Infection with oncogenic gammaherpesviruses leads to long-term viral persistence through a dynamic interplay between the virus and the host immune system. Critical for remodeling of the host cell environment after the immune responses are viral noncoding RNAs that modulate host signaling pathways without attracting adaptive immune recognition. Despite the importance of noncoding RNAs in persistent infection, the circRNA class of noncoding RNAs has only recently been identified in gammaherpesviruses. Accordingly, their roles in virus infection and associated oncogenesis are unknown. Here we report evolutionary conservation of EBV-encoded circRNAs determined by assessing the circRNAome in rLCV-infected lymphomas from an SIV-infected rhesus macaque, and we report latent and lytic circRNAs from KSHV and MHV68. These experiments demonstrate utilization of the circular RNA class of RNAs across 4 members of the gammaherpesvirus subfamily, and they identify orthologues and potential homoplastic circRNAs, implying conserved circRNA functions in virus biology and associated malignancies.
KEYWORDS: EBV, Epstein-Barr virus, KSHV, Kaposi’s sarcoma herpesvirus, MHV68, circRNA, circular RNA, lymphocryptovirus, murid herpesvirus 68, rhadinovirus
ABSTRACT
Recent studies have identified circular RNAs (circRNAs) expressed from the Epstein-Barr virus (EBV) and Kaposi’s sarcoma herpesvirus (KSHV) human DNA tumor viruses. To gain initial insights into the potential relevance of EBV circRNAs in virus biology and disease, we assessed the circRNAome of the interspecies homologue rhesus macaque lymphocryptovirus (rLCV) in a naturally occurring lymphoma from a simian immunodeficiency virus (SIV)-infected rhesus macaque. This analysis revealed rLCV orthologues of the latency-associated EBV circular RNAs circRPMS1_E4_E3a and circEBNA_U. Also identified in two samples displaying unusually high lytic gene expression was a novel rLCV circRNA that contains both conserved and rLCV-specific RPMS1 exons and whose backsplice junctions flank an rLCV lytic origin of replication (OriLyt). Analysis of a lytic infection model for the murid herpesvirus 68 (MHV68) rhadinovirus identified a cluster of circRNAs near an MHV68 lytic origin of replication, with the most abundant of these, circM11_ORF69, spanning the OriLyt. Lastly, analysis of KSHV latency and reactivation models revealed the latency associated circRNA originating from the vIRF4 gene as the predominant viral circRNA. Together, the results of this study broaden our appreciation for circRNA repertoires in the Lymphocryptovirus and Rhadinovirus genera of gammaherpesviruses and provide evolutionary support for viral circRNA functions in latency and viral replication.
IMPORTANCE Infection with oncogenic gammaherpesviruses leads to long-term viral persistence through a dynamic interplay between the virus and the host immune system. Critical for remodeling of the host cell environment after the immune responses are viral noncoding RNAs that modulate host signaling pathways without attracting adaptive immune recognition. Despite the importance of noncoding RNAs in persistent infection, the circRNA class of noncoding RNAs has only recently been identified in gammaherpesviruses. Accordingly, their roles in virus infection and associated oncogenesis are unknown. Here we report evolutionary conservation of EBV-encoded circRNAs determined by assessing the circRNAome in rLCV-infected lymphomas from an SIV-infected rhesus macaque, and we report latent and lytic circRNAs from KSHV and MHV68. These experiments demonstrate utilization of the circular RNA class of RNAs across 4 members of the gammaherpesvirus subfamily, and they identify orthologues and potential homoplastic circRNAs, implying conserved circRNA functions in virus biology and associated malignancies.
INTRODUCTION
The oncogenic properties of tumor-associated gammaherpesviruses are manifested through repertoires of viral genes expressed in their respective associated cancers. Nevertheless, expression of oncogenic Epstein-Barr virus (EBV) protein-coding latency genes is suppressed in most EBV-associated tumors and/or is variable across tumor types and patients (1–6). Differences in expression of these viral proteins are probably driven in part by the degree and mechanisms of immune privilege that are unique to tissue of origin and tumor site and between patients. More consistently expressed are EBV latency-associated noncoding RNAs, such as the small noncoding RNAs, EBER1 and EBER2, the BamHI A region microRNAs, and the BamHI A region long noncoding RNAs (referred to as RPMS1 and A73) (7–15). The pervasive expression of these noncoding RNAs across most stages of the natural EBV infection cascade presumably reflects the effectiveness of this strategy to modulate the host cell environment without eliciting a substantial adaptive immune response. The pan tissue expression of these viral noncoding RNAs is recapitulated across EBV-associated cancer types and patients, where their expression likely contributes broadly to the tumor phenotype in a relatively immunologically transparent manner. Appreciating the repertoire and functions of viral noncoding RNAs is therefore critical for efforts to understand the mechanisms of viral oncogenesis and for the development of therapeutic strategies to target virus-associated cancers.
Circular RNAs (circRNAs) are a recently appreciated class of primarily noncoding RNAs that have been detected across the 5 kingdoms of life (16–22). circRNAs are formed through backsplicing of 3′ splice donor sequences to upstream 5′ splice acceptor sequences to generate closed circular RNAs that have increased stability due to a lack of sensitivity to exonucleases. With their unique structure and increased stability, circRNAs likely play distinct functions in the cell and seem uniquely suitable for modulating pathways that require sustained effector signaling. Utilizing a series of B-cell and stomach cancer cell models representing the three major EBV latency gene expression programs (latency type I, type II, and type III) and viral reactivation, we have recently reported that EBV encodes a diverse repertoire of viral circRNAs (23). Some EBV circRNAs were uniquely associated with latency type and/or reactivation, whereas others were broadly expressed across tissue and tumor types (23). The EBV-encoded circRPMS1_E4_E3a and circRPMS1_E4_E2 RNAs, which are derived from the noncoding RPMS1 gene locus, were found to be expressed in all three latency types and in B-cell and epithelial tumor models. Further, they were found to be expressed in stomach cancer and posttransplant lymphoproliferative disease (PTLD) patient specimens (23, 24). A lower abundance of the viral circRNA circEBNA_U, derived from the Epstein-Barr nuclear antigen (EBNA) locus, was detected in B cells displaying both type I and type III latency and was detected during reactivation (23).
To gain insight into the potential significance of EBV circular RNAs, we performed a limited evolutionary study utilizing the rhesus macaque lymphocryptovirus (rLCV), model which shows nearly identical gene organization across the genome but which shares only 65% nucleotide homology with EBV (25). Utilizing three different tumor specimens from a naturally occurring simian immunodeficiency virus (SIV)-associated rhesus macaque lymphoma(s), we found high levels of rLCV gene expression, indicating a likely rLCV etiology. Assessment of the rLCV circRNAome showed low but detectable expression of the rLCV counterpart to the EBV circEBNA_U circRNA in all three samples. More-robust detection of an EBV circRPMS1_E4_E3a homologue displaying both common and rLCV-specific RPMS1 exon utilization was also observed. An additional rLCV RPMS1 circular RNA, circRPMS1_E2b_E1b, surrounds a lytic origin of replication and appears to be detected uniquely in rLCV and not in EBV. We also performed circRNA analyses of two more-distantly related gammaherpesviruses, the Kaposi’s sarcoma herpesvirus (KSHV) and murid herpesvirus 68 (MHV68) rhadinoviruses. This analysis identified a latently expressed circular RNA derived from the vIRF4 gene, consistent with previous studies (24, 26), and a lytic MHV68 circular RNA that encompasses one of MHV68’s lytic origins of replication. Together, the results of this work demonstrate the conservation of circRNA utilization as a potential mechanism to facilitate cell signaling in the absence of affecting an adaptive immune response. Further, we identified viral circRNAs and features that are conserved between the EBV and rLCV lymphocryptoviruses, with the finding of an EBV circRPMS1_E4_E3a homologue being particularly noteworthy due to the broad detection of circRPMS1_E4_E3a across latency and cell types and in the natural in vivo tumor setting (23, 24, 26).
RESULTS
Rhesus SIV/LCV lymphoma model.
