The matrix protein of the Newcastle disease virus (NDV) is one of the most abundant viral proteins that regulates the formation of progeny virions. NDV is an avian pathogen that impacts the economics of bird husbandry due to its resulting morbidity and high mortality rates. Moreover, it belongs to the Avulavirus subfamily of the Paramyxoviridae family of Mononegavirales that include dangerous representatives such as respiratory syncytial virus, human parainfluenza virus, and measles virus. Here, we investigate the solution structure and membrane binding properties of this protein at both acidic and neutral pH to distinguish between possible virus entry pathways and propose a mechanism of assembly of the viral matrix scaffold. This work is fundamental for understanding the mechanisms of viral entry as well as to inform subsequent proposals for the possible use of the virus as an adequate template for future drug or vaccine delivery.
KEYWORDS: Newcastle disease virus, ab initio modeling, atomic force microscopy, matrix protein, protein structure, small-angle X-ray scattering
ABSTRACT
Newcastle disease virus (NDV) is an enveloped paramyxovirus. The matrix protein of the virus (M-NDV) has an innate propensity to produce virus-like particles budding from the plasma membrane of the expressing cell without recruiting other viral proteins. The virus predominantly infects the host cell via fusion with the host plasma membrane or, alternatively, can use receptor-mediated endocytic pathways. The question arises as to what are the mechanisms supporting such diversity, especially concerning the assembling and membrane binding properties of the virus protein scaffold under both neutral and acidic pH conditions. Here, we suggest a novel method of M-NDV isolation in physiological ionic strength and employ a combination of small-angle X-ray scattering, atomic force microscopy with complementary structural techniques, and membrane interaction measurements to characterize the solution behavior/structure of the protein as well as its binding to lipid membranes at pH 4.0 and pH 7.0. We demonstrate that the minimal structural unit of the protein in solution is a dimer that spontaneously assembles in a neutral milieu into hollow helical oligomers by repeating the protein tetramers. Acidic pH conditions decrease the protein oligomerization state to the individual dimers, tetramers, and octamers without changing the density of the protein layer and lipid membrane affinity, thus indicating that the endocytic pathway is a possible facilitator of NDV entry into a host cell through enhanced scaffold disintegration.
IMPORTANCE The matrix protein of the Newcastle disease virus (NDV) is one of the most abundant viral proteins that regulates the formation of progeny virions. NDV is an avian pathogen that impacts the economics of bird husbandry due to its resulting morbidity and high mortality rates. Moreover, it belongs to the Avulavirus subfamily of the Paramyxoviridae family of Mononegavirales that include dangerous representatives such as respiratory syncytial virus, human parainfluenza virus, and measles virus. Here, we investigate the solution structure and membrane binding properties of this protein at both acidic and neutral pH to distinguish between possible virus entry pathways and propose a mechanism of assembly of the viral matrix scaffold. This work is fundamental for understanding the mechanisms of viral entry as well as to inform subsequent proposals for the possible use of the virus as an adequate template for future drug or vaccine delivery.
INTRODUCTION
Matrix proteins of enveloped viruses are multifunctional and commonly the most abundant viral proteins. Viral matrix proteins play an essential role in virus assembly, the budding of progeny virions, and other processes in the viral replication process (1–4). Newcastle disease virus (NDV) is a pleomorphic enveloped virus with a negative-strand, nonsegmented RNA genome belonging to the Avulavirus genus in the Paramyxoviridae family (5). The virus causes significant disease in most species of birds, resulting in burdens on agriculture and the global economy. The matrix protein of the virus, M-NDV, contains 364 amino acids (for all reported strains) and has a molecular weight (MW) of approximately 40 kDa. The M-NDV translocates into nucleus (6, 7) and plays a crucial role in virus budding (8). The majority of matrix proteins of paramyxoviruses are dimers, forming a grid-like array on the inner surface of the viral membrane (9–12) that interacts with both the cytoplasmic tails of the transmembrane dual-function hemagglutinin/neuraminidase (HN) and fusion (F) glycoproteins (8, 13) as well as the nucleocapsid (2, 14–16). The M-NDV is a highly conserved protein which has been proposed as a key marker protein to classify different NDV strains (17). The simple analysis of the protein amino acid sequence shows that the net charge of the protein is positive at pH 7.0 and that it contains a preponderance of hydrophobic residues, making the protein hydrophobic in nature. Nevertheless, there is yet no evidence that M-NDV inserts into lipid bilayers, making it a peripheral protein that is positioned between the viral lipid membrane and the nucleocapsid (6). Isolated M-NDV proteins adsorb onto phospholipid bilayers and condense into fluid-like domains, causing membrane deformation and budding of spherical vesicles (18). Recently, X-ray crystallography and electron tomography investigations (19) have shown the pseudoatomic structure of assembled M-NDV dimers. The dimers form tetrameric arrays, with the angle between dimers generating the membrane curvature necessary for virus budding. These matrix protein arrays also organize viral glycoproteins (anchored in the gaps between them) and the nucleocapsid. Thus, the matrix protein arranges the maturation and infection process in time and space, participating in the sequence of events during budding and fusion (19).
Enveloped viruses enter the cell through two main pathways: direct fusion between the viral envelope and the plasma membrane and receptor-mediated endocytosis followed by fusion (20). It is assumed that, in the case of paramyxoviruses, the membrane fusion process takes place at the host plasma membrane in a pH-independent manner. However, the fusion of NDV with cultured cells may be enhanced at acidic pH (21). Recent studies indicate that low pH can serve as a fusion trigger for some paramyxoviruses (22), and protonation of specific amino acids in their fusion proteins could promote low-pH-induced conformational changes through an electrostatic repulsion mechanism (23). Thus, the virus may also penetrate the cell via the endocytic pathway in a pH-dependent process, or, as has been suggested by Cantin et al. (24), virus entry can be a more complicated process exploiting different entry routes. The same virus may have different entry preferences, depending on the cell line. Therefore, why and how viruses are able to use exact entry pathways remain open questions, as well as the possible effects of pH on the viral matrix proteins. Answering these questions is crucial for the development of antiviral agents as well as for the use of viruses as tools in therapy, e.g., in gene therapy to deliver genes into cells or in the delivery of pharmaceuticals, in particular anticancer drugs. The use of NDV for cancer therapy is due to the fact that the virus has significant oncolytic activity. NDV appears to selectively kill cancer cells while sparing normal human cells (25, 26).
M-NDV is responsible for the viral assembly and disassembly, budding, and interactions with lipid membranes, and pH may influence not only the fusion proteins of the virus but also its protein matrix layer to facilitate virus disassembly at the endocytic route. To investigate the structural response of M-NDV at different pH values in terms of self-assembly and the dissociation of the viral scaffold, we investigated the M-NDV structure in solution using small-angle X-ray scattering (SAXS) combined with atomic force microscopy (AFM) and complementary characterization techniques, including intramembrane field compensation (IFC).
RESULTS
CD analysis of the isolated M-NDV protein.
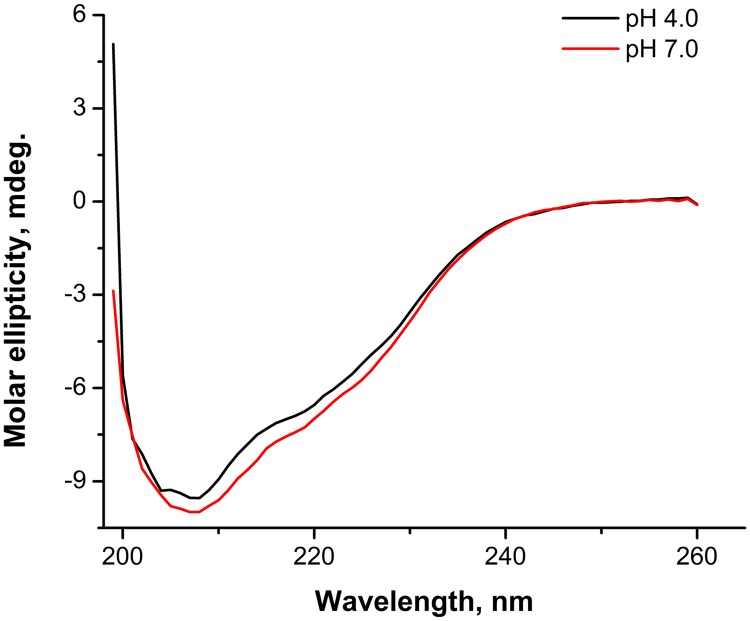
To characterize the secondary structure of the isolated matrix protein, we performed circular dichroism (CD) spectropolarimetry measurements. In the present work, we used a Chirascan dichrograph to measure the far-UV CD spectrum of isolated M-NDV in 50 mM morpholineethanesulfonic acid (MES) and 100 mM NaCl buffer, pH 4.0 and 7.0 (Fig. 1). In these spectra (with buffer value subtracted and result normalized to protein concentration), the main negative maximum (θ-max) was located at 207 nm. The position and intensity of the θ-max practically did not change with pH (−9.540 millidegrees [mdeg] at pH 4.0 and –10.000 mdeg at pH 7.0). The α-helical content of the protein determined using the Greenfield-Fasman equation (27) and K2D2 web service (28) was found to be 20% at both pH 4.0 and pH 7.0. That value is close to the 19% of α-helices obtained from high-resolution structure (PDB accession number 4G1L [19]). Thus, our results indicate little quantitative difference in the protein helical content values at acidic and neutral pH.
