Abstract
Methanol is a common solubilizing agent used to study transmembrane proteins/peptides in biological and synthetic membranes. Using small angle neutron scattering and a strategic contrast-matching scheme, we show that methanol has a major impact on lipid dynamics. Under increasing methanol concentrations, isotopically distinct 1,2-dimyristoyl-sn-glycero-3-phosphocholine large unilamellar vesicle populations exhibit increased mixing. Specifically, 1,2-dimyristoyl-sn-glycero-3-phosphocholine transfer and flip-flop kinetics display linear and exponential rate enhancements, respectively. Ultimately, methanol is capable of influencing the structure-function relationship associated with bilayer composition (e.g., lipid asymmetry). The use of methanol as a carrier solvent, despite better simulating some biological conditions (e.g., antimicrobial attack), can help misconstrue lipid scrambling as the action of proteins or peptides, when in actuality it is a combination of solvent and biological agent. As bilayer compositional stability is crucial to cell survival and protein reconstitution, these results highlight the importance of methanol, and solvents in general, in biomembrane and proteolipid studies.
Main Text
Lipid bilayers form the structural backbone of cellular membranes and possess marked lateral and transversal organization of lipids. This strict lipid organization has implications in vital cellular processes, including protein function and localization (1), vesicle fusion and budding (2), and apoptosis (3). Lipids undergo three types of spontaneous, diffusive motion: they 1) exchange between bilayers (interbilayer transfer/exchange), 2) translocate between bilayer leaflets (transverse lipid diffusion/flip-flop), and 3) laterally diffuse within the plane of the membrane surface. Herein, this study will focus on the former two because interbilayer exchange is linked to how lipids arrive, remain in, and leave cellular membranes, whereas lipid flip-flop disrupts the energy-driven maintenance of membrane asymmetry (i.e., the compositional difference between leaflets) in living cells. In essence, both dynamical actions are intrinsically linked to bilayers and their compositional stability. Previous studies have externally induced reorganization of lipids by outside factors as seen in model phospholipid membranes upon the addition of cations (4), detergents (5), and peptides (6, 7, 8, 9). Here, we focus on how common organic solvents also impact these dynamics.
As membranes do not exist in isolation, they are in constant contact with compounds or solutions. Alcohols can be commonly found in the external environment of membranes. This proximity potentiates alcohol-membrane interactions, which can cause changes in membrane composition (10). For instance, methanol, the simplest alcohol, composed of a single methyl attached to a hydroxyl group, is ubiquitously utilized as a fuel source and chemical precursor in cells and in proteolipidic studies as an organic solvent of lipids and proteins/peptides. Such alcohols have been shown to alter lipid bilayer properties (11), making their study of great importance. Despite this importance, studies on lipid dynamics under the influence of alcohols are severely lacking. Several computational studies have examined the interactions between short-chain alcohols and lipid bilayers on a physical and molecular level (12, 13) but none on lipid exchange and flip-flop in detail. To date, experimental data on these effects are also lacking, and the limited experimental reports have come into question because of the use of fluorescent probes (14, 15). These probes can disrupt physicochemical bilayer properties and have also demonstrated unreliable flip-flop rates, differing depending on the type of fluorophore used, even within the same lipid system (16). We surmise this issue to extend to interbilayer exchange as well. To our knowledge, no such dynamical study has been conducted that examines methanol and model bilayers in a probe-free manner. This study overcomes these pitfalls by using small angle neutron scattering (SANS) as a noninvasive and probe-free technique to quantify lipid dynamics in the presence of methanol. SANS can temporally and spatially monitor the molecular organization within samples and has proven to be a powerful tool in allowing the simultaneous measurement of lipid flip-flop and exchange rates (17, 18, 19, 20).
