Following primary infection by HIV-1, neutralizing antibodies (NAbs) exert selective pressure on the HIV-1 envelope glycoprotein (Env), driving the evolution of the viral population. Previous studies suggested that, as a consequence, Env has evolved at the HIV species level since the start of the epidemic so as to display greater resistance to NAbs. Here, we investigated whether the antigenic evolution of the HIV-1 Env is associated with modifications of its functional properties, focusing on cell entry efficacy and interactions with the receptor and coreceptors. Our data provide evidence that the infectious properties of the HIV-1 Env increased during the course of the epidemic. These changes may have contributed to increasing virulence of HIV-1 and an optimization of transmission between individuals.
KEYWORDS: envelope, evolution, human immunodeficiency virus, infectivity
ABSTRACT
We showed previously that during the HIV/AIDS epidemic, the envelope glycoprotein (Env) of HIV-1, and in particular, the gp120 subunit, evolved toward an increased resistance to neutralizing antibodies at a population level. Here, we considered whether the antigenic evolution of the HIV-1 Env is associated with modifications of its functional properties, focusing on cell entry efficacy and interactions with the receptor and coreceptors. We tested the infectivity of a panel of Env-pseudotyped viruses derived from patients infected by subtype B viruses at three periods of the epidemic (1987 to 1991, 1996 to 2000, and 2006 to 2010). Pseudotyped viruses harboring Env from patients infected during the most recent period were approximately 10-fold more infectious in cell culture than those from patients infected at the beginning of the epidemic. This was associated with faster viral entry kinetics: contemporary viruses entered target cells approximately twice as fast as historical viruses. Contemporary viruses were also twice as resistant as historical viruses to the fusion inhibitor enfuvirtide. Resistance to enfuvirtide correlated with a resistance to CCR5 antagonists, suggesting that contemporary viruses expanded their CCR5 usage efficiency. Viruses were equally captured by DC-SIGN, but after binding to DC-SIGN, contemporary viruses infected target cells more efficiently than historical viruses. Thus, we report evidence that the infectious properties of the envelope glycoprotein of HIV-1 increased during the course of the epidemic. It is plausible that these changes affected viral fitness during the transmission process and might have contributed to an increasing virulence of HIV-1.
IMPORTANCE Following primary infection by HIV-1, neutralizing antibodies (NAbs) exert selective pressure on the HIV-1 envelope glycoprotein (Env), driving the evolution of the viral population. Previous studies suggested that, as a consequence, Env has evolved at the HIV species level since the start of the epidemic so as to display greater resistance to NAbs. Here, we investigated whether the antigenic evolution of the HIV-1 Env is associated with modifications of its functional properties, focusing on cell entry efficacy and interactions with the receptor and coreceptors. Our data provide evidence that the infectious properties of the HIV-1 Env increased during the course of the epidemic. These changes may have contributed to increasing virulence of HIV-1 and an optimization of transmission between individuals.
INTRODUCTION
Following primary infection with human immunodeficiency virus type 1 (HIV-1), most patients develop autologous neutralizing antibodies (NAbs) directed against the gp120 and gp41 subunits of the viral envelope glycoprotein (Env) (1–6). These antibodies exert selective pressure, leading to the rapid emergence of escape Env variants and diversification of the viral population (1, 2, 7–9). HIV-1 escape from autologous neutralization involves numerous molecular mechanisms, including single amino acid substitutions, insertions/deletions, and modifications of potential N-linked glycosylation sites at the viral surface (2, 7, 8, 10, 11). During sexual transmission, a single transmitted/founder virus among the numerous viral variants present in the donor is transmitted most often, resulting in a substantial genetic bottleneck (12–15). There is preferential transmission of ancestral variants over contemporary variants circulating in the transmitting partner (16, 17), and this is expected to constrain the viral diversity at the population level, at least to some extent. Nevertheless, the evolution and spread of HIV-1 in the human population during 30 years of epidemics have led to considerable diversity (18). We and others reported evidence that the increasing diversity of the HIV-1 species has been accompanied by a drift toward increased resistance to neutralization over the course of the epidemic (19–23). Using a large panel of monoclonal NAbs, we showed that this antigenic evolution affects gp120 but not gp41 (20–23). In addition, we observed that for subtype B viruses, almost all of the major identified neutralization sites of gp120 were affected, suggesting that gp120 as a whole has progressively evolved in less than three decades (21). Interestingly, recent papers suggest that the virulence of HIV-1, estimated from the CD4 cell count at the time of primary infection and the subsequent viral set point, has increased over time, suggesting a progressive adaptation of the HIV-1 species to its human host (24–26). Here, we investigated whether the antigenic evolution of the HIV-1 Env is associated with modifications of its functional properties, focusing on cell entry efficacy and interactions with the receptor and coreceptors, properties which could impact viral fitness during the transmission process and contribute to the potential increasing virulence of HIV-1.
RESULTS
Population studied.
The HIV-1 population that we studied was described previously (20); it was derived from 40 infected patients enrolled at the time of primary infection in the ANRS SEROCO and PRIMO cohorts (27, 28) during three periods of the French epidemic: between 1987 and 1991 (historical patients [HP], n = 11), 1996 and 2000 (intermediate patients [IP], n = 15), and 2006 and 2010 (contemporary patients [CP], n = 14). They were carefully selected to be comparable for each period in the following way: all patients were Caucasian men having sex with men (MSM), infected by clade B viruses. They had similar distributions of viral loads (median values of 5.0, 5.1, and 5.2 log10 copies/ml for HP, IP, and CP, respectively) and similar distributions of CD4 T-cell counts (median values of 507, 619, and 571 cells/mm3 for HP, IP, and CP, respectively) at time of sample collection (20). Blood samples were collected shortly after infection (before 3 months postinfection, except for a few cases) (20); the variants that we analyzed were therefore considered early/transmitted viruses.
Env pseudotypes exhibit increasing infectivity.
Pseudotyped viruses expressing envelope glycoprotein (Env) variants representative of the viral quasispecies infecting each patient were generated by cotransfecting 293T cells with an env-deleted NL4-3 backbone plasmid and patient-derived env libraries (20). Infectious pseudoviruses were obtained for 11 to 15 patients from each of the three periods.
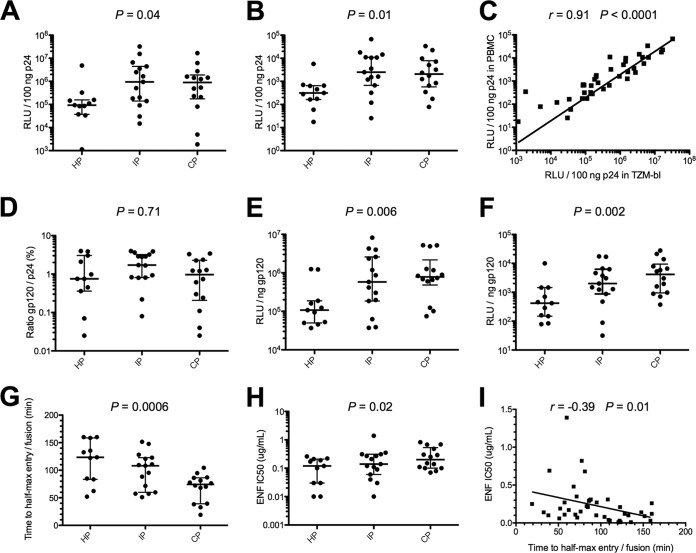
We investigated the capacity of HP, IP, and CP Env pseudotypes to infect TZM-bl cells and primary stimulated CD8-depleted peripheral blood mononuclear cells (PBMCs) in a single round of infection. The infectivity level of each pseudotyped virus, whose inputs were normalized to the p24 amounts (100 ng), was evaluated 48 h postinfection by measuring the luciferase activity (relative light units [RLU]). The results showed an increasing infectivity of approximately 10-fold in both cell types over the course of the epidemic (Fig. 1A and B). In TZM-bl cells, the median RLU increased from 9.3 × 104 for HP to 9.5 × 105 for IP and remained stable for CP (9.0 × 105) (P = 0.04) (Fig. 1A). Similarly, in PBMCs, the median RLU increased from 3.1 × 102 for HP to 2.5 × 103 for IP and remained stable for CP (2.1 × 103) (P = 0.01) (Fig. 1B). Although the luciferase activity levels were higher in TZM-bl cells than in PBMCs, there was good correlation between the cell types (r = 0.91; P < 0.0001) (Fig. 1C).