FIG 1.
CD spectra of isolated M-NDV in 50 mM MES–100 mM NaCl buffer, pH 4.0 and pH 7.0.
AUC shape characterization of the M-NDV.
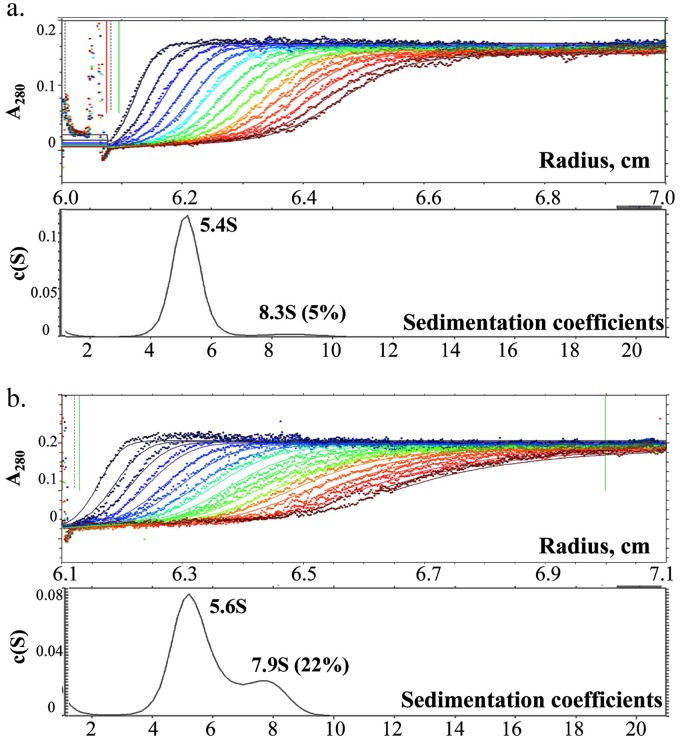
We assessed the shape of the M-NDV molecules in a solution of 0.3 mg/ml in 50 mM MES and 100 mM NaCl at room temperature, using analytical ultracentrifugation (AUC) (Fig. 2). For both pH 4.0 and pH 7.0 conditions, we obtained the major peak with almost the same sedimentation velocity. The sedimentation coefficient, s20,w, was equal to 5.4S at pH 4.0 and 5.6S at pH 7.0. The translational friction ratio f/f 0 of 1.2 ± 0.02 obtained for the M-NDV molecules indicates a slight deviation from a spherical shape. The theoretical value for the maximal possible sedimentation coefficient of the compact sphere with a molecular weight of the dimer of M-NDV is 6.2S. Thus, with regard to deviation from the spherical shape, the sedimentation coefficient values give the dimer as the basic structural unit of the M-NDV in solution. The minor peaks at 8.3S for pH 4.0 and 8.2S for pH 7.0 correspond to large protein associates, possibly tetramers. These associates are 5% (vol/vol) in acidic medium and 22% (vol/vol) in the neutral milieu, indicating the increase in concentration of self-associated M-NDV at pH 7.0.
FIG 2.
Sedimentation velocity experiment on the M-NDV protein at pH 4.0 (a) and pH 7.0 (b). Top panels show the raw data; absorbance at 280 nm was plotted as a function of the radial position, and every fifth curve is shown. Lower panels show fitted distributions of the sedimentation coefficients.
AFM of the M-NDV layers.
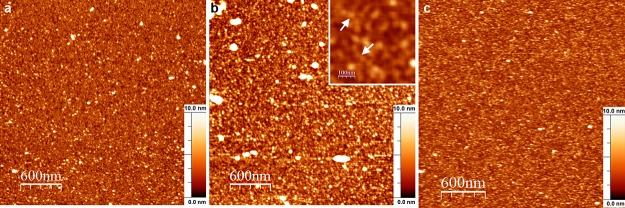
Atomic force microscopy (AFM) studies of the M-NDV adsorbed on a mica surface show the formation of a dense protein layer at protein concentrations of about 200 nM in solutions on the mica surface (Fig. 3). As the mica surface is negatively charged in aqueous solution, it provides surface charge density similar to that of the inner leaflet of the plasma membrane (29) that is necessary for the correct alignment of the matrix protein molecules (19). Moreover, the atomically flat solid mica surface allows high-resolution AFM images to be obtained. The density of the adsorbed layer was 91% ± 3% (mean ± standard deviation [SD]; n = 12) at pH 4.0 and 97% ± 2% (n = 8) at pH 7.0. The average thickness of the adsorbed layer was 2.5 nm ± 0.7 nm (n = 12) at pH 4.0 and 3.4 nm ± 0.6 nm (n = 8) at pH 7.0. We observed large protein associates with the height up to 40 nm on the surface at pH 7.0 (Fig. 3a, bright spots) occupying up to 1.5% of the surface area. The inset in Fig. 3b shows a high-resolution image of the protein layer indicating the existence of curved protein stripes with a length of about 100 nm. These structures are protein oligomers, suggesting that the protein is assembled on the mica. The observed height of the M-NDV layer at pH 7.0 was higher than the radius of the crystal dimer (PDB accession number 4G1L), with a radius of gyration (Rg) of 2.56 nm, while close to the thickness of the matrix protein layer of about 4 nm observed by cryo-electron tomography (19). The thickness of the layer at pH 4.0 was close to the Rg of the M-NDV dimer in crystal (19). Changing the pH from 7.0 to 4.0 led to disappearance of these aggregates and a decrease in the layer average thickness to 2.1 nm ± 0.7 nm (n = 5), which corresponds to the case of pH 4.0 (Fig. 3c).
FIG 3.
AFM topography images of the M-NDV layers adsorbed at freshly cleaved mica at pH 4.0 (a) and pH 7.0 (b) and the change from pH 7.0 to pH 4.0 (c). The inset in panel b shows a zoomed part of the protein layer, with white arrows indicating protein clusters. The bulk protein concentration in the solution above the surface was 200 nM. The full z-axis scale is 10 nm. The bright spots that are particularly more evident at pH 7.0 are large protein self-associates.
M-NDV interactions with lipid membranes.
