Host factors render cells susceptible to viral infection. One family of susceptibility factors, the tetraspanin proteins, facilitate enveloped virus entry by promoting virus-cell membrane fusion.
KEYWORDS: coronavirus, membrane fusion, tetraspanin, virus entry
ABSTRACT
Host factors render cells susceptible to viral infection. One family of susceptibility factors, the tetraspanin proteins, facilitate enveloped virus entry by promoting virus-cell membrane fusion. They also facilitate viral egress from infected cells. In this Gem, we discuss recent insights into how tetraspanins assemble viral entry and exit platforms on cell membranes, and we speculate that tetraspanins contribute to nonviral membrane fusions by similar mechanisms.
INTRODUCTION
For enveloped viruses, host factors determining susceptibility to infection include several “activators” which operate at particular subcellular sites to stimulate virus-cell membrane fusion. Depending on the infecting virus, activators may be cellular receptors, cations, or proteases which engage viral fusion proteins and trigger their refolding into forms that catalyze membrane coalescence. For many viruses, multiple activators are utilized in rapid succession. In these cases, activators must be in close proximity to each other, and to the incoming viral fusion proteins, to allow for rapid and efficient viral entry. Recent research identified the role of cellular proteins, including tetraspanins, in coalescing these activators. In this Gem, we discuss how tetraspanins facilitate efficient viral infection by organizing activators on host cell membranes.
TETRASPANINS
The tetraspanins are membrane proteins; they have four (tetra) transmembrane spans linked extracellularly by one large and one small extracellular loop (termed LEL and SEL, respectively) (Fig. 1, inset). They are ubiquitously present in eukaryotes (1). Their conserved structures and interacting partners suggest that they perform central roles in controlling membrane architecture. For example, they induce positive (outward) membrane curvatures (2), potentially through their cone-shaped transmembrane domains (3). Tetraspanins further influence membrane architecture by interacting more directly with lipids and with other integral membrane proteins. The tetraspanin CD81 binds cholesterol, which incorporates into a pocket between the tetraspanin transmembrane domains and alters LEL conformations (3). Several other tetraspanins associate with palmitic acid, which covalently links to cytoplasmic domains (4, 5). In conjunction with cholesterol and palmitate, tetraspanins interact with several so-called “partner” proteins. Distinct LEL- and cytoplasmic tail-dependent interactions with integrins (6, 7), adhesion molecules (8), and other tetraspanins and their associated partners (9) give rise to web-like tetraspanin-enriched microdomains (TEMs) on cellular membranes (10, 11). It has been proposed that these TEMs might be platforms for virus entry (12, 13), particularly for viruses that directly use tetraspanins as receptors (14). Yet there have been several reports that both enveloped and nonenveloped viruses preferentially enter cells through TEMs, presumably without directly binding a tetraspanin molecule (15–18). These intriguing reports stimulated our interest in the mechanisms by which viruses use TEMs for cell entry.
FIG 1.
Tetraspanins construct virus entry platforms and dictate CoV entry routes. (Inset) Tetraspanins are composed of four transmembrane spans connected by one large extracellular loop (LEL) and one small extracellular loop (SEL), with short N- and C-terminal tails protruding into the cytosol. (Left) To drive viral entry into host cells, MERS-CoV S (MERS-S) proteins (gray) bind dipeptidyl peptidase 4 (DPP4) receptors (purple) via their receptor binding domains (green). Receptor engagement exposes substrates (blue stars) susceptible to cleavage by cellular proteases, including type II transmembrane protease serine subtype 2 (TMPRSS2) (blue). The tetraspanin CD9 (red) links DPP4 to TMPRSS2 on the plasma membrane. DPP4-TMPRSS2 linkages facilitate the rapid proteolytic triggering of MERS-S proteins after DPP4 binding. Proteolytically triggered MERS-S relocates its receptor binding domains and unfolds into an extended intermediate structure that embeds hydrophobic fusion peptides into target cell membranes. Refolding of intermediates then pulls virus and cell membranes together to catalyze membrane fusion and “early” entry at or near the cell surface. (Right) In the absence of CD9, DPP4 and TMPRSS2 are not linked, and MERS-S proteins are not efficiently triggered on the cell surface. MERS-CoVs are instead endocytosed and thus encounter endosomal cathepsins (brown). At low pH (yellow), cathepsins cleave MERS-S proteins, triggering inefficient, “late” entry in the endosomal network.
