Abstract
Unlike most transmembrane proteins, phospholipids can migrate from one leaflet of the membrane to the other. Because this spontaneous lipid translocation (flip-flop) tends to be very slow, cells facilitate the process with enzymes that catalyze the transmembrane movement and thereby regulate the transbilayer lipid distribution. Nonenzymatic membrane-spanning proteins with unrelated primary functions have also been found to accelerate lipid flip-flop in a nonspecific manner and by various hypothesized mechanisms. Using deuterated phospholipids, we examined the acceleration of flip-flop by gramicidin channels, which have well-defined structures and known functions, features that make them ideal candidates for probing the protein-membrane interactions underlying lipid flip-flop. To study compositionally and isotopically asymmetric proteoliposomes containing gramicidin, we expanded a recently developed protocol for the preparation and characterization of lipid-only asymmetric vesicles. Channel incorporation, conformation, and function were examined with small angle x-ray scattering, circular dichroism, and a stopped-flow spectrofluorometric assay, respectively. As a measure of lipid scrambling, we used differential scanning calorimetry to monitor the effect of gramicidin on the melting transition temperatures of the two bilayer leaflets. The two calorimetric peaks of the individual leaflets merged into a single peak over time, suggestive of scrambling, and the effect of the channel on the transbilayer lipid distribution in both symmetric 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and asymmetric 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1,2-dimyristoyl-sn-glycero-3-phosphocholine vesicles was quantified from proton NMR measurements. Our results show that gramicidin increases lipid flip-flop in a complex, concentration-dependent manner. To determine the molecular mechanism of the process, we used molecular dynamics simulations and further computational analysis of the trajectories to estimate the extent of membrane deformation. Together, the experimental and computational approaches were found to constitute an effective means for studying the effects of transmembrane proteins on lipid distribution in both symmetric and asymmetric model membranes.
Introduction
Membranes are an essential component of all living organisms. Their structure and organization serve many functions and are tightly regulated by the cell. One prominent example is the transverse lipid distribution in cell membranes; whereas a self-assembled lipid bilayer will have the same lipid composition in its two leaflets (i.e., it is symmetric), the leaflet compositions in the plasma membranes of eukaryotic cells differ (i.e., the bilayer is asymmetric), and this difference is actively maintained by the cell. Not surprisingly, the biophysical mechanisms underlying membrane asymmetry are the subject of intense studies (1, 2, 3, 4, 5), which are rapidly increasing in number as a result of recent advances that enable the preparation and biophysical characterization of asymmetric lipid-only model membranes in vitro (6, 7, 8). Because such model membranes are not at chemical equilibrium and their asymmetry is not actively maintained, the time window for examining their properties is limited by the gradual redistribution of the lipids between the two leaflets until a symmetric lipid composition is achieved. Such unassisted interleaflet lipid movement is often referred to as passive or spontaneous lipid flip-flop (9).
Some of the key lipid and bilayer properties that determine the kinetics of spontaneous lipid flip-flop include chain length, headgroup size and charge, and cholesterol concentration, although the mechanism(s) are not fully understood (see (10) for a recent review of both experimental and computational studies on the topic). In general, flip-flop of phospholipids is many orders of magnitude slower than other lipid motions, such as rotation or lateral diffusion (11). This difference is due to the large energy barrier for moving a polar lipid headgroup from one side of the bilayer to the other, a process that requires traversing the bilayer’s hydrophobic core. In cells, the one-directional movement of lipids between the two leaflets is catalyzed by ATP-dependent enzymes; these include flippases that move phospholipids from the extracellular leaflet to the intracellular leaflet (e.g., P-type ATPases) (12) and floppases that move phospholipids in the opposite direction (e.g., ATP-binding cassette transporters) (13). An additional bidirectional and often nonspecific movement of lipids across the two leaflets is catalyzed by ATP-independent scramblases, which include the Ca2+-activated TMEM family of proteins (11). It is through a careful balance between the activity of the enzymes and scramblases that cells maintain the desired lipid compositions in their two membrane leaflets.
In addition to the flippases, floppases, and scramblases, a wide variety of proteins with nonrelated primary functions can catalyze lipid flip-flop through different proposed mechanisms that include pore-mediated scrambling (14, 15) and the so-called “credit-card” lipid movement (11). For some proteins, this secondary function has been proposed to have biological implications (16); for others, the physiological role (if any) remains unclear. Still, the ability of a variety of membrane proteins to scramble lipids has direct implications for the design and interpretation of studies using asymmetric protein-laden membranes and therefore should be carefully examined. A particularly interesting case is single-span transmembrane (TM) proteins that are often used to study protein-membrane interactions in vitro (17). Such proteins can facilitate lipid flip-flop through the so-called perturbation-mediated mechanisms, that is, lowering the energy barrier for lipid translocation between the leaflets by changing the bilayer structure and/or dynamics in the vicinity of the protein (18, 19, 20, 21, 22).
One single-span TM protein that reportedly affects lipid flip-flop only under certain conditions is the functional dimer configuration of the bacterial ion-channel gramicidin (gA) (23). To our knowledge, the channel has been shown to accelerate lipid translocation in three separate studies but to different extents: up to 30-fold in erythrocytes at moderate gA concentrations (gA/lipid ratio of ∼1:200) (24); from 2- to 10-fold in supported lipid bilayers at high gA/lipid ratios (1:50) (25); and, to a somewhat lesser extent, in 400-nm-diameter liposomes with high gA ratios (1:20) in which flip-flop was unmeasurably slow in the absence of gA (14). At the same time, flip-flop enhancement was not detected in erythrocytes at gA concentrations at which the channel performs its ion-conducting function (24). The disparate results from these studies highlight limitations in the quantification of the transbilayer lipid distribution with rather indirect experimental methodologies, including extracting the outer leaflet lipids with albumin (which in its own right could perturb the bilayer) (24) or approximating the host lipid flip-flop kinetics from the translocation rate of a fluorescent lipid analog (14). The choice of system is also important: complex and asymmetric cell membrane environments like erythrocytes present challenges both for the interpretation of results and the quantification of the gA/lipid ratio, whereas the unavoidable bilayer defects in supported lipid bilayers prepared with the Langmuir-Blodgett/Langmuir-Schaefer technique could accelerate lipid flip-flop in a manner that is difficult to control (26).
Here, we address these challenges using a new (to our knowledge) platform for measuring protein-mediated lipid flip-flop in vitro. Our approach makes use of free-floating and stress-free large unilamellar vesicles with precisely controlled symmetric and asymmetric lipid composition, both in the presence and the absence of protein. This experimental setup allows for a wide range of biophysical assays and, importantly, enables the direct measurement of the flip-flop kinetics of unlabeled lipids in chemically symmetric and asymmetric bilayers using 1H-NMR. This framework thus overcomes many of the limitations in the previous approaches by providing a robust model system for studying the effects of proteins on the transverse bilayer organization. Using this protocol, we examined the effect of gA on lipid flip-flop in both isotopically and compositionally asymmetric vesicles. Our results show that gA speeds up lipid translocation in both systems and that the rates are not directly proportional to gA concentration. Further computational analysis revealed that membrane deformations likely play a role in the observed effects at high gA mole fractions, suggesting the existence of mechanistically different regimes of gA-mediated changes in bilayer organization.
Materials and Methods
Materials
All phospholipids (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG), POPC-d31, POPC-d13, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and DMPC-d54; see Table 1 for a list of abbreviations) and gramicidin were purchased from Avanti Polar Lipids (Alabaster, AL) and Sigma-Aldrich (St. Louis, MO) respectively, as dry powders and used as supplied. The phospholipids were dissolved in high-performance liquid chromatography-grade chloroform, and gA was dissolved in high-performance liquid chromatography-grade methanol. Both phospholipids and gA were stored at −80°C until use. Methyl-β-cyclodextrin (MβCD), praseodymium(III) nitrate hexahydrate (Pr(NO3)3 6H2O), sucrose, NaCl, and hydrochloric acid were purchased from Thermo Fisher Scientific (Waltham, MA). Thallium nitrate (TlNO3) and 8-aminonaphthalene-1,3,6-trisulfonic acid disodium salt (ANTS) were purchased from Sigma-Aldrich (St. Louis, MO) and Invitrogen (Carlsbad, CA), respectively. Ultrapure H2O was obtained from a High-Q purification system (High-Q, Wilmette, IL), and D2O (99.9%) was purchased from Cambridge Isotope Laboratories (Tewksbury, MA).
Table 1.
