Abstract
Long noncoding RNAs (lncRNAs) are large RNA transcripts that do not code for proteins but exert their effects in the form of RNA. To date many thousands of lncRNAs have been identified, their molecular functions and mechanisms of action however are largely unknown. The development of high-throughput experimental technologies, such as ChIRP (Chromatin isolation by RNA purification), CHART (Capture Hybridization Analysis of RNA Targets), RAP (RNA antisense purification), RIP (RNA Immunoprecipitation), CLIP (cross-linking and immunoprecipitation) and RNA pull-down, has led to a rapid expansion of lncRNA research and resulted in many publicly-available databases. This review provides an overview of the current methodologies available for discovering and investigating functions of lncRNAs in various human diseases. A comparison and application of these methods are also included. Finally, this paper surveys current databases containing annotations, interactome networks and functions of lncRNAs. The appropriate use of these methods and databases will provide not only high-resolution functional features of lncRNAs, but also enhance our understanding of the underlying mechanisms by which lncRNAs regulate a variety of biological processes.
Keywords: lncRNA, ChIRP, CHART, RIP, RNA pull-down
1. Introduction
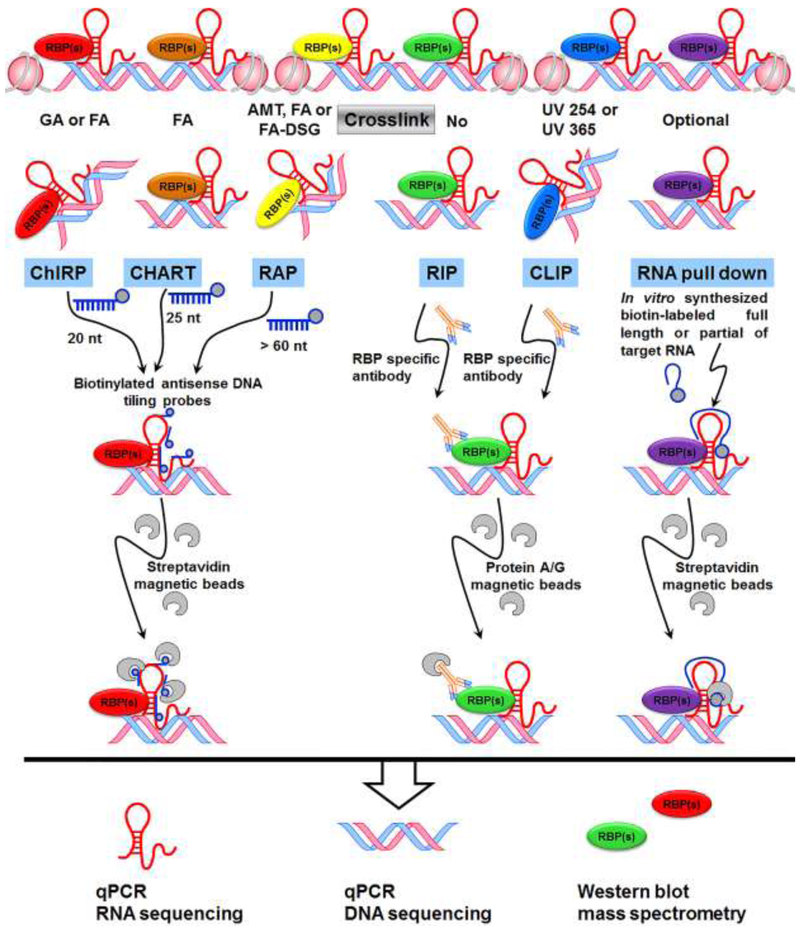
Long noncoding RNAs (lncRNAs) are non-protein coding RNA transcripts that are greater than 200 nucleotides in length [1]. lncRNAs are transcribed from various genomic regions, such as in the enhancers, promoters, introns, anti-sense coding, or intergenic regions of genes [2]. lncRNAs exhibit more highly spatial and temporal restricted expression patterns than mRNAs. Although lncRNAs are present in both the nucleus and the cytoplasm, the fact that many lncRNAs are enriched in the nucleus highlights a broader role for lncRNAs as epigenetic regulators within the nucleus [3]. Indeed, chromatin signature of actively transcribed genes has uncovered around 3,500 lncRNAs in mammals [4, 5]. More recently, over 100 thousands of lncRNAs have been defined in human genome [1, 6]. Unlike protein-coding mRNA molecules, lncRNAs are initially transcribed as primary transcripts by both RNA Polymerase II and III in the nucleus and also can be regulated by transcriptional factors [4, 5]. Multiple lines of evidence demonstrate that lncRNAs exert their functions at the level of chromatin and epigenetics via interaction with RNA binding proteins (RBPs) at specific DNA regions, such as promoters or enhancers [7–10]. Thus, to better understand the global role of lncRNAs in regulating gene expression, many genomic-context based methodologies have been developed and used for identification of lncRNA binding proteins and chromatin occupancy sites. Indeed, based on their interest, scientists can identify chromatin occupancy sites of a given lncRNA, unknown lncRNAs to which an RBP binds and interacting lncRNAs for a given lncRNA using the omic approaches followed by Next-Generation Sequencing (NGS) of RNA or DNA, and mass spectrometry (MS) (Figure 1).
