Abstract
Mutations in the GBA gene, encoding the lysosomal hydrolase glucocerebrosidase (GCase), are the most common known genetic risk factor for Parkinson’s disease (PD) and dementia with Lewy bodies (DLB). The present study aims to gain more insight into changes in lysosomal activity in different brain regions of sporadic PD and DLB patients, screened for GBA variants. Enzymatic activities of GCase, β-hexosaminidase, and cathepsin D were measured in the frontal cortex, putamen, and substantia nigra (SN) of a cohort of patients with advanced PD and DLB as well as age-matched non-demented controls (n = 15/group) using fluorometric assays. Decreased activity of GCase (− 21%) and of cathepsin D (− 15%) was found in the SN and frontal cortex of patients with PD and DLB compared to controls, respectively. Population stratification was applied based on GBA genotype, showing substantially lower GCase activity (~ − 40%) in GBA variant carriers in all regions. GCase activity was further significantly decreased in the SN of PD and DLB patients without GBA variants in comparison to controls without GBA variants. Our results show decreased GCase activity in brains of PD and DLB patients with and without GBA variants, most pronounced in the SN. The results of our study confirm findings from previous studies, suggesting a role for GCase in GBA-associated as well as sporadic PD and DLB.
Electronic supplementary material
The online version of this article (10.1007/s12035-018-1090-0) contains supplementary material, which is available to authorized users.
Keywords: GBA variants, Autophagy-lysosomal pathway, Cathepsin D, β-hexosaminidase
Introduction
Converging evidence from genetic, pathological, and experimental studies has suggested that impairment of the autophagy-lysosomal pathway (ALP) is a key pathological event in the pathogenesis of Parkinson’s disease (PD) and dementia with Lewy bodies (DLB) [1, 2]. Mutations in numerous genes encoding for ALP components have been associated with increased risk to develop PD, including Glucosidase Beta Acid 1 (GBA) [3]. Homozygous GBA mutations cause Gaucher disease (GD), and heterozygous GBA mutations form the major genetic risk factor for PD and DLB. The prevalence of GBA variants is estimated 5–10% in PD and DLB patients and higher in certain studied populations, particularly among Ashkenazi Jews (~ 20%) [4–6]. In GD, approximately 300 different GBA mutations have been described, many of which are also found in PD [7]. Recent studies showed GBA mutations impact the clinical phenotype of PD and DLB, as their presence has been associated with rapid eye movement sleep behavior disorder, a more rapid disease course, earlier age of onset, and higher risk for cognitive decline and dementia [8–11]. Moreover, a worse outcome has been reported for pathogenic mutations, for instance L444P, than for milder GBA variants such as N370S [11–14].
GBA encodes for the lysosomal hydrolase β-glucocerebrosidase (GCase), which catalyzes the conversion of glucosylceramide into glucose and ceramide [5]. Decreased protein levels and enzymatic activity of GCase were found in affected brain regions [15–18], dried blood spots [19], fibroblasts [20], peripheral blood mononuclear cells [21], and CSF [22–24] of PD patients with GBA mutations, but also in sporadic PD patients. Alterations in the activities of several other lysosomal enzymes were found in brain tissue and CSF of PD or DLB patients compared to controls, including β-hexosaminidase (β-Hex) and cathepsin D (CathD) [22–26]. In addition, decreased protein levels have been demonstrated for various other ALP components in the PD brain [27–30]. Together, these findings suggested that GCase dysfunction as well as more widespread deregulation of the lysosomal system is involved in PD and DLB pathology.
This study aims to obtain more insight into the changes in lysosomal enzymes, including GCase, in different regions of PD and DLB brains and their relation to the presence of different GBA variants. In order to do this, we measured enzymatic activities for GCase, CathD, and β-Hex in the frontal cortex (FC), putamen, and substantia nigra (SN) of a clinically and pathologically well-characterized cohort of PD and DLB patients, as well as in age-matched non-demented controls. To investigate whether fluctuations in enzymatic activities were reflected at the level of mRNA, we measured expression levels for GBA, and genes encoding CathD (CTSD), GCase’s protein interactors lysosomal integral membrane-protein 2 (LIMP-2) and saposin C (SapC) —which are important determinants for GCase activity [31]—as well as selected other components of the ALP. Our results confirm the involvement of GCase in GBA-associated as well as idiopathic PD and DLB, most pronounced in the SN, and demonstrate a stepwise decrease in GCase activity when comparing distinctive GBA genetic subgroups.
Methods
Selection of Postmortem Human Brain Tissue
Human postmortem brain tissue was collected from clinically diagnosed and neuropathologically verified advanced PD and DLB patients (15 per group) and 15 age-matched non-neurological control subjects from the Netherlands Brain Bank (NBB, Amsterdam, The Netherlands; Table 1). All donors had short postmortem delay (PMD; < 10 h), a pre-analytical factor that can potentially impact the stability of lysosomal enzymes [32]. In compliance with all local ethical and legal guidelines, informed consent for brain autopsy and the use of brain tissue and clinical information for scientific research was given by either the donor or the next of kin. Brains were dissected in compliance with standard operating protocols of the Netherlands Brain Bank (www.brainbank.nl). Frozen tissue blocks of the superior frontal cortex, putamen and SN were collected at autopsy. Tissue blocks were pulverized using a mixer mill (model MM400; Retsch, Haan, Germany) during 2 min at a frequency of 30 Hz. Pulverization took place in stainless-steel grinding jars, precooled in liquid nitrogen to prevent thawing of the tissue. The procedure was repeated, when needed, until all tissue was pulverized. Subsequently, tissue was stored at − 80 °C in aliquots.
Table 1.
