Abstract
Mucoepidermoid carcinomas are common malignant salivary gland tumors. Despite recent advances in diagnosis and treatment, there has not been much improvement in outcome of these patients, necessitating identification of novel targeted therapeutic agents. Genomic profiling of mucoepidermoid carcinomas has recently revealed aberrations in BAP1 gene. Therefore, we conducted this study to identify BAP1 loss by immunohistochemistry in these tumors. Mucoepidermoid carcinoma cases were retrieved; hematoxylin-and-eosin stained sections were reviewed. Immunohistochemistry for BAP1 was performed. Forty cases were assessed, including 25 salivary gland and 15 pulmonary mucoepidermoid carcinomas. There were 19 cases in the parotid (76%), two in submandibular gland (8%), and remaining 16% from minor salivary gland locations. Ten (40%) were low grade, nine (36%) were intermediate grade, and six (24%) were high grade mucoepidermoid carcinomas. Thirteen (86.7%) pulmonary mucoepidermoid carcinomas were tracheobronchial, while two (13.3%) were intraparenchymal; all were low grade mucoepidermoid carcinomas. On immunohistochemistry, BAP1 nuclear staining was retained in all cases (100%), irrespective of tumor location or grade. Therapeutic connotations necessitate the identification of readily applicable techniques to detect BAP1 loss in mucoepidermoid carcinomas. Using immunohistochemistry, loss of BAP1 staining was not seen in any of our cases, suggesting insensitivity of BAP1 IHC to detect aberrations at genomic level in these tumors. Analysis of BAP1 alterations by targeted sequencing may therefore be performed prior to excluding the possibility of response to BAP1-targeted therapeutics based on immunohistochemistry alone.
Keywords: BAP1, Mucoepidermoid carcinoma, Salivary gland tumor, Parotid, Bronchopulmonary tumor
Introduction
Mucoepidermoid carcinomas (MECs) account for approximately one-third of all salivary gland neoplasms [1]. They are the most common malignant salivary gland tumors, accounting for approximately 38% of adult salivary gland carcinomas [2–4]. MECs are classified into low, intermediate and high grades, based on certain histological parameters [5]. The importance of histological grade lies in the worse outcome seen in high grade MECs, which usually present at a high stage, and require adjuvant therapies following surgical resection [4]. Chen et al. [6] reported 5-year disease specific survival rates of 98.8%, 97.4%, and 67.0% for low, intermediate and high grade MECs, respectively. MECs are the commonest salivary gland-type neoplasms of the lung, usually arising in an endobronchial location in the central airways, due to their origin from submucosal glands lining the tracheobronchial tree [7, 8]. Bronchopulmonary MECs (BPMECs) are classified into low and high grades based on morphological parameters [8, 9]. While low grade BPMECs have a good outcome, high grade BPMECs are associated with aggressive behavior and significantly shorter overall survival periods [9]. Thus, despite recent advances in diagnosis and treatment, there has not been much improvement in the outcome of patients with high grade MECs, necessitating the identification of genetic alterations and novel targeted therapeutic agents for improved management of these tumors.
MECs are associated with an oncogenic recurrent genetic alteration viz. translocation t(11;19) which results in fusion of the CRTC1 and MAML2 genes. This alteration, detected by fluorescent in situ hybridization or real-time PCR, has been identified in 50–65% of MECs, and is more frequent in low and intermediate grade tumors [10–12]. More recently, comprehensive genomic profiling of clinically advanced MECs has revealed genomic aberrations in 80 unique genes, some of which include TP53, CDKN2A, BAP1 and PIK3CA [1, 4, 13]. Among these, BAP1 (BRCA1-associated protein 1) located on 3p21.1 is a relatively recently identified tumor suppressor gene which encodes for a de-ubiquinating enzyme located in the nucleus, and plays a role in regulating transcription, cell growth, cell cycle progression, cell death and DNA damage repair [14]. Its tumor suppressor role, initially described in uveal melanomas, and subsequently in malignant mesotheliomas and renal cell carcinomas, is exerted through dysregulation of these cellular processes [15–19]. Approximately 20% of MECs have recently been reported to demonstrate BAP1 truncation mutations [4]. The availability of BAP1 immunohistochemistry (IHC) has provided a simple, rapid, reliable and economical method for detection of BAP1 genetic alterations, as various tumors with BAP1 mutations show loss of BAP1 immunoexpression [19]. In view of the identification of BAP1 mutations in MEC, we conducted this study to identify loss of BAP1 by IHC in a cohort of MECs. To the best of our knowledge, this is the first study to assess BAP1 expression by IHC in MECs.
