Our results greatly expand the knowledge of EVE abundance, diversity, and function in an exceptionally wide range of arthropod species. We found that while previous findings in mosquitoes regarding the potential of EVEs to serve as sources of immunological memory via the piRNA pathway may be generalized to other arthropod species, speculation regarding the antiviral function of EVE-derived piRNAs should take into context the fact that EVEs are, in the vast majority of cases, not similar enough to currently described viruses at the nucleotide level to serve as sources of antiviral piRNAs against them.
KEYWORDS: endogenous viral element, arbovirus, arthropod, integrated viral sequences, piRNA, siRNA, small RNA
ABSTRACT
Arthropod genomes contain sequences derived from integrations of DNA and nonretroviral RNA viruses. These sequences, known as endogenous viral elements (EVEs), have been acquired over the course of evolution and have been proposed to serve as a record of past viral infections. Recent evidence indicates that EVEs can function as templates for the biogenesis of PIWI-interacting RNAs (piRNAs) in some mosquito species and cell lines, raising the possibility that EVEs may serve as a source of immunological memory in these organisms. However, whether piRNAs are derived from EVEs or serve an antiviral function in other arthropod species is unknown. Here, we used publicly available genome assemblies and small RNA sequencing data sets to characterize the repertoire and function of EVEs across 48 arthropod genomes. We found that EVEs are widespread in arthropod genomes and primarily correspond to unclassified single-stranded RNA (ssRNA) viruses and viruses belonging to the Rhabdoviridae and Parvoviridae families. Additionally, EVEs were enriched in piRNA clusters in a majority of species, and we found that production of primary piRNAs from EVEs is common, particularly for EVEs located within piRNA clusters. While the abundance of EVEs within arthropod genomes and the frequency with which EVEs give rise to primary piRNAs generally support the hypothesis that EVEs contribute to an antiviral response via the piRNA pathway, limited nucleotide identity between currently described viruses and EVEs identified here likely limits the extent to which this process plays a role during infection with known viruses in the arthropod species analyzed.
IMPORTANCE Our results greatly expand the knowledge of EVE abundance, diversity, and function in an exceptionally wide range of arthropod species. We found that while previous findings in mosquitoes regarding the potential of EVEs to serve as sources of immunological memory via the piRNA pathway may be generalized to other arthropod species, speculation regarding the antiviral function of EVE-derived piRNAs should take into context the fact that EVEs are, in the vast majority of cases, not similar enough to currently described viruses at the nucleotide level to serve as sources of antiviral piRNAs against them.
INTRODUCTION
Arthropods play key roles in terrestrial and aquatic ecosystems by pollinating plants, aiding in plant seed dispersal, controlling populations of other organisms, functioning as food sources for other organisms, and cycling nutrients (1, 2). Besides their important contributions to maintaining ecosystem stability, some arthropods are also known to serve as vectors for human, animal, and plant pathogens (3, 4). During arthropod-mediated transmission of many plant- and animal-infecting viruses, the virus replicates inside the arthropod vector, and thus, the vector serves as one of at least two possible hosts for these viruses (3, 4). Additionally, arthropods are subject to infection by arthropod-specific viruses that are not transmitted to new hosts of a different species (5). Elucidating the antiviral mechanisms arthropods use to combat viral infection is an important area of research, as a greater understanding of arthropod immunity may lead to new strategies for the control of arthropod-transmitted viruses.
RNA interference (RNAi) is the primary antiviral mechanism in arthropods and relies on three classes of small RNAs (sRNAs) (6, 7). The small interfering RNA (siRNA) pathway is the most important branch of RNAi for combating viral infection in arthropods, and this pathway relies on the production of primarily 21-nt siRNAs via cleavage of viral double-stranded RNA (7). siRNAs associate with Argonaute proteins to direct a multiprotein effector complex known as the RNA-induced silencing complex to the viral RNA, resulting in endonucleolytic cleavage of target RNA (7). The microRNA (miRNA) pathway relies primarily on inhibition of translation via imperfect base pairing between miRNAs and viral RNAs, but miRNAs can also direct cleavage of target RNA if there is sufficient complementarity between the miRNA and the target RNA (7). A third branch of RNAi, directed by PIWI-interacting RNAs (piRNAs), was discovered more recently and has been implicated as a component of antiviral defense in mosquitoes, but not in Drosophila melanogaster (8, 9).
The primary role of the piRNA pathway is control of transposable elements in animal germ cells, and studies in D. melanogaster have revealed two models for piRNA biogenesis: the primary pathway and the ping-pong cycle (secondary pathway) (10). In the primary pathway, 24- to 32-nt primary piRNAs with a strong bias for uracil as the 5′-most nucleotide (1U bias) are produced from endogenous transcripts derived from transposon sequence-rich regions of the genome denoted piRNA clusters. During the ping-pong cycle in D. melanogaster, antisense primary piRNAs guide the PIWI family Argonaute protein Aubergine to complementary RNA (cRNA), resulting in endonucleolytic cleavage of target RNA exactly 10 nt downstream from the 5′ end of the guiding primary piRNA (10). Cleaved RNA is subsequently processed into secondary piRNAs with a bias for adenine as the 10th nucleotide from the 5′ end (10A bias). The secondary piRNAs are then loaded onto Argonaute 3, another PIWI family Argonaute protein, and direct cleavage of endogenous transcripts derived from piRNA clusters, resulting in the production of additional primary piRNAs (10). Thus, in the context of defense against transposons, the ping-pong cycle serves to amplify the posttranscriptional silencing activity of the piRNA pathway in response to active transposable elements. Interestingly, the PIWI family has undergone expansion in mosquitoes, and it is now clear that the mechanisms responsible for generating virus-derived piRNAs in these organisms are distinct from the canonical piRNA pathway used to combat transposable-element activity (11, 12). Key to the novel piRNA pathway seen in mosquitoes is the biogenesis of primary piRNAs directly from exogenous viral RNA without the need for primary piRNAs derived from endogenous sequences (12).
Recent studies have revealed that the genomes of some eukaryotic species contain sequences derived from integrations of DNA and nonretroviral RNA viruses (13–17). These sequences are known as endogenous viral elements (EVEs) and are proposed to serve as a partial record of past viral infections (14). Moreover, a number of studies have demonstrated that EVEs are present within piRNA clusters and serve as sources of piRNAs in certain mosquito species and cell lines, raising the possibility that EVEs may participate in an antiviral response against exogenous viruses via the canonical piRNA pathway (13, 14, 17). While EVEs have been reported in a number of other arthropod species, their potential involvement with the piRNA pathway remains unclear. Here, we sought to expand the knowledge of EVEs and their role in the piRNA pathway beyond mosquito species. To this end, we performed a comprehensive analysis to characterize the abundance, diversity, distribution, and function of EVEs across all arthropod species with sequenced genomes for which there are corresponding publicly available sRNA sequencing data. Our results reveal that, as has been observed in mosquitoes, EVEs are abundant in arthropod genomes and many EVEs produce primary piRNAs. We found that while EVEs are widespread and commonly give rise to piRNAs, limited nucleotide identity between currently described viruses and EVEs identified here likely limits the extent to which this process plays an antiviral role during infection with known viruses.
