Abstract
Erbin has been shown to have significant effects on the development of solid tumors. However, little is known about its function and regulatory mechanism in hematological malignancies. The biological function of Erbin on cell proliferation was measured in vitro and in vivo. The predicted target of Erbin was validated by dual-luciferase reporter assay and rescue experiment. We found that overexpression of Erbin could inhibit the cell proliferation and promote the cell differentiation of acute myeloid leukemia (AML) cells, whereas depletion of Erbin could enhance the cell proliferation and block the cell differentiation in AML cells in vitro and in vivo. Besides, miR-183-5p was identified as the upstream regulator that negatively regulated the Erbin expression. The results were confirmed by dual-luciferase reporter and RNA pull-down assay. Furthermore, we found that miR-183-5p negatively regulated Erbin, resulting in enhanced cell proliferation of AML cells via activation of RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathways. The activation of RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathways was mediated by Erbin interacting with Grb2. These results were also validated by rescue experiments in vitro and in vivo. All above-mentioned findings indicated that the miR-183-5p/Erbin signaling pathway might represent a novel prognostic biomarker or therapeutic target for treatment of AML.
Keywords: Erbin, miR-183-5p, AML, proliferation, differentiation
Regulation mechanism of Erbin in hematological malignancy especially in leukemia remains unexplored. In this issue of Molecular Therapy, Zheng et al. report that the miR-183-5p/Erbin pathway modulates proliferation and differentiation of acute myeloid leukemia cells and may be served as a novel therapeutic target.
Introduction
Acute myeloid leukemia (AML) is a common hematological malignancy with higher morbidity and mortality in adults. Currently, the progress in research of AML therapy is slow, and the long-term disease-free survival for acute promyelocytic leukemia (APL) using all-trans retinoic acid (ATRA) and arsenic trioxide is the only breakthrough during the past 20 years. Despite advances in our understanding of AML biology, there is still no fundamental progress in the treatment for non-APL patients. The conventional treatment still relies on the adjustment of dosage and frequency of standard cytotoxic drug or hematopoietic stem cell transplantation techniques. Although the complete remission rate in young AML patients can reach more than 80%, its 5-year survival rate is less than 40%. Furthermore, the condition of elderly AML patients is worse, and the overall survival rate remains low even with the use of demethylation drugs.1 Leukemia stem cells with self-renewal ability are the “culprit” of malignant proliferation of leukemia cells. These stem cells are highly proliferative, showing differentiation arrest and drug resistance, which results in a high relapse rate after complete remission. The molecular characteristic is one of the important signs of AML treatment and prognosis. The extensive use of second generation sequencing technology has enabled us to have a more in-depth and accurate understanding of the molecular biology of AML. More than 95% of AML patients carry at least one somatic mutation. Unlike solid tumors, the number of mutated genes normally carried in the AML genome is very limited (an average of 13 mutated genes in each patient).2 Moreover, there are still major limitations in the use of molecular marker genes in clinical applications. Therefore, it is urgently necessary to explore new markers, as well as their functions and mechanisms of action (MOAs), in the AML environment for the diagnosis and treatment of AML.
By using a yeast two-hybrid method, Borg et al.3 have found a new PDZ (PSD95/discs large/ZO-1) protein that functions as an interacting protein of ErbB2 in epithelial cells, and named as Erbin (also called ErbB2 interacting protein). Erbin belongs to a new family of PDZ proteins, which is called the Leucine-rich repeat and PDZ domain (LAP) family. The function of Erbin is not yet fully understood, and it is abundant in brain, heart, kidney, muscle, and stomach tissues. Some studies have shown that Erbin, as a linker protein, is involved in the interaction between the scaffold protein and signal protein of the signaling pathway, and it also participates in the formation of intracellular signal transduction networks. In addition, Erbin may play an important role in the cell proliferation and differentiation, formation of organ morphology during development, and signal transduction pathway.4 The role of Erbin in the development of some solid tumors is still controversial, which is also not well characterized in AML. In the present study, we aimed to explore the role of Erbin in the pathogenesis of AML and provide new ideas for the diagnosis and treatment of AML.
Results
Endogenous Expression of Erbin and Efficiency of Lentiviral Vector in AML Cell Lines
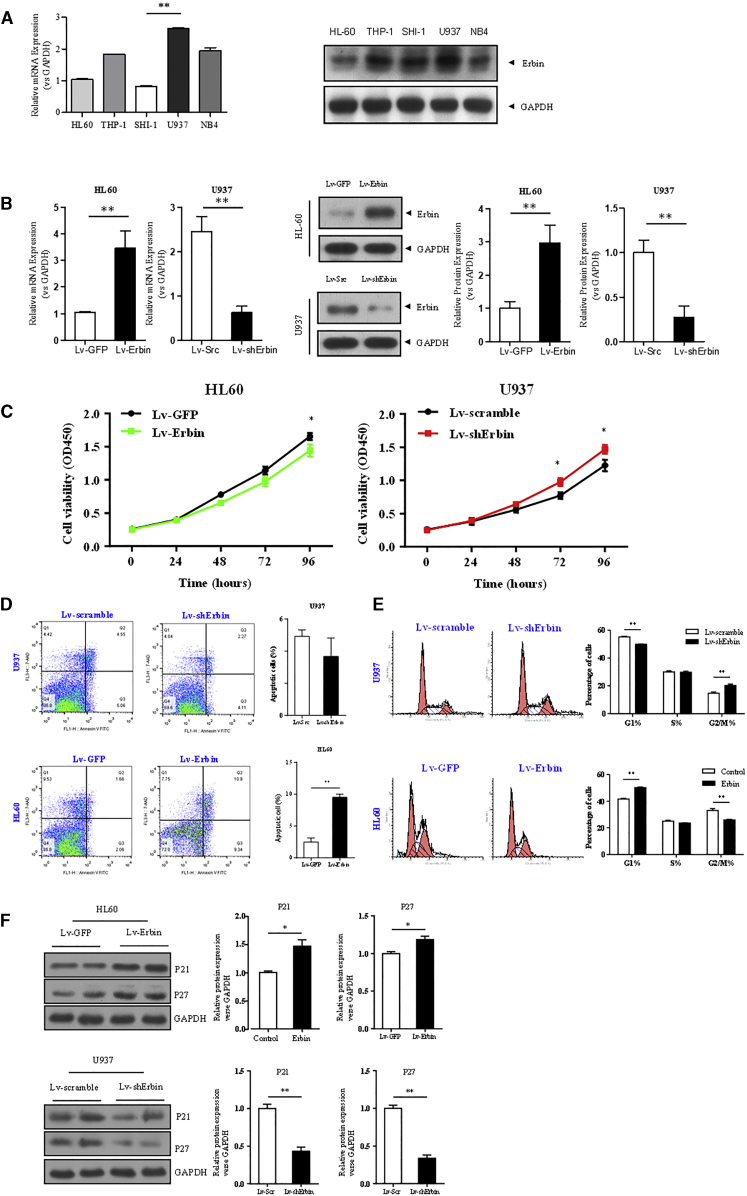
qRT-PCR and western blotting analysis showed that the expression of Erbin was relatively high in U937 cells, whereas its expression was relatively low in HL-60 and SHI-1 cells. However, the difference between HL-60 and SHI-1 cells was not statistically significant (Figure 1A). Therefore, U937 and HL-60 cell lines were selected for follow-up experiments. In addition, the expression of Erbin at the mRNA and protein levels in HL-60 cells transfected with Erbin overexpression lentiviral vector was significantly higher compared with the cells transfected with empty vector. As expected, the Erbin gene was transfected into U937 cells containing short hairpin RNA (shRNA) lentivirus (Lv). The expression of Erbin at the mRNA and protein levels in U937 cells transfected with Erbin-specific shRNA was significantly lower compared with the cells transfected with control shRNA (Figure 1B).
Figure 1.
The Impact of Erbin on Proliferation in HL-60 and U937 Cells
(A) Relative mRNA expression of Erbin was detected by qRT-PCR. Relative protein expression of Erbin was determined by western blotting analysis. (B) Efficiency of Erbin overexpression or depletion in AML cell lines. qRT-PCR and western blotting analysis were used to detect the expression of Erbin. The results were normalized to GAPDH expression and presented as relative Erbin expression. HL-60 and U937 were transfected with Erbin overexpression lentiviral vector or shRNA lentiviral vector, as well as their control vectors. (C) CCK-8 assay was used to determine cell proliferation. (D) Flow cytometry was applied to determine cell-cycle distribution. (E) Annexin V assay was used to detect cell apoptosis. (F) Expressions of p21Waf1/CIP1 and p27Kip1 were measured by western blotting analysis. **p < 0.01 versus control.
The Proliferation of AML Cells Is Regulated by Erbin
HL-60 and U937 cells were transfected with Erbin overexpression plasmid or Erbin-specific shRNA. As a control group, each cell line was also transfected with the control vector. The results of cell counting kit-8 (CCK-8) assay showed that the viability of Erbin-overexpressing HL-60 cells was moderately decreased compared with the control group. Consistently, the viability of Erbin-depleted U937 cells was measurably increased compared with the control group (Figure 1C). We further determined the cell cycle of either Erbin-overexpressing or Erbin-depleted cells, and the flow cytometry results showed that the proportion of G2/M phase cells in Erbin-overexpressing HL-60 cells was decreased compared with the cells transfected with empty vector. In contrast, the proportion of G2/M phase cells in Erbin-depleted U937 cells was increased compared with the cells transfected with empty vector (Figure 1D). Furthermore, the Annexin V assay indicated that the apoptotic rate of Erbin-overexpressing HL-60 cells was higher than that of the control group (Figure 1E). Western blotting analysis showed that the expressions of p21Waf1/CIP1 and p27Kip1 in Erbin-overexpressing HL-60 cells were upregulated compared with the control cells, whereas the expressions of p21Waf1/CIP1 and p27Kip1 in Erbin-depleted U937 cells were decreased compared with the control cells (p < 0.05) (Figure 1F). In addition, the apoptotic rate of Erbin-depleted U937 cells was lower than that of the control cells, although the difference was not statistically significant.
