Abstract
Cellular function relies on multiple pathways that are coordinated to ensure the proper execution of gene expression networks. Failure to coordinate the multiple programs active in the cell can have catastrophic consequences and lead to diseases such as cancer. At the post-transcriptional level, RNA modifications play important roles in the regulation of gene expression. N6-methyladenosine (m6A) is the most abundant internal messenger RNA (mRNA) modification and has gained increasing interest in the last few years as a dynamic regulator of RNA metabolism. Modifications regulate all stages of the RNA life cycle, from transcription to decay. Recent studies have pointed to the role of RNA methylation in cancer initiation and progression, and aberrant modification has served as a biomarker of early-stage diagnosis in several cancers. Here, we review the regulation of m6A, disruptions to methylation-dependent pathways that influence carcinogenesis, and potential avenues for m6A-related therapeutic strategies.
Keywords: m6A, epitranscriptome, gene regulation, cancer
Introduction:
Organismal development and homeostasis are fully dependent on changes in gene expression programs in response to both internal and external cues. After transcription, mRNA is subjected to multiple processing steps necessary for generating an RNA that can be translated. At each processing step, multiple pathways act to regulate how much product is generated from each mRNA, and ultimately, how a cell behaves. One of the regulatory mechanisms employed in the cell is the addition of post-transcriptional modifications to mRNAs. RNA modifications can directly influence RNA structure or modulate interactions with other molecules, contributing to complex regulatory networks. To date, over 150 modifications have been identified in RNA molecules (1) and methylation represents over 66% of known modifications (2). N6-methyladenosine (m6A), one of several methyl modifications identified in mRNAs (3–10), is the most abundant modification found internally on mRNAs and long non-coding RNAs (lncRNA). This review will focus on the mRNA modification m6A, and how disruption of m6A-dependent regulatory pathways impacts cancer.
Section 1: m6A methylation
1.1. The N6-methyladenosine modification:
Conserved throughout eukaryotic species, m6A-dependent pathways play critical roles in development (reviewed in (11)). Although m6A was first detected in mRNAs in the early 1970s (12–19), it was thought to be static and the role of this modification remained poorly understood. Recent discovery of fat mass and obesity-associated protein (FTO) as a m6A demethylase (20), and the development of methods to map sites of modification (21,22) renewed the interest in mRNA methylation. In mRNAs, the majority of m6A sites are found in a well-defined sequence context (RRACH; R can be A or G, H can be A, C or U) (23). m6A sites are enriched near the last transcribed exon (a position that often correlates with the STOP codon), in large exons, and near the 5′ end (21,22,24,25), suggesting that addition of the m6A modification is a regulated process (Figure 1A). In each cell type, while a large fraction of expressed genes can be modified, the majority of potential m6A sites (RRACH motifs) are unmethylated, and a number of m6A sites are only partially modified (26). This stoichiometry suggests a rheostat-like mechanism for fine-tuning gene regulation that can rapidly respond to cellular changes (26). The m6A modification tends to be enriched in transcript isoforms with shorter 3′ untranslated regions (UTRs), and addition of m6A modifications have been observed to impact tandem alternative polyadenylation (APA) site usage for some transcripts (24,27). Deposition of m6A is co-transcriptional (Figure 1B), with the majority of m6A sites determined by the time the mRNA is released into the nucleoplasm. A sub-optimal rate of transcription results in greater m6A deposition on mRNAs, which impacts the rate of translation (24,28,29).
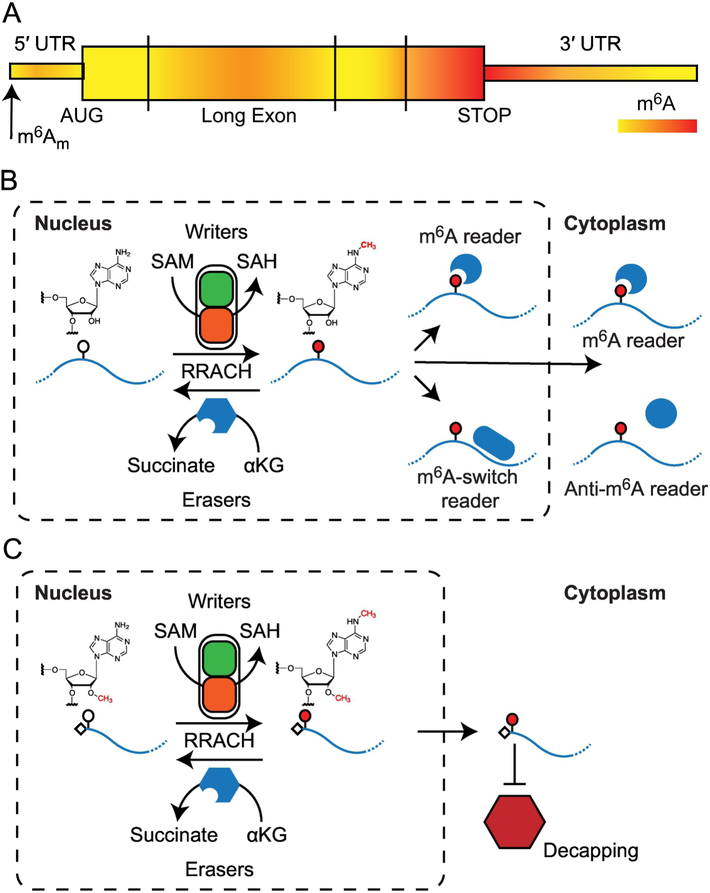
Figure 1: Cycle of methylation and demethylation.
(A) The modified bases N6-methyladenosine (m6A) and N6,2′-O-dimethyladenosine (m6Am) are present in distinct regions of mature mRNA. (B) Addition of m6A to RNA requires the co-factor SAM and is catalyzed by the methylase complex (writers). The modification is removed through the actions of the αKG-dependent demethylases (erasers). The presence or absence of m6A affects the activity of numerous RNA binding proteins (readers) that modulate the downstream processing of the RNA. (C) Formation of m6Am occurs in the nucleus. Similar to m6A, levels of m 6Am are determined by a balance between writer and eraser activity. The presence of m6Am confers resistance to decapping activity in the cytoplasm.
In addition to m6A, methylation at position 6 of adenosine also occurs in the modification N6,2′-O-dimethyladenosine (m6Am). Unlike m6A, which is an internal modification, m6Am is found at the first nucleotide adjacent to the 7-methylguanosine cap (5′ m7GpppAm) (Figure 1A) (9). The m6Am modification is also found in viral RNAs, and it is thought that this addition is catalyzed by a host enzyme (13). Formation of m6Am in mRNA follows the synthesis of the 5′ m7GpppAm and m6Am has not yet been observed in rRNA or tRNA (30). A recent report demonstrated that mRNAs modified with m6Am are more resistant to the mRNA-decapping enzyme 2 (DCP2) than those with the Am modification alone (9) (Figure 1C).
Both m6A and m6Am are regulated by the opposing activities of enzymes that introduce or remove these modifications, known as “writers” (methyltransferases) and “erasers” (demethylases), respectively. Furthermore, the presence or absence of these modifications can create or destroy binding sites for “readers” (RNA Binding Proteins). Decoding of m6A by these reader proteins leads to downstream effects in all stages of the RNA lifecycle (Figure 1B and 1C).
1.2. Writing the m6A modification - the m6A methylases:
m6A modification of mRNA is catalyzed by a nuclear RNA methyltransferase complex. Initial characterization of m6A methylation activity in HeLa cells identified two protein components required for modification: one component of approximately 200 kDa containing the enzyme methyltransferase-like 3 (METTL3), and a second unit of approximately 875 kDa (31). METTL3, the catalytic unit, forms a stable heterodimer with methyltransferase-like 14 (METTL14). This stable heterodimer interacts with an adaptor complex that includes zinc finger CCCH domain-containing protein 13 (Zc3h13); Wilms Tumor 1-associated protein (WTAP); Virilizer, Hakai and RNA binding motif protein 15 (RBM15) (32,33) (Figure 2A). The adaptor complex proteins play crucial roles in proper cellular localization of the methylase complex (34) and interactions of the METTL3/METTL14 heterodimer with mRNAs (27,32,33). Loss of function of components of the adaptor complex in multiple organisms suggests proteins in this complex may serve additional, m 6A independent roles (33).
