Insect-derived cell lines are widely used commercially for the production of vaccines and protein-based pharmaceuticals. After decades of safe and beneficial use, it was a surprise to the biotechnology industry to discover an endemic rhabdovirus in Sf9 cells. This discovery was made possible only by the substantial advancements in DNA sequencing technologies. Given the public health concerns associated with many rhabdovirus species, several initiatives were undertaken to establish that Spodoptera frugiperda rhabdovirus (Sf-RV) does not pose a threat to humans. Such actions include the generation of cell lines that have been cleared of Sf-RV. Given that Sf9 is derived from a moth whose larvae feed on human-edible foods, we explored the prevalence of Sf-RV in its wild and lab-grown populations, as well as its ability to be deposited on food items during feeding. Collectively, our data suggest that there is no overt risk from exposure to Sf-RV.
KEYWORDS: Nicotiana benthamiana, Sf21, Sf9, human-edible foods, population structure
ABSTRACT
The Sf9 and Sf21 cell lines derived from ovarian tissues of the wide-host-range phytophagous lepidopteran Spodoptera frugiperda are widely used for research and commercial-scale production of recombinant proteins. These cell lines are chronically infected with a rhabdovirus (Sf-RV) that does not cause any overt cytopathic effects. We demonstrate that wild populations of S. frugiperda in the eastern United States and Caribbean are infected with genetically diverse strains of Sf-RV and that this virus is also capable of infecting cells of Spodoptera exigua, Heliothis subflexa, and Bombyx mori. Feeding studies demonstrated the ability of S. frugiperda larvae to deposit Sf-RV onto human-consumed vegetables during feeding. Although no evidence for replication in two species of plant cells was detected, subcellular localization studies demonstrated that the Sf-RV nucleocapsid was targeted to plasmodesmata, while two forms of the accessory protein were differentiated on the basis of their ability to localize to nuclei. Collectively, the results from this study suggest that environmental exposure of humans to Sf-RV is likely to be commonplace and frequent, but its inability to replicate in plant or human cells suggests that there is no substantial risk to human health.
IMPORTANCE Insect-derived cell lines are widely used commercially for the production of vaccines and protein-based pharmaceuticals. After decades of safe and beneficial use, it was a surprise to the biotechnology industry to discover an endemic rhabdovirus in Sf9 cells. This discovery was made possible only by the substantial advancements in DNA sequencing technologies. Given the public health concerns associated with many rhabdovirus species, several initiatives were undertaken to establish that Spodoptera frugiperda rhabdovirus (Sf-RV) does not pose a threat to humans. Such actions include the generation of cell lines that have been cleared of Sf-RV. Given that Sf9 is derived from a moth whose larvae feed on human-edible foods, we explored the prevalence of Sf-RV in its wild and lab-grown populations, as well as its ability to be deposited on food items during feeding. Collectively, our data suggest that there is no overt risk from exposure to Sf-RV.
INTRODUCTION
Spodoptera frugiperda, the fall armyworm, is an extremely important agricultural pest. This polyphagous insect has a host range of more than 80 plants, many of which produce human-consumed foods such as sweet corn, tomatoes, strawberries, apples, and peanuts (1–4). In addition to S. frugiperda, at least nine Spodoptera species are native to the southern United States, where many food crops are grown. These species include Southern armyworm S. eridania (Stoll), and the beet armyworm S. exigua (Hübner) (5). Given their tropical origin, Spodoptera pests in the United States are restricted to overwintering in an area spanning from Texas to Florida. However, during the summer months, in some years, populations of these insects can rapidly expand their ranges as far as Canadian provinces (6). Effective control of S. frugiperda, particularly in corn and cotton, can be achieved through the use of transgenic plants expressing Cry proteins from Bacillus thuringiensis, to which resistance can be selected in areas of continuous use (7–9).
In addition to its significant impacts on food and fiber production, S. frugiperda, as with other lepidopterans such as Trichoplusia ni, is linked to a wide range of biotechnologies through the use of cell lines used for research and commercial-scale production of vaccines, biobetters, and other recombinant proteins (10–14). In particular, the Sf21 and Sf9 cell lines derived from ovarian tissues of S. frugiperda are used extensively in combination with baculovirus-derived vectors for recombinant protein production. The Sf9 and Sf21 cell lines have been used safely for human pharmaceutical production for a decade, and no known adventitious viruses had been found to be capable of infecting them. Recently, however, a novel rhabdovirus has been identified in all Sf9 cell lines, now called Sf-rhabdovirus (here, Sf-RV) (15–20). The discovery of Sf-RV was reported by Ma et al. (15) based on work performed at the U.S. FDA. The virus was also independently discovered by Takeda Vaccines, Inc. (then LigoCyte Pharmaceuticals) in 2012 via cloning and sequencing efforts that were intended to screen for the presence of novel insect retrovirus sequences in the Sf9 cell line. Sf-RV is noncytopathic in Sf9 cells and has a 13.5-kb negative-sense single-stranded RNA genome characteristic of viruses in the Mononegavirales (15). The genome is organized into six open reading frames (ORFs) in the order nucleocapsid protein (N), phosphoprotein (P), matrix protein, (M), glycoprotein (G), accessory protein of unknown function (F, alternatively referred to as X), and RNA-dependent RNA polymerase (L), flanked by noncoding leader and trailer regions (15). Subsequent studies have shown that the F protein is dispensable for infectivity in Sf9 cells, and deletion of 320 nucleotides from this ORF can occur when Sf9 cells are continuously passaged (16–18, 20).
To reduce the potential risk of exposure to Sf-RV, cell lines cleared of the Sf-RV have been produced, which have been shown to be of equivalent utility to Sf-RV-infected lines (17–19). However, as this study demonstrates, the environmental presence of Sf-RV and the likelihood for regular human exposure argues that there are no known human health risks warranting the need to change Sf-9 cell lines currently used in commercial applications. Importantly, Sf-RV has been shown to be noninfectious in human, mammalian, and nonlepidopteran (Drosophila-derived) cell lines (15, 20). These studies suggest that Sf-RV and the continued use of Sf-RV-infected lines already in commercial production systems do not pose adverse risks to human health. Further assurance that Sf-RV is not a significant threat to human health would be provided if there was a greater understanding of the human exposure to Sf-RV as a result of widespread occurrence in wild Spodoptera populations and if these insects have the ability to deposit this virus on food for human consumption. As such, establishing that humans have long been exposed to Sf-RV via routes of insect feeding is an important criterion in the evaluation of the risk to humans. Furthermore, given the polyphagous nature of S. frugiperda, it is important to know if plants can provide reservoirs for this virus or serve to amplify it when inoculated. To address these concerns, we surveyed wild-caught S. frugiperda from populations in seven southeastern states in the United States and in the Dominican Republic. Additionally, we examined the ability to detect Sf-RV RNA on vegetables after insect feeding and the potential for Sf-RV replication in other insect and plant cells. Finally, we determined the full-length genomes of Sf-RV that we identified in two additional insect cell lines from phytophagous species, Heliothis subflexa and Bombyx mori. Taken together, we show that Sf-RV is widely distributed in nature and has the ability to replicate in additional lepidopteran cell lines from insect species that possess the potential to deposit virus on food plants. Additionally, it was shown previously that this highly labile virus is not infectious in mammalian cells, and we demonstrate here its inability to replicate in plant cells, in which it exhibits protein localization patterns similar to those of plant-adapted species. Though further investigations are warranted, the present studies suggest that regular environmental exposure to Sf-RV is likely significant but represents negligible risk to human health.
RESULTS
Infectivity of Sf-RV in insect cell lines.
