Abstract
The oligomeric organization of the voltage-dependent anion-selective channel (VDAC) and its interactions with hexokinase play integral roles in mitochondrially mediated apoptotic signaling. Various small to large assemblies of VDAC are observed in mitochondrial outer membranes, but they do not predominate in detergent-solubilized VDAC samples. In this study, a cholesterol analog, cholesteryl-hemisuccinate (CHS), was shown to induce the formation of detergent-soluble VDAC multimers. The various oligomeric states of VDAC induced by the addition of CHS were deciphered through an integrated biophysics approach using microscale thermophoresis, analytical ultracentrifugation, and size-exclusion chromatography small angle x-ray scattering. Furthermore, CHS stabilizes the interaction between VDAC and hexokinase (Kd of 27 ± 6 μM), confirming the biological relevance of oligomers generated. Thus, sterols such as cholesterol in higher eukaryotes or ergosterol in fungi may regulate the VDAC oligomeric state and may provide a potential target for the modulation of apoptotic signaling by effecting VDAC-VDAC and VDAC-hexokinase interactions. In addition, the integrated biophysical approach described provides a powerful platform for the study of membrane protein complexes in solution.
Introduction
The characterization of integral membrane proteins in solution is often difficult relative to soluble proteins because of the necessity of adding amphipathic detergents to solubilize and stabilize them in solution (1). Nevertheless, their immense importance in biotechnology, structure-based drug design, and medicine is evident from the fact that 59% of drugs approved by the Food and Drug Administration target transmembrane proteins (2). The potentially enormous impact of these drugs, coupled with the difficulty of studying the structure-function relationships of these membrane-localized drug targets, has created a pressing need for innovative methods to study integral membrane proteins. In this study, we used a combination of existing technologies to probe the oligomeric states of an integral membrane protein, VDAC.
Voltage-dependent anion-selective channels (VDACs) are a family of mitochondrial outer membrane proteins ubiquitous in aerobic eukaryotes and have garnered interest as possible targets for anticancer drugs (3, 4, 5). VDAC family proteins form large pores in the mitochondrial outer membrane, ∼1.4 nm in diameter at the narrowest point (6, 7). The primary function of VDAC is the exchange of ions and metabolites across the mitochondrial outer membrane (8). VDAC interacts with proteins involved in apoptotic signaling, specifically through hexokinase dissociation (9, 10), interactions with bcl-2 family proteins (10, 11), and it is possibly involved in the release of cytochrome-c and apoptosis-inducing factor from mitochondria (11). The reorganization of VDAC into units larger than monomers and the disassociation of the VDAC-hexokinase complex have been linked to mitochondrial-mediated apoptosis (11).
Crystal structures of VDACs have provided insights into possible oligomeric organizations in the form of crystal packing; however, the biological relevance of these VDAC organizations is still being debated (7, 12, 13). Interestingly, many of the detergent-solubilized VDACs examined to date seem to adopt a monomeric configuration except for zebrafish VDAC2 and mouse VDAC1 (at low pH), which adopted dimeric configurations in detergent solutions (6, 7, 14, 15). Multimeric VDAC assemblies purified from native membranes have been observed using electron microscopy and atomic force microscopy; however, the components of the lipid environment that conserved the native VDAC oligomeric states were not identified (16, 17).
Here, we utilized an integrated approach combining 1) microscale thermophoresis (MST) for the determination of binding constants and relevant concentrations of proteins for oligomeric analyses, 2) analytical ultracentrifugation (AUC) to determine oligomeric state, and 3) size-exclusion chromatography (SEC) in conjunction with small angle x-ray scattering (SAXS; SEC-SAXS) to model VDAC-detergent complexes. We used this pipeline to decipher the oligomeric organization of this essential integral membrane protein and began to probe VDAC complex formation with hexokinases. The established MST-AUC-SEC-SAXS experiments allowed us to validate high-resolution structures with solution scattering data and to determine the oligomeric organizations of VDAC in detergent complexes (18, 19).
Materials and Methods
Plasmids, strains, and reagents
Native purification of Neurospora crassa VDAC was performed using a strain in which the VDAC gene was replaced via homologous recombination with a complementary DNA copy of the gene with a C-terminal 6× His tag, as described previously (20). The resulting strain was named Por-His6.
Denaturing purification of the N. crassa VDAC was conducted utilizing a codon-optimized sequence for expression in Escherichia coli. A 6× His tag was engineered onto the C-terminus of the protein sequence. The gene was synthesized and cloned into the vector pET21b via the NdeI and BamHI sites by GenScript (Piscataway, NJ). The resulting pET21b-VDAC-His6 plasmid was transformed into E. coli strain BL21 (c43) (21) for VDAC overexpression.
n-decyl-β-d-maltopyranoside (DM, Anagrade) was purchased from Anatrace, (Maumee, OH) and cholesteryl-hemisuccinate (CHS) was purchased from Sigma-Aldrich Canada (Oakville, ON, Canada). All buffer solutions used in this study were adjusted to pH 7.0 unless otherwise specified.
VDACN purification
Cultivation of N. crassa was conducted as per standard methodologies (22). 20 L of Por-His6 culture was grown at 30°C for 22 h, and mycelia were harvested via filtration using a paper filter. Mitochondria were isolated as previously described (20, 23) and diluted to a total protein concentration of 10 mg/mL. The suspension of mitochondria was mixed 1:1 with a solution of 20 mM MOPS (3-morpholinopropane-1-sulfonic acid), 300 mM NaCl, 2% DM, and 2 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride and mixed at 4°C for 1 h and then applied to 5 mL Ni-NTA resin (Thermo-Fisher Canada, Mississauga, ON, Canada) and allowed to bind in batch with mixing overnight at 4°C. The Ni-NTA resin was washed with four column volumes of 20 mM MOPS, 100 mM NaCl, and 0.3% DM followed by two column volumes of 20 mM MOPS, 100 mM NaCl, 20 mM imidazole, and 0.3% DM and then was eluted with 20 mM MOPS, 100 mM NaCl, 300 mM imidazole, and 0.3% DM. VDAC purified from N. crassa mitochondrial membranes (VDACN) was then concentrated utilizing a 50 kDa cutoff concentrator and applied to a 24 mL capacity Superdex 200 increase column (General Electric Healthcare Canada, Mississauga, ON, Canada) equilibrated with 20 mM MOPS, 100 mM NaCl, and 0.3% DM. VDAC-containing fractions (Fig. 1 A) were collected and pooled.
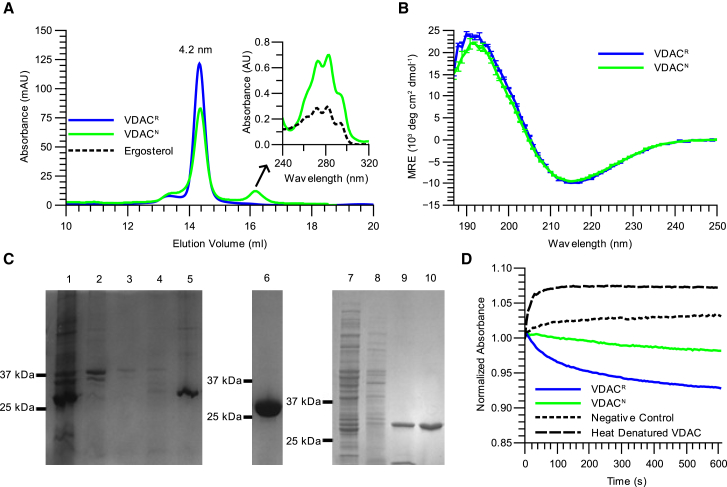
Figure 1.
VDACR retains structural integrity and functionality relative to VDACN. (A) SEC profiles of VDACN (green) and VDACR (blue) with the Rs calculated from the elution volumes of both samples displayed above the peaks are shown. The inset depicts the UV spectrum of the small second peak in the VDACN elution (green) and the UV spectrum of the fungal sterol ergosterol (black). (B) CD spectra of VDACN (green) and VDACR (blue) are shown. Averages and SDs (error bars) of three measurements are displayed. See Table 1 for secondary structure deconvolution. (C) SDS-PAGE analysis of VDAC purification is shown with cropped images of three gels displayed for clarity with labeled lanes: 1: inclusion bodies; 2–4: Ni-NTA flow through; 5: denatured VDACR elution; 6: pooled SEC fractions of refolded VDACR; 7: mitochondria; 8: DM-solubilized mitochondrial proteins; 9: Ni-NTA purified VDACN; 10: pooled SEC fractions of VDACN. Images of the full region of interest of each gel including protein Mw ladders are provided in Fig. S3, A–C. (D) Liposome reswelling assays of VDACN and VDACR are shown. Pore formation is observed through changes in the amount of scattered light that accompany reswelling of proteoliposomes. The scattered light is measured by absorbance and is displayed as values normalized to the first absorbance measurement in each curve. This figure is available in color online. To see this figure in color, go online.