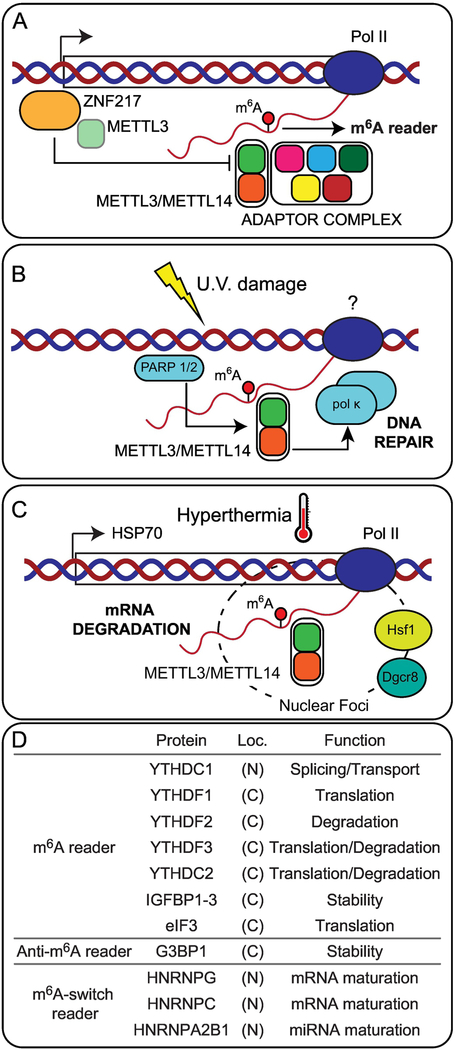
Figure 2. N6-methyladenosine regulatory complexes.
The METTL3/METTL14 proteins have been observed to interact with other cellular proteins. (A) The adaptor complex, which includes zinc finger CCCH domain-containing protein 13 (Zc3h13) Wilms Tumor 1-associated protein (WTAP) Virilizer, Hakai, and RNA binding motif protein 15 (RBM15), facilitates programed deposition of m6A during development. Additionally, the transcription factor zinc finger protein 217 can disrupt the formation of the core complex by sequestering METTL3. (B) Accumulation of m6A also occurs in response to UV damage and promotes recruitment of DNA damage repair factors. (C) Under hyperthermia, recruitment of METLL3/METTL14 to heat shock foci marks transcripts for rapid turnover. (D) Three types of RNA binding proteins that interact with RNA in a m6A-dependent manner have been described: 1) m6A readers, proteins that interact with the modified m6A base; 2) proteins repelled by the presence of m6A; and 3) proteins that interact with RNA after m6A-induced rearrangement of RNA structure. These proteins are present in both the cytoplasm and the nucleus, and determine RNA metabolism.
The core components of the methylation complex have been observed to re-localize under stress conditions (28,35). Upon ultraviolet (UV) radiation damage the heterodimer METTL3/METTL14, but not WTAP, is recruited to sites of UV damage (Figure 2B). Recruitment of METTL3/METTL14, which requires ADP-ribose polymerase (PARP), results in rapid, transient accumulation of m6A at sites of damage. Accumulation of m6A is required for early recruitment of DNA polymerase kappa (Pol κ), an enzyme implicated in both nucleotide excision repair and trans-lesion synthesis. Loss of METTL3 results in delayed repair of ultraviolet-induced cyclo-butane pyrimidine adducts and elevated sensitivity to ultraviolet light (35). In addition to UV damage, the methylation complex has also been observed to re-localize in response to thermal stress. Upon heat-shock, METTL3 and DiGeorge Syndrome Chromosomal Region 8 (DGCR8) form foci that co-localize with Heat Shock Factor 1 (HSF1) (Figure 2C). Under these conditions, binding of METTL3 is enriched at heat shock protein genes, resulting in the co-transcriptional marking of 70 kilodalton heat shock protein (Hsp70) mRNA for subsequent mRNA degradation (28). The activity of the methylation complex is controlled by transcription factor zinc finger protein 217 (ZFP217). This transcription factor binds to the promoter of m6A targets and sequesters METTL3, disrupting the formation of the methylation core complex and resulting in lower activity of the methylation complex (36) (Figure 2A).
The enzyme methyltransferase-like 16 (METTL16), which occupies a central role in the regulation of both splicing machinery and of the levels of S-adenosyl-L-methionine (SAM), is also an m6A writer. In addition to modifying the U6 snoRNA (37,38) METTL16 is also responsible for the modification and regulation of the Methionine Adenosyltransferase 2A (MAT2A) mRNA (37,39). MAT2A RNA levels are regulated in response to the availability of SAM, a common co-substrate involved in methyl group transfers. Under normal conditions, METTL16 binds to, and modifies, conserved hairpin structures in the 3′ UTR of MAT2A. When SAM is limiting, prolonged occupancy of METTL16 of a regulatory hairpin stimulates splicing of a retained intron and subsequent export and translation of the mRNA (37). An alternative model of MAT2A mRNA regulation proposes that METTL16-dependent m6A modification of the 3′-UTR of MAT2A is read by the protein YTH Domain Containing Protein 1 (YTHDC1) to control the stability of the MAT2A mRNA. Differences observed in certain cell lines in response to cycloleucine (cLEU), an inhibitor of SAM synthesis, suggests that MAT2A RNA stability is controlled through multiple mechanisms, depending on cell type (39). In addition to MAT2A, other targets of METTL16 include ncRNAs (7SK, y-RNAs, vtRNA), lncRNAs (metastasis associated lung adenocarcinoma transcript (MALAT) and X-inactive specific transcript (Xist)) and pre-mRNAs (38). Interestingly, interaction between METTL16 and MALAT occurs at the triple helix at the 3′ UTR of noncoding RNA, suggesting METTL16 can interact with triple-stranded RNAs (40). METTL16 interaction sites are enriched in introns, at sites where m6A levels are METTL16 dependent (37,38). Analysis of METTL16 homologs suggests that targeting of the MAT2A mRNA by METTL16 to regulate the levels of SAM occurred later in the evolutionary development of vertebrates (37).
1.3. Erasing m6A - The m6A demethylases:
While the m6A modification had been known for some time, the recent identification of m6A demethylases raised the possibility that this modification is dynamic and reversible. To date, two m 6A RNA demethylases have been identified: FTO and ALKB homolog 5 (ALKBH5), both members of the α-ketoglutarate (αKG)-dependent dioxygenase enzyme family. The FTO gene was first identified in genome-wide association studies and was shown to be linked to both obesity and Body Mass Index (BMI) (41–43). Glucose and amino-acid deprivation have been shown to regulate FTO expression (44). In 2011, it was demonstrated that FTO is able to demethylate m6A both in vitro an in vivo (20). Crystallographic and biochemical analyses revealed the preference of FTO for single-stranded RNA (ssRNA) (45,46). Subsequent studies revealed that FTO oxidizes m 6A in RNA to N6-hydroxymethyl adenosine (hm6A) and N6-formyl adenosine (f6A) in a step-wise manner (47). Recently, it was demonstrated that FTO preferentially demethylates m6Am over m6A (9). Altogether these studies suggest that demethylation of m6Am may lead to an increase in diversity near the 5′ cap (9). ALKBH5 was the second identified m6A demethylase (48). Like FTO, it was found to localize to nuclear speckles (20,48). Loss of ALKBH5 was shown to impact a number of RNA processing steps, including mRNA export and assembly of mRNA processing factors in nuclear speckles. ALKBH5-deficient male mice have increased m6A in mRNA and show aberrant spermatogenesis and impaired fertility (48). Finally, several crystal structures of ALKBH5 demonstrate the preference of the enzyme for a ssRNA binding model and provide evidence for why hm6A and f6A have not been observed during demethylation with ALKBH5 (49–51). Although the m6A writers show a preference for the m6A consensus sequence RRACH, this sequence is not required for substrate selectivity by FTO and ALKBH5 (52). Additionally, while FTO is expressed highly in the brain (53) ALKBH5 is primarily expressed in the testes, with lower levels in other tissues (48). It remains to be understood how these two enzymes work together.
1.4. Interpreting the presence of m6A - m6A Readers:
The functional outcome of an RNA transcript is determined by interactions between the transcript and trans-acting factors, such as RNA binding proteins and small RNAs. RNA modification by m 6A can modulate regulatory interactions through three mechanisms (Figure 1B): introduction of a binding site for proteins that preferentially interact with methylated RNA (m6A Readers), removal of a binding site of proteins that bind unmodified RNAs (Anti-m6A Readers), and alteration of RNA secondary structure, impacting how single-stranded or double-stranded RNA binding proteins interact with RNA (m6A-switch Readers). Reader proteins interpret changes in modification status, and affect a number of downstream cellular processes, including transcriptional efficiency, RNA processing, and RNA decay (Figure 2D).