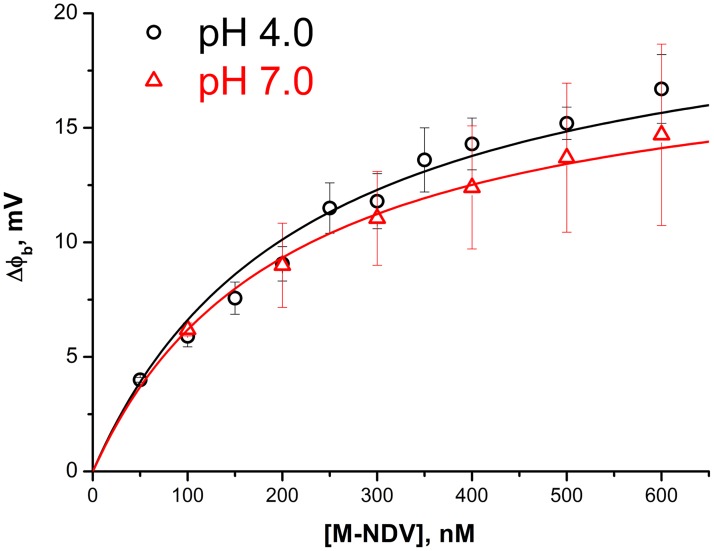
M-NDV is a positively charged protein at pH 7.0; therefore, its adsorption on the one side of the planar lipid membrane should change the transmembrane electrical field. This change can be measured by the intramembrane field compensation (IFC) technique as the shift of boundary potential difference, Δφb, providing the kinetics of protein adsorption (29). Although the M-NDV protein is overall positively charged, its membrane interactions are independent of the ionic strength (19) and are dominated by hydrophobic interactions. In addition, there is no evidence that the protein inserts into the lipid bilayer; it simply associates with the membrane surface. We constituted a diphytanoylphosphatidylserine-diphytanoylphosphatidylcholine (DPhPS-DPhPC) membrane (30:70 mol%) to have both negatively charged phosphoserine moieties for possible electrostatic interactions while providing access to the hydrophobic core of the membrane via branched phytanol lipid tails. The latter should increase the matrix protein adsorption via hydrophobic interactions (30). The addition of the M-NDV to the one side of such a freestanding planar bilayer lipid membrane (BLM) led to an increase in the boundary potential difference of the membrane, reflecting the adsorption of the protein. After the stationary level was reached, we performed perfusion with protein-free buffer that led to a decrease of the boundary potential difference to zero at both pH 7.0 and pH 4.0, meaning reversible protein adsorption (data not shown) (29). Taking the average stationary level of the potential difference at different bulk concentrations of M-NDV, we obtained adsorption isotherms at pH 4.0 and pH 7.0 (Fig. 4). Fitting the isotherms with a one-site binding model (Langmuir isotherm) resulted in M-NDV binding constants to the DPhPS-DPhPC membrane of (4.4 ± 0.4) ×106 M−1 at pH 4.0 and (4.9 ± 0.2) ×106 M−1 at pH 7.0. Therefore, M-NDV interactions with lipid membrane do not show any pH dependence.
FIG 4.
Isotherms of M-NDV adsorption on lipid bilayer membranes (DPhPS-DPhPC) obtained by IFC at pH 4.0 and pH 7.0. Each point corresponds to a stationary level for the given concentration, averaged over three independent experiments. Solid lines are the fit of the experimental data by a one-site binding model (Langmuir isotherm).
SAXS measurements of the M-NDV protein in solution.
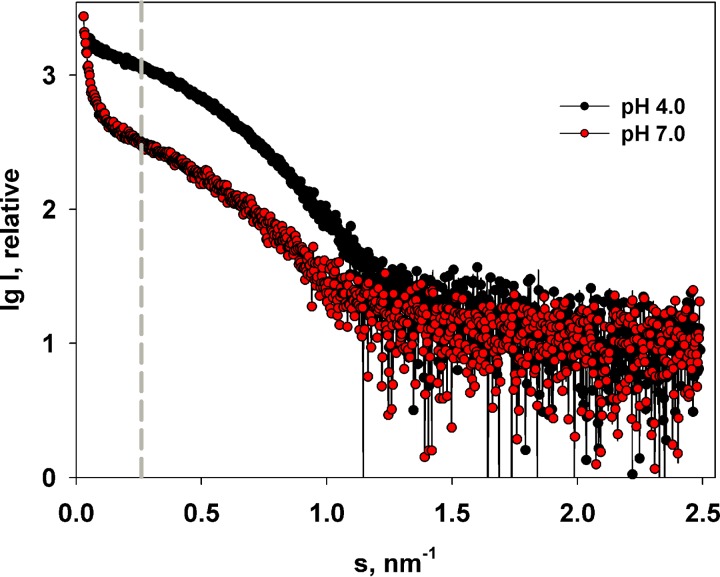
To further analyze the M-NDV structure, small-angle X-ray scattering (SAXS) measurements were performed in solutions at both pH 4.0 and 7.0 (Fig. 5) in a concentration range from 1 to 3 mg/ml. The resulting SAXS profiles can be divided into two main regions. A very low scattering-angle region (momentum transfer s of < 0.25 nm−1) that we interpret as being sensitive to the formation of higher-order self-associates and a higher scattering-angle region (momentum transfer s of >0.25 nm−1) that we interpret as scattering intensities arising from predominantly lower-order oligomeric species (dimers, tetramers, etc.). In a comparison of the higher-angle data measured from M-NDV at either pH 4.0 or pH 7.0, only minor changes in the scattering curves were observed. For example, a weak concentration dependence was detected at both pHs. This concentration dependence causes a small change in the magnitude of the radii of gyration. At pH 4.0, the Rg decreases when the protein concentration is decreased from 3.5 nm ± 0.2 nm at 3 mg/ml to 3.1 nm ± 0.2 nm at 1 mg/ml while, in a similar fashion at pH 7.0, the Rg decreases from 3.5 nm ± 0.2 nm to 3.3 nm ± 0.2 nm over the same concentration range. Given these minor concentration effects on the high-angle data, further analysis (see below) was performed from the data recorded at an average solute concentration of 2.2 mg/ml under both pH conditions.
FIG 5.
Experimental SAXS curves from 2.2 mg/ml solutions of M-NDV at pH 4.0 and pH 7.0. The gray dashed line at s = 0.25 nm−1 separates regions of the scattering profile dominated by large M-NDV self-assemblies (to the left of the line) and smaller protein oligomers, for example, dimers and tetramers (to the right of the line).
Unlike the data recorded at higher angles, a marked difference in SAXS intensities in the very-low-angle region were seen when the data measured for a momentum transfer s of <0.25 nm−1 from the M-NDV protein at low or neutral pH were compared (Fig. 5). The scattering curve at pH 7.0 shows a very strong upturn at very small angles, indicating the formation of large, soluble protein self-assemblies at pH 7.0, while their amount at pH 4.0 is rather subtle. These SAXS results are consistent with the AFM and AUC data that show the presence of larger-MW species at neutral pH, reflecting a process of pH-dependent M-NDV oligomerization.
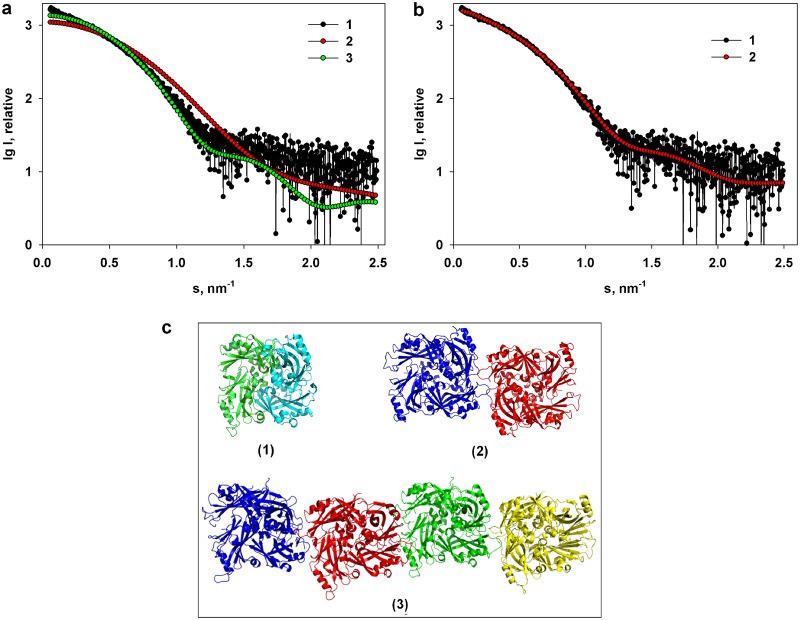
The X-ray scattering in acidic medium in the range of a momentum transfer of 0.25 < s < 2.5 nm−1 is characteristic of a solution containing a limited number of large-MW aggregates and allows for the calculation of the overall structural parameters of the M-NDV protein at pH 4.0: Rg = 3.4 nm ± 0.2 nm with a molecular weight of 110 kDa ± 10 kDa. However, these structural parameters are larger than the Rg of 2.56 nm calculated from the crystal dimer (PDB entry 4G1L) with a molecular weight of 74 kDa. Furthermore, the calculated scattering profiles computed from the dimer and monomer of the X-ray crystal structure yield a poor fit to the experimental data, giving discrepancies, χ2 values, of 3.79 for a monomer and 1.44 for the dimer (Fig. 6a). Although the fits of the monomer and the dimer structures to the pH 4.0 SAXS data are poor, the fact that the calculated dimer scattering generates a better fit to the data than the monomer may indicate that M-NDV at pH 4.0 exists as a mixture of different oligomeric species in acidic solution that tends toward a dimeric state. Consequently, to analyze the amount of these putative oligomeric forms at pH 4.0, we applied the program OLIGOMER (31) using various combinations of symmetry-related oligomers extracted from the high-resolution X-ray crystal structure of M-NDV in the PDB entry 4G1L (monomers, dimers, tetramers, hexamers, and octamers). The best fit to the experimental data (χ2 = 1.09) (Fig. 6b, curve 2) was obtained from a volume fraction-weighted mixture of the crystallographic dimers (volume fraction wi = 0.82), tetramers (wi = 0.09), and octamers (wi = 0.09). Associated volume fractions of monomers or hexamers were not required to fit the SAXS data, suggesting a negligible influence of these species on the measured SAXS intensities. The average molecular mass (∼120 kDa) evaluated by OLIGOMER of the M-NDV dimer-tetramer-octamer mixture is similar to that estimated from the experimental I(0) and protein concentration as well as the Porod volume (Vp; ∼175 to 185 nm3) obtained from the experimental SAXS profile. These results are in keeping with the observations made from the AUC; i.e., at pH 4.0, the solution state of M-NDV predominantly consists of dimers, with trace levels of higher-order oligomers in solution (Fig. 2a).
