In plants, the RNA-directed DNA methylation (RdDM) pathway is a natural antiviral defense mechanism against geminiviruses. However, how geminiviruses counter RdDM-mediated defense is largely unknown. Our findings reveal that Cotton leaf curl Multan virus V2 contributes to viral infection by interaction with NbAGO4 to suppress RNA-directed DNA methylation-mediated transcriptional gene silencing in plants. Our work provides the first evidence that a geminiviral protein is able to directly target core RdDM components to counter RdDM-mediated TGS antiviral defense in plants, which extends our current understanding of viral counters to host antiviral defense.
KEYWORDS: Argonaute 4, CLCuMuV, RdDM pathway, TGS, V2
ABSTRACT
In plants, RNA-directed DNA methylation (RdDM)-mediated transcriptional gene silencing (TGS) is a natural antiviral defense against geminiviruses. Several geminiviral proteins have been shown to target the enzymes related to the methyl cycle or histone modification; however, it remains largely unknown whether and by which mechanism geminiviruses directly inhibit RdDM-mediated TGS. In this study, we showed that Cotton leaf curl Multan virus (CLCuMuV) V2 directly interacts with Nicotiana benthamiana AGO4 (NbAGO4) and that the L76S mutation in V2 (V2L76S) abolishes such interaction. We further showed that V2, but not V2L76S, can suppresses RdDM and TGS. Silencing of NbAGO4 inhibits TGS, reduces the viral methylation level, and enhances CLCuMuV DNA accumulation. In contrast, the V2L76S substitution mutant attenuates CLCuMuV infection and enhances the viral methylation level. These findings reveal that CLCuMuV V2 contributes to viral infection by interaction with NbAGO4 to suppress RdDM-mediated TGS in plants.
IMPORTANCE In plants, the RNA-directed DNA methylation (RdDM) pathway is a natural antiviral defense mechanism against geminiviruses. However, how geminiviruses counter RdDM-mediated defense is largely unknown. Our findings reveal that Cotton leaf curl Multan virus V2 contributes to viral infection by interaction with NbAGO4 to suppress RNA-directed DNA methylation-mediated transcriptional gene silencing in plants. Our work provides the first evidence that a geminiviral protein is able to directly target core RdDM components to counter RdDM-mediated TGS antiviral defense in plants, which extends our current understanding of viral counters to host antiviral defense.
INTRODUCTION
Gene silencing is an evolutionarily conserved mechanism to regulate gene expression in eukaryotes (1, 2). In plants, transcriptional gene silencing (TGS) can be mediated by RNA-directed DNA methylation (RdDM). The establishment of RdDM entails the generation of double-stranded RNAs (dsRNAs) by RNA polymerase IV (Pol IV) and RNA-dependent RNA polymerase 2 (RDR2). dsRNAs are processed by Dicer-like 3 (DCL3) into 24-nucleotide (nt) small interfering RNAs (siRNAs), which are loaded into Argonaute 4 (AGO4) to form the core of the effector complex (3–11). Through base pairing with a scaffold transcript produced by Pol V, the AGO4/siRNA complex is recruited to target loci to direct de novo cytosine methylation (12–14).
RdDM plays a vital role in defending against DNA viruses, including geminiviruses, in plants (15, 16). AGO4 can bind virus-derived siRNAs (vsiRNAs) to mediate the methylation of viral DNA and TGS, resulting in the repression of viral transcription and replication (17). To counter the defense, geminiviruses encode suppressors of TGS. These include Tomato leaf curl Yunnan virus (TLCYNV) C4, Mungbean yellow mosaic India virus (MYMIV) AC5, Tomato yellow leaf curl Sardinia virus (TYLCSV) Rep, Tomato yellow leaf curl virus (TYLCV) V2, and Cotton leaf curl Kokhran virus (CLCKV) Rep, TrAP and βC1 (18–22). However, the mechanisms by which most geminiviral proteins suppress TGS are unknown, except for several examples. Some geminiviral proteins interfere with the methyl cycle-related enzymes to affect S-adenosylmethionine (SAM) production for TGS suppression (23–26). AL2 of Tomato golden mosaic virus (TGMV) and L2 of Beet curly top virus (BCTV) interact with and inactivate adenosine kinase (ADK) (25). Tomato yellow leaf curl China virus (TYLCCNV) βC1 interacts with S-adenosylhomocysteine hydrolase and inhibits its activity in vitro (24). BCTV C2 attenuates the degradation of SAM decarboxylase 1 (SAMDC1) (23). Cotton leaf curl Multan virus (CLCuMuV) C4 interacts with SAM synthetase (SAMS) and inhibits its activity (26). In addition, C2/TrAP from Beet severe curly top virus (BSCTV), TGMV, and Cabbage leaf curl virus (CbLCV) inhibit TGS by interacting with the H3K9 histone methyltransferase SUVH4/KYP and inhibiting its activity (27, 28). TYLCV V2 suppresses TGS by interacting with host histone deacetylase HDA6 (29).
In this study, we report that RdDM plays a role in defense against Cotton leaf curl Multan virus (CLCuMuV) and that CLCuMuV encodes V2 protein to interacts with AGO4 and suppress RdDM. Thus, we provide evidence that a DNA virus protein promotes virus infection by directly targeting an RdDM component to counter the RdDM-mediated antiviral defense in plants.
RESULTS
Identification of NbAGO4 as a CLCuMuV V2-interacting protein.
To investigate the role of V2 in CLCuMuV infection, green fluorescent protein (GFP)-Trap beads coupled with mass spectrometry were used to identify CLCuMuV V2-interacting plant proteins. We transiently expressed GFP-tagged CLCuMuV V2 (GFP-V2) via agroinfiltration in Nicotiana benthamiana leaves. N-terminally tagged V2 is functional for inhibition of TGS (see below). We then performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) and obtained an LC-MS/MS polypeptide profile. When using this profile to search for candidate V2-interacting proteins against the N. benthamiana protein databases, we found AGO4 with high matching scores (Fig. 1). In the N. benthamiana genome database (http://solgenomics.net), there are two Nicotiana benthamiana AGO4 (NbAGO4) homologs (NbAGO4-1 and NbAGO4-2) with 93% amino acid similarity. Since N. benthamiana is an allotetraploid, the two NbAGO4 homologs are likely to be alleles of a single gene of the different ancestries. Therefore, we cloned NbAGO4-2 (designated NbAGO4 here) for subsequent analysis. The amino acid alignment of NbAGO4 and the Nicotiana tabacum (NtAGO4) and Arabidopsis thaliana (AtAGO4) AGO4s shows that key amino acid domains required for siRNAs loading and RNA slicing are conserved (Fig. 2A).
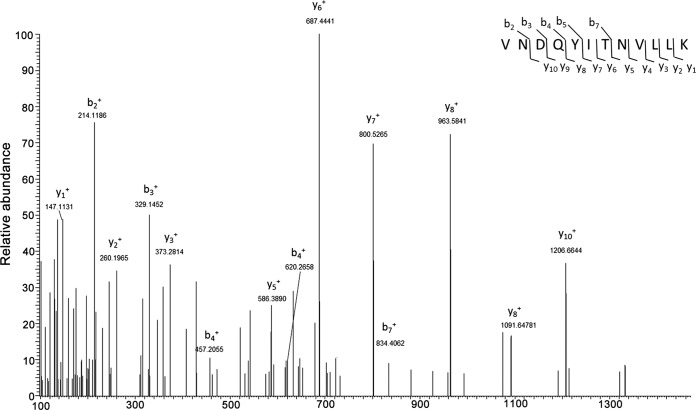
FIG 1.
Representative LC-MS/MS spectrum of the peptide in NbAGO4-2 protein (peptide sequence, VNDQYITNVLLK). GFP or GFP-tagged CLCuMuV V2 (GFP-V2) was transiently expressed through agroinfiltration in N. benthamiana leaves. The protein was isolated using GFP beads and separated by SDS-PAGE. A representative LC-MS/MS spectrum of the peptide is shown.
