Abstract
Under limited micronutrients condition, Mycobacterium tuberculosis (MTB) has to struggle for acquisition of the limited micronutrients available in the host. One such crucial micronutrient that MTB requires for the growth and sustenance is iron. The present study aimed to sequester the iron supply of MTB to control drug resistance in MTB. We found that iron restriction renders hypersensitivity to multidrug-resistant MTB strains against first-line anti-TB drugs. To decipher the effect of iron restriction on possible mechanisms of chemosensitization and altered cellular circuitry governing drug resistance and virulence of MTB, we explored MTB cellular architecture. We could identify non-intact cell envelope, tampered MTB morphology and diminished mycolic acid under iron restricted MDR-MTB cells. Deeper exploration unraveled altered lipidome profile observed through conventional TLC and advanced mass spectrometry-based LC–ESI–MS techniques. Lipidome analysis not only depicted profound alterations of various lipid classes which are crucial for pathogenecity but also exposed leads such as indispensability of iron to sustain metabolic, genotoxic and oxidative stresses. Furthermore, iron deprivation led to inhibited biofilm formation and capacity of MTB to adhere buccal epithelial cells. Lastly, we demonstrated enhanced survival of Mycobacterium-infected Caenorhabditis elegans model under iron limitation. The present study offers evidence and proposes alteration of lipidome profile and affected virulence traits upon iron chelation. Taken together, iron deprivation could be a potential strategy to rescue MDR and enhance the effectiveness of existing anti-TB drugs.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1645-4) contains supplementary material, which is available to authorized users.
Keywords: Myocbacterium, Iron, Lipids, Membrane, Lipidomics, Glyoxylate cycle, Biofilm
Introduction
Tuberculosis (TB) is a deadly disease caused by Mycobacterium tuberculosis (MTB) that affects 10.4 million of the global population annually (WHO 2018). The development of multidrug-resistant tuberculosis (MDR-TB) and extensively drug-resistant tuberculosis (XDR-TB) is an important public health concern for the treatment of TB infections. Under such circumstances, it is unavoidable to explore novel ideas that can be used either alone or in combination with current drug treatments. Targeting nutrient utilization of MTB is an efficiently adopted strategy that has been used nowadays (Vilchèze et al. 2013). Iron is one of the most significant elements required by almost all organism including humans. An estimated 30% of all enzymes need metal ions as cofactor and iron is crucial for such cellular events as the Kreb cycle, DNA biosynthesis, oxidative stress defense, and various metabolic processes (Klein and Lewinson 2011).
Iron is required for the growth and endurance of most bacterial pathogens. There are substantial body of the literature indicating the significance of iron in the growth and pathogenesis of MTB. Previous work by Kochan et al. (1969) revealed that MTB can grow in serum only when a sufficient amount of iron is present. Adding iron increases both intra- and extra-cellular in-vitro mycobacterial growth (Serafín-López et al. 2004). Moreover, it has been suggested that iron burden contributes to TB susceptibility in Africa (Gangaidzo et al. 2001), and rectification of iron burden in mice abolishes this outcome (Schaible et al. 2002). Iron, whose availability is tightly regulated in the host due to its transition nature, makes its availability limited for both the host and invading pathogen like MTB (Hameed et al. 2015). The disturbance of MTB iron metabolism has negative consequences on virulence in vitro and in vivo (Schaible et al. 2002). The mechanisms of iron acquisition in MTB have been reviewed to comprehend the potential iron-dependent candidates that are significant to establish infection (Hameed et al. 2015; Pandey et al. 2014). For instance, peptides have been designed that interfere with the iron-dependent regulator (IdeR) and thereby MTB growth (Salimizand et al. 2017). Role of iron deprivation in the persistence of MTB in human granuloma has revealed several vulnerabilities that can be exploited for anti-TB therapy (Kurthkoti et al. 2017). Lipidomics technology has enabled to identify relation between phospholipid homeostasis, virulence, and iron acquisition (Madigan et al. 2015). Pyrazolopyrimidinone and ATP exhibit antimycobacterial action only by interfering with iron acquisition (Tantano et al. 2015; Dragset et al. 2015). Thus, the significance of iron in mycobacteria is emerging and well established as apparent from a wide range of recent studies.
We formerly hypothesized the mechanisms in Mycobacterium smegmatis, whereby the antimycobacterial efficiency of known first-line anti-TB drugs (isoniazid, ethambutol, rifampicin and streptomycin) was enhanced under iron deprivation (Pal et al. 2015). To validate our hypothesis in the context of pathogenic MTB and to understand the influence of iron availability in MDR-TB clinical strains, we now report deeper insights. The study uncovers additional mechanisms which are governed by iron availability and are necessary to sustain virulence. Furthermore, we demonstrated the enhanced survival of nematode model infected with iron-deprived Mycobacterium marinum, a commonly used model for MTB due to its fast growth rate and ease of use (Shiloh et al. 2010).
Materials and methods
All media chemicals Middlebrook 7H9 broth, Middlebrook 7H10 agar, albumin/dextrose/catalase (ADC), oleic acid/albumin/dextrose/catalase (OADC) supplements were purchased from BD Biosciences (USA). Tween-80, Nitrocefin, Ethambutol (EMB), Isonizid (INH), 2′,7′ dichlorofluorescin diacetate (DCFDA) 4′,6-diamidino-2-phenylindole (DAPI), Calcoflourwhite (CFW), Vanillin (C8H8O3) were purchased from Sigma-Aldrich (St. Louis, MO, USA). 2,2, bipyridyl (2,2,-BP), ethidium bromide (EtBr), streptomycin (STP), dinitrophenol (2,4 DNP), rifampicin (RIF), and propidium iodide (PI) were purchased from Himedia (Mumbai, India). Dimethyl sulfoxide (DMSO), potassium chloride (KCl), sodium chloride (NaCl), di-sodium hydrogen orthophosphate (Na2HPO4), potassium di-hydrogen orthophosphate (KH2PO4), sodium dodecyl sulphate (SDS), glycerol, D-glucose, chloroform (CHCl3), methanol (CH3OH), n-hexane (C6H14), diethyl ether ((C2H5)2O) sulphuric acid (H2SO4), and iodine (I), were obtained from Fischer Scientific.
Bacterial strains and culture conditions
For drug susceptibility assays, all mono-resistant (RIF and EMB) and MDR isolates of MTB used in this study were derived from All India Institute of Medical Sciences and Vallabhbhai Patel Chest Institute, New Delhi, respectively. MTB cells were grown in Middlebrook 7H9 (BD Biosciences) broth supplemented with 0.05% tween-80, 10% ADC and 0.2% glycerol in 100 mL flasks (Schott Duran) and incubated at 37 °C till 12–14 days or till the exponential phase reaches. Stock cultures of log-phase cells were maintained in 30% glycerol and stored at − 80 °C.
Establishment of iron-deprived condition
To establish a suitable iron-restricted condition, MTB cells were exposed to 2,2,-BP (well-known specific iron chelator) as previously described by Pal et al. (2018a, b, c). Subinhibitory concentration of 2,2,-BP, defined as the concentration sufficient to chelate iron without causing appreciable growth defect, was further evident from reduced ferroxidase activity (Fig. S1) and downregulation of ideR transcript (Fig. S2). Thus, for subsequent biochemical and phenotypic studies on MTB under iron restriction, we used 2,2,-BP at its subinhibitory concentration of 40 µg mL− 1.
Minimum inhibitory concentration
Minimum inhibitory concentration (MIC) was done as described previously (Pal et al. 2018a, b, c) by REMA (resazurin microtiter assay) plate method. Briefly, 100 µL of Middlebrook 7H9 broth supplemented with OADC enrichment 0.5% (v/v) glycerol 0.05% (v/v) Tween 80 was placed at each well of the 96 wells plate following with the addition of the drug with the remaining media and then subsequently it was serially diluted 1:2. 100µL of cell suspension (equivalent to the McFarland standard 0.5) was added to each well of the plate (Cui et al. 2013). Plates were incubated at 37 °C for 7 days. After 7 days of incubation, 30 µL of resazurin salt was added to each well, incubated for another 2 days at 37 °C and assessed for the color development. A change from blue to red indicates reduction of resazurin and, therefore, bacterial growth. The MIC was defined as the lowest drug concentration that prevented this color change.
RT-PCR
RNA was extracted from the control and iron-deprived (2,2,-BP) MTB cells by standard TRIzol protocol with few modifications (Pal et al. 2018a, b, c). Briefly, 500 µL of TRI reagent (trizol) was added to MTB cells and sonicated. Reverse transcriptase (RT) PCR was done as described in the RevertAid H Minus kit (Invitrogen) (Pal et al. 2018a, b, c). 5 µg isolated RNA was DNase treated at 37 °C for 30 min and reaction was terminated by adding 1 µL of 25 mM EDTA and incubated at 65 °C for 60 min. RNA was subsequently primed with oligo (dT)18 for cDNA synthesis at 42 °C for 60 min. Reverse transcription reaction was terminated by heating at 70 °C for 5 min. The synthesized cDNA product (2 µL) was directly used for PCR amplification (50 µL) using gene-specific forward and reverse primers (Table S1). The amplified products were gel electrophoresed and the densities of bands (for genes of interest) were measured and quantified by normalizing to that of the constitutively expressed 16S.
PI influx assay
PI influx assay was performed as described elsewhere (Hans et al. 2017). Briefly, control and iron-deprived (2,2,-BP) MTB cells were grown at 37 °C till it reaches the exponential phase in Middlebrook 7H9 broth supplemented with 10% ADC. The cells were harvested by centrifugation at 10,000 rpm and suspended in PBS to 0.5 optical density (O.D.) units at 600 nm. 50 µg PI was added in 1 mL bacterial suspension and kept at room temperature for 15 min in the dark. 10 µL bacterial suspension was transferred to a glass slide, covered with cover-slip and examined under fluorescence microscope at 100×.
Scanning electron microscopy (SEM)
MTB cells grown in Middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (40 µg mL− 1) were observed using SEM (JEOL JEM-1011). The cells of 0.1 O.D600 were seeded to the media and were incubated for 24 h at 37 °C. Sample preparation and analysis were done using the method as described elsewhere (Pal et al. 2015). Briefly, cells were harvested in phosphate-buffered saline (PBS) fixed with 2.5% glutaraldehyde in 0.1% phosphate buffer for 1 h at room temperature (20 °C), washed with 0.1M phosphate buffer (pH 7.2), and postfixed with 1% OsO4 in 0.1Mphosphate buffer for 1 h. Cells were then dehydrated through ethanol, dried and coated with gold, and observed at magnification of 15000×.