(This article was submitted to an online preprint archive [18].)
RESULTS
EVEs are commonly found within arthropod genomes.
We began by identifying all arthropod species for which there were both publicly available genome assemblies and sRNA sequencing data sets. We then created a custom database comprised of all single-stranded DNA (ssDNA) and nonretroviral RNA virus protein sequences available in GenBank and used this database to identify putative EVEs genome wide in each arthropod genome via BLASTx. As reported previously, we found that a large number of putative EVEs could not be unambiguously classified as viral due to homology with eukaryotic, bacterial, or archaeal sequences (14). We removed the majority of putative EVEs that were homologous to eukaryotic sequences via reverse BLAST searches against the D. melanogaster proteome. The remaining putative EVEs were then filtered manually. Ultimately, we identified 4,061 EVEs within the genomes of 48 arthropod species (Table 1; Data Sets S1 and S2 in the supplemental material). With the exception of Sarcoptes scabiei, we found at least one EVE in each arthropod genome. We found that EVEs comprised a median of 0.0061% of the 48 genomes in which EVEs were identified (Table S1).
TABLE 1.
Enrichment of EVEs in piRNA clusters
| Species | Genomic region | Length (bp)a | % of genome |
No. of EVEs |
P value for EVE enrichment in piRNA clustersb |
|---|---|---|---|---|---|
| Acyrthosiphon pisum | piRNA clusters | 26,324,066 | 4.86 | 127 | <0.001 |
| Whole genome | 541,716,367 | 294 | |||
| Aedes aegypti | piRNA clusters | 43,772,915 | 3.16 | 117 | <0.001 |
| Whole genome | 1,383,978,943 | 273 | |||
| Aedes albopictus | piRNA clusters | 2,176,195 | 0.10 | 3 | <0.01 |
| Whole genome | 2,247,291,986 | 502 | |||
| Anopheles arabiensis | piRNA clusters | 1,994,683 | 0.81 | 6 | <0.001 |
| Whole genome | 246,569,081 | 16 | |||
| Anopheles gambiae | piRNA clusters | 9,472,362 | 2.88 | 7 | <0.001 |
| Whole genome | 329,012,562 | 64 | |||
| Anopheles stephensi | piRNA clusters | 2,409,359 | 1.15 | 5 | <0.001 |
| Whole genome | 209,515,279 | 23 | |||
| Apis melllifera | piRNA clusters | 1,867,492 | 0.82 | 0 | |
| Whole genome | 229,123,808 | 1 | |||
| Armadillidium vulgare | piRNA clusters | 56,355 | 0.36 | 0 | |
| Whole genome | 15,705,380 | 4 | |||
| Bactrocera dorsalis | piRNA clusters | 1,692,853 | 0.41 | 10 | <0.001 |
| Whole genome | 414,975,858 | 19 | |||
| Blattella germanica | piRNA clusters | 71,312,292 | 4.17 | 16 | <0.001 |
| Whole genome | 1,710,648,823 | 66 | |||
| Bombus terrestris | piRNA clusters | 134,793 | 0.06 | 0 | |
| Whole genome | 236,392,901 | 51 | |||
| Bombix mori | piRNA clusters | 26,961,546 | 5.86 | 26 | <0.001 |
| Whole genome | 460,334,713 | 54 | |||
| Camponotus floridanus | piRNA clusters | 24,341 | 0.01 | 0 | |
| Whole genome | 224,555,298 | 121 | |||
| Centruroides sculpturatus | piRNA clusters | 21,894,072 | 2.37 | 0 | |
| Whole genome | 925,483,296 | 13 | |||
| Ceratosolen solmsi | piRNA clusters | 1,904,847 | 0.69 | 0 | |
| Whole genome | 277,061,652 | 36 | |||
| Dermatophagoides farinae | piRNA clusters | 9,657 | 0.01 | 0 | |
| Whole genome | 91,936,773 | 18 | |||
| Diaphorina citri | piRNA clusters | 403,877 | 0.08 | 18 | <0.001 |
| Whole genome | 485,867,070 | 104 | |||
| Drosophila erecta | piRNA clusters | 227,344 | 0.16 | 0 | |
| Whole genome | 145,091,640 | 6 | |||
| Drosophila melanogaster | piRNA clusters | 489,366 | 0.34 | 0 | |
| Whole genome | 143,727,872 | 1 | |||
| Drosophila mojavensis | piRNA clusters | 6,971,121 | 3.60 | 2 | <0.001 |
| Whole genome | 193,833,151 | 5 | |||
| Drosophila persimilis | piRNA clusters | 1,924,687 | 1.02 | 0 | |
| Whole genome | 188,386,917 | 8 | |||
| Drosophila pseudoobscura | piRNA clusters | 160,414 | 0.09 | 0 | |
| Whole genome | 171,319,450 | 9 | |||
| Drosophila sechellia | piRNA clusters | 1,188,798 | 0.76 | 0 | |
| Whole genome | 157,260,000 | 20 | |||
| Drosophila simulans | piRNA clusters | 934,967 | 0.75 | 0 | |
| Whole genome | 124,956,420 | 2 | |||
| Drosophila virilis | piRNA clusters | 8,680,386 | 4.21 | 3 | <0.001 |
| Whole genome | 206,040,227 | 8 | |||
| Drosophila willistoni | piRNA clusters | 2,299,289 | 0.98 | 0 | |
| Whole genome | 235,531,186 | 31 | |||
| Drosophila yakuba | piRNA clusters | 1,964,249 | 1.21 | 10 | <0.001 |
| Whole genome | 162,595,439 | 33 | |||
| Harpegnathos saltator | piRNA clusters | 653,301 | 0.23 | 5 | <0.001 |
| Whole genome | 283,034,581 | 136 | |||
| Heliconius melpomene | piRNA clusters | 2,357,019 | 0.86 | 0 | |
| Whole genome | 275,199,408 | 67 | |||
| Helicoverpa armigera | piRNA clusters | 3,249,285 | 0.96 | 3 | <0.001 |
| Whole genome | 337,088,551 | 20 | |||
| Homalodisca vitripennis | piRNA clusters | 11,102,932 | 0.84 | 24 | <0.001 |
| Whole genome | 1,325,418,683 | 355 | |||
| Ixodes ricinus | piRNA clusters | 5,697,971 | 1.11 | 60 | <0.001 |
| Whole genome | 514,711,065 | 168 | |||
| Ixodes scapularis | piRNA clusters | 378,290 | 0.02 | 1 | <0.01 |
| Whole genome | 1,896,882,981 | 387 | |||
| Limulus polyphemus | piRNA clusters | 17,684,516 | 0.97 | 15 | <0.001 |
| Whole genome | 1,828,558,544 | 106 | |||
| Lutzomyia longipalpis | piRNA clusters | 642,293 | 0.42 | 6 | <0.001 |
| Whole genome | 154,240,798 | 41 | |||
| Musca domestica | piRNA clusters | 19,474,903 | 2.60 | 4 | <0.001 |
| Whole genome | 750,424,431 | 7 | |||
| Myzus persicae | piRNA clusters | 14,408,803 | 4.15 | 16 | <0.001 |
| Whole genome | 347,317,491 | 60 | |||
| Neobellieria bullata | piRNA clusters | 524,246 | 0.13 | 5 | <0.001 |
| Whole genome | 396,408,944 | 21 | |||
| Nicrophorus vespilloides | piRNA clusters | 3,966,609 | 2.03 | 1 | <0.001 |
| Whole genome | 195,278,032 | 2 | |||
| Oncopeltus fasciatus | piRNA clusters | 26,156,514 | 2.38 | 29 | <0.05 |
| Whole genome | 1,099,627,727 | 80 | |||
| Penaeus monodon | piRNA clusters | 14,301,335 | 0.99 | 0 | |
| Whole genome | 1,449,940,850 | 248 | |||
| Plodia interpunctella | piRNA clusters | 5,441,074 | 1.49 | 2 | <0.05 |
| Whole genome | 364,638,958 | 31 | |||
| Plutella xylostella | piRNA clusters | 5,755,516 | 1.71 | 70 | <0.001 |
| Whole genome | 336,888,803 | 171 | |||
| Spodoptera frugiperda | piRNA clusters | 9,097,455 | 1.77 | 24 | <0.001 |
| Whole genome | 514,228,299 | 241 | |||
| Tetranychus urticae | piRNA clusters | 2,545,325 | 2.84 | 0 | |
| Whole genome | 89,602,137 | 10 | |||
| Tribolium castaneum | piRNA clusters | 9,090,949 | 5.96 | 30 | <0.001 |
| Whole genome | 152,420,532 | 54 | |||
| Triops cancriformis | piRNA clusters | 4,016,357 | 3.68 | 7 | <0.01 |
| Whole genome | 109,242,312 | 62 | |||
| Varroa destructor | piRNA clusters | 35,171 | 0.01 | 0 | |
| Whole genome | 368,943,721 | 12 |
The genome or piRNA cluster size (in base pairs of DNA [bp]) is shown.