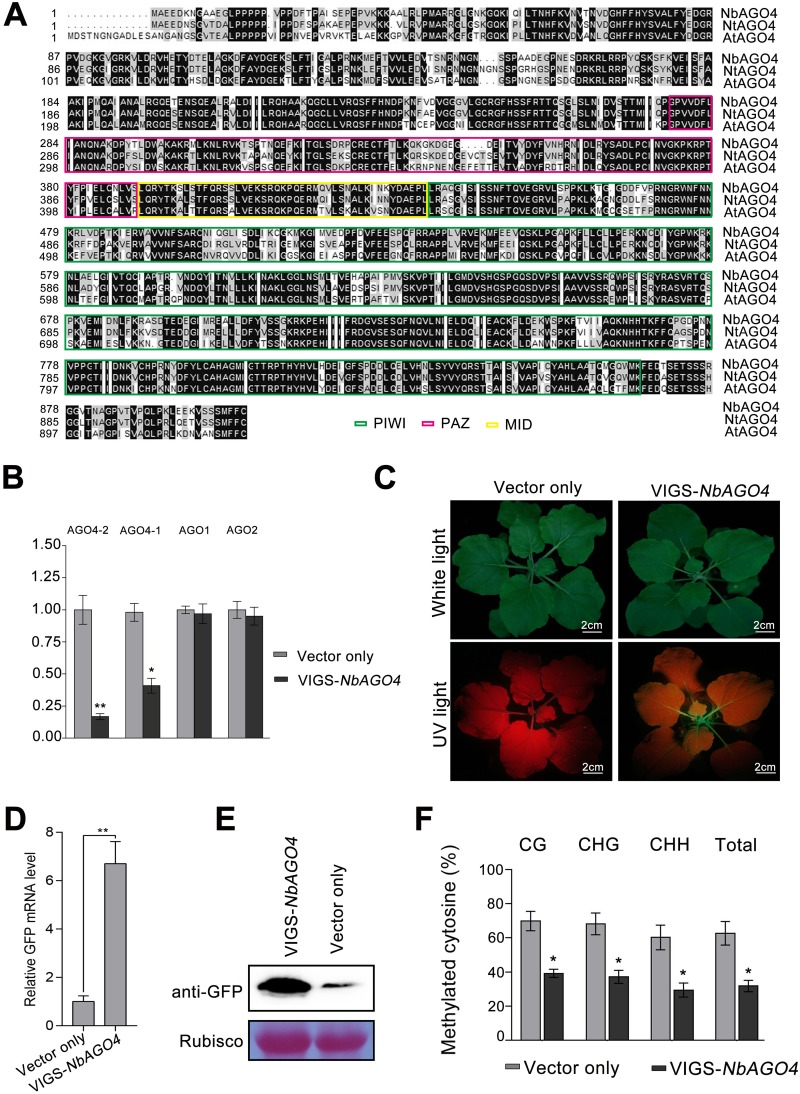
FIG 2.
Silencing of NbAGO4 reverses TGS in 16c-TGS plants. (A) Alignment of Argonaute 4 amino acid sequences from N. benthamiana, N. tabacum, and Arabidopsis thaliana. The alignment was performed with Clustal X. The black background represents 100% conserved residues across the three species. Numbers on the left indicate the positions of amino acid residues. The PIWI, PAZ, and MID domains are marked with colored rectangles. (B) RT-qPCR showing the relative mRNA levels of NbAGO4-2, NbAGO4-1, NbAGO1, and NbAGO2 in NbAGO4-silenced plants. The asterisks indicate significant differences (Student’s t test, P < 0.01). The bars represent means ± standard deviations (SD). Data were obtained from three independent experiments. (C) 16c-TGS plants were infected with TRV-NbAGO4 or TRV. Photographs were taken under white light or UV light at 14 days postinfiltration (dpi). (D) RT-qPCR showing relative GFP mRNA levels. The asterisks indicate significant differences (Student’s t test, P < 0.01). The bars represent means ± SD. Data were obtained from three independent experiments. (E) Immunoblot assay of GFP protein using anti-GFP antibody. RuBisCO was stained with Ponceau Red as a loading control. (F) Percentage of methylated cytosine in CaMV 35S promoter sites in 16c-TGS plants infected with TRV-NbAGO4 or the TRV control. The histogram represents the proportion of methylated cytosine residues in different sequence groups. The asterisks indicate significant differences (Student’s t test, P < 0.01). The bars represent means ± SD. Data were obtained from three independent experiments.
CLCuMuV V2 but not V2L76S interacts with NbAGO4 and its PAZ domain.
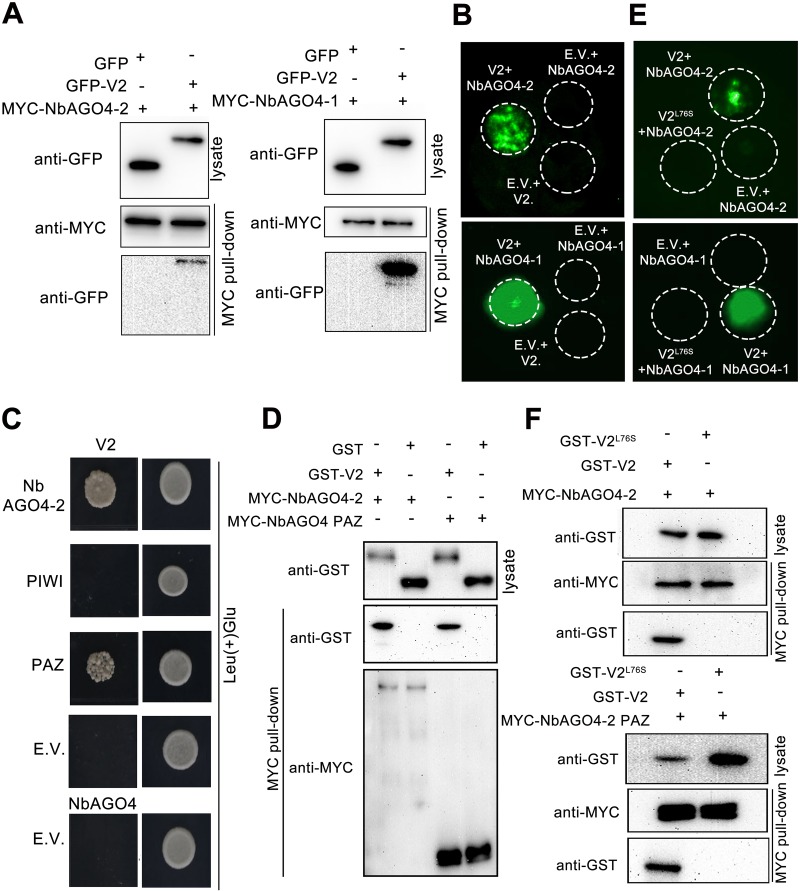
The interaction between V2 and NbAGO4 was confirmed by multiple pieces of evidence. (i) MYC-NbAGO4-2 and MYC-NbAGO4-1 were coimmunoprecipitated with GFP-V2 but not with the control protein GFP (Fig. 3A). (ii) Using a luciferase complementation imaging (LCI) assay, a strong luciferase fluorescence signal was visible only with nLUC-V2/cLUC-AGO4-2 or nLUC-V2/cLUC-AGO4-1 coagroinfiltration, whereas no such fluorescence was detectable in all control combinations (Fig. 3B). (iii) Yeast-two-hybrid assays showed that NbAGO4-2 and its PAZ domain could interact with V2 (Fig. 3C). (iv) We also confirmed the interaction using MYC pulldown assays. Because we were unable to express full-length NbAGO4-2 in Escherichia coli, we transiently expressed MYC-NbAGO4-2 and the MYC-NbAGO4-2 PAZ domain in N. benthamiana and immunoprecipitated them with anti-MYC agarose beads. The immunoprecipitates were incubated with purified GST-tagged V2 (GST-V2) or GST, which were expressed in E. coli. Our results showed that GST-V2, but not GST, was pulled down with MYC-NbAGO4 and the MYC-NbAGO4 PAZ domain, indicating that V2 interacts with NbAGO4 and its PAZ domain (Fig. 3D).