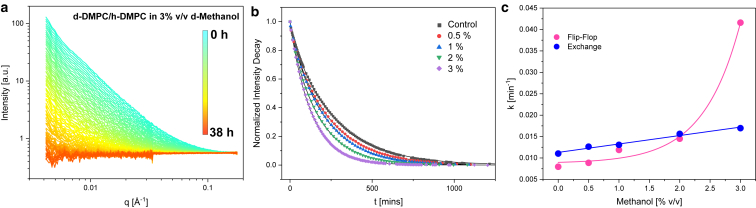
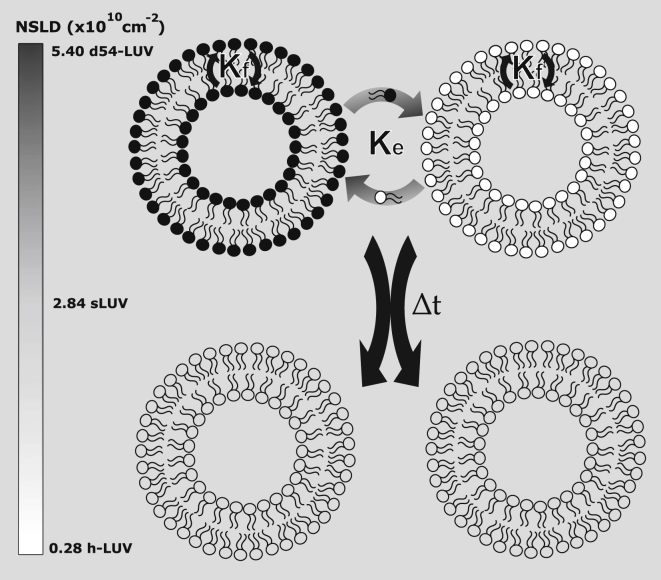
Here, we apply SANS to monitor lipid mixing of two distinct 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) populations, one chain perdeuterated DMPC (d-DMPC) and the other fully protiated DMPC (h-DMPC), in the presence of an increasing deuterated methanol (d-methanol) concentration. This measurement is achieved by setting the ratio of H2O and D2O (here 45% D2O) such that the water solvent neutron scattering length density is matched to uniformly mixed d-DMPC/h-DMPC vesicles. Unmixed vesicles will thus display contrast versus the water solvent, resulting in heightened scattering intensity, whereas fully mixed samples will display scattering intensities akin to the solvent background (i.e., a flat and featureless curve). Thus, as h-DMPC and d-DMPC large unilamellar vesicles begin to mix via lipid monomers transferring within and between bilayers, the measured intensity will decay and eventually reach an intensity baseline, corresponding to a single population of completely mixed vesicles (shown in Fig. 2 a). The experimental scheme can be seen in Fig. 1, whereas a more detailed protocol can be found in the Supporting Materials and Methods. A normalized intensity decay was calculated from the collective scattering curves of each sample and analyzed with a model for exchange/flip-flop by Nakano et al. (Fig. 2 b) (17). With this experimental setup, we are able to quantify both DMPC flip-flop (kf) and exchange (ke) rates under the influence of methanol.
Figure 2.
(a) SANS curve of d-DMPC and h-DMPC vesicles with 3% (v/v) d-methanol solvent. Periodic measurements were conducted at 37°C over 38 h. (b) Normalized contrast decay curves of increasing d-methanol presence; continuous lines indicate fitted curves used to derive flip-flop and lipid exchange rate constants. Each data point represents the normalized integrated intensity of a single SANS curve similar to those found in (a). (c) Plot of measured flip-flop and lipid exchange rate constants as a function of d-methanol percentage concentration. Solid lines represent curves of best fit.
Figure 1.
Schematic of the contrast matching scheme used. Vesicles composed solely of d-DMPC (d54-DMPC) and h-DMPC are placed together in a H2O/D2O (55/45) mixture, contrast matched to an neutron scattering length density equal to fully mixed vesicles of d-DMPC and h-DMPC. Over time, because of lipid exchange and flip-flop, intensity loss can be monitored as vesicles mix and near the contrast match point.
Increasing methanol concentrations had a profound effect on the kinetics of DMPC monomers in free-floating large unilamellar vesicles. Despite differences in vesicle size and investigative techniques, our unperturbed DMPC flip-flop and transfer rates are in excellent agreement with many values previously found (17, 21, 22, 23, 24). In a closely related study, Gerelli et al. used neutron reflectometry to measure DMPC flip-flop and exchange between vesicle dispersions and adsorbed planar bilayers (25). The exchange half-times for fluid-phase DMPC coincide with values found here (timescale of hours), whereas flip-flop was magnitudes faster (≤2.5 min). As recently shown (16), the incomplete surface coverage of planar bilayers led to microscopic defects, which can facilitate lipid flip-flop and thus result in flip-flop rates on the order of seconds to minutes. This study, with fully sealed vesicles, bypasses such issues. More significantly, our results also reveal that methanol accelerates both fluid-phase DMPC flip-flop and transfer rates (Table 1). The flip-flop rate increases exponentially (Fig. 2 c), whereas the exchange rate increases linearly under the studied concentrations. Our flip-flop finding is in line with Schwichtenhövel and co-workers, who found that radioactive and fluorescent lipid probes in human erythrocytes demonstrated exponential acceleration of inward flipping rates in the presence of 1-alkanols (C2–C8) and alkyl diols (14). Methanol has by far the weakest hydrophobic character in the short-chain alcohol group yet seems to perturb the membrane through the same or similar fashion as longer chained alcohols and alkyl diols. Although it has been shown that other short-chain alcohols affect inward flipping rates, we provide new, to our knowledge, insights with regards to both flip-flop and transfer rates in the presence of methanol. In general, at low methanol levels, DMPC undergoes slower flip-flop than transfer, but at concentrations above 2% (v/v) methanol, the situation is reversed. Interestingly, these observations suggest that methanol affects the two dynamical processes in distinct ways.
