Abstract
Currently, it reported that TAF1L gene mutation is found in a number of carcinomas, but its pathophysiological function has not been well studied. We focused on investigating expressive levels of TAF1L gene and protein in esophageal squamous cell carcinoma (ESCC) with two tissue microarrays, forty fresh paired ESCC and paracancer samples using immunohistochemistry, real-time PCR or Western blot in this study. Furthermore, we executed TAF1L silence with siRNA in ESCC cell lines to evaluate effects of TAF1L expression on cell proliferation, migration and invasion of ESCC via CCK-8, wound healing and transwell chamber assays. Moreover, key proteins related to ESCC development were also analyzed by Western blot. Results from this study showed that the expression of TAF1L mRNA and protein in ESCC tissues were significantly higher than that in matched paracancer tissues. However, its abnormal expression was not associated with other clinic features, such as the age, gender and pathological grade, except of TNM-N stage. Furthermore, the proliferation, migration and invasion of ESCC cells were inhibited after TAF1L gene silencing. As a consequence, the expression of c-Myc and phosphorylated Akt in esophageal squamous cell line after TAF1L-siRNA treatment were inversely decreased, while p53 was increased significantly, compared those to control group. Taken together, the results from this study suggest that TAF1L gene might be served as an oncogene, and its overexpression could accelerate to the tumorigenesis of ESCC via promoting the malignant cell proliferation and tumor metastasis.
Keywords: TAF1L, ESCC, tumorigenesis, cell proliferation, tumor metastasis.
Introduction
Esophageal squamous cell carcinoma (ESCC) is a major pathohistological subtype of esophageal cancer in China, which is one of the most lethal cancers, ranking 4th in mortality among all tumors globally1,2. After clinical diagnosis, main therapies of ESCC include the surgery, radiotherapy and chemotherapeutics3. Unfortunately, when ESCC patients were made definite diagnosis, approximately 90% of them had already been reached to advanced stage with poor prognosis and low survival rate4,5. Therefore, it is urgent need to find novel biomarkers for early diagnosis and precision medicine of ESCC.
TAF1L (TATA-box binding protein associated factor 1 like), also known as TAF(II)210, is located on chromosome 9p21.1, and its locus is intronless. TAF1L gene exhibits 95% amino acid identity to the homologue TAF1. A structural analysis reveals that TAF1L, as TAF1 gene, contains kinase motif with ubiquitin-conjugating activity, a HAT region and two related BDs. The products of TAF1 and TAF1L can be interchangeable in male germ cells6,7. Previous studies reported that TAF1 gene could relate with the regulation of cell growth and cell cycle8,9. A result of genome-wide RNAi screen also demonstrated that TAF1 play a role in apoptotic regulation and genotoxic stress. However, when the expression of p27Kip1 was reduced as a consequence of knocking down TAF1, those tested cells became more resistance to apoptosis10. Further information about protumorigenic roles of TAF1 gene in several tumors has been reported, i.e. uterine serous carcinoma, colorectal cancer, gastric cancer and esophageal cancer11-13. Unlike other retroposed copies of genes in the human genome without RNA translation, TAF1L gene can be normally transcribed and translated to intact protein6. Thus, TAF1L may have similar regulatory functions in cancers, as that in the homologue of TAF1 gene.
Recently, using next generation sequencing and meta-analysis, Xia J, et al found that TAF1L gene had recurrent mutation at melanomas samples14. Our previous research also demonstrated that TAF1L gene was associated with the development of human oral squamous cell carcinoma and colorectal cancer15, 16. According to that, we hypothesized that TAF1L gene may carry on special biological functions for the pathogenesis of ESCC, and then we intended to investigate whether TAF1L gene was abnormal expression in ESCC, and played important roles in disease development? Thus, via a technique of special gene silence in vitro, we analyzed TAF1L silent effects on cell proliferation, migration and invasion of ESCC, in order to further illustrate the pathophysiological effects of TAF1L gene on ESCC progress.
Material and methods
The collection and treatment of tissue specimen
Two commercial tissue microarrays include 150 cases totally were obtained from Biomax in USA. One contains 30 paired of ESCC and cancer adjacent esophagus tissue sections (total 60 sections), and another contains 120 cases of ESCC tissue, with 40 tissues of matched adjacent normal esophagus and 40 matched metastasis carcinoma tissue sections. The individual parameters (such as gender, TNM classification, clinical stage and pathology grade) of each section on the microarray were listed in Table 3. Except of 4 cases missed the information of grade, a total of 146 ESCC cases were classified based on pathological differentiation. In addition, 40 paired fresh ESCC and paracancer tissues after surgery were collected from Shantou University Cancer Hospital. The sample collection was complied with ethics agreement approved by Medical Ethics Committee of Shantou University Medical College Cancer Hospital (approved number: 2016024).
Table 3.
The association between TAF1L overexpression and clinicopathological characteristics of ESCC
| Clinicopathological characteristics | Case numbers | Overexpression | ||
|---|---|---|---|---|
| Yes (n) | No (n) | p-value | ||
| Age(years) | ||||
| <60 | 87 | 61 | 26 | 0.8615 |
| ≥60 | 63 | 45 | 18 | |
| Gender | ||||
| Female | 43 | 26 | 17 | 0.0819 |
| Male | 107 | 80 | 27 | |
| Differentiation | ||||
| Well | 49 | 33 | 16 | 0.6359 |
| Moderate | 55 | 39 | 16 | |
| Poor | 42 | 32 | 10 | |
| Stage | ||||
| Ⅰ | 12 | 8 | 4 | 0.2413 |
| Ⅱ | 78 | 51 | 27 | |
| Ⅲ | 60 | 47 | 13 | |
| TNM-T | ||||
| T1-2 | 32 | 19 | 13 | 0.1137 |
| T3 | 118 | 87 | 31 | |
| TNM-N | ||||
| N0 | 86 | 54 | 32 | 0.0141* |
| N1-2 | 64 | 52 | 12 | |
| TNM-M | ||||
| M0 | 110 | 74 | 36 | 0.7940 |
| M1 | 40 | 26 | 14 | |
* It indicated statistically significant p<0.05.