FIG 3.
CLCuMuV V2 but not V2L76S interacts with NbAGO4 and its PAZ domain (A) Coimmunoprecipitation showing the interaction between NbAGO4-1, NbAGO4-2, and CLCuMuV V2 in plants. Total protein was extracted at 60 h postagroinfiltration (hpi) and then immunoprecipitated with anti-MYC beads, followed by immunoblotting using anti-GFP or anti-MYC antibody. (B) LCI assay showing the interaction between cLUC-NbAGO4-1, cLUC-NbAGO4-2, and nLUC-V2 in plants. The agroinfiltrated leaves were monitored at 60 hpi with a low-light cooled CCD imaging apparatus. (C) Yeast two-hybrid assay showing that V2 interacts with the PAZ domain of NbAGO4-2. SKY48 yeast strains containing the indicated constructs were grown on selective plates lacking Ura, Trp, His, and Leu (−UTHL) at 28°C for 4 days. Colony formation on plates lacking leucine but containing galactose (Gal) and raffinose (Raf) indicated interaction between two proteins. In all groups, colonies were able to grow on Ura(−)Trp(−)His(−) selective plates with glucose. (D) MYC pulldown assay showing the interaction of V2 with the NbAGO4-2 PAZ domain. MYC-NbAGO4-2 and the MYC-NbAGO4-2 PAZ domain expressed in N. benthamiana leaves were immunoprecipitated with anti-MYC beads. MYC-tagged proteins in immunoprecipitates were competitively replaced by c-MYC peptide and released into buffer. MYC-NbAGO4-2 and the MYC-NbAGO4-2 PAZ domain protein in buffer were then incubated with purified GST-tagged V2 or GST from E. coli. (E) LCI assay showing that V2, but not V2L76S, interacts with NbAGO4. Inoculated leaves were monitored at 60 hpi with a low-light cooled CCD imaging apparatus. (F) MYC pulldown assays showing that V2, but not V2L76S, interacts with NbAGO4-2 and the NbAGO4-2 PAZ domain. MYC-tagged proteins corresponding to NbAGO4-2 or the NbAGO4-2 PAZ domain were expressed in plants and purified with anti-MYC beads, competitively replaced by c-MYC peptide, and released into buffer. MYC-NbAGO4-2 in buffer was then pulled down with anti-MYC beads and incubated with purified GST-tagged V2 or V2L76S.
Next, we sought to identify the key residue(s) within V2 required for its interaction with AGO4. It has been reported that amino acids 62 to 94 of Cucumber mosaic virus (CMV) 2b are essential for its interaction with AGO1 (30). Thus, we aligned the V2 amino acid sequence with CMV 2b amino acids 62 to 94 and found that they both contain a SEL peptide motif. We then changed L76 to S. Interestingly, this change in V2 (V2L76S) abolished its interaction with NbAGO4 or with NbAGO4 PAZ, as indicated by LCI assay (Fig. 3E) and MYC pulldown assay (Fig. 3F).
Taken together, our data suggest that CLCuMuV V2 interacts with NbAGO4 and its PAZ domain and that L76 of V2 is essential for the interaction.
CLCuMuV V2 suppresses TGS in N. benthamiana.
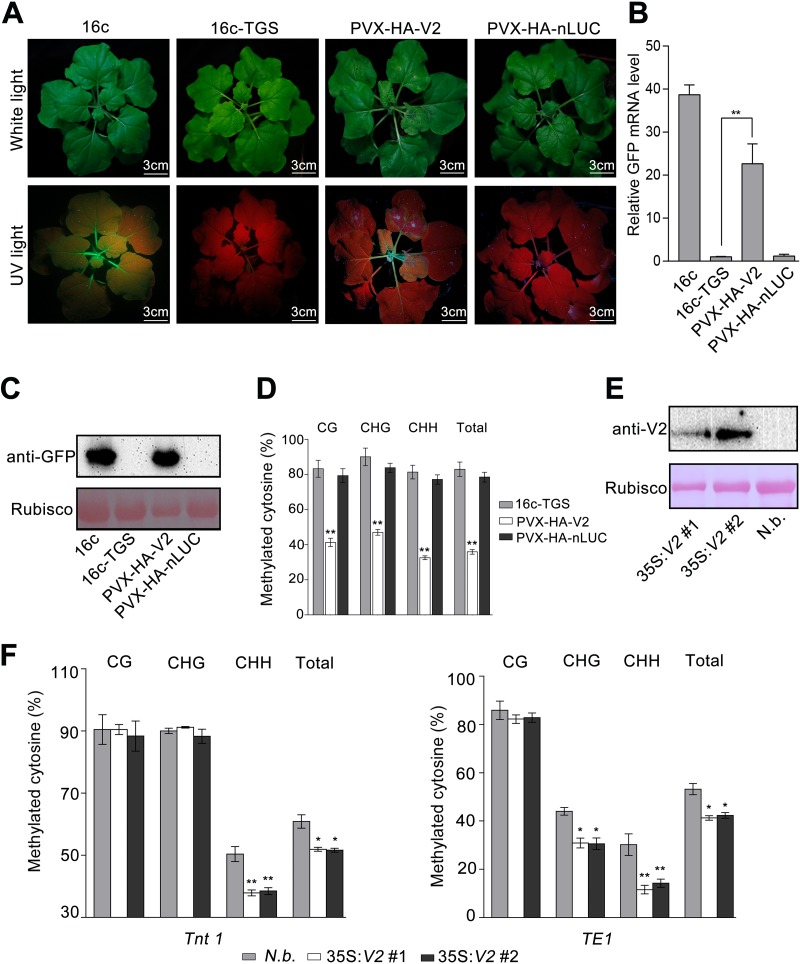
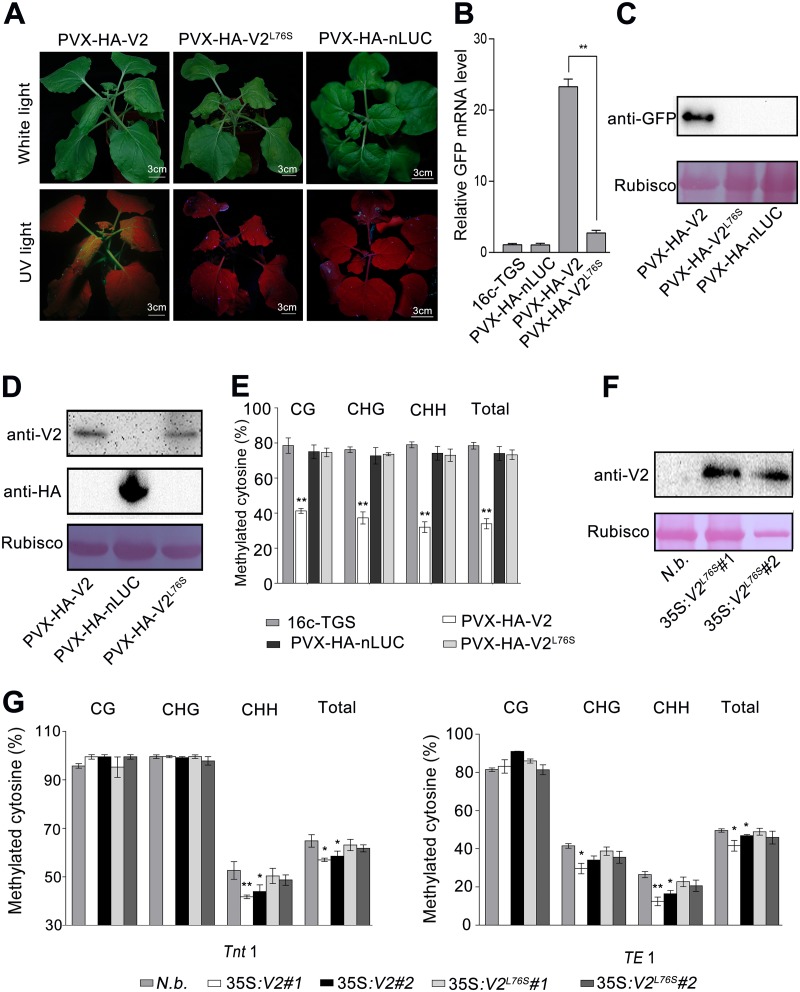
Since V2 from TYLCV is a TGS suppressor and CLCuMuV V2 interacts with AGO4, which was shown to participate in TGS, we expected that CLCuMuV V2 is also a viral TGS suppressor (20). To confirm this, we used a Potato virus X (PVX)-based vector to express hemagglutinin (HA)-tagged V2 (PVX-HA-V2) in 16c-TGS plants, which carry a transcriptionally silenced GFP transgene under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (20). Before PVX infection, no GFP fluorescence was observed in 16c-TGS plants. Plants that were infected with PVX-HA-V2 showed a severe up-curling phenotype in newly growing leaves, followed by wilting and necrosis. Seven of 30 plants showed a fast death of the apical leaf, which made it hard to observe the recovery of GFP fluorescence before plant death, and these were excluded from further analysis. The other 23 plants showed a relatively slow leaf death phenotype and were observed to have recovered GFP to different extents, especially in phloem tissue before plant death. As a control, the expression of HA-nLUC showed mild viral symptoms and no visible GFP fluorescence (Fig. 4A). Quantitative reverse transcription-PCR (RT-qPCR) and immunoblot assays indicated strong accumulation of GFP mRNA and protein in the plants expressing HA-V2 (Fig. 4B and C). These findings demonstrate that V2 can suppress TGS.