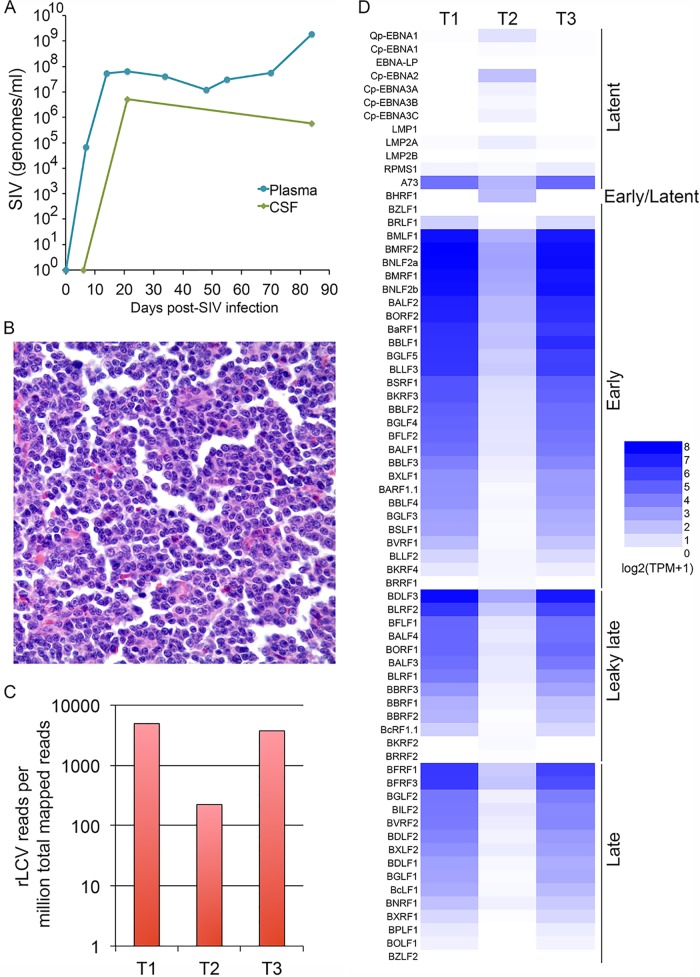
To investigate conservation of recently identified EBV circRNAs (23, 24), we utilized the rhesus lymphocryptovirus (rLCV) model, which, despite a remarkably similar genomic organization, shares only 65% nucleotide homology with EBV (25). This analysis was performed using tumor tissues from naturally occurring lymphomas in a simian immunodeficiency virus (SIV)-infected Indian rhesus macaque. This adult male macaque (14 years), which was negative for the major histocompatibility complex class I (MHC-I) alleles Mamu-A*01, Mamu B*08, and Mamu B*17, received twice-daily oral doses (60 mg and 30 mg) of dimethylfumarate for 7 days prior to intravenous inoculation with SIVmac251 (100 50% tissue culture infective doses [TCID50]). The animal then received three successive doses of the anti-CD8 antibody MT807R (10 mg/kg, 5 mg/kg, and 5 mg/kg) at days 6, 9, and 13 postinfection. Plasma and cerebrospinal fluid (CSF) samples were collected at several time points up until the day of autopsy (day 84 postinfection) for analysis of SIV RNA levels (Fig. 1A). As expected, CSF SIV RNA levels were ∼1 log lower than those in plasma, and at necropsy (day 84), the plasma SIV load was 1.9 × 109. Two effaced lymph node sections and a white nodule located next to a cut margin of the jejunum (Fig. 1B) were excised and flash frozen. RNA was isolated from snap-frozen tissues, and the RNAs were subjected to both poly(A) sequencing [poly(A)-seq] and RNase R sequencing (RNase R-seq). Mapping of the poly(A)-seq reads from each sample to the cellular plus rLCV genomes (25) demonstrated robust viral transcript detection with 224, 3,744, and 4942 viral reads per million mapped reads (Fig. 1C). These values are comparable to or higher than EBV RNA detection rates in clinical isolates of EBV-positive lymphomas and stomach cancers (2, 3, 6), indicating likely true tumor cell infection and virus etiology.
FIG 1.
(A) SIV titers in cerebrospinal fluid (CSF) and plasma through 84 days after SIV infection. (B) Hematoxylin and eosin (H&E) staining of lymphoma slides shows high tumor cell distributions. (C) rLCV reads per million mapped reads from poly(A) RNA-seq for each lymphoma specimen (T1, tumor 1; T2, tumor 2; T3, tumor 3). (D) rLCV gene expression in lymphoma samples using the lytic gene classification scheme reported by Djavadian et al. (50). Expression is plotted as log2(transcripts per million [TPM] total cellular plus viral transcripts + 1).
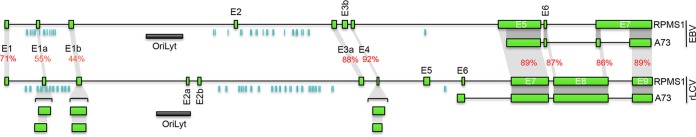
While the bulk of the reading frames across the rLCV genome have been annotated (25), rLCV transcript structures, to our knowledge, have not been globally assessed (as has been done for EBV [27]). Further, the noncoding RPMS1 and A73 genes found in EBV have not been identified or annotated in the rLCV genome build (25). Since we had previously identified EBV circRNAs derived from the EBV RPMS1 locus both in cell lines and in stomach cancer tumor tissue (23), we first utilized our poly(A)-seq and RNase R-seq coverage and splice junction data to produce a tentative working exonic structure for both of these transcripts (Fig. 2; see https://github.com/flemingtonlab/public/blob/master/annotation/rLCV_inverted.fa and https://github.com/flemingtonlab/public/blob/master/annotation/rLCV_inverted.bed for the resulting rLCV genome fasta and annotation files).
FIG 2.
Schematic comparison of the exon architecture of the RPMS1/A73 locus for EBV and rLCV. Orthologous exons are connected by gray shading. EBV and rLCV microRNAs (10, 51) are represented by aquamarine vertical bars.
Using the poly(A) sequencing data, we first assessed the viral transcription profiles for each sample. Notable in the two lymph node samples was high expression of lytic genes, with particularly high expression of many early genes such as BMLF1, BMRF2, and BNLF2a (Fig. 1D). The jejunum tumor sample displayed lower but readily detectable early lytic gene expression but was distinct in its utilization of the Cp latency promoter, expression of EBNA2, low-level expression of the other type III latency EBNAs (EBNA1, -3A, -3B, and -3C), and low levels of LMP2 (Fig. 1D). This indicated that among these three specimens, at least two distinct viral transcription programs were at our disposal for interrogating viral circRNA expression.
Detection of viral circRNAs in rLCV lymphomas.
To assess the rLCV circRNAome in these LCV-positive lymphoma samples, we analyzed the three RNase R sequencing data sets for backsplicing across the rLCV genome using find_circ (28). Backsplice reads extending from RPMS1 exon 5 to exon 3a were found to be the most abundant and were observed in all three samples (Fig. 3). Notably, two exon 5-to-exon 3a backsplice isoforms were detected that were derived from distinct exon 5 splice donor sites located only 7 bases apart (Fig. 3, denoted E5′ and E5). Backspliced reads extending from RPMS1 exon 2b to 1b were also observed in the two “lytic” tumor samples, and again two backsplice variants were detected, derived from alternative splice donor sites in exon 2b (Fig. 3). Lastly, we detected backsplice reads mapping to the EBNA U exon (29, 30), revealing an rLCV homologue of the previously reported EBV circEBNA_U circular RNA (Fig. 3) (23).
FIG 3.
rLCV circular RNAs identified in lymphoma specimens. Genome coordinates are with respect to the NC_006146 macacine gammaherpesvirus 4 genome (25), in which the genome start position was shifted to nucleotide position 102091 of NC_006146 (as described in Materials and Methods) to allow for better assessment of LMP2A transcripts that span the genome split point of NC_006146. Color intensity represents number of read counts for each backsplice junction for each lymphoma sample (T1, T2, and T3). Positions of the E5, E5′, E2b, and E2b′ splice donor junctions are marked on the respective sequences.
To validate these findings in silico, we performed alignments to a genome containing the Macaca mulatta cellular chromosomes plus conjoined backsplice exon junction sequences for each of these putative circRNAs. Junction-spanning reads with at least a 12-base overlap and a minimum of 90% homology on each side of the junction were then extracted and loaded onto the integrative genomics viewer (IGV) (31) for visualization. A staggered distribution of junction-spanning reads was detected for each backsplice call shown in Fig. 3 (data not shown), demonstrating that they represent bona fide splicing events and not random cDNA ligation chimeras generated during library preparation.
