Abstract
A small set of ribonucleoside modifications have been found in different regions of mRNA including the open reading frame. Accurate detection of these specific modifications is critical to understanding their modulatory roles in facilitating mRNA maturation, translation and degradation. While transcrip tome-wide next-generation sequencing (NGS) techniques could provide exhaustive information about the sites of one specific or class of modifications at a time, recent investigations strongly indicate cautionary interpretation due to the appearance of false positives. Therefore, it is suggested that NGS-based modification data can only be treated as predicted sites and their existence need to be validated by orthogonal methods. Liquid chromatography-tandem mass spectrometry (LCMS/MS) is an analytical technique that can yield accurate and reproducible information about the qualitative and quantitative characteristics of ribonucleoside modifications. Here, we review the recent advancements in LC-MS/MS technology that could help in securing accurate, gold-standard quality information about the resident post-transcriptional modifications of mRNA.
Keywords: Post-transcriptional modifications, LC-MS, positional isomers, nucleoside analysis, RNA modification mapping
Ribonucleic acids (RNA) play critical roles in regulating the flow of genetic information inside a cell. Ribonucleosides, the building blocks of RNA (adenosine - A, guanosine - G, cytidine - C and uridine – U), store another layer of information in the form of post-transcriptional modifications (PTMs) in almost all types of RNA, including messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), long non-coding RNA (IncRNA), and micro RNA (miRNA). These nucleoside modifications do not change the amino acid sequence of the encoded protein, but can affect the stability, localization, translational accuracy and the function of RNA [1]. More than 160 different kinds of chemically diverse PTMs have been reported in RNA [2] that can potentially impart additional cellular functions. Nucleoside modifications like 7-methylguanosine (m7G)[3], N6-methyladenosine (m6A) and 5-methylcytidine (m5C)[4] were initially reported in protein coding sequences of mRNA in the 1970s [5, 6]. Research into mRNA modifications gained attention with the discovery of specific enzymes, capable of converting adenosines to inosines[7–9]. Recent studies on mRNA and IncRNA have revealed the removable nature of certain modifications, where their functional significance is controlled by three groups of proteins: writers, readers and erasers [10, 11], While writers install these modifications, readers recognize them to determine the cellular fate of transcripts, and erasers remove the modifications. Characterization of the dynamic changes imparted by these proteins (generally referred to as the epitranscriptome[12]), demand accurate and unambiguous determination of the modification location and levels in a given RNA sequence.
1. Detection and identification of modifications
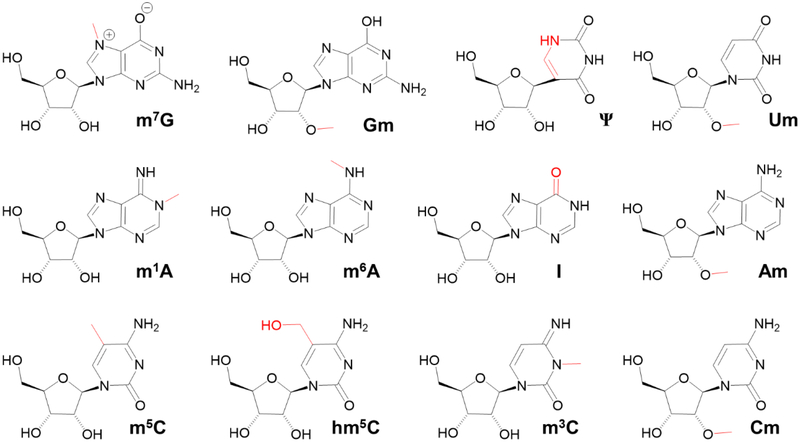
Historically, ribonucleoside modification analysis was done by thin-layer chromatography and high-performance liquid chromatography (HPLC) methods that are coupled with UV-based spectrophotometric detection [13, 14]. However, these methods exhibit low sensitivities, therefore, they are generally applied to the highly abundant modifications. Further, these methods are at best semiquantitative, and do not allow quantification of multiple modifications. The development of direct and indirect methods such as RNA mass spectrometry and next-generation sequencing (NGS) techniques, respectively, have provided powerful tools to identify and map modifications in coding and noncoding RNAs. The NGS is an indirect method operating through the synthesis of a complementary DNA (cDNA) from the transcript. In general, three strategies are combined with NGS techniques for locating modifications in mRNA - point of truncation of a cDNA product during reverse transcription, alteration of the base-pairing properties at the modification site, and enrichment of modified sequences in the transcripts [15]. These types of RNA-seq methods are high throughput but require special sample treatment to recognize the sites of modification. They include mRNA enrichment techniques such as immunoprecipitation [16] and nucleobase-specific chemical derivatization [17] or both. The repertoire of naturally occurring eukaryotic mRNA modifications (besides the 5’ cap[18, 19] and inosine) as reported by NGS technologies include N6 -methyladenosine (m6A), pseudouridine (Ψ), 5-methycytidine (m5C), 5-hydroxymethylcytidine (hm5C), 2’-O-methylated nucleosides (Nm, where N can be any of the four canonical nucleosides) and N1-methyladenosine (m1A)[17, 20–26] (Figure 1). Recently N3-methylcytidine (m3C) has been reported in mRNA of mice and humans with their corresponding writer proteins [27].
Figure 1:
Ribonucleoside modifications of mRNA. m7G – 7-methylguanosine; Gm – 2’-O-methylguanosine; Ψ – pseudouridine; Um – 2’-O-methyluridine; m1A – N1,-methyladenosine; m6A – N6-methyladenosine; I – inosine; Am – 2’-O-methyladenosine; m5C – 5-methylcytidine; hm5C – 5-hydroxymethylcytidine; m3C – 3-methylcytidine; Cm – 2’-O-methylcytidine. The modification group is indicated by red line.
Not surprisingly, die pervasive nature and reportedly higher frequency of nucleoside modifications, especially m6A, m1A, m5C and Nm at various locations of mRNA, is being challenged by other investigations [28, 29]. This is because the indirect high-throughput technologies are prone to false-positive results due to the challenges in distinguishing the signal from noise during transcriptome-wide mapping. Several Nm sites mapped to transcriptomic locations were later found to be artifacts in NGS [30]. Similarly, the number of m5C sites mapped to mRNA were highly variable exhibiting as much as 1,000-fold between studies [31]. Increased noise could also arise due to binding of antibodies to structurally similar nucleotide sequences, antibody cross reactivity (such as m6A antibody against m6Am [32]), leading to inaccurate data generation and interpretation. False positives can also arise due to incomplete derivatization, premature termination of reverse transcription (independent of modification location) and nucleotide misincorporation related artifacts [28–31]. Such data create an erroneous impression of the widespread nature of modifications in the transcriptome. Therefore, a requirement of robust statistical data analysis is suggested to distinguish the signal from noise while analyzing the NGS data [31]. Further, validation of NGS data[15, 28] by an orthogonal method of analysis is also recommended to confirm the predictions of genome-wide mapping approaches. Here, we review the current liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) approaches for identification, quantification and locating the sites of modifications in RNA sequence and their potential applicability to mRNA analysis.
1.1. LC-MS/MS approaches for qualitative analysis of ribonncleoside modifications
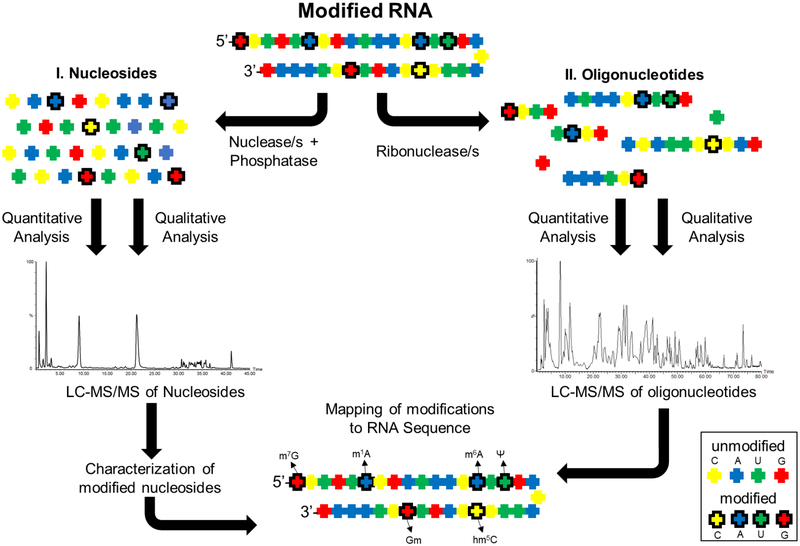
Characterization of modified nucleosides in RNA by LC-MS involve two types of analyses. Initially, the RNA is hydrolyzed to nucleosides (nucleobase linked to the sugar) and the resident modifications are catalogued. Subsequently, the RNA is digested to oligonucleotides and their nucleotide sequences are determined to locate the site of modification. A scheme that illustrates the process of characterization, quantification and mapping of RNA modifications by LC-MS/MS is shown in Figure 2. Both types of analyses involve employment of reversed-phase liquid chromatography (RP-LC). RP-LC resolves molecules based on their hydrophobicity thereby reducing the complexity of sample mixture before mass spectrometric analysis. During nucleoside analysis, modified nucleosides may exhibit varied hydrophobicity depending on the attached chemical group, therefore, they are retained for different times on a reversed-phase column. The separated modified nucleosides are detected by a mass spectrometer connected directly to the liquid chromatography column. The nucleosides are identified by their characteristic mass-to-charge (m/z) values of ionized molecules in the gas phase. The modified nucleosides display a characteristic mass shift compared to canonical nucleoside depending on the attached chemical group. Pseudouridine, which is an isomer of uridine, is an exception and is referred to as a “mass-silent” modification as it does not generate any mass shift. However, the more polar pseudouridine isomer can routinely be distinguished by its short retention time from uridine on reversed-phase liquid chromatographical column. In general, the LC-MS/MS technique presents capabilities to detect both targeted and untargeted nucleoside modifications through direct analysis of RNA without the necessity to convert it to cDNA[33]. Moreover, the LC-MS/MS based detection is mostly based on the physical properties of the molecule, thereby generating high quality and reproducible data.