Materials and Methods
Cases of MECs diagnosed between 2014 and 2017 were retrieved from the archives of Department of pathology at our institute. These included resection specimens only for salivary gland tumors, and biopsy as well as resection specimens for BPMEC. Cases with insufficient tissue for IHC were excluded. All specimens had been routinely fixed in formalin and paraffin-embedded. Hematoxylin and eosin stained slides were reviewed. Salivary gland tumors were classified as low, intermediate and high grade as per the modified Brandwein grading system; BPMECs were classified as low or high grade [5, 9]. IHC for BAP1 was performed on freshly cut 5 μ-thick formalin fixed paraffin-embedded whole tumor tissue sections using a mouse monoclonal antibody against BAP1 (Santa Cruz, Dallas, TX; clone C4) at a dilution of 1:100 with overnight incubation. Universal labeled streptavidin biotin kit was used as a detection system (Dako, Denmark). Antigen retrieval was performed in a microwave oven using citrate buffer at pH 6.0. Sections from normal testicular tissue were used as a positive control. In addition, nuclei of fibroblasts, lymphocytes, endothelial cells, adjacent normal salivary gland structures, pneumocytes, and bronchial epithelial cells served as internal positive controls [20–24]. IHC had been validated in normal salivary gland tissue and lung parenchyma. Liver biopsies were used as negative controls, as described previously [19]. Staining intensity was graded as strong, medium or weak. Tumors were categorized as having retained BAP1 when strong homogeneous nuclear staining of > 90% of tumor cells was seen, and as displaying BAP1 loss when there was no nuclear staining in tumor cells with intact expression in non-neoplastic stromal and endothelial cells or in external controls [24]. Approval was obtained from the Institute Ethics Committee (IEC-474/01.09.2017, RP37/2017), All India Institute of Medical Sciences, to conduct this observational study on archival patient tumor samples. Informed consent from patients was waived due to the retrospective nature of this study.
Results
Forty MEC cases were assessed immunohistochemically in this study. They included 25 salivary gland MECs and 15 BPMECs. The salivary gland MECs were located in the parotid gland (19 cases; 76%), submandibular gland (2 cases; 8%), and other rare sites, including two cases (8%) from palate, and one case (4%) each from tongue and pinna. Of these, 10 (40%) were low grade MEC, nine (36%) were intermediate grade, and six (24%) were high grade MECs. The mean age was 41.6 years; M:F ratio was 1.08:1. Thirteen (86.7%) BPMECs were tracheobronchial in location, while two (13.3%) were intraparenchymal. All were low grade MECs. The mean age of the BPMEC cohort was 27.3 years; M:F ratio was 2:1.
