Abstract
Hydrostatic pressure is an important environmental factor affecting the vertical distribution of marine organisms. Laboratory-based studies have shown that many extant shallow-water marine benthic invertebrates can tolerate hydrostatic pressure outside their known natural distributions. However, only a few studies have focused on the molecular mechanisms of pressure acclimatisation. In the present work, we examined the pressure tolerance of the shallow-water amphipod Eogammarus possjeticus at various temperatures (5, 10, 15, and 20 °C) and hydrostatic pressures (0.1–30 MPa) for 16 h. Six of these experimental groups were used for transcriptome analysis. We found that 100% of E. possjeticus survived under 20 MPa at all temperature conditions for 16 h. Sequence assembly resulted in 138, 304 unigenes. Results of differential expression analysis revealed that 94 well-annotated genes were up-regulated under high pressure. All these findings indicated that the pressure tolerance of E. possjeticus was related to temperature. Several biological processes including energy metabolism, antioxidation, immunity, lipid metabolism, membrane-related process, genetic information processing, and DNA repair are probably involved in the acclimatisation in deep-sea environments.
Introduction
Hydrostatic pressure is thought to be the major environmental factor that limits the vertical distribution of extent marine animals1,2. Studies in the western North Atlantic3,4 revealed a unimodal diversity-depth pattern. Diversity and biomass are relatively low at the upper bathyal and abyssal depths but peak at the intermediate depths, ranging from 1,000 m to 3,000 m5. This kind of depth zonation may have resulted from the colonisation of the deep sea by shallow-water organisms (for review see Brown & Thatje 20146).
The colonisation of the deep sea occurred throughout selection and during the slow genetic drift of species that gradually adapted to life in the deep6. However, depth range shifts, which may be as important as geographic range shifts, are observed in response to ocean warming7,8. Thus, the ability of a shallow-water animal to acclimatise to deep-sea environments during its lifetime is vital. The combined effects of temperature, pressure, and oxygen concentration, may have constrained the bathymetric migrations of marine fauna8. Substantial studies have examined the tolerance of shallow-water invertebrates to high hydrostatic pressure and low temperature (for review see Brown & Thatje 20146). These studies indicated that many extant marine benthic invertebrates can tolerate hydrostatic pressure outside their known natural distributions, and a low temperature could impede high pressure acclimatisation. A few studies have focused on gene expression, such as that of heat shock protein 70 (hsp70), in response to high pressure exposure9–11. However, the transcriptomic approach is seldom applied to relevant studies. A set of genes, which may be activated in response to high hydrostatic pressure (HHP) exposure, can be detected through comparative transcriptome analysis. Thus, the molecular mechanisms that enable shallow-water organisms to acclimatise to deep-sea environments can be studied.
Transcriptome analysis has been applied widely to discover mRNA-level responses, revealing the genetic basis of adaptation to deep-sea environments12–15. Most comparative transcriptome analyses of invertebrates were based on the comparisons of congeneric species that have different vertical distribution profiles, and common patterns of adaptation appeared in widely different taxa of deep-living organisms16. Many biological processes are possibly related to deep-sea adaptation, including alanine biosynthesis15, antioxidation13,17, energy metabolism15,18, immunity18,19, fatty acid metabolism20, and genetic information processing15. However, transcriptomic studies have seldom examined how shallow-water invertebrates acclimatise to a simulated immersion in the deep-sea. Applying transcriptome analysis to reveal deep-sea environmental acclimatisation is of great importance because both evolutionary adaptation and phenotypic acclimation are essential for high pressure adaptation21.
The impacts of hydrostatic pressure on shallow-living organisms were explored in many studies (for review see Somero 199222). Lipid bilayer is regarded as one of the most sensitive molecular assemblages of hydrostatic pressure21–25. High pressure leads to a reduction of membrane fluidity; consequently, the physiological function of membrane, including potential transmission23,26, transmembrane transportation, and cell movement27,28, are impeded. The effects of HHP and low temperature are similar29,30. Parallel effects can be detected according to membrane composition with an increase in hydrostatic pressure of 100 MPa and a reduction in temperature of 13–21 °C22. Studies on microorganisms have indicated that the increasing proportion of unsaturated fatty acid and branched-chain fatty acid can remit the reduction of membrane fluidity imposed by high pressure and low temperature27,31. The structure of protein is depolymerised under high pressure, which results in the inactivation of enzymes27,32,33. Low temperature also has a negative effect on protein structure. Therefore, it induces elevated protein chaperoning34,35, which decreases the stabilisation of the secondary structures of RNA and DNA. Moreover, high pressure can strengthen hydrogen bonds. Consequently, processes that include DNA replication, transcription and translation, are impeded36.
The amphipod Eogammarus possjeticus, belonging to the gammaridean crustaceans, is widely distributed in the coastal and estuarine areas of the Yellow Sea and Bohai Sea in northern China37. The optimal temperature and salinity of E. possjeticus are 20–25 °C and 5–35, respectively37. Amphipods are not only ubiquitous in shallow water, but are also widespread at the hadal depth38, which suggests that shallow-water amphipods could acclimatise to HHP. In the present study, we examined the hydrostatic pressure tolerance of E. possjeticus for the first time. We employed comparative transcriptome analysis on E. possjeticus to examine their molecular responses to hydrostatic pressure stress at different temperature conditions. Our findings may shed light on the mechanisms behind molecular acclimatisation to HHP at the genetic level.
Methods
Collection, maintenance, and rearing of E. possjeticus
Adult specimens of E. possjeticus were net caught from an aquaculture farm in Shandong, China on December 2016. They were maintained at a closed recirculating aquacultural system (seawater was partially refreshed once a week) in the laboratory of the Institute of Deep-sea Science and Engineering, Chinese Academy of Sciences. The amphipods were reared in aerated filtered seawater (salinity: 34.7–35.3, 1 μm filtered, natural light cycle), and were fed with brine shrimp flakes thrice a week; unconsumed food was removed after 24 h. Experimental individuals were starved for 3 d prior to pressurisation.
The experimental animals were maintained in the aquacultural system at 20 °C for 2 weeks to acclimatise them to laboratory environments and allow them to recover from collection and handling stress. The other amphipods remained in the aquacultural system at a constant temperature, while 10 individuals were chosen from the aquacultural system and used for pressure incubation. After the 20 °C experiments were finished, which took at least a week, the aquacultural system temperature was adjusted to 15 °C by a maximum of 2 °C per day. All experimental samples acclimatised to the temperature after it reached 15 °C for at least 5 d before pressurisation. The same procedure was followed for the experiments conducted at 10 °C and 5 °C. Studies on temperature acclimation of insect39–42 and amphibians43,44 indicated that nearly all studied animals can acclimatise to a temperature variation of 5 °C within 5 d. Thus, the temperature acclimatisation of experimental samples in this study is likely to be reached before pressure incubation was achieved.