Figure 2:
Characterization of ribonucleoside modification in RNA by LC-MS/MS analysis. The modified RNA is subjected to nucleosides (I) and oligonucleotide (II) analyses. Total hydrolysis of RNA leads to a mixture of both modified and unmodified nucleosides. Subsequent LC-MS/MS analysis identifies and catalogs the resident modifications. In a second analysis, the RNA is digested with nucleobase-specific ribonucleases resulting in oligonucleotides of varied length. Their nucleotide sequences are determined by different type of LC-MS/MS analysis (see the text) to identify the location of modification. The four colors represent four canonical nucleobases. The bold outline denotes the existence of modification.
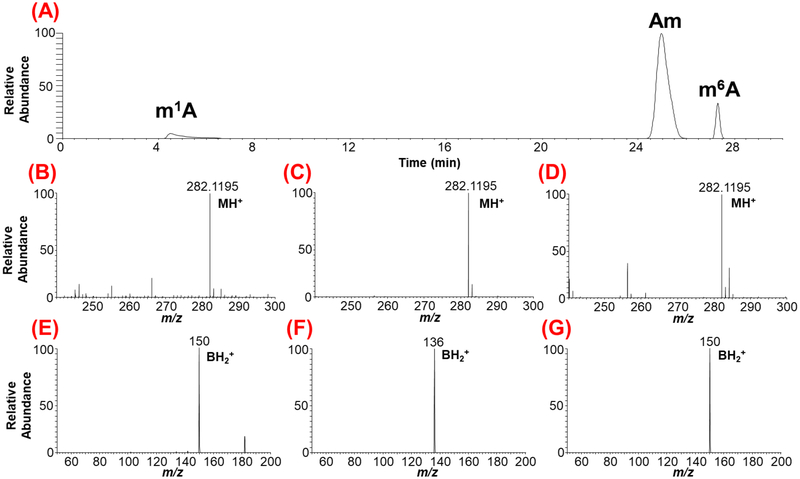
Current protocols of RNA modification detection by LC-MS/MS are largely derived from the work pioneered by the McCloskey lab[34, 35]. In short, the desired RNA is hydrolyzed to nucleosides though nonspecific enzymatic digestion (e.g., nuclease P1, phosphodiesterase I, and phosphatase) before subjecting them to LC-MS/MS analysis. As illustrated in Figure 3, a combination of retention time (RT), m/z values of the molecular (MH+) and the nucleobase (BH2+) ions are used to assign the signal to a specific ribonucleoside modification. The molecular ion refers to the nitrogenous base attached to the ribose sugar by the N-glycosidic bond, and this information is obtained in the first stage of mass spectral analysis. The nucleobase ion is the product of the molecular ion precursor following collision-induced dissociation (CID) of the N-glycosidic bond. Thus, the m/z values of ribonucleoside and nitrogenous base are recorded in the first (MS) and second stages of tandem mass spectrometry (MS/MS) analysis, respectively. In other words, identification of modified nucleosides is generally performed by two-stage mass analysis involving dissociation of the N-glycosidic bond or fragmentation of the nucleoside (Figures 3). The position of modification on nucleobase or ribose sugar can also be monitored by universal cleavage of the N-glycosidic bond that lead to neutral loss of unmodified (132 Da) or methylated (146 Da) ribose. While the majority of modified nucleosides show fragmentation at the N-glycosidic bond, other unique fragmentation patterns are possible {e.g., pseudouridine, and hypermodifications such as queuosine found in tRNA)[36].
Figure 3:
LC-MS/MS-based characterization of the methylated positional isomers of adenosine originating from yeast mRNA. (A) Extracted ion chromatogram (XIC) for m/z 282.1195 corresponding to methylated adenosine is shown. The methylated positional isomers exhibit different retention times depending on their hydrophobicity. (B), (C), (D), depict the mass spectra of chromatographic peaks with retention times at 4.8, 25.8 and 27.5 min, respectively. (E), (F), (G) represent the tandem mass spectra showing the nucleobase ion of molecular precursor ion for a given XIC. Note the differentiation of ribose methylated adenosine (Am) from base methylations (m1A and m6A) through nitrogenous base productc ion. However, the tandem mass spectra for base methylations, (E) and (G) do not distinguish the position of methylation on nitrogenous base as both exhibit identical nucleobase product ion.
The majority of LC-MS/MS-based analysis of RNA modifications have been conducted with tRNAs because of the high frequency of modifications (~ 1 for every 5 residues [37]) and high abundance (4–10%) compared to other cellular RNA [38]. Recent developments in highly sensitive and accurate LC-MS/MS methods could capture a broad range of sample levels from picogram to femtogram of modified nucleoside, thus realizing attomole levels of detection [39–42], However, the detection of a specific modified nucleoside also depends on the relative abundance of the source RNA and its population in the purified sample. Hence, purification protocols are continuously optimized to obtain a true representation of the intracellular RNA in the purified sample [43, 44].
1.2. Differentiation of the positional isomers of modified nucleosides
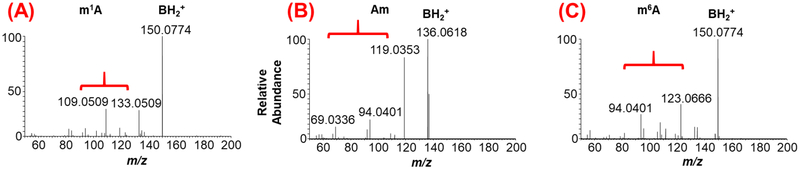
Metliylation, a common base modification, can occur at more than one location on the purine or pyrimidine ring of nucleobase leading to the occurrence of positional isomers, {e.g., m1A, m6A, m2A or m3C, m5C, m4C). These positional isomers exhibit identical m/z values for molecular and nucleobase ions at both stages (MS and MS/MS) of mass spectrometry (Figure 3). This makes it difficult to determine the exact location of the modification within the nucleoside by mass spectrometry information alone. In such cases, chromatographical retention time (RT) information becomes crucial to resolve any ambiguity for identification. RT can show a lack of reproducibility over time due to changes in experimental variables such as mobile phase composition, gradient or column aging. Such variations can be overcome by comparing the chromatographical behavior of a standard (internal or external) with the experimental sample to infer the nature of the eluting isomer. However, chromatographic reproducibility and resolving power are not always achieved for all positional isomers thus, making their accurate identification harder and challenging. To overcome this limitation, Rose et al employed ion mobility-based mass spectrometry, where differences in shape or cross-sectional area (as an additional separation tool) were employed to distinguish the isomers of methylated guanosine[45]. As these positional isomers present different shapes (or different cross-sectional areas), they could be resolved by exploiting the mobility differences of ion current in the ion mobility cell. Another recently discovered way to distinguish the modification isomers is by employing higher-energy collisional dissociation (HCD), an alternative fragmentation technique that generates more informative MS/MS spectra of nucleoside [46]. These studies indicated that the unique HCD fragmentation spectra can serve as fingerprints for each of the positional isomers tested. One such differentiation of m1A from m6A (observed in Figure 3) from yeast mRNA hydrolysate is shown in Figure 4. Because the HCD facilitates fragmentation of the modified nucleobase isomer in alternate pathways depending on the position of modification, the resulting profiles of product ions differ significantly thereby generating a fingerprint specific to the positional isomers. Such fingerprints can be used to identify the specific isomers of modification in the mixture. This type of analysis yields unambiguous isomer-specific information independently of the variations in chromatographic conditions, ion mobility differences, and is less dependent on availability of standards.