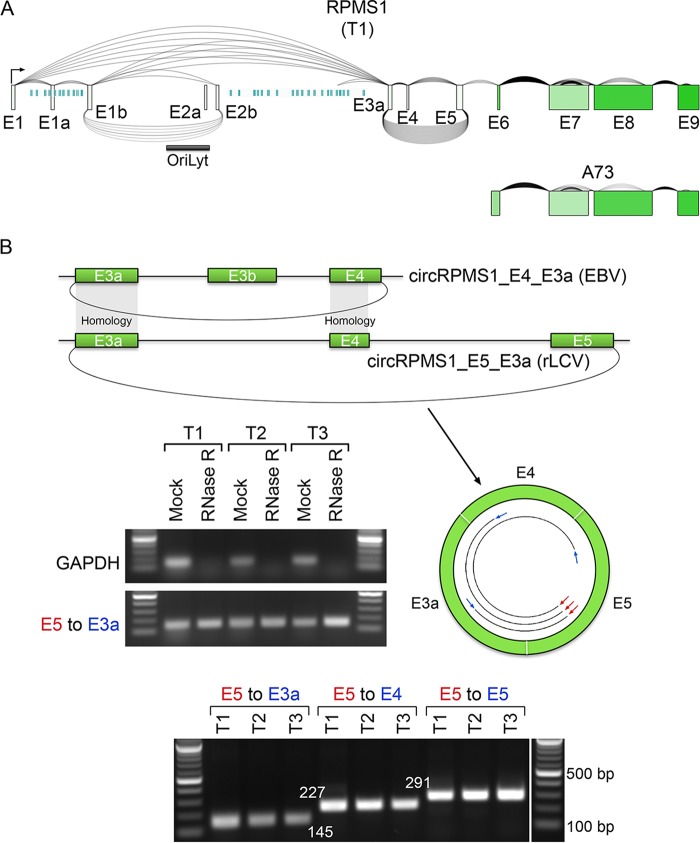
rLCV circRPMS1_E5_E3a, an rLCV orthologue of EBV circRPMS1_E4_E3a.
To validate the closed circular nature of circRPMS1_E5_E3a (Fig. 4A), RNAs from all three lymphoma samples were subjected to RNase R treatment, and exon 5- and 3a-specific divergent primers were used for PCR across the backsplice junction (Fig. 4B). While linear ACTB RNAs were found to be susceptible to RNase R digestion, no decrease in the RPMS1 exon 5-to-3a PCR signal was observed with RNase R-treated RNAs (Fig. 4B). Cloning and sequencing the RPMS1 E5-to-E3a PCR fragments revealed representation of both the RPMS1 E5′-to-E3a and the RPMS1 E5-to-E3a backsplice junctions.
FIG 4.
Validation and structure of rLCV circRPMS1_E5_E3a. (A) Graphical presentation of splicing and exon-specific coverage at the rLCV RPMS1/A73 locus for lymphoma sample T1. To provide a linear RPMS1 expression context, backsplicing read counts (underarches) derived from RNase R-seq data sets are plotted with forward splicing (overarches) and coverage data (exon color intensity) derived from poly(A)-seq data sets. The number of arches (forward and back splicing) correspond to the number of junction-spanning reads. Exon shading intensity reflects relative coverage levels across exons, and aquamarine vertical bars represent rLCV microRNAs (10, 51). (B) Validation of the circular nature and structure of rLCV circRPMS1_E5_E3a. The schematic illustrates the structural conservation between rLCV circRPMS1_E5_E3a and EBV circRPMS1_E4_E3a. Upper gels show RNase R resistance of circRPMS1_E5_E3a (E3a leftward primer and E5 rightward primer) but not linear GAPDH. The lower panel shows PCR analysis using divergent primers, exon 3a, 4, or 5 reverse primers and a common exon 5 forward primer (see schematic), demonstrating the exon 3a-to-4-to-5-to-3a configuration. PCR fragments were cloned and sequenced to validate appropriate forward- and backsplice junctions.
Visual assessment of RNase R-seq coverage across the RPMS1 exon 3a-to-exon 5 genomic region showed enriched coverage over exons 3a, 4, and 5 (data not shown). Further, forward-spliced junction reads spanning exons 3a to 4 and exons 4 to 5 were also readily detected in the RNase R-seq data (data not shown). This suggested that circRPMS1_E5′_E3a and circRPMS1_E5_E3a are composed of forward-spliced exons 3a, 4, and 5 (Fig. 4). To further assess this potential exon configuration, leftward primers specific to exons 3a, 4, and 5 and a common exon 5 rightward primer were used for reverse transcription-PCR (RT-PCR) from each of the three tumor samples (Fig. 4B). PCR fragment sizes were consistent with the circRPMS1_E5′_E3a and/or circRPMS1_E5_E3a containing consecutively forward-spliced exons 3a to 4 to 5 (Fig. 4B, bottom panel), and this was confirmed by sequencing each of the amplified fragments.
Together, these results indicate that circRPMS1_E5′_E3a and circRPMS1_E5_E3a are derived from alternative splice donors in RPMS1 exon 5, that they are expressed in all three tumor samples, and that they contain consecutively forward-spliced exons 3a, 4, and 5. While the separate evolutionary tracks of EBV and rLCV have led to species-specific circularization of some of the RPMS1 exons (for example, EBV-specific utilization of exon E3b and rLCV-specific utilization of exon 5) and distinct splice donor utilization for exon 4 (Fig. 2 and 4), the common sequences of exons 3a and 4 show high homology (88% and 92%) (Fig. 2). We therefore hypothesize that the rLCV circRPMS1_E5_E3a is a functional homologue of EBV circRPMS1_E4_E3a and that they have potentially related roles in their respective virus infection cycles.
rLCV circRPMS1_E2b_E1b spanning a lytic origin of replication.
Investigation of RPMS1 coverage and splicing information from poly(A) and RNase R sequencing data sets revealed divergent exon usage downstream from exon 1b (Fig. 2 and 5A). In rLCV, there is no evidence for the usage of the EBV RPMS1 exon 2. On the other hand, two rLCV-specific upstream “exon 2s” are detected in the two lytic SIV/rLCV lymphoma samples (Fig. 2 and 5A), with exon 2a being located within a lytic origin of replication. Whereas we found no evidence of EBV backsplicing to RPMS1 exon 1b (23), in the two lytic rLCV samples, we detected backsplicing from exons 2b to 1b (Fig. 3, 4, and 5). Using divergent primers designed against exons 2b and 1b, a product of the appropriate size was amplified, with higher levels of detection observed in the two lytic tumors (T1 and T3) (Fig. 5B). Cloning and sequencing of these fragments verified both exon 2b splice donor-to-exon 1b backsplice junctions shown in Fig. 3. The respective RNAs were resistant to RNase R digestion (Fig. 5B), indicating the circular nature of RNAs derived from RPMS1 exon 2b-to-exon 1b backsplicing.
FIG 5.
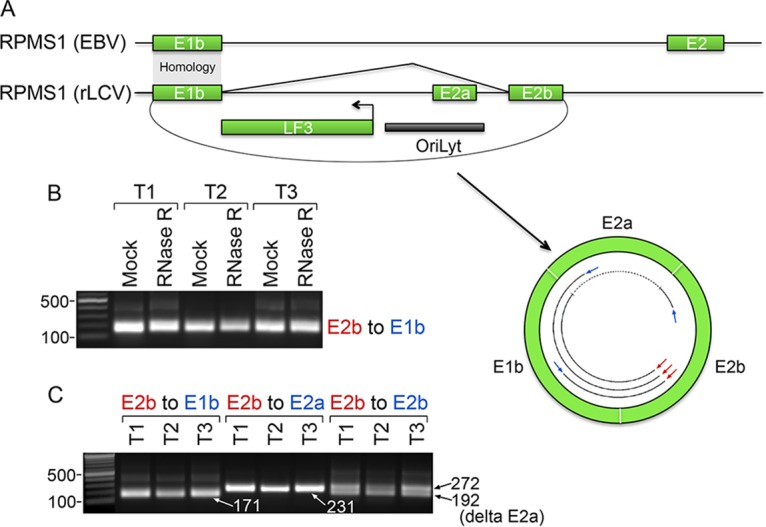
Analysis of rLCV circRPMS1_E2b_E1b. (A) Schematic illustrating the exonic structure of circRPMS1_E2b_E1b isoforms and the structural relationship with the EBV RPMS1 locus. (B) RNase R resistance of circRPMS1_E2b_E1b. PCR was performed using exon 2b and 1b divergent primers for PCR across the backsplice junction. The cDNAs used for Fig. 4B were used here, and the GAPDH PCR shown in Fig. 4B served as a control for the successful RNase R digestion of linear RNAs. (C) Divergent PCR using exon 2b, 1b, or 2a reverse primers and a common exon 2b forward primer, demonstrating the exon 2b-to-1b-to-2a-to-2b exon configuration. Also revealed in this analysis was an exon 2a skipped isoform that was observed using the E2b leftward and rightward primers (delta E2a). PCR fragments were cloned and sequenced to validate appropriate forward- and backsplice junctions.
