Capsidless mitoviruses, which are ubiquitously detected in filamentous fungi, have the simplest RNA genomes of 2.2 to 4.4 kb, encoding only RNA-dependent RNA polymerase. Despite their simple genomes, detailed biological characterization of mitoviruses has been hampered by their mitochondrial location within the cell, posing challenges to their experimental introduction and study. Here we developed a protoplast fusion-based protocol for horizontal transfer of the prototype mitovirus, Cryphonectria parasitica mitovirus 1 (CpMV1), which was isolated from strain NB631 of the chestnut blight fungus (Cryphonectria parasitica), a model filamentous fungus for studying virus-host interactions. The host range of CpMV1 has been expanded to many different strains of C. parasitica and different fungal species within and outside the Cryphonectriaceae. Comparison of CpMV1 accumulation among various RNA silencing-deficient and -competent strains showed clearly that the virus was unaffected by RNA silencing. This study provides a solid foundation for further exploration of mitovirus-host interactions.
KEYWORDS: Cryphonectria parasitica, RNA silencing, chestnut blight, mitochondria, mitovirus, mycovirus
ABSTRACT
Mitoviruses (genus Mitovirus, family Narnaviridae) are mitochondrially replicating viruses that have the simplest positive-sense RNA genomes of 2.2 to 4.4 kb with a single open reading frame (ORF) encoding an RNA-dependent RNA polymerase. Cryphonectria parasitica mitovirus 1 (CpMV1) from U.S. strain NB631 of the chestnut blight fungus, Cryphonectria parasitica, was the first virus identified as a mitochondrially replicating virus. Despite subsequent discovery of many other mitoviruses from diverse fungi, no great advances in understanding mitovirus biology have emerged, partly because of the lack of inoculation methods. Here we developed a protoplast fusion-based protocol for horizontal transmission of CpMV1 that entailed fusion of recipient and donor protoplasts, hyphal anastomosis, and single-conidium isolation. This method allowed expansion of the host range to many other C. parasitica strains. Species within and outside the family Cryphonectriaceae, Cryphonectria radicalis and Valsa ceratosperma, also supported the replication of CpMV1 at a level comparable to that in the natural host. No stable maintenance of CpMV1 was observed in Helminthosporium victoriae. PCR-based haplotyping of virus-infected fungal strains confirmed the recipient mitochondrial genetic background. Phenotypic comparison between CpMV1-free and -infected isogenic strains revealed no overt effects of the virus. Taking advantage of the infectivity to the standard strain C. parasitica EP155, accumulation levels were compared among antiviral RNA silencing-proficient and -deficient strains in the EP155 background. Comparable accumulation levels were observed among these strains, suggesting the avoidance of antiviral RNA silencing by CpMV1, which is consistent with its mitochondrial replication. Collectively, the results of study provide a foundation to further explore the biology of mitoviruses.
IMPORTANCE Capsidless mitoviruses, which are ubiquitously detected in filamentous fungi, have the simplest RNA genomes of 2.2 to 4.4 kb, encoding only RNA-dependent RNA polymerase. Despite their simple genomes, detailed biological characterization of mitoviruses has been hampered by their mitochondrial location within the cell, posing challenges to their experimental introduction and study. Here we developed a protoplast fusion-based protocol for horizontal transfer of the prototype mitovirus, Cryphonectria parasitica mitovirus 1 (CpMV1), which was isolated from strain NB631 of the chestnut blight fungus (Cryphonectria parasitica), a model filamentous fungus for studying virus-host interactions. The host range of CpMV1 has been expanded to many different strains of C. parasitica and different fungal species within and outside the Cryphonectriaceae. Comparison of CpMV1 accumulation among various RNA silencing-deficient and -competent strains showed clearly that the virus was unaffected by RNA silencing. This study provides a solid foundation for further exploration of mitovirus-host interactions.
INTRODUCTION
Mitoviruses (genus Mitovirus, family Narnaviridae) comprise a unique group of RNA viruses with small and simple positive-sense RNA genomes that are 2.2 to 4.4 kb with a single open reading frame (ORF) encoding a viral RNA-dependent RNA polymerase (RdRp) (1). Mitoviruses are the only viruses that can infect eukaryotic mitochondria, and they are characterized by their capsidless nature and possible completion of their replication cycle within host mitochondria. Most utilize the codon UGA to code for tryptophan rather than using it as a stop signal, consistent with mitochondrial codon usage in fungi. Mitoviruses are omnipresent fungal viruses (see, e.g., references 2 to 6), but they were detected in plants only recently as likely infectious entities (7). Mitovirus genome sequences are also detected in host mitochondrial and nuclear genomes in plants and fungi (6, 8–11), like other nonretrovirus RNA viruses in plants and animals (12–14). It is noteworthy that many mitovirus strains belonging to over 20 species are isolated even from one necrotrophic ascomycete fungal species, Sclerotinia sclerotiorum (15). Different mitovirus strains often coinfect single fungal strains, as exemplified by those infecting Ophiostoma novo-ulmi (Dutch elm disease fungus) or S. sclerotiorum (5, 16, 17). Phylogenetically related narnaviruses (genus Narnavirus, family Narnaviridae), ourmiaviruses (genus Ourmiavirus), and leviviruses (family Leviviridae) have been shown to replicate in the cytoplasm of yeast, plant, and bacterial cells, respectively (1, 18). A new group of ourmia-like viruses also have been reported from filamentous fungi (see, e.g., reference 19), but little information about them is available.
Despite an increasing number of mitoviruses detected, only a few mitoviruses have been biologically investigated. For example, some mitoviruses, such as Botrytis mitovirus 1 (BcMV1) (formerly Botrytis cinerea debilitation-related virus) and Sclerotinia sclerotiorum mitovirus 1, cause severe symptoms, including hypovirulence in the host fungi (9, 20, 21), while asymptomatic infections were suggested for others (3). BcMV1 induces malformation of infected mitochondria and reduces virulence of the host fungus (21). There are still many unanswered fundamental questions in general about their replication cycles, including (i) how broadly a specific mitovirus can infect and (ii) the kinds of antimitovirus host defense and counterdefense that possibly occur in mitovirus-infected cells. Initial evidence to answer the first question came when a mitovirus from the fungus Sclerotinia homeocarpa (dollar-spot fungus) was found to be conspecific with a previously characterized element, Ophiostoma novo-ulmi mitovirus 3a-Ld, from the distantly related ascomycete O. novo-ulmi (22). Similarly, the mitovirus Ophiostoma novo-ulmi mitovirus 3b was identified in the two taxonomically different fungi O. novo-ulmi and Botrytis cinerea (gray mold fungus) (21). Thus, it was clear not only that such mitochondrial elements could in some cases enter, replicate, and be maintained in heterologous hosts but also that they did so with relatively little change to their genomes. Another interesting insight into mitovirus host range was provided through a bioinformatic analysis. Nibert (23) recently showed a direct relationship between the frequency of UGA codon utilization by mitoviruses and their host mitochondria. That study implied a possibility that an absence or scarcity of mitochondrial tRNA decoding UAG for Trp restricts infection of mitovirus with internal UGA codons.
Addressing the second question, of host defense, many fungal viruses replicating in the cytosol are known to be targeted by RNA silencing, as in other eukaryotic viruses (24–27). In this regard, mitovirus-derived small RNAs were shown to be produced for a few mitoviruses, including Heterobasidion mitovirus 1 and BcMV1 (28–30). However, the important question of whether mitochondrial mitoviruses are interfered with by cytoplasmic RNA silencing has yet to be addressed. One of the constraints hampering the advancement of mitovirus research is the lack of artificial introduction methods for mitochondrial viruses, i.e., inoculation methods. This contrasts with the fact that virion or RNA transfection methods or infectious cDNA clones have increasingly been established for fungal RNA viruses (26, 27, 31–36).
Cryphonectria parasitica mitovirus 1 (CpMV1) (type member of the genus Mitovirus) was the first mitovirus characterized; it was isolated from strain NB631 of the chestnut blight fungus, Cryphonectria parasitica (order Diaporthales) (2). C. parasitica is important not only as a plant pathogen causing one of the three most destructive tree diseases in the world but also as a virus host having biological tractability (37). CpMV1 was transmitted vertically to ascospores at a rate of ∼50% in genetic crosses between NB631 and another C. parasitica strain, EP155, when infected strain NB631 was used as a female parent in the crosses (38). This is consistent with maternal inheritance of infected mitochondria but also suggests that not all mitochondria in a thallus are infected. In the current study, we developed a protocol for protoplast-based introduction of the capsidless mitovirus CpMV1 and extended its host spectrum to different fungal strains of C. parasitica and to different species within and outside the Cryphonectriaceae. The horizontal transfer of CpMV1 to the C. parasitica standard strain EP155, for which many mutant strains with knockouts of genes involved in the posttranscriptional gene silencing (PTGS) pathway are available, allowed us to suggest the avoidance of antiviral RNA silencing by CpMV1.