Pressurisation
The hydrostatic pressure system was set to desired temperature by using circulating water bath (Tianheng Instrument Factory, Zhejiang, China) at least 6 h prior to each experiment. Ten adult individuals of similar size (length: 13 ± 2 mm) were placed inside the stainless pressure chamber (volume: ~20 litres, internal diameter 20 cm, internal depth 65 cm) which is full of filtered seawater at a constant temperature (±0.8 °C), and maintained for 1 h to allow acclimatisation and recovery from handling stress. Then, the pressure vessel was pressurised at 1 MPa per minute to experimental pressure by using hydraulic pump (AILIPU Science and Technology Co., Inc., Zhejiang, China). Experimental samples would be maintained at temperature (5, 10, 15, and 20 °C) and hydrostatic pressure (5, 10, 15, 20, 25, and 30 MPa) conditions for 16 h. The pressure chamber was sealed and isolated during this time period. The pressure was released instantaneously after hydrostatic pressure exposure for 16 h, and the samples were removed from the pressure chamber and snap frozen in liquid nitrogen. The maximum time elapsed between departure from experimental pressure and flash freezing is 3 min. The flash-frozen individuals were stored at −80 °C for further use. Muscle tissues were not dissected before RNA extraction.
Dissolved oxygen, salinity and pH value were measured by using YSI Professional Plus (YSI Inc., USA) to ascertain the stabilisation of seawater quality. In addition, concentration of nitrite nitrogen (NO2-N), ammoniacal nitrogen (NH3-N) and nitric nitrogen (NO3-N) were measured by using HACH DR 1900 (HACH Company, USA) before and after each experiment. No significant difference was found between the experimental context before pressurisation (dissolved oxygen: 5.38 ± 0.3 mg.L−1, salinity: 35.0 ± 0.3, pH: 8.1 ± 0.1, NO2-N: 0.0055 ± 0.0005, NH3-N: 0.015 ± 0.005, NO3-N: 0.015 ± 0.005) and after pressurisation (dissolved oxygen: 5.26 ± 0.4 mg.L−1, salinity: 35.0 ± 0.3, pH: 8.1 ± 0.1, NO2-N: 0.007 ± 0.0005, NH3-N: 0.02 ± 0.005, NO3-N: 0.015 ± 0.005).
RNA extraction, quantification, and qualification
Approximately 50 mg pooled muscle tissues from 5 individuals of the same experimental group were used for each RNA extraction. The muscle tissues from 5 frozen samples were dissected before melted, and immediately transferred into 1 ml of QIAzol (from RNeasy Plus Universal Kit) and homogenised by T10 basic ULTRA-TURRAX (IKA, German). Total RNA was extracted by RNeasy Plus Universal Kit (QIAGEN, UK) according to the manufacturer’s protocol. All samples must meet requirements of the 260/280 ratio between 1.8 and 2.1, and the 260/230 ratio between 2.0 and 2.4, tested by using Nanodrop spectrophotometer (Thermo Fisher Scientific, USA). RNA integrity and concentration were assessed by using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA), and Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, CA, USA), respectively. Only the samples with a RIN value higher than 6.8 were further used. Moreover, RNA degradation and contamination were also monitored on 1% agarose gels. Clear bands of 28 s, 18 s and 5 s rRNA were needed.
Library preparation, Illumina sequencing, and assembly
Six of these experimental groups were chosen for further comparative transcriptome analysis. Their experimental conditions and treatment identifiers are provided in Table 1. A total of 1.5 μg RNA per sample was used for the RNA sample preparations. Sequencing libraries were generated by using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA), and index codes were added to attribute sequences to each sample. TruSeq PE Cluster Kit v3-cBot-HS (Illumina) was used for the clustering of the index-coded samples performed on a cBot Cluster Generation System. Then, the library preparations were sequenced on an Illumina Hiseq platform and paired-end reads were generated. Clean data were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. Transcriptome assembly was then accomplished based on the clean data by using Trinity45. At last, transcripts were hierarchical clustering by Corset46.
Table 1.
Experimental conditions of six experimental groups used for comparative transcriptome analysis.
| Group ID | Temperature (°C) | Pressure (MPa) |
|---|---|---|
| T20P0.1 | 20 | 0.1 |
| T20P15 | 20 | 15 |
| T15P0.1 | 15 | 0.1 |
| T15P15 | 15 | 15 |
| T10P0.1 | 10 | 0.1 |
| T10P15 | 10 | 15 |
Gene functional annotation
All unigenes were annotated in seven databases, including NCBI non-redundant protein sequences (Nr), NCBI non-redundant nucleotide sequences (Nt), a manually annotated and reviewed protein sequence database (Swiss Prot), euKaryotic Ortholog Groups (KOG), Protein family (Pfam), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO). The annotation results of all unigenes are supplied in Table S1.
Transcriptome differential expression analysis and enrichment analysis
First, FPKM and read count of six experimental groups were calculated by using RSEM47. Then, FPKM were normalised by using TMM method. The read counts and normalised FPKM of unigenes are supplied in Table S2. Second, differential expression analysis of paired-sample test was implemented by using DESeq. 2 package48 to identify differential expression genes (DEGs) involved in HHP acclimatisation. The following combinations were paired: T20P15 with T20P0.1, T15P15 with T15P0.1, and T10P15 with T10P0.1. Only genes with an adjusted p-value < 0.01 and |log2 (fold change)| > 1 were regarded as DEGs. In this study, only up-regulated genes were considered as activated genes in response to HHP exposure because only essential processes can be maintained, whereas nonessential processes are reduced outside the optimal range49–53.
The well-annotated DEGs were further analysed and they were grouped in 10 biological processes. The distance among experimental groups was calculated according to normalised FPKM of these well-annotated DEGs with vegan R package54 by using the euclidean method. Then, hierarchical clustering result was visualised by using pheatmap R package55 via the complete method. Moreover, the KEGG enrichment analysis of these up-regulated DEGs was implemented by using the KOBAS software56.
Gene expression analysis and validation by quantitative real-time reverse transcription-PCR (qPCR)
A total of 23 DEGs were employed for qPCR by StepOnePlus Real-Time PCR System (Applied Biosystems, USA) to validate the RNA-seq results. Each 25 μl reaction contained 12.5 μl of FastStart Universal SYBR Green Master (Rox) (Roche, Switzerland), and 2.5 μl of template cDNA. The primer sequences (Table S3) were designed by Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA). DNase treatment was not required because gDNA solution (from RNeasy Plus Universal Kit) was used for RNA extraction. The cDNA library was established by PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Janpan) according to the manufacturer’s standard protocol.
Relative standard curve method was used for expression level analysis with Rpl8 as internal control. The internal control was selected by GeNorm software (Primer Design, Ltd., Southampton University, Highfield Campus, Southampton Haunts, UK). Six 3-fold serial dilutions were performed on a cDNA template to ensure that each primer-set had a qPCR reaction efficiency of between 90 and 105% and a linearity greater than r2 = 0.98 across the predicted experimental cDNA concentration range. Normalised relative quantities were scaled to the control treatment (20 °C 0.1 MPa) in each comparison and converted to relative fold change (RFC). Then, log2 (RFC) was used to evaluate differential expression level. The melting curve analysis was performed at the end of each PCR to confirm only one PCR product was amplified. At last, Pearson correlation coefficients (PCC) between RNA-seq and qPCR results were calculated by corrplot R package57.