FIG 6.
(a) Comparison of the experimental scattering data from M-NDV at pH 4.0 (1) and the scattering curves calculated from the X-ray crystal structures (PDB accession number 4G1L) of the M-NDV monomer (2) and M-NDV dimer (3). (b) Analysis of the same experimental curve (1) in terms of a mixture of M-NDV X-ray crystallographic dimers, tetramers, and octamers yielding a best-fit (2) that is derived predominantly from a volume fraction-weighed mixture of dimers (∼82%) with measurable levels of tetramers (∼9%) and octamers (∼9%). (c) Different assemblies of the M-NDV protein obtained using the service PISA (protein interfaces, surfaces, and assemblies): dimer (1), tetramer (2), and octamer (3). The individual NDV monomers in the dimer (1) are shown in different colors.
Shape analysis of the M-NDV self-assemblies at neutral pH.
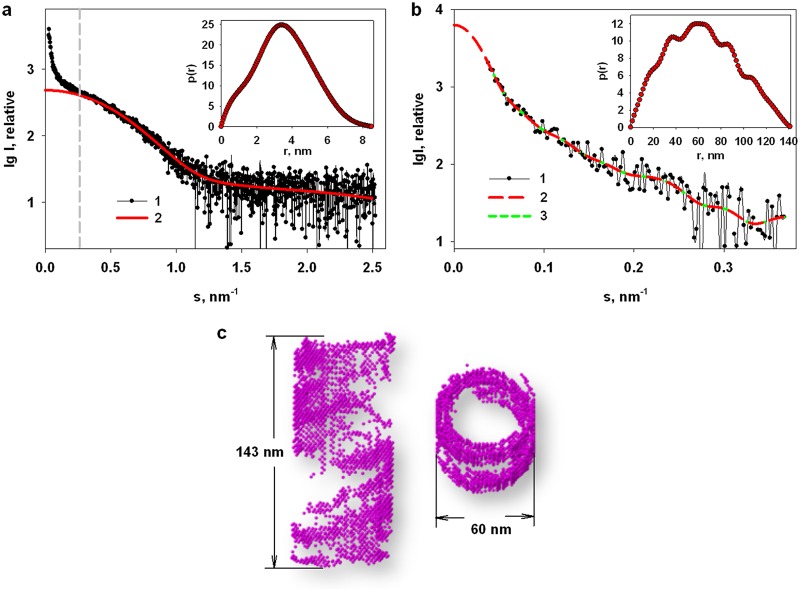
To assess the structure of the aggregates formed at pH 7.0, the following procedure was adopted. First, a distance distribution function, p(r), was calculated from the experimental data using data at an s of >0.25 nm−1 (Fig. 7a, inset), that is, in the range where the influence of large aggregates becomes negligible. These low-s data were analyzed yielding the maximum size of about 9 nm with an Rg of 3.4 nm ± 0.1 nm that is similar to the Rg of the dimer-dominated mixture observed at pH 4.0. The calculated fit to the SAXS data of the higher-angle p(r) was extrapolated to the zero angle (Fig. 7a, curve 2) and then subtracted from the experimental data at pH 7.0 across the entire s range to yield the difference SAXS profile dominated by the M-NDV aggregate scattering contributions (Fig. 7b, curve 1). The resulting SAXS profile was used to reconstruct the shape of the M-NDV clusters similar to the approach used in Shtykova et al. (32, 33).
FIG 7.
Calculation of the scattering intensity contributions from M-NDV aggregates and their shape restoration. (a) The experimental SAXS profile from 2.2 mg/ml M-NDV protein at neutral pH (1) and the modeled scattering intensities back-transformed to zero-angle from the p(r) fit to the higher-angle data for an s of >0.25 nm−1 (2). The inset shows the corresponding distance distribution function, p(r), computed using GNOM from the experimental SAXS curve from the M-NDV protein at neutral pH in the region of s > 0.25 nm−1. (b) The resulting difference scattering pattern from the M-NDV aggregates/clusters (1) and the subsequent p(r) fit to the SAXS data extrapolated to a zero scattering angle (2). The inset shows the distance distribution function, p(r), computed using GNOM from the difference curve. (c) A reconstructed dummy-atom bead model of the M-NDV clusters in two perpendicular orientations. The fit of the model to the SAXS data (χ2 =1.36) is shown in panel b (3).
For the bead model shape reconstruction, we used spherical as well as hollow-elliptical and hollow-cylindrical shapes as initial search volumes. In all cases, helical-like structures were obtained, but for the hollow-cylindrical search volume, the result of reconstruction was more compact and symmetric (Fig. 7c). The parameters of the cylindrical search volume were chosen in accordance with the parameters extracted from the p(r) function calculated from the difference SAXS profile of the M-NDV clusters (Fig. 7b, inset). The height of the cylinder, h, of 143 nm corresponds to the maximal size, Dmax, from the p(r). The outer diameter, dout, of 60 nm is equal to a cross-section of the shape while the inner diameter of the cylinder, dinn, of 46 nm was chosen so that the cylinder wall thickness, calculated as (dout – dinn)/2, of 7 nm is approximately equal to the thickness of the macromolecule of the protein. The reconstructed model of the large M-NDV clusters (Fig. 7c) appears to be characterized by a layered architecture, where the walls of the cylindrical shape are filled unevenly, forming wide, empty spaces between helical turns. The reconstructed model yields a reasonable fit to the scattering contributions attributed to the M-NDV clusters extracted from the experimental SAXS data (χ2 = 1.36).
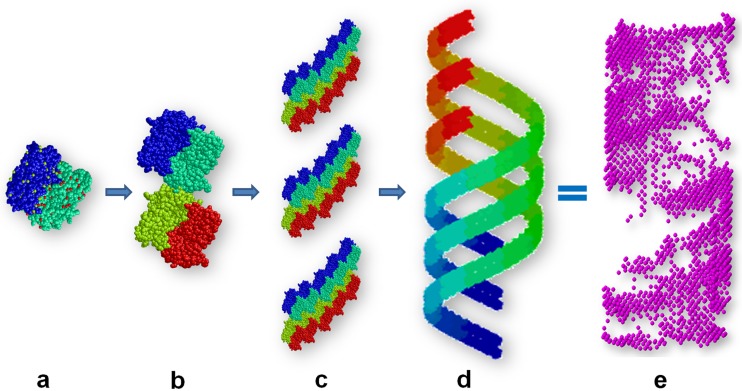
Although we present a single model to represent the M-NDV clusters, it is probably best not to view the model in isolation as the SAXS data indicate that mixtures of oligomeric components are likely present in solution and that the self-association of M-NDV is a dynamic process. Since Battisti et al. (19) revealed the NDV matrix layer as a pseudotetrameric grid-like array, our next task was to model similar constructs based on the structure of the protein (PDB entry 4G1L) and generate oligomers and helical assemblies from the high-resolution structural units. Helical assemblies mimicking the large NDV clusters were generated according to the following procedure. First, a repeating unit consisting of three spatially separated tetramers according to the ab initio model was positioned at the distance equal to the helix radius (R) of 25 nm from the origin along the x axis toward the center of the macromolecule. Each subsequent repeating unit was obtained from the previous one by rotation by angle α of 12 degrees about the Z-axis followed by the shift, Δz, of 2.5 nm along the Z-axis. The corresponding helix pitch p is equal to 360°/(αΔz), or 75 nm. The replication procedure was repeated N − 1 = 9 times yielding (Nα)/360° helical pitches (Fig. 8).