FIG 1.
Infectivity, entry kinetics, and sensitivity to enfuvirtide of Env-pseudotyped viruses. Infectivity of Env-pseudotyped viruses derived from historical patients (HP; n = 11), intermediate patients (IP; n = 15), and contemporary patients (CP; n = 14) in TZM-bl cells (A) and in CD8-depleted PBMCs (B). The infectivity of each 293T-cell-derived pseudotyped virus, with inputs normalized for amount of p24, was evaluated 48 h postinfection by assaying luciferase activity (reported as relative luminescence units [RLU]). Scatter dot plots show the distributions of infectivity values for pseudotypes of each period, expressed as RLU per 100 ng of p24. (C) Correlation between infectivity values in TZM-bl cells and CD8-depleted PBMCs. (D) Comparison of Env contents of pseudotypes derived from HP, IP, and CP. Scatter dot plots show the distributions of Env content measured as the gp120/p24 ratio. Comparison of infectivity of Env pseudotypes from HP, IP, and CP in TZM-bl cells (E) and CD8-depleted PBMCs (F) after normalization for Env content (1 ng) by dividing the infectivity RLU values (measure of infectivity) by the gp120/p24 ratio. Scatter dot plots show the distributions of infectivity values for pseudotypes of each period, expressed as RLU per 1 ng of Env. (G) Comparison of entry kinetics of Env-pseudotyped viruses from HP, IP, and CP. Env pseudotypes were added to TZM-bl cells on ice. Cells were spinoculated at 4°C to facilitate binding of viruses, and then cold medium was replaced by prewarmed medium. Saturating concentrations of ENF were then added at various times (0, 5, 10, 20, 40, 80, and 160 min), and infectivity was evaluated 72 h later by measuring luciferase activity (RLU). Scatter dot plots show the distributions of entry kinetic values for pseudotypes of each period, expressed as the time for half-max entry. (H) Comparison of ENF sensitivity of Env-pseudotyped viruses derived from HP, IP, and CP. Scatter dot plots show the distributions of IC50s of ENF for pseudotypes of each period. (I) Correlation between ENF sensitivities and entry kinetics. Differences between viruses over calendar time were evaluated using a Jonckheere-Terpstra test.
Given that Env pseudotypes were generated in an NL4-3 background, the interplay between the cytoplasmic tails of gp41 and their matching matrix proteins was disabled, possibly affecting Env incorporation. We therefore analyzed the gp120 content of each virus. Pseudotyped viruses were pelleted from viral supernatants of transfected 293T cells by centrifugation through a sucrose cushion, and virion-associated gp120 levels were quantified by enzyme-linked immunosorbent assay (ELISA). The gp120 levels differed between pseudotyped viruses of each single period but not significantly between periods (gp120/p24 ratios ranging from 0.03% to 3.97% for HP, 0.08% to 3.97% for IP, and 0.03% to 3.42% for CP; P = 0.71) (Fig. 1D). Knowing the Env content for each virus, we then assessed the infectivity of HP, IP, and CP pseudotypes normalized for Env content (1 ng) by dividing the RLU values by the gp120/p24 ratios. By this measure, infectivity increased progressively from HP to CP, both in TZM-bl cells (median RLU of 1.1 × 105, 5.8 × 105, and 7.8 × 105 for HP, IP, and CP, respectively; P = 0.006) (Fig. 1E) and in PBMCs (median RLU of 4.2 × 102, 2.0 × 103, and 4.1 × 103 for HP, IP, and CP, respectively; P = 0.002) (Fig. 1F). This indicates that the increasing infectivity of Env pseudotypes over time was linked to an intrinsic property of their Env glycoproteins and not to a potential artifactual increase in the Env content over time due to unmatched gp41 cytoplasmic tails and matrix proteins.
Env pseudotypes exhibit increasing entry kinetics and enfuvirtide resistance.
We then asked whether the increasing infectivity was associated with faster entry kinetics. We used a time-of-addition experiment with saturating concentrations of the fusion inhibitor enfuvirtide (ENF), as previously described (29). As ENF is not membrane permeative, the time to ENF escape reflects the entry rate of the virus into the cells. HP, IP, and CP Env pseudotypes were added to TZM-bl cells on ice, followed by spinoculation at 4°C to facilitate binding of viruses. Then cold medium was replaced by prewarmed medium, and saturating concentrations of ENF were added at 0, 5, 10, 20, 40, 80, and 160 min. Infectivity was evaluated 72 h later by measuring luciferase activity. The median times for half-max entry (t1/2max) decreased progressively from HP to CP (median t1/2maxs of 123.4, 108.1, and 74.2 min for HP, IP, and CP, respectively; P = 0.0006), suggesting that the rate of viral entry increased over time (Fig. 1G).
Resistance to ENF also increased from HP to CP (median 50% infective concentrations [IC50s] of 0.12, 0.14, and 0.20 μg/ml for HP, IP, and CP, respectively; P = 0.02) (Fig. 1H); however, none of the known mutations associated with resistance to ENF was present in the HR1 regions of our 40 viruses (30), and there was no specific sequence difference for this region between viruses of the three periods (Fig. 2). ENF binds to the HR1 domain of gp41, which becomes exposed after CD4/coreceptor engagement. Therefore, the sensitivity to ENF may reflect the CD4/coreceptor usage efficiency and/or the kinetics of conformational changes in Env that drive the membrane fusion reaction (31). This was further supported by a statistically significant inverse correlation between time to half-max entry and resistance to ENF (Fig. 1I).
FIG 2.
Alignment of the gp41 HR1 regions of the 40 env sequences from historical (HP), intermediate (IP), and contemporary (CP) patients with the HXB2 reference sequence. Identical residues are represented by dots. Residues G36, I37, V38, Q39, Q40, N42, and N43, whose mutations G36D/S, I37V, V38A/M/E, Q39R, Q40H, N42T, and N43D, respectively, have been shown to be associated with resistance to enfuvirtide, are highlighted in yellow.
Contemporary variants are more resistant to CCR5 antagonists than historical variants.