VDACR overexpression, purification, and refolding
A 25-mL overnight culture of BL21 (c43) E. coli cells containing the pET21b-VDAC-His6 vector was added to 500 mL of prewarmed (37°C) lysogeny broth media. The 500-mL culture was grown with shaking at 37°C for 1 h, after which time VDAC expression was induced with the addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.5 mM; the culture was grown for an additional 4 h. Cells were harvested via centrifugation for 5 min at 8000 rotations per minute (rpm) in a Sorvall SLA-3000 rotor (Thermo-Fisher Canada). Harvested cells were resuspended in 30 mL chilled (4°C) buffer (20 mM Tris, 300 mM NaCl, 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (pH 8.0)). Resuspended cells were lysed via three passes through a French pressure cell at a pressure of 20,000 psi. VDAC inclusion bodies were harvested from cell lysate via centrifugation for 15 min at 16,000 rpm in a Sorvall SS-34 rotor (Thermo-Fisher Canada). The resulting supernatant was discarded, and the pellet was resuspended in 20 mM MOPS, 150 mM NaCl, 6 M guanidine-HCl using a glass homogenizer. Insoluble material was removed via centrifugation at 20,000 rpm in a Sorvall SS-34 rotor. The supernatant was applied to 3 mL Ni-NTA resin, and binding of VDAC to the resin was done in batch with mixing overnight at 4°C. The Ni-NTA resin was washed with 10 column volumes of 20 mM MOPS, 150 mM NaCl, and 6 M guanidine-HCl followed by four column volumes of 20 mM MOPS, 150 mM NaCl, 20 mM imidazole, and 6 M guanidine-HCl. VDAC was eluted with 10 mL of 20 mM MOPS, 150 mM NaCl, 600 mM imidazole, and 6 M guanidine-HCl.
Recombinantly expressed N. crassa VDAC (VDACR) was refolded by rapid 1/10 dilution of denatured VDACR into 20 mM Tris, 300 mM NaCl, and 1% DM (pH 8.0) at 4°C. This mixture was dialyzed against 20 mM Tris and 300 mM NaCl (pH 8.0) overnight at 4°C using a dialysis membrane with a molecular weight (Mw) cutoff less than 10 kDa. Dialyzed VDACR samples were then concentrated using a concentrator with an Mw cutoff of 50 kDa. Concentrated samples were loaded onto a 24-mL capacity Superdex 200 increase column equilibrated with 20 mM MOPS, 100 mM NaCl, and 0.3% DM to separate aggregates from correctly refolded protein. For experiments containing CHS, there was an additional SEC step to exchange the buffer via a 24-mL capacity Superdex 200 increase column equilibrated with 20 mM MOPS, 100 mM NaCl, 0.3% DM, and 0.06% CHS. VDAC-containing fractions were pooled and concentrated with a 50 or 100 kDa Mw cutoff concentrator. Purified VDAC activity was assessed using liposome swelling assays as described previously (24) (Fig. 1 D).
CD
VDAC samples were exchanged into circular dichroism spectrapolarimetry (CD) buffer (either 100 mM NaCl and 0.1% DM (pH 7.0) or 20 mM MOPS, 100 mM NaCl, 0.1% DM, and 0.02% CHS (pH 7.0)) and diluted to ∼1 mg/mL in the same buffer. Final protein concentration was measured by ultraviolet (UV) absorbance at 280 nm (VDAC y0.1%: 0.79, calculated from sequence). CD measurements were taken with a Jasco J-810 (Jasco, Easton, MD) spectropolarimeter at 20°C in a 0.01-cm pathlength quartz cuvette. Raw data were converted to mean residue ellipticity, and secondary structure deconvolution was done using the CDSSTR algorithm and the SMP180 reference set on the DichroWeb server (25, 26, 27).
AUC
VDAC was diluted to appropriate concentrations in 20 mM MOPS, 100 mM NaCl, 0.3% DM or 20 mM MOPS, 100 mM NaCl, 0.3% DM, and 0.06% CHS, with the highest concentration of VDAC not exceeding 1 absorbance unit at 280 nm; final protein concentrations were determined directly from AUC absorbance data. Experiments were conducted using a Proteome XL-I analytical ultracentrifuge and an An50Ti rotor from Beckman Coulter (Brea, CA).
Sedimentation velocity experiments were conducted at a rotor speed of 30,000 rpm and a solution of 20 mM MOPS and 100 mM NaCl (ρ: 1.004 g/mL, η: 0.01006 mPa) was used as the reference solution. Other values used in the analysis: VDAC v̄ of 0.733 was determined from the protein sequence using SEDNTERP (http://www.jphilo.mailway.com/download.htm#SEDNTERP), 0.187 was used as the VDAC dn/dc (28), VDAC ε0.1%, 280 of 0.79 was determined from protein sequence, DM v̄ of 0.8 was reported by Zimmer et al. (29), and DM dn/dc of 0.1457 was experimentally determined (data not shown). Rs values were determined by SEC elution volume relative to a standard curve of proteins with known Rs values. Sedimentation velocity c(s) distribution analysis and c(s, f/f0) distribution analyses were conducted using the software SEDFIT (30). Note that c(s) assumes a single frictional ratio, f/f0, for all species producing an s distribution of measured intensity, and c(s, f/f0) makes no assumptions about frictional ratio and produces a two-dimensional (2D) distribution of the measured signal with s and f/f0 dimensions. To gain additional resolution of VDAC species with similar S values in the VDACR + CHS samples, 2D c(s, f/f0) distributions were calculated for the interference data sets (Fig. 2 E). Because of the lower signal/noise ratio in the corresponding absorbance data sets of these samples, it was not possible to produce meaningful c(s, f/f0) distributions from absorbance data. To produce comparable interference and absorbance distributions describing VDACR + CHS samples, which retained the increased resolution of VDAC species from the c(s, f/f0) distributions, we employed a Bayesian statistical analysis implemented in SEDFIT (31). This analysis was used to produce interference and absorbance c(s) distributions of VDACR + CHS samples (Fig. 2 F) using the previous knowledge of the distribution of VDAC species in these samples obtained from the c(s, f/f0) analysis of the interference data (Fig. 2 E). Four VDAC concentrations were examined in the absence of CHS and in the presence of CHS so that sedimentation coefficients could be extrapolated to a protein concentration of 0 mg/mL to account for concentration-dependent factors and to allow accurate determination of dn/dcp values for the VDAC-detergent complexes. Before use for molecular weight determination, experimentally determined sedimentation coefficients (S20, buffer: 20°C in sample buffer) were adjusted to standard conditions (S20, water: 20°C in water).
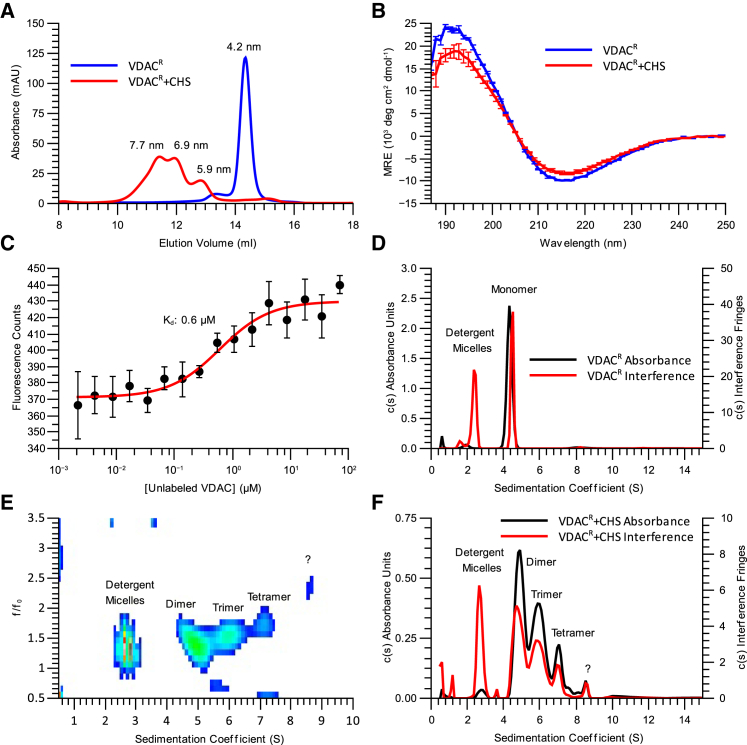
Figure 2.
