Abstract
Bacterial high-copy-number (hcn) plasmids provide an excellent model to study the underlying physical mechanisms of DNA segment segregation in an intracellular context. Using two-color fluorescent repressor-operator systems and a synthetic repressible replication origin, we tracked the motion and segregation of single hcn plasmid molecules in individual cells. The plasmid diffusion dynamics revealed between-plasmid temporal associations (clustering) as well as entropic and elastic recoiling forces in the confined intracellular spaces outside of nucleoids. These two effects could be effectively used in models to predict the heterogeneity of segregation. Additionally, the motile behaviors of hcn plasmids provide quantitative estimates of entropic exclusion strength and dynamic associations between DNA segments. Overall, this study utilizes a, to our knowledge, novel approach to predict the polymer dynamics of DNA segments in spatially confined, crowded cellular compartments as well as during bacterial chromosome segregation.
Introduction
DNA segregation, which ensures the passage of the complete genetic information from parent to daughter cells, is a critical process in cell division. Without complications from precisely scheduled and specialized transport of organelles in eukaryotic cells (1, 2), bacterial circular chromosome segregation can be accomplished in only a few minutes, alongside the other major chromosomal processes of replication and gene expression (3, 4). This high efficiency not only requires precise organization of protein networks (5, 6), but it is also influenced by the physical nature of the DNA molecule itself, including the entropic and relaxation states (7, 8). As such, the complex functions and massive size of the chromosome present major barriers to understanding the underlying physical mechanisms that determine DNA segregation. Fortunately, plasmids carrying patch sequences can be used to study a wide variety of DNA-related processes because this toolkit can be used to transfer genetic information between cells through transformation, transduction, and conjugation (9, 10). Additionally, plasmids can serve as extracellular DNA to modulate biofilm formation (11, 12). Because these molecules include only minimal biological elements and have manageable sequence lengths, plasmid DNA has been utilized as a simplified model to investigate the key factors of DNA segregation (13, 14). Among the many different types of plasmids, high-copy-number (hcn) plasmids, which lack an active motor-protein-driven partitioning system (15, 16), are widely considered to be the most applicable for studying the general physical properties of DNA segments in a cellular environment.
The mechanisms governing hcn plasmid segregation have been examined by several methodologies, which have provided increasingly detailed information about the process. For example, ColE1-derived plasmids were visualized as fluorescent foci by fluorescence in situ hybridization (17) and fluorescence repressor-operator systems (FROS) (18); the results of those experiments challenged the random distribution model that had been previously predicted from classical studies examining the loss rate (19). Recently, two single-molecule approaches have provided further insight into hcn plasmid segregation. By halting plasmid replication with genetically encoded temperature-sensitive polymerase PolIts, plasmid numbers were drastically diminished in cells, which enabled the necessary spatial resolution for tracking plasmid motion (20). The results of these single-molecule experiments suggested that hcn plasmids are freely diffusible and are only excluded by nucleoids, without other constraints on intracellular localization. Controversially, super-resolution fluorescence in situ hybridization imaging in fixed cells (21) revealed that some plasmids may be distributed within nucleoid regions, and observed plasmid clustering implies that there may be interactions between DNA segments. These seemingly inconsistent findings raise two questions regarding the motion and distribution of hcn plasmids in cells (i.e., are plasmids excluded from nucleoids, and are there interactions between plasmids?). Both of these factors would be expected to influence the dynamic behaviors of plasmids. Therefore, we endeavored to address these issues by developing a system to track single plasmids, without removing the interactions with other plasmids. We then applied our system to observe single cells, focusing on cell division and plasmid segregation.
Materials and Methods
Bacterial strains and growth conditions
Plasmid p15AA-phlFH.tq was introduced into Escherichia coli strain BW25113 to create BW25113FH. The strain BW25113FH was then transformed with pTetORK34b and pLacOIC2c for experiments testing incompatibility between plasmids with wt origin. The same strain was transformed with only pTetORK34p or additionally transformed with pLacOIC2c for experiments involving the repression of plasmid replication. Strains transformed with pZC320-tetO or pZC320-lacO, the mini-F-derived plasmids carrying FROS, were used to quantify the fluorescence background and the fluorescence intensity of a single plasmid in cells. In all experiments, bacterial cells were cultured overnight in Luria-Bertani medium (1% wild-type (wt) tryptone, 0.5% wt yeast extract, and 1% wt NaCl; BD Difco Laboratories, Detroit, MI) at 37°C. Overnight cultures were then diluted 1:50–1:100 in M9-supplemented medium (1× M9 salt, 0.5 mg/mL thiamine, 0.15 mg/mL biotin, and 0.1% casamino acids) and cultured with 0.4% glucose at 37°C until reaching an optical density of 0.4–0.6. For experiments that involved repressing replication, the culture media was M9-supplemented medium with 0.2% arabinose. Once the culture population doubled, it was diluted 1:2 to maintain the cells in log-phase growth. In all experiments, the cultures were transferred to a 2% agarose pad (SeaPlaque agarose; Lonza, Basel, Switzerland) made with M9-supplemented medium containing 0.4% glucose and covered by a microscope slide for observation. The concentrations of ampicillin, kanamycin, and chloramphenicol were 100, 50, and 34 ng/mL, respectively. However, all antibiotic selection was removed during the observation period on the 2% agarose pad. When compared to the wt strain (BW25113) under experimental conditions, the transformation of plasmids had negligible effects on the doubling time, the size of nucleoids, and the cell length for all strains used in this study.