CORONAVIRUS ENTRY
Our inroads into mechanisms by which tetraspanins facilitate virus entry came from investigations of coronaviruses (CoVs). These are enveloped viruses that cause respiratory and enteric infections in humans and animals (19); life-threatening severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV) are notable members. CoV infections are driven by spike (S) fusion glycoproteins. Extending from virions, these S proteins bind target cell receptors and then encounter cellular activators. The activators are proteases which cleave S proteins in ways that liberate domains catalyzing virus-cell fusion (20). Frequently, and in MERS-CoV infections, this proteolytic activation process takes place in stages. Furin and related proprotein convertases cleave S proteins during virus egress from producing cells (a “priming” step) (21) and then, after secreted viruses attach to target cells, serine or cysteine proteases cleave S proteins again (a “triggering” step) (22–25). Here, at the second triggering step, is where the tetraspanins come into play (Fig. 1, left panel).
TETRASPANINS PROMOTE CoV ENTRY BY LINKING CoV RECEPTORS AND PROTEASES
Since receptor-bound CoV S proteins are susceptible to triggering cleavages (26, 27), we surmised that receptors and S-cleaving proteases must be in close proximity to trigger membrane fusion. Tetraspanins appeared to be reasonable candidates for bringing receptors and proteases together. Indeed, there were good suggestions that the CoV receptors and proteases are coalesced within TEMs (28–31). Therefore, we hypothesized that tetraspanins condense CoV entry factors into localized positions whereby effective spatiotemporal activation of viral fusion takes place.
We first explored this hypothesis by using biochemical approaches. We isolated TEMs from cells containing CoV receptors and activating proteases and determined whether they localized to TEMs. Indeed, two CoV-activating proteases and the receptors of MERS-, SARS-, 229E-, and murine hepatitis virus (MHV)-CoVs were all found in TEMs (32). TEMs, while containing only ∼20% of all plasma membrane proteins, contained 50% to 90% of the CoV receptors and proteases, indicating that CoV activators are targeted to TEMs. Notably, isolated TEMs activated CoV S proteins in vitro, allowing these viruses to enter cells in a manner independent of proteases on target cells. TEM activating potential also correlated with protease abundance (32). These findings indicated that TEMs act as platforms for fusion-activating CoV S protein proteolysis.
We next used genetic manipulation to consider whether individual tetraspanins are required for fusion activation and virus entry. Given the specificity of tetraspanin-partner protein interactions (33, 34), it was likely that individual tetraspanins are responsible for ferrying particular CoV receptors into TEMs. In support of this hypothesis, the MERS-CoV receptor dipeptidyl peptidase 4 (DPP4) and the 229E-CoV receptor APN localized to TEMs only in the presence of a particular tetraspanin, CD9 (35). Furthermore, cells lacking CD9 were resistant to MERS and 229E pseudovirus entry, making it clear that these viruses needed CD9, and probably TEM-localized receptors, for their entry. In contrast, deletion of a related CD81 tetraspanin did not confer resistance to virus entry. Other CoVs, however, apparently use distinct tetraspanins, because omission of CD9 did not relocalize SARS-CoV and MHV receptors, nor did it affect susceptibility to SARS and MHV pseudoviruses. With these findings, tetraspanins were further elucidated as partners for ferrying of specific CoV receptors into TEMs.