CHS alters the oligomeric state and secondary structure of VDACR. (A) SEC of VDACR (blue) and VDACR + CHS (red) are shown, with the calculated Rs values displayed above each peak. (B) Averaged CD spectra of VDACR (blue) and VDACR + CHS (red) with SDs of three measurements are displayed as error bars. For secondary structure deconvolution, see Table 1. (C) A microscale thermophoresis (MST) isotherm of 10 nM fluorescently labeled VDACR + CHS titrated against unlabeled VDACR + CHS is shown. The Kd of the change in behavior is displayed on the plot. (D) c(s) absorbance and interference distributions of VDACR at 0.98 mg/mL are shown. Peaks corresponding to free detergent micelles and the VDACR monomer are indicated on the plot. (E) The c(s, f/f0) interference distribution of VDACR + CHS at 0.99 mg/mL used to resolve the three VDAC species present in solution is shown. Peaks corresponding to the free detergent micelles, VDAC dimer, trimer, and tetramer, and a peak of unknown composition are indicated on the plot. (F) Absorbance and interference c(s) distributions of VDACR+CHS at 0.99 mg/mL are shown. Peaks corresponding to free detergent micelles and VDAC dimers, trimers, and tetramers and a peak of unknown composition are labeled on the plot. The c(s, f/f0) distribution in (E) was reduced into a 2D c(s, ∗) distribution and was used as prior probabilities to resolve the three VDAC species in the c(s) fitting procedure displayed in (F). Peaks corresponding to free detergent micelles and VDAC dimer, trimer, tetramer and a peak of unknown composition are labeled on the plot. In all AUC distributions, detergent peaks were assigned using detergent-only control samples (data not shown). This figure is available in color online. To see this figure in color, go online.
Experimental theory and analysis of membrane protein samples and nomenclature are as previously described by Salvay et al. (32). A summary of the theory is as follows.
The mass of the protein-detergent complex (MPD) can be described by the following equation:
| (1) |
where NA is Avogadro’s number, η is the solvent viscosity, Rs is the hydrodynamic radius of the protein-detergent complex, s is the sedimentation coefficient, ρ° is the solvent density, and is the partial specific volume of the protein-detergent complex. The can be determined by the following equation:
| (2) |
where and are the partial specific volumes of the protein and detergent, respectively, and δD is the ratio of detergent/protein (g:g) in the protein-detergent complex. δD can be determined from the refractive index increment (dn/dcp, change in refractive index relative to protein concentration) of the protein-detergent complex as follows:
| (3) |
where (dn/dcp)P is the refractive index increment of the protein, and (dn/dcD)D is the refractive index increment of the detergent. Finally, the (dn/dcp)PD can be determined from the absorbance at 280 nm and interference fringe displacement of the protein-detergent complex as follows:
| (4) |
where J is the interference fringe displacement, λ is the wavelength of light used, c is the concentration of protein determined from absorbance at 280 nm, K is the magnification of the lens used, and l is the pathlength of the sample. Interference fringe displacement of a species can be directly determined from a c(s) distribution of interference fringe displacement by integrating the area under the peak of the species being examined, and the same is true for absorbance at 280 nm with the exception that a c(s) distribution of absorbance data would be used instead.
Once the mass of the protein-detergent complex has been determined from the previous equations, the mass of the protein and detergent components can be determined using δD:
| (5) |
| (6) |
where MP and MD are the masses of the protein and detergent components, respectively.
SAXS
SEC-SAXS data were collected at Beamline B21, Diamond Light Source (Didcot, UK). VDAC-detergent complexes at VDAC concentrations of ∼5–10 mg/mL in the presence and absence of CHS were applied to a 2.4 mL Superdex 200 increase column equilibrated with the appropriate buffer (20 mM MOPS and 100 mM NaCl with 0.1% DM or 0.1% DM + 0.02% CHS), and the eluate was examined in-line. Data were collected with exposure to 1 Å wavelength x-rays. ScÅtter (http://www.bioisis.net/users/sign_in/) and ATSAS (33) software packages were used to process collected SAXS data. Models were constructed using the crystal structure of mouse VDAC1 (Protein Data Bank (PDB): 3EMN) (7, 13) or the crystal structure of zebrafish VDAC2 (PDB: 4BUM) (12) to describe the protein phase of the protein-detergent complex and the Memprot algorithm (19) to model the detergent corona around the protein. Memprot allows for the geometric modeling of an elliptical detergent layer around a fixed protein phase input in the form of a PDB file (protein electron density 0.42 e−/Å2) using two different types of detergent beads corresponding to detergent headgroups (electron density 0.51e−/Å2 for maltoside detergents (34)) and detergent tails (electron density 0.28 e−/Å2 for maltoside detergents (34)).
MST
VDAC was labeled with Alexa Fluor 647 N-hydroxysuccinimide ester dye (Thermo-Fisher Canada), which labels lysine residues; VDAC samples were labeled with an efficiency of ∼1 label per protein molecule as determined by absorbance measurements. Labeled VDAC was diluted to a concentration of 100 nM for hexokinase experiments or 20 nM for VDAC self-association experiments in 20 mM MOPS, 100 mM NaCl, and 0.3% DM or 20 mM MOPS, 100 mM NaCl, 0.3% DM, and 0.06% CHS, which served as 2× working concentrations. Separated hexokinase isoforms were prepared from a mixture of Saccharomyces cerevisiae hexokinases I and II (LS002512) obtained from Worthington Biochemical (Lakewood, NJ) using a well-established method relying on the different pIs of the isoforms (35). The hexokinase mixture was immobilized on a Capto Q anion exchange column (GE Healthcare Canada), and hexokinases were separated by pI using a pH gradient formed by a linear gradient of a solution of 10 mM piperazine, 10 mM acetate, and 10 mM formate at pH 5.5 and the same buffer solution at pH 4.0 (Fig. 5, A and B). The isoforms were identified by the pH at which they eluted (Fig. 5 A) (35). Purified HK-I and HK-II were exchanged into 20 mM MOPS and 100 mM NaCl (pH 7.0) buffer via dialysis. Twofold serial dilutions (20 μL) of concentrated separated hexokinases were prepared, and each dilution was mixed 1:1 with the 100 nM stock solution of labeled VDAC for a final labeled VDAC concentration of 50 nM in each tube. For VDAC self-association experiments, twofold serial dilutions (20 μL) of concentrated unlabeled VDACR + CHS were prepared, and each dilution was mixed 1:1 with the 20 nM labeled VDACR solution resulting in a final labeled VDAC concentration of 10 nM in each tube. MST experiments were conducted with a Nanotemper Monolith NT.115 in premium treated capillaries from Nanotemper Technologies (München, Germany). MST data were recorded at 40% maximal infrared laser power. MST analysis was conducted using the MO. Analysis software from Nanotemper Technologies and Palmist (36) using the Kd model. A brief description of the model is as follows.
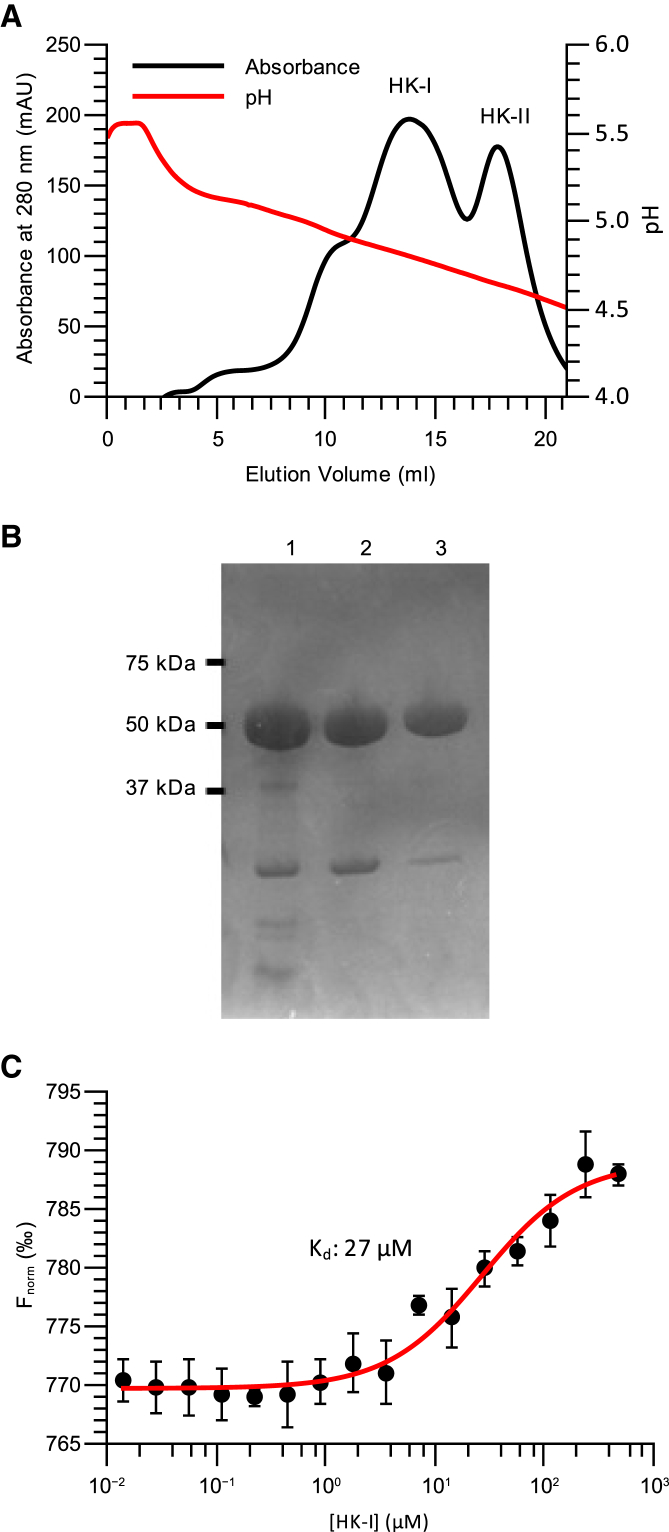
Figure 5.