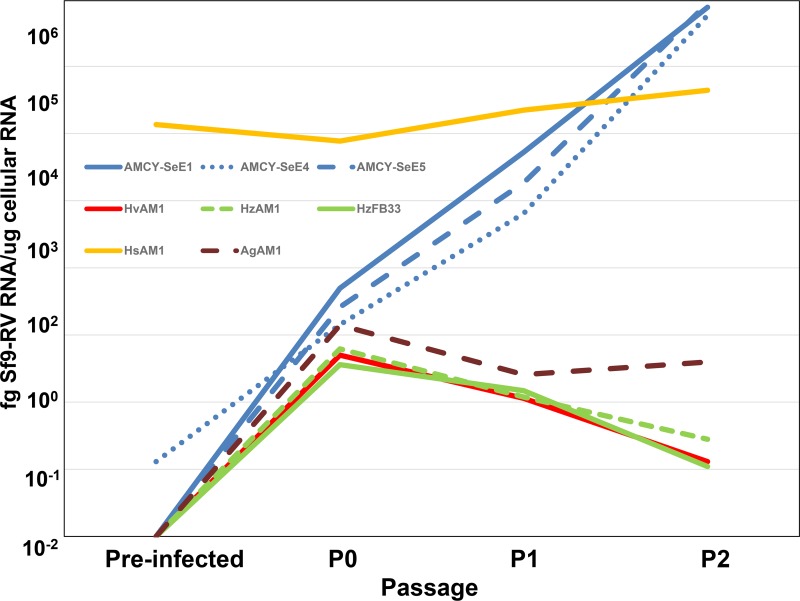
The ubiquitous presence of Sf-RV in Sf9 cells, its ability to reinfect these cells (20), and the propensity of Spodoptera sp. to exist in mixed populations raised the possibility that Sf-RV may infect or be endemic to other phytophagous lepidopterans (1, 9, 21). Therefore, we screened cell lines derived from S. exigua, Heliothis virescens, Helicoverpa zea, H. subflexa, and Anticarsia gemmatalis for Sf-RV infectivity by exposing them to filtered supernatants of Sf9 cultures. The accumulation of Sf-RV RNA was monitored over two passages by reverse transcriptase PCR (RT-PCR). Surprisingly, H. subflexa cells were positive for Sf-RV without prior addition of medium from Sf9 cultures, demonstrating that they were likely already infected with Sf-RV. The titer of Sf-RV in H. subflexa cells remained high at ∼104 fg Sf-RV RNA/μg cell RNA (Fig. 1). H. virescens, H. zea, and A. gemmatalis cells exhibited no propensity to host Sf-RV, with a rapid decline of detectable virus RNA after a single passage. In contrast, Sf-RV titers in S. exigua cell lines rose from undetectable to two orders of magnitude higher than those in H. subflexa after two passages. These observations demonstrated that Sf-RV has a host cell range beyond S. frugiperda and provided a new host cell platform (S. exigua) in which to further investigate Sf-RV infectivity. Also, it should be noted that Sf-RV infectivity in S. exigua cells does not require the presence of the intact accessory gene, since the Sf-RV found in the Takeda Sf9 cell line contains the same 320-nucleotide deletion noted by others (16–18, 20).
FIG 1.
Detection of Sf9 rhabdovirus RNA in insect cells exposed to Sf9 cell-conditioned medium. Cell lines obtained from the Agricultural Research Service (ARS) of the U.S. Department of Agriculture (USDA) were inoculated with Sf9 cell-conditioned medium and passaged three times at intervals of 72 h (P0, P1, and P2). Cell lines derived from Spodoptera exigua eggs (blue lines) were highly susceptible to Sf-RV. The ovary-derived Heliothis subflexa cell line (yellow line) was found to be endemically infected with Sf-RV at the start of the experiment (preinfected). The Helicoverpa zea ovary (dashed green line) or H. zea fat body-derived (solid green line) and H. virescens ovary (red line) cell lines did not support replication of Sf-RV.
Titrations of Sf-RV infectivity on S. exigua cells.
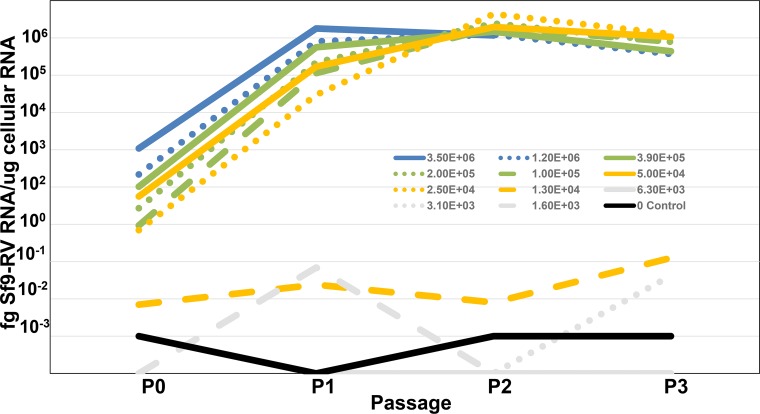
The experimental results presented in Fig. 2 are derived from a classic virus titration experiment (22) in which infectious virus in Sf9 cell lysates was quantified by inoculating susceptible S. exigua cells with decreasing quantities of virus-containing lysate and monitoring the inoculated cultures for evidence of Sf-RV infectivity by PCR. As in all infectivity studies, there will always be a greater number of total virus particles (quantified by genomic RNA copy number) than infectious particles. Our conclusions about this ratio suggest that the observation that there are many more copies of viral genomic RNA than of infectious Sf-RV viral particles is not unusual for rhabdoviruses. While the infectious-to-noninfectious particle ratio was low, it is important to point out that Sf-RV infectivity in S. exigua cells was quite robust, in that once an infection was detected, it accelerated rapidly to equilibrium.
FIG 2.
Infectivity of Sf-RV in Sf9 cells as a function of virus titer. Sf9 cell supernatants containing 106 (blue lines), 105 (green lines), 104 (yellow lines), 103 (gray lines), or no (black line) particles of Sf-RV.
Based on the observation that Sf-RV is capable of replication in Spodoptera exigua cells, two experiments were completed in which Sf-RV contained within culture supernatants of Sf9 cells was titrated for infectivity on the S. exigua cell line SeE1 to determine the ratio of infectious units to noninfectious rhabdovirus particles. The two Sf9 cell supernatants used for the titrations on SeE1 cells contained 3.5 × 107 and 5.4 × 107 particles per ml, as shown by RT-PCR. This determination was performed by quantitative RT-PCR specific for minus strand genomic RNA and following RNase treatment to eliminate any free nonencapsulated rhabdovirus RNA. Sequential flasks of SeE1 cells were inoculated with 3-fold serial dilutions of the Sf9 cell supernatants. Following inoculation, each flask was incubated until confluent (passage 0) and then passaged an additional 3 or 4 times at a 1:10 dilution. Cells not transferred into the next passage were subjected to total RNA purification. Purified RNA from each flask at each passage was subjected to RT-PCR analysis for the detection and quantification of the Sf9 rhabdovirus to determine the highest dilution that still contained at least one infectious unit. Figure 2 shows the results of the two titrations in which similar results were obtained, indicating an infectious titer of approximately 2 × 103 infectious units per ml. Based on the genomic RNA copy numbers in the original supernatants, this resulted in infectious-to-noninfectious particle ratios of approximately 1:17,000 and 1:26,000 in the two experiments. It is important to note that infectivity progressed rapidly in those SeE1 cell flasks that became infected and that infection was obvious by passage 1 in all but one flask in which infection was evident by passage 2.
Presence of Sf-RV in wild populations of S. frugiperda.