Table 1.
DMPC Flip-Flop and Exchange Half-Times and Rate Constants
| d-Methanol |
Flip-Flop |
Exchange |
||
|---|---|---|---|---|
| (v/v%) | t1/2 (min) | kf (×10−3 min−1) | t1/2 (min) | kt (×10−3 min−1) |
| 0 | 87.2 ± 1.1 | 8.0 ± 0.1 | 62.8 ± 0.3 | 11.0 ± 0.05 |
| 0.5 | 78.1 ± 2.6 | 8.9 ± 0.3 | 54.8 ± 0.4 | 12.6 ± 0.1 |
| 1.0 | 58.3 ± 1.4 | 11.9 ± 0.3 | 53.0 ± 0.4 | 13.1 ± 0.09 |
| 2.0 | 47.8 ± 1.3 | 14.5 ± 0.4 | 44.3 ± 0.3 | 15.6 ± 0.1 |
| 3.0 | 16.7 ± 1.3 | 41.6 ± 3.3 | 40.8 ± 0.2 | 17.0 ± 0.1 |
Flip-flop has a large energy barrier resulting from the transport of a polar, and often charged, lipid headgroup through the hydrophobic core. This unfavorable process can be augmented via intercalation of polar alcohol molecules within the membrane, which can cause short-lived transient pores and/or increase in the membrane’s dielectric constant; both can justify the enhanced flip-flop observed here, but the latter is unlikely to be the major driving force as shown in past work (26). During lipid transfer, it is thermodynamically unfavorable for the hydrophobic tails to pass through both the polar headgroup region and aqueous phase. As exemplified by De Cuyper et al. (27), an increase in acyl chain length reduces the transfer of lipid monomers, presumably because of a greater exposed hydrophobic moiety. They also saw that incorporation of polyalcohols into these lipid structures caused an increase in transfer. In view of these results, alcohols can play a significant role in masking a lipid’s hydrophobic character within a polar environment. Therefore, increasing methanol, an organic solvent that is fully miscible in water, should lower the energy barrier associated with lipid transfer.
Fluorescence techniques and freeze-fracture electron microscopy have demonstrated that short-chain alcohols promote vesicle hemifusion and complete fusion, hypothetically by way of outer leaflet disruption (28, 29). This fusing of individual membrane vesicles provides sites of enhanced lipid exchange via fusion pores, which allows lateral diffusion of lipids to the adjacent leaflet, yielding faster than expected lipid dynamics (30). As a result, dynamic light scattering measurements of size and polydispersity were taken before and after incubation at multiple methanol concentrations, neither of which revealed significant changes, maintaining a vesicle diameter of ∼140 nm and a polydispersity index of 0.15 ± 0.02. Significantly, these results suggest fusion events did not occur as an increase in mean particle size and a polydispersity index would have been observed.
Further biophysical studies, using elastic SANS and small angle x-ray scattering (SAXS), were applied. SANS and SAXS are complementary techniques used to probe sample structure and are known to be extremely sensitive to membrane lamellarity and lipid bilayer structure. For example, multilamellarity can be signified by the appearance of Bragg peaks in the scattering data. The fact that both SANS and SAXS yielded curves that displayed diffuse scattering (i.e., no detectable sharp Bragg peak in Fig. S1) proves that multilamellar bilayers did not evolve under the presence of d-methanol. Collectively, these qualitative findings indicate that methanol did not alter the vesicles’ morphological structure.