Extraction of total cell lipids
MTB control and 2,2-BP-treated cells at exponential phase were used for lipid extraction by modified Folch method (Pal et al. 2017, 2018a, b, c). Extracted lipids were then resolved by TLC using aluminum-backed silica gel plates (silica gel 60 F254; Merck). The lipid extract obtained from control and 2,2-BP-treated cells was loaded on TLC plate at a distance of 2 cm upward from the plate end. Chloroform/methanol/water (65:25:4; v/v/v) was used for developing the plates. Developed chromatogram was dried at room temperature for 2 min and then exposed to iodine fumes generated by iodine crystal balls placed in glass chamber to visualize the lipids.
Extraction of apolar lipids
MTB apolar lipids were extracted as described previously (Chauhan et al. 2013). Briefly, 50 mL of cultures, grown in 7H9 supplemented with ADC, were harvested at an A600nm of 1.0. Apolar lipids were extracted by adding 2 mL of methanolic solution of 0.3% sodium chloride and 1 ml of petroleum ether (60–80 °C) to the cell pellet. The cell suspension was mixed end-over-end for 30 min followed by centrifugation at 2500 rpm for 10 min. The upper layer consisting of apolar lipids was collected in a separate vial and 1 mL of petroleum ether was added to the lower layer, vortexes and mixed end-over-end for 15 min. The cell suspension was again centrifuged to recollect the upper layer. The upper layers comprising apolar lipids were pooled and dried at 60 °C. To resolve fatty acid, free mycolic acid and DG 2-D TLC were performed in chloroform:methanol (96:4 v/v) for 1st dimension and toluene:acetone (80:20 v/v) for 2nd dimension. For TG, extracted lipid was resolved in petroleum ether:diethyl ether (90:10 v/v) (Chauhan et al. 2013).
Ultra performance liquid chromatography-electrospray ionization mass spectrometry (UPLC-ESI-MS)
The samples were analyzed by LC–MS (Waters, ACQ-TQD#QBP 1152), a triple quadrupole tandem mass spectrometer in polarity switching mode as described elsewhere (Pal et al. 2017). Analysis of the mass spectral data was performed using a standalone software MS-LAMP, in which ‘Mtb LipidDB’ (http://www.mrl.colostate.edu; Sartain et al. 2011; Sabareesh and Singh 2013) and the database of Lipid Metabolites and Pathways Strategy Consortium (LIPID MAPS; http://www.lipidmaps.org) are integrated (Sartain et al. 2011). Data acquired in both positive and negative ESI mode only were considered for interpretation by ‘Mtb LipidDB’ of MS-LAMP, whereby the m/z values of the peaks in the mass spectra were assigned to singly protonated ion, i.e., [M–H]− only and by setting the mass window range for the search to 1.0. (Pal et al. 2017, 2018a, b, c) The same set of observed m/z values (that were interpreted by MS-LAMP) was analyzed using MycoMass database (Layre et al. 2011; Pal et al. 2018a, b, c) as well.
Lipase assay
MTB cells were grown in Middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (40 µg mL− 1). Whole cell protein was extracted (Lanigan et al. 2004) and protein concentration (2.5 mg mL− 1) was determined by Lowry method as previously described (Lowry et al. 1951). Lipase activity was performed by measuring the amount of p-nitrophenol (p-NP) released from p-NP ester substrate with varying lengths of fatty acids. The total lipase activity was assayed using protein extract of MTB. The standard lipase activity assays were performed in 100 µL reaction system consisting of a final concentration of 0.5 mM p-NP esters substrate in buffer (pH 8.0) of 80 mM H3BO3, 80 mM H3PO4, 300 mM NaCl, 0.3% Triton X-100 and 20% glycerol. The reaction mixture of purified protein was incubated at 37 °C for 40 min and the release of p-nitrophenol was determined spectrophotometrically (VSI-501) at 405 nm (Pal et al. 2016). Cell extract-free sample as negative control was used in the experiment.
Isocitrate lyase (ICL) and malate synthase (MS) enzymatic activity
Whole cell protein extraction and determination were carried as described elsewhere (Lanigan et al. 2004; Lowry et al. 1951). Bacterial cells were grown in Middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (iron restriction) at concentration (40 µg mL− 1). The 2 mL reaction volume (cell extract) consisted of 25 mM imidazole (solvent water, pH 6.8), 5 mM MgCl2 (solvent water), 1 mM EDTA (solvent water pH8), 4 mM phenylhydrazine HCl (solvent DMSO), 1 mM DL-isocitric acid (substrate), and cell-free extract. The reaction was started right after the addition of substrate. Glyoxylate phenylhydrazone formation was spectrophotometrically assessed at 324 nm after incubation at 30 °C for 30 min to determine the ICL activity in the reaction for the subsequent inhibition study. Similar reactions without substrate were prepared in parallel to serve as the blank (Höner Zu Bentrup et al. 1999). MS activity was determined by a coupled 4,4′-dithiodipyridine (DTP) assay at 412 nm. Reactions were carried out in 50 mM HEPES, pH 7.5 containing 15 mM MgCl2 and 200 µM DTP, substrates AcCoA and glyoxylate, to a final volume of 1 mL. All assays were performed at 25 °C and 1.5 nM MS was added to initiate the reaction (Christine et al. 2011).
Biofilm formation
MTB biofilm formation was estimated as described previously (Pal et al. 2016) on 12-well polystyrene plates. MTB cells were grown in Middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (40 µg mL− 1) till the OD600 ∼ 1.0. For quantitative assay of biofilm, 200 µl of 95% ethanol was added to each crystal violet-stained well and plates were incubated for 10 min at room temperature. Contents of each well were mixed by pipetting and then 125 µL of the crystal violet/ethanol solution was transferred from each well to a separate well of an optically clear flat bottom 12-well plate and optical densities (OD) were measured at 600 nm using spectrophotometer. Inhibition of biofilm was calculated as percentage reduction in growth. For biomasss estimation, mycobacterial biofilms were formed on coverslip in Middlebrook 7H9 broth and incubated for 28 days at 37 °C in 12 well plate. The cover-slip was rinsed with distilled water and the dry weight was measured of the matured biofilm (Hans et al. 2017).
MTB cell adherence
Epithelial cells were obtained by soft scraping of cheek mucous membrane with sterile cotton swabs from mouth cavity of the author who voluntarily agreed to donate. The cells were washed 2–3 times in PBS and the pellets were then resuspended in PBS to give approximately (0.5 OD600) by using spectrophotometer. Bacterial cells were grown in Middlebrook 7H9 broth under iron deprivation (40 µg mL− 1) and incubated overnight at 37 °C. The culture was adjusted to give an approximately OD650 of 0.5, washed twice in PBS, centrifuged for 10 min at 10,000 rpm and resuspended in PBS. The test was performed by taking equal volumes of buccal epithelial cells (OD600 of 0.5) and bacterial suspensions that were mixed and incubated under shaking (120 rpm) at 37 °C for 2–3 h. After incubation, 2–3µL of carbol fuchsin dye to stain Mycobacterium cells and crystal violet to stain epithelial cell was added to each tube and the mixture was gently shaken. 10 µL of the stained suspension was transferred to a glass slide, covered with cover slip and examined under light microscope at 40×. A total of 50 epithelial cells were counted and the mean percentage of adherence was calculated using the number of bacteria added per 50 cells as denominator. Cells were scored as adherent when attached to at least 40% of the epithelial cells, while non-adherent when attached to less than 10% of the examined epithelial cells (Pal et al. 2016).
Measurement of intracellular ROS generation
MTB cells grown in Middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (40 µg mL− 1) were seeded at a starting OD600 of 0.1. Cultures were then grown with shaking at 37 °C for 14 days and then 10 µM DCFDA was added. Bacteria continued to grow for another 30 min with shaking at 37 °C, after which cells were washed and both the OD600 and fluorescence (excitation wavelength, 488 nm, slit 5 nm; emission wavelength 540 nm, slit 10 nm) were measured every 15 min (readings at 2 h after the addition of DCFH-DA are reported) (Howell et al. 2017). H2O2 was used as a positive control and an antioxidant ascorbic acid (AA) as a negative control. To quantify ROS generation, iodonitrotetrazolium chloride (INT) assay was performed as described previously (Hans et al. 2017). Briefly, Mycobacterium cells grown in Middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (40 µg mL− 1 in MTB) were seeded at a starting OD600 of 0.1. Cultures were then grown with shaking at 37 °C till the exponential phase reached, after which cells were washed twice with PBS buffer and 0.5 mL of 0.2% INT dye was added in each set of cells. All the tubes were kept at ambient temperature for 2–3h to check the color change in the tubes. INT dye is pale yellow in color, but ROS generation changes the color from pale yellow to dark pink color which was measured spectrophotometrically within 40 min in UV–Vis double beam spectrophotometer (VSI-501) at 490 nm.
DAPI staining
MTB cells were grown in Middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (40 µg mL− 1) cells and were permeabilized by exposure to toluene (2%) for 10 min prior to staining of DNA. The MTB cells were washed, resuspended in phosphate-buffered saline (PBS; 10 mM sodium phosphate, pH 7.4, 150 mM NaCl, 15 mM KCl), and stained with DAPI (2 mg mL− 1) for 15 min at room temperature. The samples were examined under fluorescent microscope 40× (Coslab) (Hans et al. 2017).
Flow cytometry
The effect of iron deprivation on MTB cell apoptosis-like event was studied as described elsewhere (Sharma et al. 2009) with few modifications. Briefly, 0.1 OD600 of MTB cells grown in middlebrook 7H9 broth in the absence (control) and presence of 2,2-BP (40 µg mL− 1) was allowed to grow till the OD600 reaches 1.0. The cells were then harvested and fixed in 1 mL of 70% ethanol. Wash cells again in PBS and fixed with 4% paraformaldehyde. Fixed cells were stained using Annexin-V kit (sigma) and cell apoptosis-like event and necrosis were then analyzed using Fluorescence Activated Cell Sorter (FACS) (BD LSR-II). 20,000 events were counted and fluorescence intensity was compared between the control and iron-deprived cells and analyzed with BD FACS Diva 6.1.3 software.
Extraction of prenol lipids
Cells were harvested from the culture and extracted with 10 mL of ethanol at 70 °C for 30 min. Pellet the cellular debris by centrifugation at 2500 rpm for 15 min. Remove the organic extract and air dry. Resuspend the residue by the addition of 12 mL of chloroform:methanol (2:1) and partition by the addition of 3 mL water. Final ratio is chloroform:methanol:water 8:4:3. Remove the organic phase that contains the glycolipids. Evaporate to dryness and subject residue to 1 mL of 0.1 M NaOH in ethanol for 45 min at 37 °C and the partition step is repeated as described above. The organic extract contains the polyprenol-based glycolipids and can be visualized by thin-layer chromatography using solvent: (60/30/6) CHCl3/CH3OH/H2O (Slayden et al. 2001).