FIG 4.
CLCuMuV V2 suppresses TGS in plants. (A) 16c-TGS plants were agroinfected with PVX-HA-V2 or PVX-HA-nLUC. Photographs were taken under white light or UV light at 9 days postinfiltration (dpi). An HA-tagged N-terminal fragment of the firefly luciferase (HA-nLUC) was used as a negative control. (B) RT-qPCR analysis of relative GFP mRNA levels in 16c-TGS plants infected with PVX-HA-V2 or PVX-HA-nLUC. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; [Student’s t test]). The bars represent means ± SD. Data were obtained from three independent experiments. (C) Immunoblot assay of GFP protein using anti-GFP antibody in 16c-TGS plants infected with PVX-HA-V2 or PVX-HA-nLUC. RuBisCO was stained with Ponceau Red as a loading control. (D) Percentage of methylated cytosine in CaMV 35S promoter sites in 16c-TGS or 16c-TGS plants infected with PVX-V2 and PVX-HA-nLUC. The histogram represents the proportions of methylated cytosine residues in different sequence groups. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01 [Student’s t test]). (E) Detection of V2 protein in transgenic plants by immunoblotting. V2 protein was immunoblotted using an anti-V2 antibody. RuBisCO was stained with Ponceau Red as a loading control. (F) Percentage of cytosine methylation of N. benthamiana transposons Tnt1 and TE1 in 35S:HA-V2 transgenic or mock plants. The histogram represents the proportions of methylated cytosines in different sequence groups. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01 [Student’s t test]). The bars represent means ± SD. Data were obtained from three independent experiments.
To unravel the mechanism involved in V2-mediated suppression of TGS, we investigated the DNA methylation profile of the 35S promoter, which regulates expression of the GFP transgene in 16c-TGS plants. To achieve this, we measured methylation levels of cytosine within the 35S promoter sequence by bisulfite sequencing. In total, there are 69 potential methylation sites, including 8, 11, and 50 in the contexts of CG, CHG, and CHH (where H is A, T or C), respectively, across the 35S promoter (Fig. 4D). We found that PVX-based expression of HA-V2 but not HA-nLUC greatly reduced the methylation level at CG, CHG, and CHH sites. Moreover, we measured methylation level of two retrotransposons, Tnt1 (31) and TE1 (Niben101 Ctg00688, bp 1 to 514), in two transgenic N. benthamiana lines overexpressing V2 (Fig. 4E). Compared to that in the wild-type plants, CHH methylation at both loci was greatly reduced in the V2 overexpresser lines (Fig. 4F). Collectively, our results suggest that CLCuMuV V2 functions as a suppressor of RdDM-mediated TGS.
L76 is essential for the TGS suppressor activity of V2.
The requirement of L76 for the V2-AGO4 interaction prompted us to test whether L76 is important for the TGS suppressor activity of V2. HA-V2L76S was expressed by the PVX-based vector in the 16c-TGS plants. As we previously observed (Fig. 4A), plants expressing HA-V2 showed GFP fluorescence; however, plants expressing HA-V2L76S showed mild viral symptoms and no visible GFP expression (Fig. 5A), which was confirmed by RT-qPCR and immunoblot assay (Fig. 5B and C). Immunoblot assays confirmed that HA-V2L76S was expressed at a level similar to that of HA-V2 (Fig. 5D). Consistent with these results, HA-V2L76S did not cause a reduction in cytosine methylation at the 35S promoter (Fig. 5E). We also measured the DNA methylation level at Tnt1 and TE1 in transgenic N. benthamiana overexpressing HA-V2L76S. Transgenic expression of HA-V2L76S was confirmed by Western blotting (Fig. 5F). Methylation levels at Tnt1 and TE1 were not obviously changed in the HA-V2L76S-overexpressing lines (Fig. 5G). These results demonstrate that V2L76S, which loses interaction with AGO4, also loses its suppressor activity.
FIG 5.
CLCuMuV V2L76S abolishes TGS suppression function in plants. (A) 16c-TGS plants were agroinfected with PVX-HA-V2, PVX-HA-V2L76S, or PVX-HA-nLUC. Photographs were taken under white light or UV light at 9 days postinfiltration (dpi). (B) RT-qPCR analysis of relative GFP mRNA levels in 16c-TGS plants infected with PVX-HA-V2, PVX-HA-V2L76S, or PVX-HA-nLUC. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01 [Student’s t test]). The bars represent means ± SD. Data were obtained from three independent experiments. (C) Immunoblot assay of GFP protein using anti-GFP antibody in 16c-TGS plants infected with PVX-HA-V2, PVX-HA-V2L76S, or PVX-HA-nLUC. RuBisCO was stained with Ponceau Red as a loading control. (D) The protein expression of V2, V2L76S, and HA-nLUC was confirmed by immunoblot assays with anti-HA and anti-V2 polyclonal antibodies. (E) Percentage of methylated cytosine in CaMV 35S promoter sites in 16c-TGS plants infected with PVX-V2, PVX-V2L76S, and PVX-HA-nLUC. The histogram represents the proportions of methylated cytosine residues in different sequence groups. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01 [Student’s t test]). The bars represent means ± SD. Data were obtained from three independent experiments. (F) Detection of V2L76S protein in transgenic plants by immunoblotting. V2 protein was immunoblotted using an anti-V2 antibody. RuBisCO was stained with Ponceau Red as a loading control. (G) Percentage of cytosine methylation of N. benthamiana transposons Tnt1 and TE1 in 35S:HA-V2, 35S:HA-V2L76S transgenic or mock plants. The histogram represents the proportions of methylated cytosines in different sequence groups. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01 [Student’s t test]). The bars represent means ± SD. Data were obtained from three independent experiments.
AGO4-mediated RdDM plays a role in defense against CLCuMuV infection.
To confirm the role of NbAGO4 in RdDM, we silenced NbAGO4 in N. benthamiana by Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) (32). RT-qPCR indicated that gene-specific VIGS reduced the mRNA levels of NbAGO4 (NbAGO4-1 and NbAGO4-2) but not that of NbAGO1 or NbAGO2, which share 54% and 52% amino acid similarity with NbAGO4, respectively (Fig. 2B). NbAGO4-silenced plants did not show obvious developmental abnormalities (Fig. 2C). Silencing of NbAGO4 reestablished GFP expression and suppressed methylation of the 35S promoter in 16c-TGS plants (Fig. 2C to E). These results indicate that the NbAGO4 is a genuine core component of RdDM.
