Human noroviruses (NoVs) cause sporadic and epidemic gastroenteritis worldwide. They are divided into seven genogroups (GI to GVII), with each genogroup further divided into several genotypes. Human NoVs belonging to genogroup II and genotype 4 (GII.4) are the most prevalent. Currently, there are no vaccines or antiviral drugs available for NoV infection. The protease encoded by NoV is considered a valuable target because of its essential role in replication. NoV protease structures have only been determined for the GI genogroup. We show here that the structure of the GII.4 protease exhibits several significant changes from GI proteases, including a unique pairing of an arginine with the catalytic histidine that makes the proteolytic activity of GII.4 protease pH sensitive. A comparative analysis of NoV protease structures may provide a rational framework for structure-based drug design of broadly cross-reactive inhibitors targeting NoVs.
KEYWORDS: Arg-His pairing, GII.4 protease, norovirus, X-ray crystallography, antiviral agents, protease inhibitors
ABSTRACT
Human noroviruses (NoVs) are the main cause of epidemic and sporadic gastroenteritis. Phylogenetically, noroviruses are divided into seven genogroups, with each divided into multiple genotypes. NoVs belonging to genogroup II and genotype 4 (GII.4) are globally most prevalent. Genetic diversity among the NoVs and the periodic emergence of novel strains present a challenge for the development of vaccines and antivirals to treat NoV infection. NoV protease is essential for viral replication and is an attractive target for the development of antivirals. The available structure of GI.1 protease provided a basis for the design of inhibitors targeting the active site of the protease. These inhibitors, although potent against the GI proteases, poorly inhibit the GII proteases, for which structural information is lacking. To elucidate the structural basis for this difference in the inhibitor efficiency, we determined the crystal structure of a GII.4 protease. The structure revealed significant changes in the S2 substrate-binding pocket, making it noticeably smaller, and in the active site, with the catalytic triad residues showing conformational changes. Furthermore, a conserved arginine is found inserted into the active site, interacting with the catalytic histidine and restricting substrate/inhibitor access to the S2 pocket. This interaction alters the relationships between the catalytic residues and may allow for a pH-dependent regulation of protease activity. The changes we observed in the GII.4 protease structure may explain the reduced potency of the GI-specific inhibitors against the GII protease and therefore must be taken into account when designing broadly cross-reactive antivirals against NoVs.
IMPORTANCE Human noroviruses (NoVs) cause sporadic and epidemic gastroenteritis worldwide. They are divided into seven genogroups (GI to GVII), with each genogroup further divided into several genotypes. Human NoVs belonging to genogroup II and genotype 4 (GII.4) are the most prevalent. Currently, there are no vaccines or antiviral drugs available for NoV infection. The protease encoded by NoV is considered a valuable target because of its essential role in replication. NoV protease structures have only been determined for the GI genogroup. We show here that the structure of the GII.4 protease exhibits several significant changes from GI proteases, including a unique pairing of an arginine with the catalytic histidine that makes the proteolytic activity of GII.4 protease pH sensitive. A comparative analysis of NoV protease structures may provide a rational framework for structure-based drug design of broadly cross-reactive inhibitors targeting NoVs.
INTRODUCTION
Human noroviruses (NoVs) are the main cause of viral gastroenteritis worldwide (1, 2). They are responsible for 95% of nonbacterial gastroenteritis and 50% of all gastroenteritis outbreaks worldwide (3). NoVs belong to the Caliciviridae family and are divided into seven genogroups (GI to GVII), with each genogroup further divided into several genotypes. Human pathogens are restricted to genogroups GI, GII, and GIV, with genogroup II and genotype 4 (GII.4) being the most prevalent, accounting for 80% of norovirus infections worldwide (4–6). Both genetic and antigenic diversity of NoVs contribute to challenges in the development of effective treatments, and although they are much needed, there are no licensed vaccines or antiviral drugs available for human NoV infections.
The NoV genome is composed of a positive-sense, single-stranded RNA with three open reading frames (ORFs) that encode a nonstructural precursor polyprotein (ORF1), major capsid protein, VP1 (ORF2), and minor capsid protein VP2 (ORF3) (7–10). The precursor polyprotein is cleaved into six nonstructural proteins by the viral protease, which is encoded by ORF1 as part of the polyprotein. NoV protease is a typical cysteine protease similar to coronavirus 3C protease, and its cleavage of the polyprotein is an essential first step in viral replication and maturation (11–17). The NoV protease, due to its role in viral replication and relatively high homology among different genogroups, is an attractive target for the development of broadly cross-reactive antivirals. To date, NoV protease structures have only been determined for the GI genogroup, with structures available for the prototype GI.1 Norwalk virus (NV Pro) and GI.4 Chiba virus (CV Pro). Crystal structures of human rhinovirus (18), enterovirus 71 (19), poliovirus (20), Caliciviridae member rabbit hemorrhagic disease virus (12), and other viral 3C proteases revealed a common catalytic triad in the active sites with a cysteine acting as a nucleophile, histidine as a base, and glutamate or aspartate as an anion. In the NV Pro, cysteine 139 (C139), histidine 30 (H30), and glutamate 54 (E54) form a catalytic triad (21). In contrast, similar to that in hepatitis A virus 3C protease, the CV Pro active site functions as a catalytic dyad, requiring only C139 and H30, but not E54, for activity (22–24). Overlay of the two structures shows further differences in the active site, as well as in the substrate binding pockets flanking the active site, suggesting that there may be structural variability between proteases belonging to different NoV genotypes. This variability observed in the NoV proteases may be a stumbling block in the development of effective cross-reactive inhibitors. Several groups are engaged in developing structure-based NoV protease inhibitors as a potential therapeutic against NoVs (25–27). However, the inhibitors designed thus far are based on the available NV Pro structures, and although they show potent activity against GI proteases, they are not efficient in inhibiting the GII proteases, for which there is no structural information available (26). Thus, to develop a broadly cross-reactive inhibitor, it is vital to determine the protease structure from members of the GII NoVs.
In this study, we determined the crystal structure of a GII.4 protease, namely, the Houston virus protease (HOV Pro), and compared it with the available GI protease structures. Our studies show that, although the overall structure of HOV Pro is conserved, there are several significant changes in the orientation of residues comprising the active site. The side chain of an arginine residue (R112) is found inserted in the active site of the HOV Pro making interactions with the catalytic H30 residue. Our studies suggest a pH-dependent role for R112 in modulating substrate and inhibitor binding to the HOV Pro. Overall, our results provide a structural framework for structure-based drug design of broadly cross-reactive inhibitors targeting NoVs.
RESULTS
The overall structure of the HOV Pro.