RESULTS
Development of a protoplast fusion-based method for CpMV1 lateral transmission.
Because of the capsidless nature of CpMV1, potential cell-free introduction methods are limited to nonvirion transfection methods. Given the difficulty developing infectious cDNA or synthetic transcripts for mitoviruses (17), we tested a protoplast fusion protocol for CpMV1 that was reported previously for another capsidless virus, Cryphonectria hypovirus 1 (CHV1) (family Hypoviridae) (39). The protocol entailed labeling of recipient fungal strains with a hygromycin B resistance (Hygr) gene, protoplast fusion, selection of the recipient genetic backgrounds, and repeated pairing with recipient strains followed by single conidial isolation (Fig. 1). We first examined this method for the C. parasitica standard strain, EP155, which is vegetatively incompatible with NB631, the original host of CpMV1. EP155 fungal cultures were tested for the presence of CpMV1 at each step of the procedure shown in Fig. 1. Five fungal cultures obtained after protoplast fusion step 3 carried the virus when tested by reverse transcription-PCR (RT-PCR) (Fig. 2A). Two of the CpMV1-positive cultures were monitored further, and the virus was confirmed to be retained after repeated (4 rounds) anastomosis and single conidial isolation (step 4). Five single-spore isolates derived from the two cultures were subjected to double-stranded RNA (dsRNA) isolation (step 5), and 100% of them tested CpMV1 positive and carried the Hygr gene (Fig. 2B and data not shown). These results showed that the protocol is useful for lateral transfer of CpMV1 between vegetatively incompatible strains.
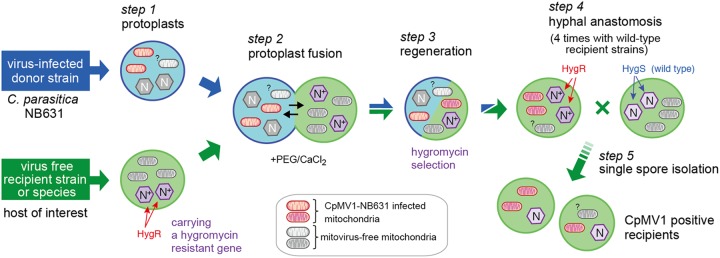
FIG 1.
Cartoon showing the experimental protocol used for the transfer of Cryphonectria parasitica mitovirus 1 (CpMV1) from NB631 to different fungal strains through protoplast fusion. Protoplasts were prepared from the naturally CpMV1-infected strain NB631 (donor) and a hygromycin B-resistant (Hygr) recipient strain (Table 1) (step 1) and fused with the aid of PEG (step 2). The resultant fusants were grown on PDA-hygromycin B plates (step 3) and anastomosed with virus-free, hygromycin B-susceptible (Hygs) recipient strains (step 4). After 1 week, mycelial plugs from the recipient were anastomosed again with virus-free recipient strains. This paring was repeated 4 times. The final step was single conidial isolation (step 5). Detailed procedures are described in Materials and Methods.
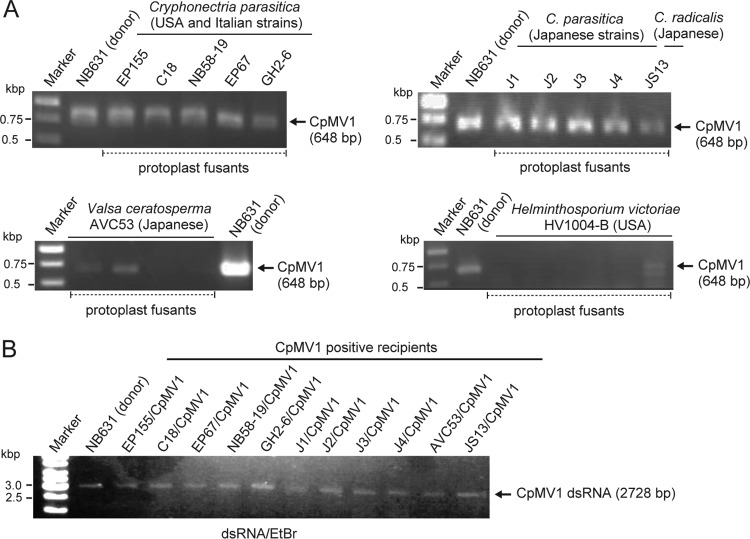
FIG 2.
RT-PCR detection of Cryphonectria parasitica mitovirus 1 (CpMV1) in recipients. (A) Fungal isolates were obtained from different batches of protoplast fusion (step 3 in Fig. 1) between the donor strain NB631 and recipients shown above the gels (Table 1). RT-PCR was designed to amplify a genomic region from map position 1290 to 1937 (see Materials and Methods for the primer sequences). (B) Fungal strains obtained from step 5 (Fig. 1) were subjected to dsRNA isolation and agarose gel electrophoresis. The 1-kb ladder DNA (Gene Ruler; Thermo Fisher Scientific, Waltham, MA) was used as a size marker in this and subsequent figures.
This technique was used for 9 other strains of C. parasitica and one strain each of Cryphonectria radicalis, Valsa ceratosperma (the fungus of apple Valsa canker, strain AVC53), and Helminthosporium victoriae (Victoria blight fungus, strain HV1004-B) (Table 1), all of which belong to the division Ascomycota. All tested strains except H. victoriae were shown to stably support CpMV1 replication. CpMV1 was detectable at each step of the procedure shown in Fig. 1 for most fungi tested (RT-PCR results at step 3 are shown in Fig. 2A). The presence of CpMV1 for recipient strains (step 5) was confirmed by RT-PCR and dsRNA agarose gel analysis (Fig. 2B and data not shown). However, after protoplast fusion with H. victoriae strain HV1000-B, CpMV1 was detected only until step 3 by RT-PCR and was undetectable at later steps. It is also noteworthy that CpMV1-infected V. ceratosperma strain AVC53 occasionally lost the virus after prolonged storage (4 to 6 weeks) (data not shown).
TABLE 1.
Fungal strains tested in this study for the ability to support CpMV1 replication
| Family | Species | Strain | Collection site or source | Strain no. or accession no. | Susceptibility to CpMV1 | vic genotyped | Reference(s) |
|---|---|---|---|---|---|---|---|
| Cryphonectriaceae | Cryphonectria parasitica | NB631 | New Jersey, USA | Yes | 1112-11 | 2 | |
| EP155 | Connecticut, USA | ATCC 38755 | Yes | 2211-22 | Standard strain | ||
| Δdcl1 mutant | EP155 derivative | NAc | Yes | NTe | 25 | ||
| Δdcl2 mutant | EP155 derivative | NA | Yes | NT | 25 | ||
| Δagl2 mutant | EP155 derivative | NA | Yes | NT | 24 | ||
| Δrdr1 mutant | EP155 derivative | NA | Yes | NT | 58 | ||
| EP67 | Italy | ATCC 38753 | Yes | 2212-22 | 62, 63 | ||
| GH2-6a | Michigan, USA | AF188515 | Yes | 1211-11 | 64 | ||
| NB58-19a | New Jersey, USA | ATCC 76221 | Yes | 2112-11 | 65 | ||
| C18-VFa | West Virginia, USA | AB073281 | Yes | 2111-11 | 66 | ||
| 1-1 (J1) | Ibaraki, Japan | MAFF 410878 | Yes | 1211-21 | 67 | ||
| J2 | Kanagawa, Japan | MAFF 305108 | Yes | 2222-21 | NA | ||
| EP48 (J3) | Hokkaido, Japan | MAFF 410557 | Yes | 2211-22 | NA | ||
| A1 (J4) | Ibaraki, Japan | MAFF 410726 | Yes | 1221-11 | NA | ||
| Cryphonectria radicalis | JS13-VFa | Kyoto, Japan | MYA-4104 | Yes | 52 | ||
| Valsaceae | Valsa ceratosperma | AVC53 | Aomori, Japan | NA | Yes | 32, 68 | |
| Massarinaceae | Helminthosporium victoriae | Hv1004-Hygrb | USA | ATCC 42020 | No | 69 |
Virus-free strains were obtained by single-conidium isolation from the originally virus-infected field isolates.
Transformant of strain B2 with an empty vector, pCB1004, carrying a Hygr gene.
NA, not available.
vic genotypes at 6 diallelic loci were determined as described by Short et al. (70), and their nomenclature is according to Choi et al. (71).
NT, not tested.
