Abstract
The nuclear lamins A, B, and C are intermediate filament proteins that form a nuclear scaffold adjacent to the inner nuclear membrane in higher eukaryotes, providing structural support for the nucleus. In the past two decades it has become evident that the final step in the biogenesis of the mature lamin A from its precursor prelamin A by the zinc metalloprotease ZMPSTE24 plays a critical role in human health. Defects in prelamin A processing by ZMPSTE24 result in premature aging disorders including Hutchinson Gilford Progeria Syndrome (HGPS) and related progeroid diseases. Additional evidence suggests that defects in prelamin A processing, due to diminished ZMPSTE24 expression or activity, may also drive normal physiological aging. Because of the important connection between prelamin A processing and human aging, there is increasing interest in how ZMPSTE24 specifically recognizes and cleaves its substrate prelamin A, encoded by LMNA. Here, we describe two humanized yeast systems we have recently developed to examine ZMPSTE24 processing of prelamin A. These systems differ from one another slightly. Version 1.0 is optimized to analyze ZMPSTE24 mutations, including disease alleles that may affect the function or stability of the protease. Using this system, we previously showed that some ZMPSTE24 disease alleles that affect stability can be rescued by the proteasome inhibitor bortezomib, which may have therapeutic implications. Version 2.0 is designed to analyze LMNA mutations at or near the ZMPSTE24 processing site to assess whether they permit or impede prelamin A processing. Together these systems offer powerful methodology to study ZMPSTE24 disease alleles and to dissect the specific residues and features of the lamin A tail that are required for recognition and cleavage by the ZMPSTE24 protease.
1. Introduction
Maturation of the nuclear scaffold protein lamin A from its precursor prelamin A by the integral membrane zinc metalloprotease ZMPSTE24 is critical for human health and longevity. This knowledge comes from understanding the molecular mechanisms of several rare diseases characterized by premature aging phenotypes. Mutations in LMNA that impede prelamin A cleavage cause the premature aging disorder Hutchinson-Gilford Progeria Syndrome (HGPS) [1–5]. The related progeroid diseases mandibuloacral dysplasia-type B (MAD-B) and restrictive dermopathy (RD) map to ZMPSTE24 and diminish or ablate its activity, respectively [6–11]. These findings have generated considerable interest in determining the specific requirements of prelamin A processing by ZMPEST24.
The complete biogenesis pathway of mature lamin A from its precursor, prelamin A, involves four enzymatic steps. Prelamin A terminates with a C-terminal CAAX motif (where “C” is cysteine, “A” is generally an aliphatic amino acid and “X” is any amino acid other than proline). The CAAX motif directs the post-translational processing steps collectively called “CAAX processing” in which the cysteine sulfhydryl is farnesylated, followed by endoproteolytic cleavage of the -AAX tripeptide, and carboxyl methylation of the farnesylated cysteine residue [8, 12–14]. What distinguishes prelamin A from other CAAX proteins is that following these processing steps, prelamin A undergoes an additional cleavage mediated by ZMPSTE24 (Fig. 1A, B) [15, 16]. Failure to perform this final cleavage results in accumulation of permanently farnesylated and carboxyl methylated prelamin A. This molecule causes aberrant nuclear morphology and is the culprit in progeroid diseases [2, 5, 17].
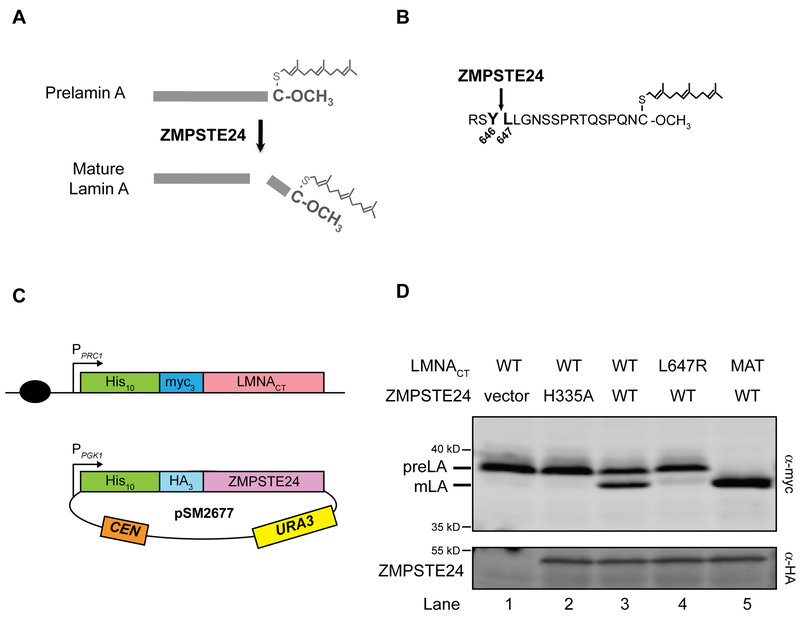
Figure 1. The “humanized yeast system” version 1.0 to assay ZMPSTE24 cleavage of prelamin A.
(A) Farnesylated and carboxylmethylated prelamin A is proteolytically cleaved by ZMPSTE24 to yield mature lamin A. ZMPSTE24 cleaves off the C-terminal 15 amino acids of prelamin A, including the modified cysteine. (B). The C-terminal residues of prelamin A are shown, including those residues flanking the cleavage site between residues 646 and 647 (bold). (C) Schematic of the “humanized yeast system”. (Top) An N-terminal 10His-3myc-tagged prelamin A substrate (LMNACT) expressed from the PRC1 promoter (pPRC1) is chromosomally integrated into the ste24Δ strain SM6158 at the TRP1 locus (black circle indicates a chromosomal centromere). (Bottom) This strain is transformed with a low-copy centromeric (CEN) plasmid expressing 10His-3HA tagged human ZMPSTE24 driven by the PGK1 promoter (pSM2677). (D) Prelamin A cleavage follows the same rules observed in mammalian cells. Lysates of cells transformed with the indicated ZMPSTE24 plasmid variant and containing the indicated chromosomally integrated LMNACT constructs were resolved by SDS-PAGE and subjected to western blotting with anti-myc and anti-HA antibodies. Prelamin A (preLA) and mature lamin A (mLA) run at ~39 and 37 kd, respectively, and ZMPSTE24 runs at ~53 kd. “MAT” is an artificially constructed version of mature lamin A, which terminates at residue Y646, and is used as a size marker. Approximately 50–70% of prelamin is cleaved to mature lamin A in this system, as exemplified in lane 3. Transformed strains in lanes 1–5 (expressed as strain name/plasmid name) and quantification of percentage of cleavage in parentheses are SM6158/pRS316 (1.5%), SM6158/pSM2673 (1.3%), SM6158/pSM2677 (53.4%), SM6177/pSM2677 (1.7%) and SM6178/pSM2677 (99.7%), respectively.
The best studied of these diseases is HGPS, which results from a dominant splicing mutation in LMNA that activates a cryptic splice donor site. This aberrant splicing results in a 50 amino acid deletion in prelamin A that includes the ZMPSTE24 processing site [3, 4]. By one year of age, children with HGPS manifest accelerated aging symptoms, including failure to thrive, lipodystrophy, hair loss, joint ailments and cardiovascular disease. Aggressive and early-onset atherosclerosis develops and these children typically die in their mid-teens from heart attack or stroke secondary to their atherosclerosis [5, 18].