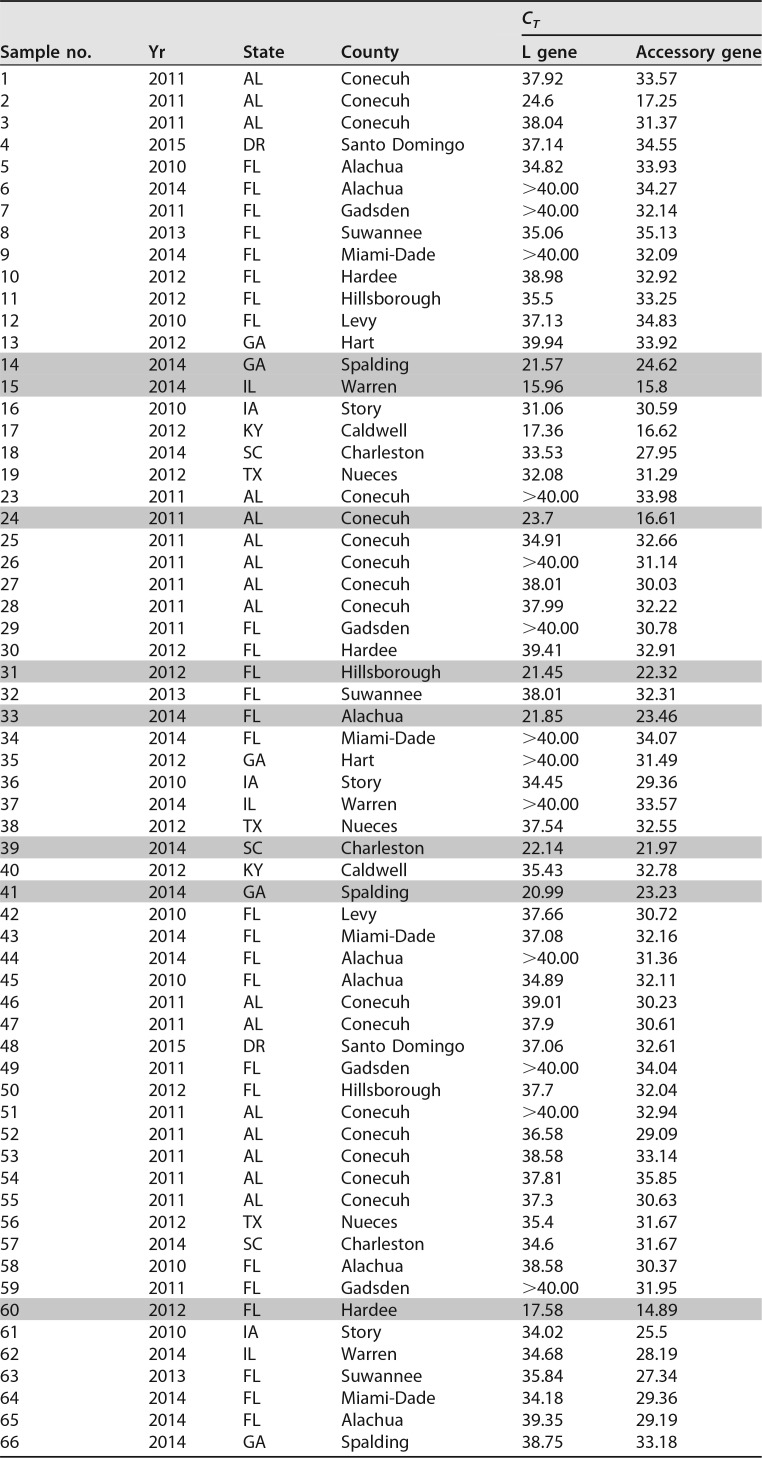
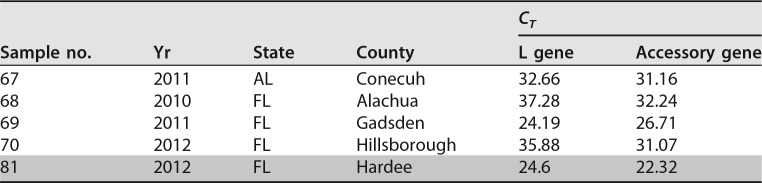
As both the Sf9 and Sf21 cell lines were derived from ovarian tissue from an S. frugiperda insect specimen, we determined whether the Sf9 rhabdovirus could be detected at a significant frequency in other wild or laboratory colony specimens of S. frugiperda. To determine the extent of the presence of this virus in the environment, we tested a number of adult S. frugiperda specimens captured in pheromone traps around the eastern region of the United States, Dominican Republic, and Puerto Rico from surveys conducted over multiple years by the USDA-ARS Center for Medical, Agricultural, and Veterinary Entomology in Gainesville, FL. Table 1 shows a list of 67 insect specimens that were collected and processed for RNA and successfully tested for the rhabdovirus by RT-PCR with appropriate negative controls. A number of the moth RNA samples exhibited very strong rhabdovirus signals (shaded in Table 1) for both the L gene and the accessory gene, providing strong evidence that these insects were infected with the same virus present in Sf9 cells. It is important to note that the accessory gene PCR assay targets a sequence that is not present in viruses containing the 320-nucleotide deletion. In the L gene assay, 54 of 67 samples (80.6%) were at least weakly positive, with 13 samples being negative (cycle threshold [CT] of ≥40). In the accessory gene assay, all 67 samples (100%) were positive, with CT values ranging from 35.85 to 14.89. For most of the moth RNA samples, the accessory gene assay was more sensitive than the L gene assay, which likely accounts for the greater number of positive samples.
TABLE 1.
Detection of Sf-RV in wild-caught S. frugiperda from counties in American states and Caribbean countriesa
Shading indicates samples that yielded full-length N gene PCR amplicons used for phylogenetic tree construction.
Variation in N gene sequences from geographically distant moth samples.
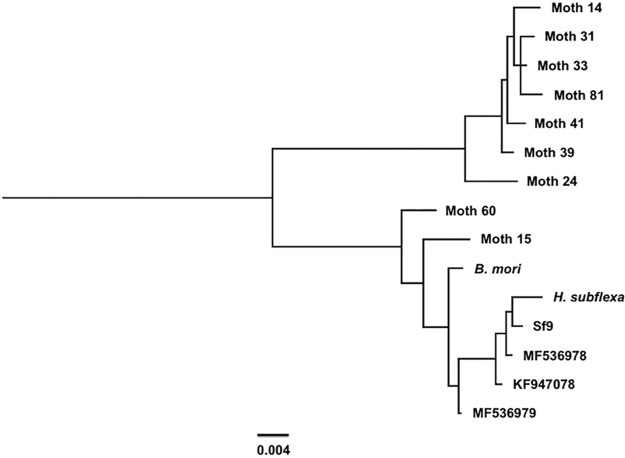
The major factor contributing to the wide variance in the ability to detect rhabdovirus signals in screens of wild-caught moths was likely due to their states of decay prior to storage and RNA isolation. That said, it is noteworthy that the accessory gene was present in every moth RNA sample collected from the environment, even though the accessory gene PCR primer and probe set used in the study targeted a sequence that is absent in the Takeda Sf-RV and which has been found to be unstable in the Sf-RV found in Sf9 cells derived from the ATCC during continuous passage (data not shown). Fortunately, 9 of the 67 moth RNA samples listed in Table 1 yielded strong RT-PCR signals, which made cloning and sequence determination of the full-length N ORF possible. These data were used to generate a phylogenetic comparison of this gene in the wild population with that of Sf-RV found in Sf9 cell lines (Fig. 3). We focused on the N gene for downstream analyses, as this gene is typically used to assess population structure in negative-strand RNA viruses (23–25). Sequence homologies between the field specimens and the published Sf9 rhabdovirus sequence from Sf9 cells ranged from 94% to 99%. The N gene sequences clustered into two distinct clades, suggesting subspecies structure in the Sf-RV population as has been noted for other negative-strand RNA viruses (26).
FIG 3.
Phylogeny of N gene sequences from wild populations of S. frugiperda and insect cell lines. Scale bar, 0.004 substitutions per site.
Sequence analysis of Sf-RV found in other insect cells.
Screening a variety of insect cell lines showed that Sf-RV is constitutively present in the H. subflexa cell line HsAM1, based on PCR data. The identification of Sf-RV in an insect line from a genus other than Spodoptera suggested that this virus might be capable of growth in a variety of insect species and be significantly more widespread in insects other than S. frugiperda in the environment. Therefore, we sequenced the version of Sf-RV found in the HsAM1 cell line to determine if it was indeed the same virus. In addition to H. subflexa, a GenBank database entry (AK377209.1) for an mRNA sequence clone derived from Bombyx mori was found to be essentially identical to one of the genes in the Sf-RV genome, implying that this virus can also be found in silk worm insects or cell lines. Indeed, acquisition of the B. mori cell line Bm-N and analysis by PCR indicated that this cell line was infected with Sf-RV similarly to the H. subflexa HsAM1 cell line. Complete sequence contigs were generated for Sf-RVs found in both the HsAM1 and Bm-N cell lines. In Table 2, the predicted proteins encoded by Sf-RV variants are compared. Notably, the size of the accessory protein in the H. subflexa strain is reduced from 12.63 kDa to 8.95 kDa due to a 320-bp deletion in the coding region of this gene. The sequence contig determined for Sf-RV from B. mori confirms the presence of the intact accessory gene region.