PCR-based mitochondrial haplotyping.
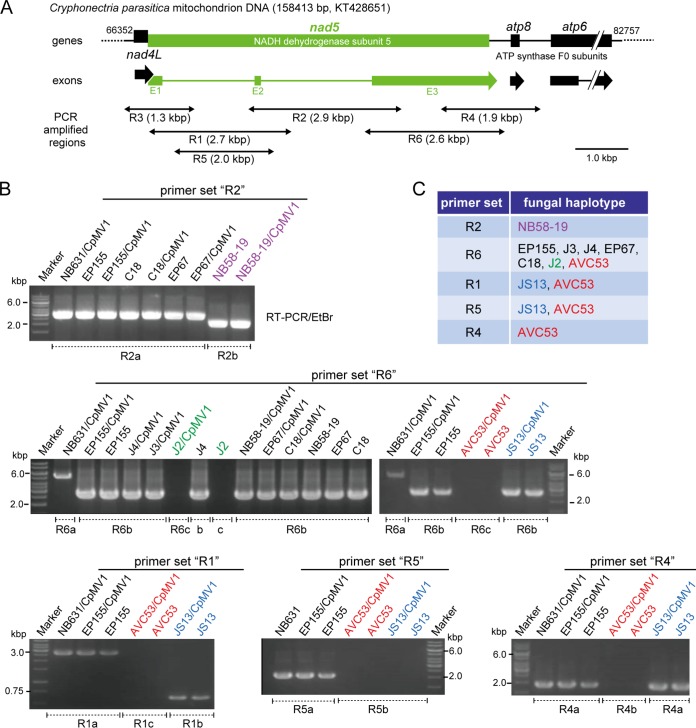
Mitochondria are believed to be prone to fusion and rearrangements, and this has been suggested for C. parasitica (38). During the process of protoplast fusion, heterokaryon formation and hybrid formation are likely to occur, and there are also chances of mixing the mitochondrial materials. Therefore, it is important to determine the mitochondrial genetic background in the CpMV1-positive recipients obtained after protoplast fusion. PCR-based mitochondrial haplotyping (mitohaplotyping) was carried out, in which a polymorphic NADH dehydrogenase subunit 5 gene (nad5) region (Fig. 3A) was targeted. Mitochondrial DNAs (mtDNAs) extracted from protoplast fusion isolates were PCR amplified using primer sets (R1 to R6) specific for the mitochondrial gene. A combination of primer pairs could distinguish five haplotypes within the order Diaporthales (Fig. 3B). Representative gel patterns are shown in Fig. 3B. For example, C. radicalis strain JS13 was differentiated from other fungal strains by primer set R1, while set R2 could distinguish NB58-19 from all other C. parasitica strains. All primer sets failed to amplify DNA fragments on mtDNA of V. ceratosperma. Note that a primer set for the mitochondrial small-subunit ribosomal gene could amplify a fragment of the expected size in all tested fungal strains (data not shown). Primer set R6 could differentiate the banding pattern of C. parasitica NB631 from those of other fungal strains (Fig. 3B). Importantly, all CpMV1-positive recipient strains possessed the same haplotypes as their original recipient strain used for protoplast fusion. These results indicated no sign of recombination within the tested mitochondrial gene region or mixed populations of mitochondria.
FIG 3.
PCR-based mitochondrial haplotyping of different fungal strains. (A) Physical map of a polymorphic region of the C. parasitica mitochondrial genome containing four genes (nad4L, nad5, atp8, and atp6). The specific primer sets R1 to R6 were adopted from reference 60. (B) Agarose gel profiles of PCR products with specific primer pairs. mtDNA was prepared from each of the CpMV1 recipient fungal strains obtained from step 5 (Fig. 1) and the original virus-free recipient strains and used as templates in PCR. Names of fungal strains are shown above the gel lanes. PCR fragments were amplified from all fungal isolates when a primer set (forward, 5′-GTGCCAGCAGTCGCGGYAANAC-3′; reverse, 5′-GGTGRARTGCTTNCACTTTCATTTATA-3′) for the mitochondrial small-subunit ribosomal gene was used (data not shown). (C) Primer sets used to distinguish mitochondrial haplotypes of CpMV1 recipients.
CpMV1 accumulation in newly established fungal hosts and their phenotypes.
A total of 12 fungal strains, including those belonging to different fungal species, were confirmed to maintain CpMV1. These newly established fungal strains were tested for virus accumulation level by Northern blot analysis. Virus accumulation levels of American, Italian, and Japanese C. parasitica strains were similar and were comparable to that observed in the original host of CpMV1, NB631, or the standard strain EP155. However, the CpMV1 accumulation level in V. ceratosperma (AVC53) appeared to be approximately 4-fold lower than that in C. radicalis (JS13) and the EP155 strain (Fig. 4A).
FIG 4.
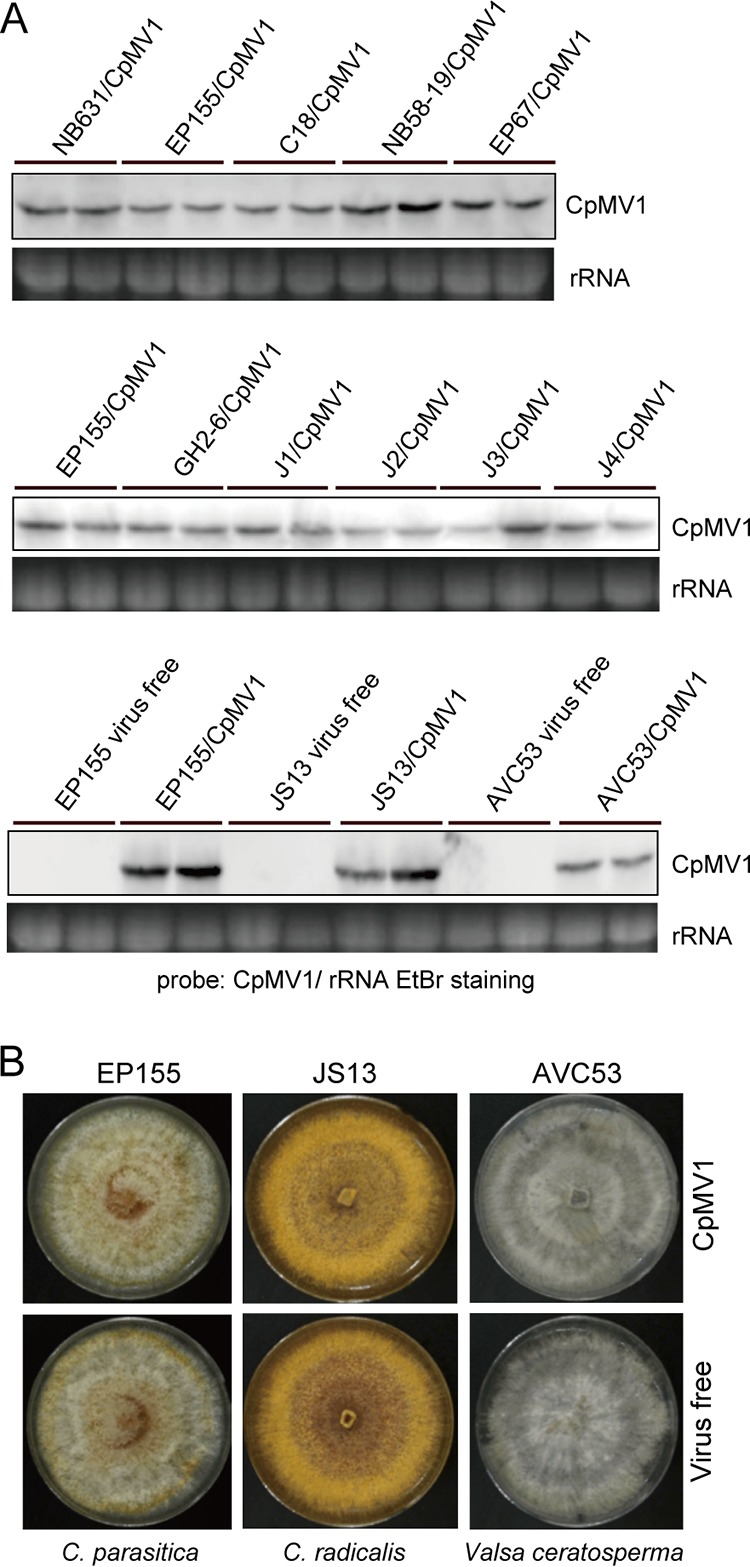
Northern blot and phenotypic analyses of Cryphonectria parasitica mitovirus 1 (CpMV1) in different fungal hosts. (A) Northern blotting of CpMV1. Total RNA was prepared from CpMV1-infected fungal strains obtained from step 5 (Fig. 1) and probed with a DIG-labeled dsDNA fragment that corresponds to the CpMV1 genomic region from position 1290 to 1937. Ethidium bromide (EtBr)-stained 28S rRNA (rRNA) served as a loading control for this and subsequent figures. (B) Colony morphologies of isogenic CpMV1-infected and -free fungal strains. Only representative fungal pairs from three different fungal species are shown.