Figure 4:
Differentiation of positional isomers of methylated adenosines (m/z 282.12) through molecular fingerprints generated by higher-energy collisional dissociation (HCD) analysis. (A) HCD of m1A indicating the presence of modified nitrogenous base and its fragments. (B) HCD of Am depicting the unmodified adenine and its fragment ions following the loss of methylated ribose. (C) HCD of m6A depicting the modified nitrogenous base and its fragment ions. The fragment ions are indicated by curved parenthesis. Note the molecular fingerprint of nitrogenous base fragment ions is different for each positional isomer.
1.3. Discovery of new modifications by LC-MS/MS
LC-MS/MS is the preferred approach for untargeted identification and discovery of new modifications [41, 47]. The appearance of new modifications can be evaluated further by stable isotope labelling (15N or 13C) of RNA, where the new modifications would exhibit a predictable mass shift due to heavy isotope incorporation [48]. By comparing the gas phase behavior of both labeled and unlabeled nucleosides during LC-MS/MS, the new modification can be confirmed by the predicted mass shift of the labeled nucleosides [49]. The position of modification on nucleobase or ribose sugar can also be monitored by universal cleavage of N-glycosidic bond, which lead to neutral loss of unmodified (132 Da) or methylated (146 Da) ribose [35] in both unlabeled and labeled (with added number of heavy isotope atoms) versions. Such an isotope labeled technique can also be used to monitor the chemical changes to the nucleoside modifications to RNA induced by stress exposure [50].
1.4. Maintaining modification integrity eluting analysis
The position of a modification on the nucleobase can undergo intramolecular alterations outside the cell during sample preparation or LC-MS/MS procedures. For example, the cyclic form of N6-threonylcarbamoyladenosine (ct6A) was observed, when E. coli tRNA hydrolysis was performed under mildly acidic and neutral conditions using nuclease P1 at pH 5.3 and bacterial alkaline phosphatase at pH 7.0[51]. This modification was not observed when hydrolysis was performed sequentially at acidic and basic conditions by the conventional method (nuclease P1 under acidic condition, pH 5.3, and bacterial alkaline phosphatase under basic condition, pH 8.2) [34]. Although such alterations have been noted with hypermodifications of tRNA [52–54], nonoptimal hydrolysis conditions can also affect the resident modifications of mRNA. For instance, under mild basic conditions (e.g., pH ≥8) m1A can be converted to m1I through deamination (our own unpublished observations), and to m6A though Dimroth rearrangement [55].
1.5. Quantification of modified ribonucleosides
Besides imparting qualitative changes, cellular regulatory pathways can alter the levels of known modified ribonucleosides. Documenting those alterations can lead to better understanding of the associated changes in gene expression, as the dynamic changes in modification levels can determine the half-life, turnover and translation efficiency of the transcripts [1, 56–60]. LC-MS/MS approaches can determine the absolute amounts of modifications that vary in response to changes in environmental cues, pathogen attack, stress responses, or any other xenobiotic insults.
Instrument:
The triple quadrupole mass spectrometer (QQQ) is ideally suited for quantification. The first quadrupole mass analyzer (Ql) selects the molecular precursor ion that correspond to the m/z; value of targeted ribonucleoside, which is transmitted to the second quadrupole (or collision cell), Q2. CID of the molecular ion precursor with the neutral gas (typically He, N2 or Ar) leads to formation of nucleobase product ions in the collision cell. Following transmission of the nucleobase ions into the third quadrupole (Q3), only predetermined nucleobase product ion/s, if present, can reach the detector to record the signal. This kind of MS approach is referred to as selected reaction monitoring (SRM) and is more sensitive and specific for a given analyte than other forms of MS-based monitoring approaches [61]. This assay provides specificity at two stages of mass analysis, i.e., selecting the specific molecular ion at Q1 and monitoring the appearance of corresponding nucleobase ion at Q3. Because the observed signal is highly specific to the analyte, the triple quadrupole-based protocol is considered as gold standard technique for quantification.
Quantification method:
The steps associated with LC-MS/MS based quantification include the determination of the important method characteristics, such as the limit of detection, lower and upper limit of quantification and the quantification range for each nucleoside standard. The limit of detection (LOD) is first determined by injecting defined amounts of a nucleoside standard until the observed signal-to-noise ratio (S/N) is at least more than three times the background noise. For quantification purposes, the signal generated by the injected standard should exhibit a S/N of ~ 10, which becomes the lower limit of quantification (LLOQ). Further, a calibration curve is generated from the detector response obtained with specified amounts of nucleoside standards. Such calibration curve defines the limits of linear response of detector for the given amount of nucleoside standard injected on the column. In general, a calibration curve prepared for RNA nucleosides spans at least three orders of magnitude. For example, if the LLOQ is 10 picogram, the upper limit of quantification (ULOQ) of the curve for quantification is around 10 nanograms. In general, 6 to 8 different concentrations of a pure standard are used to generate the calibration curve. Further, the rigor of quantification is improved with the use of stable isotope labeled compounds as internal standard during LC-MS/MS analysis. Use of internal standard reduces systematic error due to changes in injection amount, ionization efficiency and dynamic range issues. Since isotopically labeled standards are not easily available for RNA nucleosides, absolute quantification can be a challenge. One way to circumvent the lack of specific stable isotope labeled internal standard (SIL-IS) is to spike the experimental sample with the RNA derived from an organism that has been grown in media containing a heavy label [49], Although the isotope labeled internal standards provides several advantages for quantification, the relative response of the standard may differ from that of its unlabeled counterpart even though identical amounts are injected on column [62]. This is because the distribution of naturally occurring isotopes can be altered by artificial labeling. This leads to variation in measurement of the number of molecules at each isotopic state, e.g., m+0, m+1, and m+2 between unmodified analyte and manufactured internal standard [62], Such alterations can create inaccuracies as a function of the abundance, type and location of isotopic labeling, when internal standard is used as calibration standard. One way to overcome this problem is through generation of nucleoside-isotope factor for a given stable isotope labeled nucleoside internal standard (SIL-IS) [49, 63]. This factor is calculated by plotting the ratios of area under curve for light isotope and heavy isotope labeled nucleosides. The slope of the linear equation is the relative response factor for the modification of interest [49, 64]. Subsequently, the absolute amount of modified nucleoside can be computed from the ratio of modified nucleoside signal, and die product relative response factor and the SIL-IS. Here the quantification (either absolute or relative) is carried out by a combination of external calibration and sample spiking with biosynthetic SIL-IS. The validated SIL-IS can provide precise quantification with relative standard deviations ≤2%.
2. Sequencing modified nucleosides in RNA
Nucleoside analysis, either qualitative or quantitative, can provide important information about the presence (or absence) of modifications within a given RNA sample. However, what cannot be gleaned from nucleoside analysis is the exact location of that modification within the RNA sequence. Thus, nucleoside analysis alone will not provide information that could relate to changes in individual RNA sequence modification patterns. To be able to place modified nucleoside to a known RNA sequence, a different analytical methodology is required. One approach, referred to as RNase fingerprinting was developed in 1965, where the RNA sequence was determined using a combination of ribonuclease-mediated RNA digests followed by gel electrophoretic separation [65]. The gel electrophoresis technique can also be combined with the use of reverse transcriptase (RT enzyme), where the enzyme undergoes premature termination at the site of modification revealing the site of modification without necessarily identifying the resident modification. This kind of termination was expected due to the dissociation of enzyme while attempting to transcribe through chemically bulky modifications on RNA [66].
RT enzyme-based methods are recently being combined with high throughput NGS to locate specific modifications on RNA sequence at single nucleotide resolution. Though the same limitations of RT enzyme-based sequencing persist (poor quantification, false positives and single type or single class of modification-specific experimental protocols), the high-throughput nature of this technique has enabled a sensitive, transcriptome-wide analysis of PTMs [21, 23, 67]. A comprehensive description of NGS and its experimental considerations is outside the scope of this article and has been addressed elsewhere[15, 68]. However, these high-throughput detection techniques are error prone to varying degrees, which could lead to over-interpretation of data for a given biological situation. Therefore, the predicted sites in big data are postulated to be treated as candidate sites until they are confirmed by additional method to validate the predicted modification sites in a given RNA [15]. As described before, LC-MS/MS performs direct analysis of RNA without converting it into cDNA intermediate, where the modified nucleosides are detected by their inherent physicochemical properties, and thus can serve as validation method to confirm the modification sites in the mRNA species.