FIG 8.
The construction of M-NDV oligomers and higher-order helical assembly models based on the X-ray crystal structure unit cell and symmetry operators as well as previously determined pseudotetrameric grid-like arrays. (a) Dimer. (b) Tetramer. (c) Three spatially separated tetramers as repeating units to construct a pseudotetrameric grid-like array. (d) Final helical structure compared to the corresponding shape from ab initio modeling of the M-NDV helical clusters obtained from the SAXS data. The individual NDV monomers are shown in different colors. (e) Reconstruction from the SAXS data dummy-atom bead model of the M-NDV cluster.
To analyze the amount of the possible oligomeric forms of the M-NDV protein in solution, we performed OLIGOMER (31) analysis across the entire s-range of the pH 7.0 SAXS profile using the various oligomeric combinations of the high-resolution structure of the protein in combination with the pseudotetrameric grid-like structure mimicking large M-NDV helical clusters (Fig. 8e). The OLIGOMER program yields a good fit with a χ2 value of 1.02 (Fig. 9) for an equilibrium mixture consisting of crystallographic dimers (volume fraction wi = 0.81), octamers (volume fraction wi = 0.15), and trace amounts of the large helical clusters (wi = 0.04). The results reflect those obtained for the oligomeric species of M-NDV at pH 4.0, in that the main volume fraction occupied by the M-NDV protein at neutral pH are dimers (wi about 0.82), consistent with the AUC results which show a predominantly dimeric component in solution at neutral pH, with an increase in the proportion of higher-order oligomers/assemblies compared to those at pH 4.0 (Fig. 2).
FIG 9.
OLIGOMER analysis of the M-NDV protein in solution at pH 7.0. Experimental SAXS data (1) and the best fit to the experimental data (2) by an equilibrium mixture of crystallographic dimers (81%), tetramers (15%), and large helical clusters (4%).
DISCUSSION
We performed comprehensive structural analyses of the NDV matrix protein and probed its propensity to spontaneously self-associate in solution and at the mica surface as well as the effects of altering pH from neutral to acidic conditions on this self-association and interactions with lipid membranes. The CD spectropolarimetry analysis (Fig. 1) shows that the M-NDV solubilized from the intact virions has the secondary structure in solution (in terms of percentage of α-helices) close to that obtained from the X-ray crystal structure (PDB accession number 4G1L [19]) and that the CD spectra of the protein at pH 4.0 and pH 7.0 almost coincide, indicating no significant alteration to the protein secondary structure at acidic and neutral pHs.
Analytical ultracentrifugation indicates that the main structural unit of the M-NDV in solution is a dimer (Fig. 2) that is consistent with SAXS and cryo-electron tomography data (19). However, and unlike the matrix protein from influenza virus, M1 (33), we observe that a measurable portion of the M-NDV protein self-associates into oligomers even at acidic pH, where the majority of the protein is dimeric. Increasing the pH increases the proportion of higher-molecular-weight oligomers and large self-associated helical assemblies. In a neutral milieu these M-NDV dimers self-associate (octamers and above), comprising ∼20% by volume fraction of the protein molecules. This observation favors the hypothesis that a pH-triggered conformational change in the matrix proteins of paramyxoviruses could occur through an electrostatic repulsion mechanism for matrix disintegration (23), similar to M1 of influenza virus (29). Our AFM studies on mica surfaces demonstrate that the M-NDV layer densities at both pH 4.0 and pH 7.0 (Fig. 3a and b) are close, in contrast to influenza M1 that forms a much sparser layer in acidic medium (29, 32). The thickness of the M-NDV layer at the mica surface at pH 4.0 (2.5 nm ± 0.7 nm) is close to the size of crystal dimers (Rg = 2.56 nm) (19) (Fig. 3a) and lower than that observed by SAXS (Rg = 3.4 nm ± 0.2 nm), meaning that the layer in this case is a dimer thick and rather uniform. At pH 7.0 the average thickness of the layer was larger (3.4 nm ± 0.6 nm), with M-NDV aggregates above the layer (Fig. 3b). Both observations suggest that we have an increased amount of high-order oligomers, as obtained by AUC and SAXS. While even at pH 4.0 we do not detect the significant disassembly of the M-NDV layers at the mica surface, a change of pH from 7.0 to 4.0 led to disappearance of the large protein aggregates above the dimer-thick layer. Therefore, even though acidification reduces the oligomerization state of the M-NDV, electrostatic repulsion between M-NDV dimers on decreasing the pH is insufficient by itself for the complete disruption of the matrix layer. Therefore, we propose that hydrophobic interactions are the likely prevalent factor underpinning matrix layer organization under conditions of both neutral and acidic pH. This is not to say that charge interactions are unimportant as we do observe significant changes in the amount and size of the protein self-associates at pH 7.0, suggesting an enhancing role of an acidic environment in the dissociation of the M-NDV complexes. Such acidification that occurs at endocytic pathway entry could actually promote the disintegration of the protein scaffold to some extent and may be a cause of the observed diversity of virus entry pathways, depending on the cell line (24).
The analysis of the M-NDV binding to the lipid bilayer (Fig. 4) showed that the binding was reversible at both pH 4.0 and pH 7.0 with almost the same binding constants: (4.4 ± 0.4) ×106 M−1 for pH 4.0 versus (4.9 ± 0.2) ×106 M−1 for pH 7.0. The values of binding constants were close to the binding constant of VP40 from another representative of the Mononegavirales, Ebola virus (34), and some other peripheral proteins (35, 36), but an order of magnitude lower than that for the matrix protein from the influenza A virus (29). Cryoelectron tomography of NDV and influenza viruses supports these results: only 10% of Newcastle disease virions have a visible matrix layer (19) while for influenza virus the protein matrix beneath lipid membrane is detected in 90% of viral particles (37). The coincidence of M-NDV membrane binding properties under neutral and acidic conditions indicates that the protein-membrane interactions doubly change during the virus uncoating, even if it goes by the endocytic pathway, while the change in protein-protein interactions might be involved in the final disintegration of the viral protein scaffold. It means that electrostatics contribute poorly to the protein-lipid interactions, which is in line with the results of Faaberg and Peeples (38) that the M-NDV interactions with the liposomes were independent of ionic conditions. Otherwise, the change in protein net charge would influence the binding at the membrane structure, as observed for the M1 protein of influenza A virus (29).
SAXS experiments support the AUC results in concluding that the main structural unit of the M-NDV in solution is a dimer at both pH 4.0 and pH 7.0, and that increasing the pH results in the formation of larger, higher-order M-NDV oligomers and assemblies. By combining X-ray crystallography and cryoelectron tomography, Battisti et al. (19) showed that the matrix protein of Newcastle disease virus forms dimers that assemble into pseudotetrameric arrays with the angle of 6° between neighboring dimers to generate the membrane curvature for virus budding. Our results are consistent with these observations in that the oligomerization of the M-NDV protein appears to be an intrinsic biological property of the M-NDV protein that takes place even in acidic medium, with the same order of oligomerization: dimer organizes into tetramer and after that we have the helical array of repeating tetramers as a double-helix of dimers (Fig. 8). The helical structures (Fig. 7c and 8) have a layered cylindrical architecture. Moreover, the walls of the cylindrical shape are filled unevenly: there are empty spaces between the helical turns. This agrees with Battisti et al. (19) who revealed mostly partial matrix layers in the tomographic representations of the NDV virions. Moreover, the diameters of the helices obtained from our SAXS data (around 60 nm) (Fig. 7c) are close to the diameter of elongated Newcastle disease virions observed by Battisti et al. (19) by cryoelectron tomography. These structures formed in solution, beyond the virion and without participation of the other viral components, supporting the idea of an inherent property of M-NDV to self-oligomerize to form a curved viral scaffold. By comparing M-NDV helices with self-assemblies of the M1 protein of influenza A virus (32), we can say that the latter fill the search volume while the former ones form hollow structures, located mainly along the walls of the hollow cylinder. Most likely, such a difference in the scaffold structures can be explained by the difference in the structures of the individual protein macromolecules. The low-resolution structural models of influenza virus M1 built from the SAXS data reveal that the protein is a structurally anisotropic monomer consisting of compact NM domains and an extended and partially flexible C-terminal domain (32, 33) while SAXS data for M-NDV indicate that the protein is predominantly a compact dimer in solution (19), in agreement with the crystalline state.