To test possible effects of CpMV1 infection on the host phenotype, we compared the colony morphologies of isogenic pairs of CpMV1-infected and CpMV1-free fungal strains. Pictures of colonies of representative strains of each of three fungal species (C. parasitica EP155, C. radicalis JS13, and V. ceratosperma AVC53) are shown in Fig. 4B. No obvious difference in colony morphology was observed between CpMV1-infected and uninfected fungal strains (Fig. 4B).
No enhanced accumulation of CpMV1 in fungal mutants lacking RNA silencing-related genes.
RNA silencing is a primary antiviral defense mechanism in fungi (24, 25, 40) and is expected to operate in the cytosol of infected cells. Since CpMV1 is a mitochondrial virus, it is important to know whether CpMV1 is targeted by host RNA silencing. To investigate this possibility, CpMV1 was transferred to mutants of EP155 with deletions in the RNA silencing-associated genes (Δdcl2, Δdcl1, Δagl2, and Δrdr1) by hyphal fusion with CpMV1-infected EP155 as a donor (Fig. 5). Hyphal fusion with each of the RNA silencing mutants was repeated 4 times to ensure homogeneity of the nuclear genetic background (26). Comparative analyses showed no difference in CpMV1 genomic RNA accumulation between RNA silencing-competent (EP155) and mutant strains (Fig. 5A). This result suggested that CpMV1 is not affected by the RNA silencing mechanism, suggesting that CpMV1 may avoid host RNA silencing.
FIG 5.
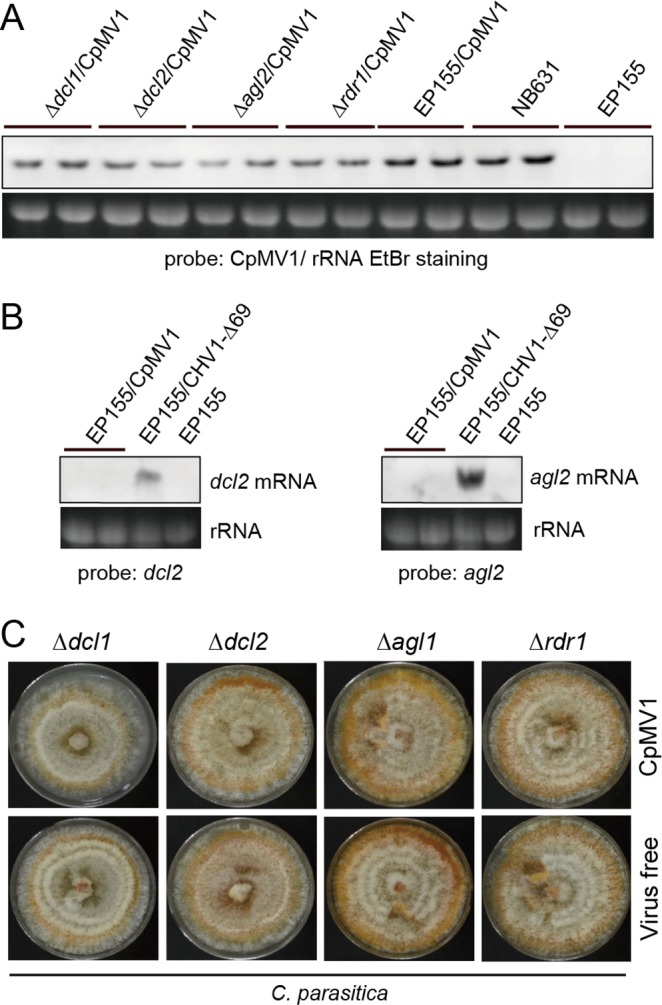
Relationship between RNA silencing and CpMV1 infection. (A) Northern blotting of CpMV1-infected fungal strains disrupted for RNA silencing-associated genes. CpMV1 was transferred from EP155 to mutants with disruptions of the dcl (Δdcl1 and Δdcl2), agl (Δagl2), and rdr (Δrdr1) genes, as reported earlier (24, 25, 58). CpMV1-infected fungal strains were then subjected to Northern blotting as described for Fig. 4. CpMV1-infected NB631 and virus-free EP155 were included as references. (B) Effects of CpMV1 infection on dcl2 and agl2 transcript levels. Total RNA was isolated from CpMV1-infected EP155, CHV1-infected EP155, and virus-free EP155. dcl2 and agl2 mRNA levels were compared among the three strains. (C) Phenotypes of fungal colonies infected and uninfected with CpMV1.
Previous studies have shown that infection of EP155 by CHV1 and several other viruses highly upregulates dcl2 and agl2 transcription (24, 40–43). To investigate this in the case of CpMV1, accumulations of dcl2 and agl2 transcripts in EP155 and CpMV1/EP155 were assessed by Northern blot analysis (Fig. 5B). The results showed that CpMV1 did not induce dcl2 and agl2 transcripts, whereas transcription levels were remarkably elevated by CHV1 Δp69, a mutant lacking the RNAi suppressor. This result suggests that CPMV1 escapes host RNA silencing by failing to induce silencing pathway genes and by avoiding their potential activity by remaining sequestered in mitochondria during replication. Moreover, phenotypic characterization of these strains showed no difference between virus-free and CpMV1-infected strains (Fig. 5C).
Production of CpMV1-derived small RNAs in infected fungal strains.
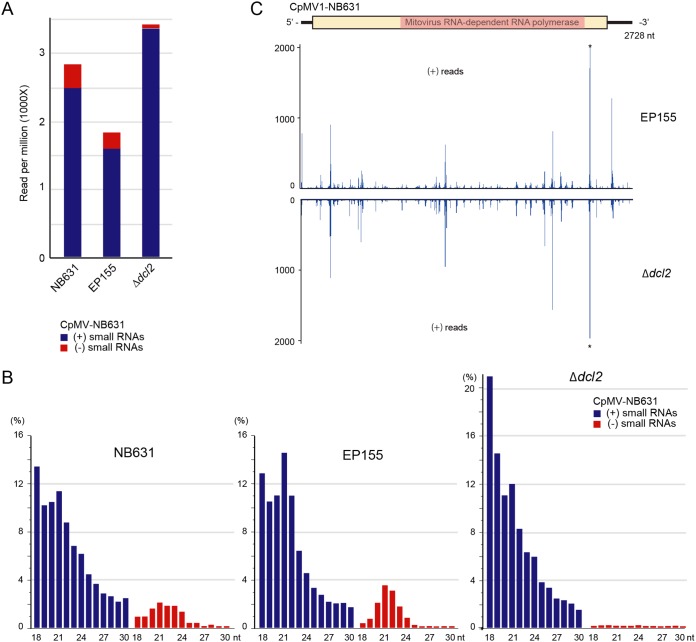
Small RNAs were prepared from four fungal strains, i.e., virus-free EP155, CpMV1-infected EP155, CpMV1-infected NB631, and the CpMV1-infected Δdcl2 mutant, and sequenced by a next-generation sequencing (NGS) method. DCL2-dependent virus-derived small RNA profiles are characterized by the production of viral minus-strand small RNAs as well as viral plus-strand small RNAs with a peak of 21 nucleotides (nt) (44, 45).
Comparison of small RNA profiles for the three fungal strains showed accumulation of CpMV1-derived small RNAs in RNA silencing-competent EP155 and NB631 and in the RNA silencing-deficient Δdcl2 mutant (Fig. 6A) but not in virus-free EP155 (data not shown). The profiles of small RNAs for CpMV1-infected NB631 and EP155 manifested a peak of 21 nt representing viral minus-strand RNA expected from the DCL2-dependent pathway, but few 21-nt viral minus-strand RNAs were detected from the CpMV1-infected Δdcl2 mutant (Fig. 6B). We recently analyzed small RNA profiles for a mutant (Δp69) of the prototype CHV1 that replicates in the nonmitochondrial cytoplasm (Andika et al., submitted). Similar patterns were observed between CpMV1-infected EP155 or Δdcl2 mutant and for CHV1-infected EP155 or Δdcl2 mutant, respectively. A notable difference was sharper peaks of 21-nt CHV1 Δp69-derived small RNAs in EP155 than of 21-nt CpMV1-derived small RNAs. This difference likely reflects a ratio of CHV1 Δp69-derived small RNAs to total small RNAs that is 4-fold greater than that of CpMV1-derived small RNAs to total small RNAs in EP155.