Plasmid constructions
All plasmids were constructed by the in-fusion method. Please see the Supporting Materials and Methods for a detailed description.
Imaging and data analysis
The plasmids were imaged in live bacteria using an epifluorescent Olympus IX71 inverted microscope (Olympus, Tokyo, Japan) with a 100× phase contrast oil objective, UPLSAPO100XOPH (numerical aperture 1.4). The fusion proteins mTurquoise, mYFP, and mCherry were excited with 405, 488, and 561 nm lasers (diode-pumped sold-state laser; TWC Opto, New Taipei City, Taiwan) using a multiband filter set (LF405/488/561/635-A-000; Semrock, Rochester, NY). An electron-multiplying charged-coupled device camera (C9100; Hamamatsu Photonics, Hamamatsu, Japan) was used for image acquisition. For time-lapse imaging, four channels, including the three different fluorescent spectrums and the phase contrast bright-field images, were recorded at each time point. The frame rates for each channel were 0.125 Hz for long-term (cell divisions, across generations) experiments and 2.5 Hz for short-term (local diffusion coefficient) experiments. Images were later extracted and processed by ImageJ- and MATLAB-based programs (The MathWorks, Natick, MA), including ND-SAFIR denoise software (22) to remove electronic noise, TrackMate particle tracking plugin (23) to acquire single-plasmid trajectory, and Schnitzcells (24), which is our home-built supplementary program to analyze the numbers of plasmids within a cell. The details regarding the calibration of fluorescence intensity and error prediction in the number of plasmids per cell are shown in the Supporting Materials and Methods.
Results and Discussion
Resolving single hcn plasmids in vivo
FROS was utilized in earlier reports (18, 20) to observe the motion of plasmids in living cells. However, because of the limitations of resolution by diffraction, it is difficult to use FROS to distinguish a single hcn plasmid molecule when all copies are uniformly labeled. We designed a strategy to overcome this issue wherein we labeled single plasmids with different fluorophores. This method allowed the plasmids to be spectrally separable and spatially resolvable from other copies in the same cell. To achieve this goal, two rationally designed elements were incorporated. One encodes a two-color fluorescent-labeling system to separate plasmids into two populations, and the other limits expression of one of the populations to allow spatial resolvability.
To generate spectrally separable labels, two repressor-operator systems (tetracycline repressor/operator and lactose repressor/operator) were utilized after confirming they do not interfere with the segregation of plasmids that have identical replication origins. Two cer-deleted ColE1-derived plasmids, pTetORK34b (P1) and pLacOIC2c (P2), respectively encoding (Ptet::tetR-myfp::tetO array) and (Plac::lacIadi-mcherry::lacO array), were constructed (Fig. 1 A). Notably, expression of the fluorescent repressors is self-regulated by the tetracycline and lactose promoters. Thus, our system stands in contrast to those that are controlled by other inducible promoters (e.g., the arabinose induction system (18)), for which the concentration of inducing agents controls the number of repressor molecules in the cells. Instead, encoding the repressor in each plasmid serves to correlate the total repressor expression level with the number of plasmids (25). This feedback circuit also automatically locks the expression level during the visualization of plasmids. The fluorescence intensities of single cassettes were resolved and quantified by inserting the cassettes into single-copy mini-F-derived plasmids and monitoring their expression in living cells (Supporting Materials and Methods; Fig. S1 B). These calibrations provided a reference that could be used to identify single hcn plasmids according to the intensities of fluorescent spots. Furthermore, the minor interaction between plasmids that arises because of the inclusion of FROS cassettes can be neglected (Supporting Materials and Methods).
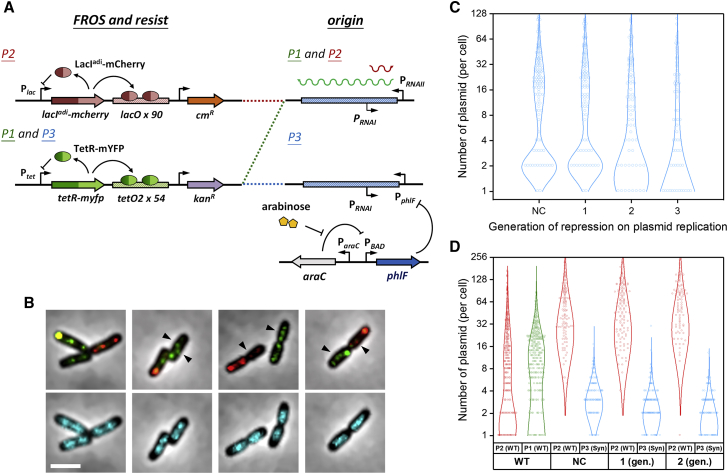
Figure 1.
Single hcn plasmid molecules can be resolved by two-color FROS and copy-number control systems. (A) The design schemes of the self-regulated TetR-YFP/tetO and LacIadi-mCherry/lacO systems are shown. Antibiotic resistance and replication origins, both wt (pTetORK34b (P1) and pLacOIC2c (P2)) and synthetic (pTetORK34p (P3)), are also shown. LacIadi is a LacI mutant that can only form dimers, not tetramers. (B) Photomicrographs show phase contrast images of cells merged with fluorescence images of cotransformed plasmids (P1 (upper panel, green) and P2 (upper panel, red)) or nucleoids (lower panel, cyan). The arrows indicate probable single plasmids that are traveling around the nucleoids. Scale bars, 3 μm. Violin plots show the estimated numbers of plasmids in cells according to generations of repression. (C) Cells were transformed with only P3 (cyan), or (D) cells were additionally transformed with the wt plasmid, P2 (red). The estimated plasmid numbers without repression are presented as the negative control (NC), and those in cells cotransformed with P1 (green) and P2 are also shown for comparison. The circles and lines are histograms and Gaussian fits, respectively. The numbers of cells in the analyzed in (C) are 151, 127, 88, and 87 from left to right. In (D), the numbers of cells are 290, 124, 136, and 103 from left to right. Of note, the asymmetry between P1 and P2 may be due to differences in antibiotic selection. To see this figure in color, go online.
