Middle East respiratory syndrome coronavirus (MERS-CoV) infections are endemic in the Middle East and a threat to public health worldwide. Rodents are not susceptible to the virus because they do not express functional receptors; therefore, we generated a new animal model of MERS-CoV infection based on transgenic mice expressing human DPP4 (hDPP4). The pattern of hDPP4 expression in this model was similar to that in human tissues (except lymphoid tissue). In addition, MERS-CoV was limited to the respiratory tract. Here, we focused on host factors involved in immunopathology in MERS-CoV infection and clarified differences in antiviral immune responses between young and adult transgenic mice. This new small-animal model could contribute to more in-depth study of the pathology of MERS-CoV infection and aid development of suitable treatments.
KEYWORDS: DPP4, MERS-CoV, animal models, immunopathology, transgenic mouse
ABSTRACT
Middle East respiratory syndrome coronavirus (MERS-CoV) infection can manifest as a mild illness, acute respiratory distress, organ failure, or death. Several animal models have been established to study disease pathogenesis and to develop vaccines and therapeutic agents. Here, we developed transgenic (Tg) mice on a C57BL/6 background; these mice expressed human CD26/dipeptidyl peptidase 4 (hDPP4), a functional receptor for MERS-CoV, under the control of an endogenous hDPP4 promoter. We then characterized this mouse model of MERS-CoV. The expression profile of hDPP4 in these mice was almost equivalent to that in human tissues, including kidney and lung; however, hDPP4 was overexpressed in murine CD3-positive cells within peripheral blood and lymphoid tissues. Intranasal inoculation of young and adult Tg mice with MERS-CoV led to infection of the lower respiratory tract and pathological evidence of acute multifocal interstitial pneumonia within 7 days, with only transient loss of body weight. However, the immunopathology in young and adult Tg mice was different. On day 5 or 7 postinoculation, lungs of adult Tg mice contained higher levels of proinflammatory cytokines and chemokines associated with migration of macrophages. These results suggest that the immunopathology of MERS-CoV infection in the Tg mouse is age dependent. The mouse model described here will increase our understanding of disease pathogenesis and host mediators that protect against MERS-CoV infection.
IMPORTANCE Middle East respiratory syndrome coronavirus (MERS-CoV) infections are endemic in the Middle East and a threat to public health worldwide. Rodents are not susceptible to the virus because they do not express functional receptors; therefore, we generated a new animal model of MERS-CoV infection based on transgenic mice expressing human DPP4 (hDPP4). The pattern of hDPP4 expression in this model was similar to that in human tissues (except lymphoid tissue). In addition, MERS-CoV was limited to the respiratory tract. Here, we focused on host factors involved in immunopathology in MERS-CoV infection and clarified differences in antiviral immune responses between young and adult transgenic mice. This new small-animal model could contribute to more in-depth study of the pathology of MERS-CoV infection and aid development of suitable treatments.
INTRODUCTION
Middle East respiratory syndrome coronavirus (MERS-CoV) was originally isolated as a novel coronavirus from a fatal case of acute respiratory distress syndrome and renal failure in 2012 (1). A human receptor for the virus, called human CD26/dipeptidyl peptidase 4 (hDPP4), was identified subsequently (2). Many epidemiological and virological investigations have been undertaken since then; however, information about the pathogenesis of MERS-CoV is limited. In addition, because MERS-CoV is endemic in the Middle East, the development of effective prophylactic and therapeutic treatment strategies remains a high priority. Therefore, appropriate animal models are needed to better understand the pathogenesis of MERS-CoV and facilitate development of effective vaccines and drugs. Some research groups experimentally infected nonhuman primates and small experimental animals with MERS-CoV (3–7). Rhesus macaques appear to develop a transient lower respiratory tract infection after a combination of intratracheal, ocular, oral, and intranasal inoculation with MERS-CoV (3), whereas the common marmoset develops progressive and severe pneumonia, which can be lethal (4). However, animal models based on nonhuman primates present both ethical and economic problems. Thus, establishing a small-animal model of MERS-CoV infection is desirable. Unfortunately, MERS-CoV does not infect or replicate in small rodents such as Syrian hamsters (8), mice (9), or rats (10) because they lack a functional MERS-CoV receptor. Zhao et al. described lung infection in a mouse model transduced with an adenovirus expressing hDPP4 (11); thus, a transgenic (Tg) mouse carrying hDPP4 should be suitable for MERS-CoV studies (5). Some research groups developed Tg mice overexpressing the hDPP4 receptor under the control of CAG or cytokeratin 18 promoters (5–7). These mice developed severe lung disease, along with infection of the brain. Autopsy data are available from only one MERS patient; therefore, it is unclear whether MERS-CoV causes a systemic infection although there is no evidence that MERS-CoV infects the human brain. Other studies describe development of an hDPP4 knock-in mouse (12–14). Although the tissue distribution and expression levels of hDPP4 in these models are largely equivalent to those of DPP4 in wild-type mice, the phenotype that determines MERS-CoV susceptibility varies from model to model. The hDPP4 knock-in mouse model described by Coleman et al. (12) succumbed to infection with wild-type MERS-CoV. In contrast, model mice described by Cockrell et al. (14) and Li et al. (13) are susceptible to infection by serially passaged MERS-CoV, which induces severe lung pathology and diffuse alveolar damage (DAD). These mice would be good models for studying pathogenesis of MERS. Here, we developed a new Tg mouse model expressing hDPP4 under the control of its endogenous promoter to better mimic physiological expression of hDPP4. These Tg mice were then backcrossed onto Th1-prone C57BL/6 mice. After evaluating susceptibility to MERS-CoV infection, we investigated age-dependent differences in disease pathogenesis because older age is one of the common factors related to MERS severity and mortality (15–20). Both young and adult Tg mice infected with MERS-CoV showed transient weight loss along with moderate pneumonia and MERS-CoV replication in the lung; however, they did not recapitulate the severe disease and lethal infection seen in humans. Young and adult Tg mice infected with MERS-CoV did, however, show different immunopathologies. Adult Tg mice showed higher levels of proinflammatory cytokine- and chemokine-mediated macrophage infiltration of the lungs than young Tg mice. Taken together, these results suggest that age affects the immunopathology of MERS-CoV infection in Tg mice. The data suggest that other factors are required to recapitulate severe human disease in these Tg mice; however, this mouse model will be useful for identifying host mediators that protect against MERS-CoV infection. This animal model will provide new insight into factors that cause severe MERS-CoV infection.
RESULTS
Expression of hDPP4 in Tg mouse tissues.
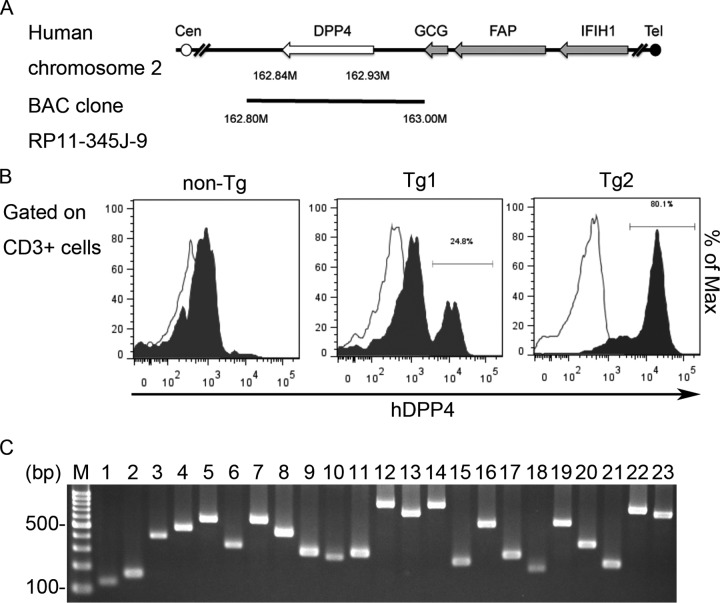
To generate Tg mice showing tissue- or cell-type-specific hDPP4 expression mimicking that in humans, we first looked at research involving Tg mice harboring human enterovirus receptors (such as the human poliovirus receptor) and SCARB2 receptor-driven endogenous promoters (21, 22). Promoter sequences, which normally include a transcriptional start site, are usually isolated from the upstream regions of endogenous mammalian genes (23). Therefore, we used a bacterial artificial chromosome (BAC) clone (RP11-345J9) containing the complete hDPP4 gene and an endogenous promoter to generate Tg mice harboring hDPP4 (Fig. 1A). To screen the Tg mice generated, we confirmed presence of the transgene by PCR genotyping using two primers sets specific for hDPP4 (Table 1, exons 3 and 10). hDPP4 is a protease expressed on the surface of cells in various organs, including T cells (24, 25). The enzyme is expressed by approximately 60% of resting T cells isolated from blood (26). Since handling of peripheral blood in a laboratory is relatively simple, we conducted flow cytometry analysis using a fluorescein isothiocyanate (FITC)-conjugated anti-human CD26/hDPP4 monoclonal antibody that does no react with murine DPP4 to detect expression of hDPP4 in mice. CD3-positive lymphocytes from 2/15 tested pups were positive for hDPP4. These mice were then crossed with C57BL/6 mice to establish two independent Tg lines (Tg1 and Tg2), which were maintained as hemizygotes carrying the hDPP4 gene. The Tg animals were born at the expected Mendelian ratio and were outwardly indistinguishable from control littermates. Because CD3-positive lymphocytes from peripheral blood of line Tg2 showed higher hDPP4 expression than those from line Tg1 (Fig. 1B), Tg2 was used for further analyses. PCR genotyping using primer sets specific for hDPP4 revealed that the complete hDPP4 gene had integrated into the genome of Tg2 mice (Fig. 1C and Table 1).
FIG 1.
Generation of transgenic mice expressing human dipeptidyl peptidase 4 (hDPP4). (A) Schematic diagram showing a bacterial artificial chromosome (BAC) clone (clone RP11-345J9) containing the hDPP4 gene used to produce the transgenic mice. The open and filled circles denote the centromere (Cen) and telomere (Tel) of human chromosome 2, respectively. The gray arrows indicate the genes located downstream of the hDPP4 gene (white arrow). The cloned region in the BAC construct is denoted by a black line. GCG, glucagon gene; FAP, fibroblast activation protein; IFIH1, interferon induced with helicase C domain 1. (B) Expression of hDPP4 on peripheral blood CD3-positive T lymphocytes from the transgenic (Tg) mice. Tg1 and Tg2, hDPP4+/− transgenic mouse lines 1 and 2; non-Tg, hDPP4−/− mouse. (C) Genomic DNA was extracted from Tg2 mice, and human DPP4 exons 1 to 26 were subjected to PCR using specific primers. Lane M, marker; lane 1, exon 1; lane 2, exon 2; lane 3, exon 3; lane 4, exon 4; lane 5, exon 5; lane 6, exons 6 and 7; lane 7, exon 8; lane 8, exon 9; lane 9, exon 10; lane 10, exon 11; lane 11, exon 12; lane 12, exons 13 and 14; lane 13, exons 15 and 16; lane 14, exons 17 and 18; lane 15, exon 19; lane 16, exon 20; lane 17, exon 21; lane 18, exon 22; lane 19, exon 23; lane 20, exon 24; lane 21, exon 25; lanes 22 and 23, exon 26.
TABLE 1.