Cumulative binomial distribution.
It should be noted that our implementation of a reverse BLAST filter against the D. melanogaster proteome could theoretically result in the exclusion of some valid EVEs, particularly in the Drosophila species analyzed. However, with the exception of D. melanogaster and Drosophila simulans, EVE density in the Drosophila species genomes was within an order of magnitude of the median for all species (Table S1). As a previous study failed to identify any EVEs within the D. melanogaster genome (13), we do not believe that our reverse BLAST filter resulted in an artificially low number of EVEs in this species.
To assess the sensitivity of our EVE identification pipeline, we compared EVEs identified in the Aedes albopictus AaloF1 assembly by Palatini et al. (13) to EVEs we identified in the same genome assembly. We found that our pipeline successfully identified 71/72 EVEs described by Palatini et al.; however, due to differences in our BLAST database, BLAST parameters, and the way in which BLAST hits in close proximity are treated, our EVEs were generally shorter and occasionally fragmented (Data Set S3). That we identified all but one of the EVEs described by Palatini et al. (13) validates the sensitivity of our approach. Here, we identified EVEs based on the similarity of endogenous sequences to a large and diverse database of viral sequences. In contrast, Palatini et al. employed a relatively small database of viral sequences restricted to only a few viral families. Thus, we identified a much larger number of EVEs.
EVEs are enriched in piRNA clusters in a majority of species.
Previous studies have pointed toward a potential role for EVE-derived piRNAs in antiviral responses, and EVEs are enriched in piRNA clusters in Aedes albopictus and Aedes aegypti (13, 14). Thus, we used publicly available sRNA data sets to define piRNA clusters in the arthropod genomes using proTRAC (19). To increase the coverage and diversity of sRNAs used for this analysis, we combined representative collections of the available sRNA data sets for each species (Table S2). We then classified the EVEs into EVEs within piRNA clusters and EVEs outside piRNA clusters (Table 1; Data Sets S2 and S3). We found that 30 of 48 arthropod genomes contained EVEs within piRNA clusters and that EVEs were enriched in piRNA clusters in 28 of these species (cumulative binomial probability of <0.05) (Table 1). The median deduced amino acid identities shared between EVEs and their closest BLASTx hit were 34.0% for EVEs in piRNA clusters and 34.3% for EVEs outside piRNA clusters. We found that deduced amino acid identity was significantly higher for piRNA cluster-resident EVEs in Acyrthosiphon pisum, Diaphorina citri, Plodia interpunctella, and Spodoptera frugiperda. Deduced amino acid identity was significantly lower for piRNA cluster-resident EVEs in Homalodisca vitripennis, Limulus polyphemus, and Oncopeltus fasciatus (Fig. 1a). Interestingly, we found that when all species are considered, EVEs in piRNA clusters are significantly longer than EVEs outside piRNA clusters (P = 0.000101, two-tailed t test). On an individual species level, EVEs were significantly longer within piRNA clusters in Anopheles stephensi, Blattella germanica, Harpegnathos saltator, H. vitripennis, L. polyphemus, Plutella xylostella, and S. frugiperda. EVEs outside piRNA clusters were significantly longer in A. pisum (Fig. 1b).
FIG 1.
(A) Distribution of amino acid identities between translated EVEs and their closest viral BLASTx hits for the respective arthropod species listed. (B) Distribution of translated EVE lengths in amino acids for the respective arthropod species listed. Blue, EVEs in piRNA clusters; red, EVEs outside piRNA clusters. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired t test).
EVEs corresponding to unclassified viruses and viruses belonging to the Rhabdoviridae and Parvoviridae families predominate both within and outside piRNA clusters.
Genome wide, we identified EVEs corresponding to viruses belonging to 54 different viral families (Data Sets S4 and S5). Both within and outside piRNA clusters, unclassified viruses and viruses belonging to the Rhabdoviridae and Parvoviridae families comprised over 70% of all EVEs (Fig. 2). Interestingly, a plurality of EVEs corresponded to viruses possessing negative-sense single-stranded RNA (ssRNA) genomes (data not shown).
FIG 2.
The most common viral families corresponding to EVEs found in arthropod genomes within piRNA clusters (A) or outside piRNA clusters (B). Complete lists of viral families corresponding to EVEs found within arthropod genomes are available in Data Sets S3 and S4 in the supplemental material.
Whitfield et al. reported the presence of EVEs corresponding to viruses belonging to the Closteroviridae and Bromoviridae families within the genome of A. aegypti-derived Aag2 cells (14). This is somewhat unexpected, as these families are comprised solely of viruses that do not infect A. aegypti but only infect plants. These viruses are transmitted by their respective insect vectors in a noncirculative manner (3). In agreement with these findings, we also identified a number of EVEs corresponding to viruses of the Closteroviridae and Bromoviridae families, as well as several other families comprised of viruses not known to replicate outside their plant hosts, including Geminiviridae, Nanoviridae, Luteoviridae, Potyviridae, Secoviridae, Tombusviridae, and Virgaviridae (Data Sets S4 and S5).
Primary piRNA production from EVEs is widespread, but nucleotide identity between EVEs and known viruses is low.