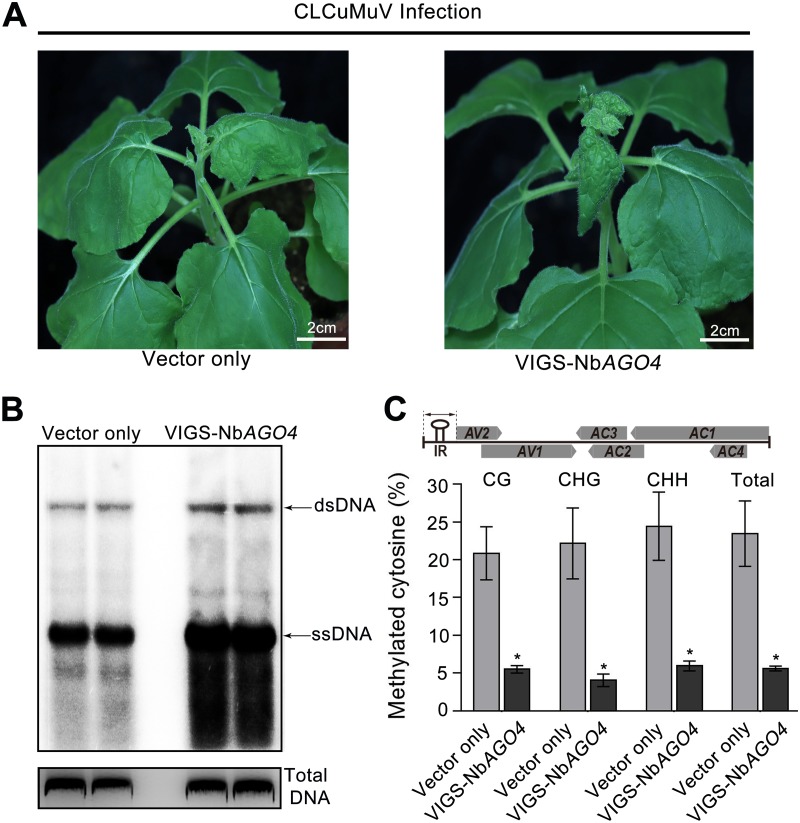
To examine whether RdDM plays a role in defense against CLCuMuV infection, we knocked down NbAGO4 in N. benthamiana by VIGS and infected the plants with CLCuMuV. We found that NbAGO4 knockdown plants showed more severe viral symptoms (Fig. 6A), which were accompanied by significantly increased viral DNA accumulation (Fig. 6B). More importantly, the methylation levels of the CLCuMuV 5′ intergenic region (IR), which contains 10 CG, 7 CNG, and 35 CHH sites, were much lower in NbAGO4 knockdown plants (Fig. 6C). These data together indicate that AGO4-mediated RdDM contributes to the antiviral defense against CLCuMuV.
FIG 6.
AGO4-mediated RdDM plays a role in defense against CLCuMuV infection. (A) Viral symptoms in NbAGO4-silenced plants infected with CLCuMuV at 18 dpi. (B) Southern blot detection showing viral DNA accumulation in NbAGO4-silenced and control plants. A partial CLCuMuV V1 DNA was used as a template to make a 32P-labeled probe. Total DNA was stained with ethidium bromide as a loading control. Single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) corresponding to CLCuMuV are indicated by arrows. (C) Detection of cytosine methylation status of the CLCuMuV 5′ intergenic region (IR) in NbAGO4-silenced or control plants. The histogram represents the proportions of methylated cytosine in different sequence groups. The asterisk indicates significant differences (*, P < 0.05, Student’s t test). The bars represent means ± SD. Data were obtained from three independent experiments.
The L76S point mutation in V2 attenuates viral infection.
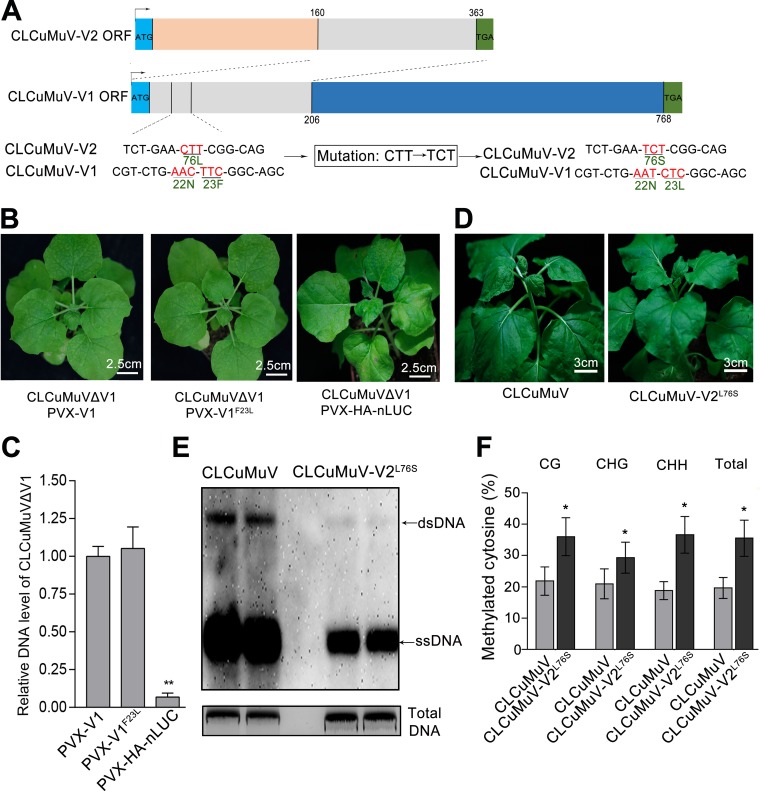
To further investigate the biological significance of V2-NbAGO4 interaction during CLCuMuV infection, we generated a CLCuMuV mutant virus (CLCuMuV-V2L76S) by replacing V2 with V2L76S. Since the V2 gene overlaps the V1 gene, the L76S substitution of the V2 protein resulted in an F23L mutation in V1 (Fig. 7A). To examine whether V1F23L impairs function during viral infection, CLCuMuVΔV1 was generated by replacing the start codon ATG with CTG to block translation of V1. PVX-based expression vectors of V1 and V1F23L were also generated. As expected, CLCuMuVΔV1 failed to infect N. benthamiana plants when it was coinfected with PVX-nLUC. However, CLCuMuVΔV1 was able to systemically infect plants when plants were coinoculated with CLCuMuVΔV1 and PVX-V1 or PVX-V1F23L. Quantitative PCR (qPCR) also indicated that CLCuMuV DNA levels in young leaves were similar between the two groups (Fig. 7B and C), indicating that F23L alteration of V1 has no effect on V1 function.
FIG 7.
Replacement of V2 with V2L76S attenuates CLCuMuV infection. (A) The V2 open reading frame (ORF) (nt 160 to 365) overlaps with the V1 ORF (nt 1 to 206) in the CLCuMuV genome. Thus, an L76S mutation in V2 (CTT-TCT) caused the F23L mutation in V1. (B) N. benthamiana was coinfected with CLCuMuVΔV1 and PVX-V1, PVX-V1F23L, or PVX-HA-nLUC. Photographs were taken under white light at 14 dpi. (C) qPCR showing the CLCuMuV DNA levels in the systemic leaves of plants coinfected with CLCuMuVΔV1 and PVX-V1, PVX-V1F23L, or PVX-HA-nLUC. Data were obtained from three independent experiments. The asterisks indicate significant differences (Student’s t test, P < 0.01). The bars represent mean ± SD. (D) Plants infected with CLCuMuV-V2L76S, which contains an L76S mutation in V2, showed weaker viral symptoms than those infected with CLCuMuV. Photographs were taken at 16 dpi. (E) Southern blot assay showing DNA accumulation for CLCuMuV and CLCuMuV-V2L76S. A partial CLCuMuV V1 DNA was used as a template to make a 32P-labeled probe. Single-stranded viral DNA (ssDNA) and double-stranded viral DNA (dsDNA) DNA are indicated by arrows. Total genomic DNA was stained with ethidium bromide as a loading control. (F) Detection of the cytosine methylation status of the viral 5′ intergenic region (IR) in CLCuMuV- or CLCuMuV-V2L76S-infected plants. The histogram shows the proportions of methylated cytosine residues in different sequence groups. The asterisk indicates significant differences (*, P < 0.05, Student’s t test). The bars represent means ± SD. Data were obtained from three independent experiments.