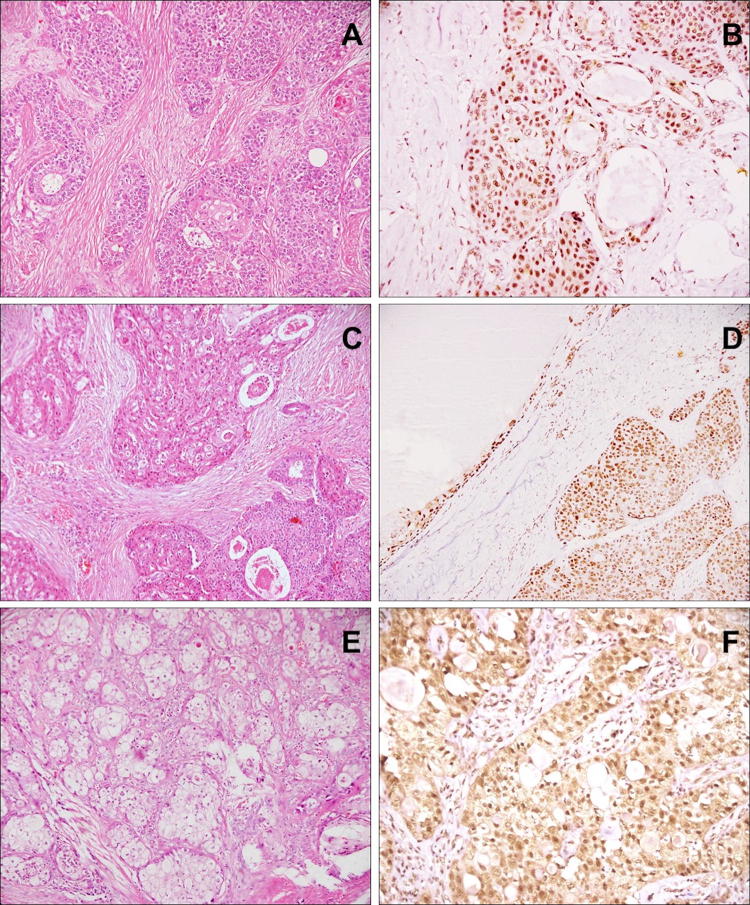
On immunohistochemistry, BAP1 nuclear staining was retained with strong intensity in all cases (100%), irrespective of tumor location or grade (Fig. 1). All types of cells, viz. epidermoid, intermediate and mucous, showed BAP1 staining. The intensity of staining was equal to that of the internal control viz. endothelial cells.
Fig. 1.
BAP1 staining in MECs: Case of submandibular high grade MEC (a HE, × 100) with retained BAP1 staining in squamoid, intermediate and mucous cells (d IHC, × 200). Case of parotid MEC, intermediate grade (c HE, × 100), with retained BAP1 expression in solid as well as cystic components (d IHC, × 100). Case of endobronchial low grade MEC (e HE, × 100) showing nuclear BAP1 positivity in tumor cells (f IHC, × 200)
Discussion
BAP1 mutations were first described in uveal melanomas, followed by malignant mesotheliomas, and renal cell carcinomas [15–18]. Germline BAP1 mutations characterize the BAP1 hereditary cancer predisposition syndrome [16]. The role of BAP1 mutations in the differential diagnosis of benign versus malignant mesothelial proliferations, and malignant mesothelioma versus pulmonary non-small cell carcinomas is now well established [25, 26]. BAP1 loss also has been found to be prognostically relevant in various malignancies such as colorectal carcinoma, lung adenocarcinoma and renal cell carcinoma, where reduced BAP1 expression was associated with poorer patient outcomes [27–29]. Identification of loss of BAP1 has therapeutic connotations as well. Currently, phase II clinical trials are under way to evaluate response of therapeutic agents like PARP inhibitors and EZH2 inhibitors in BAP1-deficient neoplasms [30, 31]. This highlights the possibility that novel targeted therapies for tumors with loss of BAP1 will be available in the near future. As such novel therapeutic options emerge, the assessment of tumors for BAP1 loss will become necessary to identify those patients that are likely to benefit from these newer drugs.
IHC for BAP1 has emerged as a simple, rapid, reliable and economical method for detection of BAP1 mutation in routine pathology practice. Koopmans et al. [32] reported a strong, significant correlation between BAP1 mutation and loss of BAP1 expression in uveal melanomas. They found a sensitivity and specificity of 88% and 97%, respectively, for the detection of BAP1 mutation by BAP1 immunostaining. Similarly, Bott et al. reported a significant association between BAP1 alterations and lack of BAP1 immunoexpression, which was corroborated by other studies [17, 19].