To reduce the population of plasmids, replication control may be the most efficient method (20, 26). Indeed, because of incompatibilities between the ColE1 derivatives in the same host cell, the two wt plasmid populations were nonevenly distributed (Fig. 1 D; Fig. S1 D). However, with this experimental design, only 10% of cells exhibited a single plasmid that could be differentiated from the majority (NP1 = 1 or NP2 = 1). To further improve our ability to track single plasmids, a repressible ColE1-derived replication origin was designed to control the numbers of the plasmid in the host cells. We replaced the native promoter, PRNAII, which regulates the synthesis of the replication initiation primer RNAII (26, 27), with the promoter, PphlF, which is modulated by the transcriptional repressor PhlF (28). Then, we expressed PhlF via an arabinose induction system, effectively suppressing the replication of plasmid pTetORK34p (P3) (Fig. 1 A). Under this system of repression, the numbers of P3 in the host cells could be reduced after dilution by cell division.
In BW25113FH cells transformed with P3, two major populations of cells were observed as determined by the number of plasmids per cell. The plasmid-rich population had roughly 20 plasmids per cell, whereas the plasmid-poor population had ∼2 plasmids per cell. In cultures with repressed replication, the plasmid-rich population (peak value ∼20 plasmids) was diminished almost entirely within only a few cell generations, and the mode value of the plasmid-poor population was simultaneously shifted from 2 to 1. Furthermore, the cells transformed with both P2 and P3 (Fig. 1 D), but without PhlF protein expression, already exhibited relatively low levels of P3, probably because of competition for replication factors. Interestingly, the mutant replication origin in P3 seemed to be relatively unstable compared to the wt; however, the plasmid P3 was found to autonomously replicate and was sustained in the host cells for multiple generations without replication repression. After repressing P3 replication for two cell generations, the estimated number of plasmids was three or less in more than half of the cells, making single-plasmid tracking much more feasible (Fig. 2 A and S2 A; Video S2). Thus, we utilized this repression process to track the motions of single P3 plasmids in all further experiments.
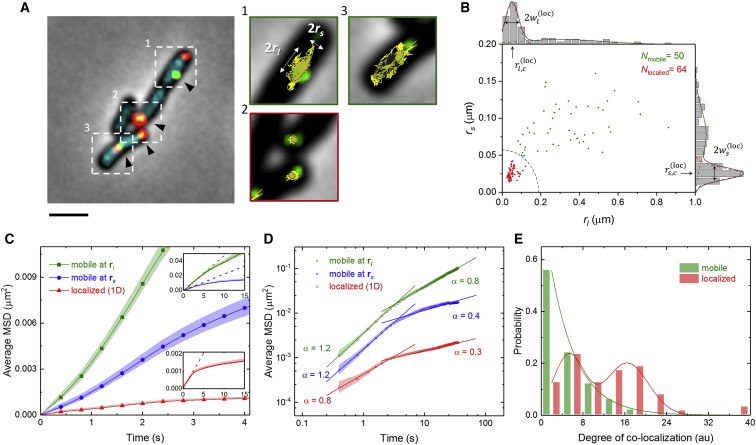
Figure 2.
The motion of single plasmids in cells. (A) A merged image shows phase contrast, and fluorescent nucleoids (cyan), single P3 plasmids (green), and P2 (red). The enlarged images correspond to the regions indicated by dashed boxes; only the fluorescence of P3 plasmids is shown along with the plasmid trajectories (yellow lines). To better visualize single plasmids, the green fluorescence intensity is 12-fold higher than that shown in Figs 1A and S1A. Scale bars, 2 μm. (B) Scatter plot and histograms show the characteristic lengths of 5-min trajectories. The dashed line indicates the cutoff criterion separating the localized (red) and mobile (green) groups. (C) and (D) are the average mean-square displacements of the trajectories of the localized (red) plasmids as well as those for mobile plasmids along the long (green) and short (blue) characteristic axes. The color bands represent the standard errors of the means. The solid and dashed lines in the insets of (C) are the experimental data and the linear fits where τ ≤ 2 s, respectively. (E) Colocalization of tracked plasmids with untracked plasmids is shown for single mobile (green) and localized (red) plasmids. The green line shows the Poisson distribution with an expected value of 4.7 arbitrary units (au), and the red line serves as a guide to the eye. To see this figure in color, go online.
Live cells: BW25113FH/pTetORK34p, pLacOIC2c. Respective colors: nucleoids (cyan), plasmids pTetORK34b (green) and pLacOIC2c (red), and cell outlines (phase contrast; gray).