CLCuMuV and CLCuMuV-V2L76S were then individually agroinoculated into N. benthamiana. Attenuated viral symptoms were observed in plants infected with the CLCuMuV-V2L76S mutant compared to the wild-type virus (Fig. 7D). CLCuMuV-V2L76S DNA accumulation was remarkably reduced compared to that of CLCuMuV, as indicated by DNA gel blot analysis (Fig. 7E). Consistently, the L76S mutation in V2 enhanced DNA cytosine methylation of the CLCuMuV IR at CG, CNG, and CHH sites in CLCuMuV-V2L76S-infected plants (Fig. 7F).
Taken together, these findings further revealed that V2 contributes to CLCuMuV infection by suppressing RdDM-mediated viral DNA methylation through its interaction with AGO4.
DISCUSSION
In plants, RdDM-mediated TGS is considered one of the major defense mechanisms against geminiviruses (15). To counterattack, geminiviruses encode viral suppressors of RNA silencing (VSRs) that are able to suppress TGS. However, it is unclear whether and by what mechanism VSRs could directly inhibit RdDM-mediated TGS, although several geminiviral proteins can suppress TGS by targeting methyl cycle-related enzymes, histone deacetylase, and H3K9 histone methyltransferase in plants (20, 23–25, 29). In addition, the 2b protein from Cucumber mosaic virus (CMV), an RNA virus, is reported to bind to AGO4-related small RNAs and impair AGO4 slicing activity (33). However, the role of TGS in RNA viral infection and whether CMV 2b affects RdDM-mediated TGS are unknown. In this study, we demonstrated for the first time that a DNA viral protein directly targets an RdDM component to counter RdDM-mediated TGS defense to facilitate virus infection.
In this study, we found that CLCuMuV V2 protein directly interacts with AGO4, a core component of the RdDM pathway, and functions as a VSR to suppress TGS. We found that CLCuMuV V2 physically interacts with NbAGO4 and its conserved PAZ domain, which is responsible for recognizing and binding to the 3′ end of single-stranded RNAs (ssRNAs) (34–37). Transient expression of V2 reversed the transcriptionally silenced status of the GFP transgene and prominently reduced the cytosine methylation level of its promoter sequence in 16c-TGS plants, while transgenic expression of V2 resulted in a reduction of cytosine methylation of endogenous transposons. An L76S mutation in V2 abolished its interaction with NbAGO4 and its TGS suppression activity. Further, silencing of NbAGO4 reversed the cytosine methylation of the CaMV 35S promoter in 16c-TGS plants, reduced viral methylation, and simultaneously enhanced viral DNA accumulation. Moreover, CLCuMuV carrying V2L76S caused much milder symptoms, reduced viral DNA accumulation, and showed increased viral DNA methylation. These results strongly suggest that CLCuMuV V2 contributes to viral infection by its interaction with NbAGO4 to interfere with the RdDM-mediated TGS antiviral defense.
In summary, we demonstrated that CLCuMuV V2 can enhance viral infection through interacting with AGO4 to counter the plant RdDM-mediated TGS antiviral defense. Our work provides direct evidence that a DNA viral protein is able to directly target the core RdDM component to counter RdDM-mediated TGS antiviral defense in plants and that AGO4 is a novel DNA viral target. In addition, the conserved PAZ domain among AGOs is responsible for recognizing and binding the 3′ end of the guide ssRNA to facilitate incorporation of sRNA into AGOs/RNA-induced silencing complex (RISC) (34–37). It is possible that CLCuMuV V2 disrupts the NbAGO4-mediated vsiRNA loading to facilitate geminiviral infection. V2 proteins from CLCuMuV and other geminiviruses have been reported to function as posttranscriptional gene silencing (PTGS) suppressors (38, 39). Considering the conservation of AGO family members, V2 could also interact with and interfere with other AGOs, such as AGO1, to suppress PTGS.
MATERIALS AND METHODS
Plant materials and growth conditions.
16c-TGS plants, which are transcriptionally silenced green fluorescent protein (GFP) transgenic N. benthamiana plants, were generated as described previously (25). All N. benthamiana plants were grown in a greenhouse at 25°C with a photoperiod cycle of 16 h light and 8 h dark.
Agroinfiltration.
Agroinfiltration was performed as described previously (40).
Plasmid construction.
The CLCuMuV infectious clone was described previously (2, 40). CLCuMuVΔV1 was generated by replacing ATG with CTG in the start codon of the V1 gene in the CLCuMuV infectious clone. CLCuMuV V2L76S was generated by replacing V2 with V2L76S in the CLCuMuV infectious clone. All constructs were confirmed by DNA sequencing. Primers used in this work are listed in Table 1.
TABLE 1.
Primers and probes used in this study
| Name | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| HindIII-virus construction fragment 1F′ | AAGCTTAGATTTGCATTTAAATTATGA | CLCuMuV infectious clone |
| XbaI-virus construction fragment 1R′ | TCTAGACAGGATCTTTCAGGAGC | |
| XbaI-virus construction fragment 2F′ | TCTAGATTTGCATTTAAATTATGAAATTG | |
| KpnI-virus construction fragment 2R′ | GGTACCAATCAAAGTACAGCACA | |
| V1 ATG-ACG mutant F′ | CGTAATTACGTCGAAGCGAGCT | CLCuMuV ΔV1 |
| V1 ATG-ACG mutant R′ | AGCTCGCTTCGACGTAATTAC | |
| V2L76S mutation F′ | GGCGTCTGAATCTCGGC | CLCuMuV V2L76S |
| V2L76S mutation R′ | GCTGCCGAGATTCAGACG | |
| Lic1 + AGO4-2 F′ | CGACGACAAGACCGTCACCATGGCTGAAGAAGATAAGAATGG | pcLUC-NbAGO4-2, pMYC-NbAGO4-2, pAD-NbAGO4-2, pBD-NbAGO4-2, pMYC-NbAGO4-1, cLUC-NbAGO4-1 |
| Lic2 + AGO4-2 R′ | GAGGAGAAGAGCCGTCGTTAACAAAAGAACATGGAACTGG | |
| Lic1 + AGO4-1 F′ | CGACGACAAGACCGTCACCATGGCTGAAGAAGACAATGGT | |
| Lic2 + AGO4-1 R′ | GAGGAGAAGAGCCGTCGCTAACAGAAGAACATGGAACTAGAAA | |
| Lic1 + AGO4 PAZ F′ | CGACGACAAGACCGTCACCATGGGGCCTGTTGTGG | pMYC-NbAGO4 PAZ domain, pAD-NbAGO4 PAZ domain, pAD-NbAGO4 PIWI domain |
| Lic2 + AGO4 PAZ R′ | GAGGAGAAGAGCCGTCGCTATGACACCAAATTGCATAGCTC | |
| Lic1 + AGO4 PIWI F′ | CGACGACAAGACCGTCACCATGCTGCTTCGTGCCTG | |
| Lic2 + AGO4 PIWI R′ | GAGGAGAAGAGCCGTCGCTTCATCCACTGTCCCATCT | |
| Lic1 + V2 F′ | CGACGACAAGACCGTCACCATGTGGGATCCATTGTTAAACGAAT | pGFP-V2, pnLUC-V2, pGST-V2, PVX-V2, pGFP-V2L76S, pnLUC-V2L76S, pGST-V2L76S, PVX-V2L76S |
| Lic2 + V2 with stop codon R′ | GAGGAGAAGAGCCGTCGTCACAACCCTTTGGAACATCCGGACT | |
| Lic1 + V1 F′ | CGACGACAAGACCGTCACCATGTCGAAGCGAGCTGC | PVX-V1, PVX-V1F23L |