Previous studies have revealed that EVEs serve as templates for piRNA production in A. aegypti, A. albopictus, and Culex quinquefasciatus (13, 14, 20). However, it is unclear whether piRNAs are produced from EVEs in nonmosquito arthropod species. We examined the sRNAs mapping to each EVE for the characteristics of primary piRNAs (i.e., a relatively higher number of 24- to 32-nt sRNAs than of other lengths and a 1U bias for 24- to 32-nt sRNAs). Some previous studies have assessed primary-piRNA production from EVEs by measuring 1U biases only for sRNAs mapping antisense with respect to the coding region of the EVE (based on comparison to the corresponding virus) (13). However, primary piRNAs could theoretically be produced from precursor transcripts derived from either genomic strand. Thus, we evaluated 1U biases for 24- to 32-nt sRNAs mapping either sense or antisense to each EVE. Biases were calculated only for EVEs displaying a significant piRNA peak, and 1U bias significance was evaluated using a binomial distribution and deemed significant when the P value was <0.001. We found that the vast majority (77.5%) of EVEs within piRNA clusters served as sources of primary piRNAs. Outside piRNA clusters, only 35.7% of EVEs served as sources of primary piRNAs. piRNA production from EVEs was particularly common in A. aegypti, A. albopictus, Anopheles stephensi, Bactrocera dorsalis, and Nicrophorus vespilloides, with over 75% of EVEs genome wide serving as templates for primary-piRNA biogenesis in these species (Fig. 3). piRNAs were not detected from EVEs in 13 species. Of these, 11 species did not possess EVEs within piRNA clusters.
FIG 3.
Percentage of EVEs producing primary piRNAs for each arthropod species. Blue, EVEs in piRNA clusters; red, EVEs outside piRNA clusters. Primary piRNA production from an EVE was defined as a significant (P < 0.001, cumulative binomial distribution) 1U bias for 24 to 32 sRNAs mapping to the EVE.
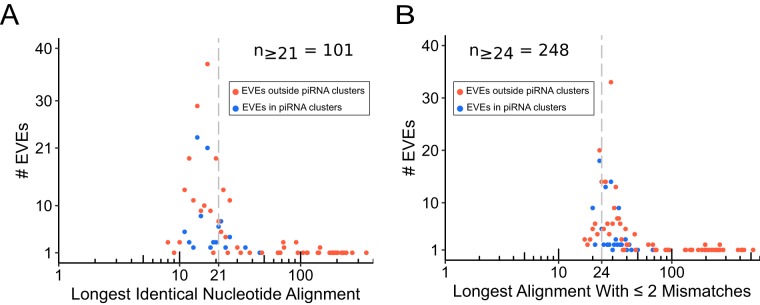
The production of piRNAs from EVEs does not imply that these piRNAs mediate antiviral responses. siRNA-directed cleavage is known to be highly dependent on the extent of base pairing between siRNAs and their targets (21). The exact complementarity requirements for piRNA-directed cleavage are unclear; however, results in the mouse model indicate that perfect base pairing between nucleotides 2 and 22 is required for efficient cleavage by Miwi (a homolog of Piwi) (22). Similarly, single-nucleotide mismatches between piRNAs and their RNA targets reduce the binding of D. melanogaster Piwi to target sites in an additive manner, with three mismatches being sufficient to abolish Piwi binding, regardless of the position of the mismatches (23). To elucidate the potential of piRNAs derived from EVEs to target known viruses, we used BLASTn to compare the nucleotide sequences of EVEs identified here to all viral nucleotide sequences available in GenBank. Based on the criteria described above, we then identified the longest stretch of identical nucleotides shared between EVEs and their BLASTn matches, as well as the longest stretch containing fewer than three mismatches and no internal gaps. We found that just 2.50% of EVEs shared aligned regions of ≥21 nt with their BLASTn hits (Fig. 4a). As a baseline for evaluating aligned regions containing fewer than three mismatches, we assumed a minimal piRNA length of 24 nt and calculated the number of EVE-virus pairs sharing a 24-nt stretch with ≤2 mismatches and no internal gaps. We found that only 6.11% of EVEs met these criteria (Fig. 4b). While these calculations are based on complementarity requirements that may not reflect the situation in species besides those in which they were experimentally determined, they provide a useful means of evaluating the nucleotide similarity between EVEs and their corresponding viruses.
FIG 4.
(A) Longest stretches of identical nucleotides shared between EVEs and their viral BLASTn matches. (B) Longest stretches of nucleotides shared between EVEs and their viral BLASTn matches containing two or fewer mismatches and/or terminal gaps.
EVE infrequently produce siRNAs.
Endogenous double-stranded RNA (dsRNA) substrates, such as those derived from bidirectional transcription of piRNA clusters (24), can give rise to endogenous siRNAs (esiRNAs), and esiRNA production from EVEs has been reported in A. albopictus (17, 24). Unlike primary piRNAs, esiRNAs are produced in similar amounts from both genomic strands (24). Thus, we determined whether EVEs identified here served as sources of siRNAs by evaluating whether sRNAs mapping to each EVE displayed a significant peak at 21 nt on both strands. The significance of 21-nt peaks is typically assessed by visual inspection of the length distribution of sRNAs mapping to a particular sequence. Given the size of our data set, we evaluated significant 21-nt peaks statistically and defined statistically significant peaks as those possessing a Z score of ≥1.96 for 21-nt reads over the range of 18 to 24 nt. We found only 27 EVEs with mapped sRNAs meeting these criteria (Data Set S6). To rule out the possibility that siRNAs could be derived from infection with exogenous viruses instead of being produced directly from EVEs, we calculated 21-nt Z scores for sRNAs mapping 1 kb upstream and downstream from each EVE. We found that only 10 EVEs resided in genomic loci for which there is strong evidence of siRNA production (Data Set S6). These results suggest that the majority of EVEs do not serve as sources of siRNAs.
sRNAs mapping to some EVEs show evidence of production via the ping-pong cycle.
While we found that nucleotide identity between EVEs and known viruses is generally low, which likely precludes induction of the ping-pong cycle by EVE-derived piRNAs upon infection with known viruses, currently described virus species are thought to represent only a small fraction of the total viral diversity, particularly for arthropod-infecting viruses (25). Thus, there is a possibility that EVE-derived piRNAs could target undescribed viruses, and the presence of ping-pong signatures in piRNAs mapping to EVEs would be one indication of the possible functionality of EVE-derived piRNAs. After defining EVEs that produced primary piRNAs (Fig. 3), we assessed whether 24- to 32-nt sRNAs mapping to these EVEs possessed significant ping-pong signatures. We defined a significant ping-pong signature as 1U and 10A biases for 24- to 32-nt sRNAs mapping to opposing strands and a ping-pong Z score of ≥3.2905. We found that sRNAs mapping to 3.4% of all EVEs displayed evidence of production via the ping-pong cycle, with 20 species possessing at least one EVE displaying evidence of ping-pong-dependent piRNA production (Table 2). This number was slightly higher for EVEs within piRNA clusters (5.37%) than for EVEs outside piRNA clusters (3.05%). While further experiments are necessary, we propose that one explanation for the observed ping-pong signatures could be infection with undescribed viruses corresponding to primary-piRNA-producing EVEs. It should be noted, however, that the observed ping-pong signatures could also be a result of bidirectional transcription of EVEs and/or transcription of duplicate copies of EVEs in opposite directions. Transcriptome-sequencing data paired with sRNA-sequencing data from the same arthropod population could clarify the contribution of such transcriptional mechanisms to the production of ping-pong-dependent EVE-derived piRNAs.