MECs are the most frequent malignant salivary gland tumors, which display aggressive clinical behavior [33]. While surgery followed by radiotherapy is the standard of care, management of patients in the setting of local recurrence or distant metastases leaves much to be desired, necessitating the identification of potentially actionable therapeutic targets, along with rapid and economical methods to identify them in routine clinical practice [1]. MECs are characterized by CRTC1/MAML2 translocation, seen in 38–70% of MECs, is more frequent in lower grades, and has also been found to be associated with prolonged survival [11, 34–38]. However, apart from this translocation, not much is known about the genomic profile of MECs, as they have mostly been included in small numbers in large studies encompassing all histological types of salivary gland carcinomas. Kato et al. included 5 MECs in their analysis of genomic landscape of 117 salivary gland tumors by targeted next-generation sequencing [1]. They identified genetic aberrations in TP53 (2 cases), PI3K pathway (2 cases), PTEN (2 cases), and BAP1 (2 cases) genes in MECs, apart from other genes. BAP1 alterations were also identified in 4/49 adenoid cystic carcinomas, 3/46 adenocarcinomas, not otherwise specified, and 1/7 acinic cell carcinomas. Subsequently, Ross et al. [13], identified BAP1 alterations in approximately 20% of 57 MECs included in their comprehensive genomic profiling of 623 salivary gland carcinomas. Almost simultaneously, the same group published their experience with genomic alterations exclusively in MECs, with BAP1 truncation mutations being seen in 10 out of 48 MECs (20.8%) [4]. However, none of these studies assessed immunoexpression of BAP1 to correlate with results of genomic analysis.
In view of the description of novel BAP1 mutations in MECs, as well as the recent identification and inclusion in clinical trials of drugs targeting tumors with BAP1 loss, we analyzed a cohort of MECs across all grades and locations for loss of BAP1 immunoexpression. In our study, mean age of patients with bronchopulmonary MEC was a decade earlier than for salivary gland MECs; a greater male preponderance was also noted in the former. None of the MEC cases showed loss of BAP1 immunoexpression, irrespective of tumor location or grade. This negative result raises several considerations. Firstly, it is possible that truncating mutations in BAP1 may have led to a qualitative but not quantitative defect in BAP1 protein, resulting in production of a functionally abnormal BAP1 protein which could be detected immunohistochemically. Next, the antibody clone used in this study detects the epitope between aa 430 and 739 of the BAP1 protein, which would detect BAP1 wild-type and mutant forms that retain the nuclear localization signals lying between aa 656–661 and aa 717–722 [19]. Any mutations outside this frame would not affect immunostaining. The third, albeit remote, possibility is that yet unknown genetic or epigenetic changes downstream of BAP1 may have led to restoration of BAP1 immunoexpression despite BAP1 mutation. Lastly, as the ethnicity and demographic profile of the patient cohort included is different from that of previous studies demonstrating BAP1 alterations, their genetic profile may also differ. It is possible that inclusion of larger number of cases may result in detection of mutations that occur at a lower frequency, and the small sample size analyzed in this preliminary study remains a pitfall.
Thus, it follows that comprehensive genetic analysis of larger numbers of MECs is required to understand the implications of our results. The urgency of this is stressed by the therapeutic connotations of identification of BAP1 loss in the near future. Our study is limited by the lack of correlation with genetic analysis and use of a single clone of BAP1 for IHC. Thus, further analysis of BAP1 genomic aberrations in MECs by targeted sequencing is recommended prior to excluding the possibility of response to BAP1-targeted therapeutics based on immunohistochemistry alone.
Compliance with Ethical Standards
Conflict of interest
The authors declared that they have no conflict of interest.