List of Abbreviations
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| POPC-d13 | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine-1,1,2,2-d4-N,N,N-trimethyl-d9 (headgroup-deuterated POPC) |
| POPC-d31 | 1-palmitoyl-d31-2-oleoyl-sn-glycero-3-phosphocholine (chain-deuterated POPC) |
| DMPC-d54 | 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine (chain-deuterated DMPC) |
| POPG | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) |
| LUV | Large unilamellar vesicle |
| sLUV | Compositionally symmetric LUV |
| s∗LUV | Compositionally symmetric but isotopically asymmetric LUV |
| aLUV | Asymmetric LUV |
| gA | Gramicidin A |
| gA-LUV | Large unilamellar vesicle with gA |
| gA-sLUV | Compositionally symmetric LUV with gA |
| gA-s∗LUV | Compositionally symmetric but isotopically asymmetric LUV with gA |
| gA-aLUV | Asymmetric LUV with gramicidin |
| MLV | Multilamellar vesicle |
| MβCD | Methyl-beta-cyclodextrin |
| GC/MS | Gas chromatography/mass spectrometry |
| SAXS | Small-angle X-ray scattering |
| CD | Circular dichroism |
| GBFA | Gramicidin-based fluorescence assay |
| DSC | Differential scanning calorimetry |
| 1H-NMR | Proton nuclear magnetic resonance |
| MD | Molecular dynamics |
| CTMD | Continuum theory of membrane deformations |
| Cout | Area of shifted peak in 1H NMR spectra after addition of shift reagent |
| Cin | Area of unshifted peak in 1H NMR spectra after addition of shift reagent |
| ΔC | Difference between Cout and Cin relative to time 0, (Cout − Cin)/( Cout (0) − Cin(0)) |
| kf | Rate of lipid flip-flop |
| t1/2 | Half time of lipid flip-flop |
Preparation of liposomes without gA
Large unilamellar vesicles (LUVs) with symmetric lipid distribution (symmetric LUVs (sLUVs)) were prepared by first mixing appropriate volumes of lipid stocks in organic solvent with a glass Hamilton syringe. The solvent was evaporated with an Argon stream followed by high vacuum overnight. The dry lipid film was hydrated at room temperature (for POPC) or 35–40°C (for mixtures with DMPC) for at least 1 h with intermittent vortexing every 15 min. The resulting multilamellar vesicle (MLV) suspension was subjected to at least five freeze/thaw cycles using a −80°C freezer and then extruded through a 100-nm pore diameter polycarbonate filter with a mini-extruder (Avanti Polar Lipids) by passing the suspension through the filter 31 times. POPG was included in all sLUVs at 5 mol% to ensure unilamellarity (27).
Asymmetric LUVs (aLUVs) were prepared following the protocol described in (28) with slight modifications. Briefly, an MLV suspension of the donor lipid (i.e., the lipid to be exchanged into the outer leaflet of the aLUVs) was prepared in a 20% w/w sucrose solution as described above. The sample was diluted 20-fold with water and centrifuged at 20°C for 30 min at 20,000 × g. The supernatant was discarded, the pellet was redissolved using a 35 mM MβCD solution in water at a nominal donor lipid/MβCD ratio of 1:8, and the MβCD/donor mixture was incubated at room temperature for 2 h with gentle stirring. sLUVs of the acceptor lipids (i.e., the lipids to be present on the inner leaflet of the aLUVs) were prepared in a 25 mM NaCl solution as described above. These were added to the MβCD/donor mixture at a nominal donor/acceptor ratio of 2:1 or 3:1 (see Table S1). The MβCD/donor/acceptor slurry was incubated for 30 min at room temperature with gentle stirring. Immediately after this incubation, the sample was filtered with a prewashed centrifugal filter device (Ultra-15, 100,000 Da molecular weight cutoff; Amicon; MilliporeSigma, Burlington, MA) for 25 min at 2.5 K × g. The retentate was diluted with water eightfold and centrifuged for 30 min at 20,000 × g and 20°C to pellet the donor MLVs. The supernatant, containing aLUVs and residual MβCD, was carefully transferred to prewashed centrifugal filtration devices (as described above), concentrated to 250–500 μL, and washed with H2O or D2O a minimum of three times to remove MβCD. The aLUVs were recovered from the final retentate and stored in a plastic centrifuge tube at room temperature until further use.
Preparation of liposomes with gA
sLUVs with gA (gA-sLUVs) were prepared as follows. The lipids were first mixed in organic solvent, and gA was added from a methanolic solution with a glass Hamilton syringe at the appropriate gA/lipid ratio. The organic solvent was evaporated on a rotary evaporator, followed by high vacuum overnight. The dry sample was gently hydrated on a rotary evaporator for 30–60 min and then incubated at room temperature until the film was fully dissolved and the solution looked uniform (typically overnight). Vortexing was performed only occasionally and at the lowest setting. The sample was subjected to at least five freeze/thaw cycles using a −80°C freezer and then extruded with a 100-nm pore diameter polycarbonate filter as described above. POPG was added to all gA-sLUVs at 5 mol% to ensure unilamellarity.
aLUVs with gA (gA-aLUVs) were prepared following the protocol described above with the only modification that gA was added to the symmetric acceptor liposomes. That is, instead of acceptor sLUVs, acceptor gA-sLUV vesicles were added to the MβCD/donor mixture after the 2-h incubation. All other steps of the protocol were the same (see Fig. S1). The vesicle size distributions measured with dynamic light scattering showed little difference in LUV radius or polydispersity index between the symmetric acceptor vesicles and the final asymmetric vesicles (Table S2).
Gas chromatography-mass spectrometry
The overall lipid composition of the aLUVs and gA-aLUVs was determined from gas chromatography-mass spectrometry (GC/MS) analysis of fatty acid methyl esters (FAMEs) generated by acid-catalyzed methanolysis. The detailed protocol is described in (28). Briefly, ∼100 μg of the sample was dried on a rotary evaporator, dissolved in 1 mL of 1 M methanolic HCl, flushed with Argon, and incubated at 85°C for 1 h in a glass culture tube sealed with a Teflon-lined cap. After allowing the sample to cool for a few minutes, 1 mL water was added, and the sample was briefly vortexed. Then, 1 mL hexane was added, and the sample was vortexed vigorously to form an emulsion and extract the FAMEs. The solution was centrifuged at a low speed (∼400 × g) for 5 min to break the emulsion, and the upper (hexane) phase containing the FAMEs was transferred to a GC autosampler vial. The total volume in the vial was brought up to 1 mL with hexane. FAME analysis was performed using an Agilent 5890A gas chromatograph (Santa Clara, CA) with a 5975C mass-sensitive detector operating in electron-impact mode and an HP-5MS capillary column (30 m × 0.25 mm, 0.25-μm film thickness). After an injection of a 5-μL aliquot into the chromatograph, a preprogrammed column temperature routine was initiated as described in (28). Total ion chromatogram peaks were assigned and integrated using GC/MSD ChemStation Enhanced Data Analysis software. The ratio of the different lipid components was determined from the ratio of the respective peak areas of the FAMEs corresponding to the lipid fatty acid chains.
Small angle x-ray scattering
LUV samples for small angle x-ray scattering (SAXS) measurements were prepared as described above and concentrated to 15–20 mg/mL with prewashed centrifugal filter devices. SAXS measurements were performed using a Rigaku BioSAXS-2000 home source system with a Pilatus 100 K two-dimensional detector and a HF007 copper rotating anode (Rigaku Americas, The Woodlands, TX). SAXS data were collected at a fixed sample-to-detector distance using a silver behenate calibration standard, with a typical data collection time of 3–4 h. The one-dimensional scattering intensity I(q) (q = 4πsin (θ)/λ, where λ is the x-ray wavelength and 2θ is the scattering angle relative to the incident beam) was obtained by radial averaging of the corrected two-dimensional image data, an operation that was performed automatically using Rigaku SAXSLab software. Data were collected in 10-min frames with each frame processed separately to assess radiation damage; there were no significant changes in the scattering curves over time. Background scattering from water collected at the same temperature was subtracted from each frame, and the background-corrected I(q) from the individual frames was then averaged with the SD taken to be the measurement uncertainty. Scattering data analysis is described in detail in the Supporting Materials and Methods.
Circular dichroism
Samples for circular dichroism (CD) were first diluted to 1 mg/mL lipid concentration for a protein concentration between 5 and 13 μM. The lipid concentration was estimated from dynamic light scattering (DLS). gA conformation was measured using a J-815 spectropolarimeter equipped with a PTC-423S Peltier temperature controller (Jasco, Easton, MD). CD spectra (raw ellipticity θ in units of millidegree versus wavelength) were collected at 25°C using a 2-mm path-length cuvette, a scan rate of 100 nm/min, and 30 accumulations. The spectrum of each gA-containing sample was first corrected for the lipid background by subtracting the spectrum of a corresponding lipid-only sample. The only exception was the gA-aLUV sample, for which the background was taken to be the spectrum of the POPC acceptor sLUVs, which were similar in size to the gA-aLUVs as determined from DLS. The background-corrected data were then converted to mean residue molar ellipticity [θ] (in units of degree cm2 dmol−1) using the relationship
| (1) |
where l is the cell path length in mm, c is the protein concentration in μM, and is the number of amino acids in the gA protein.
gA-based fluorescence assay
gA function was quantified using a fluorescence quench protocol (29). For these studies, the acceptor sLUVs were prepared with a gA/lipid ratio of 1:20,000 (corresponding to 7–8 gA monomers per 130-nm diameter vesicle) and hydrated with 25 mM Na2ANTS instead of 25 mM NaCl. After the last concentration step, the gA-aLUVs were washed once with H2O and three times with elution buffer (35 mM NaNO3 and 6 mM HEPES (pH 7.0)). The rate of quenching of the ANTS trapped inside the vesicles was measured using a stopped-flow spectrofluorometer (SX.20; Applied Photophysics, Leatherhead, UK) in a single-mixing experiment and quantified with a regular stretched exponential (29). The ANTS-loaded LUVs were mixed with either NaNO3 buffer (35 mM NaNO3 and 6 mM HEPES (pH 7.0)) or TlNO3 buffer (35 mM TlNO3 and 6 mM HEPES (pH 7.0)), in which Tl+ (thallous ion) is a gA channel-permeant quencher of the ANTS fluorescence. The samples were excited at 352 nm, and the fluorescence signal above 455 nm was recorded in the absence (four successive trials) or the presence (nine successive trials) of the quencher. As a control, gA-sLUV acceptors hydrated with ANTS were also washed three times with elution buffer, and the rate of ANTS quenching was measured as described above.
Differential scanning calorimetry
Samples for differential scanning calorimetry (DSC) were diluted to ∼5 mg/mL and measured using a Nano DSC (TA Instruments, New Castle, DE). The LUV suspension was loaded into the sample capillary cell, and degassed solvent was loaded into the reference capillary cell. The cells were pressurized to 3 atmospheres to suppress the formation of air bubbles, and a cooling scan was initiated from 30°C to −8°C at a rate of 0.2°C/min. All thermograms showed either a series of peaks or a single broad peak between ∼−5 and 20°C. A sloping background contribution was accounted for by fitting a portion of the thermogram on either side of the peaks of interest to a third-order polynomial, which was then subtracted from the raw data.