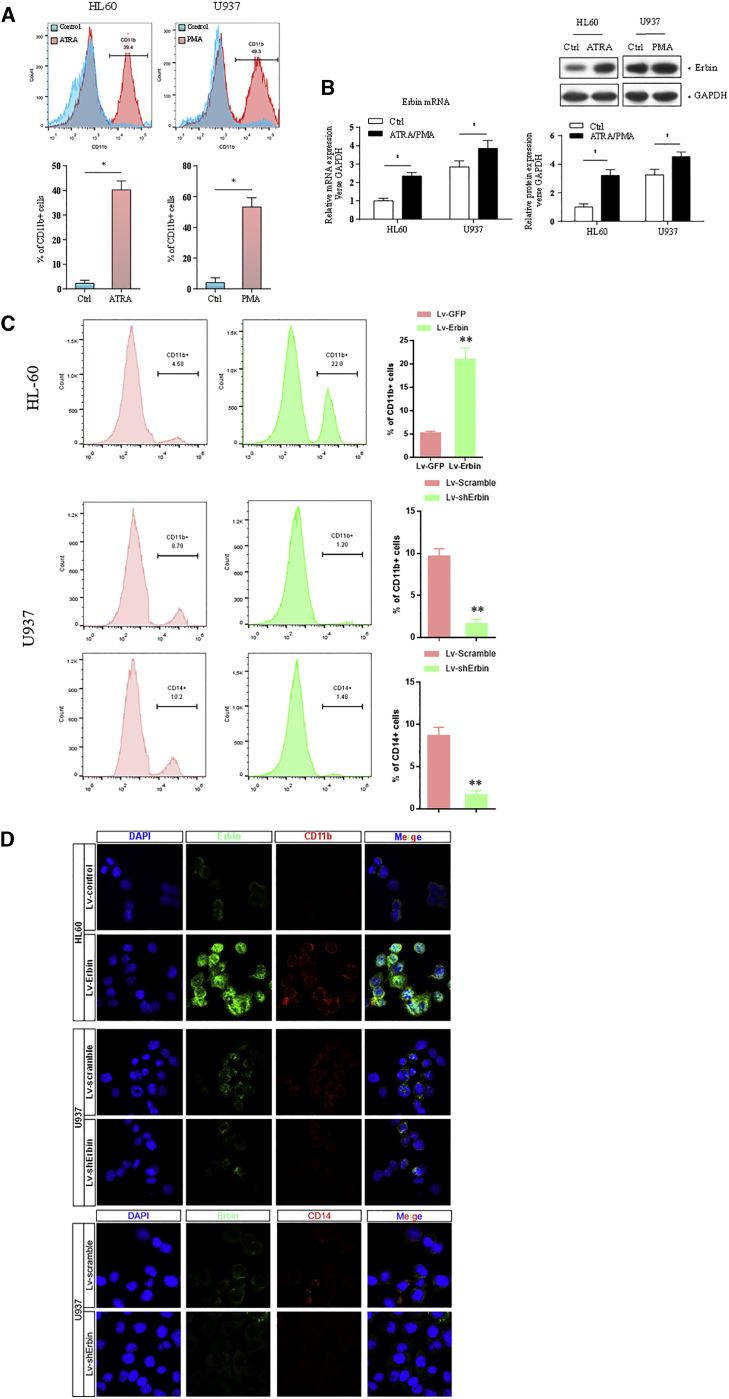
Erbin Affects the Cell Differentiation of AML Cells In Vitro
The expression of CD11b was analyzed by flow cytometry and confocal microscopy. As differentiation-inducing agents, ATRA and phorbol 12-myristate 13-acetate (PMA) could significantly induce the differentiation of HL-60 and U937 cells, leading to the upregulation of CD11b. In addition, the expression of Erbin at the mRNA and protein levels in HL-60 and U937 cells was also significantly increased compared with the untreated cells (Figures 2A and 2B). Moreover, the positive rate of CD11b in Erbin-overexpressing HL-60 cells was increased compared with that in the control cells, and the difference between Erbin-overexpressing HL-60 cells and control cells was statistically significant (p < 0.01), indicating that the cell differentiation was promoted. In contrast, the CD11b and CD14 expression in Erbin-depleted U937 cells were decreased compared with that in control cells, and such differences were statistically significant (p < 0.05), indicating that the differentiation was suppressed (Figure 2C). Immunofluorescence (IF) analysis revealed that CD11b and CD14 were localized in the membrane of cells. The expression of CD11b was stronger in Erbin-overexpressing HL-60 cells compared with the control cells, whereas such expression was weaker in Erbin-depleted U937 cells compared with the control cells, as well as the expression of CD14 (Figure 2D).
Figure 2.
The Impact of Erbin on Differentiation in HL-60 and U937 Cells
(A) The expression of CD11b in HL-60 and U937 cells was analyzed by flow cytometry after applying differentiation-inducing agents ATRA and PMA. (B) qRT-PCR and western blotting analysis were performed to determine the expression of Erbin after treating with ATRA and PMA. (C) The expression of CD11b or CD14 was analyzed by flow cytometry after transfection with the Erbin overexpression lentiviral vector or shRNA lentiviral vector in HL-60 cells or U937 cells. (D) IF staining of Erbin and CD11b or CD14 in HL-60 and U937 cells (original magnification ×400).
Erbin Affects the Proliferation and Differentiation of AML Cells In Vivo
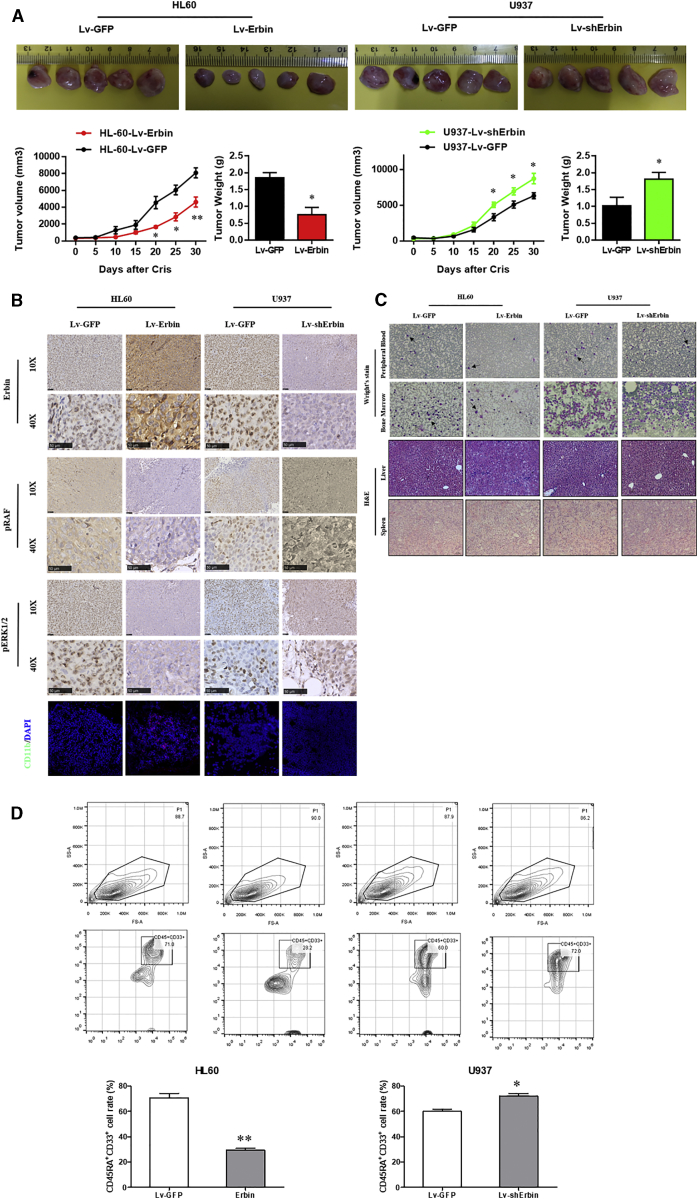
The tumor volume and mass in nude mice subcutaneously injected with Erbin-overexpressing HL-60 cells were significantly increased compared with the nude mice injected with control cells. In contrast, the tumor volume and mass in nude mice subcutaneously injected with Erbin-depleted U937 cells were significantly decreased compared with the nude mice injected with control cells (Figure 3A). The IHC analysis showed that the Erbin expression in the Erbin overexpression group was significantly higher than that in the control group, and the phosphorylation levels of RAS and ERK1/2 in tumor tissues of nude mice injected with Erbin-overexpressing HL-60 cells were significantly lower than those in the control group, suggesting that this pathway was less activated. In addition, multiple red fluorescent protein (RFP)-labeled CD11b+ cells were observed in tumors of nude mice injected with Erbin-overexpressing HL-60 cells, whereas such CD11b+ cells were not observed in the control mice, suggesting that the cell differentiation in tumor was increased in nude mice upon injection of Erbin-overexpressing HL-60 cells. The IHC analysis showed that the expression of Erbin in tumor tissue of nude mice injected with Erbin-depleted U937 cells was decreased compared with the control mice, whereas the phosphorylation levels of RAF and ERK1/2 from nude mice injected with Erbin-depleted U937 cells were significantly increased compared with the control mice, suggesting that the activation of such pathway was enhanced. In addition, the RFP-labeled CD11b+ cells were not observed in tumors of nude mice with downregulated Erbin expression, whereas the RFP-labeled CD11b+ cells were observed in tumors of control nude mice, suggesting that the cell differentiation in tumor tissues of nude mice with downregulated Erbin expression was suppressed (Figure 3B). These differences were statistically significant (p < 0.05).
Figure 3.