TABLE 2.
Percentage of EVEs with mapped 24- to 32-nt sRNAs displaying a significant ping-pong signature
| Species | Location | Total no. of EVEs |
% of EVEs with significant ping-pong signature |
|---|---|---|---|
| Aedes aegypti | Outside piRNA clusters | 156 | 13.46 |
| Inside piRNA clusters | 117 | 11.97 | |
| Aedes albopictus | Outside piRNA clusters | 499 | 7.21 |
| Anopheles gambiae | Outside piRNA clusters | 57 | 14.04 |
| Acyrthosiphon pisum | Outside piRNA clusters | 167 | 0.60 |
| Inside piRNA clusters | 127 | 0.79 | |
| Bactrocera dorsalis | Outside piRNA clusters | 9 | 11.11 |
| Blatella germanica | Outside piRNA clusters | 50 | 8.00 |
| Inside piRNA clusters | 16 | 18.75 | |
| Bombyx mori | Outside piRNA clusters | 28 | 17.86 |
| Inside piRNA clusters | 26 | 11.54 | |
| Diaphorina citri | Outside piRNA clusters | 86 | 3.49 |
| Inside piRNA clusters | 18 | 5.56 | |
| Drosophila mojavensis | Inside piRNA clusters | 2 | 50.00 |
| Drosophila virilis | Inside piRNA clusters | 3 | 33.33 |
| Helicoverpa armigera | Outside piRNA clusters | 17 | 5.88 |
| Herpegnathos saltator | Outside piRNA clusters | 131 | 7.63 |
| Musca domestica | Outside piRNA clusters | 3 | 33.33 |
| Inside piRNA clusters | 4 | 50.00 | |
| Myzus persicae | Outside piRNA clusters | 44 | 2.27 |
| Oncopeltus fasciatus | Outside piRNA clusters | 51 | 5.88 |
| Penaeus monodon | Outside piRNA clusters | 248 | 0.81 |
| Plutella xylostella | Inside piRNA clusters | 70 | 4.29 |
| Spodoptera frugiperda | Inside piRNA clusters | 24 | 4.17 |
| Triops cancriformis | Outside piRNA clusters | 55 | 12.73 |
| Inside piRNA clusters | 7 | 14.29 | |
| Tribolium castaneum | Inside piRNA clusters | 30 | 10 |
DISCUSSION
Mounting evidence points toward a role for EVEs in antiviral responses against corresponding viruses in animals, and both transcription and translation of EVEs have been hypothesized to play important roles. Indeed, some EVEs possess features of purifying selection, including maintenance of long open reading frames and low ratios of nonsynonymous/synonymous mutations (26). Moreover, experimental evidence indicating the functionality of EVE-encoded proteins has been shown in the thirteen-lined ground squirrel, the genome of which possess an EVE-encoded protein that inhibits replication of the corresponding virus in vitro (27). Proposed mechanisms of transcription-mediated EVE-based immunity include the production of primary piRNAs from EVE-derived transcripts, as well as the formation of dsRNA due to bidirectional transcription of EVEs and/or extensive secondary structure in EVE-derived transcripts (28, 29).
As reported previously for A. aegypti and A. albopictus, we found that EVEs were enriched in piRNA clusters in a majority of species analyzed. Nevertheless, EVEs were frequently found outside piRNA clusters. In general, EVEs inside piRNA clusters were not significantly different in length or identity to their closest viral BLASTx hit compared to EVEs outside piRNA clusters (Fig. 1). While the mechanisms of EVE formation are unclear, this process is thought to involve DNA chimeras consisting partly of viral sequence and partly of transposable element sequence that are formed during reverse transcription of endogenous retrotransposons and which may be capable of integrating into the host genome (30). Integration of a retrotransposon into a new genomic location and subsequent recognition of that retrotransposon by PIWI-bound piRNAs has been shown to be sufficient for the formation of a new piRNA-producing locus at the integration location (31). In this respect, it is tempting to speculate that the enrichment of EVEs within piRNA clusters may be due in part to their physical association with retrotransposons and the generation of new piRNA clusters at sites of integration of virus-transposon chimeras.
Previous research indicates that EVEs are widespread in mosquito genomes and commonly produce piRNAs (13, 14, 17). However, relatively little is known regarding the presence and functionality of EVEs in other arthropod species. Here, we examined 48 arthropod genomes representing species belonging to 16 orders. We found that, as has been demonstrated in mosquitoes, EVEs are pervasive in the genomes of species spread throughout the arthropod lineage and frequently serve as templates for the biogenesis of piRNAs. We found that EVE-derived piRNAs were common for EVEs both within and outside piRNA clusters. We note that even experimental annotation of piRNA clusters via sequencing of PIWI protein-bound piRNAs cannot place every piRNA-producing locus within a piRNA cluster (32). Thus, piRNA clusters annotated here and elsewhere represent useful and practical estimations but do not reflect the complete biological repertoire of piRNA-producing loci. In this context, the production of piRNAs from EVEs outside piRNA clusters is not surprising.
Curiously, no EVE-derived piRNAs were seen in Lutzomyia longipalpis or P. interpunctella despite the presence of EVEs within piRNA clusters in these organisms. While this may indeed reflect the biological reality, the EVE repertoire is known to vary between distinct populations of a given species and even between individuals within the same population (13, 17). This situation highlights a major difficulty of leveraging public data for analysis of EVEs. For a number of species, the available genome assemblies and sRNA data sets were derived from different strains of the organism, and in a small number of cases, sRNA data sets derived only from one sex, only from particular organs, or only from certain life stages were available. This complicates the mapping of sRNAs to host genomes, as the genomic content of the individuals from which the sRNA libraries were prepared may display important differences from that of the individuals used for genome sequencing. Moreover, most sRNA entries within the NCBI SRA database contain very little sample information (i.e., life stage, strain, sex, etc.), further complicating the interpretation of sRNA mapping results.
Besides piRNAs, there is a possibility that EVEs that are bidirectionally transcribed or that produce transcripts with sufficient secondary structure could serve as sources of dsRNA for the production of EVE-derived siRNAs. Indeed, a previous study found that some EVEs were located within regions of the A. albopictus genome that serve as sources of esiRNAs (17). We found that the production of siRNAs from EVEs identified here was extremely rare, with less than 1% of EVEs serving as sources of siRNAs.