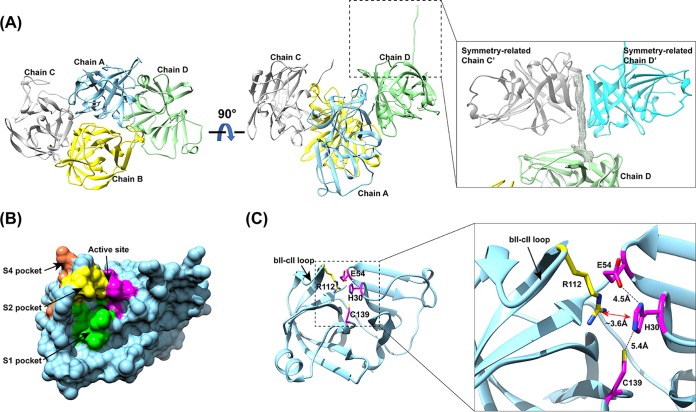
The HOV Pro crystals diffracted to 2.7 Å, and the structure was determined in the P212121 space group with four molecules in the crystallographic asymmetric unit. The phases were resolved by molecular replacement (MR) using the previously determined NV Pro structure as an initial model and refined with final Rwork and Rfree values of 20.8% and 24.7%, respectively (Table 1). The overall structure is conserved and is comprised of a chymotrypsin-like fold with an N-terminal β-barrel domain and a C-terminal twisted β-sheet domain separated by a cleft where the active site is located, similar to that in other viral cysteine proteases (Fig. 1). All four monomers (A, B, C, and D) in the asymmetric unit were similar and superimposed with a root mean square deviation (RMSD) of ∼1 Å. The C-terminal tails of molecules A, B, and C were disordered, without electron density observed, whereas that of molecule D was stabilized by the symmetry-related molecules C′ and D′ (Fig. 1A). Molecules A and D and molecules B and C form distinct tail-to-tail dimers in which the β-barrel of each HOV Pro monomer interacts along the crystallographic 2-fold symmetry to form a dimer. The significance of dimer formation by the NoV proteases is not yet established, but the dimeric forms have been reported to be important for the functions of other viral cysteine proteases, such as hepatitis A virus 3C protease and the coronavirus 3C protease (21, 28, 29).
TABLE 1.
Data processing and refinement statistics for HOV protease
| Parameter | Value(s)a |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.0 |
| Resolution range (Å) | 50.0–2.70 (2.75–2.70) |
| Space group | P212121 |
| Unit cell | |
| a, b, c (Å) | 56.76, 91.01, 141.57 |
| α, β, γ (°) | 90, 90, 90 |
| No. of reflections | 18,302 |
| Redundancy | 6.0 (5.7) |
| Completeness (%) | 99.8 (99.5) |
| Wilson B factor (Å2) | 40.14 |
| Rmerge (%) | 8 (107.6) |
| Refinement | |
| Rwork (%) | 20.8 |
| Rfree (%) | 24.7 |
| No. of atoms | 10,484 |
| RMS bonds (Å) | 0.01 |
| RMS angles (°) | 1.05 |
| Ramachandran | |
| Outliers (%) | 0 |
| Allowed | 1.74 |
| Favored | 98.26 |
| B factor (Å2) | 53.69 |
Values in parentheses refer to the highest-resolution shell.
FIG 1.
Structure of the HOV Pro. (A) Crystal packing of HOV Pro, with four molecules in the asymmetric unit shown in ribbon representation. The inset shows the C-terminal tail of chain D interacting with symmetry-related chains C′ and D′. The Fo-Fc difference electron density map of the C-terminal tail is shown in gray mesh and contoured at 3 σ. (B) Surface representation of the HOV Pro monomer with the color-coded active site (magenta), and S1 (green), S2 (yellow), and S4 (orange) substrate-binding pockets. (C) Cartoon representation of a protease monomer shown in the same orientation as in panel B. The inset shows a close view of the catalytic residues H30, E54, C139 (magenta), and R112 (yellow) of the S2 pocket rendered as sticks, with nitrogen shown in blue and oxygen shown in red. The distances between the residues are indicated by black dashed lines. The cation-π interaction between R112 and H30 is indicated by a red double arrow. The flexible bII-cII loop is indicated with black arrows.
The substrate-binding pockets.
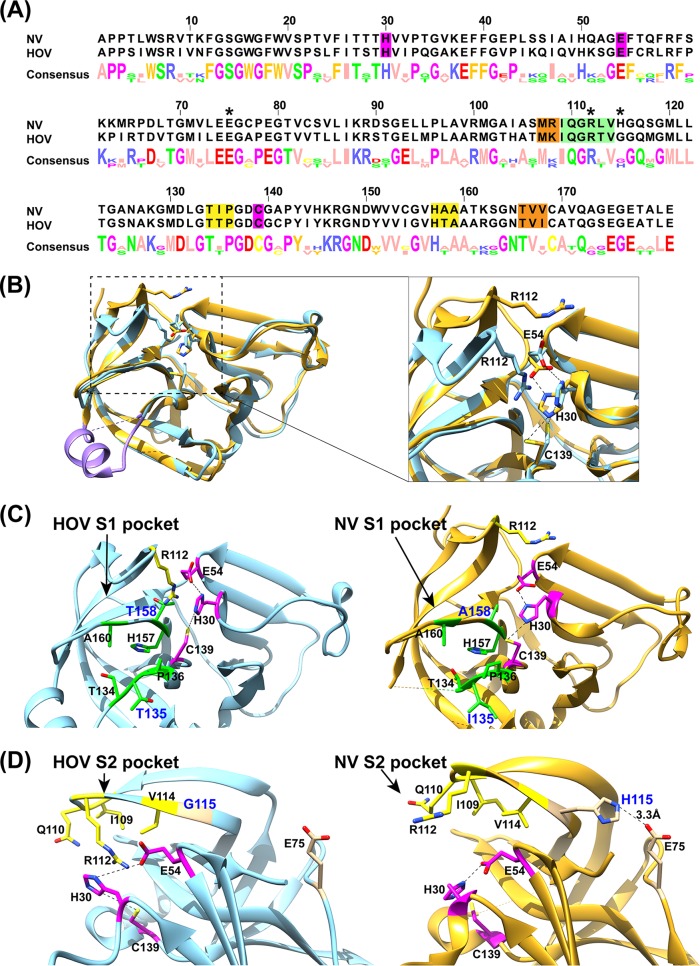
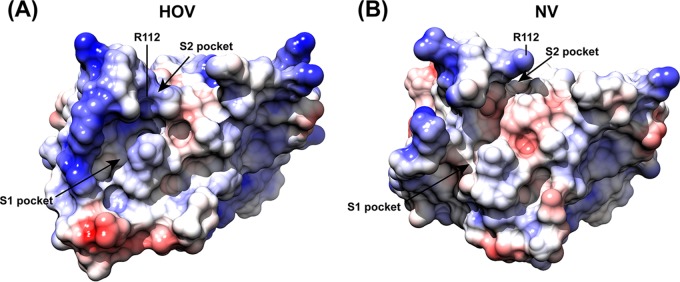
Although the GII norovirus HOV protease is homologous to the GI NV protease, with a sequence identity of 66% (Fig. 2A), the inhibitors designed thus far show more potent activity against GI proteases than against the GII proteases (26). To understand the structural basis for the inhibition of GII HOV protease, we compared the crystal structures of HOV and NV proteases (Fig. 2B to D). While the overall structure of the HOV Pro is similar to that of the NV Pro, substantial changes were observed, particularly in the S1 and S2 pockets that interact with the P1 and P2 residues of the substrate, respectively. Residues 122 to 134, which form the floor of the S1 pocket, exhibit significant changes. In the HOV Pro this stretch consists of an α-helix, whereas in the GI protease structures it is generally unstructured (Fig. 2B). The S1 pocket of the HOV Pro contains two polar residues, Thr135 and Thr158, whereas in the NV Pro these residues are replaced by Ile and Ala (Fig. 2C). Compared to the S1 pocket, the S2 pocket shows more significant structural changes. The S2 pocket of HOV is distinctly smaller than that in the NV Pro structure because of conformational changes in the bII-cII loop, with an RMSD of 4.56 Å for the matching Cα atoms between HOV and NV Pro structures. Sequence and structural comparisons reveal a key change at position 115 from His in NV Pro to Gly in HOV Pro (Fig. 2A and D). The side chain of H115 forms a hydrogen bond interaction with that of E75, which stabilizes the bII-cII loop in the structure of NV Pro. The H115G mutation in HOV Pro leads to the loss of this hydrogen bond interaction and higher flexibility of the loop, which moves closer toward the active site. As a result of this change, residues I109, Q110, R112, and V114 in the S2 pocket of HOV Pro are positioned facing the active site pocket (Fig. 2B). In addition, these changes also alter the electrostatic potential surface significantly around the active site (Fig. 3).