The idea that tetraspanins bring CoV receptors to TEM-associated proteases was further supported by the discovery that a well-known CoV-activating protease, type II transmembrane protease serine subtype 2 (TMPRSS2), was adjacent to the MERS-CoV receptor DPP4 as measured by proximity ligation assays (35). Yet in cells lacking CD9, this colocalization was not observed, and even though CD9 knockout cells retained cell surface DPP4 and TMPRSS2, they were resistant to virus entry. Only when excess TMPRSS2 was supplied through transfection was virus susceptibility restored. Therefore, these findings demonstrated that CD9 was required to coalesce DPP4 and TMPRSS2, with this juxtaposition of entry factors being necessary for robust MERS-CoV membrane fusion and infection (Fig. 1, left panel). It is conceivable that all CoVs utilize closely juxtaposed entry factors. This may explain, in part, why most known CoV receptors are transmembrane peptidases, even though their peptidase activities are not used to activate CoV fusion proteins (36, 37). The subcellular localization of transmembrane peptidases appears to be tightly controlled (38), and the CoV receptors we surveyed all localized to TEMs (32). It is possible that CoVs have selected peptidases so that virus entry is localized to protease-rich TEMs.
TETRASPANINS DICTATE CoV ENTRY ROUTES
The proteases that cleave CoV S proteins and activate virus cell entry are present at several subcellular locations. On CoV-susceptible cells, there may be serine-class TMPRs at or near plasma membranes (39, 40), and there may also be cysteine-class cathepsin proteases within endosomes (22, 39). Therefore, depending on the abundance of these proteases, a CoV can be activated for entry at different locations along the endocytic pathway, either early (TMPR mediated) or late (cathepsin mediated) after virus endocytosis (Fig. 1, right panel). Notably, in the presence of CD9, we found that MERS pseudovirus entry was early and was suppressed by serine protease inhibitors, while in the absence of CD9, entry was late and was unaffected by serine protease inhibitors but rather was blocked by cathepsin inhibitors and endosome-neutralizing agents (35). These findings fit well with the identification of CD9 as an agent of DPP4 and TMPR colocalization: by linking the receptors and TMPR proteases at or near the plasma membrane, the CD9 tetraspanin facilitated early CoV entry into target cells. Note that the early entry route is far more likely to result in productive infection, as demonstrated by Shirato and colleagues in separate studies of HCoV-229E, -OC43, and -HKU1 (41, 42). Late entry, at least for some CoVs, appears to be a lower-efficiency, last-chance infection route before virus destruction in lysosomes.
TETRASPANINS CONTRIBUTE TO CoV INFECTION AND PATHOGENESIS IN VIVO
The in vitro finding that CD9 facilitated the highly infectious early entry route suggested that CD9 and other tetraspanins might also be in vivo CoV susceptibility factors. We expected that this would be the case, as strong evidence from protease inhibitor studies indicated that early-acting TMPRs might be central for respiratory CoV infections (43). Indeed, we did find key roles for CD9 and TMPRSS2 in MERS-CoV infections. Using adenoviral vectors expressing short hairpin RNAs (shRNAs), we transiently depleted either CD9 or TMPRSS2 in mouse lungs and then determined their susceptibility to MERS-CoV infection. Depleting CD9 and TMPRSS2 reduced MERS-CoV lung titers by ∼90% and ∼80%, respectively (35). Clearly, the vast majority of MERS-CoVs enter lung epithelial cells through the early entry route, and late entry pathways cannot compensate for early entry blockade. Given that humans express 33 tetraspanins and 19 serine TMPRs, it is remarkable that MERS-CoV is so highly dependent on a single tetraspanin and a single protease to infect the lung. Further investigation is needed to determine whether other CoVs utilize particular tetraspanins and proteases to enter cells in vivo.
The importance of early entry for MERS-CoV pathogenesis was further highlighted by the mutations that occur upon adaptation of this virus to the mouse lung environment. MERS-CoVs can infect mice when the human DPP4 (hDPP4) receptor is present, but these infections usually do not cause overt respiratory disease. Serial passage of MERS-CoVs in hDPP4 mouse lungs, however, generated mouse-adapted (MA) MERS-CoVs that caused severe lung disease characteristic of acute respiratory distress in humans (44). Notably, MA viruses accumulated S protein mutations that facilitated rapid, CD9-dependent early entry (35). In contrast, the avirulent cell culture-adapted (CCA) MERS-CoVs took 3 times longer to enter cells, and their infection was not supported by CD9. It is noteworthy that Vero cells, in which CCA MERS-CoVs were adapted, have low TMPRSS2 expression levels (39), as do several other cell lines routinely used to investigate CoV entry mechanisms in vitro (27, 39). TMPR+ cell culture models may better reflect the in vivo environment of CoV entry, since CoVs utilize TMPR-mediated “early” entry routes in the lung (35, 43).