TABLE 2.
Variation in the proteins encoded by three strains of Sf-RV isolated from different species
| Strain | ORF | Function | MWa | pI | NLSa |
|---|---|---|---|---|---|
| Sf-RV (Takeda) | N | Nucleocapsid | 60.85 | 5.10 | |
| P | Phosphoprotein | 41.61 | 6.59 | 318RKPR323 | |
| M | Matrix protein | 34.97 | 9.53 | ||
| G | Glycoprotein | 69.90 | 8.38 | ||
| X | Unknown | 8.95 | 5.80 | ||
| L | Polymerase | 24.45 | 8.23 | 9RKKRP15, 554KKRH559 | |
| H. subflexa | N | Nucleocapsid | 60.78 | 5.00 | |
| P | Phosphoprotein | 42.05 | 6.82 | 321RKPR326 | |
| M | Matrix protein | 34.94 | 9.57 | ||
| G | Glycoprotein | 70.18 | 8.52 | 329RHKR323 | |
| X | Unknown | 8.95 | 5.80 | ||
| L | Polymerase | 24.41 | 8.01 | 9RKKRP15, 554KKRH559 | |
| B. mori | N | Nucleocapsid | 60.69 | 5.09 | |
| P | Phosphoprotein | 36.51 | 6.05 | 275RKPR280 | |
| M | Matrix protein | 34.90 | 9.57 | ||
| G | Glycoprotein | 70.05 | 8.39 | 329RHKR323 | |
| X | Unknown | 12.63 | 5.42 | ||
| L | Polymerase | 24.42 | 8.23 | 9RKKRP15, 554KKRH559 |
The molecular weights (MWs) of accessory proteins are in boldface font.
NLS, nuclear localization signal.
Detection of Sf-RV virus RNA on vegetables following S. frugiperda larva feeding.
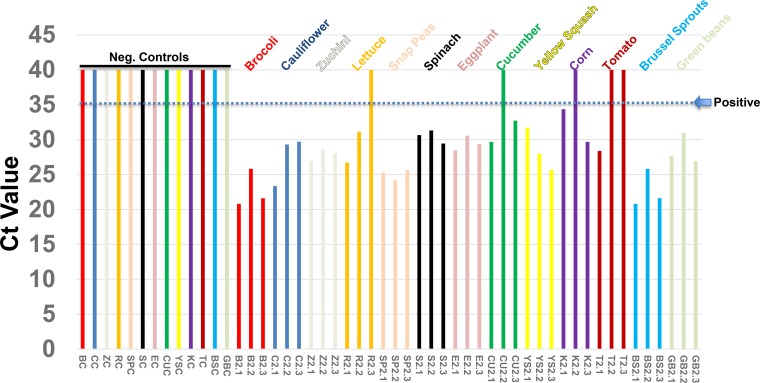
Since S. frugiperda insects are widespread in parts of North and South America, and more recently, in Africa, where they feed on important human food crops, it was of interest to determine if S. frugiperda larvae could potentially deposit Sf-RV on human food plants while feeding. To this end, S. frugiperda eggs were acquired from a commercial vendor and allowed to hatch into larvae and feed on 13 different vegetables in the laboratory for 18 h. Prior to the initiation of this study, we confirmed the presence of Sf-RV in 100% of larvae hatched from the commercially acquired eggs. Following feeding, vegetable specimens were harvested and subjected to RNA isolation, and purified RNA was tested for the presence of Sf-RV RNA. Thirty-seven of forty-two vegetable samples were shown to be positive for the presence of Sf-RV RNA, while all negative-control vegetables not exposed to S. frugiperda larvae were found to be negative. These data are shown in Fig. 4, in which 37 of 42 (88%) vegetable samples were positive (CT value, <35) for the presence of Sf-RV RNA and a few of the samples had very high titers (CT value, <25). These data are consistent with the possibility that S. frugiperda larvae deposit infectious virus on vegetables during feeding and that humans might be exposed to Sf-RV via raw vegetable handling and/or consumption. In addition to the vegetables tested in this study, S. frugiperda larvae generally feed upon members of the grass family, and the most common human-edible food plant in this category is sweet corn. Other human-edible food crops that are consumed by the larvae include apple, grape, orange, papaya, peach, and strawberry (http://entnemdept.ufl.edu/creatures/field/fall_armyworm.htm).
FIG 4.
CT values for detection of Sf-RV RNA on vegetables following overnight feeding by S. frugiperda larvae. CT values >35 were considered negative for detection of Sf-RNA. Larvae, and their frass, were removed from the vegetables prior to RNA isolation.
Characterization of Sf-RV proteins in plant cells.
Sf-RV lacks a clearly identifiable movement protein, a key mark of plant-adapted viruses, suggesting that Sf-RV is unlikely to be adapted to replicate in plants. However, its common occurrence in phytophagous insects and possible exposure to plants via feeding raised the possibility that plants could serve as reservoirs for Sf-RV and potentially amplify virus if replication was able to occur in initially infected cells. Moreover, we examined the characteristics of Sf-RV proteins in plants to determine whether their physicochemical properties might aid or inhibit adaption to plants. Protoplast experiments showed that under conditions that were conducive to the replication of a plant-adapted rhabdovirus Potato yellow dwarf virus, or plus-strand RNA Turnip crinkle virus, we were unable to detect replication of Sf-RV in protoplasts of Arabidopsis thaliana (data not shown) or Nicotiana benthamiana, plants that are known to be susceptible to a wide range of plant viruses (Fig. 5) (27). In N. benthamiana, results of Sf-RV protein localization in plant cells were consistent with that expected for rhabdoviruses, with the G and M proteins associated with membranes, particularly, perinuclear membranes. The short and long versions of the accessory protein were readily differentiated based on the ability to accumulate in nuclei. The phosphoprotein localized to the cell periphery, with no accumulation in nuclei (Fig. 6). Interestingly, the nucleocapsid protein accumulated at punctate loci on the cell periphery. These loci colocalized with the plasmodesma-associated protein PDLP1 (Fig. 7).
FIG 5.
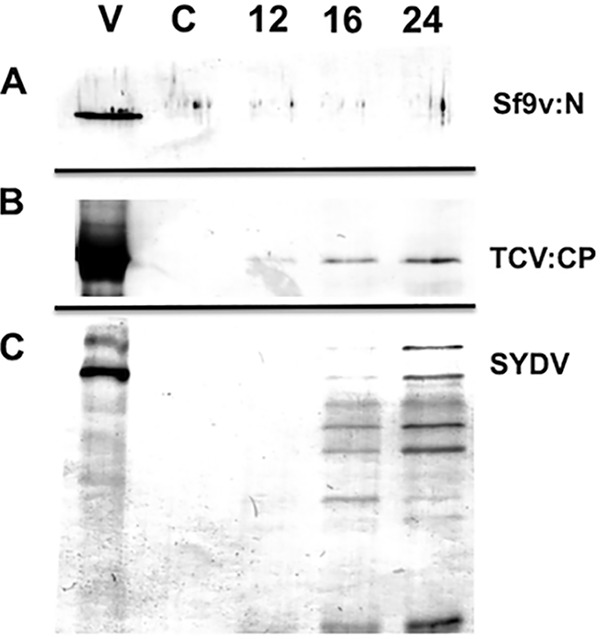
Immunodetection of viral proteins in a time course of equivalent amounts of N. benthamiana protoplasts inoculated with Sf-RV (A), Turnip crinkle virus (TCV) (B), or Potato yellow dwarf virus strain (SYDV) (C). Protoplasts were sampled at 12, 16, and 24 h after inoculation and probed via Western immunoblotting with polyclonal antibodies raised against the Sf-RV nucleocapsid (Sf-RV:N), TCV coat protein (TCV:CP), or disrupted SYDV virions, which detect a ladder of proteins corresponding to G, N, M, and P (SYDV). Purified virus (V) and uninoculated protoplasts (C) served as controls.