The first m6A readers identified were proteins belonging to the YTH domain-containing family (22), a highly conserved ssRNA binding domain in eukaryotes (54). In humans, the YTH domain family is composed of the YTH domain-containing family (YTHDF) proteins YTHDF1, YTHDF2, YTHDF3, and the YTH domain-containing (YTHDC) proteins YTHDC1 and YTHDC2 (55). These proteins interact with RNA in the presence of m6A in cell-type specific interactions. YTHDF1 plays a role in increasing translation efficiency (56) and in axon regeneration has been shown to contribute to peripheral nerve-induced global protein translation (57). Conversely, m6A recognition by YTHDF2 promotes mRNA degradation through interactions with the CCR4-NOT complex (58,59). YTHDF3 facilitates both translation and decay pathways through its interactions with YTHDF1 and YTHDF2, respectively (60). Interestingly, while YTHDF1, YTHDF2, and YTHDF3 each have their own unique target mRNAs, they also share a set of common mRNAs (56,60). The nuclear m6A reader YTHDC1 has been shown to regulate mRNA splicing through recruitment of pre-mRNA splicing factors (61) and independent of its role in splicing, can also affect the transport of mRNAs from the nucleus (62). YTHDC1 is also essential for the function of Xist, a long non-coding RNA involved in X-chromosome inactivation (63). The protein YTHDC2 localizes in the cytoplasm of germ cells, and facilitates the switch from mitosis to meiosis in both male and female gametogenesis (64–67). Several structural studies have shed light on recognition of m6A by the YTH family proteins. Analogous to the PHD and Tudor domain-containing enzymes which recognize methyl-lysine and methyl-arginine on histones tails, the YTH proteins contain a hydrophobic pocket and an “aromatic cage” to bind m6A-containing RNAs (68–71).
The insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1/2/3) recognize m6A modified RNAs and promote stability and storage of target RNAs (72). In addition to the presence of m6A, other features such as the location of the modified base can also determine interactions. The eukaryotic initiation factor 3 (eIF3) interacts with m6A-modified 5’UTR through a multi-subunit interface. This interaction recruits the 40S preinitiation complex to stimulate translation initiation, and allows for cap-independent translation of mRNA’s with m6A modified 5′ UTR (21). Conversely, the presence of m6A can also interfere with reader binding. The proteins RasGAP SH3 Domain Binding Protein 1 and 2 (G3BP1 and G3BP2) are strongly repelled by the presence of m6A and binding of G3BP1 to mRNA enhances stability (73). Interestingly, the effect of m6A on G3BP1 binding is context dependent. While m6A in the context of its consensus sequence GGACU strongly repelled G3BP1 binding, the presence of m6A in a GAACU sequence does not have a noticeable effect (73). Binding of direct m6A readers, such as Fragile X Mental Retardation 1 (FMR1), can also be context dependent (73). While the m6A modification does not affect Watson-Crick base pairing, the presence of the methyl group influences RNA secondary structure (74) and in vivo presence of m6A favors a ssRNA conformation (75). These structural alterations facilitate binding of several proteins: heterogeneous nuclear ribonucleoprotein C (HNRNPC) and heterogeneous nuclear ribonuclearprotein G (HNRNPG), proteins responsible for pre-mRNA processing and mRNA maturation; as well as HNRNPA2B1, which interacts with the microprocessor protein DGCR8 to facilitate maturation of miRNAs (24,76,77).
Several of the readers mentioned above, as well as the writer and eraser proteins mentioned earlier, have been implicated in the development of disease. Below, we discuss the expanding role m6A-dependent post-transcriptional regulation has in the development and progression of cancer.
Section 2: m6A in Cancer
Although cancer is a highly diverse disease, cancer cells share a number of traits. These “hallmarks” include factors such as dysregulated metabolism, limitless replication, and chronic inflammation (78,79). A number of m6A writers, readers, and erasers have been shown to play key roles in development and stem cell differentiation. Alterations in these pathways or dysregulation of epitranscriptomic regulators can drive tumorigenesis. While m6A modifications have been implicated in a number of cancers, it is interesting to note that while some enzymes play similar oncogenic or tumor-suppressor roles across tissue types, others function in a more tissue-specific manner (Table 1). Here, we expand upon the disruptions to methylation-dependent pathways observed in cancer, as well as the effects altered m6A-regulation have on driving cancer phenotypes.
Table 1:
m6A regulators associated with cancer.
| Cancer Type | Enzy me | Oncogene/Tumor Suppresor | Pathway(s) Affected | Hallmark of Cancer | Reference |
|---|---|---|---|---|---|
| Acute Myeloid Leukemia |
METT L3 |
oncogene | c-Myc, Blc2, PTEN | Limitless replicative potential | [120] |
| METT L3 |
oncogene | CEBPZ, c-Myc, SP1, SP2 | Limitless replicative potential | [113] | |
| METT L3 |
oncogene | MYB/MYC | Increased cell proliferation | [87] | |
| METT L14 |
oncogene | MYB/MYC | Increased cell proliferation | [87] | |
| FTO | oncogene | ASB2 and RARA | Increased cell proliferation | [86] | |
| FTO | oncogene | MYC/CEBPA | Increased cell proliferation | [106] | |
| IGF2B P |
oncogene | MYC | Increased cell proliferation | [72] | |
| Breast Cancer | FTO | unknown | unknown | Unknown | [137] |
| AlkBH 5 | oncogene | HIF | Chronic inflammation | [92] | |
| Cervical Cancer | FTO | oncogene | unknown | Increased cell proliferation | [136] |
| Colorectal Cancer | YTHD F1 |
oncogene | c-Myc | Increased cell proliferation | [127] |
| YTHD C2 |
oncogene | HIF-1α | Metastasis | [90] | |
| Gastric Cancer | FTO | oncogene | unknown | Metastasis | [133] |
| YTHD F2 |
oncogene | unknown | Increased cell proliferation | [135] | |
| Glioblastoma | METT L3 |
tumor-suppressor | ADAM19 | Cell growth and invasiveness | [130] |
| METT L14 |
oncogene | ADAM19 | Cell growth and invasiveness | [130] | |
| AlkBH 5 |
oncogene | FOXM1 | Cell-cycle regulation | [114] | |
| Hepatocellular Carcinoma | METT L3 |
tumor-suppressor | SOCS2 | Increased cell proliferation | [123] |
| METT L14 |
tumor-suppressor | miR-126 | Angiogenesis | [117] | |
| YTHD F1 |
unknown | unknown | Unknown | [134] | |
| YTHD F2 |
oncogene | miR-145 | Increased cell proliferation | [118] | |
| Lung adenocarcinoma | METT L3 |
oncogene | EGFR, TAZ | Increased cell proliferation | [124] |
| Malignant Pleural Mesothelioma | FTO | unknown | unknown | Unknown | [84] |
| Non-Small Cell Lung Cancer | METT L3 |
oncogene | miR-33-a | Increased cell proliferation | [89] |
| Pancreatic Cancer | METT L3 |
oncogene | mitogen-activated protein kinase cascades | Evading apoptosis Increased cell proliferation | [125] |
| YTHD C2 |
unknown | unknown | Unknown | [81] | |
| YTHD F2 |
oncogene tumor suppressor | EMT/YAP p-AKT/GSK3b/C yclinD1 |
Tissue invasion / metastasis Cell proliferation | [122] | |
| Renal Cell Carcinoma | METT L3 |
tumor-suppressor | EMT PI3K-Akt-mTOR |
Increased cell proliferation, tissue invasion, and metastasis | [121] |
2.1. Disruption of m6A-dependent pathways in cancer:
m 6A is a critical regulator of both self-renewal and differentiation in cells (25, 32, 36), and many of these same proteins that drive development can also become oncogenic when dysregulated. Genetic mutations, epigenetic reprogramming, and metabolic reprograming can all lead to changes in gene expression that allow cancer cells to overcome pathways that ensure tissue homeostasis. Several mechanisms are discussed below that have been shown to lead to either a loss- or gain-of-function in m6A pathways in cancer.
2.1.1. Impact of Mutations and SNP variants on m6A-dependent pathways:
Analysis of patient data from the Cancer Genome Atlas (TCGA) found that mutations and/or copy number variations (CNVs) of m6A writer, reader, and eraser genes were strongly associated with the presence of TP53 mutations in acute myeloid leukemia (AML) patients. Furthermore, these mutations were associated with poorer patient outcomes. Because TP53 is such a strong driver of cancer, additional studies may help elucidate the link between m6A writers/readers/erasers and TP53 in the progression of AML (80). A study on CNVs of patients with pancreatic adenocarcinoma identified loss of one copy of YTHDC2 as a potential candidate for pancreatic cancer susceptibility (81). While no complete loss of function has been described for METTL3 and METTL14, a highly conserved arginine of METTL14 is frequently mutated to proline in endometrial cancer. This mutation disrupts substrate recognition and reduces methylation activity (82) (Figure 3A). Genomic variations involved in the regulation of components of the m6A machinery have also been implicated in cancer. Single nucleotide polymorphisms (SNPs) in the first intron of FTO (rs7206790, rs8047395, rs9939609 and rs1477196) have been reported to be significantly associated with an increased risk of breast cancer (83) and individuals with the AT or AA genotype at rs9939609 are more likely to develop malignant pleural mesothelioma (MPM) when compared to the TT genotype (84).