To assess the exonic structure of circular RNAs containing the exon 2b-to-exon 1b backsplices, we designed exon 1b, 2a, and 2b leftward PCR primers and a common exon 2b rightward PCR primer (Fig. 5). RT-PCR with these three primer pairings resulted in fragment sizes that were consistent with a circular RNA containing exons 1b, 2a, and 2b, and sequencing of these fragments validated this assessment. This analysis indicated that circRPMS1_E2b/E2b′_E1b RNAs contain canonical forward-spliced exons 1b to 2a to 2b. With the exon 2b leftward primer, however, we also detected a lower-molecular-weight PCR fragment (Fig. 5C) that, upon sequencing, was found to result from exon 2a exon skipping. Therefore, at least two circRPMS1_E2b/E2b′_E1b isoforms were detected, differing in the inclusion or skipping of exon 2a. Lastly, while visualization of RNase R-seq coverage across this region showed elevated coverage at these exonic regions, there is relatively high intronic coverage. We therefore do not preclude the possibility of intron retention for some circRPMS1_E2b/E2b′_E1b isoforms (these products would be larger than the spliced products, and PCR amplification is expected to be less efficient than that for their spliced counterparts). Together, this analysis demonstrates the expression of a novel set of RPMS1 circular RNA isoforms spanning the lytic origin of replication that were not observed previously in our EBV circular RNAome analysis (23).
rLCV circEBNA_U.
Based on analysis of the poly(A)- and RNase R-seq splicing and coverage data, rLCV EBNA transcripts splice to a small noncoding exon with a high degree of exonic, flanking intronic, and splice junction sequence homology to the EBV noncoding EBNA U exon (data not shown). In our previous analysis of EBV circRNAs, we detected backsplicing of the EBNA U exon in most lymphoma-derived cell lines (23). Backsplicing analysis of the rLCV RNase R-seq data similarly showed evidence of backsplicing of the rLCV EBNA U exon (Fig. 3). Using divergent primers for PCR across the backsplice region, the band of the appropriate size was amplified in all three samples and was found by Sanger sequencing to validate the backspliced junction (Fig. 6). Further, RNAs containing the EBNA U backsplice were resistant to RNase R digestion, confirming the closed circular nature of the rLCV circEBNA_U (Fig. 6). Therefore, like its EBV counterpart, rLCV expresses a latency-associated circEBNA_U circular RNA in lymphomas.
FIG 6.
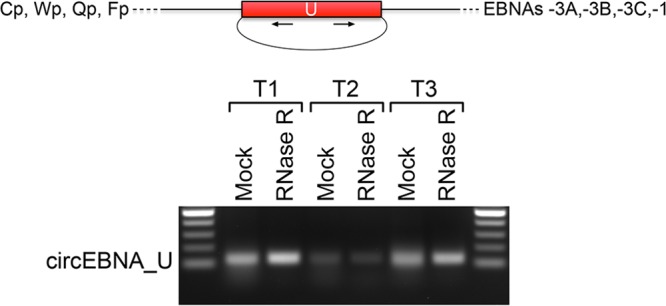
Validation of rLCV circEBNA_U. The upper schematic displays the EBNA U exon and depicts alternative promoters (Cp, Wp, Qp, and Fp) and alternative downstream EBNA coding sequences associated with EBNA U exon-containing transcripts. The gel picture shows RNase R resistance of circEBNA_U in all three tumor samples (T1, T2, and T3). The cDNAs used for Fig. 3B were used here, and the GAPDH PCR shown in Fig. 3B served as a control for the successful RNase R digestion of linear RNAs.
Detection of latently expressed circular vIRF4 transcripts in latently KSHV-infected cells of lymphoid, endothelial, and epithelial origins.
To assess whether circular RNAs are expressed from a divergent human gammaherpesvirus, we performed RNase R-seq on either untreated or tetradecanoyl phorbol acetate (TPA)-treated KSHV-positive BCBL-1 cells. Besides a cluster of low-abundance backsplice calls (5 reads or less) mapping to the PAN transcript (24, 26) and a few other sporadically distributed loci in TPA-treated BCBL1 cells, a predominant latency-associated backsplice was detected at the vIRF4 locus (32, 33), with the backsplice junctions flanking a known forward-splice junction (Fig. 7) which was also detected recently (24, 26). This backsplice was confirmed in silico through realignment of reads to conjoined backsplice junction sequences (data not shown). Coverage was restricted to the confines of the backsplice junctions, with a decrease in coverage observed within the intron (Fig. 7). Notably, although vIRF4 is defined as a lytic gene (34, 35), there is no increase in backsplice junctions observed under reactivation conditions (TPA treatment). This suggests that there is a unique transcriptional mechanism that gives rise to the observed latency expression of circvIRF4, perhaps from a previously unrecognized latency promoter.
FIG 7.
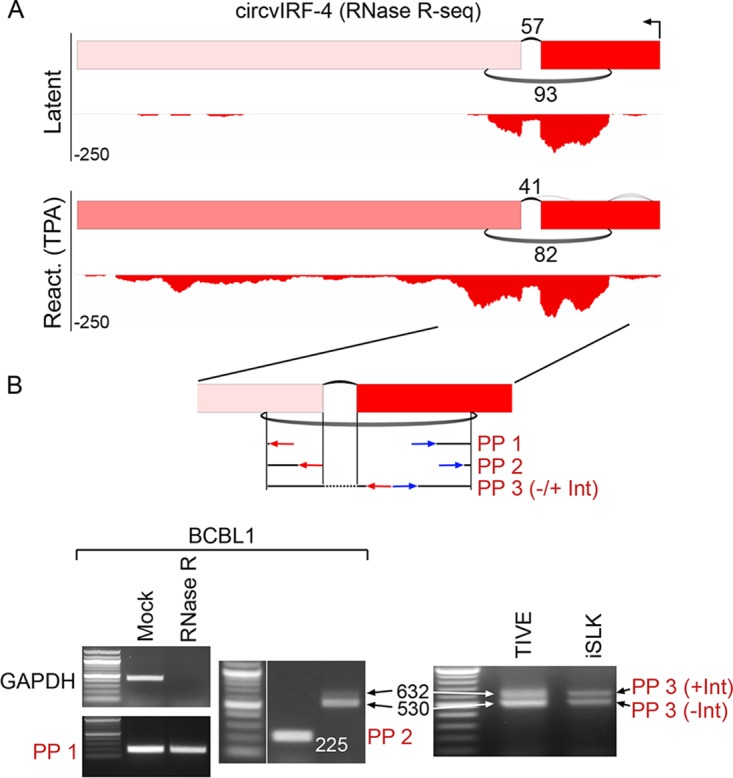
Structure of KSHV circvIRF4 isoforms. (A) Graphical presentation of RNase R-seq splicing and coverage data at the vIRF4 locus in latent and TPA-treated (reactivation) BCBL1 cells. Backsplicing read counts are represented by the number of underarches, forward splicing is displayed by overarches, and exon-level coverage data are indicated based on exon color intensity. Single-nucleotide resolution coverage data are shown below each splicing graph, with amplitudes displayed by negative numbers to represent the leftward orientation of transcription. (B) RNase R resistance and circvIRF4 isoform structure determination. Three different sets of divergent primer pairs were used, with the PP3 primer pair giving rise to two PCR fragments representing a forward-spliced version (530 bp) and an intron-retained version (632 bp) of circvIRF4. Analyses were carried out using naturally KSHV-positive BCBL1 cells and in KSHV-infected TIVE and iSLK cells.