Cohort demographics. *p < 0.05 (Pearson chi-square test)
| Controls (N = 15) | PD (N = 15) | DLB (N = 15) | p value | |
|---|---|---|---|---|
| Age of death (years ± SD) | 77 ± 6 | 77 ± 5 | 77 ± 5 | 0.67 |
| Sex (M/F) | 4/11 | 11/4 | 10/5 | 0.02* |
| Postmortem delay (hours ± SD) | 6.3 ± 1.7 | 6.0 ± 1.8 | 5.2 ± 1.2 | 0.47 |
| Braak Lewy Body Score | 0–1 (14/1) | 4–6 (3/5/7) | 4–6 (3/3/9) | < 0.01* |
| Braak score for Neurofibrillary Tangles | 0–2 (2/6/7) | 0–3 (2/9/2/2) | 0–3 (1/8/5/1) | 0.46 |
| CERAD Amyloid Plaque Score | 0-B (4/7/4) | O-C (7/5/2/1) | O-C (3/3/8/1) | 0.21 |
| Disease duration (years ± SD) | – | 15 ± 7 | 6 ± 3 | < 0.01* |
| No. of donors with GBA risk factors | 2 | 3 | 4 | – |
| No. of donors with pathogenic GBA variants | 0 | 3 | 0 | – |
Genotyping
High molecular-weight genomic DNA was extracted from frozen cerebellum specimens using the Gentra Puregene Tissue Kit (Qiagene, Hilden, Germany). The genomic region encompassing GBA open reading frame was amplified in large fragments in order to avoid amplification of the neighboring GBAP pseudogene. Subsequently, we Sanger sequenced GBA in 43 samples, as for 2 samples (1 control, 1 DLB patient), no brain tissue was available. We report variants if they are of coding effect or within 20 bp from the intron-exon boundaries, and if their minor allele frequency (MAF) is lower than 1% in Exome Aggregatium Consortium browser (ExAC, http://exac.broadinstitute.org/). Variants nomenclature is assigned according to NM_000157.3 transcript, consisting of 11 exons and encoding for a total of 536 amino acids. We also report common nomenclature attributed to GBA variants (between brackets), in which the amino acid count does not include the initial 39 amino acids corresponding to the residue signal peptide. We investigated the effect on splicing of GBA c.762-18T>A variant according to five splicing prediction tools (SpliceSiteFinder-like, MaxEntScan, NNSPLICE, GeneSplicer, and Human Splicing Finder) integrated in Alamut Visual version 4.2 (Interactive Biosoftware, Rouen, France).
Enzyme Activity Assays
Pulverized tissue (approximately 50 mg) was lysed in a 50 mM sodium/phosphate (Na/P), 150 mM NaCl buffer, pH 7.0 and homogenized using a homogenizer (Ultra-turrax, model T10B; IKA, Wilmington, NC, USA), after which 0.1% of tergitol-typenonyl phenoxypolyethoxylethanol 40 (Igepal CA630; Sigma-Aldrich, Saint Louis, MO, USA) was added. Samples were ultra-sonicated (Sonopuls, model: UW3100; Bandelin, Berlin, Germany), kept on ice for 30 min, and subsequently centrifuged at a speed of 15,000×g for 10 min in a bench centrifuge (model 5415D; Eppendorf, Hamburg, Germany). Fluorometric enzyme activity assays (EAAs) for GCase, β-Hex, and CathD were performed in accordance with the protocols previously described [16, 25]. All EAAs were performed in triplicate. Mean intra-assay coefficients of variation (CV) were lower than 5% for all enzymes, with a maximum CV of 8.5%. Enzymatic activities were normalized for total protein content of the samples to obtain the specific activity. Total protein concentration in the samples was determined according to the method described by Bradford [33].
RNA Extraction and Quantitative PCR
To determine whether fluctuations in enzymatic activities were reflected at the level of mRNA, we have measured the expression levels for different ALP-related genes using quantitative PCR (qPCR). Experiments were performed on pulverized FC and SN tissue, but not on putamen as not enough material was left. Apart from GBA, mRNA expression levels were measured for the genes that encode CathD and GCase’s protein interactors LIMP-2 (SCARB2) and saposin C (PSAP) [31]. Finally, genes encoding lysosomal membrane-associated receptors (LAMP-1; LAMP-2) and autophagy (TFEB) were measured as more general markers for the ALP. RNA extraction was performed as previously described, using a Trizol/chloroform protocol [34]. The RNA integrity number (RIN) was determined using an Agilent TM 2100 Bioanalyzer and an RNA 6000 Nano LabChip Kit (Agilent Technologies, Palo Alto, CA, USA). We applied an RIN value of 5.0 as threshold for inclusion for the qPCR analysis. Consequently, a smaller subset of 29 samples was included for the FC (11 controls/10 PD/ 8 DLB) and 27 for the SN (10 controls/9 PD/8 DLB). cDNA synthesis was done using the High-Capacity cDNA Reverse Transcription Kit (Art. No. 4368814; ThermoFischer, Waltham, MA, USA), and cDNA was stored at − 20 °C until use. For all genes of interest, intron-spanning Taqman assays were designed (Table S1). Primers were synthesized by Eurogentec (Luik, Belgium). Taqman probes were selected from the Human Universal Probe Library (UPL; Roche Applied Science, Indianapolis, IN, USA). For normalization, the housekeeping genes m-RIP, OAZ-1, and POL2RF were selected using geNorm software from a subset of candidate housekeeping genes [35]. Standard curves were generated for each assay run. The relative expression ratio of each target gene was calculated using the efficiency-corrected delta-delta Cq method [34, 36]. All assays were performed in duplicate (mean CV = 5.4%).
Statistics
Hierarchical clustering analysis was done using R software (Version 3.2.5, R Foundation for Statistical Computing, Vienna, Austria) [37], to compare lysosomal enzyme activities across different brain areas. Further statistical analyses were performed using SPSS package version 20.0 (Statistical Product and Service Solutions). As enzyme activities between brain regions were correlated within subjects, general linear models for mixed effects were used to test for differences between PD and DLB patients with controls, in which contribution of individual patient groups per brain region were further studied. In other comparisons, ANCOVA analyses were applied. In all models, age was included as a covariate. For the qPCR data, the RIN value was included as weighted least squares weight to the models. A p value < 0.05 was considered significant in all analyses. All reported p values were corrected for multiple comparisons: multiple comparisons were taken into account in the general linear model for mixed effects, while Bonferroni post hoc tests were applied in the univariate models. Graphs and figures were generated using R software and GraphPad (Version 7.0, Prism, La Jolla, CA, USA) and composed in Adobe Photoshop (version CS6, Adobe Systems Incorporated, San Jose, CA).