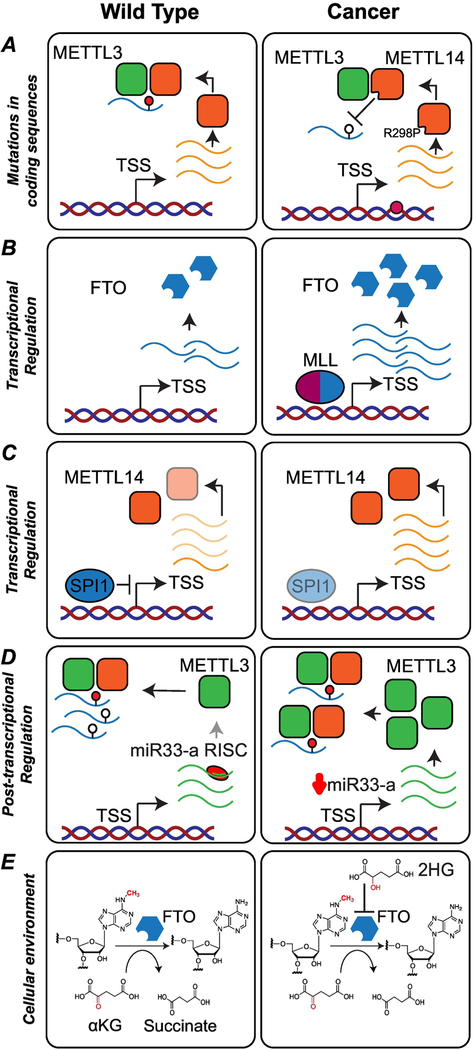
Figure 3. Cancer-induced disruptions of m6A-dependent pathways.
(A) Mutation of a highly conserved arginine in METTL14 (R298P) is frequently observed in endometrial cancer. This mutation diminishes methylation activity by disrupting substrate recognition. (B) In Mixed Lineage Leukemia (MLL)-rearranged acute myeloid leukemia (AML), FTO expression is up-regulated through binding of MLL-fusion proteins to CpG sites in the FTO locus. (C) In individuals with AML, the transcription factor SPI1, a repressor of METTL14, is suppressed, resulting in the upregulation of METTL14 and enhancement of self-renewal and proliferation. (D) Production of METTL3 can be repressed post-transcriptionally by microRNA-mediated gene silencing. In non-small cell lung cancer, miR-33a, a regulator of METTL3, is expressed at low levels, resulting in higher levels of METTL3 protein. (E) Mutations in IDH enzymes lead to the production of the onco-metabolite 2-hydroxyglutarate (2HG). 2HG is structurally similar to αKG, and acts as a competitive inhibitor of the m6A demethylases FTO and ALKBH5.
2.1.2. Transcriptional regulation of m6A pathway components:
Transcription factors are critical regulators of gene expression, and dysregulation of transcription factors is widespread in cancer (85). In Mixed Lineage Leukemia (MLL)-rearranged AML, when compared to non-MLL-rearranged AML samples or normal controls, FTO expression is upregulated through binding of MLL and particularly MLL-fusion proteins to CpG sites in the FTO locus (Figure 3B) (86). During myeloid differentiation, the transcription factor SPI1 is upregulated, repressing expression of METTL14, and indirectly of METTL14 targets that require m6A for mRNA stability. In AML cells, SPI1 expression is suppressed, resulting in the upregulation of METTL14 and enhancement of self-renewal and proliferation (87) (Figure 3C).
2.1.3. Post-transcriptional regulation:
Expression of m6A-dependent pathway components is also disrupted at the RNA level by post-transcriptional pathways. MicroRNAs (miRNAs) are single-stranded non-coding RNAs 19–25 nt in length that regulate gene expression by base-pairing to complementary sites in the 3′ UTR of mRNAs (88). In clinical samples of non-small cell lung cancer (NSCLC), levels of METTL3 are higher in tumors when compared with corresponding peri-tumor tissues (89). Interestingly, the levels of METTL3 are negatively correlated with the levels of miR-33a, a miRNA that targets the 3′ of METTL3 and is expressed at low levels in NSCLC (Figure 3D) (89). Expression of miR-33a was found to inhibit the proliferation of A549 and NCI-H460 cells, and downregulate the expression of the oncogenes EGFR, TAZ, MAPKAPK2, and DNMT3A at the protein level (89). In colon cancer, YTHDC2 is upregulated and promotes translation of HIF-1α through unwinding of the 5′ UTR and contributes to colon tumor metastasis (90).
2.1.4. Environment changes:
The tumor microenvironment influences tumor survival and progression, and m6A-dependent regulation of gene expression has been shown to respond to multiple extracellular cues. One important factor of the tumor microenvironment is oxygen availability. Under conditions of hypoxia, expression of ALKBH5 and ZNF217, two proteins that lead to reduction of m6A levels, is upregulated in breast cancer cells in a HIF-1α-dependent manner (91–93). Additionally, hypoxia has been shown to affect the splicing of YTHDC1 in gynecological tumor cell lines. Under hypoxia, changes in the splicing of YTHDC1 generate isoforms targeted by nonsense-mediated decay (NMD) which results in lower expression levels of the YTHDC1 protein. Signaling pathways also have an impact on m6A-dependent gene regulation. In human embryonic stem cells, Mothers Against Decapentaplegic Homolog 2 and Homolog 3 (SMAD2 and SMAD3), intracellular effectors of the Transforming growth factor beta (TGFβ) signaling pathway, promote binding of the methylation complex to a subset of transcripts involved in early cell fate decisions. m6A modification of these transcripts primes them for fast turn-over upon differentiation (94). While a direct link between TGFβ and m6A has not yet been described in cancer, this signaling pathway is essential for cancer progression (95). The cellular metabolic environment can also lead to changes in m6A-dependent regulation of gene expression. A powerful example is the accumulation of 2-hydroxyglutarate (2HG) in cells, a metabolite generated by mutations to isocitrate dehydrogenase (IDH). Mutations to IDH1 or IDH2 are present in approximately 80% of grade II-III gliomas (96–98), 50% of sporadic central and periosteal chondrosarcomas (99), 20–40% of angioimmunoblastic T cell lymphomas (AITL) (100), 10% to 20% of intrahepatic cholangiocarcinomas (101), and 12–16% of AML patients (102). Mutations in IDH have also been detected in hepatocellular carcinoma (HCC) (103) and prostate cancer (104). While wild type IDH enzymes catalyze the oxidative decarboxylation of isocitrate to generate α-ketoglutarate (αKG) using NADP+ as a cofactor, mutations to IDH1 (Arg132) or IDH2 (Arg140 or Arg172) confer a neomorphic, gain-of-function activity to the enzymes, leading to the production of the onco-metabolite 2HG (105). The oncometabolite 2HG, is structurally similar to αKG and functions as a competitive inhibitor of enzymes in the αKG-dependent dioxygenases family, a group that includes DNA and histone demethylases as well as the RNA demethylases FTO and ALKBH5 (Figure 3E). Interestingly, there may be cell and cancer-specific sensitivity or resistance to 2HG. In AML cells overexpressing FTO, 2HG displays anti-tumor effects. Similar sensitive and resistant phenotypes were also observed in glioblastoma cells, and taken together, may explain the responses of certain AML and glioblastoma patient populations to standard chemotherapy regimens (106). Finally, m6A-methylases have also been implicated in the process of arsenite-induced cell malignant transformation. In cells exposed to sodium arsenite, high levels of METTL3/METTL14 were shown to be correlated with the down-regulation of several miRNAs known to be involved in both small cell and non-small cell lung cancer. This suggests that external pollutants can also influence m6A-dependent pathways and play a critical role in driving carcinogenesis (107).
2.2. Dysregulation of m6A-dependent pathways contributes to the cancer phenotype
Dysregulation of m6A-dependent regulatory pathways leads to changes in expression in multiple pathways, including regulators of gene expression. Defects in such processes may contribute to cancer development and tumorigenesis. Below we describe some of the targets dysregulated as a consequence of a loss or gain of m6A regulation and their contributions to development of the cancer phenotype.
2.2.1. Transcription factors controlled through m6A-dependent pathways:
c-MYC, which lies at the crossroads of many growth-promoting signaling pathways, is under tight transcriptional control (108). MYC expression is correlated with self-renewal activity, and is considered a marker for stem cells (109). Additionally, c-MYC and N-MYC have been shown to participate in reprogramming fibroblasts into induced pluripotent stem cells (110). Because MYC drives promoter escape and the transcription of numerous genes, even small changes in c-MYC levels, including changes in mRNA half-life dependent upon regulation by m6A, may have a global impact on cellular phenotype. In fact, m6A-dependent c-MYC dysregulation has now been described in multiple cancers.
MYC expression is dysregulated in the context of tumors with IDH mutations and 2HG accumulation. In leukemia and glioma cells overexpressing FTO, 2HG displays anti-tumor effects. Mechanistically, inhibition of FTO by 2HG leads to an accumulation of m6A on MYC transcripts, destabilizing MYC mRNA and leading to downregulation of MYC signaling pathways (Figure 4A). This feedback mechanism could serve to further downregulate FTO. As 2HG is also able to inhibit the Ten-eleven translocation (TET) demethylases, this may explain why IDH and TET2 mutations are not observed together in AML (106).