FIG 2.
Sequence and structure comparisons of HOV and NV proteases. (A) Sequence alignment of HOV and NV proteases using Jalview (57). The active site (magenta), and S1 (green), S2 (yellow), and S4 (orange) substrate-binding pockets are highlighted. The residues E75, R112, and H115 are indicated with asterisks. (B) Structural overlay of HOV Pro (light blue) and NV (gold; PDB accession no. 2FYQ) proteases. The residues 122 to 134 that form the floor of the S1 pocket are colored in purple. The inset shows the structural changes of the bII-cII loop between HOV and NV proteases. Active site residues H30, E54, and C139, and the S2 substrate-binding pocket residue R112 are shown as sticks and colored as in panel A. (C and D) Detailed view of the active site and S1 (green) and S2 (yellow) substrate-binding pockets. For clarity, the overlaid structures are shown side by side. R112 in the HOV Pro structure extends into the active site and interacts with H30, while R112 in the NV protease structure is turned away from the active site.
FIG 3.
Comparison of the electrostatic potential surfaces of HOV and NV proteases. The surfaces were colored from red for negative potential to white near neutral to blue for positive potential using coulombic surface coloring in Chimera. The S1 and S2 substrate-binding pockets are indicated by black arrows. The R112 residue is labeled.
Catalytic triad or dyad?
Further notable changes were observed in the catalytic triad residues of the HOV Pro (Fig. 2B to D). In the NV Pro, the triad residues form a network of hydrogen bonds with nucleophilic C139 interacting with basic H30, which functions to deprotonate and polarize C139, and H30, in turn, forming a hydrogen bond with the acidic E54 that serves to properly align the imidazole ring of H30 with respect to C139 (21, 25). Residues H30, C139, and E54 are conserved in all GI and GII.4 proteases. However, in the HOV Pro structure, all three residues show changes in their side chain orientations compared to those in the NV Pro structure (Fig. 1C and 2B). Strikingly, instead of H30 interacting with E54, as observed in other NoV Pro structures, H30 closely interacts with the side chain of R112 via a cation-π interaction (Fig. 1C). Modeling the E54 residue in the same orientation as that observed in the NV Pro structure showed a steric clash with R112, indicating that the side chain of E54 has to be turned away from the active site to accommodate the R112 side chain in close proximity to H30. As a result of its interaction with R112, the orientation of the imidazole ring of H30 is also markedly different from that observed in previous NoV Pro structures. In that orientation, neither of the two nitrogen atoms of the imidazole are placed at a proper distance to effectively deprotonate the sulfhydryl (SH) group of C139 for the nucleophilic attack. The distance between one of the imidazole nitrogen atoms and the SH of C139 is 5.4 Å, which is significantly longer than those observed in other GI protease structures (Fig. 1C). The side chain of C139 is also oriented slightly differently compared to the NV Pro structure, with its SH group hydrogen bonded to the main chain carbonyl atom of T158. Such a hydrogen bond interaction is not observed in other GI protease structures.
HOV Pro activity is pH sensitive.
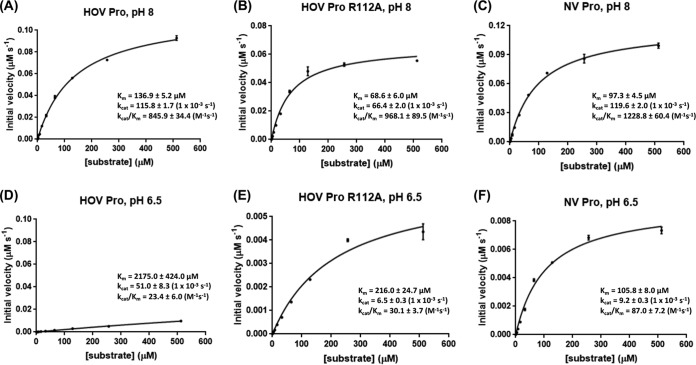
The observations of the unusual configuration of the active site residues with a unique histidine-arginine pairing, along with the consideration that the structure of the HOV Pro was determined at pH 6.5, lead us to hypothesize that R112 could influence the catalytic activity of HOV Pro in a pH-dependent manner. To directly test this hypothesis, we used a fluorescence resonance energy transfer (FRET) assay to measure the kcat/Km (enzyme efficiency) of both the HOV and NV proteases at pH values of 8, 6.5, and 5, using the decapeptide sequence containing P1 to P5 and P1′ to P5′, which correspond to the cleavage sites between p48 and p41 in their respective polyproteins. The FRET assay showed that while HOV Pro was active at pH 8, it showed extremely low activity at pH 6.5 (Fig. 4). In contrast, NV Pro, in which R112 is positioned away from the active site because of the differential changes in the bII-cII loop, was optimally active at pH 8 and retained its activity, albeit at a reduced level, at pH 6.5 (Fig. 4), indicating that the presence of the arginine in the active site is likely the reason for the greater pH sensitivity of HOV Pro. To further test this, we made an R112A mutant and performed a similar activity assay. Unlike the wild-type HOV Pro, the R112A mutant was active at pH 6.5.