Maintenance and survival of Caenorhabditis elegans
Nematodes were grown on NGM plate at 20 °C and maintained on E. coli OP50. Approximately, 30 C. elegans L4 or young adult hermaphrodites were transferred from a lawn of E.coli OP50 to the BHI medium containing 2,2,-BP. Worms were considered dead when they did not respond to touch with a platinum wire pick and was scored on daily basis. Each experimental condition was tested in triplicate. Nematode survival was plotted with the Kaplan–Meier method using MS-excel software. For the C. elegans co-infection liquid assay, methodology with few modifications was used (Brenner 1974; Hae-Eun et al. 2017). Alternatively, for infection, specific numbers of CFU of M. marinium treated with 2,2-BP bacteria from an overnight culture were directly inoculated into the liquid medium for feeding or to infect the nematodes for 7 days plates were incubated at 25 °C, animals were scored as live or dead on a daily basis and images were taken on the 7th day of infection at 10× magnification equipped with Coslab camera (Galbadage et al. 2016).
Statistical analysis
All experiments were performed in triplicates (n = 3). The results are reported as mean ± standard deviation (SD) and analyzed using Student t test wherein only p < 0.05 was considered as statistically significant.
Ethics approval
The study was approved by the Research Ethics Committee of the Amity University Haryana (reference no.AUH/EC/RP/2016/05).
Results and discussion
Iron restriction confers hypersensitivity to drug-resistant MTB
The present study was performed to elucidate the role of iron deprivation in combating drug resistance of MTB. To confirm the iron-deprived condition, we performed ferroxidase assay to measure the amount of non-oxidized ferrous iron present in the reaction mixture. MTB contains an iron storage protein known as BfrB that exhibits ferroxidase activity, where ferroxidase regulates iron by oxidizing Fe(II+) to Fe(III+) ions, to prevent the excessive accumulation of iron in MTB, thereby correlating with the iron levels. We found reduced ferroxidase activity of MTB in the presence of an iron chelator 2,2,-BP (Fig. S1). IdeR is an iron regulatory gene that principally works as a negative regulator under low iron conditions and inhibits the synthesis iron acquisition-associated genes, but works as a positive regulator for the iron storage genes bfrA and bfrB. We found downregulation of ideR gene confirming iron deprivation (Fig. S2). Next, we sought to examine if limiting iron would have any effect on drug susceptibility of sensitive and drug-resistant MTB strains against known anti-TB drugs. For this, first we tested the drug susceptibility in H37Rv strain and enhanced susceptibility was observed under iron deprivation (Table 1). Similarly, two mono-drug-resistant MTB strains (R1 and R2) were tested for EMB and RIF sensitivity. We observed that upon iron deprivation both the drug-resistant strains became hypersensitive to the same drugs, where the MIC falls from > 4 µg mL− 1 (EMBR) and > 8 µg mL− 1 (RIFR) to < 0.5 µg mL− 1, respectively (Table 1). These results prompted us to explore the effect of iron deprivation on MDR MTB strains. Interestingly, we observed that iron deprivation confers hypersensitivity to all the three tested MDR strains (Table 1). We further explored the reasons of hypersensitivity and found that most of the genes responsible for resistant phenotype in MTB, viz. rpsL, rpo, embB, katG and inhA were differentially regulated under iron deprivation as revealed by RT-PCR (Fig. S2). katG is a significant gene which is required for catalase–peroxidase activity and survival of MTB inside macrophages. In fact deletion of katG is responsible for INH drug resistance and overexpression of inhA gene confers increased INH resistance (Christine et al. 2010). Another crucial component of cell wall, arabinan encoded by the gene embB which is responsible for the resistance to EMB drug, is overexpressed in resistant MTB (Betzaida et al. 2015). Similarly, RIF and STP target β-subunit of RNA polymerase encoded by rpoB gene and 30S ribosomal protein encoded by gene rpsL, respectively, the overexpression of which makes MTB resistant to RIF and STP drugs (Telenti et al. 1993; Meier et al. 1996). Thus, the differential regulation of genes known to be responsible for drug resistance had good correlation with observed hypersensitivities of drug-resistant MTB strains. This suggested that iron plays a key role not only in MTB pathogenesis but drug resistance as well the mechanisms for which needed to be elucidated.
Table 1.
MIC values for the first-line anti-TB drugs in different MTB strains under iron deprivation
| Anti-TB | H 37 Rv | R1 | R2 | MDR1 | MDR2 | MDR3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −2,2,-BP | +2,2,-BP | −2,2,-BP | +2,2,-BP | −2,2,-BP | +2,2,-BP | −2,2,-BP | +2,2,-BP | −2,2,-BP | +2,2,-BP | −2,2,-BP | +2,2,-BP | |
| MIC Conc. (µg mL− 1) | ||||||||||||
| RIF | 0.0156 | 0.003 | N/T | – | > 8 | 0.5 | 0.5 | 0.03125 | 8 | 0.156 | 8 | 0.125 |
| EMB | 2.0 | 0.5 | > 4 | 0.5 | N/T | – | 1 | 0.0625 | 4 | 0.5 | 4 | 0.125 |
| INH | 0.031 | 0.0078 | N/T | – | N/T | – | 2 | 0.0078 | 4 | 0.5 | 0.5 | 0.125 |
| STP | 0.25 | 0.0625 | N/T | – | N/T | – | 0.0625 | 0.0078 | 0.0625 | 0.0078 | 0.125 | 0.0625 |
Values defined above were derived from the MIC obtained by REMA method. Concentration of 2,2,-BP used 40 µg mL− 1, respectively. “−2,2,-BP” indicates the absence of iron-deprived condition, whereas “+2,2,-BP” indicates the presence of iron-deprived condition
RIF rifampicin, INH isoniazid, EMB ethambutol, STP streptomycin, MIC minimum inhibitory concentration, H37Rv laboratory strain, R1 monoresistant to EMB, R2 monoresistant to RIF, MDR1, MDR2, MDR3 are multidrug resistance strains
Iron deprivation leads to loss in MTB membrane integrity
Subsequently, we used one of the MDR MTB strains to gain further mechanistic insights into iron limitation-induced chemosensitization of anti-TB drugs. First, we examined the cell membrane more closely. Fluorescence-based PI influx assay was performed, where PI can only enter and fluoresce when the cell membrane is injured. We observed fluorescence only in iron-deprived cells in contrast to the control cells (Fig. 1a). Additionally, using an uncoupler of ATP production, 2,4 DNP, we further assessed whether the observed membrane disruption is energy dependent or not. We found that in the presence of DNP (20 µg mL− 1), there was no fluorescence in the cell even under iron deprivation, confirming that the membrane disruption phenomenon is energy dependent (Fig. 1a). Non-intact membrane was also validated by nitrocefin hydrolysis, which depicts altered membrane permeability. It was observed that the rate of nitrocefin hydrolysis was greater under iron deprivation indicating enhanced membrane permeability (Fig. S3). Iron-dependent membrane perturbations were further confirmed by electron microscopy to analyze any morphological variation in size and shape of MTB cell envelope. We observed that the cells grown without any drug (control) or in the presence of anti-TB drugs (INH, EMB, RIF and STP) with subinhibitory concentrations (7.81 ng mL− 1, 62.5 ng mL− 1, 31.25 ng mL− 1 and 7.81 µg mL− 1, respectively) depicted normal morphology (Fig. 1b). On the contrary, the same cells showed roughness and a distorted morphology of the cell envelope under iron-sequestered condition. Furthermore, the cells were elongated, having disrupted cytoplasm homogeneity and showed cytoplasm shrinkage (Fig. 1b). These observations were significant from the point of view that the same drugs which were ineffective against MDR-TB strains at particular concentrations became effective under iron deprivation. Consistent with these observations, we could detect downregulation in bglS gene as revealed by RT-PCR (Fig. S2). bglS gene encodes β-glucosidase enzyme which is found in cell membrane fraction of MTB and possibly has a role in the degradation of non-reducing sugars (Ross et al. 1994). Additionally, we could observe enhanced membrane permeability under iron deprivation (Fig. S3). All these results reinforce the hypothesis that iron restriction causes alterations in membrane homeostasis of MTB.
Fig. 1.
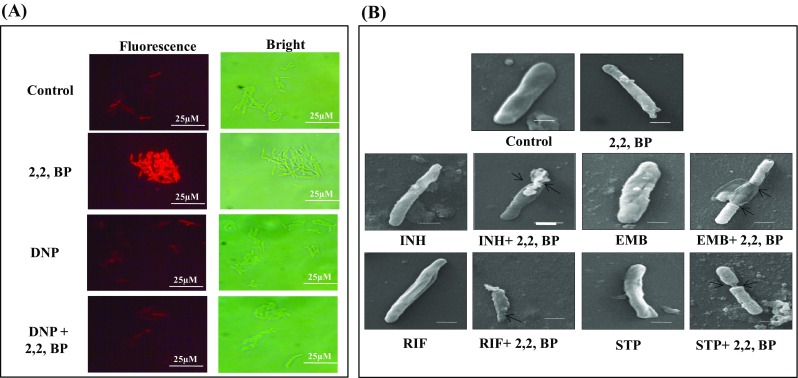
Effect of iron deprivation on membrane integrity and composition. a Fluorescence microscopy showing effect of energy depletion on PI uptake (red signal) in the absence (Control) and presence of 2,2,-BP (40 µg mL− 1) and DNP (ATP inhibitor) at ×100 magnification. Scale bar depicts 25 µm. b SEM images for MTB cells grown in the presence of drugs EMB, INH, RIF and STP with subinhibitory concentrations (0.00781 µg mL− 1, 0.0625 µg mL− 1, 0.03125 µg mL− 1 and 0.00781 µg mL− 1) alone showing smooth and filamentation of cell envelope. SEM images for MTB cells grown in the presence of drugs (EMB, INH, RIF and STP) with subinhibitory concentrations (0.00781 µg mL− 1, 0.0625 µg mL− 1, 0.03125 µg mL− 1 and 0.00781 µg mL− 1) along with 2,2,-BP (40 µg mL− 1) showing tampered morphology
Iron restriction leads to alteration in lipid profile
Further, we examined whether the observed alteration in membrane homeostasis in MTB due to iron restriction also leads to any lipid alterations. We performed TLC of cellular lipids under iron restriction and the chromatogram in control displayed various lipids classes at their respective positions contrary to the lipids under iron restriction which were not detected at similar positions (Fig. 2a). Similarly, 2D TLC also validated our result that iron restriction leads to lipid alterations (Fig. 2b). To further analyze the link between iron deprivation and lipid alteration in MTB, we performed mass spectrometric based lipidome analysis that enabled us to gain significant insights into mechanisms involved in development of lipid derived drug resistance. The data originated presents a global demonstration of varied lipids classified in MTB, facilitating a differential analysis of MTB lipid profiling in normal and iron restriction conditions. The representing mass spectra of untreated (Control) and 2,2,-BP treated samples are shown in Fig. S4 and m/z corresponding to different chromatograms obtained from control and 2,2,-BP-treated samples are shown in Supplementary Material (Excel sheets 1 and 2). As observed in chromatogram, a total of 300 m/z values were found in control and 264 m/z in 2,2,-BP-treated cells, of which 61 m/z were found to be commonly shared by both depicted in the Venn diagram (Fig. S5). The observed m/z values obtained from mass spectra were analyzed through MS-LAMP software where we found that out of 300 m/z values, 87 m/z existed in control and out of 264 m/z, 93 m/z existed in 2,2,-BP-treated cell lipids; and 9 m/z was commonly shared in both conditions (Fig. 2c). The number of lipids which corresponded to these existing 87 and 93 m/z values of control and 2,2,-BP was found to be 137 and 174, respectively (as shown in Fig. 2c heat map). Additionally, we validated the above results with another database, i.e., MycoMass reported by Layre et al. (2011), which contains 5399 molecules which is almost double the number as described by Sartain et al. (2011). Mycomass database revealed that out of 300 m/z values, 170 m/z existed in control and out of 264 m/z, 195 m/z existed in 2,2,-BP treated cell lipids, of which 52 m/z were commonly shared in both the conditions, depicted in the Venn diagram (Fig. 2d). Similarly, the number of lipids which corresponded to the 170 and 195 m/z values of control and 2,2,-BP was 224 and 252, respectively (as shown in Fig. 2d heat map).