FIG 6.
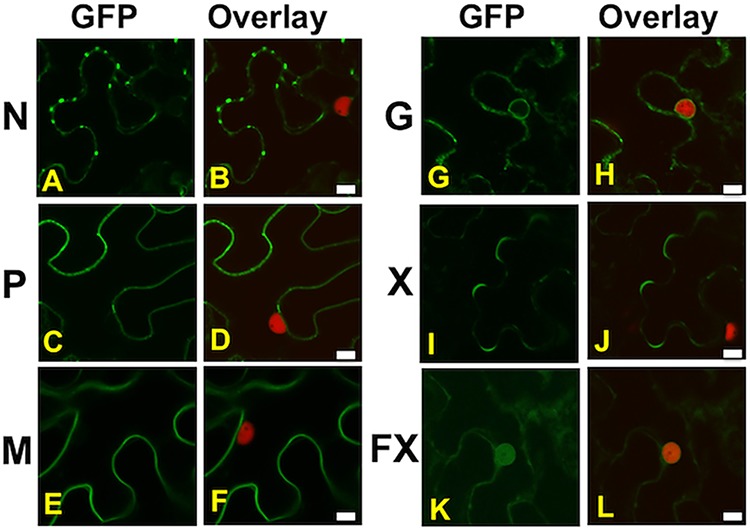
Localization of GFP:Sf-RV protein fusions in leaf epidermal cells of transgenic N. benthamiana expressing the nuclear marker fusion RFP:Histone2B by agroinfiltration. Localization patterns of GFP fusions are shown alone (GFP) or relative to the red nuclei of transgenic cells (overlay) for nucleocapsid protein (A and B), phosphoprotein (C and D), matrix protein (E and F), glycoprotein (G and H), the 9-kDa variant of the accessory protein (I and J), and the 13-kDa variant of the accessory protein (K and L). Scale bars, 10 μm.
FIG 7.
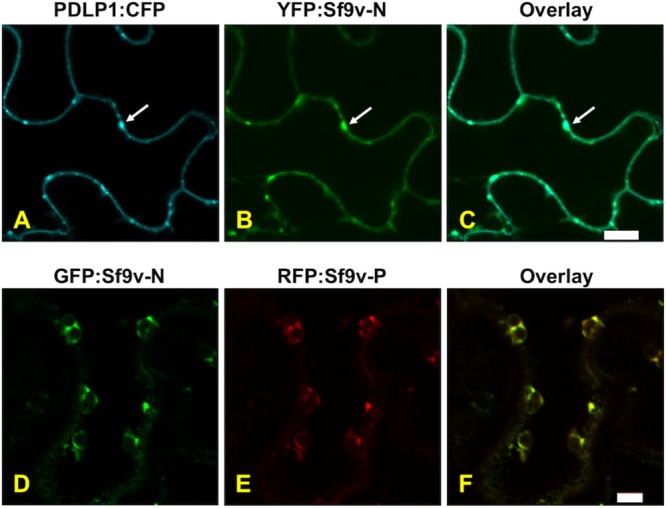
YFP:Sf-RV-N fusion has a localization pattern in leaf epidermal cells of N. benthamiana similar to that of the plasmodesmata marker fusion CFP:PDLP1. (A) CFP:PDLP1; (B) YFP:Sf-RV-N; (C) overlay of panels A and B. Scale bar, 10 μm. Coexpression of Sf-RV N and P proteins alters their expression patterns: GFP:Sf-RV-N (D), RFP:Sf-RV-P (E), overlay of panels D and E. Scale bar, 5 μm.
DISCUSSION
Virus discovery by next-generation sequencing has rapidly expanded our understanding of environs in the biosphere occupied by viruses. In many cases, these findings are surprising, as viruses are being identified in cell types that exhibit no overt cytopathic effects, as in the case of Sf9 rhabdovirus in Sf9 cell lines that have been in use for the production of vaccines and other biologicals used in humans. Cell lines cleared of Sf-RV are equivalent to virus-infected lines for the purposes of protein expression. However, established production facilities will continue to use Sf-RV-infected cell lines, which raises concerns about human exposure to this virus. In this study, we describe the first population-level studies that sought to determine how widespread Sf-RV is, and in which insect species it exists, in the wild.
Importantly, it is now well established that Sf-RV does not replicate in human cells, which is of paramount concern regarding human exposure. In contrast, our survey of S. frugiperda moths collected in hormone traps over a period of 5 years demonstrates that Sf-RV is endemic in wild populations in the southern United States. While broader surveys are warranted to gain a clearer perspective on the population structure of Sf-RV, it appears that there may be at least two lineages, based upon N gene sequences. It is expected that Sf-RV is transmitted vertically in insect populations; therefore, Sf-RV phylogenies should be congruent with the S. frugiperda population structure (4, 9, 28). Of significant interest is whether our findings for S. frugiperda may also be reflected in populations of H. subflexa and B. mori, cell lines which were found to be infected with Sf-RV. Given the plant host range of S. frugiperda, Sf-RV would have been predicted to be found in the similarly polyphagous H. virescens and not in H. subflexa, which feeds only on species in the genus Physalis (29). However, we found no evidence for Sf-RV replication in H. virescens cell lines, while the H. subflexa cell line tested was already infected with the virus. A cell line of B. mori, which also has a limited host range, was also determined to be infected with Sf-RV. A comparison of the genomes of Sf-RVs isolated from these three lepidopterans showed that the major differences lie in the sizes of the accessory proteins of unknown function. For reasons that are unclear, the accessory protein gene is unstable, and a 320-nucleotide portion is susceptible to deletion when infected cells lines are passaged in culture. In contrast, the 320-bp fragment was found to be present in all Sf-RV isolates from wild populations of S. frugiperda, suggesting that this gene is important in the context of insect infections. The full-length (12.63 kDa) accessory protein is capable of nuclear import, as shown in subcellular localization studies, whereas the truncated from (8.95 kDa) is excluded from the nucleus. Many RNA viruses with cytoplasmic sites of replication encode proteins that target the nucleus to perturb gene expression in host cells (30–34). It is possible that such regulation is required for infection of insects. Alternatively, we cannot rule out that the presence of Sf-RV in B. mori- and H. subflexa-derived cell lines is due to contamination that may have occurred when these cells were cultured in labs working with Sf-9 cells. Examination of wild populations is therefore essential to resolve this issue.
Consistent with its broad host range, we found that Sf-RV RNA was deposited on a variety of vegetables by feeding S. frugiperda larvae. This finding raises the possibility that humans are exposed on occasion to this virus via consumption of food items that overlap the S. frugiperda host range. Although we found no detectable replication of Sf-RV in two species of plants, the localization patterns of proteins exhibit some peculiarities consistent with plant-adapted species (30). Most notable among these is the ability to target the cell periphery, particularly plasmodesmata, the cell junctions in plants that provide symplastic continuity between adjacent cells separated by otherwise impenetrable cell walls (35). Sf-RV lacks a clearly identifiable movement protein, which is a hallmark of plant-adapted viruses. However, the nucleocapsid protein of Sf-RV is clearly able to target plasmodesmata, suggesting that it could provide movement protein functions. Furthermore, as for the cognate proteins of their plant-infecting counterparts, coexpression of the N and P proteins results in the formation of a complex of both proteins in spherules at the cell periphery. While plant viruses vectored by lepidopterans are rare relative to those transmitted by aphids, planthoppers, and leafhoppers (36), it is of interest to know what additional factors contribute to the inability of rhabdoviruses in these insects to fully adapt to plant cells. In the case of Sf-RV, it appears that protein stability is a key factor. While we were able to localize proteins of this virus as fusions to full-length green fluorescence protein (GFP) or red fluorescent protein (RFP), it was impossible to detect interactions between these proteins using bimolecular fluorescence complementation under conditions now standard for generating plant rhabdovirus protein interaction and localization maps (37–40) (data not shown). Mechanical inoculation of plants, and protoplast transfection experiments, did not result in evidence for replication of Sf-RV in plants.