Results
Lysosomal Enzyme Activities
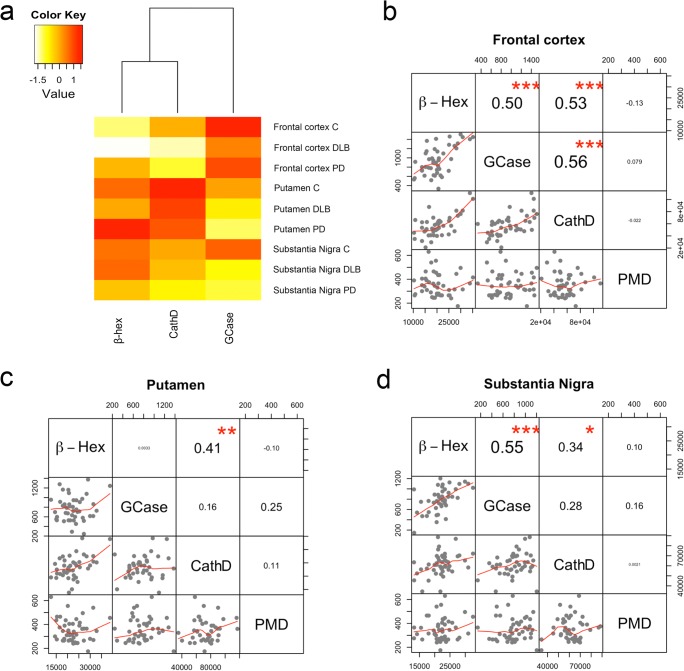
Lysosomal enzyme activities across samples revealed differences between brain regions (Fig. 1a). GCase activity was higher in the FC compared to the other regions, while CathD and β-Hex activities were highest in the putamen. Activities of different lysosomal enzymes were sometimes correlated within a brain region, but not with postmortem delay (Fig. 1b–d). GCase activity was significantly decreased in the SN of PD and DLB patients compared to controls (− 21%; p = 0.02), while this effect was not significant in other studied brain regions (Fig. 2b, d). Post hoc analyses revealed that GCase was particularly decreased in the SN of PD patients compared to controls (− 22%; p = 0.04), with a similar trend in DLB patients (− 20%; p = 0.06). No differences in GCase activity were observed between PD and DLB patient groups in any of the studied brain regions. For CathD, a significant decrease in enzymatic activity was found in the FC of PD and DLB patients compared to controls (− 15%; p = 0.05; Fig. 2c). This effect was most pronounced in DLB patients (− 18%; p = 0.04) while a similar trend did not reach significance in PD patients (− 12%; p = 0.16). For the activity of β-Hex, no between-group differences were observed in any of the measured brain regions (Fig. 2a).
Fig. 1.
Lysosomal enzymatic activities in different regions of the human postmortem brain. a Cluster analysis of average enzymatic activities across patients for GCase, β-Hex, and CathD in the different studied regions reveals that GCase activity was highest in the frontal cortex, while CathD and β-Hex activities were higher in the putamen compared to the other studied regions. b, c, and d Correlation matrices for lysosomal enzymatic activities and PMD for frontal cortex (b), putamen (c), and SN (d). High correlations were sometimes observed between activities of different lysosomal enzymes, but not with PMD. Values represent Spearman’s correlation coefficients. *p < 0.05; **p < 0.01; ***p < 0.001
Fig. 2.
Lysosomal enzyme activities in the brain of PD and DLB patients compared to controls. Group comparison of measured specific activities for β-Hex (a), GCase (b), and CathD (c) expressed in pmol/min/mg of proteins. Graphs show the average specific activity and standard deviation for non-demented controls (blue), PD patients (green), and DLB patients (blue). d. Overview of mean enzyme activities ± standard deviations. The p value indicates the between-group difference per brain region. GCase activity was significantly decreased in the SN of PD patients compared to controls, while lower CathD activity was found in the frontal cortex of DLB patients compared to controls *p < 0.05
Gene Expression
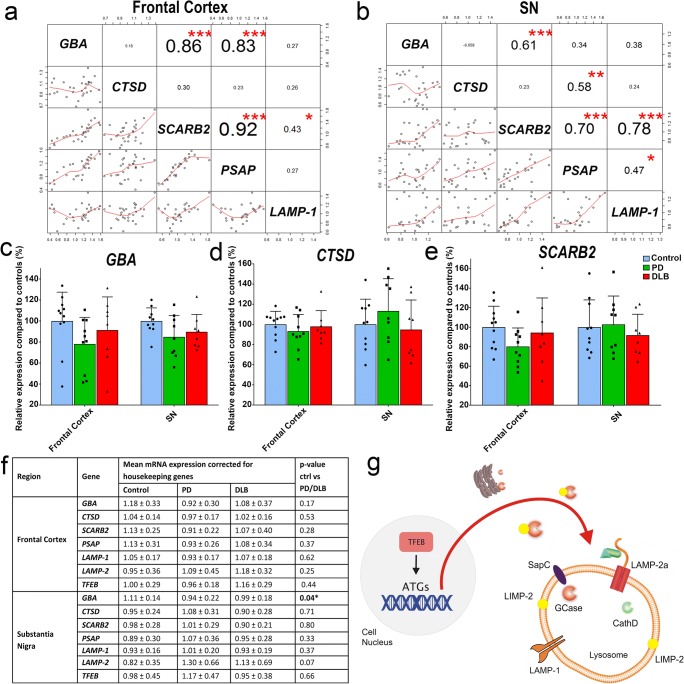
To examine whether changes in enzyme activity were reflected at the mRNA level, gene expression was assessed for GBA and CTSD. In addition, mRNA expression levels were measured for genes encoding the GCase protein interactors LIMP-2 (SCARB2) and saposin C (PSAP), which are important determinants for GCase activity [31]. Finally, gene expression levels were measured for more general markers associated with lysosomes (LAMP-1; LAMP-2) or the ALP (TFEB; Fig. 3g). GBA and CTSD expression were not correlated with GCase or CathD activity in both SN and FC (Fig. S1). However, strong correlations between mRNA expression levels were found between certain ALP genes, for instance between GBA and SCARB2 (Fig. 3a). Normalized mRNA expression levels for all measured genes showed substantial variability within groups. Still, decreased normalized GBA mRNA expression levels were observed in the SN of PD and DLB patients compared to controls (− 13%; p = 0.04; Fig. 3a, g). A similar pattern could be observed in the FC, although differences were not significant (− 16%; p = 0.17). No significant differences in the mRNA expression levels were found for CTSD or any of the other measured ALP-related genes in either FC or the SN (Fig. 3c, f).
Fig. 3.
mRNA expression levels for GBA, CTSD, and other ALP components. Correlation matrices showing association between mRNA expression levels of GBA and genes encoding CTSD, GCase’s protein interactors LIMP-2 (SCARB2) and saposin C (PSAP), and the lysosomal membrane-associated protein LAMP-1 in the frontal cortex (a) and SN (b). Normalized mRNA expression levels for GBA (c), CTSD (d), and SCARB2 (e) in PD (green) and DLB (red) patients as well as non-demented controls (blue). Expression levels are expressed as percentage of the average expression level of non-demented controls. f Schematic outline of the interplay between the measured gene products. LIMP-2 functions as a transporter molecule for GCase from the endoplasmatic reticulum, while the interaction of GCase with SapC is essential for its proper function [31]. LAMP-1 and LAMP-2a are lysosome-associated membrane proteins, while transcription factor EB (TFEB) is the master regulator of biogenesis and function of lysosomes, by activating transcription of autophagy-related genes (ATGs). *p < 0.05; **p < 0.01; ***p < 0.001
GBA Screening
According to above-mentioned criteria, six GBA variants were detected in a total of 12 out of 43 tested specimens, and specifically in 6 PD patients, 4 DLB patients and 2 non-demented controls (Table 1). Based on previous literature as well as on the predicted effect on GCase functioning, a distinction was made between putative pathogenic variants—that are causative for GD—and variants that are risk factors for PD. We identified GBA c.535G>C p.Asp179His (p.Asp140His) and GBA c.1448T>C p.Leu483Pro (p.Leu444Pro) variants in heterozygous state in ID-24 and ID-26 PD donors, respectively. In the context of recessive inheritance, both variants have been associated with GD and with PD if carried in heterozygous state [38]. In ID-18 (PD), we identified a GBA c.1073C>T p.Pro358Leu (p.Pro319Leu) variant, which has been reported before in a Dutch patient with GD type 1 [39]. As this variant affects an evolutionary-conserved amino acid, it is likely pathogenic [40].