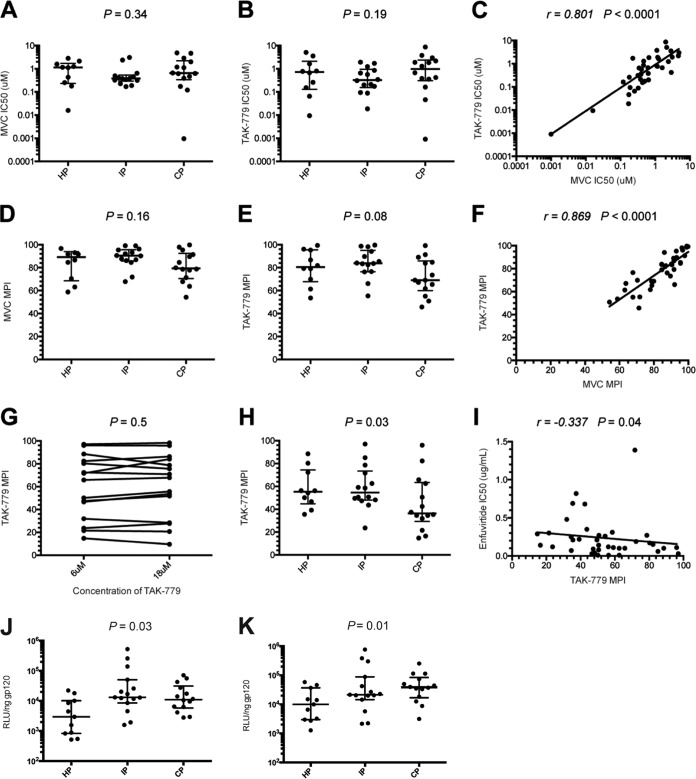
We assessed the efficiency of CCR5 usage of the HP, IP, and CP pseudotyped viruses to investigate whether the increasing resistance to ENF over time was related to a better use of the coreceptor, reducing the window period during which Env is sensitive to ENF. We first determined tropism by measuring the ability of the viruses to infect CD4+/U373 MAGI cells expressing either the CXCR4 or CCR5 coreceptor (32). All except two viruses infected exclusively CD4+/CCR5+/U373 cells (CD4+/CCR5+/U373 versus CD4+/CXCR4+/U373 RLU ratios >99%). The two other viruses (1 HP and 1 CP) were dual tropic (CD4+/CCR5+/U373 versus CD4+/CXCR4+/U373 RLU ratios of 64% and 51%, respectively). We infected TZM-bl cells with each R5 virus in the presence of various concentrations of the CCR5 antagonists TAK-779 or maraviroc (MVC). The MVC and TAK-779 IC50 values were highly correlated (r = −0.801, P < 0.0001) and did not differ between HP, IP, and CP R5 viruses (Fig. 3A to C). However, in the presence of saturating concentrations of MVC or TAK-779 (6 μM), the maximal percent inhibition (MPI) values tended to be lower for CP than for other viruses (median MVC MPIs 79.35% for CP versus 89.10% and 90.35% for HP and IP, respectively; P value, not significant [NS]; median TAK-779 MPIs 69.00% for CP versus 80.55% and 83.80% for HP and IP, respectively; P = 0.08) (Fig. 3D and E). Again, MVC and TAK MPI values were highly correlated (r = −0.869, P < 0.0001) (Fig. 3F).
FIG 3.
Sensitivity to CCR5 antagonists. Comparison of maraviroc (MVC) (A and D) and TAK-779 (B and E) sensitivity of Env-pseudotyped viruses derived from historical patients (HP; n = 10), intermediate patients (IP; n = 15), and contemporary patients (CP; n = 13) in TZM-bl cells. Scatter dot plots show the distributions of MVC and TAK-779 IC50s (A and B) and maximal percent inhibition (MPI) values (D and E) for pseudotypes of each period. (C) Correlation between MVC and TAK-779 IC50s. (F) Correlation between MVC and TAK-779 MPIs. (G) Comparison of TAK-779 MPIs in Affinofile cells in the presence of 6 μM or 12 μM of TAK-779. A panel of 15 pseudotyped viruses (5 HP, 5 IP, and 5 CP) that presented a range of sensitivities to this inhibitor was used to confirm that the concentration of TAK-779 was saturating. There was no difference in inhibition of infection between 6 μM and 12 μM, indicating that 6 μM was saturating in Affinofile cells expressing a high level of CCR5 (P = 0.50 by paired t test). (H) Comparison of TAK-779 sensitivity of Env-pseudotyped viruses derived from HP, IP, and CP in Affinofile cells. Scatter dot plots show the distributions of TAK-779 MPI values for pseudotypes of each period. Differences between viruses over calendar time were evaluated using a Jonckheere-Terpstra test. (I) Correlation between enfuvirtide IC50s and TAK-779 MPIs. Comparison of infectivity of Env pseudotypes from HP, IP, and CP in Affinofile cells expressing low levels (J) or high levels (K) of CCR5, after normalization for Env content (1 ng).
Affinofile cells expressing high levels of CCR5 can detect partial resistance to CCR5 antagonists with greater sensitivity than the other commonly used cell lines (33–36). We therefore examined the sensitivity of our pseudotyped viruses to TAK-779 on Affinofile cells expressing a high level of CCR5. Ponasterone A (PonA) treatment increased the overall expression of CCR5 on Affinofile cells from 788 to 50,178 antibody binding sites (ABS) per cell. For comparison, there were 19,032 ABS per TZM-bl cell, which was ∼2.6 times lower than on maximally induced Affinofile cells. To confirm that 6 μM of TAK-779 was saturating on Affinofile cells at the highest expression level of CCR5, we assessed the differences of MPIs in the presence of 6 μM and 18 μM of TAK-779 using a panel of 15 viruses (5 HP, 5 IP, and 5 CP) with a broad range of sensitivities to this inhibitor. The data showed that 6 μM was saturating and that further increases in TAK-779 concentrations did not have any additional effect (P = 0.50 by paired t test) (Fig. 3G). Affinofile cells expressing high levels of CCR5 were infected with each of the 38 R5 viruses in the presence or absence of 6 μM TAK-779. Consistent with previous studies (33–35), lower MPIs were observed in Affinofile cells than in cells that comparatively express lower levels of CCR5 (median TAK-779 MPIs of 50.55% in Affinofile cells versus 82.15% in TZM-bl cells). The higher resistance to TAK-779 of CP than of the other viruses observed in TZM-bl cells was more pronounced and was statistically significant in Affinofile cells (median TAK-779 MPIs of 36.40% for CP versus 55.25% and 54.63% for HP and IP, respectively; P = 0.03) (Fig. 3H). Taken together, these results showed that the most recent viruses were more resistant than older viruses to CCR5 antagonists, suggesting a greater plasticity of their Env to engage the CCR5 coreceptor. Interestingly, there was a significant inverse correlation between TAK-779 MPIs in Affinofile cells and ENF IC50 values in TZM-bl cells (r = −0.337, P = 0.04) (Fig. 3I). Possibly, increasing viral resistance to ENF over the course of the epidemic was related to expanded CCR5 usage efficiency. Nevertheless, no statistically significant correlation was observed between infectivity in PBMCs and resistance to ENF in TZM-bl cells (P = 0.36) or TAK-779 MPIs in Affinofile cells expressing high levels of CCR5 (P = 0.12), suggesting that decreasing sensitivity to ENF and CCR5 inhibitors was not the only factor involved in increasing the infectivity of viruses over time.
To further explore the relevance of our observations, we next investigated the capacity of HP, IP, and CP pseudotyped viruses to infect Affinofile cells expressing low or high levels of CCR5. In Affinofile cells with low densities of CCR5, the median RLU increased from 3.0 × 103 for HP to 1.3 × 104 for IP and remained stable for CP (1.1 × 104) (P = 0.03) (Fig. 3J). In Affinofile cells expressing high levels of CCR5, luciferase activity levels were higher; the median RLU increased progressively from HP to CP (median RLU of 9.9 × 103, 2.1 × 104, and 3.8 × 105 for HP, IP, and CP, respectively), and this evolution became more statistically significant (P = 0.01) (Fig. 3K), suggesting again a more promiscuous use of CCR5 by CP under these conditions.
Env pseudotypes do not differ in their sensitivity to soluble CD4 but exhibit increasing resistance to a CD4 mimetic miniprotein.
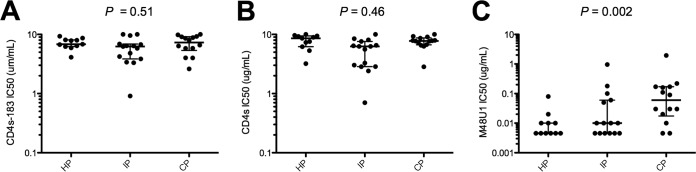
We compared CD4 receptor usage of the HP, IP, and CP pseudotyped viruses through their sensitivity to CD4 analogs. Using two soluble forms of CD4, containing either all four (sCD4) or the first two (sCD4-183) extracellular domains, we found no statistically significant differences in the IC50 ranges between HP, IP, and CP viruses (Fig. 4A and B). In contrast, resistance to M48U1, a CD4 mimetic miniprotein (miniCD4) (37, 38), increased from HP to CP viruses (median IC50s of 0.005, 0.01, and 0.06 μg/ml for HP, IP and CP pseudotypes, respectively; P = 0.002) (Fig. 4C). These detuned results may be explained by the fact that M48U1, by its flexible hydrophobic extensions, reaches deep into the gp120 Phe-43 cavity and interacts with residues that do not contact CD4 (38). This suggests that there has been evolution of the Phe-43 cavity leading to the diminishing sensitivity to M48U1.