To determine if a defect-mediated mechanism can account for the rate enhancements, we examined pertinent bilayer properties. Previous simulation and experimental studies on model membranes revealed that the area per lipid (AL) generally increases with increasing alcohol concentrations, irrespective of lipid saturation and chain length (12, 31). The in silico study, in particular, found that this led to greater transient defects that provided lipid headgroups an opportunity to traverse the bilayer core and flip-flop (12). However, because of differences in lipid and alcohol concentrations between their systems and ours, we conducted our own structural analysis. A joint refinement of SANS and SAXS data was applied to robustly derive these structural features (32). As shown in Fig. S1, the scattering profiles of pure lipid and methanol-treated samples are indiscernible. In terms of relevant bilayer structural parameters, they are essentially unchanged (shown and explained in Fig. S1) and match well with previously reported DMPC data (33). These results do not outright disagree with previous studies. For example, Klacsová et al. observed changes in AL and bilayer thickness (DB) of a diunsaturated PC system hydrated with varying concentrations of aliphatic alcohols of different chain lengths, C8–18 (34); it was generally seen that shorter chain lengths imparted fewer effects on bilayer properties. With methanol having the smallest alkyl group, it makes sense that its effect on membrane structure is more difficult to detect. A plausible mechanism of methanol perturbation could be instead due to a decrease in chain order, often seen under increasing short-chain alcohol levels (35). However, an increase in AL is also associated with such a change but, as seen here, the AL remains static. Thus, the most likely explanation must involve methanol inducing short-range and perhaps short-lived defects, which are thus difficult to discern via methods that measure a global structural average, such as the biophysical techniques used in this study.
Traditionally, protein and peptide reconstitution have relied on either solvent addition or preincorporation with lipids before thin film hydration, the choice being dependent on the folding nature of the proteins/peptides. With the former, short-chain alcohols, including methanol, are popular carrier solvents used in studies on peptide activity (36, 37), channel activity conductance (38), the critical structural motifs of proteins (39), and physical interactions between proteins and lipid bilayers (40). Our data suggest that even low concentrations of methanol, commonly used in these studies, have a profound effect on the compositional stability of membranes. In specialized cases involving lipid asymmetry, methanol-induced lipid scrambling can have unwanted consequences; lipid vesicles and likely other geometric setups exposed to methanol will have their asymmetric stability dramatically reduced manyfold. Such an effect limits the time of study and possible applicable techniques and assays. Because the response to methanol will vary depending on phospholipid composition, cholesterol content, and buffer of the studied membrane, ideal solvents that do not perturb membranes must be determined as well as the use of assays that can evaluate individual leaflet compositions and/or the degree of membrane asymmetry. These adaptations can help ensure experimental protocols have not altered the bilayer composition. Our findings further highlight an additional complication when adding proteins or peptides externally; for example, in studies on antimicrobial peptides (AMPs), though AMP attack is better simulated in this manner, these studies have the potential to incorrectly assign the cause of the enhanced lipid kinetics to the AMP, when in actuality it may be due to the carrier solvent or some combination of the two. Ultimately, this work highlights the importance of understanding the interplay between the system of interest and the carrier solvent on lipid mobility.
Author Contributions
M.H.L.N and D.M. designed the research. M.H.L.N, M.D., B.W.R., and D.M. carried out experimentation, and analyzed the data. E.G.K. and C.B.S. provided expert SANS help. M.H.L.N. wrote the article. M.H.L.N, M.D., B.W.R., C.B.S., E.G.K., and D.M. revised the manuscript.
Acknowledgments
We acknowledge the support of the National Institute of Standards and Technology, U.S. Department of Commerce in providing the neutron research facilities used in this work. Access to the VSANS instrument was provided by the Center for High Resolution Neutron Scattering, a partnership between the National Science Foundation and the National Institute of Standards and Technology under Agreement DMR-1508249. A portion of this research at Oak Ridge National Laboratory’s Spallation Neutron Source was sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy. M.D. and M.H.L.N. are both supported by Ontario Graduate Scholarships. D.M. acknowledges the support of the Natural Sciences and Engineering Research Council of Canada [funding reference number 2018-04841].
Editor: Tommy Nylander.
Footnotes
Supporting Materials and Methods and two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30057-8.
Supporting Citations
References (41, 42, 43) appear in the Supporting Material.