Apart from these GD-associated pathogenic mutations, we detected variants in our cohort for which the association with GD is less clear. First, we detected heterozygous GBA c.1093G>A p.Glu365Lys (p.Glu326Lys) and GBA c.1223C>T p.Thr408Met (p.Thr369Met) variants in eight (3 PD and 3 DLB patients, 2 controls) and two (1 PD and 1 DLB patients) brain specimens, respectively. Both of these variants are not causative for GD in homozygous carriers, although they may modify GCase activity and GD phenotype [41, 42]. In particular, p.Glu326Lys has been established as risk factor for PD, as it is the most prevalent PD-associated GBA mutation, which was demonstrated in a large meta-analysis combining data from different genome-wide association studies [43]. Although the association of p.Thr369Met variants with PD is more controversial, accumulating evidence suggests that this variant may be risk factor with a minor effect [41]. Therefore, both p.Glu326Lys and p.Thr369Met were considered as GBA risk factors in further analyses [6, 10, 41]. Finally, we found GBA c.762-18T>A variant in homozygous state in ID-16 PD patient’s brain. Yet, this variant is not reported to be associated with GD nor PD. Out of 5 addressed splicing prediction tools, only the Human Splicing Finder prediction software predicts that the alternative nucleotide introduces a weaker acceptor site, but without affecting the efficiency of the default one. Based on these observations, we have considered the c.762-18T>A as potential PD risk factor rather than pathogenic variant in our analysis. Of note, for all these detected GBA PD risk factors, although MAFs are lower than 1% in the ExAc database (p.Glu326Lys, 0.98%; p.Thr369Met, 0.66%; c.762-18T>A, 0.79%), their occurrence in the Netherlands is higher than 1%, as shown by the GoNL database (p.Glu326Lys, 2.3%; p.Thr369Met, 1.1%; c.762-18T>A, 1.2%) [44].
One PD donor (ID-24) was found to carry three GBA alterations, namely, c.535G>C p.Asp179His (p.Asp140His), c.1093G>A p.Glu365Lys (p.Glu326Lys), and c.1223C>T p.Thr408Met (p.Thr369Met) variants. Unfortunately, the phase of these variants (cis or trans) could not be established as relatives of the carrier were not available for testing. There are examples in the literature in which multiple GBA variants are located on the same chromosome as the result of complex genomic rearrangements occurring between the GBA gene and the GBAP pseudogene [7], which cannot be ruled out in this case. Moreover, the co-occurrence of p.Asp140His plus p.Glu365Lys variants on complex alleles has been reported repeatedly [12, 45, 46]. Of interest, although this patient possibly met the biochemical and genetic criteria for being diagnosed with GD [47], clinically and pathologically she was referred as affected by PD.
In our study design, it was not possible to differentiate PD/DLB patients with GBA variants (PD/DLB+GBA) from PD/DLB-GBA patients based on clinical symptoms, mainly because of the limited sample size. However, for all GBA variant carriers in our study substantial cognitive symptoms and other non-motor symptoms were reported during the disease course, including visual hallucinations. An overview of prominent clinical features of the PD and DLB patients with GBA variants in our study is provided in Table S3. All cases with GBA variants showed widespread Lewy Body pathology at autopsy, with Braak α-synuclein stages similar to non-carriers in our study.
Population Stratification Based on GBA Variants
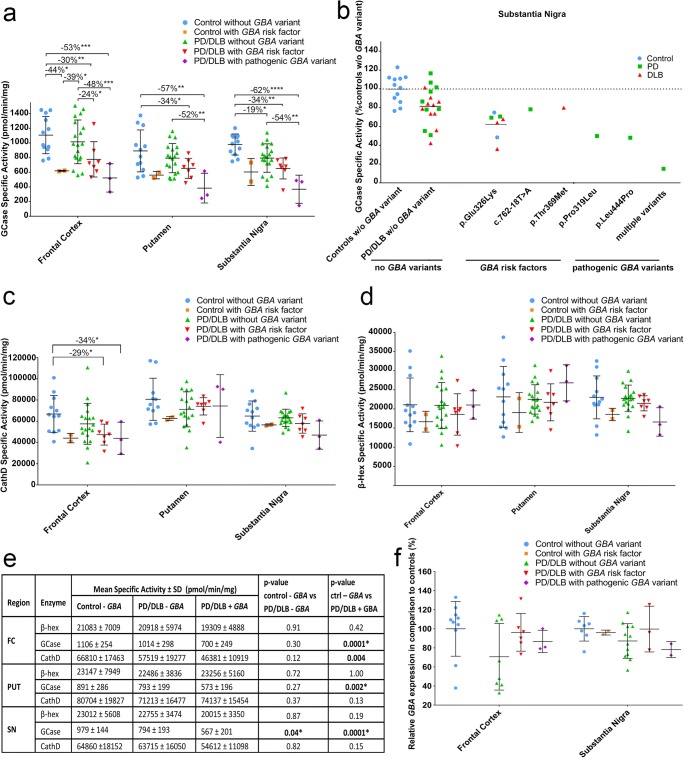
GCase activity was substantially lower in PD/DLB+GBA patients in comparison to controls without GBA variants (p < 0.0001). This effect was present in all studied brain regions (Fig. 4a–e). Moreover, GCase enzymatic activity was significantly decreased in the group of PD/DLB+GBA patients compared to PD/DLB-GBA patients, confirming that the presence of GBA variants was a major determinant for GCase activity in our cohort. However, the group of PD/DLB-GBA patient showed significantly decreased GCase activity compared to control subjects without GBA variants in the SN (19%; p = 0.04), but to lesser extent in other brain regions (Fig. 4a, b). Similar to GCase activity, CathD activity was substantially decreased in the FC of PD/DLB+GBA patients compared to controls without GBA variants (− 31%; p < 0.01; Fig. 4c). However, CathD activity was not significantly altered in the other brain regions. For β-Hex, no differences in enzymatic activity were observed between PD/DLB+GBA, PD/DLB-GBA patients, and controls in any of the studied brain regions (Fig. 4d). The differences in GCase activity between PD/DLB+GBA, PD/DLB-GBA patients, and controls were not reflected by alterations on the mRNA level in either FC or SN (Fig. 4f).