According to the results of differential expression analysis in this study, five of these genes, including glutamine synthetase (GS), phosphoenolpyruvate carboxykinase (PEPCK), peroxidase, crustin Pm5, and lysozyme, were selected and their expression patterns were examined in all experimental conditions by using qPCR. We also examined the expression level of a HHP induced gene hsp70 (not existed in the result of transcriptome differential expression analysis in this study) which has been reported in many studies. Two-way ANOVA was used to determine whether temperature and hydrostatic pressure significantly impacted the gene expression patterns, and one-way ANOVA was used to determine whether hydrostatic pressure significantly impacted the gene expression patterns at each temperature condition. The ANOVA analyses were implemented by using R software58. The expression patterns of the six genes were shown on line graph by using Graphpad Prism 7 (Graphpad software, Inc., USA).
Results
Pressure tolerance of Eogammarus possjeticus
The highest examined hydrostatic pressure condition in the present study was 30 MPa. All experimental individuals died at any temperature condition after 16 h pressure exposure under 30 MPa (Table 2). The critical pressure of E. possjeticus is 25 MPa at temperature conditions from 10 °C to 20 °C because up to 50% individuals died under these conditions. However, 25 MPa in the low temperature of 5 °C resulted in 100% mortality rate. A total of 100% survival rate was observed at all temperature conditions (5–20 °C) after 16 h pressure exposure under 20 MPa.
Table 2.
Mortality rates of Eogammarus possjeticus at different hydrostatic pressure and temperature conditions.
| 0.1 MPa | 5 MPa | 10 MPa | 15 MPa | 20 MPa | 25 MPa | 30 MPa | |
|---|---|---|---|---|---|---|---|
| 20 °C | 0% | 0% | 0% | 0% | 0% | 50% | 100% |
| 15 °C | 0% | 0% | 0% | 0% | 0% | 40% | 100% |
| 10 °C | 0% | 0% | 0% | 0% | 0% | 40% | 100% |
| 5 °C | 0% | 0% | 0% | 0% | 0% | 100% | 100% |
Ten individuals were used in each experiment.
Transcriptome sequencing, assembly and annotation
Six experimental groups were chosen for transcriptome analysis (Table 1). Qualities of sequencing are listed in Table S4. Clean reads were finally assembled into 138, 304 unigenes with a total length of 167, 172, 351 bp and an N50 length of 1, 900 bp (Table S5). A total of 60, 928 (44.05%) unigenes were annotated in at least one database (Table S6). Sequence analysis indicated that the experimental samples in the present study share 99% 16S rRNA sequence similarity with Eogammarus possjeticus according to Nt database.
DEGs involved in acclimatisation of high pressure exposure
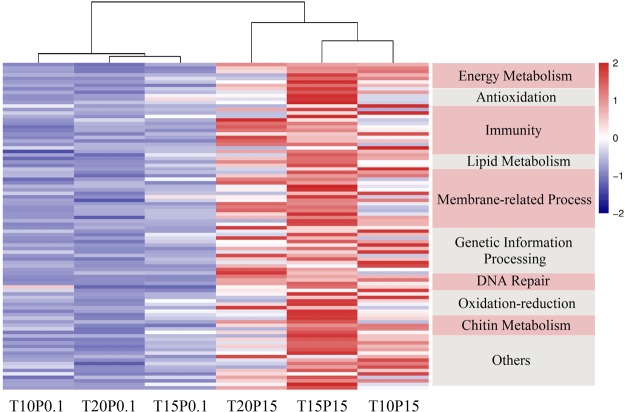
A total of 137 up-regulated genes were selected through DESeq. 2 paired test, of which 94 were well-annotated (Table S7). These genes were grouped into 10 different biological processes according to their functions as follows: energy metabolism (9 genes), antioxidation (5 genes), immunity (13 genes), lipid metabolism (4 genes), membrane-related process (18 genes), genetic information processing (13 genes), DNA repair (4 genes), oxidation-reduction (7 genes), chitin metabolism (6 genes) and others (15 genes). Their expression patterns were visualised via heatmap (Fig. 1). Results of KEGG enrichment indicated that six KEGG pathways were significant enriched (adjusted p-value < 0.01), including nitrogen metabolism, glutamatergic synapse, arginine biosynthesis, glyoxylate and dicarboxylate metabolism, GABAergic synapse,and alanine, aspartate and glutamate metabolism (Fig. S1).
Figure 1.
Expression patterns of 94 well-annotated differential expression genes. Heatmap Clustering is based on normalised FPKM of differential expression genes. Name of corresponded biological processes are listed on the right of the heatmap. T20P0.1: 20 °C, 0.1 MPa; T20P15: 20 °C, 15 MPa; T15P0.1: 15 °C, 0.1 MPa; T15P15: 15 °C, 15 MPa; T10P0.1: 10 °C, 0.1 MPa; T10P15: 10 °C, 15 MPa.
Nine genes were grouped in energy metabolism. Four of them were annotated in glutamine synthetase (GS) which catalyse the amidation of glutamate to glutamine59. Two of them were annotated in phosphoenolpyruvate carboxykinase (PEPCK) which is a key enzyme in the lyase family involved in gluconeogenesis60. One gene was annotated in carbonic anhydrase (CA) which catalyse the conversion of CO2 to the bicarbonate ion and protons61.
Five genes were grouped in antioxidation. Three of them were annotated in vitellogenin (VG) which is known as a yolk protein and was reported to act as an antioxidant to promote longevity62. The other two genes were annotated in peroxidase and catalase respectively. Both genes catalyse the decomposition of hydrogen peroxide to water and oxygen63,64.
A total of 13 genes, including crustin Pm5, lysozyme, and alpha-2-macroglobulin (α2M), were grouped in immunity. The gene crustin Pm5 and lysozyme exhibit antimicrobial activity against some gram-positive bacteria65,66, and α2M serves as humoral defense barriers against pathogens67.
Four genes were grouped in lipid metabolism. They were fatty acid desaturase, elongation of very long chain fatty acids protein (ELOVL), sphingomyelin phosphodiesterase (SMase), and fatty acid-binding protein. Fatty acid desaturase is an enzyme which creates carbon-carbon double bond. It allows the membrane to become more fluid when the temperature is decreased68. A kind of ELOVL is required for the synthesis of C28 and C30 saturated fatty acids and of C28-C38 very long chain polyunsaturated fatty acids69. SMase is involved in sphingolipid metabolism70, and its activation is suggested to be involved in the production of ceramide in response to cellular stresses71. A total of 18 genes were grouped in membrane-related process, most of which are related to ion transmembrane transportation.
A total of 13 genes were grouped in genetic information processing, including DNA replication, transcription, and translation. Four genes were grouped in DNA repair and they were annotated in C2H2 zinc finger domain, MYM-type zinc finger domain, and C4 zinc finger domain. The functions of zinc finger proteins include DNA recognition, RNA packaging, and transcriptional activation72. Studies found that genes encoding for zinc finger domains expanded in deep-sea amphipod compared with other shallow-water species genes15.