TETRASPANINS AS GENERAL PROMOTERS OF VIRUS ENTRY
Accumulating evidence indicates that tetraspanins promote the entry of multiple viruses, including influenza A virus (IAV) (15, 32), human cytomegalovirus (HCMV) (45), human papillomavirus (HPV) (17, 18), hepatitis C virus (HCV) (14, 46, 47), Lujo virus (48), and several alphaviruses (49, 50). In some of these cases, the tetraspanins may be proviral because they coalesce the multiple host factors required to catalyze membrane fusion and infection. For example, IAV hemagglutinin (HA) proteins require proteolytic cleavage to undergo fusion-catalyzing structural transitions, and these proteolytic cleavages occur in TEMs (32). Depleting the tetraspanin CD81 from target cells restricts ∼90% of IAV entry (15), suggesting that this tetraspanin may aid in localizing IAV-activating proteases with sialate receptors. Tetraspanins can also condense virus-associated membrane proteins into complexes prior to endocytosis. The tetraspanin CD151 provides an example of this, as it drives HPV endocytosis by coalescing receptor-bound viruses and integrin coreceptors targeted for endocytosis. HCV and the tetraspanin CD81 set another illustration. CD81 acts as the primary HCV receptor, binding directly to the HCV E2 glycoprotein (14), yet CD81 also coalesces the HCV coreceptors claudin-1, syndecan-1, and scavenger receptor B1 into a complex required for subsequent HCV endocytosis (46, 47, 51, 52). We speculate that many viruses have evolved to utilize entry factors that reside in close juxtaposition and that tetraspanins act to aggregate these entry factors into TEMs.
TETRASPANINS ARE TARGETED BY THE INNATE IMMUNE RESPONSE
Cells have evolved to target tetraspanins to restrict viral infection. Among the antiviral type I interferon-stimulated gene products are the interferon-induced transmembrane proteins (IFITMs), which are small alpha-helical proteins composed of ∼130 residues that inhibit several virus entry processes at the level of virus-cell membrane fusion (53, 54). Intriguingly, IFITMs intercalate into TEMs, disrupting tetraspanin interactions with their partner proteins (55). One can speculate that as these intruding IFITMs accumulate during innate immune responses, they might separate or disorganize virus entry factors on target cell membranes. Notably, it is known that IFITM1 alters HCV coreceptor interactions, in correlation with restricted HCV-cell membrane fusion (56). It is possible that TEM disruption is a common mechanism by which the IFITMs restrict viral entry and infection.
TETRASPANINS IN VIRAL EGRESS
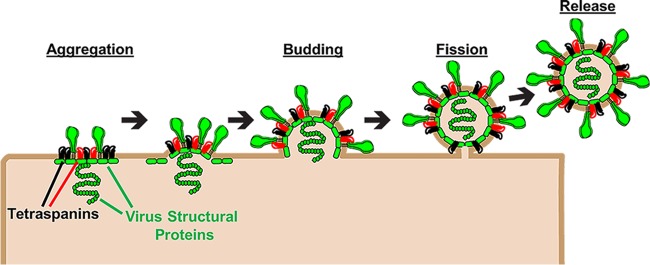
In addition to promoting virus entry into target cells, tetraspanins also facilitate viral assembly and egress (Fig. 2). Many enveloped viruses, including IAV (15), human immunodeficiency virus (HIV) (16, 57), and herpes simplex virus 1 (HSV-1) (58), bud out of TEMs. Viruses may utilize these TEMs to facilitate vesicle budding and release. One possible mechanism by which this takes place is through tetraspanin-mediated membrane remodeling. The four tetraspanin transmembrane domains form two pairs of antiparallel helices, which adopt a cone-shaped structure opening toward the outer leaflet of the lipid bilayer (3). This architecture may facilitate membrane curvatures that may promote enveloped virus vesicle formation and membrane fission (2). Investigations into IAV egress support this speculative role for tetraspanins in virus budding. IAV HA localizes to TEMs containing CD81, and this tetraspanin is required for successful virion release (15). In the absence of CD81, virions bud from the plasma membrane but do not undergo fission to be released from the cell (15). They instead become elongated, with no fission at a membrane neck. These findings suggest that tetraspanins facilitate the membrane curvatures required for membrane fission and virus release.