Our results indicate that the main structural unit of the M-NDV is a dimer that has an inherent property to oligomerize into tetramers, with these tetramers forming protein helical oligomers at neutral pH resembling the size of filamentous M-NDV virions. The geometry of the protein assemblies allows M-NDV to introduce the membrane curvature necessary for budding of virus-like particles without other viral proteins (12, 18). Although the membrane-binding propensity of the M-NDV protein did not demonstrate any pH dependence, the protein oligomerization tendency does decrease in an acidic milieu, meaning that endocytic pathways may facilitate NDV entry into the host cell (24) through the acceleration of the matrix scaffold disintegration for the nucleocapsid release.
MATERIALS AND METHODS
Newcastle disease virus preparation.
NDV (LaSota 1427 strain) was propagated in 10-day-old embryonic chicken eggs and purified by centrifugation through 20% (vol/vol) sucrose in STE buffer (100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH 7.4) at 21,000 rpm for 90 min at 8 °C in an SW 27.1 rotor of a Beckman-Spinco L5-75 centrifuge, as described in Kordyukova et al. (39).
Isolation of the matrix protein.
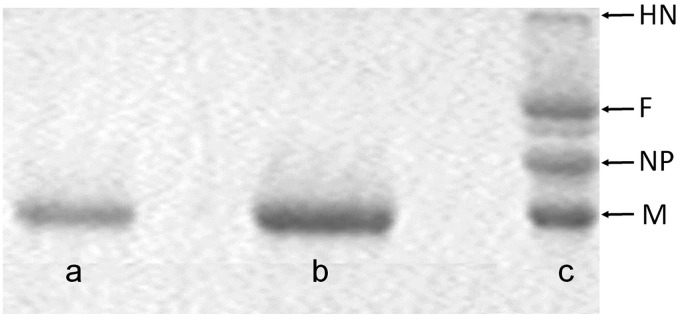
In García-Sastre et al. (40) it is demonstrated that the isolation of the M-NDV protein could be performed under high-ionic-strength conditions in a neutral milieu. Nevertheless, we decided to change the protocol based on the results obtained for the isolation of the influenza virus M1 protein (41). As for M-NDV (40), the matrix protein M1 of influenza A virus interacts electrostatically (29, 32), implying the coordination of positive and negative charges on protein molecules to form viral scaffold. Low-pH conditions will diminish all the negative charges on the protein surface, thus stabilizing the minimal possible protein unit in the solution (32). The benefit of such an approach is the possibility to further study the self-assembly of the protein in physiological ionic strength. Thus, we performed the isolation of the M-NDV protein by acid-dependent solubilization of the viral envelope with the mild nonionic detergent NP-40 (Igepal) (Sigma, USA) in 50 mM MES and 100 mM NaCl, pH 4.0 buffer. The isolated protein was dialyzed against the same buffer with Bio-Beads SM-2 Adsorbent (Bio-Rad, USA) for 18 h at 4°C using dialysis tubing cellulose membranes (14,000-molecular-weight cutoff [MWCO]) (Sigma, USA). The protein suspension/buffer ratio was 1:400 (vol/vol), and to every 200 μg of M-NDV in the suspension we added 700 mg of Bio-Beads pretreated with methanol to the buffer. The dialyzed protein was concentrated with the aid of Microcon membranes (Microcon Ultracel YM-10; regenerated cellulose 10,000 MWCO) (Millipore, USA) at 10,000 rpm for 2 h at 4°C to a final concentration up to 3 mg/ml. The purity of protein samples was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (42) (Fig. 10), and the protein content in solution was determined by quantitative amino acid analysis (Hitachi L-8800 analyzer).
FIG 10.
SDS-PAGE analysis of the protein samples. Lane a, matrix M protein isolated from purified Newcastle disease virus (NDV) virions using acid solubilization protocol (41); lane b, isolated M protein concentrated through 10,000-MWCO (Thermo Scientific) filters; lane c, purified NDV virions showing major virus proteins (HN, hemagglutinin-neuraminidase; F, fusion glycoprotein; NP, nucleoprotein; M, matrix protein).
The protein concentration was estimated using UV-visible light (UV-Vis) spectroscopy. The absorption spectra in the 240- to 338-nm range were measured in cells with an optical path of 1 cm using a Hitachi UV-2600 spectrophotometer. True absorption (E) spectra of light-scattering suspensions were calculated by an extrapolation method, using the programs of Ksenofontov et al. (27). The spectral region of 320 to 338 nm was used for the extrapolation. We used the web service Sednterp (http://bitcwiki.sr.unh.edu) to calculate extinction coefficients (E0.1% 280 nm) of the studied proteins. To determine M-NDV concentration, the coefficient E0.1% 280 nm = 0.695 was employed.
CD spectroscopy.
Circular dichroism (CD) spectra in the regions of 198 to 260 nm (far UV) were recorded on a Chiroscan CD spectropolarimeter (Applied Photophysics, United Kingdom) in 2-mm optical path length cuvettes. Protein concentrations of 0.05 to 0.15 mg/ml in 50 mM MES and 100 mM NaCl buffer, pH 4.0 and 7.0, were used for far-UV CD measurements. Far-UV spectra were calculated in ellipticity (θ) per mole of amino acids; the mean amino acid molecular weight was 110 Da. The spectra were recorded at 0.5 to 1.0 nm/s. The measured spectra were smoothed with the instrument software. Each point was measured for 1 s. The α-helical content of the protein was determined using the Greenfield-Fasman equation (43) and by the web service K2D2, which estimates protein secondary structure from CD data (28).
Analytical ultracentrifugation (AUC) measurements and data analysis.
Sedimentation velocity experiments were performed on a Beckman E analytical ultracentrifuge (USA), equipped with a scanner at a wavelength of 280 nm, at rotor speeds of 48,000 rpm at 20°C. The program SEDFIT (sedfit12p52) was applied to model sedimentation profiles using the integrated Lamm equation solutions (44). Solvent density, viscosity, and the specific partial volume of the M-NDV protein were calculated at 20°C by SEDNTERP (https://www.spinanalytical.com/auc-software.php). Intrinsic sedimentation coefficients (s20,w) corrected to water at 20°C were calculated by SEDNTERP from the experimental s values obtained in the appropriate solvent.
Scattering experiments and data analysis.
Synchrotron small-angle X-ray scattering (SAXS) measurements were performed at the European Molecular Biology Laboratory (EMBL) on the storage ring PETRA III (Deutsches Elektronen-Synchrotron [DESY], Hamburg) at the EMBL-P12 beam line equipped with a robotic sample changer and a two-dimensional (2D) photon-counting pixel X-ray detector (Pilatus-2M; Dectris, Switzerland). The scattering intensity, I(s), was recorded in the range of momentum transfer, s, of 0.05 < s < 4.5 nm−1, where s = (4πsinθ)/λ, 2θ is the scattering angle, and λ = 0.124 nm is the X-ray wavelength (45). The measurements were carried out in 100 mM NaCl and 50 mM MES buffer at pH 4.0 and in the same buffer at pH 7.0, after titration by alkali (NaOH), at 10°C using continuous-flow operation over a total exposure time of 1 s collected as 20- by 50-ms individual frames to monitor for potential radiation damage (no radiation effects were detected) (46). The data were corrected for the solvent scattering and processed using standard procedures (47, 48), with additional data analysis performed using the program PRIMUS (31). To account for interparticle interactions, we measured and compared samples of M-NDV between 1.0 and 3.0 mg/ml. A weak concentration dependence was observed and analyzed.
The values of the forward scattering and radii of gyration, Rg, were calculated from the experimental SAXS patterns using Guinier approximation:
| (1) |
which is valid in the range of sRg approximately of <1.3 (49). These parameters and the maximal diameter of the particle, Dmax, were also computed from the distance distribution function p(r). The latter was evaluated by the program GNOM (50) according to equation 2:
| (2) |
The low-resolution shapes were reconstructed ab initio with DAMMIN (51) employing a dummy atom (bead) model of the particle. Starting from a random assembly, the program utilizes simulated annealing (SA) to build models fitting the experimental data, Iexp(s), with minimal discrepancy:
| (3) |
where N is the number of experimental points, c is the scaling factor, and Icalc(sj) and σ(sj) are the calculated intensity from the model and the experimental error at the momentum transfer, sj, respectively.