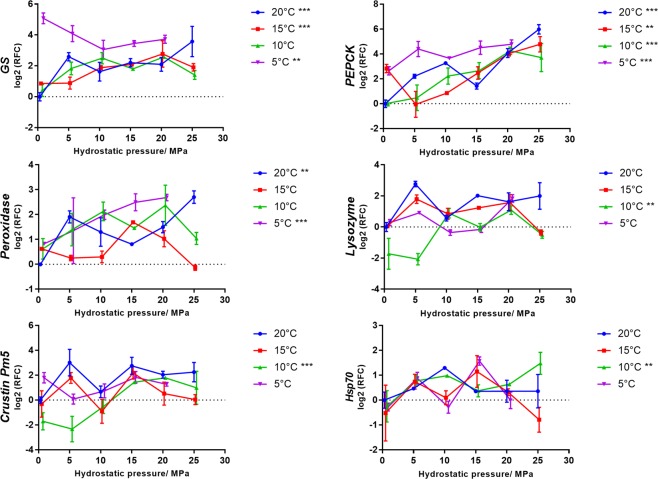
Gene expression analysis by qPCR
PCC between RNA-seq and qPCR results ranged from 0.77 to 0.99 (Table S3), which validated the reliability of the RNA-seq results. Two-way ANOVA results (Table 3) confirmed that the expression patterns of the five genes involved in pressure acclimatisation were significantly correlated with hydrostatic pressure, whereas hsp70 was not. The expression patterns of four genes, including GS, PEPCK, peroxidase, and lysozyme, were also significantly affected by temperature. Pressure and temperature had a significant interaction on the expression patterns of two genes (GS and PEPCK). One-way ANOVA analysis (Table 3) indicated that although the gene expression level was not always significantly up-regulated by hydrostatic pressure, a positive correlation can be mostly observed between gene expression patterns and hydrostatic pressure (Fig. 2). Genes involved in energy metabolism (GS and PEPCK) can also be up-regulated by low temperature, whereas genes involved in immunity (crustin Pm5 and lysozyme) did not show this trend (Fig. 2).
Table 3.
Correlations between gene expression patterns and two environmental variables tested by two-way ANOVA, and correlations between gene expression patterns and hydrostatic pressure at each temperature condition tested by one-way ANOVA.
| GS | PEPCK | Peroxidase | Crustin Pm5 | Lysozyme | Hsp70 | |
|---|---|---|---|---|---|---|
| Two-way ANOVA | ||||||
| Pressure | 0.017* | 2.44 × 10−12*** | 0.001** | 0.001** | 0.013* | 0.115 |
| Temperature | 4.31 × 10−7*** | 0.003** | 0.003** | 0.083 | 5.58 × 10−4*** | 0.606 |
| interaction | 1.81 × 10−5*** | 0.034* | 0.797 | 0.964 | 0.858 | 0.112 |
| One-way ANOVA | ||||||
| Pressure (20 °C) | 2.96 × 10−4*** | 5.93 × 10−6*** | 0.002** | 0.059 | 0.089 | 0.814 |
| Pressure (15 °C) | 1.17 × 10−4*** | 0.002** | 0.976 | 0.857 | 0.567 | 0.663 |
| Pressure (10 °C) | 0.072 | 8.48 × 10−9*** | 0.167 | 7.39 × 10−6*** | 0.009** | 0.002** |
| Pressure (5 °C) | 0.006** | 4.87 × 10−4*** | 9.11 × 10−8*** | 0.592 | 0.184 | 0.399 |
***p-value < 0.001; **p-value < 0.01; *p-value < 0.05.
Figure 2.
Effect of hydrostatic pressure on expression patterns of six genes at different temperature conditions. Normalised relative quantities were scaled to the control treatment (20 °C, 0.1 MPa) in each comparison and converted to relative fold change (RFC). Error bars represent 95% confidence intervals. Correlations between gene expression patterns and hydrostatic pressure at each temperature condition are tested by one-way ANOVA (***p-value < 0.001; **p-value < 0.01; *p-value < 0.05).
Discussion
The utilisation of a pressure vessel provided a stable and controllable experimental context allowing for pressure tolerance to be examined accurately. Although 5 d are enough for insect and amphibian to acclimate to a temperature variation of 5 °C, a longer period of acclimatisation time of more than 10 d is recommended in marine invertebrates73. We measured water quality before and after each experiment to ascertain that the experimental context only involved pressure and temperature stresses, and no significant difference was observed. Therefore, hydrostatic pressure and temperature were the only two variables.
Pressure tolerance of Eogammarus possjeticus
The shallow-water amphipod E. possjeticus can survive 100% at 20 MPa at all temperature conditions ranging from 5 °C to 20 °C for at least 16 h. The impressive tolerance of HHP was also observed among many other shallow-water invertebrates (for review see Brown & Thatje 20146). Remarkably, the pressure boundary of most studied shallow-water invertebrates ranged from 20 MPa to 25 MPa, coinciding with regions of high species turnover5,6.
Low temperatures lead to a decreasing pressure tolerance of E. possjeticus. The phenomenon of decreasing temperature resulting in a reduction of pressure tolerance was also reported among other marine animals. For example, the amphipod Stephonyx biscayensis, subjected to a 10-minute pressure exposure, can tolerate 30 MPa at 10 °C; however, its pressure tolerance fell to 20 MPa at 3 °C74. Increasing the pressure by 1 MPa every 5 min up to 30 MPa resulted in the rate of “loss of equilibrium” in ≥50% of shrimp Palaemonetes varians observed at 20 MPa at 20 °C and 15 MPa at 10 °C75.
Genes in response to high hydrostatic pressure
Most deep-sea animals are widely accepted to have originated from shallow waters as a consequence of a series of extinction events during the Phanerozoic, and the colonisation of deep sea by shallow-water organisms that fallowed76,77. Most existing studies focused on these multiple colonisation events, and the adaptation mechanisms of deep-sea species. However, the deep-sea environments acclimatisation mechanisms of shallow-water fauna are seldom studied. This question is of great importance in the present context of climate change and ocean warming, which are likely to force bathymetric migrations of marine fauna8,11.
In the present study, we explored the acclimatisation mechanisms of the shallow-water amphipod E. possjeticus via transcriptome analysis for the first time. Our results suggested that several biological processes, including energy metabolism, antioxidation, immunity, lipid metabolism, membrane-related process, genetic information processing, and DNA repair, are involved in the acclimatisation of HHP.
The HHP condition of 15 MPa is apparently beyond optimum for the shallow-water amphipod E. possjeticus. Homeostatic effort is required to maintain internal conditions within their physiological tolerance boundaries. Consequently, an increased level of energy requirement to facilitate the increased homeostatic effort is needed. Our results identified three genes are involved in energy metabolism, and two of them (PEPCK and GS) involved in energy production. PEPCK, involved in pathway of gluconeogenesis60, converts oxaloacetate into phosphoenolpyruvate, which has the highest energy phosphate bond found in living organisms. GS catalyse the amidation of glutamate to glutamine59 which is the most abundant amino acid in the plasma and plays an essential role in protein and lipid synthesis78,79. Moreover, glutamine acts as an important energy source in cells80–82, and glutaminolysis partially recruits reaction steps from the citric acid cycle. An increased oxygen demand is required to support a high level of energy requirement. Oxygen is converted into carbon dioxide through aerobic respiration, resulting in the up-regulation of CA, which converts CO2 to the bicarbonate ion and protons61.
