Abstract
Background: The oncogene DEK, which was originally identified as part of the protein product of the DEK-CAN fusion oncogene, has been shown to promote tumorigenesis in a variety of cancer cell types. However, little is known about the expression and role of DEK in pancreatic ductal adenocarcinoma (PDAC), which is one of the most refractory malignant tumors worldwide and has poor prognosis. Our study aimed to understand the role of DEK in the development and progression of pancreatic adenocarcinoma.
Materials and methods: We used western blotting and immunohistochemistry to examine the expression of DEK in pancreatic adenocarcinoma cells and tissues. We analyzed the correlation between DEK expression and clinicopathological characteristics and prognosis in 163 pancreatic adenocarcinoma patients.
Results: Protein levels of DEK in pancreatic adenocarcinoma tissues (76/136, 55.9%) were significantly higher than those in adjacent non-tumor tissues (16.2%, 22/136). A high expression level of DEK was associated with poor prognosis (P<0.001).In addition, the combination of CA19-9 and DEK expression (P<0.001) was a better prognostic indicator than CA19-9 expression alone (P=0.012).
Conclusions: DEK may play a significant role as a valuable biomarker in the development and progression of pancreatic adenocarcinoma. The combination of DEK and CA19-9 improves the prognostic prediction in patients with pancreatic adenocarcinoma.
Keywords: DEK, Pancreatic adenocarcinoma, CA19-9, prognosis
Introduction
Pancreatic cancer is a common and refractory malignant tumor with an extremely poor prognosis 1-3. The incidence of PDAC has been increasing worldwide with the growth and aging of the general population 4,5. Statistics indicate that the 5-year survival rate of PDAC is only approximately 5%, with a mortality rate of nearly 85.3% among all newly diagnosed cases of PDAC 4. Surgery remains the primary treatment for cases without distant metastasis. Since PDAC is often detected in an advanced stage with low resection rates, more effective screening techniques and a better understanding of the pathogenesis of PDAC are urgently needed.
The DEK gene is a putative proto-oncogene that encodes a 375-amino acid protein with an estimated molecular weight of 43kDa 6-8. DEK was first characterized as part of the protein product of the DEK-CAN fusion oncogene generated by a t(6;9) translocation involving breakpoints in the DEK gene on chromosome 6 and the CAN gene on chromosome 9 in a subset of patients with acute myelogenous leukemia 9,10. Many studies have demonstrated the functions of DEK, including involvement in the regulation of hematopoiesis, chromatin reconstruction, gene transcription, and cell apoptosis 11-13. Overexpression of DEK has been observed in several malignancies including breast cancer, melanoma, retinoblastoma, gastric adenocarcinoma, hepatocellular carcinoma, colorectal cancer, lung cancer, and bladder cancer 14-20, demonstrating its role in tumorigenesis and neoplastic progression 14,21-23.
The expression and role of DEK in pancreatic adenocarcinoma has rarely been reported. Through database analysis we found that DEK expression levels in PDAC tissues were much higher than those in non-tumor tissues. In this study we aimed to explore whether DEK expression is associated with clinicopathological features of pancreatic cancer and its prognostic value in this aggressive disease.
Materials and methods
Cell culture
Human pancreatic cancer cell line, including Panc-1, CFPAC-1, AsPC-1, SW1990, BxPC-3, MIAPaCa-2, and normal pancreatic ductal epithelial cells HPDE6-C7 are all obtained from Chinese Academy of Sciences (Shanghai, China). BxPC-3 and AsPC-1 cell lines were cultured in RPMI 1640 medium (Hyclone) containing 10% fetal bovine serum (FBS, Gibco, New York, USA). Panc-1, CFPAC-1, SW1990, MIAPaCa-2 and HPDE6-C7 cell lines were cultured in DMEM medium with 10% FBS.