Figure 1. Schematic representation of high-throughput strategies for lncRNA investigation.
Crosslinking enables stringent purification of target complexes. AMT specifically crosslinks direct RNA-RNA interactions; FA crosslinks RNA-protein and protein-protein interactions; FA-DSG provides stronger protein-protein crosslinking than FA alone; while UV crosslinks are more specific and only link proteins to RNAs that are in very close proximity. Also see table 1 for a detailed comparison of these methods. ChIRP, chromatin isolation by RNA purification; CHART, Capture Hybridization Analysis of RNA Targets; RAP, RNA antisense purification; RIP, RNA immunoprecipitation; CLIP, cross-linking and immunoprecipitation. FA, formaldehyde; GA, Glutaraldehyde; AMT, 4′-aminomethyltrioxalen; DSG, disuccinimidyl glutarate; UV, ultraviolet.
2. Experimental approaches for lncRNA study in the genomic context
2.1. Hybridization-based approaches (lncRNA–chromatin interactions)
To investigate lncRNA binding sites on chromatin genome-wide, two groups developed similar hybridization-based strategies independently, named ChIRP (Chromatin isolation by RNA purification) [11] and CHART (capture hybridization analysis of RNA targets) [12], and mapped the genomic binding sites for endogenous lncRNAs using those methods in 2011.
ChIRP was developed by Chu et. al., and used to map RNA occupancy at chromatin correlated with a given lncRNA using deep sequencing (ChIRP-seq) [11, 13]. In this method, a pool of 20-nt affinity-probes targeting a specific lncRNA was used to retrieve lncRNA and its binding DNA fragments followed by deep DNA sequencing, thus determining with high-resolution the genomic binding sites for a given lncRNA. To test the reliability of ChIRP, the authors performed ChIRP-seq on endogenous roX2, a lncRNA known to bind to multiple binding sites on the X chromosome in male S2 cells. The results showed that all of the identified 308 roX2 binding sites are on the X chromosome, suggesting that ChIRP-seq is highly sensitive and specific technique for mapping lncRNA occupancy genome-wide at high resolution [11]. More recently, using ChIRP, Olivier-Van Stichelen and Hanover demonstrated that the OGT (O-GlcNAc Transferase) and G6PD (Glucose-6-phosphate dehydrogenase) loci are associated with Xist RNA [14]. Additionally, the enriched RNA, and protein from ChIRP can also be isolated and subjected to RNA and protein analysis [15].
CHART, developed by Simon et. al. [12], was used to purify roX2 lncRNA, its associated proteins and DNA targets in chromatin for the purpose of mapping roX2 RNA binding sites on chromatin. A set of 25-mer desthiobiotin-conjugated-DNA oligonucleotide probes was designed and used for the enrichment of roX2 together with its DNA targets followed by DNA sequencing (CHART-seq). Consistent with the role of roX2 in dosage compensation, the results demonstrated the efficient and specific enrichment of roX2 CHART signals on chromosome X. This technique was further utilized to investigate genome-wide binding locations of Xist lncRNA [8]. CHART-seq, using oligonucleotide probes for Xist, was performed in female mouse cells at four developmental stages: before XCI (X-chromosome inactivation); early-XCI; mid-XCI; and post-XCI. The results demonstrated that Xist density on the X chromosome is increased in a developmental time-dependent manner, and high-resolution maps of Xist binding on the X chromosome across a developmental time course were generated [8].
RNA antisense purification (RAP), developed by Engreitz et. al. in 2013, is another method to map the localization of a given lncRNA across the genome [16]. In this method, a set of 120-nt oligonucleotide affinity-probes (tiled every 15 nucleotides across the sequence of a target lncRNA) was designed and used to capture the probe-RNA-chromatin complexes followed by high-throughput DNA sequencing. Using RAP, the authors demonstrated that Xist binds broadly across the X chromosome and revealed a new mechanism by which Xist orchestrates mammalian XCI by coating and silencing one X chromosome in females [16].
To avoid Type I errors, reduce background and increase specificity, it is important to evaluate the accessibility of probes to target lncRNA before performing experiments. Studies by Simon and colleagues adapted an RNase-H mapping assay strategy [8, 12]. The ability of antisense oligonucleotide probes to reach (access) their target (lncRNA) sites was determined by an RNase H sensitivity assay and only probes with high RNase-H sensitivity were selected for further CHART experiments. This is to ensure that these probes target lncRNA directly. Another strategy used in CHART was to elute probe-RNA-chromatin complexes with RNase-H (specifically hydrolyze RNA that is hybridized to DNA probes) which would release chromatin (combined with probes through lncRNA) from beads and exclude nonspecific binding complexes [12].
2.2. Immunoprecipitation-based approaches (lncRNA–protein interactions)
lncRNAs often exert their function through interaction with RBPs. To identify RNAs bound to a given RBP, immunoprecipitation-based approaches using RBP as bait have been widely used for functional study of lncRNAs. RNA Immunoprecipitation (RIP) is one of the most commonly employed techniques for protein-RNA interaction studies in the last 30 years [17, 18]. The basic principle of RIP is that protein-RNA complexes are immunoprecipitated by a specific antibody against a target protein, such as an RBP. RNAs in these protein-RNA complexes can be purified and analyzed by PCR, microarray analysis (RIP-Chip), or deep sequencing (RIP-seq).