Primers used for PCR of human DPP4 exons
| Target | Size (bp) | Sequence (5′→3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| Exon 1 | 130 | AATGTTTAACTCGGGGCCGA | CGGAAGTGAGCGTTCAGAGA |
| Exon 2 | 162 | GGACTTGATCTGCTCGGCTT | CCTGACCTGAGCTCCAACTG |
| Exon 3 | 391 | ACACACACACTCTCACACACT | TCTCAGTGCCATAAAAGCCCA |
| Exon 4 | 468 | GTGCAAAGGGAGAAAGACTGA | CCACTTTGCCATATGCTGCA |
| Exon 5 | 562 | GTGCACAGTGATGGCAATGA | CCACCATGCCCGACTTTAAC |
| Exons 6–7 | 330 | GTCTCCTATAGTGAGTGGCCA | TCTGACAACTGGAGAGACTCAC |
| Exon 8 | 565 | GGTCAGCCTTCTCGGTCTTC | GGAGACATCTGGTGCTGTGA |
| Exon 9 | 440 | AGCCCAGCAAATGCAAAGTG | GCCAGATGCTGTTGACTTCAG |
| Exon 10 | 296 | TGCAGACGTTTTTGTGCAGT | GGCTGTGATCCACTTTGCCA |
| Exon 11 | 274 | CCAAGGTCTGGCAATAGTCA | TTCACTCTCCCCAACTGCAC |
| Exon 12 | 299 | GAGCTTCCAGAAGGACCCAG | GCTGACTCATCCATAAAACCCC |
| Exons 13–14 | 848 | TGCTTGCAGCCAGAAGTCAT | CTTCTGGGCAAAGAGGGCAT |
| Exons 15–16 | 708 | CTCCGTGCACACTTAGGCTT | GGAGCTGCTTCGAAGTGAGT |
| Exons 17–18 | 847 | GCCCTGTGCCTTTCCAGTAA | GCATGTTCTCCAAATCCCTTCC |
| Exon 19 | 247 | TGCTACTGACGGACATGAGG | GAAAGGACGCATTTGGCTCC |
| Exon 20 | 549 | GGATGCATACTTCTCCACGG | AGGACATATGCCAACTCCCT |
| Exon 21 | 281 | GCAGAGAACAAATGGCAGGG | ACTGCCCAGAGACCTAAGACT |
| Exon 22 | 206 | AATGTGGAAACTGCGACTCG | TCTCTTTGTACCTGGCAGCA |
| Exon 23 | 567 | AGGTGCTTTAGCCACCCTTT | GAGAGTCTTCTGGGCTCTAAAGG |
| Exon 24 | 364 | TCCCTTCCAGTCCTGTCTCC | CAGTCTTGCCCCTCATGCTT |
| Exon 25 | 239 | CTTTCCCACCCCTTGGTACC | CCTGTCTGTGGCACTGCTAA |
| Exon 26 (1)a | 784 | TAGACCCCCTCTTTGACCCC | GAACAGCTCTTCTCCGAGGG |
| Exon 26 (2)a | 723 | AAGGGATGGCAAGATGTGGG | TCCATATGCCAGTGCGGTTT |
Exon 26 (1) and Exon 26 (2) are primer sets for the first half and the second half of exon 26.
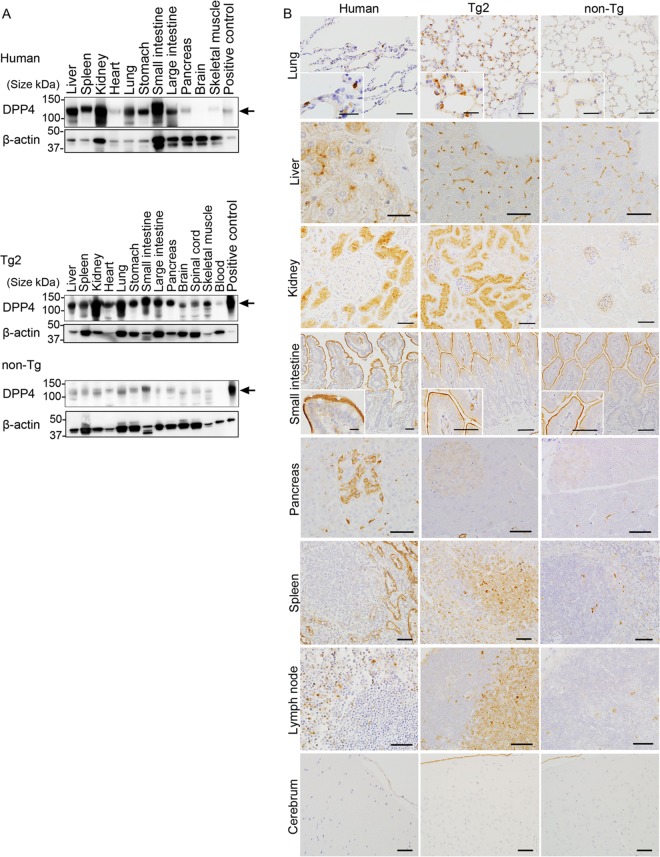
To examine hDPP4 expression in human and Tg2 tissues, we first performed Western blot analysis with a goat anti-CD26/hDPP4 polyclonal antibody (AF1180; R&D Systems), which detected hDPP4 but cross-reacted weakly with mouse DPP4. Bands of about 110 kDa (hDPP4) were detected in all tested human tissues (liver, spleen, kidney, heart, lung, stomach, small intestine, large intestine, pancreas, brain, spinal cord, and skeletal muscle), except brain (Fig. 2A). All of the tissues from hDPP4 Tg mice expressed hDPP4, including liver, spleen, kidney, heart, lung, stomach, small intestine, large intestine, pancreas, brain, spinal cord, and skeletal muscle (Fig. 2A). These results suggest that the human transgene was expressed in the majority of organs/tissues in Tg2 mice.
FIG 2.
Expression of human dipeptidyl peptidase 4 (hDPP4) in tissues from humans and mice. Tg2, hDPP4+/− transgenic mouse line 2; non-Tg, hDPP4−/− mouse. (A) Western blot analysis of homogenized human and mouse tissues with an anti-hDPP4 polyclonal antibody or an anti-β-actin polyclonal antibody (internal control). Arrows indicate the positions of hDPP4 (110 kDa). (B) Immunohistochemical analysis of hDPP4 expression in human, Tg2, and non-Tg mouse tissues stained with an anti-hDPP4 polyclonal antibody. Sections were counterstained with hematoxylin. Scale bars, 50 μm (large images of liver, kidney, small intestine, pancreas, spleen, and lymph node), 20 μm (large images of lung and brain), and 25 μm (insets).
To further determine hDPP4 distribution in tissues, immunohistochemistry (IHC) was performed (Fig. 2B). IHC using a goat anti-CD26/hDPP4 antibody detected hDPP4 antigens in pneumocytes in the lung, in bile capillaries in the liver, in renal tubular epithelium, on the surface of epithelial cells lining the small intestine, in pancreatic islets, in lymphocytes in the lymph nodes, and in several types of endothelial cell and serous membranes (Fig. 2B, left column). While hDPP4 expression was undetectable in brain tissue by Western blot analysis, IHC revealed that endothelial cells lining blood vessels and leptomeninges of the human brain were positive for hDPP4 although neurons and glia were negative. In Tg2 mice, pneumocytes and bronchial epithelial cells in the lungs, bile capillaries in the liver, the renal tubular epithelium, and the surface of epithelial cells in the small intestine were positive for hDPP4 (Fig. 2B). In addition, several types of endothelial cells and serous membranes in all tested tissues, including the central nervous system, from Tg2 mice were positive for hDPP4. Notably, most lymphocytes in the T cell zones of the spleen and lymph nodes from Tg2 mice were positive for hDPP4. Staining of tissues from non-Tg mice was very weak or absent (except for the small intestine) (Fig. 2B). These data suggest that the pattern of hDPP4 expression in Tg2 mice is similar to that in humans (except for pancreas and lymphoid tissues).
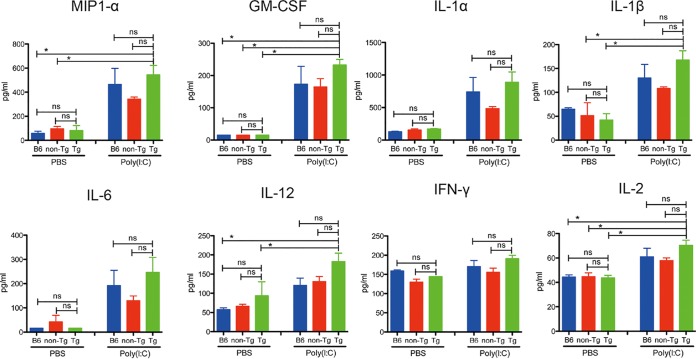
Expression of hDPP4 was higher in lymphocytes from Tg2 mice than in those from humans (Fig. 1C and Fig. 2B). Therefore, we investigated the immune response profile in Tg2 mice. To assess innate immune responses in the lungs of Tg2, non-Tg, and C57BL/6 mice, all animals received intranasal administration of PBS with or without poly(I·C), a synthetic analog of double-stranded RNA (Fig. 3). There was no statistically significant difference in cytokine expression levels between Tg2, non-Tg, and C57BL/6 mice at 24 h after inoculation with poly(I·C) or phosphate-buffered saline (PBS) (Fig. 3). However, when we set expression levels after PBS treatment as 1, we noted that expression of macrophage inflammatory protein 1α (MIP-1α), granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-1β (IL-1β), IL-12, and IL-2 in poly(I·C)-treated Tg2 mice was 0.8- to 2-fold higher than in the other two strains. These results suggest that hDPP4 expression in mice does not have a marked effect on basal innate immune responses in the three mouse strains; however, Tg2 mice show slightly stronger or earlier innate immune responses than C57BL/6 mice and non-Tg mice. Thus, when we investigated immune responses in this animal model, we made comparisons between MERS-CoV-infected and noninfected Tg mice.
FIG 3.
Innate immune responses in Tg2, non-Tg, and C57BL/6 mice. C57BL/6, non-Tg, and Tg2 mice received an intranasal inoculation of poly(I·C) or saline and were sacrificed 24 h later (n = 4/group). Expression of proinflammatory cytokines and chemokines in saline- and poly(I·C)-inoculated animals. P values were calculated using one-way ANOVA, followed by Tukey’s posttest (ns, not significant; *, P < 0.05). Error bars indicate the standard deviations.
Susceptibility of hDPP4-Tg mice to MERS-CoV infection.
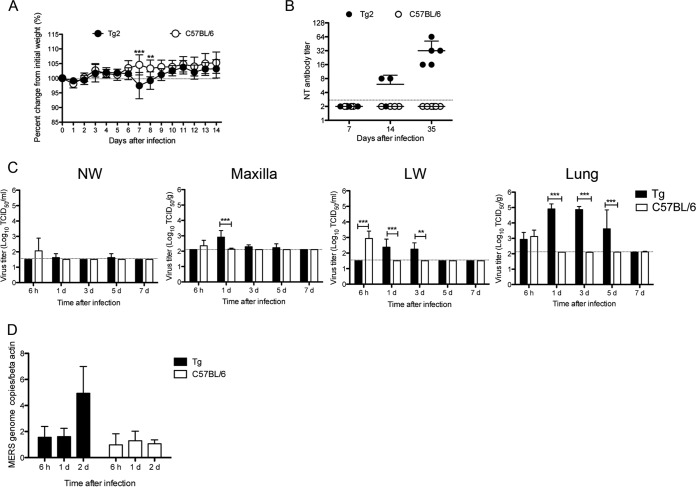
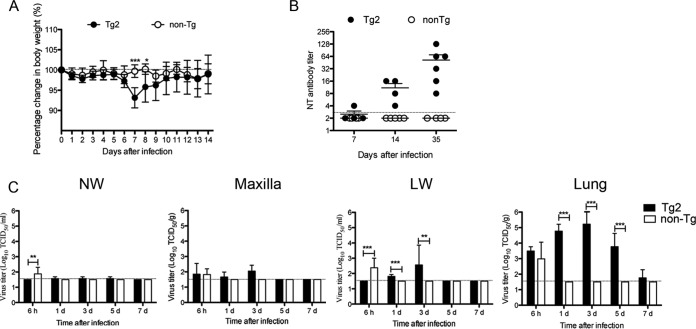
In this experiment, C57BL/6 mice were used instead of non-Tg mice because we were unable to prepare a sufficient number of non-Tg mice from littermate mice for this experiment. After intranasal inoculation of 10-week-old Tg2 and C57BL/6 mice with 105 50% tissue culture infectious doses (TCID50) of MERS-CoV, neither group was lethargic; however, Tg2 mice showed mild but transient weight loss from days 6 to 7 postinoculation (p.i.) (Tg2 mice, n = 8 [four females and four males]; C57BL/6 mice, n = 10 [five females and five males]) (Fig. 4A). Tg2 mice showed seroconversion at 35 days p.i., whereas C57BL/6 mice did not (Tg2 mice, n = 5 [three females and two males]; C57BL/6 mice, n = 6 [three females and three males]) (Fig. 4B), suggesting that Tg2 mice are susceptible to infection by MERS-CoV.