FIG 4.
Sensitivity to CD4 inhibitors. Sensitivity of Env-pseudotyped viruses derived from historical patients (HP; n = 11), intermediate patients (IP; n = 15), and contemporary patients (CP; n = 14) to sCD4-183 (A), sCD4 (B), and M48U1 (C). Scatter dot plots show the distributions of IC50 values of each inhibitor for pseudotyped viruses of each period. Differences between viruses over calendar time were evaluated using a Jonckheere-Terpstra test.
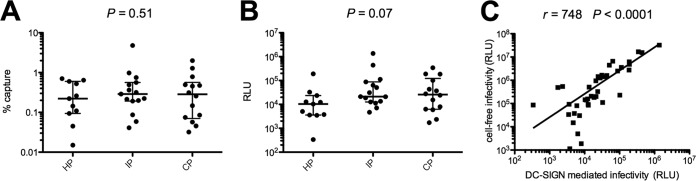
Env pseudotypes bind equally to DC-SIGN, but contemporary variants are more efficiently transferred to target cells.
HIV-1 binds and utilizes DC-SIGN, a C-type lectin expressed by dendritic cells, to promote infection of T-cells in trans (39–42). Raji/DC-SIGN cells were used to assess the binding of HP, IP, and CP viruses to DC-SIGN and their transfer to target TZM-bl cells. We found no difference in the capacity of HP, IP, and CP pseudotypes to bind to DC-SIGN (Fig. 5A). In contrast, although the data were not statistically significant, the DC–SIGN-mediated infection of TZM-bl cells tended to increase progressively from the HP to CP (median RLU of 10,313, 21,206, and 26,044 for HP, IP, and CP, respectively; P = 0.07) (Fig. 5B). DC–SIGN-mediated infection of TZM-bl cells and cell-free infection of TZM-bl cells were strongly correlated (r = 0.748, P < 0.0001) (Fig. 5C). There is thus no evidence that the increasing capacity of viruses to be transferred to target cells after binding to DC-SIGN over the course of the epidemic was due to increasing DC-SIGN binding; the change appears to involve subsequent events of the viral entry step.
FIG 5.
DC-SIGN binding and transfer to target cells. Comparison of DC-SIGN capture (A) and DC-mediated transfer to TZM-bl cells (B) of Env-pseudotyped viruses derived from historical patients (HP; n = 11), intermediate patients (IP; n = 15), and contemporary patients (CP; n = 14). Raji/DC-SIGN or Raji cells (negative control) were pulsed with each virus, whose inputs were normalized by p24 content. Samples were washed extensively to remove cell-free virions and lysed, and cell-associated virus was assayed by p24 ELISA. Percentages of capture were calculated after subtracting nonspecific binding to Raji cells. For transfer experiments, virus-bound Raji/DC-SIGN or Raji cells were cocultured with TZM-bl cells. Forty-eight hours later, cells were lysed and luciferase activity was determined. Differences between viruses over calendar time were evaluated using a Jonckheere-Terpstra test. (C) Correlation between cell-free and DC-mediated infectivities.
The increased infectious properties of Env pseudotypes are associated with the diversification of env sequences.
Full-length env nucleotidic sequences of the early/transmitted viral population infecting each subject were previously obtained by direct sequencing from bulk env PCR products (20). The deduced amino acid Env sequences from HP, IP, and CP were compared to try to detect residues that could explain the increasing infectious properties of viruses. However, using several analysis tools from the HIV sequence database, such as VESPA or AnalyzeAlign (http://www.hiv.lanl.gov/), no signature was identified, whether Env sequences were grouped according to the infectious properties of viruses or their year of isolation.
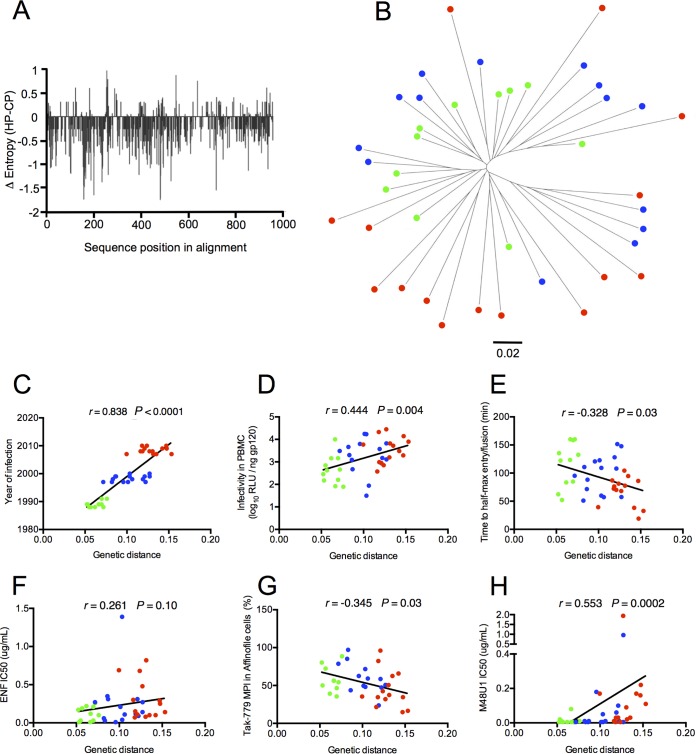
We previously reported that the mean genetic diversity among the env sequences increased gradually from HP to CP (mean genetic diversity of 8.8%, 12.4%, and 14.8% for HP, IP, and CP, respectively) (20). To determine if this genetic evolution impacts more specifically some amino acid residues, we used the Entropy tool from the HIV sequence database (http://www.hiv.lanl.gov/) to compare the Shannon entropy between HP and CP Env sequences (the two most distant groups of sequences). As shown in Fig. 6A, which represents the difference in entropy between the HP and CP Env sequences at each position in an alignment of these two sets of sequences, negative values were found at most positions all along the alignment, indicating a global diversification of CP sequences not restricted to specific areas. To complete this analysis, we measured the extent of evolution of env sequences from HP to CP by computing a maximum likelihood phylogenetic tree (Fig. 6B). The divergence represented as the genetic distance from the hypothesized most recent common ancestor (MRCA) increased gradually according to the sampling year, with historical sequences having the shortest distances and contemporaneous ones the longest distances from the root (Fig. 6B and C). Not surprisingly, this indicated that the env sequences of viruses from our study evolved over the 15-year time period. This evolution toward increasing diversification associated with the absence of a specific sequence signature suggested that the phenotypic changes of Env from CP resulted from multiple coevolutionary amino acid changes that are probably context and host dependent. To support this hypothesis, we explored whether phenotypic changes of Env pseudotypes were directly correlated to genetic distances from the MRCA. A statistically significant correlation was observed between genetic distances and most Env properties, including infectivity in PBMCs (r = 0.444, P = 0.004) (Fig. 6D), time to half-max entry (r = −0.328, P = 0.03) (Fig. 6E), TAK-779 MPIs in Affinofile cells (r = −0.345, P = 0.03) (Fig. 6G), and resistance to M48U1 (r = 0.553, P = 0.0002) (Fig. 6H). Although statistically not significant, a trend was also observed for the resistance to ENF (r = 0.261, P = 0.10) (Fig. 6F).
FIG 6.