In conclusion, this report and other studies have demonstrated that Sf-RV does not replicate in plant cells (15–20). In addition, population, infectivity, and feeding studies described here demonstrate that Sf-RV is endemic in the environment with the capability for replication in up to four insect species, S. frugiperda, S. exigua, H. subflexa, and B. mori, each with the potential to deposit virus on human-consumed foods. Indeed, S. frugiperda larva feeding studies demonstrated that significant amounts of viral RNA were deposited on a variety of vegetables. Therefore, there is increasing evidence of a history of human exposure to Sf-RV, arguing that this virus is unlikely to pose a significant health hazard to humans.
MATERIALS AND METHODS
Sf-RV genome sequence analyses.
The genomic sequence of Sf-RV found in the Takeda Vaccines Sf9 cell line was determined prior to the first published report of Sf-RV by Ma et al. (15). In experiments designed to screen for insect retroviruses, baculovirus lysates of Sf9 cells were clarified of debris and then subjected to Benzonase (Sigma-Aldrich) digestion to eliminate free (not particle-associated) nucleic acids. Following nuclease digestion, the material was centrifuged at 100,000 × g through a 20% sucrose cushion in order to collect virus particles. High-speed pelleted material was then subjected to viral nucleic acid extraction and then treated with RNase-free DNase in order to remove baculovirus and insect genomic DNA, while leaving any viral RNAs intact. A random N9 primer tagged with a Not I restriction site at its 5′ end (TATTGCGGCCGCTTCTTNNNNNNNNN) was used for both the first-strand reverse transcriptase reaction and the second-strand Klenow DNA polymerase reaction, resulting in double-stranded DNAs that could be amplified using a PCR primer consisting of the Not I sequence tag (TATTGCGGCCGCTTCTT) and then cloned into a plasmid vector. Approximately 20% of the cloned fragments were shown to be distantly homologous to the L protein of rhabdoviruses following sequence analysis (DNA sequencing facility, UC Davis). The rhabdovirus homologies were observed at the amino acid sequence level only and were in the range of 24% to 26% homology through part of the L gene. Sequence analysis of all clones showing rhabdovirus homology as well as the clones that showed no detectable homology in the GenBank database resulted in the building of two large sequence contigs of approximately 7,000 and 4,500 nucleotides, respectively, with each containing a portion of an open reading frame with homology to rhabdovirus L proteins. Suspecting that the two sequence contigs were part of the same viral genomic sequence, additional RT-PCR cloning using one primer from each contig resulted in the isolation of clones representing the sequences spanning these two contigs. Finally, rapid amplification of cDNA ends (RACE) techniques allowed for the isolation of 5′- and 3′-end clones, completing the sequence of the 13,267 nucleotide novel rhabdovirus genome. Additional RACE experiments performed at the University of Kentucky to define the 3′ and 5′ ends of the genome were performed using the BD-SMART RACE cDNA amplification kit according to the manufacturer’s instructions (Thermo Scientific). For these analyses, cDNA was synthesized by Moloney murine leukemia virus (MMLV) reverse transcriptase, and PCRs were conducted with Advantage-II DNA polymerase (Clontech).
Sequence analysis of Sf-RV in HsAM1 and Bm-N cell lines.
The H. subflexa cell line HsAM1 (gift from Cindy Goodman, USDA, ARS, Midwest Area Biological Control of Insect Research Laboratory, Columbia, MO) and the B. mori cell line BM-N. (ATCC) were found to be infected with Sf-RV by using PCR analyses. Sf-RV genome sequence analyses in these cell lines were accomplished by reverse transcriptase PCR amplification of overlapping fragments from cellular RNA followed by direct sequencing of purified PCR fragments (DNA sequencing facility, UC Davis). PCR primers were based on the known Sf9 virus genome sequence as determined by Takeda and also published by Ma et al. (15). Sequence data from the H. subflexa-derived fragments and the B. mori-derived fragments were analyzed by the ContigExpress program of the Vector NTI suite in order to assemble the individual fragment sequences into contigs.
Quantification of Sf-RV RNA by PCR.
A TaqMan quantitative RT-PCR (qRT-PCR) assay was developed in order to quantify Sf9 rhabdovirus minus-strand RNA. The primer and probe sequences are located in a 64-nucleotide target sequence within the L gene. The primer and probe sequences are as follows: forward (FWD), TCTGTATTATGGGTTTGATCAGCTAAG; reverse (REV), CTCGCTGCTGAGCGGTTT; probe, 6-carboxyfluorescein (6FAM)-AGGATTGGAGAATTATAC.
A standard curve RNA for quantification purposes was prepared by in vitro transcription of a plasmid clone containing the L gene target sequence using T7 RNA polymerase following linearization of the plasmid DNA. Template plasmid DNA was subsequently digested with DNase, and the standard curve RNA was purified (RNeasy; Qiagen). An RNA of approximately 5 kb was expected, but two RNAs of equal intensities, 3.5 kb and 5 kb, were produced as shown by glyoxal-agarose gel analysis. The smaller of the two RNAs is large enough to contain the target sequence and was likely produced as a result of premature transcription termination. For quantification purposes, the average molecular weight of the standard curve RNA was therefore 4.25 kb or 1.4 × 106 Da. Use of this RNA as a standard curve in the TaqMan assay demonstrated a reproducible limit of detection of 0.01 fg of RNA, which is equivalent to 4 molecules of standard curve RNA. Sf-RV RNA quantification was performed using the TaqMan RNA-to-Ct 1 Step kit (Thermo Fisher) and the ABI 7500 real-time PCR system. Alternatively, PCR quantification was achieved without the use of an RNA standard curve by employing the RainDrop digital PCR system (RainDance Technologies). For PCR assays of RNA derived from S. frugiperda moth or larvae specimens in which quantification of RNA copy number was not performed, cycle threshold values were reported from experiments using the L gene primer and probe set and the ABI 7500 real-time PCR system. An additional primer and probe set contained within the common 320-nucleotide accessory gene deletion region was also employed.
Sf-RV replication studies in insect cell lines.
The following insect cell lines were obtained from Cindy Goodman (USDA, ARS, Midwest Area, Biological Control of Insect Research Laboratory, Columbia, MO) (cell line name/species and tissue of origin): BCIRL/AMCY-SeE1/Spodoptera exigua eggs, BCIRL/AMCY-SeE4/S. exigua eggs, BCIRL/AMCY-SeE5/S. exigua eggs, Heliothis virescens ovaries, BCIRL/HvAM1/Heliothis virescens ovaries, BCIRL/HzAM1/Helicoverpa zea ovaries, BCIRL-HzFB33/H. zea fat bodies, BCIRL-HsAM1/Heliothis subflexa ovaries, and BCIRL-AgAM1/Anticarsia gemmatalis ovaries.
To determine the potential for Sf-RV to replicate in these cell lines, a T25 tissue culture flask of each cell line containing 10 ml of EX-Cell 420 medium plus 10% fetal bovine serum (FBS) was exposed to the Sf9 rhabdovirus by removing 5 ml of the culture medium and replacing it with 5 ml of conditioned medium from Sf9 cells that had been centrifuged and filtered through a 0.2-μm membrane in order to ensure no Sf9 cell carryover. The Sf9 conditioned medium was allowed to remain on the cell cultures for 24 h, after which the medium was removed and the insect cell lines were further cultured in EX-Cell 420 medium containing 10% FBS. Upon reaching confluence, approximately 10% of the cells of each culture were collected and stored as a frozen cell pellet (p0), while new subcultures of each were established for continued growth and passaging. In this way, two additional frozen cell pellets of each cell line were collected for passage numbers p1 and p2, respectively.
Following collection of the frozen cell samples, total cellular RNA was purified from each cell sample (RNeasy; Qiagen) as well as from control cells from each cell line that were not exposed to the Sf9 rhabdovirus (preinfected). Each RNA sample was tested in the Sf9 rhabdovirus qRT-PCR assay described above for detection and quantification of any rhabdovirus RNA signals. Data were expressed in terms of femtogram rhabdovirus RNA per microgram of total cellular RNA.