The circular nature of the RNA containing the vIRF4 backsplice junction was verified by showing resistance to RNase R digestions (Fig. 7B). Further, excising the bands from RT-PCRs using three different primer pairs (PP 1, PP 2, and PP 3 [Fig. 7B]) validated the backsplice junction. Notably, the leftward primer for the primer pair, PP3, is located to the right of the forward-splice junction, and using this PP3 primer pair, two bands were observed in BCBL1 and KSHV-infected TIVE and iSLK cells. Sequencing of both of these bands revealed that the lower band represents a circvIRF4 isoform with the intron spliced out, while the upper band represents a circvIRF4 isoform that retains this intron. Therefore, two isoforms of circvIRF4 are generated during latency in each of these cell line models.
circRNA expression at the MHV68 OriLyt locus.
We next assessed circRNA expression in an evolutionarily distinct rhadinovirus family member, MHV68. For this, we analyzed latently infected HE2.1 B cells (36), and we analyzed lytic replication through infection of NIH 3T12 cells with MHV68 (18 h after infection at a multiplicity of infection [MOI] of 5). We detected no viral backsplice junctions in latently infected HE2.1 cells, possibly due to the lack of expression of most latency genes outside the tRNA-miRNA-encoding RNAs during latency in these cells. In lytically infected NIH 3T12 cells, we detected very low backsplice read numbers scattered across the genome, and we detected a particularly high-density cluster of more than 40 backsplices detected at low levels near an MHV68 OriLyt (37–39) and a higher-abundance intergenic ca. 11-kb backsplice extending from the M11 gene to the ORF69 gene whose backsplice junctions spanned the OriLyt (Fig. 8A). The circM11_ORF69 backsplice was validated in silico through realignment to the conjoined backsplice junction (data not shown). Further, circM11_ORF69 was found to be resistant to RNase R digestion (Fig. 8B and C). Analysis of RNase R sequencing data showed good coverage across the 11-kb region spanning the backsplice junctions except for the repeat region (repeat regions commonly cause a lack of RNA-seq coverage, likely due to technical reasons). Additional nested primers within ORF69 resulted in PCR products of the appropriate sizes (Fig. 8C), indicating inclusion of these ORF69 sequences in circM11_ORF69. Forward-spliced reads were detected in the RNase R-seq data that extended into M11, suggesting that there are a number of spliced isoforms (Fig. 8A). Nevertheless, barring other undetected splice sites, these data suggest that forward-spliced and nonspliced isoforms of circM11_ORF69 could range in size from more than 8 kb to greater than 11 kb long.
FIG 8.
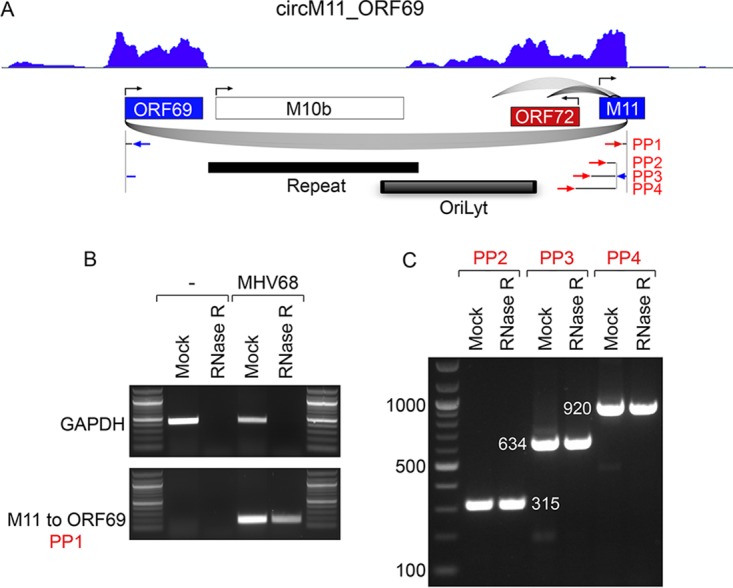
Detection and validation of lytic MHV68 circM11_ORF69. (A) Graphical representation of RNase R-seq splicing and coverage data at the ORF69 M11 locus. Backsplicing read counts are represented by the number of underarches, forward splicing is displayed by overarches, and exon-level coverage data are indicated based on exon color intensity. Single-nucleotide resolution coverage data are shown above the splicing graph, with amplitudes representing forward transcription. Primer sites used for panels B and C are indicated. (B) Uninfected (−) or infected (MHV68) NIH 3T12 cells were harvested at 18 h after infection, and RNAs were generated. The RNAs were subjected to mock or RNase R treatment, and PCR was performed using the indicated primers. (C) RNAs isolated from MHV68 infected-NIH 3T12 cells at 18 h postinfection were mock or RNase R treated, and PCR was performed using the indicated primers.
DISCUSSION
Given the recently discovered ubiquitous nature of circRNAs across all kingdoms of life, it is not unexpected to find that gammaherpesviruses of both the Lymphocryptovirus and Rhadinovirus genera carry genes that give rise to circRNAs. Despite this, however, it is important to document this previously unappreciated class of gammaherpesviral transcripts so they can be included in our efforts to understand the mechanisms of both the natural infection cycles of these viruses and their contributions to cancer. Further, the identification of individual circRNAs and our initial characterizations of their exonic structures and evolutionary conservation should provide foundational information from which to begin investigations into the functions of these RNAs in natural virus biology and disease.
From an evolutionary standpoint, our studies identify two conserved lymphocryptovirus circRNAs, circEBNA_U and circRPMS1_E4_E3a (EBV)/circRPMS1_E5/E5′_E3a (rLCV). Despite it being detected at low levels, we found EBV circEBNA_U expression in type I and type III latency B-cell models (although we have yet to detect it in any stomach cancer model tested to date) (23), and we detected the rLCV-encoded circEBNA_U in all three lymphoma samples investigated here. Its low expression could support the idea that circEBNA_U forms due to processing inefficiencies associated with upstream and/or downstream linear splicing. Even in this case, though, the evolutionary conservation between two lineages supports a possible functional significance. It is also notable that the coverage of the EBNA U exon and all other exons that make up the EBNA1 transcript is low in our poly(A)-seq data from most latency type I and type II models we have tested. The low abundance of circEBNA_U is therefore consistent with the overall expression of its parental transcript.
Visualization of the EBNA U and flanking sequence homology between EBV and rLCV shows a fairly divergent center of exon U, with greater homology near the end sequences, extending into the intronic regions (data not shown). The high homology at the intronic and exonic regions within and proximal to the splice junctions presumably maintains the structure and sequence motifs necessary to retain interactions between conserved splicing factors and splicing enhancer factors, which may in turn account for conservation of circularization. It is also notable that circular RNAs become loaded with the splicing factors involved in their genesis (40–42). As such, the observed sequence conservation of flanking introns and splice junction regions is consistent with the loading of many of the same factors onto circEBNA_U for both EBV and rLCV. Since these associated splicing factor and splicing enhancer factor cargo/effectors likely play key roles in facilitating circRNA function, this suggests conserved biological roles for circEBNA_U across these two viral relatives.
One of the notable observations arising from our analysis of the rLCV RPMS1 exon configurations was the finding of substantial variations in exon utilization between rLCV and EBV (Fig. 2). EBV utilizes two exons, E2 and E3b that we find no evidence for in any of the three rLCV lymphoma samples. Conversely, splicing analysis in the rLCV lymphoma samples supported the existence of four novel exons not detected in EBV (Fig. 2). Among RPMS1 exons that are not part of the A73 isoform (i.e., are unique to the RPMS1 isoform), it is notable that in addition to differential utilization of exons, there is relatively low homology of exons 1, 1a, and 1b. In contrast, exons 3a and 4, which are components of the EBV circRPMS1_E4_E3a and rLCV circRPMS1_E5_E3a circular RNAs, display 88% and 92% homology (Fig. 2), with homologies extending into the intronic and splice junction signaling sequences. While the high homologies between these RPMS1 exonic regions may be driven largely by selective pressure to maintain the LF2 open reading frame on the opposite strand, the homologous exonic and flanking intronic sequences important for loading of splicing factors to the respective circRNAs would nevertheless likely facilitate similar functioning of the EBV circRPMS1_E4_E3a and rLCV circRPMS1_E5_E3a circular RNAs through their bound protein cargo.