Fig. 2.
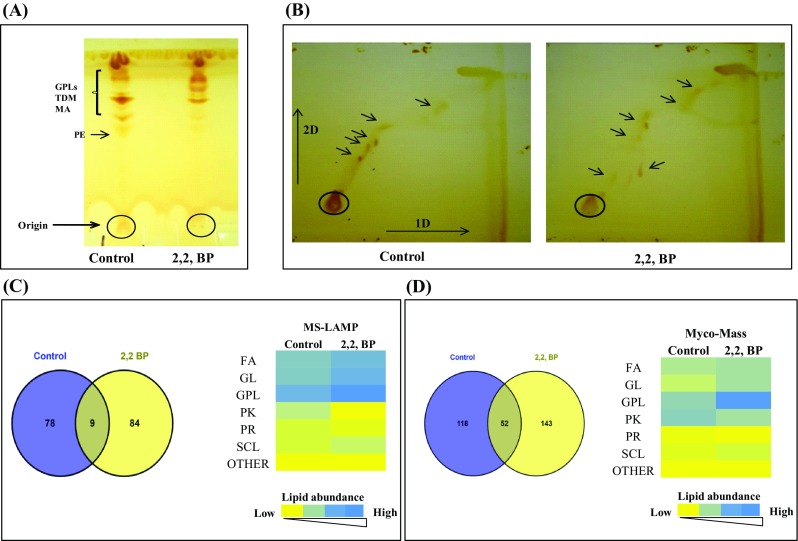
Lipid alteration under iron restriction. a Thin-layer chromatogram (TLC) showing alteration in total lipid profile under iron deprivation. b 2-D TLC of MTB (Control on left side) and 2,2,-BP (on right side) in presence of solvents as described in materials and methods. Arrows depict spots not detected under iron restriction. c Venn diagram showing the number of m/z values which were common and unique in MS-LAMP and heat map showing total lipid composition of major classes (FA, GL, GPL, PK, PR, SCL and others) in control and 2,2,-BP (treated) cells. Color bar depicts increasing number of lipid abundance from yellow to blue. d Venn diagram showing the number of m/z values which were common and unique in MycoMass and heat map showing total lipid composition of major classes (FA, GL, GPL, PK, PR, SCL and others) in control and 2,2,-BP (treated) cells. Color bar depicts increasing number of lipid abundance from yellow to blue
Iron restriction leads to increased metabolic flux towards FA and TG synthesis
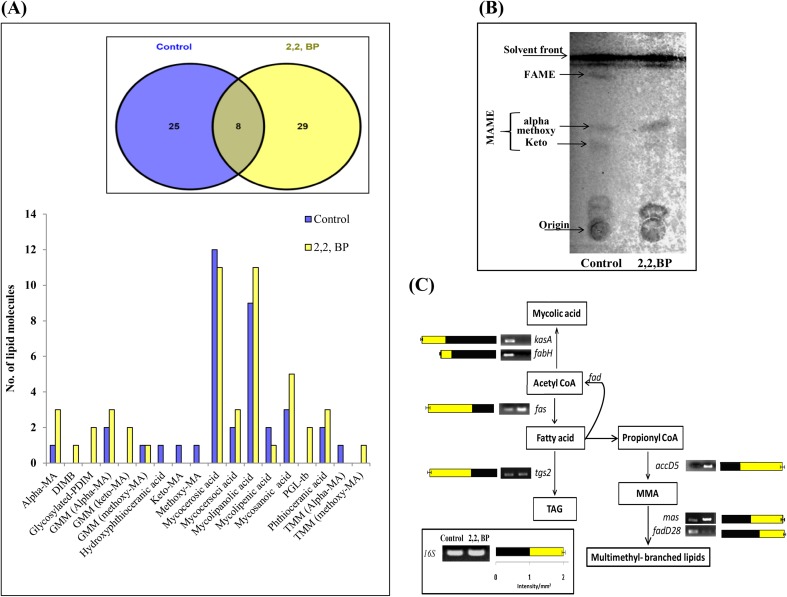
To check the effect of iron deprivation on major classes of lipids, we first monitored the FA subclasses more closely. FA, in general, was found to be elevated but pathogenic lipids, in particular, were differentially expressed. For instance, alpha MAs was increased while methoxy and keto MAs were decreased under iron deprivation. We further analyzed the mass spectrometry data through MS-LAMP and identified 25 and 29 unique m/z signals in control and 2,2,-BP, respectively, which belonged to FA subclasses. Besides these, 8 m/z signals were found to be commonly shared by both control and 2,2,-BP shown in Venn-diagram (Fig. 3a). These 25 and 29 m/z signals obtained from MS-LAMP for control and 2,2,-BP along with commonly found 8 m/z values corresponded to 38 and 49 lipid molecules of FA class shown by bar graph (Fig. 3a). The obtained results were also confirmed by TLC of MAME and FAME lipids which were found to be most affected under iron deprivation (Fig. 3b). Additionally RT-PCR of genes reveals an overexpression of fas, mas, accD5 genes and downregulation of fabH, fadD28 and kasA genes (Fig. 3c) to reinforce that FA is increased and MA is decreased under iron deprivation. Another subclass of FA which was prominently elevated was phenolic glycolipid (PGL-tb) chiefly linked with the cell wall. PGL-tb producing strains (clinical isolate HN878) cause lowered productivity of various immunological factors such as tumor necrosis factor alpha (TNF-α) and interleukin-12 (IL-12) in human monocytes (Manca et al. 2004). Further Sinsimer et al. (2008) investigated the function of PGL-tb of MTB infection in the host immune response and found that PGL-tb progressively modulates the early response of host cytokine.
Fig. 3.
Change in FA composition under iron deprivation. a Bar graph depicts number of lipid moieties identified from m/z values observed by Venn diagram at WR 1.0 through MS-LAMP. Inset (Venn diagram) showing the number of m/z values which were common and unique at WR 1.0. b TLC of MAME and FAME lipids of control and 2,2,-BP-treated cells. c RT-PCR showing transcript levels of kasA, fabH, fas, accD5, fadD28, mas, tgs2 different intermediates involved in FA synthesis and its methylated–branched lipids. Bar graph depicts the quantitation (density expressed as intensity/mm2) of transcript normalized with 16S constitutively expressed gene
Next, we analyzed glycerolipids (GL), which is another major lipid class. The data analysis was done through MS-LAMP using the same criteria as stated above. We detected 33 and 42 unique m/z signals in control and 2,2,-BP which belonged to GL subclasses, respectively. Besides these, 5 m/z signals were found to be commonly shared by both control and 2,2,-BP shown in Venn diagram (Fig. 4a). These 33 and 42 m/z signals gained from MS-LAMP of control and 2,2,-BP along with 5 m/z found in common corresponded to 39 and 53 lipid molecules of GL class shown by bar graph (Fig. 4a). It was also found that lipids belonging to the GL (DG and TG) were enhanced under iron deprivation. To confirm these results, we performed 1-D and 2-D TLC and found that the levels of DG and TG were indeed elevated under iron deprivation (Fig. 4b). Further, we examined lipase activity as there is a direct link of membrane permeability and lipase activity (Pal et al. 2016). Lipase enzymes are responsible for changes in the lipid fraction of the glycolipids membrane structure, resulting in changes in membrane permeability. Consistent with this fact, we found enhanced lipase activity under iron deprivation (Fig. 4c). Lipases in MTB play a crucial role in pathogenicity and, thus, are considered to be a virulence factor (Pal et al. 2016). These lipases hydrolyze and release fatty acid, which is provided as energy source under stress conditions (Pal et al. 2016). MTB use triacylglycerides (TG) as a major storage lipid. Interestingly, inside macrophages MTB accumulates TG using FAs secreted from host TG and this approach is vital for obtaining a dormancy characteristic (Paul et al. 2013). Remarkably, TG accumulates in MTB as they cease growth in response to stress. Gene needed for the synthesis of TG (tgs2) was also upregulated (Fig. S2) which was consistent with the results shown by Bacon et al. (2007) where iron deprivation causes elevation in TG levels. Deb et al. (2006) demonstrated that under hypoxic condition which mimics iron deprivation, MTB accumulates TG.
Fig. 4.