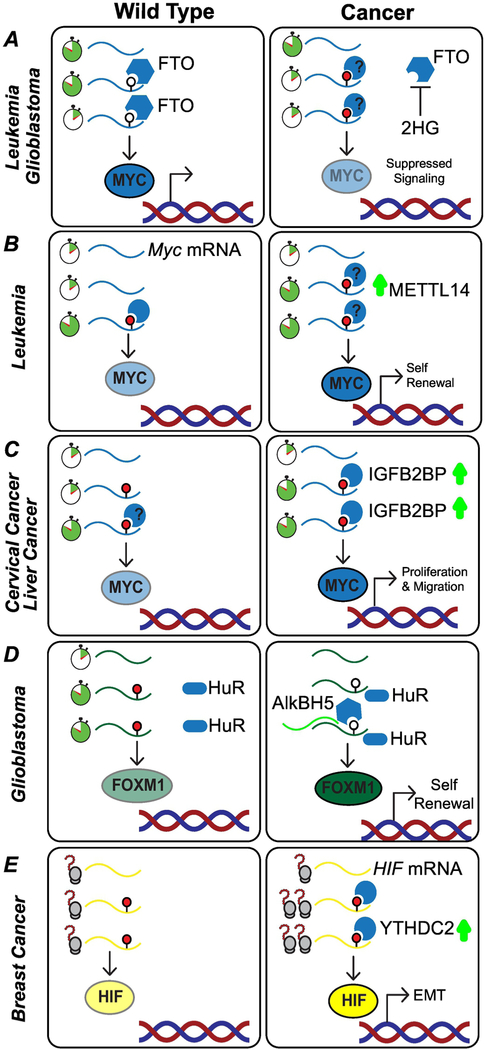
Figure 4: m6A-dependent pathways regulate control of cellular transcription factors.
(A) Mutations in IDH lead to the production of the onco-metabolite 2-hydroxyglutarate (2HG). Accumulation of 2HG inhibits the activity of the m6A demethylase FTO, leading to downregulation of MYC and suppressed MYC signaling. (B) Upregulation of METTL14 in individuals with acute myeloid leukemia (AML) leads to increased deposition of m6A. Stabilization of MYC mRNA, through interaction with an unknown reader protein, drives a self-renewal phenotype in the cancer cells; (C) In cervical cancer and hepatocellular carcinoma (HCC) cells, the m6A reader IGFB2BP stabilizes MYC mRNA, leading to increased cellular proliferation; (D) In glioblastoma cells, the lncRNA FOXM1-AS mediates the interaction between HuR and the demethylase ALKBH5, increasing expression of the transcription factor FOXM1 and driving a self-renewal phenotype. (E) Translation of the transcription factor HIF-1α is promoted by the m6A reader YTHDC2. HIF activates the transcription of genes that play a role in angiogenesis and adaptive response.
Conversely, stabilization of MYC can drive cellular proliferation and tumorigenesis. In AML, MYB and MYC, are stabilized through suppression of SP1 and over-expression of METTL14 (87). High levels of METTL14 expression correlate with increased deposition of m6A on MYB and MYC transcripts, suggesting that the increase in transcript half-life and stability is mediated through the actions of an m6A reader protein, whose identity remains unknown (Figure 4B).
An increase in MYC stability can also be driven by overexpression of ‘readers’ with a stabilizing role. The IGF2BPs are frequently amplified or highly expressed in various human cancers. Reduction in individual IGF2BP levels in vivo repressed expression of MYC, and led to phenotypes that mimic MYC silencing, such as inhibited cancer cell proliferation, colony formation ability and cell migration/invasion (72) (Figure 4C).
METTL3/METTL14 and FTO have also been shown to be co-expressed with members of the Ccaat-enhancer-binding protein (C/EBP) family. C/EBP proteins are widely expressed CCAAT box binding transcription factors that regulate a number of cellular processes, including energy metabolism, inflammation, hematopoiesis, cellular proliferation, and cellular differentiation in a context-specific manner (110). Binding sites for C/EBPs are present in the promoter regions of numerous genes that are expressed in myeloid cells (111) and are thought to be important negative regulators of cell proliferation (112). In mouse primary leukemia driven by MLL-AF9 fusions, METTL3 was found to be closely co-expressed and co-localized with CEBPZ, which is required for the recruitment of METTL3 to transcriptional start sites on chromatin (113). Additionally, expression of the transcription factor CEBPA was correlated with FTO expression. Inhibition of FTO by 2HG lead to an accumulation of m6A on CEBPA transcripts, which is recognized by the reader YTHDF2 and targeted for degradation. This feedback mechanism could serve to further downregulate FTO in AML (106).
The transcription factor Forkhead Box Protein M1 (FOXM1), an important component in cell-cycle regulation, is also an m6A target implicated in the development of cancer (114). In glioblastoma, levels of FOXM1 mRNA and protein are regulated by ALKBH5. This demethylase is highly expressed in cell lines or patient-derived primary glioblastoma cultures. High expression of ALKBH5 correlates with poor prognosis in glioblastoma patients. While knockdown of ALKBH5 had no effect on growth of non-glioma cells, loss of ALKBH5 activity inhibited growth and self-renewal of glioblastoma stem cells. Upregulation of ALKBH5 results in loss of m6A in the FOXM1 mRNA, enabling interaction with HuR, and leading to an increase in FOXM1 levels (Figure 4D). Interestingly, the interaction between FOXM1 and ALKBH5 is mediated by the lncRNA FOXM1-AS (114).
Another class of transcription factors that are frequently dysregulated in cancer are the Hypoxiainducible factor (HIF) transcription factors, which serve as regulators of cellular response to hypoxic conditions. HIF-1α activates the transcription of genes that play critical roles in angiogenesis and adaptive responses (115). Translation of HIF-1α is promoted by the m6A reader YTHDC2 (90) (Figure 4E). Finally, transcription factors that contribute to a stem cell state are also co-opted in cancer cells through dysregulation of m6A-dependent pathways. In breast cancer, hypoxia dependent over-expression of ALKBH5 leads to reduced m6A methylation of mRNAs coding for core pluripotency factors such as NANOG. Decreased m6A levels in NANOG mRNA result in increasing the stability of the mRNA, and consequent accumulation of the protein, linking the ALKBH5, and m6A-dependent pathways to the establishment of breast cancer stem cells (92).
2.2.2. Disruption of m6A-pathways impacts post-transcriptional regulation:
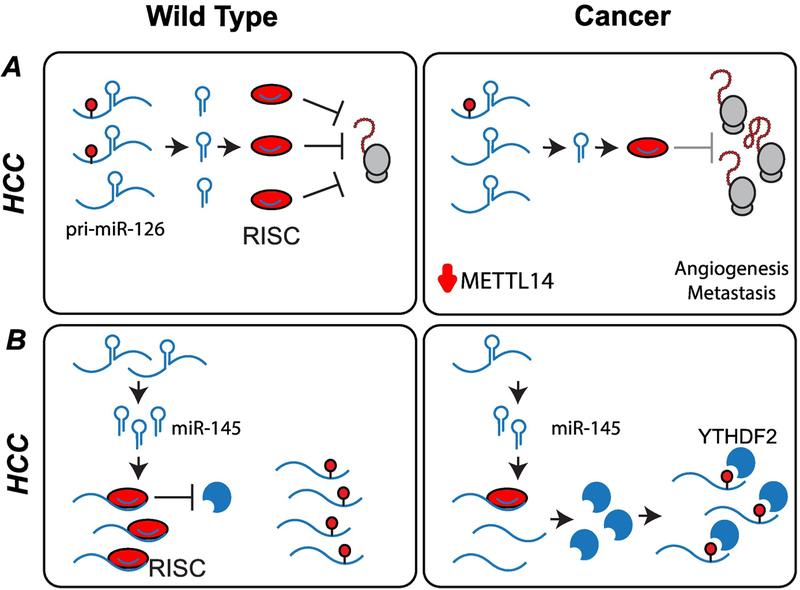
Up-regulation or down-regulation of miRNAs can have tumor-suppressor or oncogenic effects that are context- or tissue-specific (116). Patients with portal vein tumor thrombus (PVTT) (indicative of late-stage HCC) showed significantly reduced METTL14 levels compared to patients with non-metastatic tumors, or metastatic tumors without PVTT. Knockdown of METTL14 in HCC cells resulted in a decrease in migration and invasiveness. The unprocessed pri-miR126, a metastasis-associated miRNA, accumulated in METTL14 knockdown cells, while it was more rapidly processed in cells overexpressing METTL14 (Figure 5A). When m6A was immunoprecipitated directly, it was found that METTL14 overexpression significantly increased the amount of pri-miR-126 modified by m6A, indicating that this miRNA is a direct target of the methyltransferase complex (117).