1H-NMR
The interleaflet lipid distribution in the aLUVs and gA-aLUVs was quantified with a shift reagent assay performed on either a Bruker Avance III 400 MHz spectrometer (Bruker, Billerica, MA) (at Oak Ridge National Laboratory, Oak Ridge, TN) or a Bruker Avance III HD 500 MHz spectrometer (at Weill Cornell Medical College, New York, NY) as described in (26, 28). Briefly, a standard 1H pulse sequence with a 30° flip angle and a 2-s delay time was collected at 35°C. For all samples measured in H2O, the signal from the solvent was suppressed using the standard excitation sculpting sequence zgesgp. A sample aliquot was first diluted with D2O or H2O to a total volume of 600 μL and a concentration of ∼0.5 mg/mL. The diluted sample was loaded into an NMR tube, and a spectrum was recorded. A small aliquot (1–2 μL) of 20 mM Pr3+/D2O was then added to the NMR tube and mixed with its contents by inverting the tube three times ([Pr3+] ≈ 50 μM), and the spectrum was recorded immediately thereafter; another aliquot of Pr3+ was added, and the procedure was repeated. A total of at least three Pr3+ titrations were performed, and their spectra were recorded and used to establish the measurement uncertainty as described below. The typical elapsed time between the first exposure of the sample to Pr3+ and completion of the experiment was 40 min. Each Pr3+ titration resulted in a further shift of the resonance peak of the protiated choline headgroups exposed on the outer leaflet of the vesicles as shown in Fig. S2.
NMR analysis was performed by fitting each spectrum as a sum of Lorentzians using Origin or custom Mathematica scripts. The choline resonances were identified, and the fraction of exposed (i.e., outer leaflet) and protected (i.e., inner leaflet) choline headgroups was quantified from the areas of the shifted and unshifted choline peaks and used to determine the fraction of protiated-headgroup lipid in each leaflet. The measurement uncertainty was calculated as the SD of peak areas determined from the Pr3+ titration data described above. This information, together with the GC results, was used to calculate the composition of the two bilayer leaflets as described previously (28, 30).
Analysis of lipid flip-flop kinetics
The rate of lipid flip-flop, kf, was measured from the time-dependent changes in the transverse distribution of protiated-headgroup lipids in each sample as described in (26). Briefly, the NMR time traces of relative changes in the lipid distribution were expressed as
| (2) |
where C(t) is the area of the shifted (outer leaflet) and unshifted (inner leaflet) choline peaks at time t, with subscripts out and in denoting the outer and inner leaflet, respectively. The data were modeled as
| (3) |
with the corresponding half-time given by
| (4) |
All samples were kept on the bench between NMR measurements, and consequently, the flip-flop kinetics reported here correspond to sample behavior at an ambient temperature of ∼22°C.
Molecular dynamics simulations
All-atom molecular dynamics (MD) simulations of gA in the symmetric and compositionally asymmetric bilayers from the in vitro experiments were performed as described below.
For the symmetric system, a POPC bilayer with 100 lipids per leaflet (200 lipids total) was first constructed with CHARMM-GUI (31, 32, 33). The bilayer was hydrated with 70 waters per lipid and a total of 35 Na+ and 35 Cl− ions for a salt concentration of 138 mM. After a short initial equilibration with CHARMM-GUI’s protocol, the system was run for 226 ns. From the last frame of the trajectory, all water and ion atoms were removed, and a single gA dimer (Protein Data Bank [PDB]: 1JNO) was manually inserted in the bilayer by replacing ten lipids from each leaflet with a gA monomer. The system was then hydrated and ionized with VMD’s solvate and autoionize plugins, respectively, resulting in a bilayer patch with 90 lipids per leaflet (180 lipids total), 1 gA dimer, 67 waters per lipid, and 30 Na+ and 30 Cl− ions for a salt concentration of 138 mM NaCl.
For the asymmetric system, the protocol in (34) was used to identify the appropriate number of lipids in the asymmetric lipid-only bilayer with the top leaflet composed of DMPC/POPC 75/25 mol% and the bottom leaflet composed of DMPC/POPC 10/90 mol%. The resulting tension-free bilayer contained 104 and 100 lipids in the top and bottom leaflets, respectively, and was constructed and equilibrated with CHARMM-GUI’s protocols. After a production run of 445 ns, gA was manually inserted in the bilayer by removing eight lipids from each leaflet while maintaining the overall leaflet compositions. The system was hydrated with VMD’s solvate plugin, resulting in a bilayer patch with 96 lipids in the top leaflet, 92 lipids in the bottom leaflet, 1 gA dimer, and 55 waters per lipid.
All simulations were performed with the NAMD software (35) and the CHARMM36 force field for lipids (36, 37) in the NPT ensemble under constant pressure of 1 atmosphere and a temperature of 25°C. The force-field parameters for gA were kindly provided by Andrew Beaven and were based on those used in (38), made compatible with the CHARMM36 force field. Namely, the D-amino acids were treated in the same way as L-amino acids (except for their chirality), whereas the parameters for the gA terminal residues (formyl and ethanolamine) were the same as in (38) with the particular atom types renamed to conform to the CHARMM36 atom notation. For both the symmetric and asymmetric bilayers with gA, the system was first energy minimized for 104 steps and then run for 100 ps with a 1-fs time step. After this initial relaxation, the POPC/gA bilayer was simulated for 887 ns, and the asymmetric bilayer with gA was simulated for 705 ns with a 2-fs time step. Additional details of all simulated systems and the corresponding simulation parameters are found in Supporting Materials and Methods, Section S1.
Quantification of membrane deformation from simulations
To quantify the deformation of the membrane around gA, the trajectories were first centered and aligned on the backbone of gA. Because gA can tilt with respect to the bilayer normal in the course of the simulations, special care was taken to ensure that the alignment step did not result in abnormal tilting of the membrane, effectively leading to artificial changes in the bilayer thickness in the vicinity of the protein. The gA-lipid boundary was identified separately for each leaflet as the outermost layer of the time-averaged occupancy map (constructed on a 2 × 2 Å grid) of the respective gA monomer atoms projected onto the x-y plane. The leaflet thickness at the gA-lipid boundary was calculated from the interpolated z positions of the lipid atoms in the respective grid points as described in Supporting Materials and Methods, Section S2. The leaflet thickness at the gA-lipid boundary was used to obtain the optimal deformation profile around a gA monomer by a self-consistent free-energy minimization procedure as described in Supporting Materials and Methods, Section S3. The leaflet deformation around gA at distance r from the gA center was calculated as the squared deviation in the thickness averaged over the area between the grid points at distance r from the gA center and the gA-lipid boundary (the bulk leaflet thickness was taken from the corresponding lipid-only simulations). The membrane deformation was taken as the sum of the deformations of the two leaflets.
Quantification of membrane deformation in experiments
To estimate the amount of membrane deformation at each of the gA/lipid ratios used in the experiments, we first approximated the number NgA of gA dimers on a vesicle from the following: 1) the total surface area of a vesicle with diameter 130 nm (the average vesicle size measured with DLS), 2) the average area per lipid 64 Å2 (calculated from MD simulations for a POPC bilayer), and 3) the area per gA monomer 208 Å2 approximated from the occupancy map of the gA monomers described above. NgA gA dimers were then distributed uniformly on the surface of a sphere with diameter 130 nm using MATLAB (The MathWorks, Natick, MA), and the distance d between them was recorded. The membrane deformation at a given gA/lipid ratio was estimated as the membrane deformation at a distance d/2 from the gA center.
Results
gA incorporation is similar in symmetric and asymmetric liposomes
We modified a recently developed protocol for the construction of asymmetric lipid-only vesicles (28, 30) to enable the preparation of asymmetric proteoliposomes. As described in Materials and Methods, the protocol involves mixing two populations of symmetric vesicles (only one of which contains preincorporated protein) and exchanging the lipids between their outer leaflets with MβCD (Fig. S1). Using this procedure, we prepared two types of gA-containing vesicles: 1) compositionally symmetric but isotopically asymmetric liposomes (s∗LUVs) with an inner leaflet composed of POPC or its headgroup-perdeuterated variant (POPC-d13) and an outer leaflet exchanged with its chain-perdeuterated variant (POPC-d31); and 2) compositionally asymmetric vesicles (aLUV) with the same inner leaflet composition as above and an outer leaflet exchanged with the chain-perdeuterated variant of DMPC (DMPC-d54). Table S1 shows the donor mole fraction in the final samples as determined from GC/MS (see Materials and Methods), which ranged between 0.32 and 0.4 for the s∗LUVs and between 0.35 and 0.45 for the aLUVs. The size of the vesicles was measured with DLS and was on average 130 nm in diameter with polydispersity index of <0.2 (Table S2).