Although the metabolic rate of E. possjeticus was not examined in this study, existing studies on crab Lithodes maja indicated that oxygen consumption increases with increasing hydrostatic pressure and is significantly higher at 7.5–17.5 MPa than at 0.1 MPa11. The Pressure tolerance is constrained by oxygen concentration8,11. It may be because the oxygen intake of animals has its ceiling. However, the oxygen consumption of deep-sea species did not appear to be elevated compared with shallow-water congeneric species83–85, suggesting that the former are functionally adapted to high hydrostatic pressure and low temperature86. Although the oxygen consumption of deep-sea species is not significantly higher than that of shallow-water species, the genes involved in carbohydrate and energy metabolism are positively selected in amphipod Hirondellea gigas, which lives at the depth of 10, 929 m in the Challenger Deep15. In general, energy metabolism is of great important in both acclimation and adaption to the deep sea.
Increased mitochondrial activity is needed to meet the energy requirement under high pressure stress. Therefore, mitochondrial oxidative damage is elevated. Consequently, genes involved in antioxidation may be activated. Moreover, the electron transport chain of many reactions would be impeded because the enzyme activity is highly impacted by high pressure87, which consequently induces antioxidant response. In this study, five genes involved in HHP acclimatisation, including peroxidase and catalase, are grouped in antioxidation. An increasing number of evidence indicates that antioxidant defense responses can be induced by HHP and low temperature88–90. Experimental evidence indicates that the mutant of bacteria Shewanella piezotolerans, with enhanced antioxidant defense capacity by experimental evolution under H2O2 stress, has better growth ability at the high pressure of 20 MPa and low temperature of 4 °C than the wild type S. piezotolerans17.
Elevated oxidative damage leads to apoptosis91. Thus, the immune system would be activated in response to mtDNA released from apoptotic mitochondria92. Moreover, misfolded proteins that have resulted from high pressure also induce an immune response93. In the present study, a total of 13 up-regulated DEGs were grouped in immunity, including crustin Pm5 and lysozyme, both of which have antimicrobial activities and serve as part of the innate immune system65,66. Genes involved in immunity were positive selected in the deep-sea urchin Allocentrotus fragilis genome compared with shallow-water urchin Strongylocentrotus purpuratus19, and was up-regulated in the deep-sea shrimp Rimicaris sp. compared with the same species maintained at atmospheric pressure for 10 d94. However, hsp70 was not significantly induced by hydrostatic pressure exposure in the present study. Heat shock proteins play essential roles in heat shock tolerance and the refolding of denatured proteins. They also respond to a variety of stressors, such as pathogen infection, oxidative stress, heavy metals, and other xenobiotics95. A study on the gene hsp70 of shrimp Palaemonetes varians found that the expression level measured in 1 h was three times higher than that observed in shrimp maintained for 7 d under similar temperature and high pressure conditions10. Therefore, the expression level of hsp70 is related to time period under hydrostatic pressure exposure, and its up-regulation is probably a universal stress response in a short time period.
High pressure leads to a reduction of membrane fluidity21–25. The brain gangliosides of the deep-living fish are rich in mono-unsaturated fatty acids and low in saturated fatty acids96. The homeoviscous adaptation of membranes is an important component of adaptation to depth97. Moreover, studies on homeoviscous acclimation to pressure on microorganisms found that the increasing proportion of unsaturated fatty acid and branched-chain fatty acid can remit the reduction of membrane fluidity imposed by high pressure and low temperature27,31. In our studies, four genes were grouped in lipid metabolism, and two of them are involved in the synthesis of unsaturated fatty acids, including fatty acid desaturase68 and ELOVL69. Thus, homeoviscous acclimatisation is important for the deep sea acclimation of shallow-water species.
Almost all membrane-based functions are affected by high pressures98, and transmembrane ion flux is extraordinarily sensitive to pressure. The tansmembrane protein Na+-K+-adenosine triphosphatase (Na+-K+-ATPase), which plays a key role in osmoregulation, varies its activity in accordance with the fluidity of the surrounding membrane lipids99. Studies about Na+-K+-ATPase of teleost gills found that the enzyme from the deep-living fishes is far less inhibited by pressure than the homologous enzymes of shallow-living species. The evolution of Na+-K+-ATPase of deep-sea fish might be a consequence of homeoviscous adaptation of membranes. In our results, thirteen genes belong to ion transmembrane transportation genes, five of which involved in Na+ transmembrane transportation. This tendency reflects that ion transportation in E. possjeticus was impeded under high pressure, and the maintenance of its normal function is of great importance for pressure acclimatisation.
Conclusions
This study reveals that the shallow-water amphipod E. possjeticus can survive 100% under 20 MPa for at least 16 h at temperature conditions from 5 °C to 20 °C. Decreasing temperature results in a reduction of pressure tolerance of E. possjeticus. We identified several genes and biological processes involved in the acclimatisation of shallow-water invertebrates in deep-sea environments, including energy metabolism, antioxidation, immunity, lipid metabolism, membrane-related process, genetic information processing, and DNA repair.
Ethical approval and informed consent
This study did not involve any endangered or protected species and followed all relevant ethical guideline. The samples examined in this study were used as aquacultural feed in China.
Supplementary information
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2017YFC0306600), Major scientific and technological projects of Hainan Province (ZDKJ2016012), and Sanya Key Laboratory Program http://zwzx.sanya.gov.cn/ (L1501), the “Hundred Talents Program” of the Chinese Academy of Sciences (Grant No. SIDSSE-BR-201401, Y510021), and the Knowledge Innovation Program of the Chinese Academy of Sciences (Grant No. SIDSSE-201401). The author Jiawei Chen would like to acknowledge Simin Xu and Zhiyan Chen for their assistance on this study.
Author Contributions
J.C. and H.Z. designed this study. J.C. collected and analysed data, and wrote the first manuscript draft; H.L. and S.C. helped analyse data; H.Z. revised the manuscript and supervised the work.
Data Availability
Relevant data supporting this manuscript are contained within the tables of this manuscript or provided in the supplementary material. The clean data of RNA-seq were available from National Center for Biotechnology Information Sequence Read Archive database (SRA accession numbers: SRR7205161, SRR7205163, SRR7205164, SRR7205158, SRR7205159, SRR7205160).The unigenes were submitted to the Transcriptome Sequencing Assembly database (TSA accession number: SUB4070551).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-39716-y.