Supporting Material
References
- 1.Nyholm T.K. Lipid-protein interplay and lateral organization in biomembranes. Chem. Phys. Lipids. 2015;189:48–55. doi: 10.1016/j.chemphyslip.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fadok V.A., Voelker D.R., Henson P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 4.Henseleit U., Plasa G., Haest C. Effects of divalent cations on lipid flip-flop in the human erythrocyte membrane. Biochim. Biophys. Acta. 1990;1029:127–135. doi: 10.1016/0005-2736(90)90445-t. [DOI] [PubMed] [Google Scholar]
- 5.Ahyayauch H., Bennouna M., Goñi F.M. Detergent effects on membranes at subsolubilizing concentrations: transmembrane lipid motion, bilayer permeabilization, and vesicle lysis/reassembly are independent phenomena. Langmuir. 2010;26:7307–7313. doi: 10.1021/la904194a. [DOI] [PubMed] [Google Scholar]
- 6.Fattal E., Nir S., Szoka F.C., Jr. Pore-forming peptides induce rapid phospholipid flip-flop in membranes. Biochemistry. 1994;33:6721–6731. doi: 10.1021/bi00187a044. [DOI] [PubMed] [Google Scholar]
- 7.Matsuzaki K., Murase O., Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry. 1996;35:11361–11368. doi: 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- 8.Anglin T.C., Liu J., Conboy J.C. Facile lipid flip-flop in a phospholipid bilayer induced by gramicidin A measured by sum-frequency vibrational spectroscopy. Biophys. J. 2007;92:L01–L03. doi: 10.1529/biophysj.106.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor G., Nguyen M.A., Sarles S.A. Electrophysiological interrogation of asymmetric droplet interface bilayers reveals surface-bound alamethicin induces lipid flip-flop. Biochim. Biophys. Acta Biomembr. 2019;1861:335–343. doi: 10.1016/j.bbamem.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Stanley D., Bandara A., Stanley G.A. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 2010;109:13–24. doi: 10.1111/j.1365-2672.2009.04657.x. [DOI] [PubMed] [Google Scholar]
- 11.Seeman P. The membrane actions of anesthetics and tranquilizers. Pharmacol. Rev. 1972;24:583–655. [PubMed] [Google Scholar]
- 12.Dickey A.N., Faller R. How alcohol chain-length and concentration modulate hydrogen bond formation in a lipid bilayer. Biophys. J. 2007;92:2366–2376. doi: 10.1529/biophysj.106.097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patra M., Salonen E., Karttunen M. Under the influence of alcohol: the effect of ethanol and methanol on lipid bilayers. Biophys. J. 2006;90:1121–1135. doi: 10.1529/biophysj.105.062364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwichtenhövel C., Deuticke B., Haest C.W. Alcohols produce reversible and irreversible acceleration of phospholipid flip-flop in the human erythrocyte membrane. Biochim. Biophys. Acta. 1992;1111:35–44. doi: 10.1016/0005-2736(92)90271-m. [DOI] [PubMed] [Google Scholar]
- 15.Marquardt D., Geier B., Pabst G. Asymmetric lipid membranes: towards more realistic model systems. Membranes (Basel) 2015;5:180–196. doi: 10.3390/membranes5020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquardt D., Heberle F.A., Pabst G. 1H NMR shows slow phospholipid flip-flop in gel and fluid bilayers. Langmuir. 2017;33:3731–3741. doi: 10.1021/acs.langmuir.6b04485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano M., Fukuda M., Handa T. Determination of interbilayer and transbilayer lipid transfers by time-resolved small-angle neutron scattering. Phys. Rev. Lett. 2007;98:238101. doi: 10.1103/PhysRevLett.98.238101. [DOI] [PubMed] [Google Scholar]
- 18.Nakano M., Fukuda M., Handa T. Flip-flop of phospholipids in vesicles: kinetic analysis with time-resolved small-angle neutron scattering. J. Phys. Chem. B. 2009;113:6745–6748. doi: 10.1021/jp900913w. [DOI] [PubMed] [Google Scholar]
- 19.Garg S., Porcar L., Perez-Salas U. Noninvasive neutron scattering measurements reveal slower cholesterol transport in model lipid membranes. Biophys. J. 2011;101:370–377. doi: 10.1016/j.bpj.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wah B., Breidigan J.M., Perez-Salas U. Reconciling differences between lipid transfer in free-standing and solid supported membranes: a time-resolved small-angle neutron scattering study. Langmuir. 2017;33:3384–3394. doi: 10.1021/acs.langmuir.6b04013. [DOI] [PubMed] [Google Scholar]
- 21.De Cuyper M., Joniau M., Dangreau H. Intervesicular phospholipid transfer. A free-flow electrophoresis study. Biochemistry. 1983;22:415–420. doi: 10.1021/bi00271a027. [DOI] [PubMed] [Google Scholar]