As an example, Zhao et. al. identified Polycomb repressive complex 2 (PRC2)-interacting RNAs in embryonic stem cells (ESCs) using RIP-seq [19]. Antibodies against to EZH2 (one of the four core subunits of PRC2), were used to immunoprecipitate EZH2-binding RNAs followed by Illumina sequencing. This study identified 216 PRC2-interacting lncRNAs in mouse ESCs, including several well-known PRC2-binding lncRNAs (Tsix, RepA, and Xist RNAs), suggesting that the RIP-seq technique is highly sensitive and specific for the detection of protein binding lncRNAs.
Similarly, using a modified RIP-seq approach, the EZH2-associated transcriptome in human gastric cancer cells has been characterized recently by Qi and colleagues [20]. In this study, EZH2-associated RNAs were immunoprecipitated by EZH2-specific antibody from cell nuclei and reverse transcribed using a random primer flanked with a 7-nt barcode sequence. The second cDNA strand was generated using the adaptor ligation strategy instead of template switching strategy, to avoid any bias resulting from guanine preference, followed by Illumina sequencing [20]. Interestingly, the data demonstrated that most of the EZH2-interacting transcripts in gastric cancer cells are mRNAs, suggesting that mRNAs may play a regulatory role in gene expression in the nucleus, considering that cell nuclei were used for the immunoprecipitation in this study.
In another example, Dharap et. al. applied an RIP-Chip approach with antibodies against Sin3A and coREST (corepressors of the RE-1 silencing transcription factor) which are chromatin-modifying proteins (CMPs) [21]. In this work, cortical nuclear lysates from rats subjected to focal ischemia were immunoprecipitated with anti-Sin3A and anti-coREST antibodies. The precipitated RNAs were then purified and subjected to lncRNA microarray analysis. This study demonstrated that 99 Sin3A-enriched lncRNAs and 78 coREST-enriched lncRNAs were significantly increased in the ischemia group compared with the sham group. Among them, the expression of 26 Sin3A-enriched and 11 coREST-enriched lncRNAs was also up-regulated in the ischemic brains, suggesting that stroke-induced lncRNAs may play an important role in epigenetic modifications in the post-ischemic brain through interaction with CMPs [21].
Some variants of RIP have also been established over the past 10 years to precisely map the binding sites of a target protein. Ultraviolet (UV) cross-linking and immunoprecipitation (CLIP) method was originally developed by Ule et. al. in 2003 to purify protein-RNA complexes from mouse brain [22]. Brain tissue was directly irradiated with UV-B light and protein-RNA complexes were immunoprecipitated with Nova antiserum (Nova is one of the earliest identified mammalian tissue-specific splicing factor) [22]. To map the location of Nova binding sites in target RNAs, UVB- irradiated brain lysates were treated with RNase A, to digest the RNA that was not protected by protein. After immunoprecipitation with Nova antiserum, the protein protected RNA fragments were cloned with the use of linker ligation and subjected to sequencing. The results not only identified the known Nova binding sites, but also determined the role of Nova in regulating brain-specific alternative splicing. Thus, CLIP allows precise mapping of protein binding sites on RNAs [22].
HITS-CLIP (also known as CLIP-seq), is a method that combined CLIP with high-throughput sequencing for genome-wide mapping of protein-RNA binding sites in vivo [23]. Using HITS-CLIP, Licatalosi et. al. investigated Nova-binding sites in RNAs and revealed genome-wide biochemical RNA footprints of Nova in multiple mouse brains. Results from this study identified a large number of Nova - RNA interactions in 3’ untranslated regions, suggesting the role of Nova in regulating alternative polyadenylation in the brain [23]. HITS-CLIP provides a robust platform to explore transcriptome-wide binding sites of RBPs.
PAR-CLIP (photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation) was first developed by Hafner et. al. in 2010, to determine the binding sites of RBPs and miRNPs (microRNA-containing ribonucleoprotein complexes) at high resolution and transcriptome-wide [24]. In this design, 4-thiouridine (4-SU) and 6-thioguanosine (6-SG) were used to incorporate into nascent RNA transcripts by living cells. The RBP binding sites can be precisely mapped by scoring for thymidine (T) to cytidine (C) transitions in the sequenced cDNA from 4-SU-treated cells; or guanosine (G) to adenosine (A) transitions in the sequenced cDNA from 6-SU-treated cells [25]. Recently, combining PAR-CLIP and anti-m6A immunoprecipitation (MeRIP) approaches, Liu et. al. demonstrated that N6-methyladenosine (m6A) dependent mRNA and lncRNA structural remodeling affects RNA–protein interactions [26]. Furthermore, PAR-CLIP has also been employed to determine RBP binding sites in lncRNAs [24, 25].