| Lic2 + V1 with stop codon R′ | GAGGAGAAGAGCCGTCGTCAATTCGTTACAGAGTCATAAAAATATAT | |
| V1F23L mutation F′ | GGCGTCTGAATCTCGGC | PVX-V1F23L |
| V1F23L mutation R′ | GCTGCCGAGATTCAGACG | |
| Lic1 + AGO4 VIGS F′ | CGACGACAAGACCGTCACCTCGTTGGAGCTATGCATTG | pTRV-NbAGO4 |
| Lic2 + AGO4 VIGS R′ | GAGGAGAAGAGCCGTCGTGTTTGAAGAAGTCCAGTCAAA | |
| 35S BIS F′ | AAGGCAAGTAATAGAGATTGGAGT | 35S promoter bisulfate sequence |
| 35S BIS R′ | CACCTTCCTTTTCCACTATCTTCACAAT | |
| CLCuMuV IR BIS F′ | TGATTTTGGTTAATTGGAGATAA | CLCuMuV IR bisulfate sequence |
| CLCuMuV IR BIS R′ | TAAATTTAAAACTTAATACACAA | |
| Tnt1 BIS F′ | GTGATGGGGTTTAGTTTTTTTAAAT | Tnt1 bisulfate sequence |
| Tnt1 BIS R′ | TTACTCTTTATTTTCTCTCTTTTATTAATA | |
| TE1 BIS F′ | GAGGTTGTTTATTTTGGAGATTTGA | TE1 bisulfate sequence |
| TE1 BIS R′ | TATTTAACAACCTACCCACCCTAAA | |
| qeIF4a F′ | CCCAGAGAGGAAATACAGTG | RT-qPCR and qPCR |
| qeIF4a R′ | CAATAGACGGACCAGATTCG | |
| qCLCuMuV C3 F′ | GGCGCATATATCTGGGAGGT | qPCR |
| qCLCuMuV C3 R′ | GTCTGAGGGTGTAAGGTCGT | |
| qGFP F′ | ATGGGCACAAATTTTCTGTCA | RT-qPCR |
| qGFP R′ | TCCTCTCCTGCACGTATCC | |
| qNbAGO1 F′ | TGCGGAAGAAGAGAACTGATGT | |
| qNbAGO1 R′ | CACGGCCACCATGTCCTC | |
| qNbAGO2 F′ | AATCGTGGTGGTGGTGGTG | |
| qNbAGO2 R′ | GGCCTTGCTCCTGTTCGTAT | |
| qNbAGO4-2 F′ | GCTGTCTACTTGTTCGTCAGTC | |
| qNbAGO4-2 R′ | GCTATCAGAAAGTCCACAACAGG | |
| qNbAGO4-1 F′ | GCTTTGCGAGGTCAAGAGTC | |
| qNbAGO4-1 R′ | CACCTCCAACTTCCGCAAAA | |
| 32P-probe F′ | ATGTGGGATCCATTGTTAAAC | Viral Southern blot label |
| 32P-probe R′ | ACATGGGCCTTCACAACC | |
The cDNA sequences of NbAGO4 (NbAGO4-2), the PAZ domain, the PIWI domain, and NbAGO4-1 were amplified using N. benthamiana RNA as the template. For immunoprecipitation and LCI assays, MYC-NbAGO4-1, MYC-NbAGO4-2, MYC-NbAGO4 PAZ domain, GFP-V2, nLUC-V2, cLUC-NbAGO4-1, and cLUC-NbAGO4-2 fusion proteins were obtained by overlapping PCR and cloned into the pCAMBIA1300-based T-DNA vector pJG045 (41). For MYC pulldown assays, V2 and GST-V2L76S were cloned into the pGEX4T-1 vector to generate GST-V2 and GST-V2L76S for expression of glutathione S-transferase (GST)-tagged fusion proteins. For yeast two-hybrid assays, CLCuMuV V2, NbAGO4, the NbAGO4 PIWI domain, and the NbAGO4 PAZ domain were amplified and separately cloned into two yeast vectors, i.e., the LexA DNA binding domain (BD)-containing bait vector pYL302 (42) and the B42 activation domain (AD)-containing prey vector pJG4-5.
MS analysis.
Total protein was extracted from N. benthamiana leaves agroinfiltrated with GFP-V2 or GFP and immunoprecipitated using GFP-Trap_A beads (ChromoTek) as described previously (43). Immunoprecipates were denatured using β-mercaptoethanol at 98°C and separated by 12% SDS-PAGE. Silver staining of the gel and digestion of gel-embedded protein samples followed by LC-MS/MS analysis have been described previously (43).
Immunoprecipitation and immunoblotting.
MYC-NbAGO4 and GFP-V2 were transiently expressed in N. benthamiana leaves for 60 h after agroinfiltration. Immunoprecipitation was performed as described previously (43). Protein extracts were immunoprecipitated using GFP-Trap beads (Abmart, China). The precipitations were analyzed by immunoblot assays with anti-MYC (ChromoTek, Germany) and anti-GFP (Abmart, China) antibodies. Signal was detected with Pierce ECL immunoblotting substrate (Thermo Scientific, USA). Each experiment was performed three times with at least three plants each time.
LCI assay.
Luciferase complementation imaging (LCI) assays were performed as described previously (41). The nLUC-V2 group was transiently coexpressed with cLUC-AGO4 or an empty vector in N. benthamiana leaves. Leaf samples were detached at 60 h postagroinfiltration (hpi), sprayed with 1 mM luciferin, and observed with a low-light cooled charge-coupled device (CCD) imaging apparatus (iXon; Andor Technology). Photographs were taken using a 5-min exposure. The experiment was performed three times with at least 3 plants each time.
Yeast two-hybrid assay.
The yeast two-hybrid assay was performed as described previously (44). The experiment was performed three times.
MYC pulldown assay.
MYC-NbAGO4 was expressed in N. benthamiana leaves by agroinoculation. At 60 hpi, total proteins were extracted from detached leaves with native extraction buffer 1 (NB1) (40), incubated with MYC-Trap beads (Abmart, China) for 3 h at 4°C, and washed 3 or 4 times with NB1 at 4°C. Purified MYC-Trap beads were incubated with purified GST-tagged V2 and GST for 2 h at 4°C. The proteins in washed beads were analyzed by immunoblotting using anti-MYC antibody (ChromoTek, Germany) and anti-GST antibody (Abmart, China). GST-V2 and GST-V2L76S were expressed and obtained from E. coli BL21(DE3) (Transgene). The experiment was performed three times with at least 3 plants each time.
qPCR and RT-qPCR analysis.
For quantification of the CLCuMuV DNA level, total DNA was extracted from N. benthamiana leaves infected with CLCuMuV as described previously (45), and eIF4a (KX247369) was used as an internal control for normalization. All experiments were conducted three times.
For detection of NbAGO4 silencing efficiency, total RNA was extracted from N. benthamiana leaves with TRIzol reagent (Tiangen, China). RT-qPCR was performed (40), and NbeIF4a was used as an internal control for normalization. This experiment was conducted three times. Primers are listed in Table 1.
Bisulfite sequencing.
Genomic DNA was isolated with the DNAsecure plant kit (Tiangen, China). Five hundred nanograms of DNA was used for bisulfite sequencing analysis following the manufacturer’s instructions for the EZ DNA Methylation Gold kit (Zymo Research, Irvine, CA). Modified DNA was amplified and cloned into p19T vector, followed by DNA sequencing. Twenty or 21 sequences were analyzed to determine the level of viral DNA methylation each time. Analysis of the results was performed on http://geoseq.mssm.edu//kismeth/revpage.pl. Primers used for PCR are listed in Table 1. All experiments were repeated 3 times.
Southern blotting.
Total DNA was isolated from virus-infected N. benthamiana leaves. The samples were separated on a 1.2% agarose gel. DNA gel blotting was performed as described previously (46). Partial CLCuMuV V1 DNA was used as the template to generate 32P-labeled DNA probes (TaKaRa, Japan). All experiments were repeated 3 times.
Data availability.