FIG 4.
Permissiveness of transgenic mice to infection by MERS-CoV. (A) The body weight of 10-week-old mice was monitored daily after intranasal inoculation of MERS-CoV at a dose of 105 TCID50 (n = 8 for hDPP4-transgenic mouse line 2 [Tg2]; n = 10 for C57BL/6 mice). Error bars represent the standard deviation (**, P < 0.01; ***, P < 0.001, by two-way ANOVA). (B) Seroconversion of Tg2 mice inoculated with MERS-CoV. Titer of MERS-CoV-specific neutralizing (NT) antibodies in mouse serum on days 7, 14, and 35 postinoculation with MERS-CoV (Tg2, n = 3 to 5; C57BL/6, n = 4 to 6). The dotted line denotes the detection limit of the assay. Error bars represent the standard deviations. (C) Viral load in the respiratory tract of mice inoculated with MERS-CoV. NW, nasal wash fluid; maxilla, maxilla including nostril; LW, lung wash fluid. Mice were euthanized at the indicated times post-viral inoculation (n = 3 to 4 per time point). Viral titer is expressed as the means ± standard deviations. The dotted line denotes the detection limit of the assay. **, P < 0.01; ***, P < 0.001 (two-way ANOVA). (D) Quantitative real-time RT-PCR analysis of MERS-CoV viral RNA in splenocytes isolated from Tg2 and C57BL/6 mice. RNA was extracted from splenocytes infected with MERS-CoV at a multiplicity of infection of 1. RNA levels were normalized against β-actin (endogenous control).
We next examined viral replication kinetics and sites of viral replication in 10-week-old Tg2 and C57BL/6 mice (n = 3–4 [2 females and 1 to 2 males per time point]). Tg2 mice and C57BL/6 mice were inoculated intranasally with 105 TCID50 of MERS-CoV, and tissue specimens (the maxilla [including the nostril], nasal wash fluid, lung, and lung wash fluid) were collected at 6 h p.i. and on days 1, 3, 5, and 7 p.i. The nasal wash fluid from three out of four Tg2 mice contained barely detectable levels of infectious virus at days 1 and 5 p.i. (Fig. 4C). Supernatants from maxilla tissue homogenates (20%) from Tg2 mice contained 102.8 TCID50/g at day 1 p.i. although the titer fell by 5 days p.i. The viral titers in lung wash fluid and supernatants of lung tissue homogenates (20%) from Tg2 mice contained 102.3 TCID50/ml and 104.6 TCID50/g, respectively, at 1 day p.i.; these values were significantly higher than those at 6 h p.i. (P < 0.05 and P < 0.01, respectively; one-way analysis of variance [ANOVA]). The virus titers in the lungs were detectable up until 5 days p.i. Virus was undetectable in the respiratory tract at 7 days p.i. Although IHC revealed that various organs from Tg2 mice were positive for the hDPP4 antigen (Fig. 2B), no infectious virus was detected in the liver, spleen, kidney, heart, intestines, and brain up to 7 days p.i. (Table 2). In contrast, virus was detected in the respiratory tract of C57BL/6 mice at 6 h p.i. only. These observations suggest that MERS-CoV infects and replicates mainly in the lower respiratory tract of Tg2 mice and is eliminated within 7 days of infection.
TABLE 2.
Virus isolation, detection of viral antigens by immunohistochemistry, and detection of the MERS-CoV genome by real-time PCR in tissues from Tg2 and non-Tg mice inoculated with MERS-CoV
| Animala | Virus detection by sample typeb |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver |
Spleen |
Kidney |
Heart |
Lung |
Intestine |
Brain |
Blood |
|||||||||||||||||
| I | A | G | I | A | G | I | A | G | I | A | G | I | A | G | I | A | G | I | A | G | Ic | A | G | |
| Tg2 | − | − | NE | − | − | − | − | − | − | − | − | NE | + | + | NE | − | − | NE | − | − | − | − | − | +d |
| Non-Tg | − | − | NE | − | − | − | − | − | − | − | − | NE | − | − | NE | − | − | NE | − | − | − | − | − | − |
Tg2, hDPP4+/− transgenic mouse line 2; non-Tg, hDPP4−/− mouse.
Plus and minus signs indicate the presence and absence, respectively, of virus (I), antigen (A), and genome (G). NE, not examined.
Serum samples used for analysis.
Viral genome was detected in samples at 3 and 5 days p.i.
Several research groups have developed Tg mouse models of MERS-CoV infection; however, in these models, viral replication and MERS-CoV RNA were detected in the brain (5–7, 27). Thus, we next measured the amount of viral RNA in the brain of Tg2 mice at 6 h and at 1, 3, 5, and 7 days p.i. by real-time reverse transcription-PCR (RT-PCR) using primers specific for MERS-CoV; however, no MERS-CoV RNA was detected in the brain of Tg2 mice. Furthermore, another study showed that experimental infection of common marmosets with MERS-CoV resulted in viremia (4). Quantitative examination of viral RNA levels in Tg2 mice revealed very low copy numbers of viral RNA in the blood at 3 and 5 days p.i., while sera from Tg2 mice were negative for the virus (Table 2). These results suggest that although intranasal inoculation of Tg2 mice with MERS-CoV causes no neuroinvasion, it may induce viremia.
Several animal studies have identified MERS-CoV RNA in the lymphoid organs of infected animals (3, 4, 28), but no infectious virus was detected in the spleen from Tg2 mice up to 7 days p.i. (Table 2). To investigate whether lymphoid organs in Tg2 mice are susceptible to MERS-CoV infection, we harvested splenocytes from Tg2 and C57BL/6 mice and estimated infectivity and MERS-CoV RNA levels (Fig. 4D). Although the amount of infectious MERS-CoV in splenocytes was below the detection limit, viral RNA was detected in splenocytes from Tg2 mice (peaking at 2 days p.i.). Thus, lymphoid organs of Tg2 mice were as susceptible to MERS-CoV infection as those reported in other animal studies although lymphoid organs were not a major site of MERS-CoV replication in Tg2 mice after intranasal inoculation.
Acute inflammatory changes in the lungs of hDPP4-Tg mice after MERS-CoV inoculation.
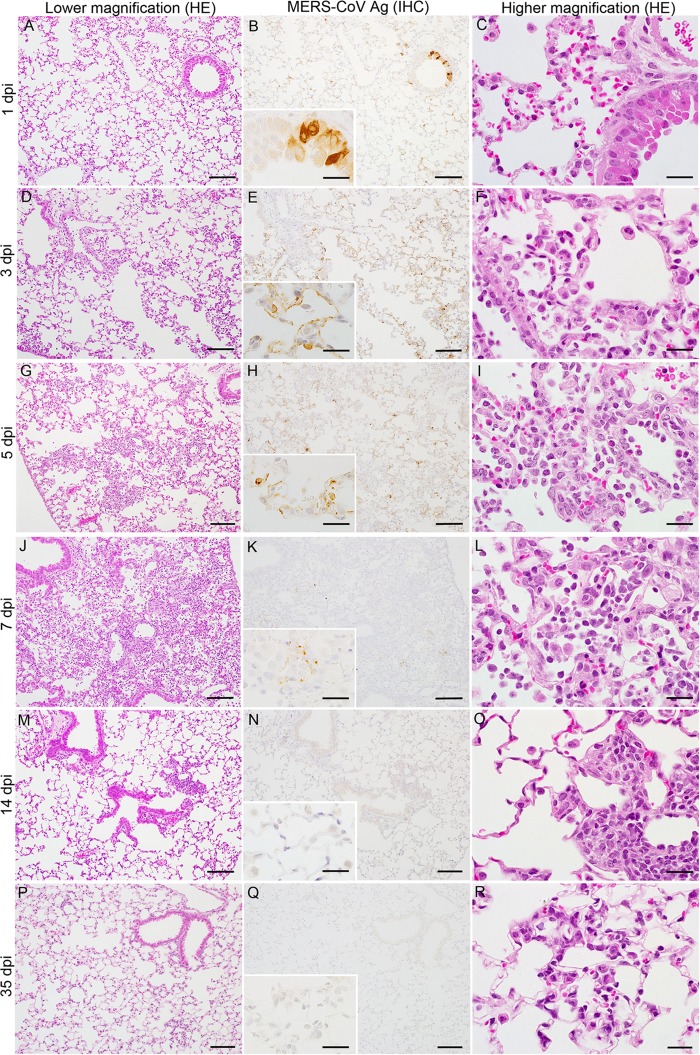
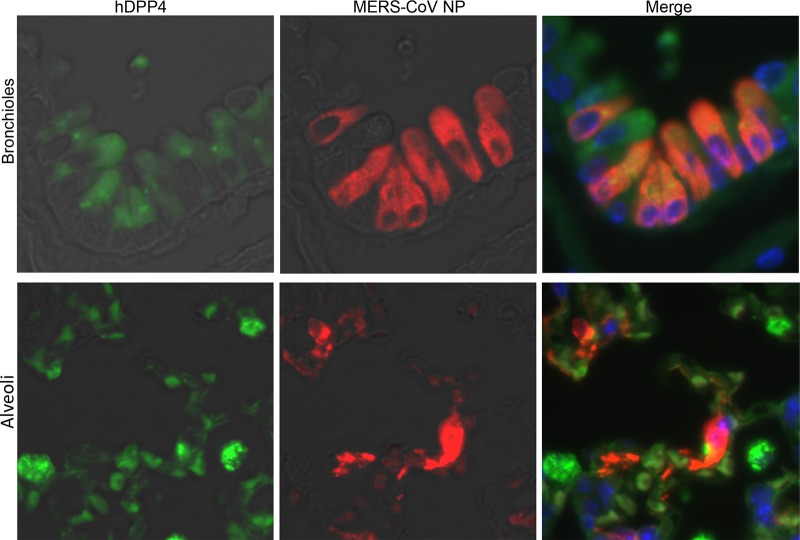
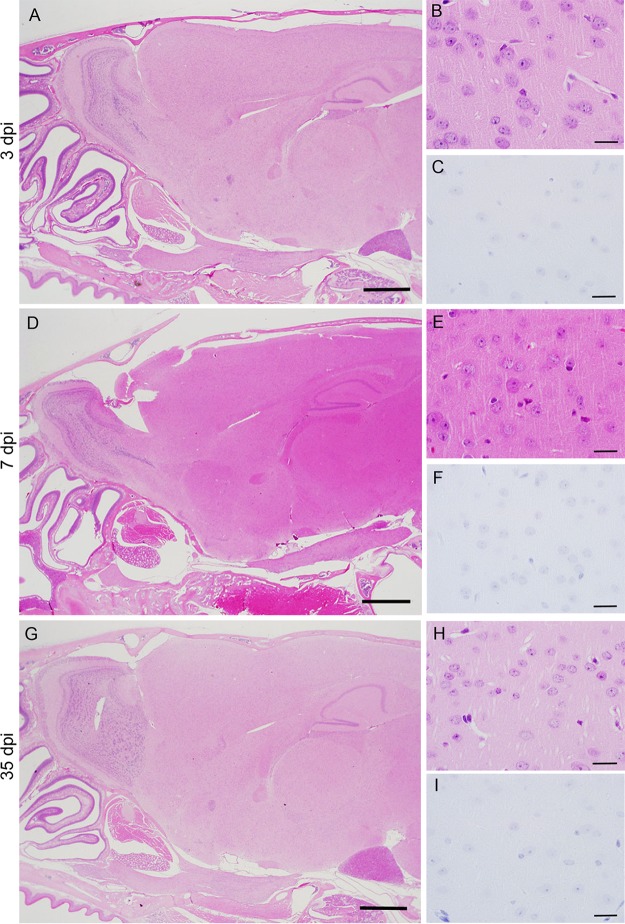
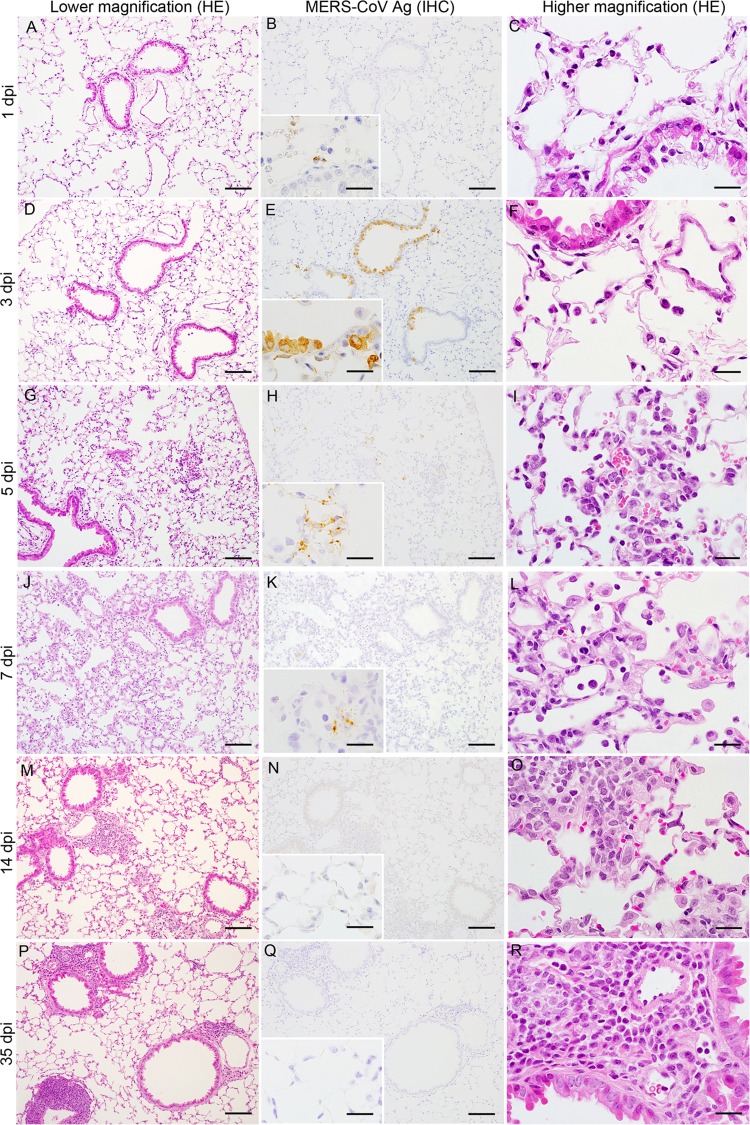
MERS-CoV infection of the nasal cavity, lungs, brain, spinal cord, liver, spleen, kidney, heart, and gastrointestinal tract of Tg2 mice was examined histopathologically on days 1, 3, 5, 7, 14, and 35 p.i (n = 3; one or two females and one or two males per time point). Histopathological investigations revealed that Tg2 mice showed progressive pulmonary inflammation associated with acute virus infection, from which they recovered (Fig. 5). On day 1 p.i., there was no obvious infiltration of the lungs in Tg2 mice; however, mild cellular degeneration and viral antigen-positive cells were seen in the bronchioles and a few alveolar areas (Fig. 5A to C). Double-immunofluorescence staining revealed that MERS-CoV antigen colocalized with hDPP4-positive bronchioles and alveolar cells on day 1 p.i. (Fig. 6). On days 3 and 5 p.i., inflammatory reactions, including partial and/or mild perivascular and peribronchiolar infiltration by mononuclear cells and eosinophils in response to viral antigens, were observed in alveolar areas of lung tissue from Tg2 mice (Fig. 5D to I). From day 3 p.i. onwards, the alveolar area was the main site of inflammatory responses to viral replication (Fig. 5F, I, and