Immunohistochemical staining (IHC)
Two tissue microarrays were deparaffinized with xylene and rehydration. Antigen retrieval was performed using 10 mM citrate buffer (pH 6.0) for 30 min in a steamer set pan (IHCWORLD, USA). Then tissue sections were incubated with 3% H2O2 for 30 min, washed with PBS, and incubated with appropriate rabbit anti-human TAF1L antibody overnight at 4°C. After washing with PBS, tissue sections were incubated with goat anti-rabbit IgG-HRP (Zymed Laboratories, USA) for 60 min at room temperature, followed by detective reaction using AEC kit (GBI, USA), and counterstained with the hematoxylin. The visualization was performed under a microscope. All resulting images were recorded and analyzed by two independent individuals. The PBS replaced primary antibody, was served as negative control. Positive controls were executed on corresponding tissue sections stained with sensitive antibodies in parallel. Five fields of each section were selected randomly under a microscopy with 400-time magnification. Intensity degrees of positive signals were determined by 0-none, 1-weak, 2-moderate, 3-intense and 4-strongly intense; and percentage degrees of positive cell number were recorded via 0: 0%, 1: 1~25%, 2: 25-50%, 3: 50-75% and 4: 75-100%. Final score of IHC staining was achieved by multiplicating above two scores. Score<4 was categorized as “negative”, and score≧4 was as “positive” for next statistical analysis.
Cell culture
Human KYSE150 cells and KYSE180 cells of ESCC, and immortalized esophageal epithelial cell line (Het-1A cells) were incubated in RPMI-1640 medium (Hyclone, USA) with 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (Hyclone, USA) at 37°C in a cell incubator with humidified atmosphere of 5% CO2.
Cell transfection
siRNAs for TAF1L gene (Sangon Biotech, China) was transfected into KYSE150 cells or KYSE180 cells with the following sequences, sense primer: 5'-GACCCAACA ACCCUUCAUTT-3' and antisense primer: 5'‑AUGAAGGGUUGUUUGGGUCTT‑3'. The transfection was conducted using Lipofectamine 2000 reagent (Invitrogen, USA), according to the manufacturer's protocol. After 6 h transfection with fresh full strength medium, cells were reincubated for growing until 24 h for mRNA detection and 48 h for protein detection.
Real-time polymerase chain reaction (real-time PCR)
Total RNAs from fresh tissues or cells were extracted using Trizol reagent (Invitrogen, USA) and each RNA sample was reversely transcribed using the cDNA synthesis kit (TaKaRa, Japan), according to the manufacturer's protocol. The primer sequences (Sangon Biotech, China) used for real-time PCR were as presented in Table 1. Real-time PCR analysis was performed using SYBR Green PCR Premix Ex TaqTM II reagents (TaKaRa, Japan) on QuantStudioTM 6 FlexI real-time system (Applied biosystems, USA). The reaction mixtures for both TAF1L and GAPDH genes were amplified at the following thermal cycling conditions: 95˚C for 30 sec; 40 cycles at 95˚C for 5 sec and 60˚C for 34 sec. Final mRNA levels of TAF1L related genes were calculated using the 2-ΔΔCt method.
Table 1.
Primer sequencing for real-time PCR
| Gene | Forward primers (5'→3') | Reverse primers (5'→3') |
|---|---|---|
| TAF1L | AAGAGTAAAGATCGGCCACG | CATCCCTGTGCGTTTGAAGT |
| GAPDH | CAATGACCCCTTCATTGACC | TTGATTTTGGAGGGATCTCG |
Western blot
Total proteins per tissue sample was extracted in RIPA lysis buffer (Biosharp, USA) with protease cocktail (Roche, Germany) for 30 min at 4°C. After a centrifugation at 14,000 xg at 4°C for 20 min, each cellular lysate was transferred to a new centrifuge tube on ice, and protein concentration was determined with a BCA assay (Thermo Scientific, USA). For Western blot analysis, 30 μg of total protein was loaded on SDS polyacrylamide gel for electrophoresis, and transferred to nitrocellulose membrane (Whatman, Germany). The nitrocellulose membrane was blocked using TBST with 5% BSA for 1 h at room temperature. As primary antibodies TAF1L (1:1000), p-Akt (Thr308, 1:1000), Akt (1:1000), p53 (1:1000), c-Myc (1:1000) and GAPDH (1:1000) were applied, and incubated at 4°C, overnight. Except TAF1L antibody was obtained from Proteintech (China), rest antibodies were purchased from Cell Signaling Technology in USA. Membranes were washed in TBST prior to addition of secondary antibody (Beyotime Biotechnology, China) at 1:1000 in blocking buffer for 1 h at room temperature. After washing with TBST, the images were exposed using chemiluminescent substrate (Pierce, USA) with a Chemiluminescent detection system (Tanon, China), and the intensity of protein band was analyzed by Tanon analysis system (Tanon), according to the manufacturer's protocol.
CCK-8 cell activity assay
After 48 h transfection, KYSE150 cells and KYSE180 cells were digested with 0.25% trypsin-EDTA to obtain the cell suspension. Cells were counted and seeded into 96-well culture plates with 5x103 cells per each well. At each indicated time point, a mixture of 100 μl fresh medium and 10 μl of CCK-8 (Dojindo, Japan) were added into each well, and incubated at 37°C for 2 h. The absorbance of CCK-8 was detected at 450 nm by a microplate reader (BioTek, USA).