Figure 5: Post-transcriptional regulation of gene expression by miRNA.
(A) Downregulation of METTL14 in hepatocellular carcinoma (HCC) cells leads to an increase in unprocessed pri-miR-126. Mature miR-126 is associated with metastasis. (B) In HCC cells, lower expression of miR-145 allows for expression of YTHDF2 protein, decreasing the levels of m6A in cells, and increasing cellular proliferation.
In HCC, the expression of YTHDF2 is negatively correlated with expression of miR-145, a microRNA that is expressed at lower levels when compared to adjacent peritumorous tissues. In a HepG2 cellular model, it was found that overexpression of miR-145 suppressed YTHDF2 at the mRNA and protein levels, thereby increasing the levels of m6A in cells, and decreasing cellular proliferation (Figure 5B). miR-145 has multiple cellular targets, and it remains to be determined if it can affect the RNA targets of other m6A writers, readers, and erasers (118).
2.2.3. Signaling pathways affected by m6A-dependent pathway dysregulation:
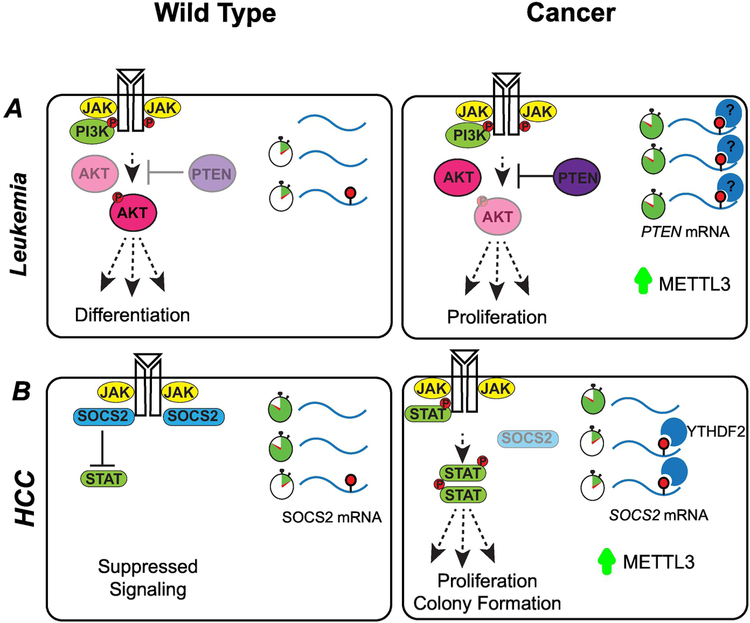
Cancer is driven by alterations that allow cells to escape mechanisms that normally control their survival and proliferation. Many of these alterations affect signaling pathways that control cell growth and division. In T cell homeostasis, the balance between the IL-7-mediated JAK–STAT and TCR-mediated ERK/AKT signaling pathways is controlled by m6A-dependent pathways (119). In multiple cancers, changes in gene expression of components of this signaling pathway have been linked to changes in m6A activity. METTL3 mRNA and protein expression was found to be elevated in multiple AML cell lines relative to normal hematopoietic cells (120). Upregulation of METTL3 results in higher levels of m6A in the PTEN mRNA, leading to an increase in PTEN protein. Increased PTEN activity reduces PI3K/AKT pathway signaling cascades and differentiation (Figure 6A). When cells were transplanted into immunodeficient recipient mice after knockdown of METTL3, animals showed a delay in leukemia development as compared to a control group.
Figure 6: m6A-dependent pathways influence signaling cascades.
(A) METTL3 is expressed more abundantly in acute myeloid leukemia (AML) cells than healthy hematopoietic stem cells. Upregulation of METTL3 results in increased mRNA methylation and higher expression levels of factors critical for the regulation of apoptosis and differentiation. For example, increased PTEN activity reduces PI3K/AKT pathway signaling, leading to proliferation and maintenance of the hematopoietic stem cell program. (B) In hepatocellular carcinoma (HCC) cells, increased METTL3 results in increased expression of SOCS2, a member of the JAK-STAT signaling pathway. Increased levels of SOCS2 lead to cellular proliferation and carcinogenesis.
In renal cell carcinoma (RCC), downregulation of METTL3 significantly promotes cell growth and colony formation. In RCC cells, expression of p-PI3K, p-AKT, p-mTOR, and p-P70 is significantly higher. Additionally, it was observed that vimentin, beta-catenin, and N-cadherin levels were significantly higher in METTL3 knockdown cells, suggesting that the epithelial-mesenchymal transition (EMT) pathway may also be altered by METTL3 knockdown (121).
In pancreatic cancers, YTHDF2 expression level is associated with poor patient prognosis, and expression increases with progression from stage I to stage IV. In several model pancreatic cancer (PC) cell lines, YTHDF2 is upregulated at both the mRNA and the protein level. Upon YTHDF2 knockdown, PC cells show reduced colony formation, lower colony density and reduced growth curves. In these cells, the protein levels for GSK3b and CyclinD1 were downregulated, leading to the hypothesis that YTHDF2 acts through the Akt/GSK3b/CyclinD1 pathway. Interestingly, YTHDF2 knockdown cells showed increased migration, invasion, and adhesion abilities in PC. This EMT upregulation occurs through the yes-associated protein (YAP) gene, although the exact link between YTHDF2 and YAP remains to be determined (122).
In the HCC-derived cell lines Huh-7 and HepG2, knockdown of METTL3, decreased cell proliferation and colony forming capacity while overexpression led to accelerated cell migration in a transwell assay. Gene expression studies in METTL3 KD cells identified 15 genes that were consistently upregulated in both cell types. Suppressor of Cytokine Signaling 2 (SOCS2), a member of the JAK-STAT signaling pathway, is modified by m6A and is a direct downstream target of METTL3. In HCC, SOCS2 down-regulation is significantly correlated to advanced TNM staging. SOCS proteins affect cellular response to cytokines and growth factors by regulating the JAK/STAT signaling pathway. Dysregulation of METTL3 may result in altered SOCS2 expression, leading to carcinogenesis (123) (Figure 6B). The EGFR signaling pathway is also regulated through proteins in the m6A pathway.
In lung adenocarcinoma (LUAD), up-regulated METTL3 directly promotes the translation of several known oncogenes, including EGFR, TAZ, MK2 and DNMT3a. METTL3 enhances the translation of its targets by interacting with eukaryotic initiation factor 3 (eIF3) in an RNA-independent manner. Most interestingly, this effect occurred independently of the binding partner METTL14, or the catalytic domain of METTL3 (124).
Section 3: Clinical Treatments and Drugability
3.1. Treatment sensitization or resistance:
Acquisition of resistance to therapeutic drugs by cancer cells represents one of the major obstacles in cancer treatment. Changes in gene expression mediated through alteration in m6A-dependent pathways can lead to changes in sensitivity to chemo and radio therapy and thus impact treatment regimens. In pancreatic cancer, knockdown of METTL3 resulted in higher sensitivity to gemcitabine, 5-fluorouracil, cisplatin, and irradiation (125). Microarray data and GO-term analysis identified a number of targets of METTL3, including genes involved in the MAPK signaling cascades, RNA splicing, and cell-cycle regulation. In glioblastoma cells, knockdown of METTL3 decreased proliferation, increased apoptosis, and increased sensitivity to temozolomide and irradiation (126). Finally, in colorectal cancer, knockdown of YTHDF1 resulted in inhibition of cell proliferation and increased sensitivity to both 5-flurouracil and oxaliplatin. Further analysis revealed that c-Myc was associated with the 5′ transcription start site of YTHDF1. Upon c-Myc knockdown, YTHDF1 was inhibited in a dose-dependent manner, but other YTH family proteins were unaffected (127). Given the important role that MYC plays in a number of cancers, further exploration of this relationship could increase our understanding of how m6A regulation plays a role in cancer. m6A-dependent pathways might also have a direct role in sensitivity to therapies that target DNA, due to its role in DNA repair (28).