Patients and clinicopathological parameters
One hundred and thirty six pancreatic ductal adenocarcinoma patients who had surgery between 2012 and 2016 were selected in Sir Run Run Shaw Hospital, affiliated with the Zhejiang University of Medicine, China. All tissue specimens were diagnosed by pathological examination after surgery. The mean age of the patients was 59.0 years (range from 35 to 82). The male to female ratio was 56:80. We retrospectively analyzed the clinical manifestations including age, gender, tumor size, differentiation, histological stage, lymph node metastasis and distant metastasis. We also reviewed the survival data carefully in all cases. The research protocol was reviewed and approved by the Research Ethics Committee of Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University. All participants or their guardians gave written consent of their tissue samples and medical information to be used for scientific research. We assessed the histological stage of PDAC patients on the basis of the newest TNM staging system by the American Joint Committee on Cancer (AJCC) in 2017.
Immunohistochemistry
Thin slices of tumor tissue from all cases were fixed in 4% formaldehyde solution (pH 7.0) for periods not exceeding 24 h. Paraffin embedding was conducted, and 4 μm thick sections were cut and placed on glass slides coated with 3-aminopropyl triethoxysilane (Seebio Biotech Inc., Shanghai, China) for immunohistochemistry. Tissue samples were stained with hematoxylin and eosin to determine histological type and grade of tumors. Briefly, pancreatic ductal adenocarcinoma tissues were cut at a thickness of 4 μm using a cryostat. The sections were mounted on microscope slides, air‑dried and then fixed in a mixture of 50% acetone and 50% methanol. The sections were then de‑waxed with xylene, gradually hydrated with gradient alcohol, and washed with phosphate-buffered saline (PBS). Sections were incubated for 60 min with the primary antibody. Following washing with PBS, sections were incubated for 30 min with the secondary biotinylated antibody (Multilink Swine anti-goat/mouse/rabbit immunoglobulin; Dako Inc., Carpinteria, CA, USA). Following washing, Avidin Biotin Complex (1:1,000 dilution, Vector Laboratories Ltd., Burlingame, CA, USA) was then applied to the sections for 30‑60 min at room temperature. The immunoreactive products were visualized by catalysis of 3, 3‑diaminobenzidine (DAB) by horseradish peroxidase in the presence of H2O2, following extensive washings. Sections were then counterstained in Gill's hematoxylin and dehydrated in ascending grades of methanol prior to clearing in xylene, and mounting under a coverslip. The sections were observed under an Olympus CX31 microscope (Olympus, Tokyo, Japan).
To score DEK as immunopositive staining, expression was classified semi-quantitatively. The staining density was scored as: 0 indicates negative, no or less than 5% positive cells, 1 indicates 5-25% positive cells, 2 indicates 26-50% positive cells and 3 indicates more than 50% positive cells. The dominant staining intensity was scored as: 0 indicates negative; 1 indicates weak; 2 indicates intermediate; and 3 indicates strong staining intensity. Expression was calculated by adding density score (0-3) and intensity score (0-3) before categorizing into low and high expression. High expression was defined as score≥3.
Western blot analysis
For western blot analysis, proteins were separated on 10% SDS-PAGE gels and transferred to a polyvinylidene difluoride membrane. After blocking, the membrane was incubated with an anti-DEK antibody (1:1,000; ab166624; Abcam, Cambridge, MA, USA) at 4˚C overnight. After washing, the membrane was incubated with the secondary goat anti-rabbit antibodies conjugated with horseradish peroxidase (1:5 000, Pierce, USA) for 1 h at room temperature. Densitometry was performed using Kodak X-OMAT LS film (Eastman Kodak, Rochester, NY).
Statistical analysis
All data was analyzed by the SPSS version 20.0. We use Kaplan—Merier method to analyze patient survival, and the differences in survival were evaluated using the log-rank test. Multivariate analysis was performed using the Cox proportional hazards model to confirm factors independently associated with survival. The paired-samples t-test was used to compare the intensity ratio between cancer tissues and matching non-cancerous livers. All p-values were based on two-sided statistical analyses and the rate of P<0.05 was considered to indicate a statistically significant difference.