Impact of Erbin on Tumor Growth In Vivo
(A) Tumors were removed and collected from nude mice injected with Erbin-overexpressing or Erbin-depleted AML cells. The tumor volume was analyzed every 5 days. The tumor weight was measured 30 days after tumor transplantation. *p < 0.05 compared with the NC group. (B) IHC detection of Erbin, pRAF, and pERK1/2 in paraffin-embedded tissue sections and IF staining of CD11b in AML cells. (C) Wright’s staining in peripheral blood and bone marrow, as well as H&E staining in liver and spleen. (D) CD45RA+CD33+ medullary cells were counted by flow cytometry.
Peripheral blood and bone marrow smears prepared from nude mice injected with HL-60 cells were stained with eosin methylene blue. Microscopic examination revealed that the amount of peripheral blood and bone marrow blast cells in nude mice injected with Erbin-overexpressing HL-60 cells was significantly decreased compared with the control group. H&E staining was performed for liver and spleen tissues. Light microscopy revealed that there was no significant difference in the infiltration of blast cells from different groups. Peripheral blood and bone marrow smears from nude mice injected with U937 cells were stained with eosin methylene blue. Light microscopic examination revealed that the amount of peripheral blood blast cells in nude mice injected with Erbin-depleted U937 cells was significantly increased compared with the control group, whereas the amount of bone marrow blast cells was slightly increased compared with the control group. However, there was no significant difference in the infiltration of blast cells in the liver and spleen tissues (Figure 3C). In addition, flow cytometry showed that the proportion of CD45RA+CD33+ bone marrow cells from nude mice injected with Erbin-overexpressing HL-60 cells was significantly lower than that of the control group, suggesting that Erbin-overexpressing HL-60 cells had lower tumorigenicity compared with the normal HL-60 cells. Consistently, the proportion of CD45RA+CD33+ bone marrow cells from nude mice injected with Erbin-depleted U937 cells was significantly higher than that of the control group, suggesting that Erbin-depleted U937 cells had an increased tumorigenicity compared with the normal U937 cells (Figure 3D). These differences were statistically significant (p < 0.05).
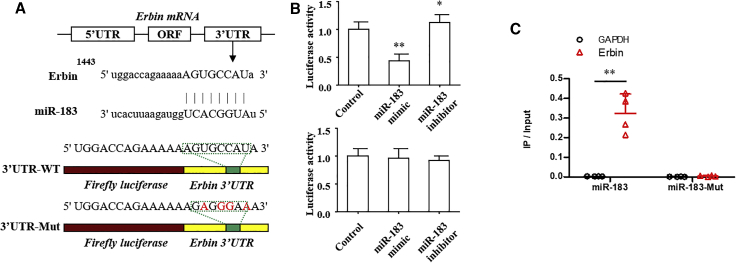
miR-183-5p Targets the 3′ UTR of Erbin and Regulates Its Expression
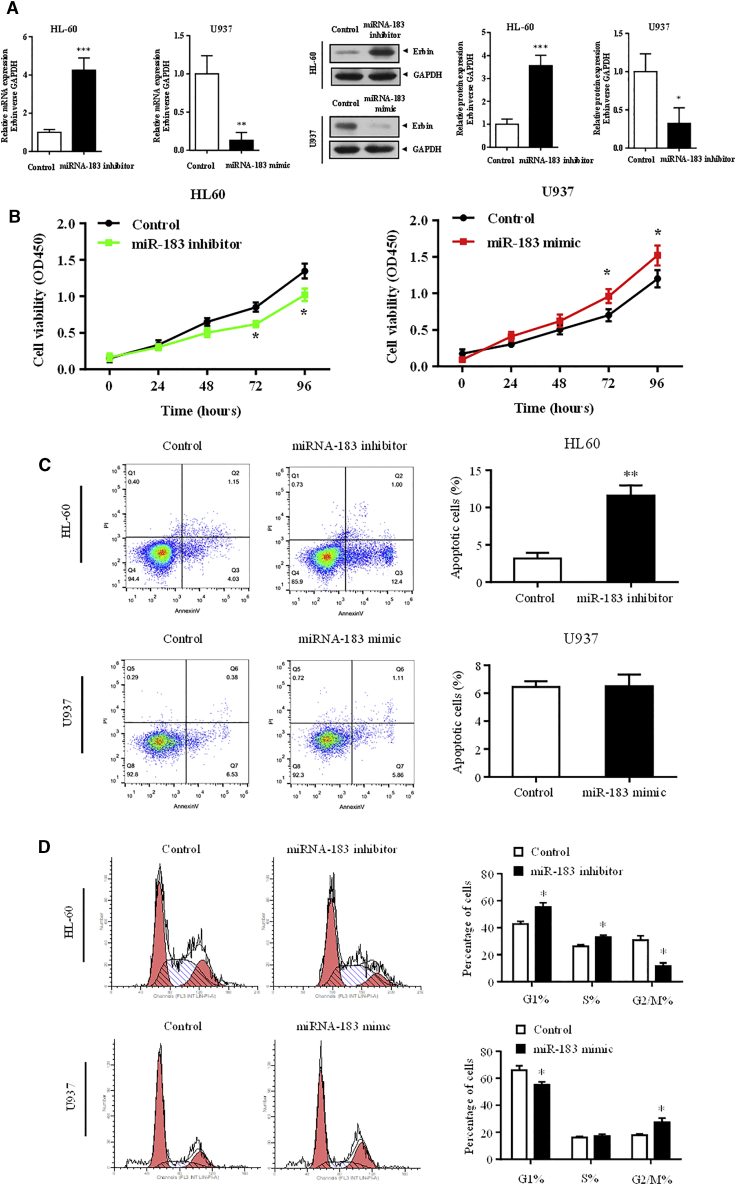
To further explore the upstream regulatory mechanism of Erbin, the possible miRNAs targeting Erbin were predicted and screened based on the bioinformatics database. We found that miR-183-5p might interact with the 3′ UTR of Erbin (Figure 4A). The Dual-Luciferase Reporter Assay System was employed to further confirm this finding. Data showed that miR-183 mimic could significantly reduce the luciferase activity of the luciferase reporter gene containing wild-type Erbin 3′ UTR, whereas the miR-183 inhibitor promoted the luciferase activity. Furthermore, miR-183 had no significant effect on the luciferase activity of the luciferase reporter gene containing the mutated Erbin 3′ UTR (Figure 4B). The results of RNA pull-down assay manifested that Erbin was more enriched in the wild-type miR-183-5p compared with that in the mutant-type miR-183-5p with broken Ebin binding site (Figure 4C). In addition, qRT-PCR and western blotting analysis indicated that miR-183-5p could negatively regulate the expression of Erbin at mRNA and protein levels (Figure 5A), and the differences from the above-mentioned results were statistically significant (p < 0.05).
Figure 4.
miR-183-5p Targets the 3′ UTR of Erbin
(A) A schematic diagram showed wild-type and mutant of miR-183-5p binding sites in the 3′ UTR of Erbin. (B) The Dual-Luciferase Reporter Assay System was used in U937 cells. Luciferase activity was measured at 48 h after transfection. (C) The biotinylated miR-183-5p or its mutant (miR-183-mut) was transfected into U937 cells. qRT-PCR was applied to quantify the RNA levels of Erbin and GAPDH. Scatterplot showed the relative ratios of the input of IP. **p < 0.01.
Figure 5.
miR-183-5p Negatively Regulates Erbin in HL-60 and U937 Cells
(A) miR-183-5p negatively regulates the expression of Erbin at the mRNA and protein levels. HL-60 cells were transfected with miR-183 inhibitor, and U937 cells were transfected with miR-330 mimic. qRT-PCR and western blotting analysis were used to determine the expression of Erbin at the mRNA and protein levels, respectively. HL-60 and U937 were transfected with miR-183 inhibitor, miR-183 mimic, or empty control. (B) CCK-8 assay was used to measure cell proliferation. (C) Flow cytometry was applied to determine cell-cycle distribution. (D) Annexin V assay was used to detect cell apoptosis. *p < 0.05 versus control.
miR-183-5p Regulates the Cell Proliferation of AML
CCK-8 assay showed that the proliferation of HL-60 cells transfected with miR-183 inhibitor was lower than that of the control cells, and the proliferation of U937 cells transfected with miR-183 mimic was higher than that of the control group (Figure 5B). The flow cytometry showed that the proportion of G2/M phase cells was decreased in HL-60 cells transfected with miR-183 inhibitor compared with the control cells, whereas the proportion of G2/M phase cells was increased in U937 cells transfected with miR-183 mimic compared with the control cells (Figure 5C). Annexin V staining showed that the apoptotic rate of HL-60 cells transfected with miR-183 inhibitor was higher than that of the control group (Figure 5D). The differences from the above-mentioned results were statistically significant (p < 0.05). There was no significant difference between the U937 cells transfected with miR-183 mimic and control cells.