Interestingly, we found that EVEs corresponding to negative-sense ssRNA viruses comprised a plurality of the EVEs identified here. This bias toward negative-sense ssRNA viruses has been noted previously (33). While the mechanisms underlying the prevalence of negative-sense ssRNA virus-derived EVEs remain unclear, it has been proposed that differences in the transcriptional strategies employed by ssRNA viruses may lead to the increased frequency of negative-sense ssRNA virus endogenization. Unlike positive-sense ssRNA viruses, which often produce long mRNAs encoding a single polyprotein, many negative-sense ssRNA viruses produce abundant short mRNAs, and thus, the predominance of negative-sense ssRNA virus endogenization may be a reflection of mRNA abundance and/or greater efficiency of reverse transcription on shorter RNA templates (33). We also identified a large number of EVEs corresponding to viruses of the family Parvoviridae. As opposed to RNA viruses, which must be reverse transcribed prior to endogenization, members of the family Parvoviridae possess ssDNA genomes and are capable of integrating into host genomes independent of retrotransposon activity, providing a potential explanation for the prevalence of parvovirus-derived EVEs (34).
Previous studies have revealed the presence of EVEs related to plant-infecting viruses within arthropod genomes (14, 35, 36). Interestingly, many of these EVEs correspond to viral families known to contain viruses that are transmitted by insect vectors but that do not replicate within insects. We identified numerous EVEs related to such plant-infecting viruses, including EVEs corresponding to the Geminiviridae, Nanoviridae, Luteoviridae, Potyviridae, Secoviridae, Tombusviridae, and Virgaviridae families. While we cannot draw conclusions regarding the origin of these EVEs related to plant viruses based on the present data, others have suggested that the phylogenetic position of such EVEs firmly within the known genetic diversity of plant viruses, as well as the fact that some plant virus-related EVEs encode putative proteins homologous to the movement proteins of plant viruses, indicates that they are derived from bona fide plant viruses rather than undocumented insect viruses (36). Yet others argue for the existence of ancient and undocumented viral families occupying phylogenetic gaps between insect- and plant-infecting viruses and that EVEs related to plant-infecting viruses may in fact be derived from novel families of insect-infecting viruses (35).
It has been proposed that EVE-derived piRNAs may play an antiviral role via the ping-pong cycle by directing posttranscriptional silencing of viral RNAs (14). Cleavage of RNA targets by primary piRNA-guided Argonaute proteins is dependent on base pairing between primary piRNAs and RNA targets (22). However, unlike siRNA-directed cleavage, piRNA-directed cleavage appears to tolerate a small number of mismatches (approximately ≤2), such that extensive but not perfect complementarity between piRNAs and their targets is required (22, 23). While we estimate that the nucleotide identity between the majority of EVEs identified here and known viruses is generally too low to permit targeting of known viruses by EVE-derived piRNAs, 24- to 32-nt sRNAs mapping to 3.4% of EVEs possessed significant ping-pong signatures. These results raise the possibility that piRNAs derived from these EVEs may play roles in responses to infection with corresponding undescribed viruses; however, further analyses are required to rule out bidirectional transcription of EVEs and/or transcription of multiple copies of EVEs in opposing directions as a source of dsRNA for induction of the ping-pong cycle. Moreover, our analysis was intended to estimate potential antiviral roles of EVE-derived piRNAs against currently known viruses based on nucleotide identity. It is possible and even likely, however, that the antiviral potential of EVE-derived piRNAs is greatest immediately following endogenization and that the antiviral effect diminishes over evolutionary time as the EVE and the corresponding virus diverge in nucleotide sequence. Thus, while the general lack of complementarity observed between EVEs and currently known viruses suggests that EVE-derived piRNAs do not play an antiviral role against these viruses, there is a possibility that the same EVE-derived piRNAs may have mediated antiviral effects in the past.
As with any homology-based method of viral or EVE discovery, our ability to identify EVEs is limited by the content of existing databases. Currently described viral species are thought to represent only a small fraction of total viral diversity, particularly for arthropod-infecting viruses, and our results must be interpreted within the context of these limitations (25). Indeed, arthropod genomes likely contain multitudes of EVEs corresponding to undescribed viruses. Moreover, the abundance and characteristics of the EVEs identified here depend entirely on the BLAST results, which themselves are a function of database content and the parameters used for the BLAST searches. Compared to EVEs that are very similar to known viruses, EVEs that are highly divergent from known viruses will be either unidentified or represented by truncated and/or fragmented BLAST output. Some previous studies have implemented curated BLAST databases containing only a subset of available viral sequences (13). In contrast, our BLAST database was comprised of all nonretroviral RNA and ssDNA virus protein sequences available in GenBank. In this respect, while our results represent a more complete picture of the EVE repertoire, the present findings must be understood to be an incomplete description of EVE abundance, diversity, function, and characteristics that is systematically biased by the content of the BLAST database.
An understanding of arthropod antiviral immunity is critical for the development of novel strategies to control vector-mediated virus transmission to animal and plant hosts. Our findings reveal that the important observations regarding the functionality of EVEs in mosquitoes apply to a wide range of other arthropod species and lend further support to the hypothesis that, in some circumstances, EVEs may constitute a form of heritable immunity against corresponding viruses. While EVEs may indeed occasionally provide the basis for an immunological response, we propose that given the lack of extensive nucleotide identity observed between EVEs identified here and currently described exogenous viruses, endogenization of viral sequences is an infrequent event and the ability of EVE-derived piRNAs to initiate a response against virus infection may decline over evolutionary time as exogenous viruses and their corresponding EVEs diverge. To gain an understanding of the general utility of the interaction between EVEs and the piRNA pathway as an antiviral mechanism, future studies should address the timescale over which acquisition of new EVEs takes places and to what extent genomic EVE content varies between geographically distinct populations of a given species.
MATERIALS AND METHODS
Data collection.
A list of currently sequenced arthropod genomes was retrieved from the 5000 Arthropod Genomes Initiative (i5K) (37). Genome sequences were then retrieved from GenBank for all species with sRNA sequencing data available in the NCBI Sequence Read Archive (SRA). The accession numbers for all genome assemblies analyzed are available in Table S3 in the supplemental material. For each arthropod species, a representative collection of available sRNA data sets was retrieved from the NCBI SRA and the data sets were combined for analysis. The accession numbers of sRNA data sets used for each species are available in Table S2.
Identification of EVEs.
To identify EVEs in arthropod genomes, we created a BLAST database containing all nonretroviral ssRNA, dsRNA, and ssDNA virus protein sequences available in GenBank. We did not include double-stranded DNA (dsDNA) viruses in our analysis due to the difficulty in unambiguously characterizing dsDNA viral sequences to be of viral origin, due to the frequency of horizontal gene transfer between dsDNA viruses and their hosts and between dsDNA viruses and transposable elements. For each arthropod species, we searched for matches to our viral-protein database genome wide with BLASTx using the default settings with the following exceptions: -evalue 0.001 -outfmt 5. The results were subsequently parsed using a custom python script (parse_xml.py). As reported previously, we found that a large number of putative EVEs identified by this process could not be unambiguously classified as viral sequences due to homology with eukaryotic, bacterial, or archaeal sequences (14). Such artifacts were initially filtered out of the data set using a custom bash script (BLAST_filter.sh) to extract the genomic nucleotide sequence corresponding to each BLASTx hit (i.e., putative EVEs) and then performing a reverse BLASTx search with these nucleotide sequences against the D. melanogaster proteome (Uniprot proteome accession number UP000000803) using the default settings with the following exceptions: -max_target_seqs 1 -max_hsps 1 -evalue 0.001 -outfmt 10. Any putative EVEs with a BLASTx hit against the D. melanogaster proteome were subsequently removed from analysis. Following this initial filter, the viral proteins corresponding to each putative EVE were compared to the nonredundant protein database with web-based BLASTp using the default settings, and the results were screened manually. If the putative EVE corresponded to a portion of the viral protein possessing a nonviral BLASTp hit or a conserved domain with a lineage not exclusive to viruses (e.g., zinc finger domains), then it was removed from the data set.