Wound healing array
KYSE150 cells and KYSE180 cells were seeded with 2×105 cells per well in 6-well plates, and transfected at following day, when cells were confluent approximately 80-90%. The wounds were gently induced by using 200 μl plastic pipette tip, and cell cultures were refed with fresh medium, and kept in a 37°C incubator with 5% CO2. After 24 h, the wound was examined under the photomicroscope (IX71 Olympus, Japan). Cell migration was evaluated by counting cell number of 5 random areas via Image-Pro Plus 6.0.
Transwell chamber assay
After 48 h transfection, KYSE150 cells and KYSE180 cells were harvested, counted and resuspended in FBS-free medium. For migration analysis, 5×104 cells per well were seeded into a upper chamber of 24‑well Costar Transwell Chamber System with 8 μm pore size insert (Corning Life Sciences, USA). The same system was also used for invasion array, when it was precoated by Matrigel-coated membrane (BD Biosciences, USA). For both assays, the lower chamber was filled by 0.5 ml of same medium containing 10% FBS, then incubated at 37°C in 5% CO2 for 24 h. After incubation, the membrane was fixed with the methanol, and remaining cells on the upper surface of filter membrane were removed with a cotton swab, and stained with 4',6-diamidino- 2-phenylindole (DAPI). The cells were recorded in 5 fields random by using IX71 photomicroscope. The number of cell migration and invasion were counted via Image-Pro Plus 6.0.
Statistical Analysis
All statistical analyses were performed via SPSS 19.0 and Graphpad prism 6, and presented as the mean ± standard error. Statistical tests were used with the student's t-test, one-way ANOVA and Chi-square test. A value of p<0.05 was considered as a statistically significant difference for each statistical analysis.
Results
TAF1L protein was overexpressed in ESCC tissues compared with in paracancer tissues
To assess TAF1L protein expression in ESCC, immunohistochemical staining was performed in two tissue microarrays (total 150 tissue sections, contained 150 cases of squamous cell carcinoma, 70 matched adjacent normal esophagus tissue and 40 metastasis carcinoma), and real-time PCR and Western blot were applied to forty paired fresh tissue samples (included 40 ESCC tissues and 40 matched paracancer tissues).
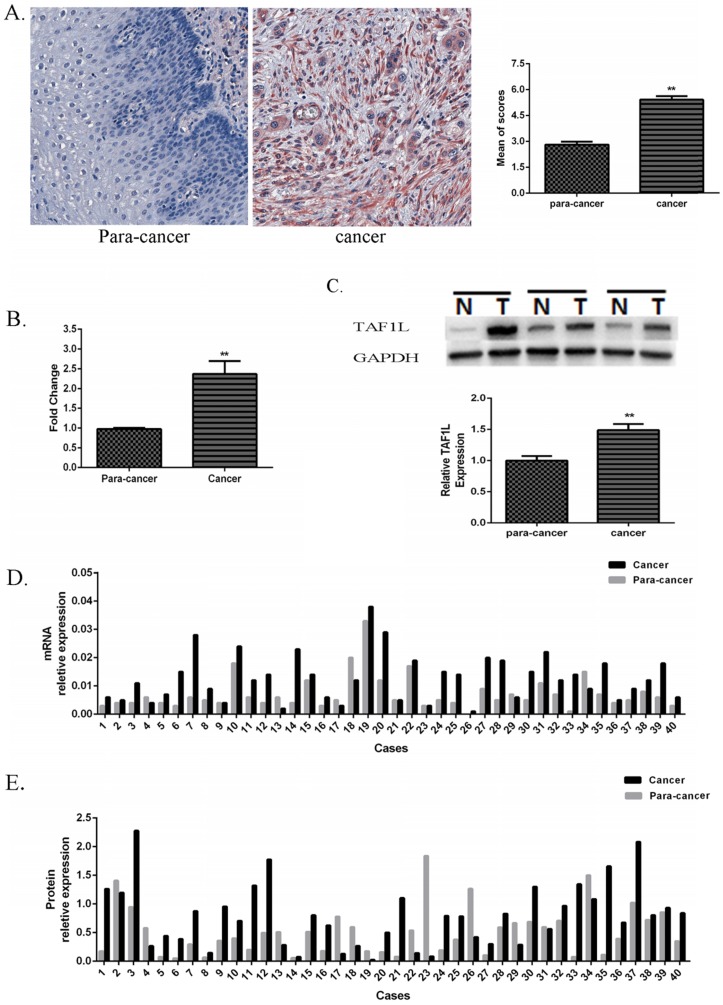
The IHC images showed that positive TAF1L signals were distributed in cell membrane and cytoplasm mainly (Figure 1A). With a comparison, its signal intensity of ESCC tissues was observed much stronger than that in paracancer tissues.
Figure 1.
Expression of TAF1L mRNA and protein in ESCC tissues and in adjacent normal esophageal epithelial tissues. (A) Higher expression of TAF1L protein in ESCC tissues (n=150) and lower expression of TAF1L in paracancer tissues (n=70) were observed by IHC (400x magnification). Red color showed positive signal. (B) mRNA expression of TAF1L in fresh ESCC tissues was detected higher than that in adjacent normal esophageal epithelial tissues with real-time PCR (paired cases=40). (C) Protein level of TAF1L in fresh ESCC tissues was showed higher expression than in adjacent normal esophageal epithelial tissues by Western blot (paired cases=40). The image of TAF1L molecular bands were calculated with triplicated samples, and the expression was also presented by mean of fold change. (D) A total of 31 of those cases exhibited elevated levels of TAF1L in ESCC by real-time PCR (paired cases=40) and (E) 29 cases in total were exhibited elevated levels of TAF1L in ESCC by Western blot (paired cases=40). N: paracancer tissues, T: ESCC tissues. *: p<0.05, **: p<0.01.