Results
DEK is over-expressed in pancreatic cancer
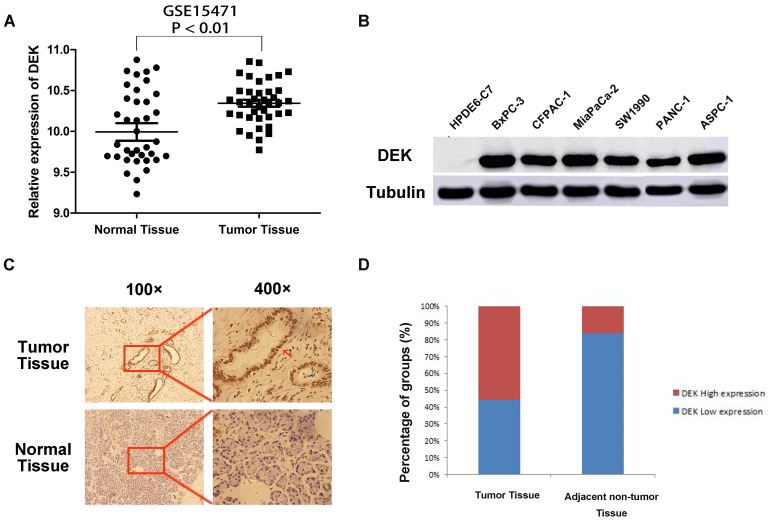
Analysis of chip results of 78 samples in the database GSE15471, which is one of the GEO database, including 39 cases of pancreatic cancer and 39 cases of corresponding non-tumor normal tissues, revealed that the expression of DEK in pancreatic cancer was significantly higher than that in corresponding non-tumor tissues (Fig. 1A). The expression of DEK in six pancreatic cancer cell lines was detected by western blotting, with normal pancreatic ductal epithelial cells HPDE6-C7 used as controls. The expression of DEK in pancreatic cancer cell lines was significantly increased relative to controls (Fig. 1B). Immunohistochemical examination showed that DEK was mainly located in the nuclei of PDAC cells, and the expression of DEK protein was significantly higher in PDAC tissues (55.9%, 76/136) compared with matched non-tumor tissues(22/136, 16.2%, Fig. 1 C,D).
Fig 1.
Expression of DEK in pancreatic ductal adenocarcinoma compared with corresponding normal pancreas. (A) Expression of DEK in pancreatic cancer and normal pancreas according to the database GSE15471, which is one of the GEO database. The mRNA expression of DEK is up-regulated in pancreatic cancer tissues (T) compared with the adjacent non-tumor tissues (N) using the GSE15471 dataset. (B) The expression of DEK in one normal pancreatic duct epithelial cell line and six pancreatic cancer cell lines detected by Western Blot. (C) Immunohistochemical detection of the DEK protein expression in PDAC tissue and corresponding normal tissue (original magnification, 100×and 400×); (D) Expression levels of DEK in pancreatic cancer tissues and the adjacent non-tumor tissues.
Correlation between DEK overexpression and clinicopathological features
We analyzed immunohistochemistry data to investigate the relationship between DEK expression and clinicopathological features. High expression of DEK was significantly associated with differentiation status (P<0.001), lymph node metastasis (P<0.001), tumor stage (P=0.002), and tumor size (P=0.038) in our 163 PDAC patients (Table 1, Fig. 2). No significant correlation was observed between high DEK expression and other clinical characteristics of patients with PDAC, such as gender (P=0.248), age (P=0.139), or serosal invasion (P=0.101, Table 1).
Table 1.