Mutations in ZMPSTE24 cause recessive diseases that share many features with HGPS. Restrictive dermopathy (RD) is a neonatal lethal disease with earlier onset and greater severity than HGPS, while mandibuloacral dysplasia-type B (MAD-B) develops later in life with milder symptoms. RD patient mutations ablate ZMPSTE24 function while ZMPSTE24 proteins with MAD-B mutations exhibit some residual function [7, 11, 13]. Together, these premature aging diseases have called attention to the lack of ZMPSTE24-mediated processing of prelamin A to mature lamin A as the basis of progeroid disorders. Importantly, additional evidence suggests that defects in prelamin A processing due to diminished ZMPSTE24 expression may also drive normal physiological aging [19], emphasizing the significance of deciphering ZMPSTE24’s proteolytic mechanism, including its prelamin A substrate specificity and the regulation of its activity and stability.
Our studies in Saccharomyces cerevisiae laid the foundation for understanding progeroid disorders. We discovered the yeast homolog of ZMPSTE24, called Ste24, in a screen for defective yeast mating, and we showed that it is required for the biogenesis of the mating pheromone a-factor. Furthermore, we demonstrated that human ZMPSTE24 can substitute for its yeast homolog to mediate a-factor processing in a ste24Δ yeast strain [7, 20, 21]. Ultimately we showed that ZMPSTE24 mediates the final step of prelamin A biogenesis in higher eukaryotes (yeast does not encode prelamin A, nor any nuclear lamins) by demonstrating that prelamin A accumulates in MEFs derived from a Zmpste24−/− mouse [15, 22]. Interestingly we have shown that yeast Ste24 and human ZMPSTE24 can also remove the -AAX from some CAAX motifs, although another integral membrane protease, RCE1, is functionally redundant with ZMPSTE24/Ste24 for CAAX cleavage [20, 21, 23]. Notably, ZMPSTE24 is unique in its ability to perform the final and critically important prelamin A maturation step (shown in Fig. 1A and B). In the humanized yeast system discussed below, yeast Ste24 is also able to mediate prelamin A cleavage [13].
Yeast and human Ste24/ZMPSTE24 define a new family of zinc metalloproteases. They are integral membrane proteins with seven transmembrane spans and localize to both the inner nuclear membrane as well as the endoplasmic reticulum membrane [24–26]. The recently solved X-ray crystal structures of human ZMPSTE24, and that of the virtually superimposable yeast Ste24, reveal a completely novel and fascinating class of protease [27–29]. The seven transmembrane spans comprise a helical barrel that surrounds a voluminous intramembrane “hollow” chamber, large enough to accommodate ~450 water molecules. Surprisingly, the HEXXH active site conserved among all zinc metalloproteases does not face the cytosol or nucleoplasm, but rather resides inside of this barrel in ZMPSTE24/Ste24. Side portals apparent in the structure are likely to provide access into the chamber interior for prelamin A and a-factor proteolytic cleavage.
Interestingly, the only known substrates of ZMPSTE24 and Ste24 are prelamin A in higher eukaryotes and a-factor in yeast. However recent studies shed light on additional roles for ZMPSTE24/Ste24. Ast et al. have shown a role for Ste24/ZMPSTE24 in a specialized type of protein quality control in the endoplasmic reticulum, involving clearance of translocationally-stalled polypeptides stuck in the Sec61 translocon [30]. Evidence for such a role was further extended by the demonstration that Ste24/ZMPSTE24 “declogging” activity may protect cells against oligomer-induced toxicity in a yeast model of pancreatic beta cell dysfunction, which occurs due to the aggregation of the human islet amyloid polypeptide (IAPP) in the pancreas of patients with Type 2 diabetes [31]. In addition, ZMPSTE24 plays a role in the defense against influenza and other pathogenic enveloped viruses, by an as yet unknown mechanism [32].
The ZMPSTE24/Ste24 structures raise a number of interesting and important questions, including how substrate specificity and access are mediated, how prelamin A is positioned for cleavages, and the role of ZMPSTE24’s large and unusual membrane-embedded hydrophilic chamber for catalysis [8, 28, 29]. Because of the importance of ZMPSTE24 in human health and disease and its novel structure, we felt it would be advantageous to have a high-throughput system to probe structure-function relationships for this protease or its prelamin A substrate. To do so, we recently have reported on a fully humanized Saccharomyces cerevisiae system in which the final cleavage of human prelamin A by human ZMPSTE24 can be assessed in yeast and the activity and in vivo stability of disease or synthetic alleles of ZMPSTE24 can be compared to WT [13]. This system (referred to as version 1.0 here) revealed that some mutant alleles of ZMPSTE24 affect solely its prelamin A processing activity, some affect mainly protein stability in vivo, and some affect both. We showed that one disease allele, P248L, is substantially correctable for prelamin A cleavage when protected from degradation by a proteasome inhibitor drug in yeast. This finding may have important therapeutic implications for individuals with this mutation if similar results are observed in patient cells.
Here we describe in detail two major variations of the humanized yeast system that can be used to analyze prelamin A cleavage. The first is version 1.0, referred to above, that is designed to analyze mutant versions of the enzyme ZMPSTE24. The other, version 2.0, is optimized to analyze mutant versions of the substrate prelamin A, encoded by LMNA. We also provide an example of how known ZMPSTE24 mutations and new mutations, along with computational modeling can begin to provide insights into a better understanding of membrane protein structure and function.
2. Methods
Strains and plasmids used are listed in Tables 1 and 2. All plasmids were constructed using standard molecular biology techniques, including NEB HiFi Assembly and verified by restriction digestion and DNA sequence analysis. When applicable, mutagenic oligonucleotides containing desired changes were used during PCR and subsequent plasmid assembly.
Table 1.