3.2. Direct Drugability:
In addition to modulating the efficacy of traditional cancer treatments, a number of studies have investigated the direct drugability of the m6A pathway. The natural product Rhein acts as a competitive inhibitor of FTO and can prevent recognition of m6A sites by FTO in cells (128). However, one drawback of Rhein is its poor selectivity. Subsequent research identified the FTO inhibitor meclofenamic acid (MA) (129). The selectivity of MA towards FTO relies in part on the interaction of MA with the FTO nucleotide recognition lid, a structural feature that is absent in other ALKBH family members, including ALKBH5. This loop provides a hydrophobic interaction surface for the inhibitor. Treatment of cells with the ethyl ester form of MA (MA2), results in increased m6A methylation of mRNA, indicating this compound is able to achieve good levels of cellular penetration. Upon FTO knockdown, m6A levels were not further affected, indicating this compound does not act through the ALKBH5 demethylation pathway. In glioblastoma cells, treatment with MA2 inhibited tumor progression (130). A third inhibitor of FTO, characterized by Toh et al., exhibits 30 to 130-fold selectivity over other members of the αKG-dependent dioxygenase family, including ALKBH5, PHD2, and KDM4A. Crystallography revealed that hydrogen-bonding interactions with the backbone of Glu234 (2.9 Å) in FTO play an important role in selectivity, as the inhibitor is unable to have similar backbone interactions with other AlkB family members (average distance ~ 4.6 Å) (131). Finally, the compound IOX3 was found to decrease protein expression of FTO in C2Cl2 cells. However, reduced expression of FTO did not appear to have a significant effect on cellular m 6A levels. In this case, it may be difficult to discern the effects of FTO inhibition from inhibition of other αKG-dependent dioxygenases, including the PHD family, which are targets of IOX3 (132). The ability to target m6A-dependent pathways might allow for the development of combination therapies. In AML cells, knockdown of FTO substantially enhanced ATRA-induced cell differentiation, suggesting that inhibiting FTO in combination with ATRA treatment represents a promising therapeutic strategy to treat leukemia (86).
Section 4. Preliminary Clinical Observations:
While our understanding of the role of m6A writers, readers, and erasers in some cancers has dramatically improved, in several clinical studies, m6A writers and erasers have been implicated in tumorigenesis, although the affected underlying pathways are still unknown. In patients with gastric cancer, it was observed that high FTO levels significantly correlated with lymph node metastasis, high TNM stage, and poor prognosis. In cell culture models, FTO overexpression led to increased cellular proliferation and migration phenotypes (133). In patients with HCC, the reader protein YTHDF1 was found to be overexpressed and was positively correlated with late-stage cancer and poor patient survival rates (134). Similarly, YTHDF2 was found to be upregulated in gastric cancer tissues and shRNA knockdown of YTHDF2 in MGC-803 cells was found to inhibit proliferation and promote apoptosis (135). In cervical cancer, it was observed that cancer tissue samples had decreased m6A levels as compared to paired normal tissue, and that these decreased levels showed strong correlation with cancer progression and patient outcomes.
In cellular models, knockdown of METTL3/METTL14 or overexpression of FTO and ALKBH5 promoted cell proliferation. Conversely, knockdown of FTO and ALKBH5 or overexpression of METTL3 and METTL14 suppressed cancer development (136). Indeed, FTO expression varies with the sub-type of breast cancer, with tumors from patients with hormone receptor (HR)-negative and (human epidermal growth factor receptor 2 (HER2) overexpression breast cancer displaying higher levels of FTO expression when compared to HR positive and HER2 negative breast cancer (137). Additionally, the levels of m6A writers, readers, and erasers, as well as the levels of m6A-modified RNA, may serve as a potential prognostic biomarkers to assist in the selection of treatment options and assess patient’s outcomes (138,139). Taken together, these clinical observations provide a number of interesting, unanswered questions. Further studies are needed to explore the underlying mechanisms.
Conclusions:
RNA post-transcriptional modifications can influence RNA structure and attract or repel RNA binding proteins. As a consequence, RNA post-transcriptional modifications are involved in all stages of the RNA life cycle, and have significant impact on the output of gene expression programs. Throughout development, and in maintaining homeostasis, several pathways are regulated in part by pathways dependent on RNA modifications. These pathways are often co-opted by the cancer cells, and disruption of RNA post-transcriptional regulation contributes to both establishment and progression of cancer. In this review, we focused on m6A, and the emerging principles of how m6A methylation contributes to cancer progression and metastasis. During development, the RNA modification m6A has a significant impact on cell identity, and in mouse embryonic stem cells, loss of m6A modification can result in different phenotypes depending on cellular identity (25,140,141). This early observation suggested that cellular identity is a key determinant of the phenotypic consequences of disrupting m6A-dependent gene regulation, a phenomenon also observed in cancer studies. Considering the heterogeneity observed in tumors, it becomes critical to understand how m6A-dependent pathways interact with both intracellular factors, such as metabolite levels or RNA binding proteins present, and extracellular factors such as oxygen availability or signaling pathways in order to effectively target these pathways in a therapeutic purpose. Future work aimed at understanding epitranscriptomic dysregulation and cancer tumorigenesis may increase our understanding of malignant transformations and aid in the design of novel therapeutics to treat cancer. (See Table 1.)
Supplementary Material
Highlights:
N6-methyladenosine (m6A) plays an important role in regulating gene expression
m6A-dependent pathways may have either a tumor-suppressive or oncogenic role
Targeting m6A-dependent pathways represents a promising new therapeutic route
Acknowledgements:
We thank George Leiman and members of the Batista Lab for discussion and critical reading of the manuscript. We apologize to colleagues whose works are not discussed due to space limitations. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, United States of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018. January 4;46(D1):D303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011. October;2(5):611–31. [DOI] [PubMed] [Google Scholar]
- 3.Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, et al. Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem. 2017. 01;292(35):14695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012. June;40(11):5023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017. 05;18(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017. May;27(5):606–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016. February 25;530(7591):441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016. May;12(5):311–6. [DOI] [PubMed] [Google Scholar]
- 9.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature. 2017. 19;541(7637):371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014. May;15(5):293–306. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Zhang S, Yuan Q. N(6)-Methyladenosine Methyltransferases and Demethylases: New Regulators of Stem Cell Pluripotency and Differentiation. Stem Cells Dev. 2016. 15;25(14):1050–9. [DOI] [PubMed] [Google Scholar]
- 12.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974. October;71(10):3971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975. April;4(4):379–86. [DOI] [PubMed] [Google Scholar]
- 14.Lavi S, Shatkin AJ. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc Natl Acad Sci U S A. 1975. June;72(6):2012–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977. April 19;16(8):1672–6. [DOI] [PubMed] [Google Scholar]
- 16.Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977. October 5;115(4):695–714. [DOI] [PubMed] [Google Scholar]
- 17.Adams JM, Cory S. Modified nucleosides and bizarre 5’-termini in mouse myeloma mRNA. Nature. 1975. May 1;255(5503):28–33. [DOI] [PubMed] [Google Scholar]
- 18.Furuichi Y, Shatkin AJ, Stavnezer E, Bishop JM. Blocked, methylated 5’-terminal sequence in avian sarcoma virus RNA. Nature. 1975. October 16;257(5527):618–20. [DOI] [PubMed] [Google Scholar]
- 19.Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci U S A. 1975. May;72(5):1904–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011. October 16;7(12):885–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012. June 22;149(7):1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012. April 29;485(7397):201–6. [DOI] [PubMed] [Google Scholar]
- 23.Harper JE, Miceli SM, Roberts RJ, Manley JL. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990. October 11;18(19):5735–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015. October 1;29(19):2037–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014. December 4;15(6):707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015. February 26;518(7540):560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knuckles P, Carl SH, Musheev M, Niehrs C, Wenger A, Bühler M. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat Struct Mol Biol. 2017. July;24(7):561–9. [DOI] [PubMed] [Google Scholar]
- 29.Slobodin B, Han R, Calderone V, Vrielink JAFO, Loayza-Puch F, Elkon R, et al. Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation. Cell. 2017. April 6;169(2):326–337.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keith JM, Ensinger MJ, Mose B. HeLa cell RNA (2’-O-methyladenosine-N6-)-methyltransferase specific for the capped 5’-end of messenger RNA. J Biol Chem. 1978. July 25;253(14):5033–9. [PubMed] [Google Scholar]
- 31.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994. July 1;269(26):17697–704. [PubMed] [Google Scholar]
- 32.Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018. March 15;69(6):1028–1038.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018. March 1;32(5–6):415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014. February;10(2):93–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang Y, Laurent B, Hsu C-H, Nachtergaele S, Lu Z, Sheng W, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017. 23;543(7646):573–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, et al. Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell. 2015. December 3;17(6):689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017. May 18;169(5):824–835.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017. November;18(11):2004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shima H, Matsumoto M, Ishigami Y, Ebina M, Muto A, Sato Y, et al. SAdenosylmethionine Synthesis Is Regulated by Selective N6-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 2017. December 19;21(12):3354–63. [DOI] [PubMed] [Google Scholar]
- 40.Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3’-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci U S A. 2016. 06;113(49):14013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scuteri A, Sanna S, Chen W-M, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007. July;3(7):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007. May 11;316(5826):889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007. June;39(6):724–6. [DOI] [PubMed] [Google Scholar]
- 44.Cheung MK, Gulati P, O’Rahilly S, Yeo GSH. FTO expression is regulated by availability of essential amino acids. Int J Obes 2005. May;37(5):744–7. [DOI] [PubMed] [Google Scholar]
- 45.Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q, et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010. April 22;464(7292):1205–9. [DOI] [PubMed] [Google Scholar]