We previously demonstrated the expression of EBV circRPMS1_E4_E3a and circRPMS1_E4_E2 in two stomach cancer patient samples (23). Assessment of 4 additional stomach cancer patient samples similarly showed expression of both circRPMS1_E4_E3a and circRPMS1_E4_E2 (Fig. 9; T3 and T5 are samples that we previously assayed [23], while T1, T2, T4, and T6 represent findings in 4 new stomach cancer specimens). In addition, Toptan et al. (24) demonstrated expression of circRPMS1_E4_E3a and circRPMS1_E4_E2 in a panel of posttransplant lymphoproliferative disease patient specimens. The findings of broad expression of circRPMS1_E4_E3a and circRPMS1_E4_E2 across a panel of type I, type II, and type III cell lines (23) and in primary stomach cancer and PTLD patient samples (Fig. 9) (23, 24), and our findings here identifying an rLCV homologue in clinical lymphoma specimens, speak to a likely conserved, ubiquitous and clinically relevant potential function for these viral circRNAs in EBV/rLCV biology and associated cancers.
FIG 9.
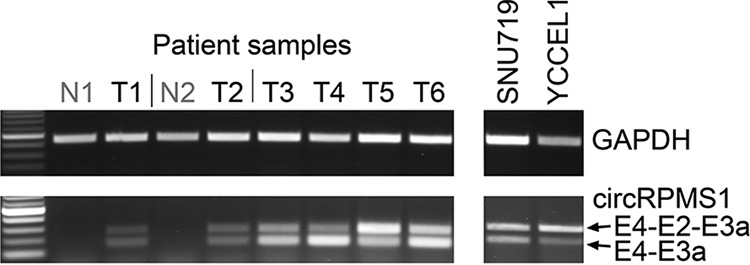
Expression of the rLCV circRPMS1_E5/E5′_E3a EBV homologues, circRPMS1_E4_E2 (upper band) and circRPMS1_E4_E3a (lower band), in primary stomach cancer specimens. N1 and N2 are normal adjacent tissue samples corresponding to the T1 and T2 tumor samples, respectively. Tumor samples T3 and T5 were assessed previously for circRPMS1_E4_E2 and circRPMS1_E4_E3a and similarly found to be positive (23). SNU719 and YCCEL1 are naturally EBV-infected stomach cancer cell lines.
We previously identified a highly expressed EBV circRNA derived from the lytic BHLF1 gene located next to one of the two EBV lytic origins of replication (23). Under reactivation conditions, this circRNA was among the top 5 most abundant circRNAs in the cell (23), and under the majority of latency conditions, low-level circBHLF1 was detected, possibly due to a small percentage of spontaneously reactivating cells among the latency population or low-level latency expression (43). Based on findings of Rennekamp et al. (44) that some undefined BHLF1 RNA isoforms bind to and activate the lytic origin of replication, we previously hypothesized that the circular BHLF1 isoforms are reasonable candidates for such a role (23). In our analysis of the rLCV circRNAome, however, we did not detect backsplice reads mapping to the corresponding BHLF1 region of rLCV in any of the three lymphoma samples assayed here despite the observation that two of these samples displayed relatively robust levels of lytic gene expression. Nevertheless, it is notable that circRPMS1_E2b_E1b spans the other rLCV lytic origin of replication and is detected uniquely in the two samples displaying lytic activity but not in the sample with the more-latent viral transcription program. In addition, we found that the predominant backsplices detected in MHV68 were a cluster arising from near one of the MHV68 lytic origins of replication during lytic infection, with the junctions of the most abundant circular RNA, circM11_ORF69, spanning this lytic origin of replication (Fig. 8). While these relationships may be coincidental or the process of replication may drive backsplicing, it also leaves open the possibility of homoplastic evolution of these circRNA/OriLyt pairings, with proximal viral circRNAs playing active roles in facilitating lytic viral DNA replication.
The KSHV vIRF4 gene is classified as a lytic gene that plays a role in regulating viral lytic gene expression (32–35, 45, 46). It is notable, however, that under latency conditions, our BCBL-1 RNase R sequencing coverage data show a strong bias for reads specifically within the confines of the circvIRF4 backsplice junctions (Fig. 7). Under reactivation conditions, however, there is an increase in coverage, presumably from inefficiently RNase R-digested linear vIRF4 transcripts, but little increase in the region within the circvIRF4 backsplices. Further, there is no observed increase in the number of backspliced reads in TPA-treated cells. This suggests that while vIRF4 linear transcripts are likely induced during reactivation, circvIRF4 is expressed as a noncoding latency transcript and may play a role in modulating the host cell environment during latency and in KSHV-associated malignancies.
Taking the results together, we have shown that both the Rhadinovirus and Lymphocryptovirus genera of gammaherpesviruses express a diverse set of circRNAs. From this limited evolutionary assessment of the lymphocryptovirus genus, we have been able to identify conservation of circEBNA_U and a variation of the EBV circRPMS1_E4_E3a circular RNA that we have detected nearly universally in our tissue culture models and in primary stomach cancers. Testing of additional rLCV samples (tissue culture and primary tissue models) as well as LCVs with other species tropisms will further address our contention of circEBNA_U and circRPMS1_E4_E3a (EBV)/circRPMS1_E5_E3a (rLCV) conservation and at the same time may reveal conservation of additional viral circRNAs detected previously for EBV (23). Further, the analysis of other rhadinoviruses that are more closely related to KSHV or MHV68 may yield insights into the possible importance of the KSHV-encoded circvIRF4 latency circRNA and the lytic-associated MHV68 circM11_ORF69 circRNA. Together, these evolutionary studies will set the stage for beginning investigations into the function and importance of these viral circRNAs in virus biology and associated cancers.
MATERIALS AND METHODS
Cell culture and infections.
BCBL1 and SNU719 (Korean Cell Line Bank) cells were grown in RPMI 1640 medium (Fisher Scientific, catalog no. SH30027) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher, catalog no. 10437). YCCEL1 cells (Korean Cell Line Bank) were cultured in Eagle’s minimum essential medium (EMEM) (ATCC, catalog no. 30-2003) supplemented with 10% FBS. All cells were cultured at 37°C in a 5% CO2 incubator. For lytic MHV68 infections, NIH 3T12 cells (grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum) were infected with MHV68 at a multiplicity of infection (MOI) of 5 and harvested at 18 h postinfection for RNA preparation.
Animal model.
An adult male (14 years) Indian rhesus macaque (MHC genotype: Mamu-A*02, Mamu-A*08, and Mamu-B*01 positive and Mamu-A*01, Mamu-A*11, Mamu B*03, Mamu B*04, Mamu B*08, and Mamu B*17 negative) received twice-daily oral doses (60 mg and 30 mg) of dimethylfumarate for 7 days prior to intravenous inoculation with SIV (SIVmac251, 100 TCID50). The animal then received three successive doses of anti-CD8 antibody (10 mg/kg, 5 mg/kg, and 5 mg/kg) at days 6, 9, and 13 postinfection. Plasma and cerebrospinal fluid (CSF) samples were collected at several time points up until the day of autopsy (day 84 postinfection) for analysis of SIV RNA levels.
At autopsy, lymphosarcoma was identified in all lymphoid tissues examined, including the tonsils, and nodes within the jejunum, cecum, and colon. Lymphocytic infiltration was observed in the kidney, liver, adrenal gland, lung, and sciatic nerve, consistent with lymphocytic inflammation. The brain demonstrated granulomatous encephalitis, and multinucleated giant cells were observed in the brain, brain stem, and cervical spinal cord. Mild to moderate amyloidosis was found in the liver and spleen. The Indian-origin rhesus macaque (Macaca mulatta) utilized in this study was housed at the Tulane National Primate Research Center (TNPRC). The TNPRC is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) International under Animal Welfare Assurance no. A3180-01. Animals were cared for in accordance with the National Research Council's Guide for the Care and Use of Laboratory Animals (52) and the Animal Welfare Act. Animal experiments were approved by the Institutional Animal Care and Use Committee of Tulane University.