FIG 4.
Rate of substrate hydrolysis by HOV Pro, HOV Pro R112A, and NV Pro at pH 8.0 and pH 6.5. Substrate hydrolysis was measured by FRET by mixing the protease at increasing concentrations with the fluorogenic peptide substrate and measuring the increase in fluorescence intensity as the substrate was cleaved. Based on these measurements, the Km and kcat values were calculated using GraphPad Prism 7. All experiments were carried out in triplicate. The standard errors of Km and kcat were generated by GraphPad software, and the standard errors of kcat/Km were calculated according to the Fenner formula (58).
The FRET assays at pH 8 further revealed that HOV Pro cleaves its substrate at a lower rate (kcat/Km = 845.9 M−1s−1) than the NV Pro (kcat/Km = 1,228.8 M−1s−1) (Fig. 4). Mutating R112 to alanine in the HOV Pro resulted in a higher affinity for the substrate and higher cleavage efficiency of the HOV Pro R112A (kcat/Km = 968.1 M−1s−1) compared to the that of wild-type HOV Pro, suggesting that R112 may regulate the rate of the catalysis by the protease by limiting the substrate access to the S2 pocket. Taken together, these results show that R112 plays an important role in substrate affinity/activity of the protease and that this regulation may be pH-dependent.
R112A mutant shows improved inhibitor potency compared to that of the wild type.
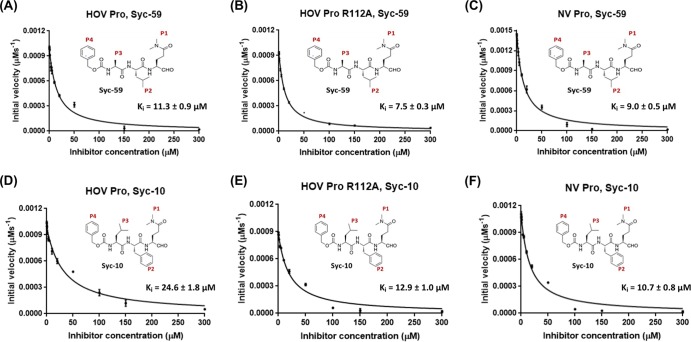
Previously, we designed a panel of substrate-based inhibitors that mimic the P1 to P4 substrate residues of the NV polyprotein with a terminal reactive group targeting the active site. Crystal structures of the NV Pro are in complex with two of the most potent inhibitors in this panel, Syc-10 and Syc-59. These inhibitors have glutamine at the P1 position but differ in their P2 position. Syc-10 has a phenylalanine at P2, whereas Syc-59 has a leucine at this position. The structures showed the P1 and P2 residues fit firmly into the S1 and S2 pockets of the NV Pro, and the reactive group forms a covalent adduct with C139, preventing the formation of the oxyanion hole in the active site, a necessary step for cleavage (25). When tested with the HOV Pro, both inhibitors showed significantly reduced potency compared to that of the NV Pro (26). For our current study, we tested Syc-10 and Syc-59 against the HOV Pro R112A mutant to examine how the absence of the arginine sidechain differentially affects inhibitor efficiency. Both inhibitors showed improved inhibition against the R112A mutant. For Syc-10, with a bulkier phenylalanine at the P2 position, Ki for the mutant was 12.9 μM, compared to 24.6 μM for the wild-type protease. For Syc-59, with leucine at the P2 position, Ki for the mutant was 7.5 μM, compared to 11.3 μM for the wild type (Fig. 5). This strongly suggests that in the absence of the arginine sidechain, the inhibitors were relatively more efficient in inhibiting the R112A mutant compared to the wild type. Taken together, these findings indicate that the presence of arginine in the active site, conformational alterations in the active site residues, and the conformation of the bII-cII loop, which narrows the S2 pocket, differentially affect both the catalytic and inhibitor efficiencies compared to those of the NV Pro, and these factors should be considered in designing GII.4-specific or broad-spectrum inhibitors.
FIG 5.
Rates of substrate hydrolysis by HOV Pro, HOV Pro R112A, and NV Pro in the presence of the Syc-10 and Syc-59 aldehyde inhibitors. Substrate hydrolysis was measured by FRET, as described above, in the presence of increasing concentrations of the Syc-10 and Syc-59 inhibitor. The Ki values were determined by fitting the initial velocities to the Morrison tight-binding equation using GraphPad Prism 7. All experiments were carried out in triplicate. The standard errors of Ki were generated by the GraphPad software.
DISCUSSION
Virus-encoded proteases have emerged as new targets for antiviral treatments and disease prevention. Inhibitors targeting the viral protease have been used effectively to treat infections caused by HIV (30–32), hepatitis C (33–37), herpesvirus (38, 39), human rhinoviruses (40), and severe acute respiratory syndrome (SARS) coronaviruses (39, 41). Advances in high-throughput screening of chemical compound libraries, together with structure-based drug design, have aided the development of these protease inhibitors. Among human NoVs, although some progress has been made in developing inhibitors against the GI proteases, these inhibitors are not as effective against the other genogroups, particularly the prevalent GII genogroup. To develop an effective broadly cross-reactive NoVs protease inhibitor, it is critical to understand the structural differences among the viral proteases of these two genogroups. To address this, we determined the protease structure of a GII.4 NoV (HOV variant). Comparison of our structure with the GI protease structure shows that although the overall polypeptide fold is conserved among the NoV proteases, substantial structural changes were observed in the vicinity of the active site, and these differences could differentially affect both catalytic and inhibitor efficacies.
HOV Pro structure shows significant differences compared to those of GI proteases.
The overall structure of the HOV Pro resembles those of the GI protease structures as well as of the other typical cysteine proteases. In contrast to the other NoV protease structures, the S2 substrate-binding pockets of the HOV Pro is markedly smaller. The bII-cII loop of the S2 pocket is highly flexible (22), and the S2 pocket of the NV Pro was shown to exist in the following three conformations: closed (unbound protease), semiopen (with bound Thr-Ala-Leu-Glu substrate residues), and open (with bound Ile-Asn-Phe-Glu substrate residues) (25). However, even in the closed conformation, the S2 pocket of the NV Pro is larger than the S2 pocket of the HOV Pro. The structure of GI.4 CV Pro also showed a smaller S2 pocket compared to the that of the NV Pro structure (22). In that study, the authors modeled the substrate-mimicking oligopeptide bound to the protease active site, showing the P1 and P2 residues of the peptide fitting into the S1 and S2 pockets without any significant clashes. The authors concluded that the smaller S2 pocket would still be able to accommodate the large P2 residue of the natural substrate. This suggests that the reduction of the inhibitor potency that was observed between the NV and the HOV Pro may not be solely attributed to the smaller S2 pocket. Most likely, changes in the substrate-binding pockets in the HOV Pro structure, together with the conformational changes of the catalytic residues and presence of the R112 side chain in the active site plus the changes in some of the amino acids surrounding the active site, contribute to the reduced inhibitor efficiencies against the HOV Pro, as discussed below.