In addition, its expression rate in ESCC tissues was up to 70.7% (106 positive tissues in total of 150 ESCC tissues), which was very higher than that in paracancer tissues (latter only 14.3%, 10 positive tissues in total of 70 matched adjacent normal esophagus tissues). A difference between those two groups was existed statistically significant (showed in Table 2, p<0.001).
Table 2.
Protein expression of TAF1L between ESCC tissues and paired adjacent normal esophageal mucosa epithelial tissues by IHC
| Group | Case numbers | Overexpression (n) | Overexpressive rate (%) | χ2 | p-value |
|---|---|---|---|---|---|
| Paracancer tissue | 70 | 10 | 14.3% | 60.87 | <0.001 |
| Cancer tissue | 150 | 106 | 70.7% |
Compared with forty pairs of matched ESCC and paracancer fresh tissues, both expressive levels of TAF1L mRNA (Figure 1B and D) and protein (Figure 1C and E) were significantly enriched in ESCC tissues, which the mean of fold change for TAF1L mRNA expression was 2.37 times (Figure 1B) and for protein expression was 1.49 times (Figure 1C), and both of them were statistically significant (p<0.01). In the same testing condition, compared with the matched paracancer sample, mRNA expression of TFA1L in 31 of 40 ESCC tissues (Figure 1D) and protein expression of TAF1L in 29 of 40 ESCC tissues (Figure 1E) were upregulated.
The association with TAF1L protein overexpression and clinic pathological characteristics of ESCC
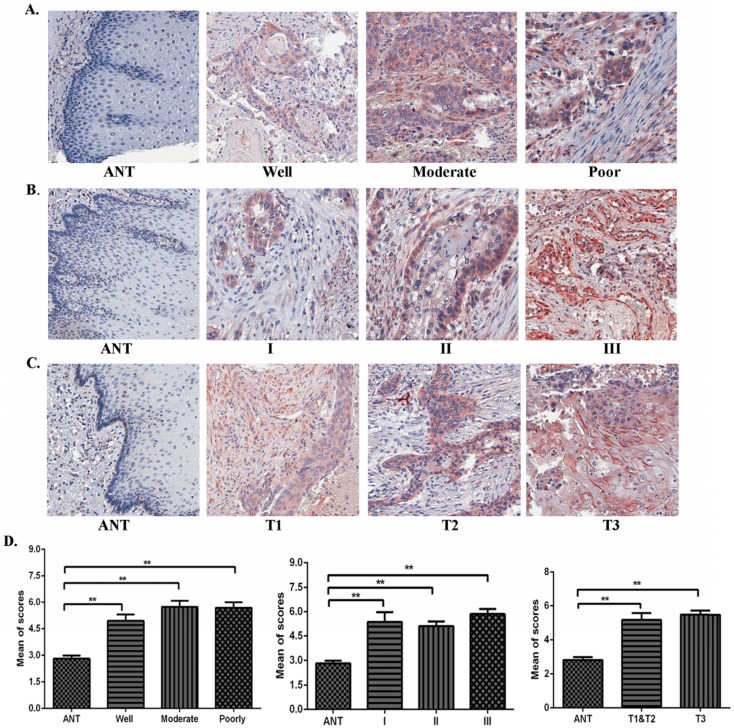
Using IHC, expression levels of TAF1L protein were further analyzed in ESCC lesions at different clinical stages, differentiation grades and TNM stages in this study. We found that TAF1L expression was significantly increased in ESCC tissues at all differentiation grades, clinical and pathological stages, compared to matched normal epithelial tissues (Figure 2A-D). Even though TAF1L expression gradually increases with the severity of ESCC, no significant difference (p>0.05) was found between any two differentiation stages of ESCC. By the chi-square test analysis, the similar results (p>0.05) were also showed in comparison of TAF1L protein expression at any two stages from I to III, and TNM-T1 to T3. In addition, the association between TAF1L expression and one of the clinical pathological features of ESCC was assayed and listed in the Table 3. There also was no significant association between the TAF1L expression and other parameters at ESCC development, except of the comparison of TNM-N0 and TNM-N1-2 stages (p=0.0141).
Figure 2.
The correlation of TAF1L expression and clinicopathological characteristics of ESCC. (A), (B) and (C): Overexpressive levels of TAF1L in ESCC tissues were detected at all of clinical factors, differentiation grades and TNM-T stages , and compared to adjacent normal esophageal epithelial tissues via IHC (400x, magnification). (D) The means of density score of TAF1L were higher at different clinical stages and pathological grades of ESCC, compared to adjacent normal esophageal mucosa epithelial tissues. *: p<0.05, **: p<0.01.