Individual-nucleotide resolution CLIP (iCLIP) was modified from CLIP by König et. al. in 2010 and was employed to investigate the RNA splicing maps of heterogeneous nuclear ribonucleoprotein C (hnRNP C) [27]. The aim of iCLIP was to map RBP binding sites at nucleotide resolution. In this study, hnRNP C-RNA complexes were excised from the nitrocellulose membrane, and treated with proteinase to release the RNAs. This also leaves a covalently bound polypeptide fragments at the RNA cross-link sites and causes cDNA premature truncation immediately before the cross-link nucleotide during the reverse transcription. cDNA molecules were then subjected to circularization, linearization, PCR-amplification and high-throughput sequencing. Results from this study revealed single-nucleotide resolution mapping of hnRNP C cross-link sites, suggesting a role for hnRNP particles in splicing regulation [27]. As with all CLIP methods, iCLIP is employed for identifying RBP binding sites on lnRNAs. This includes the interactions of lncRNAs, MALAT1 and NEAT1 with TDP-43, and MALAT1-U2AF65 interaction [28, 29].
2.3. Affinity-based approaches (RNA pull-down)
Biotinylated RNA-protein pull-down followed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) techniques are used to identify interaction proteome of a target RNA. In this method, full length or partial of the target RNA is in vitro synthesized and labeled with biotinylated uridines. After incubation with cellular lysates (or nuclear extracts), biotinylated-RNA-protein complexes are subsequently pulled down with streptavidin beads. Then, RNA-binding proteins are usually separated by SDS-PAGE, and analyzed by MS [30–32]. Owing to the use of artificially elevated levels of RNA probe, the advantage of RNA pull-down techniques is to enrich RBPs associated with low abundant target RNA. This method has been widely used for the detection of lncRNA binding proteins in the past 10 years [33–35].
Recently, a novel method, Urb-RNA immunoprecipitation (Urb-RIP), was presented by Cottrell and Djuranovic [36]. This method is based on the high-affinity interaction between the sequence-specific RNA stem loop of the MS2 bacteriophage and the RNA recognition motif 1 (RRM1) domain of the “resurrected” snRNA-binding protein Urb. Briefly, 2HA-Urb expressing cells were transfected with a tagged RNA of interest or an untagged control for 24 hours. Then, after UV-irradiation, the cell lysate was incubated with anti-HA beads to isolate 2HA-Urb- tagged RNA-protein complexes. The target RNA associated RBPs and RNAs are subjected to further analysis. The authors confirmed the interaction between polyA-binding protein (PABP) and lncRNA BC200 using Urb-RIP. Thus, Urb-RIP provides another useful tool for lncRNA study as well [36].
Although most of the methods used for investigating lncRNA are adapted from techniques originally developed for mRNA analysis, many improvements in the methods have been made to enable studies on properties and functions of lncRNA, regardless of their abundance. In addition to the modified methods mentioned above, ChIRP-ms [37] is built based on the original ChIRP-seq. Chu and Chang have provided a detailed protocol for this approach [37]. ChIRP-ms offers a comprehensive methodology for the identification of lncRNA-interacting proteins. There is no doubt that more efforts will be spent on the technologies to increase specificity or sensitivity, and to decrease false positives.
3. Employment of the methods in the discovery of disease-associated lncRNAs
The methods described above are powerful approaches for the identification of disease-associated lncRNAs as well as investigation into their acting mechanisms (Table 2). For example, using ChIRP and RIP, Jiang et al. [38] have demonstrated that CCAT1 lncRNA forms a complex with TP63 and SOX2 at the super-enhancers of EGFR, thereby increasing the expression of EGFR and activating both MEK/ERK1/2 and PI3K/AKT signaling pathways, which in turn ultimately promotes the tumorigenesis of squamous cell cancer (SCC). Importantly, knockdown of CCAT1 in the xenograft assays leads to a marked reduction in both volume and mass of the tumors [38], indicating that CCAT1 could be a rational therapeutic target. Recently, employing CHART, Gast et al. [39] have demonstrated the direct interaction between two lncRNAs, MALAT1 and NEAT1, which regulates immune genes and affects the development of atherosclerosis. Interestingly, MALAT1 deficiency causes massive immune-mediated atherosclerosis in ApoE−/− mice suggesting the underlying mechanisms of MALAT1-dependent immune regulation of the cardiovascular system as well as the potential therapeutic applications of this circuit [39]. Applying the RAP approach, Engreitz et al. [40] have demonstrated that U1 small nuclear RNA directly hybridizes to 5’ splice sites and 5’ splice site motifs throughout introns and that MALAT1 interacts with pre-mRNAs indirectly through protein intermediates, suggesting that lncRNAs may target other RNAs as part of their regulatory function. Moreover, using CLIP, Yap et al. [41] have demonstrated that ANRIL lncRNA interacts with CBX7 of the PRC1 complex at the INK4b/ARF/INK4a locus and control senescence. Subsequent studies have demonstrated ANRIL is upregulated in various human tumors, including ovarian cancer [42] and non-small cell lung cancer (NSCLC) [43] indicating that ANRIL could be a biomarker for disease progression.
Table 2.