- 46.Jia G, Yang C-G, Yang S, Jian X, Yi C, Zhou Z, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008. October 15;582(23–24):3313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013. January 10;49(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, et al. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014. April 25;289(17):11571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aik W, Scotti JS, Choi H, Gong L, Demetriades M, Schofield CJ, et al. Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014. April;42(7):4741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu C, Liu K, Tempel W, Demetriades M, Aik W, Schofield CJ, et al. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J Biol Chem. 2014. June 20;289(25):17299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou S, Toh JDW, Wong KHQ, Gao Y-G, Hong W, Woon ECY. N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci Rep. 2016. 09;6:25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerken T, Girard CA, Tung Y-CL, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007. November 30;318(5855):1469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem Sci. 2002. October;27(10):495–7. [DOI] [PubMed] [Google Scholar]
- 55.Patil DP, Pickering BF, Jaffrey SR. Reading m6A in the Transcriptome: m6A-Binding Proteins. Trends Cell Biol. 2018. February;28(2):113–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015. June 4;161(6):1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weng Y-L, Wang X, An R, Cassin J, Vissers C, Liu Y, et al. Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron. 2018. January 17;97(2):313–325.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014. January 2;505(7481):117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016. August 25;7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017. March;27(3):315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, Sun B-F, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016. February 18;61(4):507–19. [DOI] [PubMed] [Google Scholar]
- 62.Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017. October 6;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016. 15;537(7620):369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife. 2017. October 31;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017. September;27(9):1115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m6A Transcripts by the 3’→5’ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol Cell. 2017. October 19;68(2):374–387.e12. [DOI] [PubMed] [Google Scholar]
- 67.Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife. 2018. January 23;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014. November;10(11):927–9. [DOI] [PubMed] [Google Scholar]
- 69.Luo S, Tong L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc Natl Acad Sci U S A. 2014. September 23;111(38):13834–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Theler D, Dominguez C, Blatter M, Boudet J, Allain FH-T. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014. December 16;42(22):13911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014. December;24(12):1490–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018. March;20(3):285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017. October;24(10):870–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015. February 11;137(5):2107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J-W, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015. March 26;519(7544):486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015. September 10;162(6):1299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018. January 29;9(1):420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000. January 7;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 79.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011. March 4;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 80.Kwok C-T, Marshall AD, Rasko JEJ, Wong JJL. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017. February 2;10(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fanale D, Iovanna JL, Calvo EL, Berthezene P, Belleau P, Dagorn JC, et al. Germline copy number variation in the YTHDC2 gene: does it have a role in finding a novel potential molecular target involved in pancreatic adenocarcinoma susceptibility? Expert Opin Ther Targets. 2014. August;18(8):841–50. [DOI] [PubMed] [Google Scholar]
- 82.Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016. 21;63(2):306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaklamani V, Yi N, Sadim M, Siziopikou K, Zhang K, Xu Y, et al. The role of the fat mass and obesity associated gene (FTO) in breast cancer risk. BMC Med Genet. 2011. April 13;12:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khella MS, Salem AM, Abdel-Rahman O, Saad AS. The Association Between the FTO rs9939609 Variant and Malignant Pleural Mesothelioma Risk: A Case-Control Study. Genet Test Mol Biomark. 2018. February;22(2):79–84. [DOI] [PubMed] [Google Scholar]
- 85.Bhagwat AS, Vakoc CR. Targeting Transcription Factors in Cancer. Trends Cancer. 2015. September 1;1(1):53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell. 2017. January 9;31(1):127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell. 2018. February 1;22(2):191–205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017. February 26;8(1):45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017. January 22;482(4):582–9. [DOI] [PubMed] [Google Scholar]
- 90.Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016. 28;376(1):34–42. [DOI] [PubMed] [Google Scholar]
- 91.Thalhammer A, Bencokova Z, Poole R, Loenarz C, Adam J, O’Flaherty L, et al. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α (HIF-1α). PloS One. 2011. January 14;6(1):e16210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016. April 5;113(14):E2047–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016. August 31;7(40):64527–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature. 2018. March 8;555(7695):256–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pickup M, Novitskiy S, Moses HL. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer. 2013. November;13(11):788–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009. February 19;360(8):765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol (Berl). 2009. October;118(4):469–74. [DOI] [PubMed] [Google Scholar]
- 98.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009. January;30(1):7–11. [DOI] [PubMed] [Google Scholar]
- 99.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013. July;3(7):730–41. [DOI] [PubMed] [Google Scholar]
- 100.Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais J-P, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012. February 23;119(8):1901–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DiNardo CD, Propert KJ, Loren AW, Paietta E, Sun Z, Levine RL, et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood. 2013. June 13;121(24):4917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McKenney AS, Levine RL. Isocitrate dehydrogenase mutations in leukemia. J Clin Invest. 2013. September;123(9):3672–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee JH, Shin DH, Park WY, Shin N, Kim A, Lee HJ, et al. IDH1 R132C mutation is detected in clear cell hepatocellular carcinoma by pyrosequencing. World J Surg Oncol. 2017. April 12;15(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009. July 15;125(2):353–5. [DOI] [PubMed] [Google Scholar]
- 105.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010. March 16;17(3):225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell. 2018. January 11;172(1–2):90–105.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gu S, Sun D, Dai H, Zhang Z. N6-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol Lett. 2018. April 19; 292:1–11. [DOI] [PubMed] [Google Scholar]
- 108.Levens D You Don’t Muck with MYC. Genes Cancer. 2010. June 1;1(6):547–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008. December 4;3(6):611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007. November 30;131(5):861–72. [DOI] [PubMed] [Google Scholar]
- 111.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002. August 1;365(Pt 3):561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nerlov C The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007. July;17(7):318–24. [DOI] [PubMed] [Google Scholar]
- 113.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017. December 7;552(7683):126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017. April 10;31(4):591–606.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011. August 11;365(6):537–47. [DOI] [PubMed] [Google Scholar]
- 116.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ma J, Yang F, Zhou C, Liu F, Yuan J, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017. February 1;65(2):529–43. [DOI] [PubMed] [Google Scholar]
- 118.Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3’-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017. 03;292(9):3614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li H-B, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017. 17;548(7667):338–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017. November;23(11):1369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li X, Tang J, Huang W, Wang F, Li P, Qin C, et al. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget. 2017. October 10;8(56):96103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16(23):2259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen M, Wei L, Law C-T, Tsang FH-C, Shen J, Cheng CL-H, et al. RNA N6-methyladenosine methyltransferase METTL3 promotes liver cancer progression through YTHDF2 dependent post-transcriptional silencing of SOCS2. Hepatology. 2018; 67(6) 2254–2270. [DOI] [PubMed] [Google Scholar]
- 124.Lin S, Choe J, Du P, Triboulet R, Gregory RI. METTL3 promotes translation in human cancer cells. Mol Cell. 2016. May 5;62(3):335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018. February;52(2):621–9. [DOI] [PubMed] [Google Scholar]
- 126.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018. January 25;37(4):522–33. [DOI] [PubMed] [Google Scholar]
- 127.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018. January 26;9(7):7476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012. October 31;134(43):17963–71. [DOI] [PubMed] [Google Scholar]
- 129.Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015. January;43(1):373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017. March 14;18(11):2622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Toh JDW, Sun L, Lau LZM, Tan J, Low JJA, Tang CWQ, et al. A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor of N6-methyladenosine demethylase FTO. Chem Sci. 2015. January 1;6(1):112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McMurray F, Demetriades M, Aik W, Merkestein M, Kramer H, Andrew DS, et al. Pharmacological inhibition of FTO. PloS One. 2015;10(4):e0121829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017. October;38(4):2285–92. [DOI] [PubMed] [Google Scholar]
- 134.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark Sect Dis Markers. 2018;21(4):859–68. [DOI] [PubMed] [Google Scholar]
- 135.Zhang J, Pi J, Liu Y, Yu J, Feng T. [Knockdown of YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) inhibits proliferation and promotes apoptosis in MGC-803 gastric cancer cells]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin J Cell Mol Immunol. 2017. December;33(12):1628–34. [PubMed] [Google Scholar]
- 136.Wang X, Li Z, Kong B, Song C, Cong J, Hou J, et al. Reduced m6A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget. 2017. October 24;8(58):98918–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tan A, Dang Y, Chen G, Mo Z. Overexpression of the fat mass and obesity associated gene (FTO) in breast cancer and its clinical implications. Int J Clin Exp Pathol. 2015. October 1;8(10):13405–10. [PMC free article] [PubMed] [Google Scholar]
- 138.Oldenhuis CN a. M, Oosting SF, Gietema JA, de Vries EGE. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer Oxf Engl 1990. 2008. May;44(7):946–53. [DOI] [PubMed] [Google Scholar]
- 139.Goossens N, Nakagawa S, Sun X, Hoshida Y. Cancer biomarker discovery and validation. Transl Cancer Res. 2015. June;4(3):256–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015. February 27;347(6225):1002–6. [DOI] [PubMed] [Google Scholar]
- 141.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014. February;16(2):191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.