Diversification of Env pseudotypes is associated with their increased infectious properties. (A) Plot showing the difference in Shannon entropy between Env sequences derived from historical patients (HP; n = 11) and contemporary patients (C; n = 14) at each position in an alignment of these two sets of sequences. (B) Midpoint-rooted maximum likelihood phylogenetic tree of full-length env sequences. The 40 full-length env sequences of the transmitted viruses included in our study were aligned. A maximum likelihood tree was constructed using MEGA7. Env sequences from historical patients (HP; n = 11), intermediate patients (IP; n = 15), and contemporary patients (CP; n = 14) are identified by different colors (HP in green, IP in blue, and CP in red). Branch lengths correspond to nucleotide substitutions per site, as indicated in the scale. Correlation between env genetic distances from the MRCA and the infection year of patients (C), the infectivity in PBMCs (D), the time to half-max entry (E), the resistance to ENF (F), TAK-779 MPIs in Affinofile cells (G), and the resistance to M48U1 (H).
DISCUSSION
Following primary infection with HIV-1, the immune system exerts selective pressure on the viral population. In particular, NAbs arise early in the course of infection, leading to the rapid emergence of escape mutants. These NAbs constitute a major force driving the evolution of the HIV-1 envelope glycoprotein within each infected individual (1–11). As a possible consequence of this individual evolution, we and others reported evidence that the envelope glycoprotein, especially the gp120 subunit, has evolved toward increasing resistance to NAbs at a population (species) level since the start of the epidemic (19–23). The aim of the present study was to analyze whether this antigenic evolution is associated with the evolution of other functional properties and has thereby affected the relationship of HIV-1 with its human host.
We evaluated the infectivity of a panel of Env-pseudotyped viruses derived from patients infected by subtype B viruses during three periods of the epidemic (1987 to 1991, 1996 to 2000, and 2006 to 2010). The viruses from patients infected during the most recent period were approximately 10-fold more infectious in cell culture than viruses from patients infected at the beginning of the epidemic. This increase in infectivity was associated with faster viral entry kinetics, with the contemporary viruses entering the target cells approximately twice as fast as historical viruses. The changes in cell entry efficacy over time were similar in both TZM-bl cells and CD4-enriched mononuclear cells. Entry kinetics were not dependent on the Env content, because the pseudotyped viruses of the three periods had similar levels of Env incorporation. The contemporary viruses were also twice as resistant as historical viruses to enfuvirtide, a fusion inhibitor. We were unable to find particular mutations in the HR1 domain of gp41 (30), which might explain the increased resistance to ENF. Therefore, we evaluated whether this evolution might reflect differences in the CD4/coreceptor usage efficiency and/or in the transmission of conformational signals between receptor-bound gp120 and gp41, triggering the refolding of gp41 into the fusion active state (31).
The CCR5 usage efficiency was assessed indirectly by measuring the sensitivity to CCR5 antagonists in Affinofile cells expressing high levels of CCR5. These cells were previously shown to be the most relevant model to detect partial resistance to CCR5 antagonists (33–36). Under these conditions, the contemporary viruses were more resistant than historical viruses to inhibition of infection by the CCR5 antagonists. This suggests that, when expressed at high levels, a fraction of CCR5 molecules assume a conformation that CCR5 antagonists are unable to bind but that can be more efficiently used by contemporary than historic viruses. Another explanation would be that contemporary viruses were able to use a drug-bound conformation of CCR5 more efficiently that historical viruses. This coreceptor phenotype already present on cells expressing lower levels of CCR5 would become detectable only using cells expressing high levels of CCR5. These data were supported by a progressive increase of infectivity from historical viruses to that from contemporary viruses in Affinofile cells expressing high levels of CCR5. Interestingly, this evolution at the population level parallels the evolution that occurs within HIV-1 infected individuals: transmitted/founder (T/F) Envs were previously shown to be much more sensitive to MVC for entry into cells expressing high levels of CCR5 than Envs from variants present later, at the stage of chronic infection (34, 35). These observations suggest that selective pressures led the virus to expand its CCR5 usage capacity, but to what extend this evolution contributed to the increase in its infectivity remains unanswered. Further characterization of the various conformations of CCR5 expressed on CD4+ CCR5+ T-cell subsets that are targeted by historical versus contemporary viruses may reveal differences in the ability of their Env to mediate virus entry.
Contrasting with results obtained for CCR5 binding, there was no difference in CD4 binding between contemporary and historical viruses as determined by an analysis of the sensitivity to two soluble CD4 molecules (sCD4 and sCD4-183). This contrasts with our previous finding of a significant decrease in sensitivity to all broadly neutralizing antibodies (bNAbs) targeting the CD4 binding site (CD4bs) over the course of the epidemic (20, 21). Taken together, these data suggest that viruses that have been selected under pressure of NAbs targeting the CD4bs over the course of the epidemic may be either those not affected in their capacity to interact with the CD4 receptor or those that acquired compensatory mutations that restored efficient use of CD4. In favor of the second hypothesis, Lynch et al. showed recently that the VRC01 escape variants of the HIV-1 strain infecting the patient from whom VRC01 was isolated were impaired in their replication capacity initially but rapidly acquired compensatory mutations that restored a full replicative fitness (43). Interestingly, in contrast to the stable sensitivity to sCD4, we observed that resistance of our viruses to the miniCD4 protein M48U1 progressively increased over the course of the epidemic. M48U1 contains flexible hydrophobic extensions that fit into the Phe-43 cavity. It interacts with gp120 residues that do not contact with CD4 (38). This suggests that the evolution of HIV-1, possibly due in part to the selective pressure exerted by CD4bs NAbs, has involved the Phe-43 cavity and has led to diminished sensitivity to M48U1.
During sexual transmission, dendritic cells of the mucosae capture HIV-1 by attachment receptors, in particular, DC-SIGN, to transfer the virus to permissive CD4+ T cells, resulting in trans infection (39–42). We found no significant differences in the efficiency of DC-SIGN capture between viruses of the three periods of our study. However, after binding to DC-SIGN, the efficiency of virus transfer to target cells increased progressively from the oldest to the most recent period. This implicates post-DC-SIGN binding events in the evolving capacity of the viruses to infect target cells.
We tried to explore the molecular mechanisms underlying the increasing infectious properties of the envelope glycoprotein over time, but we did not identify any signature pattern in sequences from contemporary patients that could explain this phenomenon. In contrast, we observed, as expected, that Env sequences diverged and diversified over time. This genetic evolution correlated with the significant evolution of the phenotypic properties that we noted, including infectivity, entry kinetics, and resistance to CCR5 antagonists and to M48U1, suggesting that multiple amino acid changes dependent on individual selective pressures are probably involved.
In conclusion, we report evidence of increased infectiousness of the envelope glycoprotein of HIV-1 during the course of the subtype B epidemic. Our data suggest that a greater capacity of variants to engage the CCR5 coreceptor and trigger conformational changes allowing the fusion process might have contributed to this evolution, although the details of the mechanisms involved remain to be identified.
It would be valuable to determine whether these observations hold true for other viral populations, in particular, for viruses of other clades, and are common to the entire HIV-1 species. The recent papers showing that subtype C and CRF02-AG viruses also evolved toward increasing resistance to neutralization suggests that this might be the case (22, 23). From an evolutionary point of view, it might be surprising that escape from bNAbs targeting conserved regions of Env likely involved in vital functions provides a fitness advantage. One explanation would be that viruses that have been selected under pressure of NAbs represent only a minority of escape variants that were not affected in their fitness or that acquired compensatory mutations restoring or increasing their fitness. In favor of this hypothesis, we previously showed that only a minority of bNAb-resistant viruses harbored mutations in epitope key residues, suggesting that other molecular determinants should be involved in resistance, maybe without affecting vital functions (20). In addition, our findings are consistent with analyses showing that HIV-1 virulence, measured by viral loads at set points and CD4 T-cell counts at the time of primary infection, appears to have increased over the past three decades (24–26). HIV evolutionary models based on known viral load transmission rates and viral load mortality relations indicate that HIV-1 evolved to maximize virulence and overall transmission potential by optimizing the trade-off between transmission and mortality (44–46). We demonstrate here temporal changes in the function of the envelope glycoprotein of HIV-1 that might have contributed to this population-level adaptation.