FIG 6.
Small RNA profiles of RNA silencing-competent NB631and EP155 and the RNA silencing-deficient Δdcl2 mutant. Small RNA fractions were obtained from CpMV1-infected strains and analyzed (see Materials and Methods). (A) Plus-strand and minus-strand small RNAs (18 to 30 nt) derived from CpMV1 are shown on a per-million-total-small-RNA basis. Blue and red bars denote plus-strand and minus-strand small RNAs. (B) Size distributions of CpMV1 small RNAs. (C) Distributions of CpMV1 small RNAs along the CpMV1 genome. A diagram of the CpMV1 genome is shown at the top. CpMV1-derived plus-strand small RNAs from EP155 and the Δdcl2 mutant were mapped to its genome, showing very similar distribution patterns.
CpMV1-derived small RNAs detected from the Δdcl2 mutant could be products digested by DCL2 independently or degradation products during small RNA preparation. Of note is that CpMV1 positive-strand-RNA-derived small RNAs from EP155 and the Δdcl2 mutant had similar peak patterns (Fig. 6C). This observation may suggest that CpMV1 positive-strand RNA is cleaved specifically, rather than randomly, in a DCL2-independent manner and that adequate precautions are needed to interpret small RNA profiles.
DISCUSSION
Over the past few decades, virion transfection protocols have been developed for diverse mycovirus-host combinations (46, 47). This virion transfection, however, is not applicable for capsidless viruses, and instead, infectious full-length cDNA or synthetic RNA produced from it has been established for capsidless hypoviruses and yadokariviruses (calicivirus‐like positive-sense single-stranded RNA [ssRNA] viruses, proposed family Yadokariviridae) (31, 33–35). Capsidless nonsegmented mitoviruses are phylogenetically related to capsidless nonsegmented narnaviruses of Saccharomyces cerevisiae (members of the genus Narnavirus) and encapsidated, multisegmented ourmiaviruses of dicot plants (members of the genus Ourmiavirus), both of which are replicated in the nonmitochondrial cytoplasm. Infectious full-length cDNA was developed for members of the genera Narnavirus and Ourmiavirus (48–50). However, no infectious cDNA clones or synthetic transcripts are available for mitoviruses thus far, although they can be transmitted vertically via spores and horizontally via hyphal fusion only between vegetatively compatible strains within a single species (2, 38). Therefore, the protoplast fusion protocol for artificial introduction developed in this study is of great importance for studies of host range and mitovirus-host interactions, because it allows introduction of virus into any host fungus, independent of hyphal incompatibility. Note that protoplast fusion was earlier utilized for lateral transmission of CHV1 and other, unidentified viruses (39, 51, 52).
CpMV1 has been reported only from one C. parasitica strain, NB631, and not from other C. parasitica strains despite extensive surveys for fungal viruses (53, 54). The current study allowed horizontal transfer of a mitovirus experimentally to a new host and expanded its host range to an order different from the one to which C. parasitica belongs (Table 1). All nine tested C. parasitica isolates, collected from the United States, Europe, and Japan were found to be susceptible to CpMV1. Another species within the genus Cryphonectria, C. radicalis, supported the replication of CpMV1, and its accumulation level was comparable to that in C. parasitica. A slightly lower accumulation pattern was observed for V. ceratosperma, belonging to the family Valsaceae, which is different from that of C. parasitica (Cryphonectriaceae). Stable maintenance of CpMV1 was observed in these newly established hosts. This observation contrasts with the situation of cytoplasmically (nonmitochondrially) replicating RNA mycoviruses whose original hosts are C. parasitica or Rosellinia necatrix (white root rot fungus) and are experimentally introduced into V. ceratosperma. For example, mycoreovirus 1 (a reovirus [encapsidated, multisegmented dsRNA virus]), CHV1, and Rosellinia necatrix partitivirus 6 (a betapartitivirus [encapsidated, two-segmented dsRNA virus]) replicate but are not stably maintained in V. ceratosperma (41). As noted above, examples of conspecific mitovirus strains found through surveys in taxonomically distinct fungi belonging to the class Sordariomycetes (Ophiostoma) or Leotiomycetes (Sclerotinia and Botrytis) (21, 22) suggested a broader host range for mitoviruses than has been recorded for other mycoviruses. Our current study experimentally demonstrated that one single mitovirus strain, CpMV1 NB631, can infect fungi belonging to different families (Valsaceae and Cryphonectriaceae) within the order Diaporthales. Analysis performed by Nibert (23) implied that infrequency of Trp-encoding UGA codons in mitochondria of some fungi reflects the infrequency of corresponding tRNAs encoded by them and that this in turn restricts the host range of a mitovirus that contains an abundance of Trp-encoding UGA codons. The protoplast fusion technique described in this study will facilitate testing of this hypothesis.
We showed the mitochondrial and nuclear genetic backgrounds of recipients, obtained after protoplast fusion, to be the same as those of the original recipient strains before protoplast fusion, as judged or expected from mitochondrial haplotyping and repeated anastomosis and single-spore isolation (Fig. 1). A total of 5 mitochondrial haplotypes of CpMV1 recipients were distinguishable by PCR products derived from a polymorphic region (Fig. 3C). An interesting finding was that all CpMV1-positive recipients show only one single haplotype identical to that of the original recipient, and neither donor strain haplotypes nor mitochondrial DNA rearrangements, which are often reported for mitochondria resulting from protoplast fusion, were observed for the polymorphic nad5 region examined (Fig. 3B). Loss of mitochondrial genomes from one partner after protoplast fusion is not uncommon, as exemplified by somatic hybrid plants of Nicotiana spp. and Solanum spp. (55, 56). Our results suggest that there is compatibility between genotypes of mitochondria and nuclei, as expected from the fact that many genes whose products are functional in mitochondria are carried by the nuclear genome. Also, the results confirm that mitochondria of the extended hosts discussed above can support replication of CpMV1.
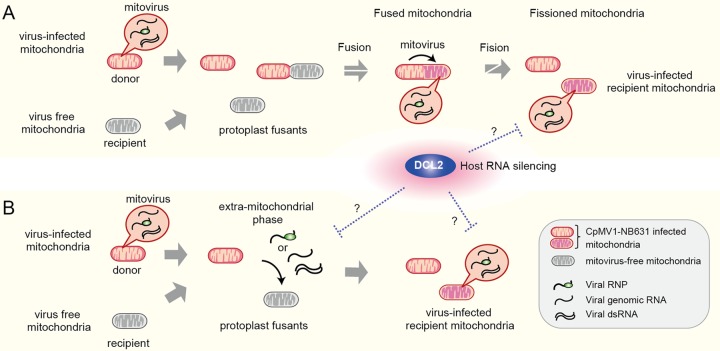
Polashock et al. (38) proposed two possibilities for the CpMV1 transmission mode with the premise that CpMV1 cannot move bidirectionally across the mitochondrial membrane: (i) partial or total replacement of uninfected mitochondria (recipient) with infected ones (donor) or (ii) fusion of infected and noninfected mitochondria, resulting in CpMV1 transmission to the latter. Our results are not consistent with the first scenario but rather favor the second one, that mitochondrial fusion is a mechanism for CpMV1 transmission to a new host while maintaining the recipient mitochondrial haplotype (Fig. 7A). We cannot completely rule out the possibility that CpMV1 has an extramitochondrial phase in the cytoplasm and reentry into uninfected recipients’ mitochondria (Fig. 7B). An extramitochondrial phase has been proposed for the presumed mitovirus M2 from Rhizoctonia solani based on initial finding of more M2 dsRNA in the nonmitochondrial cytosolic fraction than in the mitochondrial fraction (11), but follow-up subcellular localization studies have not been published, and details remain unclear. As discussed below, this idea is supported by the observation that DCL2 could produce CpMV1-derived small RNAs and by a recent report that endogenous mitochondrial dsRNA can be exported into the cytosol (57). Either scenario proposed in Fig. 7 includes open questions and needs to be validated in the future.
FIG 7.
Model of CpMV1 infection of recipient mitochondria. (A) Infection of recipient mitochondria by CpMV1 initiated via fusion with donor mitochondria. CpMV1-infected recipient mitochondria become dominant via fission. (B) Infection of recipient mitochondria by CpMV1 mediated by its extramitochondrial entities, genomic RNA-associated or nonassociated RdRp molecules, or dsRNA. The extramitochondrial dsRNA of CpMV1 could be diced by DCL2. Considering the production of CpMV1-derived small RNAs (Fig. 6), there should be extramitochondrial dsRNA of CpMV1, which is accessed in the cytosol by DCL2, even in model A. The two possibilities (A and B) are not mutually exclusive.