2.1. LC-MS based RNA modification mapping
The direct detection and sequencing of the RNA modifications by LC-MS/MS was made possible by the pioneering work of McLuckey[69, 70] and McCloskey[71, 72] in 1990s. Prior treatment of RNA with nucleobase-specific ribonuclease generates oligonucleotide lengths that are amenable to sequencing by mass spectrometer, a process referred to as RNA modification mapping [71]. The discovery of a chromatographic system suitable to oligonucleotide analysis, which uses an ion-pair reagent and pH modifier[73] in the mobile phase, dramatically improved the effectiveness and utility of the method, ushering in the next (and still current) generation of RNA modification analysis by LC-MS/MS. Tliis approach would later provide the tools for relative quantification of RNA modifications[74, 75] (see below), an application not readily achievable with RT enzyme-based methods. It should be noted that for the purposes of the current discussion, there are two major types of modified RNA samples that are routinely analyzed for modifications - RNAs prepared synthetically {e.g., small interfering RNA - siRNA), and RNAs isolated from biological origins (e.g., rRNA, tRNA, mRNA). A discussion of the analysis of synthetic (and therapeutic) oligonucleotides will not be covered here but has been recently summarized [76]. The application of LC-MS/MS to the analysis of biologically derived RNAs that participate in gene expression control will be addressed within.
2.1.1. Bottom-Up Modification Mapping
The most commonly applied RNA modification mapping approach involves isolation of the target RNA (rRNA, tRNA, etc.) and its subsequent enzymatic hydrolysis to generate oligonucleotides that are amenable to LC-MS/MS analysis. The nucleotide sequences of oligonucleotide precursor ions are deciphered from the observed product ions resulting from characteristic cleavage of the phosphodiester bond during tandem mass spectrometry. Of the four bonds available for cleavage in the phosphodiester linkage, the P-O bond is preferentially broken (in RNA) to yield a ladder of sequence informative fragment ions that share the common 5’-end (cn-ion series) or 3’-end (yn-ion series). The sequence of the oligonucleotide including the potential modification will be deciphered from this tandem mass spectrum. Thus, the intact modified RNA sequence is reconstructed from the sequences of oligonucleotide digestion products in a bottom-up approach. The most commonly used enzymes for bottom-up mapping are RNase T1 (cleaves at guanosine) and RNase A (cleaves at pyrimidines). The selection of the enzyme is an important consideration for bottom-up mapping, because generation of the fewest digestion products (highest number of oligonucleotides unique to one sequence in digested mixtures) minimizes redundant or non-sequence specific digestion products. It is ideal, if the RNA sample is subjected to more than one nucleobase-specific enzyme to strive for total sequence coverage through the generation of unique sets of complementary and overlapping digestion products. Therefore, the development of new nucleobase-specific enzymes is an important aspect of research for labs using a bottom-up approach. Recently, our lab has identified enzymes that improve sequence coverage, which include RNase U2 (unmodified purines with preference for adenosine) [77], RNase MCI (uridine specific) [78], and RNase cusativin (cytidine specific) [79]. Alternatively, a set of overlapping digestion products may be generated through partial cleavage of target RNA and subsequent sequencing in tandem mass spectrometry to reconstruct the whole sequence [80].
2.1.2. Mapping pseudourìdine locations
Though detection during nucleoside analysis has become routine, pseudouridine is isobaric (same exact mass) with uridine and is therefore “mass-silent” in LC-MS/MS-based mapping experiments. To determine the location of pseudouridine, a selective derivatization must be performed prior to the analysis. The chemical derivatization incorporates a unique mass shift that can easily be resolved by mass spectrometry. Two commonly employed approaches are the use of N-Cyclohexyl-N′-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMCT) [81] and acrylonitrile based cyanoethylation [82], which react selectively with pseudouridine (and not uridine) resulting in a detectable mass shift (+252.2 Da for carbodiimide addition and +53.0 Da for cyanoethylation) for each pseudouridine residue.
2.1.3. Chromatography of Oligonucleotides
The oligonucleotide digestion products resulting from enzymatic hydrolysis of RNA are best analyzed by a hyphenated technique involving liquid chromatography coupled with mass spectrometry (LC-MS). Such platform includes ion-pairing reversed-phase chromatography (IP-RP-LC)[73], where a hydrophobic stationary phase (e.g., octadecyl or C18 carbon chain bound to silica) is used in combination with a mobile phase containing an ion-pairing reagent (triethylamine or TEA) and volatile pH modifier (hexafluoro-2-propanol or HFIP). The presence of the ion pair reagent is necessary to obtain sufficient retention of the very polar phosphodiester containing digestion products on the hydrophobic surface of the stationary phase. The presence of HFIP aids in efficient transfer of oligonucleotides from the liquid phase to the gas phase during electrospray ionization (ESI). In spite of multiple efforts by the Bartlett group [76, 83, 84] toward further optimization, tuning of various ion-pair reagents and mobile phase modifiers for the analysis of synthetic oligonucleotides, the TEA/HFIP system remains the most commonly used approach for bottom-up mapping [41, 85]. Other types of less hydrophobic stationary phases have been evaluated [86–88] to alleviate the use of ion-pair reagents, which are known to suppress ionization efficiency during ESI process, but these chromatographic systems have yet to reach the overall performance of currently practiced IP-RP-LC.
In contrast to the enzymatic hydrolysis of RNA mixture such as total tRNA, the Suzuki group has developed a chromatographic system whereby several individual RNAs can be isolated in a series of steps. Referred to as chaplet chromatography [89], the experimental setup involves sequence-specific affinity columns containing biotinylated DNA immobilized on a solid support. To capture RNA, the mixture of RNAs is circulated through a series of columns to achieve hybridization of each target RNA to the complementary sequences at high temperature. Each individual column is then separated, and the captured sequence released from the probe. A further modification of this technique, referred to as reciprocal circulating chromatography (RCC)[90], is employed using immobilized DNA probes on pipette types to obtain similar outcome. Such a technology could be applicable to mRNA pool, where a gene-specific mRNA could be purified or enriched in a single chaplet column, eluted, digested either online or offline with a ribonuclease before subjecting it to LC-MS analysis.
2.1.4. Mass Spectrometry
As the oligonucleotides elute from the chromatographic system they are transferred from liquid phase to gas phase through ESI in the first stage of mass analysis. Here, the m/z value and charge state (number of protons lost) of oligonucleotide anion is computed. In the second stage, this oligonucleotide anion is isolated within the mass spectrometer using a quadrupole or ion trap and subsequently subjected to CID process as described above. The resulting fragment or product ions provide the tandem mass spectrum of the oligonucleotide precursor anion. This tandem mass spectrum provides a ladder of sequence informative product ions that can be assigned using the McLuckey nomenclature as c-, y-, w-, and a-B-type product ions [69, 70]. The mass differences observed between the sequential c- and y-type fragment ions (through cleavage of P-O bond in the phosphodiester backbone) will reveal the identity and location of canonical and modified nucleosides and help reconstruction of nucleotide sequence of oligomer. Since the generation of product ions from precursor ions follow a predictable and reproducible pattern for RNA[72], the MS/MS spectrum can then be interpreted manually by the analyst or through a software. A representative example of locating the RNA modification in the RNase T1 digest of E. colì total tRNA is illustrated in Figure 5. A rigorous review of the gas phase dissociation of oligonucleotides is outside of the scope of this review but has been given elsewhere [91].
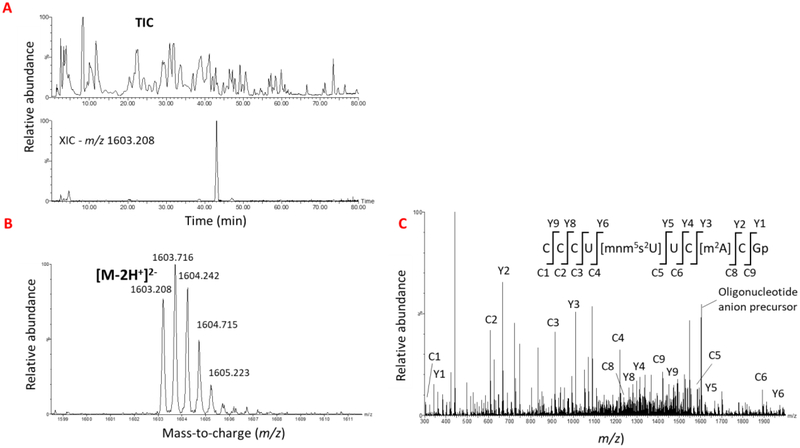
Figure 5:
A typical LC-MS/MS-based sequencing of modified oligonucleotides to identify the location of modification. (A). The total ion Chromatogram (TIC) of Rnase T1 digest of E. coli total tRNA representing the elution pattern of all the ions coming from a reverse phase column is shown. The bottom panel shows the XIC for m/z 1603.208 corresponding to oligonucleotide, CCCU[mnm5s2U]UC[m2A]CGp from tRNAGlu is shown. (B) oligonucleotide precursor ion observed in the mass spectrum of the XIC peak is depicted. (C) Tandem mass spectrum of product ions generated by CID from oligonucleotide precursor is shown. The sequence informative fragment ions that bear the common 5’-end (cn ion series) or 3’-end (yn ion series) of oligonucleotide are labeled on the sequence. The vertical lines represent the points of phosphodiester cleavage leading to formation of c and y product ion series.