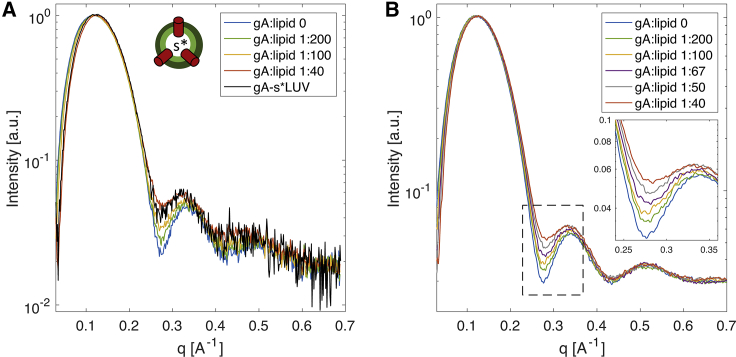
We used SAXS to examine the effect of gA incorporation on bilayer structure. Fig. 1 A shows scattering intensity versus momentum transfer vector q for POPC LUVs with an increasing concentration of gA from 0 to 2.5 mol%. The curves exhibit the typical lobes of coherent scattering separated by distinct minima that is characteristic of the core-shell structure of lipid bilayer vesicles (39). The most notable feature is a nearly linear increase in the scattering intensity near the minimum at q ∼0.28 Å−1 with increasing gA concentration. To better understand the origin of this liftoff, we used Monte Carlo simulations to calculate SAXS curves corresponding to vesicles with randomly dispersed gA TM dimers (see Supporting Materials and Methods, Section S4 for additional details of simulation methods and results). Simulated SAXS curves for vesicles with increasing gA concentration, shown in Fig. 1 B, precisely reproduced the enhanced scattering, conclusively demonstrating that this feature arises from an electron density contrast between the lipid bilayer and a TM protein. We also simulated the effect of gA aggregation within the bilayer. Fig. S11 reveals that lateral association of gA channels rapidly diminishes the liftoff, with the largest effect occurring upon lateral dimerization. Aggregates of three or more gA channels largely abrogate the liftoff, resulting in SAXS curves that are practically indistinguishable from a gA-free vesicle. A comparison of the experimental SAXS data from the gA-s∗LUVs to curves from the symmetric gA concentration series (Fig. 1 A) suggests that little if any gA was lost from acceptor vesicles during CD-mediated lipid exchange and that gA is not laterally aggregated within the vesicles.
Figure 1.
Gramicidin incorporation is similar in symmetric and asymmetric liposomes. (A) Experimental SAXS form factors of a series of POPC gA-sLUVs with an increasing concentration of gA and isotopically asymmetric LUVs, gA-s∗LUV, composed of deuterated variants of POPC and prepared with a nominal gA/lipid ratio of 1:40. The cartoon schematic next to the figure legend represents an isotopically asymmetric gA-s∗LUV. All measurements were performed at 25°C. (B) SAXS form factors calculated from Monte Carlo simulations of a POPC vesicle with different concentrations of gA (see Supporting Materials and Methods). The increase in the intensity at the minimum between the first and second scattering lobes with increasing gA concentration (shown in an expanded view in the inset) is caused by the electron density contrast between the lipid bilayer and the membrane-spanning gA dimers. To see this figure in color, go online.
Although our initial goal was to use SAXS to look for changes in bilayer structure induced by gA, we did not observe any significant shift in the positions of the scattering minima that typically accompany changes in bilayer thickness. We conducted additional Monte Carlo simulations to determine the sensitivity of SAXS to perturbations in the first shell of ∼14 lipids surrounding each gA channel. Fig. S12 shows only minor differences in SAXS curves upon changing the area per lipid of the first shell by 10 Å2 at a gA concentration of 2.5 mol%. Indeed, no significant change at all is seen for a perturbation from 64 to 70 Å2, which would correspond to a ∼3.5 Å thinning of the surrounding bilayer induced by gA. These simulations suggest that, at least for the experimental conditions used here, it is not possible to draw conclusions from SAXS data about small local bilayer perturbations induced by gA.
gA conformation and function are the same in symmetric and asymmetric liposomes
Next, we examined the structural properties of the incorporated gA. In addition to its canonical helical dimer conformation, gA can exist in other nonfunctional conformations, such as a dimeric helix in which two gA monomers are intertwined into a single extended helix (40). The dimeric helix conformation can arise from a hydrophobic mismatch (or from storing gA in some nonpolar solvents) and has a distinct CD spectrum (41). Fig. S3 shows the CD spectrum of gA in the aLUVs in comparison with the spectra of gA in sLUVs made of either just POPC or DMPCd54. The data show that most of the 2.5 mol% gA in the symmetric samples was in a helical dimer conformation and that this conformation remained unchanged during the steps of the gA-aLUV preparation protocol.
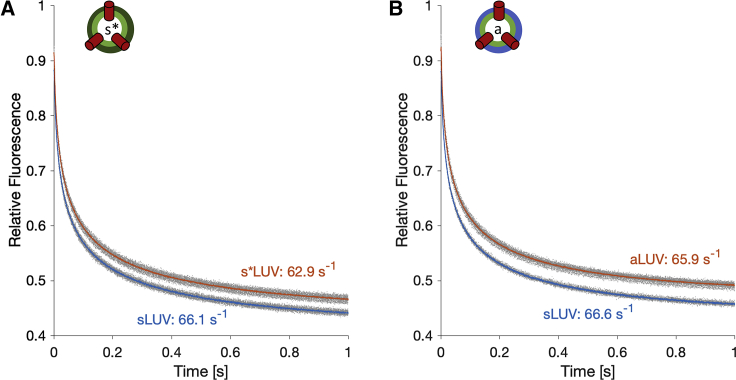
To evaluate the function of gA in the asymmetric liposomes, we measured the rate at which a fluorophore (ANTS) trapped inside the vesicles was quenched by an externally added quencher (thallium, Tl+). This assay provides a measure of the equilibrium dimer-to-monomer ratio of gA in the bilayer by taking advantage of the fact that Tl+ can enter the vesicles and quench ANTS only through functional gA dimers (29). Fig. 2 A shows the ANTS fluorescence decay as a function of time in the initially symmetric acceptor vesicles (66.1 s−1) and the final isotopically asymmetric gA-s∗LUVs (62.9 s−1). The ratio of the two rates (0.95) indicates that the dimer/monomer equilibrium of gA in the isotopically asymmetric vesicles was essentially unaffected by the cyclodextrin-mediated lipid exchange. We obtained a similar ratio of 0.99 for the compositionally asymmetric liposomes (Fig. 2 B), confirming that in both asymmetric samples, gA function remained intact.
Figure 2.
Gramicidin channel function remains intact in asymmetric liposomes. Changes in ANTS fluorescence over time for the isotopically (A) and compositionally (B) asymmetric samples (in red) and their corresponding symmetric acceptor vesicles (in blue). Replicate traces are shown in gray; their averages are shown in color. The fluorescence signal was normalized to the fluorescence measured before the addition of TI+. The average rates calculated from the traces are indicated next to each trace. The cartoon schematic in (B) represents a compositionally asymmetric gA-aLUV. All measurements were performed at an ambient temperature of ∼22°C. To see this figure in color, go online.
gA scrambles lipids in compositionally asymmetric vesicles
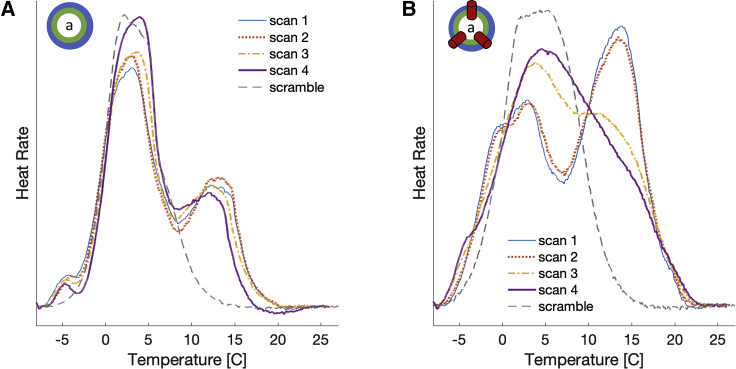
That gA can scramble lipids first became evident in DSC experiments, in which thermodynamic phase transitions (such as transitions between gel and fluid) are detected by monitoring the release of heat from a sample as a function of temperature. Fig. 3 shows a cooling temperature scan performed after sample preparation (scan 1) for compositionally asymmetric vesicles without (Fig. 3 A) and with (Fig. 3 B) gA (see Materials and Methods). Two transition peaks are visible in both samples, likely coming from the different melting temperatures of the POPC-rich inner leaflet and the DMPC-d54-rich outer leaflet (the main transition temperatures of POPC and DMPC-d54 are −2 and 19°C, respectively; see Fig. S4). After the first scan, the temperature was again brought to 30°C, and a second cooling scan (scan 2) was performed without any observable changes in the DSC spectra, indicating that cycling through the temperature range of −8 to 30°C (and consequently, through the gel-liquid crystalline transition of each leaflet) by itself did not result in any major redistribution of the lipids between the two leaflets. In the gA-aLUV sample, however, after a set of fixed temperature incubations (20°C for 12 h, followed by 45°C for 5 h), the two peaks began to merge (scan 3, Fig. 3 B), whereas those of the aLUV sample without gA remained unchanged (scan 3, Fig. 3 A). The changes in the gA-aLUV sample were even more pronounced after subjecting the samples to another set of fixed temperature incubations (scan 4). These results indicate that gA facilitated the exchange of lipids between the two leaflets, presumably through its ability to accelerate lipid flip-flop, which eventually would result in a symmetric bilayer with a single transition temperature peak (gray dashed lines in Fig. 3).
Figure 3.