Fig. 4.
Population stratification based on GBA genotype. Specific enzymatic activities for GCase (a), β-Hex (c), and CathD (d) for non-demented controls without GBA variants (blue) and with GBA risk factors (orange), for PD and DLB patients without GBA variants (green), with GBA risk factors (red), and with pathogenic GBA variants (purple). b Measured GCase activities in the SN per GBA variant for controls (blue), PD patients (green), and DLB patients (red). d Overview of mean enzyme activities ± standard deviations in different GBA genetic subgroups. p values indicate the between-group difference per brain region. e GBA mRNA expression levels for controls, PD and DLB patients without GBA variants, with GBA risk factors, and pathogenic GBA variants. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Discussion
The present study provides a comprehensive measurement of enzymatic activities as well as mRNA expression levels for GCase and other ALP components in selectively vulnerable regions of patients with advanced PD patients and DLB. We found that GCase activity is decreased in postmortem brains of PD and DLB patients with and without GBA variants compared to age-matched controls. Thereby, we confirm findings of previous studies in brain tissue from an independent cohort of clinically and pathologically well-characterized donors with PD and DLB with short PMD. Our study further provides insight in the relation between brain GCase activity and the presence of different GBA variants. In particular, population stratification based on GBA genotype showed a genotype-dependent decrease in GCase activity (Fig. 4). Finally, we observed decreased CathD activity in the FC of PD and DLB patients compared to controls, particularly in patients carrying GBA mutations.
As reported in previous studies, we find the most pronounced effects in the SN, a region associated with α-synuclein pathology and other biochemical abnormalities, including mitochondrial dysfunction, oxidative stress, and neurodegeneration [15, 16]. A contribution of dopaminergic cell-death in the SN to the reduction of GCase activity in this region cannot be excluded in our study. However, we did not observe differences in total protein content—for which we corrected enzymatic activities—or in other measured components (for instance β-Hex activity) in the SN of PD and DLB patients compared to controls. Moreover, a previous study did not detect changes in GCase activity in brain regions with massive neurodegeneration, such as the amygdala of donors with Alzheimer’s disease [15].
We found a significant 19% decrease of GCase activity in the SN of patients without GBA mutation compared to controls, which is in line with the slightly higher (33%) [15] or lower (12%) [16] differences that were previously reported in this brain region. However, in the first study, a possible contribution of GBA variants in the group of sporadic PD patients cannot be ruled out as no genetic screening was reported for this group of donors [15]. Genotyping of our cohort revealed a markedly high rate of GBA variants, with 6 carriers out of 15 screened PD donors, 4/14 DLB donors, and 2/14 screened controls. Two previously defined pathogenic variants (Asp140His and p.Leu444Pro) were detected, while one possibly pathogenic missense variant (p.Pro319Leu) was found. The presence of GBA variants resulted in a substantially lower GCase activity in all studied brain regions (~ − 40%), most pronounced in the SN, where the PD/DLB+GBA group showed no overlap with controls (Fig. 4b).
GCase activities were lowest in donors with pathogenic GBA variants. Although sample size for different genetic GBA subgroups—particularly, the number of donors with pathogenic GBA variants—was small, our data indicates a stepwise decrease in GCase activity in different GBA genetic subgroups. Interestingly, a similar “dose-effect” of GBA variants in PD has been suggested in recent clinical and genetic studies, in which differential effects of mild and severe GBA variants on PD clinical phenotype were reported. Severe GBA variants were associated with more progressive forms of PD [9, 11–13], and with higher genetic risk to develop PD [14]. Insight into the effects of different GBA variants on GCase activity in the brain can be important for patient selection and therapeutic efficacy for novel therapies that aim to increase GCase enzyme activity as disease-modifying therapy in PD/DLB, for instance using small molecule non-inhibitory chaperones [48].
The lowest GCase activity was detected in PD donor ID-24 who carried three GBA variants, whose phases (cis or trans) could not be established. Therefore, it is not clear whether these three variants contribute to the lower enzymatic activity in an independent manner, with the p.Asp140His variant acting as driving factor, or if they act in a synergic fashion. The extremely low GCase enzymatic activity prompts us to speculate that both GBA alleles are altered, with two variants being in cis and the remaining one in trans. In particular, a combination of p.Asp140His plus p.Glu365Lys variants at one allele has been reported in several studies [12, 45, 46]. Of interest is the fact that this patient, despite of very low GCase activities, was diagnosed with PD and not with GD during her life.
To investigate whether changes in enzymatic activity were reflected at the level of mRNA, we measured expression levels of GBA and CTSD. In addition, we measured expression levels for genes encoding GCase’s protein interactors LIMP-2 and SapC—which are proposed to be important determinants for GCase activity [31], to explore their interrelation. No correlation was found between GBA and CTSD mRNA expression with GCase or CathD activity in our data, while the systematic decrease in GCase activity in different pathological and genetic GBA subgroups was not observed at the mRNA level. The lack of association between mRNA levels and GCase enzymatic activity was not unexpected, as it has been reported before in studies on GD [7], and indicates that GCase activity is not simply a reflection of a GBA gene expression but also driven by other factors. In that aspect, it is interesting that we observed strong correlations between GBA and genes encoding its posttranslational regulators LIMP-2 and SapC, indicating an interrelation of these components at a transcriptional level.
Gene expression data showed large variability in patients and controls. This variability may be partially explained by differences in tissue composition. In particular, the content of cellular components in diseased tissue can be changed as a result of inflammatory and neurodegenerative processes. Despite of this variability, still, a trend for decreased GBA mRNA expression levels was observed in the SN of PD and DLB patients. Reduced GBA expression in the SN has previously been reported [16], while another study reported a trend for decreased GBA mRNA expression in the anterior cingulate cortex [17]. In other regions, including putamen [15] and occipital cortex [17], no evidence of reduced GBA mRNA expression was found. Interestingly, the results after population stratification showed that reduced GBA expression was not associated with the presence of GBA variants. Taken together, we conclude that lower GCase activities do not correspond with lower mRNA levels, while a role for altered GBA mRNA levels in sporadic PD and DLB cannot be excluded. However, at this point, the relevance of this latter observation is not clear.
We did not find differences in β-Hex activities between PD and DLB patients compared to controls. A role for β-Hex in PD pathology has been suggested, as patients affected by GM2 gangliosidosis, which results from reduced β-Hex activity, can manifest with parkinsonism [49–51]. Further, a role of β-Hex in PD pathogenesis is supported by animal studies showing that mice lacking β-Hex showed α-synuclein-positive neuronal inclusions at 4 months of age [52]. In biomarker studies, it was reported that CSF β-Hex enzyme activity is altered in PD patients compared to age-matched neurological controls [22, 23]. However, no difference in β-Hex activity was detected in brain tissue of PD and DLB patients compared to controls, in our and also other studies [15, 16, 24–26]. Together, the evidence for a role of β-Hex in sporadic PD is currently limited.