The active site of HOV Pro presents a unique configuration of the catalytic residues.
In addition to the changes in the S1 and S2 pockets, the HOV Pro structure showed significant changes in the active site, where all three catalytic residues had different orientations compared to those of the NV Pro. Unexpectedly, in the HOV Pro, the role of the E54 residue in stabilizing catalytic H30 appears to be taken up by R112. A somewhat similar observation was made in the enterovirus 71 3C protease (EV71 Pro) structure, in which, in addition to the catalytic C147, H40, and E71 residues, two other residues, arginine (R39) and asparagine (N69), were shown to be important for catalysis. R39 and N69 form hydrogen bonds with E71, connecting it via a hydrogen bond network to H40 and C147 and providing the correct environment for efficient cleavage. The mutation of these residues to alanine resulted in an inactivated EV71 Pro (19). In the HOV Pro, the removal of the R112 side chain with an alanine mutation from the active site enhanced HOV Pro activity, suggesting that, unlike any other 3C proteases, HOV Pro uses a novel catalytic site configuration that is sensitive to pH.
In another example, hepatitis A 3C protease lacks the third member of the catalytic triad, only utilizing the cysteine (C172) and histidine (H44) for the cleavage activity. The structure of the hepatitis A protease revealed a water molecule occupying the position of the third member of the triad. An additional residue, tyrosine 143 (Y143), was also found in the active site, but the distances between Y143 and H44 or Y143 and the water molecule were too great to make any hydrogen bonds (23). The authors speculated that the role of the negatively charged Y143 was to electrostatically stabilize a positive charge of the H44 imidazole ring during the rate-limiting transition state, a role that could be played by R112 in the HOV Pro.
The rotamer analysis of R112 in the NV Pro structure using the Crystallographic Object-Oriented Toolkit (COOT) showed that any of the possible side chain orientations cannot come close to the active site side chain to make a similar interaction with H30 to that observed in the HOV Pro structure because of the rather extended conformation of the bII-cII loop. The same analysis of the R112 in the CV Pro structure, which has a similar conformation of the bII-cII loop to that in the HOV Pro structure, showed that alternate orientations of the R112 side chain could engage in similar R112-H30 interactions to those observed in the HOV Pro structure. On the other hand, alternate conformations of the R112 sidechain in the HOV Pro would allow E54 to hydrogen bond with H30 and position it appropriately to interact with the C139 required for protease activation. It is possible that the role of the R112 in the HOV Pro is to regulate protease activity by disrupting the canonical triad interactions.
Unusual Arg-His pairing confers pH sensitivity to HOV Pro.
A unique feature of HOV Pro is the R112-H30 interaction. Although such a pairing between the two positively charged residues is uncommon, under certain conditions, they can participate in stabilizing interactions. Such interactions are contingent upon the protonation state of the histidine’s imidazole ring, which is highly pH dependent (42–45). Our studies showed that HOV Pro activity is significantly reduced at an acidic pH of 6.5, whereas the NV Pro still retains some of its activity at this pH. Such differential sensitivity to pH could be attributed to the presence of R112 in the catalytic pocket of HOV Pro and its close interaction with H30. At pH 6.5, the protonation state of the imidazole ring of H30 increases, shifting the equilibrium more toward the aromatic (π-system) state and allowing it to participate in a stabilizing cation-π interaction with R112, as observed in the structure, which, as noted before, was determined from crystals obtained at pH 6.5. At pH 8, however, the protonation state of the H30 decreases, and the equilibrium shifts more toward the cationic state of the imidazole ring, leading to destabilization of the H30-R112 interaction and allowing the R112 sidechain to assume a different orientation. With the sidechain of R112 moved away, the side chain of E54 can then assume an orientation conducive for hydrogen bonding with the partially protonated state of H30, allowing it to be placed appropriately to abstract proton from the SH group of C139 for the nucleophilic attack when the substrate is bound. pH sensitivity is not as pronounced in the GI protease, which is active at both pH 8 and pH 6.5. A plausible explanation is that in the absence of R112, even at this pH, some cationic nature of H30 is retained and E54 is able to hydrogen bond with H30 and position it appropriately toward the SH group of C139. Such reasoning is consistent with the observation that removing R112 from the active site restored some of the HOV Pro activity at pH 6.5. Both the NV and the HOV Pro are completely inactive at a pH of <6.0 (data not shown). At such pH values, below the pKa of histidine, the imidazole ring is expected to be fully protonated, and in such a state it will not be able to abstract the proton from the SH group of C139 for the nucleophilic attack.
A structure of the HOV Pro in complex with its native substrate would shed definitive light on the mechanism of substrate binding to the HOV Pro and R112 and the role pH may play in this binding. Regardless, the histidine-arginine pairing of the catalytic residues has not been observed in any other viral cysteine proteases and represents a significant example of viral protease evolution, also creating a unique challenge in developing inhibitors potent against GII.4 viruses.
HOV Pro exhibits lower enzyme efficiency than NV Pro.
Our biochemical studies indicate that the HOV Pro has a lower affinity for its substrate and lower proteolytic efficiency compared to those of the NV Pro. This surprising observation raises a set of interesting questions. Is the presence of the arginine in the active site a rate-limiting factor for proteolytic cleavage? Is it possible that in the infected cells, the virus uses pH to regulate proteolysis? Is the lower rate of proteolytic cleavage translating to more efficient viral replication? A study by Belliot et al. showed that the NoV RNA-dependent RNA polymerase (RdRp) may exist in two forms during NoV infection, Pol (the mature polymerase) and ProPol (a precursor comprised of both the protease and polymerase). ProPol still exhibited full protease activity and had consistently higher polymerase activity and for longer periods of time than Pol (46, 47), suggesting that the ProPol form of the polymerase would be preferred by the virus. The same observation was made for the ProPol protein of feline calicivirus (FCV), another member of the Caliciviridae, in which ProPol was a predominant form of the RdRp observed in FCV-infected cells (48, 49). Assuming that, like in the GI, both Pol and ProPol polymerase forms also exist in the GII.4 viruses, it is possible that the lower rate of polyprotein cleavage by the HOV Pro will result in the uncleaved or partially cleaved polyprotein components, among them ProPol, being present for longer periods of time, allowing the ProPol to more efficiently replicate viral RNA. Therefore, a lower rate of polyprotein cleavage could potentially be advantageous for the GII.4 NoVs.