Association and clustering of plasmids
Combining two-color FROS and replication control strategies, we were able to track a single mutant plasmid P3, successfully resolving it from the majority, wt P2, in the same host cell. According to the trajectories of the single P3 molecules, the plasmids commonly diffuse to the cell poles and midcell regions while occasionally moving across/around the nucleoids (visualized by HupA-mTurquoise2 (29)). To characterize the traveled region of a single P3 molecule, a gyration tensor describing the plasmid path during a 5 min observation period was calculated. The gyration tensor was defined as:, where m and n are the coordinates of the tracked location relative to the center of the trajectory at each time point, and N is the number of the time points. As a simplified quantification, the eigenvalues and respective eigenvectors of the gyration tensor were calculated to determine the lengths and the directions of two orthogonal characteristic axes of plasmid motion (Fig. 2 B). Based on two-peak Gaussian fittings of the rl and rS (for the long and short axes) length distributions, the motions of plasmids can be reasonably and sufficiently separated into the two groups, either “mobile” or “localized,” according to the long axis displacement. A cutoff criterion to separate these groups was defined as, which is the arc of an ellipse shown as a dashed line in Fig. 2 B. The lengths of the axes are and , where the means, and , as well as the widths, and , of the localized group were determined from the Gaussian fit shown in Fig. 2 B. According to this criterion, the portion of mobile plasmids was ∼45%, and localized plasmids accounted for the remaining 55% of 114 observed individual plasmids.
Furthermore, the diffusive properties of the mobile and localized plasmids were compared based on the mean-square displacements (MSD) of the trajectories. As shown in Fig. 2, C and D, the trajectories of the mobile plasmids can be rationally separated into components along the characteristic short and long axes of the traveled regions. Thus, the two-dimensional (2D) MSD of the localized plasmid trajectories can be divided by two to better compare the magnitudes. Using simple linear fits for MSD (τ) where τ ≤ 2 s (Fig. S2 B), we found that the mobile plasmids were more diffusive than localized plasmids and exhibited anisotropy with regard to the short and long axes. From an anomalous diffusion analysis (MSD ∼ Dappτα), we found that both the magnitude of the diffusivity and the diffusion behaviors change over different timescales. In the mobile group, a slightly superdiffusive behavior (α > 1) over a short timescale (τ < 2 s) implied that the plasmid exhibited directional motion when exiting the nucleoid region as will be discussed later. Over a long timescale (τ > 5 s), subdiffusive behavior (α < 1) indicated that the plasmid was confined, probably because of repulsion in the cellular environment (30, 31). Because the characteristic long axis only slightly deviated from the direction that defined cell length in most cases, this anisotropic diffusion may occur based on the asymmetric geometry of the intracellular space. In contrast to the mobile group, the localized group exhibited subdiffusive behavior and stronger suppression of diffusion at every timescale, implying that this population of molecules is under additional constraints. To examine if the differences in diffusion behaviors were related to plasmid-plasmid interactions, the plasmid motions were compared to those in cells only carrying P3 (Fig. S2, C and D). The comparison revealed that single plasmids exhibit similar behaviors to the mobile plasmids in cotransformed cells.
In addition, the colocalization of P2 and P3 plasmids was investigated. Though the exact numbers of colocalized plasmids cannot be directly resolved from the 2D images, the degree of colocalization provides an estimation of the number of P2 plasmids that overlap with the tracked P3 plasmid. For each frame, the area of the tracked P3 plasmid was considered to be a circle of 100-nm diameter (the Kuhn length of DNA in cells), centered at the fitted location. The degree of colocalization was then determined as the time average of the intensity of overlapping P2 plasmids divided by the intensity of a single plasmid. As shown in Fig. 2 E, the degree of colocalization between mobile plasmids and P2 was well fitted by a Poisson distribution, suggesting that the colocalization of these plasmids is likely to be a discrete, independent, and random process. This result further implies that the mobile plasmids were free from specific associations with other plasmids. On the other hand, the localized P3 plasmids were often found to be colocalized with P2 plasmids; thus, we speculate that the localized population was directly or indirectly associated with P2 plasmids in clusters. Furthermore, the localized P3 plasmids exhibited a limited region of motion (∼70 nm on average), which can be taken as a rough estimation of cluster size that is consistent with earlier estimates by super-resolution imaging (21). Additionally, the size implies that the clustering may be induced by the biological factors (e.g., replisomes (20, 32) and transcription factories (33, 34, 35), which are known to be capable of gathering DNA segments into the same machinery).
Volume exclusion effect from the nucleoids
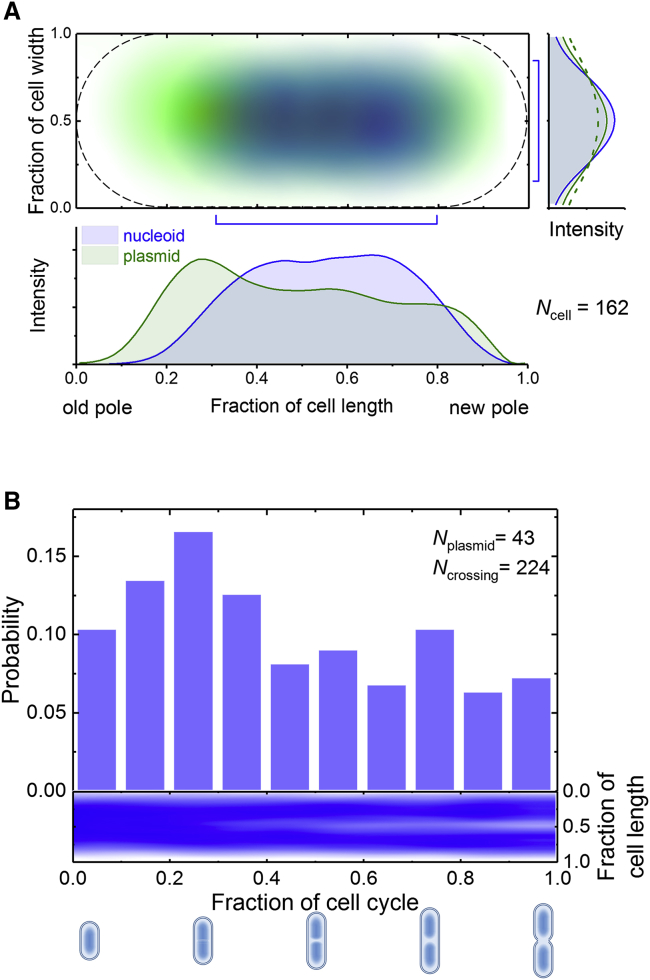
Large clusters (observed as brighter fluorescent foci) were mostly distributed in the nucleoid-free region, and only single plasmids were found to infrequently travel across the nucleoid regions (Figs. 1 B, 2 A, and S1 A; Video S1). The infrequency of nucleoid traverse by mobile plasmids suggested that the nucleoids constrain the motions of plasmids, regardless of their association with other plasmids in the cell. This exclusion effect by the nucleoid could also be observed in newly divided cells, which were quasiquantitatively identified by nucleoid fluorescence intensity, as shown in Fig. 3 A. The distribution of plasmids was higher at the old pole than the new pole and also broader in width than the nucleoid especially in the region that nucleoid locates.