Although this kind of modification mapping is quite effective, overall experimental throughput is most often limited by the complexity of the mass spectra generated in CID-based MS/MS experiments, and sequence annotation (interpretation of MS/MS spectra to the original sequence) limitations. A number of computational tools including the very first tool, simple oligonucleotide sequencer (SOS) [92], oligonucleotide mass assembler (OMA) and oligonucleotide peak analyzer (OPA) software tool box[93] and RoboOligo [94] that can simplify the MS/MS data interpretation have been developed. Subsequently, the computational platforms that can perform RNA modification mapping such as web-based Ariadne which uses MS/MS data to search against the sequence database to identify specific RNAs [95], and RNA mass mapper (RMM) that can search prokaryotic genomes or RNA FASTA sequence database [96] have been made. The recent newly developed RNAModMapper (RAMM) can accomplish both the MS/MS data interpretation and sequence annotation. Here, the CID data of oligonucleotides is interpreted, and the interpreted sequence mapped to full-length RNA [97] by a single platform. Such a computational platform will be useful to interpret MS/MS data of mRNA where the sequences are expected to be long (>1000 nt) and expected to have lower density of modifications.
2.1.5. Qualitative analysis of oligonucleotides
The primary goal of any qualitative RNA modification mapping is to obtain the maximum sequence coverage of modified RNA target. Ideally, this would include the identification of all modified and unmodified digestion products resulting in the placement of modifications detected from the nucleoside analysis onto the RNA sequence of interest (Figure 2). The power and utility of the bottom-up approach has been shown over the years for a variety of RNAs. The most significant contributions have come from a handful of research groups, particularly the labs of McCloskey, Limbach and Suzuki. Some of the selected highlights over the years include the determination of the 5’-cap structures of mRNA from T. brucei and C. fasciculata [98], the modification maps of the small subunit (SSU) rRNA of T. thermophilus [99] and T. maritima [100], tRNAs in L. lactis [101], and mitochondrial tRNAs in B. taurus [102]. Additionally, the detection and localization of more recently discovered tRNA modifications - agmatidine in H. marismortu[103], ct6A in E. coli[104] and ms2ct6A in B. subtilis and T. brucei [105] were facilitated by LC-MS/MS analysis.
An alternative approach to the characterization of modification in RNAs is the comparative analysis of one or more RNAs where modification status is probed in an unknown RNA by comparing its digest directly against the known RNA digest. This comparative approach (known as CARD or comparative analysis of RNA digests) [106] is most applicable in comparing wild-type versus mutant strains, or healthy versus diseased sample populations. Another comparative analysis approach uses metabolic isotopic labeling [107], Such an approach can help differentiating the modification status of highly similar mRNA sequences or mRNA sequences that exhibit altered modification profiles.
2.1.6. Quantitative analysis of oligonucleotides
While most studies performed to date have been qualitative, progress has also been made in the relative quantification of modified oligonucleotides. The enzymatic incorporation of stable isotope labels into RNase digestion products can be used not only to aid the identification of modifications, but also aid in the relative quantification of RNAs. The use of isotopic labels can improve and simplify the interpretation of oligonucleotide MS/MS spectra and provide relative quantification of RNAs [108], where information about changes in abundance is desired. Isotope labeling can also be introduced into the culture media, allowing for the relative quantification of modifications present in 16S rRNA [107] and during the assembly of the 16S and 23S rRNAs of E. colì [109]. More recently this technique was adapted to incorporate isotopic labels into an in vitro transcribed reference RNA which allowed detection and quantification of all PTMs in S.pombe rRNA [110]. All these approaches, if applied, can reveal information about the changes in abundance of modifications at specific locations in mRNA.
2.2. Modification mapping by top-down approaches
As an alternative to the bottom-up approach, a top-down strategy that does not involve the enzymatic hydrolysis of the RNA (analogous to top-down proteomics[111]) has seen recent use in both qualitative and quantitative mapping of modifications of various types of RNA. Top-down applications of RNA modification mapping involved samples containing single and double-stranded siRNA[112] and miRNA[113]. Significant contributions to the field of top-down RNA modification mapping have come from the Breuker group with tRNA[114] and synthetic RNA [115] and allowed to map “tat” protein binding sites in HIV-1 TAR RNA during CID-based mass spectrometric analysis [116]. This approach is typically performed without prior chromatographic separation and therefore requires a pure, single-species of RNA sequence and specialized mass spectrometers (with features of high mass accuracy, multiple gas phase dissociation techniques, and ion-ion reactions) to obtain complete sequence coverage. Nevertheless, a top down approach is limited to analysis of synthetic mRNA, where the sequence does not exceed few hundred nucleotides.
3. Challenges and potential strategies to make LC-MS applicable to mRNA modification mapping
Although LC-MS/MS has matured into an accurate and reliable means of generating high quality data for the identification and sequence mapping of RNA modifications, those studies are mostly limited to abundant cellular RNAs such as rRNA and tRNA. Such RNAs exhibit higher density of modifications. Studies on mRNA modifications, so far, have been limited to NGS-based RNA-seq approaches where RNA is converted to cDNA leading to loss of modification information. However, as described above the NGS data is mostly predictive due to the prevalence of false positives. On the other hand, the LC-MS/MS analysis require higher sample input which is a limitation considering the low percentage of mRNA (<1%) in the total pool of cellular RNA Therefore, the current detection limits afforded by LC-MS/MS require significant enrichment and purification of high quality mRNA transcripts for obtaining accurate information. To move the field of mRNA modification mapping by LC-MS/MS forward - particularly with the goal of improving sequence coverage for low abundant RNA species - a combination of enhanced sample preparation protocols, higher chromatographic peak capacities, and high mass accuracy and lowered MS detection limits are needed.
Potential strategies to mitigate the challenges could start with development of innovative sample preparation procedures. The primary difference between prokaryotic and eukaryotic mRNA is that the former lacks a poly (A) tail in the mature mRNA. This poly (A) tail provides a specific and effective capture target for enrichment of mRNA in eukaryotic organisms and is used as the primary means of isolation in several commercially available mRNA extraction kits. Technical improvements in the purification specificity, efficiency and mRNA yield need to be made for eukaryotic systems. This is because rRNA-specific modifications are still detected even after two rounds of poly (A) purification and small and large RNA depletion steps [31]. The lack of the poly(A) tail in bacterial and archaeal systems makes the isolation of mRNA in prokaryotic organisms even more difficult. Several different techniques have been demonstrated for the enrichment of mRNA from prokaryotes, including rRNA capture and removal [117], selective degradation of processed RNA (Epicenter mRNA Only Kit), selective adenylation of mRNAs [118] and immunoprecipitation using an antibody of regulatory proteins [119].
Enrichment of mRNA that harbors the modification can be one way to reduce the sample complexity and increase the signal from modified digestion product. Antibody-based immunoprecipitation of mRNA for a targeted modification such as m6A[12, 20] could pave the way for enrichment of a sub pool of mRNA for specific modification. Such a strategy can provide enough sample amount for performing LC-MS/MS analysis. Once the successful enrichment of the target sequence is obtained, the selection of appropriate nucleobase-specific ribonucleases can generate longer digestion products whose sequence can be uniquely mapped to specific mRNA.
4. Improvement of LC-MS/MS-based detection of oligonucleotides
Although it is an active and ever-maturing field of study, very little has changed fundamentally over the last 20 years in the way LC-MS/MS analysis of oligonucleotides is carried out. Therefore, technical improvements in LC-MS/MS methodology is also needed to improve the viability of mRNA modification mapping. Ion suppression due to the necessity of ion-pair reagents in liquid chromatography, signal splitting due to cation adduction and the presence of more than one charge state per oligonucleotide has a major impact on the detection limits of the technique. Improvements of an order of magnitude or more on the current limits of detection (approx. 2–5 ng or approx. 50–150 fmol for single RNA[102]) would be most welcome. The development of different electrospray and/or mobile phase additives that reduce the number of charge states, improve ion abundance, and still provide sufficient chromatographic peak capacity will be helpful. Additionally, continuing fundamental research into the development of new and novel chromatographic retention mechanisms that alleviate the necessity of ion-paif reagent use could help in reaching that goal.