Changes in the HOV Pro active site reduce the potency of GI inhibitors.
The inhibitors designed based on the GI structures were less inhibitory to the HOV Pro (50). This indicates that steric hindrance for the P2 residues of the inhibitor created by the changes in the HOV Pro S2 pocket and the active site plays a significant role in lowering inhibitor potency. Most likely, the difference observed is due not to just one factor but to the combination of all of the differences found in the HOV Pro structure. However, mutation of R112 to alanine improved inhibitor potency, indicating the importance of R112 in the inhibitor binding to the protease active site. Furthermore, the Syc-59 inhibitor that has a leucine residue in the P2 position was more potent against the wild-type HOV Pro and the R112 mutant compared to the Syc-10 inhibitor, which has a bulkier phenylalanine in the P2 position. Thus, a viable potential strategy for designing GII.4-specific inhibitors would be to design inhibitors with smaller P2 residues. Another strategy would be to design dipeptide inhibitors that will only include the reactive group and the P1 residue, which would make it possible to avoid steric clashes in the S2 pocket altogether.
In summary, our studies reported here describe the first atomic structure of the GII.4 NoV protease HOV Pro. The structure reveals unique interactions between the catalytic residues of the HOV Pro that were not observed in the previously reported structures of the GI proteases. These unusual residue interactions may be the reason for the observed differences in the inhibitor potencies between the GI.1 and GII.4 proteases and must be considered when designing broadly cross-reactive protease inhibitors.
MATERIALS AND METHODS
Protein expression and purification.
Both GI.1 NV Pro (GenBank accession no. NP_786949) and GII.4 HOV Pro (Hu/Houston/TCH186/2002/US; accession no. ABY27559) were cloned into the bacterial pET-46 Ek/LIC expression vector (Novagen, EMD Biosciences, Inc., Darmstadt, Germany) with the N-terminal 6×His tag followed by a thrombin cleavage site (LVPRGS), as described by Zeilter et al. (21). The proteins were expressed in Escherichia coli BL21(DE3) cells (Novagen, EMD Biosciences, Inc., Darmstadt, Germany) and purified using Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography resin (Qiagen). Following Ni-NTA purification, the 6×His tags were cleaved off by using thrombin (Haematologic Technologies, Inc., Essex Junction, VT) and proteins were further purified by gel filtration chromatography (HiLoad 16/60 Superdex 75; GE Healthcare, Little Chalfont, UK). Purified NV and HOV Pro were concentrated to 18 mg/ml and 20 mg/ml, respectively, and stored in 50 mM NaH2PO4 (pH 8), 100 mM NaCl, and 5 mM tris(2-carboxyethyl)phosphine) (TCEP) buffer until further use. The R112A mutant was generated by Epoch Life Science, Inc. (Sugar Land, TX) and expressed in the BL21(DE3) cells and purified as described above.
Crystallization of the HOV protease.
The HOV Pro was concentrated to ∼6 mg/ml and crystallized by the hanging drop vapor diffusion method using the Mosquito crystallization robot (TTP LabTech, Herts, Melbourn, UK) at 20°C. Final crystals were obtained in 0.2 M potassium thiocyanate, 0.1 M bis-Tris propane (pH 6.5), 20% (wt/vol) PEG 3350, placed in the cryoprotectant solution (20% glycerol), and flash-frozen in liquid nitrogen.
Data collection and processing.
A complete data set was collected to a resolution of 2.7 Å at the Argonne National Laboratory Advanced Photon Source, beamline SBC-19ID (Chicago, IL). The diffraction data were collected using a 0.5°-oscillation angle and integrated using HKL2000. HOV Pro crystallized in the space group P212121 with four molecules in the asymmetric unit. The previously determined structure of NV protease (PDB 4IN1) (25) was used for phasing, using the molecular replacement (MR) program PHASER (51) as implemented in the CCP4 suite of programs (52). Following the autobuilding and refinement using Buccaneer as implemented in the CCP4 suite, iterative cycles of refinement and further model building were carried out using PHENIX (53), Rosetta (54), and COOT (55) programs. During the course of the refinement and following the final refinement, the stereochemistry of the structures was checked using MolProbity (http://molprobity.biochem.duke.edu). Data refinement and statistics are given in Table 1. Figures were prepared using Chimera (56). Sequence alignments were carried out using Jalview (57).
Protease activity and inhibition assays.
The activity of the purified NV Pro and HOV Pro was confirmed by using a fluorescence resonance energy transfer (FRET) assay as described previously (21, 25). Fluorogenic peptides Glu(EDANS)-GDYELQGPEDLA-Lys(Dabcyl) and Glu(EDANS)-EPDFLQGPEDLAK-Lys(Dabcyl), corresponding to the natural cleavage sites between the nonstructural proteins p48 and p41 in the HOV and NV polyproteins, respectively, were synthesized by GenScript USA Inc. (Piscataway, NJ) with the fluorescent dye Edans [5-((2-aminoethyl)amino)naphthalene-1-sulfonic acid] at the N terminus of the peptide and the quencher Dabcyl [4-(dimethylaminoazo)benzene-4-carboxylic acid] at the C terminus. When the peptide is cleaved by the protease, the dye is no longer quenched, resulting in an increase in fluorescence. The assays were performed in 50 mM NaH2PO4 (pH 8, 6.5, or 5), 100 mM NaCl, and 5 mM TCEP buffer. Increasing concentrations (2, 4, 8, 16, 32, 64, 128, 256, and 512 μM) of the substrate were added to 1 μM the protease, and the fluorescence was measured at excitation/emission wavelengths of 340 nm and 490 nm using the FlexStation 3 multimode plate reader (Molecular Devices, LLC, San Jose, CA). Fluorescence signal was monitored for 120 min at 90-sec intervals at 37°C. The relative fluorescence units (RFUs) were converted to the product formed in μM, using a standard curve. Initial velocities, Michaelis constants (Km), and the catalytic constant (kcat) were calculated using nonlinear regression analysis using GraphPad Prism 7 software (GraphPad Software, Inc., La Jolla, CA). For the inhibition assays, Syc-10 and Syc-59 aldehyde inhibitors were synthesized as previously described by Deng et al. (26). HOV Pro R112A (2.5 μM) was mixed with increasing concentrations (0, 0.2, 0.5, 1, 3, 5, 10, 20, 50, 100, 150, and 300 μM) of the inhibitor and incubated at room temperature for 30 min prior to addition of the 30 μM substrate. Upon addition of the substrate, the change in the fluorescent signal was measured immediately as described above. Inhibition constants (Ki) were calculated by a nonlinear curve fit into the Morrison tight-binding equation using GraphPad software.