CHS allows for the formation of a stable VDAC-hexokinase complex. (A) The separation of yeast HK-I and HK-II isoforms via pH gradient is shown. Protein elution was monitored by absorbance at 280 nm (black), and the pH of the eluate was monitored in-line with elution (red). Peaks corresponding to the hexokinase isoforms are indicated on the plot. (B) SDS-PAGE analysis of pooled HK-I (lane 2) and HK-II (lane 3) fractions relative to starting mixture (lane 1) is shown. An image of the full region of interest of the gel including the protein Mw ladder is provided in the Fig. S3D. (C) Titration of unlabeled HK-I against a constant concentration of fluorescently labeled VDACR + CHS resulted in a change in VDAC motion because of the generated temperature gradient. Experimentally determined average values are shown by black circles with error bars representing SD. The Kd of the interaction and the fitted curve (red) used to determine the Kd are shown on the plot. This figure is available in color online. To see this figure in color, go online.
The dissociation constant of a protein-protein complex consisting of two components, A and B, can be described by the following equation:
| (7) |
where Kd is the dissociation constant, [A] is the concentration of component A, [B] is the concentration of component B, and [AB] is the concentration of the complex. For MST analysis, the fluorescence intensity of a sample after the activation of a localized heat source (Fhot) relative to the fluorescence intensity before the activation of the heat source (Fcold) is measured as follows:
| (8) |
where Fn is the resulting relative fluorescence of the sample. The measured Fn of a sample consisting of protein A, fluorescently labeled protein B, and complex AB can be described by the following equation:
| (9) |
where Btot is the total concentration of component B (bound and unbound), FB is the Fn of protein B, and FAB is the Fn of complex AB. The total concentrations of proteins A and B can be described by the following equations:
| (10) |
| (11) |
Using Eqs. 7, 9, 10, and 11, we can describe Fn as a function of Atot, which can be fitted to a series of MST measurements at different Atot values (usually a twofold serial dilution) to determine Kd:
| (12) |
Values for Atot and Btot are known because they are equivalent to the total input concentrations of each component for each measurement. Values for FB, FAB, and Kd will be determined by fitting the function to the experimental data if a full sigmoidal binding curve is observed over all measured values of Atot (36). A more thorough examination of the theory and analysis of MST data on which this brief description was based is presented by Scheuermann et al. (36).
Results
Evaluation of recombinant N. crassa VDAC
We established an isolation and characterization protocol for recombinantly expressed N. crassa VDAC (VDACR) that was folded in DM. To evaluate the validity of VDACR as a biologically equivalent model protein, we performed the following experiments on VDACR and VDACN. In a first step, detergent-solubilized VDAC was assessed through SEC (Fig. 1, A and B). Both VDACN and VDACR elute as single symmetrical peaks with identical elution volumes of 14.2 mL, indicating an Rs value of 4.2 nm from a calibration curve of proteins with known Rs values. These results suggest that VDACR and VDACN exist in the same oligomeric state when solubilized in DM. In a second step, CD analysis suggests that VDAC from both sources have identical secondary structure compositions, which largely consists of β-strands (Fig. 1 C), as observed previously (6, 7, 12). Deconvolution of the CD spectra reveals that the secondary structure compositions of VDACN and VDACR agree with the secondary structure present in the crystal structure of mouse VDAC1 (PDB: 3EMN) (Table 1) (7). Finally, liposome reswelling assays show that VDACN and VDACR can both form pores large enough to accommodate the passage of polyethylene glycol (average Mw 1000 Da) when reconstituted into lipid vesicles, indicating that they are functional (Fig. 1 D). In the assay, this is seen as a relatively slow swelling of the liposomes, accompanied by a decrease in absorbance. In addition, liposomes lacking any pores (pure lipid) as well as liposomes containing heat-denatured VDAC were examined as negative controls (Fig. 1 D). The liposomes containing heat-denatured VDAC displayed a rapid increase in absorbance relative to the pure lipid liposomes, indicating that the heat-denatured VDAC was able to facilitate the rapid transfer of water molecules across the membrane upon the addition of solute to the exterior of the liposomes. There was no subsequent reduction in absorbance in the heat-denatured VDAC liposomes, indicating that any pores formed by the misfolded VDAC were too small to allow for the passage of polyethylene glycol (average Mw 1000 Da). In summary, VDACR is advantageous for our structural and biophysical studies because it retains structural integrity and functionality in comparison to VDACN, whereas the purification and refolding procedure yields significantly higher quantities of VDAC relative to VDACN. With the methods used, the only difference noticed between the two VDAC samples observed was a small second peak that eluted after VDACN during SEC (Fig. 1 A and inset). This peak absorbed UV light at 280 nm; however, it did not contain protein as detected by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis (data not shown). The far UV spectrum of the peak as well as previous evidence of copurification (16, 37) suggests that the compound is ergosterol, a fungal sterol. Because ergosterol or cholesterol (38) was known to copurify with VDACs derived from fungal or higher eukaryotes, respectively, we became interested in investigating the effects that these sterols may have on VDAC. For this purpose, VDACR seemed to be an ideal platform because the denaturing purification from bacterial source would preclude the copurification of any sterols, which could have interfered with the investigation.
Table 1.
VDAC Secondary Structure Contributions Obtained from Deconvolution of CD Data
CHS alters the oligomeric state and secondary structure of VDACR
Attempts to introduce ergosterol or cholesterol to the sterol-free, DM-solubilized VDACR samples failed because of the insolubility of these sterols in the detergent DM (data not shown). Therefore, to study the effects of sterols on VDAC solubilized in DM, we utilized CHS, a cholesterol analog soluble in maltoside detergents (39). The addition of CHS to VDACR drastically altered the behavior of the protein. Refolded VDAC in the presence of CHS (VDACR + CHS) eluted as three overlapping peaks during SEC. All three VDACR + CHS peaks eluted at much lower volumes than the protein in the absence of CHS (Fig. 2 A). This result suggested the formation of at least three different VDAC oligomers, all of which had Rs values (5.9, 6.9, and 7.7 nm) significantly greater than that of VDACR in the absence of CHS (4.2 nm) (Table 2). In addition, the CD spectrum of VDACR + CHS was slightly altered relative to VDACR (Fig. 2 B). Deconvolution of the CD spectrum revealed a reduction of β-strand content by 7% relative to VDACR (Table 1). The change in VDACR + CHS oligomeric state was observed to be dependent on the concentration of VDACR + CHS (Fig. 2 C). The binding was detected by change in fluorescence intensity of VDACR + CHS labeled with Alexa Fluor 647 titrated against unlabeled VDACR + CHS; the Kd value of the change in state was determined to be 0.6 ± 0.2 μM using MST (Fig. 2 C). In the absence of CHS, an increase in fluorescence was also observed upon the addition of 100 μM VDACR to VDACR labeled with Alexa Fluor 647; however, an identical shift in fluorescence intensity also occurred upon the addition of 100 μM egg white lysozyme (Fig. S3 D), indicating that this change was due to the loss of fluorescent protein in the absence of a high overall protein concentration (likely caused by sticking of the labeled protein to pipette tips or sample tubes). The addition of 100 μM egg white lysozyme to VDACR labeled with Alexa Fluor 647 in the presence of CHS had no effect on fluorescence intensity, indicating that the change in fluorescence observed upon the addition of unlabeled VDACR was due to a specific interaction (Fig. S3 D).