References
- 1.Young CM, Tyler PA, Fenaux L. Potential for deep sea invasion by Mediterranean shallow water echinoids:pressure and temperature as stage-specific dispersal barriers. Marine Ecology Progress Series. 1997;154:197–209. doi: 10.3354/meps154197. [DOI] [Google Scholar]
- 2.Thatje S, Hillenbrand CD, Larter R. On the origin of Antarctic marine benthic community structure. Trends in Ecology & Evolution. 2005;20:534–540. doi: 10.1016/j.tree.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Rex MA. Community Structure in the Deep-Sea Benthos. Annual Review of Ecology & Systematics. 1981;12:331–353. doi: 10.1146/annurev.es.12.110181.001555. [DOI] [Google Scholar]
- 4.Etter RJ, Grassle JF. Patterns of species diversity in the deep sea as a function of sediment particle size diversity. Nature. 1992;360:576–578. doi: 10.1038/360576a0. [DOI] [Google Scholar]
- 5.Rex MA, Etter RK. Deep-Sea Biodiversity: Pattern and Scale. Bioscience. 2010;61:327–328. [Google Scholar]
- 6.Brown A, Thatje S. Explaining bathymetric diversity patterns in marine benthic invertebrates and demersal fishes: physiological contributions to adaptation of life at depth. Biol Rev Camb Philos Soc. 2014;89:406–426. doi: 10.1111/brv.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinsky ML, Worm B, Fogarty MJ, Sarmiento JL, Levin SA. Marine taxa track local climate velocities. Science (New York, N.Y.) 2013;341:1239–1242. doi: 10.1126/science.1239352. [DOI] [PubMed] [Google Scholar]
- 8.Brown A, Thatje S. The effects of changing climate on faunal depth distributions determine winners and losers. Global Change Biol. 2015;21:173–180. doi: 10.1111/gcb.12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris JP, et al. Acute combined pressure and temperature exposures on a shallow-water crustacean: novel insights into the stress response and high pressure neurological syndrome. Comp Biochem Physiol A Mol Integr Physiol. 2015;181:9–17. doi: 10.1016/j.cbpa.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Cottin D, et al. Sustained hydrostatic pressure tolerance of the shallow water shrimp Palaemonetes varians at different temperatures: insights into the colonisation of the deep sea. Comp Biochem Physiol A Mol Integr Physiol. 2012;162:357–363. doi: 10.1016/j.cbpa.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Brown A, et al. Metabolic costs imposed by hydrostatic pressure constrain bathymetric range in the lithodid crab Lithodes maja. J. Exp. Biol. 2017;220:3916–3926. doi: 10.1242/jeb.158543. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, et al. Adaptation and evolution of deep-sea scale worms (Annelida: Polynoidae): insights from transcriptome comparison with a shallow-water species. Sci Rep. 2017;7:46205. doi: 10.1038/srep46205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Sun QL, Luan ZD, Lian C, Sun L. Comparative transcriptome analysis of Rimicaris sp. reveals novel molecular features associated with survival in deep-sea hydrothermal vent. Sci Rep. 2017;7:2000. doi: 10.1038/s41598-017-02073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefanni S, et al. Transcriptome of the Deep-Sea Black Scabbardfish, Aphanopus carbo (Perciformes: Trichiuridae): Tissue-Specific Expression Patterns and Candidate Genes Associated to Depth Adaptation. Int J Genomics. 2014;2014:267482. doi: 10.1155/2014/267482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan, Y. et al. Molecular adaptation in the world’s deepest-living animal: Insights from transcriptome sequencing of the hadal amphipod Hirondellea gigas. Mol. Ecol., 10.1111/mec.14149 (2017). [DOI] [PubMed]
- 16.Somero GN. Biochemical ecology of deep-sea animals. Experientia. 1992;48:537–543. doi: 10.1007/bf01920236. [DOI] [PubMed] [Google Scholar]
- 17.Xie, Z., Jian, H., Jin, Z. & Xiao, X. Enhancing the Adaptability of the Deep-Sea Bacterium Shewanella piezotolerans WP3 to High Pressure and Low Temperature by Experimental Evolution under H2O2 Stress. Appl. Environ. Microbiol. 84, 10.1128/AEM.02342-17 (2018). [DOI] [PMC free article] [PubMed]
- 18.Sun J, et al. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 2017;1:121. doi: 10.1038/s41559-017-0121. [DOI] [PubMed] [Google Scholar]
- 19.Oliver TA, et al. Whole-genome positive selection and habitat-driven evolution in a shallow and a deep-sea urchin. Genome Biol Evol. 2010;2:800–814. doi: 10.1093/gbe/evq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng P, et al. Insights into deep-sea adaptations and host-symbiont interactions: A comparative transcriptome study on Bathymodiolus mussels and their coastal relatives. Mol. Ecol. 2017;26:5133–5148. doi: 10.1111/mec.14160. [DOI] [PubMed] [Google Scholar]
- 21.Delong EF, Yayanos AA. Adaptation of the Membrane Lipids of a Deep-Sea Bacterium to Changes in Hydrostatic Pressure. Science. 1985;228:1101–1103. doi: 10.1126/science.3992247. [DOI] [PubMed] [Google Scholar]
- 22.Somero GN. Adaptations to high hydrostatic pressure. Annu Rev Physiol. 1992;54:557–577. doi: 10.1146/annurev.ph.54.030192.003013. [DOI] [PubMed] [Google Scholar]
- 23.Wann KT, Macdonald AG. The effects of pressure on excitable cells. Comparative Biochemistry & Physiology Part A Physiology. 1980;66:1–12. doi: 10.1016/0300-9629(80)90353-9. [DOI] [Google Scholar]
- 24.Macdonald AG. Hydrostatic Pressure as an Environmental Factor in Life Processes. Comparative Biochemistry & Physiology Part A Physiology. 1997;116:291–297. doi: 10.1016/S0300-9629(96)00354-4. [DOI] [Google Scholar]
- 25.Winter, R. & Dzwolak, W. Exploring the temperature-pressure configurational landscape of biomolecules: from lipid membranes to proteins. Philos Trans A Math Phys Eng Sci363, 537–562; discussion 562–533, 10.1098/rsta.2004.1507 (2005). [DOI] [PubMed]
- 26.Siebenaller JF, Garrett DJ. The effects of the deep-sea environment on transmembrane signaling. Comparative Biochemistry & Physiology Part B Biochemistry & Molecular Biology. 2002;131:675. doi: 10.1016/S1096-4959(02)00027-1. [DOI] [PubMed] [Google Scholar]
- 27.Oger PM, Jebbar M. The many ways of coping with pressure. Research in Microbiology. 2010;161:799–809. doi: 10.1016/j.resmic.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Winter, R. Pressure Effects on Lipid Membranes. (Springer Berlin Heidelberg, 2013).