Yeast strains used in this study
| Straind | Genotype | Reference |
|---|---|---|
| SM4826 | ste24::KanMX met15Δ0 his3Δ1 leu2Δ0 ura3Δ0 Mata | Deletion collection |
| SM6158 | ste24::KanMX met15Δ0 his3Δ1 leu2Δ0 ura3Δ0 Mata TRP1::NatMX-PPRC1-10His-3myc-LMNA(431–664) Mata | [13] |
| SM6177 | ste24::KanMXmet15Δ0 his3Δ1 leu2Δ0 ura3Δ0 Mata TRP1::NatMX-PPRC1-10His-3myc-LMNA(431–664, L647R) Mata | [13] |
| SM6178 | ste24::KanMX met15Δ0 his3Δ1 leu2Δ0 ura3Δ0 Mata TRP1::NatMX-PPRC1-10His-3myc-LMNA(431–646) Mata | [13] |
| SM6302 | ste24::KanMX met15Δ0 his3Δ1 leu2Δ0 ura3Δ0 Mata TRP1::NatMX-PPGK1-10His-3HA-ZMPSTE24codon optimized (1X) | This study |
| SM6303 | ste24::KanMX met15Δ0 his3Δ1 leu2Δ0 ura3Δ0 Mata TRP1::NatMX-PPGK1-10His-3HA-ZMPSTE24codon optimized (2X) | This study |
Table 2.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pSM174 | pRS316 (CEN, URA3) | [48] |
| pSM2673 | pRS316::PPGK1-10His-3HA-zmpste24H335A | [7] |
| pSM2676 | pRS316::PPGK1-10His-3HA-zmpste24P248L | [7] |
| pSM2677 | pRS316::PPGK1-10His-3HA-ZMPSTE24 | [7] |
| pSM2982 | pRS316::PPGK1-10His-3HA-zmpste24L94P | [7] |
| pSM3094 | pRS316::PPGK1-10His-3HA-STE24 | [13] |
| pSM3186 | pRS316::PPGK1-10His-3HA-zmpste24Y399C | [13] |
| pSM3173 | pRS304::NatMX-PPRC1-10His-3myc-LMNA(431–664) | [13] |
| pSM3177 | pRS304::NatMX-PPRC1-10His-3myc-LMNA(431–664, L647R) | [13] |
| pSM3178 | pRS304::NatMX-PPRC1-10His-3myc-LMNA(431–646) | [13] |
| pSM3377 | pRS316::PPGK1-10His-3HA-zmpste24L94A | This study |
| pSM3378 | pRS316::PPGK1-10His-3HA-zmpste24P248A | This study |
| pSM3379 | pRS316::PPGK1-10His-3HA-zmpste24Y399A | This study |
| pSM3175 | pRS316::PPGK1-10His-3HA-Zmpste24(mouse) | This study |
| pSM3202 | pRS316::PPGK1-10His-3HA-ZMPSTE24 (human, codon-optimized) | This study |
| pSM3371 | pRS315::PPRC1-10His-3myc-LMNA(431–664) | This study |
| pSM3391 | pRS313::PPRC1-10His-3myc-LMNA(431–664) | This study |
| pSM3392 | pRS313::PPRC1-10His-3myc-LMNA(431–664, L647R) | This study |
| pSM3428 | pRS304::NatMX-PPGK1-10His-3HA-ZMPSTE24 (human, codon optimized) | This study |
| pSM3512 | pRS315::PPRC1-10His-3myc-LMNA(431–664, C661S) | This study |
| pSM3513 | pRS315::PPRC1-10His-3myc-LMNA(431–664, L647R) | This study |
As described above, two versions of the “humanized yeast system” were developed to examine prelamin A cleavage by ZMPSTE24 in yeast. For version 1.0, used to analyze ZMPSTE24 mutants introduced on plasmids, the LMNACT substrate, uncleavable (L647R), or mature (Y646X) mutant variants, were chromosomally integrated into the TRP1 locus (Fig. 1). To do so, the yeast integrating plasmids pSM3173, pSM3177 and pSM3178 were linearized by EcoRV enzyme digestion and transformed into ste24Δ (SM4826) cells by the standard lithium acetate transformation method. Transformants were selected on YPD plates containing 100 μg/ml nourseothricin. The resultant strains express chromosomally integrated WT, uncleavable, and mature 10His-3myc-LMNACT under the control of the yeast PRC1 promoter and are designated SM6158 (LMNA431–664 WT), SM6177 (LMNA431–664, L647R) and SM6178 (LMNA431–646), respectively. These strains can be transformed with low-copy CEN URA3 plasmids harboring 10His-3HA-tagged human ZMPSTE24 or yeast STE24, by selecting on SC-Uracil minimal medium. In general, strain SM6158 is used for the analysis of WT and mutant forms of ZMPSTE24 [13], and strains SM6177 and SM6178 provide controls for the migration of prelamin A and mature lamin A.
The second system, version 2.0 is used for the analysis of LMNA mutations. In this case, ZMPSTE24 is chromosomally integrated and LMNA variants are introduced on plasmids. Strains with chromosomal ZMPSTE24 were created by transforming EcoRV-digested pSM3428 into the ste24Δ (SM4826) strain, selecting transformants on nourseothricin. This transformation yielded two strains, SM6302 and SM6303, which contain respectively one or two copies of yeast codon-optimized ZMPSTE24 under control of the yeast PGK1 promoter, as determined by quantitation of ZMPSTE24. Generally for the version 2.0 system, CEN plasmids bearing WT or mutant 10His-3myc-LMNACT variants are transformed into strain SM6303, which has two copies of ZMPSTE24 integrated at the TRP1 locus, by selecting on SC-leucine or SC-histidine medium, as appropriate.
To grow cells, prepare lysates, and perform Westerns to examine steady-state cleavage of prelamin A, as well ZMPSTE24 protein levels in both versions of the humanized yeast system, the following procedures are used: Yeast strains are grown overnight in minimal synthetic complete dropout medium (SC-uracil, leucine, or histidine as appropriate to select for a plasmid), back-diluted in fresh medium to an OD600 of ~0.3–0.5, and grown for 4–6 hours at 30°C. Cells (1.5–2 OD600 units) are harvested by centrifugation in microfuge tubes at 21k × g for 2 minutes. Detergent lysates are made by the method described in Kushnirov [33], where cells are treated for 5–10 minutes in 0.1M NaOH, briefly pelleted, and then resuspended in 1X Laemmli sample buffer (2% SDS, 10% glycerol, 80mM Tris, pH6.8, 5% 2-mercaptoethanol) and heated at 65°C for 10 minutes. Lysates are vortexed and centrifuged at 21k × g for 2 minutes to pellet insoluble debris. Approximately 0.2–0.3 OD600 cell equivalents are resolved on 10% SDS polyacrylamide gels. Quantitative Western blotting is performed using an Odyssey CLx digital fluorescence scanner (LI-COR). For loading controls we probe for either hexokinase or Sec61.
Proteins are transferred to nitrocellulose (Bio-Rad Trans-blot Turbo) and the membrane is blocked using a 1:10 dilution (in phosphate buffered saline containing 0.1% Tween-20) Western Blocking Reagent (Roche). Lamin proteins are first detected using mouse anti-myc antibodies (clone 4A6, Millipore; 1:10,000 dilution) and decorated with goat anti-mouse secondary IRDye 680RD antibodies (LI-COR). Blots are re-probed using rat anti-HA (clone 3F10, Roche;1:10,000 dilution) to detect ZMPSTE24/Ste24 and rabbit anti-Sec61 (1:10,000 dilution, kindly provided by Dr. Randy Schekman, UC, Berkeley, CA), or rabbit anti-hexokinase (1:200,000 dilution) as loading controls, and then decorated with goat anti-rat IRDye 680RD and goat anti-rabbit IRDye 800CW secondary antibodies (LI-COR).
Prelamin A cleavage is calculated using ImageStudio Lite (LI-COR) by quantifying mature lamin A signal compared with total myc signal (prelamin A plus mature lamin A). ZMPSTE24 protein levels are quantified by measuring the HA signal in the entire region that contains both ZMPSTE24 bands and the intervening smear and normalizing this signal to the loading control signal.
Our computational analysis was performed using a protocol adapted from the Rosetta ΔΔG protocol with flexible backbones described by Barlow and coworkers [34,35]: Starting with the crystal structure of ZMPSTE24 (2ypt) [29], we applied the ΔΔG protocol to generate refined model of both the native and mutated proteins. Each model is an ensemble of 50 structures representing the most likely conformations. Then, we calculated the change in stability using the Rosetta all-atom energy function [36, 37]. Rosetta calculates macromolecular energies as a linear combination of terms that evaluate van der Waals, electrostatics, solvation, backbone, and side chain energies. Here, we computed the ΔΔGmut(X → Y) as the difference between the average score of the mutant conformation ensemble and the native conformation ensemble (Eq. 1).
Further, we evaluated the contribution of individual energies to the ΔΔGmut(X → Y) using PyRosetta tools described in [38].
3. Results
3.1. The humanized yeast system version 1.0 to assay ZMPSTE24 mutant alleles.
The yeast system we use to analyze the activity and stability of mutant alleles of ZMPSTE24 is schematically shown (Fig 1C). This system is designated version 1.0 and was used in our recent study of ZMPSTE24 disease alleles [13]. It features a strain with a chromosomally integrated LMNA substrate, into which WT or mutant versions of ZMPSTE24 are introduced on a low copy-number plasmid and assessed for cleavage activity. Several considerations and experimental trials helped to develop this system, as discussed below.