Molecular weight of the M-NDV protein was calculated from the SAXS data using the value of I(0) combined with protein concentration relative to a bovine serum albumin standard (52) as well as from the concentration-independent excluded Porod volume (Vp) (53).
Multiple ab initio reconstructions were performed to obtain consistent models. The DAMMIN outputs were analyzed using programs SUPCOMB (54) and DAMAVER (55) to identify the most typical models. To analyze the amounts of different associates in the M-NDV solutions, we used the program OLIGOMER (31). Given the scattering intensities of components in a mixture, Ii(s), the program fits the experimental scattering curve by their linear combination to determine their fractions, wi. The equation
| (4) |
is solved with respect to wi by nonnegative least squares to minimize the discrepancy between the experimental and calculated scattering curves. The program CRYSOL (56) was used to calculate the scattering from the crystal structure of the M-NDV protein (PDB entry 4G1L) (19) and from its different associates (dimers, tetramers, etc.) in solution.
Different assemblies of the M-NDV protein for SAXS-based modeling were obtained using the service PISA (protein interfaces, surfaces, and assemblies) (57) at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html). The crystal structure of the paramyxovirus matrix protein, PDB accession number 4G1L (19), was employed for the crystallographic interface analysis.
Docking procedure.
To model the M-NDV macromolecular assemblies, rigid-body docking simulations were conducted using the ZDOCK server (58). High-resolution structures of the M-NDV dimer obtained from the X-ray crystal structures (PDB entry 4G1L [19]) were docked against the same dimer. The top 10 models predicted by ZDOCK were visually analyzed and compared with the similar models obtained by PISA to choose the most consistent M-NDV oligomers to include in the further analysis.
AFM.
Structures formed by M-NDV protein upon adsorption were studied on a Multimode Nanoscope IV setup (Veeco Digital Instruments, USA) equipped with a J-type scanner and electrochemical fluid cell. All experiments were carried out in tapping mode at room temperature in working buffer solution (100 mM NaCl, 50 mM MES, pH 4.0 or 7.0). For scanning, SiN3 cantilevers were used with a nominal spring constant of 0.06 N/m (type SNL; Bruker, USA) with a tip radius of approximately 2 nm. After half an hour of adsorption at room temperature, the sample was placed into the atomic force microscope (AFM) cell filled with working buffer solution, and scanning commenced. Image processing was made with WSxM software (59).
IFC.
Intramembrane field compensation (IFC) measurements of M-NDV adsorption and desorption were performed as described in Batishchev et al. (29). In brief, freestanding planar bilayer lipid membranes (BLM) were formed by the Mueller-Rudin technique (60) at the small aperture (diameter of 0.8 mm) in a septum dividing two 500-μl chambers of a Teflon cell. The chambers were filled with a working buffer solution of 20 mM KCl, 0.1 mM EDTA, and 5 mM sodium citrate at pH 4.0 or 7.0. The BLM were made from a solution of 30 mol% of diphytanoylphosphatidylserine (DPhPS) and 70 mol% of diphytanoylphosphatidylcholine (DPhPC) (Avanti Polar Lipids, USA) in decane (Sigma, USA) with a total lipid concentration of 15 mg/ml. The adsorption of M-NDV protein on the one side of the BLM changed the difference of boundary potentials across the lipid bilayer, Δφb, which was measured using the IFC technique (61). The washing of the protein from the cell was performed by perfusion with protein-free buffer solution using a peristaltic pump (LKB, Sweden). To obtain the adsorption isotherm, the protein was added stepwise to change the final bulk concentration in 50 nM steps, from 50 nM to 600 nM. Each point on the isotherm corresponds to a stationary level for the given concentration, averaged over three independent experiments. Although the theory of the IFC method can explain in detail only the adsorption of small molecules, it has been demonstrated that reliable results can be obtained for large protein molecules (62–64), assuming that the measured potential difference, Δφb, is directly proportional to the amount of the protein adsorbed per unit area of the membrane. Recently, we have shown that the same approach was used successfully for the analysis of influenza virus M1 protein binding to lipid membranes (29).
ACKNOWLEDGMENTS
The work was supported in part by Russian Foundation for Basic Research (RFBR) project numbers 17-54-30022 (V.Y.T. and O.V.B.) and 16-04-00563 (A.L.K., N.V.F., and L.A.B.), Federal Agency of Scientific Organizations agreement number 007-GZ/Ch3363/26 (E.V.S., M.V.P., and L.A.D.), Ministry of Science and Higher Education of the Russian Federation project AAAA-A18-118051400023-5 (O.V.B., V.Y.T., and M.V.P.), and Presidium of Russian Academy of Sciences project AAAA-A16-116051110081-7 (E.V.S.).
REFERENCES
- 1.Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. 2010. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. 2011. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc Natl Acad Sci U S A 108:18085–18090. doi: 10.1073/pnas.1105770108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci U S A 107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorfman T, Mammano F, Haseltine WA, Gottlinger HG. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol 68:1689–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Peeples ME, Wang C, Gupta KC, Coleman N. 1992. Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J Virol 66:3263–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YE, Pernet O, Lee B. 2012. Regulation of the nucleocytoplasmic trafficking of viral and cellular proteins by ubiquitin and small ubiquitin‐related modifiers. Biol Cell 104:121–138. doi: 10.1111/boc.201100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol 80:11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell PH, Almeida JD. 1984. A regular subunit pattern seen on non-infectious Newcastle disease virus particles. J Gen Virol 65:1023–1031. doi: 10.1099/0022-1317-65-6-1023. [DOI] [PubMed] [Google Scholar]
- 10.Heggeness MH, Smith PR, Choppin PW. 1982. In vitro assembly of the nonglycosylated membrane protein (M) of Sendai virus. Proc Natl Acad Sci U S A 79:6232–6236. doi: 10.1073/pnas.79.20.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachi T. 1980. Intramembrane structural differentiation in Sendai virus maturation. Virology 106:41–49. doi: 10.1016/0042-6822(80)90219-6. [DOI] [PubMed] [Google Scholar]
- 12.Hewitt JA. 1977. Studies on the subunit composition of the M-protein of Sendai virus. FEBS Lett 81:395–397. doi: 10.1016/0014-5793(77)80562-0. [DOI] [PubMed] [Google Scholar]
- 13.Waning DL, Schmitt AP, Leser GP, Lamb RA. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J Virol 76:9284–9297. doi: 10.1128/JVI.76.18.9284-9297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison MS, Sak Aguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: Building particles that transmit infections. Int J Biochem Cell Biol 42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghildyal R, Mills J, Murray M, Vardaxis N, Meanger J. 2002. Respiratory syncytial virus matrix protein associates with nucleocapsids in infection cells. J Gen Virol 83:753–757. doi: 10.1099/0022-1317-83-4-753. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt PT, Ray G, Schmitt AP. 2010. The C-terminal end of parainfluence virus 5 NP protein is important for virus-like particle production and M–NP protein interaction. J Virol 84:12810–12823. doi: 10.1128/JVI.01885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seal BS, King DJ, Meinersmann RJ. 2000. Molecular evolution of the Newcastle disease virus matrix protein gene and phylogenetic relationships among the paramyxoviridae. Virus Res 66:1–11. doi: 10.1016/S0168-1702(99)00119-7. [DOI] [PubMed] [Google Scholar]
- 18.Shnyrova AV, Ayllon J, Mikhalyov II, Villar E, Zimmerberg J, Frolov VA. 2007. Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J Cell Biol 179:627–633. doi: 10.1083/jcb.200705062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, Steven AC, Morrison TG, Rossmann MG. 2012. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci U S A 109:13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian M, Jungerwirth S. 1990. Mechanisms of enveloped virus entry into cells. Mol Biol Med 7:17–31. [PubMed] [Google Scholar]
- 21.San Roman K, Villar E, Muñoz-Barroso I. 1999. Acidic pH enhancement of the fusion of Newcastle disease virus with cultured cells. Virology 260:329–341. doi: 10.1006/viro.1999.9841. [DOI] [PubMed] [Google Scholar]