- 29.Thatje S, Casburn L, Calcagno JA. Behavioural and respiratory response of the shallow-water hermit crab Pagurus cuanensis to hydrostatic pressure and temperature. J. Exp. Mar. Biol. Ecol. 2010;390:22–30. doi: 10.1016/j.jembe.2010.04.028. [DOI] [Google Scholar]
- 30.Calligari PA, et al. Adaptation of Extremophilic Proteins with Temperature and Pressure: Evidence from Initiation Factor 6. Journal of Physical Chemistry B. 2015;119:7860–7873. doi: 10.1021/acs.jpcb.5b02034. [DOI] [PubMed] [Google Scholar]
- 31.Michoud G, Jebbar M. High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii. Sci Rep. 2016;6:27289. doi: 10.1038/srep27289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balny C, Masson P, Heremans K. High pressure effects on biological macromolecules: from structural changes to alteration of cellular processes. Biochimica Et Biophysica Acta. 2002;1595:3. doi: 10.1016/S0167-4838(01)00331-4. [DOI] [PubMed] [Google Scholar]
- 33.Northrop DB. Effects of high pressure on enzymatic activity. Biochimica Et Biophysica Acta. 2002;1595:71–79. doi: 10.1016/S0167-4838(01)00335-1. [DOI] [PubMed] [Google Scholar]
- 34.Place SP, Hofmann GE. Constitutive expression of a stress-inducible heat shock protein gene, hsp70, in phylogenetically distant Antarctic fish. Polar Biology. 2005;28:261–267. doi: 10.1007/s00300-004-0697-y. [DOI] [Google Scholar]
- 35.Schmid B, et al. Role of Cold Shock Proteins in Growth of Listeria monocytogenes under Cold and Osmotic Stress Conditions. Appl Environ Microbiol. 2009;75:1621–1627. doi: 10.1128/AEM.02154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phadtare S, Alsina J, Inouye M. Cold-shock response and cold-shock proteins. Current Opinion in Microbiology. 1999;2:175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- 37.Suyan X, et al. Effects of temperature and salinity on the development of the amphipod crustacean Eogammarus sinensis. Chinese Journal of Oceanology and Limnology. 2013;31:1010–1017. doi: 10.1007/s00343-013-2302-0. [DOI] [Google Scholar]
- 38.Ritchie H, Jamieson AJ, Piertney SB. Phylogenetic relationships among hadal amphipods of the Superfamily Lysianassoidea: Implications for taxonomy and biogeography. Deep Sea Research Part I: Oceanographic Research Papers. 2015;105:119–131. doi: 10.1016/j.dsr.2015.08.014. [DOI] [Google Scholar]
- 39.Mellanby KL. Temperature and Insect Activity. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1939;127:473–487. doi: 10.1098/rspb.1939.0035. [DOI] [Google Scholar]
- 40.Fletcher BS, Zervas G. Acclimation of different strains of the olive fly, Dacus oleae, to low temperatures. Journal of Insect Physiology. 1977;23:649–653. doi: 10.1016/0022-1910(77)90061-0. [DOI] [Google Scholar]
- 41.Meats A. Rapid acclimatization to low temperature in the Queensland fruit fly, Dacus tryoni. Journal of Insect Physiology. 1973;19:1903–1911. doi: 10.1016/0022-1910(73)90058-9. [DOI] [Google Scholar]
- 42.Levins R. Thermal Acclimation and Heat Resistance in Drosophila Species. Amer. Natur. 1969;103:483–499. doi: 10.1086/282616. [DOI] [Google Scholar]
- 43.Dunlap DG. Influence of temperature and duration of acclimation, time of day, sex and body weight on metabolic rates in the hylid frog, Acris crepitans. Comp. Biochem. Physiol. 1969;31:555–570. doi: 10.1016/0010-406x(69)90057-7. [DOI] [PubMed] [Google Scholar]
- 44.Londos PL, Brooks RJ. Time Course of Temperature Acclimation of Locomotory Performance in the Toad, Bufo woodhousii woodhousii. Copeia. 1990;1990:827–835. doi: 10.2307/1446448. [DOI] [Google Scholar]
- 45.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29:644–U130. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidson NM, Oshlack A. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014;15:410. doi: 10.1186/s13059-014-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cossins, A. R. & Bowler, K. Temperature biology of animals. Bioscience39 (1987).
- 50.Peck, L. S. Feeding, metabolism and metabolic scope in Antarctic marine ectotherms. Society for Experimental Biology Seminar (1998).
- 51.Peck LS, Morley SA, Pörtner HO, Clark MS. Thermal limits of burrowing capacity are linked to oxygen availability and size in the Antarctic clam Laternula elliptica. Oecologia. 2007;154:479–484. doi: 10.1007/s00442-007-0858-0. [DOI] [PubMed] [Google Scholar]
- 52.Peck LS, Webb KE, Bailey DM. Extreme sensitivity of biological function to temperature in Antarctic marine species. Functional Ecology. 2004;18:625–630. doi: 10.1111/j.0269-8463.2004.00903.x. [DOI] [Google Scholar]
- 53.Peck LS, Webb KE, Miller A, Clark MS, Hill T. Temperature limits to activity, feeding and metabolism in the Antarctic starfish Odontaster validus. Marine Ecology Progress Series. 2008;358:181–189. doi: 10.3354/meps07336. [DOI] [Google Scholar]
- 54.Oksanen, J. et al. Vegan: community ecology package. v. 0.5. 2 (2013).
- 55.Kolde, R. Pheatmap: Pretty Heatmaps (2015).
- 56.Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei T. corrplot: Visualization of a correlation matrix. Mmwr Morbidity & Mortality Weekly Report. 2011;52:145–151. [Google Scholar]
- 58.Team, R. C. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2018).
- 59.Eisenberg D, et al. Some evolutionary relationships of the primary biological catalysts glutamine synthetase and RuBisCO. Cold Spring Harbor Symposia on Quantitative Biology. 1987;52:483. doi: 10.1101/SQB.1987.052.01.055. [DOI] [PubMed] [Google Scholar]
- 60.Hanson RW, Reshef L. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 1997;66:581. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 61.Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 62.Corona M, et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc Natl Acad Sci USA. 2007;104:7128–7133. doi: 10.1073/pnas.0701909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atamna H, Boyle K. Amyloid-beta peptide binds with heme to form a peroxidase: relationship to the cytopathologies of Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3381–3386. doi: 10.1073/pnas.0600134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vatanavicharn T, Supungul P, Puanglarp N, Yingvilasprasert W, Tassanakajon A. Genomic structure, expression pattern and functional characterization of crustinPm5, a unique isoform of crustin from Penaeus monodon. Comp Biochem Physiol B Biochem Mol Biol. 2009;153:244–252. doi: 10.1016/j.cbpb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 66.Ibrahim HR, Matsuzaki T, Aoki T. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett. 2001;506:27–32. doi: 10.1016/s0014-5793(01)02872-1. [DOI] [PubMed] [Google Scholar]
- 67.Borth W. Alpha 2-macroglobulin, a multifunctional binding protein with targeting characteristics. FASEB J. 1992;6:3345–3353. doi: 10.1096/fasebj.6.15.1281457. [DOI] [PubMed] [Google Scholar]
- 68.Los DA, Murata N. Structure and expression of fatty acid desaturases. Biochimica et biophysica acta. 1998;1394:3–15. doi: 10.1016/s0005-2760(98)00091-5. [DOI] [PubMed] [Google Scholar]
- 69.Agbaga M-P, et al. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12843–12848. doi: 10.1073/pnas.0802607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin X, Hengartner MO, Kolesnick R. Caenorhabditis elegans contains two distinct acid sphingomyelinases. The Journal of biological chemistry. 1998;273:14374–14379. doi: 10.1074/jbc.273.23.14374. [DOI] [PubMed] [Google Scholar]