Relationship between DEK protein expression and clinicopathological features of PDAC
| Characteristic | (n) | DEK expression | P value | |
|---|---|---|---|---|
| 136 | Low | High | ||
| Age | 0.139 | |||
| ≤58 | 72 | 28 | 44 | |
| >58 | 64 | 32 | 32 | |
| Gender | 0.248 | |||
| Man | 80 | 32 | 48 | |
| Women | 56 | 28 | 28 | |
| Tumor size | 0.038* | |||
| ≤3cm | 68 | 36 | 32 | |
| >3cm | 68 | 24 | 44 | |
| Differentiation | <0.001* | |||
| Poor | 60 | 4 | 56 | |
| Well and moderate | 76 | 56 | 20 | |
| Tumor Stage | 0.002* | |||
| Ⅰ,Ⅱ | 80 | 44 | 36 | |
| Ⅲ,Ⅳ | 56 | 16 | 40 | |
| Lymph node metastasis | 0.001* | |||
| NO | 68 | 40 | 28 | |
| YES | 68 | 20 | 48 | |
| Distant metastasis | 0.101 | |||
| M0 | 120 | 56 | 64 | |
| M1 | 16 | 4 | 12 | |
| CA19-9 | 0.099 | |||
| ≤37 IU/ml | 80 | 40 | 40 | |
| >37 IU/ml | 56 | 20 | 36 | |
* P < 0.05
Fig 2.
Expression levels of DEK in pancreatic cancer tissues with different tumor differentiation and tumor stage. (A)Typical pictures of DEK expression with different pancreatic tumor differentiation. (B) Typical pictures of DEK expression with different tumor stage.
Correlation between DEK overexpression and prognosis of PDAC patients
We also analyzed the correlation between DEK expression level and the prognosis of PDAC patients. In univariate analysis, PDAC patients with high tumor expression of DEK had significantly lower median survival time than those with low DEK expression (P<0.001, Table 2). These data suggest that DEK might be a valuable prognostic factor in PADC. We also found that tumor size (P<0.001), differentiation (P<0.001), distant metastasis (P=0.032), and high level of CA19-9(P<0.012) correlated with survival (Table 2). Multivariate Cox regression analysis identified DEK expression (P=0.002) and high level of CA19-9 (P=0.037) as independent prognostic factors of survival (Table 3).
Table 2.
Univariate survival analysis of various factors in patients with PDAC
| Characteristic | (n) | Median Survival Time | P value |
|---|---|---|---|
| Age | 0.066 | ||
| ≤58 | 72 | 12.6 | |
| >58 | 64 | 12.2 | |
| Gender | 0.09 | ||
| Man | 80 | 10 | |
| Women | 56 | 12.1 | |
| Tumor size | <0.001* | ||
| ≤3cm | 68 | 12.9 | |
| >3cm | 68 | 11.3 | |
| Differentiation | <0.001* | ||
| Poor | 60 | 14.1 | |
| Well and moderate | 76 | 10.4 | |
| Tumor Stage | 0.159 | ||
| Ⅰ,Ⅱ | 80 | 13.1 | |
| Ⅲ,Ⅳ | 56 | 11 | |
| Lymph node metastasis | 0.074 | ||
| NO | 68 | 13.1 | |
| YES | 68 | 11.6 | |
| Distant metastasis | 0.032* | ||
| M0 | 120 | 12.5 | |
| M1 | 16 | 8 | |
| DEK | <0.001* | ||
| Low expression | 60 | 15 | |
| High expression | 76 | 10.5 | |
| CA19-9 | 0.012* | ||
| ≤37 IU/ml | 76 | 14.4 | |
| >37 IU/ml | 60 | 8.7 | |
| DEK and CA19-9 | <0.001* | ||
| Low DEK and CA19-9 ≤37 IU/ml | 40 | 15.9 | |
| High DEK or CA19-9 >37 IU/ml | 60 | 12.5 | |
| High DEK and CA19-9 >37 IU/ml | 36 | 8.2 | |
* P < 0.05
Table 3.