TAF1L expression in ESCC cells was downregulated using siRNA
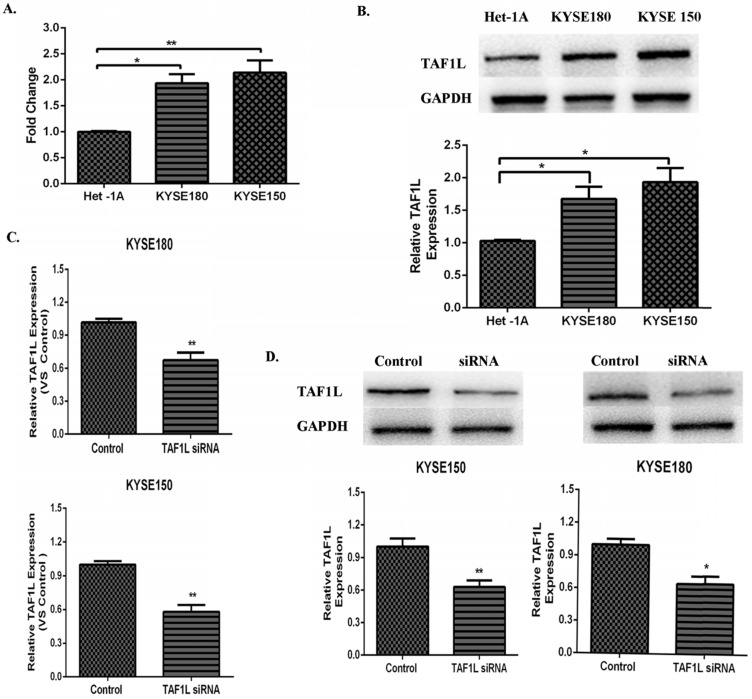
Both data from real-time PCR and Western blot were showed the higher expression levels of TAF1L mRNA and protein in KYSE150 cells and KYSE180 cells, compared that in normal human esophageal epithelial cell line (Het-1Acells), mRNA expression levels in KYSE150 cells and KYSE180 cells were raised 1.94 and 2.14 times, respectively. Expression levels of TAF1L protein were also increased 1.67 and 1.93 times in KYSE150 cells and KYSE180 cells, respectively. Both of them were statistically significant with p<0.01 and p<0.05 (Figure 3A and B). The knockdown efficiency of gene expression in KYSE150 cells and KYSE180 cells with TAF1L-siRNA treatment was tested by real-time PCR and Western blot. After transfecting with TAF1L-siRNA, the results showed that expression of TAF1L could be effectively reduced for mRNA level at 24 h and for protein level at 48 h (Figure 3C and D), compared with negative controls.
Figure 3.
The expression of TAF1L mRAN and protein in ESCC cell lines and the interference efficiency of TAF1L-siRNA. (A) TAF1L mRNA of ESCC cells was detected at higher expression, compared with that in normal human esophageal epithelial cells by real-time PCR; (B) TAF1L protein of ESCC tissues was measured with a similar trend as TAF1L mRNA, compared with that in normal human esophageal epithelial cells by Western blot; (C) decreased expression of TAF1L mRNA at 24 h post TAF1L-siRNA treatment was detected by real-time PCR; and (D) decreased expression of TAF1L protein at 48 h post TAF1L-siRNA treatment was measured by Western blot. *: p<0.05, **: p<0.01.
Suppressive effects on cell growth, migration and invasion of ESCC were observed after TAF1L gene silence
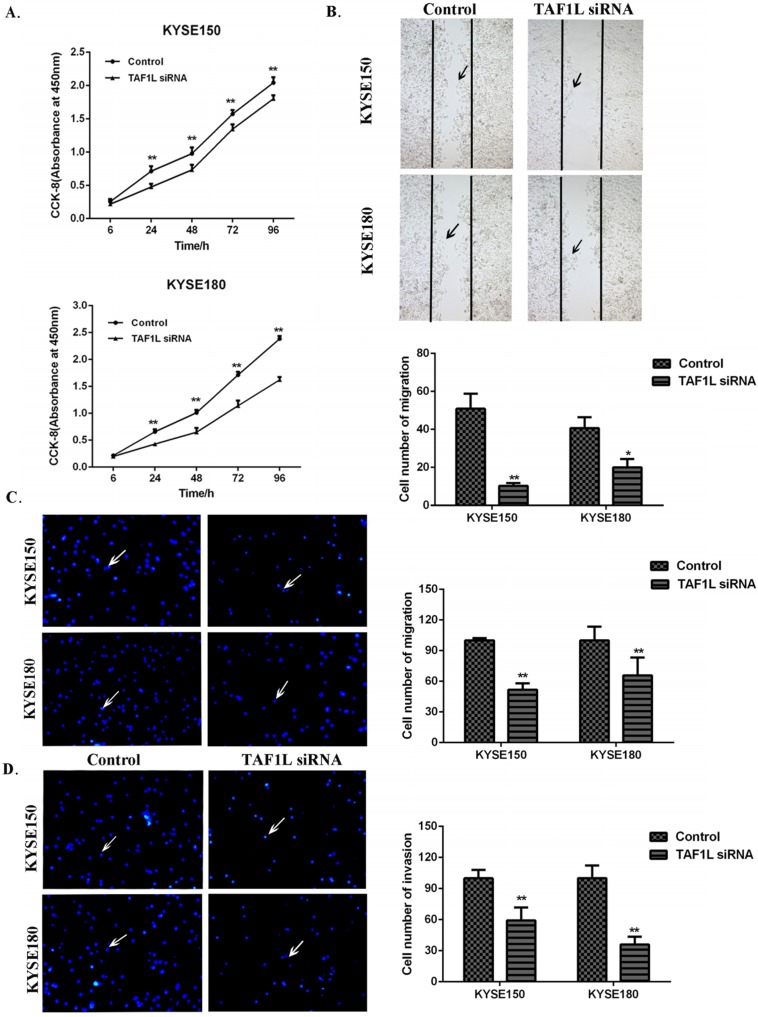
CCK-8 cell proliferation, Wound healing and Transwell chamber assays were used to assess the proliferation, migration and invasion on KYSE150 cells and KYSE180 cells. The cell proliferation of KYSE150 cells and KYSE180 cells were found significantly suppressed at 24, 48, 72 and 96 h post TAF1L-siRNA treatment, compared with control group (Figure 4A). With Wound healing assay, a comparison with the control, the cell number of the migration in KYSE150 cells and KYSE180 cells were significantly decreased after TAF1L-siRNA treatment, which both ESCC cell lines were 10.25±1.53 and 20.00±4.39. In contrast, cells in the migration in untreated control groups were 50.88±7.94 and 40.63±5.82 (p<0.05) (Figure 4B). The data of Transwell chamber assays also demonstrated that the migration of KYSE150 cells and KYSE180 cells were significantly decreased, which cell number of migration in ESCC was 51.63±2.83 and 65.73±7.79 in both KYSE150 cells and KYSE180 cells post TAF1L-siRNA treatment, compared that were 100.00±1.01 and 100.0±6.00 in untreated group of KYSE150 cells and KYSE180 cells (p<0.01) (Figure 4C). In addition, the data of Transwell chamber assays revealed the invasive cell quantities of KYSE150 cells and KYSE180 cells were significantly decreased, which was 59.34±5.00 and 36.00±3.37 in TAF1L-siRNA group, and 100.00±3.55 and 100.00±5.45 in untreated group (p<0.01) (Figure 4D).
