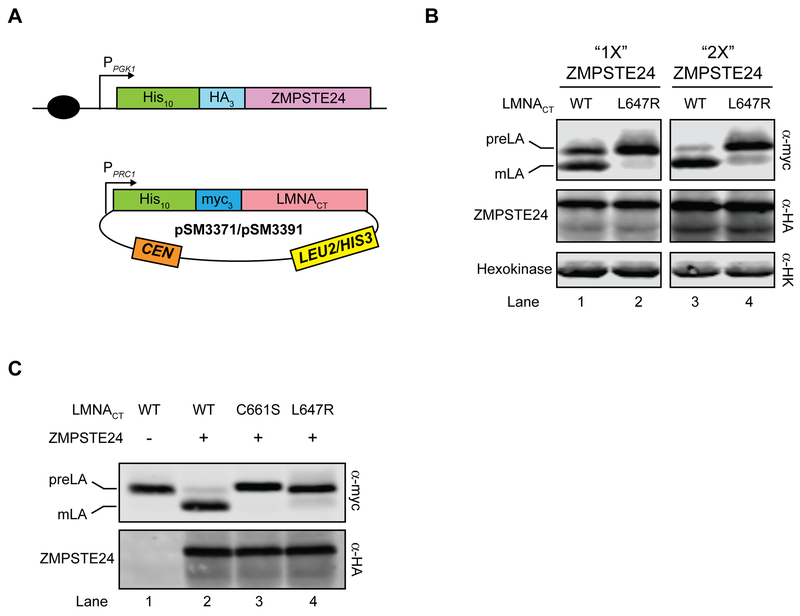
Figure 3. The “humanized yeast system” version 2.0 to study LMNA variants.
(A) Schematic of the version 2.0 system is shown. Yeast codon-optimized 10His-3HA-ZMPSTE24 is integrated into a ste24Δ strain at the TRP1 locus (SM6302/SM6303). LMNACT variants are then transformed into the yeast strain and selected with SC-leu or SC-his medium. (B). Prelamin A cleavage increases with more ZMPSTE24. SM6302 (“1X ZMP”) or SM6303 (“2X ZMP”) transformed with WT or L647R LMNACT were analyzed by western blotting with anti-myc and anti-HA antibodies. Anti-HK (hexokinase) serves as a loading control. Strains with cleavage percentage in parentheses in lanes 1–4 are SM6302/pSM3391 (54.2%), SM6302/pSM3392 (9.9%), SM6303/pSM3391 (79.6%), and SM6303/pSM3392 (15.4%), respectively. (C) Farnesylation and an intact cleavage site are necessary for prelamin A cleavage. Strain SM6303 (ste24Δ 2X 10His-3HA-ZMPSTE24) transformed with WT LMNA (pSM3371, lane 2), LMNA-C661S (pSM3512, lane 3) or LMNA-L647R (pSM3513, lane 4) were analyzed by western blotting for cleavage (anti-myc antibodies) and ZMPSTE24 level (anti-HA antibodies). Strain SM4826 (ste24Δ) transformed with WT LMNA (pSM3371, lane 1) serves as a control for no cleavage. Cleavage percentages for lanes 1–4 are 1.1%, 89.1%, 0.9% and 9.5%, respectively.