Table 2.
Parameters of VDAC-Detergent Complexes and Detergent Micelles Determined from Sedimentation Velocity Experiments
| dn/dcpa | δD (g/g)b | (mL/g)c | S20, watera | Rs (nm)d | MPD (kDa)e,f | MP (kDa)e,f | MD (kDa)e,f | |
|---|---|---|---|---|---|---|---|---|
| VDACR monomer | 0.48 ± 0.01 | 1.99 ± 0.04 | 0.78 ± 0.02 | 4.8 ± 0.05 | 4.2 ± 0.3 | 107 ± 7 | 36 ± 3 | 71 ± 5 |
| VDACR + CHS dimer | 0.43 ± 0.03 | 1.7 ± 0.1 | 0.78 ± 0.07 | 5.3 ± 0.03 | 5.9 ± 0.3 | 170 ± 30 | 60 ± 10 | 100 ± 20 |
| VDACR + CHS trimer | 0.37 ± 0.02 | 1.32 ± 0.07 | 0.77 ± 0.06 | 6.4 ± 0.07 | 6.9 ± 0.4 | 220 ± 20 | 100 ± 10 | 120 ± 20 |
| VDACR + CHS tetramer | 0.42 ± 0.02 | 1.15 ± 0.07 | 0.77 ± 0.06 | 7.3 ± 0.06 | 7.7 ± 0.4 | 290 ± 30 | 130 ± 20 | 150 ± 20 |
| DM micelles | – | – | – | 2.4 ± 0.02 | 2.8 ± 0.2 | – | – | 39 ± 3 |
| DM + CHS micelles | – | – | – | 3.0 ± 0.1 | 3.36 ± 0.02 | – | – | 59 ± 2 |
δD was determined using Eq. 3.
of the protein-detergent complex was calculated from the protein () and detergent () using Eq. 2.
Rs was determined by SEC elution volume relative to elution volumes of proteins with known Rs values for VDAC-detergent complexes and by dynamic light scattering for detergent micelles.
MPD, Mp, and MD values were calculated using Eqs. 1, 5, and 6, respectively, for VDAC-detergent complexes. MD for pure detergent micelles was calculated using Eq. 1.
The mass of VDACR monomer from sequence is 30.3 kDa.
To investigate the composition of the VDAC-detergent complexes and determine the oligomeric state of VDAC in the presence and absence of CHS, we employed AUC using both absorbance and interference optics (Fig. 2, D–F). A clear difference was observed between the VDACR and VDACR + CHS samples. The VDACR sample contained two species, which can be attributed to the monomeric VDAC-detergent complex and free detergent micelles (Fig. 2 D). In contrast, the VDACR + CHS sample contained three VDAC species as well as free detergent micelles (Fig. 2, E and F). We could clearly distinguish three distinct VDAC-detergent complexes and free detergent micelles in a c(s, f/f0) distribution (Fig. 2 E) of interference data; however, we were unable to obtain the same resolution in a c(s, f/f0) distribution of the corresponding absorbance data. Therefore, we used the interference c(s, f/f0) distributions as prior probabilities for the determination of c(s) distributions of both interference and absorbance data (Fig. 2 F). The sedimentation coefficients of the three VDACR + CHS species are all higher than the VDAC-detergent complex in the VDACR sample (Table 2). These results confirm the SEC experiments (Fig. 2 A) showing that CHS induces the formation of three different VDAC species. Determination of the composition of the VDAC-detergent complexes revealed that the VDACR sample contained exclusively monomers of VDAC, whereas the VDACR + CHS sample contained a mixture of VDAC dimers, trimers, and tetramers (Table 2).
SEC-SAXS modeling supports previously proposed VDAC multimeric arrangements
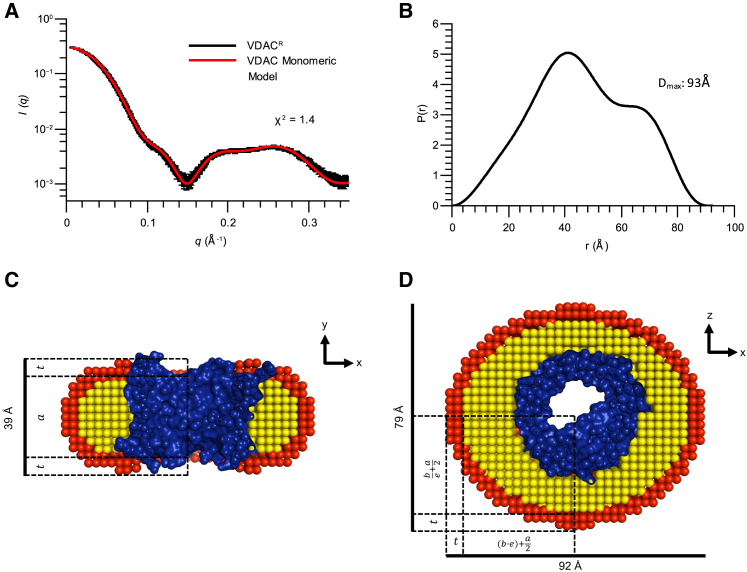
SEC-SAXS data collected of VDACR samples revealed only one VDAC-detergent complex present in solution (Rg of 35 Å) (Fig. 3; Fig. S5 A). The low χ2 value for the fit (χ2 = 1.4) of the scattering curve of the monomeric VDAC model confirms that the crystal structure of mouse VDAC1 resembles the assembly of VDACR in solution (7). The best-fitting monomeric VDAC model produced is displayed in Fig. 3, C and D.
Figure 3.
SEC-SAXS model of monomeric VDACR fits experimental scattering data. (A) Experimental scattering data of SEC eluted VDACR in 0.3% DM solution (black) and calculated scattering curve of monomeric VDAC-detergent complex (red) are shown; error bars represent the SD of averaged curves. The χ2 of the fit of the calculated curve to the experimental data is displayed on the plot. (B) The P(r) plot determined from experimental scattering data of VDACR is shown. The determined Dmax value is shown on the plot. (C) A cross section of the monomeric VDACR model through the lumen of the VDAC pore and (D) rotated 90° around the x axis is shown. Dimensions of the model are shown, and individual fitted parameters corresponding to the detergent component of the model are depicted relative to the total distances. These parameters (a, b, t, and e) are depicted to aid in visualization of their relationship to the model of the protein complex. In both panels, the protein component of the model is colored in blue, the hydrophobic layer of the detergent is colored yellow, and the hydrophilic layer of the detergent is colored red. Values for the parameters a, b, t, and e for all VDAC models are reported in Table 3. This figure is available in color online. To see this figure in color, go online.
Because of the heterogeneous nature of the VDACR + CHS sample (Fig S5, B and C), the collection of data corresponding to specific oligomeric states was more challenging. Initial SEC-SAXS experiments using a VDAC sample pre-equilibrated with CHS provided clear separation of only one VDACR + CHS complex with an Rg of 70 Å (Fig. S4 A). This species could be modeled as hexameric arrangement derived from crystal contacts of the mouse VDAC1 and produced a model with a χ2 of 1.5 (Fig. S4, B–D). The hexameric arrangement of VDACR + CHS used to construct the model is similar to previously observed organizations of VDAC isolated from native membranes (16, 17) and was previously suggested from the crystal structure of mVDAC1 as a possible biological assembly (13); however, because of the antiparallel arrangement of the molecules in the mVDAC1 crystal structure, the biological relevance of this arrangement is questionable.
