21641 [DOI] [PubMed] [Google Scholar]
- Harfe BD (2005). MicroRNAs in vertebrate development. Current Opinion in Genetics and Development. 10.1016/j.gde.2005.06.012 [DOI] [PubMed] [Google Scholar]
- Hill CG, Jabbari N, Matyunina LV, & McDonald JF (2014). Functional and Evolutionary Significance of Human MicroRNA Seed Region Mutations. PLoS ONE, 9(12), e115241 10.1371/journal.pone.0115241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.lin, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, … Ding L (2018). Pathogenic Germline Variants in 10,389 Adult Cancers. Cell. 10.1016/j.cell.2018.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, & Willoughby CE (2011). Mutation altering the miR-184 seed region causes familial keratoconus with cataract. American Journal of Human Genetics, 89(5), 628–33. 10.1016/j.ajhg.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff BW, Riazuddin SA, & Gottsch JD (2012). A Single-Base Substitution in the Seed Region of miR-184 Causes EDICT Syndrome. Investigative Opthalmology & Visual Science, 53(1), 348 10.1167/iovs.11-8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano R, Vismara MFM, Dattilo V, Trapasso F, Baudi F, & Perrotti N (2013). The role of microRNAs in cancer susceptibility. BioMed Research International, 2013, 591931 10.1155/2013/591931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan T, & Meltzer SJ (2009). MicroRNAs in Barrett’s esophagus and esophageal adenocarcinoma. Current Opinion in Pharmacology, 9(6), 727–32. 10.1016/j.coph.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, … Ding L (2013). Mutational landscape and significance across 12 major cancer types. Nature, 502(7471), 333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani A, Ha D, Schotte D, Armstrong SA, Lodish HF, Den Boer ML, … Lodish HF (2010). A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 Acute Lymphocytic Leukemia cells. Cell Cycle, 9(6), 1037–1042. http://doi.org/11011 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, & Griffiths-Jones S (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research, 39(Database issue), D152–7. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, & Griffiths-Jones S (2014). MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 42(D1), D68–73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, & James Kent W (2013). The UCSC genome browser and associated tools. Briefings in Bioinformatics, 14(2), 144–161. 10.1093/bib/bbs038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, … Jacks T (2009). Dicer1 functions as a haploinsufficient tumor suppressor. Genes & Development, 23(23), 2700–4. 10.1101/gad.1848209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwanhian W, Lenze D, Alles J, Motsch N, Barth S, Döll C, … Grässer FA (2012). MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Medicine, 1(2), 141–55. 10.1002/cam4.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, & Dutta A (2009). MicroRNAs in Cancer. Annual Review of Pathology: Mechanisms of Disease, 4(1), 199–227. 10.1146/annurev.pathol.4.110807.092222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, … MacArthur DG (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536(7616), 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Duan R, Kooy F, Sherman SL, Zhou W, & Jin P (2009). Germline mutation of microRNA-125a is associated with breast cancer. Journal of Medical Genetics, 46(5), 358–60. 10.1136/jmg.2008.063123 [DOI] [PubMed] [Google Scholar]
- Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, & Cui Q (2014). HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Research, 42(Database issue), D1070–4. 10.1093/nar/gkt1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, & Gregory RI (2015). MicroRNA biogenesis pathways in cancer. Nature Reviews Cancer, 15(6), 321–333. 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang F, Li T, Lu M, Wang L, Yue W, & Zhang D (2012). MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics, 13(1), 661 10.1186/1471-2164-13-661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, & Cui Q (2008). An Analysis of Human MicroRNA and Disease Associations. PLoS ONE, 3(10), e3420 10.1371/journal.pone.0003420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, & Lowe SW (2012). The microcosmos of cancer. Nature, 482(7385), 347–55. 10.1038/nature10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; (Baltimore, M. (2018). Online Mendelian Inheritance in Man, OMIM. Retrieved April 24, 2018, from https://omim.org/ [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, … Cunningham F (2016). The Ensembl Variant Effect Predictor. Genome Biology, 17(1), 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Nolde M, & Slack FJ (2010). OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature, 467(7311), 86–90. 10.1038/nature09284 [DOI] [PubMed] [Google Scholar]