Examples for the employment of the methods
| Method | lncRNA | Disease | Biological function (Targets) | Potential clinical application |
|---|---|---|---|---|
| ChIRP | CCAT1 | Squamous cell cancer (SCC) | Forms a complex with TP63 and SOX2 at the super-enhancers of EGFR [38]. | Therapeutic target for SCC |
| CYTOR | Colorectal cancer (CRC) | Mediates complex formation between nucleolin and Sam68 [44]. | Prognostic biomarker, therapeutic target for CRC | |
| CHART | MALAT1, NEAT1 | Atherosclerosis | MALAT1-NEAT1 interaction regulates immune genes in turn affects the development of atherosclerosis [39]. | Therapeutic targets |
| RAP | MALAT1 | Hepatocellular carcinoma (HCC) and lung cancer | Localizes to nuclear speckles and interacts with multiple serine/arginine RNA splicing proteins and is involved in mRNA processing, splicing, and the export of mRNA [40, 45]. | Biomarker for HCC [46, 47] and lung cancer [48] |
| NORAD | Pancreatic cancer and colorectal cancer (CRC) | Modulates RBMX and is essential for assembly of a topoisomerase complex and affects cell-cycle progression [49]; serve as sponge for miRNAs [50]. | Therapeutic target for pancreatic [51] cancer and CRC[50] | |
| RIP | THOR | Gastric cancer | Binding to SOX9 3’UTR and enhancing its stability [52]. | Therapeutic target for gastric cancer |
| DBH-AS1 | Hepatocellular carcinoma (HCC) | Activation of FAK/Src/ERK signaling pathway via downregulating miR-138 [53]. | Biomarker for HCC and therapeutic target | |
| FOXD2-AS1 | Osteoarthritis (OA) | Acting as a sponge of miR-206 to modulate CCND1 [54]. | Therapeutic target in the treatment of OA. | |
| CLIP | ANRIL | Epithelial ovarian cancer (EOC); Prostate Cancer; acute lymphoblastic leukemia (ALL) | Interacts with CBX7 of the PRC1 complex at the INK4b/ARF/INK4a locus [41]; Regulates P15INK4B and Bcl-2 and inhibits apoptosis and senescence [42]. | Biomarker: Prostate Cancer [41]; ALL [55]; ovarian cancer [42]; NSCLC [43] |
| Thousands of lncRNAs | KSHV- and EBV-driven cancers | lncRNA - miRNA Interactions [56]. | Therapeutic targets | |
| RNA pulldown | PINCR | human colorectal cancer cells (CRC) | Binding to Matrin 3 AT the enhancer regions of p53 target genes [57]. | Biomarker and therapeutic target |
| GAS8-AS1 | Hepatocellular carcinoma (HCC) | Recruiting the MLL1/WDR5 complex at the promoter of GAS8 [58] | Therapeutic target | |
| MANTIS | Glioblastoma | Interacts with BRG1 and promotes transcription of SOX18, SMAD6, and COUP- TFII [59]. | Biomarker and therapeutic target | |
| LINC00152 | Lung adenocarcinoma (LAD) | Interacts with EZH2 and inhibits IL24 transcription and regulates cell growth and apoptosis [60]. | Therapeutic target |
4. Conclusions and perspective
As a large number of experiments using the methods mentioned above have been conducted in the investigation of lncRNA interactome networks, millions of interactions between lncRNA, protein and chromatin have been identified. This includes transcriptome-wide binding sites of RBPs, proteome-wide interactions and genome-wide binding sites of target lncRNAs. Indeed, a variety of public databases on lncRNAs have been constructed (Table 3). DIANA-LncBase v2 contains more than 70,000 miRNA:lncRNA interactions and provides an indispensable tool for lncRNA regulation research [61]. NPInter v3.0 holds 491,416 interactions in 188 tissues (or cell lines) from 68 kinds of experimental technologies and allows researchers to predict the function of lncRNAs based on the interactions curated in the database [62]. CLIP-seq data are also annotated with database, CLIPdb [63]. Also, starBase v2.0 is designed to systematically identify RNA-RNA and protein-RNA interaction networks from 108 CLIP-seq [64]. Additionally, an excellent review on lncRNA databases has been published and could provide further insights on this subject [65].
Table 3.
lncRNA Databases
| Database | # of human lncRNAs or data size | Species | Description | Ref |
|---|---|---|---|---|
| NONCODE | 172,216 | 17 species | An integrated knowledge database of lncRNAs | [67] |
| NPInter | 491,416 interactions | 22 species | Interactions between ncRNAs and biomolecules (proteins, RNAs and DNAs) | [62] |
| lncRNAdb | 14,470 | 68 species | Comprehensive annotations of eukaryotic lncRNAs | [68] |
| NRED | 1,287 [as of 2008] | Human, Mouse | Expression data from various platforms and/or studies | [69] |
| lncRNADisease | 2,947 lncRNA-disease entries | Human | lncRNA-disease association database (support prediction) | [70] |
| LNCipedia | 107,039 | Human | A comprehensive compendium of human lncRNAs | [6] |
| ChIPBase | ~10,200 ChIP-seq datasets | Human, Mouse | T ranscription factor occupancy at lncRNA genes | [71] |
| starBase | 2.1 million protein-RNA and 1.5 million RNA-RNA interactions | Human, Mouse | RNA-RNA, and protein-RNA interaction networks from large-scale CLIP-Seq data | [64] |
| DIANA (LncBase) | 32,223 | Human, Mouse | A database of experimentally supported and in silico predicted miRNA Recognition Elements on lncRNAs | [61] |
Though the application of these methods for studying disease-associated lncRNAs is promising and has resulted in many publicly-available databases, the biological functions of these lncRNAs are still difficult to predict in comparison with protein-coding genes. By utilizing publicly-available genome-wide datasets and computational methods, Signal et al. [66] have developed a pipeline to characterize and predict the functions of lncRNAs. This pipeline is crucial not only for the functional investigation of lncRNAs in silico, but also for informing further experimental strategies.