Multivariant survival analysis of various factors in patients with PDAC
| Characteristic | SE | HR | 95%CI | P value |
|---|---|---|---|---|
| Age | 0.242 | 0.987 | 0.614-1.587 | 0.958 |
| Gender | 0.281 | 1.42 | 0.818-2.465 | 0.213 |
| Lymph node metastasis | 0.357 | 1.342 | 0.667-2.699 | 0.41 |
| Distantmetastasis | 0.6 | 0.68 | 0.209-2.204 | 0.52 |
| Tumor Stage | 0.415 | 0.32 | 0.142-0.722 | 0.006* |
| Tumor size | 0.289 | 3.07 | 1.743-5.407 | <0.001* |
| Differentiation | 0.406 | 0.344 | 0.155-0.762 | 0.009* |
| DEK | 0.317 | 0.379 | 0.204-0.706 | 0.002* |
| CA199 | 0.297 | 0.538 | 0.301-0.962 | 0.037* |
* P < 0.05
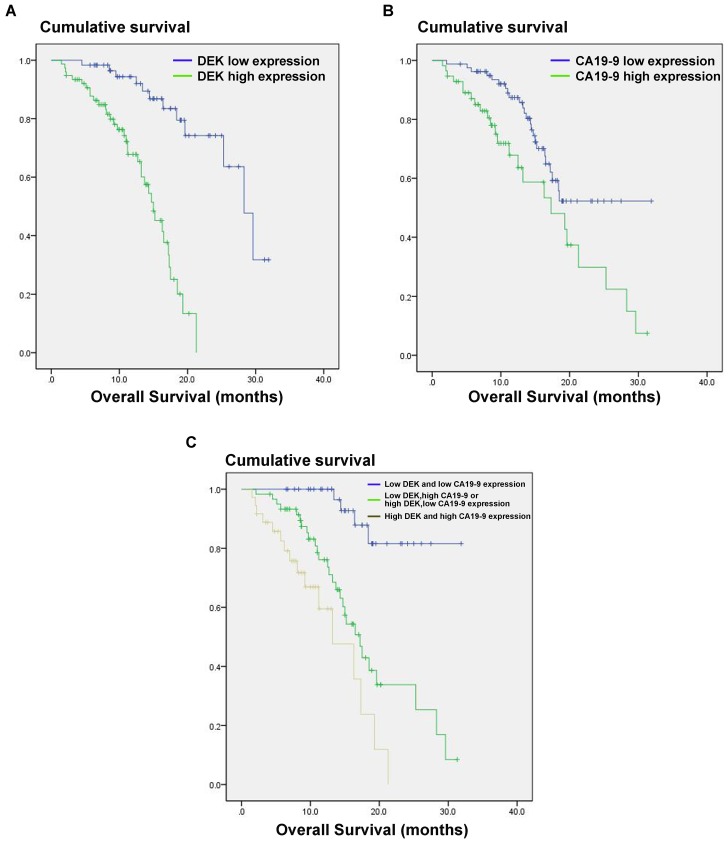
The CA19-9 expression level is increased in most cancer patients but especially in those with pancreatic cancer, and CA191-9 is the most sensitive marker in PDAC reported to date. Our results indicate that the combination of CA19-9 and DEK expression (P<0.001) is a better indicator of prognosis in PDAC patients than CA19-9 alone (P=0.012, Fig. 3).
Fig 3.
Estimated overall survival according to the expression of DEK proteinand CA19-9 protein and the combination in 136 cases of PDAC. (A) PDAC patients with high DEK expression had lower survival rates compared to those with low DEK expression (P < 0.001). (B) PDAC patients with high CA19-9 expression had lower survival rates compared to those with low CA19-9 expression (P =0.012). (C) The combination of DEK expression and CA19-9 expression make it better to distinguish the prognosis of PDAC patients (P < 0.001).
Discussion
PDAC is one of the most malignant cancers and has a 1-year survival rate of less than 20% and 5-year survival rate of less than 5% 24. It is the fourth leading cause of cancer-related death in Western societies 25. Surgery provides the only chance of cure; however, the majority of patients are diagnosed with metastasis or invasion of the superior mesenteric artery or celiac trunk and are not candidates for surgical resection of the tumor 26. Even if resection can be achieved, the survival rate is still low 27. Consequently, it is very important to find novel strategies for detection of PDAC in the early stage and thus improve the prognosis of patients.