L). On day 7 p.i., the point at which Tg2 mice showed weight loss, there was evidence of severe lung inflammation, including perivascular and alveolar septal thickening, caused by infiltrating mononuclear cells; some alveoli were positive for viral antigens (Fig. 5K). At day 14 p.i., focal cellular infiltration was still evident in the peribronchioles and alveolar septa although viral antigens and inflammatory responses had cleared from the lungs by day 35 p.i. (Fig. 5M to P). There was no evidence of diffuse alveolar damage in the lungs up to day 35 p.i. These findings suggest that progressive immunopathology occurred uniformly throughout the lungs but resolved within 14 days after infection. Neither histopathology nor IHC detected inflammatory infiltration or viral antigens in the nasal cavity through 35 days p.i. In addition, there was no inflammation or viral antigen expression in the brain through 35 days p.i. (Fig. 7). In contrast, C57BL/6 mice showed no histopathological changes or viral antigen in any organ, including the lung. These results indicate that Tg2 mice suffer acute pneumonia after infection of the lungs with MERS-CoV, which is related to expression of hDPP4 in the bronchiolar epithelium and pneumocytes. Splenocytes from Tg2 mice were susceptible to ex vivo infection with MERS-CoV (Fig. 4D); however, the spleen and other lymphoid tissues did not harbor viral antigens. Furthermore, MERS-CoV induced T cell apoptosis upon infection in vitro (28), whereas immunohistochemical staining detected no evidence of apoptosis in lymphoid tissues, including spleen and lymph nodes, of MERS-CoV-infected Tg2 mice. Similar to findings in the other mouse models in which mouse DPP4 was replaced with hDPP4 (12), MERS-CoV replication and pathology were localized in the lungs in Tg2 mice.
FIG 5.
Histopathological changes in the lungs of human dipeptidyl peptidase 4 (hDPP4)-transgenic mice inoculated with MERS-CoV. Representative images of lungs from hDPP4+/− transgenic mouse line 2 on days 1, 3, 5, 7, 14, and 35 postinoculation. Mild but progressive interstitial infiltration was seen within 7 days postinoculation (dpi) (left column, A, D, G, J, M, and P). IHC staining of sequential sections revealed abundant MERS-CoV antigen-positive cells in affected areas (middle column, B, E, H, K, N, and Q). Severe inflammation, with many mononuclear cells in the alveolar spaces and regenerated type II pneumocytes in the alveolar wall, was observed within 7 days p.i. (right column, C, F, I, L, O, and R). Scale bars: 100 μm (left and middle columns), 50 μm (right column), and 20 μm (insets of middle column). HE, hematoxylin and eosin staining; IHC, immunohistochemistry using an anti-MERS-CoV nucleocapsid protein polyclonal antibody; Ag, antigen.
FIG 6.
Double-immunofluorescence images taken at 1 day p.i. showing human dipeptidyl peptidase 4 (hDPP4) (green) and MERS-CoV antigen (red) in the lungs of Tg2 mice infected with MERS-CoV. Viral antigen-positive cells in the lungs were hDPP4-positive bronchiolar epithelial cells (upper panels) and alveolar epithelial cells (lower panels). Original magnification, ×600.
FIG 7.
Histopathological changes in the brain of human dipeptidyl peptidase 4 (hDPP4)-transgenic mice inoculated with MERS-CoV. (A, D, and G) Sagittal sections of the head, including the nasal cavity, olfactory bulb, and brain, of a Tg2 mouse infected with MERS-CoV (images taken at 3, 7, and 35 days p.i. [dpi]). Right panels show the brain cortex from samples from panels A, D, and G, respectively, with hematoxylin and eosin staining (B, E, and F) and immunohistochemical analysis of MERS-CoV antigen (C, F, I). Neither lesions nor MERS-CoV antigen-positive cells were detected in the brain. Scale bars, 1 mm (A, D, and G) and 20 μm (B, C, E, F, H, and I).
Differences in the immunopathology of MERS-CoV infection between young and adult hDPP4-Tg mice.
According to an epidemiological study (29), age (>45 years) is considered to be one of the risk factors for MERS-CoV infection in humans. Therefore, we infected 25-week-old mice with MERS-CoV. Tg2 mice showed significant weight loss from days 7 and 8 p.i. before recovering by day 14 p.i. (Tg2 mice, n = 6 [one female and five males]; non-Tg mice, n = 7 [three females and four males]) (Fig. 8A). However, 25-week-old Tg2 mice showed no obvious clinical signs (such as respiratory illness and mortality). The 25-week-old Tg2 mice had seroconverted by 35 days p.i., whereas the C57BL/6 mice had not (Fig. 8B). Next, we examined viral replication kinetics and sites of viral replication in 25-week-old Tg2 and non-Tg mice (n = 4; two females and two males per time point). The nasal wash fluid from one out of four Tg2 mice contained barely detectable levels of infectious virus at 1, 3, and 5 days p.i. (Fig. 8C). Supernatants from maxilla tissue homogenates (20%) from Tg2 mice contained 101.7 and 102.0 TCID50/g at 1 and 3 days p.i., respectively, although the titer was undetectable at 5 days p.i. The viral titers in lung wash fluid from Tg2 mice were detectable up until 3 days p.i., while the supernatants from lung tissue homogenates (20%) from Tg2 mice showed a high viral load from 1 to 5 days p.i.; infectious virus was detectable up until 7 days p.i. Viral loads in the respiratory tract peaked at 3 days p.i. Although no virus was detectable in the respiratory tract of 10-week-old Tg mice at 7 days p.i., infectious virus was detected in the lungs of 25-week-old Tg mice up until 5 days p.i. In addition, infectious virus was detected in the lungs of one of four 25-week-old Tg mice (102.5 TCID50/g) even at 7 days p.i. We also found that viral titers in the nasal wash fluid, maxilla (including nostril), lung wash fluid, and lungs of 25-week-old Tg2 mice on day 3 p.i. (102.6 TCID50/ml, 103.3 TCID50/ml, 102.3 TCID50/ml, and 104.9 TCID50/ml, respectively) were slightly higher than those in 10-week-old Tg2 mice (101.6 TCID50/ml, 102.9 TCID50/ml, 101.8 TCID50/ml, and 104.5 TCID50/ml, respectively) (P = 0.04, Student's t test with Welch’s correction).
FIG 8.
Susceptibility of adult human dipeptidyl peptidase 4 (hDPP4)-transgenic mice to MERS-CoV infection. Tg2, hDPP4+/− transgenic mouse line 2; non-Tg, hDPP4−/− mouse. (A) The body weight of 25-week-old mice was monitored daily after intranasal inoculation with MERS-CoV (n = 6 Tg2 mice and n = 7 non-Tg mice). *, P < 0.05; ***, P < 0.001 (two-way ANOVA). (B) Seroconversion of Tg2 mice inoculated with MERS-CoV. Titer of MERS-CoV-specific neutralizing (NT) antibodies in mouse serum on days 7, 14, and 35 postinoculation with MERS-CoV (Tg2, n = 4 to 6; non-Tg, n = 4). The dotted line denotes the detection limit of the assay. Error bars represent the standard deviation. (C) Viral titer in nasal wash fluid (NW), maxilla (including nostril), lung wash fluid (LW), and lungs of 25-week-old Tg2 and non-Tg mice at 3 days postinoculation (p.i.) (n = 4 mice per group). Viral titer is expressed as the mean ± standard deviation. The dotted line denotes the detection limit of the assay. **, P < 0.01; ***, P < 0.001 (two-way ANOVA).