Figure 3.
Plasmid exclusion from the nucleoid region as shown by the spatial distribution and motion of plasmids. (A) The average fluorescence intensity of the nucleoids (blue) and all plasmids (green) in newly divided cells is presented in a 2D map (upper panel). Intensities are shown along the long (lower panel) and short (right panel) axes of the cells. The solid and dashed lines indicate the respective distribution profiles of the whole cell and the nucleoid region indicated on the 2D map. (B) The probability of the nucleoid-crossing events (upper panel) and the nucleoid distribution along the direction of the cell length (lower panel) is shown according to the cell cycle. The illustrations below the x axis represent the states of the nucleoid segregation in cells. To see this figure in color, go online.
Live cells: BW25113FH/pTetORK34b, pLacOIC2c. Respective colors: nucleoids (blue), plasmids pTetORK34b (green) and pLacOIC2c (red), and cell outlines (phase contrast; gray).
We also performed long-term tracking of single plasmids and simultaneously recorded the nucleoid profiles along the long axis of the cell. The movement of plasmid DNA across the nucleoid was observed at every stage of the cell cycle, but the probability of nucleoid transit was not uniform throughout the cell cycle (Video S3). This lack of uniformity suggests a relationship between the motility of the plasmids and the physiological state of the nucleoids (Fig. 3 B). Interestingly, the highest probability of nucleoid-crossing events was roughly at the end of the first quarter of a cell cycle when the nucleoid initiated segregation. It can be speculated that the nucleoid blocks diffusion of plasmids, and its obstructive strength is related to the density of the chromosome, which is lowest during segregation and highest just after the completion of replication. An additional discovery from our long-term tracking experiment was that although roughly the half of the plasmids in the population should have been prevented from crossing the nucleoid because of clustering, only 3 out of 43 plasmids exhibited a complete lack of nucleoid crossing along an entire cell cycle. This result implies that although the association between plasmids is common, it is not enduring, and the strength may be weak. Thus, most plasmids seem to dissociate from the clusters at least once within a cell cycle. This speculation is further supported by the short temporal profiles of replication and transcription (36).
Live cells: BW25113FH/pTetORK34p, pLacOIC2c. Respective colors: nucleoids (cyan), plasmids pTetORK34p (green), and cell outlines (phase contrast; gray).
Based on our measurements, the space between the cell boundary and the nucleoid is ∼150 nm (Fig. 3 A), which is smaller than the full-spread size of an unconfined supercoiled plasmid (more than 200 nm) as estimated by twice the radius of gyration (37). This geometry can be modeled as an entropic barrier between the nucleoid and nucleoid-free regions. Because of this spatial confinement, a plasmid as a polymer will experience entropic and elastic recoiling forces (38, 39, 40) when exiting the nucleoid region, which potentially explains the superdiffusive motions recorded in short timescales (Fig. 2 D). On the other hand, according to our observations, a plasmid is expected to undertake an average of five crossing events during one whole 50-min cell cycle, with each crossing event averaging around 80 s. Thus, the probability for a single plasmid to be found in the nucleoid region was 13%. Also, only half of the plasmids were free from clusters, so the probability of observing a free plasmid in nucleoid-free region Pfr was calculated to be ∼3-fold higher than that in the nucleoid region Pnuc. Because the volumes of these two regions are roughly equal (according to the width and length of nucleoids, Fig. 3, A and B), the probability difference does not require normalization to volume. Therefore, equipartition theorem and the calculated probabilities of observing plasmids in the nucleoid and nucleoid-free regions can provide a rough estimation of the barrier height between the compartments as ΔE/kBT∗ ∼ ln (Pfr/Pnuc) ∼ 1, where kBT∗ is the effective thermal energy in cells. Notably, the activity of ATP-dependent enzymes will affect the fluctuating motions of DNA in cells (41). According to the diffusion behaviors of chromosome loci, the effective temperature T∗ should be two- to sevenfold higher than that of a room-temperature culture environment T, depending on the phase in the cell cycle (8). Although plasmids may not exhibit protein interactions as frequently as chromosomes, entanglement and repulsion from the chromosome are likely to alter the motion of the plasmids. Based on these and other uncertainties, this calculation represents a simplified order-of-magnitude estimation of the barrier height.