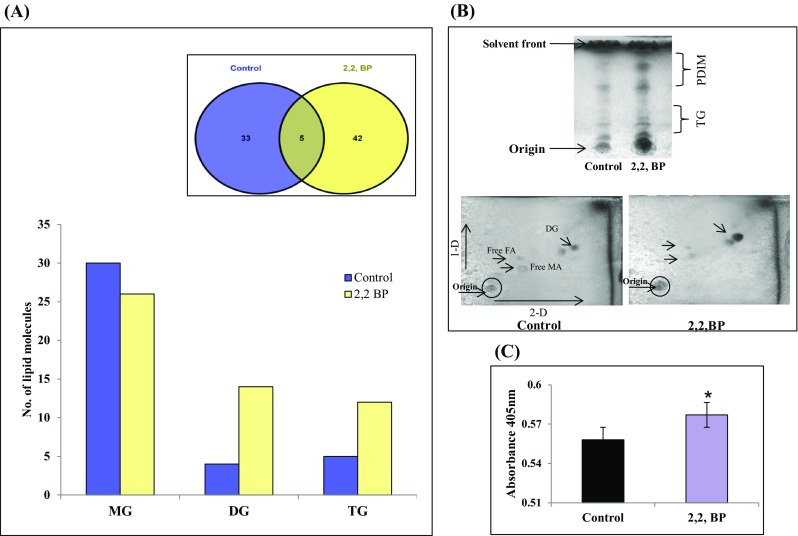
Changes in GL composition under iron deprivation. a Bar graph depicts number of lipid moieties identified from m/z values observed by Venn diagram at WR 1.0 through MS-LAMP. Inset (Venn diagram) showing the number of m/z values which were common and unique at WR 1.0. b Upper panel shows TLC of apolar lipids TG and PDIM extracted from control and 2,2,-BP cells. Lower panel shows 2D TLC of apolar lipids extracted from control and 2,2,-BP-treated cells. c Lipase activity depicted as bar graph in the absence (control) and presence of 2,2,-BP. Mean of O.D405 ± SD of three independent sets of experiments is depicted on Y-axis and *p value < 0.05
Iron deprivation inhibits glyoxylate cycle
Iron deprivation is known to elicit changes in central carbon metabolism where the cell prioritizes the metabolic pathways in such a manner that iron-dependent pathways are downregulated while iron-independent pathways are activated (Gil et al. 2013). Metabolic flexibility under stress conditions is one of the crucial attribute for pathogen survival. One such metabolic pathway that is prevalent in invading pathogens, including MTB, is glyoxylate cycle, as it involves a bypass step preventing carbon loss when low carbon sources are prevailing in hostile niche. Thus, functional GC is required by MTB for its persistence, maintenance and virulence, and the two key enzymes of this pathway ICL and MS represent an attractive target for anti-TB therapy. Increased metabolic flux towards FA synthesis as observed above prompted us to study GC under low carbon conditions such as citrate, acetate or ethanol. ICL1 and MS are two key enzymes of GC required to cope metabolic stress during infection. As a positive control, we also used a serine threonine kinase mutant (ΔPknG) responsible for nutrient utilization and also known to govern virulence and metabolism in MTB (Rieck et al. 2017). We explored that iron deprivation confers hypersensitivity under tested low carbon utilizing conditions such as citrate and ethanol and phenocopied serine threonine protein kinase G (ΔPknG) mutant (Fig. 5a). Biochemical assays further revealed reduced ICL1 and MS activities under iron deprivation (Fig. 5b, c). Subsequent validation by RT-PCR indicated that only the icl1 expression but not ms was considerably reduced under iron deprivation (Fig. S2). Thus, our observations indicate that iron is indispensible to cope with metabolic stress during MTB infection.
Fig. 5.
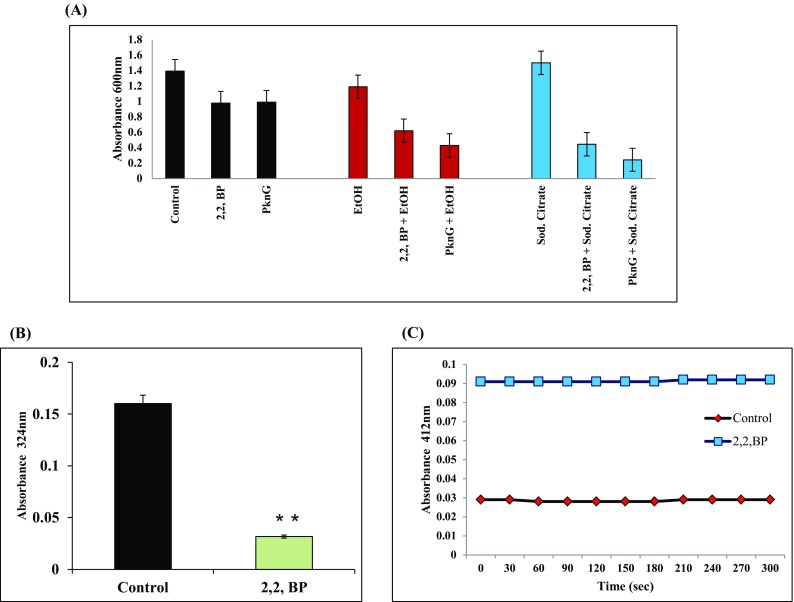
Effect of iron deprivation on metabolic stress (a). Effect of low carbon sources (0.2% of ethanol and 0.2% of sodium citrate) on MTB growth in iron-deprived condition and ΔPknG mutant. Mean O.D600 of three independent sets of experiments is depicted on Y-axis. b ICL activity depicted as bar graph. Mean of absorbance O.D324 ± SD of three independent sets of experiments is depicted on Y-axis and *p value < 0.05. c MS activity is depicted as graph. Mean of absorbance O.D412 ± SD of three independent sets of experiments is depicted on Y-axis
Iron deprivation inhibits biofilm formation and cell adherence
MAs are uniquely present in MTB and play critical roles in the biofilm formation (Ojha et al. 2008) which is another virulence marker which resist bacteria to anti-TB drugs (Trivedi et al. 2016). Biofilm formation under iron deprivation was studied by both qualitative and quantitative methods. CV and CFW staining for qualitative analysis showed that biofilm formation was inhibited under iron-deprived condition in contrast to control (Fig. 6a). For quantitation of biofilms, metabolic activity of CV stained MTB cells was spectrophotometrically measured. It was observed that metabolic activity in iron-deprived biofilms was considerably less in comparison to control cells (Fig. 6b). We also quantified the dry weight of biofilm biomass and found that the biomass was also significantly decreased under iron deprivation (Fig. 6b). These results were also consistent with the fact that RT-PCR showed downregulation of pks-1 gene under iron deprivation (Fig. S2). Recent studies have shown that free MAs, keto MAs and polyketide synthase 1 (PKS-1) (Pang et al. 2012) are some of the factors which are crucial in the formation of the MTB biofilms. Since the present study reveals alteration in lipid profile under iron deprivation, it is only fitting to further study the intricate lipid alterations in response to iron deprivation.
Fig. 6.
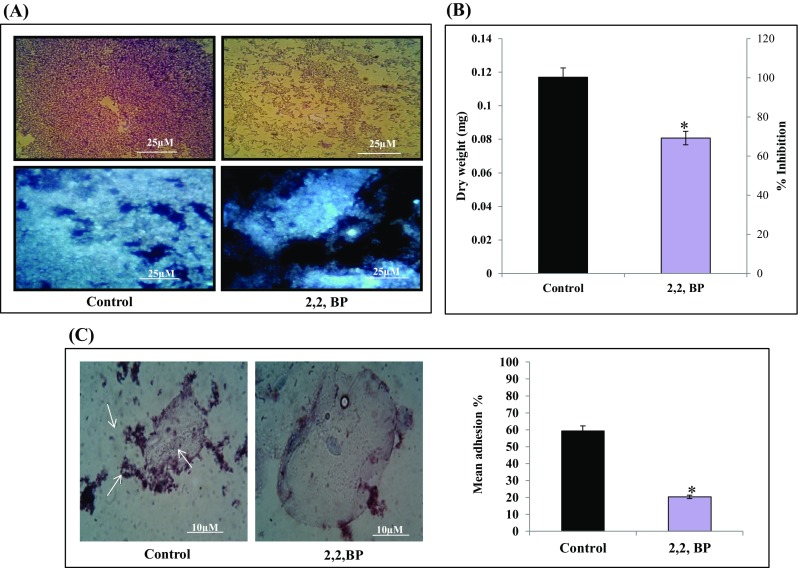
Effect of iron deprivation on MTB biofilm formation. a Biofilm formation depicted by CV staining (upper panel) and CFW staining (lower panel), respectively. b Biofilm biomass (dry weight) and metabolic activity are depicted by bar graph. Mean of biomass in mg ± SD and percentage inhibition, respectively, of three independent sets of experiments is depicted on Y-axis and *p value < 0.05. c Left panel showing Control (untreated) cells appeared adhered (depicted by arrow) to human buccal epithelial cells while iron-deprived cells (2,2,-BP) are not adhered to the epithelial cells (magnification ×40). Right panel showing mean percentage adherence of MTB cells depicted as bar graph showing O.D600 ± SD of three independent sets of experiments on Y-axis and *p value < 0.05
Inhibited biofilm formation under iron deprivation also prompted us to study the adherence of MTB on human buccal epithelial cells as this is known to be a critical step prior to biofilm formation. Interestingly, we found that iron deprivation resulted in the inhibition of the adherence of MTB cells on epithelial cells as compared to control cells (Fig. 6c). To our expectation, the control cells showed mean percent adherence of 59.34% in comparison to the iron-deprived cells which only showed mean percent adherence of 20.34% (Fig. 6c) Together, iron deprivation inhibits potential virulence traits of MTB resulting in the inhibition of biofilm formation and cell adherence.
GPL profiling reveals oxidative and genotoxic vulnerability under iron deprivation
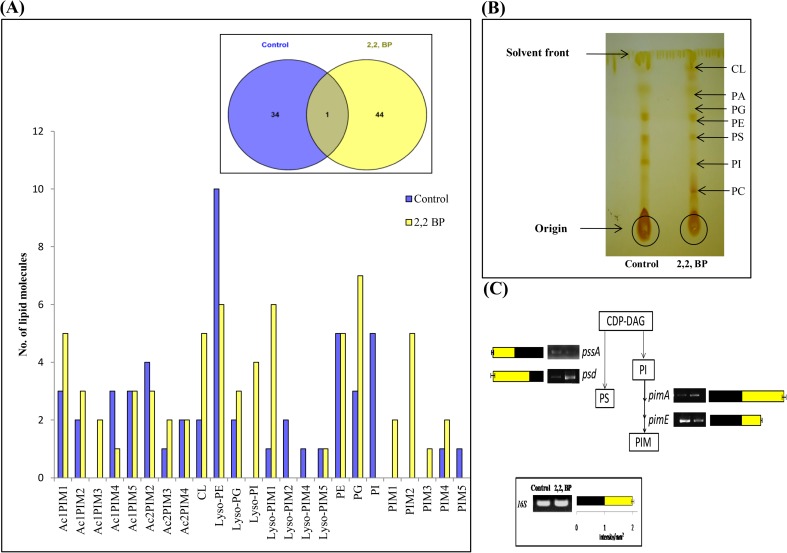
Next, we also analyzed another major class i.e., GPL. While analyzing with MS-LAMP, 34 and 44 unique m/z signals in control and 2,2,-BP were detected which belonged to GPL subclasses, respectively. Besides these, only 1 m/z signal was found to be commonly shared by both control and 2,2,-BP shown in Venn diagram (Fig. 7a). These 34 and 44 m/z signals gained from MS-LAMP of control and 2,2,-BP along with 1 m/z found in common corresponded to 52 and 68 lipid molecules of GPL category shown by bar graph (Fig. 7a). It was also found that almost all the lipid subclasses of GPL were enhanced in iron deprivation (2,2,-BP) as compared to control. Furthermore, TLC revealed that except PE, all the subclasses of GPL were elevated (Fig. 7b). Concomitant with these findings, we could also observe an increased expression of psd and pimA genes and downregulation of pssA and pimE genes (Fig. 7c). PI is a crucial phospholipid in MTB and needed for the synthesis of PIMs, which are the substrates for substantial LMs/LAMs encoded by the gene PgsA (Rv2612c) (Paul et al. 2013) and are required for viability. PIs are further modified by conjugation with glucose residues. The sequential outcomes are PIMs, LMs, and LAMs that are key components of mycobacterial cell wall. Previous study revealed that substantial amounts of PI/PIMs were recognized in the outer membrane. The significance of PIMs is still unclear but high ranking PIMs (PIM1 < PIM2 < PIM3 < PIM4 < PIM5 < PIM6) may be involved in nurturing of membrane integrity. Similarly, our results correlate with the above fact that iron deprivation inhibits the higher ranking PIMs (PIM4 and PIM5) as compared to (PIM1, PIM2, PIM3) (Fig. 7a, b).