Data availability.
The atomic structure coordinates of the norovirus GII.4 protease have been deposited in the Protein Data Bank under accession number 6NIR.
ACKNOWLEDGMENTS
We acknowledge the use of the Argonne National Laboratory Advanced Photon Source, beamline SBC-19ID (Chicago, IL) for diffraction data collection. We thank the staff of this institution for the excellent help.
The SBC-CAT 19ID beamline at Advanced Photon Source is supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38. This research is supported by National Institutes of Health Grant NIH PO1 AI057788 (to M.K.E., Y.S., T.P., and B.V.V.P.) and by Robert Welch Foundation Grant Q1279 (to B.V.V.P.).
ADDENDUM IN PROOF
Six days before the present paper was published, the X-ray structure of a catalytically inactive C139A mutant of another GII.4 protease (Minerva strain) was reported. Reference 59 was added in proof.
REFERENCES
- 1.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. 2014. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green KY, Ando T, Balayan MS, Berke T, Clarke IN, Estes MK, Matson DO, Nakata S, Neill JD, Studdert MJ, Thiel HJ. 2000. Taxonomy of the caliciviruses. J Infect Dis 181:S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- 3.Atmar RL, Estes MK. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol Clin North Am 35:275–290. viii. doi: 10.1016/j.gtc.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Vinje J. 2015. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 53:373–381. doi: 10.1128/JCM.01535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramani S, Atmar RL, Estes MK. 2014. Epidemiology of human noroviruses and updates on vaccine development. Curr Opin Gastroenterol 30:25–33. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindesmith LC, Donaldson EF, Baric RS. 2011. Norovirus GII.4 strain antigenic variation. J Virol 85:231–242. doi: 10.1128/JVI.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy ME, Estes MK. 1996. Completion of the Norwalk virus genome sequence. Virus Genes 12:287–290. [DOI] [PubMed] [Google Scholar]
- 8.Glass PJ, White LJ, Ball JM, Leparc-Goffart I, Hardy ME, Estes MK. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J Virol 74:6581–6591. doi: 10.1128/JVI.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, Wang M, Wang K, Estes MK. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 10.Vongpunsawad S, Venkataram Prasad BV, Estes MK. 2013. Norwalk virus minor capsid protein VP2 associates within the VP1 shell domain. J Virol 87:4818–4825. doi: 10.1128/JVI.03508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazan JF, Fletterick RJ. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A 85:7872–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boniotti B, Wirblich C, Sibilia M, Meyers G, Thiel HJ, Rossi C. 1994. Identification and characterization of a 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J Virol 68:6487–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belliot G, Sosnovtsev SV, Mitra T, Hammer C, Garfield M, Green KY. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J Virol 77:10957–10974. doi: 10.1128/JVI.77.20.10957-10974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blakeney SJ, Cahill A, Reilly PA. 2003. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology 308:216–224. [DOI] [PubMed] [Google Scholar]
- 15.Hardy ME, Crone TJ, Brower JE, Ettayebi K. 2002. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res 89:29–39. [DOI] [PubMed] [Google Scholar]
- 16.Someya Y, Takeda N, Miyamura T. 2005. Characterization of the norovirus 3C-like protease. Virus Res 110:91–97. doi: 10.1016/j.virusres.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sosnovtsev SV, Garfield M, Green KY. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J Virol 76:7060–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews DA, Smith WW, Ferre RA, Condon B, Budahazi G, Sisson W, Villafranca JE, Janson CA, McElroy HE, Gribskov CL. 1994. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77:761–771. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Fan T, Yao X, Wu Z, Guo L, Lei X, Wang J, Wang M, Jin Q, Cui S. 2011. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J Virol 85:10021–10030. doi: 10.1128/JVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosimann SC, Cherney MM, Sia S, Plotch S, James MN. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol 273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 21.Zeitler CE, Estes MK, Venkataram Prasad BV. 2006. X-ray crystallographic structure of the Norwalk virus protease at 1.5-Å resolution. J Virol 80:5050–5058. doi: 10.1128/JVI.80.10.5050-5058.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura K, Someya Y, Kumasaka T, Ueno G, Yamamoto M, Sato T, Takeda N, Miyamura T, Tanaka N. 2005. A norovirus protease structure provides insights into active and substrate binding site integrity. J Virol 79:13685–13693. doi: 10.1128/JVI.79.21.13685-13693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmann EM, Mosimann SC, Chernaia MM, Malcolm BA, James MN. 1997. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J Virol 71:2436–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Someya Y, Takeda N, Miyamura T. 2002. Identification of active-site amino acid residues in the Chiba virus 3C-like protease. J Virol 76:5949–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muhaxhiri Z, Deng L, Shanker S, Sankaran B, Estes MK, Palzkill T, Song Y, Prasad BV. 2013. Structural basis of substrate specificity and protease inhibition in Norwalk virus. J Virol 87:4281–4292. doi: 10.1128/JVI.02869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng L, Muhaxhiri Z, Estes MK, Palzkill T, Prasad BV, Song Y. 2013. Synthesis, activity and structure-activity relationship of noroviral protease inhibitors. Medchemcomm 4. doi: 10.1039/C3MD00219E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damalanka VC, Kim Y, Galasiti Kankanamalage AC, Lushington GH, Mehzabeen N, Battaile KP, Lovell S, Chang KO, Groutas WC. 2017. Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease. Eur J Med Chem 127:41–61. doi: 10.1016/j.ejmech.2016.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 29.Peters H, Kusov YY, Meyer S, Benie AJ, Bauml E, Wolff M, Rademacher C, Peters T, Gauss MV. 2005. Hepatitis A virus proteinase 3C binding to viral RNA: correlation with substrate binding and enzyme dimerization. Biochem J 385:363–370. doi: 10.1042/BJ20041153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanne I, Piliero P, Squires K, Thiry A, Schnittman S, Group AICT. 2003. Results of a phase 2 clinical trial at 48 weeks (AI424-007): a dose-ranging, safety, and efficacy comparative trial of atazanavir at three doses in combination with didanosine and stavudine in antiretroviral-naive subjects. J Acquir Immune Defic Syndr 32:18–29. doi: 10.1097/00126334-200301010-00004. [DOI] [PubMed] [Google Scholar]