MATERIALS AND METHODS
Ethics statements.
National ethics committee approvals were obtained for the two ANRS cohorts(SEROCO: Commission Nationale de l'Informatique et des Libertés [CNIL]; PRIMO: Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale [CCPPRB] Paris-Cochin and Comité de Protection des Personnes [CPP] Ile de France III), and all patients gave written informed consent to participate in the cohort. PBMCs were separated from the buffy coat collected from HIV-1-negative blood donors of the regional blood transfusion center (Etablissement Français du Sang, Tours, France) according to the ethics rules described in the official agreement (CA-PLER-2015 162).
Study population.
The HIV-1 population that we studied was described previously (20). It was derived from 40 Caucasian men having sex with men with primary infection, infected by clade B viruses at three periods of the French epidemic: between 1987 and 1991 (historical patients [HP], enrolled in the ANRS SEROCO cohort), 1996 and 2000 (intermediate patients [IP]), and 2006 and 2010 (contemporary patients [CP]), these latter were enrolled in the ANRS PRIMO cohort. The estimated date of infection was defined as the date of symptom onset minus 15 days for patients with symptomatic primary infection, or, for asymptomatic patients, the date of the incomplete Western blot (presence of antibodies to gp160 and P24) minus 1 month or the midpoint between a negative and a positive ELISA result (47).
Production of pseudotyped viruses and analysis of their Env content.
Pseudotyped viruses expressing the Env variants representative of the viral quasispecies infecting each patient were generated from cells as previously described (20). Briefly, HIV-1 RNA was extracted from plasma using the QIAamp viral RNA minikit (Qiagen). Full-length (gp160) env genes were amplified by nested reverse transcriptase PCR (RT-PCR) using group M env-specific degenerated primers and cloned into the mammalian expression vector pCI. The resulting pCI-env plasmids representing the amplified virus populations were propagated by transformation of Electromax DH5α electrocompetent Escherichia coli (Invitrogen). Env-pseudotyped viruses were produced by cotransfecting 3 × 106 293T cells with 4 μg of each patient-derived pCI-env library and 8 μg of pNL4.3.LUC.R-E- (48) using FuGene-6 transfection reagent (Promega). Viruses contained in supernatants of 293T cell cultures were harvested 72 h later, purified by filtration (0.45-μm filter), and stored as aliquots at −80°C. For the analysis of the Env content, supernatants of 293T cell cultures were overlaid on a 20% sucrose cushion, and viral particles were pelleted at 87,000 × g for 1.5 h at 4°C. Viral pellets were solubilized overnight at 4°C in 100 μl of phosphate-buffered saline (PBS) supplemented with 1% Triton X-100 and an antiprotease cocktail. p24 antigen content was determined by ELISA (INNOTEST HIV Antigen MAb; Innogenetics). The Env ELISA was performed in Nunc Maxisorp plates (Dutscher) as previously described (49). A pool of serum samples from 60 chronically infected patients enrolled between 1987 and 2007 in the PRIMO or SEROCO cohorts (but different from patients selected in our study) was used for the detection of Env captured on D7324-coated (Aalto Bioreagents Ltd., Dublin, Ireland) microplates. Dilutions of purified gp120IIIB (Advanced Bioscience Laboratories) were used to construct a standard curve.
Viral infectivity in TZM-bl cells, peripheral blood mononuclear cells, and Affinofile cells.
Viral infectivity was determined in quadruplicates in TZM-bl cells and PBMCs. Samples of 100 μl of virus stock normalized to 100 ng of p24 were added to 100 μl of culture medium. Aliquots of 1 × 104 TZM-bl cells, 1 × 105 PBMCs (pool of PBMCs issued from 5 HIV-negative donors), or 4 × 105 Affinofile cells (ponasterone A treated or not, as described below) were added to viruses in the presence of 30 μg/ml DEAE-dextran. Infection levels were determined after 48 h by measuring the luciferase activity of cell lysates using the Bright-Glo luciferase assay (Promega) and a Centro LB 960 luminometer (Berthold Technologies) (50).
Enfuvirtide time-of-addition assay.
To assess the entry kinetics, 8 × 103 TZM-bl cells per well were plated the day prior to infection. Aliquots of 50 μl of pseudotyped viruses (400 50% tissue culture infective dose [TCID50]/ml) were chilled to 4°C and added to TZM-bl cells as previously described (29). Briefly, cells were spinoculated at 300 × g for 90 min at 4°C to enhance viral binding. Immediately postspinoculation, cold supernatant was aspirated off and all wells were flooded with 180 μl of prewarmed 37°C medium and transferred to a 37°C incubator. Enfuvirtide (ENF; NIBSC), 20 μl of 10 μg/ml (final concentration, 1 μg/ml), was then added at 0, 5, 10, 20, 40, 80, or 160 min postwarming. A no-drug control was also included to normalize percent infection. Cells were then incubated for 72 h, and luciferase activity was measured. Three wells per virus per time point were included in each experiment, and all Envs were examined in at least two independent experiments. Data were analyzed with Prism 6.0 (GraphPad Software, Inc.) by fitting a best-fit sigmoidal line to the results from each independent experiment and then averaging the Hill slopes and time to half-maximal entry/fusion.
Determination of coreceptor usage.
Coreceptor usage was determined using the U373 MAGI cells stably expressing CD4 and either CCR5 (CD4+/U373/CCR5) or CXCR4 (CD4+/U373/CXCR4). Aliquots of 1.5 × 104 cells were plated the day prior to infection. Cells were infected with 25 μl of a p24 normalized amount (25 ng) of pseudotyped viruses for 2 h at 37°C. Then, 175 μl of Dulbecco’s modified Eagle medium (DMEM) supplemented with 20 μg/ml of DEAE-dextran and 5% fetal bovine serum (FBS) was added; 48 h after infection, the luciferase activity was measured and the viral tropism was determined.
Inhibition of entry by enfuvirtide, CCR5 antagonists, and CD4 analogs.
TZM-bl cells were employed in duplicate experiments to assess the sensitivity of pseudotyped viruses to the fusion inhibitor ENF (NIBSC), CCR5 antagonists MVC and TAK-779, and CD4 inhibitors sCD4, sCD4-183, and M48U1 (L. Martin, CEA, Gif-sur-Yvette, France). Aliquots of 50 μl of pseudotyped virus stocks (400 TCID50/ml) were incubated for 1 h at 37°C with 50 μl of 3-fold serial dilutions of ENF, sCD4, sCD4-183, or M48U1 (10 μg/ml to 0.0046 μg/ml). The virus-inhibitor mixture was then used to infect 1 × 104 TZM-bl cells in the presence of 30 μg/ml DEAE-dextran. Infection levels were determined after 48 h by measuring the luciferase activities of cell lysates. IC50 values were calculated as the reciprocal of the inhibitor concentration reducing RLU values by 50%. Results are expressed as mean values from at least two independent experiments performed in triplicates.
For MVC and TAK-779 inhibition, 8 × 103 TZM-bl cells per well were prepared the day prior to infection. Cells were first treated for 1 h at 37°C with 150 μl of 3-fold serial dilutions of TAK-779 or MVC (6 μM to 0.3 nM) before adding 50 μl of pseudotyped viruses (400 TCID50/ml); 100 μl of DMEM supplemented with 30 μg/ml DEAE-dextran was then added to cells. Luciferase activity was measured 48 h after infection. CCR5 antagonist susceptibility is expressed as maximal percent inhibition (MPI) and IC50 values. Presented values are mean values from at least two independent experiments performed in triplicates.
To measure inhibition of entry at high concentrations of CCR5 at the cell surface, 4 × 105 Affinofile cells were seeded in triplicates into 96-well plates (36). Ponasterone A (ponA; Invitrogen) was added 24 h later at 8 μM to induce the expression of CCR5. The drug was removed 18 h later, and receptor densities were quantified by fluorescence-activated cell sorting (FACS) as described below. Affinofile cells were then treated for 1 h at 37°C with 150 μl of DMEM supplemented with 6 μM TAK-779 and 40 μg/ml DEAE-dextran before infecting the cells with 50 μl of pseudotyped viruses (400 TCID50/ml) using spinoculation, as described previously (34); 48 h later, the luciferase activity in cell lysates was measured.