FIG 2.
Tetraspanins construct virus exit platforms. Tetraspanins (black and red) associate with viral structural proteins (green), leading to their aggregation into TEMs. Tetraspanins also induce positive membrane curvatures that may promote nascent virus budding. Finally, tetraspanins facilitate membrane fission, allowing for virus release.
Another way in which tetraspanins facilitate viral egress is by aggregating viral structural proteins. For example, the HSV-1 capsid protein VP26 interacts with the tetraspanin TSPAN7, leading to its accumulation on the nuclear membrane (58). This interaction is required for viral budding and egress from the nucleus. In the absence of TSPAN7 or upon mutation of VP26, HSV-1 capsids are sequestered in the nucleus, unable to exit (58). In addition, filamentous influenza virus strains accumulate CD81 at the ends of budding virions (15), along with the viral M2 protein, which mediates scission of IAV membranes from host cells (59). It is conceivable that CD81 facilitates M2 accumulation at the site of virus-cell membrane scission (15). These investigations highlight the ability of tetraspanins to facilitate multiple stages of the viral life cycle.
During viral biogenesis, tetraspanins incorporate into nascent virions (15, 57, 60, 61), suggesting that they may function in the context of the extracellular virion, perhaps at the virion cell entry stage. Intriguingly, virus-incorporated tetraspanins suppress rather than promote membrane fusion and virus entry (62, 63). In the context of HIV, overexpressing the tetraspanins CD9, CD63, and CD81 in virus producer cells inhibits HIV envelope-mediated membrane fusion (62–65), while depleting tetraspanins by use of small interfering RNAs (siRNAs) promotes membrane fusion (63, 65). We speculate that the high abundance of tetraspanins in HIV membranes may lead to these inhibitory effects. HIV Gag actively recruits tetraspanins to viral assembly sites, leading to the aggregation of large TEMs in HIV membranes (16). Notably, overexpressed tetraspanins inhibit HIV membrane fusion only in the presence of Gag (63). It is possible that Gag concentrates tetraspanins and cholesterols to rigidify HIV membranes, rendering them incompatible with membrane merger. Antibody-mediated tetraspanin cross-linking aggregates TEMs and suppresses CoV entry (32), supporting this notion. Further investigation is needed to determine whether the relative abundance of tetraspanins may dictate their pro- or antifusion activities.
TETRASPANINS IN NONVIRAL MEMBRANE FUSIONS
Tetraspanins and assembled TEMs contribute to a wide variety of biological processes, including those involved in cell metabolism, signal transduction, and intercellular adhesion. One of their additional critical roles is in controlling nonviral cellular membrane fusions. Cell-cell fusions generate many biologically important structures, including fertilized zygotes (66), skeletal muscles (67), placentae (68), and multinucleated macrophages (69). Notably, tetraspanins are known regulators of many cell-cell fusion reactions (70–73). Their role is most striking during fertilization, which absolutely requires the tetraspanin CD9 (70). CD9 knockout mice are infertile, and eggs lacking CD9 are unable to fuse with sperm (70). CD9-laden extracellular vesicles, however, can compensate for loss of CD9 by bridging sperm and egg cells and subsequently fusing them together, in a process known as “fusion from without” (71). CD9 overexpression stimulates monocyte cell-cell fusions (72), which lead to the formation of multinucleated osteoclasts that resorb bone. Similarly, antibodies targeting CD9 suppress the fusion of myoblasts into the large syncytia required to form skeletal muscle (73). While the mechanisms behind tetraspanin-facilitated cell-cell fusions are not fully elucidated, we speculate that tetraspanins generally regulate membrane fusion events through their ability to bend membranes and cluster the factors required for membrane fusion catalysis. Surprisingly, many eukaryotic cell-cell fusion catalysts remain unidentified. Proteomic analysis of TEMs from fusogenic cells, along with molecular genetic approaches, may identify these elusive endogenous cellular fusogens.