Two additional Sf-RV replication experiments were performed in S. exigua cells in which the Sf9 cell-conditioned medium was serially diluted to reach the endpoint of infectivity in order to determine the number of infectious units per total number of Sf-RV genomic RNA copies. Using the Spearman-Karber method of 50% tissue culture infective dose (TCID50), the calculations for both virus preparations showed a similar rhabdovirus titer of 2 × 103 infectious units per ml (22). Based on the original rhabdovirus particle count determined by qRT-PCR, the virus for which the titer was determined in experiment 602-132 had an infectious-to-noninfectious ratio of 1:17,000, whereas the virus for which the titer was determined in experiment 602-170 had an infectious-to-noninfectious ratio of 1:26,000.
Measurement of Sf-RV in adult S. frugiperda specimens collected from the environment.
RNA samples isolated from the collected moth specimens were analyzed for the presence of Sf9 rhabdovirus RNA by 2 different TaqMan RT-PCR assays (see above) specific for a target sequence in the L (polymerase) gene and a second target sequence in the accessory gene within the boundaries of the common 320-nucleotide deletion. The accessory gene target sequence is in an unstable area of the Sf9 rhabdovirus genome in a 320-nucleotide (nt) region that is sometimes deleted. Negative-control samples produced a CT value of ≥40, consistent with the absence of contaminating rhabdovirus sequences producing false-positive results.
Insect feeding assay.
S. frugiperda eggs were acquired from Benzon Research, Inc. (Carlisle, PA, USA) and were hatched under optimal controlled environmental conditions in the presence of an artificial lepidopteran diet acquired from the insect supplier. At approximately day 14 posthatching and slightly before the insects were to begin pupating, the larvae were placed on a variety of vegetables and allowed to feed for 18 h (overnight). The vegetables included broccoli, cauliflower, zucchini, romaine, snap peas, spinach, eggplant, cucumber, yellow squash, corn, tomato, Brussels sprouts, and green beans. After overnight feeding, samples of the vegetables where insect feeding had taken place were excised and subjected to RNA extraction and RT-PCR analysis to assay for the presence of Sf-RV viral RNA.
Protoplasts and immunoblotting.
Protoplasts were isolated from 1 g of N. benthamiana leaves from young plants at the 4- to 6-leaf stage of growth, as described by Panaviene et al. (41). Experiments were always conducted in quadruplets, with each lot of protoplasts, in a final suspension of 1 ml, being split into 250-μl aliquots. One aliquot served as the uninoculated control, while the remaining three were inoculated with Sf-RV (using 5.5 × 107 particles), while the other two were inoculated with Potato yellow dwarf virus (PYDV) or Turnip crinkle virus (TCV) by the addition of 100 μl of clarified supernatant after grinding 0.1 g of infected leaf tissue in 1 ml of protoplast isolation buffer. At 12, 16, and 24 h postinoculation, 50 μl of protoplast suspension was sampled, mixed with equal volumes of SDS-PAGE loading buffer, and processed for Western immunoblotting, essentially as described previously (42).
Protein localization in plant cells.
To our knowledge, the subcellular localization patterns of Sf9-RV proteins have not been determined. Heterologous expression systems are generally valid for localization studies given the conservation of subcellular localization signals (i.e., the same nuclear localization signal functions correctly if expressed in yeast, plant, or mammalian cells) (43–45). As such, the plant-based system used is appropriate for these studies (39, 46). One of the aims of this study was to determine the replication competency of Sf9-RV in plants given the opportunity for transmission by its phytophagous host. We chose to use N. benthamiana for protein localization studies, which is widely used for such experiments (27, 43). Furthermore, initial phylogenetic comparisons suggested that Sf9-RV is related to lettuce necrotic yellows virus, a plant-adapted rhabdovirus (data not shown). Therefore, plant-based expression was necessary to determine if Sf9-RV encoded a protein that could target the virus to the cell periphery, in lieu of a clearly identifiable movement of protein. To accomplish the expression of GFP protein fusions in plant cells, full-length ORFs of Sf-RV were amplified by PCR, cloned, and sequenced verified. These clones were used to express fusions to fluorescent proteins in leaf epidermal cells, essentially as described previously for the construction of rhabdovirus protein interaction and localization maps (30, 39, 46, 47).
Accession number(s).
The Sf-RV genome sequences from B. mori, H. subflexa, and Sf9 cells (Takeda cell culture), were deposited in GenBank as accessions MH926029, MH926030, and MH926031.
ACKNOWLEDGMENTS
We thank John Shaw for critical readings of the manuscript prior to submission. We also thank Robert L. Meagher, Jr. and Rodney Nagoshi at the USDA-ARS CMAVE (Gainesville, FL) for the generous contributions of wild-caught Spodoptera sp. moths.
REFERENCES
- 1.Westbrook JK, Nagoshi RN, Meagher RL, Fleischer SJ, Jairam S. 2016. Modeling seasonal migration of fall armyworm moths. Int J Biometeorol 60:255–267. doi: 10.1007/s00484-015-1022-x. [DOI] [PubMed] [Google Scholar]
- 2.Ray S, Basu S, Rivera-Vega LJ, Acevedo FE, Louis J, Felton GW, Luthe DS. 2016. Lessons from the far end: caterpillar FRASS-induced defenses in maize, rice, cabbage, and tomato. J Chem Ecol 42:1130–1141. doi: 10.1007/s10886-016-0776-x. [DOI] [PubMed] [Google Scholar]
- 3.Bano A, Muqarab R. 2017. Plant defence induced by PGPR against Spodoptera litura in tomato (Solanum lycopersicum L.). Plant Biol J 19:406–412. doi: 10.1111/plb.12535. [DOI] [PubMed] [Google Scholar]
- 4.Acevedo FE, Peiffer M, Ray S, Meagher R, Luthe DS, Felton GW. 2018. Intraspecific differences in plant defense induction by fall armyworm strains. New Phytol 218:310–321. doi: 10.1111/nph.14981. [DOI] [PubMed] [Google Scholar]
- 5.Heppner JB. 1998. Spodoptera armyworms in Florida (Lepidoptera: Noctuidae), p 1–5. no. 390 Florida Department of Agriculture Entomology Circular. Florida Department of Agriculture and Consumer Services, Tallahassee, FL. [Google Scholar]
- 6.Luginbill P. 1928. The fall army worm. U.S. Department of Agriculture, Washington, DC. [Google Scholar]
- 7.Banerjee R, Hasler J, Meagher R, Nagoshi R, Hietala L, Huang F, Narva K, Jurat-Fuentes JL. 2017. Mechanism and DNA-based detection of field-evolved resistance to transgenic Bt corn in fall armyworm (Spodoptera frugiperda). Sci Rep 7:10877. doi: 10.1038/s41598-017-09866-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrasena DI, Signorini AM, Abratti G, Storer NP, Olaciregui ML, Alves AP, Pilcher CD. 2018. Characterization of field-evolved resistance to Bacillus thuringiensis-derived Cry1F delta-endotoxin in Spodoptera frugiperda populations from Argentina. Pest Manag Sci 74:746–754. doi: 10.1002/ps.4776. [DOI] [PubMed] [Google Scholar]
- 9.Nagoshi RN, Koffi D, Agboka K, Tounou KA, Banerjee R, Jurat-Fuentes JL, Meagher RL. 2017. Comparative molecular analyses of invasive fall armyworm in Togo reveal strong similarities to populations from the eastern United States and the Greater Antilles. PLoS One 12:e0181982. doi: 10.1371/journal.pone.0181982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith GE, Summers MD, Fraser MJ. 1983. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol 3:2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller LK. 1989. Insect baculoviruses: powerful gene expression vectors. Bioessays 11:91–95. doi: 10.1002/bies.950110404. [DOI] [PubMed] [Google Scholar]
- 12.Vlak JM, Keus RJ. 1990. Baculovirus expression vector system for production of viral vaccines. Adv Biotechnol Processes 14:91–128. [PubMed] [Google Scholar]
- 13.Fath-Goodin A, Kroemer J, Martin S, Reeves K, Webb BA. 2006. Polydnavirus genes that enhance the baculovirus expression vector system. Adv Virus Res 68:75–90. doi: 10.1016/S0065-3527(06)68002-0. [DOI] [PubMed] [Google Scholar]