Heterogeneity of hcn plasmid segregation
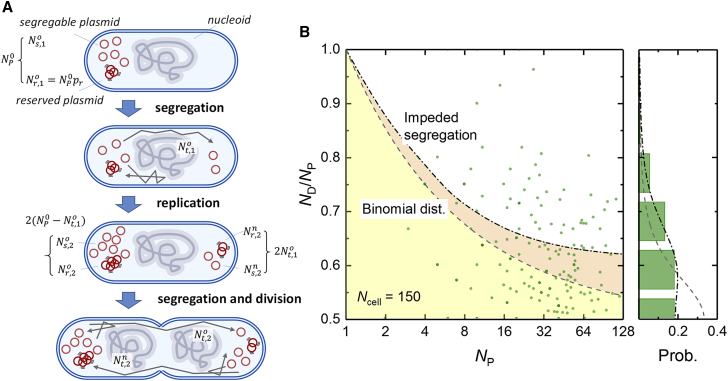
Because of the effects of association between plasmids and volume exclusion from the nucleoids, we speculated that the heterogeneity of hcn plasmid segregation should be high. An added benefit of the self-regulated FROS cassette is that it is possible to quantify the distributions of parental and inherited plasmids (NP and ND) in a cell division as shown in Fig. 4 B. We clearly observed a broader distribution of inherited plasmid copies than that would be predicted by a random segregation model, which is represented by a binomial distribution with an equal probability of distribution to each daughter cell. Therefore, we applied a simplified segregation model (Fig. 4 A), which includes two segregation steps and an intervening replication step, to further investigate the effects of impeded plasmid motility on segregation. In the model, the original plasmid, (= NP/2), is initially localized entirely in the old half of the cell. In the first segregation step, a portion of the plasmids, , are transferred from the old to the new half. Then, in the replication step, the plasmid copies in both halves are simply assumed to be doubled. Finally, after another exchange of and plasmids between the old and new halves in a second segregation event, the numbers of the plasmids in each half are fixed in the respective daughter cells. The final number of plasmids segregated to the old half can be calculated as . The numbers and the respective probability of the transferred plasmids in the segregation steps were determined as follows. Among all N plasmids found in the original half, a portion, Nr, determined by the proportionality constant pr, are clustered and retained. In the rest of the segregable plasmids, Ns (= N − Nr), the probability of transferring Nt plasmids to the other half can be determined by a binomial distribution Pb (Nt; Ns, pt), where the transfer probability pt < 0.5, because of nucleoid exclusion. Therefore, the probability of a combination of the segregable and transferred plasmid numbers in the two segregation steps can be represented as , where , and are the segregable plasmid numbers in the segregation steps. Because more than one combination of , , and may produce a particular , all possible combinations should be summed to calculate the respective probability . Notably, the probability that a number of inherited copies, ND, will be found in either daughter cell can be determined as . By fitting the probability distribution of the proportion of inheritance ND/NP as a function of the all measured NP values, we found that pr = 0.05 and pt = 0.3. These results agree with the observation that nucleoid crossing occurs throughout the whole cell cycle as well as the limited number of crossing events and short time spent in the nucleoid region. Also, 55% of the events were within the deviation of the distribution predicted by this simplified impeded segregation model. Although the model predicts this value should be 62%, it still performs much better than the simple random segregation model, in which only 33% of the experimental events were within the deviation, which was predicted to be 68%. Additionally, the effectiveness of our impeded segregation model was compared to the random segregation model using the Akaike information criterion (42), which can estimate the relative information lost between two test models based on the fitting residuals and the number of the parameters in each model. According to this comparison, the probability of the null hypothesis that the random segregation model was better able to explain the information in the experiment is less than 10−6. Therefore, the better fit of the simplified impeded segregation model was confirmed to be an informative improvement over the random segregation model and not simply the overfitting of the data. Further, the predictive nature of the simplified impeded segregation model also supports the idea that plasmid association and nucleoid exclusion are determinants of DNA segment motion in vivo.
Figure 4.
The heterogeneity of hcn plasmid segregation. (A) A scheme showing the simplified impeded model of plasmid segregation, which includes two segregation steps and an intervening replication step. This model represents the processes of plasmid transfer between cell halves and the doubling of the plasmid copies after replication. (B) The proportion of inheritance (ND/NP) of plasmids (green spots) with respect to the copy number in parent cells (left panel) is shown. Each spot indicates a single division event. The upper boundaries of the yellow (gray dashed line) and orange (black dot-dashed line) regions are predicted from the deviations of the probability distributions following the simple random segregation model (analytical solution, 0.5 + Np−1/2) and the impeded segregation model (numerical solution), respectively. The right panel shows the probability distribution of the proportion of inheritance from the experimental results (green bars) as well as the prediction from the random segregation scenario (gray dashed line) and the impeded segregation model (black dot-dashed line). Because of symmetry and the fact that all plasmids should be eventually segregated into either one of the daughter cells, only the half region of the proportion of inheritance (ND/NP ≥ 0.5) is shown. To see this figure in color, go online.