Investigation of antiviral RNA silencing versus mitoviruses is of great interest from several perspectives. If mitoviruses have a cytosolic phase in their replication cycles, though seemingly unlikely, they would be targeted by RNA silencing. In this context, some recent studies have reported mitovirus-derived small RNAs produced in infected fungi (28, 30), suggesting the accessibility of Dicer to mitovirus RNAs. DCL2-dependent CpMV1-derived small RNAs were also detectable in infected EP155 (Fig. 6). The replicative form (dsRNA) of CpMV1 may be exposed in the cytosol at an unknown step of replication or by accidental release of dsRNAs from mitochondria. In support of this notion, long mitochondrial dsRNAs (endogenous, bidirectional transcription products) were recently shown to be excreted into the cytoplasm and to induce a dsRNA sensor-mediated interferon type I antiviral response in humans (57). The next question is whether CpMV1 is really targeted by RNA silencing. This study allowed us to address this question, because CpMV1 could be moved laterally to the standard EP155 strain of C. parasitica, for which many mutant strains are available. Many cytoplasmically (nonmitochondrially) replicating RNA viruses have been shown to be targeted by antiviral RNA silencing and to transcriptionally induce RNA silencing genes in C. parasitica. Only a few viruses, such as Rosellinia necatrix victorivirus 1 (RnVV1) (a victorivirus [nonsegmented dsRNA virus]), fail to induce dcl2 and agl2 (26, 40). Like RnVV1, CpMV1 induced neither dcl2 nor agl2 (Fig. 5B). Figure 5A and C show comparable accumulation levels of CpMV1 between RNAi-proficient (EP155) and -deficient (Δdcl2 and Δagl2 mutant) strains, and no phenotypic alteration was observed in these strains. These results contrast with the situation of other cytoplasmically (nonmitochondrially) replicating viruses, including RnVV1, that usually show higher levels of virus accumulation and more severe symptoms in RNAi-deficient strains. Collectively, this study suggests the possible avoidance of host antiviral RNA silencing, consistent with its mitochondrial replication.
MATERIALS AND METHODS
Fungal strains.
The CpMV1-infected U.S. strain NB631 of C. parasitica was described earlier (2). The fungal strains used in this study (Table 1) included the standard C. parasitica strain EP155, which is used extensively in many laboratories, EPI55-derived deletion mutants, and field-collected strains of C. parasitica from the United States (EP155, C18, GH2, and NB58-19), Italy (EP67), and Japan (J1, J2, J3, J4). Strains of some other ascomycetous fungi, such as C. radicalis (JS13), V. ceratosperma (AVC53), and H. victoriae (HV1004-B), were also used. C. parasitica and C. radicalis share the same niche, where both thrive in chestnut trees. V. ceratosperma belongs to the family Valsaceae, whereas Cryphonectria spp. belong to the family Cryphonectriaceae, but they are within the same order Diaporthales. H. victoriae belongs to the order Pleosporales.
Protoplast fusion.
All tested fungal strains (Table 1) were transformed with pCPXHY2 carrying a hygromycin B phosphotransferase gene (hph) to obtain hygromycin B-resistant (Hygr) strains. An equal number of protoplasts (approximately 2 × 107) from the CpMV1 donor strain NB631 and a virus-free Hygr recipient strain were fused in the presence of polyethylene glycol (PEG)-CaCl2. The protoplast fusants were regenerated on the regeneration medium and were allowed to grow overnight. The next day, top agar containing 40 μg/ml hygromycin B was overlaid on regeneration medium for screening the recipient (host of interest). Protoplast regenerants were then allowed to grow over top agar for several days. Hygr mycelia growing over the top agar were transferred into new potato dextrose agar (PDA) plates containing 80 μg/ml hygromycin B and incubated for 3 to 5 days before detection of CpMV1. After confirming the presence of CpMV1, CpMV1-positive colonies were anastomosed with the original nontransformed recipients that were hygromycin B susceptible (Hygs) and CpMV1 free. Mycelial plugs taken from the recipient’s side were fused again with the original CpMV1-free recipient. Karyons are known to be transferred between donors and recipients (26). A total of four rounds of this hyphal fusion procedure were carried out for each of the fungal strains to avoid carryover of karyons from donors. Finally, single conidial isolates from each of 11 CpMV1-infected fungal hosts were prepared to obtain monokaryons and were used in subsequent characterization (Fig. 1). Steps 1 and 2 shown in Fig. 1 are commonly used for protoplast fusion experiments. However, the objective of this procedure is to follow the mitochondrial lineage from CpMV1-infected monokaryotic recipients, not from CpMV1-infected heterokaryotic fusants.
RT-PCR detection of CpMV1.
cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific, Invitrogen, Waltham, MA) and a primer, 5′-GGAATTACCTCCAAAC-3′, that corresponds to the RdRp-coding region (positions 1951 to 1966) of CpMV1 NB631 (GenBank accession no. NC_00404). PCR amplification was carried out using Quick Taq (Toyobo, Osaka, Japan) along with primer set 5′-GCAACGGATAGACTTC-3′ (map positions 1290 to 1305) and 5′-AAATAGCCTGATGCCA-3′ (map positions 1922 to 1937).
Northern blotting.
Northern blotting was performed as described by Eusebio-Cope and Suzuki (57). The heat-denatured ssRNA fraction (2 μg) was electrophoresed in a 1% agarose gel under denaturing conditions. The separated RNAs were capillary transferred onto a Hybond-N+ nylon membrane (GE Healthcare Life Sciences, Pittsburgh, PA) and fixed by a UV cross-linker, CL-1000 (UVP). The membrane was then treated with digoxigenin (DIG)-labeled DNA probes amplified from cDNA by PCR (PCR DIG labeling mix; Roche, Risch-Rotkreuz, Switzerland). The primer set is described above for CpMV1 and was described earlier for the dcl2 and agl2 mutants (40). Prehybridization and hybridization steps were performed based on the information provided by the supplier (Roche, Risch-Rotkreuz, Switzerland). Two low- and high-stringency washes were carried out in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1× SSC, respectively. Hybridized nucleic acids were detected with ready-to-use CDP-Star (Roche) through digital imaging in the ImageQuant LAS 4000 system (GE Healthcare Life Sciences).
Mitochondrial haplotyping.
Mitochondrial DNA extraction was performed as described by Yamada et al. (59) with slight modification. Fungal mycelia grown on liquid medium (potato dextrose broth [PDB]) were harvested after 3 days of inoculation on the benchtop. Mycelia were ground in the presence of liquid nitrogen with the help of pestle and mortar and were homogenized with extraction buffer (100 mM Tris HCl [pH 8.0], 1% SDS, 2% Triton X-100, 10 mM EDTA, 100 mM NaCl), followed by phenol-isoamyl alcohol and chloroform extraction. The supernatant was then precipitated by adding an equal amount of isopropanol and incubating the sample at room temperature for 1 h. The pellet was then washed twice with 70% ethanol and vacuum dried. Finally, the pellet was dissolved in MilliQ water and stored at −20°C until use.
Based on the paper published by Gobbi et al. (60), a highly polymorphic region, i.e., region 2 from mtDNA of EP155, was utilized to make different sets of primers within it (Fig. 3A) to distinguish recipient mitochondria from donor mitochondria. Various amounts of mtDNA (0.5 ng, 1 ng, 5 ng, and 10 ng) were used as templates, and conventional PCR was performed.
Analysis of small RNA sequences.
Total RNA samples from three C. parasitica strains (NB631, EP155, and the Δdcl2 mutant) infected with CpMV1 NB631 were subjected to small RNA sequencing. Small RNA cDNA library preparation and subsequent deep sequencing with the Illumina platform (HiSeq 2500; 50-bp single-end reads) were done by Macrogen Inc. (Tokyo, Japan). After trimming adapters and low-quality bases and size filtering (18 to 30 nt in length), the retained read sequences in each library were mapped to the CpMV1 NB631 genome (GenBank accession no. L31849) using CLC Genomics Workbench (version 11; CLC Bio-Qiagen). The virus-derived small RNA reads were used for in-depth analysis with the program MISIS-2 (61).
ACKNOWLEDGMENTS
This study was supported in part by Yomogi Inc., the Uehara Memorial Foundation, by Grants-in-Aid for Scientific Research on Innovative Areas from the Japanese Ministry of Education, Culture, Sports, Science and Technology (KAKENHI 25252011, 15K14663, 16H06436, 16H06429, and 16K21723 to H.K. and N.S.), and by the New Jersey Agricultural Experiment Station and USDA-NIFA (to B.I.H.).