- 14.Steele KH, Stone BJ, Franklin KM, Fath-Goodin A, Zhang X, Jiang H, Webb BA, Geisler C. 2017. Improving the baculovirus expression vector system with vankyrin-enhanced technology. Biotechnol Prog 33:1496–1507. doi: 10.1002/btpr.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H, Galvin TA, Glasner DR, Shaheduzzaman S, Khan AS. 2014. Identification of a novel rhabdovirus in Spodoptera frugiperda cell lines. J Virol 88:6576–6585. doi: 10.1128/JVI.00780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisler C, Jarvis DL. 2016. Rhabdovirus-like endogenous viral elements in the genome of Spodoptera frugiperda insect cells are actively transcribed: implications for adventitious virus detection. Biologicals 44:219–225. doi: 10.1016/j.biologicals.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maghodia AB, Geisler C, Jarvis DL. 2016. Characterization of an Sf-rhabdovirus-negative Spodoptera frugiperda cell line as an alternative host for recombinant protein production in the baculovirus-insect cell system. Protein Expr Purif 122:45–55. doi: 10.1016/j.pep.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geisler C, Jarvis DL. 2018. Adventitious viruses in insect cell lines used for recombinant protein expression. Protein Expr Purif 144:25–32. doi: 10.1016/j.pep.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto Y, Macri D, Srivastava I, McPherson C, Felberbaum R, Post P, Cox M. 2017. Complete study demonstrating the absence of rhabdovirus in a distinct Sf9 cell line. PLoS One 12:e0175633. doi: 10.1371/journal.pone.0175633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maghodia AB, Jarvis DL. 2017. Infectivity of Sf-rhabdovirus variants in insect and mammalian cell lines. Virology 512:234–245. doi: 10.1016/j.virol.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia AG, Godoy WAC, Thomas JMG, Nagoshi RN, Meagher RL. 2018. Delimiting strategic zones for the development of fall armyworm (Lepidoptera: Noctuidae) on corn in the state of Florida. J Econ Entomol 111:120–126. doi: 10.1093/jee/tox329. [DOI] [PubMed] [Google Scholar]
- 22.Hierholzer J, Killington RA. 1996. Virus isolation and quantitation, p 25–46. In Kangro H, Mahy B (ed), Virology methods manual. Academic Press, San Diego, CA. [Google Scholar]
- 23.Higgins CM, Chang WL, Khan S, Tang J, Elliott C, Dietzgen RG. 2016. Diversity and evolutionary history of lettuce necrotic yellows virus in Australia and New Zealand. Arch Virol 161:269–277. doi: 10.1007/s00705-015-2626-5. [DOI] [PubMed] [Google Scholar]
- 24.Talbi C, Cabon J, Baud M, Bourjaily M, de Boisséson C, Castric J, Bigarré L. 2011. Genetic diversity of perch rhabdoviruses isolates based on the nucleoprotein and glycoprotein genes. Arch Virol 156:2133–2144. doi: 10.1007/s00705-011-1103-z. [DOI] [PubMed] [Google Scholar]
- 25.Carnieli P Jr, Ruthner Batista HB, de Novaes Oliveira R, Castilho JG, Vieira LF. 2013. Phylogeographic dispersion and diversification of rabies virus lineages associated with dogs and crab-eating foxes (Cerdocyon thous) in Brazil. Arch Virol 158:2307–2313. doi: 10.1007/s00705-013-1755-y. [DOI] [PubMed] [Google Scholar]
- 26.Kondo H, Hirota K, Maruyama K, Andika IB, Suzuki N. 2017. A possible occurrence of genome reassortment among bipartite rhabdoviruses. Virology 508:18–25. doi: 10.1016/j.virol.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 27.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. 2008. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 21:1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 28.Venkatesan M, Rasgon JL. 2010. Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Mol Ecol 19:1573–1584. doi: 10.1111/j.1365-294X.2010.04577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groot AT, Classen A, Inglis O, Blanco CA, López J, Téran Vargas A, Schal C, Heckel DG, Schöfl G. 2011. Genetic differentiation across North America in the generalist moth Heliothis virescens and the specialist H. subflexa. Mol Ecol 20:2676–2692. doi: 10.1111/j.1365-294X.2011.05129.x. [DOI] [PubMed] [Google Scholar]
- 30.Min BE, Martin K, Wang R, Tafelmeyer P, Bridges M, Goodin M. 2010. A host-factor interaction and localization map for a plant-adapted rhabdovirus implicates cytoplasm-tethered transcription activators in cell-to-cell movement. Mol Plant Microbe Interact 23:1420–1432. doi: 10.1094/MPMI-04-10-0097. [DOI] [PubMed] [Google Scholar]
- 31.Redondo N, Madan V, Alvarez E, Carrasco L. 2015. Impact of vesicular stomatitis virus M proteins on different cellular functions. PLoS One 10:e0131137. doi: 10.1371/journal.pone.0131137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettit Kneller EL, Connor JH, Lyles DS. 2009. hnRNPs Relocalize to the cytoplasm following infection with vesicular stomatitis virus. J Virol 83:770–780. doi: 10.1128/JVI.01279-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen JM, Her LS, Varvel V, Lund E, Dahlberg JE. 2000. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol Cell Biol 20:8590–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarbrough ML, Mata MA, Sakthivel R, Fontoura BM. 2014. Viral subversion of nucleocytoplasmic trafficking. Traffic 15:127–140. doi: 10.1111/tra.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. 2008. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6:e7. doi: 10.1371/journal.pbio.0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ossiannilsson F. 1966. Insects in the epidemiology of plant viruses. Annu Rev Entomol 11:213–232. doi: 10.1146/annurev.en.11.010166.001241. [DOI] [PubMed] [Google Scholar]
- 37.Jang C, Wang R, Wells J, Leon F, Farman M, Hammond J, Goodin MM. 2017. Genome sequence variation in the constricta strain dramatically alters the protein interaction and localization map of Potato yellow dwarf virus. J Gen Virol 98:1526–1536. doi: 10.1099/jgv.0.000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi D, Raikhy G, Pappu HR. 2015. Movement and nucleocapsid proteins coded by two tospovirus species interact through multiple binding regions in mixed infections. Virology 478:137–147. doi: 10.1016/j.virol.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- 40.Citovsky V, Gafni Y, Tzfira T. 2008. Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods 45:196–206. doi: 10.1016/j.ymeth.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Panaviene Z, Baker JM, Nagy PD. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191–205. [DOI] [PubMed] [Google Scholar]
- 42.Ghosh D, Brooks RE, Wang R, Lesnaw J, Goodin MM. 2008. Cloning and subcellular localization of the phosphoprotein and nucleocapsid proteins of Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virus Res 135:26–35. doi: 10.1016/j.virusres.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Bally J, Jung H, Mortimer C, Naim F, Philips JG, Hellens R, Bombarely A, Goodin MM, Waterhouse PM. 2018. The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annu Rev Phytopathol 56:405–426. doi: 10.1146/annurev-phyto-080417-050141. [DOI] [PubMed] [Google Scholar]
- 44.Lamm CE, Link K, Wagner S, Milbradt J, Marschall M, Sonnewald U. 2016. Human cytomegalovirus nuclear egress proteins ectopically expressed in the heterologous environment of plant cells are strictly targeted to the nuclear envelope. Viruses 8:73. doi: 10.3390/v8030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodin MM, Austin J, Tobias R, Fujita M, Morales C, Jackson AO. 2001. Interactions and nuclear import of the N and P proteins of sonchus yellow net virus, a plant nucleorhabdovirus. J Virol 75:9393–9406. doi: 10.1128/JVI.75.19.9393-9406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakrabarty R, Banerjee R, Chung SM, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M. 2007. PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20:740–750. doi: 10.1094/MPMI-20-7-0740. [DOI] [PubMed] [Google Scholar]
- 47.Bandyopadhyay A, Kopperud K, Anderson G, Martin K, Goodin M. 2010. An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology 402:61–71. doi: 10.1016/j.virol.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]