Based on the heterogeneity of inherited hcn plasmid copy numbers, it may be predicted that some rate of plasmid loss would occur in the host cells. However, with regard to population fitness, this loss of genetic material may be compensated by improvements in the diversity of the population. As the carrier for horizontal gene transfer, plasmids serve as genomic patches that often play crucial roles in the survival of host cells. In diverse and variable environments, the appropriate population distribution of plasmids is determined by a balance between the burden of carrying the plasmid and the total fitness provided by the genes encoded in the plasmids (43, 44). Higher population diversity provides a better opportunity to optimize the number of plasmid copies across the population of host cells. In other words, heterogeneity of plasmid copy numbers may increase the persistence of bacterial cells in different conditions as well as the difficulty to control pathogens and bacteria in biofilm states.
Conclusions
Leveraging single-molecule and single-cell observations, we found that a few dominant factors determine plasmid motion (i.e., the association between plasmids and exclusion by nucleoids) and can sufficiently explain heterogeneity of segregation. Moreover, we were able to quantify the influence of each factor on segregation. The principles underlying the dynamics of plasmid motion may be generalizable to the physical behaviors of many types of DNA segments transiting in cellular environments. We found non-negligible entropic differences between the nucleoid and non-nucleoid space, which led to the confinement of DNA segments to crowded and complex cellular compartments. Quantification of these entropic differences allowed us to estimate the strength of the DNA segment exclusion by chromosomes. Additionally, the coexistence of a large population of plasmids in clusters and the high probability of observing plasmids transiting across nucleoids suggest that interactions between DNA segments are both common and variable. These interactions have been suggested to be a driving force of bacterial chromosome organization and segregation (34, 45) and appear to depend on the state of the chromosomes. The insights gained herein will be helpful for further understanding the physics of bacterial chromosome segregation.
Author Contributions
Y.-R.C. conceived and designed the experiments. T.-M.H. performed the experiments. Y.-R.C. and T.-M.H. analyzed the data. Y.-R.C. derived the model and wrote the article.
Acknowledgments
We thank Dr. Chia-Fu Chou (Academia Sinica, Taiwan) and Dr. Jung-Ren Huang (Academia Sinica, Taiwan) for their beneficial discussion. We appreciate Dr. Wen-Chen Tsai’s support on the calibration of plasmid numbers in cells by quantitative PCR. We are also grateful to Dr. Chia-Fu Chou and Dr. Kenn Gerdes (University of Copenhagen, Danmark) for their kind donations of plasmids pSOT37 and pJMJ178, respectively.
This work was supported by the Ministry of Science and Technology, Taiwan (NSC 102-2112-M-003-019-MY3 and MOST 106-2112-M-003-012).
Editor: Julie Biteen.
Footnotes
Supporting Materials and Methods, three figures, one table, and three videos are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30055-4.
Supporting Material
References
- 1.Murray A.W. Creative blocks: cell-cycle checkpoints and feedback controls. Nature. 1992;359:599–604. doi: 10.1038/359599a0. [DOI] [PubMed] [Google Scholar]
- 2.Ruchaud S., Carmena M., Earnshaw W.C. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 3.Wang J.D., Levin P.A. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyes-Lamothe R., Nicolas E., Sherratt D.J. Chromosome replication and segregation in bacteria. Annu. Rev. Genet. 2012;46:121–143. doi: 10.1146/annurev-genet-110711-155421. [DOI] [PubMed] [Google Scholar]
- 5.Kleckner N., Fisher J.K., Witz G. The bacterial nucleoid: nature, dynamics and sister segregation. Curr. Opin. Microbiol. 2014;22:127–137. doi: 10.1016/j.mib.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badrinarayanan A., Le T.B., Laub M.T. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015;31:171–199. doi: 10.1146/annurev-cellbio-100814-125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun S., Wright A. Entropy as the driver of chromosome segregation. Nat. Rev. Microbiol. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lampo T.J., Kuwada N.J., Spakowitz A.J. Physical modeling of chromosome segregation in Escherichia coli reveals impact of force and DNA relaxation. Biophys. J. 2015;108:146–153. doi: 10.1016/j.bpj.2014.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 10.Johnsen A.R., Kroer N. Effects of stress and other environmental factors on horizontal plasmid transfer assessed by direct quantification of discrete transfer events. FEMS Microbiol. Ecol. 2007;59:718–728. doi: 10.1111/j.1574-6941.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- 11.Mann E.E., Rice K.C., Bayles K.W. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madsen J.S., Burmølle M., Sørensen S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 13.Gerdes K., Møller-Jensen J., Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S.K., Hajra S., Jayaram M. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 15.Dmowski M., Jagura-Burdzy G. Active stable maintenance functions in low copy-number plasmids of Gram-positive bacteria I. Partition systems. Pol. J. Microbiol. 2013;62:3–16. [PubMed] [Google Scholar]
- 16.Baxter J.C., Funnell B.E. Plasmid partition mechanisms. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.PLAS-0023-2014. Published online November 7, 2014. [DOI] [PubMed] [Google Scholar]
- 17.Pogliano J., Ho T.Q., Helinski D.R. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2001;98:4486–4491. doi: 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao S., Helinski D.R., Toukdarian A. Localization of the naturally occurring plasmid ColE1 at the cell pole. J. Bacteriol. 2007;189:1946–1953. doi: 10.1128/JB.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durkacz B.W., Sherratt D.J. Segregation kinetics of colicinogenic factor col E1 from a bacterial population temperature sensitive for DNA polymerase I. Mol. Gen. Genet. 1973;121:71–75. doi: 10.1007/BF00353694. [DOI] [PubMed] [Google Scholar]