We are grateful to Donald L. Nuss, Said A. Ghabrial, and Satoko Kanematsu for generous gifts of C. parasitica strains EP155 and the Δdcl2 and Δagl2 mutants, H. victoriae strain Hv1004-Hygr, and V. ceratosperma strain AVC53 and to Sotaro Chiba and Ida Bagus Andika for fruitful discussions.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Hillman BI, Cai G. 2013. The family Narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176. doi: 10.1016/B978-0-12-394315-6.00006-4. [DOI] [PubMed] [Google Scholar]
- 2.Polashock JJ, Hillman BI. 1994. A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc Natl Acad Sci U S A 91:8680–8684. doi: 10.1073/pnas.91.18.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu M, Deng Y, Zhou Z, He G, Chen W, Li G. 2016. Characterization of three mycoviruses co-infecting the plant pathogenic fungus Sclerotinia nivalis. Virus Res 223:28–38. doi: 10.1016/j.virusres.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Osaki H, Nakamura H, Nomura K, Matsumoto N, Yoshida K. 2005. Nucleotide sequence of a mitochondrial RNA virus from the plant pathogenic fungus, Helicobasidium mompa Tanaka. Virus Res 107:39–46. doi: 10.1016/j.virusres.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Hong YG, Dover SL, Cole TE, Brasier CM, Buck KW. 1999. Multiple mitochondrial viruses in an isolate of the Dutch elm disease fungus Ophiostoma novo-ulmi. Virology 258:118–127. doi: 10.1006/viro.1999.9691. [DOI] [PubMed] [Google Scholar]
- 6.Turina M, Ghignone S, Astolfi N, Silvestri A, Bonfante P, Lanfranco L. 2018. The virome of the arbuscular mycorrhizal fungus Gigaspora margarita reveals the first report of DNA fragments corresponding to replicating non-retroviral RNA viruses in fungi. Environ Microbiol doi: 10.1111/1462-2920.14060. [DOI] [PubMed] [Google Scholar]
- 7.Nibert ML, Vong M, Fugate KK, Debat HJ. 2018. Evidence for contemporary plant mitoviruses. Virology 518:14–24. doi: 10.1016/j.virol.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruenn JA, Warner BE, Yerramsetty P. 2015. Widespread narnavirus sequences in plant genomes. Peer J 3:e876. doi: 10.7717/peerj.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Wu S, Liu L, Cheng J, Fu Y, Jiang D, Xie J. 2015. A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res 197:127–136. doi: 10.1016/j.virusres.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Hong YG, Cole TE, Brasier CM, Buck KW. 1998. Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 246:158–169. doi: 10.1006/viro.1998.9178. [DOI] [PubMed] [Google Scholar]
- 11.Lakshman DK, Jian JH, Tavantzis SM. 1998. A double-stranded RNA element from a hypovirulent strain of Rhizoctonia solani occurs in DNA form and is genetically related to the pentafunctional AROM protein of the shikimate pathway. Proc Natl Acad Sci U S A 95:6425–6429. doi: 10.1073/pnas.95.11.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, Suzuki N. 2011. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog 7:e1002146. doi: 10.1371/journal.ppat.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Fu Y, Jiang D, Li G, Xie J, Cheng J, Peng Y, Ghabrial SA, Yi X. 2010. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol 84:11876–11887. doi: 10.1128/JVI.00955-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, Tomonaga K. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu F, Xie J, Cheng S, You MP, Barbetti MJ, Jia J, Wang Q, Cheng J, Fu Y, Chen T, Jiang D. 2017. Virome characterization of a collection of S. sclerotiorum from Australia. Front Microbiol 8:2540. doi: 10.3389/fmicb.2017.02540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalifa ME, Pearson MN. 2013. Molecular characterization of three mitoviruses co-infecting a hypovirulent isolate of Sclerotinia sclerotiorum fungus. Virology 441:22–30. doi: 10.1016/j.virol.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Hillman BI, Annisa A, Suzuki N. 2018. Viruses of plant-interacting fungi. Adv Virus Res 100:99–116. doi: 10.1016/bs.aivir.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Turina M, Hillman BI, Izadpanah K, Rastgou M, Rosa C, ICTV Report Consortium. 2017. ICTV virus taxonomy profile: Ourmiavirus. J Gen Virol 98:129–130. doi: 10.1099/jgv.0.000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrabakova L, Koloniuk I, Petrzik K. 2017. Phomopsis longicolla RNA virus 1—novel virus at the edge of myco- and plant viruses. Virology 506:14–18. doi: 10.1016/j.virol.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Wu MD, Zhang L, Li GQ, Jiang DH, Hou MS, Huang HC. 2007. Hypovirulence and double-stranded RNA in Botrytis cinerea. Phytopathology 97:1590–1599. doi: 10.1094/PHYTO-97-12-1590. [DOI] [PubMed] [Google Scholar]
- 21.Wu M, Zhang L, Li G, Jiang D, Ghabrial SA. 2010. Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 406:117–126. doi: 10.1016/j.virol.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Deng F, Xu R, Boland GJ. 2003. Hypovirulence-associated double-stranded RNA from Sclerotinia homoeocarpa Is conspecific with Ophiostoma novo-ulmi mitovirus 3a-Ld. Phytopathology 93:1407–1414. doi: 10.1094/PHYTO.2003.93.11.1407. [DOI] [PubMed] [Google Scholar]
- 23.Nibert ML. 2017. Mitovirus UGA(Trp) codon usage parallels that of host mitochondria. Virology 507:96–100. doi: 10.1016/j.virol.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Q, Choi GH, Nuss DL. 2009. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc Natl Acad Sci U S A 106:17927–17932. doi: 10.1073/pnas.0907552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci U S A 104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiba S, Lin YH, Kondo H, Kanematsu S, Suzuki N. 2013. A novel victorivirus from a phytopathogenic fungus, Rosellinia necatrix is infectious as particles and targeted by RNA silencing. J Virol 87:6727–6738. doi: 10.1128/JVI.00557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salaipeth L, Chiba S, Eusebio-Cope A, Kanematsu S, Suzuki N. 2014. Biological properties and expression strategy of Rosellinia necatrix megabirnavirus 1 analyzed in an experimental host, Cryphonectria parasitica. J Gen Virol 95:740–750. doi: 10.1099/vir.0.058164-0. [DOI] [PubMed] [Google Scholar]
- 28.Vainio EJ, Jurvansuu J, Streng J, Rajamaki ML, Hantula J, Valkonen JPT. 2015. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J Gen Virol 96:714–725. doi: 10.1099/jgv.0.000003. [DOI] [PubMed] [Google Scholar]
- 29.Donaire L, Ayllon MA. 2017. Deep sequencing of mycovirus-derived small RNAs from Botrytis species. Mol Plant Pathol 18:1127–1137. doi: 10.1111/mpp.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz-Adalia EJ, Diez JJ, Fernandez MM, Hantula J, Vainio EJ. 2018. Characterization of small RNAs originating from mitoviruses infecting the conifer pathogen Fusarium circinatum. Arch Virol 163:1009–1018. doi: 10.1007/s00705-018-3712-2. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R, Hisano S, Tani A, Kondo H, Kanematsu S, Suzuki N. 2016. A capsidless ssRNA virus hosted by an unrelated dsRNA virus. Nat Microbiol 1:15001. doi: 10.1038/nmicrobiol.2015.1. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki A, Onoue M, Kanematsu S, Suzaki K, Miyanishi M, Suzuki N, Nuss DL, Yoshida K. 2002. Extending chestnut blight hypovirus host range within diaporthales by biolistic delivery of viral cDNA. Mol Plant Microbe Interact 15:780–789. doi: 10.1094/MPMI.2002.15.8.780. [DOI] [PubMed] [Google Scholar]
- 33.Chen B, Choi GH, Nuss DL. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762–1764. [DOI] [PubMed] [Google Scholar]
- 34.Choi GH, Nuss DL. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800–803. [DOI] [PubMed] [Google Scholar]
- 35.Marzano SYL, Hobbs HA, Nelson BD, Hartman GL, Eastburn DM, McCoppin NK, Domier LL. 2015. Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus. J Virol 89:5060–5071. doi: 10.1128/JVI.03199-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillman BI, Supyani S, Kondo H, Suzuki N. 2004. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the Coltivirus genus of animal pathogens. J Virol 78:892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eusebio-Cope A, Sun L, Tanaka T, Chiba S, Kasahara S, Suzuki N. 2015. The chestnut blight fungus for studies on virus/host and virus/virus interactions: from a natural to a model host. Virology 477:164–175. doi: 10.1016/j.virol.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 38.Polashock JJ, Bedker PJ, Hillman BI. 1997. Movement of a small mitochondrial double-stranded RNA element of Cryphonectria parasitica: ascospore inheritance and implications for mitochondrial recombination. Mol Gen Genet 256:566–571. [DOI] [PubMed] [Google Scholar]
