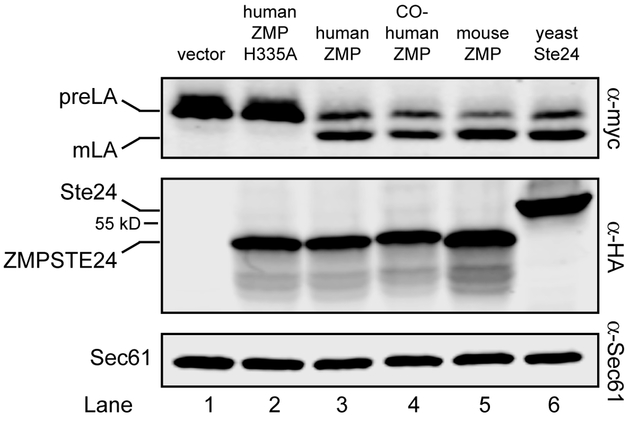
Figure 2. ZMPSTE24 from other species process the LMNACT substrate.
Transformants of strain SM6158 (ste24Δ 10His-3myc-LMNACT) containing the indicated protease constructs were analyzed by western blotting as described in Figure 1. Yeast codon-optimized human ZMPSTE24 (lane 4), mouse cDNA Zmpste24 (lane 5), and yeast Ste24 (lane 6) all process the prelamin A substrate to the same degree as human cDNA ZMPSTE24 (lane 3), but strains with vector only (no ZMPSTE24) or the catalytically dead mutant H335A) are processing deficient (lanes 1 and 2). Proteases were detected with anti-HA antibodies, using anti-Sec61 as a loading control. Plasmids with cleavage percentages (in parentheses) used here for lanes 1 –6 are pRS316 (1.8%), pSM2673 (1.8%), pSM2677 (59.5%), pSM3202 (58.2%), pSM3175 (74.0%) and pSM3094 (65.3%), respectively.