- 31.Pulido F, Arribas JR, Delgado R, Cabrero E, Gonzalez-Garcia J, Perez-Elias MJ, Arranz A, Portilla J, Pasquau J, Iribarren JA, Rubio R, Norton M, Group OKS. 2008. Lopinavir-ritonavir monotherapy versus lopinavir-ritonavir and two nucleosides for maintenance therapy of HIV. AIDS 22:F1–F9. doi: 10.1097/QAD.0b013e3282f4243b. [DOI] [PubMed] [Google Scholar]
- 32.Kempf DJ, Rode RA, Xu Y, Sun E, Heath-Chiozzi ME, Valdes J, Japour AJ, Danner S, Boucher C, Molla A, Leonard JM. 1998. The duration of viral suppression during protease inhibitor therapy for HIV-1 infection is predicted by plasma HIV-1 RNA at the nadir. AIDS 12:F9–14. [DOI] [PubMed] [Google Scholar]
- 33.Pause A, Kukolj G, Bailey M, Brault M, Do F, Halmos T, Lagace L, Maurice R, Marquis M, McKercher G, Pellerin C, Pilote L, Thibeault D, Lamarre D. 2003. An NS3 serine protease inhibitor abrogates replication of subgenomic hepatitis C virus RNA. J Biol Chem 278:20374–20380. doi: 10.1074/jbc.M210785200. [DOI] [PubMed] [Google Scholar]
- 34.Lamarre D, Anderson PC, Bailey M, Beaulieu P, Bolger G, Bonneau P, Bös M, Cameron DR, Cartier M, Cordingley MG, Faucher A-M, Goudreau N, Kawai SH, Kukolj G, Lagacé L, LaPlante SR, Narjes H, Poupart M-A, Rancourt J, Sentjens RE, St George R, Simoneau B, Steinmann G, Thibeault D, Tsantrizos YS, Weldon SM, Yong C-L, Llinàs-Brunet M. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 35.Reesink HW, Zeuzem S, Weegink CJ, Forestier N, van Vliet A, van de Wetering de Rooij J, McNair L, Purdy S, Kauffman R, Alam J, Jansen PL. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997–1002. doi: 10.1053/j.gastro.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 36.Sarrazin C, Rouzier R, Wagner F, Forestier N, Larrey D, Gupta SK, Hussain M, Shah A, Cutler D, Zhang J, Zeuzem S. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270–1278. doi: 10.1053/j.gastro.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 37.Lin TI, Lenz O, Fanning G, Verbinnen T, Delouvroy F, Scholliers A, Vermeiren K, Rosenquist A, Edlund M, Samuelsson B, Vrang L, de Kock H, Wigerinck P, Raboisson P, Simmen K. 2009. In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob Agents Chemother 53:1377–1385. doi: 10.1128/AAC.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimba N, Nomura AM, Marnett AB, Craik CS. 2004. Herpesvirus protease inhibition by dimer disruption. J Virol 78:6657–6665. doi: 10.1128/JVI.78.12.6657-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson J, Schiffer C, Lee SK, Swanstrom R. 2009. Viral protease inhibitors. Handb Exp Pharmacol. 4:85–110. doi: 10.1007/978-3-540-79086-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayden FG, Turner RB, Gwaltney JM, Chi-Burris K, Gersten M, Hsyu P, Patick AK, Smith GJ 3rd, Zalman LS. 2003. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob Agents Chemother 47:3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fear G, Komarnytsky S, Raskin I. 2007. Protease inhibitors and their peptidomimetic derivatives as potential drugs. Pharmacol Ther 113:354–368. doi: 10.1016/j.pharmthera.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Browne CA, Campbell ID, Kiener PA, Phillips DC, Waley SG, Wilson IA. 1976. Studies of the histidine residues of triose phosphate isomerase by proton magnetic resonance and X-ray crystallography. J Mol Biol 100:319–343. [DOI] [PubMed] [Google Scholar]
- 43.Khandogin J, Chen J, Brooks CL. 3rd, 2006. Exploring atomistic details of pH-dependent peptide folding. Proc Natl Acad Sci U S A 103:18546–18550. doi: 10.1073/pnas.0605216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyagi M, Nakazawa T. 2008. Determination of pKa values of individual histidine residues in proteins using mass spectrometry. Anal Chem 80:6481–6487. doi: 10.1021/ac8009643. [DOI] [PubMed] [Google Scholar]
- 45.Heyda J, Mason PE, Jungwirth P. 2010. Attractive interactions between side chains of histidine-histidine and histidine-arginine-based cationic dipeptides in water. J Phys Chem B 114:8744–8749. doi: 10.1021/jp101031v. [DOI] [PubMed] [Google Scholar]
- 46.Belliot G, Sosnovtsev SV, Chang KO, Babu V, Uche U, Arnold JJ, Cameron CE, Green KY. 2005. Norovirus proteinase-polymerase and polymerase are both active forms of RNA-dependent RNA polymerase. J Virol 79:2393–2403. doi: 10.1128/JVI.79.4.2393-2403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheffler U, Rudolph W, Gebhardt J, Rohayem J. 2007. Differential cleavage of the norovirus polyprotein precursor by two active forms of the viral protease. J Gen Virol 88:2013–2018. doi: 10.1099/vir.0.82797-0. [DOI] [PubMed] [Google Scholar]
- 48.Sosnovtseva SA, Sosnovtsev SV, Green KY. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J Virol 73:6626–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei L, Huhn JS, Mory A, Pathak HB, Sosnovtsev SV, Green KY, Cameron CE. 2001. Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J Virol 75:1211–1219. doi: 10.1128/JVI.75.3.1211-1219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng TFF, Zhang W, Sachsenröder J, Kondov NO, da Costa AC, Vega E, Holtz LR, Wu G, Wang D, Stine CO, Antonio M, Mulvaney US, Muench MO, Deng X, Ambert-Balay K, Pothier P, Vinjé J, Delwart E. 2015. A diverse group of small circular ssDNA viral genomes in human and non-human primate stools. Virus Evol 1:vev017. doi: 10.1093/ve/vev017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project N. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. [DOI] [PubMed] [Google Scholar]
- 53.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 54.DiMaio F, Echols N, Headd JJ, Terwilliger TC, Adams PD, Baker D. 2013. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods 10:1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenner G. 1931. Das Genauigkeitsmass von Summen, Differenzen, Produkten und Quotienten der Beobachtungsreihen. Naturwissenschaften 19:310–315. doi: 10.1007/BF01520425. [DOI] [Google Scholar]
- 59.Muzzarelli KM, Kuiper B, Spellmon N, Brunzelle J, Hackett J, Amblard F, Zhou S, Liu P, Kovari IA, Yang Z, Schinazi RF, Kovari LC. 2019. Structural and antiviral studies of the human norovirus GII.4 2 protease. Biochemistry doi: 10.1021/acs.biochem.8b01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The atomic structure coordinates of the norovirus GII.4 protease have been deposited in the Protein Data Bank under accession number 6NIR.