References
- 1.Kato S, Elkin SK, Schwaederle M, et al. Genomic landscape of salivary gland tumors. Oncotarget. 2015;6:25631–25645. doi: 10.18632/oncotarget.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahlberg P, Anderson H, Biorklund A, et al. Carcinoma of the parotid and submandibular glands—a study of survival in 2465 patients. Oral Oncol. 2002;38:706–713. doi: 10.1016/s1368-8375(02)00007-6. [DOI] [PubMed] [Google Scholar]
- 3.Jones AV, Craig GT, Speight PM, et al. The range and demo-graphics of salivary gland tumours diagnosed in a UK population. Oral Oncol. 2008;44:407–417. doi: 10.1016/j.oraloncology.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, McDermott JD, Schrock AB, et al. Comprehensive genomic profiling of salivary mucoepidermoid carcinomas reveals frequent BAP1, PIK3CA, and other actionable genomic alterations. Ann Oncol. 2017;28:748–753. doi: 10.1093/annonc/mdw689. [DOI] [PubMed] [Google Scholar]
- 5.Brandwein MS, Ivanov K, Wallace DI, et al. Mucoepidermoid carcinoma: a clinicopathologic study of 80 patients with special reference to histological grading. Am J Surg Pathol. 2001;25:835–845. doi: 10.1097/00000478-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Chen MM, Roman SA, Sosa JA, et al. Histologic grade as prognostic indicator for mucoepidermoid carcinoma: a population-level analysis of 2400 patients. Head Neck. 2014;36:158–163. doi: 10.1002/hed.23256. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh CC, Sun YH, Lin SW, et al. Surgical outcomes of pulmonary mucoepidermoid carcinoma: a review of 41 cases. PLoS ONE. 2017;12:e0176918. doi: 10.1371/journal.pone.0176918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalhor N, Moran CA. Pulmonary mucoepidermoid carcinoma: diagnosis and treatment. Expert Rev Respir Med. 2018;12:249–255. doi: 10.1080/17476348.2018.1428563. [DOI] [PubMed] [Google Scholar]
- 9.Salem A, Bell D, Sepesi B, et al. Clinicopathologic and genetic features of primary bronchopulmonary mucoepidermoid carcinoma: the MD Anderson Cancer Center experience and comprehensive review of the literature. Virchows Arch. 2017;470:619–626. doi: 10.1007/s00428-017-2104-4. [DOI] [PubMed] [Google Scholar]
- 10.Saade RE, Bell D, Garcia J, et al. Role of CRTC1/MAML2 translocation in the prognosis and clinical outcomes of mucoepidermoid carcinoma. JAMA Otolaryngol Head Neck Surg. 2016;142:234–240. doi: 10.1001/jamaoto.2015.3270. [DOI] [PubMed] [Google Scholar]
- 11.Jee KJ, Persson M, Heikinheimo K, et al. Genomic profiles and CRTC1-MAML2 fusion distinguish different subtypes of mucoepidermoid carcinoma. Mod Pathol. 2013;26:213–222. doi: 10.1038/modpathol.2012.154. [DOI] [PubMed] [Google Scholar]
- 12.Kang H, Tan M, Bishop JA, et al. Whole-exome sequencing of salivary gland mucoepidermoid carcinoma. Clin Cancer Res. 2017;23:283–288. doi: 10.1158/1078-0432.CCR-16-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross JS, Gay LM, Wang K, et al. Comprehensive genomic profiles of metastatic and relapsed salivary gland carcinomas are associated with tumor type and reveal new routes to targeted therapies. Ann Oncol. 2017;28:2539–2546. doi: 10.1093/annonc/mdx399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White AE, Harper JW. Cancer Emerging anatomy of the BAP1 tumor suppressor system. Science. 2012;337:1463–1464. doi: 10.1126/science.1228463. [DOI] [PubMed] [Google Scholar]
- 15.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasu M, Emi M, Pastorino S, et al. High incidence of somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol. 2015;10:565–576. doi: 10.1097/JTO.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankar GM, Abedalthagafi M, Vaubel RA, et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017;19:535–545. doi: 10.1093/neuonc/now235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piris A, Mihm MC, Jr, Hoang MP. BAP1 and BRAFV600E expression in benign and malignant melanocytic proliferations. Hum Pathol. 2015;46:239–245. doi: 10.1016/j.humpath.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Loeser H, Waldschmidt D, Kuetting F, et al. Somatic BRCA1-associated protein 1 (BAP1) loss is an early and rare event in esophageal adenocarcinoma. Mol Clin Oncol. 2017;7:225–228. doi: 10.3892/mco.2017.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cigognetti M, Lonardi S, Fisogni S, et al. BAP1 (BRCA1-associated protein 1) is a highly specific marker for differentiating mesothelioma from reactive mesothelial proliferations. Mod Pathol. 2015;28:1043–1057. doi: 10.1038/modpathol.2015.65. [DOI] [PubMed] [Google Scholar]