Histopathological analysis revealed that 25-week-old Tg2 mice showed delayed and prolonged inflammatory responses in the lung compared with those in 10-week-old Tg2 mice (Fig. 9). Interestingly, viral antigen-positive cells were seen in the alveolar area on day 1 p.i. and then in the bronchi on day 3 p.i., along with a sparse cellular infiltrate (Fig. 9A to F). Cellular infiltration (which included mononuclear cells and polynuclear cells) was observed in the alveoli from 5 days p.i.; this expanded on day 7 p.i. (Fig. 9G to L). Focal cell infiltration was seen in the alveoli on day 14 p.i., and lymphoid cell aggregates were seen around bronchioles and blood vessels on day 35 p.i. (Fig. 9M to R).
FIG 9.
Histopathological changes in the lungs of human dipeptidyl peptidase 4 (hDPP4)-transgenic mice inoculated with MERS-CoV. Representative histopathological images of the lungs from 25-week-old Tg2 mice at 1, 3, 5, 7, 14, and 35 days post-MERS-CoV infection. Images in the left (A, D, G, J, M, and P) and right (C, F, I, L, O, and R) columns show time-dependent recruitment of inflammatory cells to the lung. Marked inflammatory cell infiltration was noted at 7 days postinoculation (dpi) in panels J and L. Images in the middle column (B, E, H, K, N, and Q) show immunohistochemical staining for MERS-CoV antigen (Ag). Scale bars: 100 μm (left and middle columns), 50 μm (right column), and 20 μm (insets of middle column). HE, hematoxylin and eosin staining; IHC, immunohistochemistry using an anti-MERS-CoV nucleocapsid protein polyclonal antibody.
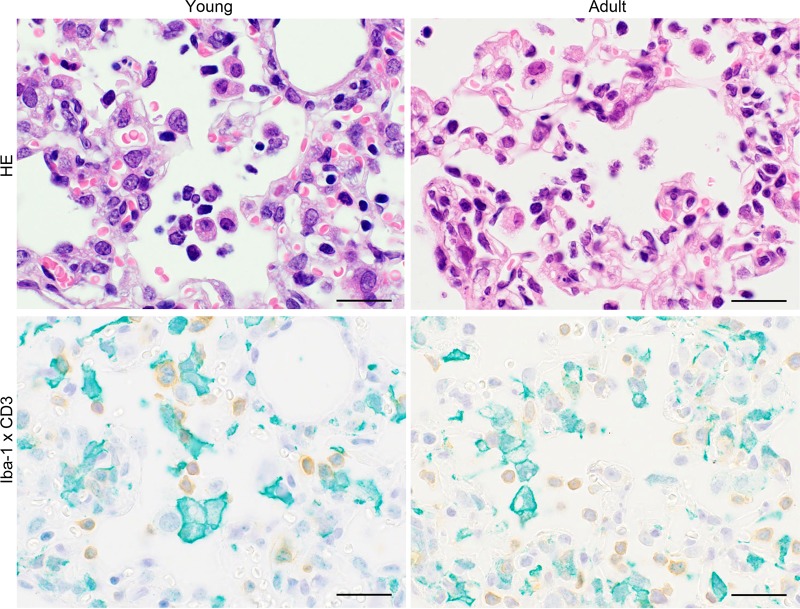
Next, we compared inflammation of the alveoli in 10-week-old Tg mice and 25-week-old Tg mice on day 7 p.i. Ionized calcium binding adaptor molecule 1 (Iba-1) is expressed specifically by monocytes/macrophages and is upregulated when these cells are activated. The predominant inflammatory infiltrate within the lungs of both 10-week-old and 25-week-old Tg mice comprised Iba-1-positive large cells and CD3-positive mononuclear cells (Fig. 10). Phagocytic vacuoles were prominent in large Iba-1-positive cells from 25-week-old Tg mice.
FIG 10.
Identification of cells infiltrating the lung of Tg2 mice infected with MERS-CoV. Representative images of lungs from 10-week-old (young) and 25-week-old (adult) hDPP4+/− transgenic mice (line 2) on day 7 postinoculation (p.i.). Infiltrating cells were positive for Iba-1 (green) or CD3 (brown). Bar, 20 μm.: Hematoxylin and eosin staining (HE) was used for the images in the upper panels, and anti-Iba-1 polyclonal antibody and anti-CD3 monoclonal antibody were used for IHC in the lower panels.
Expression of proinflammatory cytokines and chemokines in hDPP4-Tg mice after MERS-CoV infection.
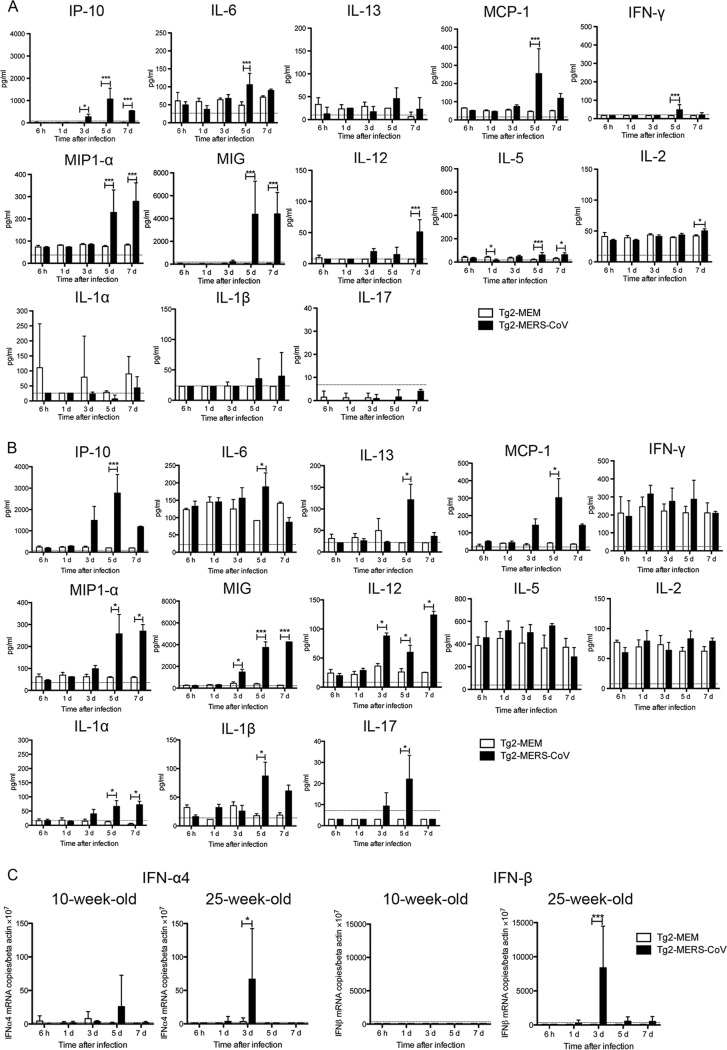
Dysregulated cytokine and chemokine expression is observed in MERS-CoV-infected patients (30). Therefore, we measured the levels of 20 cytokines and chemokines in lung samples from both 10-week-old and 25-week-old Tg2 mice inoculated with either MERS-CoV or minimal essential medium (MEM). Measurements were made at 6 h and at 1, 3, 5, and 7 days p.i. (n = 3 to 4 [2 females and 1 to 2 males per time point] and n = 4 [2 females and 2 males per time point]) (Fig. 11A and B, for 10-week-old and 25-week-old mice, respectively). On day 3 p.i., we observed early expression of gamma interferon (IFN-γ)-induced protein 10 (IP-10) in the lungs of both 10- and 25-week-old Tg2 mice, a level which was significantly higher than that in control mice; this increase lasted through day 7. This was followed by a transient increase in expression of interleukin-6 (IL-6), IL-13, and monocyte chemotactic protein-1 (MCP-1) in lungs from both 10- and 25-week-old Tg2 mice at day 5 p.i. High levels of macrophage inflammatory protein 1α (MIP-1α) and monokine induced by IFN-γ (MIG) were detected in the lungs of both groups of Tg2 mice from days 3 or 5 to 7 p.i., whereas IL-12 levels increased at day 7 p.i. IFN-γ production in the lungs of 10-week-old Tg2 mice peaked significantly on day 5 p.i. while high values were seen in both infected and noninfected 25-week-old mice during the observation period. In contrast, expression of IP-10, IL-12, and IL-1β in 25-week-old Tg mice was higher than that in 10-week-old Tg2 mice. Interestingly, IL-1α and IL-17 were detected only in 25-week-old Tg2 mice infected with MERS-CoV. IL-1α, a potent proinflammatory cytokine associated with inflammation and fever, was detected from 3 days p.i. and remained significantly elevated until 7 days p.i.; IL-17 (a proinflammatory cytokine that recruits monocytes and neutrophils) was detected from 3 days p.i. and peaked at 5 days p.i. before falling at 7 days p.i. These results indicate that MERS-CoV infection induces production of acute inflammatory chemokines and cytokines in the lungs. In addition, aging causes more severe immunopathology; this means that young and adult Tg mice show different pathologies in the lung after MERS-CoV infection.
FIG 11.
Cytokine and chemokine levels and expression of type I interferon (IFN) genes in the lungs of Tg2 mice infected with MERS-CoV. Cytokine and chemokine levels in lung samples from 10-week-old (A) and 25-week-old (B) mice are shown. Tg2 mice were inoculated with MERS-CoV or cell culture medium containing 2% FBS. Lungs were collected at the indicated times post-viral inoculation (n = 3 to 4 mice per time point). Data represent the means ± standard deviations. A dotted line denotes the detection limit of the assay. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-way ANOVA). (C) Quantitative real-time RT-PCR analysis of type I IFN gene expression in lung homogenates from Tg2 mice inoculated with MERS-CoV or cell culture medium containing 2% FBS. RNA levels were normalized against those of β-actin (endogenous control). Data represent the means ± standard deviations. A dotted line denotes the detection limit of the assay. *, P < 0.05; ***, P < 0.001 (two-way ANOVA).
Furthermore, we measured expression of mRNA encoding IFN-α4 and IFN-β (type I IFN with antiviral activity) in the lungs of 10- and 25-week-old mice at 6 h and at 1, 3, 5, and 7 days p.i. by real-time reverse transcription-PCR (RT-PCR) (31). We did not find any evidence of IFN-α4 or IFN-β in the lungs from 10-week-old Tg2 mice at early times postinfection; however, we observed a transient increase in IFN-α4 expression in the lungs of two out of four Tg2 mice at 5 days p.i.; this level fell by 7 days p.i. (Fig. 11C). Day 5 p.i. was the time point at which the amount of virus in the lungs of Tg2 mice began to fall. No IFN-β mRNA was detectable in lung samples from either group. On the other hand, the lungs of 25-week-old Tg2 mice showed a transient increase in IFN-α4 and IFN-β mRNA expression at 3 days p.i. (Fig. 11C). In addition, IFN-α4 and IFN-β mRNA expression was higher than that in 10-week-old Tg2 mice. Taken together, these results indicate that both type I and type II IFN contribute to the immunopathology of the lungs of 25-week-old Tg2 mice infected with MERS-CoV.
DISCUSSION
Here, we describe a new hDPP4-Tg mouse expressing the human gene under the control of an endogenous human promoter. This mouse model shows a pattern of hDPP4 expression that closely mimics that in human tissues and is similar to that in other recent models (5–7). This mouse model also shows susceptibility to infection by MERS-CoV; this mimics the nonlethal observations in other mouse models (13, 14). After intranasal inoculation with a human isolate of MERS-CoV, the Tg mice developed acute and mild interstitial pneumonia; however, the infection was nonlethal and so did not mimic severe cases seen in human MERS-CoV patients. MERS-CoV infection can cause severe illness, resulting in acute respiratory distress syndrome, although a large number of MERS-CoV infections follow a mild or asymptomatic course in healthy individuals (32–35). Thus, this Tg mouse model reflects the natural course of a mild MERS-CoV infection.