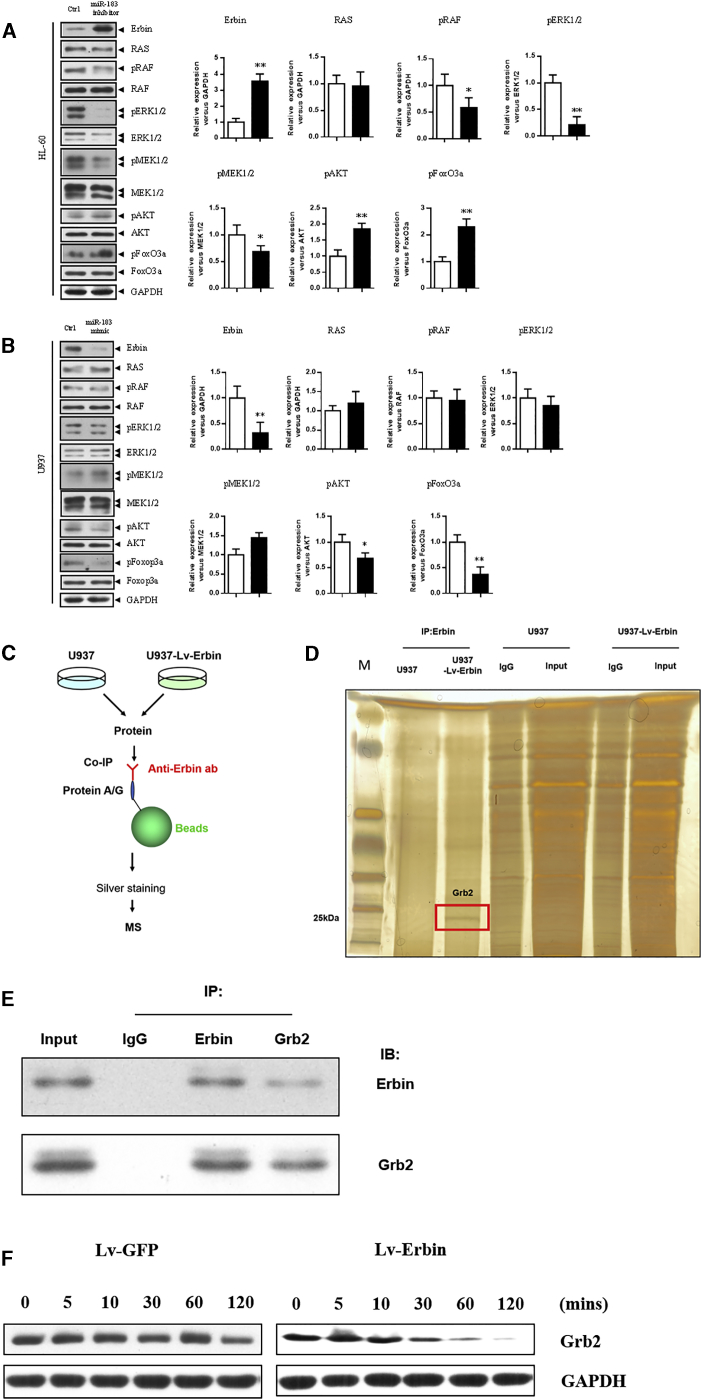
Western blotting analysis showed that the expression of Erbin at the protein level in HL-60 cells transfected with miR-183 inhibitor was higher than that in the control cells. Moreover, the phosphorylation level of the RAS/RAF/MEK/ERK pathway in HL-60 cells transfected with miR-183 inhibitor was significantly decreased, whereas the phosphorylation level of the PI3K/AKT/FoxO3a pathway was significantly increased compared with the control cells. In addition, the expression of Erbin at the protein level in U937 cells transfected with miR-183 mimic was lower than that in the control cells, and the phosphorylation level of the PI3K/AKT/FoxO3a pathway was also significantly decreased compared with the control cells. All of these differences were statistically significant (p < 0.05). However, the phosphorylation level of the RAS/RAF/MEK/ERK pathway in U937 cells transfected with miR-183 mimic was not significantly changed (Figures 6A and 6B).
Figure 6.
Regulatory Mechanism of Erbin in HL-60 and U937 Cells
Western blotting analysis was used to measure the phosphorylation levels of the RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathway, as well as the expression of Erbin in HL-60 (A) and U937 (B) cells after transfection with miR-183 inhibitor and miR-183 mimic. (C) Flow diagram of co-IP and mass spectrum assays was shown as a schematic diagram. (D) Co-immunoprecipitation was performed to identify potential interactive candidate of Erbin in U937 and U937-Lv-Erbin (transfected with Lv-Erbin) cells. The enriched products were eluted and separated by SDS-PAGE and silver staining. The differential band appearing in U937-Lv-Erbin lane was analyzed by mass spectrum. (E) Co-immunoprecipitation (IP) showed Erbin complexed with Grb2. (F) Western blotting analysis showed the expression of Erbin and Grb2 in U937 cells that transfected with Lv-Erbin and control vector. *p < 0.05.
To further explore the mechanism that Erbin affect RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathway, we performed co-immunoprecipitation (co-IP) and mass spectrum to identify potential interactive proteins in U937 cells. Grb2, a well-known upstream regulatory protein of the RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathway, was identified as a potential candidate (Figures 6C and 6D). Then we confirmed the interaction between Erbin and Grb2 by co-IP assays (Figure 6E). The result of western blotting assay revealed that the degradation of Grb2 protein could be obviously accelerated when Erbin was exogenously overexpressed in U937 cells (Figure 6F). These results indicated that Erbin may modulate the RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathway by directly affecting activity of Grb2.
The Rescue Experiment of the miR-183-5p/Erbin Pathway In Vitro and In Vivo
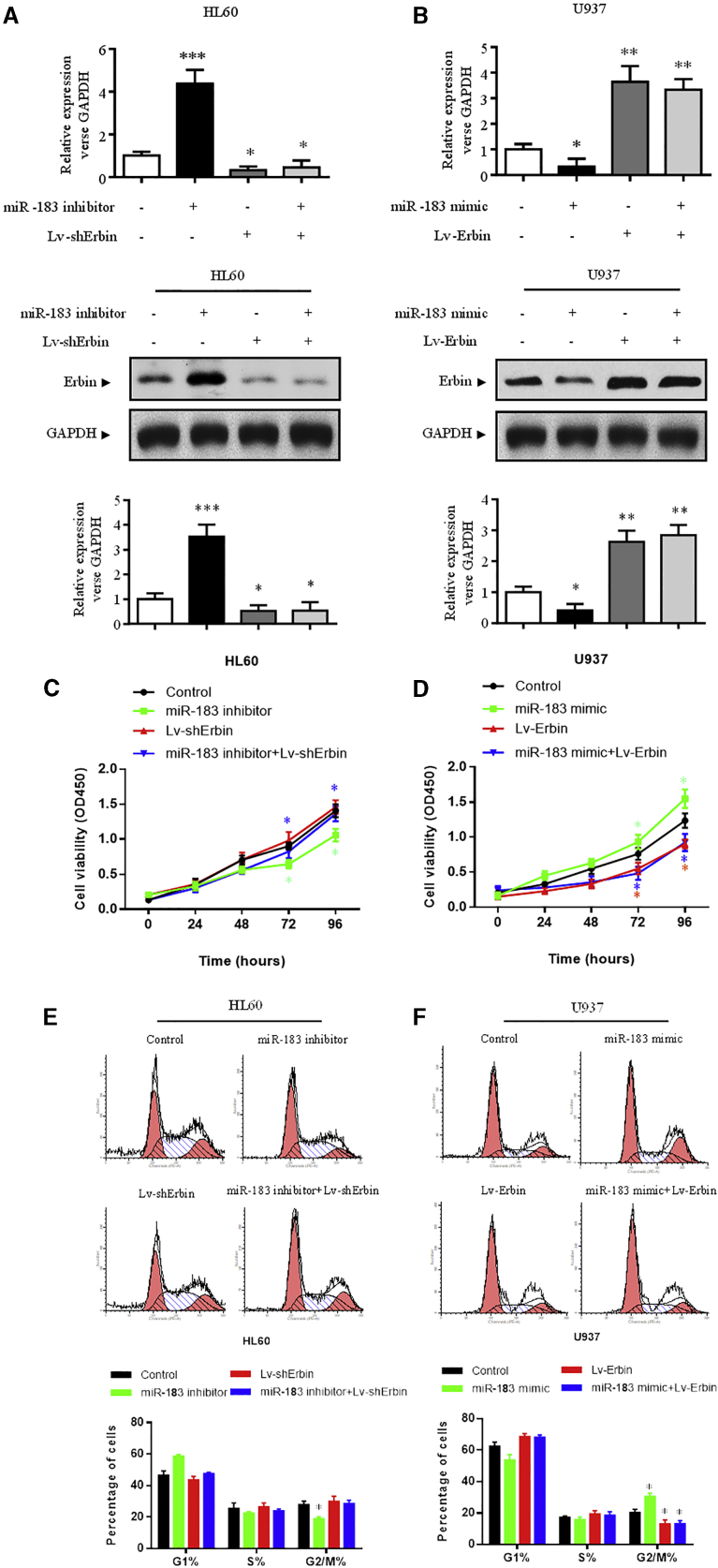
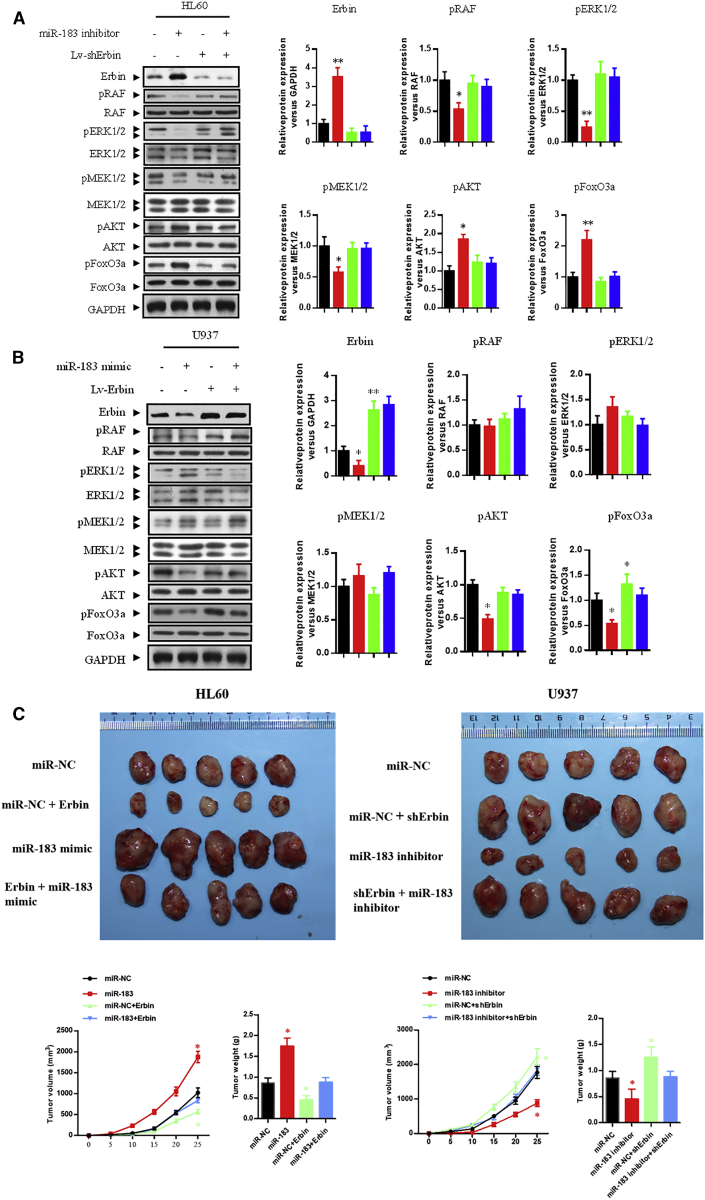
In HL-60 cells, interference of Erbin expression could significantly abolish miR-183-5p inhibitor-mediated elevation of Erbin expression (Figure 7A), inhibition of cell proliferation (Figures 7C and 7E), decrease of the RAS/RAF/MEK/ERK phosphorylation, and increase of the PI3K/AKT/FoxO3a phosphorylation (Figure 8A) (p < 0.05). In U937 cells, overexpression of Erbin could abrogate miR-183-5p mimic-mediated Erbin expression (Figure 7B), promotion of cellular proliferation (Figures 7D and 7F), and decrease of PI3K/AKT/FoxO3a phosphorylation (Figure 8B) (p < 0.05). Because miR-183 mimic showed little effect on the RAS/RAF/MEK/ERK phosphorylation in U937 cells, overexpression of Erbin had less significant effect on the RAS/RAF/MEK/ERK phosphorylation (Figure 8B).