RNA preparations.
Whole-cell RNA preparations were carried out using TRIzol reagent (Thermo Fisher, catalog no. 15596) according to the vendor’s recommended protocol. For tumor and normal tissue, pieces were first ground finely using a mortar and pestle in liquid nitrogen prior to disruption with TRIzol reagent. Nuclear and cytoplasmic RNAs were isolated using the Cytoplasmic & Nuclear RNA purification kit from Norgen Biotek Corp. (catalog no. 21000) according to the vendor’s protocol. All RNA preparations were subjected to DNase treatment twice using the DNA-free kit (Thermo Fisher, catalog no. AM1906).
RNA sequencing.
RNA sequencing was performed at the Beijing Genomics Institute (BGI). For poly(A)-seq, RNAs were selected using a poly(dT) column, and for RNase R-seq, RNAs were subjected to DNA and rRNA depletion, followed by linear RNA depletion using RNase R. For all sequencing experiments, TruSeq stranded libraries were generated and sequenced using 2 × 100-base sequencing on a HiSeq 4000 system.
Backsplice junction analysis.
Backsplicing was analyzed using find_circ (28) with default parameters. For analysis of BCBL1 cell RNase R-seq data, alignments were carried out using the human hg38 genome build plus the NC_009333 human herpesvirus 8 genome (47). Alignment for backsplicing analysis of rhesus lymphoma samples was performed using the Macaca mulatta mmul8.01 genome build plus the NC_006146 macacine gammaherpesvirus 4 genome (25), which was split at nucleotide position 102091 rather than the beginning of the terminal repeats (position 1) to allow for assessment of splicing across the LMP2 locus that spans the terminal repeats. For MHV68 backsplicing analyses, RNase R-seq data were aligned to the mouse mm10 genome build plus the MHV68 genome (48). For in silico validations, STAR (49) genome indices were generated containing the human hg38 (BCBL1), Macaca mulatta (rhesus lymphomas), or murine mm10 genome builds plus the respective conjoined backsplice junctions for KSHV, LCV, or MHV68 identified by find_circ. Raw fastq files were aligned to the respective genome indices using STAR (-outFilterMultimapNmax 20 -outSAMtype BAM SortedByCoordinate -outWigType wiggle -outWigNorm None), reads spanning backsplice junctions with a minimum of a 12-base overlap (minimum of 90% homology) on each side of the junction were pulled out for visualization on the Integrative Genomics Viewer (IGV) (31), and the number of reads mapping to each junction was quantified for reporting.
Splice junction and expression exon structure plots.
For canonical splicing and coverage display, RNA-seq data from poly(A)+ or RNase R sequencing libraries were analyzed by STAR alignment against the respective cellular plus viral genomes using STAR -outFilterMultimapNmax 20 -outSAMtype BAM SortedByCoordinate -outWigType wiggle -outWigNorm None). Splice junction data from .SJ files and wiggle output files were used to generate junction read numbers and coverage information. Backsplice read counts were extracted from .bed junction count files derived from find_circ output. Forward splicing, coverage, and backsplicing were visualized together using the software SpliceV (53). For these plots (Fig. 4A, 7A, and 8A), exon-level coverage is represented by color intensity, with canonical and backsplice junction curves plotted above and below the exon diagram, respectively.
RNase R resistance analysis.
Five micrograms of total RNA was incubated with or without 60 units of RNase R (Lucigen, catalog no. RNR07250). Briefly, no RNase R (control) or 1.5 μl RNase R (30 units) and 3 μl of 10× RNase R buffer were added to 5 μg of RNA in a total volume of 30 μl and incubated in a 37°C water bath for 30 min. Either no RNase R (control) or 1.5 μl (30 units) more RNase R was added to the reaction mixtures and incubated for an additional 1.5 h in a 37°C water bath. RNAs were then cleaned and concentrated using the RNA Clean & Concentrator-5 kit (Zymo Research, catalog no. R1015) and eluted in 10 μl H2O, 9 μl of which was used for cDNA preparations (see below).
RT-PCR.
cDNA was synthesized using the SuperScript IV first-strand synthesis system (Thermo Fisher, catalog no. 18091) or ProtoScript II reverse transcriptase (New England Biolabs, MA, USA), and the cDNAs were amplified by taq-PCR (Thermo Fisher, catalog no. 11304) following the vendor’s protocol. PCR products were run on a 1% agarose gel at room temperature. PCR products were cut out and purified using the NucleoSpin Gel & PCR Clean-up kit (Clontech, catalog no. 740609). The resulting PCR fragments were cloned into the pCR4-TOPO vector (Thermo Fisher, catalog no. 450030), and the inserts were Sanger sequenced.
The following PCR primers were used: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for (human/mouse), 5′-ACCACAGTCCATGCCATCAC; GAPDH for (rhesus), 5′-TGGCCAAGGTCATCCATGACA; GAPDH rev (human/mouse/rhesus), 5′-TCCACCACCCTGTTGCTGTA; rhesus RPMS1 E5 for, 5′-GAGCACCAGGGCAAAGAC; rhesus RPMS1 E3a rev, 5′-CACGACTCCGTTCCTGAAGT; rhesus RPMS1 E4 rev, 5′-AGGAGCCCATGCAGCACTA; rhesus RPMS1 E5 rev, 5′-GCTATCTCCTGGCGGGTATC; rhesus RPMS1 E1b for, 5′-AAGCACCACAGACACGAGA; rhesus RPMS1 E2a for, 5′-GGAAGCGTGGACCCAGA; rhesus RPMS1 E2b for, 5′-GCCAGGACTGGTACCTGAGA; rhesus RPMS1 E1b rev, 5′-GGTTGGGCCGTTTCCTAC; rhesus EBNA-U for, 5′-AGACCGTCGCGTCGTAGA; rhesus EBNA-U rev, 5′-GCAGAATCAGCTCTCCCAGA; KSHV circvIRF4-PP1-for, 5′-CTCCGTGTGGATACCAGTGA; KSHV circvIRF4-PP1-rev, 5′-TGGTTCCACGCAACAGTCT; KSHV circvIRF4-PP2-for, 5′-AGAACAAAGCTACGAGGAGGCA; KSHV circvIRF4-PP2-rev, 5′-GAATACCAGCCAGGCGGGATA; KSHV circvIRF4-PP3-for, 5′-AACCACGGCTACGCGACG; KSHV circvIRF4-PP3-rev, 5′-TGCATTGGGGGGGACAAC; MHV68 circM11_ORF69-PP1-for, 5′-GGCACTATGACAGCGTTTACC; MHV68 circM11_ORF69-PP1-rev, 5′-CTCTCGCCAGAGCAGGAT; MHV68 circM11_ORF69-PP2-for, 5′-ATGAGTCATAAGAAAAGCGGGA; MHV68 circM11_ORF69-PP3-for, 5′-TCGCTGCGATAGATCATCTG; MHV68 circM11_ORF69-PP4-for, 5′-TGCTCCTCCACAAAGGTATG; MHV68 circM11_ORF69-PP2,3,4-rev, 5′-ATGGAGCAGAGCCTCCTCACACA; EBV circRPMS1 E4 forward, 5′-CTAGTGCTGCATGGGCTCCT; and EBV circRPMS1 E3a reverse, 5′-GTCATACGCCCGTATTCACA.
Data availability.
All RNA sequencing data have been deposited in the NCBI GEO public database (accession number GSE124711).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01AI101046 (E.K.F.), R01AI106676 (E.K.F.), R01CA188609 (Y.D.), R00GM112877 (W.N.M.), R01AI108407 (S.A.T.), R01CA119917 (R.R.), and P01CA214091 (R.R., S.A.T., and E.K.F.) and Department of Defense grants W81XWH-16-1-0318 (E.K.F.) and W81XWH-16-1-0317 (Y.D.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Terri Lehman and Michael Seddon are currently employed by Reprocell Inc. Reprocell Inc. provided funds, resources, and equipment to conduct some of the studies and provided salary and benefits for Terri Lehman and Michael Seddon. This does not alter our adherence to all Journal of Virology policies on sharing data and materials.