CCR5 expression levels were checked by flow cytometry. Eighteen hours after induction with 8 μM ponA, Affinofile cells were processed for quantitative FACS (qFACS) analysis with a phycoerythrin-conjugated mouse anti-human CCR5 antibody (clone 2D7; BD Biosciences) as previously described (35). Receptor expression levels were quantified with a QuantiBRITE fluorescence quantitation system (BD Biosciences). Regression curves were generated with Prism 6.0 (GraphPad Software, Inc.), such that ponA concentrations could be converted to cell surface concentrations of CCR5 in units of ABS/cell.
Virus capture and transfer by Raji/DC-SIGN cells.
For the DC-SIGN capture experiment, 100 μl of Raji or Raji/DC-SIGN cells (2 × 105 cells/well) were seeded in a V-bottom 96-well tissue culture plate (Corning) by centrifugation at 300 × g for 10 min. Cells were resuspended in 100 μl containing pseudotyped virus equivalent to 50 ng of p24 and incubated at 37°C for 2 h. The cells were then washed four times with a wash buffer (ice-cold PBS, 2.5% fetal bovine serum, and 1 mM CaCl2), lysed, and assayed for p24 content by ELISA (INNOTEST HIV Antigen MAb; Innogenetics). For the DC-SIGN transfer experiment, washed cells were used to infect 100 μl of TZM-bl (104 cells). After 48 h, the TZM-bl cells were lysed and luciferase activity was measured.
Sequence analysis.
Full-length env sequences of the viral quasispecies infecting each patient were previously obtained by direct sequencing bulk env PCR products using the Sanger method according to the Dye Terminator cycle sequencing protocol (Applied Biosystems, Foster City, CA) (20).
Phylogenetic analysis of env sequences.
Nucleotide sequences were aligned using CLUSTALW and manually edited. A maximum likelihood tree was computed with MEGA7, using the GTRGAMMA model of nucleotide substitutions with bootstrap analysis to assess branch support (1,000 replicates).
Statistical analysis.
Differences between results for viruses from HP (1987 to 1991), IP (1996 to 2000), and CP (2006 to 2010) from the various assays were evaluated using a Jonckheere-Terpstra test for trend. The Jonckheere-Terpstra trend test is a nonparametric test for ordered differences among classes. It is used to test for differences between more than two populations when the samples are ordered. Correlations between continuous variables were examined with the Spearman’s correlation test.
Data availability.
All sequences have been submitted to GenBank and assigned accession numbers KC699001 to KC699040.
ACKNOWLEDGMENTS
This work was supported by the Agence Nationale de Recherche sur le SIDA et les hépatites (ANRS, Paris, France). Mélanie Bouvin-Pley was supported by doctoral fellowships from the Région Centre and Sidaction (France). Maxime Beretta was supported by doctoral fellowships from INSERM, the Région Centre, and Sidaction (France).
We thank the patients and clinicians who participated in the ANRS SEROCO CO2 and PRIMO CO6 cohorts. We thank B. Giraudeau (University of Tours, France) for his kind help with statistical analyses. We thank B. Lee and K. Chikere (University of California, Los Angeles) for the generous gift of Affinofile cells. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4.3.LUC.R-E- from N. Landau; TZM-bl cells from J. C. Kappes, X. Wu, and Tranzyme Inc.; Raji/DC-SIGN cells from L. Wu and V. N. KewalRamani; maraviroc (catalog no. 11580); TAK-779 from M. Baba; sCD4-183 from Pharmacia, Inc.; sCD4 from Progenics; U373-MAGI-CXCR4 and U373-MAGI-CCR5 cells from Michael Emerman.
REFERENCES
- 1.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A 100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 3.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alter G, Moody MA. 2010. The humoral response to HIV-1: new insights, renewed focus. J Infect Dis 202:S315–S322. doi: 10.1086/655654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mouquet H. 2014. Antibody B cell responses in HIV-1 infection. Trends Immunol 35:549–561. doi: 10.1016/j.it.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Burton DR, Mascola JR. 2015. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A 102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagar M, Wu X, Lee S, Overbaugh J. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol 80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahalanabis M, Jayaraman P, Miura T, Pereyra F, Chester EM, Richardson B, Walker B, Haigwood NL. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J Virol 83:662–672. doi: 10.1128/JVI.01328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 Infection. J Virol 82:7932–7941. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, Allen SA, Pinter A, Shaw GM, Hunter E, Robinson JE, Gnanakaran S, Derdeyn CA. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog 5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 13.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C, CAPRISA Acute Infection Study Team, Center for HIV-AIDS Vaccine Immunology Consortium . 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, Hahn BH, Allen SA, Derdeyn CA, Hunter E. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog 5:e10000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer MJ, Gray RH, Serwadda D, Sewankambo NK, Shepherd JC, Toma J, Huang W, Quinn TC. 2009. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis 199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redd AD, Collinson-Streng AN, Chatziandreou N, Mullis CE, Laeyendecker O, Martens C, Ricklefs S, Kiwanuka N, Nyein PH, Lutalo T, Grabowski MK, Kong X, Manucci J, Sewankambo N, Wawer MJ, Gray RH, Porcella SF, Fauci AS, Sagar M, Serwadda D, Quinn TC. 2012. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J Infect Dis 206:1433–1442. doi: 10.1093/infdis/jis503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peeters M, Jung M, Ayouba A. 2013. The origin and molecular epidemiology of HIV. Expert Rev Anti Infect Ther 11:885–896. doi: 10.1586/14787210.2013.825443. [DOI] [PubMed] [Google Scholar]
- 19.Bunnik EM, Euler Z, Welkers MR, Boeser-Nunnink BD, Grijsen ML, Prins JM, Schuitemaker H. 2010. Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat Med 16:995–997. doi: 10.1038/nm.2203. [DOI] [PubMed] [Google Scholar]
- 20.Bouvin-Pley M, Morgand M, Moreau A, Jestin P, Simonnet C, Tran L, Goujard C, Meyer L, Barin F, Braibant M. 2013. Evidence for a continuous drift of the HIV-1 species towards higher resistance to neutralizing antibodies over the course of the epidemic. PLoS Pathog 9:e1003477. doi: 10.1371/journal.ppat.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouvin-Pley M, Morgand M, Meyer L, Goujard C, Moreau A, Mouquet H, Nussenzweig M, Pace C, Ho D, Bjorkman PJ, Baty D, Chames P, Pancera M, Kwong PD, Poignard P, Barin F, Braibant M. 2014. Drift of the HIV-1 envelope glycoprotein gp120 toward increased neutralization resistance over the course of the epidemic: a comprehensive study using the most potent and broadly neutralizing monoclonal antibodies. J Virol 88:13910–13917. doi: 10.1128/JVI.02083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rademeyer C, Korber B, Seaman MS, Giorgi EE, Thebus R, Robles A, Sheward DJ, Wagh K, Garrity J, Carey BR, Gao H, Greene KM, Tang H, Bandawe GP, Marais JC, Diphoko TE, Hraber P, Tumba N, Moore PL, Gray GE, Kublin J, McElrath MJ, Vermeulen M, Middelkoop K, Bekker LG, Hoelscher M, Maboko L, Makhema J, Robb ML, Abdool Karim S, Abdool Karim Q, Kim JH, Hahn BH, Gao F, Swanstrom R, Morris L, Montefiori DC, Williamson C. 2016. Features of recently transmitted HIV-1 clade C viruses that impact antibody recognition: implications for active and passive immunizations. PLoS Pathog 12:e1005742. doi: 10.1371/journal.ppat.1005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefic K, Bouvin-Pley M, Essat A, Visdeloup C, Moreau A, Goujard C, Chaix ML, Braibant M, Meyer L, Barin F, ANRS PRIMO Cohort Study Group . 7 November 2018. Sensitivity to broadly neutralizing antibodies of recently transmitted HIV-1 clade CRF02_AG viruses with a focus on evolution over time. J Virol. doi: 10.1128/JVI.01492-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbeck JT, Müller V, Maust BS, Ledergerber B, Torti C, Di Giambenedetto S, Gras L, Günthard HF, Jacobson LP, Mullins JI, Gottlieb GS. 2012. Is the virulence of HIV changing? A meta-analysis of trends in prognostic markers of HIV disease progression and transmission. AIDS 26:193–205. doi: 10.1097/QAD.0b013e32834db418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantazis N, Porter K, Costagliola D, De Luca A, Ghosn J, Guiguet M, Johnson AM, Kelleher AD, Morrison C, Thiebaut R, Wittkop L, Touloumi G, CASCADE Collaboration in EuroCoord . 2014. Temporal trends in prognostic markers of HIV-1 virulence and transmissibility: an observational cohort study. Lancet HIV 1:e119. doi: 10.1016/S2352-3018(14)00002-2. [DOI] [PubMed] [Google Scholar]