- 39.Lee KM, Yu J, Son M, Lee YW, Kim KH. 2011. Transmission of Fusarium boothii mycovirus via protoplast fusion causes hypovirulence in other phytopathogenic fungi. PLoS One 6:e21629. doi: 10.1371/journal.pone.0021629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiba S, Suzuki N. 2015. Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc Natl Acad Sci U S A 112:E4911–E4918. doi: 10.1073/pnas.1509151112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiba S, Lin YH, Kondo H, Kanematsu S, Suzuki N. 2016. A novel betapartitivirus RnPV6 from Rosellinia necatrix tolerates host RNA silencing but is interfered by its defective RNAs. Virus Res 219:62–72. doi: 10.1016/j.virusres.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Andika IB, Jamal A, Kondo H, Suzuki N. 2017. SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc Natl Acad Sci U S A 114:E3499–E3506. doi: 10.1073/pnas.1701196114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Shi D, Nuss DL. 2012. Variations in hypovirus interactions with the fungal-host RNA-silencing antiviral-defense response. J Virol 86:12933–12939. doi: 10.1128/JVI.00961-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. 2008. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J Virol 82:2613–2619. doi: 10.1128/JVI.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andika IB, Kondo H, Suzuki N. 23 January 2019. Dicer functions transcriptionally and post-transcriptionally in a multilayer antiviral defense. Proc Natl Acad U S A doi: 10.1073/pnas.1812407116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kondo H, Kanematsu S, Suzuki N. 2013. Viruses of the white root rot fungus, Rosellinia necatrix. Adv Virus Res 86:177–214. doi: 10.1016/B978-0-12-394315-6.00007-6. [DOI] [PubMed] [Google Scholar]
- 47.Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, Jiang D. 2014. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci U S A 111:12205–12210. doi: 10.1073/pnas.1401786111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crivelli G, Ciuffo M, Genre A, Masenga V, Turina M. 2011. Reverse genetic analysis of Ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation. J Virol 85:5091–5104. doi: 10.1128/JVI.02565-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esteban R, Fujimura T. 2003. Launching the yeast 23S RNA Narnavirus shows 5' and 3' cis-acting signals for replication. Proc Natl Acad Sci U S A 100:2568–2573. doi: 10.1073/pnas.0530167100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esteban R, Vega L, Fujimura T. 2005. Launching of the yeast 20 s RNA narnavirus by expressing the genomic or antigenomic viral RNA in vivo. J Biol Chem 280:33725–33734. doi: 10.1074/jbc.M506546200. [DOI] [PubMed] [Google Scholar]
- 51.van Diepeningen AD, Debets AJ, Hoekstra RF. 1998. Intra- and interspecies virus transfer in Aspergilli via protoplast fusion. Fungal Genet Biol 25:171–180. doi: 10.1006/fgbi.1998.1096. [DOI] [PubMed] [Google Scholar]
- 52.van Diepeningen AD, Debets AJ, Slakhorst SM, Fekete C, Hornok L, Hoekstra RF. 2000. Interspecies virus transfer via protoplast fusions between Fusarium poae and black Aspergillus strains. Fungal Genet Newsl 47:99–100. doi: 10.4148/1941-4765.1216. [DOI] [Google Scholar]
- 53.Liu YC, Dynek JN, Hillman BI, Milgroom MG. 2007. Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol Res 111:433–442. doi: 10.1016/j.mycres.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Peever TL, Liu YC, Wang KR, Hillman BI, Foglia R, Milgroom MG. 1998. Incidence and diversity of double-stranded RNAs occurring in the chestnut blight fungus, Cryphonectria parasitica, in China and Japan. Phytopathology 88:811–817. doi: 10.1094/PHYTO.1998.88.8.811. [DOI] [PubMed] [Google Scholar]
- 55.Patel D, Power JB, Anthony P, Badakshi F, Pat Heslop-Harrison JS, Davey MR. 2011. Somatic hybrid plants of Nicotiana x sanderae (+) N. debneyi with fungal resistance to Peronospora tabacina. Ann Bot 108:809–819. doi: 10.1093/aob/mcr197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iovene M, Savarese S, Cardi T, Frusciante L, Scotti N, Simon PW, Carputo D. 2007. Nuclear and cytoplasmic genome composition of Solanum bulbocastanum (+) S. tuberosum somatic hybrids. Genome 50:443–450. doi: 10.1139/g07-024. [DOI] [PubMed] [Google Scholar]
- 57.Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, Crow YJ, Rice GI, Duffy D, Tamby C, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, Proudfoot NJ. 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560:238–242. doi: 10.1038/s41586-018-0363-0:10.1038/s41586-018-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eusebio-Cope A, Suzuki N. 2015. Mycoreovirus genome rearrangements associated with RNA silencing deficiency. Nucleic Acids Res 43:3802–3813. doi: 10.1093/nar/gkv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamada Y, Makimura K, Merhendi H, Ueda K, Nishiyama Y, Yamaguchi H, Osumi M. 2002. Comparison of different methods for extraction of mitochondrial DNA from human pathogenic yeasts. Jpn J Infect Dis 55:122–125. [PubMed] [Google Scholar]
- 60.Gobbi E, Firrao G, Carpanelli A, Locci R, Van Alfen NK. 2003. Mapping and characterization of polymorphism in mtDNA of Cryphonectria parasitica: evidence of the presence of an optional intron. Fungal Genet Biol 40:215–224. [DOI] [PubMed] [Google Scholar]
- 61.Seguin J, Otten P, Baerlocher L, Farinelli L, Pooggin MM. 2016. MISIS-2: a bioinformatics tool for in-depth analysis of small RNAs and representation of consensus master genome in viral quasispecies. J Virol Methods 233:37–40. doi: 10.1016/j.jviromet.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 62.Anagnostakis SL. 1984. Nuclear gene-mutations in Endothia (Cryphonectria) parasitica that affect morphology and virulence. Phytopathology 74:561–565. doi: 10.1094/Phyto-74-561. [DOI] [Google Scholar]
- 63.Milgroom MG, Lipari SE, Powell WA. 1992. DNA fingerprinting and nalysis of population structure in the chestnut blight fungus, Cryphonectria parasitica. Genetics 131:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smart CD, Yuan W, Foglia R, Nuss DL, Fulbright DW, Hillman BI. 1999. Cryphonectria hypovirus 3, a virus species in the family Hypoviridae with a single open reading frame. Virology 265:66–73. doi: 10.1006/viro.1999.0039. [DOI] [PubMed] [Google Scholar]
- 65.Hillman BI, Tian Y, Bedker PJ, Brown MP. 1992. A North American hypovirulent isolate of the chestnut blight fungus with European isolate-related dsRNA. J Gen Virol 73:681–686. doi: 10.1099/0022-1317-73-3-681. [DOI] [PubMed] [Google Scholar]
- 66.Enebak SA, Hillman BI, Macdonald WL. 1994. A hypovirulent isolate of Cryphonectria parasitica with multiple, genetically unique dsRNA segments. MPMI 7:590–595. doi: 10.1094/MPMI-7-0590. [DOI] [Google Scholar]
- 67.Eusebio-Cope A, Suzuki N, Sadeghi-Garmaroodi H, Taga M. 2009. Cytological and electrophoretic karyotyping of the chestnut blight fungus Cryphonectria parasitica. Fungal Genet Biol 46:342–351. doi: 10.1016/j.fgb.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 68.Suzaki K, Yoshida K, Ito T. 1997. Pathogenicity to apple branch and phloridzin degrading activity of Valsa ceratosperma isolates from some broad leaf trees including apple tree. Annu Rep Soc Plant Prot N Jpn 48:145–157. [Google Scholar]
- 69.Ghabrial SA, Dunn SE, Li H, Xie J, Baker TS. 2013. Viruses of Helminthosporium (Cochlioblus) victoriae. Adv Virus Res 86:289–325. doi: 10.1016/B978-0-12-394315-6.00011-8. [DOI] [PubMed] [Google Scholar]
- 70.Short DP, Double M, Nuss DL, Stauder CM, MacDonald W, Kasson MT. 2015. Multilocus PCR assays elucidate vegetative incompatibility gene profiles of Cryphonectria parasitica in the United States. Appl Environ Microbiol 81:5736–5742. doi: 10.1128/AEM.00926-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi GH, Dawe AL, Churbanov A, Smith ML, Milgroom MG, Nuss DL. 2012. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190:113–127. doi: 10.1534/genetics.111.133983. [DOI] [PMC free article] [PubMed] [Google Scholar]