To continue to improve the accuracy and throughput of oligonucleotide data processing, the use of state-of-the-art high mass-accuracy and maximal duty cycle (parallel instrument operations for efficient analysis) mass spectrometers (FT-ICR and Orbitrap configurations in particular) is a very important experimental consideration. As the accuracy and precision of measured masses for both precursor and product ions increase, the number of possible elemental compositions and thereby the oligonucleotides that correspond to the measured mass decrease — which can reduce computational time and false positive rates during data processing. High duty cycle of the mass spectrometer can conduct parallel operations to meet with continuously changing oligonucleotide analyte population of the chromatographic eluent.
Due to the low density of modifications in mRNA, bottom-up modification mapping would generate many unmodified and redundant digestion products that could interfere with the detection of the modified digestion products. Because of their sheer numbers, these unmodified oligonucleotides will predominate and overwhelm the MS analysis. Therefore, the mass spectrometer would end up spending more time on acquiring spectra of less useful unmodified oligonucleotides than spending time on the oligonucleotides that contain the modifications. By excluding these unmodified oligonucleotides from sequence-informative fragmentation pathways, the mass spectrometer can be tuned to spend more time on modified oligonucleotides to obtain high quality sequence information and provide high sequence coverage. Thus, the use of an exclusion list approach[120], where the unmodified digestion products are excluded from MS/MS analysis, can increase the efficiency of analysis of modified oligonucleotides thereby improving the coverage in mRNA modification mapping experiments. The introduction of another, preferably orthogonal, separation medium (e.g., two dimensional LC and ion mobility) could also provide an important means of improving analytical peak capacity with a positive impact on total sequence coverage. Such an exclusionary approach and capacity to acquire direct information of modified oligonucleotides based on the physicochemical properties of resident modifications is not available with RNA-seq type of NGS approaches. Thus, the LC-MS can be a unique, ideal option and an appropriate orthogonal method for validation of the predicted NGS sites of modifications.
In summary, recent improvements in LC-MS/MS based nucleoside analysis make an ideal platform for unambiguous detection of nucleoside modifications in mRNA. LC-MS/MS-based mRNA modification mapping, however, require further improvements in methodology associated with mRNA purification, sample preparation, liquid chromatography techniques. Combining the improved methodologies with the available state-of-the-art mass spectrometers that provide high mass accuracy and maximal duty cycle can facilitate accurate qualitative and quantitative analysis besides validating the NGS predicted sites of modified mRNA oligonucleotides.
Acknowledgements
The authors appreciate the critical reading and valuable suggestions offered by Prof. Patrick Limbach during manuscript preparation, and thank the financial support extended by National Institutes of Health (NIGMS R01 058843 and OD OD018485 to Patrick Limbach), REU program award (to B Williams of NSF) and the University of Cincinnati.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest with this manuscript.
References
- [1].Hoernes TP, Huttenhofer A, Erlacher MD, mRNA modifications: Dynamic regulators of gene expression?, RNA Biol, 13 (2016) 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM, MODOMICS: a database of RNA modification pathways. 2017 update, Nucleic Acids Research, 46 (2018) D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shatkin AJ, Capping of eucaryotic mRNAs, Cell, 9 (1976) 645–653. [DOI] [PubMed] [Google Scholar]
- [4].Dubin DT, Stollar V, Methylation of Sindbis virus “26S” messenger RNA, Biochemical and biophysical research communications, 66 (1975) 1373–1379. [DOI] [PubMed] [Google Scholar]
- [5].Desrosiers R, Friderici K, Rottman F, Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells, Proceedings of the National Academy of Sciences of the United States of America, 71 (1974) 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dubin DT, Taylor RH, The methylation state of poly A-containing messenger RNA from cultured hamster cells, Nucleic Acids Research, 2 (1975) 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K, Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing, Proceedings of the National Academy of Sciences of the United States of America, 91 (1994) 11457–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Benne R, RNA editing. The long and the short of it, Nature, 380 (1996) 391–392. [DOI] [PubMed] [Google Scholar]
- [9].Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M, A mammalian RNA editing enzyme, Nature, 379 (1996) 460–464. [DOI] [PubMed] [Google Scholar]
- [10].Roignant JY, Soller M, m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression, Trends in genetics: TIG, 33 (2017) 380–390. [DOI] [PubMed] [Google Scholar]
- [11].Meyer KD, Jaffrey SR, The dynamic epitranscriptome: N6-methyladenosine and gene expression control, Nature reviews. Molecular cell biology, 15 (2014) 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR, Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons, Cell, 149 (2012) 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Grosjean H, Droogmans L, Roovers M, Keith G, Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates, Methods in enzymology, 425 (2007) 55–101. [DOI] [PubMed] [Google Scholar]
- [14].Kohrer C, Rajbhandary UL, The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases, Methods, 44 (2008) 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Helm M, Motorin Y, Detecting RNA modifications in the epitranscriptome: predict and validate, Nature reviews. Genetics, 18 (2017) 275–291. [DOI] [PubMed] [Google Scholar]
- [16].Wang CY, Lin MH, Su HT, A Method for Measuring RNA N 6-methyladenosine Modifications in Cells and Tissues, Journal of visualized experiments: JoVE, (2016) 54672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV, Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells, Nature, 515 (2014) 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramanathan A, Robb GB, Chan SH, mRNA capping: biological functions and applications, Nucleic Acids Res, 44 (2016) 7511–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Warminski M, Sikorski PJ, Kowalska J, Jemielity J, Applications of Phosphate Modification and Labeling to Study (m)RNA Caps, Topics in current chemistry (Cham), 375 (2017) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G, Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq, Nature, 485 (2012) 201–206. [DOI] [PubMed] [Google Scholar]
- [21].Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T, Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA, Nucleic Acids Res, 40 (2012) 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Dore LC, Amariglio N, Rechavi G, He C, The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA, Nature, 530 (2016) 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C, Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome, Nat Chem Biol, 12 (2016) 311–316. [DOI] [PubMed] [Google Scholar]
- [24].Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R, Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs, PLoS genetics, 9 (2013) e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, Wetzel C, Kreher J, Soin R, Creppe C, Limbach PA, Gueydan C, Kruys V, Brehm A, Minakhina S, Defrance M, Steward R, Fuks F, RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine, Science (New York, N.Y.), 351 (2016) 282–285. [DOI] [PubMed] [Google Scholar]
- [26].Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, Dominissini D, He C, Nmseq maps 2’-O-methylation sites in human mRNA with base precision, Nat Methods, 14 (2017) 695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, Fu XY, Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans, The Journal of biological chemistry, 292 (2017) 14695–14703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Grozhik AV, Jaffrey SR, Distinguishing RNA modifications from noise in epitranscriptome maps, Nat Chem Biol, 14 (2018) 215–225. [DOI] [PubMed] [Google Scholar]
- [29].Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S, The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution, Nature, 551 (2017) 251–255. [DOI] [PubMed] [Google Scholar]
- [30].Gillen AE, Yamamoto TM, Kline E, Hesselberth JR, Kabos P, Improvements to the HITS-CLIP protocol eliminate widespread mispriming artifacts, BMC genomics, 17 (2016) 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M, Lyko F, Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs, Genome research, 27 (2017) 1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Munns TW, Oberst RJ, Sims HF, Liszewski MK, Antibody-nucleic acid complexes. Immunospecific recognition of 7-methylguanine- and N6-methyladenine-containing 5’-terminal oligonucleotides of mRNA, The Journal of biological chemistry, 254 (1979) 4327–4330. [PubMed] [Google Scholar]
- [33].Alfonzo JD, Post-transcriptional modifications are very important after all, RNA Biology, 11 (2014) 1481–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Crain PF, [42] Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry, Methods in Enzymology, Academic Press, Place Published, 1990, pp. 782–790. [DOI] [PubMed] [Google Scholar]