We observed decreased CathD activity in the FC of DLB patients compared to controls, while a similar non-significant effect was observed in PD patients. Of interest, CathD activity was particularly decreased in patients with GBA variants. A role for CathD has been proposed in different neurodegenerative diseases [53], including Alzheimer’s disease, Huntington’s disease, and PD [54–57]. Our results are in line with a previous study, which also showed lower CathD activity in the FC of patients with PD and DLB [28], while reduced expression levels of CathD have also been reported in the SN of PD patients [27]. CathD is a lysosomal protease involved in the posttranslational cleavage of prosaposin (encoded by PSAP), leading to the production of saposin C, a GCase activator [31, 58]. Thus, interestingly, although we did not find changes in the mRNA expression of PSAP, the activity of the enzyme responsible for its posttranslational regulation was altered.
The within-subject design of our study allowed to demonstrate the differential regulation of lysosomal enzymatic activity in different brain regions, as has been suggested previously in the literature [16]. However, in contrast to this study [16], enzyme activities were not generally higher in the SN. Rather, this effect seems to depend on the specific lysosomal enzyme, as CathD activity was significantly higher in the putamen than in SN and FC. A systematic mapping of lysosomal enzyme gene/protein expression and activities in the brain could provide more insight into the differential regulation of lysosomal enzymes between brain regions, and may allow better understanding of region-specific effects of lysosomal proteins.
In conclusion, our results show decreased GCase activity in the brain of PD and DLB patients with and without GBA variants. GCase activities in brain tissue were related to the presence as well as the pathogenicity of GBA variants. The results of our study confirm findings from previous studies, and provide important insights into the role of GCase in GBA-associated as well as sporadic PD and DLB.
Electronic Supplementary Material
(DOCX 185 KB)
(DOCX 25.0 KB)
(DOCX 18.7 KB)
(DOCX 18.9 KB)
(XLSB 34 kb)
Acknowledgements
We are very grateful to all patients who donated their brains to the Netherlands Brain Bank (NBB; www.brainbank.nl), and we thank the team of the NBB, in particular Michiel Kooreman, for their cooperation and their help in the selection of brain tissue.
Author Contributions
Project was initiated by WB, LB, and TB. TM, SP, AT, TB, and AI were involved in processing of the tissue samples, and design and execution of biochemical assays, while VB, MQ, and GB performed GBA genotyping. Statistical analyses were set up and performed by TM, PE, DC, and GD. Figures were composed by TM and PE. TM, TK, LP, TB, WB, VB, and MQ contributed to the interpretation of the data, and TM and WB wrote the first draft of the manuscript. After receiving significant contributions of co-authors (SP, AT, MQ, TK, GD, VB, PC, VB, LP, and TB) for finalizing the manuscript, the manuscript was edited and finalized by TM and WB.
Funding Information
This work was supported by Roche Pharma and grants from the Stichting Parkinson Fonds to WB, from Fondazione Cassa di Risparmio di Perugia to TB, and from the Stichting Parkinson Fonds to VB.
Compliance with Ethical Standards
Competing Interests
All authors declare that they have no competing interests.
References
- 1.Dehay B, Martinez-Vicente M, Caldwell GA, Caldwell KA, Yue Z, Cookson MR, Klein C, Vila M, Bezard E. Lysosomal impairment in Parkinson’s disease. Mov Disord. 2013;28:725–732. doi: 10.1002/mds.25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xilouri M, Brekk OR, Stefanis L. Autophagy and alpha-synuclein: relevance to Parkinson’s disease and related synucleopathies. Mov Disord. 2016;31:178–192. doi: 10.1002/mds.26477. [DOI] [PubMed] [Google Scholar]
- 3.Gan-Or Z, Dion PA, Rouleau GA. Genetic perspective on the role of the autophagy-lysosome pathway in Parkinson disease. Autophagy. 2015;11:1443–1457. doi: 10.1080/15548627.2015.1067364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sidransky E, Lopez G. The link between the GBA gene and parkinsonism. Lancet Neurol. 2012;11:986–998. doi: 10.1016/S1474-4422(12)70190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schapira AH. Glucocerebrosidase and Parkinson disease: recent advances. Mol Cell Neurosci. 2015;66:37–42. doi: 10.1016/j.mcn.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen CM, Clark LN, Condroyer C, de Marco EV, Dürr A, Eblan MJ, Fahn S, Farrer MJ, Fung HC, Gan-Or Z, Gasser T, Gershoni-Baruch R, Giladi N, Griffith A, Gurevich T, Januario C, Kropp P, Lang AE, Lee-Chen GJ, Lesage S, Marder K, Mata IF, Mirelman A, Mitsui J, Mizuta I, Nicoletti G, Oliveira C, Ottman R, Orr-Urtreger A, Pereira LV, Quattrone A, Rogaeva E, Rolfs A, Rosenbaum H, Rozenberg R, Samii A, Samaddar T, Schulte C, Sharma M, Singleton A, Spitz M, Tan EK, Tayebi N, Toda T, Troiano AR, Tsuji S, Wittstock M, Wolfsberg TG, Wu YR, Zabetian CP, Zhao Y, Ziegler SG. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 8.Gan-Or Z, Mirelman A, Postuma RB, Arnulf I, Bar-Shira A, Dauvilliers Y, Desautels A, Gagnon JF, Leblond CS, Frauscher B, Alcalay RN, Saunders-Pullman R, Bressman SB, Marder K, Monaca C, Högl B, Orr-Urtreger A, Dion PA, Montplaisir JY, Giladi N, Rouleau GA. GBA mutations are associated with rapid eye movement sleep behavior disorder. Ann Clin Transl Neurol. 2015;2:941–945. doi: 10.1002/acn3.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamez-Valero A, Prada-Dacasa P, Santos C, Adame-Castillo C, Campdelacreu J, Rene R, Gascon-Bayarri J, Ispierto L, Alvarez R, Ariza A, Beyer K. GBA mutations are associated with earlier onset and male sex in dementia with Lewy bodies. Mov Disord. 2016;31:1066–1070. doi: 10.1002/mds.26593. [DOI] [PubMed] [Google Scholar]