Fig. 7.
Changes in GPL composition under iron deprivation. a Bar graph depicts number of lipid moieties identified from m/z values observed by Venn diagram at WR 1.0 through MS-LAMP. Inset (Venn diagram) showing the number of m/z values which were common and unique at WR 1.0. b TLC showing alteration in GPL profile under iron deprivation. c RT-PCR showing transcript levels of pssA, psd, pimA, and pimE different intermediates involved in GPL (PS and PIM) synthesis. Bar graph depicts the quantitation (density expressed as Intensity/mm2) of transcript normalized with 16S constitutively expressed gene
Iron deprivation leads to enhanced generation of reactive oxygen species
Lipids are the most considerable target of oxidative damage. Nambi et al. (2015) showed that membrane-associated oxidoreductase complex (MRC), a thiol-deficient mutant (DMsSseA, DMsDoxX, DMtSseA, and DMtDoxX), in MTB produces more lipid peroxidation as compared to control implying the role of phospholipid synthetic genes in the oxidative stress system (Nambi et al. 2015). Hence, we investigated the link between supra-physiologic elevations in the levels of reactive oxygen species (ROS) coupled to an iron-starvation response, which in turn could foster the eventual cell death. One of the antimycobacterial responses of RIF, which is triggered after target binding, is ROS generation (Piccaro et al. 2014).We determined intracellular ROS generation through membrane-permeating fluorescent dye 2′,7′-dichlorofluorescin diacetate (DCFDA), which fluoresces upon interacting with ROS. We observed that in comparison to the RIF-treated cells, more fluorescence was observed when similar treated cells were deprived of iron (Fig. 8a). Interestingly, the ROS generation could be reverted by the addition of AA (ascorbic acid), a known antioxidant (Fig. 8a). This result was further validated by quantification of ROS generation using INT dye described in methods. As expected, we observed enhanced pink color solution under iron deprivation along with RIF which confirms the ROS generation (Fig. 8b). We could also detect downregulation in oxyS gene, an oxidative stress-regulated protein which corroborates with our findings (Fig. S2).
Fig. 8.
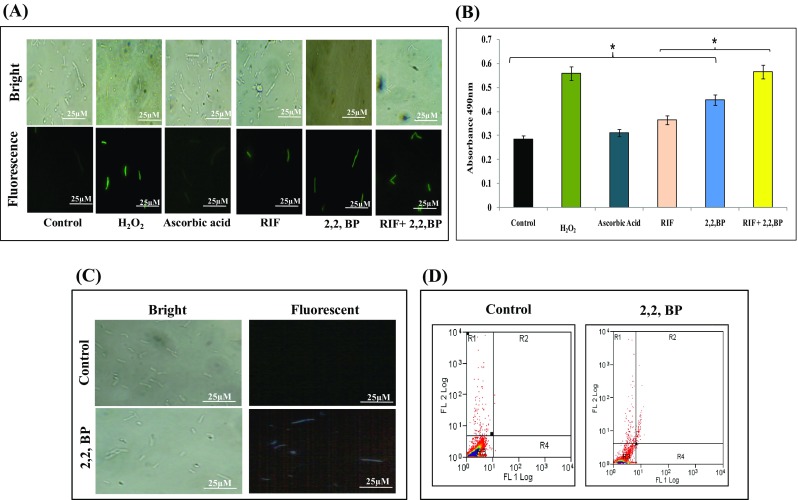
Effect of iron deprivation on oxidative stress. a Estimation of ROS generation with DCFDA observed by fluorescence microscope at ×100 magnification. Scale bar depicts 25 µm. b ROS quantification under iron restriction with INT dye. Mean of absorbance O.D490 ± SD of three independent sets of experiments is depicted on Y-axis and *p value < 0.05. c Fluorescence microscopy of DAPI staining for the detection of DNA damage in the presence of 2,2,-BP at ×100 magnification. Scale bar depicts 25 µm. d Flow cytometry of control and 2,2,-BP (40 µg mL− 1)-treated MTB cells depicting necrosis and apoptosis-like events
Enhanced ROS generation observed in this study is necessitated to study the DNA damage response under iron deprivation. MTB cells were stained with DAPI (4′,6-Diamidine- 2′-phenylindole dihydrochloride) which is a popular nuclear counter stain that preferentially stains A + T region of the damaged dsDNA. Our results depicted that under iron deprivation, blue color fluorescence was observed in contrast to untreated cells (control) where we observed no fluorescence (Fig. 8c). recA responsible for DNA excision repair was also downregulated (Fig. S2). recA gene is crucial for the repair mechanism of damaged DNA and it was already elucidated that deletion in the recA gene makes mycobacteria susceptible to DNA damage (Papavinasasundaram et al. 1998). We further determined whether ROS generation under iron deprivation leads to apoptosis-like cell death in MTB cells (Bayles 2014). FACS analysis indicates that apoptosis-like event was pronounced in iron-deprived cells as compared to the control cells (Fig. S6).
Iron deprivation leads to accumulation of PR and PK sub-species
Subtle changes were also noted when minor lipid classes like PR, PK and SCL were studied under iron deprivation. We found that more number of lipid molecules of PR and PK categories were present under iron deprivation (2,2,-BP) as compared to control (Fig. 9a). Pol-P (Poly-Prenol) is required for bacterial growth and cell wall components’ biosynthesis and is needed for the viability of MTB (Crick et al. 2000). Our lipidomic data were supported by earlier study (Bacon et al. 2007) where gene required for the PR (idsBB) and PK (ppsC, ppsD and pks2) syntheses was upregulated under iron limitation. However, for SCL we could not detect any clear trend as represented by heat map (Fig. 9a). Furthermore, TLC displayed more prenols in iron deprivation as revealed by their denser cone-shaped spots (Fig. 9b). We could also detect upregulation in pds (Prenyl diphosphate synthases) gene which is required for the synthesis of prenols (Fig. S2).
Fig. 9.
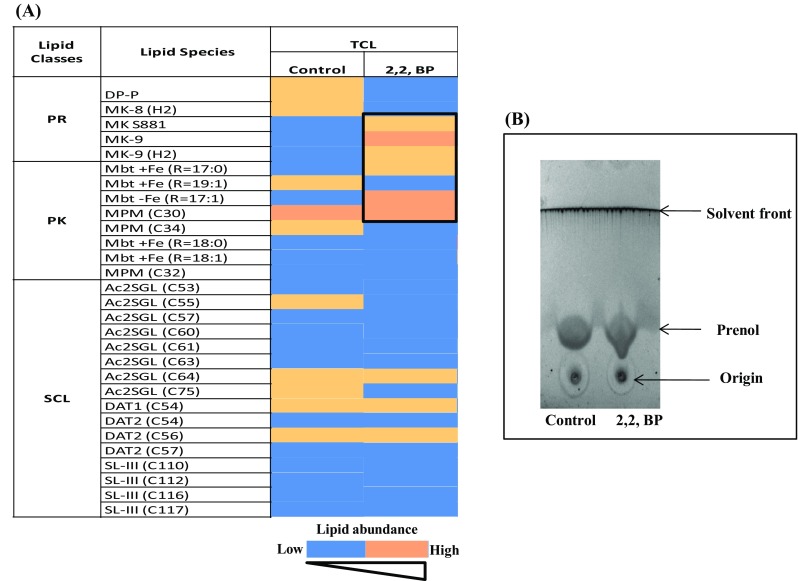
Changes in minor lipid classes under iron deprivation. a Heat map representing composition of (PR, PK and SCL) of MTB control and 2,2,-BP-treated cells present at WR 1.0, through MS-LAMP. Data are shown by color bar depicting increasing number of lipid abundance from yellow to blue. b TLC showing lipid profiling of prenol under iron deprivation
Iron restriction inhibited the virulence of M. marinum in C. elegans
C. elegans lacks both an adaptive immune system and some crucial features of the mammalian innate immune response which includes Toll-like receptor (TLR) adaptor protein MYD88 or NF-κB. Additionally, homolog of TLR in C. elegans does not exhibit a key role in activating the innate immune response either directly or indirectly as a receptor for pathogen-associated molecular pattern (PAMP) molecules. Moreover, also it does not synthesize homolog of cytokines. This obviously limits its relevance for the understanding of human immunity (Pukkila-Worley and Ausubel 2012). Despite of the absence of these immune responses nematode produce an immune response that use some evolutionarily perpetuating signaling pathways which includes, β-catenin, p38 mitogen-activated protein kinase (MAPK) and FOXO transcription factors, altogether function parallelly to activate at least partly overlay sets of effectors genes. With these factors, now it can be argued that nematodes offer an admirable opportunity to recognize TLR-independent and NF-κB-independent characters of the metazoan innate immune response that may be not easy to recognize in vertebrate models (Pukkila-Worley and Ausubel 2012). On the other hand, its relative simplicity and the extensive range of available experimental methods facilitate its use in biology. The major molecular mechanisms involved in the epidermal innate immune response in C. elegans have been identified. As in humans, they involve the upregulation of AMP expression and wound healing, governed by a combination of highly conserved and nematode-specific pathways. A common theme that has emerged is the importance of damage recognition as a trigger for the induction of defense genes.
To evaluate the consequences of iron deficiency in MTB, the colonization of C. elegans, an established infection model for mycobacteria (Galbadage et al. 2016), was tested. Antimycobacterial effects of iron restriction were validated in C. elegans nematode model infected with M. marinum. Firstly, we examined the toxicity of 2,2,-BP (iron restriction) by treating worms with same concentration (40 µg mL− 1) of 2,2,-BP that was used in the present study for 6 days in the absence of M. marinum infection. 2,2,-BP at this concentration did not affect nematode viability (Fig. 10a), suggesting that iron restriction does not have toxic effect on the nematode. Further, microscopic observations of infected nematodes revealed that untreated M. marinum caused death in C. elegans. However, worm survival was increased when infected with M. marinum grown under iron-deprived condition (40 µg mL− 1 of 2,2,-BP) (Fig. 10b). In C. elegans, we found significant decreased viability when infected with non-treated controls (Fig. 10b), whereas treated M. marinum infection increased the nematode survival. In agreement with this finding, we hypothesized that both pharmacological and genetic inhibition of iron are effective for nematode survival.