Gramicidin scrambles lipids in compositionally asymmetric vesicles. DSC data of compositionally asymmetric LUVs with DMPC-d54 and POPC and without (A) or with (B) gramicidin at gA/lipid ratio of 1:40 are shown. Four consecutive cooling scans were performed as follows: after the asymmetric vesicle preparation (scan 1, thin blue line), after scan 1 (scan 2, dotted red line), after subsequent incubation at 20°C for 12 h followed by incubation at 45°C for 5 h (scan 3, dash-dotted yellow line), and after another set of incubations at 20°C for 12 h and 45°C for 5 h (scan 4, thick purple line). Also shown for comparison with gray dashed lines are data for the symmetric samples (scramble) with the same overall composition as the asymmetric vesicles (Table S1). To see this figure in color, go online.
gA increases the rate of lipid flip-flop in compositionally asymmetric vesicles
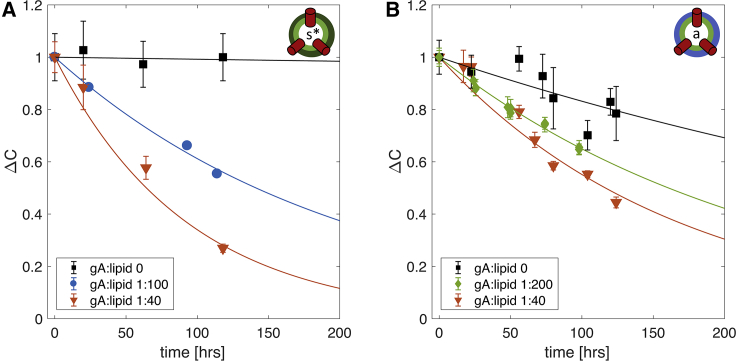
To quantify the effect of gA on lipid flip-flop, we used 1H-NMR to measure the rate of lipid translocation in the presence and the absence of the channel (see Materials and Methods). The nine equivalent protons on a protiated choline headgroup produce a clearly defined resonance peak at 3.1–3.6 ppm. The shift reagent Pr3+, when added externally to the sample, interacts only with the lipid headgroups exposed on the outer leaflet of the vesicles and shifts their resonance downfield (26, 42). (We further confirmed that Pr3+ does not alter bilayer properties using the fluorescence quenching assay mentioned above; see Table S3.) After exchanging lipids with protiated choline headgroups (POPC-d31 or DMPC-d54) into acceptor vesicles composed of the headgroup-deuterated POPC-d13, the only detectable resonance comes from the exchanged donor lipids that are initially in the outer leaflet. The distribution of the protiated-headgroup lipids across the two bilayer leaflets thus can be determined from the ratio of the areas of the shifted and unshifted choline peaks in the NMR spectrum. Repeating the shift experiment on different aliquots of the sample over time allows for direct quantification of the lipid flip-flop rate (26). To minimize the exposure of the LUVs to Pr3+, the sample was incubated at room temperature in the absence of Pr3+; the shift reagent was added to a sample aliquot immediately before the NMR measurement, and the aliquot containing Pr3+ was discarded after measurement as described in Materials and Methods. The main transition temperature TM of DMPC-d54 (the DMPC variant used in these experiments) is less than 20°C (see Fig. S5) in contrast to fully protiated DMPC (TM = 24°C). As a result, the asymmetric samples both with and without gA were fully in the fluid phase at room temperature (Fig. 3).
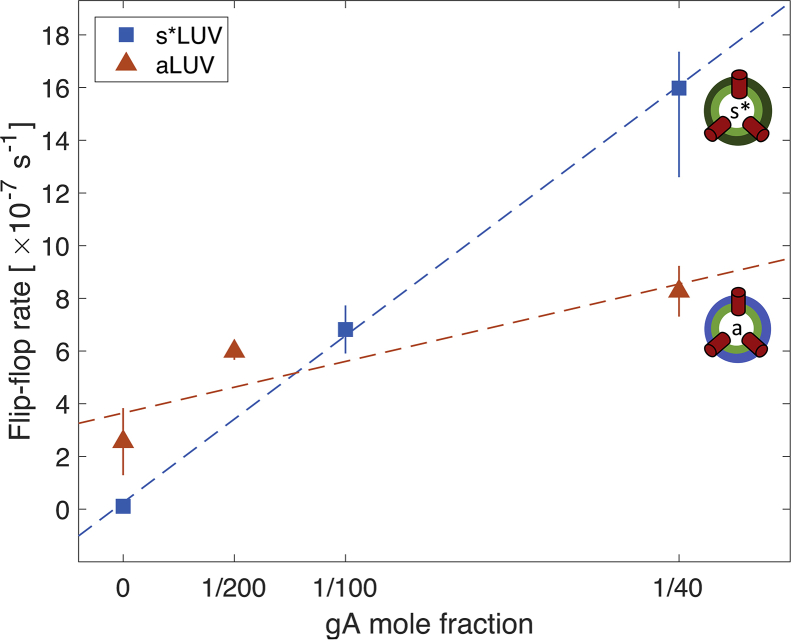
Fig. 4 shows the relative changes in transverse lipid distribution ΔC (see Eq. 2) as a function of time for both the chemically symmetric (Fig. 4 A) and asymmetric (Fig. 4 B) liposomes with different amounts of gA (see Materials and Methods). Table 2 lists the corresponding lipid flip-flop rates and half times calculated as described in Materials and Methods. Whereas the spontaneous lipid flip-flop rate in the absence of gA is immeasurably slow in the s∗LUVs, it is markedly increased when DMPC is exchanged into the outer leaflet of POPC (Fig. 4, A and B, black squares and curves). Importantly, the rate of lipid translocation in both samples increases in the presence of the channel in a manner that depends on gA concentration.
Figure 4.
Gramicidin increases the rate of lipid flip-flop in isotopically and compositionally asymmetric vesicles. The time evolution of the interleaflet distribution of POPC-d31 in s∗LUVs (A) and DMPC-d54 in aLUVs (B) either without gA (black squares) or with gA at a gA/lipid ratio of 1:40 (red triangles), 1:100 (blue circles), and 1:200 (green diamonds). Both plots show the time-dependent changes in the distribution of POPC-31 between the outer and inner leaflets, relative to the first time point measured after vesicle preparation (ΔC). See text for details. Error bars represent SDs of at least three consecutive Pr3+ additions. Each time trace in (A) is from a single sample. The time traces of the compositionally asymmetric vesicles in (B) are from one (gA/lipid 1:40), two (no gA), and three (gA/lipid 1:200) separately prepared samples, respectively. The kinetics reported here corresponds to sample behavior at an ambient temperature of ∼22°C. To see this figure in color, go online.
Table 2.
Translocation Kinetics in the Examined Systems
| Outer Leaflet | gA/lipid | t1/2 [h] | kf/10−7 s−1 |
|---|---|---|---|
| POPC-d31/POPC-d13 | 0 | ∼8900 | ∼0.1 |
| 1:100 | 141 [125; 163] | 6.8 [5.9; 7.7] | |
| 1:40 | 64 (55; 76) | 16.0 [12.6; 17.4] | |
| DMPC-d54/POPC-d13 | 0 | 376 [251; 748] | 2.6 [1.3; 3.8] |
| 1:200 | 160 [153; 168] | 6.0 [5.7; 6.3] | |
| 1:40 | 116 [104; 132] | 8.3 [7.3; 9.2] |
Shown are the corresponding half times and the rates of lipid flip-flop calculated from the NMR data shown in Fig. 4 as described in Materials and Methods. The numbers in brackets represent 95% confidence intervals.
Models for gA-mediated lipid flip-flop
If each additional gA dimer accelerated the movement of a constant, fixed number of lipids between the leaflets, the rate of lipid flip-flop would vary linearly with gA mole fraction (20). Fig. 5 shows the flip-flop rates calculated from the NMR data (Table 2) as a function of the gA/lipid ratio. Indeed, the data for the s∗LUV samples (shown in blue) are consistent with the model of single gA channel-mediated lipid translocation. However, in the aLUV samples (shown in red), the linear relationship predicted from the model is not as clear, and the uncertainty in the data precludes any firm conclusions. Considered together, the two data sets introduce a conundrum: in the absence of gA, the flip-flop rate in the compositionally asymmetric bilayers is clearly faster than the rate in the symmetric membranes, yet in the presence of gA at a gA/lipid ratio of 1:40, this trend is reversed and the flip-flop kinetics in the aLUVs are much slower than those in the s∗LUVs. This result suggests a mechanism different from a single channel-induced perturbation, by which gA catalyzes lipid translocation, based on the following considerations: 1) because the gA channel is relatively short, it is likely to cause thickness deformations in the membrane; and 2) the thickness deformations in a POPC bilayer could be alleviated in the presence of a shorter-chain lipid like DMPC. Thus, gA-induced bilayer stress is likely a contributing factor to the observed trends.
Figure 5.
The rate of lipid flip-flop shows a complex relationship with gA mole fraction. Flip-flop rates were calculated from the data in Fig. 4 for the compositionally symmetric (blue squares) and asymmetric (red triangles) LUVs (see Table 2) as a function of the nominal gA mole fraction in the samples. Error bars represent 95% confidence intervals. To see this figure in color, go online.
Membrane deformations are likely to play a role in gA-mediated lipid flip-flop
gA has been routinely used as a model peptide to study local bilayer deformations that result from protein-membrane interactions (43, 44). In this context, it is important that the gA channel’s helical pitch is invariant with respect to changes in the channel-bilayer hydrophobic mismatch, meaning that the bilayer will adjust to the channel rather than the channel adjusting to the bilayer (in bilayers having a hydrophobic thickness greater than the channel length) (45). gA has been shown to induce thinning in a pure DMPC bilayer (46), and one would expect the same effect in bilayers with a hydrophobic thickness greater than DMPC, such as a membrane composed of POPC (47). Such local deformations produce stress on the membrane that will increase with an increasing gA concentration and could potentially affect lipid flip-flop. We therefore investigated the response of the bilayer to the presence of a single gA channel in the two experimentally studied systems.