- 71.Hannun YA, Obeid LM. The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. The Journal of biological chemistry. 2002;277:25847–25850. doi: 10.1074/jbc.r200008200. [DOI] [PubMed] [Google Scholar]
- 72.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Current Opinion in Structural Biology. 2001;11:39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 73.New P, et al. The effects of temperature and pressure acclimation on the temperature and pressure tolerance of the shallow-water shrimp Palaemonetes varians. Mar. Biol. 2014;161:697–709. doi: 10.1007/s00227-013-2371-9. [DOI] [Google Scholar]
- 74.Brown, A. & Thatje, S. Respiratory Response of the Deep-Sea Amphipod Stephonyx biscayensis Indicates Bathymetric Range Limitation by Temperature and Hydrostatic Pressure. PLoS One6, 10.1371/journal.pone.0028562 (2011). [DOI] [PMC free article] [PubMed]
- 75.Oliphant A, et al. Pressure tolerance of the shallow-water caridean shrimp Palaemonetes varians across its thermal tolerance window. The Journal of Experimental Biology. 2011;214:1109–1117. doi: 10.1242/jeb.048058. [DOI] [PubMed] [Google Scholar]
- 76.Harnik PG, et al. Extinctions in ancient and modern seas. Trends in Ecology & Evolution. 2012;27:608–617. doi: 10.1016/j.tree.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 77.Raup DM, Sepkoski JJ. Mass Extinctions in the Marine Fossil Record. Science. 1982;215:1501–1503. doi: 10.1126/science.215.4539.1501. [DOI] [PubMed] [Google Scholar]
- 78.Corbet C, Feron O. Metabolic and mind shifts: from glucose to glutamine and acetate addictions in cancer. Current Opinion in Clinical Nutrition & Metabolic Care. 2015;18:346–353. doi: 10.1097/MCO.0000000000000178. [DOI] [PubMed] [Google Scholar]
- 79.Gouw AM, Toal GG, Felsher DW. Metabolic vulnerabilities of MYC-induced cancer. Oncotarget. 2016;7:29879–29880. doi: 10.18632/oncotarget.7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 81.Mckeehan WL. Glycolysis, glutaminolysis and cell proliferation. Cell Biology International Reports. 1982;6:635–650. doi: 10.1016/0309-1651(82)90125-4. [DOI] [PubMed] [Google Scholar]
- 82.Goossens V, Grooten J, Fiers W. The oxidative metabolism of glutamine. A modulator of reactive oxygen intermediate-mediated cytotoxicity of tumor necrosis factor in L929 fibrosarcoma cells. J. Biol. Chem. 1996;271:192–196. doi: 10.1074/jbc.271.1.192. [DOI] [PubMed] [Google Scholar]
- 83.Treude T, Janßen F, Queisser W, Witte U. Metabolism and decompression tolerance of scavenging lysianassoid deep-sea amphipods. Deep Sea Research Part I: Oceanographic Research Papers. 2002;49:1281–1289. doi: 10.1016/s0967-0637(02)00023-7. [DOI] [Google Scholar]
- 84.Childress JJ, Cowles DL, Favuzzi JA, Mickel TJ. Metabolic rates of benthic deep-sea decapod crustaceans decline with increasing depth primarily due to the decline in temperature. Deep Sea Research Part A Oceanographic Research Papers. 1990;37:929–949. doi: 10.1016/0198-0149(90)90104-4. [DOI] [Google Scholar]
- 85.Seibel BA, Drazen JC. The rate of metabolism in marine animals: environmental constraints, ecological demands and energetic opportunities. Philos Trans R Soc Lond B Biol Sci. 2007;362:2061–2078. doi: 10.1098/rstb.2007.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Childress JJ. Are there physiological and biochemical adaptations of metabolism in deep-sea animals? Trends in Ecology & Evolution. 1995;10:30–36. doi: 10.1016/S0169-5347(00)88957-0. [DOI] [PubMed] [Google Scholar]
- 87.Somero GN. Protein adaptations to temperature and pressure: complementary roles of adaptive changes in amino acid sequence and internal milieu. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 2003;136:577–591. doi: 10.1016/s1096-4959(03)00215-x. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Li X, Bartlett DH, Xiao X. Current developments in marine microbiology: high-pressure biotechnology and the genetic engineering of piezophiles. Curr Opin Biotechnol. 2015;33:157–164. doi: 10.1016/j.copbio.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 89.Ambily Nath IV, Loka Bharathi PA. Diversity in transcripts and translational pattern of stress proteins in marine extremophiles. Extremophiles: life under extreme conditions. 2011;15:129–153. doi: 10.1007/s00792-010-0348-x. [DOI] [PubMed] [Google Scholar]
- 90.Shah J, Desai PT, Chen D, Stevens JR, Weimer BC. Preadaptation to cold stress in Salmonella enterica serovar Typhimurium increases survival during subsequent acid stress exposure. Appl. Environ. Microbiol. 2013;79:7281–7289. doi: 10.1128/AEM.02621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anuradha CD, Kanno S, Hirano S. Oxidative damage to mitochondria is a preliminary step to caspase-3 activation in fluoride-induced apoptosis in HL-60 cells. Free Radic Biol Med. 2001;31:367–373. doi: 10.1016/s0891-5849(01)00591-3. [DOI] [PubMed] [Google Scholar]
- 92.Mcarthur K, et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science. 2018;359:eaao6047. doi: 10.1126/science.aao6047. [DOI] [PubMed] [Google Scholar]
- 93.Kovac A, et al. Misfolded truncated protein τ induces innate immune response via MAPK pathway. Journal of Immunology. 2011;187:2732. doi: 10.4049/jimmunol.1100216. [DOI] [PubMed] [Google Scholar]
- 94.Zhang, J., Sun, Q., Luan, Z., Lian, C. & Sun, L. Comparative transcriptome analysis ofRimicarissp. reveals novel molecular features associated with survival in deep-sea hydrothermal vent. Scientific Reports7 (2017). [DOI] [PMC free article] [PubMed]
- 95.Young DB. Stress proteins and the immune response. Antonie Van Leeuwenhoek. 1990;58:203–208. doi: 10.1007/bf00548934. [DOI] [PubMed] [Google Scholar]
- 96.Avrova NF. The Effect of Natural Adaptations of Fishes to Environmental-Temperature on Brain Ganglioside Fatty-Acid and Long-Chain Base Composition. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 1984;78:903–909. doi: 10.1016/0305-0491(84)90207-4. [DOI] [Google Scholar]
- 97.Macdonald AG, Cossins AR. The theory of homeoviscous adaptation of membranes applied to deep-sea animals. Symp Soc Exp Biol. 1985;39:301–322. [PubMed] [Google Scholar]
- 98.Macdonald AG. The effects of pressure on the molecular structure and physiological functions of cell membranes. Philos Trans R Soc Lond B Biol Sci. 1984;304:47–68. doi: 10.1098/rstb.1984.0008. [DOI] [PubMed] [Google Scholar]
- 99.Chong PL, Fortes PA, Jameson DM. Mechanisms of inhibition of (Na,K)-ATPase by hydrostatic pressure studied with fluorescent probes. The Journal of biological chemistry. 1985;260:14484–14490. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relevant data supporting this manuscript are contained within the tables of this manuscript or provided in the supplementary material. The clean data of RNA-seq were available from National Center for Biotechnology Information Sequence Read Archive database (SRA accession numbers: SRR7205161, SRR7205163, SRR7205164, SRR7205158, SRR7205159, SRR7205160).The unigenes were submitted to the Transcriptome Sequencing Assembly database (TSA accession number: SUB4070551).