In this system, an epitope-tagged C-terminal segment of LMNA (10His-3myc-LMNACT; referred to as LMNACT) is chromosomally integrated in a strain deleted for the yeast ZMPSTE24 homolog STE24 (ste24Δ), generating strain SM6158 (Fig. 1C). The LMNA construct contains the C-terminal “lamin A tail” (amino acids 431–664) [39, 40]. It lacks the coiled-coil region of lamin A that mediates dimer and polymer formation and the nuclear localization signal, since the presence of these domains causes a yeast growth defect and they are not needed for ZMPSTE24 processing (unpublished observations and [41]). The LMNACT construct is expressed from the PRC1 promoter. While many yeast studies utilize strong promoters, including the inducible GAL1 or constitutive TDH3 promoters, to drive expression of heterologous proteins, we found that use of the weaker PRC1 promoter allowed for the most optimal cleavage of LMNACT, ensuring that ZMPSTE24 activity is in excess of the substrate. For the protease, epitope-tagged human ZMPSTE24, constitutively expressed from the strong yeast PGK1 promoter, is present on a low copy-number centromeric (CEN) plasmid (Fig. 1C).
To assay cleavage, plasmids containing WT (pSM2677) or mutant versions of ZMPSTE24 are transformed into strain SM6158, purified transformants are grown in liquid media that selects for plasmid retention, extracts are prepared, and then analyzed by 10% SDS-PAGE and Western blotting using a digital fluorescence scanner. Anti-Myc antibodies detect the precursor and mature forms of lamin A and anti-HA antibodies detect ZMPSTE24, as shown in Figure 1, panel D. (See Materials and Methods for details).
Prelamin A processing in this humanized yeast system version 1.0 follows the same rules as in mammalian cells [13]. Notably, we observe between 50–70% cleavage of prelamin A to lamin A using plasmid-encoded WT human ZMPSTE24 (Fig .1D, lane 3). The steady-state processing efficiency for WT and mutant forms of ZMPSTE24 is defined as the amount of mature lamin A divided by the total (prelamin A and mature lamin A). As in mammalian cells, processing does not occur when ZMPSTE24 is absent or harbors a catalytically dead mutation H335A (Fig. 1D, lanes 1 and 2, respectively). Nor can ZMPSTE24 in yeast efficiently cleave a mutant form of LMNA, L647R, with a mutation just C-terminal to the cleavage site in prelamin A known to render it uncleavable in mammalian cells (Fig. 1D, lane 4) [41–44].
3.2. ZMPSTE24 proteins from several species process the human prelamin A substrate encoded by LMNACT in our yeast system.
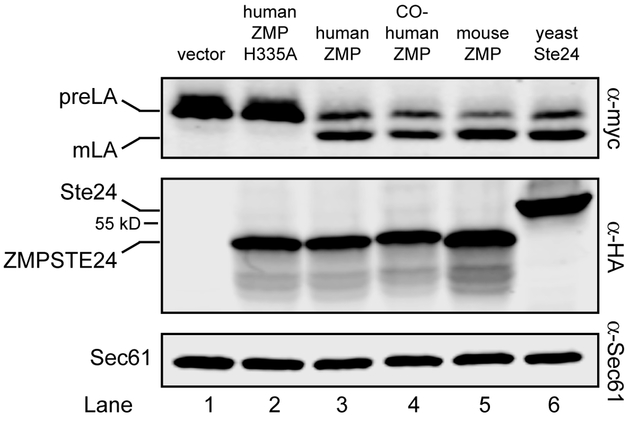
The human ZMPSTE24 gene on pSM2677 used in this version 1.0 system contains the codons corresponding to human cDNA. We replaced the human ZMPSTE24 in this plasmid with mouse Zmpste24, yeast STE24, and a version of human ZMPSTE24 that is codon-optimized for expression in yeast [27] (kindly provided by M. Dumont, University of Rochester) (Fig. 2). Cells containing no ZMPSTE24 (vector only) or a catalytically dead version (H355A) do not process prelamin A (Fig. 2, lanes 1 and 2, top panel). Not surprisingly, the mouse Zmpste24 protein, which is 93% identical (and 96% similar) to human ZMPSTE24 processed prelamin A to the same extent as its human homolog (Fig. 2, compare lanes 3 and 5). Yeast Ste24, which is 35% identical to ZMPSTE24 (and 51% similar) and has a remarkably similar structure [28, 29] also is proficient in human prelamin A cleavage (Fig. 2, compare lanes 3 and 6). We also found that the codon-optimized (co) version of human ZMPSTE24 does not appear to significantly enhance ZMPSTE24 protein levels nor prelamin A cleavage (Fig. 2, compare lanes 3 and 4).
Figure 2. ZMPSTE24 from other species process the LMNACT substrate.
Transformants of strain SM6158 (ste24Δ 10His-3myc-LMNACT) containing the indicated protease constructs were analyzed by western blotting as described in Figure 1. Yeast codon-optimized human ZMPSTE24 (lane 4), mouse cDNA Zmpste24 (lane 5), and yeast Ste24 (lane 6) all process the prelamin A substrate to the same degree as human cDNA ZMPSTE24 (lane 3), but strains with vector only (no ZMPSTE24) or the catalytically dead mutant H335A) are processing deficient (lanes 1 and 2). Proteases were detected with anti-HA antibodies, using anti-Sec61 as a loading control. Plasmids with cleavage percentages (in parentheses) used here for lanes 1 –6 are pRS316 (1.8%), pSM2673 (1.8%), pSM2677 (59.5%), pSM3202 (58.2%), pSM3175 (74.0%) and pSM3094 (65.3%), respectively.
It is worth pointing out that yeast Ste24, which is 22/21 amino acids shorter than human/mouse ZMPSTE24 (453 versus 475/474 amino acids, respectively; 498 and 517/516 amino acids for their epitope-tagged versions) has a significantly slower mobility in SDS-PAGE than its mammalian homologs (Fig. 2, middle panel, compare lanes 3 and 6). However, anomalous migration is not uncommon for membrane proteins, whose hydrophobic spans may influence SDS binding and thus migration [45, 46]. In addition, human and mouse ZMPSTE24 are detected as two bands (e.g. Fig. 3B) or a major band with underlying faster migrating smear (Fig. 2), as also observed by others [27]. This is not specific to yeast, since two bands are also detected for ZMPSTE24 in the mouse [16].
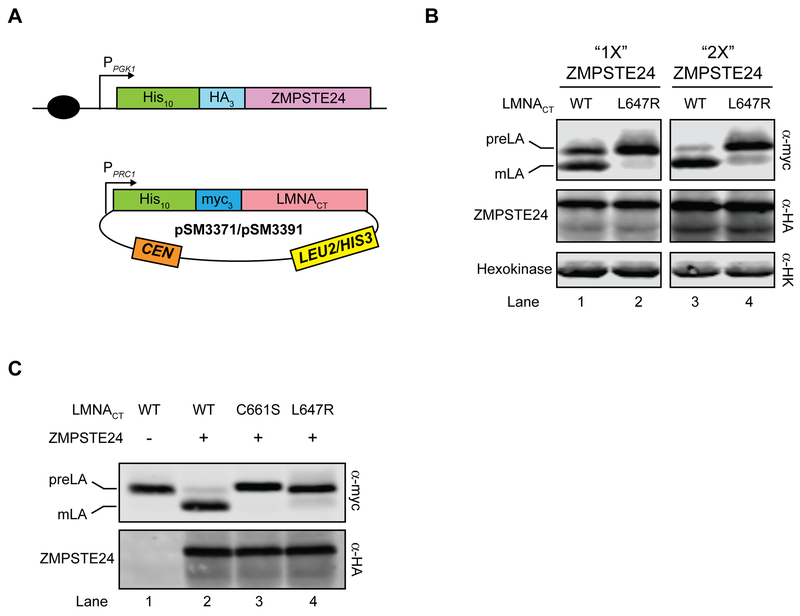
Figure 3. The “humanized yeast system” version 2.0 to study LMNA variants.