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: NbAGO4, MH476459; CLCuMuV, GQ924756; eIF4a, KX247369; GFP, U87973; NbTE1, MH476460; NbTnt1, MH476461; NbAGO4-1, DQ321490.1; and NbAGO4-2, DQ321491.1. Accession numbers MH476459 to MH476461 were newly determined in the present study.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31421001, 31530059, and 31470254) and the National R&D Project of Transgenic Crops of the Ministry of Science and Technology of China (2016ZX08005-001, 2016ZX08009-003, and 2016ZX08009001-004).
Y. Wang and Y. Liu initiated the project and designed the experiments. Y. Wang, Y. Wu, Q. Gong, A. Ismayil, Y. Yuan, B. Lian, Q. Jia, M. Han, and H. Deng performed the experiments. Y. Wang, Y. Hong, L. Hanley-Bowdoin, Y. Qi, and Y. Liu analyzed the data. H. Deng, Y. Qi, and Y. Liu contributed reagents, materials, and analysis. Y. Wang, Y. Hong, L. Hanley-Bowdoin, Y. Qi, and Y. Liu wrote and revised the paper.
REFERENCES
- 1.Heo I, Kim VN. 2009. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell 139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. 2008. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 3.Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. 2004. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 4.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. 2005. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 5.Qi Y, Denli AM, Hannon GJ. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. 2006. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 7.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. 2006. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. 2007. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A 104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. 2005. RNA polymerase IV directs silencing of endogenous DNA. Science 308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 11.Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. 2009. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 13.Wierzbicki AT, Haag JR, Pikaard CS. 2008. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. 2009. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raja P, Sanville BC, Buchmann RC, Bisaro DM. 2008. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol 82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coursey T, Regedanz E, Bisaro DM. 2018. Arabidopsis RNA polymerase V mediates enhanced compaction and silencing of geminivirus and transposon chromatin during host recovery from infection. J Virol 92:e01320-17. doi: 10.1128/JVI.01320-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol 10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 18.Saeed M, Briddon RW, Dalakouras A, Krczal G, Wassenegger M. 2015. Functional analysis of cotton leaf curl Kokhran virus/cotton leaf curl Multan betasatellite RNA silencing suppressors. Biology (Basel) 4:697–714. doi: 10.3390/biology4040697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Xu X, Huang C, Gu Z, Cao L, Hu T, Ding M, Li Z, Zhou X. 2015. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol 208:555–569. doi: 10.1111/nph.13473. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Li F, Huang C, Yang X, Qian Y, Xie Y, Zhou X. 2014. V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J Gen Virol 95:225–230. doi: 10.1099/vir.0.055798-0. [DOI] [PubMed] [Google Scholar]
- 21.Xie Y, Zhao L, Jiao X, Jiang T, Gong H, Wang B, Briddon RW, Zhou X. 2013. A recombinant begomovirus resulting from exchange of the C4 gene. J Gen Virol 94:1896–1907. doi: 10.1099/vir.0.053181-0. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Negrete E, Lozano-Duran R, Piedra-Aguilera A, Cruzado L, Bejarano ER, Castillo AG. 2013. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol 199:464–475. doi: 10.1111/nph.12286. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, Zhou X, Guo H, Xie Q. 2011. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23:273–288. doi: 10.1105/tpc.110.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, Bisaro DM, Zhou X. 2011. Suppression of methylation-mediated transcriptional gene silencing by betaC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog 7:e1002329. doi: 10.1371/journal.ppat.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. 2009. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol 83:5005–5013. doi: 10.1128/JVI.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ismayil A, Haxim Y, Wang Y, Li H, Qian L, Han T, Chen T, Jia Q, Yihao Liu A, Zhu S, Deng H, Gorovits R, Hong Y, Hanley-Bowdoin L, Liu Y. 2018. Cotton leaf curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog 14:e1007282. doi: 10.1371/journal.ppat.1007282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castillo-Gonzalez C, Liu X, Huang C, Zhao C, Ma Z, Hu T, Sun F, Zhou Y, Zhou X, Wang XJ, Zhang X. 2015. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. Elife 4:e06671. doi: 10.7554/eLife.06671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun YW, Tee CS, Ma YH, Wang G, Yao XM, Ye J. 2015. Attenuation of histone methyltransferase KRYPTONITE-mediated transcriptional gene silencing by Geminivirus. Sci Rep 5:16476. doi: 10.1038/srep16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B, Yang X, Wang Y, Xie Y, Zhou X. 2018. Tomato yellow leaf curl virus V2 interacts with host histone deacetylase 6 to suppress methylation-mediated transcriptional gene silencing in plants. J Virol 92:e00036-18. doi: 10.1128/JVI.00036-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan CG, Fang YY, Zhou BJ, Zhao JH, Hou WN, Zhu H, Ding SW, Guo HS. 2012. Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24:259–274. doi: 10.1105/tpc.111.092718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernhettes S, Grandbastien MA, Casacuberta JM. 1998. The evolutionary analysis of the Tnt1 retrotransposon in Nicotiana species reveals the high variability of its regulatory sequences. Mol Biol Evol 15:827–836. doi: 10.1093/oxfordjournals.molbev.a025988. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Schiff M, Dinesh-Kumar SP. 2002. Virus-induced gene silencing in tomato. Plant J 31:777–786. [DOI] [PubMed] [Google Scholar]
- 33.Hamera S, Song X, Su L, Chen X, Fang R. 2012. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J 69:104–115. doi: 10.1111/j.1365-313X.2011.04774.x. [DOI] [PubMed] [Google Scholar]
- 34.Lingel A, Simon B, Izaurralde E, Sattler M. 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 35.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 36.Lingel A, Simon B, Izaurralde E, Sattler M. 2004. Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol 11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 37.Gu S, Jin L, Huang Y, Zhang F, Kay MA. 2012. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr Biol 22:1536–1542. doi: 10.1016/j.cub.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y. 2008. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci U S A 105:157–161. doi: 10.1073/pnas.0709036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Dong J, Xu Y, Wu J. 2012. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res 163:51–58. doi: 10.1016/j.virusres.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Jia Q, Liu N, Xie K, Dai Y, Han S, Zhao X, Qian L, Wang Y, Zhao J, Gorovits R, Xie D, Hong Y, Liu Y. 2016. CLCuMuB betaC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog 12:e1005668. doi: 10.1371/journal.ppat.1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du Y, Zhao J, Chen T, Liu Q, Zhang H, Wang Y, Hong Y, Xiao F, Zhang L, Shen Q, Liu Y. 2013. Type I J-domain NbMIP1 proteins are required for both Tobacco mosaic virus infection and plant innate immunity. PLoS Pathog 9:e1003659. doi: 10.1371/journal.ppat.1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Liu Q, Zhang H, Jia Q, Hong Y, Liu Y. 2013. The Rubisco small subunit is involved in tobamovirus movement and Tm-2(2)-mediated extreme resistance. Plant Physiol 161:374–383. doi: 10.1104/pp.112.209213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y, Liu Y. 2015. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27:1316–1331. doi: 10.1105/tpc.114.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Chen Y, Liu Y, Zhou X, Fan D. 2002. Local cytokines profile in gastric cancer lesions. Zhonghua Zhong Liu Za Zhi 24:14–16. [PubMed] [Google Scholar]
- 45.Haxim Y, Ismayil A, Jia Q, Wang Y, Zheng X, Chen T, Qian L, Liu N, Wang Y, Han S, Cheng J, Qi Y, Hong Y, Liu Y. 2017. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife 6:e23897. doi: 10.7554/eLife.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paprotka T, Deuschle K, Metzler V, Jeske H. 2011. Conformation-selective methylation of geminivirus DNA. J Virol 85:12001–12012. doi: 10.1128/JVI.05567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: NbAGO4, MH476459; CLCuMuV, GQ924756; eIF4a, KX247369; GFP, U87973; NbTE1, MH476460; NbTnt1, MH476461; NbAGO4-1, DQ321490.1; and NbAGO4-2, DQ321491.1. Accession numbers MH476459 to MH476461 were newly determined in the present study.