Custom python scripts were used to remove overlapping EVEs (parse_xml.py, remove_duplicates.py, and remove_in_frame.py). When two EVEs had any degree of overlap, the EVE with the higher BLASTx score was retained (13). An EVE was defined as one continuous BLASTx hit. Custom python scripts (viral_families.py, count_names.py, compare_tax_and_names.py, species_family.py, and count_families.py) were then used to assign a viral family to each EVE by searching the NCBI taxonomy database (ftp://ftp.ncbi.nih.gov/pub/taxonomy/) using as a query the closest viral match to each EVE (as identified by BLASTx) (14, 15).
Identification of EVEs in piRNA clusters.
Adapter sequences were removed from the sRNA data sets with Cutadapt (version 1.16), using the default settings with the exception that reads as short as 18 nt were retained (38). After trimming, all the sRNA data sets for each species were concatenated into one data set per species. These concatenated sRNA data sets were used for all further analysis. piRNA clusters were defined with proTRAC (version 2.3.1) using the default settings with the following exceptions: sliding window size = 1,000, sliding window increment = 500, threshold cluster size = 1,500, and threshold-density P value = 0.1 (19). We identified EVEs within piRNA cluster sequences obtained with proTRAC as described above for the identification of EVEs genome wide. Despite the publication of an alternative and equally flexible piRNA cluster detection algorithm during preparation of the manuscript (39), we chose to use proTRAC to maintain consistency with previous results (14). A custom python script (remove_picluster.py) was then used to remove any EVEs from the genome-wide EVE list that were present in the piRNA cluster EVE list. If an EVE was partially inside and partially outside a piRNA cluster, it was marked as residing outside the piRNA cluster.
To determine whether EVEs were enriched within piRNA clusters, we estimated the probability of our observed EVE counts within piRNA clusters using a cumulative binomial distribution in which the probability of integration was assumed to equal the total number of EVEs genome wide (i.e., the number of EVEs inside and outside piRNA clusters) divided by the total length of the genome in base pairs (13). A cumulative binomial probability of <0.05 was deemed to be evidence of significant enrichment of EVEs within piRNA clusters (13).
Small RNA mapping, piRNA identification, and siRNA identification.
Concatenated sRNA reads were mapped to arthropod genomes with Bowtie (version 1.1.2), using the default settings (40). Individual BAM files corresponding to each EVE were then generated using SAMtools based on the genomic coordinates of each EVE, and sRNAs mapping to each EVE were extracted from these BAM files using BEDTools (41, 42). Custom scripts were used to calculate whether an EVE served as a source of primary piRNAs. EVEs were marked as producing primary piRNAs if sRNAs mapping to that EVE displayed a significant peak within the piRNA length range (determined with sign_piRNA_peak.py) and sRNAs within the piRNA length range possessed a significant 1U bias on one or both strands (determined with nt_bias.sh and python scripts executed therein). To identify significant piRNA peaks, we used a binomial test to calculate whether 24- to 32-nt sRNAs were enriched compared to all sRNAs within the 18- to 36-nt length range (excluding sRNAs with a length of 21 nt), assuming a null hypothesis that sRNAs are evenly distributed across the 18- to 36-nt length range. Enrichment within the 24- to 32-nt piRNA length range was defined as a P value of <0.001. To assess 1U biases, we calculated the percentage of 24- to 32-nt sRNAs mapping to each EVE that began with uridine and determined the significance of these percentages using a binomial distribution, assuming that the probability of an sRNA beginning with a uridine was 25%. A significant 1U bias was defined as a P value of <0.001 for 24- to 32-nt sRNAs mapping to one strand of the EVE. Unlike some other previously described approaches, our analysis examined 1U biases on either strand individually and did not require primary piRNAs to be derived from the antisense strand with respect to the coding potential of the EVEs.
To determine whether sRNAs mapping to each EVE possessed a significant ping-pong signature, we first used custom scripts to calculate whether 24- to 32-nt sRNAs mapping to each EVE possessed a significant 1U bias as described above. If a 1U bias was observed for sRNAs mapping to one strand, we determined whether 24- to 32-nt sRNAs mapping to the opposite strand possessed a significant 10A bias (P < 0.001, binomial distribution). We then calculated a ping-pong Z score for 24- to 32-nt sRNAs mapping to each EVE using previously published and custom scripts (signature.py [43], sig_ping_pong.sh, and python scripts executed therein). sRNAs mapping to each EVE were classified as possessing a significant ping-pong signature if we observed significant 1U and 10A biases for 24- to 32-nt sRNAs mapping to opposing strands and if the ping-pong Z score was ≥3.2905 (which corresponds to a P value of 0.001 for a two-tailed hypothesis).
To determine whether EVEs served as sources of siRNAs, we first used a custom script (Z_score_21.py) to evaluate whether sRNAs mapping to each EVE displayed a significant peak at 21 nt on both strands. A significant 21-nt peak was defined as a Z score of ≥1.96 for the number of 21-nt reads compared to the number of reads of other lengths within the range of 18 to 24 nt. To exclude the possibility that putative EVE-derived siRNAs were produced from corresponding exogenous viruses as a result of viral infection, we also calculated Z scores at 21 nt for sRNAs mapping 1 kb upstream and downstream from each EVE. Only siRNA-producing EVEs located within siRNA-producing genomic loci were marked as producing siRNAs.
Calculation of nucleotide identities.
To calculate the longest identical nucleotide stretches shared between EVEs and their BLASTn matches, we compared each EVE nucleotide sequence to all nonretroviral ssRNA, dsRNA, and ssDNA viral nucleotide sequences available in GenBank with BLASTn, using the default parameters with the following exceptions: -evalue .001 -word_size 7 -outfmt “6 pident qcovs btop” -parse_deflines. A custom bash script (highest_nt_stretch.sh) was used to extract the longest stretch of identical nucleotides from each alignment. To prevent spurious alignments, we required query coverage to be >39.9%. Using these same BLASTn alignments, we calculated the longest stretches of shared nucleotides between EVEs and their BLASTn hits with ≤2 mismatches and no internal gaps using a custom python script (calculate_string_value.py). Up to two total terminal subject gaps (EVE query contains additional nucleotides compared to the viral subject) were permitted at either end of each calculated nucleotide stretch. Internal subject gaps were not permitted. Neither internal nor external query gaps (viral subject contains additional nucleotides compared to the EVE query) were permitted in calculated nucleotide stretches.
Data availability.
The custom scripts produced during the current study are available on GitHub (https://github.com/AnneliektH/EVEs_arthropod). Data sets not provided within the supplemental material are available from the corresponding author upon request.