The majority of severe MERS-CoV cases occur in middle-aged and older males (36, 37). Therefore, we infected Tg2 mice aged 25 weeks with MERS-CoV. The mice showed prominent proinflammatory responses and prolonged pulmonary inflammation compared with responses in Tg2 mice aged 10 weeks. However, none of the infected Tg2 mice had a severe outcome, such as respiratory distress, that led to death. Epidemiologically, patients with diabetes, kidney failure, or chronic lung disease, all of which might weaken the immune system, tend to have a poor outcome after infection by MERS-CoV (http://www.who.int/csr/don/23-september-2015-mers-kuwait/en/). Thus, it is presumed that a combination of older age and underlying disease might also increase mortality in this animal model.
This hDPP4-Tg mouse model, which lacks the clinical signs and mortality associated with severe MERS-CoV infection, is likely to be less advantageous than other lethal mouse models with respect to development of novel vaccines or antiviral agents (12–14). When we asked why the Tg2 mice showed nonlethal responses to infection, we could not ignore the fact that virus levels in lungs were lower than those reported for other MERS mouse models (5, 12, 13, 38, 39). One reason for this is that the transgene used in this study is a hemizygote, meaning that the copy number or expression level may be lower than that in mice homozygous for hDPP4. In addition, the DPP4 protein is active as a dimer (40), but the Tg2 mice harbor both murine and hDPP4. We presume that the viral yield in the lungs of Tg2 mice was low because of heterodimers formed by hDPP4 and murine DPP4. To address this, we constructed structural models of homo- and heterodimers comprising human and mouse DPP4. Notably, the estimated interaction energy of the DPP4 heterodimers was greater than that of murine and hDPP4 homodimers (−358.2, −347.6, and −344.6 kcal/mol, respectively). These results suggest that DPP4 heterodimers are as stable as DPP4 homodimers. Cockrell et al. reported that mouse DPP4 does not support MERS-CoV entry (38). Thus, the presence of stable DPP4 heterodimers may be a reason for the lower levels of infection in our mouse model. Further study is necessary to clarify this.
Some research groups generated a mouse-adapted MERS-CoV for use in severe/lethal MERS-CoV infection mouse models (13, 14). This may be one way to establish severe MERS infection in our Tg2 mice.
While the Tg2 mice expressed hDPP4 protein in the liver, spleen, kidney, heart, lung, gastrointestinal tract, pancreas, and brain, viral infection and replication were limited (mainly) to the lower respiratory tract, with little upper respiratory tract involvement, after intranasal inoculation of MERS-CoV. Tg2 mice developed interstitial pneumonia, and MERS-CoV antigens were detected in the lungs. Virus yields in the lung were up to 100-fold higher than those in the upper respiratory tract. Most MERS patients exhibit a severe lower respiratory tract infection, with little involvement of the upper respiratory tract (36). This suggests that MERS-CoV infection in Tg2 mice mimics mild infection in humans.
In vitro analysis of MERS-CoV suggests that the virus also infects human T cells and macrophages (28, 41, 42). We detected MERS-CoV RNA in serum and spleen cells from Tg2 mice. These data are similar to those generated from another Tg mouse model in which mouse DPP4 was replaced with hDPP4 under the control of the endogenous mouse DPP4 promoter (12). MERS-CoV infection of T cells might affect immunopathology or induction of apoptosis in Tg mice, but we found no clear evidence of this. Although Tg2 mice showed systemic viremia, infection of organs (except lung) did not lead to secondary complications. The disease phenotype (including clinical symptoms, viral titer in the lung, and acute pneumonia) appeared to be driven by infiltration by macrophages and lymphocytes; this is similar to the phenotype observed in another Tg mouse model harboring hDPP4 under the control of the endogenous mouse DPP4 promoter (12).
The Tg2 mice described herein did not show any brain or renal lesions after MERS-CoV infection. Other Tg mouse models in which hDPP4 is expressed under a strong ubiquitous promoter show high levels of viral RNA and inflammation in the lungs, which are accompanied by brain lesions (5, 7, 11). A fatal case of human MERS-CoV infection published by Ng et al. showed no sign of MERS-CoV infection in the brain (43). To date, no reports suggest that MERS-CoV shows tropism for brain tissue. The primary target of MERS-CoV is the lower respiratory tract; however, patients with MERS often show signs of acute kidney failure (1, 43). In addition, MERS-CoV was identified in the urine of MERS patients (44, 45). Data from the first autopsy case did find pathological signs in the patient’s kidneys although IHC revealed no evidence of MERS-CoV replication in the kidneys (43). In addition, acute renal failure in this patient was not caused by MERS-CoV directly; rather, it was caused by hypotension (43) and/or acute respiratory distress syndrome (46).
Histopathological analysis identified CD3-positive T cells and Iba-1-positive macrophages in the lungs of Tg2 mice on day 7 p.i., which correlated with expression of inflammatory cytokines and inflammatory infiltrates in the lung. Tg2 mice aged 10 and 25 weeks showed increased expression of cytokines and chemokines associated with migration of T cells and activation of macrophages, including IP-10, IL-6, IL-13, MCP-1, IFN-γ, MIP-1α, MIG, and IL-12, in the lungs at day 5 and/or 7 p.i. This result is the same as that observed in a hDPP4 knock-in mouse model reported by Coleman et al. (12). In this hDPP4 knock-in mouse model, CD8+ T cells and macrophages affected the course of MERS-CoV-induced disease (12). In addition, Tg2 mice expressed mRNA encoding the type I IFN, IFN-α4, during the early phase of the MERS-CoV infection. Importantly, the pathogenic and immune response data from the knock-in mouse model and our own model are similar. Thus, an acute inflammatory reaction (including production of type I and type II IFNs) and infiltration by macrophages might clear the virus from the lung, thereby preventing progression to MERS. Interestingly, we detected IL-1α and IL-17 in the lungs of 25-week-old Tg2 mice, but not those of 10-week-old Tg2 mice, after MERS-CoV infection. Both IL-1α and IL-17 are proinflammatory cytokines that attract monocytes and neutrophils; therefore, they may exacerbate immunopathology after infection. These findings support the notion that the severity of MERS-CoV infection is age dependent. Age is one of the most common factors related to severity and mortality of MERS infection; however, the underlying pathology is unclear (47).
Many studies have examined immune responses of MERS patients (30, 48–52). Indeed, elevated serum levels of IL-6, IL-12, IP-10, and IFN-γ are observed in patients during the early period after severe infection (48–51). In addition, a prominent proinflammatory Th1 and Th17 response, including production of IFN-γ, tumor necrosis factor alpha (TNF-α), IL-15, and IL-17, is seen in patients during the acute phase of MERS-CoV infection (52). In contrast, we found that administration of poly(I·C) to Tg2 mice induced a mild increase (or earlier induction) in innate immune responses compared with those in C57BL/6 mice and non-Tg mice. This suggests that overexpression of hDPP4 might influence immune responses in Tg2 mice. Thus, we must exercise caution when assessing the relationship between immunopathology and outcome in patients and animal models of MERS-CoV; however, proinflammatory responses seem to contribute to immunopathology during the acute phase of MERS-CoV infection in both patients and mouse models.
In summary, we generated an hDPP4-Tg mouse model showing mild respiratory infection by MERS-CoV. While this Tg mouse has limitations as a model for human MERS (i.e., lower virus titer in the lungs and mild disease), the immunopathology seems to resemble a mild and early stage of infection in humans. Even though it has limitations, this Tg mouse model will increase our understanding of the mechanisms underlying MERS-CoV infection. Indeed, we recently used this mouse model to confirm a role for transmembrane protease serine type 2 (TMPRSS2) during MERS-CoV infection (53). This animal model may provide new insight into disease pathogenesis and guide development of therapeutic interventions that mitigate MERS.
MATERIALS AND METHODS
Ethics statements.
Experiments using recombinant DNA and pathogens were approved by the Committee for Experiments using Recombinant DNA and Pathogens at the National Institute of Infectious Diseases, Tokyo, Japan. All animal experiments were approved by the Animal Care and Use Committee of the National Institute of Infectious Diseases and the National Center for Global Health and Medicine (NCGM) Research Institute and were conducted in accordance with institutional Guidelines for the Care and Use of Animals. All animals were housed in a Japan Health Sciences Foundation-certified facility. All human samples used in this study were obtained from US Biomax, Inc., GeneTex, Inc., Alpha Diagnostics International, or Protein Biotechnologies. The protocols were approved by the Health Insurance Portability and Accountability Act (HIPAA) or Institutional Review Board (IRB).
Cells and viruses.
MERS-CoV, HCoV-EMC 2012 strain, was kindly provided by Bart Haagmans and Ron Fochier (Erasmus Medical Center, Rotterdam, the Netherlands) and was used throughout the study. Vero E6 cells, purchased from the American Type Cell Collection (Manassas, VA), were cultured in Eagle’s MEM containing 5% fetal bovine serum (FBS), 50 IU/ml penicillin G, and 50 μg/ml streptomycin (5% FBS-MEM). Stocks of MERS-CoV were propagated and titrated on Vero E6 cells and cryopreserved at −80°C. Viral infectivity titers are expressed as the TCID50/milliliter on Vero E6 cells and were calculated according to the Behrens-Kärber method. Work with infectious MERS-CoV was performed under biosafety level 3 conditions.
Virus isolation and titration.
Lung wash fluids and liver, kidney, heart, spleen, intestine, and brain tissue samples from Tg2, non-Tg, and C57BL/6 mice were collected at the time of postmortem examination and stored at −80°C. Tissue homogenates (20%, wt/vol) were prepared in 2% FBS-MEM, and samples were inoculated onto Vero E6 cell cultures, which were then examined for cytopathic effects (CPE) for 5 days. Blind passage was performed after freezing and thawing cells from the first- or second-round passages. If MERS-CoV-specific CPE were not observed in the first-, second-, or third-round cultures, the samples were deemed negative for infectious virus. Viral infectivity titers were determined in Vero E6 cell cultures using the microtitration assay described above.
MERS-CoV neutralizing assay.
Blood was obtained from each mouse and allowed to clot. Sera were then obtained by centrifugation and inactivated by incubation at 56°C for 30 min. One hundred TCID50 aliquots of MERS-CoV were incubated for 1 h in the presence or absence of mouse serum (serially diluted 2-fold) and then added to confluent Vero E6 cell cultures in 96-well microtiter plates. The presence of viral CPE was determined on day 5, and the titers of neutralizing antibody were determined as the reciprocal of the highest dilution at which no CPE were observed. The lowest and highest serum dilutions tested were 1:2 and 1:512, respectively.
Generation of hDPP4-Tg mice.
To generate Tg mice expressing hDPP4, a BAC vector carrying the hDPP4 gene (clone RP11-345J9) was purchased from Advanced GenoTechs Co., Japan. The BAC DNA was purified using a Large-Construct kit (Qiagen) according to the manufacturer’s instructions and suspended in TE buffer (10 mM Tris-HCl and 0.1 mM EDTA, pH 7.5) at a concentration of 4 ng/μl. Tg mice were generated using standard procedures (23). The purified BAC clones were microinjected into the pronuclei of fertilized eggs from BDF1×C57BL/6NCr mice (SLC, Inc., Hamamatsu, Japan) and then transplanted into pseudopregnant ICR mice (SLC Inc.). Expression of the transgene was assessed by fluorescence-activated cell sorter (FACS) analysis as described below. The Tg mice were then backcrossed onto C57BL/6NCr for five generations. After weaning, the mice were tested for Tg integration by PCR and FACS analysis. Briefly, genomic DNA isolated from ear punch tissues was subjected to PCR using hDPP4-specific primers (Table 2), and lymphocytes were isolated from blood taken from the tail vein. Lymphocytes were screened for hDPP4 protein expression by flow cytometry analysis. Tg mice and their non-Tg littermates were used for the MERS-CoV infection study.
Flow cytometry analysis.