Figure 7.
Rescue Experiment with miR-183-5p/Erbin in HL-60 and U937 Cells
The expression of Erbin at the mRNA (A) and protein (B) levels was measured by qRT-PCR and western blotting analysis, respectively. The viability (C, HL-60; D, U937) and cell-cycle distribution (E, HL-60; F, U937) were measured by CCK-8 assay and flow cytometry, respectively. These experiments were conducted after transfection with control vector, miR-183 mimic or miR-183 inhibitor, Lv-Erbin or Lv-shErbin, and miR-183 mimic+Lv-Erbin or miR-183 inhibitor+Lv-shErbin. *p < 0.05 versus control.
Figure 8.
Rescue Experiment with miR-183-5p/Erbin
The expression of Erbin and the phosphorylation levels of RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathways were detected by western blotting analysis in HL-60 (A) and U937 (B) cells after transfection with control vector, miR-183 mimic or miR-183 inhibitor, Lv-Erbin or Lv-shErbin, and miR-183 mimic+Lv-Erbin or miR-183 inhibitor+Lv-shErbin. *p < 0.05 versus control. (C) Tumors were collected from nude mice injected with AML cells transfected with miR-NC, miR-183 mimic or miR-183 inhibitor, Lv-Erbin+miR-NC or Lv-shErbin+miR-NC, and miR-183 mimic+Lv-Erbin or miR-183 inhibitor+Lv-shErbin. The tumor volume was analyzed every 5 days. The tumor weight was measured 30 days after tumor transplantation. *p < 0.05 compared with the NC group.
In nude mouse xenografts, the tumor volume and mass could be significantly affected by modulation of miR-183-5p in AML cells, which subcutaneously injected to nude mice (p < 0.05). The differences of tumor volume and mass in nude mice between the co-transfecting HL-60 group (Lv-Erbin + miR-183-5p mimic) and control HL-60 group were not statistically significant. Similarly, the differences between the co-transfecting U937 group (Lv-shErbin + miR-183-5p inhibitor) and control U937 group were also not statistically significant (Figure 8C). All of these results suggested that the Erbin/miR-183-5p pathway could eventually modulate the proliferation in AML cells.
The Expressions of Erbin and miR-183-5p in Clinical Samples
To further validate the in vitro results, seven bone marrow samples and 30 peripheral blood samples were collected from initially diagnosed AML patients without treatment. Clinical characteristics were summarized in Table S1. In addition, 10 peripheral blood samples from healthy individuals were used as controls. qRT-PCR showed that the expression of Erbin at the mRNA level in the peripheral blood of healthy individuals was significantly lower than that in the bone marrow and peripheral blood of AML patients (Figure S1A). The expressions of pre-miR-183-5p and mature-miR-183-5p at the mRNA level in peripheral blood and bone marrow from AML patients were significantly higher than those in the healthy individuals (Figures S1B and S1C), suggesting that the increased expression of miR-183-5p in bone marrow and peripheral blood of AML patients could be attributed to the increased synthesis at the transcription level, instead of reduced degradation. These differences were statistically significant (p < 0.05). Additionally, the expression of Erbin was not correlated with the expression of pre-miR-183-5p or mature-miR-183-5p in clinical samples, which might mainly attribute to small sample size (Figure S1D).
The Expression of Erbin and miR-183-5p in Different Hematopoietic Cells
Different hematopoietic cells including granulocytes, monocytes, erythrocytes, megakaryocytes, T lymphocytes, B lymphocytes, and NK cells were collected from healthy people by flow cytometer. qRT-PCR was used to measure the level of Erbin and miR-183-5p. The result indicated that the expression of Erbin was comparatively high in granulocytes, monocytes, erythrocytes, and megakaryocytes and comparatively low in T lymphocytes, B lymphocytes, and NK cells. The difference among granulocytes, monocytes, erythrocytes, and megakaryocytes was not statistically significant. Also, the difference among T lymphocytes, B lymphocytes, and NK cells was not statistically significant. The expression of miR-183-5p in hematopoietic cells was fairly low. The difference among these cells was not statistically significant (Figure S2).
Discussion
The function of Erbin in tumors has not been fully elucidated. It has been reported that Erbin expression in human breast cancer tissue is significantly reduced, and the inhibition of Erbin expression in breast cancer cells can trigger the resistance to the targeted therapeutic drug Herceptin (trastuzumab), promoting the invasion and metastasis of cancer cells.5 However, some other studies have shown that Erbin expression in ErbB2-positive breast cancer cells is increased.6 Inhibition of Erbin expression can enhance the sensitivity of MCF-7 cancer cells to tumor necrosis factor-related apoptosis-inducing ligand TNF superfamily member 10 (TRAIL), thereby promoting cell apoptosis.7 Hu et al.8 have demonstrated that the reduced or absent expression of Erbin can induce tumor cell proliferation, drug resistance, and migration in cervical cancer, suggesting that the underlying mechanism may be related to the absence of Erbin-mediated incapability of inhibiting STAT3 signaling, which results in the enhanced resistance of cervical cancer cells to apoptosis, thereby promoting tumor development and metastasis. In the study of rectal cancer, Erbin expression is upregulated, the expression level of Erbin is associated with the stage and progression of the tumor, and it can regulate the expression of epidermal growth factor receptor (EGFR),9 thereby exhibiting the characteristics of oncogenes. Additionally, some studies have shown that Erbin reduces the expression of the tumor suppressor gene Melin by inhibiting the activity of protein kinase PAK2 and indirectly participates in the development of neurofibroma.10
Current studies suggest that Erbin is associated with the development of multiple tumors. However, its function and mechanism in hematological malignancies still remain largely unexplored. Hematological malignancies and solid tumors have distinct characteristics in terms of pathological types, clinical features, and prognosis. AML is the most common type of leukemia in adults. This study found that Erbin expression negatively regulated the proliferation and differentiation of AML cells, and such ability of Erbin might be related to the activation of tumor suppressor genes p21Waf1/CIP1 and p27Kip1. In the tumor-bearing mouse model, the tumor volume and mass of nude mice injected with Erbin-overexpressing cells were lower than those of mice injected with control cells. Moreover, Erbin might inhibit the proliferation of AML cells in vivo by decreasing the RAS/RAF/mitogen-activated protein kinase (MAPK) phosphorylation and decrease the tumorigenicity of AML cells by promoting the differentiation of naive cells.
To further elucidate the regulatory mechanism of Erbin in AML, a bioinformatics method was employed to predict and screen the possible targets of Erbin, and miR-183-5p was found to bind directly to the 3′ UTR of Erbin. The function of miR-183 in tumors has been intensively reported; it often shows high expression in tumors, accompanied by allelic imbalance, chromosome breakage, and increased copy number,11, 12, 13, 14, 15, 16, 17 and most of these phenotypes are manifested as carcinogenic. Some phenotypes also show tumor suppressor activity, suggesting that its tumorigenicity may be dependent on the environment and cell type.18 Moreover, ectopic overexpression of miR-183 in medulloblastoma cells cultured in Ptch1+/−/PtenFloxp−/+ transgenic mice exhibits enhanced proliferative capacity.19 In addition, miR-183 is highly expressed in colorectal cancer and hepatocellular carcinoma, and its tumorigenicity is mainly achieved by suppressing tumor suppressor mRNA.20, 21, 22, 23, 24 However, overexpression of miR-183 inhibits the invasion and migration of 801D lung cancer cells and regulates the metastasis-related genes.25 The ectopic expression of miR-183 can inhibit the migration, invasion, and activity of ovarian cancer cells in vitro.26 In normal breast tissue, estrogen therapy increases miR-183 levels.27 Paradoxically, miR-183 expression is highest in estrogen receptor (ER)-negative breast cancer.28 Similarly, overexpression of miR-183 in breast cancer cell line T47D, which already possesses a high expression level of miR-183, inhibits the cell migration,29 displaying a similar effect as high-level expression of miR-183 in lung cancer cells. Moreover, miR-183 often shows differences among different tumor cells, which may be attributed to different transcriptional environments. Our study further showed that miR-183-5p regulated the expression of Erbin at the mRNA and protein levels in AML cells. Meanwhile, miR-183-5p could also regulate the proliferative capacity of AML cells. The luciferase reporter assay and RNA pull-down assay confirmed the direct binding of miR-183-5p to Erbin. In-depth study found that the miR-183-5p/Erbin pathway played a role in regulating cell proliferation and differentiation by mediating the phosphorylation levels of RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathways. The modulation on phosphorylation of RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathways was not direct but secondary and was mediated by Grb2, which was confirmed by co-IP, mass spectrometry, and western blotting assays. Grb2 was a famous upstream regulator of such pathways that was previously proven by several studies.30, 31, 32 Furthermore, a rescue experiment was carried out to prove the reliability of the miR-183-5p/Erbin pathway. The results showed that negative regulation of Erbin expression in AML cells could restore the proliferation of AML cells affected by miR-183-5p, suggesting that miR-183-5p, as a regulatory factor upstream of Erbin, could directly bind to and inhibit Erbin expression and modulate the phosphorylation levels of RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathways, thereby being involved in regulation of cellular proliferation and differentiation, and playing an important role in occurrence and development of AML. Rescue experiment in vivo further validated these conclusions. Moreover, we also showed that the expression of the miR-183-5p/Erbin pathway was significantly different between AML patients and healthy individuals, and such difference could be attributed to the increased expression of miR-183-5p at the transcription level. For healthy individuals, hematopoietic cells of lymphatic origin expressed less Erbin than hematopoietic cells of myelocytic, erythrocytic, and megakaryocytic origin. All hematopoietic cells expressed only very little miR-183-5p. The functional phenotype of Erbin in AML was effective against the characteristics of high proliferation, low apoptosis, and differentiation arrest of AML cells, making it not only a marker for the development, efficacy, and prognosis of AML but also a potential therapeutic target. The finds in AML were different from those in several solid tumors, which reflected functional diversity of Erbin in cancers.