- 10.Davis MY, Johnson CO, Leverenz JB, Weintraub D, Trojanowski JQ, Chen-Plotkin A, Van Deerlin VM, Quinn JF, Chung KA, Peterson-Hiller AL, et al. Association of GBA mutations and the E326K polymorphism with motor and cognitive progression in Parkinson disease. JAMA Neurol. 2016;73:1217–1224. doi: 10.1001/jamaneurol.2016.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cilia R, Tunesi S, Marotta G, Cereda E, Siri C, Tesei S, Zecchinelli AL, Canesi M, Mariani CB, Meucci N, Sacilotto G, Zini M, Barichella M, Magnani C, Duga S, Asselta R, Soldà G, Seresini A, Seia M, Pezzoli G, Goldwurm S. Survival and dementia in GBA-associated Parkinson’s disease: the mutation matters. Ann Neurol. 2016;80:662–673. doi: 10.1002/ana.24777. [DOI] [PubMed] [Google Scholar]
- 12.Liu G, Boot B, Locascio JJ, Jansen IE, Winder-Rhodes S, Eberly S, Elbaz A, Brice A, Ravina B, van Hilten JJ, Cormier-Dequaire F, Corvol JC, Barker RA, Heutink P, Marinus J, Williams-Gray CH, Scherzer CR, for the International Genetics of Parkinson Disease Progression (IGPP) Consortium Specifically neuropathic Gaucher’s mutations accelerate cognitive decline in Parkinson’s. Ann Neurol. 2016;80:674–685. doi: 10.1002/ana.24781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thaler A, Gurevich T, Bar Shira A, Gana Weisz M, Ash E, Shiner T, Orr-Urtreger A, Giladi N et al (2016) A “dose” effect of mutations in the GBA gene on Parkinson’s disease phenotype. Parkinsonism Relat Disord [DOI] [PubMed]
- 14.Gan-Or Z, Amshalom I, Kilarski LL, Bar-Shira A, Gana-Weisz M, Mirelman A, Marder K, Bressman S, Giladi N, Orr-Urtreger A. Differential effects of severe vs mild GBA mutations on Parkinson disease. Neurology. 2015;84:880–887. doi: 10.1212/WNL.0000000000001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gegg ME, Burke D, Heales SJ, Cooper JM, Hardy J, Wood NW, Schapira AH. Glucocerebrosidase deficiency in substantia nigra of Parkinson disease brains. Ann Neurol. 2012;72:455–463. doi: 10.1002/ana.23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiasserini D, Paciotti S, Eusebi P, Persichetti E, Tasegian A, Kurzawa-Akanbi M, Chinnery PF, Morris CM, Calabresi P, Parnetti L, Beccari T. Selective loss of glucocerebrosidase activity in sporadic Parkinson’s disease and dementia with Lewy bodies. Mol Neurodegener. 2015;10:15. doi: 10.1186/s13024-015-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy KE, Gysbers AM, Abbott SK, Tayebi N, Kim WS, Sidransky E, Cooper A, Garner B, Halliday GM. Reduced glucocerebrosidase is associated with increased alpha-synuclein in sporadic Parkinson’s disease. Brain. 2014;137:834–848. doi: 10.1093/brain/awt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocha EM, Smith GA, Park E, Cao H, Brown E, Hallett P, Isacson O. Progressive decline of glucocerebrosidase in aging and Parkinson’s disease. Ann Clin Transl Neurol. 2015;2:433–438. doi: 10.1002/acn3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcalay RN, Levy OA, Waters CC, Fahn S, Ford B, Kuo SH, Mazzoni P, Pauciulo MW, Nichols WC, Gan-Or Z, et al. Glucocerebrosidase activity in Parkinson’s disease with and without GBA mutations. Brain. 2015;138:2648–2658. doi: 10.1093/brain/awv179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNeill A, Magalhaes J, Shen C, Chau KY, Hughes D, Mehta A, Foltynie T, Cooper JM, Abramov AY, Gegg M, Schapira AH. Ambroxol improves lysosomal biochemistry in glucocerebrosidase mutation-linked Parkinson disease cells. Brain. 2014;137:1481–1495. doi: 10.1093/brain/awu020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papagiannakis N, Xilouri M, Koros C, Stamelou M, Antonelou R, Maniati M, Papadimitriou D, Moraitou M, Michelakakis H, Stefanis L. Lysosomal alterations in peripheral blood mononuclear cells of Parkinson’s disease patients. Mov Disord. 2015;30:1830–1834. doi: 10.1002/mds.26433. [DOI] [PubMed] [Google Scholar]
- 22.Balducci C, Pierguidi L, Persichetti E, Parnetti L, Sbaragli M, Tassi C, Orlacchio A, Calabresi P, Beccari T, Rossi A. Lysosomal hydrolases in cerebrospinal fluid from subjects with Parkinson’s disease. Mov Disord. 2007;22:1481–1484. doi: 10.1002/mds.21399. [DOI] [PubMed] [Google Scholar]
- 23.Parnetti L, Chiasserini D, Persichetti E, Eusebi P, Varghese S, Qureshi MM, Dardis A, Deganuto M, De Carlo C, Castrioto A, et al. Cerebrospinal fluid lysosomal enzymes and alpha-synuclein in Parkinson’s disease. Mov Disord. 2014;29:1019–1027. doi: 10.1002/mds.25772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parnetti L, Paciotti S, Eusebi P, Dardis A, Zampieri S, Chiasserini D, Tasegian A, Tambasco N, Bembi B, Calabresi P, Beccari T. Cerebrospinal fluid beta-glucocerebrosidase activity is reduced in Parkinson’s disease patients. Mov Disord. 2017;32:1423–1431. doi: 10.1002/mds.27136. [DOI] [PubMed] [Google Scholar]
- 25.van Dijk KD, Persichetti E, Chiasserini D, Eusebi P, Beccari T, Calabresi P, Berendse HW, Parnetti L, van de Berg WD. Changes in endolysosomal enzyme activities in cerebrospinal fluid of patients with Parkinson’s disease. Mov Disord. 2013;28:747–754. doi: 10.1002/mds.25495. [DOI] [PubMed] [Google Scholar]
- 26.Parnetti L, Balducci C, Pierguidi L, De Carlo C, Peducci M, D’Amore C, Padiglioni C, Mastrocola S, Persichetti E, Paciotti S, et al. Cerebrospinal fluid beta-glucocerebrosidase activity is reduced in dementia with Lewy bodies. Neurobiol Dis. 2009;34:484–486. doi: 10.1016/j.nbd.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Chu Y, Dodiya H, Aebischer P, Olanow CW, Kordower JH. Alterations in lysosomal and proteasomal markers in Parkinson’s disease: relationship to alpha-synuclein inclusions. Neurobiol Dis. 2009;35:385–398. doi: 10.1016/j.nbd.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Kurzawa-Akanbi M, Hanson PS, Blain PG, Lett DJ, McKeith IG, Chinnery PF, Morris CM. Glucocerebrosidase mutations alter the endoplasmic reticulum and lysosomes in Lewy body disease. J Neurochem. 2012;123:298–309. doi: 10.1111/j.1471-4159.2012.07879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, Boya P, Vila M. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30:12535–12544. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, Schapira AH. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol. 2010;67:1464–1472. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- 31.Siebert M, Sidransky E, Westbroek W. Glucocerebrosidase is shaking up the synucleinopathies. Brain. 2014;137:1304–1322. doi: 10.1093/brain/awu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persichetti E, Chiasserini D, Parnetti L, Eusebi P, Paciotti S, De Carlo C, Codini M, Tambasco N, Rossi A, El-Agnaf OM, et al. Factors influencing the measurement of lysosomal enzymes activity in human cerebrospinal fluid. PLoS One. 2014;9:e101453. doi: 10.1371/journal.pone.0101453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Dijkstra AA, Ingrassia A, de Menezes RX, van Kesteren RE, Rozemuller AJ, Heutink P, van de Berg WD. Evidence for immune response, axonal dysfunction and reduced endocytosis in the Substantia Nigra in early stage Parkinson’s disease. PLoS One. 2015;10:e0128651. doi: 10.1371/journal.pone.0128651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Development Core R. Team: R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 38.Beutler E, Gelbart T, Scott CR. Hematologically important mutations: Gaucher disease. Blood Cells Mol Dis. 2005;35:355–364. doi: 10.1016/j.bcmd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Smid BE, Ferraz MJ, Verhoek M, Mirzaian M, Wisse P, Overkleeft HS, Hollak CE, Aerts JM. Biochemical response to substrate reduction therapy versus enzyme replacement therapy in Gaucher disease type 1 patients. Orphanet J Rare Dis. 2016;11:28. doi: 10.1186/s13023-016-0413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallett V, Ross JP, Alcalay RN, Ambalavanan A, Sidransky E, Dion PA, Rouleau GA, Gan-Or Z. GBA p.T369M substitution in Parkinson disease: polymorphism or association? A meta-analysis. Neurol Genet. 2016;2:e104. doi: 10.1212/NXG.0000000000000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pastores GM, Hughes DA (1993) Gaucher disease. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K (eds) GeneReviews(R). Seattle
- 43.Pankratz N, Beecham GW, DeStefano AL, Dawson TM, Doheny KF, Factor SA, Hamza TH, Hung AY, Hyman BT, Ivinson AJ, et al. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann Neurol. 2012;71:370–384. doi: 10.1002/ana.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genome of the Netherlands C Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 45.Eyal N, Firon N, Wilder S, Kolodny EH, Horowitz M. Three unique base pair changes in a family with Gaucher disease. Hum Genet. 1991;87:328–332. doi: 10.1007/BF00200914. [DOI] [PubMed] [Google Scholar]
- 46.Grace ME, Ashton-Prolla P, Pastores GM, Soni A, Desnick RJ. Non-pseudogene-derived complex acid beta-glucosidase mutations causing mild type 1 and severe type 2 Gaucher disease. J Clin Invest. 1999;103:817–823. doi: 10.1172/JCI5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabowski GA. Phenotype, diagnosis, and treatment of Gaucher’s disease. Lancet. 2008;372:1263–1271. doi: 10.1016/S0140-6736(08)61522-6. [DOI] [PubMed] [Google Scholar]
- 48.Moors TE, Hoozemans JJ, Ingrassia A, Beccari T, Parnetti L, Chartier-Harlin MC, van de Berg WD. Therapeutic potential of autophagy-enhancing agents in Parkinson’s disease. Mol Neurodegener. 2017;12:11. doi: 10.1186/s13024-017-0154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moors T, Paciotti S, Chiasserini D, Calabresi P, Parnetti L, Beccari T, van de Berg WD. Lysosomal dysfunction and alpha-synuclein aggregation in Parkinson’s disease: diagnostic links. Mov Disord. 2016;31:791–801. doi: 10.1002/mds.26562. [DOI] [PubMed] [Google Scholar]
- 50.Argov Z, Navon R. Clinical and genetic variations in the syndrome of adult GM2 gangliosidosis resulting from hexosaminidase A deficiency. Ann Neurol. 1984;16:14–20. doi: 10.1002/ana.410160105. [DOI] [PubMed] [Google Scholar]
- 51.Inzelberg R, Korczyn AD. Parkinsonism in adult-onset GM2 gangliosidosis. Mov Disord. 1994;9:375–377. doi: 10.1002/mds.870090325. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki K, Iseki E, Katsuse O, Yamaguchi A, Katsuyama K, Aoki I, Yamanaka S, Kosaka K. Neuronal accumulation of alpha- and beta-synucleins in the brain of a GM2 gangliosidosis mouse model. Neuroreport. 2003;14:551–554. doi: 10.1097/00001756-200303240-00004. [DOI] [PubMed] [Google Scholar]
- 53.Vidoni C, Follo C, Savino M, Melone MA, Isidoro C. The role of cathepsin D in the pathogenesis of human neurodegenerative disorders. Med Res Rev. 2016;36:845–870. doi: 10.1002/med.21394. [DOI] [PubMed] [Google Scholar]
- 54.Cullen V, Lindfors M, Ng J, Paetau A, Swinton E, Kolodziej P, Boston H, Saftig P, Woulfe J, Feany MB, et al. Cathepsin D expression level affects alpha-synuclein processing, aggregation, and toxicity in vivo. Mol Brain. 2009;2:5. doi: 10.1186/1756-6606-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiao L, Hamamichi S, Caldwell KA, Caldwell GA, Yacoubian TA, Wilson S, Xie ZL, Speake LD, Parks R, Crabtree D, Liang Q, Crimmins S, Schneider L, Uchiyama Y, Iwatsubo T, Zhou Y, Peng L, Lu YM, Standaert DG, Walls KC, Shacka JJ, Roth KA, Zhang J. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol Brain. 2008;1:17. doi: 10.1186/1756-6606-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sevlever D, Jiang P, Yen SH. Cathepsin D is the main lysosomal enzyme involved in the degradation of alpha-synuclein and generation of its carboxy-terminally truncated species. Biochemistry. 2008;47:9678–9687. doi: 10.1021/bi800699v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi M, Ko LW, Kulathingal J, Jiang P, Sevlever D, Yen SH. Oxidative stress-induced phosphorylation, degradation and aggregation of alpha-synuclein are linked to upregulated CK2 and cathepsin D. Eur J Neurosci. 2007;26:863–874. doi: 10.1111/j.1460-9568.2007.05736.x. [DOI] [PubMed] [Google Scholar]
- 58.Ambrosi G, Ghezzi C, Zangaglia R, Levandis G, Pacchetti C, Blandini F. Ambroxol-induced rescue of defective glucocerebrosidase is associated with increased LIMP-2 and saposin C levels in GBA1 mutant Parkinson’s disease cells. Neurobiol Dis. 2015;82:235–242. doi: 10.1016/j.nbd.2015.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 185 KB)
(DOCX 25.0 KB)
(DOCX 18.7 KB)
(DOCX 18.9 KB)
(XLSB 34 kb)