- 22.Schowalter RM, Smith SE, Dutch RE. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J Virol 80:10931–10941. doi: 10.1128/JVI.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. 2009. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J Virol 83:1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantín C, Holguera J, Ferrerira L, Villar E, Muñoz-Barroso I. 2007. Newcastle disease virus may enter cells by caveolae-mediated endocytosis. J Gen Virol 88:559–569. doi: 10.1099/vir.0.82150-0. [DOI] [PubMed] [Google Scholar]
- 25.Reichard KW, Lorence RM, Cascino CJ, Peeples ME, Walter RJ, Fernando MB, Reyes HM, Greager JA. 1992. Newcastle disease virus selectively kills human tumor cells. J Surg Res 52:448–453. doi: 10.1016/0022-4804(92)90310-V. [DOI] [PubMed] [Google Scholar]
- 26.Zamarin D, Palese P. 2012. Oncolytic Newcastle Disease Virus for cancer therapy: old challenges and new directions. Future Microbiol 7:347–367. doi: 10.2217/fmb.12.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ksenofontov AL, Kozlovskii VS, Kordyukova LV, Radyukhin VA, Timofeeva AV, Dobrov EN. 2006. Determination of concentration and aggregate size in influenza virus preparations using the true UV-absorption spectra. Mol Biol 40:152–158. doi: 10.1134/S0026893306010201. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Iratxeta C, Andrade-Navarro MA. 2008. K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct Biol 8:25. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batishchev OV, Shilova LA, Kachala MV, Tashkin VY, Sokolov VS, Fedorova NV, Baratova LA, Knyazev DG, Zimmerberg J, Chizmadzhev YA. 2016. pH-dependent formation and disintegration of the influenza A virus protein scaffold to provide tension for membrane fusion. J Virol 90:575–585. doi: 10.1128/JVI.01539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kordyukova LV, Shtykova EV, Baratova LA, Svergun DI, Batishchev OV. 2018. Matrix proteins of enveloped viruses: a case study of Influenza A virus M1 protein. J Biomol Struct Dyn 13:1–46. doi: 10.1080/07391102.2018.1436089. [DOI] [PubMed] [Google Scholar]
- 31.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 36:1277–1282. doi: 10.1107/S0021889803012779. [DOI] [Google Scholar]
- 32.Shtykova EV, Dadinova LA, Fedorova NV, Golanikov AE, Bogacheva EN, Ksenofontov AL, Baratova LA, Shilova LA, Tashkin VY, Galimzyanov TR, Jeffries CM, Svergun DI, Batishchev OV. 2017. Influenza virus matrix protein M1 preserves its conformation with pH, changing multimerization state at the priming stage due to electrostatics. Sci Rep 7:16793. doi: 10.1038/s41598-017-16986-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shtykova EV, Baratova LA, Fedorova NV, Radyukhin VA, Ksenofontov AL, Volkov VV, Shishkov AV, Dolgov AA, Shilova LA, Batishchev OV, Jeffries CM, Svergun DI. 2013. Structural analysis of influenza А virus matrix protein M1 and its self-assemblies at low pH. PLoS One 8:e82431. doi: 10.1371/journal.pone.0082431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soni SP, Adu-Gyamfi E, Yong SS, Jee CS, Stahelin RV. 2013. The Ebola virus matrix protein deeply penetrates the plasma membrane: an important step in viral egress. Biophys J 104:1940–1949. doi: 10.1016/j.bpj.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahelin RV. 2013. Surface plasmon resonance: a useful technique for cell biologists to characterize biomolecular interactions. Mol Biol Cell 24:883–886. doi: 10.1091/mbc.E12-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minton AP. 2010. Analysis of membrane binding equilibria of peripheral proteins: allowance for excluded area of bound protein. Anal Biochem 397:247–249. doi: 10.1016/j.ab.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontana J, Steven AC. 2013. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J Virol 87:5621–5628. doi: 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faaberg KS, Peeples ME. 1988. Association of soluble matrix protein of Newcastle disease virus with liposomes is independent of ionic conditions. Virology 166:123–132. doi: 10.1016/0042-6822(88)90153-5. [DOI] [PubMed] [Google Scholar]
- 39.Kordyukova LV, Serebryakova MV, Polyakov VY, Ovchinnikova TV, Smirnova YA, Fedorova NV, Baratova LA. 2008. Influenza A virus M1 protein structure probed by in situ limited proteolysis with bromelain. Protein Pept Lett 15:922–930. doi: 10.2174/092986608785849254. [DOI] [PubMed] [Google Scholar]
- 40.García-Sastre A, Cabezas J, Villar E. 1989. Proteins of Newcastle disease virus envelope: interaction between the outer hemagglutinin-neuraminidase glycoprotein and the inner non-glycosylated matrix protein. Bioch Biophys Acta 999:171–175. doi: 10.1016/0167-4838(89)90214-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhirnov O. 1992. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology 186:324–330. doi: 10.1016/0042-6822(92)90090-C. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Greenfield N, Fasman GD. 1969. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 44.Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanchet CE, Spilotros A, Schwemmer F, Graewert MA, Kikhney AG, Jeffries CM, Franke D, Mark D, Zengerle R, Cipriani F, Fiedler S, Roessle M, Svergun DI. 2015. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J Appl Crystallogr 48:431–443. doi: 10.1107/S160057671500254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeffries CM, Graewert MA, Svergun DI, Blanchet CE. 2015. Limiting radiation damage for high brilliance biological solution scattering: practical experience at the EMBL P12 beam line, PETRAIII. J Synchrotron Rad 22:273–279. doi: 10.1107/S1600577515000375. [DOI] [PubMed] [Google Scholar]
- 47.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. 2012. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr 45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, Svergun DI. 2017. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr 50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feigin LA, Svergun DI. 1987. Structure analysis by small-angle X-ray and neutron scattering, Plenum Press, New York, NY. [Google Scholar]
- 50.Svergun DI. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25:495–505. doi: 10.1107/S0021889892001663. [DOI] [Google Scholar]
- 51.Svergun DI. 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mylonas E, Svergun DI. 2007. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J App Cryst 40:245–249. [Google Scholar]
- 53.Porod G. 1982. Small-angle X-ray scattering, p 17–51. In Glatter O, Kratky O (ed). Academic Press, London, United Kingdom. [Google Scholar]
- 54.Kozin MV, Svergun DI. 2001. Automated matching of high- and low-resolution structural models. J Appl Crystallogr 34:33–41. doi: 10.1107/S0021889800014126. [DOI] [Google Scholar]
- 55.Volkov VV, Svergun DI. 2003. Uniqueness of ab initio shape determination in small angle scattering. J Appl Crystallogr 36:860–864. doi: 10.1107/S0021889803000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svergun DI, Barberato C, Koch MHJ. 1995. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28:768–773. doi: 10.1107/S0021889895007047. [DOI] [Google Scholar]
- 57.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 58.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. 2014. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horcas I, Fernández R, Gómez-Rodríguez JM, Colchero J, Gómez-Herrero J, Baro AM. 2007. WSxM: a software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum 78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 60.Mueller P, Rudin DO, Tien HT, Wescott WC. 1963. Methods for the formation of single bimolecular lipid membranes in aqueous solution. J Phys Chem 67:534–535. doi: 10.1021/j100796a529. [DOI] [Google Scholar]
- 61.Sokolov VS, Kuz'min VG. 1980. Measurement of the difference in the surface potentials of bilayer membranes from the second harmonic of the capacitance current. Biofizika 25:174–177. [PubMed] [Google Scholar]
- 62.Bahr G, Diederich A, Vergeres G, Winterhalter M. 1998. Interaction of the effector domain of MARCKS and MARCKS-related protein with lipid membranes revealed by electric potential measurements. Biochemistry 37:16252–16261. doi: 10.1021/bi981765a. [DOI] [PubMed] [Google Scholar]
- 63.Sokolov VS, Sokolenko EA, Filinsky DV, Ermakov YA, Lopina OD, Apell HJ. 2007. Electrostatic potentials arising due to adsorption of Na+, K+-ATPase-containing membrane fragments on bilayer lipid membrane. Biol Membrany 24:259–272. [Google Scholar]
- 64.Sokolov VS, Sokolenko EA, Sokolov AV, Dontsov AE, Chizmadzhev YA, Ostrovsky MA. 2007. Interaction of pyridinium bis-retinoid (A2E) with bilayer lipid membranes. J Photochem Photobiol B 86:177–185. doi: 10.1016/j.jphotobiol.2006.09.006. [DOI] [PubMed] [Google Scholar]