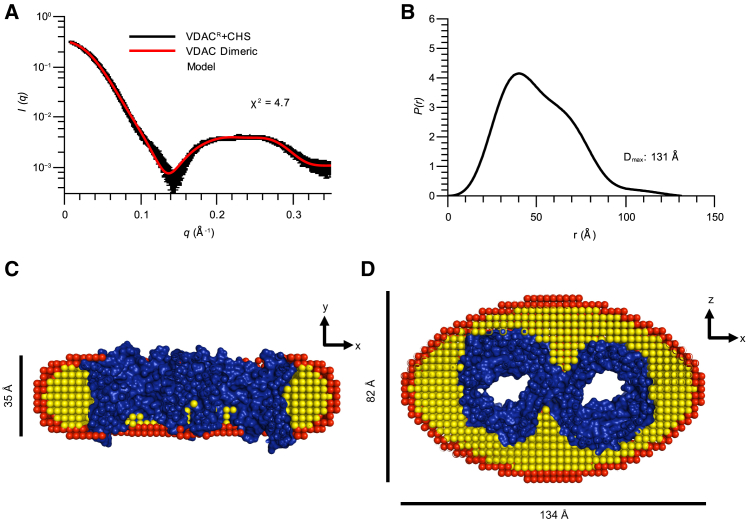
We obtained a clearer separation of the VDAC dimer species when CHS was introduced to the VDAC sample during the SEC step of the SEC-SAXS experiment. This VDACR + CHS species had an Rg of 40 Å (Fig. 4 A). The P(r) distribution of the protein-detergent complex clearly demonstrates a Dmax value of 131 Å (Fig. 4 B). The best-fitting model (χ2 value of 4.7) was a dimeric zebrafish VDAC2 (PDB: 4BUM) (Fig. 4, C and D). Parameters of the detergent layer of each model are summarized in Table 3.
Figure 4.
SEC-SAXS model of dimeric VDACR + CHS fits experimental scattering data. (A) Experimental SAXS data (black) and calculated scattering curve of a dimeric VDAC-detergent complex (red) are shown; error bars represent the SD of averaged curves. The χ2 of the fit of the model to the experimental data is shown on the plot. (B) P(r) function calculated from experimental data of VDACR + CHS dimer is shown. Determined Dmax value is shown on the plot. (C) A cross section of the dimeric VDACR + CHS model through the lumen of the pore and (D) rotated 90° around the x axis is shown. Dimensions of the model and the orientation relative to the real space axes are displayed in each panel. The protein component of the model is colored in blue, the hydrophobic layer of the detergent is colored yellow, and the hydrophilic layer of detergent is colored in red. Best-fitting parameters of the detergent layer of the models are reported in Table 3, and a visualization of how each parameter relates to the model can be seen in Fig. 3. This figure is available in color online. To see this figure in color, go online.
Table 3.
Parameters of Best-Fitting VDAC-Detergent Complex Models from SEC-SAX Analysis
| Rg (Å) (Guinier/Model)a | Dmax (Å) (P(r)/Model)b | a (Å)c | b (Å)c | t (Å)c | e (°)c | |
|---|---|---|---|---|---|---|
| VDACR (monomer) | 36/35 | 93/92 | 29 | 23 | 5 | 1.16 |
| VDACR + CHS (dimer) | 40/40 | 131/134 | 27 | 34 | 4 | 1.45 |
| VDACR + CHS (hexamer) | 70/71 | 243/242 | 27 | 77 | 4 | 1.35 |
CHS allows for a stable VDAC-hexokinase interaction
Previous reports link apoptosis to the oligomeric state of VDAC and to its interactions with hexokinase (9, 11). To gain insight into the onset of apoptosis triggered by the disassociation of the VDAC-hexokinase complex, we performed MST (Fig. 5). Our established experimental setup provides an excellent opportunity to study integral membrane protein complexes while also providing novel information on the role VDAC plays in apoptosis (10).
To determine the binding behavior, we used full-length native-source hexokinase isoforms I and II from S. cerevisiae (HK-I and HK-II, respectively) (Fig. 5, A and B). Whereas HK-II did not show any binding (Fig. S2), it was determined that the stable interaction of HK-I with VDACR is strictly dependent upon addition of CHS to the sample. VDACR-HK-I mixtures showed signs of aggregation when the MST infrared laser was activated, and the rate of aggregation seemed to be dependent on the HK-I concentration, a behavior which does not occur in the presence of other proteins such as egg white lysozyme (Fig. S2 D). Because this behavior was not observed in the absence of HK-I or in the presence of egg white lysozyme, this result suggests that a thermally unstable VDAC-HKI complex was formed. In VDACR + CHS + HK-I mixtures, aggregation was not observed. A binding constant for VDACR + CHS + HK-I of 27 ± 6 μM was determined (Fig. 5 C) from the thermophoresis profiles of a HK-I titration.
Discussion
The methods discussed in this study were used to gain an understanding of the self-association of VDAC induced by the cholesterol analog CHS and to generate a platform for exploring the formation of the protein-protein complexes that include integral membrane proteins and drive essential biological processes. VDAC NMR (6) and crystal structures (7, 12, 14) focused on the arrangement of the VDAC polypeptide, which was a critical first step in understanding the structure-function relationships of VDAC and formed the basis of future work probing the protein function. However, the isolated protein cannot be studied in aqueous environments, and studies of the function of VDAC are carried out in artificial membranes (8, 40) and membrane-mimetic systems (14, 16). Evidence suggests that VDAC interacts with many proteins, including proteins involved in apoptotic signaling, and thorough studies of these interactions will be most readily achieved using refolded VDAC solubilized in detergent; therefore, a detailed understanding of the complete protein-detergent system is critical to the interpretation of structure-function relationships (9, 11, 41).
Previous works have adapted AUC and SAXS to the analysis of detergent-solubilized membrane proteins (19, 32). AUC provides information pertaining to molecular weight, shape, composition of oligomers and complexes, and the relative abundance of distinct species present in a mixture (42). The analysis of membrane proteins via AUC sedimentation velocity can be achieved via the solubilization of the membrane protein in detergent; however, the interpretation of the data obtained from these experiments is complicated by the presence of detergent bound to the solubilized membrane protein, adding mass and obscuring the shape and composition of oligomers of the protein (32). SAXS allows for the low-resolution modeling of macromolecules in solution from x-ray scattering intensities. The main complications in the analysis of detergent-solubilized membrane proteins via SAXS are the presence of free detergent micelles in the solution and the detergent layer surrounding the membrane protein (19).
To overcome these obstacles, we have established a new, to our knowledge, combination of 1) MST to investigate the concentration dependence of VDAC oligomerization and the interaction of VDAC with hexokinase, 2) AUC sedimentation experiments to investigate the composition of the VDAC-detergent complexes, and 3) SEC-SAXS studies to create models of detergent micelles and VDAC-detergent complexes. Previous studies have shown that VDAC function is altered by the surrounding lipid environment through the modulation of its oligomeric state, which can influence apoptotic signaling (16, 43). There is some previous evidence for VDAC multimers in detergent solutions; however, evidence toward a detailed understanding of the regulation of these assemblies was not obtained (12). In this study, the oligomerization of VDAC was shown to be dependent on VDACR concentration in the presence of CHS via MST analysis, and a Kd value of 0.6 ± 0.2 μM (∼0.02 mg/mL) was determined, implying that the protein readily forms larger than monomer units in the presence of CHS. This contrasts with the behavior of the protein in the absence of CHS in that only monomers of VDAC were observed. It should be noted that the Kd model used in this work to describe the self-association of VDAC is likely too simple of a model to accurately describe the self-association of an integral membrane protein in detergent solution and also is unlikely to completely represent the self-association of VDAC in a lipid bilayer. This is due to several complications, with the main issue being the confinement of VDAC in the “detergent micelle phase” of the solution, drastically reducing the volume of the solution that VDAC molecules can possibly occupy. A more suitable model may be suggested by incorporating the volume of available detergent micelles into the model; however, this approach itself would be too simplistic because the AUC results presented in this work (Table 2) as well as information presented by others (32, 44) have demonstrated that integral membrane proteins do not associate with the same mass of detergent as is in a single detergent micelle. In the case of VDAC, a further complication arises with different oligomers of VDAC associating with different quantities of detergent (Table 2). Nevertheless, the Kd model used in this work can describe a relative binding constant for the comparison of VDAC self-association in the same detergent environment, establishing a baseline for the examination of the self-association of VDAC mutants or VDAC sequences from other organisms.
Dimeric organizations of VDAC have been suggested from previous crystal packing arrangement of mouse VDAC1 (antiparallel molecules) and zebrafish VDAC2 (parallel molecules) (7, 12, 13) and furthered by the examination of these two molecules in solution by electron paramagnetic resonance (EPR) revealing two different dimeric interfaces (12, 15). The previously mentioned EPR experiments revealed that a parallel mouse VDAC1 dimer was formed at low pH and depended on the protonation state of residue E73 in contrast to the antiparallel mouse VDAC1 crystal packing arrangement (15). The EPR studies of zebrafish VDAC2 supported the parallel arrangement observed in the zebrafish VDAC2 crystal asymmetric unit (12). In this study, we observed dimers, trimers, tetramers, and possibly hexamers of VDAC in the presence of CHS via AUC and collected SEC-SAXS data corresponding to a VDAC dimer in the presence of CHS. The dimeric VDAC model produced from SEC-SAXS data in this study fit best utilizing the dimeric zebrafish VDAC2 organization for the protein phase of the model, implying that the N. crassa VDACR dimer is organized in this way in detergent solution in the presence of CHS. The two alternative models for VDAC dimer arrangements, which come from mouse VDAC1 data (7, 15), are unlikely arrangements of the N. crassa VDACR dimer observed in this study, considering that one arrangement contains antiparallel VDAC molecules, which is likely a crystal packing artifact (7, 13), and the other relies on the protonation state of E73, which corresponds to a proline residue in the aligned N. crassa VDAC sequence (8).