Figure 4.
The effects of TAF1L-siRNA on the proliferation, migration and invasion of ESCC cells. (A) Via CCK-8 proliferation array, the inhibition of growth effects were detected in both of KYSE150 cells and KYSE180 cells post TAF1L-siRNA treatment at 6, 24, 48, 72 and 96 h time points. (B) Via Wound healing array, migration decrease of ESCC cell was detected post TAF1L-siRNA treatment (100x magnification). (C) and (D) By Transwell chamber array, the down-regulative migration and invasion in ESCC cells were detected at 48 h post TAF1L-siRNA treatment (100x magnification). All the data were compared with control group. *: p<0.05, **: p<0.01.
Possible regulatory mechanisms of TAF1L effect on ESCC cells
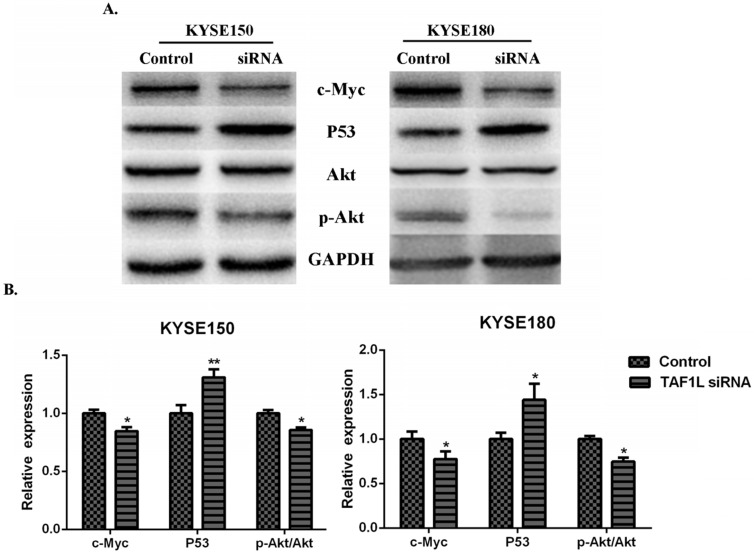
In order to investigate the regulatory mechanisms of TAF1L gene expression to ESCC, several key proteins related to tumorgenesis, such as Akt, c-Myc, p53 and etc., were detected for further research. The results showed that the expression downregulation of TAF1L post TAF1L-siRNA treatment had no effect on the total Akt expression in ESCC cells, compared with untreated group. However, under same condition, expression levels of c-Myc and phosphorylated Akt (Thr308) were significantly decreased, while the expression of p53 was inversely increased, all p<0.05 (Figure 5A and B).
Figure 5.
The effects of TAF1L-siRNA on several key proteins that related to tumorgenesis of ESCC cells. (A) Western blot was used to measure p53, c-Myc, Akt and p-Akt in ESCC cells; (B) semi-quantitative analysis of Western blot was performed to analyze above same markers in ESCC cells. p53 was showed a increase expression, while c-Myc and p-Akt were showed decrease expression between either ESCC cells post TAF1L-siRNA treatment. All the data were compared with control group. *: p<0.05, **: p<0.01.
Discussion
Recently, increasing bioinformatic data indicate that TAF1L gene may have a closed relationship with tumorigenesis in several tumors12,15-17. While as a gene regulator, expressive features and potential roles of TAF1L on carcinoma are almost unknown. Mutative biofunctions of some oncogenes (e.g. GRP78 and PBK/TOPK) in ESCC with their abnormal expression have been reported, which the most of them are closely associated with ESCC tumorigenesis18-21, and can lead us a journey to explore the potential pathogenesis and possible biomarkers for interfering prognosis and making early diagnosis. In this study, we reported that TAF1L is significantly upregulated in human ESCC tissues and cells (Figure 1). IHC staining analysis revealed that TAF1L expression is significantly correlated with the malignancy of ESCC.
With abnormal numbers of lymph node in metastasis were also observed in ESCC patients. As strongest prognostic factors, the ratio of regional lymph node is closely related to risk of locoregional recurrence in many cancers, such as breast cancer22, gallbladder cancer23 and esophageal squamous cell carcinoma24. The current study discovered that TAF1L overexpression was significantly related to TNM-N stage (p=0.0141), which indicated that TAF1L may serve as a valuable biomarker for early diagnosis, therapeutic evaluation, and prognostic intervention of ESCC patients.