- 22.McLean L.R., Phillips M.C. Kinetics of phosphatidylcholine and lysophosphatidylcholine exchange between unilamellar vesicles. Biochemistry. 1984;23:4624–4630. doi: 10.1021/bi00315a017. [DOI] [PubMed] [Google Scholar]
- 23.Drazenovic J., Ahmed S., Wunder S.L. Lipid exchange and transfer on nanoparticle supported lipid bilayers: effect of defects, ionic strength, and size. Langmuir. 2015;31:721–731. doi: 10.1021/la503967m. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg R.D., McConnell H.M. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971;10:1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- 25.Gerelli Y., Porcar L., Fragneto G. Lipid exchange and flip-flop in solid supported bilayers. Langmuir. 2013;29:12762–12769. doi: 10.1021/la402708u. [DOI] [PubMed] [Google Scholar]
- 26.Barchfeld G.L., Deamer D.W. Alcohol effects on lipid bilayer permeability to protons and potassium: relation to the action of general anesthetics. Biochim. Biophys. Acta. 1988;944:40–48. doi: 10.1016/0005-2736(88)90314-8. [DOI] [PubMed] [Google Scholar]
- 27.De Cuyper M., Joniau M. Spontaneous intervesicular transfer of anionic phospholipids differing in the nature of their polar headgroup. Biochim. Biophys. Acta. 1985;814:374–380. [Google Scholar]
- 28.Chanturiya A., Leikina E., Chernomordik L.V. Short-chain alcohols promote an early stage of membrane hemifusion. Biophys. J. 1999;77:2035–2045. doi: 10.1016/S0006-3495(99)77044-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondal Roy S., Sarkar M. Membrane fusion induced by small molecules and ions. J. Lipids. 2011;2011:528784. doi: 10.1155/2011/528784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jahn R., Lang T., Südhof T.C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 31.Ly H.V., Longo M.L. The influence of short-chain alcohols on interfacial tension, mechanical properties, area/molecule, and permeability of fluid lipid bilayers. Biophys. J. 2004;87:1013–1033. doi: 10.1529/biophysj.103.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kučerka N., Nagle J.F., Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and X-ray scattering data. Biophys. J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kučerka N., Nieh M.P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Klacsová M., Bulacu M., Balgavý P. The effect of aliphatic alcohols on fluid bilayers in unilamellar DOPC vesicles--a small-angle neutron scattering and molecular dynamics study. Biochim. Biophys. Acta. 2011;1808:2136–2146. doi: 10.1016/j.bbamem.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Gawrisch K., Holte L.L. NMR investigations of non-lamellar phase promoters in the lamellar phase state. Chem. Phys. Lipids. 1996;81:105–116. [Google Scholar]
- 36.Epand R.F., Epand R.M., Toniolo C. Analogs of the antimicrobial peptide trichogin having opposite membrane properties. Eur. J. Biochem. 2001;268:703–712. doi: 10.1046/j.1432-1327.2001.01922.x. [DOI] [PubMed] [Google Scholar]
- 37.Hu X., Tan J., Ye S. Reversible activation of pH-responsive cell-penetrating peptides in model cell membrane relies on the nature of lipid. J. Phys. Chem. C. 2017;121:15181–15187. [Google Scholar]
- 38.Rokitskaya T.I., Kotova E.A., Antonenko Y.N. Membrane dipole potential modulates proton conductance through gramicidin channel: movement of negative ionic defects inside the channel. Biophys. J. 2002;82:865–873. doi: 10.1016/S0006-3495(02)75448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serrano A.G., Ryan M., Pérez-Gil J. Critical structure-function determinants within the N-terminal region of pulmonary surfactant protein SP-B. Biophys. J. 2006;90:238–249. doi: 10.1529/biophysj.105.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz A., Casals C., Pérez-Gil J. Different modes of interaction of pulmonary surfactant protein SP-B in phosphatidylcholine bilayers. Biochem. J. 1997;327:133–138. doi: 10.1042/bj3270133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eicher B., Heberle F.A., Pabst G. Joint small-angle X-ray and neutron scattering data analysis of asymmetric lipid vesicles. J. Appl. Cryst. 2017;50:419–429. doi: 10.1107/S1600576717000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J.K., Gao C.Y., Liu D. The extended Q-range small-angle neutron scattering diffractometer at the SNS. J. Appl. Cryst. 2010;43:1068–1077. [Google Scholar]
- 43.Petoukhov M.V., Franke D., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.