Supplementary Material
ACKNOWLEDGMENTS
A.M.T.H., J.C.N., and F.M.D. wrote the scripts. A.M.T.H. and J.C.N. performed the analyses. J.C.N. conceived the study. A.M.T.H., J.C.N., and B.W.F. analyzed the data and wrote the manuscript.
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under grant 1650042 and by grants from the U.S. Department of Agriculture (grants 13-002NU-781 and 2015-70016-23011) and the University of California.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02124-18.
REFERENCES
- 1.Joern A, Laws AN. 2013. Ecological mechanisms underlying arthropod species diversity in grasslands. Annu Rev Entomol 58:19–36. doi: 10.1146/annurev-ento-120811-153540. [DOI] [PubMed] [Google Scholar]
- 2.Momot WT. 1995. Redefining the role of crayfish in aquatic ecosystems. Rev Fish Sci 3:33–63. doi: 10.1080/10641269509388566. [DOI] [Google Scholar]
- 3.Whitfield AE, Falk BW, Rotenberg D. 2015. Insect vector-mediated transmission of plant viruses. Virology 479:278–289. doi: 10.1016/j.virol.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Gray SM, Banerjee N. 1999. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol Mol Biol Rev 63:128–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calisher CH, Higgs S. 2018. The discovery of arthropod-specific viruses in hematophagous arthropods: an open door to understanding the mechanisms of arbovirus and arthropod evolution? Annu Rev Entomol 63:87–103. doi: 10.1146/annurev-ento-020117-043033. [DOI] [PubMed] [Google Scholar]
- 6.Palmer WH, Varghese FS, Van Rij RP. 2018. Natural variation in resistance to virus infection in dipteran insects. Viruses 10:E118. doi: 10.3390/v10030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obbard DJ, Gordon KH, Buck AH, Jiggins FM. 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci 364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miesen P, Joosten J, van Rij RP. 2016. PIWIs go viral: arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog 12:e1006017. doi: 10.1371/journal.ppat.1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petit M, Mongelli V, Frangeul L, Blanc H, Jiggins F, Saleh M-C. 2016. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc Natl Acad Sci U S A 113:E4218–E4227. doi: 10.1073/pnas.1607952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 41:324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell CL, Black WC, Hess AM, Foy BD. 2008. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics 9:425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miesen P, Girardi E, van Rij RP. 2015. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res 43:6545–6556. doi: 10.1093/nar/gkv590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palatini U, Miesen P, Carballar-Lejarazu R, Ometto L, Rizzo E, Tu Z, van Rij RP. 2017. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics 18:512. doi: 10.1186/s12864-017-3903-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitfield ZJ, Dolan PT, Kunitomi M, Tassetto M, Seetin MG, Oh S, Heiner C, Paxinos E, Andino R. 2017. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr Biol 27:3511–3519. doi: 10.1016/j.cub.2017.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet 6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.François S, Filloux D, Roumagnac P, Bigot D, Gayral P, Martin DP, Froissart R, Ogliastro M. 2016. Discovery of parvovirus-related sequences in an unexpected broad range of animals. Sci Rep 6:30880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki Y, Frangeul L, Dickson LB, Blanc H, Verdier Y, Vinh J, Lambrechts L, Saleh M-C. 2017. Uncovering the repertoire of endogenous flaviviral elements in Aedes mosquito genomes. J Virol 91:e00571-17. doi: 10.1128/JVI.00571-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ter Horst AM, Nigg JC, Falk BW. 2018. Endogenous viral elements are widespread in arthropod genomes and commonly give rise to piRNAs. bioRxiv doi: 10.1101/396382. [DOI] [PMC free article] [PubMed]
- 19.Rosenkranz D, Zischler H. 2012. proTRAC—a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformatics 13:5. doi: 10.1186/1471-2105-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lourenço-de-Oliveira R, Marques JT, Sreenu VB, Atyame Nten C, Aguiar ERGR, Varjak M, Kohl A, Failloux A-B. 2018. Culex quinquefasciatus mosquitoes do not support replication of Zika virus. J Gen Virol 99:258–264. doi: 10.1099/jgv.0.000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena S, Jónsson ZO, Dutta A. 2003. Small RNAs with imperfect match to endogenous mRNA repress translation implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem 278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 22.Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 23.Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X, Qin X-C, Li J, Cao J-P, Eden J-S, Buchmann J, Wang W, Xu J, Holmes EC, Zhang Y-Z. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 26.Aswad A, Katzourakis A. 2012. Paleovirology and virally derived immunity. Trends Ecol Evol 27:627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Fujino K, Horie M, Honda T, Merriman DK, Tomonaga K. 2014. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc Natl Acad Sci U S A 111:13175–13180. doi: 10.1073/pnas.1407046111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh M-C. 2013. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol 14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 29.Parrish NF, Fujino K, Shiromoto Y, Iwasaki YW, Ha H, Xing J, Makino A, Kuramochi-Miyagawa S, Nakano T, Siomi H, Honda T, Tomonaga K. 2015. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA 21:1691–1703. doi: 10.1261/rna.052092.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirier EZ, Goic B, Tomé-Poderti L, Frangeul L, Boussier J, Gausson V, Blanc H, Vallet T, Loyd H, Levi LI, Lanciano S, Baron C, Merkling SH, Lambrechts L, Mirouze M, Carpenter S, Vignuzzi M, Saleh M-C. 2018. Dicer-2-dependent generation of viral DNA from defective genomes of RNA viruses modulates antiviral immunity in insects. Cell Host Microbe 23:353–365.E8. doi: 10.1016/j.chom.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olovnikov I, Ryazansky S, Shpiz S, Lavrov S, Abramov Y, Vaury C, Jensen S, Kalmykova A. 2013. De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment. Nucleic Acids Res 41:5757–5768. doi: 10.1093/nar/gkt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 33.Holmes EC. 2011. The evolution of endogenous viral elements. Cell Host Microbe 10:368–377. doi: 10.1016/j.chom.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapoor A, Simmonds P, Lipkin WI. 2010. Discovery and characterization of mammalian endogenous parvoviruses. J Virol 84:12628–12635. doi: 10.1128/JVI.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo H, Chiba S, Maruyama K, Andika IB, Suzuki N. 2017. A novel insect-infecting virga/nege-like virus group and its pervasive endogenization into insect genomes. Virus Res 2017:S0168-1702(17)30701-3. doi: 10.1016/j.virusres.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Cui J, Holmes EC. 2012. Endogenous RNA viruses of plants in insect genomes. Virology 427:77–79. doi: 10.1016/j.virol.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.i5K Consortium. 2013. The i5K initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered 104:595–600. doi: 10.1093/jhered/est050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 39.Ray R, Pandey P. 2018. piRNA analysis framework from small RNA-Seq data by a novel cluster prediction tool—PILFER. Genomics 110:355–365. doi: 10.1016/j.ygeno.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antoniewski C. 2014. Computing siRNA and piRNA overlap signatures, p 135–146. In Werner A. (ed), Animal endo-siRNAs: methods and protocols. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The custom scripts produced during the current study are available on GitHub (https://github.com/AnneliektH/EVEs_arthropod). Data sets not provided within the supplemental material are available from the corresponding author upon request.