(A) Schematic of the version 2.0 system is shown. Yeast codon-optimized 10His-3HA-ZMPSTE24 is integrated into a ste24Δ strain at the TRP1 locus (SM6302/SM6303). LMNACT variants are then transformed into the yeast strain and selected with SC-leu or SC-his medium. (B). Prelamin A cleavage increases with more ZMPSTE24. SM6302 (“1X ZMP”) or SM6303 (“2X ZMP”) transformed with WT or L647R LMNACT were analyzed by western blotting with anti-myc and anti-HA antibodies. Anti-HK (hexokinase) serves as a loading control. Strains with cleavage percentage in parentheses in lanes 1–4 are SM6302/pSM3391 (54.2%), SM6302/pSM3392 (9.9%), SM6303/pSM3391 (79.6%), and SM6303/pSM3392 (15.4%), respectively. (C) Farnesylation and an intact cleavage site are necessary for prelamin A cleavage. Strain SM6303 (ste24Δ 2X 10His-3HA-ZMPSTE24) transformed with WT LMNA (pSM3371, lane 2), LMNA-C661S (pSM3512, lane 3) or LMNA-L647R (pSM3513, lane 4) were analyzed by western blotting for cleavage (anti-myc antibodies) and ZMPSTE24 level (anti-HA antibodies). Strain SM4826 (ste24Δ) transformed with WT LMNA (pSM3371, lane 1) serves as a control for no cleavage. Cleavage percentages for lanes 1–4 are 1.1%, 89.1%, 0.9% and 9.5%, respectively.
Taken together, the version 1.0 system discussed above provides a functional tool to compare specific human disease alleles of ZMPSTE24, and assay specific mutations derived from the known crystal structure to try to elucidate the mechanism of substrate cleavage; see also Fig. 4). In addition, ZMPSTE24 variants from different species can be compared (Fig. 2). Since the LMNA gene is integrated into a ste24Δ background, the only variable is the processing enzyme supplied on a single-copy CEN plasmid.
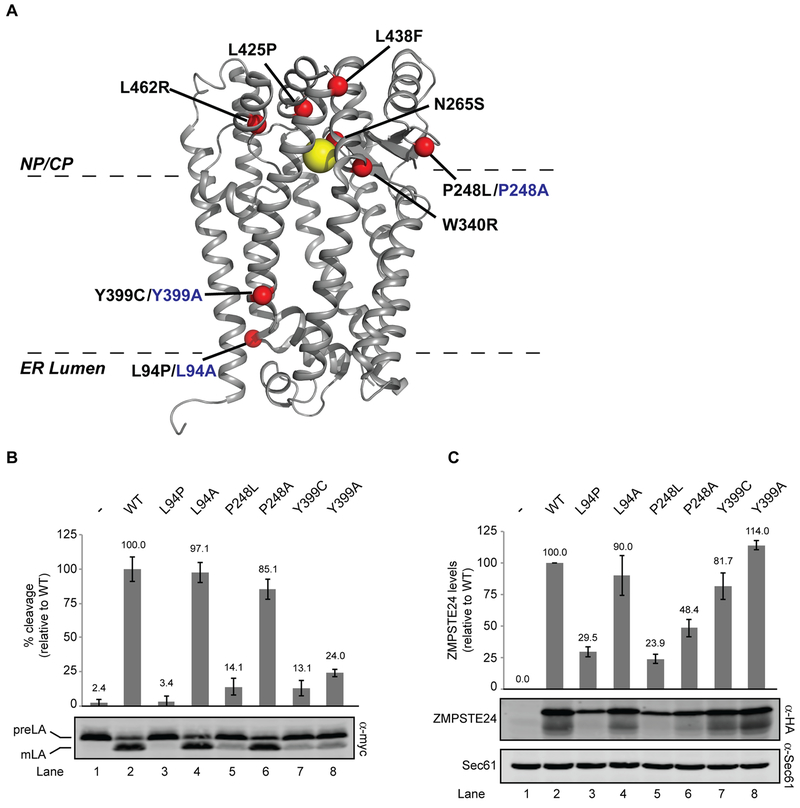
Figure 4. Structure-function studies of ZMPSTE24.
(A) X-ray crystal structure of human ZMPSTE24 (PDB entry 2ypt) with zinc indicated by the yellow ball and disease residues indicated with red balls. Disease alleles are labeled in black, with alanine changes in blue. The predicted placement of ZMPSTE24 in the membrane bilayer is shown (B). SM6158 (ste24Δ 10His-3myc-LMNACT) transformed with the indicated ZMPSTE24 mutants were analyzed by western blotting to determine prelamin A cleavage activity, which was calculated as a ratio of mature lamin A to total myc signal (prelamin A + mature lamin A), as described in Methods section. Average cleavage and standard deviation of the mean for three independent experiments is shown, with wild-type ZMPSTE24 activity set to 100% for comparison. (C) ZMPSTE24 protein levels were analyzed by western blotting using anti-HA (normalized to the loading control Sec61). The average and standard deviation of the mean is shown for the same three experiments as in (B). Wild-type ZMPSTE24 protein levels are set to 100% for comparison.
3.3. The humanized yeast system version 2.0 to assay ZMPSTE24 mutant alleles
We designed a second system (version 2.0) whereby ZMPSTE24 is integrated into the genome of a ste24Δ strain, which can be transformed with plasmid-encoded WT or mutant LMNA constructs (Fig. 3A). We had previously demonstrated that prelamin A cleavage could be increased (from ~60% to over 90%) in our version 1.0 system when two differently tagged plasmid-borne versions of ZMPSTE24 are present [13]. Here, we constructed a pair of yeast strains, SM6302 and SM6303, that have either one copy or two copies of ZMPSTE24, integrated into the genome at the chromosomal TRP1 locus and expressed from the strong PGK1 promoter. Consistent with previous results, we observe an increase in prelamin A cleavage to 80–90%, concomitant with an increase in ZMPSTE24 level (Fig. 3B, compare lanes 1 and 3). Version 2.0 thus uses strain SM6303 which contains two integrated copies of ZMPSTE24 to maximize cleavage. In this system, prelamin A cleavage remains dependent on its farnesylation and an intact cleavage site, since cleavage fails to occur in the C661S LMNACT mutant in which farnesylation is blocked and in the uncleavable LMNA mutant L647R (Fig. 3C, compare lane 2 with lanes 3 and 4, respectively). Overall, the version 2.0 system is optimal to query prelamin A substrates from different species, or test whether disease-causing or otherwise mutated alleles of LMNA affect ZMPSTE24-dependent proteolysis.
3.4. Application of humanized yeast system version 1.0 to study ZMPSTE24 function and structure: Comparison of in vivo findings for ZMPSTE24 mutants with computational models.
Our humanized yeast systems were developed with the goal of fully understanding the mechanism of ZMPSTE24-dependent substrate cleavage, as well as testing the effects of specific ZMPSTE24 mutations, including disease alleles (Fig. 4A). Our published work has shown that ZMPSTE24 missense patient mutations were all defective in prelamin A cleavage [13]. However the mechanistic basis for the observed processing defects appear to differ in vivo since the disease alleles defined 3 classes: those with an in vivo stability defect due to degradation by the ubiquitin-proteasome system, those mainly affected for processing, and those mutants which show both defects. These three classes are exemplified by the disease alleles P248L, Y399C, and L94P, respectively.