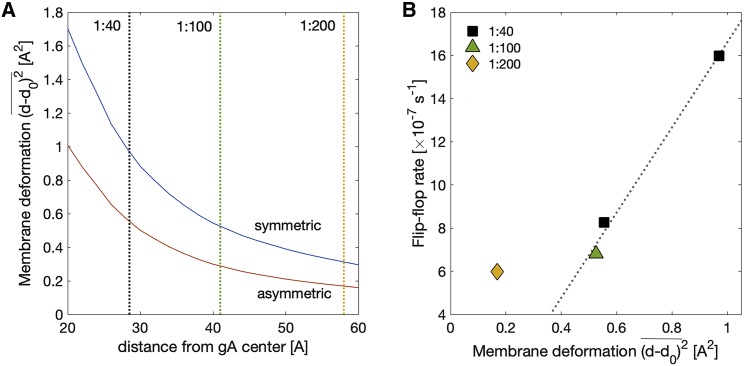
MD simulations, in combination with the continuum theory of membrane deformations (CTMD) (48), were used to estimate the amount of membrane deformation at the different gA mole fractions from the experiments. This was achieved by first quantifying the changes in membrane thickness as a function of distance from the channel and then relating them to the average distances between channels in the samples. To enable these calculations, MD simulations were performed on a system containing a single gA channel embedded in a symmetric POPC bilayer or in an asymmetric membrane with a top leaflet of DMPC/POPC 0.75/0.25 and a bottom leaflet of DMPC/POPC 0.10/0.90. The compositions of the two leaflets were based on a different set of samples prepared for small-angle neutron scattering experiments (unpublished data) and were similar to the leaflet compositions of the samples examined with NMR in this work (Table S1). At the end of the simulations, the thickness of each leaflet at the gA-lipid boundary was analyzed and used to calculate the optimal thickness deformation profile around gA using the CTMD formalism as described in Materials and Methods and Supporting Materials and Methods, Section S3. Because the membrane deformation energy varies with the square of the hydrophobic mismatch (Eq. S3), Fig. 6 A shows the resulting average squared deviations in thickness as a function of distance from the gA center in both systems. The amount of membrane deformation decreases gradually further away from the protein in the two bilayers, and it is less in the asymmetric membrane, indicating that gA is able to adapt more easily to the asymmetric bilayer environment.
Figure 6.
The gA-mediated lipid flip-flop rate at high gA concentrations correlates with membrane deformation. (A) Average squared deviations in bilayer thickness as a function of distance from gA center for a symmetric POPC bilayer and an asymmetric bilayer as in the NMR experiments in Fig. 4. The thickness deviations were calculated from the membrane deformation profiles around a single gA channel obtained by a CTMD-guided free-energy minimization utilizing information from MD simulations (see Materials and Methods). Dotted lines indicate the approximated half distances between gA channels on the surface of the LUVs at three different gA/lipid ratios: 1:40 (black), 1:100 (green), and 1:200 (yellow). (B) gA-mediated lipid flip-flop rate from Fig. 5 as a function of the corresponding membrane deformation from (A). The data points denote different gA/lipid mole ratios: 1:40 (black square), 1:100 (green triangle), and 1:200 (yellow diamond). To see this figure in color, go online.
To examine whether membrane deformations could be involved in the ability of gA to scramble lipids, we approximated the packing density of gA dimers within the experimentally prepared LUVs. We estimated the distance between the channels at each gA mole fraction by assuming that they are uniformly distributed on the surface of the vesicles (see Materials and Methods). The dotted lines on Fig. 6 A indicate the respective half distances between the gA centers. The compositionally symmetric sample with gA at a gA/lipid ratio of 1:40, in which the rate of lipid flip-flop was highest (Table 2), also seemed to have the largest membrane deformation (as determined from the MD simulations) among the examined asymmetric proteoliposomes. Fig. 6 B shows the relationship between the measured rates of lipid flip-flop in the four gA-aLUVs and the corresponding amounts of membrane deformation as quantified by the analysis in Fig. 6 A. Whereas the correlation between gA-mediated lipid scrambling and membrane deformation at gA mole fractions ≥0.01 is very strong (0.998), the sample with a gA/lipid ratio of 1:200 did not follow the same relationship and appeared as an outlier (Fig. 6 B). This result suggests that the stress induced in the membrane as a result of gA-membrane interactions is an important contributor to the observed effects of gA on lipid scrambling and that at high mole fractions, the channels accelerate lipid flip-flop in a mechanistically different way than at low gA concentrations. We note that the same qualitative results for the gA-induced flip-flop rate and membrane deformation are obtained, even assuming a 20% loss of gA during the preparation of asymmetric vesicles (an upper limit estimated from the SAXS data).
Discussion
In studies of protein-membrane interactions, the effects of the membrane (e.g., its composition and structure) on protein function and oligomerization have been examined extensively. It is equally important, however, to account for the protein’s effect on the membrane because a protein embedded in a bilayer is prone to introduce defects that can perturb the bilayer structure and affect its transverse and lateral organization. We have, therefore, developed, to our knowledge, new protocols for the systematic investigation of protein-mediated changes in the lipid compositions of the two bilayer leaflets by utilizing model systems that allow for fine tuning of various membrane parameters. As discussed below, these protocols have been devised to exploit biophysical properties that bring to light specific elements of the complex interplay between the protein and the membrane and to quantify the consequences.
In the following, we first discuss the preparation and properties of the gA-containing asymmetric lipid vesicles; we then turn to the effect of gA on lipid flip-flop; and, finally, we present a possible mechanism for the gA-induced lipid scrambling observed in the experiments.
Preparation and biophysical characterization of asymmetric proteoliposomes
Asymmetric membranes containing TM proteins have been successfully prepared in the past using two general approaches. In one, the protein (either soluble or micelle stabilized) is added externally to preformed asymmetric membranes, including LUVs filled with sucrose (49), droplet interface bilayers (50), and planar supported bilayers (51). In the other (also used here), the protein is first reconstituted into symmetric vesicles, after which the outer leaflet lipids are exchanged with different lipids. For example, using the latter approach, Vitrac et al. prepared sonicated small unilamellar vesicles (SUVs) with the 12 TM protein LacY from Escherichia coli and examined the effect of asymmetric charge distribution on the topology of the protein (4, 52). In a different study, asymmetric SUVs containing the nicotinic acetylcholine receptor were prepared by using MβCD-loaded lipid complexes to exchange the lipids on the outer leaflet of symmetric SUVs containing nicotinic acetylcholine receptor with sphingomyelin (3). Although these examples illustrate how a variety of TM proteins can be incorporated into asymmetric model membranes of different geometries, the effect of the protein (and protein incorporation) on the lipid compositions of the two bilayer leaflets can vary and therefore has to be examined explicitly.
To this end, the protocols and assays presented here offer an advantageous platform for biophysical characterization of the sample, much improved by utilizing large tensionless proteoliposomes that are free from the potential effects of curvature, residual organic solvent, or polymer cushion supports. Indeed, we found only minimal effects of the preparation protocol on the incorporation, conformation, and function of gA (Figs. 1, 2, and S3). Still, these effects are likely to depend on the types of lipids in the vesicles, and their negligible magnitude cannot be taken for granted. For example, the energetic cost of gA dimer formation increases with increasing bilayer thickness, resulting in a shift of the gA monomer/dimer equilibrium toward the monomeric state (41, 53); in such cases, the gA monomers have been shown to more easily exchange between vesicles (54). This could result in a partial loss of the protein during the CD-mediated lipid exchange between donor and acceptor vesicles. Furthermore, a preference of a gA monomer for the composition of one leaflet versus the other in the gA-aLUVs may require additional tests of the interleaflet gA localization and vesicle stability. The thermodynamic phase properties of the bilayer in the presence of gA (whether in dimeric or monomeric state) should also be considered. For example, we found that gA broadens and slightly lowers the main transition temperature of DMPC-d54 (Fig. S5), similar to previous reports for DMPC (55). This effect is likely to depend on the channel concentration in the membrane as well as the bilayer lipid composition and is likely to differ among proteins. A gel environment, for example, could affect not only the protein dynamics but also the efficiency of lipid exchange during the aLUV preparation protocol (see (28)).
Rates of lipid flip-flop measured with NMR
The slow rate of spontaneous lipid flip-flop that we measured in the chemically symmetric vesicles in the absence of gA is consistent with previous reports (14, 56). Flip-flop kinetics in the compositionally asymmetric membranes, however, was significantly faster (Fig. 5). Because both POPC and DMPC have phosphocholine headgroups, this increase is likely due to differences in the chain lengths of the lipids (16 and 18 carbons for POPC and 14 carbons for DMPC) and the corresponding leaflet thicknesses (Table S4). This explanation is consistent with results from small-angle neutron scattering experiments showing a faster flip-flop rate in DMPC LUVs compared to POPC LUVs (56) and the corresponding free energy for flip-flop in the two bilayers, quantified with MD simulations (57).
Our results for the gA-mediated half time of lipid translocation in the POPC liposomes (t1/2 ≈ 64 h at 22°C and a gA/lipid ratio of 1:40) are comparable to those of Fattal et al. who used the chain-labeled fluorescent lipid analog 1-oleoyl-2-[12-[(7-nitro-1,2,3-benzoxadiazol-4-yl)amino]dodecanoyl] phosphatidylcholine (NBD-PC) to determine a half time t1/2 > 96 h in POPC vesicles at room temperature and a gA/lipid ratio of 1:30 (14). This similarity is surprising in light of the different chemical structures of the acyl chains of NBD-PC relative to POPC and suggests that—in agreement with earlier experimental observations (58)—the nature of the lipid headgroup is the dominant factor in determining the flip-flop rate. Yet, the chain structure also has been shown to strongly affect flip-flop for lipids with multiple double bonds (59). In contrast, the reported kinetics of lipid translocation in the presence of gA in erythrocytes (24) and supported lipid bilayers (25) are much faster, likely because of the specific experimental conditions in the studies as mentioned in the Introduction.