- 26.Brey F, Seybold U, Kollan C, Bogner J, ClinSurv-HIV Study Group . 2016. Accelerated CD4+ cell count decline in untreated HIV-1 patients points toward increasing virulence over the course of the epidemic. AIDS 30:1995–1997. doi: 10.1097/QAD.0000000000001165. [DOI] [PubMed] [Google Scholar]
- 27.Desquilbet L, Goujard C, Rouzioux C, Sinet M, Deveau C, Chaix ML, Séréni D, Boufassa F, Delfraissy JF, Meyer L, PRIMO and SEROCO Study Groups . 2004. Does transient HAART during primary HIV-1 infection lower the virological set-point? AIDS 18:2361–2369. [PubMed] [Google Scholar]
- 28.Laanani M, Ghosn J, Essat A, Melard A, Seng R, Gousset M, Panjo H, Mortier E, Girard PM, Goujard C, Meyer L, Rouzioux C, Agence Nationale de Recherche sur le Sida PRIMO Cohort Study Group . 2015. Impact of the timing of initiation of antiretroviral therapy during primary HIV-1 infection on the decay of cell-associated HIV-DNA. Clin Infect Dis 60:1715–1721. doi: 10.1093/cid/civ171. [DOI] [PubMed] [Google Scholar]
- 29.Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, Haim H, Petersen JE, Wojcechowskyj JA, Sodroski J, Haynes BF, Montefiori DC, Tilton JC, Shaw GM, Hahn BH, Doms RW. 2011. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol 85:8514–8527. doi: 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wensing AM, Calvez V, Günthard HF, Johnson VA, Paredes R, Pillay D, Shafer RW, Richman DD. 2014. 2014 update of the drug resistance mutations in HIV-1. Top Antivir Med 22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 31.Miyauchi K, Kozlov MM, Melikyan GB. 2009. Early steps of HIV-1 fusion define the sensitivity to inhibitory peptides that block 6-helix bundle formation. PLoS Pathog 5:e1000585. doi: 10.1371/journal.ppat.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 33.Roche M, Jakobsen MR, Ellett A, Salimiseyedabad H, Jubb B, Westby M, Lee B, Lewin SR, Churchill MJ, Gorry PR. 2011. HIV-1 predisposed to acquiring resistance to maraviroc (MVC) and other CCR5 antagonists in vitro has an inherent, low-level ability to utilize MVC-bound CCR5 for entry. Retrovirology 8:89. doi: 10.1186/1742-4690-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker ZF, Iyer SS, Wilen CB, Parrish NF, Chikere KC, Lee FH, Didigu CA, Berro R, Klasse PJ, Lee B, Moore JP, Shaw GM, Hahn BH, Doms RW. 2013. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol 87:2401–2411. doi: 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ping LH, Joseph SB, Anderson JA, Abrahams MR, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, Keys J, Potter EL, Chu H, Moore P, Salazar MG, Iyer S, Jabara C, Kirchherr J, Mapanje C, Ngandu N, Seoighe C, Hoffman I, Gao F, Tang Y, Labranche C, Lee B, Saville A, Vermeulen M, Fiscus S, Morris L, Karim SA, Haynes BF, Shaw GM, Korber BT, Hahn BH, Cohen MS, Montefiori D, Williamson C, Swanstrom R, CAPRISA Acute Infection Study and the Center for HIV-AIDS Vaccine Immunology Consortium . 2013. Comparison of viral Env proteins from acute and chronic infections of subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol 87:7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnston SH, Lobritz MA, Nguyen S, Lassen K, Delair S, Posta F, Bryson YJ, Arts EJ, Chou T, Lee B. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J Virol 83:11016–11026. doi: 10.1128/JVI.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin L, Stricher F, Missé D, Sironi F, Pugnière M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, Ménez A, Lusso P, Veas F, Vita C. 2003. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol 21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- 38.Acharya P, Luongo TS, Louder MK, McKee K, Yang Y, Kwon YD, Mascola JR, Kessler P, Martin L, Kwong PD. 2013. Structural basis for highly effective HIV-1 neutralization by CD4-mimetic miniproteins revealed by 1.5 Å cocrystal structure gp120 and M48U1. Structure 21:1018–1029. doi: 10.1016/j.str.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, Figdor CG, van Kooyk Y. 2000. DC-SIGN, a dendritic cell-specific HIV-1 binding protein that enhances trans-infection of T cells. Cell 100:587–597. doi: 10.1016/S0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 40.Jameson B, Baribaud F, Pöhlmann S, Ghavimi D, Mortari F, Doms RW, Iwasaki A. 2002. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J Virol 76:1866–1875. doi: 10.1128/JVI.76.4.1866-1875.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turville SG, Cameron PU, Handley A, Lin G, Pöhlmann S, Doms RW, Cunningham AL. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat Immunol 3:975–983. doi: 10.1038/ni841. [DOI] [PubMed] [Google Scholar]
- 42.van Kooyk Y, Geijtenbeek TB. 2003. DC-SIGN: escape mechanism for pathogens. Nat Rev Immunol 3:697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 43.Lynch RM, Wong P, Tran L, O'Dell S, Nason MC, Li Y, Wu X, Mascola JR. 2015. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J Virol 89:4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirreff G, Pellis L, Laeyendecker O, Fraser C. 2011. Transmission selects for HIV-1 strains of intermediate virulence: a modeling approach. PLoS Comput Biol 7:e1002185. doi: 10.1371/journal.pcbi.1002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraser C, Lythgoe K, Leventhal GE, Shirreff G, Hollingsworth TD, Alizon S, Bonhoeffer S. 2014. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343:1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herbeck JT, Mittler JE, Gottlieb GS, Mullins JI. 2014. An HIV epidemic model based on viral load dynamics: value in assessing empirical trends in HIV virulence and community viral load. PLoS Comput Biol 10:e1003673. doi: 10.1371/journal.pcbi.1003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goujard C, Chaix ML, Lambotte O, Deveau C, Sinet M, Guergnon J, Courgnaud V, Rouzioux C, Delfraissy JF, Venet A, Meyer L. 2009. Spontaneous control of viral replication during primary HIV infection: when is “HIV controller” status established? Clin Infect Dis 49:982–986. doi: 10.1086/605504. [DOI] [PubMed] [Google Scholar]
- 48.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 49.Lambelé M, Labrosse B, Roch E, Moreau A, Verrier B, Barin F, Roingeard P, Mammano F, Brand D. 2006. Impact of natural polymorphism within the gp41 cytoplasmic tail of human immunodeficiency virus type 1 on the intracellular distribution of envelope glycoproteins and viral assembly. J Virol 81:125–140. doi: 10.1128/JVI.01659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences have been submitted to GenBank and assigned accession numbers KC699001 to KC699040.