- Mencía Á, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, … Moreno-Pelayo MÁ (2009). Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nature Genetics, 41(5), 609–613. 10.1038/ng.355 [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Jo MH, Choi Y-G, Park J, Kwon SC, Hohng S, … Woo J-S (2015). Functional Anatomy of the Human Microprocessor. Cell, 161(6), 1374–1387. 10.1016/j.cell.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, & Rubin EM (2001). Genomic strategies to identify mammalian regulatory sequences. Nature Reviews Genetics, 2(2), 100–109. 10.1038/35052548 [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, & Siepel A (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Research, 20(1), 110–121. 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR (2014). BEDTools: The Swiss-Army tool for genome feature analysis. Current Protocols in Bioinformatics, 2014(1), 11.12.1–11.12.34. 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rie D. de, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, … de Hoon MJL (2017). An integrated expression atlas of miRNAs and their promoters in human and mouse. Nature Biotechnology, 35(Table 2), 872–878. 10.1038/nbt.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden C, Gaillard J, Kanoria S, Rennie W, Barish S, Cheng J, … Lu J (2017). Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Research, 27(3), 374–384. 10.1101/gr.208900.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, Dunger I, … Theis FJ (2010). PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes. Genome Biology, 11(1), R6 10.1186/gb-2010-11-1-r6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan B, Robles A, & Harris C (2010). Genetic variation in microRNA networks: the implications for cancer research. Nature Reviews Cancer, 10(6), 389–402. 10.1038/nrc2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, … Daly MJ (2014). A framework for the interpretation of de novo mutation in human disease. Nature Genetics, 46(9), 944–950. 10.1038/ng.3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Ambrosone CB, & Zhao H (2009). Novel genetic variants in microRNA genes and familial breast cancer. International Journal of Cancer. Journal International Du Cancer, 124(5), 1178–82. 10.1002/ijc.24008 [DOI] [PubMed] [Google Scholar]
- Shen J, DiCioccio R, Odunsi K, Lele SB, & Zhao H (2010). Novel genetic variants in miR-191 gene and familial ovarian cancer. BMC Cancer, 10, 47 10.1186/1471-2407-10-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, … Haussler D (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Research, 15(8), 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Pollard KS, & Haussler D (2006). New methods for detecting lineage-specific selection In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) (Vol. 3909 LNBI, pp. 190–205). Springer; Berlin Heidelberg: 10.1007/11732990_17 [DOI] [Google Scholar]
- Silva J, Wylie T, Hundal J, McGrath S, Magrini V, Ramsingh G, … Link DC (2013). Dysregulation and Recurrent Mutation Of miRNA-142 In De Novo AML. Blood, 122(21). Retrieved from http://www.bloodjournal.org.ezproxyhost.library.tmc.edu/content/122/21/472?sso-checked=true [Google Scholar]
- Smith C-M, Watson DI, Michael MZ, & Hussey DJ (2010). MicroRNAs, development of Barrett’s esophagus, and progression to esophageal adenocarcinoma. World Journal of Gastroenterology, 16(5), 531–7. 10.3748/WJG.V16.I5.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, & Meltzer SJ (2012). MicroRNAs in Pathogenesis, Diagnosis, and Treatment of Gastroesophageal Cancers. Gastroenterology, 143(1), 35–47.e2. 10.1053/j.gastro.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Croce CM, & Calin G. a. (2009). SnapShot: MicroRNAs in Cancer. Cell, 137(3), 586–586.e1 10.1016/j.cell.2009.04.040 [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, … Rajewsky K (2007). Regulation of the germinal center response by MicroRNA-155. Science, 316(5824), 604–608. 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- Tuna M, Machado AS, & Calin GA (2016). Genetic and epigenetic alterations of microRNAs and implications for human cancers and other diseases. Genes Chromosomes and Cancer, 55(3), 193–214. 10.1002/gcc.22332 [DOI] [PubMed] [Google Scholar]
- Vergoulis T, Kanellos I, Kostoulas N, Georgakilas G, Sellis T, Hatzigeorgiou A, & Dalamagas T (2015). MirPub: A database for searching microRNA publications. Bioinformatics, 31(9), 1502–1504. 10.1093/bioinformatics/btu819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, … Hatzigeorgiou AG (2015). DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Research, 43(Database issue), D153–9. 10.1093/nar/gku1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, … Zhang W (2015). The UK10K project identifies rare variants in health and disease. Nature, 526(7571), 82–89. 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lu M, Qiu C, & Cui Q (2010). TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Research, 38(Database issue), D119–22. 10.1093/nar/gkp803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, & Hakonarson H (2010). ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research, 38(16), 1–7. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth CL, Gong S, & Blundell TL (2009). Structural and functional constraints in the evolution of protein families. Nature Reviews Molecular Cell Biology, 10(10), 709 10.1038/nrm2762 [DOI] [PubMed] [Google Scholar]
- Yang Z (1995). A space-time process model for the evolution of DNA sequences. Genetics, 139(2), 993–1005. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7713447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J-C, Wong TN, Trissal M, Ramaswamy R, Sun Y, Ley TJ, … Link DC (2016). MIR142 Loss-of-Function Mutations Promote Leukemogenesis through Derepression of ASH1L Resulting in Increased HOX Gene Expression. Blood, 128(22). Retrieved from http://www.bloodjournal.org.ezproxyhost.library.tmc.edu/content/128/22/2718?sso-checked=true [Google Scholar]
- Zeng Y, & Cullen BR (2003). Sequence requirements for micro RNA processing and function in human cells. RNA (New York, N.Y.), 9(1), 112–23. 10.1261/rna.2780503.known [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Gao W, Qian Z, & Miao Y (2009). Genetic variation of miRNA sequence in pancreatic cancer. Acta Biochimica et Biophysica Sinica, 41(5), 407–413. 10.1093/abbs/gmp023.Advance [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.