The hexameric VDACR in the presence of CHS modeled from SEC-SAXS data was not observed by AUC sedimentation velocity experiments; however, a possible explanation for this is the low concentrations of VDACR used in AUC (0.5–1 mg/mL) relative to SEC-SAXS (5–10 mg/mL) experiments. Interestingly, there is a small peak in the AUC c(s) and c(s, f/f0) distributions of VDACR + CHS samples at a larger S value than the VDACR tetramer (Fig. 2 d); however, the concentration of this species was too low to accurately determine its composition. Ultimately, the data describing the hexameric species was limited relative to the other species examined in this study; in addition, the only available crystallographic model for a VDAC hexamer arrangement (mouse VDAC1; PDB: 3EMN) is composed of antiparallel barrels leading to the suggestion that this is a biologically irrelevant model. However, it is possible that because of the low resolution of models produced by SAXS analysis, the data represent a similarly arranged parallel-barrel hexamer as similar VDAC arrangements have been observed by atomic force microscopy of mitochondrial membranes (17). The hexamer model has been included in Fig. S4. Future work in this area should focus on establishing the orientation of N. crassa VDAC molecules in these oligomers; this could be accomplished using the aforementioned EPR methods used to analyze the arrangement of mouse VDAC1 (15) and zebrafish VDAC2 (12) in solution.
The change in oligomeric organization of VDACR induced by the addition of CHS could be attributed to the direct binding of the sterol to VDAC. Direct binding of cholesterol to human VDAC1 has been described using NMR spectroscopy (6) and is supported by evidence of copurification of VDAC proteins with ergosterol or cholesterol (16, 38). The binding of sterols to VDAC could therefore be a possible target for drugs that could influence the oligomeric state of VDAC if direct binding of CHS to VDAC is the cause of the change in behavior. Such a drug could be designed to block or promote the binding of sterols to VDAC to influence apoptosis. However, biological systems are more complex, precluding direct extrapolation of our in vitro results to biological systems. The accumulation of cholesterol in mitochondria is observed in some cancer cell lines (45), and the accumulation of cholesterol in mitochondria has been shown to inhibit the activity of the proapoptotic protein BAX (46), suggesting that increased cholesterol content has an anti-apoptotic effect. The ability of CHS to promote the formation of VDAC oligomers is interesting as oligomerization is thought to have a pro-apoptotic effect (11). The relevance of the effect of cholesterol on mitochondrial-mediated apoptosis is further enhanced by evidence suggesting that some VDACs also interact directly with BAX (47). These data suggest an intricate system with many factors at play. Overall, the roles of VDAC and mitochondrial cholesterol content in apoptotic signaling warrant future study.
An alternative explanation for the change in VDAC behavior is that CHS may impact the detergent components of the system. CHS is known to alter the size and shape of n-dodecyl-β-D-maltopyranoside micelles, and CHS addition to maltoside detergents has been shown to stabilize solubilized G-protein-coupled receptors (GPCRs) (39, 48). CHS has also been shown to be required for the retention of ligand recognition by detergent solubilized and purified GPCRs such as the adenosine A2a receptor (48). This contrasts with VDAC in that in isolation, this protein showed no signs of instability in DM solutions. The VDAC-hexokinase complex, however, was stabilized by the presence of CHS (Fig. 5; Fig. S2). In addition, the restoration of the ability for VDAC to form oligomers by the introduction of CHS provides another interesting parallel with the effect of cholesterol on some GPCRs in that works using molecular dynamic simulations of the β2-adrenergic receptor (49) and the chemokine receptor type 4 (50) have shown that the dimerization of these proteins is modulated by cholesterol. It is possible that the change in size and shape of the detergent micelles is affecting the oligomeric reorganization of VDAC through changes to the structure of the molecule. Indeed, a change to the secondary structure was observed by our CD measurements, which could be due to the change in surrounding detergent environment.
The localization of hexokinase to the surface of mitochondria has been observed in mammalian and yeast cells (51, 52). Interestingly, the hexokinase could be detached from the mitochondrial surface by the introduction of a peptide consisting of the first 15 amino acids of the hexokinase, identifying the N-terminus as a possible VDAC binding site (10, 51). In this work, the binding of full-length S. cerevisiae HK-I with VDAC was reported, and an interaction between S. cerevisiae HK-II and VDAC was not detected. The relevance of this isoform specificity is supported by the upregulation of HK-I and downregulation of HK-II during respiration relative to fermentative growth in S. cerevisiae (53). However, because both S. cerevisiae hexokinases have identical N-termini, the differential binding behavior of the two isoforms would imply that the N-terminus of hexokinase is required but not sufficient for binding of full-length hexokinase to VDAC and that there is likely at least one other site of contact between the two proteins that is required for specific binding. Subcellular localization data for S. cerevisiae HK-I and II under fermentative growth conditions place both proteins in the cytoplasm; however, HK-I does associate with the mitochondria after treatment with rapamycin, which causes an increase in HK-I expression (52).
A combination of MST, AUC, and SEC-SAXS have revealed the most detailed picture of VDAC-detergent complexes and the effect of CHS on VDAC oligomeric organization. The identification of CHS as an effector of VDAC oligomeric organization and the detection of a VDAC-HK-I binding event will provide a platform for the further investigation of this system as well as lending the use of these methods in the examination of other membrane protein complexes. All data and models presented in this work are available upon reasonable request to the corresponding author, J.S.
Author Contributions
F.G.F. developed the biophysics pipeline. F.G.F., T.R.P., D.A.C., and J.S. designed the experiments. F.G.F., T.R.P., and G.O. performed the experiments. F.G.F. analyzed the data. F.G.F., T.R.P., D.A.C., and J.S. prepared the manuscript.
Acknowledgments
We thank the staff of Beamline B21 for their assistance with SAXS data collection and advice. T.R.P. is a Canada Research Chair in RNA & Protein Biophysics. SEC-SAXS data were collected at Diamond Light Source at Beamline B21 under the proposal SM16028.
This work was supported by the Natural Science and Engineering Council of Canada (RGPIN-004954-2017 to J.S. and RGPIN-05930-2016 to D.A.C.). F.G.F. was partially supported by the Faculty of Science and the Graduate Enhancement of Tri-Council Stipends of the Faculty of Graduate Studies at the University of Manitoba. J.S. is a Canada Research Chair in Structural Biology and Biophysics.
Editor: Andreas Engel.
Footnotes
Five figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30081-5.