Fig. 10.
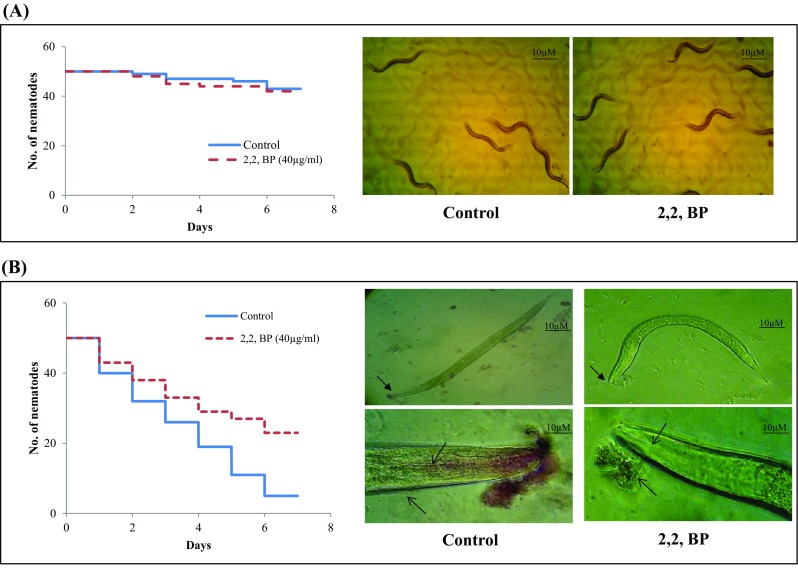
M. marinum infected nematode model of C. elegans under iron deprivation. a The toxicity of 2,2,-BP was studied on non-infected nematodes by determining survival rates after 7 days depicted by Kaplan–Meier graph. b Left panel depicting Kaplan–Meier survival graph of C. elegans on exposure of infected nematodes under iron deprivation (2,2,-BP). Right panel depicts microscopic images showing survival (upper image; magnification ×10) and persistence (lower image; magnification ×40) by carbol fuchsin stain of M. marinum infected nematodes under iron deprivation (2,2,-BP). Worm survival was determined based on the movement. At least, three independent experiments were conducted
Conclusion
The increasing appreciation that metals do have impact on microbial resistance and virulence is gaining prominence. The present lipidomics approach adopted in this study unveils susceptible iron-dependent mechanisms in MTB which can be exploited for the design of new anti-TB therapies (Fig. 11). Sequestering Fe supply as a treatment option should be carefully evaluated considering the fact that the host also needs iron and there is iron overload in phagosomes. In fact, this is one of the suggested mechanisms that macrophages employ to kill intracellular pathogens. Therefore, although it may be important for extracellular bacteria in necrotic granulomas, the response to Fe depletion should be further scrutinized for intracellular bacteria. Thus, dissecting the mechanisms that regulate cellular circuitry governing drug resistance and other crucial virulence attributes although holds great promise for elucidating new anti-TB therapeutic strategies requires further clinical validation.
Fig. 11.
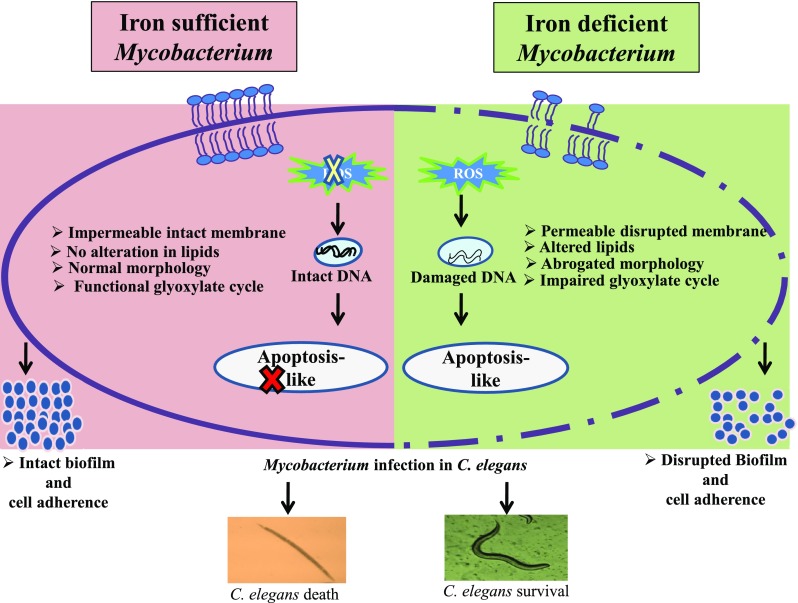
Model showing summary of vulnerable targets affected in MTB under iron deprivation
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 Excel sheet showing the data obtained from MS-LAMP in untreated cells (XLS 147 KB)
Supplementary material 3 Excel sheet showing the data obtained from MS-LAMP in 2, 2,-BP treated cells (XLS 179 KB)
Acknowledgements
Z. F. thanks Board of Research in Nuclear Sciences (BRNS), Mumbai (2013/37B/45/BRNS/1903) for the financial assistance. We thank Anindya Ghosh for providing wild-type C. elegans (N2) and Escherichia coli OP50 strains as generous gift. We are grateful to Mandira Varma-Basil, Pramod Mehta and Yossef Av-Gay for providing MTB MDR strains, M. marinium and ΔPknG mutant as generous gifts, respectively. We thank Sanjeev Kanojiya for assisting us in mass spectrometry experiments. We thank Varatharajan Sabareesh for his intellectual support in lipidome data analysis.
Abbreviations
- MTB
Mycobacterium tuberculosis
- MDR
Multidrug resistance
- ADC
Albumin dextrose catalase
- OADC
Oleic albumin dextrose catalase
- 2,4 DNP
2,4 dinitrophenol
- CFW
Calcoflour white
- CV
Crystal violet
- INT
Iodonitrotetrazolium chloride
- SEM
Scanning electron microscopy
- PI
Propidium iodide
- DCFDA
2′,7′-dichlorofluorescin diacetate
- DAPI
4′,6-diamidino-2-phenylindole
- MS
Malate synthase
- ICLI
Isocitrate lyase
- ROS
Reactive oxygen species
- EMB
Ethambutol
- RIF
Rifampicin
- INH
Isoniazid
- STP
Streptomycin
- 2,2,-BP
2,2, Bipyridyl
- FA
Fatty acid
- GL
Glycerolipid
- GPL
Glycerophospholipid
- PK
Polykedide
- PR
Prenol
- SCL
Saccharolipide
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Contributor Information
Saif Hameed, Phone: +91-124-2337015, Email: saifhameed@yahoo.co.in.
Zeeshan Fatima, Email: drzeeshanfatima@gmail.com.
References
- Bacon J, Dover LG, Hatch KA, Zhang Y, Gomes JM, Kendall S, Wernisch L, Stoker NG, Butcher PD, Besra GS, Marsh PD. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: identification of a novel wax ester. Microbiology. 2007;153(Pt 5):1435–1444. doi: 10.1099/mic.0.2006/004317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KW. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol. 2014;12(1):63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzaida CC, Dulce MJE, Raquel AV, Muñiz-Salazar R, Laniado-Laborin R, Zenteno-Cuevas R. Mutation at embB Codon 306, a potential marker for the identification of multidrug resistance associated with ethambutol in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59(9):5455–5462. doi: 10.1128/AAC.00117-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetic. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan P, Reddy PV, Singh R, Jaisinghani N, Gandotra S, Tyagi AK. Secretory phosphatases deficient mutant of Mycobacterium tuberculosis imparts protection at the primary site of infection in guinea pigs. PLoS One. 2013;8(10):e77930. doi: 10.1371/journal.pone.0077930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christine EQ, John SB. Kinetic and chemical mechanism of malate synthase from Mycobacterium tuberculosis. Biochemistry. 2011;50(32):6879–6887. doi: 10.1021/bi2007299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christine EC, Adrienne CD, Katalin FM, Salas-Castillo SP, Ghiladi RA. Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: catalase, peroxidase, and INH-NADH adduct formation activities. Protein Sci. 2010;19(3):458–474. doi: 10.1002/pro.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick DC, Schulbach MC, Zink EE, Macchia M, Barontini S, Besra GS, Brennan PJ. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol. 2000;182(20):5771–5778. doi: 10.1128/jb.182.20.5771-5778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Wang J, Lu J, Huang X, Zheng R, Hu Z. Evaluation of methods for testing the susceptibility of clinical Mycobacterium tuberculosis isolates to pyrazinamide. J Clin Microbiol. 2013;51(5):1374–1380. doi: 10.1128/JCM.03197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, Kolattukudy PE. A novel lipase belonging to the hormonesensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J Biol Chem. 2006;281(7):3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragset MS, Poce G, Alfonso S, Padilla-Benavides T, Ioerger TR, Kaneko T, Sacchettini JC, Biava M, Parish T, Argüello JM, Steigedal M, Rubin EJ. A novel antimycobacterial compound acts as an intracellular iron chelator. Antimicrob Agents Chemother. 2015;59(4):2256–2264. doi: 10.1128/AAC.05114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbadage T, Shepherd TF, Cirillo SL, Gumienny TL, Cirillo JD. Caenorhabditis elegans p38 MAPK gene plays a key role in protection from mycobacteria. Microbiologyopen. 2016;5(3):436–452. doi: 10.1002/mbo3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaidzo IT, Moyo VM, Mvundura E, Aggrey G, Murphree NL, Khumalo H, Saungweme T, Kasvosve I, Gomo ZA, Rouault T, Boelaert JR, Gordeuk VR. Association of pulmonary tuberculosis with increased dietary iron. J Infect Dis. 2001;184(7):936–939. doi: 10.1086/323203. [DOI] [PubMed] [Google Scholar]
- Gil M, Graña M, Schopfer FJ, Wagner T, Denicola A, Freeman BA, Alzari PM, Batthyány C, Durán R. Inhibition of Mycobacterium tuberculosis ∆PknG by non-catalytic rubredoxin domain specific modification: reaction of an electrophilic nitro-fatty acid with the Fe–S center. Free Radic Biol Med. 2013;65:150–161. doi: 10.1016/j.freeradbiomed.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hae-Eun HP, Yoonji J, Lee Seung-Jae V. Survival assays using Caenorhabditis elegans. Mol Cells. 2017;40(2):90–99. doi: 10.14348/molcells.2017.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed S, Pal R, Fatima Z. Iron acquisition mechanisms: promising target against Mycobacterium tuberculosis. Open Microbiol J. 2015;9:91–97. doi: 10.2174/1874285801509010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Sharma S, Hameed S, Fatima Z. Sesamol exhibits potent antimycobacterial activity: underlying mechanisms and impact on virulence traits. J Glob Antimicrob Resist. 2017;10:228–237. doi: 10.1016/j.jgar.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Höner Zu Bentrup K. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J Bacteriol. 1999;181(23):7161–7167. doi: 10.1128/jb.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell Wescott HA, Roberts DM, Allebach CL, Kokoczka R, Parish T. Imidazoles induce reactive oxygen species in Mycobacterium tuberculosis which is not associated with cell death. ACS Omega. 2017;2(1):41–51. doi: 10.1021/acsomega.6b00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Lewinson O. Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics. 2011;3:1098–1108. doi: 10.1039/c1mt00073j. [DOI] [PubMed] [Google Scholar]
- Kochan I, Golden CA, Bukovic JA. Mechanism of tuberculostasis in mammalian serum. II. Induction of serum tuberculostasis in guinea pigs. J Bacteriol. 1969;100(1):64–70. doi: 10.1128/jb.100.1.64-70.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurthkoti K, Amin H, Marakalala MJ, Ghanny S, Subbian S, Sakatos A, Livny J, Fortune SM, Berney M, Rodriguez GM. The capacity of Mycobacterium tuberculosis to survive iron starvation might enable it to persist in iron-deprived. Microenviron Hum Granulomas MBio. 2017;8(4):e01092–e01017. doi: 10.1128/mBio.01092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan MD, Vaughan JA, Shiell BJ, Beddome GJ, Michalski WP. Mycobacterial proteome extraction: comparison of disruption methods. Proteomics. 2004;4(4):1094–1100. doi: 10.1002/pmic.200300672. [DOI] [PubMed] [Google Scholar]