REFERENCES
- 1.Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, Xie X, Yu Y, Yu X, Chen S, Zhang S, Pontarotti P, Xu A. 2005. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86:674–684. doi: 10.1016/j.ygeno.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Bari R, Guo Q, Xia B, Zhang YH, Giesert EE, Levy S, Zheng JJ, Zhang XA. 2011. Tetraspanins regulate the protrusive activities of cell membrane. Biochem Biophys Res Commun 415:619–626. doi: 10.1016/j.bbrc.2011.10.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, Blacklow SC. 2016. Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167:1041.e11–1051.e11. doi: 10.1016/j.cell.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Claas C, Kraeft S-K, Chen LB, Wang Z, Kreidberg JA, Hemler ME. 2002. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol Biol Cell 13:767–781. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charrin S, Manie S, Oualid M, Billard M, Boucheix C, Rubinstein E. 2002. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett 516:139–144. [DOI] [PubMed] [Google Scholar]
- 6.Slupsky JR, Seehafer JG, Tang SC, Masellis-Smith A, Shaw AR. 1989. Evidence that monoclonal antibodies against CD9 antigen induce specific association between CD9 and the platelet glycoprotein IIb-IIIa complex. J Biol Chem 264:12289–12293. [PubMed] [Google Scholar]
- 7.Serru V, Le Naour F, Billard M, Azorsa DO, Lanza F, Boucheix C, Rubinstein E. 1999. Selective tetraspan-integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem J 340:103–111. doi: 10.1042/bj3400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barreiro O, Zamai M, Yáñez-Mó M, Tejera E, López-Romero P, Monk PN, Gratton E, Caiolfa VR, Sánchez-Madrid F. 2008. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol 183:527–542. doi: 10.1083/jcb.200805076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stipp CS, Kolesnikova TV, Hemler ME. 2003. Functional domains in tetraspanin proteins. Trends Biochem Sci 28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 10.Boucheix C, Rubinstein E. 2001. Tetraspanins. Cell Mol Life Sci 58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemler ME. 2005. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 12.Martin F, Roth DM, Jans DA, Pouton CW, Partridge LJ, Monk PN, Moseley GW. 2005. Tetraspanins in viral infections: a fundamental role in viral biology? J Virol 79:10839–10851. doi: 10.1128/JVI.79.17.10839-10851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charrin S, Le Naour F, Silvie O, Milhiet P-E, Boucheix C, Rubinstein E. 2009. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J 420:133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 14.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. [DOI] [PubMed] [Google Scholar]
- 15.He J, Sun E, Bujny MV, Kim D, Davidson MW, Zhuang X. 2013. Dual function of CD81 in influenza virus uncoating and budding. PLoS Pathog 9:e1003701. doi: 10.1371/journal.ppat.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol 173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, Florin L. 2008. Clathrin- and caveolin-independent entry of human papillomavirus type 16—involvement of tetraspanin-enriched microdomains (TEMs). PLoS One 3:e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. 2013. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J Virol 87:3435–3446. doi: 10.1128/JVI.02906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Haan CAM, Stadler K, Godeke G-J, Bosch BJ, Rottier PJM. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J Virol 78:6048–6054. doi: 10.1128/JVI.78.11.6048-6054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A 102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wicht O, Burkard C, de Haan CAM, van Kuppeveld FJM, Rottier PJM, Bosch BJ. 2014. Identification and characterization of a proteolytically primed form of the murine coronavirus spike proteins after fusion with the target cell. J Virol 88:4943–4952. doi: 10.1128/JVI.03451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada Y, Liu DX. 2009. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J Virol 83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belouzard S, Chu VC, Whittaker GR. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuyama S, Taguchi F. 2009. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J Virol 83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J-E, Li K, Barlan A, Fehr AR, Perlman S, McCray PB, Gallagher T. 2016. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A 113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rana S, Claas C, Kretz CC, Nazarenko I, Zoeller M. 2011. Activation-induced internalization differs for the tetraspanins CD9 and Tspan8: impact on tumor cell motility. Int J Biochem Cell Biol 43:106–119. doi: 10.1016/j.biocel.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto T, Iwata S, Yamazaki H, Hatano R, Komiya E, Dang NH, Ohnuma K, Morimoto C. 2014. CD9 negatively regulates CD26 expression and inhibits CD26-mediated enhancement of invasive potential of malignant mesothelioma cells. PLoS One 9:e86671. doi: 10.1371/journal.pone.0086671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Naour F, Andre M, Boucheix C, Rubinstein E. 2006. Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics 6:6447–6454. doi: 10.1002/pmic.200600282. [DOI] [PubMed] [Google Scholar]
- 31.Andre M, Le Caer J-P, Greco C, Planchon S, El Nemer W, Boucheix C, Rubinstein E, Chamot-Rooke J, Le Naour F. 2006. Proteomic analysis of the tetraspanin web using LC-ESI-MS/MS and MALDI-FTICR-MS. Proteomics 6:1437–1449. doi: 10.1002/pmic.200500180. [DOI] [PubMed] [Google Scholar]
- 32.Earnest JT, Hantak MP, Park J-E, Gallagher T. 2015. Coronavirus and influenza virus proteolytic priming takes place in tetraspanin-enriched membrane microdomains. J Virol 89:6093–6104. doi: 10.1128/JVI.00543-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charrin S, Manié S, Billard M, Ashman L, Gerlier D, Boucheix C, Rubinstein E. 2003. Multiple levels of interactions within the tetraspanin web. Biochem Biophys Res Commun 304:107–112. [DOI] [PubMed] [Google Scholar]
- 34.Yauch RL, Berditchevski F, Harler MB, Reichner J, Hemler ME. 1998. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol Biol Cell 9:2751–2765. doi: 10.1091/mbc.9.10.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earnest JT, Hantak MP, Li K, McCray PB, Perlman S, Gallagher T. 2017. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog 13:e1006546. doi: 10.1371/journal.ppat.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tellier E, Canault M, Rebsomen L, Bonardo B, Juhan-Vague I, Nalbone G, Peiretti F. 2006. The shedding activity of ADAM17 is sequestered in lipid rafts. Exp Cell Res 312:3969–3980. doi: 10.1016/j.yexcr.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 39.Shirato K, Kawase M, Matsuyama S. 2013. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pohlmann S. 2011. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirato K, Kanou K, Kawase M, Matsuyama S. 2016. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J Virol 91:e01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirato K, Kawase M, Matsuyama S. 2018. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Renslo AR, Simmons G. 2015. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li K, Wohlford-Lenane CL, Channappanavar R, Park J-E, Earnest JT, Bair TB, Bates AM, Brogden KA, Flaherty HA, Gallagher T, Meyerholz DK, Perlman S, McCray PBJ. 2017. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A 114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hochdorfer D, Florin L, Sinzger C, Lieber D. 2016. Tetraspanin CD151 promotes initial events in human cytomegalovirus infection. J Virol 90:6430–6442. doi: 10.1128/JVI.00145-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset F-L. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem 278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 47.Grigorov B, Reungoat E, Gentil Dit Maurin A, Varbanov M, Blaising J, Michelet M, Manuel R, Parent R, Bartosch B, Zoulim F, Ruggiero F, Pecheur E-I. 2017. Hepatitis C virus infection propagates through interactions between Syndecan-1 and CD81 and impacts the hepatocyte glycocalyx. Cell Microbiol 19:12711. doi: 10.1111/cmi.12711. [DOI] [PubMed] [Google Scholar]
- 48.Raaben M, Jae LT, Herbert AS, Kuehne AI, Stubbs SH, Chou Y-Y, Blomen VA, Kirchhausen T, Dye JM, Brummelkamp TR, Whelan SP. 2017. NRP2 and CD63 are host factors for Lujo virus cell entry. Cell Host Microbe 22:688.e5–696.e5. doi: 10.1016/j.chom.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. 2013. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog 9:e1003835. doi: 10.1371/journal.ppat.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stiles KM, Kielian M. 2016. Role of TSPAN9 in alphavirus entry and early endosomes. J Virol 90:4289–4297. doi: 10.1128/JVI.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krieger SE, Zeisel MB, Davis C, Thumann C, Harris HJ, Schnober EK, Mee C, Soulier E, Royer C, Lambotin M, Grunert F, Dao Thi VL, Dreux M, Cosset F-L, McKeating JA, Schuster C, Baumert TF. 2010. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology 51:1144–1157. doi: 10.1002/hep.23445. [DOI] [PubMed] [Google Scholar]