Taken together, data generated from genomic-context based methodologies will provide not only high-resolution functional features of lncRNAs, but also enhance our understanding of the underlying mechanisms by which lncRNAs regulate a variety of biological processes.
Table 1.
Comparison of the methods
| Method | Input (Bait) | Output (Prey) | Endpoint detection | Advantage | Disadvantage |
|---|---|---|---|---|---|
| ChIRP | DNA probes (20nt each) for a target lncRNA | lncRNA-binding RNAs, genomic elements and proteins | RNA/DNA- sequencing and Mass Spectrometry | Requires no prior knowledge of the RNA’s structure or functional domains | lncRNA sequence information is required and large enough for probe design |
| CHART | DNA probes (25nt each) for a target lncRNA | lncRNA-binding RNAs and genomic elements | RNA/DNA- sequencing | Requires no prior knowledge of the RNA’s structure or functional domains | lncRNA sequence information is required and large enough for probe design |
| RAP | >60nt antisense RNA probes for a target lncRNA | lncRNA-binding RNAs, proteins and genomic elements | RNA/DNA- sequencing and Mass Spectrometry | High resolution, specificity, and sensitivity | Requires overlapping probes tiled across the entire length of the target RNA to ensure capture even in the case of extensive protein-RNA interactions, RNA secondary structure, or partial RNA degradation |
| RIP | Antibody against a target protein | Protein-binding RNAs | RNA- sequencing | Requires no specialized equipment or reagents | Lack of actually protein binding site identification, nonspecific RNA interaction identification, and high signal-to-noise ratio |
| CLIP | Antibody against a target protein | Protein-binding RNAs | RNA- sequencing | Low background noise and high resolution | Low sensitivity and possible mutations caused by UV crosslink |
| RNA pulldown | In vitro synthesized biotin-labeled full length or partial of a target RNA | lncRNA-binding proteins and RNAs | Mass Spectrometry and RNA- sequencing | Higher chance to identify weak/transient bindings | Artificially elevated levels of lncRNA may result in false positives; difficult to synthesize large lncRNAs (e.g. > 5kb) |
Acknowledgements
This work was supported by the National Institutes of Health (DA042704 and DA046831 to GH) and the National Natural Science Foundation of China (31270175 to JZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflicts of interest.
References
- [1].Quinn JJ, Chang HY, Unique features of long non-coding RNA biogenesis and function, Nat Rev Genet 17(1) (2016) 47–62. [DOI] [PubMed] [Google Scholar]
- [2].Kung JT, Colognori D, Lee JT, Long noncoding RNAs: past, present, and future, Genetics 193(3) (2013) 651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kopp F, Mendell JT, Functional Classification and Experimental Dissection of Long Noncoding RNAs, Cell 172(3) (2018) 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL, Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression, Proc Natl Acad Sci U S A 106(28) (2009) 11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES, Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals, Nature 458(7235) (2009) 223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P, An update on LNCipedia: a database for annotated human lncRNA sequences, Nucleic Acids Res 43(8) (2015) 4363–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL, Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression, Proc Natl Acad Sci U S A 106(28) (2009) 11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT, High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation, Nature 504(7480) (2013) 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].He R, Zhang FH, Shen N, LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC), Biomed Pharmacother 95 (2017) 331–338. [DOI] [PubMed] [Google Scholar]
- [10].Werner MS, Sullivan MA, Shah RN, Nadadur RD, Grzybowski AT, Galat V, Moskowitz IP, Ruthenburg AJ, Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription, Nat Struct Mol Biol 24(7) (2017) 596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chu C, Qu K, Zhong FL, Artandi SE, Chang HY, Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions, Mol Cell 44(4) (2011) 667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE, The genomic binding sites of a noncoding RNA, Proc Natl Acad Sci U S A 108(51) (2011) 20497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chu C, Quinn J, Chang HY, Chromatin isolation by RNA purification (ChIRP), J Vis Exp (61) (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Olivier-Van Stichelen S, Hanover JA, X-inactivation normalizes O-GlcNAc transferase levels and generates an O-GlcNAc-depleted Barr body, Frontiers in genetics 5 (2014) 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chu C, Spitale RC, Chang HY, Technologies to probe functions and mechanisms of long noncoding RNAs, Nat Struct Mol Biol 22(1) (2015) 29–35. [DOI] [PubMed] [Google Scholar]