Blood was collected from the retro-orbital venous plexus under isoflurane anesthesia using heparinized capillary tubes. The samples were then treated with red blood cell lysis buffer (Sigma-Aldrich, St. Louis, MO) to remove erythroid cells. For immunofluorescence staining, cells were resuspended in staining buffer (PBS containing 2.5% FBS, 0.5 mM EDTA, 0.05% NaN3), and Fc receptors were blocked by incubation for 20 min on ice with an unlabeled anti-mouse CD16/CD32 monoclonal antibody (clone 2.4G2; Bay Bioscience Co., Ltd., Kobe, Japan). After a wash step, the cells were stained for 30 min with an FITC-labeled anti-hDPP4 antibody (clone BA5b) or with a control antibody against mouse IgG2a (clone MOPC-173; BioLegend, San Diego, CA) and an allophycocyanin (APC)-labeled anti-human CD3 antibody (clone 145-2C11; BioLegend). Cells were then washed and analyzed on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Data were processed using Cell Quest software (BD Biosciences).
Western blot analysis.
To examine expression of DPP4 in human tissues, human tissue lysates prepared from the liver, spleen, kidney, heart, lung, stomach, small intestine, large intestine, pancreas, and brain were purchased from Alpha Diagnostics International (San Antonio, TX). Human skeletal muscle lysates were purchased from Protein Biotechnologies (Ramona, CA) and used under IRB-approved protocols. To prepare protein samples from the Tg and non-Tg mouse organs, tissues were homogenized in 0.5 ml of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris/HCl [pH 7.5], 150 mM NaCl, 1% [vol/vol] Nonidet P40 [NP-40], 0.5% sodium deoxycholate, 0.1% SDS) containing a protease inhibitor mixture (Complete Mini; Roche Diagnostics, Basel, Switzerland), and the protein concentrations were measured using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific Inc., Waltham, MA). Homogenates (equivalent to 5 μg of protein) were subjected to SDS/PAGE on 4 to 12% Bis-Tris Protein Gels (Thermo Fisher Scientific), followed by transfer to a polyvinylidene difluoride (PVDF) membrane (Millipore Corp., Billerica, MA). After being blocked, the membranes were incubated for 1 h with a goat anti-hDPP4 antibody (0.1 μg/ml) (AF1180; R&D Systems, Inc., Minneapolis, MN) or a rabbit anti-β-actin antibody (1 μg/ml) (ab8227; Abcam, Cambridge, UK), followed by incubation with a donkey anti-goat horseradish peroxidase (HRP)-conjugated antibody (Abcam) and an anti-rabbit HRP-conjugated antibody (Abcam). The bands were detected by an Immobilon Western Chemiluminescent HRP substrate (Millipore) and an LAS-3000 apparatus (Fujifilm, Tokyo, Japan).
Intranasal administration of poly(I·C).
Tg2, non-Tg, and C57BL/6 mice (10 weeks old) were anesthetized and received 20 μg of poly(I·C) (Invitrogen, San Diego, CA) in 20 μl of saline intranasally. The dose of poly(I·C) was determined from previous studies (54). Control mice received saline alone. To evaluate production of cytokines and chemokines in the lung, mice were sacrificed at 1 day postadministration (n = 4 per group; two females and two males), and lungs were collected. Inflammatory cytokine profiles in 20% (wt/vol) lung homogenates were detected using a commercial Mouse Cytokine 20-Plex antibody bead kit (Thermo Fisher Scientific), as described by the manufacturer.
Inoculation of mice with MERS-CoV.
Tg2 and non-Tg mice (9 to 10 weeks or 25 weeks old) and Tg2-BALB mice (12 to 22 weeks old) were anesthetized and inoculated intranasally with 1 × 105 TCID50 (30 μl) of MERS-CoV. Body weight was measured daily for 14 days (n = 4 to 8 per group), and animals were sacrificed at 6 h and at 1, 3, 5, 7, 14, and 35 days p.i. to analyze virus replication, hematological parameters, cytokine expression, and disease pathology (n = 3 to 6 per group). Clinical signs were observed up until 14 days p.i. All mock-infected mice were inoculated with 2% FBS-MEM and used as controls for all analyses involving mice aged 9 to 10 weeks.
Histopathology and IHC.
Formalin-fixed paraffin-embedded normal human tissue sections were purchased from separate sources: liver, spleen, lung, trachea, small intestine, colon, pancreas, cerebrum, cerebellum, and skeletal muscle were obtained from US Biomax, Inc. (Rockville, MD), whereas kidney, heart, spinal cord, stomach, and lymph node were obtained from GeneTex, Inc. (Irvine, CA). These human tissues were collected under HIPAA-approved protocols. To obtain animal tissues, mice were anesthetized and perfused with 2 ml of 10% phosphate-buffered formalin, and the lungs, liver, spleen, kidney, heart, gastrointestinal tract, salivary glands, and brain tissue were harvested and fixed. Fixed tissues were routinely embedded in paraffin, sectioned, and stained with hematoxylin and eosin. For IHC, antigen retrieval of formalin-fixed mouse tissue sections was performed by autoclaving at 121°C for 10 min in retrieval solution at pH 6.0 (Nichirei, Tokyo, Japan). hDPP4 and MERS-CoV antigens were detected using a standard immunoperoxidase method and a goat anti-hDPP4 antibody (R&D Systems) and a rabbit anti-MERS-CoV nucleocapsid antibody (40068-RP01; Sino Biological, Inc., Beijing, China).
For double staining of CD3 (T cells) and Iba-1 (macrophages) antigen, we used a rabbit anti-human CD3 antibody (790-4341; Ventana Medical System, Inc., Tucson, AZ) and a rabbit anti-human Iba-1 antibody (019-19741; Wako Pure Chemical Industries, Ltd., Osaka, Japan). Diaminobenzidine (DAB; Sigma-Aldrich Co., MO, USA) and a Vina Green Chromogen kit (Biocare Medical, CA, USA) were used as chromogens for HRP visualization. Following the first staining of CD3 using the polymer-based detection system with DAB, denaturing was performed by hydrolytic autoclaving in citrate buffer (pH 6.0) for 10 min at 121°C. The second staining was performed for Iba-1 with Vina Green. Nuclei were counterstained with hematoxylin for 10 s. To detect apoptosis, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) was performed using an In Situ Cell Death Detection kit (Roche).
Quantitative real-time RT-PCR.
To measure the levels of type I IFN mRNA expression and the number of viral genome copies, RNA was extracted from 20% (wt/vol) lung and brain tissue homogenates and from the blood of Tg2 and non-Tg mice infected with MERS-CoV using RNeasy minikits (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. mRNAs encoding IFN-α, IFN-β, and the E gene of MERS-CoV were examined by real-time RT-PCR using an ABI Prism 7900HT Fast real-time PCR system (Applied Biosystems, Foster City, CA). The TaqMan probes and primers and the reaction conditions have been described previously (31). Expression of each gene was normalized to that of β-actin.
Detection of inflammatory cytokines and chemokines.
Cytokines and chemokines in mouse lung homogenates (10% wt/vol) were measured using a commercial Mouse Cytokine 20-Plex antibody bead kit (Thermo Fisher Scientific). A panel of inflammatory cytokines and chemokines (basic fibroblast growth factor [bFGF], granulocyte-macrophage colony-stimulating factor [GM-CSF], IFN-γ, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12p40/p70, IL-13, IL-17, IP-10, keratinocyte chemoattractant [KC], MCP-1, MIG, MIP-1α, TNF-α, and vascular endothelial growth factor) was measured according to the manufacturer’s protocols.
Isolation of splenocytes and infection with MERS-CoV.
Spleens were removed aseptically from Tg2 and C57BL/6 mice (n = 3 each), dissociated in RPMI 1640 medium, and pressed gently through a 40-μm-pore-size nylon mesh filter. The cell suspension was centrifuged at 400 × g for 10 min, and red blood cells were lysed with blood cell lysis buffer (final concentrations of 155 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA, pH 7.3) at room temperature for 5 min. The cells were washed twice with RPMI 1640 medium and centrifuged at 1,000 × g for 10 min. hDPP4-expressing CD3+ T cells within the splenocyte population were detected by flow cytometry analysis. The percentage of CD3+ T cells was 39.11% ± 3.8%, and that of hDPP4-expressing CD3+ T cells was 26.46% ± 1.9%. The cells were resuspended in the medium and infected with MERS-CoV (multiplicity of infection [MOI] of 1). Viral replication was determined after 1 and 2 days of culture. Viral infectivity titers were measured in Vero E6 cell cultures using a microtitration assay. To detect the MERS-CoV genome in splenocytes, RNA from splenocytes infected with MERS-CoV was extracted at 1 and 2 days p.i. and subjected to quantitative real-time RT-PCR (10).
Molecular modeling of DPP4 homo- and heterodimers.
DPP4 dimer models were constructed using the Molecular Operating Environment (MOE) (Chemical Computing Group, Inc., Montreal, QC, Canada) based on the crystal structure of hDPP4 at a resolution of 2.55 Å (PDB accession number 2ONC). Stereochemical quality was assessed using the Ramachandran plot and atom clashes applications in MOE. Interaction energy, which is an indicator of the affinity of the dimer, was calculated using the potential energy application in MOE.
Statistical analysis.
Data are expressed as the means and standard errors of the means. Statistical analyses were performed using GraphPad Prism, version 5, software (GraphPad Software, Inc., La Jolla, CA). Intergroup comparisons were performed using one-way and two-way ANOVA or Student's t test with Welch’s correction. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Bart Haagmans and Ron Fouchier for providing MERS-CoV (isolate HCoV-EMC/2012) and Satoshi Koike, Ken Fujii (Tokyo Metropolitan Institute of Medical Science), and Shutoku Matsuyama (National Institute of Infectious Diseases) for helpful discussions. We also thank Ayako Harashima and Midori Ozaki for technical assistance.
This work was supported by the following: a grant-in-aid for research (H25-Shinko-Wakate-004) from the Ministry of Health, Labor, and Welfare, Japan; the Research Program on Emerging and Reemerging Infectious Diseases (grants JP17fk0108313, JP18fk0108058, and JP18fk0108072) from the Japan Agency for Medical Research and Development; grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (16K09951 and 18H02665), and a grant from the National Center for Global Health and Medicine (27A1102).
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JVI.01815-18.