Here we have generated alanine replacements at these same residues (L94A, P248A, and Y399A) and compared their in vivo activity and stability to the corresponding disease alleles. The position of ZMPSTE24 disease mutants along with the new alanine mutants, is indicated on the structure (Fig. 4A) and the results of the in vivo activity and stability assays are shown in Figs. 4B and 4C. Interestingly, in contrast to the disease alleles L94P and P248L, the corresponding alanine substitutions L94A and P248A were significantly more proficient in activity and stability (Fig. 4B, C compare lanes 3 to 4 and 5 to 6). The functionality of the alanine mutants suggests that the wild-type residues are not absolutely needed for function and proper folding. In contrast, for the disease allele Y399C and its corresponding alanine allele Y399A, both show decreased prelamin A cleavage, although both variants were as essentially as stable as the wild-type protein (Fig. 4B, C compare lanes 7 and 8 to lane 2).
The medium resolution structure of human ZMPSTE24 at 3.4 Å resolution shown in Fig. 4A [29] provides an opportunity to attempt to understand the role of specific ZMPSTE24 residues at the molecular level. Here, we compared the above experimental data for ZMPSTE24 mutations at positions L94, P248, and Y399 with computational modeling to begin to see if we could generate structure-based hypotheses of each variant’s role in stability and activity. We used the Rosetta macromolecular modeling suite [47] to predict the structural and energetic effects of the disease alleles or alanine substitutions. Our approach is adapted from the flexible-backbone ΔΔG protocol described in Barlow and coworkers [34]. The goal was to calculate the free-energy change for each mutant as compared to WT ZMPSTE24 (ΔΔGmut), since the predicted thermodynamic cost of a mutation might provide insight into the observed changes in in vivo stability. Our procedures for the ΔΔGmut calculations are described in the Materials and Methods.
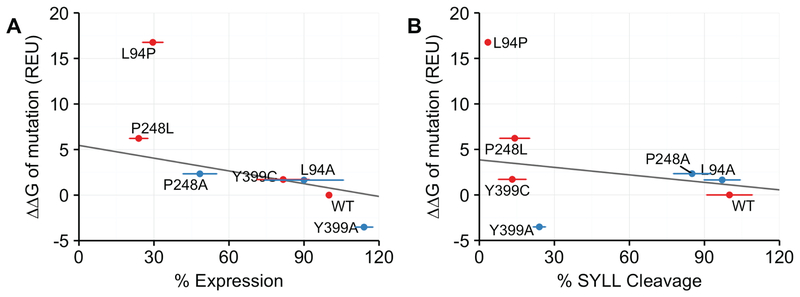
We compared our ΔΔGmut calculations to the experimental in vivo stability (from Fig. 4C) and cleavage measurements (from Fig. 4B) for each variant. The calculated and experimental data are graphed in Fig 5. In general, for this panel of mutants, we observed that the calculated ΔΔGmut values (y axis), expressed as Rosetta Energy Units (REU), exhibit a strong negative correlation with protein stability (x axis) (R = −0.843) (Fig 5A.). As the magnitude of ΔΔGmut decreases, protein expression, which reflects in vivo stability, increases. For instance, the P248L is predicted by Rosetta to be much less thermodynamically stable than WT and Y399A is predicted to be more stable than WT, which roughly match the experimental data. The ΔΔGmut of L94P is an outlier in our dataset, which is unsurprising because in the Rosetta modeling program it is challenging to accommodate a proline while only moderately sampling backbone flexibility. These results indicate that the thermodynamic stability measurements calculated by Rosetta correlate reasonably well with in vivo stability and suggest that what the cellular machinery detects as misfolding is reflected in the Rosetta energy calculations of thermodynamic stability.
Figure 5. Comparison of calculated ΔΔGmut values for mutant ZMPSTE24 alleles with experimental data for ZMPSTE24 stability and prelamin A cleavage activity from Fig. 4B and Fig.4C.
ΔΔGmut is the predicted difference in free energy of folding between a WT and mutant protein. A) The calculated ΔΔGmut values in Rosetta Energy Units (REU) (y axis) for the indicated mutant alleles of ZMPSTE24 is compared with the level of ZMPSTE24 stability (denoted as expression; × axis) from the assay shown in Fig 4B. When the proline mutation L94P is excluded, the correlation coefficient is −0.843. (B) Comparison of ΔΔGmut calculations (y axis) with percentage of prelamin A cleavage determined in Fig. 4A (x axis). When the proline mutation L94P is excluded, the correlation coefficient is −0.207. Disease variants are in red and additional variants are in blue. Horizontal error bars are the standard deviation of the mean taken from Fig 4.
In contrast to the stability measurements, the prelamin A cleavage results do not correlate well with the Rosetta ΔΔGmut predictions (Fig. 5B). For instance, the calculated ΔΔGmut of Y399C is 1.711 REU, while that of Y399A is strikingly low at −3.512 REU, yet both show strong cleavage defects. This result is not unexpected, because the Rosetta energy function is designed to capture thermodynamic and not catalytic effects.
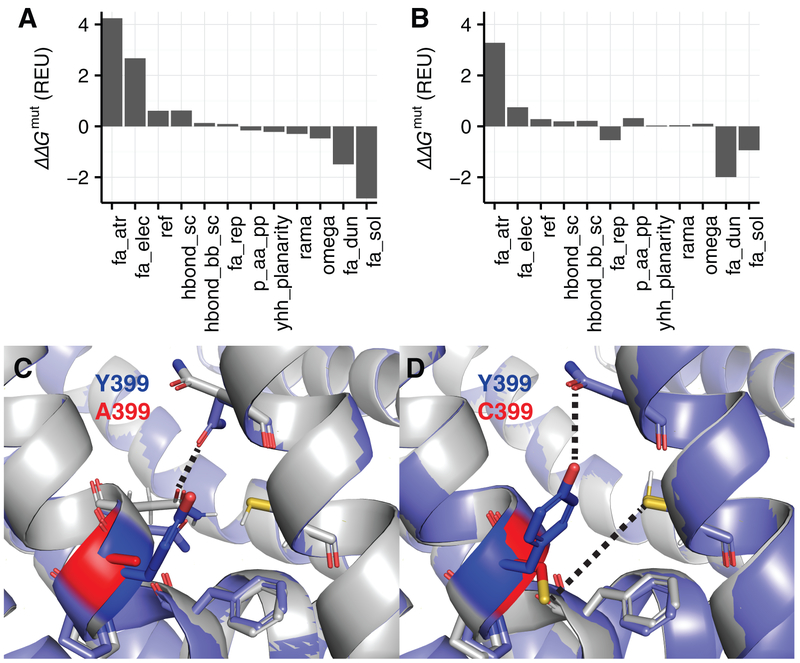
To further investigate how the calculated ΔΔGmut values relate to predicted structural changes for these two mutations, we used energy decomposition and structure visualization. The comparison of the energy breakdown and structures for the Y399A/Y399C variants is shown in Fig.6. In the Y399A variant, the conformation loses favorable van der Waals attractive and electrostatic interactions, likely due to the loss of a hydroxyl group capable of hydrogen bonding to nearby side chains (Fig. 6A). However, this mutation is rescued by favorable side-chain conformations and solvation energies-define these terms briefly. The rotamer score improves because alanine is very small and is easy to fit into a pocket. The solvation score improves because position 399 is buried, thereby minimizing exposure of the hydrophobic alanine to solvent (Fig. 6C). In contrast, Y399C also loses favorable van der Waals and electrostatic interactions (Fig. 6B). This variant is not rescued by favorable solvation because cysteine is not as hydrophobic as traditional nonpolar residues. For Y399C, the structure is particularly revealing (Fig. 2D). There is a cysteine nearby capable of forming a disulfide bond with position 399, further deforming the aqueous cavity and potentially impacting catalytic activity. Overall, this structure-based analysis of Y399A and Y399C provides insight into the mechanism of stability loss and may ultimately be useful to suggest sites for potential therapeutics that rescue activity.