- [16].Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M, The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome, Science 341(6147) (2013) 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ule J, Hwang HW, Darnell RB, The Future of Cross-Linking and Immunoprecipitation (CLIP), Cold Spring Harb Perspect Biol 10(8) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lerner MR, Steitz JA, Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus, Proc Natl Acad Sci U S A 76(11) (1979) 5495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT, Genome-wide identification of polycomb-associated RNAs by RIP-seq, Mol Cell 40(6) (2010) 939–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan S, Yu Q, Li YY, Kang Y, Li H, Xiong Z, Zhu T, Liu B, Shao Z, Zhao X, MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10, Oncotarget 7(11) (2016) 12693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dharap A, Pokrzywa C, Vemuganti R, Increased binding of stroke-induced long non-coding RNAs to the transcriptional corepressors Sin3A and coREST, ASN Neuro 5(4) (2013) 283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB, CLIP identifies Nova-regulated RNA networks in the brain, Science 302(5648) (2003) 1212–5. [DOI] [PubMed] [Google Scholar]
- [23].Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB, HITS-CLIP yields genome-wide insights into brain alternative RNA processing, Nature 456(7221) (2008) 464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr., Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T, Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP, Cell 141(1) (2010) 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T, PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins, J Vis Exp (41) (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T, N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions, Nature 518(7540) (2015) 560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J, iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution, Nature structural & molecular biology 17(7) (2010) 909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J, Characterizing the RNA targets and position-dependent splicing regulation by TDP-43, Nature neuroscience 14(4) (2011) 452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schor IE, Lleres D, Risso GJ, Pawellek A, Ule J, Lamond AI, Kornblihtt AR, Perturbation of chromatin structure globally affects localization and recruitment of splicing factors, PloS one 7(11) (2012) e48084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marin-Bejar O, Huarte M, RNA pulldown protocol for in vitro detection and identification of RNA-associated proteins, Methods in molecular biology 1206 (2015) 87–95. [DOI] [PubMed] [Google Scholar]
- [31].Jazurek M, Ciesiolka A, Starega-Roslan J, Bilinska K, Krzyzosiak WJ, Identifying proteins that bind to specific RNAs - focus on simple repeat expansion diseases, Nucleic acids research 44(19) (2016) 9050–9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bai Q, Bai Z, Sun L, Detection of RNA-binding Proteins by In Vitro RNA Pull-down in Adipocyte Culture, J Vis Exp (113) (2016). [DOI] [PubMed] [Google Scholar]
- [33].Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY, Long noncoding RNA as modular scaffold of histone modification complexes, Science 329(5992) (2010) 689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, Kim J, Curtis J, Moad CA, Wohler CM, Indig FE, de Paula W, Dudekula DB, De S, Piao Y, Yang X, Martindale JL, de Cabo R, Gorospe M, HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP, Genes & development 30(10) (2016) 1224–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, Wang Z, Sun M, Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with LSD1 and EZH2 in Gastric Cancer, Molecular therapy : the journal of the American Society of Gene Therapy (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [36].Cottrell KA, Djuranovic S, Urb-RIP - An Adaptable and Efficient Approach for Immunoprecipitation of RNAs and Associated RNAs/Proteins, PloS one 11(12) (2016) e0167877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chu C, Chang HY, ChIRP-MS: RNA-Directed Proteomic Discovery, Methods Mol Biol 1861 (2018) 37–45. [DOI] [PubMed] [Google Scholar]
- [38].Jiang Y, Jiang YY, Xie JJ, Mayakonda A, Hazawa M, Chen L, Xiao JF, Li CQ, Huang ML, Ding LW, Sun QY, Xu L, Kanojia D, Jeitany M, Deng JW, Liao LD, Soukiasian HJ, Berman BP, Hao JJ, Xu LY, Li EM, Wang MR, Bi XG, Lin DC, Koeffler HP, Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression, Nat Commun 9(1) (2018) 3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gast M, Rauch BH, Nakagawa S, Haghikia A, Jasina A, Haas J, Nath N, Jensen L, Stroux A, Bohm A, Friebel J, Rauch U, Skurk C, Blankenberg S, Zeller T, Prasanth KV, Meder B, Kuss A, Landmesser U, Poller W, Immune system-mediated atherosclerosis caused by deficiency of long noncoding RNA MALAT1 in ApoE−/− mice, Cardiovasc Res (2018). [DOI] [PubMed] [Google Scholar]
- [40].Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES, RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites, Cell 159(1) (2014) 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM, Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a, Mol Cell 38(5) (2010) 662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Qiu JJ, Wang Y, Liu YL, Zhang Y, Ding JX, Hua KQ, The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer, Oncotarget 7(22) (2016) 32478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ, Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression, Mol Cancer Ther 14(1) (2015) 268–77. [DOI] [PubMed] [Google Scholar]
- [44].Wang X, Yu H, Sun W, Kong J, Zhang L, Tang J, Wang J, Xu E, Lai M, Zhang H, The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68, Mol Cancer 17(1) (2018) 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A, A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains, BMC Genomics 8 (2007) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lin R, Maeda S, Liu C, Karin M, Edgington TS, A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas, Oncogene 26(6) (2007) 851–8. [DOI] [PubMed] [Google Scholar]