DEK was identified as a putative proto-oncogene that can be linked to cancer through multiple mechanisms including cell division, DNA repair, aging, apoptosis, differentiation, or oncogenic transformation 28-30. Many studies have reported the expression of DEK in different cancers. Khodadoust et al. suggested that the expression level of DEK is a useful tool for distinguishing benign nevi from malignant melanomas 31. Ying et al. recently reported that DEK expression correlated with chemotherapy resistance and may be an independent prognostic factor for breast cancer, as well as a potential therapeutic target 14. Moreover, studies in retinoblastoma patients identified DEK as a potential target on chromosome 6p22 and suggested that a gain of DEK function enhances tumor development, with similar findings in patients with bladder cancer 1. Increasing evidence of a relationship between tumor type and DEK protein expression indicates the potential of DEK as a tumor marker 32.
A previous report by Sun et al. showed correlations between DEK expression and clinical pathological parameters and prognosis 33.The overall goal of our study was to further determine the effect of DEK expression on the prognosis of pancreatic cancer. We performed immunohistochemical staining of DEK protein and survival data analysis using 136 PDAC tissues and their adjacent normal tissue counterparts. DEK protein was mainly expressed in the nucleus, and the expression of DEK protein was significantly higher in cancer tissues than adjacent tumor tissues. Sun's research showed that high expression of DEK was significantly related to differentiation status, tumor stage, and tumor size.In addition to that, our study also found the relationship between the high expression of DEK and the lymph node metastasis. Consistent with the results of sun's study, our study also indicated that patients with high expression of DEK had a significantly lower median survival time, which validated DEK as a valuable prognostic factor in PDAC. Besides, we also showed that the expression level of DEK was significantly related to the CA19-9 expression level, and might be used as an important supplement to CA19-9 in the diagnosis of PDAC.
In summary, overexpression of DEK correlated with tumor size, differentiation, tumor stage, and lymph node metastasis, indicating the important role of DEK protein in the progression of PDAC. The combination of DEK expression and CA19-9 expression levels represents a better prognostic index than CA19-9 alone. DEK may potentially be used as a biomarker for prognostic evaluation and a molecular therapeutic target in PADC patients.
Acknowledgments
This research was supported by the Major Research Project of Science Technology Department of Zhejiang Province (2015C03G2010160), A Project Supported by Scientific Research Fund of Zhejiang Provincial Education Department (Y201534615), and the Foundation Project for Medical Science and Technology of Zhejiang province (2015KYB218).
References
- 1.Wise-Draper TM, Allen HV, Jones EE. et al. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26(20):7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Löhr JM. Medical treatment of pancreatic cancer. Expert Rev Anticancer Ther. 2007;7(4):533–544. doi: 10.1586/14737140.7.4.533. [DOI] [PubMed] [Google Scholar]
- 3.Yeh JJ, Der CJ. Targeting signal transduction in pancreatic cancer treatment. Expert Opin Ther Targets. 2007;11(5):673–694. doi: 10.1517/14728222.11.5.673. [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Ferlay J, Shin HR, Bray F. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 6.Riveiro-Falkenbach E, Soengas MS. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin Cancer Res. 2010;16(11):2932–2938. doi: 10.1158/1078-0432.CCR-09-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu HG, Scholten I, Gruss C. et al. The distribution of the DEK protein in mammalian chromatin. Biochem Biophys Res Commun. 2007;358(4):1008–1014. doi: 10.1016/j.bbrc.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Privette Vinnedge LM, Ho SM, Wikenheiser-Brokamp KA. et al. The DEK oncogene is a target of steroid hormone receptor signaling in breast cancer. PLoS One. 2012;7(10):e46985. doi: 10.1371/journal.pone.0046985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Lindern M, Fornerod M, van Baal S. et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12(4):1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornerod M, Boer J, van Baal S. et al. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10(9):1739–1748. [PubMed] [Google Scholar]