The lipid flip-flop rates accessible to NMR measurements are limited by the time needed to perform a single experiment with the shift reagent. Depending on the type of instrument (i.e., the strength of the magnet) and sample concentration, one measurement in the absence of shift reagent, followed by 3 Pr3+ titrations (needed to obtain error bars, for example), could take anywhere from 15 min to 1 h. Thus, this technique can be used to measure only slower time-dependent translocation processes. However, because the lipid flip-flop rate would be expected to increase with increasing temperatures, systems with faster rates can be compared if incubated at lower temperatures (see also (60)). All samples in this study were incubated on the benchtop at an ambient temperature of ∼22°C.
Mechanisms of gA-induced lipid scrambling
It has previously been proposed that the ability of gA to scramble lipid analogs is not due to bilayer perturbations induced by individual gA channels because of the following (23, 24): 1) gA increases the translocation rate of lysophosphatidylcholine only at concentrations much higher than those in which gA performed its normal function as a conducting channel, but in which gA produces nonspecific solute leakage; and 2) formylation of gA’s tryptophan residues, which prevents the clustering of gA, abolished the gA-catalyzed lyso-lipid scrambling. Instead, it was proposed that gA at gA/lipid mole fractions of 1:2000 or higher forms some sort of lateral aggregate(s) in the erythrocyte membrane that may be intermediates for the formation of hexagonal phases. The proposed clusters would induce transient defects in the bilayer with a subsequent formation of aqueous leaks that allow for the passage of molecules as large as choline and sucrose across the cell membrane as well as the translocation of lipid analogs (24). The proposed formation of gA clusters would depend on channel-bilayer hydrophobic mismatch because gA did not cause detectable aggregates in DMPC bilayers, in which there is minimal channel-bilayer hydrophobic mismatch (46) and little accumulation of stress in the membrane.
The lamellar SAXS form factors of POPC liposomes used in this study (Fig. 1 A) definitively exclude the presence of nonlamellar phases known to form at high gA concentrations under different experimental conditions (19). We can also exclude changes in the lateral association of gA in the asymmetric liposomes because aggregation to even as few as a 2-mer (two gA dimers) is expected to abrogate the liftoff effect of the gA 1-mer in the SAXS form factor, which is not seen in the experimental data (see Fig. 1; Supporting Materials and Methods). We further observe that gA broadens, but does not shift, the main transition temperature of the aLUVs (Fig. S6 A) as well as the sLUVs made of a binary mixture of DMPC-d54 and POPC (Fig. S6 B). According to the model in (55), the lack of a shift is consistent with the gA exhibiting ideal mixing in both phases, though nonideal mixing would be expected at larger channel-bilayer hydrophobic mismatches.
In this context, our NMR and computational analysis further illuminates the importance of the frustration energy in the bilayer: at high channel densities, the bilayer thickness is not able to relax to its unperturbed state between the channels (Fig. 6), leading to bilayer-deformation-induced stress. This stress would increase the probability of transient clusters of bilayer-spanning gA channels (61, 62, 63), which could serve to decrease the energy barrier for lipid translocation and thereby increase the lipid flip-flop rate (64). The deviation of the gA-aLUVs at a gA/lipid ratio of 1:200 from the other samples in Fig. 6 B further suggests that at this lower gA mole fraction, the role of the frustration energy is different, resulting in a mechanistically distinct route for the observed gA effect on lipid flip-flop.
Conclusion
Applying, to our knowledge, new methodologies and protocols for preparing and characterizing asymmetric proteoliposomes to the system of gA channels, we have demonstrated the ability of gA to accelerate interleaflet lipid translocation in both chemically symmetric and asymmetric membranes. The mechanistic analysis of the results shows that the channel-induced bilayer deformation likely contributes to the rate of lipid flip-flop. The ability to characterize and quantify the interplay between TM proteins and their solvating lipid environment allows us to determine the properties of the protein-laden bilayers. If such properties are considered properly (e.g., with the type of methodology illustrated here), the mechanistic understanding of much more complex biomimetic systems becomes feasible and practical.
Author Contributions
M.D., F.A.H., H.W., G.W.F., and O.S.A. designed the research. M.D., F.A.H., R.R., R.L.S., and T.A.P. performed the experiments. M.D., F.A.H., D.M., and T.A.P. analyzed the experimental data. M.D. performed all MD simulations and computational analysis. M.D., F.A.H., D.M., J.K., G.W.F., H.W., and O.S.A. wrote the manuscript.
Acknowledgments
We thank Haden Scott for technical assistance with CD measurements, Andrew Beaven for providing the MD force-field parameters for gA, and W. Clay Bracken for help with identifying the water suppression conditions for the NMR measurements at Weill Cornell Medical College.
This work was supported by National Science Foundation grant MCB-1817929 (to F.A.H.), National Institutes of Health grants P01 DA012408 (to H.W.) and R01 GM021342 (to O.S.A.), and by a grant from Natural Sciences and Engineering Research Council of Canada (funding reference number 2018-04841) (to D.M.). GC/MS, DSC, and DLS measurements were supported by the Biophysical Characterization Laboratory suite of the Shull Wollan Center at Oak Ridge National Laboratory. SAXS measurements were supported by Department of Energy scientific user facilities at Oak Ridge National Laboratory.
Editor: Charles Deber.
Footnotes
Milka Doktorova's present address is Department of Integrative Biology and Pharmacology, University of Texas Health Science Center, Houston, Texas.
Supporting Materials and Methods, 12 figures, and six tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30051-7.
Supporting Material
References
- 1.Collins M.D., Keller S.L. Tuning lipid mixtures to induce or suppress domain formation across leaflets of unsupported asymmetric bilayers. Proc. Natl. Acad. Sci. USA. 2008;105:124–128. doi: 10.1073/pnas.0702970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiessling V., Wan C., Tamm L.K. Domain coupling in asymmetric lipid bilayers. Biochim. Biophys. Acta. 2009;1788:64–71. doi: 10.1016/j.bbamem.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perillo V.L., Peñalva D.A., Antollini S.S. Transbilayer asymmetry and sphingomyelin composition modulate the preferential membrane partitioning of the nicotinic acetylcholine receptor in Lo domains. Arch. Biochem. Biophys. 2016;591:76–86. doi: 10.1016/j.abb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Vitrac H., MacLean D.M., Dowhan W. Dynamic membrane protein topological switching upon changes in phospholipid environment. Proc. Natl. Acad. Sci. USA. 2015;112:13874–13879. doi: 10.1073/pnas.1512994112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlmutter J.D., Sachs J.N. Interleaflet interaction and asymmetry in phase separated lipid bilayers: molecular dynamics simulations. J. Am. Chem. Soc. 2011;133:6563–6577. doi: 10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- 6.St. Clair J.R., Wang Q., London E. Preparation and physical properties of asymmetric model membrane vesicles. In: Epand R., Ruysschaert J.M., editors. The Biophysics of Cell Membranes. Springer; 2017. pp. 1–27. [Google Scholar]
- 7.Markones M., Drechsler C., Fiedler S. Engineering asymmetric lipid vesicles: accurate and convenient control of the outer leaflet lipid composition. Langmuir. 2018;34:1999–2005. doi: 10.1021/acs.langmuir.7b03189. [DOI] [PubMed] [Google Scholar]
- 8.Takaoka R., Kurosaki H., Nakano M. Formation of asymmetric vesicles via phospholipase D-mediated transphosphatidylation. Biochim. Biophys. Acta. Biomembr. 2018;1860:245–249. doi: 10.1016/j.bbamem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Contreras F.X., Sánchez-Magraner L., Goñi F.M. Transbilayer (flip-flop) lipid motion and lipid scrambling in membranes. FEBS Lett. 2010;584:1779–1786. doi: 10.1016/j.febslet.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 10.Sperotto M.M., Ferrarini A. Spontaneous lipid flip-flop in membranes: a still unsettled picture from experiments and simulations. In: Epand R., Ruysschaert J.M., editors. The Biophysics of Cell Membranes. Springer; 2017. pp. 29–60. [Google Scholar]
- 11.Pomorski T.G., Menon A.K. Lipid somersaults: uncovering the mechanisms of protein-mediated lipid flipping. Prog. Lipid Res. 2016;64:69–84. doi: 10.1016/j.plipres.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebastian T.T., Baldridge R.D., Graham T.R. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta. 2012;1821:1068–1077. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aye I.L., Singh A.T., Keelan J.A. Transport of lipids by ABC proteins: interactions and implications for cellular toxicity, viability and function. Chem. Biol. Interact. 2009;180:327–339. doi: 10.1016/j.cbi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Fattal E., Nir S., Szoka F.C., Jr. Pore-forming peptides induce rapid phospholipid flip-flop in membranes. Biochemistry. 1994;33:6721–6731. doi: 10.1021/bi00187a044. [DOI] [PubMed] [Google Scholar]
- 15.Tieleman D.P., Marrink S.J. Lipids out of equilibrium: energetics of desorption and pore mediated flip-flop. J. Am. Chem. Soc. 2006;128:12462–12467. doi: 10.1021/ja0624321. [DOI] [PubMed] [Google Scholar]