REFERENCES
- 1.Strong MJ, Laskow T, Nakhoul H, Blanchard E, Liu Y, Wang X, Baddoo M, Lin Z, Yin Q, Flemington EK. 2015. Latency expression of the Epstein-Barr virus-encoded MHC class I TAP inhibitor, BNLF2a in EBV-positive gastric carcinomas. J Virol 89:10110–10114. doi: 10.1128/JVI.01110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strong MJ, O'Grady T, Lin Z, Xu G, Baddoo M, Parsons C, Zhang K, Taylor CM, Flemington EK. 2013. Epstein-Barr virus and human herpesvirus 6 detection in a non-Hodgkin's diffuse large B-cell lymphoma cohort by using RNA sequencing. J Virol 87:13059–13062. doi: 10.1128/JVI.02380-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR, Strong AL, Lehman TA, Seddon MB, Lin Z, Concha M, Baddoo M, Ferris M, Swan KF, Sullivan DE, Burow ME, Taylor CM, Flemington EK. 2013. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog 9:e1003341. doi: 10.1371/journal.ppat.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu L, Lin Z, Wu Y, Dong J, Zhao B, Cheng Y, Huang P, Xu L, Xia T, Xiong D, Wang H, Li M, Guo L, Kieff E, Zeng Y, Zhong Q, Zeng M. 2016. Comprehensive profiling of EBV gene expression in nasopharyngeal carcinoma through paired-end transcriptome sequencing. Front Med 10:61–75. doi: 10.1007/s11684-016-0436-0. [DOI] [PubMed] [Google Scholar]
- 5.Arvey A, Ojesina AI, Pedamallu CS, Ballon G, Jung J, Duke F, Leoncini L, De Falco G, Bressman E, Tam W, Chadburn A, Meyerson M, Cesarman E. 2015. The tumor virus landscape of AIDS-related lymphomas. Blood 125:e14. doi: 10.1182/blood-2014-11-599951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR. 2012. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog 8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 10.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HL, Lung MM, Sham JS, Choy DT, Griffin BE, Ng MH. 1992. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology 191:193–201. doi: 10.1016/0042-6822(92)90181-N. [DOI] [PubMed] [Google Scholar]
- 12.Marquitz AR, Mathur A, Edwards RH, Raab-Traub N. 2015. Host gene expression is regulated by two types of noncoding RNAs transcribed from the Epstein-Barr virus BamHI A rightward transcript region. J Virol 89:11256–11268. doi: 10.1128/JVI.01492-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe JG, Steitz JA. 1986. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc Natl Acad Sci U S A 83:9006–9010. doi: 10.1073/pnas.83.23.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lerner MR, Andrews NC, Miller G, Steitz JA. 1981. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A 78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss WN, Lee N, Pimienta G, Steitz JA. 2014. RNA families in Epstein-Barr virus. RNA Biol 11:10–17. doi: 10.4161/rna.27488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolakofsky D. 1976. Isolation and characterization of Sendai virus DI-RNAs. Cell 8:547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- 17.Hsu MT, Coca-Prados M. 1979. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 18.Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. 1980. Some yeast mitochondrial RNAs are circular. Cell 19:313–319. doi: 10.1016/0092-8674(80)90505-X. [DOI] [PubMed] [Google Scholar]
- 19.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. 2015. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Kristensen LS, Okholm TLH, Veno MT, Kjems J. 2018. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol 15:280–291. doi: 10.1080/15476286.2017.1409931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M, Lehman T, Tibbetts S, Renne R, Dong Y, Flemington EK. 2018. The Epstein Barr virus circRNAome. PLoS Pathog 14:e1007206. doi: 10.1371/journal.ppat.1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, Shair KH, Moore PS, Chang Y. 2018. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1811728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivailler P, Jiang H, Cho YG, Quink C, Wang F. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J Virol 76:421–426. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, Serquina AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan R, Ziegelbauer JM. 2018. Discovery of Kaposi's sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1816183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Grady T, Wang X, Bentrup KHZ, Baddoo M, Concha M, Flemington EK. 2016. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res 44:e145. doi: 10.1093/nar/gkw629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 29.Sample J, Brooks L, Sample C, Young L, Rowe M, Gregory C, Rickinson A, Kieff E. 1991. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc Natl Acad Sci U S A 88:6343–6347. doi: 10.1073/pnas.88.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speck SH, Strominger JL. 1985. Analysis of the transcript encoding the latent Epstein-Barr virus nuclear antigen I: a potentially polycistronic message generated by long-range splicing of several exons. Proc Natl Acad Sci U S A 82:8305–8309. doi: 10.1073/pnas.82.24.8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham C, Barnard S, Blackbourn DJ, Davison AJ. 2003. Transcription mapping of human herpesvirus 8 genes encoding viral interferon regulatory factors. J Gen Virol 84:1471–1483. doi: 10.1099/vir.0.19015-0. [DOI] [PubMed] [Google Scholar]
- 33.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanno T, Sato Y, Sata T, Katano H. 2006. Expression of Kaposi's sarcoma-associated herpesvirus-encoded K10/10.1 protein in tissues and its interaction with poly(A)-binding protein. Virology 352:100–109. doi: 10.1016/j.virol.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Katano H, Sato Y, Kurata T, Mori S, Sata T. 2000. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology 269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 36.Forrest JC, Speck SH. 2008. Establishment of B-cell lines latently infected with reactivation-competent murine gammaherpesvirus 68 provides evidence for viral alteration of a DNA damage-signaling cascade. J Virol 82:7688–7699. doi: 10.1128/JVI.02689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adler H, Steer B, Freimuller K, Haas J. 2007. Murine gammaherpesvirus 68 contains two functional lytic origins of replication. J Virol 81:7300–7305. doi: 10.1128/JVI.02406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng H, Chu JT, Park NH, Sun R. 2004. Identification of cis sequences required for lytic DNA replication and packaging of murine gammaherpesvirus 68. J Virol 78:9123–9131. doi: 10.1128/JVI.78.17.9123-9131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong D, Qi J, Arumugaswami V, Sun R, Deng H. 2009. Identification and functional characterization of the left origin of lytic replication of murine gammaherpesvirus 68. Virology 387:285–295. doi: 10.1016/j.virol.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY. 2017. Sensing self and foreign circular RNAs by intron identity. Mol Cell 67:228–238.e225. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. 2017. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell 67:214–227.e217. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 42.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes DJ, Dickerson CA, Shaner MS, Sample CE, Sample JT. 2011. trans-repression of protein expression dependent on the Epstein-Barr virus promoter Wp during latency. J Virol 85:11435–11447. doi: 10.1128/JVI.05158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rennekamp AJ, Lieberman PM. 2011. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J Virol 85:2837–2850. doi: 10.1128/JVI.02175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinzelmann K, Scholz BA, Nowak A, Fossum E, Kremmer E, Haas J, Frank R, Kempkes B. 2010. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4/K10) is a novel interaction partner of CSL/CBF1, the major downstream effector of Notch signaling. J Virol 84:12255–12264. doi: 10.1128/JVI.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xi X, Persson LM, O'Brien MW, Mohr I, Wilson AC. 2012. Cooperation between viral interferon regulatory factor 4 and RTA to activate a subset of Kaposi's sarcoma-associated herpesvirus lytic promoters. J Virol 86:1021–1033. doi: 10.1128/JVI.00694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezaee SA, Cunningham C, Davison AJ, Blackbourn DJ. 2006. Kaposi's sarcoma-associated herpesvirus immune modulation: an overview. J Gen Virol 87:1781–1804. doi: 10.1099/vir.0.81919-0. [DOI] [PubMed] [Google Scholar]
- 48.Virgin HWt, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71:5894–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Djavadian R, Hayes M, Johannsen E. 2018. CAGE-seq analysis of Epstein-Barr virus lytic gene transcription: 3 kinetic classes from 2 mechanisms. PLoS Pathog 14:e1007114. doi: 10.1371/journal.ppat.1007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skalsky RL, Kang D, Linnstaedt SD, Cullen BR. 2014. Evolutionary conservation of primate lymphocryptovirus microRNA targets. J Virol 88:1617–1635. doi: 10.1128/JVI.02071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 53.Ungerleider NA, Flemington EK. 2019. SpliceV: analysis and publication quality printing of linear and circular RNA splicing, expression and regulation. bioRxiv doi: 10.1101/509661. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All RNA sequencing data have been deposited in the NCBI GEO public database (accession number GSE124711).