In the present study, we also found that downregulation of Erbin in U937 cells did not significantly inhibit the apoptosis, which could possibly be explained by that AML cells generally had anti-apoptotic properties. Therefore, downregulation of Erbin exerted only a limited impact on the further decrease of apoptotic rate. Collectively, it was not reasonable to make a conclusion that Erbin could not affect the apoptosis of AML cells. In addition, the phosphorylation level of the RAS/RAF/MEK/ERK pathway in U937 cells was not significantly altered, whereas the PI3K/AKT/FoxO3a pathway had normal feedback after the expression of Erbin was decreased by miR-183-5p, which might be attributed to several reasons. The first reason is that the increased cell proliferation caused by downregulation of Erbin in U937 cells was not mediated by the phosphorylation of the RAS/RAF/MEK/ERK pathway, although it was mediated by PI3K/AKT/FoxO3 and other signaling pathways. The second reason is that the U937 cell line had high proliferative and low apoptotic characteristics; thus, there was an upper limit or a bottleneck for the proliferative capacity even when the Erbin expression was downregulated. Moreover, there might be other negative feedback mechanisms, such as the presence of a relevant threshold, which could trigger a negative feedback mechanism counteracting the phosphorylation of the RAS/RAF/MEK/ERK pathway to achieve negative regulatory function. The above-mentioned hypothesis needs to be further determined. Actually, we also observed that further downregulation of Erbin in HL-60 cells did not result in a significant increase in proliferative capacity, which might be attributed to the low level of endogenous Erbin in HL-60 cells, as well as its weak ability to regulate cell proliferation. In the tumor model, cell infiltration of AML was not detected in the liver and spleen. However, given the fact that clinical AML patients possessed a low proportion of extramedullary invasiveness, we believed that the large difference in invasiveness between AML cells might be the main cause of this finding.
It was possible that Erbin-targeted therapy might benefit AML patients. Various types of differentiation-inducing agents, such as ATRA, have been widely used in the treatment of APL and myelodysplastic syndromes (MDSs), showing satisfactory efficacy.33, 34, 35 However, there are still some limitations, such as how to maintain an effective blood concentration of oral drugs at low doses with low toxicity and side effects, how to accurately localize intravenous drugs to the bone marrow, and the presence of ATRA-insensitive fusion gene mutations, such as PLZF-RARα.36, 37, 38, 39 Gene-level intervention allows Erbin to directly affect the differentiation of AML cells, and it has an advantage over conventional drugs in terms of its MOA. In combination with gene-editing technology, Erbin could possibly be used as a good therapeutic target.
In the present study, we described the mechanism by which the miR-183-5p/Erbin pathway affected the proliferation and differentiation of AML cells. Our findings enriched the pathogenesis of AML and provided a theoretical basis for the study of disease progression and treatment. However, several questions still remain unanswered. For example, it is necessary to explore the downstream coding proteins of Erbin and the regulatory long noncoding RNAs (lncRNAs), circ-RNAs, or transcription factors existing upstream of miR-183-5p. In addition, the clinical efficiency of Erbin-targeted therapy needs to be further verified by large-sample clinical trials. It is foreseeable that the miR-183-5p/Erbin pathway had good potential in the treatment of hematological malignancies, and Erbin-targeted treatment might improve the therapeutic scheme for AML patients.
Materials and Methods
Cell Culture
Human leukemia cell lines, THP-1 (monocytic leukemia), HL-60 (promyelocytic leukemia), and U937 (monocytic leukemia/lymphoma), were obtained from the Chinese Academy of Sciences, Shanghai Institutes for Biological Sciences (Shanghai, China). Human leukemia cell lines, NB-4 (promyelocytic leukemia) and SHI-1 (monocytic leukemia), were kindly provided by Prof. Suning Chen (Blood Research Institute of Soochow University). All tested cells were maintained in RPMI-1640 medium supplemented with 10% fetal calf serum (HyClone Laboratories, Logan, UT, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Lentiviral Transduction and Generation of Stable Cell Lines
Human Erbin Lv (Lv-Erbin), shRNA Lv targeting Erbin (Lv-shErbin), negative control (Lv-GFP), and scrambled shRNA Lv (Lv-scramble) were constructed by Genelily (Shanghai, China). EGFP and puromycin were labeled to all lentiviruses. The target sequences of Erbin were 5′-GGAAGTGCCTGAAGTACTTGA-3′ for shErbin and 5′-TTCTCCCCGAACAACAACGTG-3′ for shRNA-NC. After 2 days of lentiviral transduction, stable cell lines were selected with 2 μg/mL puromycin for at least 2 weeks. The overexpression or depletion efficiency of cells was determined by qRT-PCR and western blotting analysis.
Transfection
Transient transfection was respectively conducted with 50 nm miRNA-183 mimic, miRNA-183 inhibitor, and miRNA-NC using Lipofectamine 2000 (Invitrogen). miRNA-183 mimic, miRNA-183 inhibitor, and miRNA-NC were generated by GenePharma (GenePharma Corporation, Shanghai, China).
Cell Viability Assay
Cell growth was monitored by CCK-8 assay. In brief, cells (5 × 103 per well) were incubated in 96-well plates for 0, 24, 48, or 72 h. Subsequently, CCK-8 solution (Dojindo, Japan) was added to each well, followed by 4-h incubation. Cell viability was detected at a wavelength of 450 nm. All experiments were performed in triplicate.
qRT-PCR
Total RNA was extracted from exponentially growing cells by TRIzol reagent (Invitrogen, USA). Purified RNA was reverse transcribed into cDNA using M-MLV (Promega, USA). qRT-PCR was performed on an Applied Biosystems vii7 system using SYBR qPCR mix and has-miRNA-183 qPCR kit (Thermo Fisher). The primers were synthesized as follows: Erbin, 5′-ATCTCACCAAACGACCGACT-3′ and 5′-TCCTGGCATCATTGGAGGAG-3′; GAPDH, 5′-CACCATCTTCCAGGAGCGAG-3′ and 5′-AAATGAGCCCCAGCCTTCTC-3′. The relative expression of the target gene was calculated using the 2−ΔΔCt method. All experiments were performed in triplicate.
Western Blotting Analysis
Cells of logarithmic growth phase were collected. Total proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer; then protein contents were quantified. Equal amounts of proteins were subjected to SDS-PAGE. Western blotting analysis was performed according to a previously established method. Antibodies against p21, p27, phospho-ERK1/2, ERK1/2, phospho-MEK1/2, MEK1/2, Akt, phospho-Akt, FOXO3a, phospho-FOXO3a, RAS, and phospho-RAS were purchased from Cell Signaling Technology (USA), whereas antibodies against Erbin, Grb2, and GAPDH were obtained from Abcam (Cambridge, MA, USA). GAPDH was used as a protein loading control.
Differentiation Assay
Flow cytometric analysis was employed to assess the differentiation of AML cells. Phycoerythrin (PE-anti CD11b; BD) was conjugated to CD11b antibody. Cells (20 μL of antibody/106 cells) were washed and incubated with conjugated antibodies at 4°C for 1 h. Subsequently, cells were washed and analyzed by FACSCalibur flow cytometer (BD).
Cell-Cycle Analysis
Cells were fixed in 70% ice-cold ethanol at 4°C overnight and then stained in a mixed solution (200 μg/mL PI, 0.1% sodium azide, 0.1% Triton X-100, and 10 μg/mL RNase) in the dark at 23°C for 4 h. Cell-cycle distribution was analyzed by FACSCalibur flow cytometer using the Modfit software.
Apoptosis Assay
Allophycocyanin (APC)-Annexin V apoptosis detection kit (BD Biosciences Pharmingen, San Diego, CA, USA) was used to examine the apoptosis-mediated cell death. In brief, cells (1 × 106) were harvested and incubated with Annexin V and propidium iodide (PI) solutions. The apoptotic cells were detected by a FACSCalibur flow cytometer and quantified by BD CellQuest Pro software according to the manufacturer’s instructions.