Figure 6. Structural and energetic effects of Y399A vs. Y399C in ZMPSTE24.
We compared the Y399A and Y399C variants by analyzing the decomposed Rosetta energies and structural ensembles. (A) Decomposed ΔΔGmut of the Y399A ZMPSTE24 variant. Energies with a contribution to the ΔΔGmut > 0.1 REU are shown. The energy terms are as follows: fa_atr = Van der Waals attractive energy, fa_rep = repulsive energy, fa_elec = Coulomb electrostatics energy, ref = Rosetta reference energy, fa_dun = Dunbrack rotamer energy, hbond_sc = side-chain to side-chain hydrogen-bonding energy, hbond_bb_sc = backbone to side-chain hydrogen-bonding energy, fa_sol = Lazaridis and Karplus implicit solvation energy, p_aa_pp = amino acid propensity, ramachandran backbone ϕ,ψ score, omega = backbone ω score [36]. (B) Decomposed ΔΔGmut of the Y399C ZMPSTE24 variant. (C) Lowest scoring structure in the native ensemble (gray) superimposed onto the lowest scoring structure in the Y399A ensemble (purple). The native tyrosine is highlighted in blue and the mutant alanine is highlighted in red. Side chains within 3.0 Å of the mutation are shown. (D) Lowest scoring structure in the native ensemble (gray) superimposed onto the lowest scoring structure in the Y399C ensemble (purple). The native tyrosine is highlighted in blue and the mutant cysteine is highlighted in red. Side chains within 3.0 Å of the mutation are shown.
DISCUSSION
The cleavage of prelamin A by ZMPSTE24 to yield mature a-factor is clearly critical for human health, since mutations in either the LMNA or ZMPSTE24 genes that affect processing result in progeroid diseases. In addition, provocative evidence suggests that diminished ZMPSTE24 expression or activity may contribute to normal physiological aging, particularly in the vasculature [19]. Thus, understanding the features of this integral membrane protease and its farnesylated substrate that are important for cleavage is likely to have far-reaching implications.
Here we describe two versions of a “humanized yeast system” we have recently developed to study the cleavage of human prelamin A by human ZMPSTE24 in S. cerevisiae. The two versions are optimized to allow direct comparison of either ZMPSTE24-encoded variants (version 1.0) or LMNA- encoded prelamin A variants (version 2.0), by introducing the WT or mutant versions of the respective genes on plasmids. Our recent analysis of ZMPSTE24 missense disease alleles using version 1.0 demonstrates the power of the “humanized yeast” approach [13]. We showed that some disease alleles are defective solely for activity, while other alleles (i.e. P248L) mainly affect in vivo stability, thus likely affecting mainly folding. Together these results suggest distinct classes of mechanistic defects for the disease alleles. The ability to readily make use of a yeast ER-associated degradation (ERAD) ubiquitin E3 ligase mutant was beneficial for this analysis, as it was immediately informative about the role of the ubiquitin-proteasome ERAD pathway for the degradation of P248L and other unstable mutant proteins. Tests are underway in our laboratory to determine whether the same fate occurs for the P248L ZMPSTE24 mutant protein in patient cells. The version 2.0 system is equally as useful. We are currently using this latter system to analyze hundreds of LMNA mutants generated by site-directed mutagenesis. This will allow us to assess the requirements for prelamin A cleavage, including evaluating residues surrounding the cleavage site and alterations of the amino acid composition and length of the cleaved portion of prelamin A.
In addition to the ease of analyzing site-directed mutations, the model organism yeast affords an ideal system for developing high throughput screens and selections to identify particular types of mutants. Using deep mutational scanning coupled with next generation sequencing technology it will be possible to score tens of thousands of variants in parallel in a single screening or selection experiment, making it feasible, for instance, to generate and assess all possible substitution mutations of ZMPSTE24. This approach, coupled with computational analysis described here that predicts protein stability, could help address the question of how the protein quality control machinery selects and degrades some misfolded variants, while ignoring others. Overall, we expect that the humanized yeast systems presented here have the versatility to answer fundamental questions about how ZMPSTE24 recognizes its substrate prelamin A and how disease alleles affect protein structure, function, and stability. Ultimately understanding these issues could lead to improved pharmacological approaches for certain forms of progeria and for optimizing healthy physiological aging.
ACKNOWLEDGEMENTS
We thank Mark Dumont for the kind gift of human ZMPSTE24 that was codon-optimized for expression in yeast [27]. This work was funded by grants to SM from the NIH (R01 GM041223, R35 GM127073, and R21 AG058032), to JJG from the NIH (R01 GM078221), and to RFA from the Hertz Foundation.
REFERENCES
- [1].Capell BC, Collins FS, Human laminopathies: nuclei gone genetically awry, Nat Rev Genet 7(12) (2006) 940–52. [DOI] [PubMed] [Google Scholar]
- [2].Davies BS, Fong LG, Yang SH, Coffinier C, Young SG, The posttranslational processing of prelamin A and disease, Annu Rev Genomics Hum Genet 10 (2009) 153–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, Levy N, Lamin a truncation in Hutchinson-Gilford progeria, Science 300(5628) (2003) 2055. [DOI] [PubMed] [Google Scholar]
- [4].Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS, Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome, Nature 423(6937) (2003) 293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gordon LB, Rothman FG, Lopez-Otin C, Misteli T, Progeria: a paradigm for translational medicine, Cell 156(3) (2014) 400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Agarwal AK, Fryns JP, Auchus RJ, Garg A, Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia, Hum Mol Genet 12(16) (2003) 1995–2001. [DOI] [PubMed] [Google Scholar]
- [7].Barrowman J, Wiley PA, Hudon-Miller SE, Hrycyna CA, Michaelis S, Human ZMPSTE24 disease mutations: residual proteolytic activity correlates with disease severity, Hum Mol Genet 21(18) (2012) 4084–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Michaelis S, Hrycyna CA, Biochemistry. A protease for the ages, Science 339(6127) (2013) 1529–30. [DOI] [PubMed] [Google Scholar]
- [9].Moulson CL, Go G, Gardner JM, van der Wal AC, Smitt JH, van Hagen JM, Miner JH, Homozygous and compound heterozygous mutations in ZMPSTE24 cause the laminopathy restrictive dermopathy, J Invest Dermatol 125(5) (2005) 913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Navarro CL, Cadinanos J, De Sandre-Giovannoli A, Bernard R, Courrier S, Boccaccio I, Boyer A, Kleijer WJ, Wagner A, Giuliano F, Beemer FA, Freije JM, Cau P, Hennekam RC, Lopez-Otin C, Badens C, Levy N, Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of Lamin A precursors, Hum Mol Genet 14(11) (2005) 1503–13. [DOI] [PubMed] [Google Scholar]