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, Fischer ER, Martellaro C, Okumura A, Chang J, Scott D, Benecke AG, Katze MG, Feldmann H, Munster VJ. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci U S A 110:16598–16603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falzarano D, de Wit E, Feldmann F, Rasmussen AL, Okumura A, Peng X, Thomas MJ, van Doremalen N, Haddock E, Nagy L, LaCasse R, Liu T, Zhu J, McLellan JS, Scott DP, Katze MG, Feldmann H, Munster VJ. 2014. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog 10:e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal AS, Garron T, Tao X, Peng BH, Wakamiya M, Chan TS, Couch RB, Tseng CT. 2015. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol 89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao G, Jiang Y, Qiu H, Gao T, Zeng Y, Guo Y, Yu H, Li J, Kou Z, Du L, Tan W, Jiang S, Sun S, Zhou Y. 2015. Multi-organ damage in human dipeptidyl peptidase 4 transgenic mice infected with Middle East respiratory syndrome-coronavirus. PLoS One 10:e0145561. doi: 10.1371/journal.pone.0145561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K, Wohlford-Lenane C, Perlman S, Zhao J, Jewell AK, Reznikov LR, Gibson-Corley KN, Meyerholz DK, McCray PB. Jr, 2016. Middle East respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis 213:712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit E, Prescott J, Baseler L, Bushmaker T, Thomas T, Lackemeyer MG, Martellaro C, Milne-Price S, Haddock E, Haagmans BL, Feldmann H, Munster VJ. 2013. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS One 8:e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman CM, Matthews KL, Goicochea L, Frieman MB. 2013. Wild type and innate immune deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol 95:408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwata-Yoshikawa N, Fukushi S, Fukuma A, Suzuki T, Takeda M, Tashiro M, Hasegawa H, Nagata N. 2016. No susceptibility of neonatal and adult rats against the Middle East respiratory syndrome coronavirus. Jpn J Infect Dis 69:510–516. [DOI] [PubMed] [Google Scholar]
- 11.Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ Jr, Baric RS, Enjuanes L, Gallagher T, McCray PB Jr, Perlman S. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A 111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman CM, Sisk JM, Halasz G, Zhong J, Beck SE, Matthews KL, Venkataraman T, Rajagopalan S, Kyratsous CA, Frieman MB. 2017. CD8+ T cells and macrophages regulate pathogenesis in a mouse model of Middle East respiratory syndrome. J Virol 91:e01825-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K, Wohlford-Lenane CL, Channappanavar R, Park JE, Earnest JT, Bair TB, Bates AM, Brogden KA, Flaherty HA, Gallagher T, Meyerholz DK, Perlman S, McCray PB. Jr, 2017. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A 114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cockrell AS, Yount BL, Scobey T, Jensen K, Douglas M, Beall A, Tang XC, Marasco WA, Heise MT, Baric RS. 2016. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol 2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korea Centers for Disease Control and Prevention. 2015. Middle East respiratory syndrome coronavirus outbreak in the republic of Korea, 2015. Osong Public Health Res Perspect 6:269–278. doi: 10.1016/j.phrp.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saad M, Omrani AS, Baig K, Bahloul A, Elzein F, Matin MA, Selim MA, Al Mutairi M, Al Nakhli D, Al Aidaroos AY, Al Sherbeeni N, Al-Khashan HI, Memish ZA, Albarrak AM. 2014. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int J Infect Dis 29:301–306. doi: 10.1016/j.ijid.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed AE. 2017. The predictors of 3- and 30-day mortality in 660 MERS-CoV patients. BMC Infect Dis 17:615. doi: 10.1186/s12879-017-2712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsahafi AJ, Cheng AC. 2016. The epidemiology of Middle East respiratory syndrome coronavirus in the Kingdom of Saudi Arabia, 2012–2015. Int J Infect Dis 45:1–4. doi: 10.1016/j.ijid.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majumder MS, Kluberg SA, Mekaru SR, Brownstein JS. 2015. Mortality risk factors for Middle East respiratory syndrome outbreak, South Korea, 2015. Emerg Infect Dis 21:2088–2090. doi: 10.3201/eid2111.151231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi WS, Kang CI, Kim Y, Choi JP, Joh JS, Shin HS, Kim G, Peck KR, Chung DR, Kim HO, Song SH, Kim YR, Sohn KM, Jung Y, Bang JH, Kim NJ, Lee KS, Jeong HW, Rhee JY, Kim ES, Woo H, Oh WS, Huh K, Lee YH, Song JY, Lee J, Lee CS, Kim BN, Choi YH, Jeong SJ, Lee JS, Yoon JH, Wi YM, Joung MK, Park SY, Lee SH, Jung SI, Kim SW, Lee JH, Lee H, Ki HK, Kim YS. 2016. Clinical presentation and outcomes of Middle East respiratory syndrome in the Republic of Korea. Infect Chemother 48:118–126. doi: 10.3947/ic.2016.48.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koike S, Taya C, Kurata T, Abe S, Ise I, Yonekawa H, Nomoto A. 1991. Transgenic mice susceptible to poliovirus. Proc Natl Acad Sci U S A 88:951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii K, Nagata N, Sato Y, Ong KC, Wong KT, Yamayoshi S, Shimanuki M, Shitara H, Taya C, Koike S. 2013. Transgenic mouse model for the study of enterovirus 71 neuropathogenesis. Proc Natl Acad Sci U S A 110:14753–14758. doi: 10.1073/pnas.1217563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haruyama N, Cho A, Kulkarni AB. 2009. Overview: engineering transgenic constructs and mice. Curr Protoc Cell Biol Chapter 19:Unit 19.10. doi: 10.1002/0471143030.cb1910s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geppert TD, Davis LS, Gur H, Wacholtz MC, Lipsky PE. 1990. Accessory cell signals involved in T-cell activation. Immunol Rev 117:5–66. [DOI] [PubMed] [Google Scholar]
- 25.Fleischer B. 1994. CD26: a surface protease involved in T-cell activation. Immunol Today 15:180–184. doi: 10.1016/0167-5699(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 26.Hegen M, Niedobitek G, Klein CE, Stein H, Fleischer B. 1990. The T cell triggering molecule Tp103 is associated with dipeptidyl aminopeptidase IV activity. J Immunol 144:2908–2914. [PubMed] [Google Scholar]
- 27.Fan C, Wu X, Liu Q, Li Q, Liu S, Lu J, Yang Y, Cao Y, Huang W, Liang C, Ying T, Jiang S, Wang Y. 2018. A human DPP4-knockin mouse's susceptibility to infection by authentic and pseudotyped MERS-CoV. Viruses 10:E448. doi: 10.3390/v10090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu H, Zhou J, Wong BH, Li C, Chan JF, Cheng ZS, Yang D, Wang D, Lee AC, Li C, Yeung ML, Cai JP, Chan IH, Ho WK, To KK, Zheng BJ, Yao Y, Qin C, Yuen KY. 2016. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alghamdi IG, Hussain II, Almalki SS, Alghamdi MS, Alghamdi MM, El-Sheemy MA. 2014. The pattern of Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive epidemiological analysis of data from the Saudi Ministry of Health. Int J Gen Med 7:417–423. doi: 10.2147/IJGM.S67061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Brand JM, Smits SL, Haagmans BL. 2015. Pathogenesis of Middle East respiratory syndrome coronavirus. J Pathol 235:175–184. doi: 10.1002/path.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwata-Yoshikawa N, Uda A, Suzuki T, Tsunetsugu-Yokota Y, Sato Y, Morikawa S, Tashiro M, Sata T, Hasegawa H, Nagata N. 2014. Effects of Toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-inactivated severe acute respiratory syndrome-related coronavirus vaccine. J Virol 88:8597–8614. doi: 10.1128/JVI.00983-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memish ZA, Zumla AI, Assiri A. 2013. Middle East respiratory syndrome coronavirus infections in health care workers. N Engl J Med 369:884–886. doi: 10.1056/NEJMc1308698. [DOI] [PubMed] [Google Scholar]
- 33.The WHO MERS-CoV Research Group. 2013. State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Curr Outbreaks 1:23143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Hagan JJ, Carias C, Rudd JM, Pham HT, Haber Y, Pesik N, Cetron MS, Gambhir M, Gerber SI, Swerdlow DL. 2016. Estimation of severe Middle East respiratory syndrome cases in the Middle East, 2012–2016. Emerg Infect Dis 22:1797–1799. doi: 10.3201/eid2210.151121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omrani AS, Matin MA, Haddad Q, Al-Nakhli D, Memish ZA, Albarrak AM. 2013. A family cluster of Middle East respiratory syndrome coronavirus infections related to a likely unrecognized asymptomatic or mild case. Int J Infect Dis 17:e668–e672. doi: 10.1016/j.ijid.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CDC. 2013. Update: severe respiratory illness associated with Middle East respiratory syndrome coronavirus (MERS-CoV)—worldwide, 2012–2013. MMWR Morb Mortal Wkly Rep 62:480–483. [PMC free article] [PubMed] [Google Scholar]
- 37.Assiri A, Abedi GR, Bin Saeed AA, Abdalla MA, Al-Masry M, Choudhry AJ, Lu X, Erdman DD, Tatti K, Binder AM, Rudd J, Tokars J, Miao C, Alarbash H, Nooh R, Pallansch M, Gerber SI, Watson JT. 2016. Multifacility outbreak of Middle East respiratory syndrome in Taif, Saudi Arabia. Emerg Infect Dis 22:32–40. doi: 10.3201/eid2201.151370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cockrell AS, Peck KM, Yount BL, Agnihothram SS, Scobey T, Curnes NR, Baric RS, Heise MT. 2014. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J Virol 88:5195–5199. doi: 10.1128/JVI.03764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascal KE, Coleman CM, Mujica AO, Kamat V, Badithe A, Fairhurst J, Hunt C, Strein J, Berrebi A, Sisk JM, Matthews KL, Babb R, Chen G, Lai KM, Huang TT, Olson W, Yancopoulos GD, Stahl N, Frieman MB, Kyratsous CA. 2015. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A 112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chien CH, Huang LH, Chou CY, Chen YS, Han YS, Chang GG, Liang PH, Chen X. 2004. One site mutation disrupts dimer formation in human DPP-IV proteins. J Biol Chem 279:52338–52345. doi: 10.1074/jbc.M406185200. [DOI] [PubMed] [Google Scholar]
- 41.Tynell J, Westenius V, Ronkko E, Munster VJ, Melen K, Osterlund P, Julkunen I. 2016. Middle East respiratory syndrome coronavirus shows poor replication but significant induction of antiviral responses in human monocyte-derived macrophages and dendritic cells. J Gen Virol 97:344–355. doi: 10.1099/jgv.0.000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Chu H, Li C, Wong BH, Cheng ZS, Poon VK, Sun T, Lau CC, Wong KK, Chan JY, Chan JF, To KK, Chan KH, Zheng BJ, Yuen KY. 2014. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis 209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng DL, Al Hosani F, Keating MK, Gerber SI, Jones TL, Metcalfe MG, Tong S, Tao Y, Alami NN, Haynes LM, Mutei MA, Abdel-Wareth L, Uyeki TM, Swerdlow DL, Barakat M, Zaki SR. 2016. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol 186:652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poissy J, Goffard A, Parmentier-Decrucq E, Favory R, Kauv M, Kipnis E, Mathieu D, van der Werf S, Guery B. 2014. Kinetics and pattern of viral excretion in biological specimens of two MERS-CoV cases. J Clin Virol: 61:275–278. doi: 10.1016/j.jcv.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drosten C, Seilmaier M, Corman VM, Hartmann W, Scheible G, Sack S, Guggemos W, Kallies R, Muth D, Junglen S, Muller MA, Haas W, Guberina H, Rohnisch T, Schmid-Wendtner M, Aldabbagh S, Dittmer U, Gold H, Graf P, Bonin F, Rambaut A, Wendtner CM. 2013. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis 13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeley EJ. 2013. Updates in the management of acute lung injury: a focus on the overlap between AKI and ARDS. Adv Chronic Kidney Dis 20:14–20. doi: 10.1053/j.ackd.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Park JE, Jung S, Kim A, Park JE. 2018. MERS transmission and risk factors: a systematic review. BMC Public Health 18:574. doi: 10.1186/s12889-018-5484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faure E, Poissy J, Goffard A, Fournier C, Kipnis E, Titecat M, Bortolotti P, Martinez L, Dubucquoi S, Dessein R, Gosset P, Mathieu D, Guery B. 2014. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One 9:e88716. doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan WD, Mok CK, Chen ZL, Feng LQ, Li ZT, Huang JC, Ke CW, Deng X, Ling Y, Wu SG, Niu XF, Perera RA, Da Xu Y, Zhao J, Zhang LQ, Li YM, Chen RC, Peiris M, Chen L, Zhong NS. 2015. Characteristics of traveler with Middle East respiratory syndrome, China, 2015. Emerg Infect Dis 21:2278–2280. doi: 10.3201/eid2112.151232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Min CK, Cheon S, Ha NY, Sohn KM, Kim Y, Aigerim A, Shin HM, Choi JY, Inn KS, Kim JH, Moon JY, Choi MS, Cho NH, Kim YS. 2016. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep 6:25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin HS, Kim Y, Kim G, Lee JY, Jeong I, Joh JS, Kim H, Chang E, Sim SY, Park JS, Lim DG. 27 July 2018. Immune responses to MERS coronavirus during the acute and convalescent phases of human infection. Clin Infect Dis doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. 2018. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. 2019. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol 93:e01815-18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ichinohe T, Watanabe I, Ito S, Fujii H, Moriyama M, Tamura S, Takahashi H, Sawa H, Chiba J, Kurata T, Sata T, Hasegawa H. 2005. Synthetic double-stranded RNA poly(I:C) combined with mucosal vaccine protects against influenza virus infection. J Virol 79:2910–2919. doi: 10.1128/JVI.79.5.2910-2919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]