- [35].Pomerantz SC, McCloskey JA, [44] Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry, Methods in Enzymology, Academic Press, Place Published, 1990, pp. 796–824. [DOI] [PubMed] [Google Scholar]
- [36].Ross R, Cao X, Yu N, Limbach PA, Sequence mapping of transfer RNA chemical modifications by liquid chromatography tandem mass spectrometry, Methods (San Diego, Calif.), 107 (2016) 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jackman JE, Alfonzo JD, Transfer RNA modifications: Nature’s combinatorial chemistry playground, Wiley interdisciplinary reviews. RNA, 4 (2013) 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kirchner S, Ignatova Z, Emerging roles of tRNA in adaptive translation, signalling dynamics and disease, Nature reviews. Genetics, 16 (2015) 98–112. [DOI] [PubMed] [Google Scholar]
- [39].Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, Ng CS, Pang YLJ, Prestwich EG, Lim KS, Ramesh Babu I, Begley TJ, Dedon PC, Chapter Three - A Platform for Discovery and Quantification of Modified Ribonucleosides in RNA: Application to Stress-Induced Reprogramming of tRNA Modifications, in: He C (Ed.) Methods in Enzymology, Academic Press, Place Published, 2015, pp. 29–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dudley E, Bond L, Mass spectrometry analysis of nucleosides and nucleotides, Mass Spectrometry Reviews, 33 (2013) 302–331. [DOI] [PubMed] [Google Scholar]
- [41].Wetzel C, Limbach PA, Mass spectrometry of modified RNAs: recent developments, Analyst, 141 (2016) 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Basanta-Sanchez M, Temple S, Ansari SA, D’Amico A, Agris PF, Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells, Nucleic Acids Research, 44 (2016) e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chionh YH, Ho C-H, Pruksakorn D, Ramesh Babu I, Ng CS, Hia F, McBee ME, Su D, Pang YLJ, Gu C, Dong H, Prestwich EG, Shi P-Y, Preiser PR, Alonso S, Dedon PC, A multidimensional platform for the purification of non-coding RNA species, Nucleic Acids Research, 41 (2013) e168–e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M, Lyko F, Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs, Genome Research, 27 (2017) 1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rose RE, Quinn R, Sayre JL, Fabris D, Profiling ribonucleotide modifications at full-transcriptome level: a step toward MS-based epitranscriptomics, RNA, 21 (2015) 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jora M, Burns AP, Ross RL, Lobue PA, Zhao R, Palumbo CM, Beal PA, Addepalli B, Limbach PA, Differentiating Positional Isomers of Nucleoside Modifications by Higher-Energy Collisional Dissociation Mass Spectrometry (HCD MS), J Am Soc Mass Spectrom, 29 (2018) 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Thuring K, Schmid K, Keller P, Helm M, Analysis of RNA modifications by liquid chromatography-tandem mass spectrometry, Methods, 107 (2016) 48–56. [DOI] [PubMed] [Google Scholar]
- [48].Bruckl T, Globisch D, Wagner M, Muller M, Carell T, Parallel isotope-based quantification of modified tRNA nucleosides, Angew Chem Int Ed Engl, 48 (2009) 7932–7934. [DOI] [PubMed] [Google Scholar]
- [49].Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M, Profiling of RNA modifications by multiplexed stable isotope labelling, Chemical Communications, 50 (2014) 3516–3518. [DOI] [PubMed] [Google Scholar]
- [50].Sun C, Jora M, Solivio B, Limbach PA, Addepalli B, The Effects of Ultraviolet Radiation on Nucleoside Modifications in RNA, ACS Chem Biol, 13 (2018) 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Miyauchi K, Kimura S, Suzuki T, A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification, Nature Chemical Biology, 9 (2012) 105. [DOI] [PubMed] [Google Scholar]
- [52].Matuszewski M, Wojciechowski J, Miyauchi K, Gdaniec Z, Wolf WM, Suzuki T, Sochacka E, A hydantoin isoform of cyclic N(6)-threonylcarbamoyladenosine (ct(6)A) is present in tRNAs, Nucleic Acids Research, 45 (2017) 2137–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kang B.-i., Miyauchi K, Matuszewski M, D’Almeida GS, Rubio Mary Anne T., Alfonzo JD, Inoue K, Sakaguchi Y, Suzuki T, Sochacka E, Suzuki T, Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs, Nucleic Acids Research, 45 (2017) 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nagao A, Ohara M, Miyauchi K, Yokobori S.-i., Yamagishi A, Watanabe K, Suzuki T, Hydroxylation of a conserved tRNA modification establishes non-universal genetic code in echinoderm mitochondria, Nature Structural &Amp; Molecular Biology, 24 (2017) 778. [DOI] [PubMed] [Google Scholar]
- [55].Macon JB, Wolfenden R, 1-Methyladenosine. Dimroth rearrangement and reversible reduction, Biochemistry, 7 (1968) 3453–3458. [DOI] [PubMed] [Google Scholar]
- [56].Agris PF, Narendran A, Sarachan K, Vare VYP, Eruysal E, The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity, The Enzymes, 41 (2017) 1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen K, Zhao Boxuan S., He C, Nucleic Acid Modifications in Regulation of Gene Expression, Cell Chemical Biology, 23 (2016) 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Engel M, Chen A, The emerging role of mRNA methylation in normal and pathological behavior, Genes, brain, and behavior, 17 (2018) e12428. [DOI] [PubMed] [Google Scholar]
- [59].Harcourt EM, Kietrys AM, Kool ET, Chemical and structural effects of base modifications in messenger RNA, Nature, 541 (2017) 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hoernes TP, Erlacher MD, Methylated mRNA Nucleotides as Regulators for Ribosomal Translation, Methods in molecular biology (Clifton, N.J.), 1562 (2017) 283–294. [DOI] [PubMed] [Google Scholar]
- [61].Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham A-JL, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander Å, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA, Multi-site assessment of the precision and reproducibility of multiple reaction monitoring–based measurements of proteins in plasma, Nature Biotechnology, 27 (2009) 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rappold BA, Hoofnagle AN, Bias due to isotopic incorporation in both relative and absolute protein quantitation with carbon-13 and nitrogen-15 labeled peptides, Clinical Mass Spectrometry, 3 (2017) 13–21. [Google Scholar]
- [63].Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M, Absolute and relative quantification of RNA modifications via biosynthetic isotopomers, Nucleic Acids Res, 42 (2014) e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thuring K, Schmid K, Keller P, Helm M, LC-MS Analysis of Methylated RNA, Methods in molecular biology (Clifton, N.J.), 1562 (2017) 3–18. [DOI] [PubMed] [Google Scholar]
- [65].Sanger F, Brownlee GG, Barrell BG, A two-dimensional fractionation procedure for radioactive nucleotides, J Mol Biol, 13 (1965) 373–398. [DOI] [PubMed] [Google Scholar]
- [66].Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR, Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses, Proceedings of the National Academy of Sciences of the United States of America, 82 (1985) 6955–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, Regev A, Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA, Cell, 159 (2014) 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Schwartz S, Motorin Y, Next-generation sequencing technologies for detection of modified nucleotides in RNAs, RNA Biol, 14 (2017) 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].McLuckey SA, Habibi-Goudarzi S, Ion trap tandem mass spectrometry applied to small multiply charged oligonucleotides with a modified base, J Am Soc Mass Spectrom, 5 (1994) 740–747. [DOI] [PubMed] [Google Scholar]
- [70].McLuckey SA, Van Berkel GJ, Glish GL, Tandem mass spectrometry of small, multiply charged oligonucleotides, J Am Soc Mass Spectrom, 3 (1992) 60–70. [DOI] [PubMed] [Google Scholar]
- [71].Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA, A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry, Nucleic Acids Research, 21 (1993) 4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ni J, Pomerantz C, Rozenski J, Zhang Y, McCloskey JA, Interpretation of oligonucleotide mass spectra for determination of sequence using electrospray ionization and tandem mass spectrometry, Anal Chem, 68 (1996) 1989–1999. [DOI] [PubMed] [Google Scholar]
- [73].Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS, Analysis of Oligonucleotides by HPLCElectrospray Ionization Mass Spectrometry, Anal Chem, 69 (1997) 1320–1325. [DOI] [PubMed] [Google Scholar]
- [74].Gaston KW, Limbach PA, The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry, RNA Biology, 11 (2014) 1568–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Paulines MJ, Limbach PA, Stable Isotope Labeling for Improved Comparative Analysis of RNA Digests by Mass Spectrometry, J Am Soc Mass Spectrom, 28 (2017) 551–561. [DOI] [PubMed] [Google Scholar]
- [76].Basiri B, Bartlett MG, LC-MS of oligonucleotides: applications in biomedical research, Bioanalysis, 6 (2014) 1525–1542. [DOI] [PubMed] [Google Scholar]
- [77].Houser WM, Butterer A, Addepalli B, Limbach PA, Combining recombinant ribonuclease U2 and protein phosphatase for RNA modification mapping by liquid chromatography-mass spectrometry, Anal Biochem, 478 (2015) 52–58. [DOI] [PubMed] [Google Scholar]