Supporting Material
References
- 1.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Uhlén M., Fagerberg L., Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 3.Shoshan-Barmatz V., Ben-Hail D. VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochondrion. 2012;12:24–34. doi: 10.1016/j.mito.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Shoshan-Barmatz V., Krelin Y., Arif T. Voltage-dependent anion channel 1 as an emerging drug target for novel anti-cancer therapeutics. Front. Oncol. 2017;7:154. doi: 10.3389/fonc.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simamura E., Shimada H., Hirai K. Mitochondrial voltage-dependent anion channels (VDACs) as novel pharmacological targets for anti-cancer agents. J. Bioenerg. Biomembr. 2008;40:213–217. doi: 10.1007/s10863-008-9158-6. [DOI] [PubMed] [Google Scholar]
- 6.Hiller S., Garces R.G., Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ujwal R., Cascio D., Abramson J. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summers W.A., Court D.A. Origami in outer membrane mimetics: correlating the first detailed images of refolded VDAC with over 20 years of biochemical data. Biochem. Cell Biol. 2010;88:425–438. doi: 10.1139/o09-115. [DOI] [PubMed] [Google Scholar]
- 9.Arzoine L., Zilberberg N., Shoshan-Barmatz V. Voltage-dependent anion channel 1-based peptides interact with hexokinase to prevent its anti-apoptotic activity. J. Biol. Chem. 2009;284:3946–3955. doi: 10.1074/jbc.M803614200. [DOI] [PubMed] [Google Scholar]
- 10.Bryan N., Raisch K.P. Identification of a mitochondrial-binding site on the N-terminal end of hexokinase II. Biosci. Rep. 2015;35:e00205. doi: 10.1042/BSR20150047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoshan-Barmatz V., Keinan N., Aram L. Apoptosis is regulated by the VDAC1 N-terminal region and by VDAC oligomerization: release of cytochrome c, AIF and Smac/Diablo. Biochim. Biophys. Acta. 2010;1797:1281–1291. doi: 10.1016/j.bbabio.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Schredelseker J., Paz A., Abramson J. High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J. Biol. Chem. 2014;289:12566–12577. doi: 10.1074/jbc.M113.497438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ujwal R., Cascio D., Abramson J. Crystal packing analysis of murine VDAC1 crystals in a lipidic environment reveals novel insights on oligomerization and orientation. Channels (Austin) 2009;3:167–170. doi: 10.4161/chan.3.3.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayrhuber M., Meins T., Zeth K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergdoll L.A., Lerch M.T., Abramson J. Protonation state of glutamate 73 regulates the formation of a specific dimeric association of mVDAC1. Proc. Natl. Acad. Sci. USA. 2018;115:E172–E179. doi: 10.1073/pnas.1715464115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clémençon B., Fine M., Hediger M.A. Conservation of the oligomeric state of native VDAC1 in detergent micelles. Biochimie. 2016;127:163–172. doi: 10.1016/j.biochi.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Gonçalves R.P., Buzhynskyy N., Scheuring S. Supramolecular assembly of VDAC in native mitochondrial outer membranes. J. Mol. Biol. 2007;369:413–418. doi: 10.1016/j.jmb.2007.03.063. [DOI] [PubMed] [Google Scholar]
- 18.Koutsioubas A., Berthaud A., Pérez J. Ab initio and all-atom modeling of detergent organization around Aquaporin-0 based on SAXS data. J. Phys. Chem. B. 2013;117:13588–13594. doi: 10.1021/jp407688x. [DOI] [PubMed] [Google Scholar]
- 19.Pérez J., Koutsioubas A. Memprot: a program to model the detergent corona around a membrane protein based on SEC-SAXS data. Acta Crystallogr. D Biol. Crystallogr. 2015;71:86–93. doi: 10.1107/S1399004714016678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferens F.G., Spicer V., Court D.A. A deletion variant partially complements a porin-less strain of Neurospora crassa. Biochem. Cell Biol. 2017;95:318–327. doi: 10.1139/bcb-2016-0166. [DOI] [PubMed] [Google Scholar]
- 21.Miroux B., Walker J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 22.Davis R.H., de Serres F.J. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- 23.Summers W.A.T. University of Manitoba; 2010. An in vivio approach to elucidating the function of mitochondrial porin by the characterization of neurospora crassa strains deficient in porin. Ph.D. thesis. [Google Scholar]
- 24.Bay D.C., O’Neil J.D., Court D.A. Two-step folding of recombinant mitochondrial porin in detergent. Biophys. J. 2008;94:457–468. doi: 10.1529/biophysj.107.115196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdul-Gader A., Miles A.J., Wallace B.A. A reference dataset for the analyses of membrane protein secondary structures and transmembrane residues using circular dichroism spectroscopy. Bioinformatics. 2011;27:1630–1636. doi: 10.1093/bioinformatics/btr234. [DOI] [PubMed] [Google Scholar]
- 26.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H., Brown P.H., Schuck P. On the distribution of protein refractive index increments. Biophys. J. 2011;100:2309–2317. doi: 10.1016/j.bpj.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmer J., Doyle D.A., Grossmann J.G. Structural characterization and pH-induced conformational transition of full-length KcsA. Biophys. J. 2006;90:1752–1766. doi: 10.1529/biophysj.105.071175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown P.H., Balbo A., Schuck P. Using prior knowledge in the determination of macromolecular size-distributions by analytical ultracentrifugation. Biomacromolecules. 2007;8:2011–2024. doi: 10.1021/bm070193j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvay A.G., Santamaria M., Ebel C. Analytical ultracentrifugation sedimentation velocity for the characterization of detergent-solubilized membrane proteins Ca++-ATPase and ExbB. J. Biol. Phys. 2007;33:399–419. doi: 10.1007/s10867-008-9058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petoukhov M.V., Franke D., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipfert J., Columbus L., Doniach S. Size and shape of detergent micelles determined by small-angle X-ray scattering. J. Phys. Chem. B. 2007;111:12427–12438. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- 35.Kopetzki E., Entian K.D. Purification of yeast hexokinase isoenzymes using affinity chromatography and chromatofocusing. Anal. Biochem. 1982;121:181–185. doi: 10.1016/0003-2697(82)90573-5. [DOI] [PubMed] [Google Scholar]
- 36.Scheuermann T.H., Padrick S.B., Brautigam C.A. On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 2016;496:79–93. doi: 10.1016/j.ab.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freitag H., Neupert W., Benz R. Purification and characterisation of a pore protein of the outer mitochondrial membrane from Neurospora crassa. Eur. J. Biochem. 1982;123:629–636. doi: 10.1111/j.1432-1033.1982.tb06578.x. [DOI] [PubMed] [Google Scholar]
- 38.De Pinto V., Benz R., Palmieri F. Interaction of non-classical detergents with the mitochondrial porin. A new purification procedure and characterization of the pore-forming unit. Eur. J. Biochem. 1989;183:179–187. doi: 10.1111/j.1432-1033.1989.tb14911.x. [DOI] [PubMed] [Google Scholar]
- 39.Thompson A.A., Liu J.J., Stevens R.C. GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods. 2011;55:310–317. doi: 10.1016/j.ymeth.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Runke G., Maier E., Court D.A. Deletion variants of Neurospora mitochondrial porin: electrophysiological and spectroscopic analysis. Biophys. J. 2006;90:3155–3164. doi: 10.1529/biophysj.105.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roman I., Figys J., Zizi M. Hunting interactomes of a membrane protein: obtaining the largest set of voltage-dependent anion channel-interacting protein epitopes. Mol. Cell. Proteomics. 2006;5:1667–1680. doi: 10.1074/mcp.T600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Cole J.L., Lary J.W., Laue T.M. Methods Cell. Biol. Vol. 84. 2008. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium; pp. 143–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betaneli V., Petrov E.P., Schwille P. The role of lipids in VDAC oligomerization. Biophys. J. 2012;102:523–531. doi: 10.1016/j.bpj.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gimpl K., Klement J., Keller S. Characterising protein/detergent complexes by triple-detection size-exclusion chromatography. Biol. Proced. Online. 2016;18:4. doi: 10.1186/s12575-015-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Ruiz C., Mari M., Fernández-Checa J.C. Mitochondrial cholesterol in health and disease. Histol. Histopathol. 2009;24:117–132. doi: 10.14670/HH-24.117. [DOI] [PubMed] [Google Scholar]
- 46.Lucken-Ardjomande S., Montessuit S., Martinou J.C. Bax activation and stress-induced apoptosis delayed by the accumulation of cholesterol in mitochondrial membranes. Cell Death Differ. 2008;15:484–493. doi: 10.1038/sj.cdd.4402280. [DOI] [PubMed] [Google Scholar]
- 47.Lauterwasser J., Todt F., Edlich F. The porin VDAC2 is the mitochondrial platform for Bax retrotranslocation. Sci. Rep. 2016;6:32994. doi: 10.1038/srep32994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss H.M., Grisshammer R. Purification and characterization of the human adenosine A(2a) receptor functionally expressed in Escherichia coli. Eur. J. Biochem. 2002;269:82–92. doi: 10.1046/j.0014-2956.2002.02618.x. [DOI] [PubMed] [Google Scholar]
- 49.Prasanna X., Chattopadhyay A., Sengupta D. Cholesterol modulates the dimer interface of the β2-adrenergic receptor via cholesterol occupancy sites. Biophys. J. 2014;106:1290–1300. doi: 10.1016/j.bpj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pluhackova K., Gahbauer S., Böckmann R.A. Dynamic cholesterol-conditioned dimerization of the G protein coupled chemokine receptor type 4. PLoS Comput. Biol. 2016;12:e1005169. doi: 10.1371/journal.pcbi.1005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiara F., Castellaro D., Rasola A. Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PLoS One. 2008;3:e1852. doi: 10.1371/journal.pone.0001852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chong Y.T., Koh J.L., Andrews B.J. Yeast proteome dynamics from single cell imaging and automated analysis. Cell. 2015;161:1413–1424. doi: 10.1016/j.cell.2015.04.051. [DOI] [PubMed] [Google Scholar]
- 53.Roberts G.G., Hudson A.P. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol. Genet. Genomics. 2006;276:170–186. doi: 10.1007/s00438-006-0133-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.