- 11.Broxmeyer HE, Mor-Vaknin N, Kappes F. et al. Concise review: role of DEK in stem/progenitor cell biology. Stem Cells. 2013;31(8):1447–1453. doi: 10.1002/stem.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang DM, Liu L, Fan L. et al. Expression level of DEK in chronic lymphocytic leukemia is regulated by fludarabine and Nutlin-3 depending on p53 status. Cancer Biol Ther. 2012;13(14):1522–1528. doi: 10.4161/cbt.22252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Privette Vinnedge LM, Benight NM, Wagh PK. et al. The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers. Oncogene. 2015;34(18):2325–2336. doi: 10.1038/onc.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying G, Wu Y. DEK: A novel early screening and prognostic marker for breast cancer. Mol Med Rep. 2015;12(5):7491–7495. doi: 10.3892/mmr.2015.4380. [DOI] [PubMed] [Google Scholar]
- 15.Piao J, Shang Y, Liu S. et al. High expression of DEK predicts poor prognosis of gastric adenocarcinoma. Diagn Pathol. 2014;9:67. doi: 10.1186/1746-1596-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin L, Piao J, Gao W. et al. DEK over expression as an independent biomarker for poor prognosis in colorectal cancer. BMC Cancer. 2013;13:366. doi: 10.1186/1471-2407-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Zhang W, Zhou L. et al. MicroRNA-592 targets DEK oncogene and suppresses cell growth in the hepatocellular carcinoma cell line HepG2. Int J Clin Exp Pathol. 2015;8(10):12455–12463. [PMC free article] [PubMed] [Google Scholar]
- 18.Datta A, Adelson ME, Mogilevkin Y. et al. Oncoprotein DEK as a tissue and urinary biomarker for bladder cancer. BMC Cancer. 2011;11:234. doi: 10.1186/1471-2407-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carro MS, Spiga FM, Quarto M. et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5(11):1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 20.Kroes RA, Jastrow A, McLone MG. et al. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156(2):191–198. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- 21.Alexiadis V, Waldmann T, Andersen J. et al. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14(11):1308–1312. [PMC free article] [PubMed] [Google Scholar]
- 22.Vance EB, Wagner NN. Written descriptions of orgasm: a study of sex differences. Arch Sex Behav. 1976;5(1):87–98. doi: 10.1007/BF01542242. [DOI] [PubMed] [Google Scholar]
- 23.Soares LM, Zanier K, Mackereth C. et al. Intron removal requires proofreading of U2AF/3' splice site recognition by DEK. Science. 2006;312(5782):1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 24.German RR, Fink AK, Heron M. et al. The accuracy of cancer mortality statistics based on death certificates in the United States. Cancer Epidemiol. 2011;35(2):126–131. doi: 10.1016/j.canep.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 26.Sener SF, Fremgen A, Menck HR. et al. Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985-1995, using the National Cancer Database. J Am Coll Surg. 1999;189(1):1–7. doi: 10.1016/s1072-7515(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 27.Neoptolemos JP, Stocken DD, Friess H. et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350(12):1200–1210. doi: 10.1056/NEJMoa032295. [DOI] [PubMed] [Google Scholar]
- 28.Kavanaugh GM, Wise-Draper TM, Morreale RJ. et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39(17):7465–7476. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brázda V, Laister RC, Jagelská EB. et al. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol Biol. 2011;12:33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fahrer J, Popp O, Malanga M. et al. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49(33):7119–7130. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khodadoust MS, Verhaegen M, Kappes F. et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69(16):6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng J, Kohler ME, Chen Q. et al. Serum from mice immunized in the context of Treg inhibition identifies DEK as a neuroblastoma tumor antigen. BMC Immunol. 2007;8:4. doi: 10.1186/1471-2172-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Bi F, Yang Y. et al. DEK protein overexpression predicts poor prognosis in pancreatic ductal adenocarcinoma. Oncol Rep. 2017;37(2):857–864. doi: 10.3892/or.2016.5302. [DOI] [PubMed] [Google Scholar]