- [78].Addepalli B, Lesner NP, Limbach PA, Detection of RNA nucleoside modifications with the uridine-specific ribonuclease MC1 from Momordica charantia, Rna, 21 (2015) 1746–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Addepalli B, Venus S, Thakur P, Limbach PA, Novel ribonuclease activity of cusativin from Cucumis sativus for mapping nucleoside modifications in RNA, Anal Bioanal Chem, 409 (2017) 5645–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Solivio B, Yu N, Addepalli B, Limbach PA, Improving RNA modification mapping sequence coverage by LC-MS through a nonspecific RNase U2-E49A mutant, Analytica Chimica Acta, (2018) (In Press) 10.1016/j.aca.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Patteson KG, Rodicio LP, Limbach PA, Identification of the mass-silent post-transcriptionally modified nucleoside pseudouridine in RNA by matrix-assisted laser desorption/ionization mass spectrometry, Nucleic Acids Res, 29 (2001) E49–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mengel-Jorgensen J, Kirpekar F, Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry, Nucleic Acids Res, 30 (2002) e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Basiri B, Murph MM, Bartlett MG, Assessing the Interplay between the Physicochemical Parameters of Ion-Pairing Reagents and the Analyte Sequence on the Electrospray Desorption Process for Oligonucleotides, J Am Soc Mass Spectrom, 28 (2017) 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Basiri B, van Hattum H, van Dongen WD, Murph MM, Bartlett MG, The Role of Fluorinated Alcohols as Mobile Phase Modifiers for LC-MS Analysis of Oligonucleotides, J Am Soc Mass Spectrom, 28 (2017) 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ross RL, Cao X, Limbach PA, Mapping Post-Transcriptional Modifications onto Transfer Ribonucleic Acid Sequences by Liquid Chromatography Tandem Mass Spectrometry, Biomolecules, 7 (2017) 21. [Google Scholar]
- [86].Gong L, McCullagh JS, Analysis of oligonucleotides by hydrophilic interaction liquid chromatography coupled to negative ion electrospray ionization mass spectrometry, J Chromatogr A, 1218 (2011) 5480–5486. [DOI] [PubMed] [Google Scholar]
- [87].Li L, Leone T, Foley JP, Welch CJ, Separation of small interfering RNA stereoisomers using reversed-phase ion-pairing chromatography, J Chromatogr A, 1500 (2017) 84–88. [DOI] [PubMed] [Google Scholar]
- [88].Studzinska S, Lobodzinski F, Buszewski B, Application of hydrophilic interaction liquid chromatography coupled with mass spectrometry in the analysis of phosphorothioate oligonucleotides in serum, J Chromatogr B Analyt Technol Biomed Life Sci, 1040 (2017) 282–288. [DOI] [PubMed] [Google Scholar]
- [89].Suzuki T, Suzuki T, Chaplet column chromatography: isolation of a large set of individual RNAs in a single step, Methods in enzymology, 425 (2007) 231–239. [DOI] [PubMed] [Google Scholar]
- [90].Miyauchi K, Ohara T, Suzuki T, Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method, Nucleic Acids Res, 35 (2007) e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Schurch S, Characterization of nucleic acids by tandem mass spectrometry - The second decade (2004–2013): From DNA to RNA and modified sequences, Mass Spectrom Rev, 35 (2016) 483–523. [DOI] [PubMed] [Google Scholar]
- [92].Rozenski J, McCloskey JA, SOS: a simple interactive program for ab initio oligonucleotide sequencing by mass spectrometry, J Am Soc Mass Spectrom, 13 (2002) 200–203. [DOI] [PubMed] [Google Scholar]
- [93].Nyakas A, Blum LC, Stucki SR, Reymond JL, Schurch S, OMA and OPA--software-supported mass spectra analysis of native and modified nucleic acids, J Am Soc Mass Spectrom, 24 (2013) 249–256. [DOI] [PubMed] [Google Scholar]
- [94].Sample PJ, Gaston KW, Alfonzo JD, Limbach PA, RoboOligo: software for mass spectrometry data to support manual and de novo sequencing of post-transcriptionally modified ribonucleic acids, Nucleic Acids Res, 43 (2015) e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Nakayama H, Akiyama M, Taoka M, Yamauchi Y, Nobe Y, Ishikawa H, Takahashi N, Isobe T, Ariadne: a database search engine for identification and chemical analysis of RNA using tandem mass spectrometry data, Nucleic Acids Res, 37 (2009) e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Matthiesen R, Kirpekar F, Identification of RNA molecules by specific enzyme digestion and mass spectrometry: software for and implementation of RNA mass mapping, Nucleic Acids Res, 37 (2009) e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Yu N, Lobue PA, Cao X, Limbach PA, RNAModMapper: RNA Modification Mapping Software for Analysis of Liquid Chromatography Tandem Mass Spectrometry Data, Anal Chem, 89 (2017) 10744–10752. [DOI] [PubMed] [Google Scholar]
- [98].Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC, Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides, The Journal of biological chemistry, 267 (1992) 9805–9815. [PubMed] [Google Scholar]
- [99].Guymon R, Pomerantz SC, Crain PF, McCloskey JA, Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus, Biochemistry, 45 (2006) 4888–4899. [DOI] [PubMed] [Google Scholar]
- [100].Guymon R, Pomerantz SC, Ison JN, Crain PF, McCloskey JA, Post-transcriptional modifications in the small subunit ribosomal RNA from Thermotoga maritima, including presence of a novel modified cytidine, RNA, 13 (2007) 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Puri P, Wetzel C, Saffert P, Gaston KW, Russell SP, Cordero Varela JA, van der Vlies P, Zhang G, Limbach PA, Ignatova Z, Poolman B, Systematic identification of tRNAome and its dynamics in Lactococcus lactis, Mol Microbiol, 93 (2014) 944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Suzuki T, Suzuki T, A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs, Nucleic Acids Res, 42 (2014) 7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Mandal D, Kohrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Soll D, RajBhandary UL, Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine, Proceedings of the National Academy of Sciences of the United States of America, 107 (2010) 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Miyauchi K, Kimura S, Suzuki T, A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification, Nat Chem Biol, 9 (2013) 105–111. [DOI] [PubMed] [Google Scholar]
- [105].Kang B, Miyauchi K, Matuszewski M, D’Almeida GS, Rubio MAT, Alfonzo JD, Inoue K, Sakaguchi Y, Suzuki T, Sochacka E, Suzuki T, Identification of 2-methylthio cyclic N-6-threonylcarbamoyladenosine (ms(2)ct(6)A) as a novel RNA modification at position 37 of tRNAs, Nucleic Acids Research, 45 (2017) 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Li S, Limbach PA, Mass spectrometry sequencing of transfer ribonucleic acids by the comparative analysis of RNA digests (CARD) approach, Analyst, 138 (2013) 1386–1394. [DOI] [PubMed] [Google Scholar]
- [107].Waghmare SP, Dickman MJ, Characterization and quantification of RNA post-transcriptional modifications using stable isotope labeling of RNA in conjunction with mass spectrometry analysis, Anal Chem, 83 (2011) 4894–4901. [DOI] [PubMed] [Google Scholar]
- [108].Castleberry CM, Limbach PA, Relative quantitation of transfer RNAs using liquid chromatography mass spectrometry and signature digestion products, Nucleic Acids Res, 38 (2010) e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Popova AM, Williamson JR, Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry, J Am Chem Soc, 136 (2014) 2058–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Taoka M, Nobe Y, Hori M, Takeuchi A, Masaki S, Yamauchi Y, Nakayama H, Takahashi N, Isobe T, A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: the complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs, Nucleic Acids Res, 43 (2015) e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Cui W, Rohrs HW, Gross ML, Top-down mass spectrometry: recent developments, applications and perspectives, Analyst, 136 (2011) 3854–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Huang TY, Liu J, Liang X, Hodges BD, McLuckey SA, Collision-induced dissociation of intact duplex and single-stranded siRNA anions, Anal Chem, 80 (2008) 8501–8508. [DOI] [PubMed] [Google Scholar]
- [113].Nakayama H, Yamauchi Y, Taoka M, Isobe T, Direct identification of human cellular microRNAs by nanoflow liquid chromatography-high-resolution tandem mass spectrometry and database searching, Anal Chem, 87 (2015) 2884–2891. [DOI] [PubMed] [Google Scholar]
- [114].Taucher M, Breuker K, Characterization of modified RNA by top-down mass spectrometry, Angew Chem Int Ed Engl, 51 (2012) 11289–11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Glasner H, Riml C, Micura R, Breuker K, Label-free, direct localization and relative quantitation of the RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass spectrometry, Nucleic Acids Res, 45 (2017) 8014–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Schneeberger EM, Breuker K, Native Top-Down Mass Spectrometry of TAR RNA in Complexes with a Wild-Type tat Peptide for Binding Site Mapping, Angew Chem Int Ed Engl, 56 (2017) 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, Joint I, Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities, PLoS One, 3 (2008) e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Wendisch VF, Zimmer DP, Khodursky A, Peter B, Cozzarelli N, Kustu S, Isolation of Escherichia coli mRNA and comparison of expression using mRNA and total RNA on DNA microarrays, Anal Biochem, 290 (2001) 205–213. [DOI] [PubMed] [Google Scholar]
- [119].Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J, Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq, PLoS genetics, 4 (2008) e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Cao X, Limbach PA, Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LC-MS/MS, Anal Chem, 87 (2015) 8433–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]