Luciferase miRNA Target Reporter Assay
PsiCheck2 (Promega, USA) was cloned by the 3′ UTR of Erbin mRNA containing wild-type or mutant miRNA-183 binding sites. Then HEK293T cells were co-transfected with psiCheck2-Erbin-wild-typeormutant plasmids and miRNA-183 or miRNA-NC using Lipofectamine 2000 (Invitrogen). Cells were collected after 48 h. Glomax 96 luminometer (Promega) was used to determine the luciferase activity. In addition, β-galactosidase activity was applied to normalize the transfection efficiency.
Xenograft Model
About 4- to 5-week-old female nude mice (BALB/c) were randomly divided into two groups according to random number table and subcutaneously injected with approximately 107 of shRNA-infected and mock-infected HL60 and U937 cells at the dorsal region. No blinding was done. Mice were examined every 5 days 2 weeks post-injection. The formed tumor was measured with a micrometer. All mice were sacrificed after 1 month. The tumors were then excised and weighed. Micro-hematocrit capillary tubes were used to collect blood samples from the orbital sinus. Blood and bone marrow smears were performed and stained with the May-Grünwald-Giemsa stain using approximately 0.1 mL of blood or bone marrow. The leukemic infiltration was evaluated by optical microscope observation. Tumor, spleen, and liver were collected for further immunohistochemistry (IHC) and IF analyses. For rescue experiments, 4- to 5-week-old female BALB/c nude mice were randomly divided into eight groups according to random number table and subcutaneously injected with approximately 107 of transfected AML cells and control cells. The experiments were carried out according to the ethical guidelines for laboratory animal use and approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University.
IHC
IHC staining was performed as previously described.40 In brief, the tumor tissues were cut into 4-mm-thick sections, dewaxed in xylene, and rehydrated in a graded series of alcohols. Antigen was retrieved by heating the tissue sections at 100°C for 30 min in EDTA solution (1 mM, pH 9.0). Cooled tissue sections were immersed in 0.3% hydrogen peroxide solution for 15 min to block endogenous peroxidase activity, rinsed with PBS for 5 min, and blocked with 3% BSA solution at room temperature for 30 min. Subsequently, the sections were incubated with mouse monoclonal antibody against human Erbin (1:100), phospho-ERK1/2 (1:200), and phospho-RAF (1:200) at 4°C overnight, followed by incubation with HRP-conjugated goat anti-rabbit secondary antibody. Diaminobenzene was used as the chromogen, and hematoxylin was used as the nuclear counterstain. For IF analysis of CD11b and CD14 expression, the sections were incubated with mouse monoclonal antibody against human CD11b (1:100) and CD14 (1:100) at 4°C overnight and then incubated with Alexa 488-conjugated goat anti-rabbit secondary antibody (Thermo Fisher, CA, USA). The nuclear stain Hoechst 34580 (5 μg/mL; Molecular Probes, Thermo Fisher, CA, USA) was added prior to final washes after incubation of the secondary antibody. Finally, the sections were dehydrated, cleared, and mounted using both wide-field microscope Leica AF7000 and Zeiss confocal microscope. The resulting area and cell measurement were quantified using ImageJ software analysis.
RNA Pull-Down Assay
In brief, U937 cells were transfected with the 3′ end biotinylated miR-183 or miR-183-mut for 24 h at a final concentration of 20 nmol/L. Then the cells were incubated in the cell lysate with streptavidin-coated magnetic beads (Ambion, Life Technologies). The biotin-coupled RNA complex was pulled down, and analysis of the abundance of Erbin in bound fractions was then conducted by qRT-PCR. The pull-down assay was performed as previously described.41
co-IP
The soluble cortical extract was diluted and pre-cleared by using BSA and protein A-Sepharose (Zymed, San Francisco, CA, USA) or A/G agarose (Thermo Scientific, Waltham, MA, USA) beads. Pre-cleared samples were incubated first with antibodies and then with fresh beads. The co-immunoprecipitated proteins were eluted from the beads, resolved by SDS-PAGE, and analyzed by mass spectrometry. Also, western blotting was used for analysis as previously described.
Clinical Samples
A total of seven bone marrow samples and 30 peripheral blood samples were collected from initially diagnosed AML patients who did not undergo the treatment at the Third Affiliated Hospital of Soochow University. Another 10 peripheral blood samples of healthy controls were obtained from the Physical Examination Center. Two bone marrow samples of healthy donors were used for different kinds of hematopoietic cells sorting. All the samples were stored in liquid nitrogen (−80°C). Informed consent was obtained from each participant. The use of human tissues was approved by the Institutional Human Experiment and Ethics Committee of the Third Affiliated Hospital of Soochow University, and the experiments were strictly conducted according to the Declaration of Helsinki.
FACS Analysis and Purification
Cells were harvested at the immature stage by mechanical selection and at later stages using 0.05% trypsin/EDTA or TrypLE Express (GIBCO, Grand Island, NY, USA; http://www.thermofisher.com/). Cells were filtered through cell strainer caps (35-m mesh) to obtain a single-cell suspension after gentle trituration. Surface antigens (granulocytes: CD11/CD13/CD33, monocytes: CD14/CD15, erythrocytes: CD36/CD47/CD59/CD71, megakaryocytes: CD41/CD42/CD61, T lymphocytes: CD2/CD3/CD7, B lymphocytes: CD19/CD20/CD21, and NK cells: CD25/CD56) were labeled by incubating with the primary antibodies described above for 30 min in the dark at 4°C to prevent internalization of antibodies, then incubated for 20–30 min with the appropriate secondary antibodies (Alexa Fluor 488 or Alexa Fluor 647 fluorescent). After washing, the stained cells were analyzed and sorted on a fluorescence-activated cell sorter FACSAria (BD Biosciences, San Diego, CA, USA; http://www.bdbiosciences.com) using FACSDiva software (BD Biosciences); FlowJo software (Tree Star, Ashland, OR, USA; http://www.treestar.com) was used for data analysis. The fluorochromes were excited with this instrument’s standard 488-nm and 633-nm lasers, and green fluorescence was detected using 490 LP and 510/20 filters, and far red fluorescence using 660/20 filters. All analyses and sorts were repeated three times. Purity of sorted fractions was checked visually and by FACS reanalysis.
Statistical Analysis
Statistical analyses were performed using the SPSS version 21.0 (SPSS, Chicago, IL, USA). Other data were assessed by two-tailed Student’s t tests using GraphPad Prism 5.0 software package (GraphPad Software, San Diego, CA, USA). A p value <0.05 was considered statistically significant.
Author Contributions
Z.Z. wrote the manuscript. X.Z., X.G., and W.H. prepared figures. X.X., W.G., and J.J. edited the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Prof. Suning Chen, Dr. Jinlong Liu, and all authors collaborated in the collection and interpretation of the data and contributed to the manuscript. Special thanks for bone marrow healthy donors. This work was supported by the National Key Technology Research and Development Program of China (grant 2015BAI12B12), the Joint Research Fund for Overseas Chinese, Hong Kong and Macao Scholars (grant 31729001), the General Program of National Natural Science Foundation of China (grants 81301960, 31570877, 31570908, and 81171653), the Engineering Research Center for Tumor Immunotherapy of Jiangsu (grant BM2014404), and the Science and Technology Bureau foundation application project of Changzhou (grant CJ20179025).
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at https://doi.org/10.1016/j.ymthe.2019.01.016.
Contributor Information
Weiying Gu, Email: guweiying2001@163.com.
Wenwei Hu, Email: viphuwenwei@vip.163.com.
Jingting Jiang, Email: jiangjingting@suda.edu.cn.