- 24.Joseph NM, Chen YY, Nasr A, et al. Genomic profiling of malignant peritoneal mesothelioma reveals recurrent alterations in epigenetic regulatory genes BAP1, SETD2, and DDX3X. Mod Pathol. 2017;30:246–254. doi: 10.1038/modpathol.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Churg A, Sheffield BS, Galateau-Salle F. New markers for separating benign from malignant mesothelial proliferations: Are we there yet? Arch Pathol Lab Med. 2016;140:318–321. doi: 10.5858/arpa.2015-0240-SA. [DOI] [PubMed] [Google Scholar]
- 26.Carbone M, Shimizu D, Napolitano A, et al. Positive nuclear BAP1 immunostaining helps differentiate non-small cell lung carcinomas from malignant mesothelioma. Oncotarget. 2016;7:59314–59321. doi: 10.18632/oncotarget.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang J, Xi S, Wang G, et al. Prognostic significance of BRCA1-associated protein 1 in colorectal cancer. Med Oncol. 2013;30:541. doi: 10.1007/s12032-013-0541-8. [DOI] [PubMed] [Google Scholar]
- 28.Shen C, Wang Y, Wei P, et al. BRCA1-associated protein 1 deficiency in lung adenocarcinoma predicts poor outcome and increased tumor invasion. BMC Cancer. 2016;16:670. doi: 10.1186/s12885-016-2670-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minardi D, Lucarini G, Milanese G, et al. Loss of nuclear BAP1 protein expression is a marker of poor prognosis in patients with clear cell renal cell carcinoma. Urol Oncol. 2016;34:11–18. doi: 10.1016/j.urolonc.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 30.A Trial of Niraparib in BAP1 and Other DNA Damage Response (DDR) Deficient Neoplasms (2017). Retrieved from http://clinicaltrials.gov/ct2. Identification No. NCT03207347
- 31.Study of the EZH2 Inhibitor Tazemetostat in Malignant Mesothelioma (2016). Retrieved from http://clinicaltrials.gov/ct2. Identification No. NCT02860286
- 32.Koopmans AE, Verdijk RM, Brouwer RW, et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Pathol. 2014;27:1321–1330. doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 33.Badlani J, Gupta R, Balasubramanian D, et al. Primary salivary gland malignancies: a review of clinicopathological evolution, molecular mechanisms and management. ANZ J Surg. 2018;88:152–157. doi: 10.1111/ans.14201. [DOI] [PubMed] [Google Scholar]
- 34.Martins C, Cavaco B, Tonon G, et al. A study of MECT1-MAML2 in mucoepidermoid carcinoma and Warthin’s tumor of salivary glands. J Mol Diagn. 2004;6:205–210. doi: 10.1016/S1525-1578(10)60511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonon G, Modi S, Wu L, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33:208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 36.Okabe M, Miyabe S, Nagatsuka H, et al. MECT1-MAML2 fusion transcript defines a favorable subset of mucoepidermoid carcinoma. Clin Cancer Res. 2006;12:3902–3907. doi: 10.1158/1078-0432.CCR-05-2376. [DOI] [PubMed] [Google Scholar]
- 37.Schwarz S, Stiegler C, Müller M, et al. Salivary gland mucoepidermoid carcinoma is a clinically, morphologically and genetically heterogeneous entity: a clinicopathological study of 40 cases with emphasis on grading, histological variants and presence of the t(11;19) translocation. Histopathology. 2011;58:557–570. doi: 10.1111/j.1365-2559.2011.03777.x. [DOI] [PubMed] [Google Scholar]
- 38.Behboudi A, Enlund F, Winnes M, et al. Molecular classification of mucoepidermoid carcinomas-prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer. 2006;45:470–481. doi: 10.1002/gcc.20306. [DOI] [PubMed] [Google Scholar]