- [47].Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, Ranganathan S, Michalopoulos GK, Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas, Hepatology 44(4) (2006) 1012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C, MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer, Oncogene 22(39) (2003) 8031–41. [DOI] [PubMed] [Google Scholar]
- [49].Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP, Subramanian V, Chen J, Schenone M, Guttman M, Carr SA, Lander ES, The NORAD lncRNA assembles a topoisomerase complex critical for genome stability, Nature 561(7721) (2018) 132–136. [DOI] [PubMed] [Google Scholar]
- [50].Zhang J, Li XY, Hu P, Ding YS, LncRNA NORAD contributes to colorectal cancer progression by inhibition of miR-202–5p, Oncol Res (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, Cheng D, Chen H, Deng X, Peng C, Shen B, Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer, Mol Cancer 16(1) (2017) 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Song H, Xu Y, Shi L, Xu T, Fan R, Cao M, Xu W, Song J, LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability, Biomed Pharmacother 108 (2018) 338–346. [DOI] [PubMed] [Google Scholar]
- [53].Bao J, Chen X, Hou Y, Kang G, Li Q, Xu Y, LncRNA DBH-AS1 facilitates the tumorigenesis of hepatocellular carcinoma by targeting miR-138 via FAK/Src/ERK pathway, Biomed Pharmacother 107 (2018) 824–833. [DOI] [PubMed] [Google Scholar]
- [54].Cao L, Wang Y, Wang Q, Huang J, LncRNA FOXD2-AS1 regulates chondrocyte proliferation in osteoarthritis by acting as a sponge of miR-206 to modulate CCND1 expression, Biomed Pharmacother 106 (2018) 1220–1226. [DOI] [PubMed] [Google Scholar]
- [55].Iacobucci I, Sazzini M, Garagnani P, Ferrari A, Boattini A, Lonetti A, Papayannidis C, Mantovani V, Marasco E, Ottaviani E, Soverini S, Girelli D, Luiselli D, Vignetti M, Baccarani M, Martinelli G, A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia, Leuk Res 35(8) (2011) 1052–9. [DOI] [PubMed] [Google Scholar]
- [56].Sethuraman S, Thomas M, Gay LA, Renne R, Computational analysis of ribonomics datasets identifies long non-coding RNA targets of gamma-herpesviral miRNAs, Nucleic Acids Res 46(16) (2018) 8574–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chaudhary R, Gryder B, Woods WS, Subramanian M, Jones MF, Li XL, Jenkins LM, Shabalina SA, Mo M, Dasso M, Yang Y, Wakefield LM, Zhu Y, Frier SM, Moriarity BS, Prasanth KV, Perez-Pinera P, Lal A, Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pan W, Zhang N, Liu W, Liu J, Zhou L, Liu Y, Yang M, The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8, J Biol Chem (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, Miller MJ, Epah J, Hofmann P, Gunther S, Moll F, Valasarajan C, Heidler J, Ponomareva Y, Freiman TM, Maegdefessel L, Plate KH, Mittelbronn M, Uchida S, Kunne C, Stellos K, Schermuly RT, Weissmann N, Devraj K, Wittig I, Boon RA, Dimmeler S, Pullamsetti SS, Looso M, Miller FJ Jr., Brandes RP, Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function, Circulation 136(1) (2017) 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chen QN, Chen X, Chen ZY, Nie FQ, Wei CC, Ma HW, Wan L, Yan S, Ren SN, Wang ZX, Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression, Mol Cancer 16(1) (2017) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, Hatzigeorgiou AG, DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts, Nucleic Acids Res 44(D1) (2016) D231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hao Y, Wu W, Li H, Yuan J, Luo J, Zhao Y, Chen R, NPInter v3.0: an upgraded database of noncoding RNA-associated interactions, Database (Oxford) 2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yang YC, Di C, Hu B, Zhou M, Liu Y, Song N, Li Y, Umetsu J, Lu ZJ, CLIPdb: a CLIP-seq database for protein-RNA interactions, BMC Genomics 16 (2015) 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li JH, Liu S, Zhou H, Qu LH, Yang JH, starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data, Nucleic Acids Res 42(Database issue) (2014) D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ruhle F, Stoll M, Long non-coding RNA Databases in Cardiovascular Research, Genomics Proteomics Bioinformatics 14(4) (2016) 191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Signal B, Gloss BS, Dinger ME, Computational Approaches for Functional Prediction and Characterisation of Long Noncoding RNAs, Trends Genet 32(10) (2016) 620–637. [DOI] [PubMed] [Google Scholar]
- [67].Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, Zhao L, Li X, Teng X, Sun X, Sun L, Zhang MQ, Chen R, Zhao Y, NONCODEV5: a comprehensive annotation database for long non-coding RNAs, Nucleic Acids Res 46(D1) (2018) D308–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME, lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs, Nucleic Acids Res 43(Database issue) (2015) D168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS, NRED: a database of long noncoding RNA expression, Nucleic Acids Res 37(Database issue) (2009) D122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q, LncRNADisease: a database for long-non-coding RNA-associated diseases, Nucleic Acids Res 41(Database issue) (2013) D983–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhou KR, Liu S, Sun WJ, Zheng LL, Zhou H, Yang JH, Qu LH, ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data, Nucleic Acids Res 45(D1) (2017) D43–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]