- 16.Ernst O.P., Menon A.K. Phospholipid scrambling by rhodopsin. Photochem. Photobiol. Sci. 2015;14:1922–1931. doi: 10.1039/c5pp00195a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen O.S., Koeppe R.E., II Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 18.de Kruijff B., van Zoelen E.J., van Deenen L.L. Glycophorin facilitates the transbilayer movement of phosphatidylcholine in vesicles. Biochim. Biophys. Acta. 1978;509:537–542. doi: 10.1016/0005-2736(78)90246-8. [DOI] [PubMed] [Google Scholar]
- 19.Tournois H., Leunissen-Bijvelt J., de Kruijff B. Gramicidin-induced hexagonal HII phase formation in erythrocyte membranes. Biochemistry. 1987;26:6613–6621. doi: 10.1021/bi00395a008. [DOI] [PubMed] [Google Scholar]
- 20.Kol M.A., de Kroon A.I., de Kruijff B. Membrane-spanning peptides induce phospholipid flop: a model for phospholipid translocation across the inner membrane of E. coli. Biochemistry. 2001;40:10500–10506. doi: 10.1021/bi010627+. [DOI] [PubMed] [Google Scholar]
- 21.Kol M.A., van Laak A.N., de Kruijff B. Phospholipid flop induced by transmembrane peptides in model membranes is modulated by lipid composition. Biochemistry. 2003;42:231–237. doi: 10.1021/bi0268403. [DOI] [PubMed] [Google Scholar]
- 22.Anglin T.C., Brown K.L., Conboy J.C. Phospholipid flip-flop modulated by transmembrane peptides WALP and melittin. J. Struct. Biol. 2009;168:37–52. doi: 10.1016/j.jsb.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killian J.A. Gramicidin and gramicidin-lipid interactions. Biochim. Biophys. Acta. 1992;1113:391–425. doi: 10.1016/0304-4157(92)90008-x. [DOI] [PubMed] [Google Scholar]
- 24.Classen J., Haest C.W., Deuticke B. Gramicidin-induced enhancement of transbilayer reorientation of lipids in the erythrocyte membrane. Biochemistry. 1987;26:6604–6612. doi: 10.1021/bi00395a007. [DOI] [PubMed] [Google Scholar]
- 25.Anglin T.C., Liu J., Conboy J.C. Facile lipid flip-flop in a phospholipid bilayer induced by gramicidin A measured by sum-frequency vibrational spectroscopy. Biophys. J. 2007;92:L01–L03. doi: 10.1529/biophysj.106.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquardt D., Heberle F.A., Pabst G. 1H NMR shows slow phospholipid flip-flop in gel and fluid bilayers. Langmuir. 2017;33:3731–3741. doi: 10.1021/acs.langmuir.6b04485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kucerka N., Pencer J., Katsaras J. Curvature effect on the structure of phospholipid bilayers. Langmuir. 2007;23:1292–1299. doi: 10.1021/la062455t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doktorova M., Heberle F.A., Marquardt D. Preparation of asymmetric phospholipid vesicles for use as cell membrane models. Nat. Protoc. 2018;13:2086–2101. doi: 10.1038/s41596-018-0033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingólfsson H.I., Andersen O.S. Screening for small molecules’ bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay Drug Dev. Technol. 2010;8:427–436. doi: 10.1089/adt.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heberle F.A., Marquardt D., Pabst G. Subnanometer structure of an asymmetric model membrane: interleaflet coupling influences domain properties. Langmuir. 2016;32:5195–5200. doi: 10.1021/acs.langmuir.5b04562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 32.Jo S., Lim J.B., Im W. CHARMM-GUI membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doktorova M., Weinstein H. Accurate in silico modeling of asymmetric bilayers based on biophysical principles. Biophys. J. 2018;115:1638–1643. doi: 10.1016/j.bpj.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klauda J.B., Monje V., Im W. Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B. 2012;116:9424–9431. doi: 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- 37.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaven A.H., Maer A.M., Im W. Gramicidin A channel formation induces local lipid redistribution I: experiment and simulation. Biophys. J. 2017;112:1185–1197. doi: 10.1016/j.bpj.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heberle F.A., Pan J., Katsaras J. Model-based approaches for the determination of lipid bilayer structure from small-angle neutron and X-ray scattering data. Eur. Biophys. J. 2012;41:875–890. doi: 10.1007/s00249-012-0817-5. [DOI] [PubMed] [Google Scholar]
- 40.Bystrov V.F., Arsenev A.S. Diversity of the gramicidin A spatial structure: two-dimensional 1H NMR study in solution. Tetrahedron. 1988;44:925–940. [Google Scholar]
- 41.Galbraith T.P., Wallace B.A. Phospholipid chain length alters the equilibrium between pore and channel forms of gramicidin. Faraday Discuss. 1999;11:159–164. doi: 10.1039/a808270g. discussion 225–246. [DOI] [PubMed] [Google Scholar]
- 42.Perly B., Smith I.C., Ingold K.U. Estimation of the location of natural alpha-tocopherol in lipid bilayers by 13C-NMR spectroscopy. Biochim. Biophys. Acta. 1985;819:131–135. doi: 10.1016/0005-2736(85)90203-2. [DOI] [PubMed] [Google Scholar]
- 43.Andersen O.S., Bruno M.J., Koeppe R.E., II Single-molecule methods for monitoring changes in bilayer elastic properties. Methods Mol. Biol. 2007;400:543–570. doi: 10.1007/978-1-59745-519-0_37. [DOI] [PubMed] [Google Scholar]
- 44.Lundbaek J.A., Collingwood S.A., Andersen O.S. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. J. R. Soc. Interface. 2010;7:373–395. doi: 10.1098/rsif.2009.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katsaras J., Prosser R.S., Davis J.H. Constant helical pitch of the gramicidin channel in phospholipid bilayers. Biophys. J. 1992;61:827–830. doi: 10.1016/S0006-3495(92)81888-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harroun T.A., Heller W.T., Huang H.W. Experimental evidence for hydrophobic matching and membrane-mediated interactions in lipid bilayers containing gramicidin. Biophys. J. 1999;76:937–945. doi: 10.1016/S0006-3495(99)77257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kučerka N., Nieh M.P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 48.Mondal S., Khelashvili G., Weinstein H. Quantitative modeling of membrane deformations by multihelical membrane proteins: application to G-protein coupled receptors. Biophys. J. 2011;101:2092–2101. doi: 10.1016/j.bpj.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Q., London E. The influence of natural lipid asymmetry upon the conformation of a membrane-inserted protein (perfringolysin O) J. Biol. Chem. 2014;289:5467–5478. doi: 10.1074/jbc.M113.533943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang W.L., Chen M., Bayley H. Asymmetric droplet interface bilayers. J. Am. Chem. Soc. 2008;130:5878–5879. doi: 10.1021/ja802089s. [DOI] [PubMed] [Google Scholar]
- 51.Hussain N.F., Siegel A.P., Naumann C.A. Bilayer asymmetry influences integrin sequestering in raft-mimicking lipid mixtures. Biophys. J. 2013;104:2212–2221. doi: 10.1016/j.bpj.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vitrac H., Bogdanov M., Dowhan W. In vitro reconstitution of lipid-dependent dual topology and postassembly topological switching of a membrane protein. Proc. Natl. Acad. Sci. USA. 2013;110:9338–9343. doi: 10.1073/pnas.1304375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mobashery N., Nielsen C., Andersen O.S. The conformational preference of gramicidin channels is a function of lipid bilayer thickness. FEBS Lett. 1997;412:15–20. doi: 10.1016/s0014-5793(97)00709-6. [DOI] [PubMed] [Google Scholar]
- 54.Lum K., Ingólfsson H.I., Andersen O.S. Exchange of gramicidin between lipid bilayers: implications for the mechanism of channel formation. Biophys. J. 2017;113:1757–1767. doi: 10.1016/j.bpj.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivanova V.P., Makarov I.M., Heimburg T. Analyzing heat capacity profiles of peptide-containing membranes: cluster formation of gramicidin A. Biophys. J. 2003;84:2427–2439. doi: 10.1016/S0006-3495(03)75047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano M., Fukuda M., Handa T. Flip-flop of phospholipids in vesicles: kinetic analysis with time-resolved small-angle neutron scattering. J. Phys. Chem. B. 2009;113:6745–6748. doi: 10.1021/jp900913w. [DOI] [PubMed] [Google Scholar]
- 57.Sapay N., Bennett W.F.D., Tieleman D.P. Thermodynamics of flip-flop and desorption for a systematic series of phosphatidylcholine lipids. Soft Matter. 2009;5:3295–3302. [Google Scholar]
- 58.Homan R., Pownall H.J. Transbilayer diffusion of phospholipids: dependence on headgroup structure and acyl chain length. Biochim. Biophys. Acta. 1988;938:155–166. doi: 10.1016/0005-2736(88)90155-1. [DOI] [PubMed] [Google Scholar]
- 59.Renooij W., Van Golde L.M. The transposition of molecular classes of phosphatidylcholine across the rat erythrocyte membrane and their exchange between the red cell membrane and plasma lipoproteins. Biochim. Biophys. Acta. 1977;470:465–474. doi: 10.1016/0005-2736(77)90137-7. [DOI] [PubMed] [Google Scholar]
- 60.Wolfenden R., Snider M.J. The depth of chemical time and the power of enzymes as catalysts. Acc. Chem. Res. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 61.Goforth R.L., Chi A.K., Andersen O.S. Hydrophobic coupling of lipid bilayer energetics to channel function. J. Gen. Physiol. 2003;121:477–493. doi: 10.1085/jgp.200308797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rokitskaya T.I., Kotova E.A., Antonenko Y.N. Tandem gramicidin channels cross-linked by streptavidin. J. Gen. Physiol. 2003;121:463–476. doi: 10.1085/jgp.200208780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis B.A., Engelman D.M. Bacteriorhodopsin remains dispersed in fluid phospholipid bilayers over a wide range of bilayer thicknesses. J. Mol. Biol. 1983;166:203–210. doi: 10.1016/s0022-2836(83)80006-0. [DOI] [PubMed] [Google Scholar]
- 64.Gurtovenko A.A., Vattulainen I. Molecular mechanism for lipid flip-flops. J. Phys. Chem. B. 2007;111:13554–13559. doi: 10.1021/jp077094k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.