Oh, et al reported that TAF1L has frame shift mutations in gastric and colorectal cancers due to the mononucleotide repeats12. Glaser, et al found that a variant of TAF1L in lymphatic endothelial cells was derived from lymphatic malformations, using whole exome sequencing17. Based on the references12,17, mentioned SNP mutation sites (rs10971047, rs10758145, rs10971046 and rs199701622) were examined by sequencing and analyzing in this study. The data showed that DNA samples from both of ESCC and paired paracancer tissues (40 cases per group) were with lower mutative ratio for all above four SNP sites, and there was no significant difference between two groups. The results from this study suggest that enlarge sample numbers for further verification may be needed.
Via checking cellular functions, cell proliferation, migration and invasion of ESCC were found all effectively weaken by the inhibition of TAF1L expression with siRNA technique. It is well-known fact that Akt signal pathways can play critical roles in the regulation of cell proliferation, differentiation, invasion and metastasis during tumor development25-27. As a serine/threonine kinase, Akt is an important mediator in some signal transduction pathways. In addition, a variety of growth factors28, hormones29 and cytokines30 can also stimulate Akt activation. Phosphorylation of Akt and its downstream molecules are involved in inflammation process and other regulatory pathways, as well as tumor development. As for ESCC, phosphorylated Akt phosphorylation was reported to be elevated in tumor tissues compared with paired normal tissues31, and genetic variations of Akt might predict increased recurrence risk after chemoradiotherapy32. Results obtained in this study identified that blocking TAF1L gene could reduce expressive level of downstream phosphorylated Akt, suggesting that TAF1L may have potential roles in the malignant proliferation and metastasis of ESCC cells. p53, as a tumor suppressor gene, can derive many mutations in more than 50% of malignancies, and promote tumorigenesis via repressing downstream target genes33. As a protooncogene, c-Myc is negatively regulated by p5334, which is involved in cell proliferation and apoptosis induction35. Moreover, previous studies showed that the P13K/Akt signaling pathway could immoderately lead to tumor cell proliferation by downregulating tumor suppressor gene p5336, and upregulating protooncogene gene c-Myc37. Taken together, it indicates that downregulation of TAF1L protein expression could increase P53 expression, decreased c-Myc expression, and enhance cancer proliferation and metastasis of ESCC via Akt signal pathway.
Conclusion
This study has found that TAF1L may act as an oncogene to participate the ESCC development, and more supporting data indicate that it has possibilities as a new biomarker of ESCC for early diagnosis, therapeutic evaluation and prognostic intervention.
Acknowledgments
This study was supported by Science and Technology Program: JCYJ20160520174247 964, the Science and Technology Innovation Committee of Shenzhen. In addition, this study was also supported by Instrumental Analysis Center of Shenzhen University for providing research instruments.
Ethics approval and consent to participate
Human Research Ethics Committee of Shantou University Medical College Cancer Hospital (approved number: 2016024).
Authors' contributions
SZ participated in the project as a major performer and wrote this draft. HY collected tissue samples and worked as a counselor of clinical pathology. ZC, YS and YW participated in data analysis in part. YL cultured cells in part. LL performed Western blot in part. SS participated in experimental plan in part. YW designed and organized this study, and edited this manuscript. All authors have read and approved this final manuscript. Note: Two YW as authors' initials were appeared in this manuscript, first one is Yongyu Wang and last YW is Yun Wang.
References
- 1.Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Guo M, Zhao YD, Yang HJ. et al. Analysis of clinicopathological characteristics for 5406 cases of esophageal neoplasm. Chin J Cancer Prev Treat. 2008;15:54–6. [Google Scholar]
- 3.Ando N, Ozawa S, Kitagawa Y. et al. Improvement in the results of surgical treatment of advanced squamous esophageal carcinoma during 15 consecutive years. Annals of surgery. 2000;232:225–32. doi: 10.1097/00000658-200008000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford HE. Gefitinib for oesophageal cancer: a cog in need of a wheel? The Lancet Oncology. 2014;15:790–1. doi: 10.1016/S1470-2045(14)70271-2. [DOI] [PubMed] [Google Scholar]
- 5.Rustgi AK, El-Serag HB. Esophageal carcinoma. New England Journal of Medicine. 2014;371:2499–509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 6.Wassarman DA, Sauer F. TAFII250. Journal of cell science. 2001;114:2895–902. doi: 10.1242/jcs.114.16.2895. [DOI] [PubMed] [Google Scholar]
- 7.Vanin EF. Processed pseudogenes: characteristics and evolution. Annual review of genetics. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 8.Lee DH, Gershenzon N, Gupta M. et al. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Molecular and cellular biology. 2005;25:9674–86. doi: 10.1128/MCB.25.21.9674-9686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Developmental biology. 2010;339:225–9. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura J, Nguyen ST, Liu H. et al. A functional genome-wide RNAi screen identifies TAF1 as a regulator for apoptosis in response to genotoxic stress. Nucleic acids research. 2008;36:5250–9. doi: 10.1093/nar/gkn506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao S, Choi M, Overton JD. et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proceedings of the National Academy of Sciences. 2013;110:2916–21. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh HR, An CH, Yoo NJ. et al. Frameshift mutations in the mononucleotide repeats of TAF1 and TAF1L genes in gastric and colorectal cancers with regional heterogeneity. Pathology & Oncology Research. 2017;23:125–30. doi: 10.1007/s12253-016-0107-0. [DOI] [PubMed] [Google Scholar]