Supplemental Information
References
- 1.Dombret H., Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127:53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kantarjian H. Acute myeloid leukemia—major progress over four decades and glimpses into the future. Am. J. Hematol. 2016;91:131–145. doi: 10.1002/ajh.24246. [DOI] [PubMed] [Google Scholar]
- 3.Borg J.P., Marchetto S., Le Bivic A., Ollendorff V., Jaulin-Bastard F., Saito H., Fournier E., Adélaïde J., Margolis B., Birnbaum D. ERBIN: a basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat. Cell Biol. 2000;2:407–414. doi: 10.1038/35017038. [DOI] [PubMed] [Google Scholar]
- 4.Kolch W. Erbin: sorting out ErbB2 receptors or giving Ras a break? Sci. STKE. 2003;2003:pe37. doi: 10.1126/stke.2003.199.pe37. [DOI] [PubMed] [Google Scholar]
- 5.Liu D., Shi M., Duan C., Chen H., Hu Y., Yang Z., Duan H., Guo N. Downregulation of Erbin in Her2-overexpressing breast cancer cells promotes cell migration and induces trastuzumab resistance. Mol. Immunol. 2013;56:104–112. doi: 10.1016/j.molimm.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Tao Y., Shen C., Luo S., Traoré W., Marchetto S., Santoni M.J., Xu L., Wu B., Shi C., Mei J. Role of Erbin in ErbB2-dependent breast tumor growth. Proc. Natl. Acad. Sci. USA. 2014;111:E4429–E4438. doi: 10.1073/pnas.1407139111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu N., Zhang J., Zhang J., Liu S., Liu Y., Zheng D. Erbin-regulated sensitivity of MCF-7 breast cancer cells to TRAIL via ErbB2/AKT/NF-kappaB pathway. J. Biochem. 2008;143:793–801. doi: 10.1093/jb/mvn032. [DOI] [PubMed] [Google Scholar]
- 8.Hu Y., Chen H., Duan C., Liu D., Qian L., Yang Z., Guo L., Song L., Yu M., Hu M. Deficiency of Erbin induces resistance of cervical cancer cells to anoikis in a STAT3-dependent manner. Oncogenesis. 2013;2:e52. doi: 10.1038/oncsis.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao S., Zheng P., Wu H., Song L.M., Ying X.F., Xing C., Li Y., Xiao Z.Q., Zhou X.N., Shen T. Erbin interacts with c-Cbl and promotes tumourigenesis and tumour growth in colorectal cancer by preventing c-Cbl-mediated ubiquitination and down-regulation of EGFR. J. Pathol. 2015;236:65–77. doi: 10.1002/path.4502. [DOI] [PubMed] [Google Scholar]
- 10.Wilkes M.C., Repellin C.E., Hong M., Bracamonte M., Penheiter S.G., Borg J.P., Leof E.B. Erbin and the NF2 tumor suppressor Merlin cooperatively regulate cell-type-specific activation of PAK2 by TGF-beta. Dev. Cell. 2009;16:433–444. doi: 10.1016/j.devcel.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neville P.J., Conti D.V., Krumroy L.M., Catalona W.J., Suarez B.K., Witte J.S., Casey G. Prostate cancer aggressiveness locus on chromosome segment 19q12-q13.1 identified by linkage and allelic imbalance studies. Genes Chromosomes Cancer. 2003;36:332–339. doi: 10.1002/gcc.10165. [DOI] [PubMed] [Google Scholar]
- 12.Lange E.M., Ho L.A., Beebe-Dimmer J.L., Wang Y., Gillanders E.M., Trent J.M., Lange L.A., Wood D.P., Cooney K.A. Genome-wide linkage scan for prostate cancer susceptibility genes in men with aggressive disease: significant evidence for linkage at chromosome 15q12. Hum. Genet. 2006;119:400–407. doi: 10.1007/s00439-006-0149-6. [DOI] [PubMed] [Google Scholar]
- 13.Greshock J., Nathanson K., Medina A., Ward M.R., Herlyn M., Weber B.L., Zaks T.Z. Distinct patterns of DNA copy number alterations associate with BRAF mutations in melanomas and melanoma-derived cell lines. Genes Chromosomes Cancer. 2009;48:419–428. doi: 10.1002/gcc.20651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dambal S., Baumann B., McCray T., Williams L., Richards Z., Deaton R., Prins G.S., Nonn L. The miR-183 family cluster alters zinc homeostasis in benign prostate cells, organoids and prostate cancer xenografts. Sci. Rep. 2017;7:7704. doi: 10.1038/s41598-017-07979-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., Wang S., Li C. MiR-183 overexpression inhibits tumorigenesis and enhances DDP-induced cytotoxicity by targeting MTA1 in nasopharyngeal carcinoma. Tumour Biol. 2017;39 doi: 10.1177/1010428317703825. 1010428317703825. [DOI] [PubMed] [Google Scholar]
- 16.Supic G., Zeljic K., Rankov A.D., Kozomara R., Nikolic A., Radojkovic D., Magic Z. miR-183 and miR-21 expression as biomarkers of progression and survival in tongue carcinoma patients. Clin. Oral Investig. 2018;22:401–409. doi: 10.1007/s00784-017-2126-y. [DOI] [PubMed] [Google Scholar]
- 17.Peng C., Li L., Zhang M.D., Bengtsson Gonzales C., Parisien M., Belfer I., Usoskin D., Abdo H., Furlan A., Häring M. miR-183 cluster scales mechanical pain sensitivity by regulating basal and neuropathic pain genes. Science. 2017;356:1168–1171. doi: 10.1126/science.aam7671. [DOI] [PubMed] [Google Scholar]
- 18.Chandel R., Saxena R., Das A., Kaur J. Association of rno-miR-183-96-182 cluster with diethyinitrosamine induced liver fibrosis in Wistar rats. J. Cell. Biochem. 2018;119:4072–4084. doi: 10.1002/jcb.26583. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z., Li S., Cheng S.Y. The miR-183∼96∼182 cluster promotes tumorigenesis in a mouse model of medulloblastoma. J. Biomed. Res. 2013;27:486–494. doi: 10.7555/JBR.27.20130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi D.P., Yin C.H., Zhang X.Y., Yang N.N., Xu J.Y. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol. Rep. 2016;35:2873–2879. doi: 10.3892/or.2016.4631. [DOI] [PubMed] [Google Scholar]
- 21.Anwar S.L., Krech T., Hasemeier B., Schipper E., Schweitzer N., Vogel A., Kreipe H., Buurman R., Skawran B., Lehmann U. hsa-mir-183 is frequently methylated and related to poor survival in human hepatocellular carcinoma. World J. Gastroenterol. 2017;23:1568–1575. doi: 10.3748/wjg.v23.i9.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan D., Li K., Zhu K., Yan R., Dang C. Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer. Cancer Biol. Ther. 2015;16:268–275. doi: 10.1080/15384047.2014.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z.B., Li Z.Z., Li L., Chu H.T., Jia M. MiR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. Eur. Rev. Med. Pharmacol. Sci. 2015;19:3208–3217. [PubMed] [Google Scholar]
- 24.Sarver A.L., French A.J., Borralho P.M., Thayanithy V., Oberg A.L., Silverstein K.A., Morlan B.W., Riska S.M., Boardman L.A., Cunningham J.M. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Z., Pan L., Niu Z., Li W., Dang X., Wan L., Zhang R., Yang S. Identification of microRNAs as potential biomarkers for lung adenocarcinoma using integrating genomics analysis. Oncotarget. 2017;8:64143–64156. doi: 10.18632/oncotarget.19358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H., Zhang L., Zhang L., Du J., Wang H., Wang B. MicroRNA-183 correlates cancer prognosis, regulates cancer proliferation and bufalin sensitivity in epithelial ovarian caner. Am. J. Transl. Res. 2016;8:1748–1755. [PMC free article] [PubMed] [Google Scholar]
- 27.Kovalchuk A., Ilnytskyy Y., Rodriguez-Juarez R., Katz A., Sidransky D., Kolb B., Kovalchuk O. Growth of malignant extracranial tumors alters microRNAome in the prefrontal cortex of TumorGraft mice. Oncotarget. 2017;8:88276–88293. doi: 10.18632/oncotarget.19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song C., Zhang L., Wang J., Huang Z., Li X., Wu M., Li S., Tang H., Xie X. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci. Rep. 2016;6:24502. doi: 10.1038/srep24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowery A.J., Miller N., Dwyer R.M., Kerin M.J. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ijaz M., Wang F., Shahbaz M., Jiang W., Fathy A.H., Nesa E.U. The role of Grb2 in cancer and peptides as Grb2 antagonists. Protein Pept. Lett. 2018;24:1084–1095. doi: 10.2174/0929866525666171123213148. [DOI] [PubMed] [Google Scholar]
- 31.Xu L.J., Wang Y.C., Lan H.W., Li J., Xia T. Grb2-associated binder-2 gene promotes migration of non-small cell lung cancer cells via Akt signaling pathway. Am. J. Transl. Res. 2016;8:1208–1217. [PMC free article] [PubMed] [Google Scholar]
- 32.Giubellino A., Burke T.R., Jr., Bottaro D.P. Grb2 signaling in cell motility and cancer. Expert Opin. Ther. Targets. 2008;12:1021–1033. doi: 10.1517/14728222.12.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravandi F., Stone R. Acute promyelocytic leukemia: a perspective. Clin. Lymphoma Myeloma Leuk. 2017;17:543–544. doi: 10.1016/j.clml.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 34.Su J., Veillon D., Shackelford R., Cotelingam J., El-Osta H., Mills G., Munker R., Devarakonda S. Acute promyelocytic leukemia and chronic lymphocytic leukemia: concomitant presentation of two molecularly distinct entities. J. La. State Med. Soc. 2017;169:68–70. [PubMed] [Google Scholar]
- 35.Itzykson R., Ayari S., Vassilief D., Berger E., Slama B., Vey N., Suarez F., Beyne-Rauzy O., Guerci A., Cheze S., Groupe Francophone des Myelodysplasies (GFM) Is there a role for all-trans retinoic acid in combination with recombinant erythropoetin in myelodysplastic syndromes? A report on 59 cases. Leukemia. 2009;23:673–678. doi: 10.1038/leu.2008.362. [DOI] [PubMed] [Google Scholar]
- 36.Noack K., Mahendrarajah N., Hennig D., Schmidt L., Grebien F., Hildebrand D., Christmann M., Kaina B., Sellmer A., Mahboobi S. Analysis of the interplay between all-trans retinoic acid and histone deacetylase inhibitors in leukemic cells. Arch. Toxicol. 2017;91:2191–2208. doi: 10.1007/s00204-016-1878-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platzbecker U., Avvisati G., Cicconi L., Thiede C., Paoloni F., Vignetti M., Ferrara F., Divona M., Albano F., Efficace F. Improved outcomes with retinoic acid and arsenic trioxide compared with retinoic acid and chemotherapy in non-high-risk acute promyelocytic leukemia: final results of the randomized Italian-German APL0406 Trial. J. Clin. Oncol. 2017;35:605–612. doi: 10.1200/JCO.2016.67.1982. [DOI] [PubMed] [Google Scholar]
- 38.Rohr S.S., Pelloso L.A., Borgo A., De Nadai L.C., Yamamoto M., Rego E.M., de Lourdes L F Chauffaille M. Acute promyelocytic leukemia associated with the PLZF-RARA fusion gene: two additional cases with clinical and laboratorial peculiar presentations. Med. Oncol. 2012;29:2345–2347. doi: 10.1007/s12032-011-0147-y. [DOI] [PubMed] [Google Scholar]
- 39.Spicuglia S., Vincent-Fabert C., Benoukraf T., Tibéri G., Saurin A.J., Zacarias-Cabeza J., Grimwade D., Mills K., Calmels B., Bertucci F. Characterisation of genome-wide PLZF/RARA target genes. PLoS ONE. 2011;6:e24176. doi: 10.1371/journal.pone.0024176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong J., Wang R., Ren G., Li X., Wang J., Sun Y., Liang J., Nie Y., Wu K., Feng B. HMGA2-FOXL2 axis regulates metastases and epithelial-to-mesenchymal transition of chemoresistant gastric cancer. Clin. Cancer Res. 2017;23:3461–3473. doi: 10.1158/1078-0432.CCR-16-2180. [DOI] [PubMed] [Google Scholar]
- 41.Cheng X., Zhang L., Zhang K., Zhang G., Hu Y., Sun X., Zhao C., Li H., Li Y.M., Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 2018;77:770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.