- [11].Navarro CL, Esteves-Vieira V, Courrier S, Boyer A, Duong Nguyen T, Huong le TT, Meinke P, Schroder W, Cormier-Daire V, Sznajer Y, Amor DJ, Lagerstedt K, Biervliet M, van den Akker PC, Cau P, Roll P, Levy N, Badens C, Wehnert M, De Sandre-Giovannoli A, New ZMPSTE24 (FACE1) mutations in patients affected with restrictive dermopathy or related progeroid syndromes and mutation update, Eur J Hum Genet 22(8) (2014) 1002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Michaelis S, Barrowman J, Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease, Microbiol Mol Biol Rev 76(3) (2012) 626–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Spear ED, Hsu ET, Nie L, Carpenter EP, Hrycyna CA, Michaelis S, ZMPSTE24 missense mutations that cause progeroid diseases decrease prelamin A cleavage activity and/or protein stability, Dis Model Mech 11(7) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Worman HJ, Michaelis S, Permanently Farnesylated Prelamin A, Progeria, and Atherosclerosis, Circulation 138(3) (2018) 283–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, Mohr A, Meta M, Genant H, Jiang Y, Wisner ER, Van Bruggen N, Carano RA, Michaelis S, Griffey SM, Young SG, Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect, Proc Natl Acad Sci U S A 99(20) (2002) 13049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, Lopez-Otin C, Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice, Nat Genet 31(1) (2002) 94–9. [DOI] [PubMed] [Google Scholar]
- [17].Worman HJ, Fong LG, Muchir A, Young SG, Laminopathies and the long strange trip from basic cell biology to therapy, J Clin Invest 119(7) (2009) 1825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO 3rd, Gahl WA, Introne WJ, Phenotype and course of Hutchinson-Gilford progeria syndrome, N Engl J Med 358(6) (2008) 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ragnauth CD, Warren DT, Liu Y, McNair R, Tajsic T, Figg N, Shroff R, Skepper J, Shanahan CM, Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging, Circulation 121(20) (2010) 2200–10. [DOI] [PubMed] [Google Scholar]
- [20].Fujimura-Kamada K, Nouvet FJ, Michaelis S, A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor, J Cell Biol 136(2) (1997) 271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tam A, Nouvet FJ, Fujimura-Kamada K, Slunt H, Sisodia SS, Michaelis S, Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing, J Cell Biol 142(3) (1998) 635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Young SG, Fong LG, Michaelis S, Prelamin A, Zmpste24, misshapen cell nuclei, and progeria--new evidence suggesting that protein farnesylation could be important for disease pathogenesis, J Lipid Res 46(12) (2005) 2531–58. [DOI] [PubMed] [Google Scholar]
- [23].Boyartchuk VL, Rine J, Roles of prenyl protein proteases in maturation of Saccharomyces cerevisiae a-factor, Genetics 150(1) (1998) 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Barrowman J, Hamblet C, George CM, Michaelis S, Analysis of prelamin A biogenesis reveals the nucleus to be a CaaX processing compartment, Mol Biol Cell 19(12) (2008) 5398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Barrowman J, Michaelis S, ZMPSTE24, an integral membrane zinc metalloprotease with a connection to progeroid disorders, Biol Chem 390(8) (2009) 761–73. [DOI] [PubMed] [Google Scholar]
- [26].Tam A, Schmidt WK, Michaelis S, The multispanning membrane protein Ste24p catalyzes CAAX proteolysis and NH2-terminal processing of the yeast a-factor precursor, J Biol Chem 276(50) (2001) 46798–806. [DOI] [PubMed] [Google Scholar]
- [27].Clark KM, Jenkins JL, Fedoriw N, Dumont ME, Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors, Protein Sci 26(2) (2017) 242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pryor EE Jr., Horanyi PS, Clark KM, Fedoriw N, Connelly SM, Koszelak-Rosenblum M, Zhu G, Malkowski MG, Wiener MC, Dumont ME, Structure of the integral membrane protein CAAX protease Ste24p, Science 339(6127) (2013) 1600–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Quigley A, Dong YY, Pike AC, Dong L, Shrestha L, Berridge G, Stansfeld PJ, Sansom MS, Edwards AM, Bountra C, von Delft F, Bullock AN, Burgess-Brown NA, Carpenter EP, The structural basis of ZMPSTE24-dependent laminopathies, Science 339(6127) (2013) 1604–7. [DOI] [PubMed] [Google Scholar]
- [30].Ast T, Michaelis S, Schuldiner M, The Protease Ste24 Clears Clogged Translocons, Cell 164(1–2) (2016) 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kayatekin C, Amasino A, Gaglia G, Flannick J, Bonner JM, Fanning S, Narayan P, Barrasa MI, Pincus D, Landgraf D, Nelson J, Hesse WR, Costanzo M, Consortium ATDG, Myers CL, Boone C, Florez JC, Lindquist S, Translocon Declogger Ste24 Protects against IAPP Oligomer-Induced Proteotoxicity, Cell 173(1) (2018) 62–73 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fu B, Wang L, Li S, Dorf ME, ZMPSTE24 defends against influenza and other pathogenic viruses, J Exp Med 214(4) (2017) 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kushnirov VV, Rapid and reliable protein extraction from yeast, Yeast 16(9) (2000) 857–60. [DOI] [PubMed] [Google Scholar]
- [34].Barlow KA, S OC, Thompson S, Suresh P, Lucas JE, Heinonen M, Kortemme T, Flex ddG: Rosetta Ensemble-Based Estimation of Changes in Protein-Protein Binding Affinity upon Mutation, J Phys Chem B 122(21) (2018) 5389–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, Baker D, RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite, PLoS One 6(6) (2011) e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Alford RF, Leaver-Fay A, Jeliazkov JR, O’Meara MJ, DiMaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K, Labonte JW, Pacella MS, Bonneau R, Bradley P, Dunbrack RL Jr., Das R, Baker D, Kuhlman B, Kortemme T, Gray JJ, The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design, J Chem Theory Comput 13(6) (2017) 3031–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].O’Meara MJ, Leaver-Fay A, Tyka MD, Stein A, Houlihan K, DiMaio F, Bradley P, Kortemme T, Baker D, Snoeyink J, Kuhlman B, Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta, J Chem Theory Comput 11(2) (2015) 609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chaudhury S, Lyskov S, Gray JJ, PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta, Bioinformatics 26(5) (2010) 689–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dittmer TA, Misteli T, The lamin protein family, Genome Biol 12(5) (2011) 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Simon DN, Wilson KL, Partners and post-translational modifications of nuclear lamins, Chromosoma 122(1–2) (2013) 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Barrowman J, Hamblet C, Kane MS, Michaelis S, Requirements for efficient proteolytic cleavage of prelamin A by ZMPSTE24, PLoS One 7(2) (2012) e32120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hennekes H, Nigg EA, The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties, J Cell Sci 107(Pt 4) (1994) 1019–29. [DOI] [PubMed] [Google Scholar]
- [43].Mallampalli MP, Huyer G, Bendale P, Gelb MH, Michaelis S, Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson-Gilford progeria syndrome, Proc Natl Acad Sci U S A 102(40) (2005) 14416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang Y, Lichter-Konecki U, Anyane-Yeboa K, Shaw JE, Lu JT, Ostlund C, Shin JY, Clark LN, Gundersen GG, Nagy PL, Worman HJ, A mutation abolishing the ZMPSTE24 cleavage site in prelamin A causes a progeroid disorder, J Cell Sci 129(10) (2016) 1975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Newman MJ, Foster DL, Wilson TH, Kaback HR, Purification and reconstitution of functional lactose carrier from Escherichia coli, J Biol Chem 256(22) (1981) 11804–8. [PubMed] [Google Scholar]
- [46].Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM, Detergent binding explains anomalous SDS-PAGE migration of membrane proteins, Proc Natl Acad Sci U S A 106(6) (2009) 1760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P, ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules, Methods Enzymol 487 (2011) 545–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sikorski RS, Hieter P, A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae, Genetics 122(1) (1989) 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]