- 52.Harris HJ, Davis C, Mullins JGL, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, Balfe P, McKeating JA. 2010. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen L-M, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, Brass AL. 2011. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. 2014. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanagata N, Li X. 2011. Osteoblast-enriched membrane protein IFITM5 regulates the association of CD9 with an FKBP11-CD81-FPRP complex and stimulates expression of interferon-induced genes. Biochem Biophys Res Commun 409:378–384. doi: 10.1016/j.bbrc.2011.04.136. [DOI] [PubMed] [Google Scholar]
- 56.Wilkins C, Woodward J, Lau DT-Y, Barnes A, Joyce M, McFarlane N, McKeating JA, Tyrrell DL, Gale M. 2013. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jolly C, Sattentau QJ. 2007. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol 81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Liu L, Che Y, Wang L, Jiang L, Dong C, Zhang Y, Li Q. 2010. Egress of HSV-1 capsid requires the interaction of VP26 and a cellular tetraspanin membrane protein. Virol J 7:156. doi: 10.1186/1743-422X-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. 2008. Cellular proteins in influenza virus particles. PLoS Pathog 4:e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orentas RJ, Hildreth JE. 1993. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res Hum Retroviruses 9:1157–1165. doi: 10.1089/aid.1993.9.1157. [DOI] [PubMed] [Google Scholar]
- 62.Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, Koyanagi Y. 2008. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol 82:1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weng J, Krementsov DN, Khurana S, Roy NH, Thali M. 2009. Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J Virol 83:7467–7474. doi: 10.1128/JVI.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Symeonides M, Lambele M, Roy NH, Thali M. 2014. Evidence showing that tetraspanins inhibit HIV-1-induced cell-cell fusion at a post-hemifusion stage. Viruses 6:1078–1090. doi: 10.3390/v6031078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gordón-Alonso M, Yañez MM, Barreiro O, Álvarez S, Muñoz FMÁ, Valenzuela FA, Sánchez-Madrid F. 2006. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol 177:5129–5137. doi: 10.4049/jimmunol.177.8.5129. [DOI] [PubMed] [Google Scholar]
- 66.Okabe M. 2013. The cell biology of mammalian fertilization. Development 140:4471–4479. doi: 10.1242/dev.090613. [DOI] [PubMed] [Google Scholar]
- 67.Kim JH, Jin P, Duan R, Chen EH. 2015. Mechanisms of myoblast fusion during muscle development. Curr Opin Genet Dev 32:162–170. doi: 10.1016/j.gde.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerbaud P, Pidoux G. 2015. Review: an overview of molecular events occurring in human trophoblast fusion. Placenta 36:S35–S42. doi: 10.1016/j.placenta.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 69.Vignery A. 2000. Osteoclasts and giant cells: macrophage-macrophage fusion mechanism. Int J Exp Pathol 81:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. 2000. The gamete fusion process is defective in eggs of CD9-deficient mice. Nat Genet 24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 71.Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito C, Toshimori K, Nakamura A, Ito M, Miyado M, Mekada E, Umezawa A. 2008. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc Natl Acad Sci U S A 105:12921–12926. doi: 10.1073/pnas.0710608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishii M, Iwai K, Koike M, Ohshima S, Kudo-Tanaka E, Ishii T, Mima T, Katada Y, Miyatake K, Uchiyama Y, Saeki Y. 2006. RANKL-induced expression of tetraspanin CD9 in lipid raft membrane microdomain is essential for cell fusion during osteoclastogenesis. J Bone Miner Res 21:965–976. doi: 10.1359/jbmr.060308. [DOI] [PubMed] [Google Scholar]
- 73.Tachibana I, Hemler ME. 1999. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J Cell Biol 146:893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]