- 20.Reyes-Lamothe R., Tran T., Tolmasky M.E. High-copy bacterial plasmids diffuse in the nucleoid-free space, replicate stochastically and are randomly partitioned at cell division. Nucleic Acids Res. 2014;42:1042–1051. doi: 10.1093/nar/gkt918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Penkul P., Milstein J.N. Quantitative localization microscopy reveals a novel organization of a high-copy number plasmid. Biophys. J. 2016;111:467–479. doi: 10.1016/j.bpj.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulanger J., Kervrann C., Salamero J. Patch-based nonlocal functional for denoising fluorescence microscopy image sequences. IEEE Trans. Med. Imaging. 2010;29:442–454. doi: 10.1109/TMI.2009.2033991. [DOI] [PubMed] [Google Scholar]
- 23.Tinevez J.Y., Perry N., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Young J.W., Locke J.C., Elowitz M.B. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat. Protoc. 2011;7:80–88. doi: 10.1038/nprot.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong Ng J., Chatenay D., Poirier M.G. Plasmid copy number noise in monoclonal populations of bacteria. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;81:011909. doi: 10.1103/PhysRevE.81.011909. [DOI] [PubMed] [Google Scholar]
- 26.Panayotatos N. DNA replication regulated by the priming promoter. Nucleic Acids Res. 1984;12:2641–2648. doi: 10.1093/nar/12.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eguchi Y., Itoh T., Tomizawa J. Antisense RNA. Annu. Rev. Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- 28.Stanton B.C., Nielsen A.A., Voigt C.A. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu F., Van Rijn E., Dekker C. Multi-color imaging of the bacterial nucleoid and division proteins with blue, orange, and near-infrared fluorescent proteins. Front. Microbiol. 2015;6:607. doi: 10.3389/fmicb.2015.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber S.C., Spakowitz A.J., Theriot J.A. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys. Rev. Lett. 2010;104:238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber S.C., Theriot J.A., Spakowitz A.J. Subdiffusive motion of a polymer composed of subdiffusive monomers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:011913. doi: 10.1103/PhysRevE.82.011913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cagliero C., Zhou Y.N., Jin D.J. Spatial organization of transcription machinery and its segregation from the replisome in fast-growing bacterial cells. Nucleic Acids Res. 2014;42:13696–13705. doi: 10.1093/nar/gku1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Romero M.A., Lee D.J., Busby S.J. Location and dynamics of an active promoter in Escherichia coli K-12. Biochem. J. 2012;441:481–485. doi: 10.1042/BJ20111258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin D.J., Cagliero C., Zhou Y.N. Role of RNA polymerase and transcription in the organization of the bacterial nucleoid. Chem. Rev. 2013;113:8662–8682. doi: 10.1021/cr4001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stracy M., Lesterlin C., Kapanidis A.N. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc. Natl. Acad. Sci. USA. 2015;112:E4390–E4399. doi: 10.1073/pnas.1507592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin D.J., Cagliero C., Zhou Y.N. The dynamic nature and territory of transcriptional machinery in the bacterial chromosome. Front. Microbiol. 2015;6:497. doi: 10.3389/fmicb.2015.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latulippe D.R., Zydney A.L. Radius of gyration of plasmid DNA isoforms from static light scattering. Biotechnol. Bioeng. 2010;107:134–142. doi: 10.1002/bit.22787. [DOI] [PubMed] [Google Scholar]
- 38.Yeh J.W., Taloni A., Chou C.F. Entropy-driven single molecule tug-of-war of DNA at micro-nanofluidic interfaces. Nano Lett. 2012;12:1597–1602. doi: 10.1021/nl2045292. [DOI] [PubMed] [Google Scholar]
- 39.Tang J., Trahan D.W., Doyle P.S. Coil-stretch transition of DNA molecules in slit-like confinement. Macromolecules. 2010;43:3081–3089. doi: 10.1021/ma902689c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin P.-k., Hsieh C.-C., Chou C.-F. Effects of topology and ionic strength on double-stranded DNA confined in nanoslits. Macromolecules. 2012;45:2920–2927. [Google Scholar]
- 41.Weber S.C., Spakowitz A.J., Theriot J.A. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc. Natl. Acad. Sci. USA. 2012;109:7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burnham K.P., Anderson D.R. Multimodel inference: understanding AIC and BIC in model selection. Sociol. Methods Res. 2004;33:261–304. [Google Scholar]
- 43.Paulsson J., Ehrenberg M. Trade-off between segregational stability and metabolic burden: a mathematical model of plasmid ColE1 replication control. J. Mol. Biol. 1998;279:73–88. doi: 10.1006/jmbi.1998.1751. [DOI] [PubMed] [Google Scholar]
- 44.Ghozzi S., Wong Ng J., Robert J. Inference of plasmid-copy-number mean and noise from single-cell gene expression data. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:051916. doi: 10.1103/PhysRevE.82.051916. [DOI] [PubMed] [Google Scholar]
- 45.Bakshi S., Choi H., Weisshaar J.C. The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol. 2015;6:636. doi: 10.3389/fmicb.2015.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live cells: BW25113FH/pTetORK34p, pLacOIC2c. Respective colors: nucleoids (cyan), plasmids pTetORK34b (green) and pLacOIC2c (red), and cell outlines (phase contrast; gray).
Live cells: BW25113FH/pTetORK34b, pLacOIC2c. Respective colors: nucleoids (blue), plasmids pTetORK34b (green) and pLacOIC2c (red), and cell outlines (phase contrast; gray).
Live cells: BW25113FH/pTetORK34p, pLacOIC2c. Respective colors: nucleoids (cyan), plasmids pTetORK34p (green), and cell outlines (phase contrast; gray).