- Layre E, Sweet L, Hong S, Madigan CA, Desjardins D, Young DC, Cheng TY, Annand JW, Kim K, Shamputa IC, McConnell MJ, Debono CA, Behar SM, Minnaard AJ, Murray M, Barry CE, Matsunaga I, Moody DB. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol. 2011;18(12):1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- Madigan CA, Martinot AJ, Wei JR, Madduri A, Cheng TY, Young DC, Layre E, Murry JP, Rubin EJ, Moody DB. Lipidomic analysis links mycobactin synthase K to iron uptake and virulence in M. tuberculosis. PLoS Pathog. 2015;11(3):e1004792. doi: 10.1371/journal.ppat.1004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C, Reed MB, Freeman S, Mathema B, Kreiswirth B, Barry CE, Kaplan G. Differential monocyte activation underlies strainspecific Mycobacterium tuberculosis pathogenesis. Infect Immun. 2004;72(9):5511–5514. doi: 10.1128/IAI.72.9.5511-5514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A, Sander P, Schaper KJ, Scholz M, Böttger EC. Correlation of molecular resistance mechanisms and phenotypic resistance levels in streptomycin-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1996;40(11):2452–2454. doi: 10.1128/aac.40.11.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin DE, Alshamaony L, Goodfellow M. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole-organism methanolysates. J Gen Microbiol. 1975;88(1):200–204. doi: 10.1099/00221287-88-1-200. [DOI] [PubMed] [Google Scholar]
- Murphy RC, Axelsen PH. Mass spectrometric analysis of long-chain lipids. Mass Spectrom Rev. 2011;30(4):579–599. doi: 10.1002/mas.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambi S, Long JE, Mishra BB, Baker R, Murphy KC, Olive AJ, Nguyen HP, Shaffer SA, Sassetti CM. The oxidative stress network of Mycobacterium tuberculosis reveals coordination between radical detoxification systems. Cell Host Microbe. 2015;17(6):829–837. doi: 10.1016/j.chom.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR, Jr, Hatfull GF. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69(1):164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Hameed S, Fatima Z (2015) Iron deprivation affects drug susceptibilities of mycobacteria targeting membrane integrity. J Pathog 2015:938523 [DOI] [PMC free article] [PubMed]
- Pal R, Hameed S, Sharma S, Fatima Z. Influence of iron deprivation on virulence traits of mycobacteria. Braz J Infect Dis. 2016;20(6):585–591. doi: 10.1016/j.bjid.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Hameed S, Kumar P, Singh S, Fatima Z. Comparative lipidomics of drug sensitive and resistant Mycobacterium tuberculosis reveals altered lipid imprints. 3 Biotech. 2017;7(5):325. doi: 10.1007/s13205-017-0972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Ansari MA, Saibabu V, Das S, Fatima Z, Hameed S. Nonphotodynamic roles of methylene blue: display of distinct antimycobacterial and anticandidal mode of actions. J Pathog. 2018;2018:3759704. doi: 10.1155/2018/3759704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Hameed S, Sabareesh V, Kumar P, Singh S, Fatima Z. Investigations into isoniazid treated Mycobacterium tuberculosis by electrospray mass spectrometry reveals new insights into its lipid composition. J Pathog. 2018;2018:1454316. doi: 10.1155/2018/1454316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Hameed S, Fatima Z. Altered drug efflux under iron deprivation unveils MmpL3 driven abrogated mycolic acid transport and fluidity in mycobacteria. Biometals. 2018 doi: 10.1007/s10534-018-0157-8. [DOI] [PubMed] [Google Scholar]
- Pandey R, Rodriguez GM. IdeR is required for iron homeostasis and virulence in Mycobacterium tuberculosis. Mol Microbiol. 2014;91(1):98–109. doi: 10.1111/mmi.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JM, Layre E, Sweet L, Sherrid A, Moody DB, Ojha A, Sherman DR. The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. J Bacteriol. 2012;194(3):715–721. doi: 10.1128/JB.06304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavinasasundaram KG, Colston MJ, Davis EO. Construction and complementation of a recA deletion mutant of Mycobacterium smegmatis reveals that the intein in Mycobacterium tuberculosis recA does not affect RecA function. Mol Microbiol. 1998;30(3):525–534. doi: 10.1046/j.1365-2958.1998.01083.x. [DOI] [PubMed] [Google Scholar]
- Paul KC, Chu-Yuan L, Yasu SM. In: Metabolism of plasma membrane lipids in Mycobacteria and Corynebacteria, lipid metabolism. Baez RV, editor. New York: InTech; 2013. pp. 119–148. [Google Scholar]
- Piccaro G, Pietraforte D, Giannoni F, Mustazzolu A, Fattorini L. Rifampin induces hydroxyl radical formation in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58(12):7527–7533. doi: 10.1128/AAC.03169-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol. 2012;24(1):3–9. doi: 10.1016/j.coi.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieck B, Degiacomi G, Zimmermann M, Cascioferro A, Boldrin F, Lazar-Adler NR, Bottrill AR, le Chevalier F, Frigui W, Bellinzoni M, Lisa MN, Alzari PM, Nguyen L, Brosch R, Sauer U, Manganelli R, O'Hare HM. PknG senses amino acid availability to control metabolism and virulence of Mycobacterium tuberculosis. PLoS Pathog. 2017;13(5):e1006399. doi: 10.1371/journal.ppat.1006399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross GS, Wegrzyn T, MacRae EA, Redgwell RJ. Apple beta-galactosidase. Activity against cell wall polysaccharides and characterization of a related cDNA clone. Plant Physiol. 1994;106(2):521–528. doi: 10.1104/pp.106.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabareesh V, Singh G. Mass spectrometry based lipid(ome) analyzer and molecular platform: a new software to interpret and analyze electrospray and/or matrix-assisted laser desorption/ ionization mass spectrometric data of lipids: a case study from Mycobacterium tuberculosis. J Mass Spectrom. 2013;48(4):465–477. doi: 10.1002/jms.3163. [DOI] [PubMed] [Google Scholar]
- Salimizand H, Jamehdar SA, Nik LB, Sadeghian H. Design of peptides interfering with iron-dependent regulator (IdeR) and evaluation of Mycobacterium tuberculosis growth inhibition. Iran J Basic Med Sci. 2017;20(6):722–728. doi: 10.22038/IJBMS.2017.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain MJ, Dick DL, Rithner CD, Crick DC, Belisle JT. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J Lipid Res. 2011;52(5):861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, Collins HL, Priem F, Kaufmann SH. Correction of the iron overload defect in beta-2-microglobulin knockout mice by lactoferrin abolishes their increased susceptibility to tuberculosis. J Exp Med. 2002;196(11):1507–1513. doi: 10.1084/jem.20020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafín-López J, Chacón-Salinas R, Muñoz-Cruz S, Enciso-Moreno JA, Estrada-Parra SA, Estrada-García I. The effect of iron on the expression of cytokines in macrophages infected with Mycobacterium tuberculosis. Scand J Immunol. 2004;60(4):329–337. doi: 10.1111/j.0300-9475.2004.01482.x. [DOI] [PubMed] [Google Scholar]
- Sharma S, Sharma M, Bose M. Mycobacterium tuberculosis infection of human monocyte-derived macrophages leads to apoptosis of T cells. Immunol Cell Biol. 2009;87(3):226–234. doi: 10.1038/icb.2008.97. [DOI] [PubMed] [Google Scholar]
- Shiloh MU, Champion PA. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol. 2010;13(1):86–92. doi: 10.1016/j.mib.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsimer D, Huet G, Manca C, Tsenova L, Koo MS, Kurepina N, Kana B, Mathema B, Marras SA, Kreiswirth BN, Guilhot C, Kaplan G. The phenolic glycolipid of Mycobacterium tuberculosis differentially modulates the early host cytokineresponse but does not in itself confer hypervirulence. Infect Immun. 2008;76(7):3027–3036. doi: 10.1128/IAI.01663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden RA, Barry CE. Analysis of the lipids of Mycobacterium tuberculosis. Methods Mol Med. 2001;54:229–245. doi: 10.1385/1-59259-147-7:229. [DOI] [PubMed] [Google Scholar]
- Tatano Y, Kanehiro Y, Sano C, Shimizu T, Tomioka H. ATP exhibits antimicrobial action by inhibiting bacterial utilization of ferric ions. Sci Rep. 2015;5:1–8. doi: 10.1038/srep08610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341(8846):647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- Trivedi A, Mavi PS, Bhatt D, Kumar A. Thiol reductive stress induces cellulose-anchored biofilm formation in Mycobacterium tuberculosis. Nat Commun. 2016;7:11392. doi: 10.1038/ncomms11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchèze C, Hartman T, Weinrick B, Jacobs WR., Jr Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat Commun. 2013;4:1881. doi: 10.1038/ncomms2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Global tuberculosis report. Geneva: WHO; 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 2 Excel sheet showing the data obtained from MS-LAMP in untreated cells (XLS 147 KB)
Supplementary material 3 Excel sheet showing the data obtained from MS-LAMP in 2, 2,-BP treated cells (XLS 179 KB)