- 13.Findlay JM, Castro-Giner F, Makino S. et al. Differential clonal evolution in oesophageal cancers in response to neo-adjuvant chemotherapy. Nature communications. 2016;7:11111–3. doi: 10.1038/ncomms11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia J, Jia P, Hutchinson KE. et al. A meta-analysis of somatic mutations from next generation sequencing of 241 melanomas: a road map for the study of genes with potential clinical relevance. Molecular cancer therapeutics. 2014;13:1918–28. doi: 10.1158/1535-7163.MCT-13-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, Zhang J, Jin H. et al. Whole transcriptome sequencing identifies tumor-specific mutations in human oral squamous cell carcinoma. BMC medical genomics. 2013;6:28–35. doi: 10.1186/1755-8794-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu T, Wang Y, Zhao S. et al. Two new molecular biomarkers in human colorectal cancer. Journal of Shenzhen University Science and engineering. 2018;35:529–35. [Google Scholar]
- 17.Glaser K, Dickie P, Neilson D. et al. Linkage of Metabolic Defects to Activated PIK3CA Alleles in Endothelial Cells Derived from Lymphatic Malformation. Lymphatic research and biology. 2018;16:43–55. doi: 10.1089/lrb.2017.0033. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Kwong DL, Cao T. et al. Esophageal squamous cell carcinoma (ESCC): advance in genomics and molecular genetics. Diseases of the Esophagus. 2015;28:84–9. doi: 10.1111/dote.12088. [DOI] [PubMed] [Google Scholar]
- 19.Ren P, Chen C, Yue J. et al. High expression of glucose-regulated protein 78 (GRP78) is associated with metastasis and poor prognosis in patients with esophageal squamous cell carcinoma. OncoTargets and therapy. 2017;10:617–25. doi: 10.2147/OTT.S123494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Li Q, Wang C. et al. Prognostic and diagnostic potential of isocitrate dehydrogenase 1 in esophageal squamous cell carcinoma. Oncotarget. 2016;7:86148–60. doi: 10.18632/oncotarget.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohashi T, Komatsu S, Ichikawa D. et al. Overexpression of PBK/TOPK Contributes to Tumor Development and Poor Outcome of Esophageal Squamous Cell Carcinoma. Anticancer research. 2016;36:6457–66. doi: 10.21873/anticanres.11244. [DOI] [PubMed] [Google Scholar]
- 22.Liao GS, Chou YC, Golshan M. et al. Prognostic value of the lymph node ratio in breast cancer subtypes. The American Journal of Surgery. 2015;210:749–54. doi: 10.1016/j.amjsurg.2014.12.054. [DOI] [PubMed] [Google Scholar]
- 23.Birnbaum DJ, Viganò L, Russolillo N. et al. Lymph node metastases in patients undergoing surgery for a gallbladder cancer. Extension of the lymph node dissection and prognostic value of the lymph node ratio. Annals of surgical oncology. 2015;22:811–8. doi: 10.1245/s10434-014-4044-4. [DOI] [PubMed] [Google Scholar]
- 24.Chen SB, Weng HR, Wang G. et al. Lymph node ratio-based staging system for esophageal squamous cell carcinoma. World Journal of Gastroenterology: WJG. 2015;21:7514–21. doi: 10.3748/wjg.v21.i24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo KT, Chen CL, Chou TY. et al. Nm23H1 mediates tumor invasion in esophageal squamous cell carcinoma by regulation of CLDN1 through the AKT signaling. Oncogenesis. 2016;5:239–48. doi: 10.1038/oncsis.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urakawa N, Utsunomiya S, Nishio M. et al. GDF15 derived from both tumor-associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression via Akt and Erk pathways. Laboratory investigation. 2015;95:491–503. doi: 10.1038/labinvest.2015.36. [DOI] [PubMed] [Google Scholar]
- 28.Magner NL, Jung Y, Wu J. et al. Insulin and IGFs enhance hepatocyte differentiation from human embryonic stem cells via the PI3K/AKT pathway. Stem Cells. 2013;31:2095–103. doi: 10.1002/stem.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirabo A, Ryzhov S, Gupte M. et al. Neuregulin-1β induces proliferation, survival and paracrine signaling in normal human cardiac ventricular fibroblasts. Journal of molecular and cellular cardiology. 2017;105:59–69. doi: 10.1016/j.yjmcc.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner NA, Das A, Warburton P. et al. Interleukin-1α stimulates proinflammatory cytokine expression in human cardiac myofibroblasts. American Journal of Physiology-Heart and Circulatory Physiology. 2009;297:1117–27. doi: 10.1152/ajpheart.00372.2009. [DOI] [PubMed] [Google Scholar]
- 31.Li B, Li J, Xu WW. et al. Suppression of esophageal tumor growth and chemoresistance by directly targeting the PI3K/AKT pathway. Oncotarget. 2014;5:11576–87. doi: 10.18632/oncotarget.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hildebrandt MA, Yang H, Hung MC. et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. Journal of clinical oncology. 2009;27:857–71. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 34.Levy N, Yonish-Rouach E, Oren M. et al. Complementation by wild-type p53 of interleukin-6 effects on M1 cells: induction of cell cycle exit and cooperativity with c-myc suppression. Molecular and cellular biology. 1993;13:7942–52. doi: 10.1128/mcb.13.12.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nature Reviews Cancer. 2002;2:764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 36.Maurya AK, Vinayak M. Quercetin regresses Dalton's lymphoma growth via suppression of PI3K/AKT signaling leading to upregulation of p53 and decrease in energy metabolism. Nutrition and cancer. 2015;67:354–63. doi: 10.1080/01635581.2015.990574. [DOI] [PubMed] [Google Scholar]
- 37.Galmozzi E, Casalini P, Iorio MV. et al. HER2 signaling enhances 5′ UTR-mediated translation of c-Myc mRNA. Journal of cellular physiology. 2004;200:82–8. doi: 10.1002/jcp.20012. [DOI] [PubMed] [Google Scholar]